Abstract
Aims
Arrhythmogenic cardiomyopathy (ACM) is a primary myocardial disease that typically manifests with cardiac arrhythmias, progressive heart failure, and sudden cardiac death (SCD). ACM is mainly caused by mutations in genes encoding desmosome proteins. Desmosomes are cell–cell adhesion structures and hubs for mechanosensing and mechanotransduction. The objective was to identify the dysregulated molecular and biological pathways in human ACM in the absence of overt heart failure.
Methods and results
Transcriptomes in the right ventricular endomyocardial biopsy samples from three independent individuals carrying truncating mutations in the DSP gene and five control samples were analysed by RNA-Seq (discovery group). These cases presented with cardiac arrhythmias and had a normal right ventricular function. The RNA-Seq analysis identified ∼5000 differentially expressed genes (DEGs), which predicted suppression of the Hippo and canonical WNT pathways, among others. Dysregulated genes and pathways, identified by RNA-Seq, were tested for validation in the right and left ventricular tissues from five independent autopsy-confirmed ACM cases with defined mutations (validation group), who were victims of SCD and had no history of heart failure. Protein levels and nuclear localization of the cWNT and Hippo pathway transcriptional regulators were reduced in the right and left ventricular validation samples. In contrast, levels of acetyltransferase EP300, known to suppress the Hippo and canonical WNT pathways, were increased and its bona fide target TP53 was acetylated. RNA-Seq data identified apical junction, reflective of cell–cell attachment, as the most disrupted biological pathway, which were corroborated by disrupted desmosomes and intermediate filament structures. Moreover, the DEGs also predicted dysregulation of over a dozen canonical signal transduction pathways, including the Tec kinase and integrin signalling pathways. The changes were associated with increased apoptosis and fibro-adipogenesis in the ACM hearts.
Conclusion
Altered apical junction structures are associated with activation of the EP300-TP53 and suppression of the Hippo/cWNT pathways in human ACM caused by defined mutations in the absence of an overt heart failure. The findings implicate altered mechanotransduction in the pathogenesis of ACM.
Keywords: Cardiomyopathy, Hippo pathway, WNT pathway, TP53, RNA-sequencing, Gene expression
Graphical Abstract
1. Introduction
Arrhythmogenic cardiomyopathy (ACM) comprises a group of enigmatic hereditary myocardial diseases whose cardinal manifestation is ventricular arrhythmia occurring early and prior to the onset of overt cardiac dysfunction.1,2 Cardiac dysfunction, manifested as an increased ventricular diameter and decreased ejection fraction, commonly manifests later in the course of the disease, is progressive, and leads to refractory heart failure as well as secondary cardiac arrhythmias.1,2 ACM is an important cause of sudden cardiac death (SCD) in the young, which is often the first manifestation of the disease.3–5
The classic form of ACM predominantly involves the right ventricle, particularly in the early stages, and is referred to as arrhythmogenic right ventricular cardiomyopathy (ARVC). The pathological hallmark of ARVC is fibrofatty infiltration of the myocardium, which typically starts from the epicardium of the right ventricle.4 A subtype of ACM predominantly involves the left ventricle and presents with left ventricular dilatation and failure, overlapping with the diagnosis of primary dilated cardiomyopathy.6
The molecular genetic basis of ACM has been partially elucidated upon identification of mutations in genes encoding protein constituents of the intercalated discs (IDs), particularly the desmosomes.7 PKP2 encoding plakophilin 2 is the most common causal gene for ACM followed by DSP, DSC2, DSG2, and JUP, encoding desmoplakin, desmocollin 2, desmoglein 2, and junction protein plakoglobin, respectively.7–9 The genetic spectrum of ACM has been recently expanded to include genes coding for proteins other than the ID proteins, such as the TMEM43 and LMNA genes, coding for nuclear membrane proteins transmembrane protein 43 and lamin A/C, respectively. Moreover, mutations in PLN, encoding calcium homoeostasis protein phospholamban; FLNC and DES encoding cytoskeletal protein filamin C and desmin, respectively; and TTN, which codes for the giant sarcomere protein titin have been identified as causes of ACM.7 Other implicated genes include TGFB3, CDH2, and CTNNA3, encoding transforming growth factor β3, cadherin 2, and αT-catenin, respectively.2 Thus far, genetic studies have led to identification of the causal genes in approximately half of the ACM families/cases.7
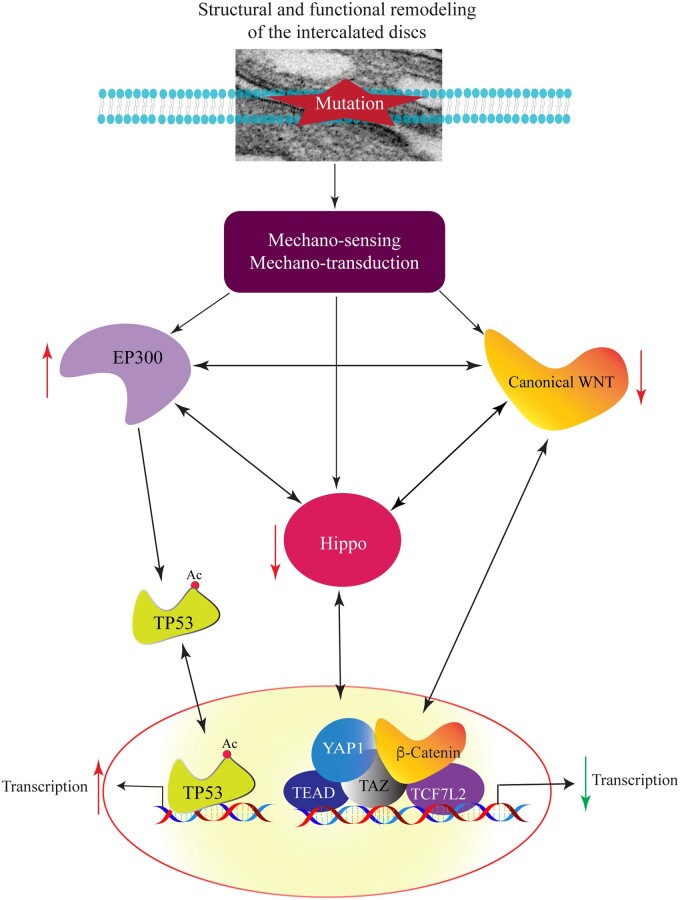
Desmosomes, abundantly expressed in cardiac myocytes and epithelial cells, are cell–cell attachment structures, often referred to as the apical junction complex, whose primary function in the heart is to maintain mechanical integrity of the myocardium.10,11 The desmosomes are also signalling hubs, responsible, in part, for sensing the mechanical stress, connecting with the cell cytoskeleton, and regulating the mechanosensing and mechanotransduction signalling pathways, including the Hippo and the canonical WNT (cWNT) pathways.10,12,13 Accordingly, the Hippo pathway is activated and gene expression through its co-effectors TEAD, YAP1, and TAZ is suppressed in the context of heart failure in the advanced stages of ACM.13 Likewise, gene expression through the cWNT downstream co-effector, namely β-catenin (CTNNB1) and TCF7L2, is also suppressed in advanced ACM.12 It is unknown, however, whether these changes are secondary to cardiac dysfunction or are early preceding the onset of overt heart failure in ACM. The significance of identification of the early molecular changes in ACM is self-evident, as the early changes would be free of the confounding effects of heart failure and might imply a pathogenic role. Thus, the purpose of this study was to identify the early molecular changes that occur in the heart prior to the development of overt heart failure in genetically defined ACM.
2. Methods
2.1 Ethical statements
The studies with human heart samples were approved by the institutional review board of the University of Texas Health Science Center at Houston (HSC-IMM-07-0016) and Hospital Universitario y Politécnico La Fe de Valencia, Spain. All participants signed informed consents prior to participation in the study and the investigation conformed to the principles outlined in the Declaration of Helsinki.
2.2 Clinical studies
ACM probands and family members underwent clinical evaluation, as per the conventional medical care of ACM patients, which included electrocardiography (ECG), Holter monitoring, echocardiography, and cardiac magnetic resonance (CMR) imaging, in addition to history and physical examination.
2.3 Molecular genetic studies
Genetic testing was performed in the probands by candidate gene Sanger sequencing or whole exome sequencing with a focused analysis of cardiomyopathy panel, followed by cascade genetic testing of the related family members by Sanger sequencing or multiplex ligatioin-dependent probe amplification (MLPA), as appropriate.
2.4 Discovery and validation study populations
The discovery samples were from three independent patients with familial ACM caused by three different truncating DSP mutations. They presented with ventricular arrhythmias in the setting of family screening and were evaluated for ACM per the 2010 revised Task Force guidelines, which included ECG, CMR, and echocardiography, as well as genetic testing. The patients underwent endomyocardial biopsy per the conventional procedure and samples from the right ventricle near the apical septum were obtained. Given that endomyocardial biopsy in control healthy individuals is not justifiable, the right ventricular tissues from five donor explanted hearts with no apparent cardiovascular diseases that were not used for heart transplantation were included as controls. The endomyocardial samples from the patients and the control samples were used for whole heart RNA-sequencing (RNA-Seq).
Right and left ventricular tissues from five victims of SCD who had no known history of heart failure but were found to have ACM on autopsy were used as the validation samples. They carried pathogenic variants (PVs) in genes known to cause ACM.14 Right and left ventricular samples from five explanted hearts with no documented history of cardiovascular disease were used as control heart.
2.5 RNA-Seq
Whole heart RNA-Seq was performed, as published with some modifications.15–18 Total RNA was extracted from flash-frozen samples using the miRNAeasy mini kit (CAT#217004). RNA quality was assessed using the Agilent bioanalyzer, and only samples with RNA Integrity Number (RIN) of >7 were used. Strand-specific libraries were prepared following depletion of ribosomal RNA and sequenced on an Illumina HiSeq 4000 instrument using the paired-end sequencing chemistry.
Raw RNA sequencing reads were aligned to the human reference genome (build GRCh38/hg38) using HISAT2.19 The number of the uniquely mapped reads per gene was determined using the featureCounts tool, as the sum of reads associated with each of the exons in the gene. The read data were normalized using the Remove Unwanted Variation (RUVr) method.20 Differentially expressed genes (DEGs) were defined as genes whose transcript levels differed between ACM and control samples at the Benjamini–Hochberg adjusted P value (i.e. q value) of <0.05, as determined using the DESeq2 software in the R package.21 Heat maps and volcano plots were generated for data visualization in Rstudio using normalized count per million (CPM) values (www.rstudio.com). Circos maps were plotted using the GOCHORD function in R.
2.6 Predicted transcriptional regulators and biological pathways
The RNA-Seq data were analysed as published with minor modifications.15–18 To obtain the biological functions associated with the DEGs, Gene Set Enrichment (GSEA, version 2.2.3, http://software.broadinstitute.org/gsea/) was performed. Significance level was determined by the permutation-based statistical inference and only gene sets with a q < 0.05 were presented. Strength of the enrichment was ranked by Normalized Enrichment Score (NES) using the Molecular signature database (MSigDB) 3.0, curated gene sets for the Hallmark canonical pathways. GSEA for YAP1 targets was also performed using the following Gene Set: CORDENONSI_YAP_CONSERVED_SIGNATURE. The transcriptional regulators (TRs) responsible for differential gene expression were predicted using the Upstream regulator analysis function of the Ingenuity Pathway Analysis software (IPA®, QIAGEN Redwood City). Those showing an overlap P-value of <0.05 and a predicted Z score of < −2 or >2 were considered significant. Targets of the specific TRs were obtained from IPA.
2.7 Immunofluorescence
Immunofluorescence (IF) was performed as previously described.16,17 Briefly, fresh-frozen right ventricular tissues were embedded into OCT containing compound and frozen using 2-Methyl butane in liquid nitrogen, cut into 5 µm sections, fixed in 4% formaldehyde for 10 min at room temperature. Following fixation sections were washed, washed twice with PBS for 5 min each, and blocked using 10% goat serum in PBS containing 0.3% Triton X-100 solution for 1 h at room temperature. The myocardial sections were then incubated with the primary antibodies in the presence of 1% bovine serum albumin (BSA) with 5% goat serum in PBS containing 0.5% Triton X-100 solution overnight at 4°C. All the primary antibodies along with dilutions are listed in Supplementary material online, Table S1. The corresponding secondary antibody conjugates were added the next day and the nuclei were counter-stained with 4′,6-diamidino-2-phenylindole (DAPI). The percentage of nuclei that stained for each protein was determined by counting at least 13 000–40 000 nuclei in each group and was compared.
Immuno-stained myocardial sections were imaged using a Zeiss microscope and Axiovision software at 40X magnification. Approximately 20–30 fields per each section were imaged and the images were converted to 8-bit grayscale. Stained pixel area was calculated for each image using Image J software by adjusting the threshold to the background. The distribution of pixel area was compared between control and ACM samples using density plots in R-Studio after normalizing for the sample size.
2.8 Immunoblotting
Immunoblotting was performed as published.16,17 In brief, 5–10 mg aliquots of right and left ventricular tissue samples from ACM and control hearts were homogenized in a RIPA buffer containing 0.5–1% SDS in the presence of protease and phosphatase inhibitors (cat #4693116001 and cat #49068459001, Roche Molecular Biochemicals, respectively). Protein extracts were quantitated by the detergent compatible protein assay kit (Biorad Cat# 500-0114) using a spectrophotometer set at 750 nm. Approximately 50–100 µg aliquots of protein lysates were denatured in a Laemmli sample loading buffer, loaded onto an SDS-PAGE gel, and transferred to a nitrocellulose membrane. Expressions of the proteins of interest were detected by probing the membranes with the specific primary antibodies and the corresponding HRP conjugated secondary antibodies. The list of all primary and secondary antibodies used in the study was reported in Supplementary material online, Table S1. The signal for each protein was normalized to the endogenous loading control protein GAPDH signal. Quantitation of western blot signal was performed using Image J software.
2.9 TUNEL assay
Apoptosis was detected in the control and ACM hearts as described previously.22 Briefly, thin myocardial sections were deparaffinized in xylene, rehydration in a series of decreasing ethanol gradients, washed in PBS, and treated with proteinase K (20 μg/mL) in 10 mM Tris pH 7.5 and 1 mg/mL BSA for 20 min at room temperature. Sections were incubated with terminal deoxynucleotidyl transferase dUTP nick end labelling (TUNEL) assay by In-Situ cell death detection Fluorescein kit (Roche Cat#11684795910) as per the manufacturer’s instructions for one hour at 37°C. Nuclei were counter-stained with 1 μg/mL 4′,6 Diamidino-2-phenylindole dihydrochloride (DAPI, Sigma-Aldrich St Louis, MO; Cat# D8417). Approximately 5000–10 000 nuclei were counted per sample and the percentage of nuclei stained for TUNEL was quantified using image J software.
2.10 Conventional statistical methods
Parametric variables were presented as mean and SD and the differences between the two groups were comparted by unpaired t test. Categorical and non-parametric variables were compared by Fisher’s exact and Mann–Whitney U test, respectively. Cumulative distributions were plotted by Kernel density plots and were compared by Kolmogorov–Smirnov test.
3. Results
3.1 Phenotypic characteristics
The discovery population, used for RNA-Seq, was comprised of members of three independent families with defined truncating mutations in the DSP gene, who were found to have ventricular arrhythmias in the setting of family screening triggered by the diagnosis of ACM in a first-degree relative. These individuals fulfilled the 2010 Task Force Criteria for the diagnosis of ACM (Supplementary material online, Table S2).23 None had symptoms or signs of heart failure and each had a normal right ventricle size and function on the echocardiogram and/or CMR imaging. The characteristics of one patient who had a mildly enlarged left ventricle and a mildly reduced LVEF but a normal right ventricular function and no clinical evidence of heart failure have been published.24 The other two individuals had normal left ventricular size and ejection fraction. Detailed characteristics of the patients are listed in Supplementary material online, Tables S2 and S3. Representative phenotypic data are shown in Supplementary material online, Figures S1 and S2.
Right and left ventricular samples were obtained from hearts of five victims of SCD and used in the validation studies. All were Caucasian male individuals with a mean (±SD) age of 37.4 ± 12.3 years. One case had a known diagnosis of ACM but four were diagnosed for the first time at autopsy. Phenotypic characteristics of these cases have been described in Supplementary material online, Figures S3–S6. None of the cases had a known history of heart failure and all were asymptomatic, except for one individual who had an episode of syncope due to ventricular arrhythmias (Supplementary material online, Figures S3–S6). Phenotypic data in one case have been published and therefore, were not presented.14
Right and left ventricular samples from five explanted hearts from individuals who had died from non-cardiovascular causes and were not known to have cardiovascular diseases or chronic health conditions, such as cancer, were included as controls. The mean age of the group was 31.6 ± 12.8 years. Three were of Caucasian ethnicity (two male and one female) and two were Hispanic (both male).
3.2 Mutations
Cases in the discovery population had three distinct truncating variants in the DSP gene (Supplementary material online, Table S2). Likewise, all five cases in the validation population carried PVs in genes known to cause ACM, including DSP (c.2878-46_6599del, large deletion including exons 21–23), PKP2 (p.Lys672ArgfsTer12 and p. Arg79Ter), TMEM43 (p.Ser358Leu), and FLNC (p.Arg1370Ter), as listed in Supplementary material online, Table S3. Two cases carried additional variants, which were categorized as variants of unknown significance, in genes encoding desmosome proteins (DSC2: p. Thr358Ile; DSP:IVS2 + 5G>A and DSG2:c.523 + 2_523 + 3insT). These variants are listed in each pedigree.
3.3 RNA-Seq
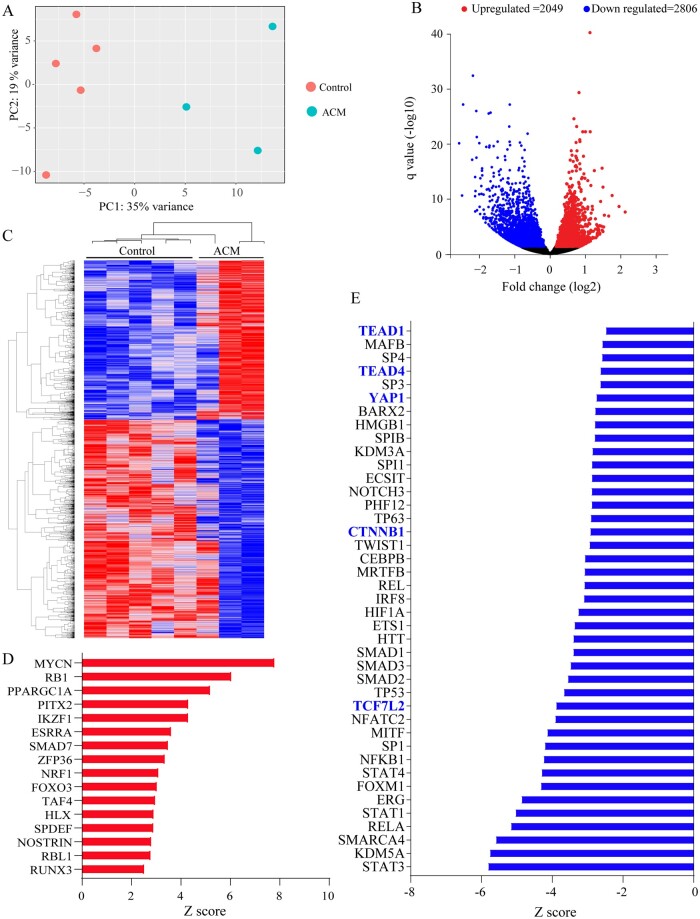
Strand-specific RNA-Seq was performed on ribosome-depleted RNA extracts from three ACM and five control samples. PCA plot showed distinct segregation of transcripts in the ACM and control samples (Figure 1A). There were 4855 differentially expressed genes (DEGs), comprised of 2806 down-regulated and 2049 up-regulated genes, between the ACM and control samples. The corresponding volcano plot and the heat map of the DEGs are depicted in Figure 1B and C, respectively. To assess the effects of the truncating mutations on DSP expression, transcript levels of DSP were compared between control and ACM hearts, which showed ∼60% reduction in the ACM hearts (Control: 67 362 ± 5006 cpm vs. ACM: 25 735 ± 5157, P = 0.0008).
Figure 1.
RNA-Seq of endomyocardial biopsy samples from ACM and control hearts showing differentially expressed genes and dysregulated biological pathways. (A) Principle component analysis (PCA) plots showing distinct separation of the ACM and control samples. Each dot represents one sample. (B) Volcano plot showing differentially expressed genes (DEGs, q < 0.05). Down-regulated genes are shown in blue and up-regulated ones in red. Those shown in black reflect genes whose expression levels were unchanged. (C) Heat map of the DEGs, blue indicated low and red high transcript levels. (D) Transcriptional regulators (TRs), which were predicted to be activated upon IPA analysis of the DEGs. Only TRs that showed an enrichment P value of <0.05 and a Z score of > 2 are shown. (E) Panel depicts TRs that were predicted to be suppressed (enrichment P value of <0.05 and a Z score of <−2). Those highlighted in blue are major transcriptional effector of the Hippo pathway (YAP1, TEAD1, and TEAD4) or canonical WNT signalling (TCF7L2 and CTNNB1) pathways.
3.4 Dysregulated TRs
The DEGs were analysed to identify TRs that were responsible for differential gene expression in the ACM samples. The up-regulated genes predicted activation of MYCN, RB1, and PPARGC1A among others, whereas the down-regulated genes predicted suppression of STAT3, KDM5A, and SMARCA4 among others (Figure 1D and E). Pertinent to the pathways implicated in the pathogenesis of ACM, the DEGs predicted suppression of gene expression through YAP1, TEAD1, and TEAD4, TRs of the Hippo pathway (Figure 1E). Likewise, gene expression through TCF7L2 and CTNNB1, the TRs of the cWNT pathway, was predicted to be suppressed (Figure 1E).
The DEGs comprised 642 genes whose protein products are expected to be secreted, i.e. secretome (Supplementary material online, Figure S7). Notable among the differentially expressed secretome were cytokines PF4 (11.5-fold increase, q < 0.0001), CXCL5 (2.82-fold increase, q < 0.0001) and CCL5 (2.2-fold increase, q = 0.002), as well as growth and mitotic factor FGF18 (2.7-fold increase, q < 0.0001). The DEGs coding for the secretome predicted activation of PPARGC1A, SMAD7, and KLF2 and suppression of STATs, TEADs, YAP1, and CTNNB1, among others (Supplementary material online, Figure S7B). Clustering of the genes coding for the secretome based on their biological functions identified those involved in the regulation of cell migration and motility, cytokine-mediated signalling, and mitochondrial respiratory chain complex among the largest clusters (Supplementary material online, Figure S7C).
3.4.1 Suppression of gene expression through the Hippo pathway
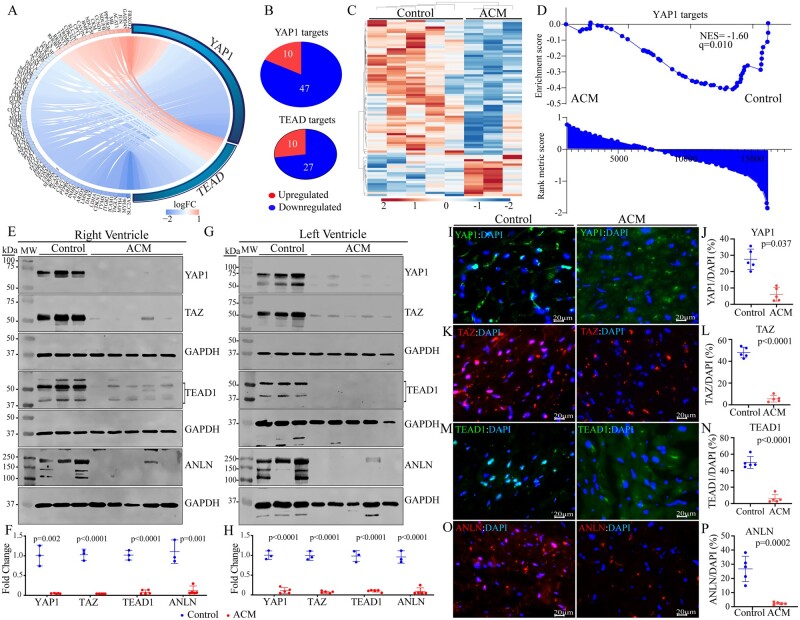
To expand on the RNA-Seq data predicting suppression of the YAP1/TEAD transcriptional activities, transcript levels of DEGs that are known to be regulated by Hippo pathway were compared between the ACM and control hearts, which showed reduced levels in the ACM hearts (Figure 2A). Among the DEGs, expression levels of 74 YAP1/TEADs target genes were reduced, whereas transcript levels of 20 target genes were increased (Figure 2B, P < 0.0001). A heat map of the Hippo pathway target genes showing suppression of the majority of the transcripts in the endomyocardial biopsy samples in ACM is presented in Figure 2C. Likewise, GSEA predicted suppression of gene expression through YAP1 in the ACM hearts (Figure 2D).
Figure 2.
Suppression of the Hippo pathway, predicted from the RNA-Seq data. (A) Circos map of the genes regulated by the YAP1 and TEAD TRs of the Hippo pathway. Connecting lines show common and unique genes and are coloured based on the status of fold change in gene expression in ACM vs Control samples. Blue indicates suppression and red activation of gene expression. (B) Pie charts showing the number of suppressed and activated genes for each TR obtained from IPA. (C) Heat map of the YAP-TEAD targets showing reduced transcript levels (blue) of the majority of the genes in this pathway, as opposed to increased transcript levels (red) in ACM as compared to control hearts. Each lane represents one sample. (D) Gene set enrichment analysis (GSEA) plot showing enrichment of the YAP1 targets among the suppressed genes. GSEA plots are shown with corresponding normalized enrichment score (NES) and the q value. (E–H) Immunoblotting and the corresponding quantitative data (normalized to GAPDH) showing reduced levels of YAP1, TAZ, TEAD1 isoforms, and ANLN proteins in the right (E and F) and left (G and H) ventricular ACM heart samples as compared to control hearts. P-values were calculated by unpaired t test. (I–P) Immunofluorescence staining of thin myocardial sections for YAP1 (I), TAZ (K), TEAD1 (M), and ANLN (O) of the Hippo pathway in the control and ACM hearts. Quantitative data showing the percentage of nuclei stained for the selected Hippo pathway proteins in the five control and five ACM hearts (J, L, N, and P). Between 13 000 and 40 000 nuclei per group were counted for this analysis and P values were determined by unpaired t test.
To test for validation of the RNA-Seq findings, expression levels of selected Hippo pathway proteins were assessed by immunoblotting in right and left ventricular samples from five independent victims of SCD. Expression levels of YAP1, TAZ (WWTR1), and TEAD1, key downstream effectors of the Hippo pathway, were reduced in the right and left ventricular samples from the ACM hearts, as compared to the corresponding ventricular tissues from the control hearts (Figure 2E–H). Similarly, expression levels of ANLN, a Hippo pathway target involved in regulation of cytoskeleton, were reduced (Figure 2E–H). The selected Hippo pathway proteins are known to be localized to the nucleus, enabling unequivocal detection of their expression and localization. Therefore, nuclear localization of YAP1, TAZ, TEAD1, and ANLN was analysed by immunofluorescence staining of thin myocardial sections. The number of cells stained positive for the expression of each Hippo pathway protein was reduced markedly in the ACM as compared to control heart samples (Figure 2I–P). Thus, the findings of RNA-Seq, immunoblotting, and immunofluorescence staining were concordant in showing suppression of the transcriptional activity of the Hippo pathway and were present in both ventricles, irrespective of chamber-specific histological involvement.
3.4.2 Suppression of gene expression through the cWNT pathway
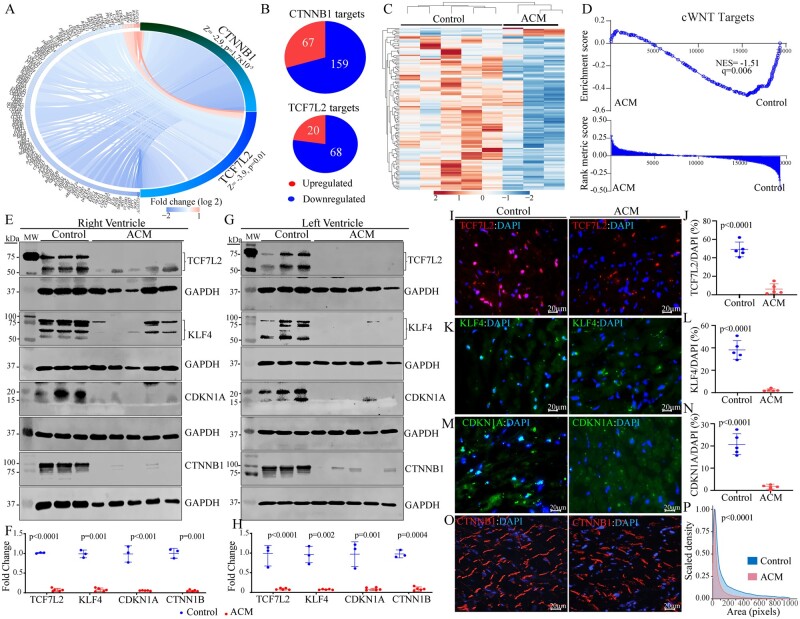
Given that the DEGs predicted suppression of transcriptional activities of the CTNNB1 and TCF7L2, the co-effector of the cWNT pathway (Figure 1E), transcript levels of CTNNB1 and TCF7L2 target genes were compared in the control and ACM hearts. A circos map of the cWNT pathway target genes regulated by the CTNNB1 and TCF7L2 transcriptional co-effectors illustrated significant down-regulation of the transcript levels in the ACM hearts (Figure 3A). Of the 226 known CTNNB1 target genes, as listed in the IPA dataset, 159 (70%) had lower expression levels in the ACM hearts (Figure 3B, P = 0.0002). Likewise, 68/88 (77%) of the TCF7L2 target genes were suppressed in the ACM hearts (Figure 3B, P = 0.0002). A heat map of the cWNT pathway target genes is depicted in Figure 3C, which illustrated down-regulation of the transcript levels of the majority of the cWNT target genes. Finally, the DEGs were analysed by GSEA, which showed enrichment of the cWNT target genes among the down-regulated genes in the ACM hearts (Figure 3D).
Figure 3.
Suppression of the canonical WNT (cWNT) pathway, predicted from the RNA-Seq data. (A) Circos map of the genes regulated by the CTNNB1 and TCF7L2 TRs of the cWNT pathway. Blue indicates suppression and red activation of gene expression. (B) Pie charts showing the number of suppressed and activated genes obtained from IPA for each TR. (C) Heat map of the cWNT pathway targets showing reduced transcript levels (blue) of the majority of the genes in this pathway, as opposed to increased transcript levels (red) in ACM as compared to control hearts. Each lane represents one sample. (D) Gene set enrichment analysis (GSEA) plot showing enrichment of the cWNT targets among the suppressed genes. (E–H) Immunoblotting showing reduced levels of both isoforms of TCF7L2 and KLF4 proteins CDKN1A, and CTNNB1 in the right (E and F) and left (G and H) ventricular ACM heart samples as compared to control hearts. Quantitative data corresponding to immunoblots in panels F and H (normalized to GAPDH). The p value was determined by unpaired t test. (I–P) Immunofluorescence staining of thin myocardial sections for TCF7L2 (I), KFL4 (K), CDKN1A (M), and CTNNB1 (O) of the cWNT pathway in the control and ACM hearts. Quantitative data showing the percentage of nuclei stained for the selected cWNT pathway proteins in the control and ACM hearts (J, L, and N). Between 13 000 and 40 000 nuclei per heart were counted to obtain % positive nuclei. The P value was determined by unpaired t test. (P) Kernel density map showing quantitative data on the mean cross-sectional areas stained for CTNNB1 protein in the ACM and control hearts. Approximately 10 000 CTNNB1-stained areas per sample were counted and Kernel size distribution density maps were compared by Kolmogorov–Smirnov test.
To test for validation of suppressed cWNT pathway in ACM, as predicted by the RNA-Seq data, expression levels of selected cWNT signalling pathway molecules, namely TCF7L2, KLF4, CDKN1A, and CTNNB1, were examined by immunoblotting in the right and left ventricular samples. Levels of TCF7L2 isoforms, KLF4 isoforms, CDKN1A, and CTNNB1 were markedly reduced in the right and left ventricular tissues in the ACM hearts as compared to controls (Figure 3E–H). Immunostaining of thin myocardial sections corroborated the findings by showing reduced number of cells with nuclear localization of TCF7L2, KLF4, and CDKN1A (Figure 3I–N) and altered localization and reduced size density of CTNNB1 (Figure 3O and P) in the ACM hearts.
3.5 Dysregulated biological pathways
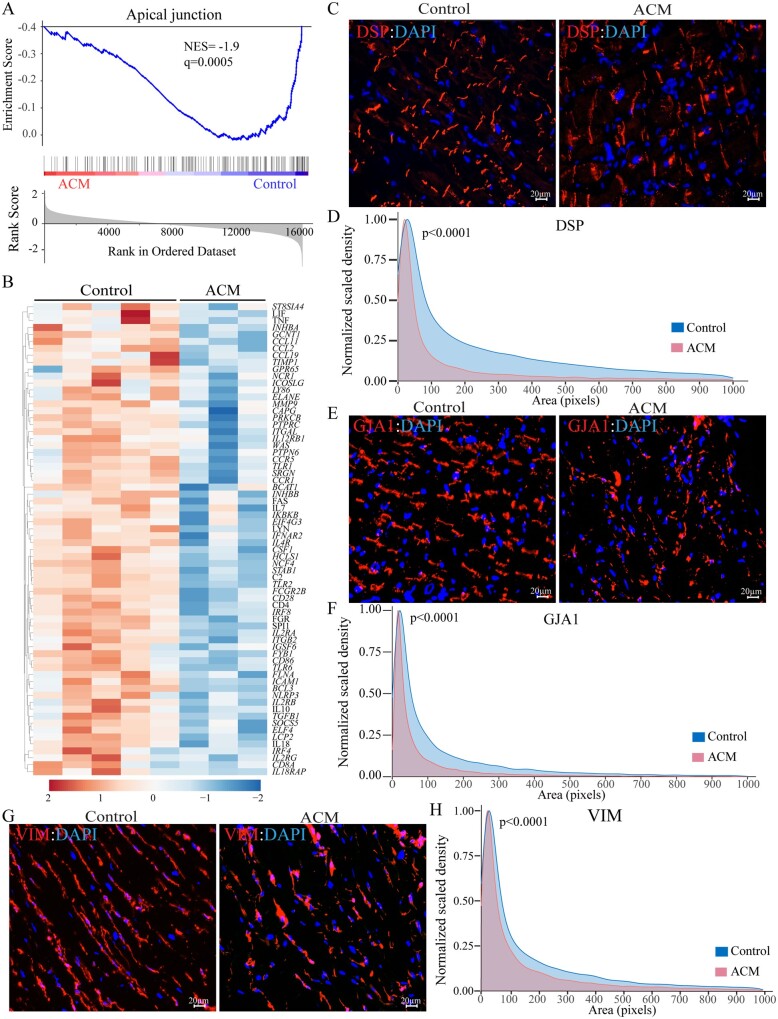
The DEGs predicted adipogenesis as the most activated biological pathway and apical junction, representing cell–cell attachment, as the most suppressed pathways in the ACM hearts (Supplementary material online, Figure S8 and Figure 4A). The predicted dysregulated biological pathways also included fatty acid metabolism and oxidative phosphorylation, which were activated, whereas inflammatory responses and IL6/JAK/STATs were predicted to be suppressed (Supplementary material online, Figure S8). In accord with truncating DSP mutations in the ACM patients, the apical junction pathway was suppressed in the ACM as compared to control hearts, depicted in the GSEA plot, showing depletion of the DEGs in the ACM group (Figure 4A). Similarly, the heat map showed reduced transcript levels of DEGs representing the apical junction pathway (Figure 4B).
Figure 4.
Deranged structure of the intercalated disc and intermediary filaments. (A) GSEA plot of apical junction, which was the most suppressed biological pathway in ACM hearts. (B) Heat map corresponding to genes involved in apical junction, which shows predominance of the suppressed genes (blue). (C) Myocardial areas stained for the expression of DSP, representing structure of the desmosomes, in the ACM hearts. (D) Kernel density map showing quantitative data on the mean cross-sectional areas stained for DSP protein in the ACM and control hearts. Approximately 10 000 desmosomes per sample were counted and Kernel size distribution density maps were compared by Kolmogorov–Smirnov test. (E) Myocardial areas stained for the expression of GJA1 protein, representing gap junction at the cell–cell junction, in the ACM hearts. (F) Kernel density map of GJA1 distribution depicting the mean cross-sectional myocardial areas stained for the GJA1 protein expression in the ACM and control hearts. Statistical analysis was as in D. (G) Myocardial areas stained for the expression of VIM protein, representing the intermediate filaments structure in the ACM hearts. (H) Kernel size density map of VIM-stained myocardial areas in the ACM and control hearts. Statistical analysis was as in D.
The DEGs were also analysed by IPA to predict the dysregulated canonical signalling pathways, as listed in the IPA database, in the human hearts with ACM. The results, shown in Supplementary material online, Figure S9, are notable for suppression of over a dozen and activation of a few pathways in the hearts from patients with ACM.
3.6 Altered ID and cytoskeletal structures
The RNA-Seq data predicted suppression of the ‘apical junction’, a term used in the GSEA parlance to describe cell–cell adhesion, which in cardiac myocytes includes the IDs (Figure 4A). To assess the effects of the PVs on the ‘apical junction’, thin myocardial sections were stained with antibodies against selected proteins, as representative of the IDs, cytoskeletal, and intermediate filament proteins. There were marked differences in the size of the areas stained for the expression of DSP in the ACM as compared to control hearts, which was depicted as a leftward shift in the size-density plot, indicating smaller sizes in the ACM tissues (Figure 4C and D). Likewise, size-density distribution of GJA1 (connexin 43), which localizes to the IDs, was significantly altered in the ACM hearts (Figure 4E and F). In accord with the structural changes in the IDs, orderly organization of the cytoskeletal and intermediate filament protein VIM (vimentin), which is known to be connected to the IDs, was altered in the ACM hearts, as reflected in a shift toward smaller sizes in the kernel density plots (Figure 4G and H).
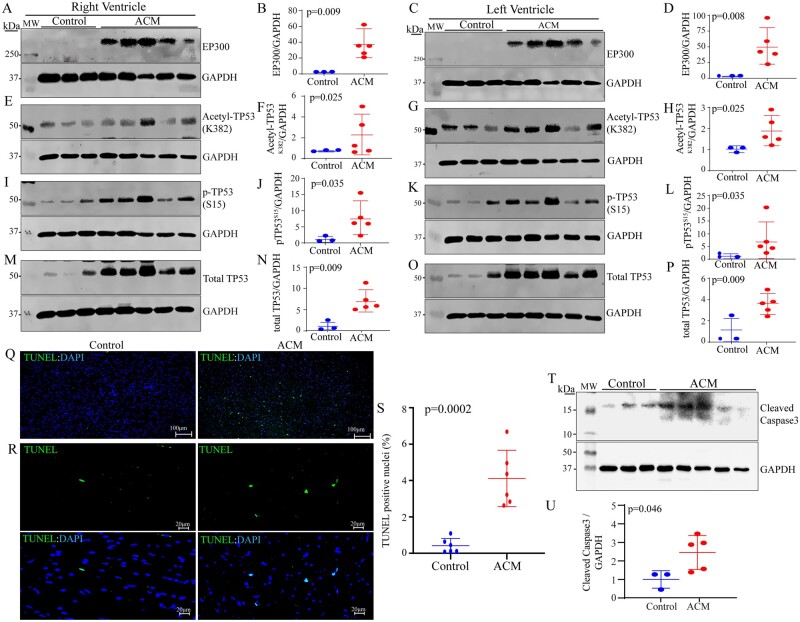
3.7 Activation of EP300-TP53 pathway
In view of the role of the cell junction in regulating the mechanosensitive signalling pathways, including the Hippo and cWNT pathways, expression of acetyltransferase EP300, which is regulated by the mechanical stress and known to interact with the Hippo and cWNT pathways, was analysed in the right and left ventricular ACM and control samples.25–27 Levels of the EP300 protein were markedly increased in the right as well as left ventricular myocardium in the ACM hearts as compared to controls (Figure 5A–D). Given that EP300 acetylates its target proteins, levels of acetylated TP53 (acetyl-TP53K382), the best-established target of EP300, were analysed by immunoblotting, which showed increased levels in the right and left ventricular samples (Figure 5E–H). In accord with increased levels of acetyl-TP53K382, levels of phosphorylated TP53 at serine residue 15 (pTP53S15) were also increased in the samples from both ventricles in the ACM hearts, as compared to the controls (Figure 5I–L). Furthermore, levels of total TP53 were also increased in the ACM tissues from both ventricles (Figure 5M–P).
Figure 5.
Activation of the EP300-TP53 pathway in the ACM hearts. (A–P) Immunoblots showing increased levels of EP300 proteins, acetyl-TP53 (K382), phospho (S15)-TP53, and total TP53 in the right and left ventricular samples in the ACM hearts as compared to controls. The corresponding quantitative data, normalized to GAPDH, are shown next to each panel. (Q–R) Increased myocardial apoptosis in the ACM hearts as compared to controls (Q: low magnification and R: high magnification). The corresponding quantitative data are shown in S. (T–U) Immunoblots showing increased levels of the cleaved (activated) Caspase 3 (CASP3) and the corresponding loading control GAPDH in the ACM hearts (T). Quantitative data are shown in U.
Given the well-established role of TP53 in apoptosis, thin myocardial sections were analysed by the TUNEL assay, which showed increased number of nuclei staining positive for the assay in the ACM hearts (Figure 5Q–S). To complement the findings of the TUNEL assay, levels of cleaved Caspase 3 (CASP3) were analysed by immunoblotting, which corroborated the findings by showing increased levels in the ACM hearts (Figure 5T and U).
4. Discussion
Analysis of the transcripts by RNA-Seq in the endomyocardial biopsy samples from human patients with defined truncating mutations in the DSP gene and normal right ventricular size and function, led to identification of over two-dozen dysregulated TRs, including the Hippo/cWNT pathways. The findings of the RNA-Seq were corroborated by immunoblotting and immunofluorescence studies in independent autopsy-proven ACM samples obtained from the right and left ventricles of five victims of SCD who carried PVs in ACM genes but had no prior history of heart failure. A novel finding was activation of the EP300 protein, which is an acetyltransferase known to interact with and suppress the cWNT/Hippo pathways.26,27 In conjunction with the activation of EP300, the TP53 protein, a bona fide target of EP300, was acetylated and its phosphorylated levels were increased. Activation of EP300-TP53 pathway and suppression of the Hippo/cWNT pathways were detected in the right and left ventricular ACM samples, irrespective of the ventricle showing fibroadiposis, and occurred in the absence of heart failure. Given the well-established role of these regulators of gene expression in various biological processes pertaining to the phenotype in ACM, such as apoptosis, cell size, cell cycle regulation, and fibro-adipogenesis, the findings suggest their pathogenic involvements in ACM.
Whereas identification of activation of the EP300-TP53 pathway in ACM is novel, the findings on suppression of the Hippo/cWNT transcriptional activities, in the absence of overt heart failure, are also novel and extend the previous findings in ACM patients with advanced heart failure.13 The findings were validated by complementary methods at the RNA and proteins levels, the latter in the right and left ventricular ACM samples, and were corroborated by detection of nuclear localization of the selected proteins. Both the discovery samples, used for RNA-Seq and the validation samples were obtained from individuals with defined PVs in the ACM genes, which were shown to co-segregate with the phenotype in the families or are published.24 The biological effects of the PVs were evident by ∼50% reduction in the DSP mRNA levels, consistent with haplo-insufficiency. The cases were members of families who exhibited either left-dominant ACM or isolated involvement of the left ventricle, which further expand the role of the dysregulated TRs to the broader spectrum of ACM. Collectively, the data suggest that activation of the EP300/TP53 and suppression of gene expression through the Hippo/cWNT pathways are independent of the confounding effects of heart failure and therefore, likely pathogenic in ACM. Moreover, identification of over two dozen dysregulated TRs in the absence of overt heart failure in ACM indicates the complexity of the molecular pathways that are directly or indirectly regulated by altered cell–cell junctions in ACM.
The putative mechanisms responsible for activation of the EP300-TP53 and suppression of gene expression through the Hippo/cWNT pathways were not explored but likely pertain to involvement of their components in the apical junctions, which encompass the desmosomes among other cell–cell junction structures.13 One might posit that mutations in genes encoding the desmosome proteins by affecting integrity of the apical junction in sensing the mechanical stress of myocardial contraction and relaxation, dysregulate these mechanosensitive pathways. The notion was supported by the RNA-Seq data, which identified the apical junction pathway, a Hallmark term for GSEA, as the most suppressed biological pathway in the ACM hearts. Indeed, ACM, mainly a disease of apical junction, is the prime model of dysregulation of mechanosensing and mechanotransduction in the heart.13 In agreement with this notion, experimental data point to impaired mechanosensing in cells defective in the desmosome proteins.28 Moreover, the upstream molecules of the Hippo/cWNT pathways are known to be localized to the apical junctions, including the IDs, and are dysregulated in ACM.13,29
Impaired signal transduction at the apical junction is further complemented with the complex multi-stage interactions among the components of these pathways and consequent suppressive effects of activation of the EP300/TP53 on the Hippo/cWNT signalling pathways.26,27,30,31 For example, TAZ, whose levels were decreased in the ACM hearts, is known to inhibit the EP300-mediated acetylation of TP53 and EP300 is known to inhibit the cWNT pathway.26,27 Likewise, TAZ inhibits phosphorylation of dishevelled (DVL), resulting in suppression of the cWNT pathway.31 Moreover, MST1/2, a key kinase in the Hippo pathway, inactivates DVL2 and suppresses the cWNT pathway.32 Furthermore, LATS1/2, downstream targets of MAST1/2 in the Hippo pathway, are essential for transcriptional activity of the cWNT pathway.33 In addition, YAP1 and CTNNB1 are known to interact both in the cytoplasm and in the nucleus.13 Whereas the cytoplasmic interaction results in sequestration of YAP1 and CTNNB1 and suppression of gene expression through the Hippo/cWNT pathways, the nuclear interaction has the opposite effects, as it leads to increased transcription activities of the Hippo and cWNT pathways.30,34,35 The net effect of complex interactions among the mechanosensing pathways in ACM is activation of EP300/TP53 and suppression of transcriptional activities of the Hippo/cWNT pathways.
The study has a number of limitations. The sample size of study population is small (a total of eight ACM cases), which was constrained partly because of rarity of the disease and the inherent challenges in obtaining endomyocardial biopsy in patients with ACM, given the potential risk associated with performing an invasive procedure. In addition, it is important to note that the RNA-Seq was performed in the RNA extracted from the right ventricular endomyocardial biopsy, whereas the control and validation samples were from the right and left ventricular transmural tissues. Consequently, differences in the myocardial regional gene expressions could confound the findings, even though the findings in the discovery and validation studies were concordant. Likewise, the discovery cases had internal defibrillator/cardioverter implanted, which could cause local fibrosis and potentially affect gene expression in the endomyocardial samples. Mutations in the DSP gene are known to cause left-dominant ACM, often resembling dilated cardiomyopathy.9,36–40 Despite the chamber-specific differences in the phenotypic expression, DSP and other desmosome proteins are expressed and desmosomes are formed in both ventricles. The concordant findings in the right and left ventricles provide further credence to independence of these changes from cardiac dysfunction, in keeping with the findings on CMR and echocardiography.
Some of the validation samples exhibited pathological features of ACM in one of both ventricles, including fibro-adipocytic infiltration of the myocardium, which could also confound the RNA-Seq findings. To reduce such effects tissues from the apparently unaffected ventricles were used in the validation studies. Nevertheless, microscopic histological changes could remain undetected and affect gene expression. Tissue obtained by the endomyocardial biopsy is scant in amount and typically insufficient for complementary molecular studies. Despite these drawbacks, good quality RNA and a high number of unique sequence reads were obtained in the RNA-Seq studies, which enabled a reliable identification of the DEGs and prediction of the dysregulated TRs and biological pathways. Furthermore, the disease commonly starts from the epicardium of the right ventricle, which might differ in gene expression from the endocardial region. Epicardial and left ventricular tissues, however, are seldom available from ACM patients, unless at autopsy or in the explanted hearts of patients who have undergone heart transplantation.
Whereas the endomyocardial biopsy samples were fresh and yielded excellent quality RNA, the right and left ventricular tissues, which were used for the validation studies, were obtained post-mortem. Consequently, partial post-mortem protein degradation might have occurred, which could confound the findings. Post-translational protein modifications, such as phosphorylation, often affect protein stability, consistent with increased levels of TP53-K382Ac and pTP53S15 as well as levels of the EP300 acetyltransferase. In addition, protein levels were presented as normalized values to the house-keeping proteins. While some level of post-mortem degradation could not be excluded, considering the above, it is unlikely that the observed changes in the levels of the Hippo and cWNT pathway proteins are mainly due to post-mortem degradation.
It is important to recognize that the findings are descriptive and the links among various components of the EP300/TP53 and Hippo/cWNT pathways are associative, mainly supported by the existing data.26,27,30,31 The small sample size of the study population hinders correlation between changes in the gene expression with the histological, genetic, or clinical phenotypes in patients with ACM. Moreover, two of the validation cases had PVs in FLNC and TMEM43 gene, which might involve different or additional sets of mechanisms from those pertaining to PVs in DSP gene.
The phenotype in ACM, comprised mainly of cardiac arrhythmias, heart failure, and fibrofatty infiltration of the myocardium, is complex, involving a diverse array of biological pathways and mechanisms in its pathogenesis. The RNA-Seq data in this study also predicted dysregulation of over a dozen transcriptional regulators and canonical signalling pathways in the human hearts with ACM. The findings as well as the previous data implicate involvement of a large number of upstream pathways, in addition to the EP300/TP53/Hippo/cWNT pathways, such as integrins, receptor tyrosine kinases, and the transforming growth factor β in the pathogenesis of the phenotype in ACM.28,41–43
In conclusion, the findings, validated in independent samples and by complementary methods, indicate activation of the EP300/TP53 and suppression of gene expression through the Hippo/cWNT pathways in human hearts with ACM caused by defined PVs in the absence of overt heart failure. The findings support the pathogenic roles of the EP300/TP53 and the Hippo/cWNT pathways in ACM, including in left dominant ACM.
Data availability
RNA-Seq data have been deposited in the public database GEO (GSE156869). Additional data pertaining to the present article are available from the corresponding author upon request.
Supplementary material
Supplementary material is available at Cardiovascular Research online.
Authors’ contributions
L.R. performed the western blotting and immunofluorescent studies. S.F. extracted RNA and protein from the tissue samples and performed the initial experiments. S.M.C. performed some of the western blotting and immunofluorescent studies. A.B.-B., M.S.M., Y.Y., J.R.G., P.M., and E.Z. performed clinical and pathological evaluation of the patients, genetic studies, and edited the manuscript. C.C. and M.J.R. performed the bioinformatics analysis of the RNA-sequencing data. P.G. performed secondary bioinformatics analysis of RNA-Sequencing data, contributed to data analysis, and edited the manuscript. A.J.M. conceived the studies, supervised the experiments, and wrote the manuscript.
Supplementary Material
Acknowledgements
The authors wish to acknowledge Dr Nuria Mancheño, MD, PhD from the Pathology Department at Hospital Universitario y Politécnico La Fe de Valencia (Spain); Dr Joaquina Belchi MD, PhD from the Cardiology Department at Hospital General Universitario de Valencia (Spain); and Dr Ana Bel, MD from the Cardiovascular Surgery Department at Hospital Universitario y Politécnico La Fe de Valencia (Spain) for their assistance in procuring samples, clinical evaluations, and genetic testing.
Conflict of interest: All authors declares that they have no personal, professional, or financial matter related by the authors that might be construed as a conflict of interest.
Contributor Information
Leila Rouhi, Department of Medicine, Center for Cardiovascular Genetics, Institute of Molecular Medicine, University of Texas Health Sciences Center at Houston, 6770 Bertner Ave, Suite C900A, Houston, TX 77030, USA.
Siyang Fan, Department of Medicine, Center for Cardiovascular Genetics, Institute of Molecular Medicine, University of Texas Health Sciences Center at Houston, 6770 Bertner Ave, Suite C900A, Houston, TX 77030, USA.
Sirisha M Cheedipudi, Department of Medicine, Center for Cardiovascular Genetics, Institute of Molecular Medicine, University of Texas Health Sciences Center at Houston, 6770 Bertner Ave, Suite C900A, Houston, TX 77030, USA.
Aitana Braza-Boïls, Unidad de Cardiopatías Familiares, Muerte Súbita y Mecanismos de Enfermedad (CaFaMuSMe), Instituto de Investigación Sanitaria La Fe, Valencia, Spain; Center for Biomedical Network Research on Cardiovascular Diseases (CIBERCV), Madrid, Spain.
Maria Sabater Molina, Department of Genetics and Microbiology, Cardiogenetic Laboratory, Instituto Murciano de Investigación Biosanitaria, Murcia, Spain.
Yan Yao, Department of Cardiology, Fuwai Hospital, Peking Union Medical College, Beijing, China.
Matthew J Robertson, Department of Cell Biology, Baylor College of Medicine, Houston, TX 77030, USA.
Cristian Coarfa, Department of Cell Biology, Baylor College of Medicine, Houston, TX 77030, USA.
Juan R Gimeno, Center for Biomedical Network Research on Cardiovascular Diseases (CIBERCV), Madrid, Spain; Department of Cardiology, Unidad CSUR Cardiopatias Familiares, Hospital Universitario Virgen de la Arrixaca, Murcia, Spain.
Pilar Molina, Unidad de Cardiopatías Familiares, Muerte Súbita y Mecanismos de Enfermedad (CaFaMuSMe), Instituto de Investigación Sanitaria La Fe, Valencia, Spain; Servicio de Patologia, Instituto de Medicina Legal y Ciencias Forenses de Valencia, Histology Unit at the Universitat de València, Valencian, Spain.
Priyatansh Gurha, Department of Medicine, Center for Cardiovascular Genetics, Institute of Molecular Medicine, University of Texas Health Sciences Center at Houston, 6770 Bertner Ave, Suite C900A, Houston, TX 77030, USA.
Esther Zorio, Unidad de Cardiopatías Familiares, Muerte Súbita y Mecanismos de Enfermedad (CaFaMuSMe), Instituto de Investigación Sanitaria La Fe, Valencia, Spain; Center for Biomedical Network Research on Cardiovascular Diseases (CIBERCV), Madrid, Spain; Department of Cardiology, Unidad de Cardiopatías Familiares, Servicio de Cardiología, Hospital Universitario y Politécnico La Fe, Valencia, Spain.
Ali J Marian, Department of Medicine, Center for Cardiovascular Genetics, Institute of Molecular Medicine, University of Texas Health Sciences Center at Houston, 6770 Bertner Ave, Suite C900A, Houston, TX 77030, USA.
Funding
The work was in part supported by grants from the Instituto de Salud Carlos III and FEDER Union Europea, Una forma de hacer Europa (PI18/01231 and PI18/01582) and (PI18/01231); FEDER Union Europea, Una forma de hacer Europa (PI18/01582) and La Fe Biobank (PT17/0015/0043). Memorial Nacho Barberá. National Institutes of Health (NIH), National Heart, Lung and Blood Institute (NHLBI: 1R01HL132401 and R01HL151737). NIH (S10 OD018135). Leducq Foundation (14 CVD 03). NIA (R21AG060413-01). The Ewing Halsell Foundation, George and Mary Josephine Hamman Foundation, and TexGen Fund from Greater Houston Community Foundation.
Translational perspective
The findings suggest that altered mechanosensing at the cell–cell junction instigates a cascade of molecular events through the activation of acetyltransferase EP300/TP53 and suppression of gene expression through the Hippo/canonical WNT pathways in human arrhythmogenic cardiomyopathy (ACM) caused by defined mutations. These molecular changes occur early and in the absence of overt heart failure. Consequently, one may envision cell type-specific interventions to target the dysregulated transcriptional, mechanosensing, and mechanotransduction pathways to prevent the evolving phenotype in human ACM.
References
- 1. Corrado D, Link MS, Calkins H. Arrhythmogenic right ventricular cardiomyopathy. N Engl J Med 2017;376:61–72. [DOI] [PubMed] [Google Scholar]
- 2. Gandjbakhch E, Redheuil A, Pousset F, Charron P, Frank R. Clinical diagnosis, imaging, and genetics of arrhythmogenic right ventricular cardiomyopathy/dysplasia: JACC State-of-the-Art Review. J Am Coll Cardiol 2018;72:784–804. [DOI] [PubMed] [Google Scholar]
- 3. Groeneweg JA, Bhonsale A, James CA, Te Riele AS, Dooijes D, Tichnell C, Murray B, Wiesfeld AC, Sawant AC, Kassamali B, Atsma DE, Volders PG, de Groot NM, de Boer K, Zimmerman SL, Kamel IR, van der Heijden JF, Russell SD, Jan Cramer M, Tedford RJ, Doevendans PA, van Veen TA, Tandri H, Wilde AA, Judge DP, van Tintelen JP, Hauer RN, Calkins H. Clinical presentation, long-term follow-up, and outcomes of 1001 arrhythmogenic right ventricular dysplasia/cardiomyopathy patients and family members. Circ Cardiovasc Genet 2015;8:437–446. [DOI] [PubMed] [Google Scholar]
- 4. Thiene G. The research venture in arrhythmogenic right ventricular cardiomyopathy: a paradigm of translational medicine. Eur Heart J 2015;36:837–846. [DOI] [PubMed] [Google Scholar]
- 5. Corrado D, Basso C, Schiavon M, Thiene G. Screening for hypertrophic cardiomyopathy in young athletes. N Engl J Med 1998;339:364–369. [DOI] [PubMed] [Google Scholar]
- 6. Mast TP, Teske AJ, Vd Heijden JF, Groeneweg JA, Te Riele AS, Velthuis BK, Hauer RN, Doevendans PA, Cramer MJ. Left ventricular involvement in arrhythmogenic right ventricular dysplasia/cardiomyopathy assessed by echocardiography predicts adverse clinical outcome. J Am Soc Echocardiogr 2015;28:1103–1113.e1109. [DOI] [PubMed] [Google Scholar]
- 7. Corrado D, Basso C, Judge DP. Arrhythmogenic cardiomyopathy. Circ Res 2017;121:784–802. [DOI] [PubMed] [Google Scholar]
- 8. van Tintelen JP, Entius MM, Bhuiyan ZA, Jongbloed R, Wiesfeld AC, Wilde AA, van der Smagt J, Boven LG, Mannens MM, van Langen IM, Hofstra RM, Otterspoor LC, Doevendans PA, Rodriguez LM, van Gelder IC, Hauer RN. Plakophilin-2 mutations are the major determinant of familial arrhythmogenic right ventricular dysplasia/cardiomyopathy. Circulation 2006;113:1650–1658. [DOI] [PubMed] [Google Scholar]
- 9. Castelletti S, Vischer AS, Syrris P, Crotti L, Spazzolini C, Ghidoni A, Parati G, Jenkins S, Kotta MC, McKenna WJ, Schwartz PJ, Pantazis A. Desmoplakin missense and non-missense mutations in arrhythmogenic right ventricular cardiomyopathy: genotype-phenotype correlation. Int J Cardiol 2017;249:268–273. [DOI] [PubMed] [Google Scholar]
- 10. Broussard JA, Getsios S, Green KJ. Desmosome regulation and signaling in disease. Cell Tissue Res 2015;360:501–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Nitoiu D, Etheridge SL, Kelsell DP. Insights into desmosome biology from inherited human skin disease and cardiocutaneous syndromes. Cell Commun Adhes 2014;21:129–140. [DOI] [PubMed] [Google Scholar]
- 12. Garcia-Gras E, Lombardi R, Giocondo MJ, Willerson JT, Schneider MD, Khoury DS, Marian AJ. Suppression of canonical Wnt/beta-catenin signaling by nuclear plakoglobin recapitulates phenotype of arrhythmogenic right ventricular cardiomyopathy. J Clin Invest 2006;116:2012–2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chen SN, Gurha P, Lombardi R, Ruggiero A, Willerson JT, Marian AJ. The hippo pathway is activated and is a causal mechanism for adipogenesis in arrhythmogenic cardiomyopathy. Circ Res 2014;114:454–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Molina P, Sanz-Sanchez J, Fenollosa M, Martinez-Matilla M, Giner J, Zorio E. Arrhythmogenic cardiomyopathy with left ventricular involvement versus ischemic heart disease: lessons learned from the family study and the reviewed autopsy of a young male. Forensic Sci Res 2019;4:274–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Cheedipudi SM, Hu J, Fan S, Yuan P, Karmouch J, Czernuszewicz G, Robertson MJ, Coarfa C, Hong K, Yao Y, Campbell H, Wehrens X, Gurha P, Marian AJ. Exercise restores dysregulated gene expression in a mouse model of arrhythmogenic cardiomyopathy. Cardiovasc Res 2020;116:1199–1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Auguste G, Rouhi L, Matkovich SJ, Coarfa C, Robertson MJ, Czernuszewicz G, Gurha P, Marian AJ. BET bromodomain inhibition attenuates cardiac phenotype in myocyte-specific Lamin A/C-deficient mice. J Clin Invest 2020;130:4740–4758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Chen SN, Lombardi R, Karmouch J, Tsai JY, Czernuszewicz G, Taylor MRG, Mestroni L, Coarfa C, Gurha P, Marian AJ. DNA damage response/TP53 pathway is activated and contributes to the pathogenesis of dilated cardiomyopathy associated with LMNA (Lamin A/C) mutations. Circ Res 2019;124:856–873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Auguste G, Gurha P, Lombardi R, Coarfa C, Willerson JT, Marian AJ. Suppression of activated FOXO transcription factors in the heart prolongs survival in a mouse model of laminopathies. Circ Res 2018;122:678–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kim D, Paggi JM, Park C, Bennett C, Salzberg SL. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat Biotechnol 2019;37:907–915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Risso D, Ngai J, Speed TP, Dudoit S. Normalization of RNA-seq data using factor analysis of control genes or samples. Nat Biotechnol 2014;32:896–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 2014;15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Rouhi L, Cheedipudi SM, Chen SN, Fan S, Lombardi R, Chen X, Coarfa C, Robertson MJ, Gurha P, Marian AJ. Haplo-insufficiency of Tmem43 in cardiac myocytes activates the DNA damage response pathway leading to a late-onset senescence-associated pro-fibrotic cardiomyopathy. Cardiovasc Res 2020;doi: 10.1093/cvr/cvaa300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Marcus FI, McKenna WJ, Sherrill D, Basso C, Bauce B, Bluemke DA, Calkins H, Corrado D, Cox MG, Daubert JP, Fontaine G, Gear K, Hauer R, Nava A, Picard MH, Protonotarios N, Saffitz JE, Sanborn DM, Steinberg JS, Tandri H, Thiene G, Towbin JA, Tsatsopoulou A, Wichter T, Zareba W. Diagnosis of arrhythmogenic right ventricular cardiomyopathy/dysplasia: proposed modification of the Task Force Criteria. Eur Heart J 2010;31:806–814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. López-Ayala JM, Gómez-Milanés I, Sánchez Muñoz JJ, Ruiz-Espejo F, Ortíz M, González-Carrillo J, López-Cuenca D, Oliva-Sandoval MJ, Monserrat L, Valdés M, Gimeno JR. Desmoplakin truncations and arrhythmogenic left ventricular cardiomyopathy: characterizing a phenotype. Europace 2014;16:1838–1846. [DOI] [PubMed] [Google Scholar]
- 25. Ma Z, Huebsch N, Koo S, Mandegar MA, Siemons B, Boggess S, Conklin BR, Grigoropoulos CP, Healy KE. Contractile deficits in engineered cardiac microtissues as a result of MYBPC3 deficiency and mechanical overload. Nat Biomed Eng 2018;2:955–967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Li J, Sutter C, Parker DS, Blauwkamp T, Fang M, Cadigan KM. CBP/p300 are bimodal regulators of Wnt signaling. EMBO J 2007;26:2284–2294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Miyajima C, Kawarada Y, Inoue Y, Suzuki C, Mitamura K, Morishita D, Ohoka N, Imamura T, Hayashi H. Transcriptional coactivator TAZ negatively regulates tumor suppressor p53 activity and cellular senescence. Cells 2020;9:171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Puzzi L, Borin D, Gurha P, Lombardi R, Martinelli V, Weiss M, Andolfi L, Lazzarino M, Mestroni L, Marian AJ, Sbaizero O. Knock down of plakophillin 2 dysregulates adhesion pathway through upregulation of miR200b and alters the mechanical properties in cardiac cells. Cells 2019;8:1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Nelson WJ, Nusse R. Convergence of Wnt, beta-catenin, and cadherin pathways. Science 2004;303:1483–1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Heallen T, Zhang M, Wang J, Bonilla-Claudio M, Klysik E, Johnson RL, Martin JF. Hippo pathway inhibits Wnt signaling to restrain cardiomyocyte proliferation and heart size. Science 2011;332:458–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Varelas X, Miller BW, Sopko R, Song S, Gregorieff A, Fellouse FA, Sakuma R, Pawson T, Hunziker W, McNeill H, Wrana JL, Attisano L. The Hippo pathway regulates Wnt/beta-catenin signaling. Dev Cell 2010;18:579–591. [DOI] [PubMed] [Google Scholar]
- 32. Xu F, Wang YL, Chang JJ, Du SC, Diao L, Jiang N, Wang HJ, Ma D, Zhang J. Mammalian sterile 20-like kinase 1/2 inhibits the Wnt/beta-catenin signalling pathway by directly binding casein kinase 1epsilon. Biochem J 2014;458:159–169. [DOI] [PubMed] [Google Scholar]
- 33. Li Q, Sun Y, Jarugumilli GK, Liu S, Dang K, Cotton JL, Xiol J, Chan PY, DeRan M, Ma L, Li R, Zhu LJ, Li JH, Leiter AB, Ip YT, Camargo FD, Luo X, Johnson RL, Wu X, Mao J. Lats1/2 sustain intestinal stem cells and Wnt activation through TEAD-dependent and independent transcription. Cell Stem Cell 2020;26:675–692.e678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Imajo M, Miyatake K, Iimura A, Miyamoto A, Nishida E. A molecular mechanism that links Hippo signalling to the inhibition of Wnt/beta-catenin signalling. EMBO J 2012;31:1109–1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Park HW, Kim YC, Yu B, Moroishi T, Mo JS, Plouffe SW, Meng Z, Lin KC, Yu FX, Alexander CM, Wang CY, Guan KL. Alternative Wnt signaling activates YAP/TAZ. Cell 2015;162:780–794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Bhonsale A, Groeneweg JA, James CA, Dooijes D, Tichnell C, Jongbloed JD, Murray B, Te Riele AS, van den Berg MP, Bikker H, Atsma DE, de Groot NM, Houweling AC, van der Heijden JF, Russell SD, Doevendans PA, van Veen TA, Tandri H, Wilde AA, Judge DP, van Tintelen JP, Calkins H, Hauer RN. Impact of genotype on clinical course in arrhythmogenic right ventricular dysplasia/cardiomyopathy-associated mutation carriers. Eur Heart J 2015;36:847–855. [DOI] [PubMed] [Google Scholar]
- 37. Augusto JB, Eiros R, Nakou E, Moura-Ferreira S, Treibel TA, Captur G, Akhtar MM, Protonotarios A, Gossios TD, Savvatis K, Syrris P, Mohiddin S, Moon JC, Elliott PM, Lopes LR. Dilated cardiomyopathy and arrhythmogenic left ventricular cardiomyopathy: a comprehensive genotype-imaging phenotype study. Eur Heart J Cardiovasc Imaging 2020;21:326–336. [DOI] [PubMed] [Google Scholar]
- 38. Smith ED, Lakdawala NK, Papoutsidakis N, Aubert G, Mazzanti A, McCanta AC, Agarwal PP, Arscott P, Dellefave-Castillo LM, Vorovich EE, Nutakki K, Wilsbacher LD, Priori SG, Jacoby DL, McNally EM, Helms AS. Desmoplakin cardiomyopathy, a fibrotic and inflammatory form of cardiomyopathy distinct from typical dilated or arrhythmogenic right ventricular cardiomyopathy. Circulation 2020;141:1872–1884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Cipriani A, Bauce BD, Lazzari M, Rigato I, Bariani R, Meneghin S, Pilichou K, Motta R, Aliberti C, Thiene G, McKenna WJ, Zorzi A, Iliceto S, Basso CP, Marra M, Corrado D. Arrhythmogenic right ventricular cardiomyopathy: characterization of left ventricular phenotype and differential diagnosis with dilated cardiomyopathy. J Am Heart Assoc 2020;9:e014628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Haas J, Frese KS, Peil B, Kloos W, Keller A, Nietsch R, Feng Z, Müller S, Kayvanpour E, Vogel B, Sedaghat-Hamedani F, Lim W-K, Zhao X, Fradkin D, Köhler D, Fischer S, Franke J, Marquart S, Barb I, Li DT, Amr A, Ehlermann P, Mereles D, Weis T, Hassel S, Kremer A, King V, Wirsz E, Isnard R, Komajda M, Serio A, Grasso M, Syrris P, Wicks E, Plagnol V, Lopes L, Gadgaard T, Eiskjær H, Jørgensen M, Garcia-Giustiniani D, Ortiz-Genga M, Crespo-Leiro MG, Deprez RHLD, Christiaans I, van Rijsingen IA, Wilde AA, Waldenstrom A, Bolognesi M, Bellazzi R, Mörner S, Bermejo JL, Monserrat L, Villard E, Mogensen J, Pinto YM, Charron P, Elliott P, Arbustini E, Katus HA, Meder B. Atlas of the clinical genetics of human dilated cardiomyopathy. Eur Heart J 2015;36:1123–1135a. [DOI] [PubMed] [Google Scholar]
- 41. Dubash AD, Kam CY, Aguado BA, Patel DM, Delmar M, Shea LD, Green KJ. Plakophilin-2 loss promotes TGF-beta1/p38 MAPK-dependent fibrotic gene expression in cardiomyocytes. J Cell Biol 2016;212:425–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Shoykhet M, Trenz S, Kempf E, Williams T, Gerull B, Schinner C, Yeruva S, Waschke J. Cardiomyocyte adhesion and hyperadhesion differentially require ERK1/2 and plakoglobin. JCI Insight 2020;5: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Hall CL, Gurha P, Sabater-Molina M, Asimaki A, Futema M, Lovering RC, Suarez MP, Aguilera B, Molina P, Zorio E, Coarfa C, Robertson MJ, Cheedipudi SM, Ng KE, Delaney P, Hernandez JP, Pastor F, Gimeno JR, McKenna WJ, Marian AJ, Syrris P. RNA sequencing-based transcriptome profiling of cardiac tissue implicates novel putative disease mechanisms in FLNC-associated arrhythmogenic cardiomyopathy. Int J Cardiol 2020;302:124–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA-Seq data have been deposited in the public database GEO (GSE156869). Additional data pertaining to the present article are available from the corresponding author upon request.