INTRODUCTION
Breast cancer is the most commonly diagnosed cancer and the second leading cause of cancer death among women in the United States, with over 281,000 estimated new cases and 43,000 estimated deaths in 2021.1 Owing to its high prevalence, the advancement of clinical practice and basic research to predict the risk, detect and diagnose the disease, and predict response to therapy has a high potential impact. Over the course of many decades, medical imaging modalities have been developed and used in routine clinical practice for these purposes in several capacities, including detection through screening programs, staging when a cancer is found, and planning and monitoring treatment. Screening with mammography is associated with a 20% – 40% reduction in breast cancer deaths.2 However, screening with mammography alone may be insufficient for women at high risk of breast cancer.3 For example, cancers can be missed at mammography in women with dense breasts because of the camouflaging effect.4 The need for more effective assessment strategies has led to the emergence of newer imaging techniques for supplemental screening and diagnostic, prognostic, and treatment purposes, including full-field digital mammography (FFDM), multiparametric magnetic resonance imaging (MRI), digital breast tomosynthesis (DBT), and automated breast ultrasound (ABUS).2,5
While imaging technologies have expanding roles in breast cancer and have provided radiologists with multimodality diagnostic tools applied to various clinical scenarios, they have also led to an increased need for interpretation expertise and reading time. The desire to improve the efficacy and efficiency of clinical care continues to drive innovations, including artificial intelligence (AI). AI offers the opportunity to optimize and streamline the clinical workflow as well as aid in many of the clinical decision-making tasks in image interpretations. AI’s capacity to recognize complex patterns in images, even those that are not noticeable or detectable by human experts, transforms image interpretation into a more quantitative and objective process. AI also excels at processing the sheer amount of information in multimodal data, giving it the potential to integrate not only multiple radiographic imaging modalities but also genomics, pathology, and electronic health records to perform comprehensive analyses and predictions.
AI-assisted systems, such as computer-aided detection (CADe), diagnosis (CADx), and triaging (CADt), have been under development and deployment for clinical use for decades and have accelerated in recent years with the advancement in computing power and modern algorithms.6–10 These AI methods extract and analyze large volumes of quantitative information from image data, assisting radiologists in image interpretation as a concurrent, secondary, or autonomous reader at various steps of the clinical workflow. It is worth noting that while AI systems hold promising prospects in breast cancer image analysis, they also bring along challenges and should be developed and used with abundant caution.
It is important to note that in AI development, two major aspects need to be considered: (1) development of the AI algorithm and (2) evaluation of how it will be eventually used in practice. Currently, most AI systems are being developed and cleared by US Food and Drug Administration (FDA) to augment the interpretation of the medical image, as opposed to autonomous use. These computer-aided methods of implementation include a second reader, a concurrent reader, means to triage cases for reading prioritization, and methods to rule out cases that might not require a human read (a partial autonomous use). In the evaluation of such methods, the human needs to be involved as in dedicated reader studies to demonstrate effectiveness and safety. Table 1 provides a list of AI algorithms cleared by the FDA for various use cases in breast imaging.11
Table 1.
FDA-cleared AI algorithm for breast imaging
| Product | Company | Modality | Use Case | Date Cleared |
|---|---|---|---|---|
| ClearView cCAD | ClearView Diagnostics Inc | US | Diagnosis | 12/28/16 |
| QuantX | Quantitative Insights, Inc | MRI | Diagnosis | 7/19/17 |
| Insight BD | Siemens Healthineers | FFDM, DBT | Breast density | 2/6/18 |
| DM-Density | Densitas, Inc | FFDM | Breast density | 2/23/18 |
| PowerLook Density Assessment Software | ICAD Inc | DBT | Breast density | 4/5/18 |
| DenSeeMammo | Statlife | FFDM | Breast density | 6/26/18 |
| Volpara Imaging Software | Volpara Health Technologies Limited | FFDM, DBT | Breast density | 9/21/18 |
| cmTriage | CureMetrix, Inc | FFDM | Triage | 3/8/19 |
| Koios DS | Koios Medical, Inc | US | Diagnosis | 7/3/19 |
| ProFound AI Software V2.1 | ICAD Inc | DBT | Detection, Diagnosis | 10/4/19 |
| densitas densityai | Densitas, Inc | FFDM, DBT | Breast density | 2/19/20 |
| Transpara | ScreenPoint Medical B.V. | FFDM, DBT | Detection | 3/5/20 |
| MammoScreen | Therapixel | FFDM | Detection | 3/25/20 |
| HealthMammo | Zebra Medical Vision Ltd | FFDM | Triage | 7/16/20 |
| WRDensity | Whiterabbit.ai Inc | FFDM, DBT | Breast density | 10/30/20 |
| Genius AI Detection | Hologic, Inc | DBT | Detection, Diagnosis | 11/18/20 |
| Visage Breast Density | Visage Imaging GmbH | FFDM, DBT | Breast density | 1/29/21 |
Abbreviations: US, ultrasound.
Data from FDA Cleared AI Algorithm. https://models.acrdsi.org.
This article focuses on the research and development of AI systems for clinical breast cancer image analysis, covering the role of AI in the clinical tasks of risk assessment, detection, diagnosis, prognosis, as well as treatment response monitoring and risk of recurrence. In addition to presenting applications by task, the article will start with an introduction to human-engineered radiomics and deep learning techniques and conclude with a discussion on current challenges in the field and future directions.
HUMAN-ENGINEERED ANALYTICS AND DEEP LEARNING TECHNIQUES
AI algorithms often use either human-engineered, analytical, or deep learning methods in the development of machine intelligence tasks. Human-engineered features are mathematical descriptors/model-driven analytics developed to characterize lesions or tissue in medical images. These features quantify visually discernible characteristics, such as size, shape, texture, and morphology, collectively describing the phenotypes of the anatomy imaged. They can be automatically extracted from images of lesions using computer algorithms with analytical expressions encoded, and machine learning models, such as linear discriminant analysis and support vector machines, can be trained on the extracted features to produce predictions for clinical questions. The extraction of human-engineered features often requires a prior segmentation of the lesion from the parenchyma background. Such extraction of features has been conducted on mammography, tomosynthesis, ultrasound, and MRI.8,12 For example, Fig. 1 presents a CADx pipeline that automatically segments breast lesions and extracts six categories of human-engineered radiomic features from dynamic contrast-enhanced (DCE) MRI on a workstation.13–18 Note that the extraction and interpretation of features depend on the imaging modality and the clinical task required.
Fig. 1.
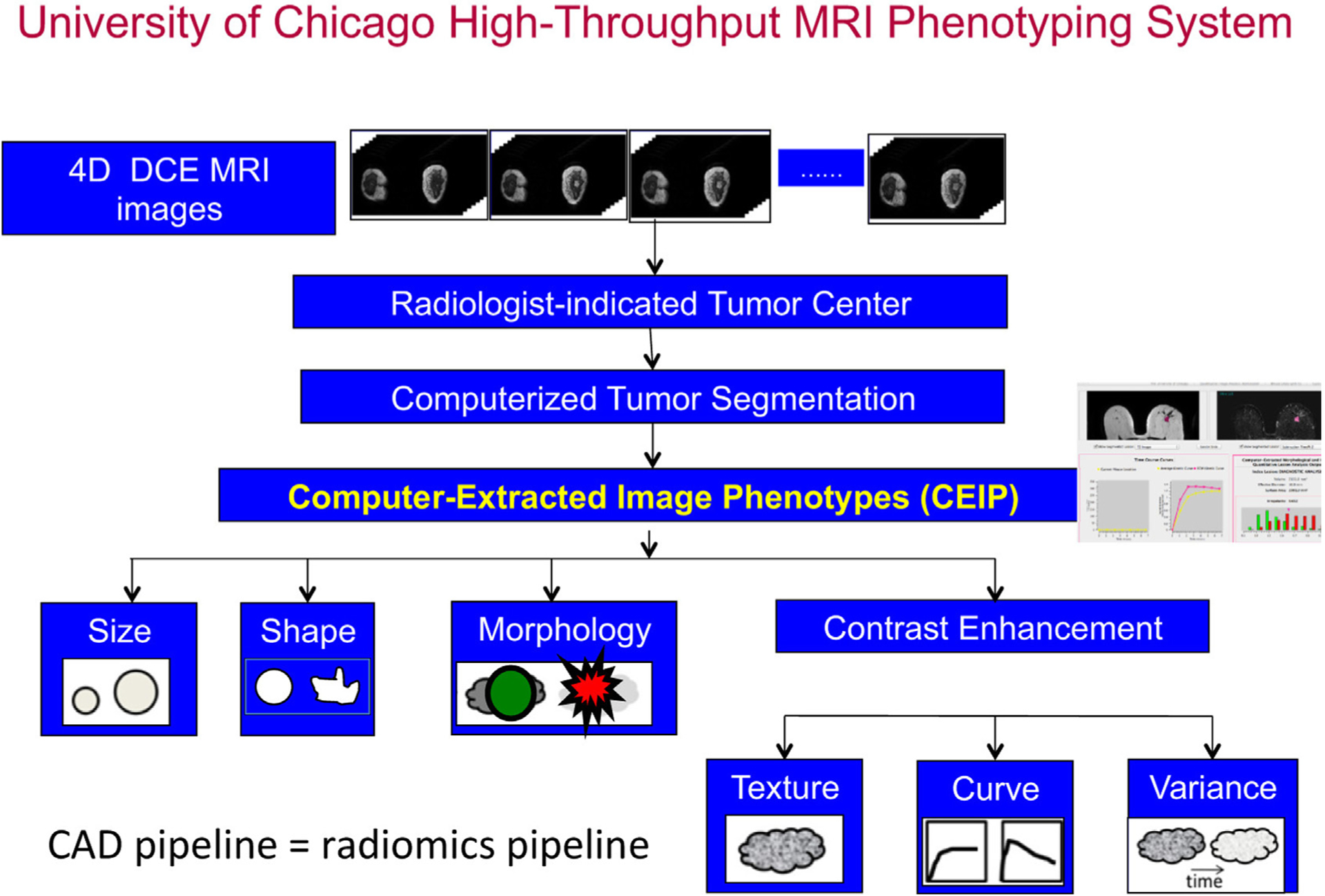
Schematic flowchart of a computerized tumor phenotyping system for breast cancers on DCE-MRI. The CAD radiomics pipeline includes computer segmentation of the tumor from the local parenchyma and computer-extraction of human-engineered radiomic features covering six phenotypic categories: (1) size (measuring tumor dimensions), (2) shape (quantifying the 3D geometry), (3) morphology (characterizing tumor margin), (4) enhancement texture (describing the heterogeneity within the texture of the contrast uptake in the tumor on the first postcontrast MRIs), (5) kinetic curve assessment (describing the shape of the kinetic curve and assessing the physiologic process of the uptake and washout of the contrast agent in the tumor during the dynamic imaging series, and (6) enhancement-variance kinetics (characterizing the time course of the spatial variance of the enhancement within the tumor). CAD, computer-aided diagnosis; DCE-MRI, dynamic contrast-enhanced MRI. (From Giger ML. Machine learning in medical imaging. J Am Coll Radiol. 2018;15(3):512–520; with permission.)
In addition, AI systems that use deep learning algorithms have been increasingly developed for health care applications in recent years.19 Deep learning is a subfield of machine learning and has seen a dramatic resurgence recently, largely driven by increases in computational power and the availability of large data sets. Some of the greatest successes of deep learning have been in computer vision, which considerably accelerated AI applications of medical imaging. Numerous types of models, including convolutional neural networks (CNNs), recurrent neural networks (RNNs), autoencoders, generative adversarial networks, and reinforcement learning, have been developed for medical imaging applications, where they automatically learn features that optimally represent the data for a given task during the training process.20,21 Medical images, nevertheless, pose a set of unique challenges to deep-learning-based computer vision methods, and breast imaging is no exception. For one, the high-dimensionality and large size of medical images allow them to contain a wealth of clinically useful information but make them not suitable for naive applications of standard CNNs developed for image recognition and object detection tasks in natural images. Furthermore, medical imaging data sets are usually relatively small in size and can have incomplete or noisy labels. The lack of interpretability is another hurdle in adopting deep-learning-based AI systems for clinical use. Transfer learning, fusion and aggregation methods, multiple-instance learning, and explainable AI methods continue to be developed to address these challenges in using deep learning algorithms for medical image analysis.22–25
Human-engineered radiomics and deep learning methods for breast imaging analysis both have advantages and disadvantages regarding computation efficiency, amount of data required, preprocessing, interpretability, and prediction accuracy. They should be chosen based on the specific tasks and can be potentially integrated to maximize the benefits of each.26,27 The following sections will cover both of them for each clinical task when applicable.
ARTIFICIAL INTELLIGENCE IN BREAST CANCER RISK ASSESSMENT AND PREVENTION
Computer vision techniques have been developed to extract quantitative biomarkers from normal tissue that are related to cancer risk factors in the task of breast cancer risk assessment in AI-assisted breast image analysis. To improve upon the current one-size-fits-all screening programs, computerized risk assessment can potentially help estimate a woman’s lifetime risk of breast cancer and, thus, recommend risk-stratified screening protocols and/or preventative therapies to reduce the overall risk. Risk models consider risk factors, including demographics, personal history, family history, hormonal status, and hormonal therapy, as well as image-based characteristics such as breast density and parenchymal pattern.
Breast density and parenchymal patterns have been shown to be strong indicators in breast cancer risk estimation.28 Breast density refers to the amount of fibroglandular tissue relative to the amount of fatty tissue. These tissue types are distinguishable on FFDM because fibroglandular tissue attenuates x-ray beams much more than fatty tissue. Breast density has been assessed by radiologists using the four-category Breast Imaging Reporting and Data System (BI-RADS) density ratings, proposed by the American College of Radiology.29 Computerized methods for assessing breast density include calculating the skewness of the gray-level histograms of FFDMs, as well as estimating volumetric density from the 2D projections on FFDMs.8,30,31 Automated assessment of breast density on mammograms is now routinely performed using FDA-cleared clinical systems in breast cancer screening.
On a mammogram, the parenchymal pattern indicates the spatial distribution of dense tissue. To quantitatively evaluate the parenchymal pattern of the breast, various texture-based approaches have been investigated to characterize the spatial distribution of gray levels in FFDMs.8 Such radiomic texture analyses have been conducted using data sets from high-risk groups (eg, BRCA1/BRCA2 gene mutation carriers and women with contralateral cancer) and data sets from low or average risk groups (eg, routine screening populations). Results have shown that women at high risk of breast cancer tend to have dense breasts with parenchymal patterns that are coarse and low in contrast.32–34 Texture analysis on DBT images have also been conducted for risk assessment, with early results showing that texture features correlated with breast density better on DBT than on FFDM.35
Beyond FFDM and DBT, investigators have also assessed the background parenchymal enhancement (BPE) on DCE-MRI. It has been shown that quantitative measures of BPE are associated with the presence of breast cancer, and relative changes in BPE percentages are predictive of breast cancer development after risk-reducing salpingo-oophorectomy.36 A more recent study shows that BPE is associated with an increased risk of breast cancer, and the risk is independent of breast density.37
Various deep learning methods for breast cancer risk assessment have been reported.38,39 One of these methods has shown strong agreement with BI-RADS density assessments by radiologists.40 Another deep learning approach has demonstrated superior performance to methods based on human-engineered features in assessing breast density on FFDMs, as deep learning algorithms can potentially extract additional information contained in FFDMs beyond features defined by human-engineered analytical expressions.41 Moreover, studies have also compared and merged radiomic texture analysis and deep learning approaches in characterizing parenchymal patterns on FFDMs, showing that the combination yield improved results in predicting risk of breast cancer (Fig. 2).39 Besides analyses of FFDM, a deep learning method based on U-Net has been developed to segment fibroglandular tissue on MRI to calculate breast density.42
Fig. 2.
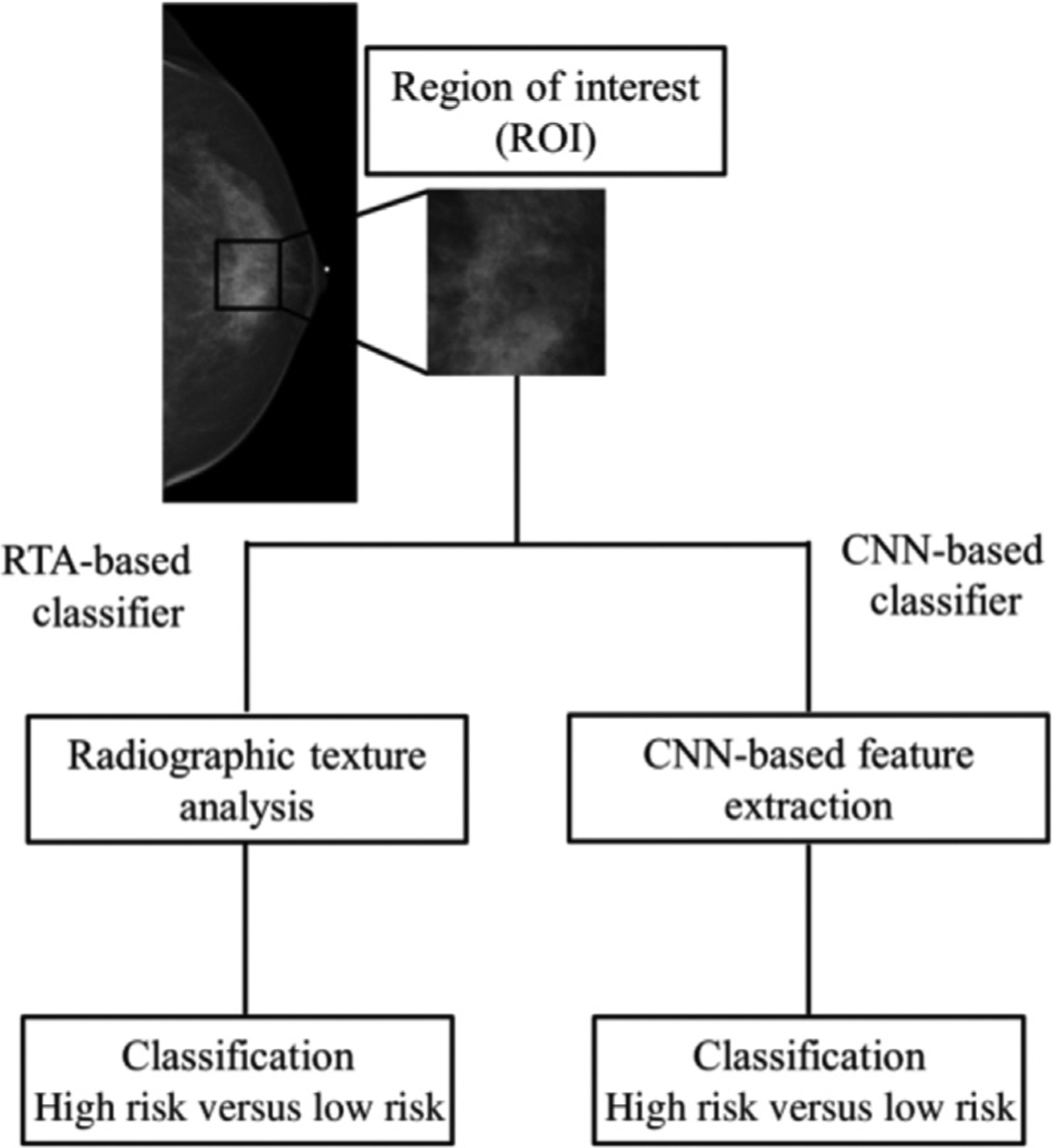
Schematic of methods for the classification of ROIs using human-engineered texture feature analysis and deep convolutional neural network methods. ROI, region of interest. (From Li H. et al. Deep learning in breast cancer risk assessment: evaluation of convolutional neural networks on a clinical dataset of full-field digital mammograms. J Med Imaging 2017;4; with permission.)
ARTIFICIAL INTELLIGENCE IN BREAST CANCER SCREENING AND DETECTION
Detection of abnormalities in breast imaging is a common task for radiologists when reading screening images. Detection task refers to the localization of a lesion, including mass lesion, clustered microcalcifications, and architectural distortion, within the breast. One challenge when detecting abnormalities is that dense tissue can mask the presence of an underlying lesion at mammogram, resulting in missed cancers during breast cancer screening. In addition, radiologists’ ability to detect lesions is also limited by inaccurate assessment of subtle or complex patterns, suboptimal image quality, and fatigue. Therefore, although screening programs have contributed to a reduction in breast cancer–related mortality,43 this process tends to be costly, time-consuming, and error-prone. As a result, CADe methods have been in development for decades to serve a reader besides the radiologists in the task of finding suspicious lesions within images.
In the 1980s, CADe for clustered microcalcifications in digitized mammography was developed using a difference-image technique in which a signal-suppressed image was subtracted from a signal-enhanced image to remove the structured background.44 Human-engineered features were extracted based on the understanding of the signal presentation on mammograms. With the introduction of FFDM, various radiomics methods have evolved and progressed over the years.6–9,12 In 1994, a shift-invariant artificial neural network was used for computerized detection of clustered microcalcifications in breast cancer screening, which was the first journal publication on the use of CNN in medical image analysis (Fig. 3).45
Fig. 3.
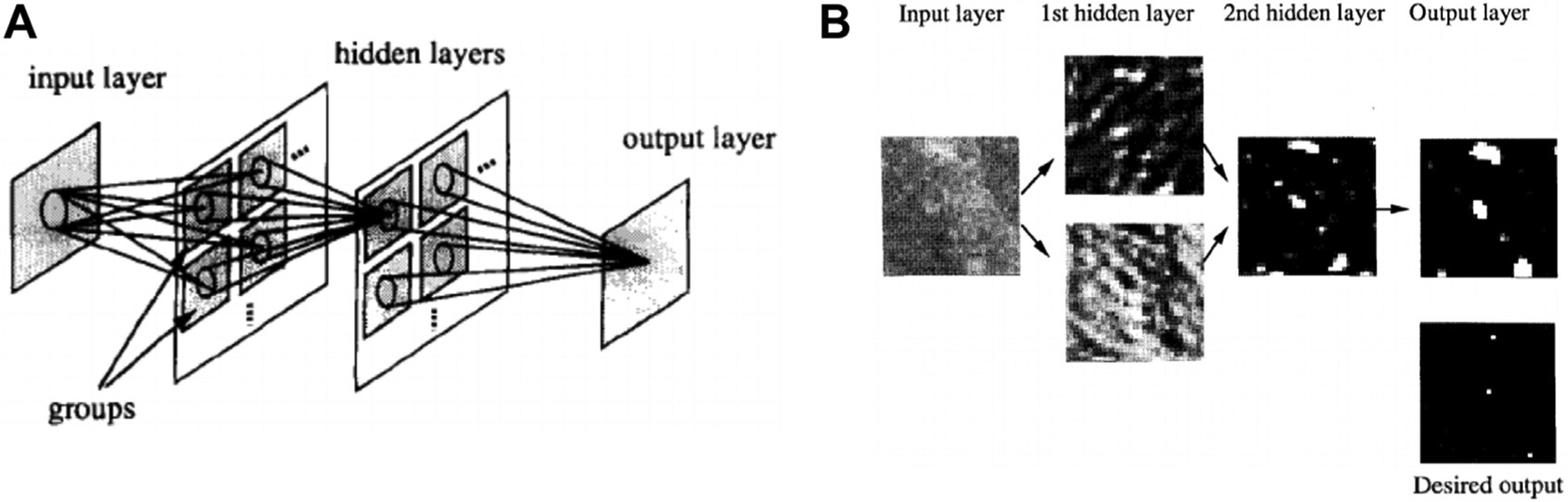
Illustration of the first journal publication on the use of a convolutional neural network (CNN), that is, a shift-invariant neural network, in medical image analysis. The CNN was used in a computer-aided detection system to detect microcalcification on digitized mammograms and later on full-field digital mammograms. (A) A 2D shift-invariant neural network. (B) Illustration of input testing image, output image, desired output image, and responses in hidden layers. (From Zhang W. et al. Computerized detection of clustered microcalcifications in digital mammograms using a shift-invariant artificial neural network. Med Phys. 1994;21(4):517–524; with permission.)
The ImageChecker M1000 system (version 1.2; R2 Technology, Los Altos, CA) was approved by the FDA in 1998, which marked the first clinical translation of mammographic CADe. The system was approved for use as a second reader, where the radiologist would first perform their own interpretation of the mammogram and would only view the CADe system output afterward. A potential lesion indicated by the radiologist but not by the computer output would not be eliminated, ensuring that the sensitivity would not be reduced with the use of CADe. Clinical adoption increased as CADe systems continued to improve. By 2008, CADe systems were used in 70% of screening mammography studies in hospital-based facilities and 81% of private offices46 and stabilized at over 90% of digital mammography screening facilities in the US from 2008 to 2016.47
With the adoption of DBT in screening programs, the development of CADe methods for DBT images accelerated, first as a second reader and more recently as a concurrent reader.48 A multireader, multi-case reader study evaluated a deep learning system developed to detect suspicious soft-tissue and calcified lesions in DBT images and found that the concurrent use of AI improved cancer detection accuracy and efficiency as shown by the increased area under the receiver operating characteristic curve (AUC), sensitivity, and specificity, as well as the reduced recall rate and reading time.49
The recommendation for high-risk patients to receive additional screening with ABUS and/or MRI has motivated the development of CADe on these imaging modalities.50–54 In a reader study, an FDA-approved AI system for detecting lesions on 3D ABUS images showed that when used as a concurrent reader, the system was able to reduce the interpretation time while maintaining diagnostic accuracy.55 Another study developed a CNN-based method that was able to detect breast lesions on the early-phase images in DCE-MRI examinations, suggesting its potential use in screening programs with abbreviated MRI protocols.
Investigators continue working toward the ultimate goal of using AI as an autonomous reader in breast cancer screening and have delivered promising results.56 A recent study demonstrated that their deep learning system yielded a higher AUC than the average AUC of six human readers and was noninferior to radiologists’ double-reading consensus opinion. They also showed through simulation that the system could obviate double reading in 88% of UK screening cases while maintaining a similar level of accuracy to the standard protocol.57 Another recent study proposed a deep learning model whose AUC was greater than the average AUC of 14 human readers, reducing the error approximately by half, and combining radiologists’ assessment and model prediction improved the average specificity by 6.3% compared to human readers alone (Fig. 4).58 It is worth noting that similar to their predecessors, the new CADe systems require additional studies, especially prospective ones, to gauge their real-world performance, robustness, and generalizability before being introduced into the clinical workflow.
Fig. 4.
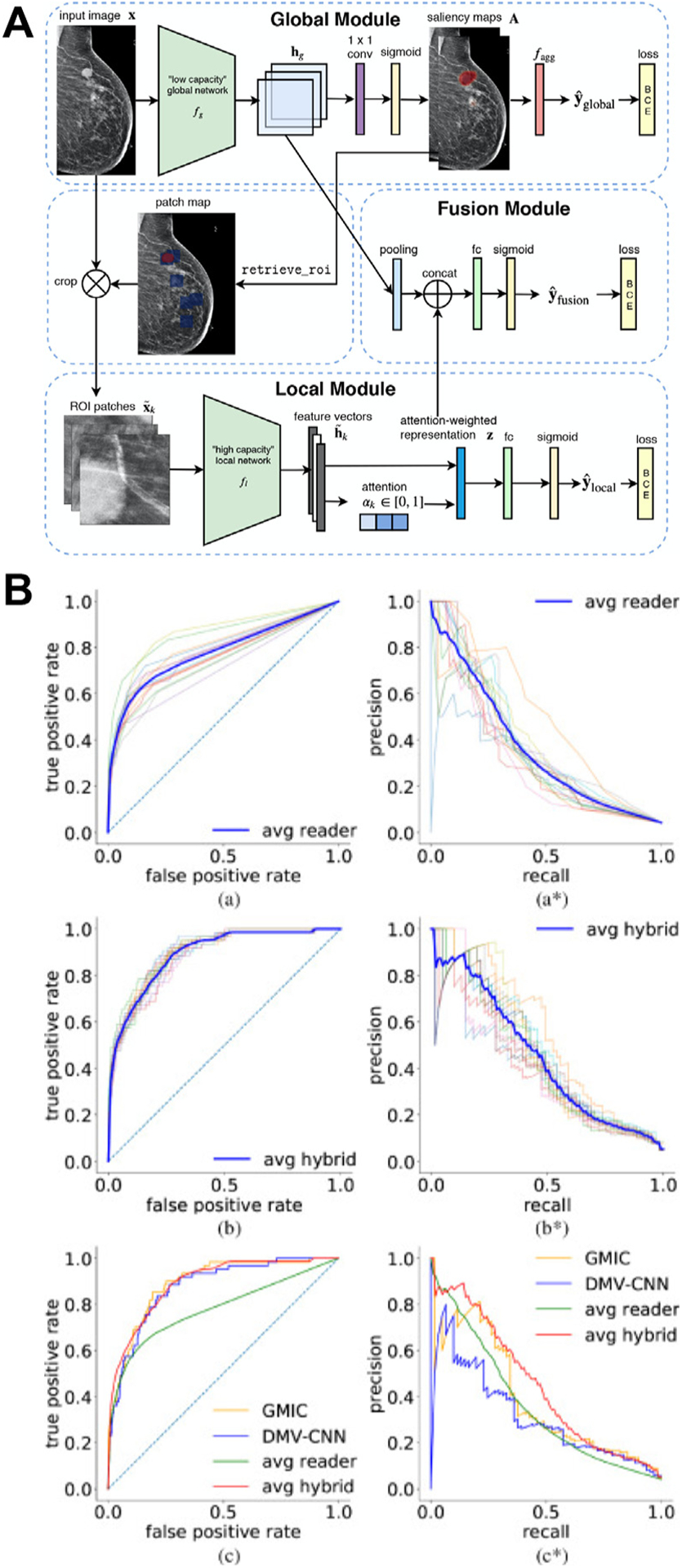
(A) Overall architecture of the globally aware multiple instance classifier (GMIC), in which the patch map indicates positions of the region of interest patches (blue squares) on the input. (B) The receiver operating characteristic curves and precision-recall curves computed on the reader study set. (a, a*): Curves for all 14 readers. (b, b*): Curves for hybrid models with each single reader. The curve highlighted in blue indicates the average performance of all hybrids. (c, c*): Comparison among the GMIC, deep multiview convolutional neural network (DMV-CNN), the average reader, and average hybrid. (From Shen Y. et al. An interpretable classifier for high-resolution breast cancer screening images utilizing weakly supervised localization. Med Image Anal. 2021;68:101908; with permission. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.))
Furthermore, researchers are also investigating the use of AI for triaging and rule out in CADt systems. In a screening program with a CADt system implemented, a certain percentage of FFDMs would be deemed negative by the algorithm without having a radiologist read their mammograms, and those patients would be asked to return for their next screening after the regular screening interval (Fig. 5). In a simulation study, a deep learning approach was used to triage 20% of screening mammograms, and the results showed improvement in radiologist efficiency and specificity without sacrificing sensitivity.59
Fig. 5.
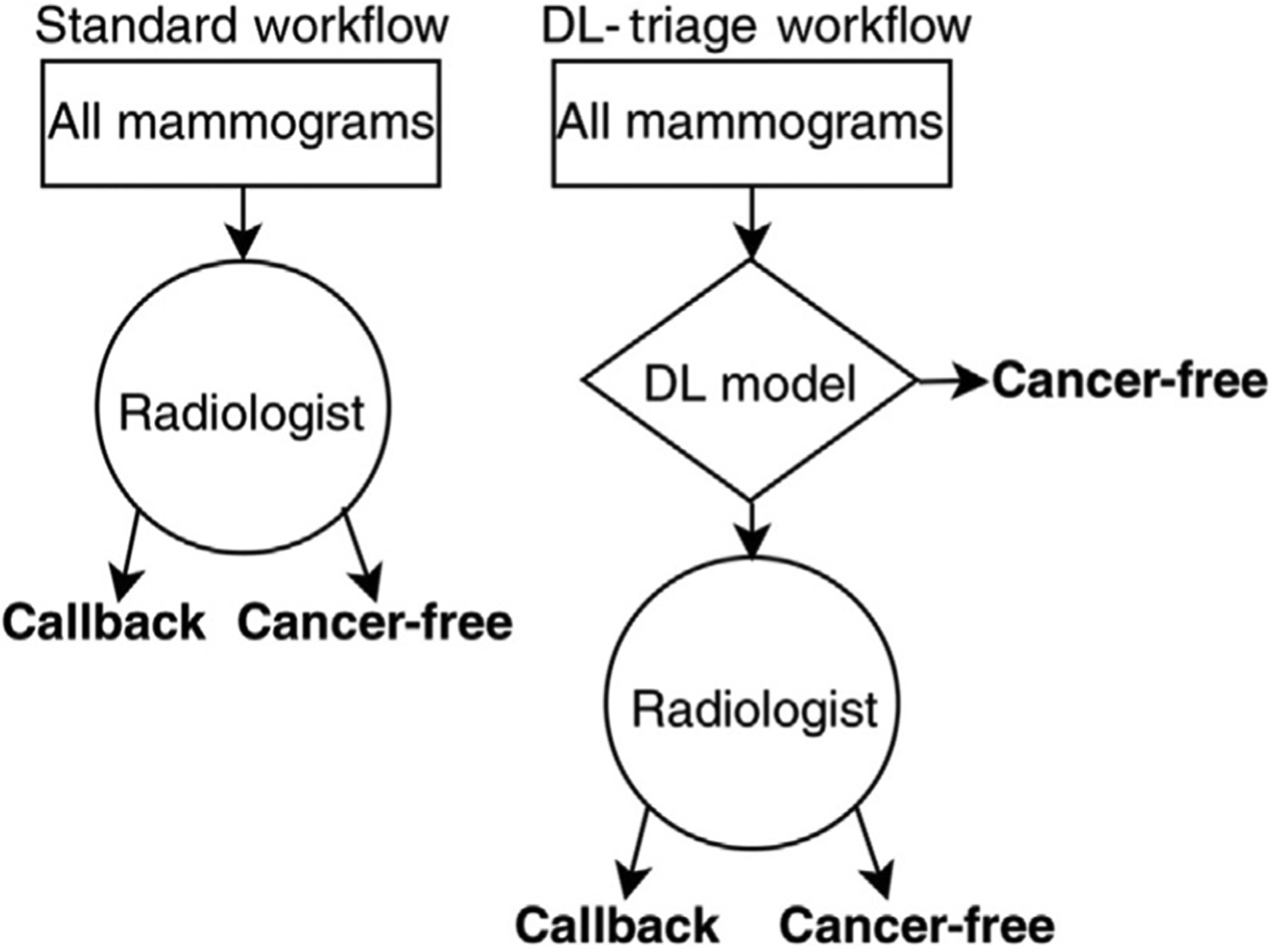
Diagram illustrating the experimental setup for triage analysis (CADt). In the standard scenario, radiologists read all mammograms. In CADt (or rule-out), radiologists only read mammograms above the model’s cancer-free threshold. (From Yala A. et al. A deep learning model to triage screening mammograms: a simulation study. Radiology 2019;293:38–46; with permission.)
ARTIFICIAL INTELLIGENCE IN BREAST CANCER DIAGNOSIS AND PROGNOSIS
During the workup of a breast lesion, diagnosis and prognosis occur after the lesion has been detected by either screening mammography or other examinations. Lesion characterization occurs at this step, and thus, it is a classification task, leaving the radiologist to further assess the likelihood that the lesion is cancerous and determine if the patient should proceed to biopsy for pathologic confirmation. Oftentimes, multiple imaging modalities, including additional mammography, ultrasound, or MRI, are involved in this diagnostic step to better characterize the suspect lesion. When a cancerous tumor is diagnosed, additional imaging is usually conducted to assess the extent of the disease and determine patient management. Given its ability to quantitatively analyze complex patterns in images and process large amounts of information, AI is well suited for the tasks of breast cancer diagnosis and prognosis using image data.
Since the 1980s, investigators have been developing machine learning techniques for CADx in the task of distinguishing between malignant and benign breast lesions.8 From the input image of a lesion, the AI algorithm either extracts human-engineered radiomic features or automatically learns predictive features in the case of deep learning and then outputs a probability of malignancy of the lesion. Algorithms should be trained using pathologically confirmed ground truth to ensure the quality of the data and, in turn, the predictions.
Over the decades, investigators have developed CADx methods that merge features into a tumor signature.9,12,18 Although some radiomic features, such as size, shape, and morphology, can be extracted across various imaging modalities, others are dependent on the modality. For example, spiculation may be extracted from mammographic images of lesions with high spatial resolution, while kinetics-based features are special for DCE-MRI, which contains a temporal sequence of images that visualize the uptake and washout of the contrast agent in the breast.13–17 Moreover, when a feature defined by the same analytical expression is extracted from different imaging modalities, the phenotypes being quantified can be different. For example, texture features can be extracted from the enhancement patterns in DCE-MRI to assess the effects of angiogenesis, but these mathematical texture descriptors characterize different properties of the lesion when extracted from T2-weighted MRI or diffusion-weighted MRI because of the underlying differences between these MRI sequences.60–62 There is also evidence that radiomic analysis on contralateral parenchyma, in addition to the lesion itself, may add value to breast lesion classification.63
Deep learning–based methods have also been developed and proven promising to differentiate benign and cancerous lesions on multiple imaging modalities, including FFDM, DBT, ABUS, and MRI.64–67 Owing to the limited size of data sets in the medical imaging domain, transfer learning is often used where the deep network is initialized with weights pretrained on millions of natural images. As natural images are 2D color images, various approaches have been proposed to maximally use the information in high-dimensional (3D or 4D), gray-level medical images with the pretrained model architectures. For example, the maximum intensity projection of image slices or feature space as well as 3D CNNs has been used to incorporate volumetric information (Fig. 6), and subtraction images, RGB channels in pretrained CNNs, and RNNs have been used to incorporate temporal information (Fig. 7).68–73
Fig. 6.
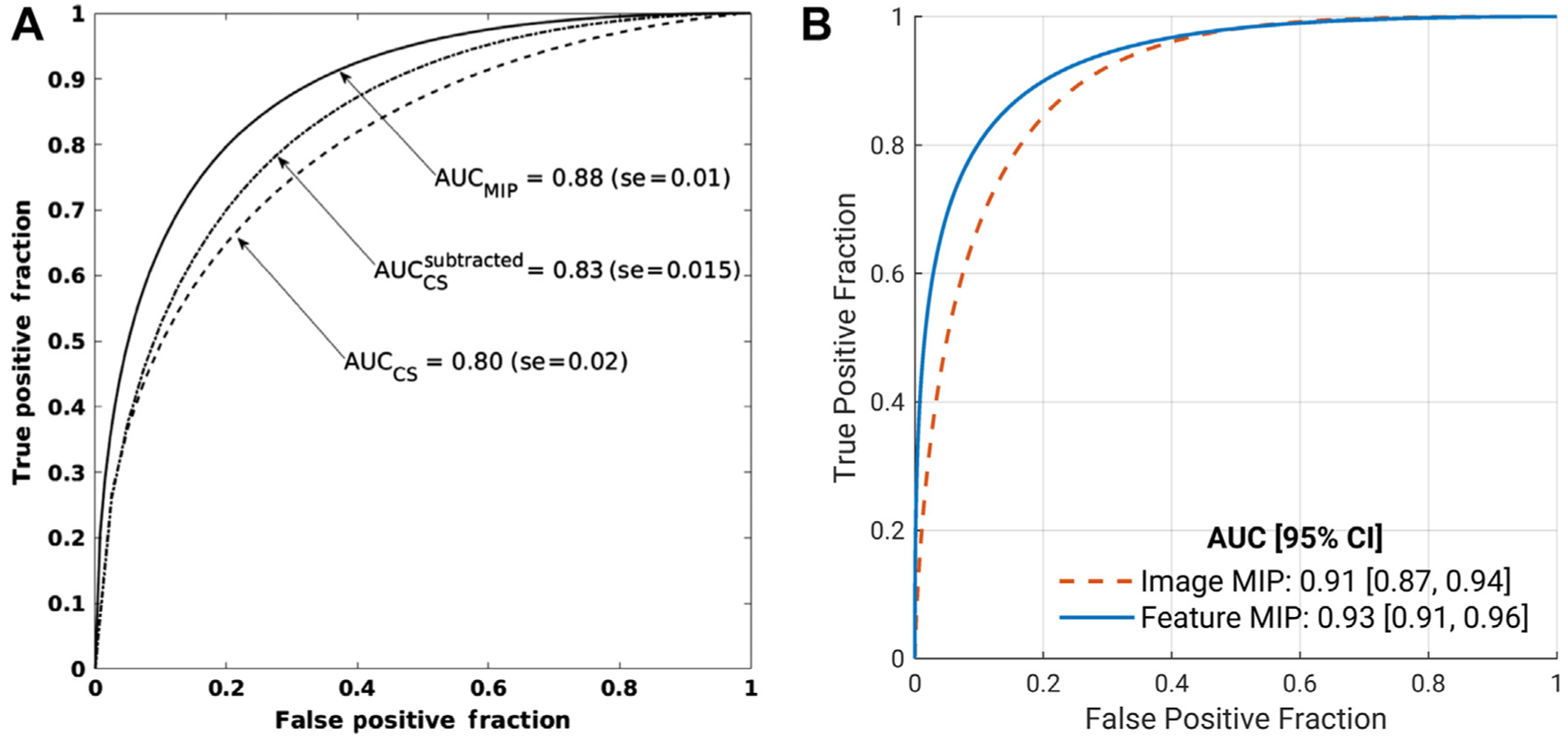
Comparisons of classifier performance in distinguishing between benign and malignant breast lesions when different methods are used to incorporate volumetric information of breast lesions on DCE-MRI. (A) Using the maximum intensity projection (MIP) of the second postcontrast subtraction image outperformed using the central slice of both the second postcontrast subtraction images and the second-post contrast images. CS, central slice. (B) Feature MIP, that is, max pooling the feature space of all slices along the axial dimension, outperformed using image MIP at the input. (From [A] Antropova N, Abe H, Giger ML. Use of clinical MRI maximum intensity projections for improved breast lesion classification with deep convolutional neural networks. J Med Imaging 2018;5(1):14503; with permission. [B] Hu Q, et al. Improved Classification of Benign and Malignant Breast Lesions using Deep Feature Maximum Intensity Projection MRI in Breast Cancer Diagnosis using Dynamic Contrast-Enhanced MRI. Radiol Artif Intell. 2021:e200159; with permission.)
Fig. 7.
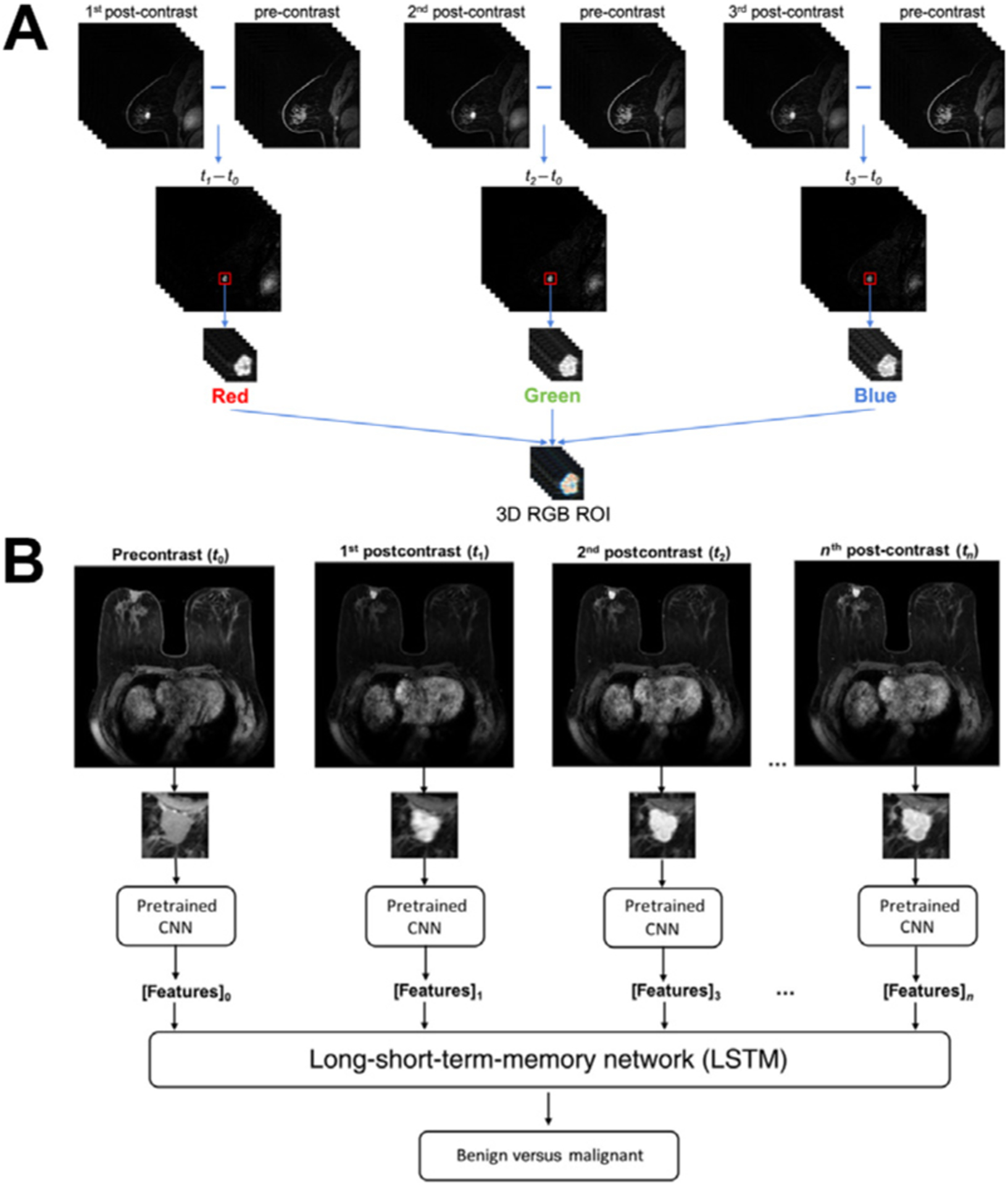
Illustrations of image two approaches of using the temporal sequence of images in DCE-MRI in deep learning–based computer-aided diagnosis methods in distinguishing between benign and malignant breast lesions. (A) The same region of interest (ROI) is cropped from the first, second, and third postcontrast subtraction images and combined in the red, green, and blue (RGB) channels to form an RGB ROI. (B) Features extracted using a pretrained convolutional neural network (CNN) from all time points in DCE-MRI sequences are analyzed by a long short-term memory (LSTM) network to predict the probability of malignancy. (From [A] Hu Q. et al. Improved Classification of Benign and Malignant Breast Lesions using Deep Feature Maximum Intensity Projection MRI in Breast Cancer Diagnosis using Dynamic Contrast-Enhanced MRI. Radiol Artif Intell. 2021:e200159; with permission. [B] Antropova N, et al. Breast lesion classification based on dynamic contrast-enhanced magnetic resonance images sequences with long short-term memory networks. J Med Imaging 2018;6(1):1–7; with permission.)
-With the advancements in breast imaging technology, CADx methods continue to evolve to use the increasingly rich phenotypical information provided in the images and improve the diagnostic performance. For example, multiparametric MRI has been adopted for routine clinical use and has proven to improve clinical diagnostic performance for breast cancer because the different sequences in a multiparametric MRI examination provide complementary information.5,74 AI methods that incorporate the multiple MRI sequences in an examination have been developed in recent years and have demonstrated improved diagnostic performance compared with using DCE-MRI alone (Fig. 8).60,61,67,71 One of these studies investigated fusion strategies at various levels of the classification pipeline and found that latent feature fusion was superior to image fusion and classifier output fusion.67 Notable disagreement was observed between the predictions from classifiers based on different MRI sequences, suggesting that incorporating multiple sequences would be valuable in predicting the probability of malignancy of a lesion (Fig. 9).61,67
Fig. 8.
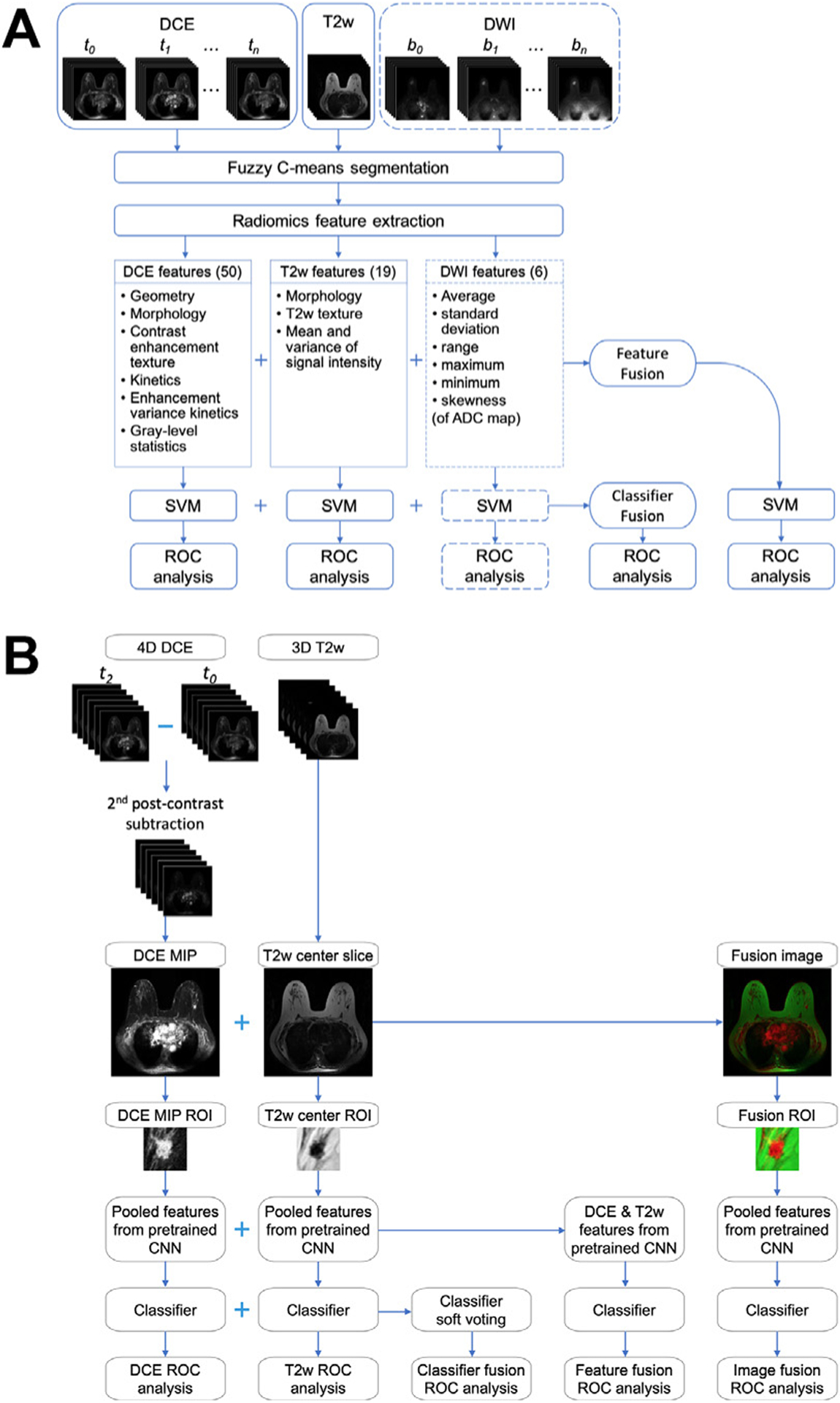
Breast lesion classification pipeline using multiparametric MRI exams using (A) human-engineered radiomics and (B) deep learning. (A) Radiomic features are extracted from dynamic contrast-enhanced (DCE), T2-weighted (T2w), and diffusion-weighted (DWI) MRI sequences. Information from the three sequences is integrated using two fusion strategies: feature fusion, that is, concatenating features extracted from all sequences to train a classifier, and classifier fusion, that is, aggregating the probability of malignancy output from all single-parametric classifiers via soft voting. Parentheses contain the numbers of features extracted from each sequence. The dashed lines for DWI indicate that the DWI sequence is not available in all cases and is included in the classification process when it is available. (B) Information DCE and T2w MRI sequences are integrated using three fusion strategies: image fusion, that is, fusing DCE and T2w images to create RGB composite image, feature fusion as defined in (A), and classifier fusion as defined in (A). ADC, apparent diffusion coefficient; CNN, convolutional neural network; MIP, maximum intensity projection; ROI, region of interest; ROC, receiver operating characteristic; SVM, support vector machine. ([A] From Hu Q, Whitney HM, Giger ML. Radiomics methodology for breast cancer diagnosis using multiparametric magnetic resonance imaging. J Med Imaging. 2020;7(4):44502; with permission. [B] Adapted from Hu Q, Whitney HM, Giger ML. A deep learning methodology for improved breast cancer diagnosis using multiparametric MRI. Sci Rep. 2020;10(1):1–11; with permission. This article is licensed under a Creative Commons Attribution 4.0 International License: http://creativecommons.org/licenses/by/4.0/.)
Fig. 9.
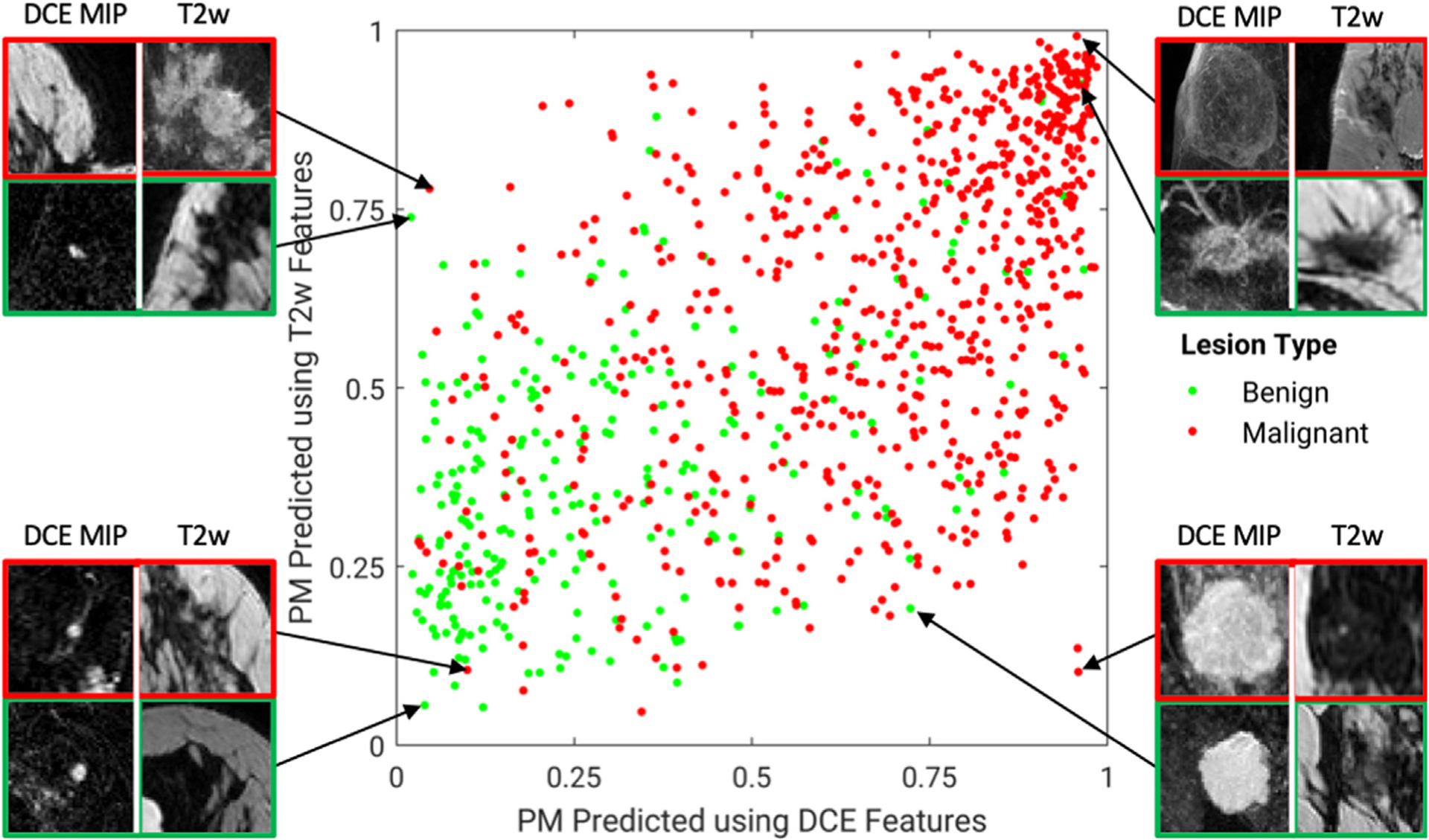
A diagonal classifier agreement plot between the T2-weighted (T2w) and dynamic contrast-enhanced (DCE)-MRI single-sequence deep learning–based classifiers. The x-axis and y-axis denote the probability of malignancy (PM) scores predicted by the DCE classifier and the T2w classifier, respectively. Each point represents a lesion for which predictions were made. Points along or near the diagonal from bottom left to top right indicate high classifier agreement; points far from the diagonal indicate low agreement. A notable disagreement between the two classifiers is observed, suggesting that features extracted from the two MRI sequences provide complementary information, and it is likely valuable to incorporate multiple sequences in multiparametric MRI when making a computer-aided diagnosis prediction. Examples of lesions on which the two classifiers are in extreme agreement/disagreement are also included. (From Hu Q, Whitney HM, Giger ML. A deep learning methodology for improved breast cancer diagnosis using multiparametric MRI. Sci Rep. 2020;10(1):1–11; with permission. This article is licensed under a Creative Commons Attribution 4.0 International License: http://creativecommons.org/licenses/by/4.0/.)
CADx systems based on human-engineered radiomics and deep learning models have been compared and combined. It is worth noting that these two types of machine intelligence have achieved comparable performances and have shown synergistic improvements when combined across multiple modalities (Fig. 10).26,65,75
Fig. 10.
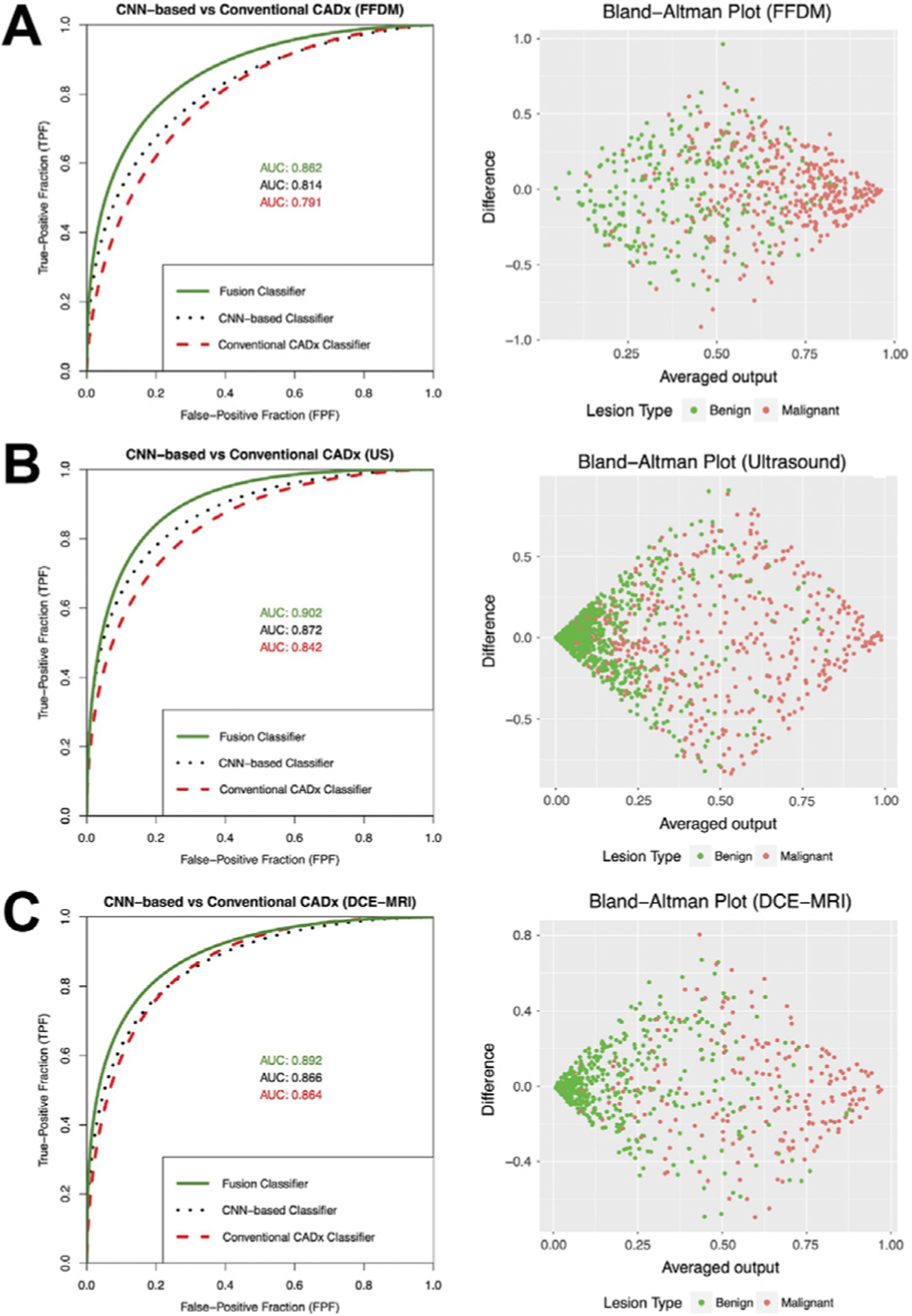
Left column: Fitted binormal ROC curves comparing the performances of classifiers based on human-engineered radiomics, convolutional neural network (CNN), and fusion of the two on three imaging modalities. Right column: Associated Bland-Altman plots illustrating agreement between the classifiers based on human-engineered radiomics and CNN. Since the averaged output is used in the fusion classifier, these plots also help visualize potential decision boundaries for the fusion classifier. (From Antropova N, Huynh BQ, Giger ML. A deep feature fusion methodology for breast cancer diagnosis demonstrated on three imaging modality datasets. Med Phys 2017;44(10):5162–71; with permission.)
The FDA cleared the first commercial breast CADx system (QuantX from Quantitative Insights, Chicago, IL; now Qlarity Imaging) for clinical translation in 2017,76,77 and others have followed for various breast imaging modalities for use as secondary or concurrent readers. In addition to evaluating the diagnostic performance of the AI algorithms themselves, CADx systems have also been evaluated in reader studies when their predictions are incorporated into the radiology workflow as an aid. Improvement in radiologists’ performance has been demonstrated on multiple imaging modalities in the task of distinguishing between benign and malignant breast lesions.77–80
Once a cancer is identified, further workup through biopsies provides information on the stage, molecular subtypes, and other histopathological factors to yield information on prognosis and treatment options. Beyond diagnosis, AI algorithms can also further characterize cancerous lesions to assist in prognosis and subsequent patient management decisions. Many studies have proposed methods to assess the tumor grade, tumor extent, tumor subtype, and molecular subtypes and other histopathological information of breast lesions using various imaging modalities and shown promising results (Fig. 11).81–93 AI methods can relate imaging-based characteristics to clinical, histopathology, or genomic data, contributing to precision medicine for breast cancer. In a collaborative effort through the National Cancer Institute The Cancer Genome Atlas Breast Phenotype Research Group, for example, investigators studied mappings from image-based information of breast tumors extracted to clinical, molecular, and genomic markers. Statistically significant associations were observed between enhancement texture features on DCE-MRI and molecular subtypes.84 Associations between MRI and genomic data were also reported, shedding light on the genetic mechanisms that govern the development of tumor phenotypes, which formed a basis for the future development of noninvasive imaging-based techniques for accurate cancer diagnosis and prognosis (Fig. 12).94
Fig. 11.
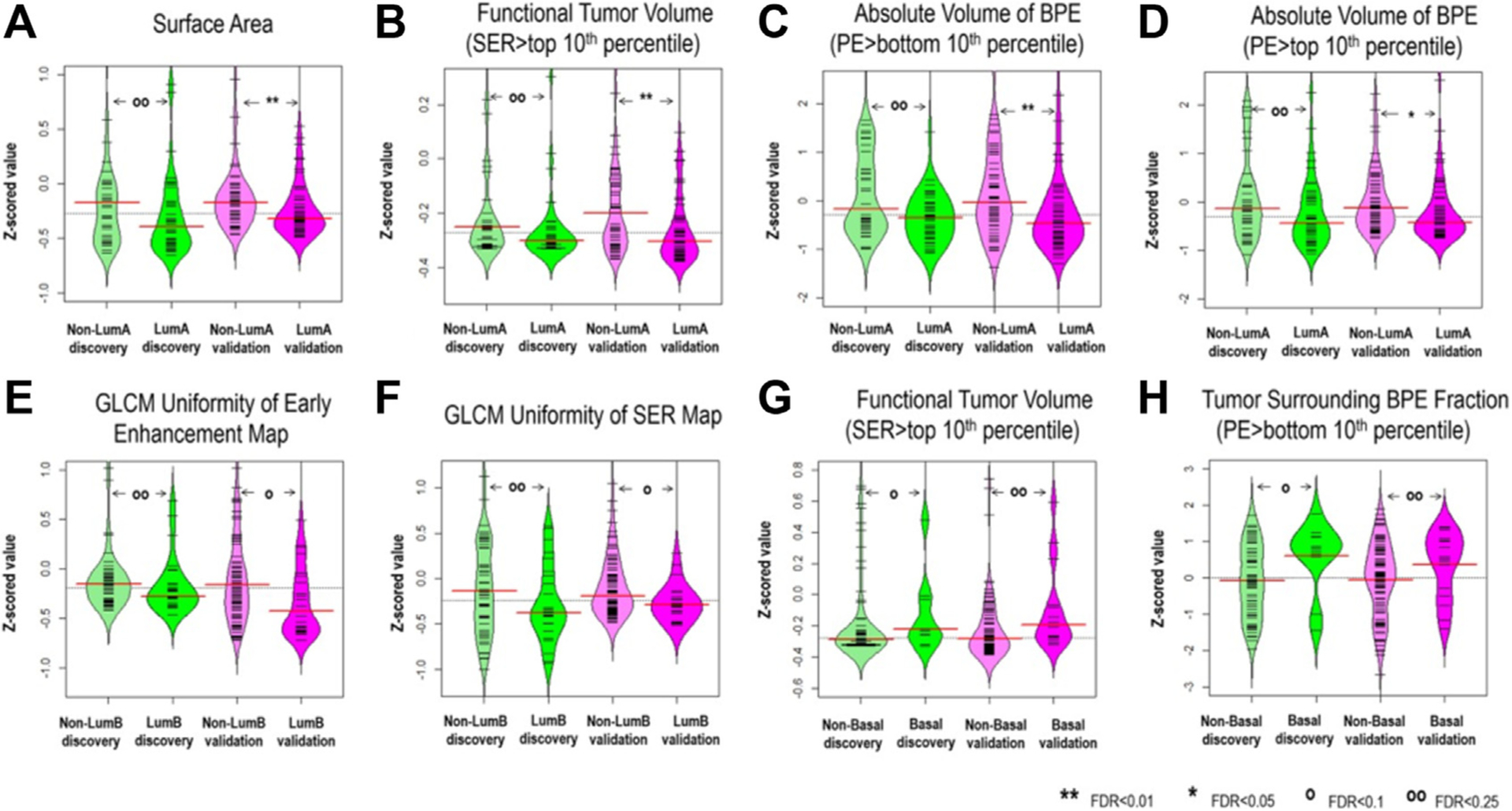
Imaging features significantly associated with molecular subtypes (after correction for multiple testing) in both discovery and validation cohorts, (A–D) four features for distinguishing luminal A versus nonluminal A; (E, F) two features for distinguishing luminal B versus nonluminal B; and (G, H) two features for distinguishing basal-like versus nonbasal-like. Wilcoxon rank-sum test was implemented to investigate pairwise differences. Also, the FDR adjusted for multiple testing is reported. (From Wu J, Sun X, Wang J, et al. Identifying relations between imaging phenotypes and molecular subtypes of breast cancer: Model discovery and external validation. J Magn Reson Imaging. 2017;46(4):1017–1027; with permission.)
Fig. 12.
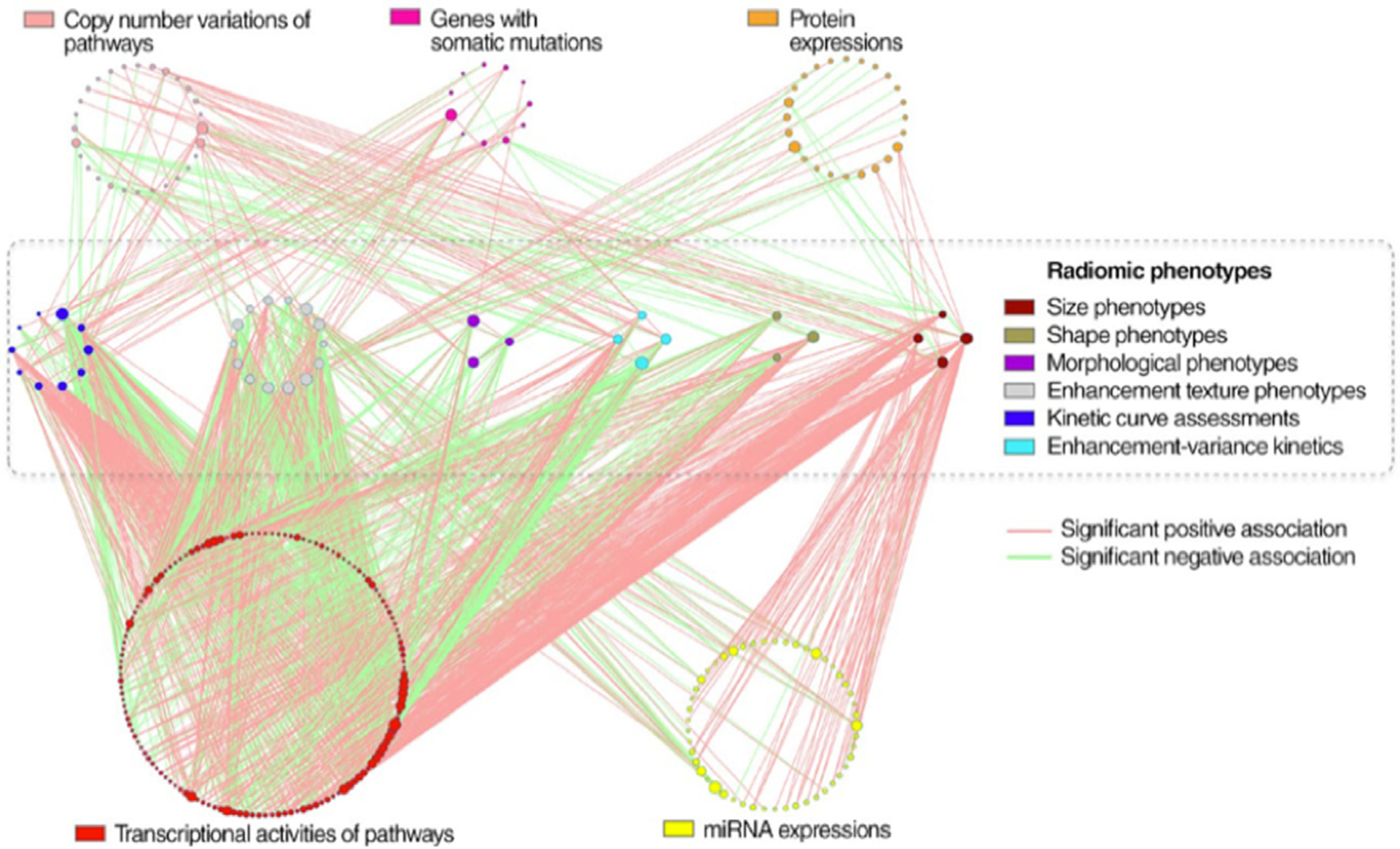
Statistically significant associations between genomic features and radiomic features on MRI in breast carcinoma. Genomic features are organized into circles by data platform and indicated by different node colors. Genomic features without statistically significant associations are not shown. Radiomic phenotypes in six categories are also indicated by different node colors. The node size is proportional to its connectivity relative to other nodes in the category. (From Zhu Y. et al. Deciphering genomic underpinnings of quantitative MRI-based radiomic phenotypes of invasive breast carcinoma. Sci Rep 2015;42(6):3603; with permission.)
Besides its noninvasiveness, such an imaging-based “virtual biopsy” can also provide the advantage of examining a tumor in its entirety, rather than only evaluating the biopsy samples that constitute small parts of a tumor. Given the important role that tumor heterogeneity plays in the prognosis of cancerous lesions, AI-assisted analysis of images presents a strong potential impact on breast cancer prognosis.
ARTIFICIAL INTELLIGENCE IN BREAST CANCER TREATMENT RESPONSE AND RISK OF RECURRENCE
AI algorithms can also be used to assess tumor response to therapy and the risk of recurrence during the treatment of breast cancer. In one example, the size of the most enhancing voxels within the tumor, which was initially extracted from DCE-MRI using a fuzzy c-means method for CADx, has been found useful in assessing recurrence-free survival before or early on during neoadjuvant chemotherapy (NAC) and yielded a comparable performance as another semi-manual method on cases in the I-SPY1 trial.95–97 Another recent study found that while pre-NAC tumor features generally appear uninformative in predicting response to therapy, some pre-NAC lymph node features are predictive.98 Others have used CNNs to predict pathologic complete response (pCR) using the I-SPY1 database and yielded probability heatmaps that indicate regions within the tumors most strongly associated with pCR.99 In another study, a long short-term memory network was able to predict recurrence-free survival in patients with breast cancer only using MRIs acquired early on during the NAC treatment.99 Furthermore, in a study by the TCGA Breast Phenotype Research Group, breast MRIs were quantitatively mapped to research versions of gene assays and showed a significant association between the radiomics signatures and the multigene assay recurrence scores, demonstrating the potential of MRI-based biomarkers for predicting the risk of recurrence.100
Using AI to predict treatment response effectively serves as a “virtual biopsy” that can be conducted during multiple rounds of therapy to track the tumor over time when an actual biopsy is not practical. Such image-based signatures have the potential for increasing the precision in individualized patient management. Moreover, currently unknown correlations between observed phenotypes and genotypes may be discovered through the mappings between imaging data and genomic data, and the unveiled image-based biomarkers can be used in routine screening, prognosis, and monitoring in the future, providing possibilities to improve early detection and better management of the disease.
CHALLENGES IN THE FIELD AND FUTURE DIRECTIONS
Although many breast imaging AI publications are published each year, there are still only a few algorithms that are being translated to clinical care, hindered often by lack of large data sets, which are diverse and well-curated, for training and independent testing. Creating such large data sets will require a change in data-sharing culture allowing for medical institutions to contribute their medical images and data to common image repositories for the public good.
Furthermore, for AI methods to be translated ultimately to clinical care, the algorithms need to demonstrate high performance over a large range of radiological presentations of various breast cancer states on user-friendly interfaces.
In addition, AI methods can be perceived as a “black box” for medical tasks. More research is needed on the explainability of AI methods to help the developer and on the interpretability of AI methods to aid the user. It is interesting to contemplate when might AI be acceptable without giving reasons for its output. To reach such a stage will require high performance in the given clinical task along with high reproducibility and repeatability.
AI in breast imaging is expected to impact both interpretation efficacy and workflow efficiency in radiology as it is applied to imaging examinations being routinely obtained in clinical practice. Although there are more and more novel machine intelligence methods being developed, currently the main use will be in decision support, that is, computers will augment human decision-making as opposed to replacing radiologist decision-making.
KEY POINTS.
AI in breast imaging is expected to impact both interpretation efficacy and workflow efficiency in radiology as it is applied to imaging examinations being routinely obtained in clinical practice.
AI algorithms, including human-engineered radiomics algorithms and deep learning methods, have been under development for multiple decades.
AI can have a role in improving breast cancer risk assessment, detection, diagnosis, prognosis, assessing response to treatment, and predicting recurrence.
Currently the main use of AI algorithms is in decision support, where computers augment human decision-making as opposed to replacing radiologists.
CLINICS CARE POINTS.
Radiologists need to understand the performance level and correct use of AI applications in medical imaging.
FUNDING INFORMATION
The authors are partially supported by NIH QIN Grant U01CA195564, NIH S10 OD025081 Shared Instrument Grant, the University of Chicago Comprehensive Cancer Center, and an RSNA/AAPM Graduate Fellowship.
Funded by:
NIHHYB. Grant number(s): U01CA195564.
Footnotes
DISCLOSURE
Q.Hu declares no competing interests. M.L. Giger is a stockholder in R2 technology/Hologic and QView; receives royalties from Hologic, GE Medical Systems, MEDIAN Technologies, Riverain Medical, Mitsubishi, and Toshiba; and is a cofounder of Quantitative Insights (now Qlarity Imaging). Following the University of Chicago Conflict of Interest Policy, the investigators disclose publicly actual or potential significant financial interest that would reasonably appear to be directly and significantly affected by the research activities.
REFERENCE
- 1.Siegel RL, Miller KD, Fuchs HE, et al. Cancer statistics, 2021. CA Cancer J Clin 2021;71(1):7–33. [DOI] [PubMed] [Google Scholar]
- 2.Niell BL, Freer PE, Weinfurtner RJ, et al. Screening for breast cancer. Radiol Clin North Am 2017;55(6): 1145–62. [DOI] [PubMed] [Google Scholar]
- 3.ACR appropriateness criteria: breast cancer screening. American College of Radiology; 2017. Available at: https://acsearch.acr.org/docs/70910/Narrative. Accessed January 17, 2021.
- 4.Nelson HD, O’Meara ES, Kerlikowske K, et al. Factors associated with rates of false-positive and false-negative results from digital mammography screening: an analysis of registry data. Ann Intern Med 2016;164(4):226–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mann RM, Cho N, Moy L. Breast MRI: state of the art. Radiology 2019;292(3):520–36. [DOI] [PubMed] [Google Scholar]
- 6.Vyborny CJ, Giger ML. Computer vision and artificial intelligence in mammography. Am J Roentgenol 1994;162(3):699–708. [DOI] [PubMed] [Google Scholar]
- 7.Vyborny CJ, Giger ML, Nishikawa RM. Computer-aided detection and diagnosis of breast cancer. Radiol Clin North Am 2000;38(4):725–40. [DOI] [PubMed] [Google Scholar]
- 8.Giger ML, Karssemeijer N, Schnabel JA. Breast image analysis for risk assessment, detection, diagnosis, and treatment of cancer. Annu Rev Biomed Eng 2013;15:327–57. [DOI] [PubMed] [Google Scholar]
- 9.El Naqa I, Haider MA, Giger ML, et al. Artificial Intelligence: reshaping the practice of radiological sciences in the 21st century. Br J Radiol 2020; 93(1106):20190855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li H, Giger ML. Chapter 15 - Artificial intelligence and interpretations in breast cancer imaging. In: Xing L, Giger ML, Min JK, editors. Artificial intelligence in medicine. Cambridge (MA): Academic Press; 2021.p. 291–308. [Google Scholar]
- 11.FDA cleared AI algorithm. Available at: https://models.acrdsi.org. Accessed March 7, 2021.
- 12.Giger ML, Chan H-P, Boone J. Anniversary paper: history and status of CAD and quantitative image analysis: the role of medical physics and AAPM. Med Phys 2008;35(12):5799–820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gilhuijs KGA, Giger ML, Bick U. Computerized analysis of breast lesions in three dimensions using dynamic magnetic-resonance imaging. Med Phys 1998;25(9):1647–54. [DOI] [PubMed] [Google Scholar]
- 14.Chen W, Giger ML, Lan L, et al. Computerized interpretation of breast MRI: investigation of enhancement-variance dynamics. Med Phys 2004;31:1076–82. [DOI] [PubMed] [Google Scholar]
- 15.Chen W, Giger ML, Bick U, et al. Automatic identification and classification of characteristic kinetic curves of breast lesions on DCE-MRI. Med Phys 2006;33(8):2878–87. [DOI] [PubMed] [Google Scholar]
- 16.Chen W, Giger ML, Li H, et al. Volumetric texture analysis of breast lesions on contrast-enhanced magnetic resonance images. Magn Reson Med 2007;58(3):562–71. [DOI] [PubMed] [Google Scholar]
- 17.Chen W, Giger ML, Newstead GM, et al. Computerized assessment of breast lesion malignancy using DCE-MRI: robustness study on two independent clinical datasets from two manufacturers. Acad Radiol 2010;17(7):822–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Giger ML. Machine learning in medical imaging. J Am Coll Radiol 2018;15(3):512–20. [DOI] [PubMed] [Google Scholar]
- 19.Esteva A, Robicquet A, Ramsundar B, et al. A guide to deep learning in healthcare. Nat Med 2019;25(1):24–9. [DOI] [PubMed] [Google Scholar]
- 20.Litjens G, Kooi T, Bejnordi BE, et al. A survey on deep learning in medical image analysis. Med Image Anal 2017;42:60–88. [DOI] [PubMed] [Google Scholar]
- 21.Sahiner B, Pezeshk A, Hadjiiski LM, et al. Deep learning in medical imaging and radiation therapy. Med Phys 2019;46(1):e1–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maron O, Lozano-Pérez T. A framework for multiple-instance learning. In: Kearns MJ, Solla SA, Cohn DA, editors. Advances in neural information processing systems. Cambridge (MA): The MIT Press; 1998. p. 570–6. [Google Scholar]
- 23.Tajbakhsh N, Shin JY, Gurudu SR, et al. Convolutional neural networks for medical image analysis: Full training or fine tuning? IEEE Trans Med Imaging 2016;35(5):1299–312. [DOI] [PubMed] [Google Scholar]
- 24.Shin H-C, Roth HR, Gao M, et al. Deep convolutional neural networks for computer-aided detection: CNN architectures, dataset characteristics and transfer learning. IEEE Trans Med Imaging 2016;35(5):1285–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang Q, Zhu S-C. Visual interpretability for deep learning: a survey. Front Inf Technol Electron Eng 2018;19(1):27–39. [Google Scholar]
- 26.Whitney HM, Li H, Ji Y, et al. Comparison of breast MRI tumor classification using human-engineered radiomics, transfer learning from deep convolutional neural networks, and fusion method. Proc IEEE 2019;108(1):163–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Truhn D, Schrading S, Haarburger C, et al. Radiomic versus convolutional neural networks analysis for classification of contrast-enhancing lesions at multiparametric breast MRI. Radiology 2019; 290(2):290–7. [DOI] [PubMed] [Google Scholar]
- 28.Freer PE. Mammographic breast density: impact on breast cancer risk and implications for screening. RadioGraphics 2015;35(2):302–15. [DOI] [PubMed] [Google Scholar]
- 29.D’Orsi C, Sickles E, Mendelson E, et al. ACR BI-RADS® atlas, breast imaging reporting and data system. Reston, VA, American College of Radiology; 2013. [Google Scholar]
- 30.Byng JW, Yaffe MJ, Lockwood GA, et al. Automated analysis of mammographic densities and breast carcinoma risk. Cancer 1997;80(1):66–74. [DOI] [PubMed] [Google Scholar]
- 31.van Engeland S, Snoeren PR, Huisman H, et al. Volumetric breast density estimation from full-field digital mammograms. IEEE Trans Med Imaging 2006;25(3):273–82. [DOI] [PubMed] [Google Scholar]
- 32.Li H, Giger ML, Olopade OI, et al. Power spectral analysis of mammographic parenchymal patterns for breast cancer risk assessment. J Digit Imaging 2008;21(2):145–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li H, Giger ML, Olopade OI, et al. Fractal analysis of mammographic parenchymal patterns in breast cancer risk assessment. Acad Radiol 2007;14(5):513–21. [DOI] [PubMed] [Google Scholar]
- 34.Huo Z, Giger ML, Olopade OI, et al. Computerized analysis of digitized mammograms of BRCA1 and BRCA2 gene mutation carriers. Radiology 2002; 225(2):519–26. [DOI] [PubMed] [Google Scholar]
- 35.Kontos D, Bakic PR, Carton A-K, et al. Parenchymal texture analysis in digital breast tomosynthesis for breast cancer risk estimation: a preliminary study. Acad Radiol 2009;16(3): 283–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wu S, Weinstein SP, DeLeo MJ, et al. Quantitative assessment of background parenchymal enhancement in breast MRI predicts response to risk-reducing salpingo-oophorectomy: preliminary evaluation in a cohort of BRCA1/2 mutation carriers. Breast Cancer Res 2015;17(1):67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Arasu VA, Miglioretti DL, Sprague BL, et al. Population-based assessment of the association between magnetic resonance imaging background parenchymal enhancement and future primary breast cancer risk. J Clin Oncol 2019;37(12):954–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gastounioti A, Oustimov A, Hsieh M-K, et al. Using convolutional neural networks for enhanced capture of breast parenchymal complexity patterns associated with breast cancer risk. Acad Radiol 2018;25(8):977–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li H, Giger ML, Huynh BQ, et al. Deep learning in breast cancer risk assessment: evaluation of convolutional neural networks on a clinical dataset of full-field digital mammograms. J Med Imaging 2017;4(4):41304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lee J, Nishikawa RM. Automated mammographic breast density estimation using a fully convolutional network. Med Phys 2018;45(3):1178–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li S, Wei J, Chan H-P, et al. Computer-aided assessment of breast density: comparison of supervised deep learning and feature-based statistical learning. Phys Med Biol 2018;63(2):25005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dalmış MU, Litjens G, Holland K, et al. Using deep learning to segment breast and fibroglandular tissue in MRI volumes. Med Phys 2017;44(2):533–46. [DOI] [PubMed] [Google Scholar]
- 43.Lauby-Secretan B, Scoccianti C, Loomis D, et al. Breast-cancer screening — viewpoint of the IARC Working Group. N Engl J Med 2015;372(24):2353–8. [DOI] [PubMed] [Google Scholar]
- 44.Chan H-P, Doi K, Galhotra S, et al. Image feature analysis and computer-aided diagnosis in digital radiography. I. Automated detection of microcalcifications in mammography. Med Phys 1987;14(4):538–48. [DOI] [PubMed] [Google Scholar]
- 45.Zhang W, Doi K, Giger ML, et al. Computerized detection of clustered microcalcifications in digital mammograms using a shift-invariant artificial neural network. Med Phys 1994;21(4):517–24. [DOI] [PubMed] [Google Scholar]
- 46.Rao VM, Levin DC, Parker L, et al. How widely is computer-aided detection used in screening and diagnostic mammography? J Am Coll Radiol 2010;7(10):802–5. [DOI] [PubMed] [Google Scholar]
- 47.Keen JD, Keen JM, Keen JE. Utilization of computer-aided detection for digital screening mammography in the United States, 2008 to 2016. J Am Coll Radiol 2018;15(1, Part A):44–8. [DOI] [PubMed] [Google Scholar]
- 48.Geras KJ, Mann RM, Moy L. Artificial intelligence for mammography and digital breast tomosynthesis: current concepts and future perspectives. Radiology 2019;293(2):246–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Conant EF, Toledano AY, Periaswamy S, et al. Improving accuracy and efficiency with concurrent use of artificial intelligence for digital breast tomosynthesis. Radiol Artif Intell 2019;1(4):e180096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kuhl CK. Abbreviated breast MRI for screening women with dense breast: the EA1141 trial. Br J Radiol 2018;91(1090):20170441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Supplemental screening for breast cancer in women with dense breasts: a systematic review for the U.S. preventive services task force. Ann Intern Med 2016;164(4):268–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Giger ML, Inciardi MF, Edwards A, et al. Automated breast ultrasound in breast cancer screening of women with dense breasts: reader study of mammography-negative and mammography-positive cancers. Am J Roentgenol 2016;206(6): 1341–50. [DOI] [PubMed] [Google Scholar]
- 53.Comstock CE, Gatsonis C, Newstead GM, et al. Comparison of abbreviated breast MRI vs digital breast tomosynthesis for breast cancer detection among women with dense breasts undergoing screening. JAMA 2020;323(8):746–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.van Zelst JCM, Tan T, Clauser P, et al. Dedicated computer-aided detection software for automated 3D breast ultrasound; an efficient tool for the radiologist in supplemental screening of women with dense breasts. Eur Radiol 2018; 28(7):2996–3006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jiang Y, Inciardi MF, Edwards AV, et al. Interpretation time using a concurrent-read computer-aided detection system for automated breast ultrasound in breast cancer screening of women with dense breast tissue. Am J Roentgenol 2018;211(2): 452–61. [DOI] [PubMed] [Google Scholar]
- 56.Sechopoulos I, Mann RM. Stand-alone artificial intelligence - The future of breast cancer screening? Breast 2020;49:254–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.McKinney SM, Sieniek M, Godbole V, et al. International evaluation of an AI system for breast cancer screening. Nature 2020;577(7788):89–94. [DOI] [PubMed] [Google Scholar]
- 58.Shen Y, Wu N, Phang J, et al. An interpretable classifier for high-resolution breast cancer screening images utilizing weakly supervised localization. Med Image Anal 2021;68:101908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yala A, Schuster T, Miles R, et al. A deep learning model to triage screening mammograms: a simulation study. Radiology 2019;293(1):38–46. [DOI] [PubMed] [Google Scholar]
- 60.Bhooshan N, Giger M, Lan L, et al. Combined use of T2-weighted MRI and T1-weighted dynamic contrast-enhanced MRI in the automated analysis of breast lesions. Magn Reson Med 2011;66(2):555–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hu Q, Whitney HM, Giger ML. Radiomics methodology for breast cancer diagnosis using multiparametric magnetic resonance imaging. J Med Imaging 2020;7(4):44502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Parekh VS, Jacobs MA. Integrated radiomic framework for breast cancer and tumor biology using advanced machine learning and multiparametric MRI. NPJ Breast Cancer 2017;3(1):43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Li H, Mendel KR, Lan L, et al. Digital mammography in breast cancer: additive value of radiomics of breast parenchyma. Radiology 2019;291(1):15–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Huynh BQ, Li H, Giger ML. Digital mammographic tumor classification using transfer learning from deep convolutional neural networks. J Med Imaging 2016;3(3):34501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Antropova N, Huynh BQ, Giger ML. A deep feature fusion methodology for breast cancer diagnosis demonstrated on three imaging modality datasets. Med Phys 2017;44(10):5162–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Samala RK, Chan H-P, Hadjiiski LM, et al. Evolutionary pruning of transfer learned deep convolutional neural network for breast cancer diagnosis in digital breast tomosynthesis. Phys Med Biol 2018;63(9):95005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hu Q, Whitney HM, Giger ML. A deep learning methodology for improved breast cancer diagnosis using multiparametric MRI. Sci Rep 2020;10(1):1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Antropova N, Abe H, Giger ML. Use of clinical MRI maximum intensity projections for improved breast lesion classification with deep convolutional neural networks. J Med Imaging 2018;5(1):14503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hu Q, Whitney HM, Giger ML. Transfer learning in 4D for breast cancer diagnosis using dynamic contrast-enhanced magnetic resonance imaging. arXiv Prepr arXiv191103022. 2019. [Google Scholar]
- 70.Hu Q, Whitney HM, Li H, et al. Improved classification of benign and malignant breast lesions using deep feature maximum intensity projection MRI in breast cancer diagnosis using dynamic contrast-enhanced MRI. Radiol Artif Intell 2021;3(3):e200159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Dalmis MU, Gubern-Mérida A, Vreemann S, et al. Artificial intelligence–based classification of breast lesions imaged with a multiparametric breast MRI protocol with ultrafast DCE-MRI, T2, and DWI. Invest Radiol 2019;54(6):325–32. [DOI] [PubMed] [Google Scholar]
- 72.Li J, Fan M, Zhang J, et al. Discriminating between benign and malignant breast tumors using 3D convolutional neural network in dynamic contrast enhanced-MR images. In: Cook TS, Zhang J, editors. medical imaging 2017: imaging Informatics for healthcare, research, and applications. vol. 10138. Bellingham (WA): SPIE Press; 2017: 1013808. [Google Scholar]
- 73.Antropova N, Huynh B, Li H, et al. Breast lesion classification based on dynamic contrast-enhanced magnetic resonance images sequences with long short-term memory networks. J Med Imaging 2018;6(1):1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Leithner D, Wengert GJ, Helbich TH, et al. Clinical role of breast MRI now and going forward. Clin Radiol 2018;73(8):700–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hu Q, Whitney HM, Edwards A, et al. Radiomics and deep learning of diffusion-weighted MRI in the diagnosis of breast cancer. In: Mori K, Hahn HK, editors. Medical imaging 2019: computer-aided diagnosis. vol. 10950. Bellingham (WA): SPIE Press; 2019:109504A. [Google Scholar]
- 76.Evaluation of Automatic Class III Designation (De Novo) Summaries. The Food and Drug Administration. [Google Scholar]
- 77.Jiang Y, Edwards AV, Newstead GM. Artificial intelligence applied to breast MRI for improved diagnosis. Radiology 2021;298(1):38–46. [DOI] [PubMed] [Google Scholar]
- 78.Huo Z, Giger ML, Vyborny CJ, et al. Breast cancer: effectiveness of computer-aided diagnosis—observer study with independent database of mammograms. Radiology 2002;224(2):560–8. [DOI] [PubMed] [Google Scholar]
- 79.Horsch K, Giger ML, Vyborny CJ, et al. Performance of computer-aided diagnosis in the interpretation of lesions on breast sonography. Acad Radiol 2004;11(3):272–80. [DOI] [PubMed] [Google Scholar]
- 80.Shimauchi A, Giger ML, Bhooshan N, et al. Evaluation of clinical breast MR imaging performed with prototype computer-aided diagnosis breast MR imaging workstation: reader study. Radiology 2011; 258(3):696–704. [DOI] [PubMed] [Google Scholar]
- 81.Loiselle C, Eby PR, Kim JN, et al. Preoperative MRI improves prediction of extensive occult axillary lymph node metastases in breast cancer patients with a positive sentinel lymph node biopsy. Acad Radiol 2014;21(1):92–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Schacht DV, Drukker K, Pak I, et al. Using quantitative image analysis to classify axillary lymph nodes on breast MRI: a new application for the Z 0011 Era. Eur J Radiol 2015;84(3):392–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Guo W, Li H, Zhu Y, et al. Prediction of clinical phenotypes in invasive breast carcinomas from the integration of radiomics and genomics data. J Med Imaging 2015;2(4):1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Li H, Zhu Y, Burnside ES, et al. Quantitative MRI radiomics in the prediction of molecular classifications of breast cancer subtypes in the TCGA/TCIA data set. npj Breast Cancer 2016;2(1):16012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Bhooshan N, Giger ML, Jansen SA, et al. Cancerous breast lesions on dynamic contrast-enhanced MR images. Breast Imaging 2010;254(3):680–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Liu W, Cheng Y, Liu Z, et al. Preoperative prediction of Ki-67 status in breast cancer with multiparametric MRI using transfer learning. Acad Radiol 2021; 28(2):e44–53. [DOI] [PubMed] [Google Scholar]
- 87.Liang C, Cheng Z, Huang Y, et al. An MRI-based radiomics classifier for preoperative prediction of Ki-67 status in breast cancer. Acad Radiol 2018; 25(9):1111–7. [DOI] [PubMed] [Google Scholar]
- 88.Ma W, Zhao Y, Ji Y, et al. Breast cancer molecular subtype prediction by mammographic radiomic features. Acad Radiol 2019;26(2):196–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Leithner D, Mayerhoefer ME, Martinez DF, et al. Non-invasive assessment of breast cancer molecular subtypes with multiparametric magnetic resonance imaging radiomics. J Clin Med 2020;9(6):1853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Son J, Lee SE, Kim E-K, et al. Prediction of breast cancer molecular subtypes using radiomics signatures of synthetic mammography from digital breast tomosynthesis. Sci Rep 2020;10(1):21566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Grimm LJ, Zhang J, Baker JA, et al. Relationships between MRI Breast Imaging-Reporting and Data System (BI-RADS) lexicon descriptors and breast cancer molecular subtypes: internal enhancement is associated with luminal B subtype. Breast J 2017;23(5):579–82. [DOI] [PubMed] [Google Scholar]
- 92.Wu J, Sun X, Wang J, et al. Identifying relations between imaging phenotypes and molecular subtypes of breast cancer: model discovery and external validation. J Magn Reson Imaging 2017; 46(4):1017–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Burnside ES, Drukker K, Li H, et al. Using computer-extracted image phenotypes from tumors on breast magnetic resonance imaging to predict breast cancer pathologic stage. Cancer 2016;122(5):748–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Zhu Y, Li H, Guo W, et al. TU-CD-BRB-06: deciphering genomic underpinnings of quantitative MRI-based radiomic phenotypes of invasive breast carcinoma. Med Phys 2015;42(6Part32):3603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Chen W, Giger ML, Bick U. A fuzzy c-means (FCM)-based approach for computerized segmentation of breast lesions in dynamic contrast-enhanced MR images. Acad Radiol 2006;13(1):63–72. [DOI] [PubMed] [Google Scholar]
- 96.Hylton NM, Gatsonis CA, Rosen MA, et al. Neoadjuvant chemotherapy for breast cancer: functional tumor volume by MR imaging predicts recurrence-free survival—results from the ACRIN 6657/CALGB 150007 I-SPY 1 TRIAL. Radiology 2016;279(1):44–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Drukker K, Li H, Antropova N, et al. Most-enhancing tumor volume by MRI radiomics predicts recurrence-free survival “early on” in neoadjuvant treatment of breast cancer. Cancer Imaging 2018;18(1):12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Drukker K, Edwards AV, Doyle C, et al. Breast MRI radiomics for the pretreatment prediction of response to neoadjuvant chemotherapy in node-positive breast cancer patients. J Med Imaging 2019;6(3):34502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Ravichandran K, Braman N, Janowczyk A, et al. A deep learning classifier for prediction of pathological complete response to neoadjuvant chemotherapy from baseline breast DCE-MRI. In: Petrick N, Mori K, editors. Medical imaging 2018: computer-aided diagnosis, vol. 10575. Bellingham (WA): SPIE; 2018. p. 79–88. [Google Scholar]
- 100.Li H, Zhu Y, Burnside ES, et al. Mr imaging radiomics signatures for predicting the risk of breast cancer recurrence as given by research versions of mammaprint, oncotype DX, and PAM50 gene assays. Radiology 2016;281(2):382–91. [DOI] [PMC free article] [PubMed] [Google Scholar]


