Abstract
Background
The conversion of fibroblasts into induced cardiomyocytes may regenerate myocardial tissue from cardiac scar through in situ cell transdifferentiation. The efficiency transdifferentiation is low, especially for human cells. We explored the leveraging of Hippo pathway intermediates to enhance induced cardiomyocyte generation.
Methods and Results
We screened Hippo effectors Yap (yes‐associated protein), Taz (transcriptional activator binding domain), and Tead1 (TEA domain transcription factor 1; Td) for their reprogramming efficacy with cardio‐differentiating factors Gata4, Mef2C, and Tbx5 (GMT). Td induced nearly 3‐fold increased expression of cardiomyocyte marker cTnT (cardiac troponin T) by mouse embryonic and adult rat fibroblasts versus GMT administration alone (P<0.0001), while Yap and Taz failed to enhance cTnT expression. Serial substitution demonstrated that Td replacement of TBX5 induced the greatest cTnT expression enhancement and sarcomere organization in rat fibroblasts treated with all GMT substitutions (GMTd versus GMT: 17±1.2% versus 5.4±0.3%, P<0.0001). Cell contractility (beating) was seen in 6% of GMTd‐treated cells by 4 weeks after treatment, whereas no beating GMT‐treated cells were observed. Human cardiac fibroblasts likewise demonstrated increased cTnT expression with GMTd versus GMT treatment (7.5±0.3% versus 3.0±0.3%, P<0.01). Mechanistically, GMTd administration increased expression of the trimethylated lysine 4 of histone 3 (H3K4me3) mark at the promoter regions of cardio‐differentiation genes and mitochondrial biogenesis regulator genes in rat and human fibroblast, compared with GMT.
Conclusions
These data suggest that the Hippo pathway intermediate Tead1 is an important regulator of cardiac reprogramming that increases the efficiency of maturate induced cardiomyocytes generation and may be a vital component of human cardiodifferentiation strategies.
Keywords: cardiomyocytes, direct reprogramming, regeneration
Subject Categories: Cellular Reprogramming, Gene Therapy, Myocardial Regeneration
Nonstandard Abbreviations and Acronyms
- cTnT
cardiac troponin T
- GFP
green fluorescent protein
- GMT
Gata4, Mef2c, and Tbx5
- iCMs
induced cardiomyocytes
- TAZ
transcriptional activator binding domain
- TEAD
transcriptional enhanced associate domain
- Tead 1
TEA domain transcription factor 1
- YAP
yes‐associated protein
Clinical Perspective
What Is New?
Hippo pathway effector Tead1 in combination with Gata4 and Mef2c enhances the efficiency and improves the structural and functional transdifferentiation of reprogrammed rat and human cardiac fibroblasts compared with use of a standard Gata4, Mef2C, and Tbx5 cocktail.
Tead1 enhance cardiac reprogramming by regulating mitochondrial biogenesis through peroxisome proliferator‐activated receptor‐γ coactivator PGC (peroxisome proliferator‐activated receptor‐γ coactivator)‐1A/1B.
What Are the Clinical Implications?
Tead1 may provide an important transcriptional regulatory role in cardiac fibroblast transdifferentiation essential for induced cardiomyocyte generation, providing a cocktail of genetic factors that have potential for further applications in in vivo cardiac regeneration.
Heart failure remains a leading cause of cardiac death worldwide, for which current interventional treatments remain suboptimal. 1 While cardiac regeneration strategies hold great theoretical promise as a treatment for heart failure, they are hindered by the limited innate regenerative capacity of the adult human heart. 2 The in situ reprogramming of cardiac fibroblasts into induced cardiomyocytes (iCMs) thus represents a promising new myocardial regeneration strategy. 3 , 4 We and others have shown that a wide variety of cardiac reprogramming cocktails based upon the core cardiac transcription factors Gata4, Mef2c and Tbx5 (GMT) can convert fibroblasts into iCMs, albeit with low efficiency in inducing contractile cells in vitro, especially as applied to human cells. 5 , 6 , 7 , 8 , 9 , 10 , 11 , 12 , 13 , 14 , 15 , 16
The TEAD (transcriptional enhance associate domain) transcription factor family is a component of the Hippo signaling pathway, the inactivation of which has been implicated in cell replication, growth, and development. 17 , 18 , 19 , 20 The role of TEAD in the Hippo pathway is broadly believed to be regulated by the presence or the absence of nuclear YAP (yes‐associated protein) and TAZ (transcriptional activator binding domain). 21 Recent studies suggest, however, that TEAD itself is actively regulated and may be an active effector in the Hippo pathway. 22
Of the 4 members of the TEAD family, Tead1 (also known as transcriptional enhancer factor 1 [TEF‐1]) is the most abundant in the heart, is a transcriptional regulator of myogenic genes, and is required for heart development. 17 , 23 A recent report also suggests that Tead1 is an integral component of the cardiac transcriptional regulatory network and that it regulates core cardiomyocyte‐specific functions including contraction and metabolism, and shares binding domains with key cardio‐differentiation factors such as p300, Gata4, Nkx2.5, and Mef2a. 24 , 25 , 26 , 27 These observations led us to hypothesize that Tead1 may provide an important transcriptional regulatory role in cardiac fibroblast transdifferentiation essential for iCMs generation. We consequently sought to test Tead1 (Td) as a potential factor that could increase the efficiency of cardiac reprogramming by enhancing cardio‐differentiating gene activation.
METHODS
Per the Transparency and Openness Promotion Guidelines, the data, analytic methods, and study materials will be available to other researchers on request to the corresponding author.
Tissue Collection and Isolation of Cardiac Cells
All animal experiments were approved by Institutional Animal Care and Use Committee at Baylor College of Medicine and all methods were carried out in accordance with the NIH guidelines (Guide for the care and use of laboratory animals) and under protocol AN‐6223, approved by the Institutional Animal Care and Use Committee. Cardiac fibroblasts were isolated from adult rat cardiac tissues harvested from 6‐ to 8‐week‐old male Sprague‐Dawley rats (Envigo International Holding Inc., Hackensack, NJ) using standard cell isolation techniques. 8 Adult human cardiac fibroblasts were harvested from ventricular tissue obtained from heart failure pateints undergoing cardiac transplantation at Baylor St. Luke's Medical Center and under a protocol approved by the Baylor College of Medicine Institutional Review Board (IRB H‐33421). 9 Human tissues were manually sharply miced and placed in fibroblast growth medium (Dulbecco modified eagle medium (DMEM), 10% fetal bovine serum (FBS), and 1% penicillin/streptomycin), as previously described. 10
Cell Reprogramming
Lentivirus vectors encoding Gata4 (G), Mef2c (M), Tbx5 (T), Tead1 (Td), YAP, TAZ, PGC (peroxisome proliferator‐activated receptor‐γ coactivator)‐1A, PGC‐1B and GFP (green fluorescent protein) were prepared from relevant plasmids by the Baylor College of Medicine Gene Vector Core, as previously described. 8 , 9 , 15 A combination of 4 factors (GMTTd) was used for reprogramming rat and human cardiac fibroblasts reprogramming. Rat and human cardiac fibroblasts were seeded at a density of 5×104 cells per well of a 24‐well plate in Iscove modified dulbecco media medium containing 20% FBS, and 1% penicillin/streptomycin. Lentivirus vectors with a multiplicity of infection: 20 was added with polybrene (5 μg/μL) after 24 hours. Two days later, medium was replaced with induction medium (iCM media), containing DMEM/Medium‐199 (4:1), 10% FBS, 5% horse serum, 1% penicillin/streptomycin, 1% non‐essential amino acids, 1% essential amino acids, 1% GlutaMAX, and 1% sodium pyruvate. 11 Induction media was changed twice a week until cells were harvested.
For rat contractility studies, iCM medium was changed on day 14 of culture to beating media containing RPMI1640/2% B27/2%FBS/0.05%BSA/50 μg/mL ascorbic acid/0.2 mmol/L GlutaMAX, 1×NEAA/1% insulin‐selenium‐transferrin/1% vitamin mixture/10% conditioned medium obtained from neonatal rat/mouse cardiomyocyte culture. Medium was changed every 3 to 4 days.
For mitochondrial induction studies, media was replaced with induction media after 48 hours of lentival transduction and the compounds PGC‐1A activator (ZLN005, MCE, MedChem Express, # HY‐17538, 1 μmol/L) PGC‐1A inhibitor (SR‐18292, MCE, MedChem Express, # HY‐101491, 0.1 μmol/L) were added and maintained in culture throughout the duration of the experiment for 14 days.
For human reprogramming studies using co‐culture techniques, neonatal rat pups were used to isolate cardiomyocytes under protocol (AN‐6223), as previously described. 9 Human fibroblasts were coinfected with lentiviral GMT or GMTd. After 1 week of transduction, the infected fibroblasts were harvested and re‐plated onto neonatal rat cardiomyocytes at a ratio of 1:10 and cultured in DMEM/M‐199/10% FBS medium. 12
High Throughput Screening
Induced murine embryonic fibroblast cell line were used for high throughput screening for iCMs that overexpressed GMT in response to doxycycline treatment and expressed a GFP reporter gene driven by a promoter sequence responsive to the cardiomyocyte marker alpha muscle heavy chain‐GFP. 28 In brief, the alpha muscle heavy chain‐GFP cardiac reporter in the induced murine embryonic fibroblast cell line indicates reprogramming with GFP reporter expression. Real‐time quantification of GFP+ events were captured by IncucyteG3 imaging system (IncuCyte Live‐Cell Analysis System; Sartorius) at 37°C for 7 days. Five images (1.75×1.29 mm FOV) per well at ×10 magnification were acquired every 4 hours using phase contrast and a GFP filter (excitation: 460 nm with passband [440 480 nm]; emission: 524 nm with passband [504, 544 nm]). Images were exported as 8 bit TIFF and 16 bit TIFF for phase and GFP, respectively, for further analysis.
Flow Cytometry
Cells were trypsinized and fixed with fixation buffer (BD Biosciences, San Jose, CA) for 15 minutes at room temperature. Fixed cells were washed with Perm/Wash buffer (BD Biosciences). 8 , 9 Then, cells were incubated with primary mouse anti‐cTnT (cardiac troponin T) antibody (ab8295) in Perm/Wash buffer for 90 minutes at room temperature followed by incubation with donkey anti‐mouse Alexa Fluor 647 (ab150107). Cells were washed with Perm/Wash buffer, and then analyzed for cTnT expression using an LSR Fortessa cell sorter (BD Biosciences, Franklin Lakes, NJ) with FlowJo software (Flowjo, LLC, Ashland, Ore).
Immunocytochemistry
Immunofluorescence staining was performed as previously described. 9 Cells were fixed in 4% paraformaldehyde and permeabilized with 0.5% Triton‐X solution. Cells were then blocked with 10% goat serum and incubated with primary antibodies against cTnT (1:300 dilution; Thermo Fisher Scientific), Gja 1 (1:500 dilution; Abcam), cTnI (1:200 dilution; Abcam), or α‐actinin (1:300 dilution; Sigma‐Aldrich, St. Louis, MO), followed by the appropriate Alexa fluorogenic secondary antibodies (Invitrogen), and finally with DAPI (Invitrogen). 8 , 9 , 11 Proliferation assay was performed using anti‐Ki‐67 (1:250 dilution; Abcam). After being washed with PBS 3 times, cells were incubated with secondary antibodies conjugated with Alexa Fluor 555 (Invitrogen) for 1 hour at room temperature in the dark. DAPI (1:1000; Sigma) was used to stain nuclei. iCM proliferation was calculated by counting Ki‐67+, cTnT+, and DAPI+ cells in every image.
Real‐Time Polymerase Chain Reaction
Total RNA was extracted using the TRIzol method (Invitrogen), after which relative quantification was performed using SYBR‐green detection of polymerase chain reaction (PCR) products in real time with the ABI ViiA 7 (Applied Biosystems Inc., Foster City, CA). The following rat and human primers were used for these studies: CTNT (cardiac troponin T), NKX‐2.5 (homebox protein 2.5), MYL7 (myosin light chain 7), ACTC1 (actin alpha cardiac muscle 1), RYR2 (ryanodine receptor 2), GJA1 (gap junction protein alpha 1), ATP2A2 (sarcoplasmic reticulum calcium‐ATPase 2), SCN5A (sodium channel protein 5), HCN4 (hyperpolarization‐activated cyclic nucleotide‐gated channel), KCNA5 (Potassium Voltage‐Gated Channel 5), COL1A1 (Collagen type 1 alpha 1), COL1A2 (Collagen type 1 alpha 2), S100A4 (S100 Calcium‐Binding Protein A4), POSTN (periostin), PGC1A/B (Peroxisome proliferator‐activated receptor gamma coactivator 1‐alpha and beta), and TFAM (mitochondrial transcription factor). Sequences of the primers are listed in Table S1. mRNA levels were normalized by comparison to GAPDH. Briefly, the cycle threshold (Ct) values of each gene of interest and of GAPDH were calculated for each sample and then the normalized value was derived by subtracting the Ct value of GAPDH from that of the gene of interest (∆Ct). Data are shown as mRNA fold change (2−∆∆Ct) relative to the mRNA level of the corresponding transcript in the control samples as indicated.
Western Analysis
To perform Western analyses, cell lysates were prepared by homogenization of cells in cell lysis buffer and run on SDS‐PAGE. After seperation, the protein bands were transferred to nitrocellulose membrane (Invitrogen Catalog number: IB301001), immune detection was performed with the following primary antibodies: PGC‐1A (Abcam: ab106814), PGC‐1B (Abcam: ab176328), TFAM (Abcam: ab131607), and actin (Sigma‐Aldrich), followed by treatment with appropriate horseradish peroxidase‐conjugated secondary antibodies (Millipore, Billerica, MA). Membranes were visualized by chemiluminescence detection (Thermo Scientific catalog no: 34580).
Measurements of Contractility, Calcium Transient, and Beating
To measure cell shortening and calcium transients iCMs were placed to a perfusion chamber mounted on the stage of an inverted epifluorescence microscope (Nikon Diaphot 200). iCMs were continuously perfused with 1.8 mmol/L Ca2+‐Tyrode solution containing (in mmol/L): NaCl 140, KCl 5.4, MgCl2 1, CaCl21.8, HEPES 5, and glucose 10, pH 7.4. When required, cells were field stimulated by means of platinum electrodes within the chamber, with a suprathreshold voltage applied over a range of frequencies. Contractions of iCMs was assessed using a video‐based edge detection system (IonOptix, Dublin, Ireland). Ca2+ transients measurements was measured with 2 μmol/L Fura‐2/AM (Life Technologies) together with 0.1% Pluronic F‐127 (Invitrogen) according to the manufacturer’s instruction. [Ca2+]i was detected by exciting Fura‐2 alternately at 340 and 380 nm at 0.5 Hz, and recording emitted fluorescences at 510 nm. Both edge detection and Fura‐2 fluorescence measurement were made simultaneously from the same contracting region of the cell monolayer. Data analysis was performed using IonWizard software (IonOptix). 9 , 11
For beating cell counting, we seeded 50 000 fibroblasts per well on 12‐well plates, performed cell transductions as detailed above and then analyzed cell contraction by light microscopy performed at room temperature. Spontaneous contracting cells were manually counted in 10 randomly selected high‐power fields per well in at least 3 independent experiments. We identified the individual beating iCMs based on differences in beating frequency, cell‐membrane boundary, and GFP fluorescence expression. The cardiac reprogramming efficiency was determined as the number of beating cells per 50 000 starting fibroblasts.
Chromatin Immunoprecipitation Assay
Chromatin immunoprecipitation assays were performed on murine embryonic fibroblasts (ATCC) using ChromaFlash Chromatin Extraction kits (p‐2001, Epigenetek, Farmingdale, NY) as described previously. 29 Cells were seeded onto 150 mm dishes at density of 106, treated 24 hours later with lentivirus encoding GFP or Tead1 (20 moi) with 5 μg/μL polybrene. Cells were harvested for chromatin immunoprecipitation assay‐quantitative polymerase chain reaction (qPCR) 7 days later. Chromatin immunoprecipitations were conducted with a rabbit polyclonal antibody anti‐Histone H3 (tri methyl K4) CHIP Grade (ab8580), and normal rabbit immunoglobulin G (AB‐105‐C) was obtained from R&D systems. All primers for chromatin immunoprecipitation assay‐qPCR assays were synthesized by Sigma. These sequences are listed in Table S2. Fold enrichment of PCR product was calculated after normalization with input of all 3 types of infected cells.
Statistical Analysis
Statistical analysis was performed using SAS, version 9.4. Results are presented as mean±SEM. Three independent experiments with triplicate determinations were performed. The non‐parametric Mann–Whitney U test (Wilcoxon rank‐sum test) was used when comparing 2 groups, and the Kruskal–Wallis test, followed by Boneferroni corrections, was used when comparing >2 groups to analyze statistical significance. A P<0.05 was regarded as significant.
RESULTS
Hippo Pathway Effector Screening Demonstrating That Tead1 Enhances Cardiodifferentiation Marker Expression
We found that Tead1 in combination with GMT administration increased the expression of the cardiomyocyte marker cTnT in mouse embryonic and adult rat cardiac fibroblasts by ≈3‐fold compared with GMT administration alone (P<0.0001), whereas 2 other Hippo pathway effectors YAP and TAZ did not increase cTnT expression above GMT treatment levels (Figure 1A). We then used our GMT‐ (doxycycline) inducible alpha muscle heavy chain‐GFP murine embryonic fibroblast high throughput cardio‐differentiated cell assay to demonstrate that lentiviral‐mediated administration of Tead1 to GMT overexpressing cells increased cardiac reprogramming efficiency compared with cells overexpressing GMT without addition of Tead1, as indicated by the percentage of GFP+ cells, (P<0.0001). In comparison, we observed that lentiviral‐mediated administration of YAP or TAZ failed to enhance cardiac reprogramming efficiency above doxycycline‐induced GMT overexpression (Figure 1B).
Figure 1. Hippo pathway effector screening in mouse embryonic and adult rat cardiac fibroblasts.

A, Quantitative reverse transcription polymerase chain reaction (qRT‐PCR) analysis of mouse embryonic fibroblasts (MEFs; top) and adult rat cardiac fibroblasts (RCFs; bottom) demonstrating increased cTnT (cardiac troponin T) gene expression 7 days after combined treatment with Gata4, Mef2c, and Tbx5 (GMT) and Tead1 (TEA domain transcription factor 1) compared with GMT treatment with other indicated effectors (n=3 biological replicates, each performed in triplicate; *P<0.001 vs GFP (green fluorescent protein), **P<0.0001 vs GMT; Wilcoxon rank‐sum test). B, Real‐time high throughput cardiomyocyte marker cell screening using an induced murine embryonic fibroblast cell line and “IncucyteG3” imaging system (see Methods). Left: Representative immunocytochemistry images of GFP)‐positive cells (indicating activation of α‐muscle heavy chain‐ GFP reporter expression). Right: Quantification of the percentage of GFP‐positive cells 7 days after treatment with indicated effectors (n=3 biological replicates, each performed in triplicate. cTnT indicates cardiac troponin T; GFP, green fluorescent protein; GMT, Gata4, Mef2C, and Tbx5; icMEF, induced murine embryonic fibroblast cell line; MEF, mouse embryonic fibroblasts; RCFs, rat cardiac fibroblasts; TAZ, transcriptional activator binding domain; and YAP, yes‐associated protein. *P<0.0001 vs doxycycline only; Wilcoxon rank‐sum test). Scale bar: 400 μm.
Tead1 Enhances the Efficiency and Improves the Cardiomyocyte‐Like Maturation of Reprogrammed Rat Cardiac Fibroblasts
In serial substitution studies using GMT as our baseline transdifferentiation strategy, we demonstrated that TBX5 was dispensable in the presence of Tead1 and that the GMTd yielded the highest levels of cTnT expression compared with cells treated with GMT alone (GMTd versus GMT: 17±1.2% versus 5.4±0.3%; P<0.0001; Figure 2A and 2B). Interestingly, replacement of Gata4 and Mef2c with Tead1 in a GMT cocktail did not increase cardiac reprogramming compared with GMT administration alone, suggesting these genes are not dispensable during reprogramming (Figure 2A and 2B).
Figure 2. Tead1 enhances expression of a portfolio of cardiomyocyte marker genes in adult rat cardiac fibroblasts.

A, Representative flow cytometry plots of rat cardiac fibroblasts 14 days after transduction demonstrating that the combination of Gata4, Mef2c and Tead1 (GMTd) enhances the expression of depicted cardiomyocyte marker genes compared with cell transduction with GFP (green fluorescent protein),GMT, or other Td substitutions of GMT (MTTd, GTTd). B, Quantification of the percentage of cTnT+ cells treated as above, as assessed by flow cytometry (n=3 biological replicates, each performed in triplicate; *P<0.01 vs GFP, **P<0.0001 vs GMT; Wilcoxon rank‐sum test). C, Quantitative reverse transcription polymerase chain reaction (qRT‐PCR) analysis of rat cardiac fibroblasts demonstrating increased expression of depicted cardiomyocyte marker genes (CTNT, MYL7, ACTC1, NKX2.5, RYR2) and ion channel genes (GJA1, ATP2A2, SCN5A, HCN4, KCNA5) and decreased expression of fibroblast marker gene (COL1A1, COL1A2, S100A4, POSTN) 14 days after reprogramming factor administration (n=3 biological replicates, each performed in triplicate. ACTC1, actin alpha cardiac muscle 1; ATP2A2, sarco (endo)plasmic reticulum calcium‐ATPase 2; COL1A1, Collagen type 1 alpha 1; COL1A2, Collagen type 1 alpha 2; cTnT indicates cardiac troponin T; FSC, forward scatter; GFP, green fluorescent protein; GMT, Gata4, Mef2C, and Tbx5; GMTd, Gata4, Mef2C, and Tead1; GJA1, gap junction protein alpha 1; HCN4, hyperpolarization‐activated cyclic nucleotide‐gated channel; KCNA5, Potassium Voltage‐Gated Channel 5; MYL7, myosin light chain 7; Nkx‐2.5, homebox protein 2.5; POSTN, periostin; RCFs, rat cardiac fibroblasts; RYR2, ryanodine receptor 2; SCN5A, sodium channel protein 5; and S100A4, S100 Calcium‐Binding Protein A4. *P<0.01, **P<0.001 vs GFP; Kruskal–Wallis test).
To assess transdifferentiated cell maturity, we examined the expression of a panel of genes relevant to cardiomyocyte functional structure (CTNT, MYL7, ACTC1, NKX2.5, RYR2), and ion channels (GJA1, ATP2A2, SCN5A, HCN4, KCNA5). We observed by qRT‐PCR that GMTd treatment increased the expression of these genes in rat cardiac fibroblasts compared with treatment with GFP alone (Figure 2C). Consistent with these findings, we observed that GMTd also decreased the expression of the fibroblast marker gene COL1A1, COL1A2, S100A4, POSTN versus GFP treatment alone (Figure 2C).
Immunofluorescence analyses likewise demonstrated greater numbers of cells expressing the cardiomyocyte markers cTnT and α‐ sarcomeric actinin after 2 weeks of reprogramming with GMTd compared with treatment with GMT alone; however, sarcomeric structures was not obvious at this early stage (Figure 3A and 3B). The GMTd‐induced iCMs showed clear cardiac TNT, TNI3, phalloidin and, α‐sarcomeric actinin with advanced sarcomere organization after 4 weeks of reprogramming (Figure 3C). In addition, GMTd treatment was seen to induce a ≈4‐fold increase in the number of α‐sarcomeric actinin+ cells with assembled sarcomeres after 4 weeks compared with cells treated with GMT alone (Figure 3D). We also observed formation of gap‐junction protein Gja1 channels after GMTd but not after GMT treatment (Figure 3E). In the context of the known proliferative properties of Tead1, Ki67 proliferation assays revealed that the increase in these cardio‐differentiated GMTd‐treated cells was not attributable to cell proliferation (Figure S1).
Figure 3. Tead1 enhances cardiomyocyte marker protein expression and ultrastructure.

Immunofluorescence analyses of rat cardiac fibroblasts 14 days after transduction with GFP (green fluorescent protein), GMT, or GMTd (n=3/group). DAPI nuclear marker: blue; GFP: green; cardiomyocyte markers: red. A, Representative images of immunofluorescence staining for cTnT (left top) and for α‐actinin (left bottom) and high magnification views for cTnT (cardiac troponin T) and α‐actinin (right), after 2 weeks. Scale bars: 100 μm (left images) and 25 μm (right [high magnification] images). B, Quantification of cells expressing cardiomyocyte markers cTnT and α‐actinin after 2 weeks by immunofluorescence labelling (*P<0.001 vs GMT; Wilcoxon rank‐sum test). C, Representative images of immunofluorescence staining for cTnT, CTnI3, and phalloidin (left) and α‐actinin (miidle) after 4 weeks of GMTd transduction demonstrating sarcomeric structures, most clearly visible in α‐actinin labeled cells (middle). Scale bars: 25 μm. D, Quantification of cells with well‐developed sarcomeres as a percentage of total α‐actinin+ cells after 4 weeks of GMT or GMTd transduction (*P<0.001 vs GMT; Wilcoxon rank‐sum test). E, Representative images of immunofluorescence staining for GJA1 (gap junction alpha‐1) protein demonstrating gap junction channels formed between cells treated with GMTd. GFP indicates green fluorescent protein; and GMT, Gata4, Mef2C, and Tbx5. Scale bars: 100 μm (left images); 25 μm (right [high magnification] images).
Tead1 Enhances the Generation of Functional iCMs
Spontaneous contraction was observed in 6% of rat cardiac fibroblasts as early as 4 weeks after GMTd treatment but was not observed in untreated cells or GMT‐treated cells for up 8 weeks after cell transduction. (Figure 4A and Videos S1 through S3). All contracting GMTd‐treated cells were also observed to demonstrate calcium transients on electrical stimulation, but calcium transients were not seen in untreated or GMT‐treated cells (Figure 4B). The number of these beating GMTd‐treated cells increased over time (Figure 4C).
Figure 4. Tead1 enhances contractile function of reprogrammed cells.

A, Representative immunofluorescence staining 4 weeks after rat cardiac fibroblast transfection with lentivirus encoding GFP (green fluorescent protein), Gata4, Mef2C, and Tbx5 (GMT), or GMTd, as depicted by expression of encoded (green) GFP marker protein. Scale bar: 100 μm. B, Representative peaks from GFP+ rat cells after 4 weeks in culture reflecting cell contraction (top row) and Ca2+ transients (bottom row) in cells treated with GMTd, but not in cells treated with GMT or GFP alone. Scale bar: 0.5 seconds. C, Quantification of the number of spontaneously beating GMTd‐treated cells treated as a function of time (n=3 biological replicates, each performed in triplicate). GFP indicates green fluorescent protein; GMT, Gata4, Mef2C, and Tbx5; GMTd, Gata4, Mef2C, Tead1, and iCMs, induced cardiomyocytes.
Tead1 Enhances Human Cardiac Fibroblast Reprogramming
Human cardiac fibroblasts treated with GMTd were shown by fluorescence‐activated cell sorting to demonstrate increased cTnT expression compared with cells treated with GMT alone (7.5±0.3% versus 3±0.3%; P<0.01; Figure 5A and 5B). Quantitative reverse transcription polymerase chain reaction (qRT‐PCR) analysis likewise demonstrated increased expression of other cardiomyocyte transdifferentiation genes (CTNT, MYH6, ACTC1) and ion channel genes (SCN5A, HCN4) and downregulation of the fibroblast marker gene (COL1A1, COL1A2, POSTN) after GMTd treatment (Figure 5C). Increased expression of cTnT and α‐sarcomeric actinin in GMTd versus GMT‐treated human cells was also demonstrated by immunofluorescence studies after 2 weeks of reprogramming; however, sarcomeric structures was not clear at this early stage (Figure 5D and 5E). The GMTd‐induced iCMs showed clear cardiac TNT and α‐sarcomeric actinin with advanced sarcomere organization after 4 weeks of reprogramming (Figure 5F). GMTd‐treated human cells demonstrated a 4‐fold increase in the number of α‐sarcomeric actinin+ cells with assembled sarcomeres after 4 weeks compared with human cells treated with GMT (Figure 5G).
Figure 5. GMTd induces cardio‐differentiation of human cardiac fibroblasts.

A, Representative flow cytometry plots of human cardiac fibroblasts 14 days after transduction demonstrating that GMTd enhances the expression of depicted cardiomyocytes marker genes compared with cell transduction with GFP (green fluorescent protein), Gata4, Mef2C, and Tbx5 (GMT), or other Td substitutions of GMT (MTTd, GTTd). B, Quantification of the percentage of cTnT+ cells treated as above, as assessed by flow cytometry (n=3 biological replicates, each performed in triplicate; *P<0.01 vs GMT; Wilcoxon rank‐sum test). C, Quantitative reverse transcription polymerase chain reaction (qRT‐PCR) analysis of human cardiac fibroblasts demonstrating increased expression of depicted cardiomyocyte marker genes (CTNT, MYH6, ACTC1) and ion channel genes (SCN5A, HCN4) and decreased expression of fibroblast marker gene (COL1A1, COL1A2 and POSTN) 14 days after reprogramming factor administration (n=3 biological replicates, each performed in triplicate; *P<0.05, **P<0.001 vs GFP; Kruskal–Wallis test). Data were normalized against GFP values. Immunofluorescence analyses of human cardiac fibroblasts 14 days after transduction with GFP, GMT, or GMTd (n=3/group). DAPI nuclear marker: blue; GFP: green; cardiomyocyte markers: red. D, Representative images of immunofluorescence staining for cTnT (left top) and for α‐actinin (left bottom) and high magnification views for cTnT and α‐actinin (right) after 2 weeks. Scale bar: 100 μm (left images) and 25 μm (right [high magnification] images). E, Quantification of cells expressing cardiomyocytes markers cTnT and α‐actinin after 2 weeks by immunofluorescence labelling (*P<0.01 vs GMT; Wilcoxon rank‐sum test). F, Representative images of immunofluorescence staining for cTnT (top) and α‐actinin (bottom) after 4 weeks of GMTd transduction demonstrating sarcomeric structures, most clearly visible in α‐actinin labeled cells. Scale bar: 25 µm. G, Quantification of cells with well‐developed sarcomeres as a percentage of total α‐actinin+ cells after 4 weeks of GMT or GMTd transduction (*P<0.001 vs GMT; Wilcoxon rank‐sum test). H, Representative immunofluorescence staining demonstrating (green) GFP expression by adult human cardiac fibroblasts treated with GMT (left) or lentivirus expressing GMTd (right) after 4 weeks in co‐culture with (non‐transduced) neonatal rat cardiomyocytes (negative for GFP). Scale bar: 100 μm. I, Representative peaks from GFP+ cells treated with GMTd, reflecting contraction (top row) and Ca2+ transients (bottom row; n=3 independent experiment). Contractility parameters were not observed in cells treated with GMT alone. Scale bar: 0.5 seconds. cTnT indicates cardiac troponin T; GMT, Gata4, Mef2C, and Tbx5; FSC, forward scatter; GMTd, Gata4, Mef2C, and Tead1 and HCFs, human cardiac fibroblasts.
Although human cardiac fibroblasts treated with GMTd were not observed to contract independently after up to 6 weeks in culture, ≈5% of human cardiac fibroblasts treated with GMTd, as verified by their GFP expression (Figure 5H), contracted synchronously with surrounding neonatal rat cardiomyocytes after 4 weeks in co‐culture. In comparison, cells treated with GMT alone failed to contract in co‐culture experiments (Figure 5H and Videos S4, S5). All contracting GMTd‐treated human cells were also observed to demonstrate calcium transients on electrical stimulation, but calcium transients were not seen in cells treated with GMT alone (Figure 5I). Verification that the contracting cells were transfected fibroblasts, rather than the neonatal cardiomyocytes, was provided by the uniform expression of GFP by these contracting cells, reflecting the transfection of the fibroblast culture in isolation before their reseeding into the co‐culture plates.
Mechanisms of Action of Tead1 in Cardiac Reprogramming
Enrichment of the transcriptionally active chromatin mark trimethylated lysine 4 of histone 3 (H3K4me3) was demonstrated in MEFs (mouse embryonic fibroblasts) 1 week after these cells were treated with GMTd compared with GMT‐treated cells (Figure 6A). In the GMTd‐treated group, H3K4me3 was significantly enriched at the promoters for TNNT2 (2.3x), GATA4 (2.4x), RYR2 (2.6x), and MEF2C (2.5x). These findings suggest that Tead1 may enhance cell cardio‐differentiation by reversing chromatin depression of the expression of relevant cardio‐differentiation factors.
Figure 6. Tead 1 mechanisms of action in cardiac fibroblast reprogramming.
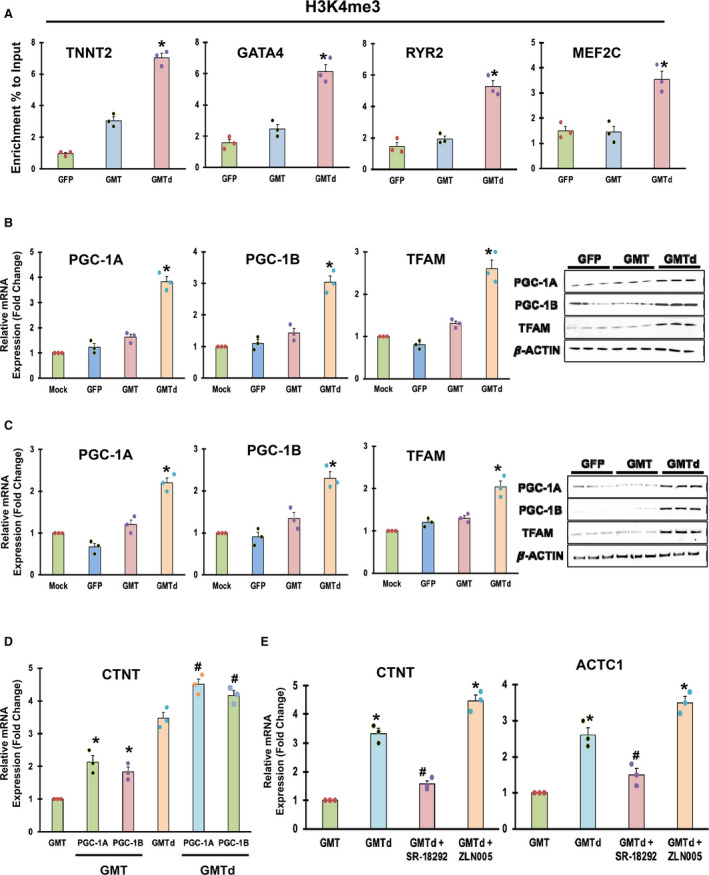
A, Chromatin immunoprecipitation assay analysis of mouse embryonic fibroblasts treated with GFP (green fluorescent protein), Gata4, Mef2C, and Tbx5 (GMT), and GMTd demonstrating the trimethylated status of histone H3 of lysine 4 for the promoters of Tnnt2, Gata4, Ryr2, and, Mef2c (n=3 biological replicates, each performed in triplicate; *P<0.01 vs GMT; Wilcoxon rank‐sum test). B and C, Quantitative reverse transcription polymerase chain reaction (qRT‐PCR) [left] and Western analysis (right) of rat cardiac fibroblasts and human cardiac fibroblasts, respectively, demonstrating increased expression of depicted mitochondrial biogenesis regulator gene mRNA and protein expression 14 days after reprogramming factor administration (n=3 biological replicates, each performed in triplicate; *P<0.01 vs GMT [Wilcoxon rank‐sum test] for qRT‐PCR, n=3 independent experiments for western blotting. Beta actin is shown as a loading control). D, qRT‐PCR analysis of cTnT (cardiac troponin T) in rat cardiac fibroblasts transduced with GMT and GMTd administration with or without PGC (peroxisome proliferator‐activated receptor‐γ coactivator)‐1A and PGC‐1B, as indicated (n=3 biological replicates, each performed in triplicate; *P<0.05 vs GMT, # P<0.05 vs GMTd; Kruskal–Wallis test). E, qRT‐PCR analysis of cTnT (left) and Actc1 (right) in rat cardiac fibroblasts after GMT and GMTd administration with or without addition of PGC‐1A activator or inhibitor, as indicated (n=3 biological replicates, each performed in triplicate. CTNT indicates cardiac troponin T; GFP, green fluorescent protein; GMTd, Gata4, Mef2C, and Tead1 and GMT, Gata4, Mef2C, and Tbx5. *P<0.01 vs GMT, # P<0.05 vs GMTd; Kruskal–Wallis test).
Increased peroxisome PGC‐1A and ‐1B and TFAM expression in rat and human cardiac fibroblast treated with GMTd was also demonstrated by qRT‐PCR and protein expression compared with the treatment of these cells with GMT (Figure 6B and 6C). These findings suggest that Tead1 may enhance cardio‐differentiation efficiency by upregulating these energy metabolism and mitochondrial biogenesis regulator factors thought to be relevant to cardiomyocyte viability. 30
We accordingly investigated whether PGC‐1A and ‐1B could also induce cardiac reprogramming in rat cardiac fibroblasts. In these studies, qRT‐PCR analysis demonstrated further increased cTnT expression after overexpression of PGC‐1A and PGC‐1B together with GMT and GMTd treatment compared with treatment with GMT and GMTd alone (Figure 6D). We then demonstrated that administration of PGC‐1A activator (ZLN005) to GMTd‐treated cells further increased cTnT and Actc1 expression compared with treatment with GMT or GMTd alone, as detected by qRT‐PCR, while PGC‐1A inhibitor (SR‐18292) administration completely reversed GMTd‐mediated upregulation of cTnT and Actc1 expression (Figure 6E). These results suggest that PGC‐1A and 1B is necessary for cardiomyocyte reprogramming and that Tead1 may enhance cardiac reprogramming by regulating mitochondrial biogenesis through PGC‐1A.
DISCUSSION
Our present data demonstrate that the Hippo pathway effector Tead1 in combination with Gata4 and Mef2c increases the efficiency of the structural and functional transdifferentiation of rat and human cardiac fibroblasts into mature (contractile) iCMs compared with use of a standard GMT cocktail. We and others have previously demonstrated that other combinations of cardio‐differentiating transcription factors, growth factors, kinases, small molecules, microRNAs, and epigenetic factors can also be used improve GMT‐induced cardiac reprogramming in vitro. 9 , 31 , 32 , 33 , 34 , 35 , 36 , 37 , 38 , 39 , 40 , 41 Most of these reports, however, used murine or embryonic/juvenile fibroblasts as a substrate for their studies. In comparison our study of adult rat and human cardiac fibroblasts represents the targeting of cells more resistant to reprogramming and less conducive to the generation of functional (contractile) iCMs. 10 , 42
One potential explanation of this enhanced reprogramming efficacy is our finding of increased Tead1‐mediated upregulation of the expression of the gene activation H3K4me3 histone mark at the promoter regions of key cardio‐differentiation genes. These include cTnT, Gata4, Mef2c, and Ryr2 and the mitochondrial biogenesis regulator genes PGC‐1A/1B and TFAM, thought to play an important role in cardiomyocyte development. This latter supposition in particular is supported by our observation that PGC‐1A and PGC‐1B further increase GMT and GMTd mediated reprogramming and antagonists of Tead1 effects on mitochondrial biogenesis regulator genes leads to the loss of Tead1‐mediated improvements in reprogramming efficacy. These findings of the epigenetic effects of Tead1 is consistent with the observations of the mechanism of action of other potent reprogramming formulations. 25 , 27 , 30 , 33 , 43
The efficacy of Tead1 in enhancing cardio‐differentiation may be reflective of its significant role in enhancing myocyte development through its binding to MCAT (malonyl‐coA‐acyl carrier protein transacylase) elements and its motif found at muscle gene promoters. 44 Tead1 and its upstream regulators have likewise been shown to be critical to cardiomyocyte proliferation during embryonic and postnatal periods, with recent studies also demonstrating the role of the mammalian Hippo‐Tead1 pathway in cardiomyocyte regeneration after injury. 45 , 46 , 47 , 48 These effects are thought to be mediated via the physical interactions of Tead1 with the cardiac reprogramming factors Nkx2.5, Mef2c, Srf, Gata4, and p300. 26 , 27 An earlier study also demonstrated that Tead1 along with Mef2 and FoxO directly target myocardin during cardiovascular development. 49 Recent studies have also demonstrated that Tead1 is also required for maintenance of adult cardiomyocyte mitochondrial and overall heart function, with its loss associated with lethal dilated cardiomyopathy. 24 , 25 While these data support Tead1 as a potentially potent cardio‐differentiation factor, the use of Tead1 for this purpose has not to our knowledge been previously reported.
Our observation of significantly enhanced GMTd‐mediated reprogramming is particularly noteworthy for its early and robust transdifferentiation of adult rat cell into mature induced cardiomyocytes, as evidenced by our observation of well‐organized sarcomeres in 50% of cells within 4 weeks of reprogramming factor administration and onset of cell contractility in 6% of cells within 4 weeks of reprogramming. In comparison, Miyamota et al demonstrated transdifferentiation of 10% of cells into beating iCMs, but used more “plastic” mouse embryonic fibroblasts as their substrate target cell. 12 Likewise, Song et al reported that addition of Hand2, to GMT yielded an “optimized” rate of 0.2% of spontaneously beating adult mouse cardiac fibroblasts, while Addis et al reported that administering HAND2 and NKX2.5 with GMT yielded only rare spontaneous beating cells. 5 , 6 The beating percentage reported in the current study is on the other hand consistent with the previous reports of Wang et al 35 ; who demonstrated 1% to 6% beating cells after MGT and shBecn1 transduction using murine cardiac fibroblasts. Likewise, Mohamed et al 40 ; demonstrated that 5% to 40% of cells were beating after treatment with GMT and GMT plus TGF‐beta/WNT inhibitor using adult mouse cardiac fibroblsts. Recently, Kurotsu et al 50 ; likewise reported 6% to 15% beating iCMs after Sev‐GMT treatment in mouse cardiac fibroblats.
Mitochondrial Biogenesis
PGC‐1α is a multi‐functional transcriptional coactivator that regulates the activities of multiple nuclear receptors and transcriptional factors involved in mitochondrial biogenesis through TFAM. 51 Our results suggest that Tead1 may enhance cardiac reprogramming by regulating mitochondrial biogenesis through expression of PGC‐1A and mitochondrial transcription factor.
Although mitochondria primarily regulate cellular metabolism, they are also key participants in programmed cellular death (apoptosis) since they (1) contain several proapoptotic proteins (ie, cytochrome c) that, on release, can lead to cell death; and (2) produce reactive oxygen species (ROS) that can initiate apoptotic signaling. 52 Other studies have also correlated reduced PGC‐1α expression levels with decreased mitochondrial function and increased vulnerability to apoptosis. 53 In view of mitochondria as an energy center and important for cellular homeostasis, it would be interesting to explore the roles of mitochondrial biogenesis via the PGC‐1α/TFAM pathway which may play crucial roles on apoptotic activity in Tead1 induced cardiac reprogramming.
Study Limitations
Despite our encouraging observations regarding Tead1‐mediated cardiomyocyte reprogramming, it is possible that other mechanisms might also be involved in the Tead1‐mediated cardiac reprogramming, including alteration of signaling pathways, metabolic switch from glycolysis to fatty acid oxidation and epigenetic changes other than those that we have reported. Identification of such pathways will provide new insights into the mechanism of cardiac reprogramming and help to improve reprogramming efficiency.
Our premise that PGC‐1A and PGC‐1B are necessary for cardiac reprogramming is supported both by our PGC overexpression and inhibition studies, including those in the presence of GMTd. Additional PGC‐1A and ‐1B gene knock down studies performed in the presence of Tead1 are planned to confirm these findings. Defining the types or size of iCMs that can be obtained from GMTd‐induced reprogramming is essential for further research using this model system and for any potential utilization of these cells for gene‐based therapies. A variety of techniques can be used to discriminate between different types of CMs including gene expression studies, immunochemistry (for the measurement of size and change in sarcomeric length), and perhaps most importantly, functional studies. The cardiac action potential is the result of multiple ion channels and Ca2+ cycling proteins interacting in concert, and so the action potential measurement will provides a functional signature for the given type of iCM compared with neonatal/adult heart ventricle, atria, or pacemaker cells. Charecterization of these iCMs on the basis of their functional property is the subject of current investigations in our laboratory. Finally, further studies are also needed to clarify the relationship between G, M, and Td binding, epigenetic repatterning, and transcriptional activation of cardiac genes during iCMs reprogramming to further leverage of our current findings into optimized reprogramming strategies.
Sources of Funding
This study was funded by the National Heart, Lung, and Blood Institute (1R01HL152280‐01 [Rosengart]) and supported in part by an NIH/NHLBI Research Training Program in Cardiovascular Surgery (T32 HL139430 [Ryan]) and by the BCM Cytometry and Cell Sorting Core (National Institutes of Health grants P30AI036211, P30CA125123, and S10RR024574; National Center for Research Resources grant S10RR024574; National Institute of Allergy and Infectious Diseases grant AI036211, and National Cancer Institute grant P30CA125123).
Disclosures
Rosengart is a board member, paid consultant and holds a significant financial interest with XyloCor Therapeutics, LLC and receives royalty payments relevant to this work for intellectual property held by Cornell University. Mathison receives royalty payments relevant to this work for intellectual property held by Cornell University. Martin is a cofounder of and owns shares in Yap Therapeutics. The remaining authors have no disclosures to report.
Supporting information
Tables S1–S2
Figure S1
Videos S1
Videos S2
Videos S3
Videos S4
Videos S5
Acknowledgments
We thank Dr Kazuhiro Oka from the Gene Vector Core at Baylor College of Medicine for the preparation of viral vectors. We thank Dr Li for induced murine embryonic fibroblast cell line. In addition, the authors thanks BCM core facilities, including the Integrated Microscopy and Flow Cytometry Cores.
Supplementary Material for this article is available at https://www.ahajournals.org/doi/suppl/10.1161/JAHA.121.022659
For Sources of Funding and Disclosures, see page 14.
REFERENCES
- 1. Liu L, Eisen HJ. Epidemiology of heart failure and scope of the problem. Cardiol Clin. 2014;32:1–8, vii. doi: 10.1016/j.ccl.2013.09.009 [DOI] [PubMed] [Google Scholar]
- 2. Prabhu SD, Frangogiannis NG. The biological basis for cardiac repair after myocardial infarction: from inflammation to fibrosis. Circ Res. 2016;119:91–112. doi: 10.1161/CIRCRESAHA.116.303577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ieda M, Fu JD, Delgado‐Olguin P, Vedantham V, Hayashi Y, Bruneau BG, Srivastava D. Direct reprogramming of fibroblasts into functional cardiomyocytes by defined factors. Cell. 2010;142:375–386. doi: 10.1016/j.cell.2010.07.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Patel V, Mathison M, Singh VP, Yang J, Rosengart TK. Direct cardiac cellular reprogramming for cardiac regeneration. Curr Treat Options Cardiovasc Med. 2016;18:58. doi: 10.1007/s11936-016-0480-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Addis RC, Ifkovits JL, Pinto F, Kellam LD, Esteso P, Rentschler S, Christoforou N, Epstein JA, Gearhart JD. Optimization of direct fibroblast reprogramming to cardiomyocytes using calcium activity as a functional measure of success. J Mol Cell Cardiol. 2013;60:97–106. doi: 10.1016/j.yjmcc.2013.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Song K, Nam Y‐J, Luo X, Qi X, Tan W, Huang GN, Acharya A, Smith CL, Tallquist MD, Neilson EG, et al. Heart repair by reprogramming non‐myocytes with cardiac transcription factors. Nature. 2012;485:599–604. doi: 10.1038/nature11139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Qian L, Huang Y, Spencer CI, Foley A, Vedantham V, Liu L, Conway SJ, Fu JD, Srivastava D, Qian L. In vivo reprogramming of murine cardiac fibroblasts into induced cardiomyocytes. Nature. 2012;485:593–598. doi: 10.1038/nature11044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Mathison M, Singh VP, Gersch RP, Ramirez MO, Cooney A, Kaminsky SM, Chiuchiolo MJ, Nasser A, Yang J, Crystal RG, et al. Triplet polycistronic vectors encoding Gata4, Mef2c, and Tbx5 enhances postinfarct ventricular functional improvement compared with singlet vectors. J Thorac Cardiovasc Surg. 2014;148:1656–1664. doi: 10.1016/j.jtcvs.2014.03.033 [DOI] [PubMed] [Google Scholar]
- 9. Singh VP, Mathison M, Patel V, Sanagasetti D, Gibson BW, Yang J, Rosengart TK. MiR‐590 Promotes transdifferentiation of porcine and human fibroblasts toward a cardiomyocyte‐like fate by directly repressing specificity protein 1. J Am Heart Assoc. 2016;5:e003922. doi: 10.1161/JAHA.116.003922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Nam YJ, Song K, Luo X, Daniel E, Lambeth K, West K, Hill JA, DiMaio JM, Baker LA, Bassel‐Duby R, et al. Reprogramming of human fibroblasts toward a cardiac fate. Proc Natl Acad Sci USA. 2013;110:5588–5593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Patel V, Singh VP, Pinnamaneni JP, Sanagasetti D, Olive J, Mathison M, Cooney A, Flores ER, Crystal RG, Yang J, et al. p63 Silencing induces reprogramming of cardiac fibroblasts into cardiomyocyte‐like cells. J Thorac Cardiovasc Surg. 2018;156:556–565.e1. doi: 10.1016/j.jtcvs.2018.03.162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Miyamoto K, Akiyama M, Tamura F, Isomi M, Yamakawa H, Sadahiro T, Muraoka N, Kojima H, Haginiwa S, Kurotsu S, et al. Direct in vivo reprogramming with sendai virus vectors improves cardiac function after myocardial infarction. Cell Stem Cell. 2018;22:91–103.e5. [DOI] [PubMed] [Google Scholar]
- 13. Fu J‐D, Stone Nicole R, Liu L, Spencer CI, Qian L, Hayashi Y, Delgado‐Olguin P, Ding S, Bruneau Benoit G, Srivastava D. Direct reprogramming of human fibroblasts toward a cardiomyocyte‐like state. Stem Cell Reports. 2013;1:235–247. doi: 10.1016/j.stemcr.2013.07.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Zhang Z, Zhang W, Nam YJ. Stoichiometric optimization of Gata4, Hand2, Mef2c, and Tbx5 expression for contractile cardiomyocyte reprogramming. Sci Rep. 2019;9:14970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Mathison M, Singh VP, Chiuchiolo MJ, Sanagasetti D, Mao Y, Patel VB, Yang J, Kaminsky SM, Crystal RG, Rosengart TK. In situ reprogramming to transdifferentiate fibroblasts into cardiomyocytes using adenoviral vectors: implications for clinical myocardial regeneration. J Thorac Cardiovasc Surg. 2017;153:329–339.e3. doi: 10.1016/j.jtcvs.2016.09.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wang L, Liu Z, Yin C, Asfour H, Chen O, Li Y, Bursac N, Liu J, Qian L. Stoichiometry of Gata4, Mef2c, and Tbx5 influences the efficiency and quality of induced cardiac myocyte reprogramming. Circ Res. 2015;116:237–244. doi: 10.1161/CIRCRESAHA.116.305547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Chen Z, Friedrich GA, Soriano P. Transcriptional enhancer factor 1 disruption by a retroviral gene trap leads to heart defects and embryonic lethality in mice. Genes Dev. 1994;8:2293–2301. doi: 10.1101/gad.8.19.2293 [DOI] [PubMed] [Google Scholar]
- 18. Ota M, Sasaki H. Mammalian Tead proteins regulate cell proliferation and contact inhibition as transcriptional mediators of Hippo signaling. Development. 2008;135:4059–4069. doi: 10.1242/dev.027151 [DOI] [PubMed] [Google Scholar]
- 19. Yu FX, Zhao B, Guan KL. Hippo pathway in organ size control, tissue homeostasis, and cancer. Cell. 2015;163:811–828. doi: 10.1016/j.cell.2015.10.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Plouffe SW, Hong AW, Guan KL. Disease implications of the Hippo/YAP pathway. Trends Mol Med. 2015;21:212–222. doi: 10.1016/j.molmed.2015.01.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Meng Z, Moroishi T, Guan KL. Mechanisms of Hippo pathway regulation. Genes Dev. 2016;30:1–17. doi: 10.1101/gad.274027.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lin KC, Park HW, Guan KL. Regulation of the hippo pathway transcription factor TEAD. Trends Biochem Sci. 2017;42:862–872. doi: 10.1016/j.tibs.2017.09.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Yoshida T. MCAT elements and the TEF‐1 family of transcription factors in muscle development and disease. Arterioscler Thromb Vasc Biol. 2008;28:8–17. doi: 10.1161/ATVBAHA.107.155788 [DOI] [PubMed] [Google Scholar]
- 24. Liu R, Lee J, Kim BS, Wang Q, Buxton SK, Balasubramanyam N, Kim JJ, Dong J, Zhang A, Li S, et al. Tead1 is required for maintaining adult cardiomyocyte function, and its loss results in lethal dilated cardiomyopathy. JCI Insight. 2017;2:e93343. doi: 10.1172/jci.insight.93343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Liu R, Jagannathan R, Sun L, Li F, Yang P, Lee J, Negi V, Perez‐Garcia EM, Shiva S, Yechoor VK, et al. Tead1 is essential for mitochondrial function in cardiomyocytes. Am J Physiol Heart Circ Physiol. 2020;319:H89–H99. doi: 10.1152/ajpheart.00732.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. He A, Kong SW, Ma Q, Pu WT. Co‐occupancy by multiple cardiac transcription factors identifies transcriptional enhancers active in heart. Proc Natl Acad Sci USA. 2011;108:5632–5637. doi: 10.1073/pnas.1016959108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Akerberg BN, Gu F, VanDusen NJ, Zhang X, Dong R, Li K, Zhang B, Zhou B, Sethi I, Ma Q, et al. A reference map of murine cardiac transcription factor chromatin occupancy identifies dynamic and conserved enhancers. Nat Commun. 2019;10:4907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Vaseghi HR, Yin C, Zhou Y, Wang L, Liu J, Qian L. Generation of an inducible fibroblast cell line for studying direct cardiac reprogramming. Genesis. 2016;54:398–406. doi: 10.1002/dvg.22947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Mathison M, Singh VP, Sanagasetti D, Yang L, Pinnamaneni JP, Yang J, Rosengart TK. Cardiac reprogramming factor Gata4 reduces postinfarct cardiac fibrosis through direct repression of the profibrotic mediator snail. J Thorac Cardiovasc Surg. 2017;154:1601–1610.e3. doi: 10.1016/j.jtcvs.2017.06.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Liu J, Wen T, Dong K, He X, Zhou H, Shen J, Fu Z, Hu G, Ma W, Li J, et al. TEAD1 protects against necroptosis in postmitotic cardiomyocytes through regulation of nuclear DNA‐encoded mitochondrial genes. Cell Death Differ. 2021;28:2045–2059. doi: 10.1038/s41418-020-00732-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Mathison M, Gersch RP, Nasser A, Lilo S, Korman M, Fourman M, Hackett N, Shroyer K, Yang J, Ma Y, et al. In vivo cardiac cellular reprogramming efficacy is enhanced by angiogenic preconditioning of the infarcted myocardium with vascular endothelial growth factor. J Am Heart Assoc. 2012;1:e005652. doi: 10.1161/JAHA.112.005652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Singh VP, Pinnamaneni JP, Pugazenthi A, Sanagasetti D, Mathison M, Wang K, Yang J, Rosengart TK. Enhanced generation of induced cardiomyocytes using a small‐molecule cocktail to overcome barriers to cardiac cellular reprogramming. J Am Heart Assoc. 2020;9:e015686. doi: 10.1161/JAHA.119.015686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Zhou Y, Wang LI, Vaseghi H, Liu Z, Lu R, Alimohamadi S, Yin C, Fu J‐D, Wang G, Liu J, et al. Bmi1 is a key epigenetic barrier to direct cardiac reprogramming. Cell Stem Cell. 2016;18:382–395. doi: 10.1016/j.stem.2016.02.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Jayawardena TM, Egemnazarov B, Finch EA, Zhang L, Payne JA, Pandya K, Zhang Z, Rosenberg P, Mirotsou M, Dzau VJ. MicroRNA‐mediated in vitro and in vivo direct reprogramming of cardiac fibroblasts to cardiomyocytes. Circ Res. 2012;110:1465–1473. doi: 10.1161/CIRCRESAHA.112.269035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wang LI, Ma H, Huang P, Xie Y, Near D, Wang H, Xu J, Yang Y, Xu Y, Garbutt T, et al. Down‐regulation of Beclin1 promotes direct cardiac reprogramming. Sci Transl Med. 2020;12:eaay7856. doi: 10.1126/scitranslmed.aay7856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Zhao Y, Londono P, Cao Y, Sharpe EJ, Proenza C, O’Rourke R, Jones KL, Jeong MY, Walker LA, Buttrick PM. High‐efficiency reprogramming of fibroblasts into cardiomyocytes requires suppression of pro‐fibrotic signaling. Nat Commun. 2015;6:8243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Abad M, Hashimoto H, Zhou H, Morales MG, Chen B, Bassel‐Duby R, Olson EN. Notch inhibition enhances cardiac reprogramming by increasing MEF2C transcriptional activity. Stem Cell Reports. 2017;8:548–560. doi: 10.1016/j.stemcr.2017.01.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Zhou H, Dickson ME, Kim MS, Bassel‐Duby R, Olson EN. Akt1/protein kinase B enhances transcriptional reprogramming of fibroblasts to functional cardiomyocytes. Proc Natl Acad Sci USA. 2015;112:11864–11869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Christoforou N, Chakraborty S, Kirkton RD, Adler AF, Addis RC, Leong KW. Core transcription factors, microRNAs, and small molecules drive transdifferentiation of human fibroblasts towards the cardiac cell lineage. Sci Rep. 2017;7:40285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Mohamed TMA, Stone NR, Berry EC, Radzinsky E, Huang YU, Pratt K, Ang Y‐S, Yu P, Wang H, Tang S, et al. Chemical enhancement of in vitro and in vivo direct cardiac reprogramming. Circulation. 2017;135:978–995. doi: 10.1161/CIRCULATIONAHA.116.024692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Garry GA, Bezprozvannaya S, Chen K, Zhou H, Hashimoto H, Morales MG, Liu N, Bassel‐Duby R, Olson EN. The histone reader PHF7 cooperates with the SWI/SNF complex at cardiacsuper enhancers to promote direct reprogramming. Nat Cell Biol. 2021;23:467–475. doi: 10.1038/s41556-021-00668-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Huang P, Zhang L, Gao Y, He Z, Yao D, Wu Z, Cen J, Chen X, Liu C, Hu Y, et al. Direct reprogramming of human fibroblasts to functional and expandable hepatocytes. Cell Stem Cell. 2014;14:370–384. doi: 10.1016/j.stem.2014.01.003 [DOI] [PubMed] [Google Scholar]
- 43. Mammoto A, Muyleart M, Kadlec A, Gutterman D, Mammoto T. YAP1‐TEAD1 signaling controls angiogenesis and mitochondrial biogenesis through PGC1alpha. Microvasc Res. 2018;119:73–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Jiang SW, Desai D, Khan S, Eberhardt NL. Cooperative binding of TEF‐1 to repeated GGAATG‐related consensus elements with restricted spatial separation and orientation. DNA Cell Biol. 2000;19:507–514. doi: 10.1089/10445490050128430 [DOI] [PubMed] [Google Scholar]
- 45. Xin M, Kim Y, Sutherland LB, Murakami M, Qi X, McAnally J, Porrello ER, Mahmoud AI, Tan W, Shelton JM, et al. Hippo pathway effector Yap promotes cardiac regeneration. Proc Natl Acad Sci USA. 2013;110:13839–13844. doi: 10.1073/pnas.1313192110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Liu R, Jagannathan R, Li F, Lee J, Balasubramanyam N, Kim BS, Yang P, Yechoor VK, Moulik M. Tead1 is required for perinatal cardiomyocyte proliferation. PLoS One. 2019;14:e0212017. doi: 10.1371/journal.pone.0212017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Heallen T, Zhang M, Wang J, Bonilla‐Claudio M, Klysik E, Johnson RL, Martin JF. Hippo pathway inhibits Wnt signaling to restrain cardiomyocyte proliferation and heart size. Science. 2011;332:458–461. doi: 10.1126/science.1199010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Zhou Q, Li L, Zhao B, Guan KL. The hippo pathway in heart development, regeneration, and diseases. Circ Res. 2015;116:1431–1447. doi: 10.1161/CIRCRESAHA.116.303311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Schlesinger J, Schueler M, Grunert M, Fischer JJ, Zhang Q, Krueger T, Lange M, Tönjes M, Dunkel I, Sperling SR. The cardiac transcription network modulated by Gata4, Mef2a, Nkx2.5, Srf, histone modifications, and microRNAs. PLoS Genet. 2011;7:e1001313. doi: 10.1371/journal.pgen.1001313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Kurotsu S, Sadahiro T, Fujita R, Tani H, Yamakawa H, Tamura F, Isomi M, Kojima H, Yamada YU, Abe Y, et al. Soft matrix promotes cardiac reprogramming via inhibition of YAP/TAZ and suppression of fibroblast signatures. Stem Cell Reports. 2020;15:612–628. doi: 10.1016/j.stemcr.2020.07.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Adhihetty PJ, Irrcher I, Joseph AM, Ljubicic V, Hood DA. Plasticity of skeletal muscle mitochondria in response to contractile activity. Exp Physiol. 2003;88:99–107. [DOI] [PubMed] [Google Scholar]
- 52. Hickson‐Bick DL, Jones C, Buja LM. Stimulation of mitochondrial biogenesis and autophagy by lipopolysaccharide in the neonatal rat cardiomyocyte protects against programmed cell death. J Mol Cell Cardiol. 2008;44:411–418. doi: 10.1016/j.yjmcc.2007.10.013 [DOI] [PubMed] [Google Scholar]
- 53. Puigserver P, Spiegelman BM. Peroxisome proliferator‐activated receptor‐gamma coactivator 1 alpha (PGC‐1 alpha): transcriptional coactivator and metabolic regulator. Endocr Rev. 2003;24:78–90. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Tables S1–S2
Figure S1
Videos S1
Videos S2
Videos S3
Videos S4
Videos S5


