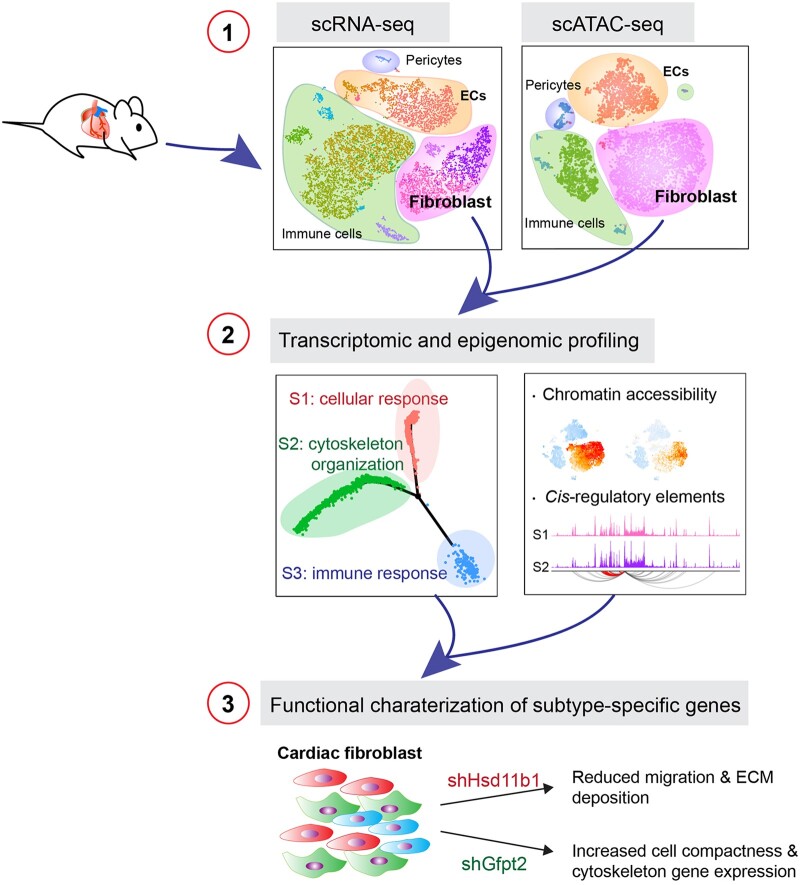
Abstract
Aims
The precise cellular identity and molecular features of non-myocytes (non-CMs) in a mammalian heart at a single-cell level remain elusive. Depiction of epigenetic landscape with transcriptomic signatures using the latest single-cell multi-omics has the potential to unravel the molecular programs underlying the cellular diversity of cardiac non-myocytes. Here, we characterized the molecular and cellular features of cardiac non-CM populations in the adult murine heart at the single-cell level.
Methods and results
Through single-cell dual omics analysis, we mapped the epigenetic landscapes, characterized the transcriptomic profiles and delineated the molecular signatures of cardiac non-CMs in the adult murine heart. Distinct cis-regulatory elements and trans-acting factors for the individual major non-CM cell types (endothelial cells, fibroblast, pericytes, and immune cells) were identified. In particular, unbiased sub-clustering and functional annotation of cardiac fibroblasts (FBs) revealed extensive FB heterogeneity and identified FB sub-types with functional states related to the cellular response to stimuli, cytoskeleton organization, and immune regulation, respectively. We further explored the function of marker genes Hsd11b1 and Gfpt2 that label major FB subpopulations and determined the distribution of Hsd11b1+ and Gfp2+ FBs in murine healthy and diseased hearts.
Conclusions
In summary, we characterized the non-CM cellular identity at the transcriptome and epigenome levels using single-cell omics approaches and discovered previously unrecognized cardiac fibroblast subpopulations with unique functional states.
Keywords: Non-myocytes, Single-cell ATAC-seq, Single-cell transcriptomics, Murine adult heart, Fibroblast
Graphical Abstract
1. Introduction
Cardiac non-myocytes (non-CMs) are a critical constituent of the heart and have been implicated in regulating many important aspects of heart development, physiology, and pathology.1 Among the non-CMs, cardiac fibroblast (FB) is the major cell type that produces extracellular matrix, which, under cardiac homeostasis, provides a basic structural scaffold for the heart.2 Under pathological conditions, however, FBs become activated and transdifferentiated into myofibroblasts that secrete an excessive amount of ECM components, leading to the formation of fibrotic scar. Endothelial cells (ECs) form the innermost layer of blood and lymphatic vessels that serve to supply nutrients/oxygen and maintain tissue fluid levels, respectively. Mural cells (pericytes and vascular smooth muscle cells) intimately envelop the endothelial layer and function to maintain the vascular integrity and stability.3,4 Immune cells are also major cardiac non-CM populations that contribute to cardiac homeostasis in a healthy heart and function to remove dead cells and promote cardiac repair after tissue damage.5 Non-CMs dysfunction often leads to the pathogenesis of cardiac hypertrophy, fibrosis, and heart failure,6,7 yet, a lack of complete dissection of the heterogeneity and annotation of the cellular identity of the non-CM populations in health and disease greatly hindered our understanding of the underlying mechanisms associates with various heart diseases.
Single-cell RNA sequencing (scRNA-seq) has emerged as a powerful tool to dissect the transcriptional profiles of complex tissues at single-cell resolution. This allows for the identification and classification of cell sub-types, detection of rare cell populations and characterization of intermediate cellular states for trajectory reconstruction.8,9 Evolving insights have been provided by scRNA-seq studies into cardiac biology at embryonic, postnatal, and adult stages, revealing an unprecedented molecular details and intercellular communications during heart development and cardiac repair.10–21 Recent studies using adult murine heart depicted the cellular heterogeneity of non-CMs and identified disease-associated factors and regulatory pathways.18,20,22 Expression of the context-specific genes is tightly controlled by cis-regulatory elements such as promoters and enhancers, and trans-regulatory factors including transcription factors. Recent advances of single-cell Assay for Transposase Accessible Chromatin using sequencing (scATAC-seq) has emerged as an accurate proxy to investigate the epigenomic landscape and uncover the regulatory elements in single cells. In this study, we performed single-cell dual-omics analysis of cardiac non-CMs from murine adult hearts to dissect the heterogeneity and functionality of non-CMs at transcriptomic and epigenetic levels. We characterized gene expression signatures of major non-CM cell types and investigated the specific activity of cis-regulatory elements (CREs) and trans-regulatory factors controlling the cellular identity. Using sub-clustering and trajectory analysis, we found that cardiac FBs comprise three distinct subpopulations featuring functional states relating to cellular response, cytoskeleton organization, and immune response. Gene enrichment analysis identified Hsd11b1 and Gfpt2 as the representative marker genes for corresponding FB states. Consistently, in vivo immunohistochemistry (IHC) staining validated that Hsd11b1 and Gfpt2 marked distinct FB subpopulations on heart sections. In vitro experiments demonstrated that knockdown Gfpt2 altered cell compactness whereas knockdown Hsd11b1 reduced FB migration and ECM deposition. Our results altogether indicate that the cardiac non-CM subpopulations can be defined by their functional states at the single-cell level and thus could provide molecular insights on the role of non-CMs during cardiac haemostasis or disease progression.
2. Methods
2.1 Mice and myocardial infarction model
Animal studies were approved by the Institutional Animal Care and Use Committee (IACUC) of the University of North Caroline at Chapel Hill and conform to the National Institutes of Health (NIH) Guidelines for the care and use of laboratory animals. Male mice (C57BL/6J strain) aged at 10–12 weeks were used in all experiments unless otherwise stated. Mice were anaesthetized with intraperitoneal injection of 100 mg/kg body weight (BW) ketamine and 10 mg/kg BW xylazine for one time. Myocardial infarction (MI) was then performed by permanent ligation of the left anterior descending artery (LAD) with a 7-0 prolene suture when anaesthetized as described previously.23,24 Heart was harvested for IHC at 1 week and 4 weeks after injury. In accordance with UNC-Chapel Hill LARC guidelines, all animals were euthanized using CO2 asphyxiation, followed by cervical dislocation. This method is consistent with the recommendations of the Panel on Euthanasia of the American Veterinary Medical Association and is acceptable under the DLAM guidelines and IUCAC.
2.2 Single-cell transcriptome library preparation and sequencing
Single live cells from both ventricles were loaded to 10× Genomics Chromium chip per factory recommendations (details in Supplementary material online). Reverse transcription and library preparation were conducted using Chromium Single Cell 3′ Library and Gel Bead Kit v.2 and Chromium i7 Multiplex kit. Sequencing was performed on Illumina NextSeq 500 with a high output kit.
2.3 Single-cell ATAC library preparation and sequencing
The single nuclei were processed for library preparation (Chromium Single Cell ATAC v1.0) following manufactural instructions from 10× Chromium. Sequencing was performed on Illumina NOVAseq-S1 platform.
2.4 scRNAseq data analysis
Raw sequencing reads were processed with Cell Ranger v.2.2.0 pipeline. We first performed analysis on each of the samples separately and merged samples following default pipelines in Seurat v3.25 In this study, we mainly focus on non-CMs, thus we carried out one more pre-processing step that filters out the clusters expressing a high level of cardiomyocyte markers. All the immune clusters were further validated by W plot using the online tools of Immunological Genome Project (ImmGen).26 For FBs, ECs, and macrophages/monocytes (MCs), differentially expressed genes (DEGs) among sub-clusters were detected, respectively, using FindMarkers function of Seurat package. All the identified DEGs were functionally annotated in DAVID Bioinformatics Resources v. 6.8.27 Gene ontology (GO) terms were considered as significantly enriched if P-value < 0.05.
To unveil the underlying cell transition among different functional states within fibroblasts, we implemented single-cell trajectory analysis in all identified fibroblasts in both sample 1 and 2, respectively, using Monocle v. 2.8.0.28 The high-dimensional data are reduced into a two-dimensional subspace using DDRTree algorithm, and the pseudotime and state were defined by orderCells function. Genes were considered to be differentially expressed among three states if their UMI counts in the corresponding state > 50 and log (Fold-change) > 0.25 with an adjusted P-value < 0.01.
2.5 scATAC-seq data analysis
Sequencing reads of scATAC-seq were processed using Cell Ranger ATAC v. 1.1.0 software. The outputs of the two samples were first aggregated together using cellranger-atac aggr pipeline and analysed using SnapATAC package.29 Only high-quality barcodes, whose log10(UMI) ≥ 3.5 and ≤ 5 and promoter ration ≥ 0.15 and ≤ 0.7, were retained for downstream analysis. Then, the genome regions were segmented into 5-kb bins and the features within each bin were counted and binarized. The bins overlapped with the ENCODE blacklist or located on mitochondrial fragment were filtered out to ensure data quality. Moreover, the top 5% bins, which were considered as invariant features, were also removed. Diffusion maps were computed to reduce data’s dimensionality.
To annotate the nuclei from scATAC-seq data, we projected scATAC-seq data to the integrative scRNA-seq data following the SnapATAC and Seurat v3 pipelines with minor modifications. To ensure the prediction accuracy, only the nuclei with a prediction score > 0.5 were retained for further analysis. Furthermore, because only a few nuclei were annotated as FB3, MC3, or MC4 (<20), our downstream analysis only focus on the major subpopulations of FBs and MCs. MACS230 was employed for peak calling for the clusters with > 100 cells, and the differentially accessible regions (DARs) among cell type/sub-type were identified using findDAR function of SnapATAC package. The peaks with FDR < 0.05 and log (Fold-change) > 0 were considered as significant DARs. For the cell type/sub-type containing < 2000 DARs, we ranked the peaks according to their P-values and selected the top 2000 most significant peaks as representative DARs. The findMotifsGenome.pl program of Homer v. 4.1131 was applied to discover the enriched motifs in the DARs of each cell type/sub-type, and the scanMotifGenomeWide.pl program was employed to scan all the potential motifs within each DAR. If two motifs were overlapped, only the motif with a higher log-odds score was retained. We also inferred motif variability among cells using chromVAR,32 where the profiles of 746 non-redundant vertebrate motifs were downloaded from JASPAR database (the latest 8th release (2020)).33 Furthermore, we predicted promoter-enhancer pairs for the feature genes identified in two FB sub-types (FB1 and 2) via predictGenePeakPair function. The genome tracks of peaks and/or predicted promoter–enhancer pairs were visualized by the WashU Epigenome Browser v52.0.0.34
2.6 Other assays
Single-cell preparation, immuno-cytochemistry (ICC), and -histochemistry (IHC), wound healing assays are provided in Supplementary material online.
2.7 Statistical analysis
Unless otherwise stated, values are expressed as average ± standard mean of error (SEM) of three biologically independent samples and were statistically analysed by GraphPad Prism 7 and R software. Generally, a P-value of < 0.05 was considered statistically significant (*), a P-value of < 0.01 was considered highly significant (**), and a P-value of < 0.001 was considered strongly significant (***).
3. Results
3.1 Single-cell RNA-seq and ATAC-seq revealed heterogeneity of non-myocyte populations
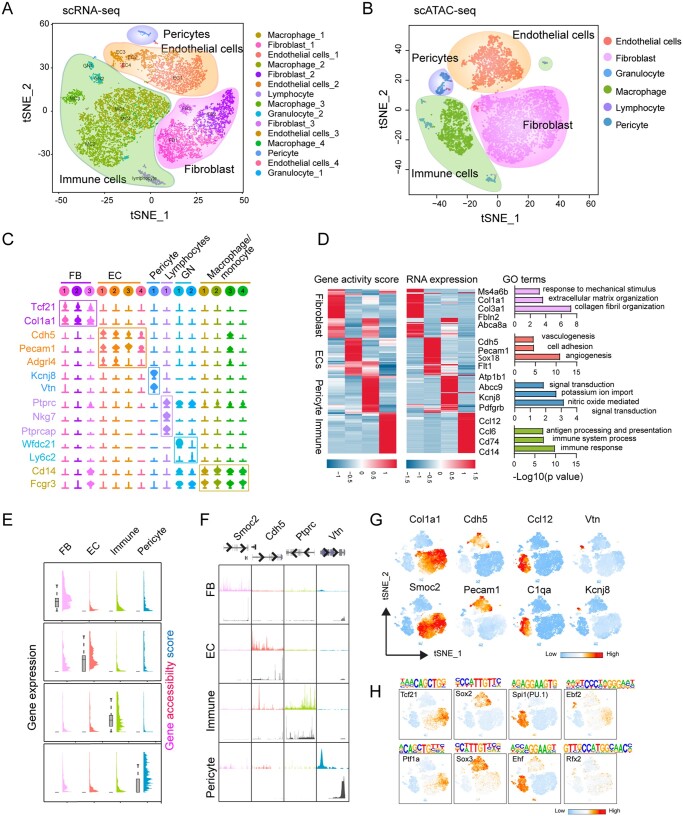
Non-CMs from ventricle were harvested and sorted single live cells (∼80%) were subjected to scRNA-seq and scATAC-seq using 10× Genomics platform (see Section 2 for details) (Supplementary material online, Figure S1A and B). scRNA-seq data from biological duplicates were integrated for downstream analysis if otherwise mentioned (Supplementary material online, Figure S1C–F). Following stringent quality control steps, we performed unsupervised clustering analysis and identified 15 distinct cell clusters shown on the t-SNE (t-distributed stochastic neighbour embedding) plot, including the major non-CM cell types FBs (three sub-clusters, 26.6 ± 4.0%), ECs (four sub-clusters, 24.6 ± 6.8%), pericytes (1.0 ± 0.3%), and immune cells (MCs, 44.1 ± 1.7%; lymphocytes, 2.9 ± 0.4%) (Figure 1A). Each cell type preferentially expresses known marker genes (Figure 1C), for example, FBs express Tcf21, Col1a1 and Pdgfrα, whereas ECs express Pecam1 and Cdh5. We further validated the identity of immune cells based on the annotations from ImmGen,26 and we observed clear differences among lymphocytes, granulocytes (GN) and MCs (Supplementary material online, Figure S1G). Analysis with individual dataset achieves similar clustering results (Supplementary material online, Figure S2A–F).
Figure 1.
Integrated single-cell transcriptomic and epigenomic analysis of non-cardiac (non-CM) cells. (A) Unsupervised clustering demonstrates 15 distinct cell types shown in a t-SNE plot. Non-CMs (n = 12 779) were obtained from two adult hearts. (B) t-SNE plot of major cell types captured by scATAC-seq (n = 9524). (C) Violin plots showing the relative expression levels of representative marker genes across the 15 main clusters. (D) Heatmaps showing the promoter accessibility and the corresponding gene expression of differentially expressed genes (DEGs) in four major non-CM cell populations. Representative gene ontology (GO) terms of the DEGs obtained from scRNA-seq were shown at the right side. (E) Plots showing the gene accessibility score (coloured peaks) and expression levels (grey boxes) of the scRNA-seq derived cell-type signature genes for major non-CM clusters including fibroblasts (FBs), endothelial cells (ECs), pericytes and immune cells. (F) Aggregate chromatin accessibility (colour peaks) and gene expression (grey peaks) profiles for each cell cluster at representative marker gene loci (Smoc2 for FBs, Cdh5 for ECs, Ptprc for immune cells and Vtn for pericytes). (G) t-SNE plot showing the differential accessibilities of representative marker genes for major non-CM clusters. (H) t-SNE plot showing the differentially enriched transcription factor (TF) binding motifs among major non-CM clusters.
We next interrogated the epigenetic landscape of non-CMs by measuring chromatin accessibility across 9524 single nuclei which had a median of 16 602 fragments per cell. Nuclei were annotated by the integrative analysis of both scRNA-seq and scATAC-seq data. In scATAC-seq data, we recovered major non-CM populations consistently comprising of FBs, ECs, pericytes, and immune cells (Figure 1B), and the chromatin accessibility of canonical markers in each cell population are well correlated to their expression levels measured by scRNA-seq (Figure 1D and E). Known signature genes for each cell type, including Col1a1 and Smoc2 for FBs, Cdh5 and Pecam1 for ECs, Vtn for pericytes and Ptptrc (encoding CD45) for immune cells, have been marked with elevated gene accessibility score (the accessibility level at the gene promoter and the peaks nearby) (Figure 1F and G).
Next, we determined DEGs and DARs in each cell type and found that both DEGs and DARs exhibited highly cell-type specificities (Figure 1D). GO enrichment analysis on the genes nearest to the DARs showed that the DARs detected in each cell populations were closely related to the cellular functions (Supplementary material online, Figure S1H). Especially, the genes associated with DARs in FBs were involved in adhesion and Wnt signalling pathway, consistent with the functions of FBs revealed by our own scRNA-seq analysis (Supplementary material online, Figure S1H) and those from others.18
We further performed motif enrichment analysis using the JASPAR database as the source of DNA binding profiles33 to identify potential transcription factors (TFs) binding sites in each cell population. We analysed accessibility variations in binding sites for cell-type-specific TFs. Indeed, the binding motif for Tcf21, the well-known basic helix–loop–helix (bHLH) family members regulating cardiac FB identity, was overrepresented in FB population. The motifs for key regulatory TFs, including Sox2/Sox3, and Spi1 (PU.1)/Ehf, and Ebf2/Rbx2 were enriched in ECs, immune cells, and pericytes, respectively (Figure 1H). We inferred motif enrichment scores of all 746 vertebrate TF motifs from JASPAR and discovered cell-type-specific enriched TFs that can be grouped into 2–5 families (Supplementary material online, Figure S3A–F). For example, the EC cluster was highly enriched in binding motifs for high-mobility group (HMG) domain factor family members such as SRY-Box (Sox) TFs, and for C2H2 zinc finger factors including Sp1 family members (Supplementary material online, Figure S3A and B). Several TFs from these two families have been shown to regulate EC development, vessel formation, and function.35–37 TF activity of FBs will be further depicted in another section (see below). Collectively, these results revealed the general transcriptomic and epigenomic features of non-CMs, indicating that dual-omics mapping of non-CMs broadly captured major non-CM cell types and the clustering outcome is concordant using scRNA-seq and scATAC-seq.
3.2 Four cardiac EC subpopulations have distinct functional profiles
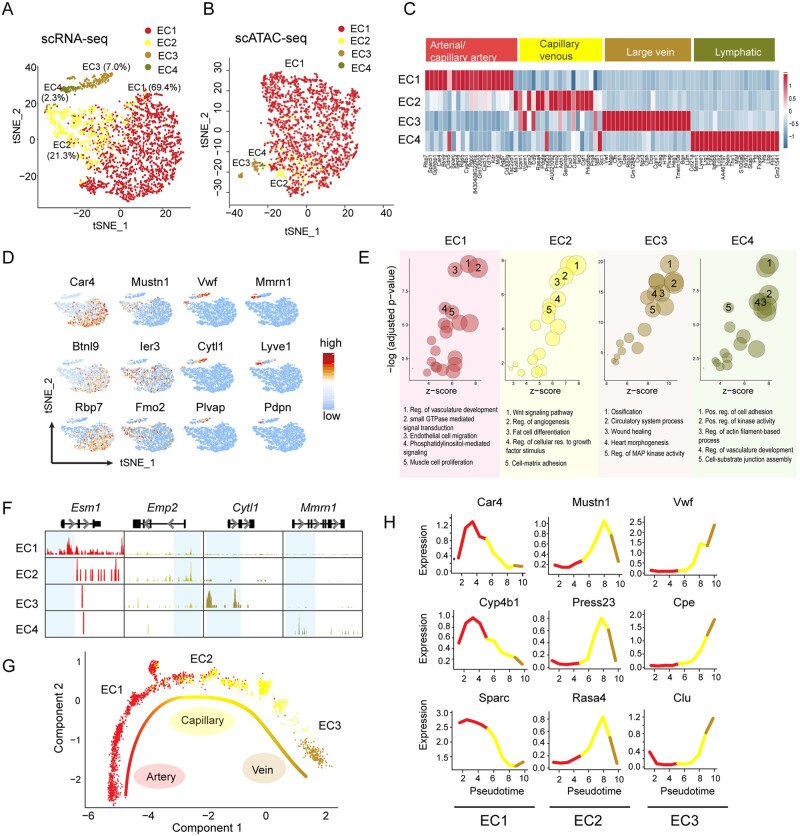
Using our transcriptomic and chromatin profiles of healthy non-CMs, we next investigated the cellular identities of the non-CMs in more detail. ECs, MCs, and FBs were further analysed, respectively. ECs are one of the most abundant cell types in the heart and display discrete functional states.1,10,38 In our study, ECs were characterized by the expression of Cdh5 with Pecam1 (encoding for CD31) and were clustered into four subpopulations (EC1–EC4) (Figure 2A and B). To infer the putative functions of the EC sub-clusters, we determined DEGs in each EC sub-type, by comparing each EC subpopulation with the rest EC groups. Based on the differential expression profiles of the established EC lineage-specific marker genes,35 we classified EC1–EC4 as artery/capillary artery, capillary venous, large vein, and lymphatic ECs, respectively (Figure 2C and D). For instance, EC4 population uniquely expressed Mmrn1, Lyve1, and Pdpn, all known markers for lymphatic endothelial cells.38 Consistently, DNA accessibility score for sub-type-specific marker genes loci were also specifically higher in the corresponding EC sub-type (Figure 2E). GO analysis using DAR nearby genes indicated distinct functions of individual EC sub-types (Figure 2F).
Figure 2.
Characterization of endothelial cell subpopulations from cardiac non-myocytes. (A) t-SNE plot showing subpopulations of ECs from scRNA-seq. EC cells (n = 2939) were subclustered into four subsets and the percentage of each subcluster was shown. (B) t-SNE plot showing subpopulations of ECs from scATAC-seq (n = 2287). (C) Heatmap of top 20 DEGs in each EC subcluster that re-define ECs into arterial/capillary artery ECs, capillary venous, large vein and lymphatic ECs. (D) t-SNE plot showing expression of representative markers in individual EC sub-clusters. (E) Genome track showing the chromatin accessibility profiles of the representative markers in each EC subcluster. EC subcluster specific peaks were highlighted in blue. (F) Enriched GO terms of each EC sub-types using DAR nearby genes. Top five non-redundant GO terms were shown as representatives. Reg, regulation; pos, positive; res, response. (G) Trajectory analysis of the ECs forming blood vessels. (H) Expression profile of marker genes differentially expressed along the trajectory in EC sub-clusters forming blood vessels.
At the tissue level, blood vessels organize themselves into tree-like hierarchical networks comprised of arteries, veins, and the interconnected capillaries.39 To investigate the putative topography of the blood vessel-forming EC clusters within the vascular tree, we performed the differentiation trajectory of EC1–EC3 sub-clusters and discovered a molecular trajectory from artery, capillary, to vein ECs (Figure 2G). Expression of known artery marker genes (e.g., Car4) was enriched at the arterial end of the pseudotime trajectory, whereas expression of capillary marker (Mustn1) was detected in the middle part, and large vein marker Vwf was identified at the other end of the trajectory. This analysis also identified Cyp4b1 and Sparc as artery markers, Press23 and Rasa4 as capillary markers, and Cpe and Clu as venous markers (Figure 2H). Altogether, our analysis of ECs indicates that cardiac ECs exhibited heterogeneity and differences among sub-types with distinct gene expression patterns representing its unique functionality.
3.3 Gene set enrichment profiles identify distinct cardiac macrophage subpopulations
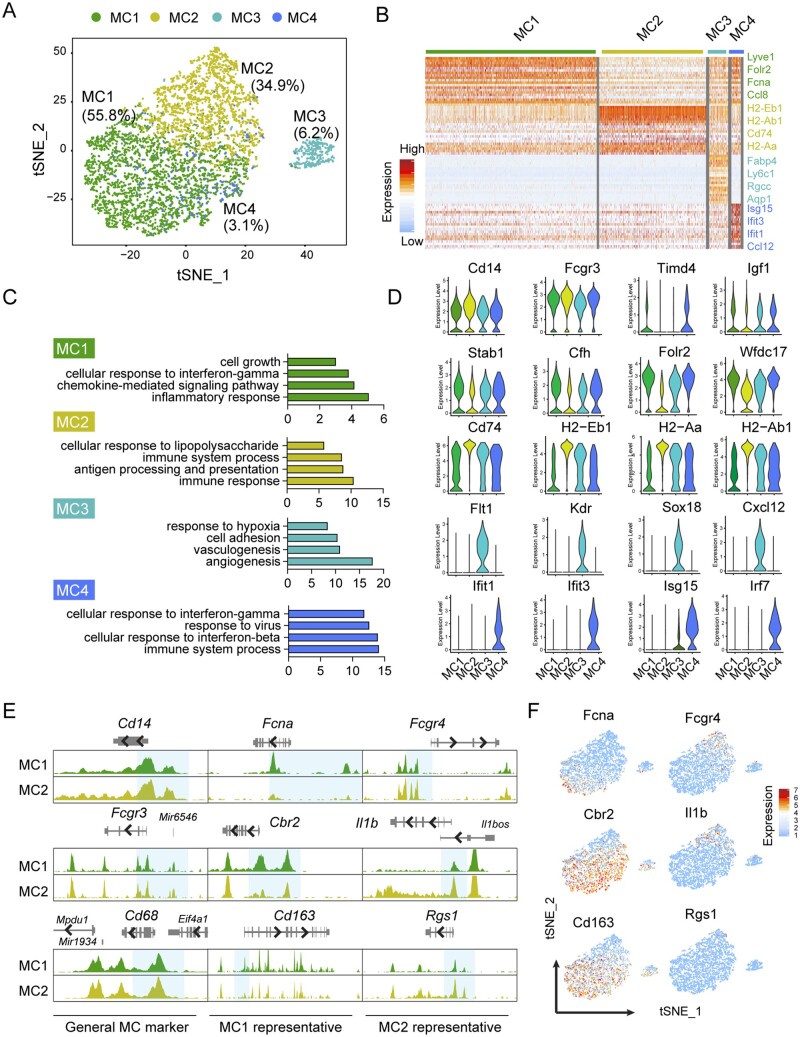
Among cardiac immune cells, MCs represented the most abundant cell population. We identified four sub-clusters of MCs that exhibited similar levels of RNA expression and chromatin accessibility for the common MC markers Cd14, Fcgr3, and Cd68 (Figure 3A–E). To extrapolate information regarding the identity and function of these sub-types, we plotted the top 30 DEGs that differentiate each cluster in a heat map (Figure 3B), evaluated representative DEGs expression (Figure 3D), confirmed cell identity using ImmGen data portal26 (Supplementary material online, Figure S1G), and interrogated significant GO terms using all DEGs identified in each cluster (Figure 3C).40,41
Figure 3.
Characterization of macrophages/monocytes from cardiac non-myocytes. (A) t-SNE plot showing 4 macrophage/monocyte (MC) sub-types (n = 5532) from non-CMs. (B) Heatmap showing expression of top 30 DEGs in each MC sub-type. (C) GO terms enriched for each MC sub-type. (D) Violin plot showing the expression of the representative genes in each MC sub-type. (E) Genome tracks showing the chromatin accessibility profiles of the representative marker genes for the two major MC sub-type (MC1 and MC2). Differentially accessible regions near promoter were highlighted in blue. (F) t-SNE plot showing the expressions of representative marker genes for different MC sub-types.
Both MC1 and MC2 demonstrated enriched functional pathways related to inflammatory response. MC1 showed high expression of various genes previously described in M2-like macrophages (Folr2, Cbr2, and Mrc1), chemokines (Ccl8 and Ccl24), and arterial resident macrophages (e.g. Lyve1 and F13a1).30,31 Whereas, MC2 had relatively increased expression of genes (such as H2-Eb1, Ha-Aa, and Ha-Ab1) that contribute to antigen processing and presentation, suggesting these cells may play a role in immunosurveillance.42 Interestingly, we identified MC3 with endothelial-like features as reported by other groups.18 MC4 expressed a higher level of genes (Ifit1, Ifit2, and Isf15) associated with innate immune response to type I interferon, and interferon-γ (Figure 3D).
Consistent with the above-mentioned gene expression from scRNAseq, we observed a higher gene activity score of MC1 marker genes including Fcna, Cbr2, and Cd163 in MC1 but not MC2 population. Meanwhile, genes specifically expressed in MC2 (Fcgr4, Il1b, and Rgs1) were more accessible in MC2 cluster than MC1 cluster (Figure 3E and F). These results indicate a positive correlation of MC sub-type specific marker gene expression with its chromatin landscape, which further suggests the functionality of MC sub-groups.
3.4 Single-cell chromatin landscape of cardiac fibroblasts
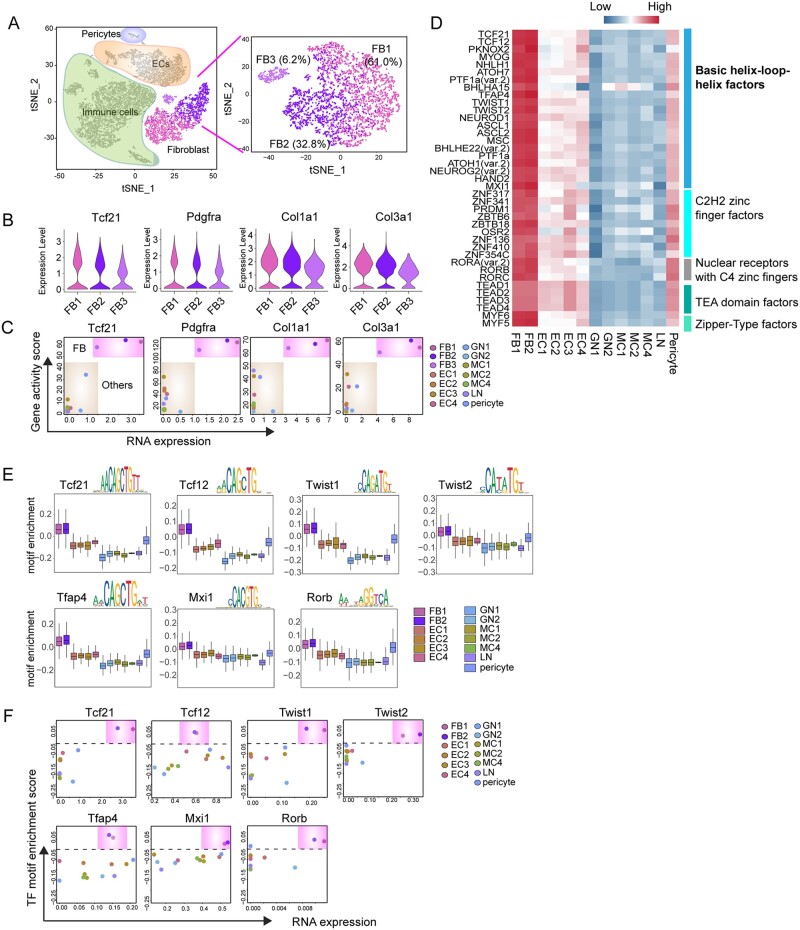
Fibroblast is one of the major cell types that constitute the cardiac microenvironment and can rapidly respond to various stresses. Recent studies have begun to define the transcriptome of FB at the single-cell level.18,22 However, the epigenetic regulation that co-ordinately determines FB identity and function is largely unknown. Thus, we sought to perform a joint analysis of transcriptomic and chromatin landscapes to delineate cardiac FB identity and function. Unbiased cell clustering using the transcriptomic data allowed us to identify three FB sub-types: namely the FB1, FB2, and FB3 (Figure 4A), which accounted for 57.2%, 36.6%, and 6.2% of the total FB population, respectively. We found that previously known FB markers, including Tcf21, Pdgfrα, Col1a1, and Col3a1,43 expressed at comparable levels in all three FB sub-types (Figure 4B). Consistently, scATAC profiling indicated that these loci exhibited a highly accessible chromatin state (Figure 4C).
Figure 4.
Cis- and trans-regulatory elements of cardiac fibroblast. (A) Unsupervised clustering of FBs (n = 3459) by Seurat demonstrating three FB sub-types. (B) Violin plot showing the relative expression of canonical FB marker genes in three FB sub-types. (A) X–Y plots showing the gene expression levels (X-axis) and the gene accessibility score (Y-axis) of canonical FB marker genes. (D) Heatmap showing the motif enrichment scores for TF motifs highly enriched in FB sub-types. (E) Box plot showing the enrichment scores for seven representative TF motifs that are specifically enriched and highly expressed in FB population. (F) X–Y plots showing the gene expression levels and TF motif enrichment scores of FB-specific TFs.
To understand the regulatory mechanism in the control of FB cellular identity, we determined the promoter accessibility of the TFs curated in the JASPR database (746 in total) across the major non-CM cell types. This analysis identified putative binding motifs for 42 TFs enriched specifically in FB populations (Figure 4D). Approximately 50% of the corresponding TFs belonged to bHLH family, and other factors are zinc finger factors, TEA domain factors, and zipper-type factors, respectively (Figure 4D). Among the 42 TFs with enriched binding motifs in FBs, we found that 7 of them exhibited high expression levels in FBs, including 6 bHLH family members (Figure 4E and F). The bHLH family TFs share common features including recognition to E-box motif (a canonical consensus sequence 5′-CANNTG-3′) and the HLH domain that dimerizes to facilitate DNA binding. Interestingly, Tcf21, the first reported bHLH TF essential for the development of cardiac FBs,44 showed in our results, together with Twist1, another TF reported to regulate epithelial‐to‐mesenchymal transition (EMT) of cardiac FBs.45 These results indicate that we have identified known and new TFs together with cis-regulatory elements that potentially regulate FB cellular and molecular features.
3.5 Cardiac fibroblast sub-types defined by their functional states
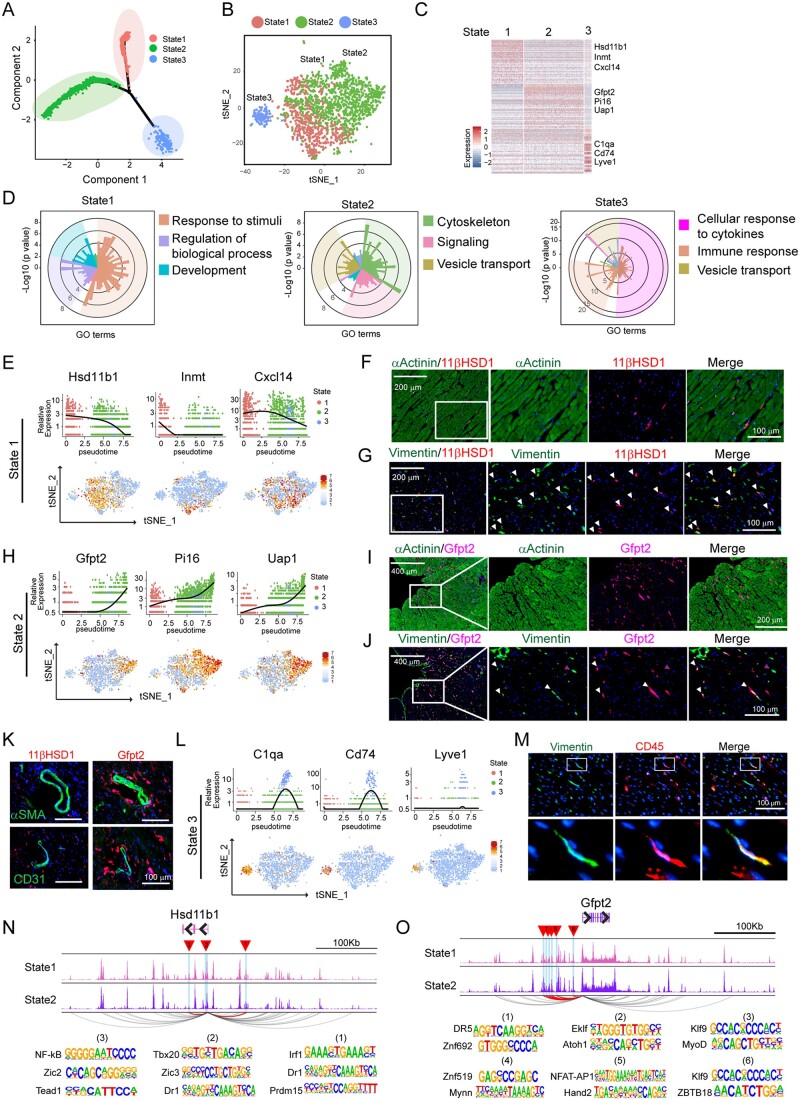
To investigate distinct signatures of each fibroblast subpopulation, we applied single-cell trajectory analysis on three FB subpopulations to order the cells along the pseudotime. By doing so, we identified three FB states, each of which occupied a separate trajectory branch (Figure 5A and Supplementary material online, Figure S4A). Interestingly, three states of FBs were highly correlated with the three subpopulations. State 1 subpopulation mainly composed of FB2 cells, while State 2 and State 3 corresponded FB1 and FB3, respectively (Figure 5B and Supplementary material online, Figure S4B–G). The first branch (State 1, in red) was occupied by more than half of FB1, the second branch (State 2, in green) was occupied mainly by FB2, whereas the last branch (State 3, in blue) mainly comprised of FB3. Cell cycle status and gene expression remained similar among all states (Supplementary material online, Figure S4C and D). Next, we performed DEG analysis by comparing fibroblast sub-type in each state with other clusters, followed by pathway annotation (Figure 5C and D and Supplementary material online, Figure S4H). Interestingly, we found that each FB subset corresponds to a major functional state that distinguishes their cellular identities. The top three highly enriched categories of GO terms were shown (Supplementary material online, Figure S4H). The State 1 FB pathways were primarily associated with cellular response to various stimuli including drug, hormone, cytokines etc. The State 2 FBs were involved in cytoskeleton organization and cellular signalling, whereas classic immune response pathways were enriched in State 3 FBs. Similar findings were also discovered using the DEGs identified by Seurat package (Supplementary material online, Figure S4I). These analyses reveal the existence of three major subpopulations of FBs that could be responsible for distinct biological functions during homeostatic stage.
Figure 5.
Heterogeneity of fibroblast reflected by three subpopulations in distinct functional states. (A) Ordering single fibroblast cells along a cell state trajectory using Monocle (n = 2048). (B) Projection of FBs in three state into t-SNE plot clustered by Seurat. (C) Heat map showing a subset of transcripts that are enriched in each functional state. Three representative genes were listed. (D) Top 3 enriched GO categories analysed from DEGs of each cell state. In each panel, each histogram represents a GO term, the height of the bar indicates the P-value, and the colour indicates similar functional GO categories. GO terms of similar biological function were gathered as a function group, representative GO terms were shown and followed by a P-value. (E) Representative gene expression in State 1 FB shown in total fibroblast population along pseudotime trajectory (upper panel) and their distribution in major cell populations in t-SNE plot (bottom panel). (F) Representative immunohistochemistry (IHC) images showing 11βHDS1+αActinin-State 1 FBs. (G) Representative IHC images showing the identification of State 1 FB positive for both 11βHDS1 and Vimentin (white arrowhead). (H) Representative gene expression in State 2 FB shown in total fibroblast population along pseudotime trajectory (upper panel) and their distribution in major cell populations in t-SNE plot (bottom panel). (I) Representative IHC images showing Gfpt2+α-Actinin-State 2 FBs. (J) Representative IHC images showing the identification of State 2 FB positive for both Gfpt2 and Vimentin (white arrowhead). (K) Representative IHC images showing the co-staining of 11βHDS1+ and Gfpt2+ cells with either αSMA or CD31. (L) Representative gene expression in State 3 FBs shown in total FB population along pseudotime trajectory (upper panel) and in major cell populations in tSNE plot (bottom panel). (M) Representative IHC images showingV the identification of FB (state 3) positive for both CD45 and Vimentin. (N,O) Genome track of the Hsd11b1 (N) and Gfpt2 (O) loci. Predicted promoter-enhancer interactions of statistical significance (P-value < 0.05) are shown in arcs, where the interactions involving differentially accessible regions (DARs; highlighted in blue, as well as red triangles) are highlighted in purple, and the others are in gray. For panels F, G, I, J, K, and M, n = 3 hearts were used, and n = 4 sections at different positions from each heart were stained for indicated protein.
3.6 Identification of novel cardiac fibroblast markers associated with functional states
Having identified three subpopulations of FBs that correlated with distinct functional status, we next determined specific markers for each FB subpopulation. From the top 50 DEGs among three states (Figure 5C and Supplementary material online, Table), we chose three representative genes that were highly enriched in each state (Figure 5E, H,and L and Supplementary material online, Figure S5A). FBs from State 1 were characterized by the relatively high level of Hsd11b1, Inmt, and Cxcl14 expression (Figure 5E). Hsd11b1 encodes 11β-hydroxysteroid dehydrogenase type 1 (11β-HSD1), an enzyme that regenerates glucocorticoids within cells.46 By IHC, we found that 11β-HSD1+ cells localized at interstitial space between cardiomyocytes (αActinin+) (Figure 5F), and barely co-localized with the endothelial cell marker CD31 or smooth muscle marker αSMA (Figure 5K). Vimentin has been widely used as a pan FB marker.47,48 We discovered that around 23% of the Vimentin positive cells were also positive for 11β-HSD1, and this percentage of the Vimentin+ 11β-HSD1+ population was consistent with our single-cell clustering result (Figure 5G). FBs from State 2 were characterized by high level of Gfpt2, pi16, and Uap1 expression. Gfpt2 encodes glutamine-fructose-6-phosphate transaminase2 (GFPT2), the rate-limiting enzyme of the hexosamine biosynthesis pathway.49 Recent study demonstrated a role of Gftp2 in metabolic programming of the cancer-associated fibroblasts.50 Its function in the heart, especially in cardiac FB has not been characterized. We found that GFPT2+ cells were well aligned between the bundles of cardiomyocytes (Figure 5I). Around 60% of Vimentin+ cells expressed GFPT2. These cells exhibited fibre-like shape, morphologically distinct from the 11β-HSD1+ dot-like cells (Figure 5J). GFPT2 expression was undetectable in the endothelial cells and smooth muscle cells (Figure 5K). State 3 FBs consisted of only 10% of the total FB population, and exhibited characteristics of the previously characterized ‘fibrocytes’ as evidenced by the expression of immune cell markers, such as Lyve1 and Cd7418 (Figure 5L). We also confirmed the existence of this FB subpopulation by co-staining of Vimentin and CD45 (pan immune marker) (Figure 5M).
As CREs can regulate gene expression from a large genomic distance,51 we asked if the difference in the use of the distal CREs could contribute to differential gene expression among the FB sub-types. We analysed the interaction networks linking the distal regulatory elements [within a 500 kb range of transcription start site (TSS)] to the promoter of sub-type representative genes. Forty-seven candidate regulatory regions were identified to be linked to HSD11b1, three of which showed significantly higher accessibility in State 1 than State 2 (P-value < 0.05). Within the three regions, we identified 71 TF binding motifs, where the top 3 TFs showing the highest log-odds score (the probability for the nucleotide in each position of a certain motif) in each region were Irf1 (12.88), Dr1 (11.39) and Prdm15 (11.03) in region 1, Tbx20 (11.59), Zic3 (9.60), and Dr1 (7.62) for region 2, and NF-kB (12.52), Zic2(11.63), and Tead1 (10.34) for region 3, respectively (Figure 5N). Similarly, 6 out of 41 regulatory regions linked to Gfpt2 were found to be more accessible in State 2 FBs than that in State 1 FBs (Figure 5O). Predicted interactions between DARs and other representative genes were also explored (Supplementary material online, Figure S5B and C).
Together, we discovered representative markers for FBs in each functional state that helps to re-define FB heterogeneity and sub-type function. Further motif analysis has identified differential accessibilities of the CREs among different FB populations that potentially regulate the expression of marker genes in each FB subpopulation.
3.7 Knockdown of Hsd11b1 and Gfpt2 differentially affects cardiac fibroblast functions
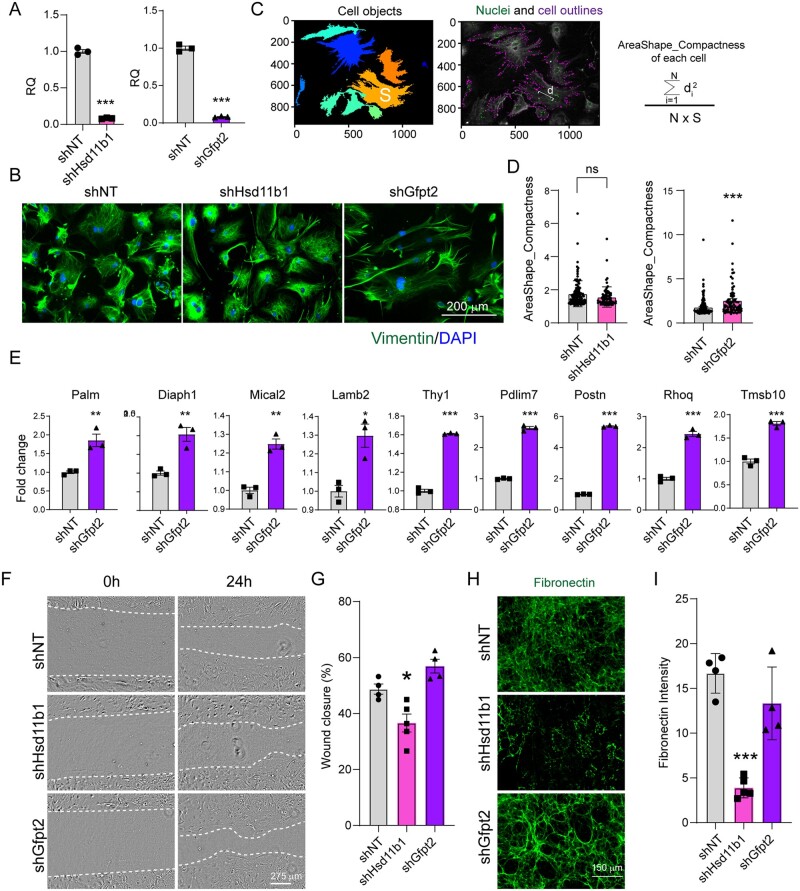
To explore the biological function of major FB sub-types, we introduced short hairpin RNAs (shRNAs) targeting Hsd11b1 and Gfpt2 into FBs freshly dissociated from the adult heart. Over 95% knockdown efficiency was achieved compared with non-targeting (shNT) control (Figure 6A). Intriguingly, morphology of FBs upon Gfpt2 depletion was dramatically changed (with a higher area/shape compactness score), while Hsd11b1 knockdown minimally altered cell morphology (Figure 6B–D). Furthermore, knockdown Gfpt2 significantly upregulated the expression of genes related to actin cytoskeleton organization pathway (GO: 0030036) such as Diaph1,52 Mical2,53 and Lamb2.54 This result is consistent with our single-cell omics data analysis and suggests a possible role of Gfpt2 in controlling FB cytoskeleton organization (State 2 FBs). Interestingly, Gfpt2 knockdown increased periostin (Postn) expression, suggesting a potential transition to pathological conditions upon Gfpt2 deficiency.
Figure 6.
Effects of Hsd11b1 and Gfpt2 knockdown on cardiac fibroblasts in vitro. (A) Knockdown efficiency of Hsd11b1 and Gfpt2 evaluated by RT-qPCR. Lenti-viral shRNAs targeting Hsd11b1, Gfpt2 and non-targeting control (shNT) were individually introduced to FBs isolated from 3 m adult hearts. Three days post-viral infection, cells were collected for qPCR. This experiment was repeated three times with n = 5 hearts and averaged numbers from technical triplicates were used for statistics. Error bars indicate mean ± s.e.m; *** P < 0.001. (B) Representative ICC images showing the morphology of cultured FBs upon shHsd11b1 and shGfpt2 treatment. (C) Representative images and formula showing the parameters of area (S) and distance (d) for measuring cell compactness using Cellprofiler. (D) Histogram showing the cell compactness changes upon shHsd11b1 and shGfpt2 treatment in comparison with shNT control. In left panel, n = 112 for shNT, and n = 73 for shHsd11b1; in right panel, n = 146 for shNT and n = 76 for shGfpt2. (E) Quantification of the expression of genes involved in cytoskeleton organization in FBs treated with shNT or shGfpt2 by qPCR. This experiment was repeated three times with n = 5 hearts each time. Averaged numbers from technical triplicates were used for statistics. Error bars indicate mean ± s.e.m; * P < 0.05, ** P < 0.01, ***P < 0.001. (F,G) Representative light microscopic images (F) and quantification (G) of cardiac FB migration at 0 h and 24 h after scratching. (H,I) Representative ICC images (H) and quantification (I) showing the deposition of fibronectin in FBs treated with shRNAs targeting Hsd11b1, Gfpt2, and the non-targeting control. For panels (F–I), the experiments were repeated three times in technical duplicates using n = 3 hearts each time. In each replicate, n = 4–5 individual fields were taken and calculated for statistics. Error bars indicate mean ± s.e.m; * P < 0.05, ** P < 0.01, *** P < 0.001.
We next determined if and how other functions of cardiac FBs, including migration and extracellular matrix (ECM) deposition, could be influenced by a deficiency in Hsd11b1 and Gfpt2. Surprisingly, Hsd11b1 knockdown in cultured adult FBs reduced migration in wound healing assay (Figure 6F and G) and decreased ECM fibronectin deposition (Figure 6H and I). Whereas, Gfpt2 deficiency had minimal effect in both assays compared to shNT controls. These results were also in line with our observation of State 2 FB functions from single-cell data analysis, indicating the potential involvement of cellular response by Hsd11b1. Overall, we observed different effects of Hsd11b1 and Gfpt2 knockdown on FB function in vitro, suggesting that integrated data analysis may faithfully identify FB functional sub-types.
3.8 Activation of fibroblast sub-types upon cardiac injury
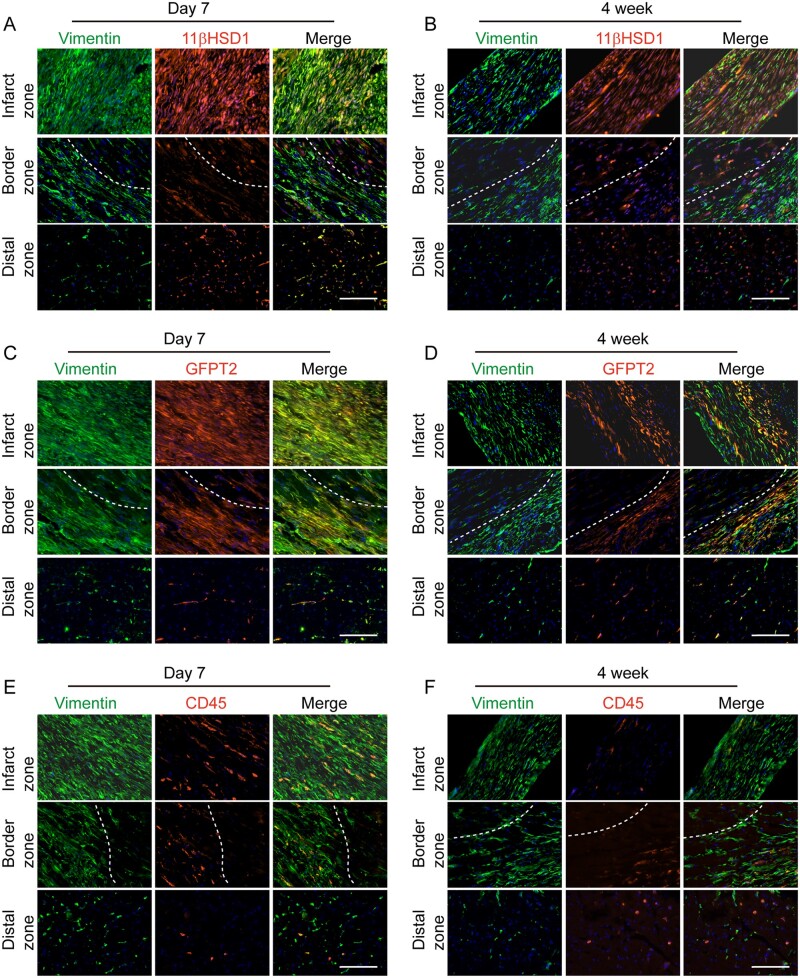
Previous work has demonstrated that by 3–7 days after acute MI, cardiac FBs are rapidly activated to become Postn+ myofibroblast, and are then further transformed into matrifibrocyte 10 days after MI forming a stable fibrotic scar.47,55 To assess the dynamic responses of FB sub-types identified from our study, we analysed the expression of sub-type specific marker expression (Hsd11b1, Gfpt2, and CD45) in FBs (identified by Vim staining) at 1 week, and 4 weeks post-MI. Myocardial injury caused massive expansion of FBs in the injured area and border zone (a region between injured and uninjured tissue) while FBs residing at the distal region were minimally changed. Majority of the Vim+ cells located in the infarcted region and border zone were positive for 11-βHSD1 and GFPT2 at 1- and 4-weeks post-MI (Figure 7A–D). In comparison, the spatial localization pattern of each marker with Vim was similar to that from healthy mice (Figure 5G, J,and M) at the distal region. The number of CD45+Vim+ fibrocytes in and near the infarct region dramatically increased at Day 7 post-MI compared to that in healthy hearts (Figure 5M), but reduced sharply at 4 weeks post-MI (Figure 7E and F).
Figure 7.
Expression of Hsd11b1 and Gfpt2 in cardiac fibroblast upon myocardial injury. (A,B) Representative IHC images showing the expression of Hsd11b1 and its colocalization with Vimentin in infarcted region, border zone and distal zone of heart 7 days (A) and 4 weeks (B) post-MI. (C,D) Representative IHC images showing the expression of Gfpt2 and its colocalization with Vimentin in infarcted region, border zone and distal zone of heart 7 days (C) and 4 weeks (D) post-MI. (E,F) Representative IHC images showing the expression of Gfpt2 expression and its colocalization with Vimentin in infarcted region, border zone and distal zone of heart 7 days (E) and 4 weeks (F) post-MI. Dashed lines indicate the borderline between injured and uninjured or less injured area. The experiment was performed with n = 5 mice per each group. In each heart, n = 4 sections at different layers of heart were used.
To further explore the potential biological function of the Hsd11b1+ and Gfpt2+ FBs, we re-analysed previously published Pdgfα-GFP+ FBs single-cell dataset18 and recapitulated the sub-cluster classification (Supplementary material online, Figure S6A and B). We then mapped the expression of Hsd11b1+ and Gfpt2+ onto this scRNA-seq dataset (Supplementary material online, Figure S6C). We found that, under sham condition, Hsd11b1 and Gfpt2 were mostly expressed in the Fibroblast-Sca1-low (F-SL) and Fibroblast-Sca1-high (F-SH) sub-clusters, respectively, but not in the Postn positive activated MI sub-clusters (F-Act and MYO) (Supplementary material online, Figure S6A–C). At Day 3 post-MI, the majority of Hsd11b1+ and Gfpt2+ FBs transited to become the activated fibroblast (F-Act). At Day 7 post-MI, however, Hsd11b1 expression appeared to be reduced in the F-Act sub-cluster yet regained its high level of expression in the F-SL sub-cluster, while increased expression of Gfpt2 was found in F-SH and myoblast (MYO) sub-clusters, indicating the Hsd11b1+ and Gfpt2+ fibroblast subsets function differently upon injury (Supplementary material online, Figure S6C).
4. Discussion
The mammalian heart harbours diverse cell types with distinct molecular signatures and functions. Although heterogeneity of CMs and non-CMs have been interrogated at developmental stages or under pathological conditions,10,13,14,18–22,56,57 the association between the identity of cell/sub-cell type and functional profile, remains unclear. Here, we applied a single-cell dual-omics approach to investigate the chromatin accessibility and transcriptome of major non-CM populations including fibroblast, endothelial cells, and immune cells. We further studied the putative association between cis- and trans-regulatory elements with gene expression that shapes cell identity. For FBs, we further defined three subpopulations based on their functional status corresponding to cellular response, cytoskeleton organization and immune function. Novel markers such as Hsd11b1 and Gfpt2 for fibroblast subpopulation were identified and experimentally validated. Knocking down Hsd11b1 and Gfpt2 affected FB migration and cytoskeleton organization, respectively. Together, this work reveals a global map of gene expression and chromatin accessibility landscape in adult non-CMs at single-cell resolution and provides additional new clues and information for future work to deconstruct molecular events underlying cardiac homeostasis and disease progression.
Cardiac FBs have been recognized as a critical cell type for maintaining structural and mechanical homeostasis of the heart.58 The identity and function of FBs have long been a focus in the field. Previous studies investigated general roles of cardiac FBs under physiological and pathological conditions based on the markers that were somewhat non-specific to this cell type.43 For example, the surface protein Thy1 is also expressed in most immune and endothelial cells. With the development of lineage tracing technique, fibroblast specific inducible Cre mouse lines offer useful platforms to elucidate the origin and identity of FBs, and to genetically trace and manipulate FB during healthy or diseased stage.47 Under these scenarios, fibroblasts were considered relatively homogenous and were universally labelled with markers such as TCF21 and PDGFRa. Recent advances in scRNA-seq technology provide unique opportunities to further study FB molecular signature at single-cell resolution. Recent comparative scRNA-seq analysis revealed the heterogeneity of the FB population and identified novel marker gene expression.18,20,22 Similar to our research, three sub-types of FB population were identified while further characterization of specific signatures is limited.22 In our study, we discovered that the canonical FB marker expression remains similar among subpopulations, consistent with the previous lineage-tracing results.43 We further annotated the open chromatin landscape of FBs and identified enriched transcription factors as well as enriched binding motifs, thus providing new insights into how FB identity is generally regulated in the heart. Next, based on unsupervised clustering and analysis of DEGs, we discovered that the three FB subpopulations were function-associated sub-types that are mainly responsible for cellular response, cytoskeleton organization and immune response, respectively. We propose that multiple functions of cardiac fibroblasts were separately executed by subpopulations of FBs that are distinct in their morphologies, molecular signatures, and cellular functions. Thus, we defined exclusive markers for each population, evaluated their distal regulatory elements, and experimentally validated representative marker expression including Hsd11b1 and Gfpt2 in healthy and injured hearts. Our work further expanded our understanding of cardiac FB identity, plasticity, and possible functional states. This work may also provide an insight for elucidating other cellular atlas by defining the cell identity based on functional status.
While this work delineates cardiac cell gene expression and chromatin accessibility, several key points require further functional validation. For example, it remains to be explored if FB subpopulations respond differently to cardiac insults as one can speculate from their functional status. Future work is needed to establish animal models in which each FB subpopulation could be exclusively labelled and genetically tranced, and/or functionally ablated under healthy and diseased conditions. Recent study also suggested the functional importance of Wnt expressing fibroblasts,18 which we also identified through scATAC-seq, suggesting consistent findings.
Our study has several limitations. For example, using only male adult murine hearts for single-cell analysis excludes us from discovering sex-related molecular features. We performed Langendorff perfusion to isolate non-CM cells from the hearts, which led to the relative high abundance of immune cells and low enrichment of smooth muscle cells. We also discovered that cellular composition differed between our scRNA-seq and scATAC-seq studies. This may be caused by the inherent technical limitations such as additional cell lysis step to obtain nuclei for scATAC-seq, and different quality control procedures for both omics analyses. In addition, future studies shall analyse the functional relevance of non-myocyte sub-types under various physiological and pathological conditions in the human heart.
In summary, we comprehensively studied the cellular identity of cardiac non-CMs at the single-cell level. We further defined fibroblast subpopulations based on their functional states. Our study thus provides a different perspective for interpreting the heterogeneity of cardiac fibroblast and new insights into the basic biology of cardiac cells.
Supplementary material
Supplementary material is available at Cardiovascular Research online.
Authors’ contributions
L.W. designed and performed experiments, analysed data, and wrote the manuscript. Y.Y analysed the scRNA-seq and scATAC-seq data. H.M. performed heart perfusion with Langendorff system. Y.X, J.X, D.N, and T.G prepared frozen samples for cryosection, performed IHC and quantification. J.X and H.W performed Cell profiler analysis. Y.L. supervised the single-cell metadata analysis. J.L. supervised the work and wrote the manuscript. L.Q. supervised the work, designed the experiments, analysed data, and wrote the manuscript.
Supplementary Material
Acknowledgements
We thank the UNC Lineberger Translational Genomics Lab for all sequencing work and assistance on bioinformatics analysis, the UNC FLOW Core for assistance on flow cytometry. We thank members of the Qian and Liu laboratories for helpful discussions and critical reviews of the manuscript.
Conflict of interest: The authors declare no competing interests.
Funding
This work was supported by American Heart Association (18CDA34110340 to L.W., 15GRNT25530005 to J.L., and 18TPA34180058 to L.Q.); and National Institutes of Health (R01HL129132, R01GM105785 to Y.L., R01HL139880, R01HL139976 to J.L., and R01HL128331, R01HL144551 to L.Q.).
Data availability
The scRNA-seq and scATAC-seq datasets have been deposited the Gene Expression Ominbus (GSE157446). Codes used in this study are freely available from the corresponding author upon reasonable request.
Contributor Information
Li Wang, Department of Pathology and Laboratory Medicine, University of North Carolina, 111 Mason Farm Rd, University of North Carolina, Chapel Hill, Chapel Hill, NC 27599, USA; McAllister Heart Institute, University of North Carolina, 111 Mason Farm Rd, University of North Carolina, Chapel Hill, Chapel Hill, NC 27599, USA.
Yuchen Yang, Department of Pathology and Laboratory Medicine, University of North Carolina, 111 Mason Farm Rd, University of North Carolina, Chapel Hill, Chapel Hill, NC 27599, USA; McAllister Heart Institute, University of North Carolina, 111 Mason Farm Rd, University of North Carolina, Chapel Hill, Chapel Hill, NC 27599, USA; Department of Genetics, University of North Carolina, Chapel Hill, NC 27599, USA.
Hong Ma, Department of Pathology and Laboratory Medicine, University of North Carolina, 111 Mason Farm Rd, University of North Carolina, Chapel Hill, Chapel Hill, NC 27599, USA; McAllister Heart Institute, University of North Carolina, 111 Mason Farm Rd, University of North Carolina, Chapel Hill, Chapel Hill, NC 27599, USA.
Yifang Xie, Department of Pathology and Laboratory Medicine, University of North Carolina, 111 Mason Farm Rd, University of North Carolina, Chapel Hill, Chapel Hill, NC 27599, USA; McAllister Heart Institute, University of North Carolina, 111 Mason Farm Rd, University of North Carolina, Chapel Hill, Chapel Hill, NC 27599, USA.
Jun Xu, Department of Pathology and Laboratory Medicine, University of North Carolina, 111 Mason Farm Rd, University of North Carolina, Chapel Hill, Chapel Hill, NC 27599, USA; McAllister Heart Institute, University of North Carolina, 111 Mason Farm Rd, University of North Carolina, Chapel Hill, Chapel Hill, NC 27599, USA.
David Near, Department of Pathology and Laboratory Medicine, University of North Carolina, 111 Mason Farm Rd, University of North Carolina, Chapel Hill, Chapel Hill, NC 27599, USA; McAllister Heart Institute, University of North Carolina, 111 Mason Farm Rd, University of North Carolina, Chapel Hill, Chapel Hill, NC 27599, USA.
Haofei Wang, Department of Pathology and Laboratory Medicine, University of North Carolina, 111 Mason Farm Rd, University of North Carolina, Chapel Hill, Chapel Hill, NC 27599, USA; McAllister Heart Institute, University of North Carolina, 111 Mason Farm Rd, University of North Carolina, Chapel Hill, Chapel Hill, NC 27599, USA.
Tiffany Garbutt, Department of Pathology and Laboratory Medicine, University of North Carolina, 111 Mason Farm Rd, University of North Carolina, Chapel Hill, Chapel Hill, NC 27599, USA; McAllister Heart Institute, University of North Carolina, 111 Mason Farm Rd, University of North Carolina, Chapel Hill, Chapel Hill, NC 27599, USA.
Yun Li, Department of Genetics, University of North Carolina, Chapel Hill, NC 27599, USA; Department of Biostatistics, University of North Carolina, Chapel Hill, NC 27599, USA; Department of Computer Science, University of North Carolina, Chapel Hill, NC 27599, USA.
Jiandong Liu, Department of Pathology and Laboratory Medicine, University of North Carolina, 111 Mason Farm Rd, University of North Carolina, Chapel Hill, Chapel Hill, NC 27599, USA; McAllister Heart Institute, University of North Carolina, 111 Mason Farm Rd, University of North Carolina, Chapel Hill, Chapel Hill, NC 27599, USA.
Li Qian, Department of Pathology and Laboratory Medicine, University of North Carolina, 111 Mason Farm Rd, University of North Carolina, Chapel Hill, Chapel Hill, NC 27599, USA; McAllister Heart Institute, University of North Carolina, 111 Mason Farm Rd, University of North Carolina, Chapel Hill, Chapel Hill, NC 27599, USA.
Translational perspective
Our research identified discrete cell types of non-CM in the heart and differentially expressed genes with regulatory factors. Unveiling the heterogeneity of non-CMs and molecular signatures of each cell type or sub-types allows for study, precise capture and manipulation of specific cell type(s) in heart and will provide insights into the development of therapeutics for cardiovascular diseases.
References
- 1. Pinto AR, Ilinykh A, Ivey MJ, Kuwabara JT, D'Antoni ML, Debuque R, Chandran A, Wang L, Arora K, Rosenthal NA, Tallquist MD. Revisiting cardiac cellular composition. Circ Res 2016;118:400–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Tallquist MD, Molkentin JD. Redefining the identity of cardiac fibroblasts. Nat Rev Cardiol 2017;14:484–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Armulik A, Abramsson A, Betsholtz C. Endothelial/pericyte interactions. Circ Res 2005;97:512–523. [DOI] [PubMed] [Google Scholar]
- 4. Lee LL, Chintalgattu V. Pericytes in the heart. Adv Exp Med Biol 2019;1122:187–210. [DOI] [PubMed] [Google Scholar]
- 5. Prabhu SD, Frangogiannis NG. The biological basis for cardiac repair after myocardial infarction: from inflammation to fibrosis. Circ Res 2016;119:91–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Tian Y, Morrisey EE. Importance of myocyte-nonmyocyte interactions in cardiac development and disease. Circ Res 2012;110:1023–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lajiness JD, Conway SJ. The dynamic role of cardiac fibroblasts in development and disease. J Cardiovasc Transl Res 2012;5:739–748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Stuart T, Satija R. Integrative single-cell analysis. Nat Rev Genet 2019;20:257–272. [DOI] [PubMed] [Google Scholar]
- 9. Birnbaum KD. Power in numbers: single-cell RNA-seq strategies to dissect complex tissues. Annu Rev Genet 2018;52:203–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Li Z, Solomonidis EG, Meloni M, Taylor RS, Duffin R, Dobie R, Magalhaes MS, Henderson BEP, Louwe PA, D'Amico G, Hodivala-Dilke KM, Shah AM, Mills NL, Simons BD, Gray GA, Henderson NC, Baker AH, Brittan M. Single-cell transcriptome analyses reveal novel targets modulating cardiac neovascularization by resident endothelial cells following myocardial infarction. Eur Heart J 2019;40:2507–2520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hill MC, Kadow ZA, Li L, Tran TT, Wythe JD, Martin JF. A cellular atlas of Pitx2-dependent cardiac development. Development 2019;146:dev180398-180410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Xiong H, Luo Y, Yue Y, Zhang J, Li AS, Wang X, Zhang X, Wei YL, Li Y, Hu H, Li HX, He CA. Single-cell transcriptomics reveals chemotaxis-mediated intraorgan crosstalk during cardiogenesis. Circ Res 2019;125:398–410. [DOI] [PubMed] [Google Scholar]
- 13. Lescroart F, Wang X, Lin X, Swedlund B, Gargouri S, Sanchez-Danes A, Moignard V, Dubois C, Paulissen C, Kinston S, Gottgens B, Blanpain C. Defining the earliest step of cardiovascular lineage segregation by single-cell RNA-seq. Science 2018;359:1177–1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. de Soysa TY, Ranade SS, Okawa S, Ravichandran S, Huang Y, Salunga HT, Schricker A, Del SA, Gifford CA, Srivastava D. Single-cell analysis of cardiogenesis reveals basis for organ-level developmental defects. Nature 2019;572:120–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ackers-Johnson M, Tan WLW, Foo RS. Following hearts, one cell at a time: recent applications of single-cell RNA sequencing to the understanding of heart disease. Nat Commun 2018;9:4434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hulin A, Hortells L, Gomez-Stallons MV, O'Donnell A, Chetal K, Adam M, Lancellotti P, Oury C, Potter SS, Salomonis N, Yutzey KE. Maturation of heart valve cell populations during postnatal remodeling. Development 2019;146:dev173047-173059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Cui Y, Zheng Y, Liu X, Yan L, Fan X, Yong J, Hu Y, Dong J, Li Q, Wu X, Gao S, Li J, Wen L, Qiao J, Tang F. Single-cell transcriptome analysis maps the developmental track of the human heart. Cell Rep 2019;26:1934–1950. e1935. [DOI] [PubMed] [Google Scholar]
- 18. Farbehi N, Patrick R, Dorison A, Xaymardan M, Janbandhu V, Wystub-Lis K, Ho JW, Nordon RE, Harvey RP. Single-cell expression profiling reveals dynamic flux of cardiac stromal, vascular and immune cells in health and injury. Elife 2019;8:e43882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Dick SA, Macklin JA, Nejat S, Momen A, Clemente-Casares X, Althagafi MG, Chen J, Kantores C, Hosseinzadeh S, Aronoff L, Wong A, Zaman R, Barbu I, Besla R, Lavine KJ, Razani B, Ginhoux F, Husain M, Cybulsky MI, Robbins CS, Epelman S. Self-renewing resident cardiac macrophages limit adverse remodeling following myocardial infarction. Nat Immunol 2019;20:29–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gladka MM, Molenaar B, de Ruiter H, van der Elst S, Tsui H, Versteeg D, Lacraz GPA, Huibers MMH, van Oudenaarden A, van Rooij E. Single-cell sequencing of the healthy and diseased heart reveals cytoskeleton-associated protein 4 as a new modulator of fibroblasts activation. Circulation 2018;138:166–180. [DOI] [PubMed] [Google Scholar]
- 21. Schafer S, Viswanathan S, Widjaja AA, Lim WW, Moreno-Moral A, DeLaughter DM, Ng B, Patone G, Chow K, Khin E, Tan J, Chothani SP, Ye L, Rackham OJL, Ko NSJ, Sahib NE, Pua CJ, Zhen NTG, Xie C, Wang M, Maatz H, Lim S, Saar K, Blachut S, Petretto E, Schmidt S, Putoczki T, Guimaraes-Camboa N, Wakimoto H, van Heesch S, Sigmundsson K, Lim SL, Soon JL, Chao VTT, Chua YL, Tan TE, Evans SM, Loh YJ, Jamal MH, Ong KK, Chua KC, Ong BH, Chakaramakkil MJ, Seidman JG, Seidman CE, Hubner N, Sin KYK, Cook SA. IL-11 is a crucial determinant of cardiovascular fibrosis. Nature 2017;552:110–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Skelly DA, Squiers GT, McLellan MA, Bolisetty MT, Robson P, Rosenthal NA, Pinto AR. Single-cell transcriptional profiling reveals cellular diversity and intercommunication in the mouse heart. Cell Rep 2018;22:600–610. [DOI] [PubMed] [Google Scholar]
- 23. Qian L, Huang Y, Spencer CI, Foley A, Vedantham V, Liu L, Conway SJ, Fu JD, Srivastava D. In vivo reprogramming of murine cardiac fibroblasts into induced cardiomyocytes. Nature 2012;485:593–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ma H, Wang L, Yin C, Liu J, Qian L. In vivo cardiac reprogramming using an optimal single polycistronic construct. Cardiovasc Res 2015;108:217–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Satija R, Farrell JA, Gennert D, Schier AF, Regev A. Spatial reconstruction of single-cell gene expression data. Nat Biotechnol 2015;33:495–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Shay T, Kang J. Immunological Genome Project and systems immunology. Trends Immunol 2013;34:602–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 2009;4:44–57. [DOI] [PubMed] [Google Scholar]
- 28. Qiu X, Hill A, Packer J, Lin D, Ma YA, Trapnell C. Single-cell mRNA quantification and differential analysis with census. Nat Methods 2017;14:309–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Fang R, Preissl S, Hou X, Lucero J, Wang X, Motamedi A, Shiau AK, Mukamel EA, Zhang Y, Behrens MM. Fast and accurate clustering of single cell epigenomes reveals cis-regulatory elements in rare cell types. bioRxiv 2019; 615179.
- 30. Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, Liu XS. Model-based analysis of ChIP-Seq (MACS). Genome Biol 2008;9:R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, Glass CK. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell 2010;38:576–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Schep AN, Wu B, Buenrostro JD, Greenleaf WJ. chromVAR: inferring transcription-factor-associated accessibility from single-cell epigenomic data. Nat Methods 2017;14:975–978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Fornes O, Castro-Mondragon JA, Khan A, van der Lee R, Zhang X, Richmond PA, Modi BP, Correard S, Gheorghe M, Baranasic D, Santana-Garcia W, Tan G, Cheneby J, Ballester B, Parcy F, Sandelin A, Lenhard B, Wasserman WW, Mathelier A. JASPAR 2020: update of the open-access database of transcription factor binding profiles. Nucleic Acids Res 2020;48:D87–D92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Li D, Hsu S, Purushotham D, Sears RL, Wang T. WashU Epigenome Browser update 2019. Nucleic Acids Res 2019;47:W158–W165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Park C, Kim TM, Malik AB. Transcriptional regulation of endothelial cell and vascular development. Circ Res 2013;112:1380–1400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Shi Q, Le X, Abbruzzese JL, Peng Z, Qian CN, Tang H, Xiong Q, Wang B, Li XC, Xie K. Constitutive Sp1 activity is essential for differential constitutive expression of vascular endothelial growth factor in human pancreatic adenocarcinoma. Cancer Res 2001;61:4143–4154. [PubMed] [Google Scholar]
- 37. Gory S, Dalmon J, Prandini MH, Kortulewski T, de Launoit Y, Huber P. Requirement of a GT box (Sp1 site) and two Ets binding sites for vascular endothelial cadherin gene transcription. J Biol Chem 1998;273:6750–6755. [DOI] [PubMed] [Google Scholar]
- 38. Lother A, Bergemann S, Deng L, Moser M, Bode C, Hein L. Cardiac endothelial cell transcriptome. Arterioscler Thromb Vasc Biol 2018;38:566–574. [DOI] [PubMed] [Google Scholar]
- 39. Potente M, Makinen T. Vascular heterogeneity and specialization in development and disease. Nat Rev Mol Cell Biol 2017;18:477–494. [DOI] [PubMed] [Google Scholar]
- 40. Ensan S, Li A, Besla R, Degousee N, Cosme J, Roufaiel M, Shikatani EA, El-Maklizi M, Williams JW, Robins L, Li C, Lewis B, Yun TJ, Lee JS, Wieghofer P, Khattar R, Farrokhi K, Byrne J, Ouzounian M, Zavitz CC, Levy GA, Bauer CM, Libby P, Husain M, Swirski FK, Cheong C, Prinz M, Hilgendorf I, Randolph GJ, Epelman S, Gramolini AO, Cybulsky MI, Rubin BB, Robbins CS. Self-renewing resident arterial macrophages arise from embryonic CX3CR1(+) precursors and circulating monocytes immediately after birth. Nat Immunol 2016;17:159–168. [DOI] [PubMed] [Google Scholar]
- 41. Cochain C, Vafadarnejad E, Arampatzi P, Pelisek J, Winkels H, Ley K, Wolf D, Saliba AE, Zernecke A. Single-cell RNA-Seq reveals the transcriptional landscape and heterogeneity of aortic macrophages in murine atherosclerosis. Circ Res 2018;122:1661–1674. [DOI] [PubMed] [Google Scholar]
- 42. Epelman S, Lavine KJ, Beaudin AE, Sojka DK, Carrero JA, Calderon B, Brija T, Gautier EL, Ivanov S, Satpathy AT, Schilling JD, Schwendener R, Sergin I, Razani B, Forsberg EC, Yokoyama WM, Unanue ER, Colonna M, Randolph GJ, Mann DL. Embryonic and adult-derived resident cardiac macrophages are maintained through distinct mechanisms at steady state and during inflammation. Immunity 2014;40:91–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Ivey MJ, Tallquist MD. Defining the cardiac fibroblast. Circulation Journal 2016;80:2269–2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Acharya A, Baek ST, Huang G, Eskiocak B, Goetsch S, Sung CY, Banfi S, Sauer MF, Olsen GS, Duffield JS, Olson EN, Tallquist MD. The bHLH transcription factor Tcf21 is required for lineage-specific EMT of cardiac fibroblast progenitors. Development 2012;139:2139–2149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Al-Hattab DS, Safi HA, Nagalingam RS, Bagchi RA, Stecy MT, Czubryt MP. Scleraxis regulates Twist1 and Snai1 expression in the epithelial-to-mesenchymal transition. Am J Physiol Heart Circ Physiol 2018;315:H658–H668. [DOI] [PubMed] [Google Scholar]
- 46. Chapman K, Holmes M, Seckl J. 11beta-hydroxysteroid dehydrogenases: intracellular gate-keepers of tissue glucocorticoid action. Physiol Rev 2013;93:1139–1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Fu X, Khalil H, Kanisicak O, Boyer JG, Vagnozzi RJ, Maliken BD, Sargent MA, Prasad V, Valiente-Alandi I, Blaxall BC, Molkentin JD. Specialized fibroblast differentiated states underlie scar formation in the infarcted mouse heart. J Clin Invest 2018;128:2127–2143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Snider P, Standley KN, Wang J, Azhar M, Doetschman T, Conway SJ. Origin of cardiac fibroblasts and the role of periostin. Circ Res 2009;105:934–947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Yamazaki K, Mizui Y, Oki T, Okada M, Tanaka I. Cloning and characterization of mouse glutamine:fructose-6-phosphate amidotransferase 2 gene promoter. Gene 2000;261:329–336. [DOI] [PubMed] [Google Scholar]
- 50. Zhang W, Bouchard G, Yu A, Shafiq M, Jamali M, Shrager JB, Ayers K, Bakr S, Gentles AJ, Diehn M, Quon A, West RB, Nair V, van de Rijn M, Napel S, Plevritis SK. GFPT2-expressing cancer-associated fibroblasts mediate metabolic reprogramming in human lung adenocarcinoma. Cancer Res 2018;78:3445–3457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Dekker J, Marti-Renom MA, Mirny LA. Exploring the three-dimensional organization of genomes: interpreting chromatin interaction data. Nat Rev Genet 2013;14:390–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Tominaga T, Sahai E, Chardin P, McCormick F, Courtneidge SA, Alberts AS. Diaphanous-related formins bridge Rho GTPase and Src tyrosine kinase signaling. Mol Cell 2000;5:13–25. [DOI] [PubMed] [Google Scholar]
- 53. McDonald CA, Liu YY, Palfey BA. Actin stimulates reduction of the MICAL-2 monooxygenase domain. Biochemistry 2013;52:6076–6084. [DOI] [PubMed] [Google Scholar]
- 54. Noakes PG, Gautam M, Mudd J, Sanes JR, Merlie JP. Aberrant differentiation of neuromuscular junctions in mice lacking s-laminin/laminin beta 2. Nature 1995;374:258–262. [DOI] [PubMed] [Google Scholar]
- 55. Kanisicak O, Khalil H, Ivey MJ, Karch J, Maliken BD, Correll RN, Brody MJ, Jl SC, Aronow BJ, Tallquist MD, Molkentin JD. Genetic lineage tracing defines myofibroblast origin and function in the injured heart. Nat Commun 2016;7:12260–12274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. See K, Tan WLW, Lim EH, Tiang Z, Lee LT, Li PYQ, Luu TDA, Ackers-Johnson M, Foo RS. Single cardiomyocyte nuclear transcriptomes reveal a lincRNA-regulated de-differentiation and cell cycle stress-response in vivo. Nat Commun 2017;8:225–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Nomura S, Satoh M, Fujita T, Higo T, Sumida T, Ko T, Yamaguchi T, Tobita T, Naito AT, Ito M, Fujita K, Harada M, Toko H, Kobayashi Y, Ito K, Takimoto E, Akazawa H, Morita H, Aburatani H, Komuro I. Cardiomyocyte gene programs encoding morphological and functional signatures in cardiac hypertrophy and failure. Nat Commun 2018;9:4435–4452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Tallquist MD. Cardiac fibroblasts: from origin to injury. Curr Opin Physiol 2018;1:75–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The scRNA-seq and scATAC-seq datasets have been deposited the Gene Expression Ominbus (GSE157446). Codes used in this study are freely available from the corresponding author upon reasonable request.