Abstract
Background
Conventional prognostic scores usually require predefined clinical variables to predict outcome. The advancement of natural language processing has made it feasible to derive meaning from unstructured data. We aimed to test whether using unstructured text in electronic health records can improve the prediction of functional outcome after acute ischemic stroke.
Methods and Results
Patients hospitalized for acute ischemic stroke were identified from 2 hospital stroke registries (3847 and 2668 patients, respectively). Prediction models developed using the first cohort were externally validated using the second cohort, and vice versa. Free text in the history of present illness and computed tomography reports was used to build machine learning models using natural language processing to predict poor functional outcome at 90 days poststroke. Four conventional prognostic models were used as baseline models. The area under the receiver operating characteristic curves of the model using history of present illness in the internal and external validation sets were 0.820 and 0.792, respectively, which were comparable to the National Institutes of Health Stroke Scale score (0.811 and 0.807). The model using computed tomography reports achieved area under the receiver operating characteristic curves of 0.758 and 0.658. Adding information from clinical text significantly improved the predictive performance of each baseline model in terms of area under the receiver operating characteristic curves, net reclassification improvement, and integrated discrimination improvement indices (all P<0.001). Swapping the study cohorts led to similar results.
Conclusions
By using natural language processing, unstructured text in electronic health records can provide an alternative tool for stroke prognostication, and even enhance the performance of existing prognostic scores.
Keywords: acute ischemic stroke, machine learning, natural language processing, outcome prediction, risk score
Subject Categories: Ischemic Stroke, Machine Learning, Prognosis
Nonstandard Abbreviations and Acronyms
- AIS
acute ischemic stroke
- ASTRAL
Acute Stroke Registry and Analysis of Lausanne
- AUC
area under the receiver operating characteristic curve
- BERT
bidirectional encoder representations from transformers
- BOW
bag‐of‐words
- CYCH
Chia‐Yi Christian Hospital
- EHR
electronic health record
- HPI
history of present illness
- IDI
integrated discrimination improvement
- ML
machine learning
- NIHSS
National Institutes of Health Stroke Scale
- NRI
net reclassification improvement
- NTUH
National Taiwan University Hospital
- PLAN
preadmission comorbidities, level of consciousness, age, and neurological deficit
Clinical Perspective
What Is New?
Using natural language processing, it is feasible to develop machine learning models for predicting functional outcome after acute ischemic stroke based on unstructured clinical text stored in electronic health records.
Machine learning models using deep learning techniques outperformed those based on the traditional “bag‐of‐words” text representation model.
The machine learning model based on the “history of present illness” in the admission note performed nearly as well as the National Institutes of Health Stroke Scale score and achieved an adequate discriminatory ability in both within‐site and across‐site validations.
What Are the Clinical Implications?
The developed machine learning models could not only provide an alternative method of stroke prognostication but also could enhance the predictive performance of conventional risk models.
The process of model development can be reproduced in individual hospitals to build customized versions of similar prognostic models.
Stroke is a common disabling neurologic condition. About one quarter of adults aged ≥25 years will develop a stroke in their lifetime. 1 Even though the acute treatment of strokes has advanced substantially, more than half of patients who have had strokes still have poor outcomes such as permanent functional dependence or even death. 2 Thus far, several prognostic risk models have been developed to predict functional outcomes following an acute stroke. Most of them use similar input variables to make predictions, such as age, initial stroke severity, and comorbidities. While most of the risk models were validated to have reasonable prognostic accuracy, they still are not widely adopted into clinical practice, probably because of implementation issues. 3 Hence, having a readily available digital tool that provides automated prognostication is beneficial for clinical decision‐making and resource allocation.
Artificial intelligence–aided prediction has been introduced to improve diagnostic precision and streamline clinical decision‐making. 4 With the advances in machine learning (ML) and deep learning, it has become feasible to integrate various types of structured data for the data‐driven prediction of clinically meaningful outcomes in patients with stroke. 5 , 6 Furthermore, by using natural language processing (NLP) to extract hidden but valuable information stored in textual data, it is possible to automate the detection of acute ischemic stroke (AIS) or the classification of stroke subtypes from neuroimaging reports, 7 , 8 , 9 and even improve the prognostication of patients with critical illness using clinical notes. 10 , 11
Supposedly, all hospitalized patients with stroke would have their corresponding admission note, in which the history of present illness (HPI) comprises the most essential textual data regarding the clinical features of the index stroke event. Furthermore, patients with stroke would also undergo baseline neuroimaging, especially a head computed tomography (CT) scan. The textual component of the CT report may disclose relevant information about the extent of cerebrovascular diseases. Considering the points above, we aimed to develop and validate ML models to investigate whether unstructured clinical text in the HPI and CT report can improve the prediction of functional outcome at an early stage after AIS.
METHODS
Data Source
Data that support the study findings are available from the corresponding author on reasonable request. The Ditmanson Medical Foundation Chia‐Yi Christian Hospital (CYCH) is a 1000‐bed teaching hospital in southern Taiwan. The National Taiwan University Hospital (NTUH) is a university‐affiliated medical center with a capacity of >2000 beds in northern Taiwan. The study protocol was independently approved by the CYCH Institutional Review Board (CYCH‐IRB No. 2020090) and Research Ethics Committee B of NTUH (202104028RINB). Study data were maintained with confidentiality to ensure the privacy of all participants.
The stroke centers of both hospitals have maintained their stroke registries since 2007 and 1995, respectively. The stroke registries prospectively registered all cases of stroke by daily screening of all patients receiving head CT or those with a diagnosis of stroke at the emergency department or during hospitalization, as well as screening for a diagnosis at discharge using the International Classification of Diseases, Ninth and Tenth Revisions (ICD ‐9 and ICD‐ 10 revisions with clinical modification) codes. Data regarding the demographics, cause, risk factor profiles, intervention, and outcomes of patients with stroke were collected. Stroke severity was assessed using the National Institutes of Health Stroke Scale (NIHSS) and functional outcome was assessed by the modified Rankin Scale.
Study Design
We followed the Transparent Reporting of a Multivariable Prediction Model for Individual Prognosis or Diagnosis guidelines for the methods and reporting of prediction models. 12 All consecutive adult patients with first‐ever AIS admitted to CYCH from October 2007 to December 2019 and those admitted to NTUH from January 2016 to December 2020 were identified using the institutional stroke registries. Patients who had an in‐hospital stroke or whose clinical data included a missing admission NIHSS score were excluded. Those who did not provide consent for follow‐up or who were lost to follow‐up at 90 days were also eliminated. For each patient, we retrieved the HPI upon admission and the initial CT report from the electronic health record (EHR) database. Both types of documents were written in English in the study hospitals. Patients whose EHRs were unavailable were excluded.
The CYCH cohort (Cohort A in Figure 1) were randomly split into a derivation set that consisted of 75% of the patients, and an internal validation set comprising the remaining 25% of the patients, who were withheld from all ML models during the training process. The NTUH cohort (Cohort B in Figure 1) comprised the external validation set. In addition, to test the generalizability of the ML approach, we did another experiment where the NTUH cohort was used for derivation and internal validation and the CYCH cohort was used for external validation.
Figure 1. Process of model development and validation.
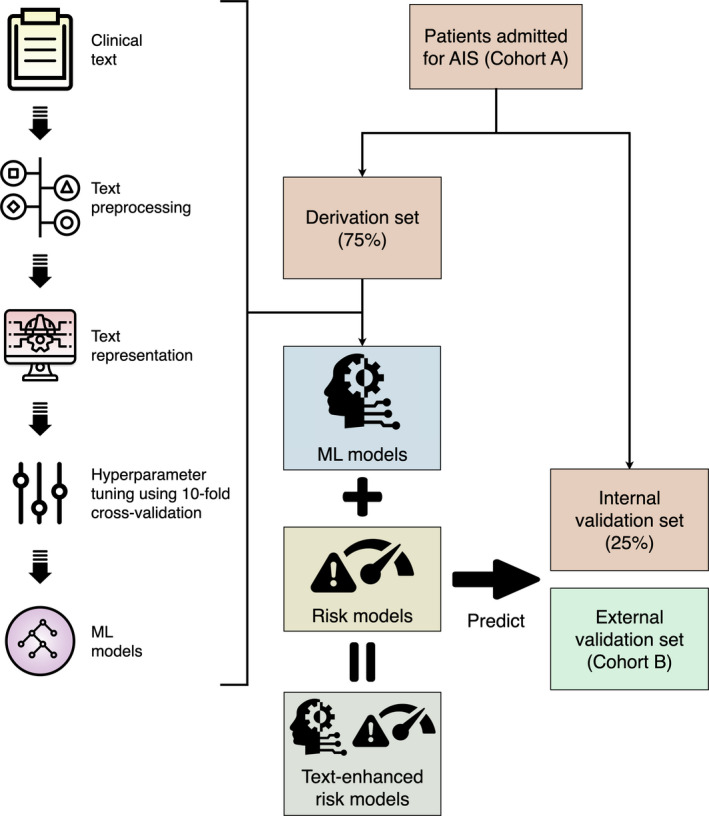
AIS indicates acute ischemic stroke; and ML, machine learning.
Outcome Variable
The outcome variable was a poor functional outcome at 90 days poststroke, which was defined as a modified Rankin Scale score of 3 to 6.
Baseline Risk Models
We used 4 prognostic models that used clinical variables available upon admission as the baseline risk models for comparison. The first risk model was the NIHSS score because stroke severity is the most important determinant for poststroke functional outcome. 13 The second model, consisting of age and NIHSS score within the first 6 hours of the onset of AIS, was useful in predicting 3‐month mortality and functional outcome. 13 The third model was preadmission comorbidities, level of consciousness, age, and neurologic deficit (PLAN) score, 14 which was developed to predict 30‐day and 1‐year mortality and a modified Rankin Scale score of 5 to 6 at discharge. In the PLAN score, preadmission comorbidities refer to preadmission dependence, cancer, congestive heart failure, and atrial fibrillation, whereas neurologic focal deficits indicate weakness of the leg or arm, aphasia, and neglect. The fourth model was derived using a cohort of patients from the Acute Stroke Registry and Analysis of Lausanne (ASTRAL). 15 The ASTRAL score, comprising age, NIHSS score, time from stroke onset to admission, range of visual fields, acute glucose level, and level of consciousness, was designed to predict 3‐month unfavorable outcome (modified Rankin Scale >2) poststroke upon hospital admission.
NLP and ML Models
Figure 1 illustrates the process of model development and validation. ML models were trained separately using HPI (model HPI) and CT reports (model CT). Experiments were done with 2 approaches of text representation: a simple “bag‐of‐words” (BOW) approach and a more sophisticated deep learning approach using the bidirectional encoder representations from transformers (BERT).
Free text was preprocessed as follows: (1) words were spell‐checked, and misspelled words were automatically corrected using the Jazzy spell checker (https://github.com/kinow/jazzy); (2) acronyms and abbreviations were expanded to their full forms by looking up a list of common clinical acronyms and abbreviations used locally; (3) non‐ASCII characters and nonword special characters were deleted; (4) words were converted to lowercase; (5) words were lemmatized to their root forms; and (6) stop words were removed. Only step 1 through step 3 were needed for the BERT approach.
In the BOW approach, we built a document‐term matrix, where each column stood for a unique word from the text corpus, the rows represented each document (HPI or CT report for each patient), and the cells indicated whether each word appeared within each document. The random forest algorithm was used to build the ML models. 16 After fitting the ML models, the permutation importance method was used to assess the impact of each word on the prediction results, 16 thus aiding in interpreting the ML models.
Clinical notes are generally lengthy, and their words are usually dependent on each other. However, the BOW approach does not consider the sequence of words and cannot capture the meaning of words in their context. Even conventional word‐embedding methods such as Word2Vec disregard the long‐range dependency and may not fully capture clinical meaning from clinical notes. 11 Therefore, the BERT approach was used for representation of clinical notes. BERT is a deep neural network–based contextualized word‐embedding model that is pre‐trained using bidirectional transformers based on masked language modeling and next sentence prediction. 17 The original BERT is pre‐trained using the general BooksCorpus and English Wikipedia corpus. 17
For the current study, a clinical domain‐specific BERT model (ie, ClinicalBERT) was used. ClinicalBERT is pre‐trained on the Medical Information Mart for Intensive Care III clinical notes, 18 which is a large collection of clinical notes from >40 000 patients who stayed in the intensive care unit. 19 The embeddings learned from the ClinicalBERT model were directly fed to a feed‐forward neural network with a softmax function for classification. The weights of the pre‐trained CliniclBERT model along with the classification layer were updated simultaneously during the fine‐tuning stage of the model. The preprocessed text was split into BERT tokens. Because the BERT model can only deal with 512 tokens and most of the documents contained <512 tokens (Table 1), input documents were truncated to 512 tokens to simplify the analysis.
Table 1.
Characteristics of the Study Cohorts
| CYCH (n=3847) | NTUH (n=2668) | P value | |
|---|---|---|---|
| Age, mean (SD) | 69.5 (12.3) | 69.8 (13.9) | 0.528 |
| Female | 1583 (41.1) | 1118 (41.9) | 0.543 |
| Hypertension | 3098 (80.5) | 2090 (78.3) | 0.031 |
| Diabetes | 1602 (41.6) | 1024 (38.4) | 0.008 |
| Hyperlipidemia | 2195 (57.1) | 1369 (51.3) | <0.001 |
| Atrial fibrillation | 684 (17.8) | 790 (29.6) | <0.001 |
| Congestive heart failure | 196 (5.1) | 223 (8.4) | <0.001 |
| Cancer | 249 (6.5) | 424 (15.9) | <0.001 |
| Preadmission dependence (mRS >2) | 419 (10.9) | 407 (15.3) | <0.001 |
| Onset‐to‐admission delay >3 h | 2763 (71.8) | 1913 (71.7) | 0.915 |
| NIHSS, median (IQR) | 5 (3–10) | 5 (2–13) | 0.267 |
| Glucose, mean (SD), mg/dL | 163.2 (82.6) | 146.8 (67.7) | <0.001 |
| PLAN score, median (IQR) | 8 (6–12) | 9 (7–12) | 0.001 |
| ASTRAL score, median (IQR) | 21 (18–27) | 22 (18–30) | 0.178 |
| Word count in HPI, median (IQR) | 132 (109–161) | 268 (209–342) | <0.001 |
| BERT tokens in HPI, median (IQR) | 192 (156–240) | 420 (329–535) | <0.001 |
| Word count in CT reports, median (IQR) | 127 (93–189) | 42 (34–52) | <0.001 |
| BERT tokens in CT reports, median (IQR) | 225 (164–351) | 86 (68–106) | <0.001 |
| Poor outcome (mRS >2) | 1674 (43.5) | 1118 (41.9) | 0.196 |
Data are expressed in number (percentage) unless specified otherwise. ASTRAL indicates Acute Stroke Registry and Analysis of Lausanne; BERT, bidirectional encoder representations from transformers; CT, computed tomography; CYCH, Chia‐Yi Christian Hospital; HPI, history of present illness; IQR, interquartile range; mRS, modified Rankin Scale; NIHSS, National Institutes of Health Stroke Scale; NTUH, National Taiwan University Hospital; and PLAN, preadmission comorbidities, level of consciousness, age, and neurological deficit.
Only data from the derivation set were used in the training (fine‐tuning) process, which was implemented using Python 3.8.5 on a Windows 10 system with graphics processing unit. Hyperparameter optimization was performed using 10‐fold cross‐validation. The number of trees for the random forest classifier was varied from 10 to 200 with an increment of 10. During the fine‐tuning of the ClinicalBERT model for the prediction task, the batch size of the neural network was set at 16. The learning rate of the Adam optimizer was varied from 1×10−5 to 5×10−5 with an increment of 1×10−5, and the number of epochs from 2 to 4 with an increment of 1. Model error was minimized in terms of the area under the receiver operating characteristic curve (AUC). Once the optimal hyperparameters were determined, the ML models were fitted with the full derivation set.
Statistical Analysis
Categorical variables were expressed as counts (percentages) while continuous variables were expressed as means (SDs) or medians (interquartile ranges). Differences between 2 groups were tested by χ2 tests for categorical variables and t tests or Mann–Whitney U tests for continuous variables, as appropriate.
Model performance was evaluated on the internal and external validation sets separately. For each patient in the validation sets, the probability of a poor functional outcome was estimated using the ML models. Model discrimination was assessed with AUCs. The AUCs between the BOW and BERT approaches were compared using the DeLong method. 20 The approach that resulted in higher AUCs was used in the following analysis.
For each baseline model, a logistic regression model was fitted by entering the risk score as a continuous variable. To construct a “text‐only” risk model, named model HPI+CT, a logistic regression model was fitted by entering the probabilities of a poor functional outcome predicted separately by model HPI and model CT as continuous variables. To assess the incremental value of adding information from clinical text to the baseline models, the probabilities of a poor functional outcome predicted by model HPI and model CT were introduced to the logistic regression model as continuous variables to construct “text‐enhanced” risk models. Model discrimination was assessed with AUCs. Model calibration was evaluated by visual inspection of the calibration plot, 21 which depicts the observed risk versus the predicted risk. In addition, the added predictive ability of clinical text was evaluated by calculating the continuous net reclassification improvement (NRI) and integrated discrimination improvement (IDI) indices. 22 , 23 Unlike categorical NRI, the continuous NRI does not require established risk categories. It quantifies upward and downward changes in the predicted probabilities of an event. The IDI is equivalent to the difference in discrimination slopes, which measure the difference between mean predicted probabilities of an event for those with events and the corresponding mean for those without events. 22 , 23 Higher values of NRI and IDI indicate superior discrimination.
All statistical analyses were performed using Stata 15.1 (StataCorp, College Station, TX) and R version 4.0.5 (R Foundation for Statistical Computing, Vienna, Austria). The calibration plot and the analysis of NRI and IDI were performed using R package “PredictABEL”. Two‐tailed P values of 0.05 were considered significant.
RESULTS
A total of 3847 eligible patients with AIS were recruited from CYCH. The derivation and internal validation sets consisted of 2885 and 962 patients, respectively. The external validation set comprised 2668 patients with AIS from NTUH. The characteristics of the study cohorts are listed in Table 1. The CYCH and NTUH cohorts were similar in age, sex, onset‐to‐admission delay, NIHSS, ASTRAL score, and the proportion of a poor functional outcome. However, they significantly differed in the prevalence of comorbidities and preadmission dependence, glucose level, PLAN score, and word counts in the HPI and CT reports.
In the internal validation set, the AUC of model HPI of the BERT approach was not significantly different from that of the BOW approach (0.820 versus 0.802, P=0.111), whereas model CT of the BERT approach achieved a higher AUC than that of the BOW approach (0.758 versus 685, P<0.001). Model HPI+CT of the BERT approach yielded a higher AUC than that of the BOW approach (0.840 versus 0.819, P=0.042). In the external validation set, model HPI of the BERT approach had a significantly higher AUC than that of the BOW approach (0.792 versus 0.761, P<0.001), whereas the AUCs of model CT of both approaches were similar (0.658 versus 662, P=0.690). Model HPI+CT of the BERT approach achieved a higher AUC than that of the BOW approach (0.798 versus 0.778, P=0.009). Therefore, ML models built by the BERT approach were used in the following analysis. Figure S1 shows the top 20 most influential words for model HPI and model CT.
Figure 2 shows the AUCs of the baseline and ML models. In the internal validation set, model HPI+CT yielded an AUC of 0.840, which was comparable to those of NIHSS (0.811, P=0.062), age and NIHSS (0.841, P=0.935), PLAN score (0.837, P=0.830), and ASTRAL score (0.840, P=0.995). Model HPI achieved an AUC of 0.820, which was comparable to those of NIHSS (P=0.581), age and NIHSS (P=0.111), PLAN score (P=0.194), and ASTRAL score (P=0.134). Model CT yielded an AUC of 0.758, which was significantly lower than those of NIHSS (P=0.005) and the other 3 baseline models (all P<0.001). In the external validation set, the AUCs of model HPI+CT, model HPI, and model CT were 0.798, 0.792, and 0.658, respectively. Model HPI+CT and model HPI performed equally well with NIHSS (0.807, P=0.359 and 0.134, respectively), but were inferior to the other 3 baseline models (all P<0.001). Model CT had a significantly lower AUC than the 4 baseline models (all P<0.001).
Figure 2. Receiver operating characteristic curves for predicting a poor functional outcome in the internal (A) and external (B) validation sets.
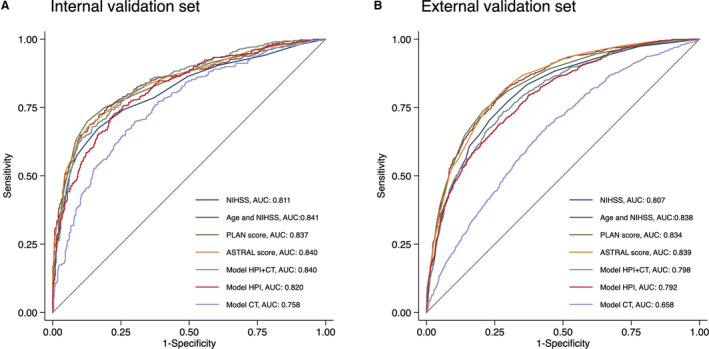
ASTRAL indicates Acute Stroke Registry and Analysis of Lausanne; AUC, area under the receiver operating characteristic curve; CT, computed tomography; HPI, history of present illness; NIHSS, National Institutes of Health Stroke Scale; and PLAN, preadmission comorbidities, level of consciousness, age, and neurological deficit.
Table 2 lists the comparison of performance between the baseline and text‐enhanced risk models. The AUCs of the text‐enhanced models were significantly higher than those of the baseline models in both the internal and external validation sets (all P<0.001). The NRI and IDI indices also indicated a statistically significant improvement (all P<0.001) in predictive performance when the baseline models were enhanced by the information from the clinical text. The calibration plots of the baseline and text‐enhanced models are displayed in Figure 3. It shows that the text‐enhanced models were generally well calibrated over the entire risk range because all points lie close to the 45‐degree line.
Table 2.
Comparison of the Predictive Ability of Baseline Models With or Without Adding Information From Clinical Text
| Baseline AUC (95% CI) | Text‐enhanced AUC (95% CI) | P value | NRI (95% CI) | P value | IDI (95% CI) | P value | |
|---|---|---|---|---|---|---|---|
| Internal validation | |||||||
| NIHSS | 0.811 (0.783–0.839) | 0.869 (0.846–0.891) | <0.001 | 0.766 (0.648–0.884) | <0.001 | 0.109 (0.089–0.129) | <0.001 |
| Age and NIHSS | 0.841 (0.815–0.866) | 0.872 (0.850–0.895) | <0.001 | 0.514 (0.391–0.637) | <0.001 | 0.065 (0.049–0.080) | <0.001 |
| PLAN score | 0.837 (0.811–0.863) | 0.870 (0.847–0.893) | <0.001 | 0.593 (0.471–0.715) | <0.001 | 0.061 (0.046–0.077) | <0.001 |
| ASTRAL score | 0.840 (0.814–0.866) | 0.871 (0.849–0.894) | <0.001 | 0.527 (0.405–0.650) | <0.001 | 0.070 (0.054–0.086) | <0.001 |
| External validation | |||||||
| NIHSS | 0.807 (0.790–0.823) | 0.843 (0.828–0.858) | <0.001 | 0.719 (0.648–0.791) | <0.001 | 0.089 (0.078–0.100) | <0.001 |
| Age and NIHSS | 0.838 (0.823–0.853) | 0.854 (0.840–0.868) | <0.001 | 0.556 (0.482–0.630) | <0.001 | 0.043 (0.035–0.052) | <0.001 |
| PLAN score | 0.834 (0.818–0.849) | 0.852 (0.838–0.867) | <0.001 | 0.561 (0.488–0.635) | <0.001 | 0.045 (0.037–0.054) | <0.001 |
| ASTRAL score | 0.839 (0.824–0.854) | 0.854 (0.840–0.868) | <0.001 | 0.572 (0.499–0.646) | <0.001 | 0.052 (0.043–0.061) | <0.001 |
ASTRAL indicates Acute Stroke Registry and Analysis of Lausanne; AUC, area under the receiver operating characteristic curve; IDI, integrated discrimination improvement; NIHSS, National Institutes of Health Stroke Scale; NRI, net reclassification improvement; and PLAN, preadmission comorbidities, level of consciousness, age, and neurological deficit.
Figure 3. Calibration plots of the baseline and text‐enhanced models.
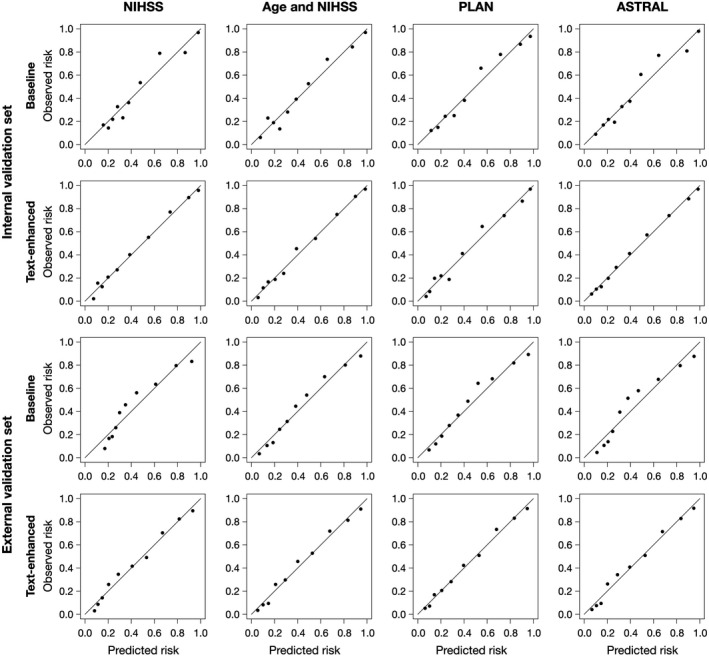
ASTRAL indicates Acute Stroke Registry and Analysis of Lausanne; NIHSS, National Institutes of Health Stroke Scale; and PLAN, preadmission comorbidities, level of consciousness, age, and neurological deficit.
In the additional experiment where the 2 study cohorts were exchanged, similar results were obtained. Figure S2 shows the AUCs of the baseline and ML models. In the internal validation cohort, model HPI+CT achieved an AUC of 0.818, which was comparable to those of NIHSS (0.815, P=0.867), age and NIHSS (0.842, P=0.115), PLAN score (0.837, P=0.214), and ASTRAL score (0.847, P=0.056). Model HPI yielded an AUC of 0.818, which was comparable to those of NIHSS (0.815, P=0.889), age and NIHSS (0.842, P=0.134), PLAN score (0.837, P=0.234), and ASTRAL score (0.847, P=0.065). Model CT achieved an AUC of 0.674, which was significantly lower than the 4 baseline models (all P<0.001). In the external validation set, the AUCs of model HPI+CT, model HPI, and model CT were 0.778, 0.772, and 0.662, respectively. However, all of the 3 models had significantly lower AUCs than the 4 baseline models (all P<0.001). The calibration plots of the baseline and text‐enhanced models are displayed in Figure S3. Table S1 gives the comparison of performance between the baseline and text‐enhanced risk models. Significant improvements in AUCs, NRI, and IDI indices (all P<0.001) were observed in the text‐enhanced models versus the baseline models.
DISCUSSION
This study demonstrates that ML models trained from clinical text could not only provide an alternative method of stroke prognostication but also enhance the predictive performance of conventional risk models in terms of the prediction of 90‐day functional outcome. In general, the deep learning NLP approach (BERT) outperformed the simpler NLP approach (BOW paired with random forest classifiers). Based on the BERT approach, the text‐only model based on HPI and CT reports and the model based on HPI alone both achieved an adequate discriminatory ability in within‐site and across‐site validations and they performed nearly as well as the NIHSS score. Moreover, the text‐enhanced risk models demonstrated a considerably higher discriminatory ability than the baseline risk models as well as acceptable model calibration.
The functional outcome of AIS is largely determined by stroke severity, 13 which is closely related to the location and size of ischemic brain lesions. Therefore, the free‐form text in the HPI and CT reports, which may implicitly contain information regarding stroke severity, can be used to predict stroke outcomes. However, the predictive performance of model CT was worse than model HPI. One possible explanation might be that the initial unenhanced CT scan generally does not reflect the final extent of brain ischemia. 24 In this regard, magnetic resonance imaging studies are more sensitive than CT studies for detection of acute ischemia, 25 and magnetic resonance imaging reports seemed to be promising for predicting outcome after AIS. 26 Nevertheless, magnetic resonance imaging is not as widespread and readily available for emergency situations as CT.
On the other hand, clinical text may contain much richer information than that captured by conventional stroke prognostic models. Most of the existing models tended to base their predictions on the same concepts, such as demographics, initial stroke severity, pre‐stroke functional status, and comorbidities, and thus shared a similar degree of prognostic accuracy. 3 It might reasonably be expected that incorporating other less traditional factors, such as the degree of frailty, emotional wellness, strength of social support, or even the clinician’s clinical gestalt could improve the utility of prognostic models. The text in the HPI could complement such information and thereby enhance the predictive performance of the baseline models. Even though model HPI based on the BERT approach performed and generalized well across hospitals, such deep learning NLP models are often considered a “black box” model lacking interpretability. In situations where model interpretability is given a high priority, simpler NLP approaches such as the BOW approach may be reasonable alternatives despite their lower predictive ability. 9 , 26 Furthermore, influential features identified from free text might be collected and used to develop new prognostic models. 10
ML methods have been applied to develop models for prognostication of AIS. 6 , 27 , 28 , 29 , 30 , 31 , 32 The ML models in the existing studies generally had a comparable or even higher discriminatory ability than conventional logistic regression models. 27 , 28 , 30 One of the reasons may be that ML algorithms can handle potential nonlinear relationships and model complex interactions between variables. 6 , 33 , 34 However, these ML models were seldom externally validated, 31 undermining their utility in other populations or health care settings. Like any other diagnostic or prognostic tool, a ML model should be validated in an independent data set by assessing its discrimination and calibration. 35 , 36 In particular, in order to improve model performance, ML models usually undergo hyperparameter optimization where the hyperparameters are tuned on a tuning set independent of the validation set. 35 , 36 This study followed these recommendations by tuning the hyperparameters by cross‐validation within the derivation set and validating the ML models on both a holdout test set (within‐site validation) and a completely independent data set (across‐site validation).
Although ML methods are gaining popularity, textual data have rarely been analyzed or used in previous ML prognostic models of stroke. Despite this, NLP has been applied in the field of stroke medicine, such as building ML models to identify AIS 7 , 9 , 37 or automating AIS subtype classification. 38 , 39 The merit of using textual data is that clinical notes are generated within EHRs in the process of medical care, thus saving the extra effort required for data collection and coding. Furthermore, the nuances of symptoms across patients are more likely to be preserved in unstructured textual data. 40 The study findings supported the incremental value of unstructured clinical text over the conventional prognostic models.
With the routine use of EHRs in clinical practices, a large amount of health care data, either structured or unstructured, has not only accumulated rapidly but also has become more available for downstream use. Big data analytics is now increasingly used in diverse health care applications such as disease surveillance, health management, and clinical decision support. 41 In addition, by directly drawing data from EHRs, conventional prognostic models can be integrated into the EHR system to provide automated outcome prediction. 42 , 43 Nevertheless, this approach would miss the opportunity to capture meaningful information embedded in clinical notes. By contrast, the methods used in this study harnessed both unstructured and structured data to generate prognostic models, which can be easily implemented as an electronic decision support tool to help health care professionals to establish a prognosis.
This study has some limitations that need to be addressed. First, although data‐driven ML methods have their own advantages, the relationships discovered from the data do not mean any causal inference, and prediction accuracy should not be interpreted in any way as causal validity. 44 Second, the vocabulary and style used in clinical documentation may vary across hospitals and regions, thereby affecting the performance of the ClinicalBERT model, which was pre‐trained on clinical notes from a US hospital. 18 A BERT model pre‐trained on clinical text from the local health system is likely to further improve the predictive performance of NLP models but requires a larger computational cost. Despite this, the study results showed that the developed models generalized well in the external validation group from a geographically distant hospital. Therefore, we believe that the influence by the variation in clinical documentation is not substantial. Furthermore, the process of model development can be reproduced in individual hospitals to build customized versions of similar prognostic models. Third, ML models based on features directly derived from neuroimaging data are promising for predicting various stroke outcomes. 45 , 46 Although this issue is out of the scope of the current study, future studies may explore the value of alternative sources of unstructured data such as imaging data in the prediction of poststroke functional outcome.
CONCLUSIONS
By using NLP and ML methods, information derived from clinical text has the potential to prognosticate patients with AIS. This study developed and validated text‐enhanced prognostic models to aid in the early prediction of functional outcome after AIS. However, further studies are needed to confirm the generalizability of this approach and the clinical usefulness in routine practice.
Sources of Funding
This research was funded by the Ditmanson Medical Foundation Chia‐Yi Christian Hospital (grant number R109‐37‐1). The funder of the research had no role in the design and conduct of the study, interpretation of the data, or decision to submit for publication.
Disclosures
None.
Supporting information
Table S1
Figures S1–S3
Acknowledgments
The authors would like to thank Li‐Ying Sung for English language editing.
Supplementary Material for this article is available at https://www.ahajournals.org/doi/suppl/10.1161/JAHA.121.023486
For Sources of Funding and Disclosures, see page 9.
REFERENCES
- 1. GBD 2016 Lifetime Risk of Stroke Collaborators . Global, regional, and country‐specific lifetime risks of stroke, 1990 and 2016. N Engl J Med. 2018;379:2429–2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Campbell BCV, Khatri P. Stroke. Lancet. 2020;396:129–142. doi: 10.1016/S0140-6736(20)31179-X [DOI] [PubMed] [Google Scholar]
- 3. Drozdowska BA, Singh S, Quinn TJ. Thinking about the future: a review of prognostic scales used in acute stroke. Front Neurol. 2019;10:274. doi: 10.3389/fneur.2019.00274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Rasmy L, Xiang Y, Xie Z, Tao C, Zhi D. Med‐BERT: pretrained contextualized embeddings on large‐scale structured electronic health records for disease prediction. NPJ Digit Med. 2021;4:86. doi: 10.1038/s41746-021-00455-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ding L, Liu C, Li Z, Wang Y. Incorporating artificial intelligence into stroke care and research. Stroke. 2020;51:e351–e354. doi: 10.1161/STROKEAHA.120.031295 [DOI] [PubMed] [Google Scholar]
- 6. Matsumoto K, Nohara Y, Soejima H, Yonehara T, Nakashima N, Kamouchi M. Stroke prognostic scores and data‐driven prediction of clinical outcomes after acute ischemic stroke. Stroke. 2020;51:1477–1483. doi: 10.1161/STROKEAHA.119.027300 [DOI] [PubMed] [Google Scholar]
- 7. Kim C, Zhu V, Obeid J, Lenert L. Natural language processing and machine learning algorithm to identify brain MRI reports with acute ischemic stroke. PLoS One. 2019;14:e0212778. doi: 10.1371/journal.pone.0212778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Rannikmäe K, Wu H, Tominey S, Whiteley W, Allen N, Sudlow C, Biobank U. Developing automated methods for disease subtyping in UK Biobank: an exemplar study on stroke. BMC Med Inform Decis Mak. 2021;21:191. doi: 10.1186/s12911-021-01556-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ong CJ, Orfanoudaki A, Zhang R, Caprasse FPM, Hutch M, Ma L, Fard D, Balogun O, Miller MI, Minnig M, et al. Machine learning and natural language processing methods to identify ischemic stroke, acuity and location from radiology reports. PLoS One. 2020;15:e0234908. doi: 10.1371/journal.pone.0234908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Weissman GE, Hubbard RA, Ungar LH, Harhay MO, Greene CS, Himes BE, Halpern SD. Inclusion of unstructured clinical text improves early prediction of death or prolonged ICU stay. Crit Care Med. 2018;46:1125–1132. doi: 10.1097/CCM.0000000000003148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Huang K, Altosaar J & Ranganath R ClinicalBERT: modeling clinical notes and predicting hospital readmission. Paper presented at: CHIL ’20: ACM Conference on Health, Inference, and Learning; Workshop Track. April 02–04, 2020; Toronto, Ontario, Canada. Available at: https://arxiv.org/abs/1904.05342. Accessed June 27, 2021
- 12. Moons KGM, Altman DG, Reitsma JB, Ioannidis JPA, Macaskill P, Steyerberg EW, Vickers AJ, Ransohoff DF, Collins GS. Transparent Reporting of a multivariable prediction model for Individual Prognosis or Diagnosis (TRIPOD): explanation and elaboration. Ann Intern Med. 2015;162:W1–W73. doi: 10.7326/M14-0698 [DOI] [PubMed] [Google Scholar]
- 13. Weimar C, König IR, Kraywinkel K, Ziegler A, Diener HC. Age and National Institutes of Health Stroke Scale Score within 6 hours after onset are accurate predictors of outcome after cerebral ischemia. Stroke. 2004;35:158–162. doi: 10.1161/01.STR.0000106761.94985.8B [DOI] [PubMed] [Google Scholar]
- 14. O’Donnell MJ, Fang J, D’Uva C, Saposnik G, Gould L, McGrath E, Kapral MK; Investigators of the Registry of the Canadian Stroke Network . The PLAN score: a bedside prediction rule for death and severe disability following acute ischemic stroke. Arch Intern Med. 2012;172:1548–1556. doi: 10.1001/2013.jamainternmed.30 [DOI] [PubMed] [Google Scholar]
- 15. Ntaios G, Faouzi M, Ferrari J, Lang W, Vemmos K, Michel P. An integer‐based score to predict functional outcome in acute ischemic stroke. Neurology. 2012;78:1916–1922. [DOI] [PubMed] [Google Scholar]
- 16. Breiman L. Random forests. Mach Learn. 2001;45:5–32. [Google Scholar]
- 17. Devlin J, Chang M‐W, Lee K, Toutanova K. BERT: pre‐training of deep bidirectional transformers for language understanding. Paper presented at: Proceedings of the 2019 Conference of the North American Chapter of the Association for Computational Linguistics: Human Language Technologies, Volume 1 (Long and Short Papers). June 2019; Minneapolis, MN. Available at: https://arxiv.org/abs/1810.04805. Accessed June 27, 2021 [Google Scholar]
- 18. Alsentzer E, Murphy J, Boag W, Weng W‐H, Jindi D, Naumann T, McDermott M. Publicly available clinical BERT Embeddings. Paper presented at: Proceedings of the 2nd Clinical Natural Language Processing Workshop, pages 72‐78. June 7, 2019; Minneapolis, MN. Available at: https://arxiv.org/abs/1904.03323. Accessed June 27, 2021 [Google Scholar]
- 19. Johnson AEW, Pollard TJ, Shen L, Lehman LH, Feng M, Ghassemi M, Moody B, Szolovits P, Celi LA, Mark RG. MIMIC‐III, a freely accessible critical care database. Sci Data. 2016;3:160035. doi: 10.1038/sdata.2016.35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. DeLong ER, DeLong DM, Clarke‐Pearson DL. Comparing the areas under two or more correlated receiver operating characteristic curves: a nonparametric approach. Biometrics. 1988;44:837–845. doi: 10.2307/2531595 [DOI] [PubMed] [Google Scholar]
- 21. Steyerberg EW, Vickers AJ, Cook NR, Gerds T, Gonen M, Obuchowski N, Pencina MJ, Kattan MW. Assessing the performance of prediction models: a framework for traditional and novel measures. Epidemiology. 2010;21:128–138. doi: 10.1097/EDE.0b013e3181c30fb2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Pencina MJ, D' Agostino RB, D' Agostino RB, Vasan RS. Evaluating the added predictive ability of a new marker: from area under the ROC curve to reclassification and beyond. Stat Med. 2008;27:157–172. doi: 10.1002/sim.2929 [DOI] [PubMed] [Google Scholar]
- 23. Pencina MJ, D’Agostino RB, Steyerberg EW. Extensions of net reclassification improvement calculations to measure usefulness of new biomarkers. Stat Med. 2011;30:11–21. doi: 10.1002/sim.4085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kloska SP, Nabavi DG, Gaus C, Nam E‐M, Klotz E, Ringelstein EB, Heindel W. Acute stroke assessment with CT: do we need multimodal evaluation? Radiology. 2004;233:79–86. doi: 10.1148/radiol.2331030028 [DOI] [PubMed] [Google Scholar]
- 25. Vilela P, Rowley HA. Brain ischemia: CT and MRI techniques in acute ischemic stroke. Eur J Radiol. 2017;96:162–172. doi: 10.1016/j.ejrad.2017.08.014 [DOI] [PubMed] [Google Scholar]
- 26. Heo TS, Kim YS, Choi JM, Jeong YS, Seo SY, Lee JH, Jeon JP, Kim C. Prediction of stroke outcome using natural language processing‐based machine learning of radiology report of brain MRI. J Pers Med. 2020;10:286. doi: 10.3390/jpm10040286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Monteiro M, Fonseca AC, Freitas AT, Melo TPE, Francisco AP, Ferro JM, Oliveira AL. Using machine learning to improve the prediction of functional outcome in ischemic stroke patients. IEEE/ACM Trans Comput Biol Bioinform. 2018;15:1953–1959. doi: 10.1109/TCBB.2018.2811471 [DOI] [PubMed] [Google Scholar]
- 28. Heo J, Yoon JG, Park H, Kim YD, Nam HS, Heo JH. Machine learning‐based model for prediction of outcomes in acute stroke. Stroke. 2019;50:1263–1265. doi: 10.1161/STROKEAHA.118.024293 [DOI] [PubMed] [Google Scholar]
- 29. Xie Y, Jiang B, Gong E, Li Y, Zhu G, Michel P, Wintermark M, Zaharchuk G. Use of gradient boosting machine learning to predict patient outcome in acute ischemic stroke on the basis of imaging, demographic, and clinical information. AJR Am J Roentgenol. 2019;212:44–51. [DOI] [PubMed] [Google Scholar]
- 30. Li X, Pan X, Jiang C, Wu M, Liu Y, Wang F, Zheng X, Yang J, Sun C, Zhu Y, et al. Predicting 6‐month unfavorable outcome of acute ischemic stroke using machine learning. Front Neurol. 2020;11:539509. doi: 10.3389/fneur.2020.539509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Alaka SA, Menon BK, Brobbey A, Williamson T, Goyal M, Demchuk AM, Hill MD, Sajobi TT. Functional outcome prediction in ischemic stroke: a comparison of machine learning algorithms and regression models. Front Neurol. 2020;11:889. doi: 10.3389/fneur.2020.00889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lin C‐H, Hsu K‐C, Johnson KR, Fann YC, Tsai C‐H, Sun YU, Lien L‐M, Chang W‐L, Chen P‐L, Lin C‐L, et al.; Taiwan Stroke Registry Investigators . Evaluation of machine learning methods to stroke outcome prediction using a nationwide disease registry. Comput Methods Programs Biomed. 2020;190:105381. doi: 10.1016/j.cmpb.2020.105381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Orfanoudaki A, Chesley E, Cadisch C, Stein B, Nouh A, Alberts MJ, Bertsimas D. Machine learning provides evidence that stroke risk is not linear: the non‐linear Framingham stroke risk score. PLoS One. 2020;15:e0232414. doi: 10.1371/journal.pone.0232414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. van Os HJA, Ramos LA, Hilbert A, van Leeuwen M, van Walderveen MAA, Kruyt ND, Dippel DWJ, Steyerberg EW, van der Schaaf IC, Lingsma HF, et al. Predicting outcome of endovascular treatment for acute ischemic stroke: potential value of machine learning algorithms. Front Neurol. 2018;9:784. doi: 10.3389/fneur.2018.00784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Liu Y, Chen P‐HC, Krause J, Peng L. How to read articles that use machine learning: users’ guides to the medical literature. JAMA. 2019;322:1806–1816. doi: 10.1001/jama.2019.16489 [DOI] [PubMed] [Google Scholar]
- 36. Obermeyer Z, Emanuel EJ. Predicting the future — big data, machine learning, and clinical medicine. N Engl J Med. 2016;375:1216–1219. doi: 10.1056/NEJMp1606181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Sedghi E, Weber JH, Thomo A, Bibok M, Penn AMW. Mining clinical text for stroke prediction. Netw Model Anal Health Inform Bioinform. 2015;4:688. doi: 10.1007/s13721-015-0090-5 [DOI] [Google Scholar]
- 38. Garg R, Oh E, Naidech A, Kording K, Prabhakaran S. Automating ischemic stroke subtype classification using machine learning and natural language processing. J Stroke Cerebrovasc Dis. 2019;28:2045–2051. doi: 10.1016/j.jstrokecerebrovasdis.2019.02.004 [DOI] [PubMed] [Google Scholar]
- 39. Sung S‐F, Lin C‐Y, Hu Y‐H. EMR‐based phenotyping of ischemic stroke using supervised machine learning and text mining techniques. IEEE J Biomed Health Inform. 2020;24:2922–2931. doi: 10.1109/JBHI.2020.2976931 [DOI] [PubMed] [Google Scholar]
- 40. Kuhn T, Basch P, Barr M, Yackel T; Medical Informatics Committee of the American College of Physicians . Clinical documentation in the 21st century: executive summary of a policy position paper from the American College of Physicians. Ann Intern Med. 2015;162:301. doi: 10.7326/M14-2128 [DOI] [PubMed] [Google Scholar]
- 41. Khanra S, Dhir A, Islam N, Mäntymäki M. Big data analytics in healthcare: a systematic literature review. Enterp Inf Syst. 2020;14:878–912. doi: 10.1080/17517575.2020.1812005 [DOI] [Google Scholar]
- 42. Aakre C, Franco PM, Ferreyra M, Kitson J, Li M, Herasevich V. Prospective validation of a near real‐time EHR‐integrated automated SOFA score calculator. Int J Med Inform. 2017;103:1–6. doi: 10.1016/j.ijmedinf.2017.04.001 [DOI] [PubMed] [Google Scholar]
- 43. Osborne TF, Veigulis ZP, Arreola DM, Röösli E, Curtin CM. Automated EHR score to predict COVID‐19 outcomes at US Department of Veterans Affairs. PLoS One. 2020;15:e0236554. doi: 10.1371/journal.pone.0236554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Li J, Liu L, Le TD, Liu J. Accurate data‐driven prediction does not mean high reproducibility. Nat Mach Intell. 2020;2:13–15. doi: 10.1038/s42256-019-0140-2 [DOI] [Google Scholar]
- 45. Hope TMH, Seghier ML, Leff AP, Price CJ. Predicting outcome and recovery after stroke with lesions extracted from MRI images. Neuroimage Clin. 2013;2:424–433. doi: 10.1016/j.nicl.2013.03.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Yang HE, Kyeong S, Kang H, Kim DH. Multimodal magnetic resonance imaging correlates of motor outcome after stroke using machine learning. Neurosci Lett. 2020;741:135451. doi: 10.1016/j.neulet.2020.135451 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1
Figures S1–S3


