Summary
RNA transfer via extracellular vesicles (EVs) influences cell phenotypes; however, lack of information regarding biogenesis of RNA-containing EVs has limited progress in the field. Here, we identify endoplasmic reticulum membrane contact sites (ER MCS) as platforms for generation of RNA-containing EVs. We identify a subpopulation of small EVs that is highly enriched in RNA and regulated by the ER MCS linker protein VAP-A. Functionally, VAP-A-regulated EVs are critical for miR-100 transfer between cells and in vivo tumor formation. Lipid analysis of VAP-A-knockdown EVs revealed reductions in the EV biogenesis lipid ceramide. Knockdown of the VAP-A-binding ceramide transfer protein CERT led to similar defects in EV RNA content. Imaging experiments revealed that VAP-A promotes luminal filling of multivesicular bodies (MVBs), CERT localizes to MVBs, and the ceramide-generating enzyme neutral sphingomyelinase 2 colocalizes with VAP-A-positive ER. We propose that ceramide transfer via VAP-A-CERT linkages drives biogenesis of a select RNA-containing EV population.
Graphical Abstract
eTOC blurb:
Biogenesis of RNA-containing extracellular vesicles (EVs) is poorly understood. Barman et al. delineate a pathway where endoplasmic reticulum membrane contact sites (ER MCS), the ER MCS linker VAP-A, and the ceramide transporter CERT are essential for biogenesis of a select subpopulation of RNA-containing EVs.
Introduction
Extracellular vesicles (EVs) are small lipid-bound carriers of bioactive cargoes that are released from diverse cell types to promote cellular communication. Multiple biogenesis mechanisms can promote EV formation and cargo selection, including budding from the plasma membrane as microvesicles and intraluminal budding in endosomes to form exosomes. Recently, it has become apparent that EVs are more heterogeneous than previously appreciated and that diverse cargoes may utilize distinct and potentially non-classical mechanisms for incorporation into EVs (Raposo and Stoorvogel, 2013; van Niel et al., 2018).
In addition to proteins and lipids, EVs contain diverse types of RNA, including miRNAs, lncRNAs, snoRNAs, tRNAs, rRNAs, Y RNAs, and mRNAs (Chow et al., 2019; Crescitelli et al., 2013; Driedonks et al., 2018; Hinger et al., 2018; Lasser et al., 2017; Skog et al., 2008; Valadi et al., 2007). EV-carried RNAs can affect gene expression and the phenotype of recipient cells, which may be important for a variety of diseases (Bell and Taylor, 2017; de Candia et al., 2016; Falcone et al., 2015; O'Brien et al., 2020). EV-enclosed RNA is also being studied for potential use as therapeutics and biomarkers. While extracellular RNA (exRNA) can also be present in a non-vesicular form, encapsulation of RNA in EVs protects it from degradation and allows it to be delivered directly to the cytoplasm of recipient cells via membrane fusion (O'Brien et al., 2020).
Although certain exRNAs are known to be selectively enriched in EVs (Cha et al., 2015; Lee et al., 2019; Lin et al., 2019; Lu et al., 2017), the mechanisms by which this packaging occurs is poorly understood. RNA-binding proteins play a major role in this process, controlling both stability and sorting of the RNAs (Deng et al., 2020; Leidal et al., 2020; Li et al., 2019; McKenzie et al., 2016; Mukherjee et al., 2016; Santangelo et al., 2016; Shurtleff et al., 2017; Temoche-Diaz et al., 2019; Villarroya-Beltri et al., 2013; Wozniak et al., 2020; Zietzer et al., 2020). We and others identified Argonaute 2 (Ago2) and other RISC complex proteins as potent mediators of miRNA sorting into EVs (Bukong et al., 2014; Clancy et al., 2019; Mantel et al., 2016; McKenzie et al., 2016; Melo et al., 2014). Other studies have shown that miRNAs with specific sequence motifs (i.e., GGAG and GGCU) are selectively sorted into EVs by heterogenous nuclear ribonucleoprotein A2B1 (hnRNPA2B1) and synaptotagmin binding cytoplasmic RNA-interacting protein SYNCRIP (Santangelo et al., 2016; Villarroya-Beltri et al., 2013). In addition, the RNA-binding protein Y-box I (YBX-1) is suggested to be involved in the packaging of mRNAs, miRNAs and other exRNAs into EVs (Kossinova et al., 2017; Shurtleff et al., 2016; Shurtleff et al., 2017).
Despite the accumulating evidence of a role for RNA binding Proteins (RBPs) in determining the RNA content of EVs, it is unclear how these RBP-RNA complexes are trafficked to and selected for incorporation into newly forming EVs at multivesicular bodies (MVB) and the plasma membrane. One clue may come from the typical cellular location of these known RBPs and their activities. hnRNPA2B1 and SYNCRIP are both hnRNPs that mediate RNA processing and translation, with functions in both the nucleus and at the endoplasmic reticulum (ER) (Hong et al., 2017; Kamma et al., 1999; Quaresma et al., 2009). Likewise, YBX1 affects translation of select mRNAs, localizing to ribosomes and the ER (Matsumoto et al., 2005) and miRNA-loaded Ago2 was shown to be physically associated with rough ER membranes (Barman and Bhattacharyya, 2015; Stalder et al., 2013) before moving to MVBs (Bose et al., 2017; Gibbings et al., 2009).
Membrane contact sites with the endoplasmic reticulum (ER MCS) are areas of close apposition between the ER and other organelles (Phillips and Voeltz, 2016; Wu et al., 2018). Key described functions of ER MCS include calcium and lipid exchange between the organelles, and organelle fission. However, the major physiological functions of MCS and the underlying mechanisms are still under investigation. A number of tether proteins have been identified that mediate these contacts and control molecular signaling and exchange at contact points by binding to additional proteins (Phillips and Voeltz, 2016; Wu et al., 2018), including vesicle-associated membrane protein-associated protein-A (VAP-A), VAP-B, and Motile sperm domain-containing protein 2 (Alpy et al., 2013; De Vos et al., 2012; Di Mattia et al., 2018; Rocha et al., 2009). Of these tether proteins, VAP-A and VAP-B are the most well studied and undergo both homo- and heterodimerization (James and Kehlenbach, 2021; Neefjes and Cabukusta, 2021). While VAP-B mutations are associated with mitochondrial defects and neurodegenerative diseases (Chen et al., 2010; Nishimura et al., 2004), VAP-A is known most for its binding to multiple endosome-localized lipid transport proteins and function in lipid transfer from the ER to endosomes and other organelles (Alpy et al., 2013; Jansen et al., 2011; Kirmiz et al., 2019; Neefjes and Cabukusta, 2021; Weber-Boyvat et al., 2015).
A key VAP-A-binding lipid transporter is ceramide transfer protein (CERT), which mediates ceramide transfer from the ER to the Golgi at ER-Golgi MCSs (Hanada et al., 2003; Peretti et al., 2008). Recently, CERT was also shown to mediate ceramide transport at ER-endosome MCS to affect EV secretion from palmitate-stimulated hepatocytes (Fukushima et al., 2018). Transfer of ceramide via MCS could potentially provide an alternative mechanism to in situ ceramide synthesis, which is known to promote nonclassical exosome biogenesis (Trajkovic et al., 2008); however, its general relevance and impact on specific cargoes has not been determined.
To test the role of ER MCS in the biogenesis of RNA-containing EVs, we knocked down (KD) or overexpressed VAP-A in colon cancer cells. VAP-A was chosen as the most well-defined MCS tether that is associated with endosomes and the plasma membrane, the two sites of EV biogenesis (Alpy et al., 2013; Kirmiz et al., 2019; Rocha et al., 2009; Weber-Boyvat et al., 2015). We identified multiple small RNAs and RBPs that are differentially enriched in both small and large EVs compared to control cells. In addition, confocal microscopy analysis of control and VAP-A KD cells revealed a strong defect in intraluminal filling of MVBs with RNA and RBP cargoes. In-depth analysis of alterations in small EVs revealed that VAP-A promotes formation of a select subpopulation of small EVs that carries the majority of RNA and is enriched in RBPs, the EV marker flotillin-1, and the autophagy protein LC3B. Moreover, VAP-A-regulated EV biogenesis controls the ability of colon cancer cells to transfer miR-100 to recipient cells and to grow tumors in xenograft mouse experiments. Investigation of the molecular mechanism revealed that VAP-A regulates the ceramide content of EVs and intraluminal filling of MVB with the VAP-A binding partner CERT. Likewise, KD of CERT leads to a decrease in the RNA content of both small and large EVs. Immunofluorescence experiments revealed strong colocalization of the ceramide generating enzyme neutral sphingomyelinase 2 (nSMase2) with VAP-A-positive ER but little colocalization with MVBs, suggesting that ceramide generated via nSMase2 may depend on CERT for transfer to MVBs. Altogether, these data suggest a model in which VAP-A-CERT linkages at ER MCS drive biogenesis of a unique subset of RNA-containing EVs.
Results
To explore whether the ER may be associated with RBPs trafficked into EVs, we mined publicly available EV proteomics data. Analysis of the human RNA Binding Proteome (Hentze et al., 2018), EV proteome (Kalra et al., 2012; Pathan et al., 2019) and ER proteome (Thul et al., 2017) revealed that 52% (809 RBPs out of 1542 RBPs) of RBPs are secreted in EVs. Among them, 8% are ER–associated proteins (61 RBPs out of 809 RBPs) (Figure 1A, Table S1). We also examined a recent report in which the most highly represented RBPs across the EV proteomics datasets in the online database EVpedia were manually identified (Mateescu et al., 2017). Of these 80 RBPs, 28% are ER-associated ribosomal proteins (22 RBPs out of 80 RBPs), and an additional 18% are non-ER-associated ribosomal proteins (Figure 1B, Table S1). Together with previous reports showing that Ago2-miRNA complexes are assembled at ER-associated ribosomes (Barman and Bhattacharyya, 2015; Maroney et al., 2006; Nottrott et al., 2006) these data led us to hypothesize that a significant portion of EV-incorporated RNAs and RBPs are associated with the ER.
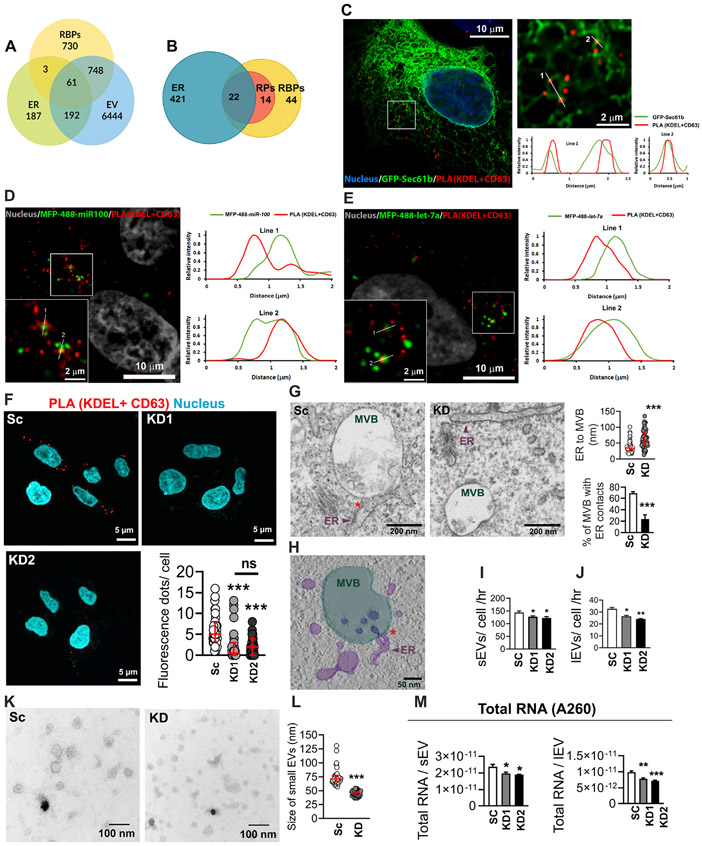
Figure 1: VAP-A regulates ER-MVB contact sites and EV characteristics.
(A) Venn diagram shows the overlap of human RBPs (1542) (Hentze et al., 2018), endoplasmic reticulum (ER) proteins (443) (Thul et al., 2017), and extracellular vesicle (EV) proteins (7445) (from Vesiclepedia (Kalra et al., 2012; Pathan et al., 2019)).
(B) Representation of previously published top 80 EV associated RBPs (Mateescu et al., 2017) present on ER membranes (Thul et al., 2017). Venn diagram shows 22 RBPs (28%) are ER associated and an additional 14 RBPs (18%) are ribosomal proteins (RPs).
(C) Representative merged image for proximity ligation assay (PLA) analysis for ER MCS in GFP-Sec61b-expressing DKs-8 cell. PLA reaction was performed with KDEL (ER marker) and CD63 (late endosome/MVB marker) and appears as red fluorescent dots. DAPI (blue) was used to stain the nuclei. The selected area is enlarged at the right side. Numbered lines were scanned for the intensity of each fluorescence channel of the image and plotted at the right side.
(D and E) Representative merged images for PLA analysis of ER MCS in MFP488-miR100 (D) or MFP-488-let-7a (E)-transfected DKs-8 cells. DAPI (gray) was used to stain nuclei. Selected areas are enlarged in the left bottom corner of each image. Numbered lines were scanned for the intensity of each fluorescence channel of the images and plotted at the right side.
(F) Representative merged images of PLA analysis of ER MCS in scrambled control (Sc) and VAP-A knockdown (KD1 and KD2) DKs-8 cells. DAPI, blue. Fluorescence dots per cell were calculated and plotted from sixty cells per condition from three independent experiments.
(G) Representative TEM images of control (Sc) and VAP-A KD2 (KD) DKs-8 cells. ER-endosome MCS are indicated by red asterisks. Quantification shows distance of ER to MVBs and percentage of MVBs with ER contacts (defined as MVBs with ≤40 nm distance from ER). Each circle represents an MVB. n = 61 and 59 MVBs from Sc and KD respectively from 10 (Sc) and 8 (KD) sections. Data were taken from three independent experiments.
(H) Tomographic reconstruction of an MVB/ER contact site observed in a DKs-8 (Sc) cell (see also Video S1). Three-dimensional segmentations of organelles depict MVB (light green), intraluminal vesicles (ILVs) (dark blue), and ER tubules (purple). Note the presence of an ILV still connected to the MVB limiting membrane at the ER contact site.
(I and J) Graphs of EV release rate from control and KD cells quantitated from NTA data and normalized based on final cell number and conditioned media collection time. Data from five independent experiments.
(K and L) Representative TEM images and size analysis for small EVs purified from DKs-8 control (Sc) and VAP-A KD2 (KD) cells. Quantification of a total of 150 vesicles per condition (control or KD) from three independent experiments.
(M) Graphs of total RNA concentration measured by NanoDrop (A260) for small and large EVs isolated from control (Sc) and VAP-A KD DKs-8 cells. Data from five independent experiments.
Bar graphs indicate mean +/− S.E.M. Scatter plots indicate median and interquartile range. *p<0.05, **p<0.01, ***p<0.001, ns, not significant. See also Figures S1 and S2, and Table S1.
To assess whether RNAs known to be selectively incorporated into EVs localize to ER MCS, we identified ER-endosome MCS with a proximity ligation assay (PLA) using antibodies against KDEL and CD63 in wild type KRAS DKs-8 colorectal carcinoma cells. DKs-8 was chosen as a good model cell line based on previous studies showing selective export of RNAs and RBPs in their EVs, including miR-100 and let-7a miRNAs (Demory Beckler et al., 2013; McKenzie et al., 2016). Confocal microscopy identified localization of the PLA signal in close proximity with the general ER marker Sec61b (Fig 1C, note overlap of signals in line scans). We also localized miR-100 and let-7a to the identified MCS and found that both displayed overlap with the PLA signal (Figures 1D, 1E).
The MCS tether protein VAP-A controls the number, size and cargo content of EVs
To determine whether ER MCS may affect the biogenesis of RNA-containing EVs, we knocked down (KD) the ER MCS linker protein VAP-A in DKs-8 colon cancer cells. (Figure S1A). Both proximity ligation and transmission electron microscopy (TEM) analyses confirmed a reduction in ER-endosome MCS in VAP-A KD cells compared with controls (Figures 1F and 1G, with PLA controls and full TEM images shown in Figures S1B, S1C and S1D). As a part of the TEM analysis, we also observed events in which intraluminal vesicles within MVBs appeared to be forming at ER MCS in control cells. A tomogram of one of these events is shown in Fig 1H and Supplemental Video 1. In contrast to the effect on ER MCS with MVBs, loss of VAP-A had no discemable effect on cell viability, apoptosis, or ER stress markers, suggesting that loss of VAP-A did not generally disrupt cell functions (Figures S1E-S1G).
To assess the effect of VAP-A on EV number and cargo content, EVs were purified from conditioned media (see methods) by serial ultracentrifugation to pellet cells, debris, and large EVs followed by cushion density gradient method to purify small EVs (Li et al., 2018) (Fig S1H) from DKs-8 control or VAP-A KD cells. While we expect large EV fractions to contain mostly microvesicles and small EV fractions to contain mostly exosomes, we cannot identify them as such from these biochemical purifications. Thus, we will use the terms small and large EVs to describe our EV preparations, as is the convention in the field (Thery et al., 2018). Nanoparticle tracking analysis (NTA) of the EVs revealed a small but significant decrease in the number of small and large EVs purified from VAP-A KD cells (Figures S1I, 1I and 1J). From this analysis, we also observed an apparent decrease in the size of small EVs purified from VAP-A KD cells (note different peak sizes in Fig S1I), which we validated by analysis of TEM images of negatively stained small EVs (Figures 1K and 1L). Western blot analysis of our small and large EV preparations confirmed the presence of typical EV marker proteins and the absence of the negative marker GM130 (Thery et al., 2018) (Figure S1H). We also knocked down VAP-A in a second colon cancer cell line, DKO-1, and found similar alterations in the number of EVs as assessed by NTA (Figures S2A-S2C).
To determine whether VAP-A affects the RNA content of EVs, total RNA was extracted and analyzed. Assessment of the total RNA content of small and large EVs by A260 reading with a NanoDrop indicated that VAP-A KD EVs contained significantly less RNA than control EVs for both DKs-8 and DKO-1 cells (Figures 1M, and S2D). To identify specific small RNAs that are dependent on MCS for trafficking into EVs, we performed next generation sequencing on equal amounts of small RNA purified from control and VAP-A KD DKs-8 cells and EVs (Supplementary Datasheets 1-3). Principal component analysis of the data revealed that VAP-A KD alters the small RNA profiles of small EVs, large EVs, and cells (graphs for miRNA shown in Figures 2A, S3A and S3B). To identify individual miRNAs whose secretion was altered by VAP-A knockdown, we normalized the miRNA levels in EVs to the levels in the corresponding cells of origin. Using a criterion of ≤ 0.5 or ≥ 2-fold change and FDR ≤ 0.05, we identified 82 miRNAs that were differentially exported into VAP-A KD EVs compared to control EVs (Figure 2B). Of these, 26 were common to both small EVs and large EVs (Figure 2C). To validate our sequencing results, we performed qRT-PCR analysis for specific miRNAs taken from our sequencing dataset (miR-371a, miR-372) that were downregulated in VAP-A KD EVs. We also analyzed 4 miRNAs known to be selectively exported in DKs-8 EVs: let-7a, miR-100, miR-320, miR-125b ((McKenzie et al., 2016) and unpublished data). These miRNAs were also decreased in KD EVs in our dataset but did not reach the criteria of FDR ≤ 0.05 (Data S1). The levels of candidate RNAs were normalized to U6, which is exported in EVs but not affected by VAP-A. We found that all of the candidate miRNAs were decreased in both small and large EVs (Figures 2D-2E). Consistent with a specific role for VAP-A in RNA sorting into EVs, there was either no change or an increase in the cell levels of the same RNAs (Fig 2F). Similar results were found in DKO-1 cells, with a decrease in candidate miRNAs in VAP-A-KD EVs and an increase in VAP-A-KD cells (Figures S2E-G). We also validated several miRNAs predicted to be upregulated in VAP-A KD DKs-8 EVs, miR-30a, miR-129, and miR-99, and found that they were indeed present at higher levels in KD EVs while there was no change in KD cells (Fig S2H).
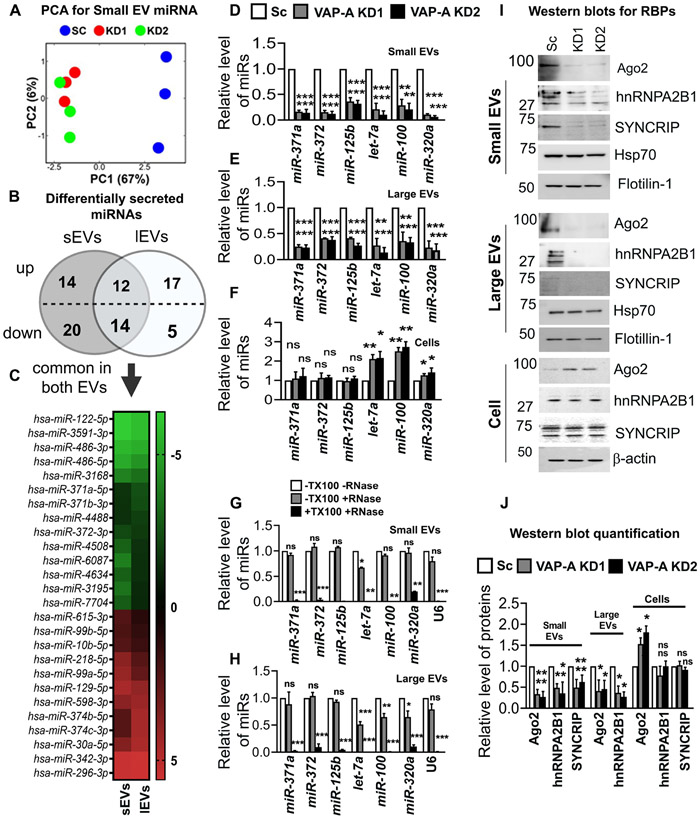
Figure 2: VAP-A regulates the miRNA composition of small and large EVs.
(A) Principal component analysis (PCA) of miRNA composition in small EVs showing segregation of KD (KD1 and KD2) from control (SC) data.
(B) Venn diagram depicts numbers of up- or down-regulated miRNAs in small and large EVs upon VAP-A knockdown. Levels (log 2-fold change) of miRNAs in small and large EVs were normalized to levels in their parental cells and then compared between control and VAP-A KD. miRNAs were considered significantly changed if either ≥ 2-fold or ≤ 0.5-fold enriched with a FDR value ≤ 0.05.
(C) Heat map represents levels (log 2-fold change) of differentially secreted 29 miRNAs in small and large EVs compared to their parental cells upon VAP-A KD. Green indicates downregulation in VAP-A KD EVs whereas red indicates upregulation.
(D-F) Relative levels of miR-371a, miR-372, miR-125b, let-7a, miR-100, miR-320a in control and VAP-A KD small EVs, large EVs and cells. Quantitative RT PCR was performed with 10 ng of total RNA. All experiments were from three biological replicates with three technical replicates. U6 snRNA was used to normalize Ct values.
(G-H) Relative levels of miRNAs (miR-371a, miR-372, miR-125b, let-7a, miR-100, miR-320a) in small and large EVs purified from control DKs-8 cells. Small and large EVs were treated with (+) or without (−) RNase in absence (−) or presence (+) of 1% Triton-X-100 (TX100) followed by total RNA isolation and qRT-PCR. All experiments were done in three biological replicates with three technical replicates
(I and J) Immunoblots show levels of RBPs (Ago2, hnRNPA2B1, SYNCRIP) and EV markers (flotillin-1, HSP70) in control (Sc) and VAP-KD EVs and cells. Quantification of immunoblot from three independent experiments were shown. Values were normalized by HSP70 (for small and large EVs) and by beta-actin (for cell lysates).
Data were plotted as mean +/− S.E.M. *p<0.05, **p<0.01, ***p<0.001. ns, not significant.
See also Figures S2 and S3.
We also analyzed snoRNA levels in EVs and cells from our RNA-seq dataset. Using our previous criteria, we found alterations in secretion of 14 snoRNAs (11 reduced and 3 increased) in small EVs with VAP-A KD, but no alterations in snoRNAs in large EVs (Fig S3C, Data S2). qRT-PCR for specific snoRNAs (snoRD105, snoRA40, snoRA42 and snoRD45) taken from our dataset revealed that all four snoRNAs were reduced in VAP-A KD small EVs but unchanged in KD cells (Figures S3D and S3F). In addition, snoRA42 and snoRD45 levels were reduced slightly in VAP-A KD large EVs (Figure S3E). Analysis of the RNA-Seq dataset also revealed alterations in the levels of tRNA fragments in VAP-A KD EVs (Figures S3G and S3H, Data S3).
To further test our hypothesis that VAP-A is a positive regulator of EV number and cargo content, we overexpressed VAP-A (Figure S4A). Consistent with that hypothesis, we found that overexpression of VAP-A in DKs-8 cells increased the number of small and large EVs per cell, the total level of RNA per EV, and the levels of specific miRNAs in small and large EVs (Figures S4B-S4G). Interestingly, the levels of those same miRNAs in VAP-A-OE cells were significantly decreased, suggesting that export of miRNAs into EVs may impact their levels in cells (Chiou et al., 2018).
Since non-vesicular RNAs can associate with the outside of EVs in a nonspecific manner and could theoretically contaminate our assays, we analyzed whether the miRNAs associated with our EVs were sensitive to RNase in the absence or presence of detergent. For small EVs, we found that five out of six candidate miRNAs along with U6 are completely unaffected by RNase treatment in the absence of detergent but are almost fully depleted by RNase in the presence of detergent (Figures 2G and 2H). For let-7a detection in small EVs, there was a small amount of depletion with RNase in the absence of detergent, but the majority was protected. For large EVs, there was some sensitivity to RNase in the absence of detergent for three of the six miRNAs tested whereas the other three miRNAs and U6 were fully protected. The origin of the extravesicular RNA on large EVs is unclear since no serum (a source of nonvesicular RNA contamination) was used during the conditioning of the media. While it is possible that some contaminants remain associated with the plasma membrane (the likely source of large EVs) even after removal of serum, specific association of RNA to the outside of the cell (Flynn et al., 2021) cannot be ruled out. Overall, these data are consistent with the candidate RNAs being on the inside of the EVs, as would be expected for a selective biogenesis mechanism.
Previous reports have shown that RBPs such as Ago2, hnRNPA2B1, and SYNCRIP, are involved in RNA sorting to EVs (McKenzie et al., 2016; Santangelo et al., 2016; Villarroya-Beltri et al., 2013). Western blot analysis revealed that Ago2 and hnRNPA2B1 are reduced in both small and large EVs isolated from VAP-A KD cells while SYNCRIP is reduced in small EVs from KD cells and undetectable in large EVs (Figures 2I and 2J). To test whether the RBPs we detect in our Western blots are on the inside or outside of EVs in our preparations, we used a previously published dot blot method (Lai et al., 2015; McKenzie et al., 2016; Patel and Weaver, 2021; Sung and Weaver, 2017). Serially diluted EV samples were dotted onto nitrocellulose membranes and immunoblotted for Ago2, hnRNPA2B1, CD63, or flotillin-1 in the presence or absence of 0.1% Tween-20 detergent to permeabilize the EVs. As the antibody to CD63 was to an extracellular epitope, it served as a positive control for small EVs and was detected in both the presence and absence of detergent (Fig S5A). Flotillin-1 was used as a control for large EVs, as they do not have detectable CD63 (Fig S1C). As expected for a protein that binds the cytosolic leaflet of the plasma membrane, flotillin-1 was mostly detected on the inside of EVs (Fig S5B). Likewise, Ago2 and hnRNPA2B1 were detected only in the presence of detergent for both small and large EVs, indicating that they are present inside the vesicles (Figures S5A and S5B) and unlikely to represent protein aggregate contamination of our EV preparations.
A subpopulation of small EVs is highly enriched in RNA and is regulated by VAP-A.
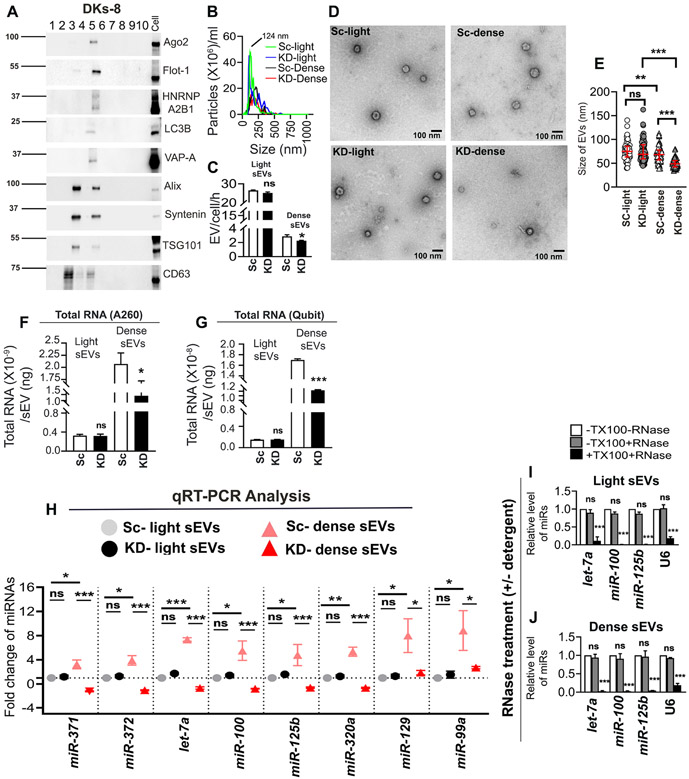
A central question in the field has been whether RNA is primarily present in a small subset of EVs or is uniformly distributed at low levels in most EVs (Chevillet et al., 2014). To address this question and determine whether VAP-A regulates biogenesis of a subset of cellular EVs containing RNAs, we used a previously published density gradient protocol (Kowal et al., 2016) to isolate “light” and “dense” subpopulations of small EVs from control and VAP-A KD cells. Consistent with the previous publication, we found two peaks of EVs on the density gradient, a peak at fraction 3 that represents less dense material and is enriched for the EV markers Alix, Syntenin, TSG101 and CD63 and a peak at fraction 5 that contains more dense material and is enriched for the EV marker Flotillin-1, the RBPs Ago2 and hnRNPA2B1, as well as VAP-A (Figure 3A). As the autophagy protein LC3B has recently been shown to induce formation of exosomes containing RNAs and RBPs (Leidal et al., 2020), we also probed for LC3B and found that indeed it was enriched in the dense fraction (Figure 3 A). We validated these Western blot findings in a second cell line, HT1080 fibrosarcoma cells (Fig S6A). Nanoparticle tracking analysis revealed that the majority of the EVs are found in the light fraction (Figures 3B and 3C (DKs-8) and S6B, S6C, S6G, and S6H (HT1080 and DKO-1). In addition, VAP-A KD led to a reduction in the number of dense EVs secreted over time but no change in the number of light EVs (Figures 3C and S6H). To further characterize the dense and light EV populations, we performed transmission electron microscopy on negative stained EVs purified from control and VAP-A KD DKs-8 cells. As shown in Figure 3D, small EVs with similar morphology were observed in all samples. Quantitation of the diameter of EVs in the four different samples revealed that dense EVs had a small but significant reduction in size compared to light EVs. More striking was the reduction in the diameter of dense EVs purified from VAP-A KD cells (Figure 3E). Estimation of the total RNA found in each EV population revealed that the dense EVs are highly enriched in RNA, compared to light EVs (Figures 3F and 3G, 6.3-fold and 11.3-fold enrichment comparing control dense to light EVs by NanoDrop and Qubit methods, respectively). In addition, VAP-A KD significantly decreased the amount of total RNA in the dense EVs, but not in light EVs (Figures 3F and 3G). Similar results were found for HT1080 and DKO-1 light and dense EVs (Figures S6D, S6E, S6I and S6J). QRT-PCR analysis further revealed that miRNAs are enriched in dense EVs (Figures 3H and S6F) and that VAP-A KD selectively decreased the levels of eight miRNAs in dense but not light small EVs (Figure 3H). Surprisingly, two of these miRNAs – miR-129 and miR-99a – were predicted to be upregulated in our RNA-Seq dataset and indeed validated that way when comparing control and KD EVs from our standard cushion gradient method. By contrast, these miRNAs were significantly less abundant in KD dense EVs compared to controls yet unchanged in light EVs. It is unclear at this point why these RNAs gave inconsistent results between the two preparations. Furthermore, quantitation of total RNA/EV gives higher numbers for both light and dense EVs as compared to EVs purified by cushion density gradient (compare Y-axis scales between Fig 1M and 3F). This does not appear to be due to contamination with extravesicular RNA since, similar to our other method of EV preparation (Figure 2), candidate small RNAs associated with dense and light EVs are depleted by RNase treatment only in the presence of detergent (Figures 3I and 3J). Since the PCR analyses were all done using equal amounts of RNA for the PCR reactions, we also checked whether analyzing RNA based on equal vesicle number gives similar results. Indeed, it does, with total RNA, as well as specific miRNAs, greatly enriched in DKs-8 dense EVs compared to light EVs. (Figs S6K and S6L). Overall, these data indicate that VAP-A regulates a subpopulation of EVs that is enriched in RNA.
Fig 3: A subpopulation of small EVs is highly enriched in RNA and is regulated by VAP-A.
(A) Representative immunoblot of different EV cargo and marker proteins (Ago2, flotillin-1, hnRNPA2B1, LC3B, VAP-A, Alix, syntenin, TSG101 and CD63) shows segregation in fractions 3 and 5 of “light” and “dense” EVs purified from control DKs-8 cells. Representative of n=3 independent experiments.
(B) NTA traces for “light” and “dense” EVs purified from control (Sc) and VAP-A KD2 (KD) DKs-8 cells. Median values from three independent experiments were plotted.
(C) EV concentrations were calculated from NTA data and plotted from three independent experiments.
(D and E). Representative TEM images of “light” and “dense” EVs purified from control (Sc) and VAP-A KD2 (KD) DKs-8 cells are shown. Graphs show size of the “light” and “dense” EVs calculated from a total of 150 vesicles per condition from three independent experiments.
(F and G) Total RNA quantity measured by NanoDrop (A260) or Qubit (fluorescence) per “light” and “dense” small EVs isolated from control and KD cells. Data from three independent experiments. Note the enrichment of RNA in dense EVs compared to light EVs, regardless of the method of measurement.
(H) Fold change values of specific miRNAs in control and KD light and dense small EVs show VAP-A KD selectively affects the dense EV population. Data from three independent experiments and candidate miRNA Ct values were normalized by respective U6 values which is unchanged.
(I and J) Specific miRNAs (let-7a, miR-100, miR-320a and U6) are present inside light and dense small EVs, as they are only susceptible to RNase treatment in the presence of Triton-X-100 (TX100). Data from three independent experiments.
Data plotted as mean +/− S.E.M. Scatter plots indicate median and interquartile range.
*p<0.05, **p<0.01, ***p<0.001. ns, not significant.
VAP-A controls intraluminal filling of Rab5Q79L-positive MVBs with RNA and RBP cargoes.
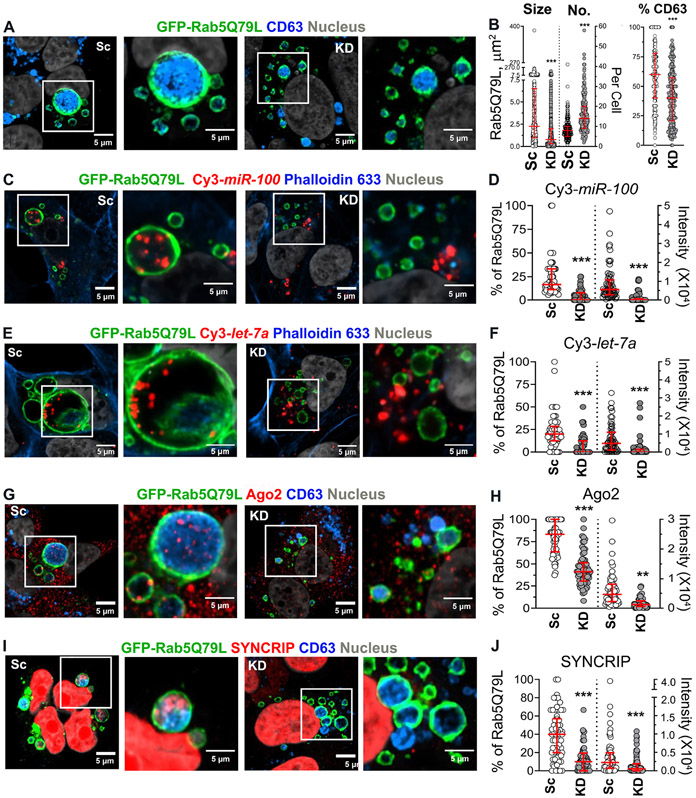
Based on our findings that VAP-A affects a select subpopulation of EVs enriched for RNA and RBP cargoes, we hypothesized that VAP-A controls biogenesis of EVs containing those cargoes. To test that hypothesis for exosomes, we expressed in cells a constitutively active mutant of Rab, Rab5Q79L, that leads to enlarged multivesicular endosomes (Baietti et al., 2012; Ghossoub et al., 2014; Mercier et al., 2020; Roberts et al., 1999; Sinha et al., 2016; Stenmark et al., 1994; Wegner et al., 2010) and greatly facilitates visualization and quantitation of intraluminal vesicle formation (Baietti et al., 2012; Ghossoub et al., 2014) by confocal microscopy. Indeed, we verified that the canonical exosome marker CD63 fills the lumen of GFP-Rab5Q79L-positive endosomes in control cells. We found that the size of GFP-Rab5Q79L-positive endosomes is decreased in VAP-A KD DKs-8 cells compared to controls while the number is increased (Figures 4A and 4B). In addition, there is a small but significant decrease in the luminal filling of GFP-Rab5Q79L-positive endosomes with CD63 in KD cells. These data suggest that CD63 is present on numerous types of intraluminal vesicles, including those regulated by VAP-A, and is consistent with our previous finding that Ago2 is present in EVs that are immunoprecipitated using a CD63 antibody (McKenzie et al., 2016).
Figure 4. VAP-A regulates intraluminal filling of GFP-Rab5Q79L-positive MVBs.
(A) Representative merged images of GFP-Rab5Q79L-transfected DKs-8 cells with CD63 immunofluorescence staining. Selected areas are enlarged at the right. Sc, scrambled control; KD, knockdown.
(B) Quantitation of size and number/cell (No.) of GFP-Rab5Q79L rings and the percentage of CD63-positive GFP-Rab5Q79L rings. Each circle for size represents a single GFP-Rab5Q79L ring whereas each circle for number/cell represents a single cell. Each circle for the percentage of CD63-positive rings represents a single cell. n = 2328 and 4342 rings from 12 independent experiments for Sc and KD for the size measurement. n = 273 and 267 cells from 12 independent experiments for Sc and KD for the number of GFP-Rab5Q79L rings and the percentage of CD63-positive rings measurement.
(C and E) Representative merged images of GFP-Rab5Q79L and Cy3-miR-100 (C) or Cy3-let-7a (E)-co-transfected DKs-8 cells with Alexa633-conjugated phalloidin staining. Selected areas are enlarged at the right.
(D and F) Quantitation of the percentage of Cy3-miR-100 (D)- or Cy3-let-7a (F)-positive GFP-Rab5Q79L rings and the intensity of Cy3-miR-100 (D) or Cy3-let-7a (F) presented in GFP-Rab5Q79L rings. Each circle represents a cell. n = 63 and 64 cells (D) or 62 and 79 cells (F) from 3 independent experiments for Sc and KD, respectively.
(G and I) Representative merged images of GFP-Rab5Q79L-transfected DKs-8 cells with Ago2 (G) or Syncrip (I) and CD63 immunofluorescence staining. Selected areas are enlarged at the right.
(H and J) Quantitation of the percentage of Ago2 (H)- or Syncrip (J)-positive GFP-Rab5Q79L rings and the intensity of Ago2 (H) or Syncrip (J) presented in GFP-Rab5Q79L rings. Each circle represents a cell. n = 63 and 65 cells (H) or 67 and 63 cells (J) from 3 independent experiments for Sc and KD, respectively. All data were plotted with median and interquartile range. ** P < 0.01; *** P < 0.001 by the two-sided unpaired Mann-Whitney test.
We leveraged this assay to test whether VAP-A regulates intraluminal filling of MVB with two candidate miRNAs and two candidate RBPs (Figures 4C-J). To visualize the miRNAs by fluorescence, miR-100 and let-7a were labelled with Cy3 dye and co-transfected into cells with GFP-Rab5Q79L and then stained with Fluor™ 633-conjugated Phalloidin to visualize actin filaments and cell boundaries. For RBPs, cells transfected with GFP-Rab5Q79L were immunostained for CD63 and either Ago2 or SYNCRIP. In all cases, RNAs and RBPs were present in a sparse punctate distribution across many MVBs in control cells. Consistent with a key role for VAP-A in biogenesis of RNA/RBP-containing exosomes, there was a large decrease in the percent of MVBs containing miR-100, let-7a, Ago2, and SYNCRIP. There was also a decrease in the intensity of those RNAs and RBPs inside of MVBs (Figures 4C-J).
VAP-A expression controls the function of EVs.
To test whether VAP-A affects the function of EVs, we leveraged our previous work, in which we showed that miR-100 can be transferred in a coculture from donor cells grown on Transwell filters to recipient cells present in culture wells below (Cha et al., 2015). Since mutant KRAS-expressing DKO-1 cells secrete more miR-100 in SEVs compared to matched isogenic wild type KRAS DKs-8 cells (Cha et al., 2015), we used control and VAP-A KD DKO-1 cells (Figure S2) as donor cells.
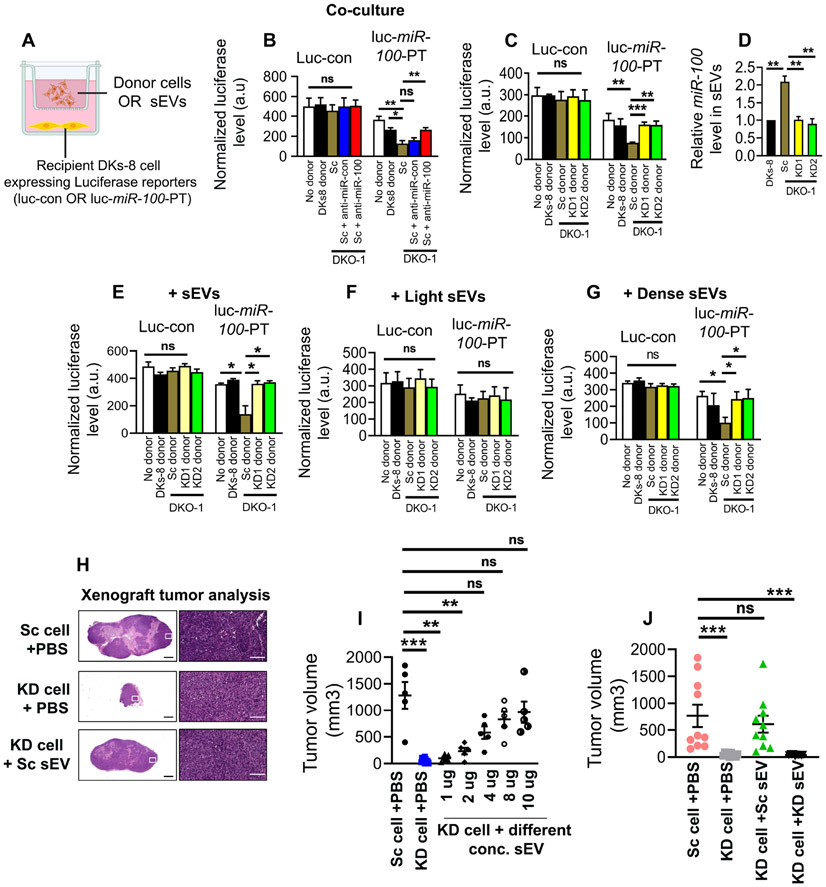
To perform the Transwell co-culture assay, DKs-8 recipient cells were seeded in culture wells and transiently transfected with luciferase reporters containing either 3 artificial miR-100 binding sites in the 3’ UTR (luc-miR-100-PT) or control scrambled sites (luc-con) (Cha et al., 2015) (Figure 5A). Scrambled control (Sc) or VAP-A KD DKO1 cells, or parental DKs-8 cells were used as donors. Consistent with our previous data (Cha et al., 2015), luciferase expression from the miR-100-PT reporter was significantly reduced in recipient DKs-8 cells when co-cultured with control DKO-1 cells in comparison to either the DKs-8 donor cells or the no donor condition (Figure 5B). This decrease in luciferase was reversed when DKO-1 donor cells were co-transfected with an antagomir to miR-100 but not with a control antagomir (Figure 5B), demonstrating that the effect on luciferase was due to miR-100 originating in the donor cells. VAP-A KD in DKO-1 donor cells also reversed this reduction in luciferase, bringing it back to the levels found in the no donor or Dks-8 donor conditions (Figure 5C). There were no alterations in luciferase expression from the control reporter under any of the conditions. To confirm that the effects of VAP-A KD in the coculture system were due to EV transfer, we purified small EVs from control or VAP-A KD DKO-1 cells or from DKs-8 cells. As expected, control DKO-1 EVs contained ~2-fold more miR-100 than did KD DKO-1 EVs or DKs-8 EVs (Figure 5D). When added to recipient cells expressing miR-100-PT luciferase, the control DKO-1 EVs - but not the VAP-A KD EVs - reduced luciferase expression, similar to the co-culture results (Figure 5E).
Figure 5: VAP-A controls miR-100 transfer and tumorigenic functions of EVs.
(A) Illustration of co-culture setup. Control (luc-control: luciferase with scrambled sites in the 3’ UTR) or miR-100 expressing luciferase reporters (luc-miR-100-PT: luciferase with three perfect miR-100 sites in the 3’ UTR) (Cha et al., 2015) were expressed in recipient DKs-8 cells that were plated in the bottom of a Transwell plate. Different donor cells (DKs-8, or control or VAP-A KD DKO-1 cells) were cultured in Transwell inserts. Alternatively, sometimes purified EVs were added instead of donor cells.
(B) Graph shows luciferase expression levels, normalized by co-expressed beta galactosidase, after lysis of recipient control or miR-100 reporter-expressing DKs-8 cells that were co-cultured with the indicated donor cells. miR-100 transfer was confirmed by expressing an antagomir to miR-100 (anti-miR-100) in DKO-1 cells, which blocked the reduction in luciferase caused by control DKO1 donors (anti-miR-control or Sc). Data from three independent experiments.
(C) Graph shows luciferase expression levels, normalized by co-expressed beta galactosidase, after lysis of recipient control or miR-100 reporter-expressing DKs-8 cells that were co-cultured with the indicated donor cells. Data from three independent experiments.
(D) Relative miR-100 levels in small EVs isolated from different donor cells, quantified by qRT-PCR. Data from three independent experiments.
(E-G) Relative luciferase expression in recipient DKs-8 cells after addition of small EVs purified from donor cells, as indicated. (E), small EVs purified by cushion density gradient. (F and G) Light EVs or Dense EVs purified as in (Kowal et al., 2016).
Luciferase data (B-G) from three independent experiments with three technical replicates per condition each time. Luciferase data were analyzed by unpaired Mann-Whitney test. *p<0.05, **p<0.01, ***p<0.001. ns, not significant.
(H-J) Control (Sc) and VAP-A KD2 (KD) DKO-1 cells were mixed with PBS (+PBS) or small EVs (+sEV) and injected subcutaneously in nude mice and allowed to grow for 3 weeks, with injection of PBS or EVs twice a week. (H) Representative images of Hematoxylin and Eosin stained sections of tumors. (I) Tumor volume after injecting PBS or different concentrations of small EVs purified from control (Sc) DKO-1 cells. Each circle represents an animal (n≥5 per condition). (J) Tumor volume for control and KD tumors injected with PBS, or 10 μg control or KD sEVs, as indicated. Each condition from ten animals. Some data points in Figure H and I are in common. Tumor data were compared by unpaired Mann-Whitney test.
*p<0.05, **p<0.01, ***p<0.001. ns, not significant.
Our EV fractionation analysis in Fig 3 showed that the dense subpopulation of small EVs is enriched in RNA, including miR-100, and is regulated by VAP-A. To further validate that finding, we added light or dense small EVs purified from control or VAP-A KD DKO-1 cells, or from DKs-8 cells. Indeed, only the dense small EVs purified from control DKO-1 cells reduced luciferase expression in miR-100-PT-luciferase-expressing recipient cells, dependent on VAP-A (Figures 5F and 5G).
Previous reports showed that mutant KRAS DKO-1 cells are tumorigenic when grafted into mice (Shirasawa et al., 1993). To test whether VAP-A-mediated EV production promotes tumor growth, we injected control and VAP-A KD DKO-1 cells into the flanks of nude mice and allowed tumors to grow for 21 days. Compared with control tumors, VAP-A KD tumors were much smaller or absent at the time of harvest (Figures 5H and 5I). To determine whether the defect in VAP-A KD growth was due to alterations in EV secretion, we performed a reconstitution experiment in which purified small EVs were mixed with VAP-A KD cells. Indeed, purified small EVs from control DKO-1 cells rescued the growth of KD tumors in a concentration dependent manner (Figure 5I). However, an equal amount of the highest concentration (10 μg) of EVs purified from VAP-A KD cells was not able to rescue VAP-A KD tumor growth (Figure 5J). These data suggest that VAP-A controls a specific subpopulation of EVs that promotes DKO-1 tumor growth.
VAP-A controls the lipid content of EVs.
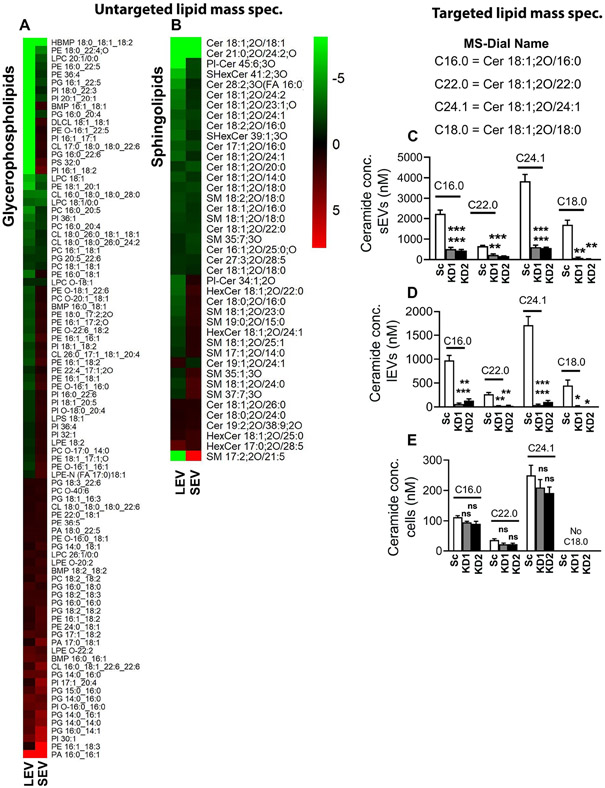
VAP-A is known to promote efflux of lipids from the ER to diverse organelles by binding to lipid transporters, including oxysterol binding proteins (OSBPs) and ceramide transporters (Hanada et al., 2003; Mesmin et al., 2013; Perry and Ridgway, 2006). As ceramides and potentially other lipids are thought to be involved in the biogenesis of EVs, we hypothesized that VAP-A-mediated lipid transfer may be a critical component of the mechanism by which VAP-A promotes biogenesis of RNA-containing EVs. To determine whether VAP-A affects the lipid composition of EVs, we carried out an untargeted discovery lipidomics analysis of control and VAP-A KD small EVs, large EVs, and cells. We found a variety of lipids predicted to be altered in KD EVs and cells, including glycerophospholipids and sphingolipids (Data S4, Figures 6A and B). Notably, compared to controls, multiple ceramide species were decreased in both small and large KD EVs (Figure 6B). We validated these findings for multiple 18:1;2O ceramide species using targeted mass spectrometry with calibrated lipid standards. While Cer18:1;2O/18:1 was not detected in the targeted mass spec in either cells or EVs (not shown), Cer18:1;2O/16:0, Cer18:1;2O/18:0, Cer18:1;2O/22:0, and Cer18:1;2O/24:1 ceramide were significantly reduced in VAP-A KD small and large EVs and unchanged or undetectable in cells (Figures 6C-6E).
Figure 6: Ceramide levels are reduced in VAP-A KD EVs.
(A and B) Levels (log 2-fold change) of lipids in small and large EVs were normalized to levels in their parental cells and then compared between control and VAP-A KD. Heat maps show differentially secreted glycerosphingolipids (A) or sphingolipids (B) upon VAP-A KD. Green indicates downregulation in VAP-A KD EVs whereas red indicates upregulation. Heat map scale is from −8 to 8, any values outside of these do not show a further increase in green or red on the heatmap.
(C-E) Ceramide levels are reduced in VAP-A KD small and large EVs, but not in cells. Equal numbers of control and VAP-A KD small or large EVs, or cells were taken for ceramide (C16.0, C22.0, C24.1) measurements by targeted mass spectrometry. Data from three biological replicates.
Graphs were plotted as mean ±S.E.M. *p<0.05, **p<0.01, ***p<0.001. ns, not significant.
The VAP-A binding partner CERT is critical for biogenesis of RNA-containing EVs.
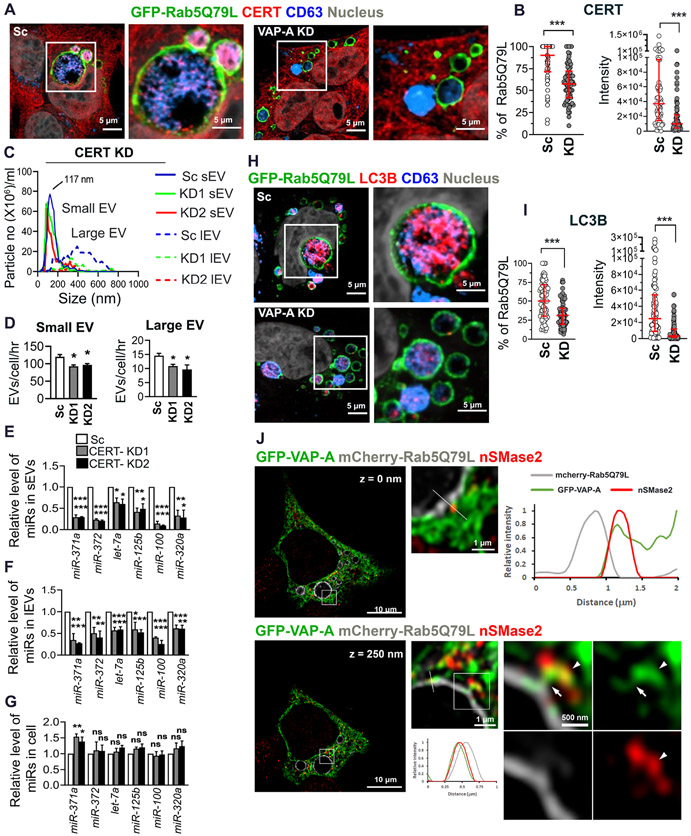
Since VAP-A interacts with the ceramide transporter CERT/STARD11 (Hanada et al., 2003) and VAP-A KD EVs have reduced ceramide levels, we hypothesized that CERT located at MVB (Fukushima et al., 2018) may interact with VAP-A and transfer ceramide to promote biogenesis of RNA-containing EVs. To determine whether CERT is present at MVB in our cells, we immunolocalized CERT to GFP-Rab5Q79L MVB, along with CD63. Interestingly, in control cells, CERT was present not only at the limiting membrane of MVBs, but also inside of MVBs indicating association with intraluminal vesicles (Fig 7A). By contrast, intraluminal filling of MVBs with CERT was greatly diminished in VAP-A KD cells (Figures 7A and 7B). We also tested whether CERT affects the RNA content of EVs by KD of CERT in DKs-8 cells. Similar to the VAP-A KD phenotype, CERT-KD significantly reduced the number of EVs released from cells (Figure 7C and 7D). There was also a significant effect of CERT-KD on total RNA content of small and large EVs (Figures S7A and S7B). In addition, CERT-KD led to large reductions in the levels of candidate miRNAs in EVs but either no change or an increase in the levels of those same miRNAs in cells (Figures 7E-G).
Figure 7: CERT controls the number and RNA content of EVs.
(A) Representative merged images of GFP-Rab5Q79L-transfected DKs-8 cells (scrambled control (Sc) and VAP-A-knockdown (KD)) with CERT and CD63 immunofluorescence staining. Selected areas are enlarged at the right.
(B) Quantitation of the percentage of CERT-positive GFP-Rab5Q79L rings and the intensity of CERT presented in GFP-Rab5Q79L rings. Each circle represents a cell. n = 68 and 70 cells from 3 independent experiments for Sc and KD, respectively.
(C) Graph shows NTA traces of small and large EVs purified from control (Sc) and CERT-KD (KD1, KD2) DKs-8 cells. Median values were plotted from three independent experiments.
(D) Small and large EV release rates from control and CERT-KD cells calculated from three independent NTA datasets.
(E-G) qRT PCR analysis of miRNA levels in control and CERT-KD small and large EVs, and their parental cells, normalized to U6 snRNA. Data from three independent experiments.
(H) Representative merged images of GFP-Rab5Q79L-transfected DKs-8 cells (scrambled control (Sc) and VAP-A-knockdown (KD)) with LC3 and CD63 immunofluorescence staining. Selected areas are enlarged at the right.
(I) Quantitation of the percentage of LC3-positive GFP-Rab5Q79L rings and the intensity of LC3 presented in GFP-Rab5Q79L rings. Each circle represents a cell. n = 75 and 69 cells from 3 independent experiments for Sc and KD, respectively.
(J) Representative deconvolved and merged images of GFP-VAP-A and mCherry-Rab5Q79L-co-transfected DKs-8 cell with nSMase2 immunofluorescence staining. Selected areas are enlarged at the right. Lines were scanned for the intensity of each fluorescence channel of the images and plotted at the right for 0 nm and at the bottom for 250 nm Z-step images. Arrows indicate a bridge of GFP-VAP-A between the limiting membrane of an mCherry-Rab5Q79L ring and nSMase2. Arrowheads indicate nSMase2 association with the ER.
Data plotted as mean +/− S.E.M for bar graphs and as median and interquartile range for scatter plots. *p<0.05, **p<0.01, ***p<0.001. ns, not significant.
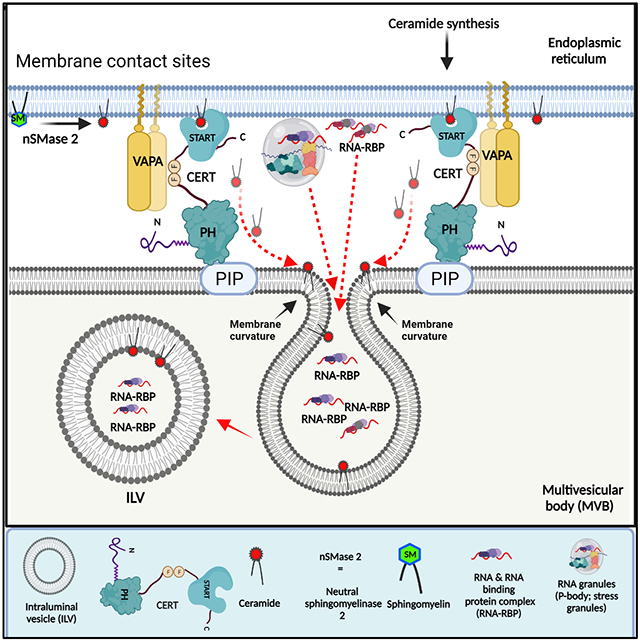
Ceramide synthesis is known to induce EV biogenesis, and the predominant model suggests that ceramide is generated on site in endosomes by the enzymatic action of neutral sphingomyelinase 2 (nSMase 2) on sphingomyelin (Trajkovic et al., 2008). A recent manuscript described a mechanism in which the autophagy protein LC3B together with its binding partner FAN activates nSMase2 to promote biogenesis of exosomes containing snoRNAs and RBPs (Leidal et al., 2020). Since we found that LC3B is present in the dense small EVs that are regulated by VAP-A (Figures 3A and S6A), we tested whether LC3B is present at MVBs and dependent on VAP-A for its incorporation into intraluminal vesicles. Similar to CERT, we found that luminal filling of GFP-Rab5Q79L-positive MVBs with LC3B depends on VAP-A (Figures 7H and 7I). Our data showing that VAP-A controls luminal filling of MVB with LC3B suggest that VAP-A and LC3B may act to control biogenesis of the same population of RNA-containing exosomes. However, if ceramides are generated directly on site at MVB limiting membranes by nSMase2 downstream of LC3B-FAN complexes, then there would presumably be no need to transfer ceramide via VAP-A-CERT linkages. To test whether nSMase2 is more highly associated with MVB or with the ER, we performed immunostaining for nSMase2 in cells expressing GFP-VAP-A to mark the ER and mCherry-Rab5Q79L to mark MVB. Analysis of single plane confocal images revealed that nSMase2 is highly associated with the ER but very little associated with MVBs (Figures S7C and S7D). We did observe a few punctate nSMase2 structures that touched or overlapped with the limiting membrane of the mCherry-Rab5Q79L-positive endosomes. Line scans of such puncta revealed that VAP-A was also present (Figures S7C and S7E-G). To obtain further resolution of the relationship between the nSMase2- and VAP-A-positive structures, we acquired high resolution confocal Z-stacks of the triple stained cells and deconvolved the images. These images revealed that VAP-A-positive ER appears to serve as a bridge between nSMase2-positive structures and MVBs (Figure 7J and Video S2). These data indicate that nSMase2 is closely associated with the ER and suggest that ceramide generated either by the action of nSMase2 or by de novo or salvage synthesis in the ER could be transferred to MVBs by CERT (see model in Graphical Abstract).
Discussion
Currently the models of how RNA is trafficked into vesicles are extraordinarily rudimentary, focusing on select recruitment of RNA by their partner RBPs and lacking an overall picture of how the RBPs themselves connect to the membranes at which the EVs are made. We found that ER MCS are key platforms for this process, impacting the number of both small and large EVs and explicitly controlling the biogenesis of a specific subset of small EVs. While this EV subset accounts for the minority of the small EVs released from cells, it is highly enriched in RNA. Furthermore, this EV population is critical for transfer of miR-100 to recipient cells and growth of DKO-1 tumors in mice. Mechanistically, this biogenesis process depends on VAP-A and its lipid transfer partner CERT, suggesting a model whereby transfer of ceramide from the ER mediates vesicle formation and is coupled to selection of ER-associated RNA-RBP cargoes (Graphical Abstract).
Recent studies have shown that EVs are released from cells as a heterogeneous population containing diverse protein cargoes (Jeppesen et al., 2019; Kowal et al., 2016; Zhang et al., 2018). Leveraging a recently published method that sub fractionates small EVs into light and dense populations (Kowal et al., 2016), we demonstrated that the dense population contains the minority of the small EVs (~10%) but is greatly enriched in RNA (~9-fold/EV) compared to light EVs. Likewise, our imaging data showed sparse punctate distribution of specific miRNAs and RBPs in GFP-Rab5Q79L-positive MVBs. These data suggest that RNA-containing EVs are relatively rare in a general EV population, which may explain why previous calculations of RNA copies/EV are so low (Chevillet et al., 2014) despite their ability to transfer functional RNA to recipient cells (Abels et al., 2019; Chen et al., 2019; Ghamloush et al., 2019; Lucero et al., 2020; Shen et al., 2019; Ying et al., 2017). This subset of EVs is dependent on VAP-A expression in cells, as only dense EVs are diminished in number, size, and RNA content with VAP-A KD. Our data further show that biogenesis of this EV population can be boosted, since VAP-A overexpression greatly increased the number and RNA content of EVs released from cells. Furthermore, since it is dependent on ceramide transfer at ER MCS, one could anticipate regulation by metabolic and signaling alterations that impact ER MCS and/or sphingolipid metabolism, such as occurs in a variety of disorders, such as obesity, metabolic syndrome, and cancer (Holland and Summers, 2008; Ogretmen, 2018).
Although RBPs are known to be important for the transport of RNAs into EVs (Leidal et at., 2020; Lin et al., 2019; Santangelo et al., 2016; Shurtleff et al., 2016; Villarroya-Beltri et al., 2013; Zietzer et al., 2020), it has been unclear how the RBP-RNA complexes are recruited to membranes for incorporation into EVs. Recent work has shown that two membraneless organelles that are comprised of RBP-RNA complexes – processing bodies and stress granules – form contacts with the ER (Lee et al., 2020). Furthermore, both biogenesis and fission of these organelles was shown to occur at these ER contact sites. Also, the ER is associated with additional RNA-RBP complexes, including ribosomes and TIGER domains (Ma and Mayr, 2018). Consistent with our localization of miR-100 and let-7a to ER-endosome contacts and our data that formation of RNA- and RBP-containing EVs depends on ER MCS proteins, one possibility is that RNA-containing membraneless organelles contact the ER at sites of EV biogenesis and contribute material to newly forming EVs.
A major function of VAP-A at ER MCS is to promote lipid transport from one organelle to another. Indeed, we found that EVs purified from VAP-A KD cells had reductions in ceramide, and other lipids and that the ceramide transporter CERT is present at MVB and is critical for biogenesis of RNA-containing EVs. Ceramide is known to be important for biogenesis of exosomes through induction of membrane curvature (Trajkovic et al., 2008) and the major source of ceramide generation for EV biogenesis is thought to be hydrolysis of sphingomyelin by nSMase2 (Maas et al., 2017; Trajkovic et al., 2008). nSMase2 has also been shown to regulate RNA trafficking into small EVs (Kosaka et al., 2010; Leidal et al., 2020). To understand the relationship of nSMase2 to the mechanism we describe involving ceramide transfer via VAP-A-CERT linkages, we localized nSMase2 in cells expressing GFP-VAP-A and Rab5Q79L-marked MVB. Our finding that nSMase2 is highly associated with VAP-A-positive ER and poorly associated with MVBs suggests that CERT could transfer ceramides generated not only via de novo synthesis in the ER but potentially also via nSMase2 activity on associated membranes (see model in Graphical Abstract).
Several recent studies have shown that the early autophagic machinery is involved in exosome biogenesis (Guo et al., 2017; Leidal et al., 2020). By conjugation of a biotin ligase to the key autophagy protein LC3B, Leidal et al (2020) showed that lipidated LC3 (LC3B-II) induces formation of exosomes that contain snoRNAs and a number of RNA binding proteins. They also found that LC3B-II binds to an activator of nSMase2, FAN, that is critical for formation of the subpopulation of EVs regulated by LC3B. As LC3 conjugation takes place at ER-associated membranes, especially the ER-Golgi intermediate compartment (Dikic and Elazar, 2018; Ge et al., 2015; Ge et al., 2014), it seems likely that LC3B-II may recruit FAN and nSMase2 to membranes in close proximity to the ER. Indeed, our findings that nSMase2 is on a structure closely associated with VAP-A-positive ER and that VAP-A ER may bridge those structures to MVB (Figure 7J and Video S2) are consistent with that model. An important future direction is to clearly identify the nSMase2 compartment associated with the ER.
In our subpopulation analysis of small EVs, we found that dense small EVs are enriched for both RNA and LC3B and that candidate miRNAs are on the inside of the same dense small EV population (Figures 3 and S6). We also found that VAP-A-KD reduces intraluminal filling of MVB with LC3B. These data suggest strongly that LC3B and VAP-A act together at ER MCS to promote biogenesis of RNA-containing small EVs. A recent paper showed that a double depletion of VAP-A and VAP-B leads to a defect in transition from the autophagic isolation membrane stage, with lipidated LC3B, to the phagophore stage (Zhao and Zhang, 2019). Since the induction of EV biogenesis by LC3B was shown to be independent of phagophore formation (Leidal et al., 2020), we favor a model in which VAPs interact with early stage LC3B-positive isolation membranes and can mediate either EV biogenesis or phagophore membrane formation depending on the metabolic and signaling state of the cell. Consistent with this model, induction of autophagy with rapamycin was shown to decrease biogenesis of EVs downstream of LC3B (Leidal et al., 2020), suggesting a diversion of lipidated LC3B away from sites of EV biogenesis.
The selective EV biogenesis mechanism that we describe suggests rethinking several related biological processes, including RNA virus assembly and RNAi machinery functions. Indeed, viruses frequently remodel the ER and some also hijack the EV biogenesis machinery for their assembly (Ghosh et al., 2020; Nolte-'t Hoen et al., 2016; Romero-Brey and Bartenschlager, 2016). With regard to RNAi, we found that up- or down-regulation of the ER MCS linker machinery and lipid transfer affects the levels of miRNAs and Argonaute 2, not only in EVs, but also in many cases causing changes in the opposite direction in the parental cells (Figs 2, 7, S2, and S4). Thus, the sorting of miRNAs and Ago2 to EVs, via ER MCS, reduces their levels in cells. These data are in line with previous publications showing regulation of RISC function by MVBs (Bose et al., 2017; Gibbings et al., 2009) and indicate that our mechanism may broadly regulate the miRNA repertoire of cells via selective localization and extracellular sorting of the RISC machinery and associated RNAs.
Our RNA sequencing data suggest that not all EV-associated RNAs are regulated by VAP-A mediated lipid transport. Indeed, U6, which is a small nuclear RNA commonly used to normalize miRNA levels in EVs (Cha et al., 2015; McKenzie et al., 2016) was not altered with VAP-A- or CERT-KD. We do not believe it is a contaminant, because it was predominantly present inside of EVs, based on RNase sensitivity tests. Thus, it seems likely that there may be additional mechanisms that incorporate distinct RNAs into both small and large EVs. Nonetheless, biogenesis of RNA-containing EVs at ER MCS appears to be a major mechanism that controls specific sorting of miRNAs and other small noncoding RNAs.
Several lines of evidence support the premise that VAP-A controls biogenesis of a functionally important subset of small EVs. In miRNA transfer experiments, we found that VAP-A expression in donor DKO-1 colon cancer cells was critical for functional transfer of miR-100 to recipient DKs-8 colon cancer cells in both a co-culture setting and by direct addition of purified small EVs. Furthermore, dense but not light EV subfractions mediated functional transfer of miR-100 to DKs-8 cells. We also carried out xenograft tumor experiments, in which we observed that VAP-A KD DKO-1 colon cancer cells had a defect in tumor growth. The rescue of tumor growth defects by the addition of control but not VAP-A KD EVs indicates that the subpopulation of EVs controlled by VAP-A has important functional properties for tumor survival. The precise VAP-A-regulated EV cargo that mediates tumor survival is yet to be defined and could include RNA, lipid, or protein. Indeed, VAP-A was recently shown to mediate biogenesis of small EVs carrying the ECM protein Tenascin C (Albacete-Albacete et al., 2020), which could mediate tumor cell survival (Yoshida et al., 2015). Identifying key cargoes regulated by VAP-A that promote tumor aggressiveness is an important topic for future research. Regardless, these data establish EV biogenesis as an important function of VAP-A in cancer cells.
Although our mechanistic investigation primarily focused on small EVs/exosomes, we found that VAP-A and CERT also control the number and RNA content of large EVs, which presumably represent microvesicles originating from the plasma membrane. While acid sphingomyelinases are present at the outer leaflet of the plasma membrane and could induce formation of some types of microvesicles in response to stimuli (Bianco et al., 2009), our data indicate that VAP-A-CERT linkages are important for generation of RNA- and ceramide-containing microvesicles. Given that the ER membrane is continuous with the outer nuclear membrane and a recent study found that export of pre-miRNA into microvesicles involved handoff from nuclear export proteins (Clancy et al., 2019), it seems likely that ER contact sites with the plasma membrane may function similarly to ER-endosome linkages to promote ceramide transfer and RNA-RBP sorting. Future studies should investigate the molecular details of how RNA-containing microvesicles are generated.
In summary, we identified a novel biogenesis mechanism for RNA-containing small and large EVs that takes place at ER MCS. Our findings identify a new function for ER MCS, elucidate a poorly understood area of RNA and EV biology, and suggest pathways that could be leveraged for production of RNA-containing therapeutic EVs.
Limitations of the study:
To visualize EV cargoes in MVBs, we utilized expression of a dominant active mutant of Rab5 (GFP-Rab5Q79L) that leads to enlarged endosomes filled with ILVs. The increased size and distinctly labeled boundary of these endosomes greatly facilitates identification of sorted exosome cargoes (Baietti et al., 2012). Using this system, we observed CERT, LC3B, RBPs, and RNAs inside of MVBs, dependent on VAP-A expression. One limitation is that the Rab5Q79L-positive endosomes are not exactly the same as MVBs present in unperturbed cells (Wegner et al., 2010). Nonetheless, previous studies have shown that CERT, LC3, the RBP Ago2, and the miRNA Let7a all colocalize with CD63-positive endosomes in unperturbed cells and connected these localizations to exosome biogenesis (CERT, LC3) or cargo sorting into exosomes (Ago2, Let-7a) (Fukushima et al., 2018; Leidal et al., 2020; McKenzie et al., 2016). Another limitation is that we did not assess the frequency with which ER MCS-induced ILV formation occurs. While other studies have shown that the ER increases its contact with endosomes as they mature (Friedman et al., 2013) and that ER MCS contributes to ILV formation in other contexts (Albacete-Albacete et al., 2020; Eden et al., 2016; Eden et al., 2010; Fukushima et al., 2018), fast super-resolution live imaging would be required to directly assess the frequency at which these events take place.
STAR*METHODS
RESOURCE AVAILABILITY
Lead contact and materials availability
Further information and requests for reagent and resources should be addressed to and will be met by the Lead Contact, Alissa Weaver (alissa.weaver@vanderbilt.edu). All unique/stable reagents generated in this study are available from the Lead Contact with a completed Materials Transfer Agreement.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Cell Lines:
WT KRAS DKs-8 and Isogenic KRAS Mut DKO-1 were cultured in DMEM (Corning) supplemented with 10% Fetal bovine serum (FBS), non-essential amino acids (Sigma), and L-glutamine. HEK293FT cells were cultured in DMEM supplemented with 10% FBS and sodium pyruvate as per the manufacturer’s instructions. HT1080 cells were cultured in DMEM with 10% bovine growth serum (BGS). HEK 293FT lentiviral packaging cells were cultured in DMEM supplemented with 10% FBS and 0.5mg/ml G418 Sulfate (Corning). Stable shRNA scrambled control and shRNA VAP-A or CERT knockdown cell lines were produced using the ViraPower Lentiviral expression system (Thermo Fisher Scientific). The shRNA constructs for VAP-A or CERT in pLKO.l lentiviral shRNA expression system were purchased from Dharmacon. The scrambled control construct was acquired from Addgene.
Animal subjects:
7-12 weeks old female athymic nude mice were purchased from Charles River Laboratory and kept in a pathogen-free facility approved by the American Association for the Accreditation of Laboratory Animal Care that met all current regulations and standards of the U.S. Department of Agriculture, U.S. Department of Health and Human Services, and the National Institutes of Health. Mice were fed irradiated standard mouse chow (LabDiet) and autoclaved, reverse osmosis treated water.
Non-orthotopic nude mouse model for tumor cell xenograft.
Subconfluent cultures were harvested by trypsinization and washed with PBS. Subcutaneous tumors were established by injecting cells (7 X 106 control or VAP-A-KD DKO-1 cells) suspended in 150 μL of serum-free DMEM into the flanks of nude mice. In some cases, small EVs or PBS was mixed with the cells before implantation and small EVs or PBS was injected twice a week until tumor harvest. Mice were examined twice a week for tumor size and weight loss. Subcutaneous tumor size was measured with micro calipers. Tumor volume was calculated as (A) X (B2) X 0.52 where A is the longest dimension of the tumor and B is the dimension of the tumor perpendicular to A. Mice were sacrificed after 3 weeks and tumors were fixed, sectioned, and stained with haematoxylin and eosin (H&E). Imaging of H&E-stained tumor sections was performed using an Aperio Versa 200 scanner (Leica) in the Vanderbilt Digital Histology Shared Resource.
METHOD DETAILS
Extracellular vesicle isolation and nanoparticle tracking analysis
For cushion density gradient method, cells were cultured at 80% confluence in serum-free DMEM. After 48 hours, the conditioned medium was collected from the cells and the EVs were isolated via serial centrifugation. Floating live cells, dead cell debris, and large EVs were respectively collected from the conditioned medium by centrifugation at 300 × g for 10 min, 2,000 × g for 25 min, and 10,000 × g Ti45 rotor, Beckman Coulter)) for 30 min. The supernatant was then overlaid onto a 2 ml 60% iodixanol cushion and centrifuged at 100,000 × g (SW32 rotor, Beckman Coulter) for 18h. The bottom 3 ml, including the 1 ml of collected EVs + 2 ml iodixanol (40% iodixanol final concentration) were transferred to the bottom of another tube and then 20%, 10% and 5% iodixanol were layered successively on top. These iodixanol dilutions were prepared by diluting OptiPrep (60% aqueous iodixanol) with 0.25 M sucrose/10 mM Tris, pH 7.5. After an 18-hour centrifugation step at 100,000 × g, 12 density gradient fractions were collected, diluted in PBS and centrifuged at 100,000 × g for 3 hours. EVs from fractions 6 and 7 were combined and used as small EVs. To quantitate the size and concentration of EVs, nanoparticle tracking analysis (NTA) was performed using a Particle Metrix ZetaView PMX 110.
For “light” and “dense” small EV purification, floating live cells, dead cell debris, and large EVs were removed from the conditioned medium by centrifugation at 300 × g for 10 min, 2,000 × g for 25 min, and 10,000 × g Ti45 rotor, Beckman Coulter)) for 30 min. Small EV-containing pellets were then obtained by ultracentrifugation of the supernatant (100,000 x g in Ti45 Beckman Coulter rotor for 70 min. at 4 ° C). Pellets were then washed and resuspended in 1.25 ml buffer [0.25 M sucrose, 10 mM Tris pH 8.0, 1 mM EDTA (pH 7.4)], transferred to a SW55Ti rotor tube (Beckman Coulter) and mixed with 60% (wt/vol) stock solution of iodixanol (1:1). Next, 1.1 ml 20% (wt/vol) iodixanol and 1 ml of 10% (wt/vol) iodixanol successively layered on top of the vesicles suspension and tubes were centrifuged for 1h at 4 °C at 350,000 x g in SW55Ti rotor; Ten fractions of 460 ul were collected from the top. Fractions were diluted and washed in PBS for 1h at 100,000 x g in a TLA 110 rotor (Beckman). Fractions were resuspended in 35 ul of PBS. This method was from a previously published report (Kowal et al., 2016).
Cell Labeling for fluorescence microscopy
Cells on coverslips coated with poly-D-Lysine (100 μg/ml) were fixed with 4% paraformaldehyde in PBS then permeabilized with 0.5% Triton X-100 in PBS. Cells were stained for proximity ligation assay (PLA) using a Duolink® kit according to the manufacturer’s protocol (DUO92102-1KT, Millipore Sigma). Briefly, cells were blocked by Duolink® Blocking Solution for 60 minutes in a 37° C humid chamber. Primary antibodies were diluted (KDEL 1:100 and CD63 1:100) in the Duolink® Antibody Diluent and incubated overnight at 4° C in a humid chamber. After washing, the cover slips were incubated with PLA probes for lh in a 37° C humid chamber. The ligation reaction was performed for 30 minutes at 37° C followed by washing and amplification at 37 degrees Celsius for 100 minutes. Cover slips were washed and mounted with antifade gold mounting media with DAPI. For colocalization of EV cargoes with Rab5Q79L, cells were transfected with 100 ng of GFP-Rab5Q79L or 150 ng mCherryRab5Q79L or 150 ng EGFP-VAP-A in a 12 well plate for 5h. Cells were reseeded after 24h on PDL coated coverslips and grown for 18h and then changed to media with serum free DMEM and grown for 24h before fixed with 4% paraformaldehyde in PBS. Cells were permeabilized in 0.2% saponin in PBS followed by blocking with 5% BSA and 0.1% saponin in PBS. Primary antibodies were diluted in blocking buffer (CD63 1:100, Ago2 1:100, SYNCRIP 1:100, CERT 1:100, nSMase 1:100) and incubated overnight at 4° C in a humid chamber. After washing, the cover slips were incubated with Alexa Fluor 647™ and/or Alexa Fluor 546™ -conjugated secondary antibodies for 1h at RT in a humid chamber. Cover slips were washed and mounted with antifade gold mounting media with DAPI. For let-7a and miR-100 colocalization, cells were transfected with 100 ng of GFP-Rab5Q79L and 10 pg of MFP488 labelled (Mirus-bio) let-7a or miR-100 and grown as mentioned above before fixation. Cells were permeabilized in 0.2% saponin in PBS followed by blocking. Cells were stained with Phalloidin 633 for 1h at RT in a humid chamber. Cover slips were washed and mounted with antifade gold mounting media with DAPI.
Confocal Microscopy and Post-Acquisition Deconvolution
After mounting coverslips on glass slides, single Z-slice images were acquired with a Nikon A1R-HD25 confocal microscope (run by NIS-Elements) equipped with an Apo TIRF 60x/1.49 oil immersion lens using 1 Airy unit for pinhole. For images used for deconvolution, multi-channel image stacks (as well as single optical sections) were acquired via point scanning confocal microscopy (A1R-HD25, Nikon Instruments, Inc.) on an inverted fluorescence microscope stand (Ti2, Nikon Instrument, Inc.) equipped with an Apo TIRF 60x 1.49 NA oil immersion lens. At least 7 slices per stack with 50 nm Z-step intervals were imaged using the Nikon A1 Piezo Z Drive with the pinhole set to 0.9 Airy unit. The theoretical resolution afforded by this lens was slightly oversampled (~1.75x laterally, resulting in 50 nm pixels) to optimize anticipated downstream deconvolution of resultant data, in combination with a pinhole set to 0.9 Airy units. Likewise, the axial step size chosen (50 nm/step) oversampled the resolution via integrated piezo stage (Mad City Labs), as well as provided cubic voxels for ease of downstream processing and visualization. Excitation lasers in this microscope configuration were comprised of 405 nm, 488nm, 561 nm, and 647 nm lines. Acquisition of said data was managed by NIS-Elements software package (Nikon Instruments, Inc.). Post-acquisition, image stacks were deconvolved using 10 iterations of Richardson-Lucy deconvolution (via NIS-Elements software) in order to quantitatively improve image contrast, and thus potentially enhance resolving capability. Both the method of deconvolution, as well as number of iterations, were empirically chosen such that introduction of processing artifact was minimized.
Transmission electron microscopy (TEM)
For negative staining of regular small EVs, Formvar carbon film–coated grids (FCF-200-Cu; Electron Microscopy Sciences) were washed in double distilled water and then washed by 100% ethanol. 10-μl samples were added to grids overnight at 4 ° C. Grids were then incubated with 2% phosphotungstic acid, pH 6.1 for 30 s and followed by immediate blotting. For negative staining of purified “light” and “dense” EVs, Formvar carbon film–coated grids (FCF-200-Cu; Electron Microscopy Sciences) were freshly glow discharged before use. Grids were incubated with samples for 20 seconds, followed by brief washes in ddH2O and stained with uranyl acetate for 5 seconds and immediately blotting.
For TEM of the cells, cells were grown on Matrigel-coated Transwells (Corning) for 48h before fixing in 2.5% glutaraldehyde in 0.1 M cacodylate for 1 hour at room temperature followed by 48 hours at 4 ° C. Samples were post-fixed in 1% tannic acid, followed by 1% OsO4, and en bloc stained with 1% uranyl acetate. Samples were dehydrated with a graded ethanol series, infiltrated with Quetol 651 based Spurr’s resin (Electron Microscopy Sciences) using propylene oxide as a transition solvent, and polymerized at 60 ° C for 48 hours. Samples were sectioned on a UC7 ultramicrotome (Leica) at a nominal thickness of 70 nm and stained with 2% uranyl acetate and lead citrate.
All TEM samples were imaged using a Tecnai T12 operating at 100 kV with an AMT NanoSprint CMOS camera using AMT imaging software for single images. Quantification of TEM data was performed in Fiji. Tilt series acquisition for tomography was performed using SerialEM. Tomographic reconstructions were performed in the IMOD software suite using patch tracking and back projection. MVB, ER, and ILVs were manually segmented in Dragonfly ORS to paint organelles in every Z-plane of the tomograms Z-stacks, these ROIs were used to generate three-dimensional contours. Images and movies depicting the segmented organelles were generated in Dragonfly ORS, movies were reformatted in FIJI.
Image Analyses
Analysis of GFP-Rab5Q79L data: GFP-Rab5Q79L-transfected cells from all-related immunofluorescence staining experiments were used to analyze Rab5Q79L size and number/cell, the percentage of cargo-positive Rab5Q79L-endosomes, and the intensity of cargoes within Rab5Q79L rings. Each ring of GFP-Rab5Q79L was outlined using the oval selections tool or the freehand selections tool in Fiji and the number of rings each cell was counted for the total number (Analyze/Analyze Particles). The size was measured using Fiji (Analyze/Analyze Particles). To measure the percentage of GFP-Rab5Q79L rings which are positive for specific cargoes, each cargo-positive GFP-Rab5Q79L ring was manually counted, divided by the total number of GFP-Rab5Q79L rings for each cell, and multiplied by 100. The intensity for each cargo located in GFP-Rab5Q79L rings was measured using Fiji (Analyze/Measure) after thresholding.
Fluorescence colocalization analyses:
Colocalization was measured using Fiji (Analyze/Colocalization/ Colocalization Threshold) after background subtraction (Process/Subtract Background) of each fluorescence channel. All datasets were exported to and plotted using GraphPad Prism 9.2.0. Line scanning to show intensity distribution of multiple channels was done using Fiji (Analyze/Plot Profile), then the datasets were exported to an Excel sheet. The relative intensity of each channel was calculated and plotted using Excel.
PLA analysis:
For fluorescent dots quantification, images were segmented from the background by thresholding and particle number per cell was calculated by Fiji (Analyze/analyze particles).
3D reconstruction of deconvolved images:
The 3D structure shown in Video S2 was reconstructed from deconvolved images in a Z-stack and rotated using NIS-Elements. The zooming 2D effect in the movie was recorded using Adobe Photoshop 2020 (Window/Timeline function). The zooming 2D movie and 3D rotating movie were concatenated and annotated using Fiji.
Analyses of TEM images:
Diameter of EVs from TEM images were measured by Fiji plugin (Analyze/measure). The ER and MVB contacts were manually identified and distance between ER and MVB was analyzed by Fiji plugin (Analyze/measure). Analysis of the TEM data was performed in Fiji plugin (Analyze/measure).
Western blot analysis
The protein concentrations of total cell lysates were determined utilizing Pierce BCA Assay (Cat. 23225, Thermo Fisher). The protein concentrations of the EVs were determined utilizing Pierce Micro BCA Assay (Cat. 23235, Thermo Fisher). For Western blots, 15 μg of TCLs, small EVs, large EVS or an equal volume of resuspended vesicles from density gradient fractions (for control markers blots) were boiled in SDS-Page sample buffer for 5 min and loaded on 10-well or 15-well 8% or 10% polyacrylamide gels. Proteins were transferred to nitrocellulose membranes for 1 h at 100 volts or 25 volts for overnight at 4°C. Membranes were blocked in 3% BSA diluted in Tris-buffered saline with 0.5% Tween 20 (TBST) for 4h at room temperature. Primary antibodies were diluted in 3% BSA -TBST (Ago2, 1:1000; hnRNPA2/B1,1:1000; Hsp70, 1:1000; CD63, 1:1000; Flotillin, 1:1000; TSG101, 1:1000; GM130, 1:2000; LC3B 1:1000; Syntenin 1:1000; Alix 1:1000; GAPDH 1:5000 and beta actin, 1:10000) and incubated overnight at 4°C. Membranes were washed 3 times for 15 min in TBST and subsequently incubated with species-specific HRP-conjugated secondary antibodies (1:10000; Promega) in 3% BSA -TBST for 1h at room temp. All membranes were washed 3 times for 15 min in TBST and incubated with an enhanced chemiluminescence (ECL) reagent (Thermo Scientific) for 1 min before being exposed to film or using a ChemiDoc Imager (BioRad) or Amersham 680 imager (GE). Multiple exposures were taken for each blot to have the complete dynamic range for densitometry measurements. The densitometry measurements for the protein bands were done using the Analysis Gels feature of ImageJ (NIH).
Dot blot analysis
Dot blotting of EV preparations was performed as described previously (Lai et al., 2015). Different concentrations of sEVs or lEVs were collected from conditioned media of DKs-8 cells were dotted onto nitrocellulose membranes and allowed to dry at room temperature for 1 h. The membrane was - blocked with 3% BSA in Tris-buffered saline (TBS) in the absence or presence of 0.1% (v/v) Tween 20 (TBS-T) at room temperature for 1h, followed by incubation with anti-Ago2, anti-hnRNPA2/B1, anti-flotillin-1 or antiCD63 antibody in TBS or TBS-T overnight at 4 °C.
RNA purification
Total RNA from cell, small and large EVs was purified using the miRNeasy kit (Qiagen Inc., Valencia, CA, USA) according to the manufacturer’s protocol. Final RNAs were eluted with two rounds of 35 ul of Nuclease free water extraction.
miRNA library preparation and sequencing
Total RNA from each sample was used for small RNA library preparation using NEBNext. Small RNA Library Prep Set from Illumina (New England BioLabs Inc., Ipswich, MA, USA). Briefly, 3′ adapters were ligated to total input RNA followed by hybridization of multiplex single read (SR) reverse transcription (RT) primers and ligation of multiplex 5′ SR adapters. RT was performed using ProtoScript II RT for 1 hr at 50°C. Immediately after RT reactions, PCR amplification was performed for 15 cycles using LongAmp Taq 2× master mix. Illumina-indexed primers were added to uniquely barcode each sample. Post-PCR material was purified using QIAquick PCR purification kits (Qiagen Inc.). Post-PCR yield and concentration of the prepared libraries were assessed using Qubit 2.0 Fluorometer (Invitrogen, Carlsbad, California, CA, USA) and DNA 1000 chip on Agilent 2100 Bioanalyzer (Applied Biosystems, Carlsbad, CA, USA), respectively. Size selection of small RNA with a target size range of approximately 146–148 bp was performed using 3% dye free agarose gel cassettes on a Pippin Prep instrument (Sage Science Inc., Beverly, MA, USA). Post-size selection yield and concentration of libraries were assessed using Qubit 2.0 Fluorometer and DNA high-sensitivity chip on an Agilent 2100 Bioanalyzer, respectively. Accurate quantification for sequencing applications was performed using qPCR-based KAPA Biosystems Library Quantification kits (Kapa Biosystems, Inc., Woburn, MA, USA). Each library was diluted to a final concentration of 1.25 nM and pooled in equimolar ratios prior to clustering. Single-end sequencing was performed to generate at least 15 million reads per sample on an Illumina HiSeq2500 v4 using a 50-cycle kit.
Small RNA sequencing analysis
Small RNA-seq reads were trimmed using cutadapt v1.18 (https://github.com/marcelm/cutadapt). After trimming, reads longer than 15 nucleotides were retained. ncPRO-seq (version 1.5.1) (Chen et al., 2012) was used to map reads to the reference genome hg19 and quantitate small RNA. The miRBase v18, ACA_snoRNA and CD_snoRNA from Rfam v11.0, and tRNA from UCSC (hg19) were employed for reads annotation to miRNA, snoRNA, and tRNA. miRNA annotation was extended in both upstream and downstream regions by using miRNA_e_+2_+2. Principal component analysis was performed to assess the similarity between samples. DESeq2 (Love et al., 2014) was used to identify small RNAs differentially exported from Cell to EVs in VAP-A KD compared to SC. For identifying miRNAs that were upregulated or downregulated in KD EVs or cells, all KD samples were compared to control in one group whether from KD1 or KD2 cells. Small RNAs with fold change≥2 or ≤0.5 and a false discovery rate (FDR) ≤ 0.05 were considered to be significantly differentially expressed.
qRT-PCR for miRNA
Total RNA was isolated from small EVs, large EVs, and cells using the miRNeasy kit (Qiagen Inc., Valencia, CA, USA), which isolates all small RNAs <200 nt, including miRNAs. Total RNA amount of sEVs, lEVs and Cells were measured by Nanodrop. Taqman small RNA assays (ThermoFisher Scientific) were performed for small EVs, large EVs and cellular RNAs according to the manufacturer’s protocol; U6 snRNA: 001973; hsa-let7a-5p: 000377; hsa-miR-100-5p:000437; hsa-miR320a: 002277; has-miR-371a: 002124; has-miR-372: 000560. Individual reverse transcription reactions were performed using 10 ng RNA from each sample per Taqman miRNA primer in a final reaction volume of 10 μl. After transcription, 0.34 ng (0.67 μl) cDNA was used as the template together with the corresponding Taqman miRNA probe for qPCR in a final reaction volume of 10 μl. Each Taqman miRNA qPCR was performed with technical triplicates on a Bio-Rad CFX96. C(t) values were averaged for each technical triplicate. U6 snRNA was used as a normalization control for each biological sample. To calculate fold changes (FC), the ΔΔC(t) method was used (Schmittgen and Livak, 2008). Briefly, ΔC(t) values were calculated for each biological sample, where ΔC(t) = C(t)miRNA - C(t)U6 snRNA. Relative fold changes were determined by Fold change = 2-ΔΔC(t), where ΔΔC(t) = ΔC(t)-ΔC(t)control. For ΔΔC(t) values < 0 (signifying a negative fold change), the negative reciprocal Fold Change formula was used (−1/(2-ΔΔC(t)). Statistical analyses were performed from three independent biological replicates
RNase protection assay for EV samples
EV pellets resuspended in PBS were mixed with 10 Units RNase A (Thermo) in the presence or absence of Triton-X-100 (TX-100) (final concentration 1%) in 100 μl and incubated for 30 min at 37° C. Enzyme was inactivated at 95° C for 10 min and 700 μl Trizol was added followed immediately by RNA extraction using the miRNeasy kit (Qiagen Inc., Valencia, CA, USA).
Co-culture and Luciferase reporter assay
Recipient DKs-8 cells were plated in six-well plates at a density of ~2.5 × 105 cells and cultured in DMEM supplemented with 10% FBS for 24 hr. The media was replaced with serum-free Opti-MEM and the cells were co-transfected with 1.5 μg of Luc-reporter plasmid and 1.5 μg β-gal plasmid DNA/well. Donor cells were plated in 0.4-μm pore Transwell filters (Corning, 3450, Corning, NY, USA) at ~2.5 × 105 cells/well for 24 hr. The media from donor Transwells and recipient 6-well plates were removed and replaced with serum-free DMEM. Co-culture of donor and recipient cells was then conducted for 48 hr before recipient cells were harvested. In some cases, purified small EVs were added instead of co-culturing with donor cells (8X109 per well for Fig 5E, 2X109 per well for Fig 5F, G). The number of EVs to add was estimated by the EV/cell/hour secretion rate of parent DKO-1 cells x number of cells x number of hours of assay then refined in pilot experiments. Lysates were prepared in 1× Reporter lysis buffer (Promega, E2510), and Luciferase assays were performed according to the manufacturer's protocol (Promega, E2510). β-gal expression was simultaneously determined from the lysates according to the manufacturer's protocol (Promega, E2000). Differences in transfection efficiency were accounted for by normalizing Luc expression to β-Gal expression (Luc/β-Gal). All assays were performed on three biological replicates, each with three technical replicates.
Lipid mass spectrometry
Untargeted Lipidomics.
Discovery lipidomics data were acquired using a Vanquish ultrahigh performance liquid chromatography (UHPLC) system interfaced to a Q Exactive HF quadrupole/orbitrap mass spectrometer (Thermo Fisher Scientific). Exosomes, microvesicles, and cell pellets were resuspended in 250 μL aqueous 20 mM ammonium acetate and spiked with a mixture of C12:0 ceramide and SPLASH lipidomics MS standards (Avanti). For lipid extraction, 1 mL of MeOH/MTBE/CHCl3 (1.3:1:1) was added, briefly vortexed and shaken gently for 20 min, followed by centrifugation at 3,000 x g for 15 min at room temperature. The supernatant was transferred to a clean glass vial, evaporated under a gentle stream of N2 gas, and resuspended in 100 μL HPLC-grade methanol for LC-MS analysis. Lipid extracts were injected a total of four times. Two injections were made in positive ESI mode followed by two injections in negative mode. Pooled QCs were injected to assess the performance of the LC and MS instruments at the beginning and at the end of each sequence.
Chromatographic separation was performed with a reverse-phase Acquity BEH C18 column (1.7 μm, 2.1x150mm, Waters, Milford, MA) at a flow rate of 300 μl/min. Mobile phases were made up of 10 mM ammonium acetate in (A) H2O/CH3CN (1:1) and in (B) CH3CN/iPrOH (1:1). Gradient conditions were as follows: 0–1 min, B = 20 %; 1–8 min, B = 20– 100 %; 8–10 min, B = 100 %; 10–10.5 min, B = 100–20 %; 10.5–15 min, B = 20%. The total chromatographic run time was 20 min; the sample injection volume was 10 μL. Mass spectra were acquired over a precursorion scan range of m/z 100 to 1,200 at a resolving power of 30,000 using the following ESI source parameters: spray voltage 5 kV (3 kV in negative mode); capillary temperature 300 °C; S-lens RF level 60 V; N2 sheath gas 40; N2 auxiliary gas 10; auxiliary gas temperature 100 °C. MS/MS spectra were acquired for the top-five most abundant precursor ions with an MS/MS AGC target of 1e5, a maximum MS/MS injection time of 100 ms, and a normalized collision energy of 30. High resolution mass spectrometry data were processed with MS-DIAL version 4.70 in lipidomics mode (Tsugawa et al., 2020). MS1, and MS2 tolerances were set to 0.01 and 0.025 Da respectively. Minimum peak height was set to 30000 to decrease the number of false positive hits. Peaks were aligned on a quality control (QC) reference file with a RT tolerance of 0.1 min and a mass tolerance of 0.015 Da. Default lipid library was used (Msp20210527163602_converted.lbm2), solvent type was set to CH3COONH4 to match the solvent used for separation, and the identification score cut off was set to 80%. All lipid classes were made available for the search. MS-DIAL results were cleaned after identification was completed using blank sample as a template and all peak areas were exported into Excel for further processing. Differentially expressed lipids from cells and EVs in SC compared to KD were identified with an interaction model using the package Limma version 3.48.1 (Ritchie et al., 2015). Lipids with a fold change > 2 and false discovery rate (FDR) < 0.05 were considered significantly different and were plotted in heatmaps with package pheatmap version 1.0.12 (pheatmap: Pretty Heatmaps version 1.0.12 from CRAN (rdrr.io)).
Targeted ceramide quantification by LC-MS/MS.
Tandem mass spectrometric detection was performed using a TSQ Quantum Ultra triple quadrupole mass spectrometer (Thermo Scientific, San Jose, CA) equipped with an Ion Max API source, a standard ESI probe, and a 50 μm ID stainless steel high voltage capillary. The mass spectrometer was operated in positive ion mode. Quantitation was based on single reaction monitoring detection. The following optimized source parameters were used for the detection of analyte and internal standards. N2 sheath gas 30 psi; N2 auxiliary gas 15 psi; spray voltage 5 kV; ion transfer tube temp 300 °C; declustering voltage 10 V.
For calibrating the instrument response, milligram quantities of C12:0 (IS), C16:0, C18:0, C18:1, C22:0, C24:0, and C24:1 ceramide standards (Avanti) were weighed out in aluminum weigh boats using a UMT2 microbalance (Mettler Toledo, Columbus, OH), dissolved in an appropriate volume of EtOH/CHCl3 (3:1) to produce primary stock solutions at a concentration of 0.1–0.2 mg/mL, and stored in the dark at −20 °C. Working stocks were prepared by serial dilution of primary stocks in ethanol and stored in the dark at 2–8 °C for up to 4 weeks before use. Calibration samples (PBS) were spiked with the appropriate working stocks of C16:0, C18:0, C18:1, C22:0, C24:0, and C24:1 ceramides and internal standard C12:0 ceramide. Exosomes, microvesicles, and cell pellets were extracted and reconstituted in methanol as described for untargeted lipidomics (see above). Sample analyses were carried out using a Waters Acquity UPLC system (Waters, Milford, MA), made up of a binary solvent manager, refrigerated sample manager, and a heated column manager. A Kinetix C8 analytical column (2.1 mm x 100 mm, 1.7 μm particle size, Phenomenex, Torrance, CA) was used for all chromatographic separations. The autosampler tray temperature was maintained at 5 °C; the column compartment was not thermostatted. Mobile phases were made up of 0.2% HCOOH in (A) H2O/CH3CN/CH3OH (3:2:2) and in (B) CE3CN/iPrOH (1:1). Seven ceramides were resolved in less than five minutes using isocratic elution (A/B 80:20) at a flow rate of 0.3 mL/min. The sample injection volume (partial loop) was 10 μL. Calibration curves were constructed by plotting peak area ratios (analyte / internal standard) against analyte concentrations for a series of ten calibrants, ranging in concentration from 10 ng/mL to 20 μg/mL. A weighting factor of 1/Ct2 was applied in the linear least-squares regression analysis to maintain homogeneity of variance across the concentration range (% error ≤ 20% for at least four out of every five standards). Data acquisition and analysis were carried out using Xcalibur v.2.1.0, and LCOuan v.2.7.0 software (Thermo).
Animal subjects:
7-12 weeks old female athymic nude mice were purchased from Charles River Laboratory and kept in a pathogen-free facility approved by the American Association for the Accreditation of Laboratory Animal Care that met all current regulations and standards of the U.S. Department of Agriculture, U.S. Department of Health and Human Services, and the National Institutes of Health. Mice were fed irradiated standard mouse chow (LabDiet) and autoclaved, reverse osmosis treated water.
Non-orthotopic nude mouse model for tumor cell xenograft.
Subconfluent cultures were harvested by trypsinization and washed with PBS. Subcutaneous tumors were established by injecting cells (7 X 106 control or VAP-A-KD DKO-1 cells) suspended in 150 μL of serum-free DMEM into the flanks of nude mice. In some cases, small EVs (1X1011 to 10X1011 EVs) or PBS were mixed with the cells before implantation and small EVs or PBS were injected twice in a week until tumor harvest. The number of EVs to add was first estimated from the EV secretion rate x number of cells x hours before next injection then converted to protein, for ~4 μg. Pilot experiments then tested 1-10 μg protein concentrations (Fig 4H). Mice were examined twice a week for tumor size and weight loss. Subcutaneous tumor size was measured with micro calipers. Tumor volume was calculated as (A) X (B2) X 0.52 where A is the longest dimension of the tumor and B is the dimension of the tumor perpendicular to A. Mice were sacrificed after 3 weeks and tumors were fixed, sectioned, and stained with haematoxylin and eosin (H&E). Imaging of H&E-stained tumor sections was performed using an Aperio Versa 200 scanner (Leica) in the Vanderbilt Digital Histology Shared Resource.
Statistics
Experimental data were acquired from at least three independent experiments. Data plotted by bar graph were compared using student’s t test and plotted as mean and standard error of the mean using GraphPad Prism 9. Tumor data were compared by non-parametric Mann-Whitney test. All datasets from imaging analyses were analyzed as non-parametric data groups and were compared by the two-sided unpaired Mann-Whitney test and plotted with median and interquartile range.
Supplementary Material
Data S1: microRNA data from small RNA-Seq data for control, VAP-A KD1, VAP-A KD2 cells, small EVs and large EVs. Related to Figure 2.
Data S2: snoRNA data from small RNA-Seq data for control, VAP-A KD1, VAP-A KD2 cells, small EVs and large EVs. Related to Figure 2 and Figure S3.
Data S3: tRNA data from small RNA-Seq data for control, VAP-A KD1, VAP-A KD2 cells, small EVs and large EVs. Related to Figure 2 and Figure S3.
Data S4: Untargeted lipid mass spectrometry data for control, VAP-A KD1, VAP-A KD2 cells, small EVs and large EVs. Related to Figure 6.
Table S1: Curated data for RNA binding proteins (RBPs). Related to Figure 1.
List of 61 RBPs shown in Figure 1A overlapped with endoplasmic reticulum and EV proteomes; 22 RBPs shown in Figure 1B overlapped with endoplasmic reticulum proteome.
Video S1. Intraluminal vesicle formation at an ER membrane contact site. Related to Figure 1.
Video shows a tomographic reconstruction of an MVB/ER contact site observed in a DKs-8 (Sc) cell (see also Figure 1H). Three-dimensional segmentations of organelles depict MVB (light green), intraluminal vesicles (ILVs) (dark blue), and ER tubules (purple). Note the presence of an ILV still connected to the MVB limiting membrane at the ER contact site.
Video S2. VAP-A-positive ER connects nSMase2-positive vesicles to an MVB. Related to Figure 7.
GFP-VAP-A- and mCherry-Rab5Q79L-cotransfected DKs-8 cell with nSMase2 immunofluorescence staining was deconvolved. Zooming effect is shown at 250 nm Z-step (see Figure 7J). The selected area is converted to a 3D structure reconstructed with 650 nm thickness. An arrow indicates GFP-VAP-A penetration into the lumen of mCherry-Rab5Q79L-labeled MVB. An arrowhead indicates the presence of nSMase2 at the ER-MVB contact site.
Key Resources Table
| REAGENT OR RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Mouse monoclonal anti-VAPA | Novus biologicals | Cat no # H00009218 |
| Rabbit polyclonal anti-CERT | Abeam | Cat no # ab72536 |
| Mouse anti-Flotillin-1 | BD Biosciences | Cat no # 610820 |
| Mouse monoclonal anti-HSP70 | SCBT | Cat no # sc-66048 |
| Rabbit polyclonal anti-Tsg101 | Abcam | Cat no # ab30871 |
| Rabbit monoclonal anti-CD63 (for WB) | Abcam | Cat no # ab134045 |
| Mouse anti-CD63 (for IF) | Abcam | Cat no #ab8219 |
| Rabbit monoclonal anti-Ago2 | Cell Signaling | Cat no # 2897 |
| Mouse monoclonal anti-hnRNPA2B1 | Cell Signaling | Cat no # 9304 |
| Rabbit polyclonal anti-hnRNPQ | Abcam | Cat no # ab189405 |
| Mouse monoclonal anti-KDEL | Abcam | Cat no # ab |
| Mouse monoclonal anti-Beta actin | Cell Signaling | Cat no# 58169 |
| Rabbit monoclonal anti-Bip-1 | Cell Signaling | Cat no# 3177 |
| Rabbit monoclonal anti-IRE1a | Cell Signaling | Cat no # 3294 |
| Rabbit anti-Cleaved caspase 3 | Abcam | Cat no # ab32042 |
| Rabbit anti-nSMase 2 | Abcam | Cat no # ab85017 |
| Rabbit anti-hnRNPQ (SYNCRIP) | Abcam | Cat no# abl 84946 |
| Rabbit anti-KDEL | Abcam | Cat no # ab176333 |
| Rabbit anti-Syntenin | Abcam | Cat no # ab133267 |
| Rabbit anti-CERT | Abcam | Cat no # ab151285 |
| Mouse anti-Alix | Cell Signaling | Cat no # 2171A |
| Rabbit anti-LC3B | Cell signaling | Cat no # 3868 |
| Mouse anti-GM130 | BD Biosciences | Cat no # BD610822 |
| Rabbit monoclonal anti-GAPDH | Cell Signaling | Cat no #5174 |
| Anti-Mouse IgG (H+L), HRP Conjugate | Promega | Cat no # W4021 |
| Anti-Rabbit IgG (H+L), HRP Conjugate | Promega | Cat no # W4011 |
| Chemicals | ||
| Thapsigargin | Millipore-Sigma | Cat no # T9033 |
| Staurosporine | Cell Signaling | Cat no #9953 |
| Puromycin Dihydrochloride | Sigma Aldrich | Cat no # P8333 |
| Critical Commercial Assays | ||
| Micro BCA Protein Assay Kit | Thermo Fisher Scientific | Cat no # 23235 |
| BCA protein assay kit | Thermo Fisher Scientific | Cat no # 23225 |
| Micro Rneasy Mini Kit | Qiagen | Cat no # 217004 |
| Steady-Glo® Luciferase Assay System | Promega | Cat no # E2510 |
| β-Galactosidase Enzyme Assay System with Reporter Lysis Buffer | Promega | Cat no # E2000 |
| TaqMan™ MicroRNA Reverse Transcription Kit | Thermo Fisher Scientific | Cat no # 4366597 |
| TaqMan™ Universal Master Mix II, no UNG | Thermo Fisher Scientific | Cat no # 4440049 |
| iScript™ cDNA Synthesis Kit | Bio Rad | Cat no # 1708890 |
| SsoAdvanced Universal SYBR Green Supermix | Bio Rad | Cat no # 1725270 |
| Qubit™ RNA HS Assay Kit | Thermo Fisher Scientific | Cat No #Q32852 |
| Duolink™ In Situ Orange Starter Kit Mouse/Rabbit | Millipore Sigma | Cat no #DUO92102-1KT |
| Experimental Models: Organism/Strain | ||
| BLAB/c female mice | Charles River Laboratory | |
| Oligonucleotides | ||
| snoRA42 Forward 5’TGGATTTATGGTGGGTCCTTCTCTG3’ | MilliporeSigma/Genosys | |
| snoRA42 Reverse 5’CAGGTAAGGGGACTGGGCAATGGTT3’ | MilliporeSigma/Genosys | |
| snoRD45 Forward 5’CATCTATAATGGCTGAATTGGAA3’ | MilliporeSigma/Genosys | |
| snoRD45 Reverse 5’ATGAACTTTCCAACAAATGTTGTT3’ | MilliporeSigma/Genosys | |
| snoRA40 Forward 5’ ATGTATGTTTTTGTTTAACG 3’ | MilliporeSigma/Genosys | |
| snoRA40 Reverse 5’ CAAAACTCATACTGAACAATG 3’ | MilliporeSigma/Genosys | |
| snoRD105 forward 5’ ATCTCTCATGATGAACACATATG3’ | MilliporeSigma/Genosys | |
| snoRD105 Reverse 5’ CCATCTCTTCTTCAGAGCG 3’ | MilliporeSigma/Genosys | |
| TaqMan™ MicroRNA Assay | Thermo Fisher Scientific | Cat no # 4427975 |
| U6 snRNA | Thermo Fisher Scientific | Cat no # 001973 |
| hsa-miR-371a | Thermo Fisher Scientific | Cat no # 002124 |
| hsa-miR-372 | Thermo Fisher Scientific | Cat no # 000560 |
| hsa-let-7a | Thermo Fisher Scientific | Cat no # 000377 |
| hsa-miR-100 | Thermo Fisher Scientific | Cat no # 000437 |
| hsa-miR-125b | Thermo Fisher Scientific | Cat no # 000449 |
| hsa-miR-320a | Thermo Fisher Scientific | Cat no # 002277 |
| hsa-miR-30a | Thermo Fisher Scientific | Cat no #000417 |
| hsa-miR-129 | Thermo Fisher Scientific | Cat no #000590 |
| hsa-miR-99a | Thermo Fisher Scientific | Cat no #000435 |
| TRC Lentiviral shRNA -VAP-A | Dharmacon | Cat no #RHS3979-201759439 Cat no #RHS3979-201759438 |
| TRC Fentiviral shRNA -CERT | Dharmacon | Cat no #RHS3979-201738486 Cat no #RHS3979-201738485 |
| pLKO.1 scrambled control construct | Addgene | Plasmid no #26701 |
| Pre-miR-let-7a | Thermo Scientific | Cat no #AM17100 (ID PM10050) |
| Pre-miR-100 | Thermo Scientific | Cat no #AM17100 (ID PM10188) |
| Anti-miR-100 | Thermo Scientific | Cat no #AM17000 (ID AM10188) |
| Anti-miR control | Thermo Scientific | Cat no #AM17010 |
| RNase A | Thermo Scientific | Cat no #EN0531 |
| Recombinant DNA | ||
| EGFP-Rab5A Q79F | Addgene | Cat no # 28046 |
| mCherry-Rab5CA(Q79L) | Addgene | Cat no # 35138 |
| pEGFPC1-hVAP-A | Addgene | Cat no #104447 |
| Tissue culture reagents | ||
| DMEM | Corning | Cat no #10-013-CV |
| Fetal bovine serum | Sigma | Cat no #F0926 |
| Bovine growth serum | Hyclone | Cat no #SH30073.03 |
| Lipofectamine 2000 | Thermo Scientific | Cat no #11668-019 |
| TransITX2 | Mirus Bio | Cat no #MIR 6004 |
| MFP-488 | Mirus Bio | Cat no #MIR7125 |
| Cy3 | Mirus Bio | Cat no #MIR3625 |
| Software and algorithms | ||
| GraphPad Prism 9.2.1 | https://www.graphpad.com/scientific-software/prism/ | |
| ImageJ / Fiji | NIH | |
| NIS Elements | Nikon Instruments, Inc | |
| Dragonfly ORS | http://www.theobjects.com/dragonfly | |
| IMOD | ||
| cutadapt v1.18 | (https://github.com/marcelm/cutadapt) | |
| ncPRO-seq (version 1.5.1) | ||
| DESeq2 | ||
| Msp20210527163602_converted.lbm2 | ||
| pheatmap version 1.0.12 | pheatmap: Pretty Heatmaps version 1.0.12 from CRAN (rdrr.io) | |
| Xcalibur v.2.1.0 software | Thermo | |
| LCQuan v.2.7.0 software | Thermo | |
| Limma version 3.48.1 | https://bioconductor.org/packages/release/bioc/html/limma.html | |
| MS-DIAL ver4 | http://prime.psc.riken.jp/ | |
| Adobe Photoshop 2020 | Adobe | |
Highlights.
VAP-A-CERT linkages at ER membrane contact sites drive biogenesis of RNA-containing EVs
A distinct subset of small EVs is enriched in RNA and controlled by VAP-A
VAP-A controls functional transfer of microRNAs to recipient cells
Ceramide transporter CERT localizes to MVBs and controls biogenesis of RNA-containing EVs
ACKNOWLEDGMENTS
We gratefully acknowledge the P01 group and Weaver laboratory for feedback, including Renee Dawson and Andrew Dixson. Funding was provided by NIH grants P01CA229123, U19CA179514, and NSF-2036809, and the Chan Zuckerberg Initiative Imaging Scientists Program. Core facility usage (electron microscopy (Vanderbilt Cell Imaging Shared Resource) and lipid mass spectrometry (Vanderbilt Mass Spectrometry Core)) was supported in part by vouchers from Vanderbilt CTSA grant UL1 RR024975 and UL1 TR002243 and by NIH support of those facilities.
Footnotes
DECLARATION OF INTERESTS: The authors declare no competing interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Abels ER, Maas SLN, Nieland L, Wei Z, Cheah PS, Tai E, Kolsteeg CJ, Dusoswa SA, Ting DT, Hickman S, et al. (2019). Glioblastoma-Associated Microglia Reprogramming Is Mediated by Functional Transfer of Extracellular miR-21. Cell Rep 28, 3105–3119 e3107. 10.1016/j.celrep.2019.08.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albacete-Albacete L, Navarro-Lerida I, Lopez JA, Martin-Padura I, Astudillo AM, Ferrarini A, Van-Der-Heyden M, Balsinde J, Orend G, Vazquez J, and Del Pozo MA (2020). ECM deposition is driven by caveolin-1-dependent regulation of exosomal biogenesis and cargo sorting. J Cell Biol 219. 10.1083/jcb.202006178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alpy F, Rousseau A, Schwab Y, Legueux F, Stoll I, Wendling C, Spiegelhalter C, Kessler P, Mathelin C, Rio MC, et al. (2013). STARD3 or STARD3NL and VAP form a novel molecular tether between late endosomes and the ER. J Cell Sci 126, 5500–5512. 10.1242/jcs.139295. [DOI] [PubMed] [Google Scholar]
- Baietti MF, Zhang Z, Mortier E, Melchior A, Degeest G, Geeraerts A, Ivarsson Y, Depoortere F, Coomans C, Vermeiren E, et al. (2012). Syndecan-syntenin-ALIX regulates the biogenesis of exosomes. Nat Cell Biol 14, 677–685. 10.1038/ncb2502. [DOI] [PubMed] [Google Scholar]
- Barman B, and Bhattacharyya SN (2015). mRNA Targeting to Endoplasmic Reticulum Precedes Ago Protein Interaction and MicroRNA (miRNA)-mediated Translation Repression in Mammalian Cells. The Journal of biological chemistry 290, 24650–24656. 10.1074/jbc.C115.661868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell E, and Taylor MA (2017). Functional Roles for Exosomal MicroRNAs in the Tumour Microenvironment. Comput Struct Biotechnol J 15, 8–13. 10.1016/j.csbj.2016.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianco F, Perrotta C, Novellino L, Francolini M, Riganti L, Menna E, Saglietti L, Schuchman EH, Furlan R, Clementi E, et al. (2009). Acid sphingomyelinase activity triggers microparticle release from glial cells. EMBO J 28, 1043–1054. 10.1038/emboj.2009.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bose M, Barman B, Goswami A, and Bhattacharyya SN (2017). Spatiotemporal Uncoupling of MicroRNA-Mediated Translational Repression and Target RNA Degradation Controls MicroRNP Recycling in Mammalian Cells. Mol Cell Biol 37. 10.1128/MCB.00464-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bukong TN, Momen-Heravi F, Kodys K, Bala S, and Szabo G (2014). Exosomes from hepatitis C infected patients transmit HCV infection and contain replication competent viral RNA in complex with Ago2-miR122-HSP90. PLoS Pathog 10, e1004424. 10.1371/journal.ppat.1004424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cha DJ, Franklin JL, Dou Y, Liu Q, Higginbotham JN, Demory Beckler M, Weaver AM, Vickers K, Prasad N, Levy S, et al. (2015). KRAS-dependent sorting of miRNA to exosomes. Elife 4, e07197. 10.7554/eLife.07197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CJ, Servant N, Toedling J, Sarazin A, Marchais A, Duvernois-Berthet E, Cognat V, Colot V, Voinnet O, Heard E, et al. (2012). ncPRO-seq: a tool for annotation and profiling of ncRNAs in sRNA-seq data. Bioinformatics 28, 3147–3149. 10.1093/bioinformatics/bts587. [DOI] [PubMed] [Google Scholar]
- Chen HJ, Anagnostou G, Chai A, Withers J, Morris A, Adhikaree J, Pennetta G, and de Belleroche JS (2010). Characterization of the properties of a novel mutation in VAPB in familial amyotrophic lateral sclerosis. The Journal of biological chemistry 285, 40266–40281. 10.1074/jbc.M110.161398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Tang Y, Liu Y, Zhang P, Lv L, Zhang X, Jia L, and Zhou Y (2019). Exosomes derived from miR-375-overexpressing human adipose mesenchymal stem cells promote bone regeneration. Cell Prolif 52, e12669. 10.1111/cpr.12669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chevillet JR, Kang Q, Ruf IK, Briggs HA, Vojtech LN, Hughes SM, Cheng HH, Arroyo JD, Meredith EK, Gallichotte EN, et al. (2014). Quantitative and stoichiometric analysis of the microRNA content of exosomes. Proc Natl Acad Sci U S A 111, 14888–14893. 10.1073/pnas.1408301111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiou NT, Kageyama R, and Ansel KM (2018). Selective Export into Extracellular Vesicles and Function of tRNA Fragments during T Cell Activation. Cell Rep 25, 3356–3370 e3354. 10.1016/j.celrep.2018.11.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow FW, Koutsovoulos G, Ovando-Vazquez C, Neophytou K, Bermudez-Barrientos JR, Laetsch DR, Robertson E, Kumar S, Claycomb JM, Blaxter M, et al. (2019). Secretion of an Argonaute protein by a parasitic nematode and the evolution of its siRNA guides. Nucleic Acids Res 47, 3594–3606. 10.1093/nar/gkz142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clancy JW, Zhang Y, Sheehan C, and D'Souza-Schorey C (2019). An ARF6-Exportin-5 axis delivers pre-miRNA cargo to tumour microvesicles. Nat Cell Biol 21, 856–866. 10.1038/s41556-019-0345-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crescitelli R, Lasser C, Szabo TG, Kittel A, Eldh M, Dianzani I, Buzas EI, and Lotvall J (2013). Distinct RNA profiles in subpopulations of extracellular vesicles: apoptotic bodies, microvesicles and exosomes. J Extracell Vesicles 2. 10.3402/jev.v2i0.20677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Candia P, De Rosa V, Casiraghi M, and Matarese G (2016). Extracellular RNAs: A Secret Arm of Immune System Regulation. The Journal of biological chemistry 291, 7221–7228. 10.1074/jbc.R115.708842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Vos KJ, Morotz GM, Stoica R, Tudor EL, Lau KF, Ackerley S, Warley A, Shaw CE, and Miller CC (2012). VAPB interacts with the mitochondrial protein PTPIP51 to regulate calcium homeostasis. Hum Mol Genet 21, 1299–1311. 10.1093/hmg/ddr559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demory Beckler M, Higginbotham JN, Franklin JL, Ham AJ, Halvey PJ, Imasuen IE, Whitwell C, Li M, Liebler DC, and Coffey RJ (2013). Proteomic analysis of exosomes from mutant KRAS colon cancer cells identifies intercellular transfer of mutant KRAS. Mol Cell Proteomics 12, 343–355. 10.1074/mcp.M112.022806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng X, Ruan H, Zhang X, Xu X, Zhu Y, Peng H, Zhang X, Kong F, and Guan M (2020). Long noncoding RNA CCAL transferred from fibroblasts by exosomes promotes chemoresistance of colorectal cancer cells. Int J Cancer 146, 1700–1716. 10.1002/ijc.32608. [DOI] [PubMed] [Google Scholar]
- Di Mattia T, Wilhelm LP, Ikhlef S, Wendling C, Spehner D, Nomine Y, Giordano F, Mathelin C, Drin G, Tomasetto C, and Alpy F (2018). Identification of MOSPD2, a novel scaffold for endoplasmic reticulum membrane contact sites. EMBO Rep 19. 10.15252/embr.201745453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dikic I, and Elazar Z (2018). Mechanism and medical implications of mammalian autophagy. Nat Rev Mol Cell Biol 19, 349–364. 10.1038/s41580-018-0003-4. [DOI] [PubMed] [Google Scholar]
- Driedonks TAP, van der Grein SG, Ariyurek Y, Buermans HPJ, Jekel H, Chow FWN, Wauben MHM, Buck AH, t Hoen PAC, and Nolte-'t Hoen ENM (2018). Immune stimuli shape the small noncoding transcriptome of extracellular vesicles released by dendritic cells. Cell Mol Life Sci 75, 3857–3875. 10.1007/s00018-018-2842-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eden ER, Sanchez-Heras E, Tsapara A, Sobota A, Levine TP, and Futter CE (2016). Annexin A1 Tethers Membrane Contact Sites that Mediate ER to Endosome Cholesterol Transport. Dev Cell 37, 473–483. 10.1016/j.devcel.2016.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eden ER, White IJ, Tsapara A, and Futter CE (2010). Membrane contacts between endosomes and ER provide sites for PTP1B-epidermal growth factor receptor interaction. Nat Cell Biol 12, 267–272. 10.1038/ncb2026. [DOI] [PubMed] [Google Scholar]
- Falcone G, Felsani A, and D'Agnano I (2015). Signaling by exosomal microRNAs in cancer. Journal of experimental & clinical cancer research : CR 34, 32. 10.1186/s13046-015-0148-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flynn RA, Pedram K, Malaker SA, Batista PJ, Smith BAH, Johnson AG, George BM, Majzoub K, Villalta PW, Carette JE, and Bertozzi CR (2021). Small RNAs are modified with N-glycans and displayed on the surface of living cells. Cell 184, 3109–3124 e3122. 10.1016/j.cell.2021.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman JR, Dibenedetto JR, West M, Rowland AA, and Voeltz GK (2013). Endoplasmic reticulum-endosome contact increases as endosomes traffic and mature. Mol Biol Cell 24, 1030–1040. 10.1091/mbc.E12-10-0733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukushima M, Dasgupta D, Mauer AS, Kakazu E, Nakao K, and Malhi H (2018). StAR-related lipid transfer domain 11 (STARD11)-mediated ceramide transport mediates extracellular vesicle biogenesis. The Journal of biological chemistry 293, 15277–15289. 10.1074/jbc.RA118.002587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge L, Wilz L, and Schekman R (2015). Biogenesis of autophagosomal precursors for LC3 lipidation from the ER-Golgi intermediate compartment. Autophagy 11, 2372–2374. 10.1080/15548627.2015.1105422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge L, Zhang M, and Schekman R (2014). Phosphatidylinositol 3-kinase and COPII generate LC3 lipidation vesicles from the ER-Golgi intermediate compartment. Elife 3, e04135. 10.7554/eLife.04135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghamloush F, Ghayad SE, Rammal G, Fahs A, Ayoub AJ, Merabi Z, Harajly M, Zalzali H, and Saab R (2019). The PAX3-FOXO1 oncogene alters exosome miRNA content and leads to paracrine effects mediated by exosomal miR-486. Sci Rep 9, 14242. 10.1038/s41598-019-50592-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh S, Dellibovi-Ragheb TA, Kerviel A, Pak E, Qiu Q, Fisher M, Takvorian PM, Bleck C, Hsu VW, Fehr AR, et al. (2020). beta-Coronaviruses Use Lysosomes for Egress Instead of the Biosynthetic Secretory Pathway. Cell 183, 1520–1535 e1514. 10.1016/j.cell.2020.10.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghossoub R, Lembo F, Rubio A, Gaillard CB, Bouchet J, Vitale N, Slavik J, Machala M, and Zimmermann P (2014). Syntenin-ALIX exosome biogenesis and budding into multivesicular bodies are controlled by ARF6 and PLD2. Nat Commun 5, 3477. 10.1038/ncomms4477. [DOI] [PubMed] [Google Scholar]
- Gibbings DJ, Ciaudo C, Erhardt M, and Voinnet O (2009). Multivesicular bodies associate with components of miRNA effector complexes and modulate miRNA activity. Nat Cell Biol 11, 1143–1149. 10.1038/ncb1929. [DOI] [PubMed] [Google Scholar]
- Guo H, Chitiprolu M, Roncevic L, Javalet C, Hemming FJ, Trung MT, Meng L, Latreille E, Tanese de Souza C, McCulloch D, et al. (2017). Atg5 Disassociates the V1V0-ATPase to Promote Exosome Production and Tumor Metastasis Independent of Canonical Macroautophagy. Dev Cell 43, 716–730 e717. 10.1016/j.devcel.2017.11.018. [DOI] [PubMed] [Google Scholar]
- Hanada K, Kumagai K, Yasuda S, Miura Y, Kawano M, Fukasawa M, and Nishijima M (2003). Molecular machinery for non-vesicular trafficking of ceramide. Nature 426, 803–809. 10.1038/nature02188. [DOI] [PubMed] [Google Scholar]
- Hentze MW, Castello A, Schwarzl T, and Preiss T (2018). A brave new world of RNA-binding proteins. Nat Rev Mol Cell Biol 19, 327–341. 10.1038/nrm.2017.130. [DOI] [PubMed] [Google Scholar]
- Hinger SA, Cha DJ, Franklin JL, Higginbotham JN, Dou Y, Ping J, Shu L, Prasad N, Levy S, Zhang B, et al. (2018). Diverse Long RNAs Are Differentially Sorted into Extracellular Vesicles Secreted by Colorectal Cancer Cells. Cell Rep 25, 715–725 e714. 10.1016/j.celrep.2018.09.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland WL, and Summers SA (2008). Sphingolipids, insulin resistance, and metabolic disease: new insights from in vivo manipulation of sphingolipid metabolism. Endocr Rev 29, 381–402. 10.1210/er.2007-0025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong F, Mohammad Rachidi S, Lundgren D, Han D, Huang X, Zhao H, Kimura Y, Hirano H, Ohara O, Udono H, et al. (2017). Mapping the Interactome of a Major Mammalian Endoplasmic Reticulum Heat Shock Protein 90. PLoS One 12, e0169260. 10.1371/journal.pone.0169260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James C, and Kehlenbach RH (2021). The Interactome of the VAP Family of Proteins: An Overview. Cells 10. 10.3390/cells10071780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen M, Ohsaki Y, Rega LR, Bittman R, Olkkonen VM, and Ikonen E (2011). Role of ORPs in sterol transport from plasma membrane to ER and lipid droplets in mammalian cells. Traffic 12, 218–231. 10.1111/j.1600-0854.2010.01142.x. [DOI] [PubMed] [Google Scholar]
- Jeppesen DK, Fenix AM, Franklin JL, Higginbotham JN, Zhang Q, Zimmerman LJ, Liebler DC, Ping J, Liu Q, Evans R, et al. (2019). Reassessment of Exosome Composition. Cell 177, 428–445 e418. 10.1016/j.cell.2019.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalra H, Simpson RJ, Ji H, Aikawa E, Altevogt P, Askenase P, Bond VC, Borras FE, Breakefield X, Budnik V, et al. (2012). Vesiclepedia: a compendium for extracellular vesicles with continuous community annotation. PLoS Biol 10, e1001450. 10.1371/journal.pbio.1001450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamma H, Horiguchi H, Wan L, Matsui M, Fujiwara M, Fujimoto M, Yazawa T, and Dreyfuss G (1999). Molecular characterization of the hnRNP A2/B1 proteins: tissue-specific expression and novel isoforms. Exp Cell Res 246, 399–411. 10.1006/excr.1998.4323. [DOI] [PubMed] [Google Scholar]
- Kirmiz M, Gillies TE, Dickson EJ, and Trimmer JS (2019). Neuronal ER-plasma membrane junctions organized by Kv2-VAP pairing recruit Nir proteins and affect phosphoinositide homeostasis. The Journal of biological chemistry 294, 17735–17757. 10.1074/jbc.RA119.007635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosaka N, Iguchi H, Yoshioka Y, Takeshita F, Matsuki Y, and Ochiya T (2010). Secretory mechanisms and intercellular transfer of microRNAs in living cells. The Journal of biological chemistry 285, 17442–17452. 10.1074/jbc.M110.107821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kossinova OA, Gopanenko AV, Tamkovich SN, Krasheninina OA, Tupikin AE, Kiseleva E, Yanshina DD, Malygin AA, Ven'yaminova AG, Kabilov MR, and Karpova GG (2017). Cytosolic YB-1 and NSUN2 are the only proteins recognizing specific motifs present in mRNAs enriched in exosomes. Biochim Biophys Acta Proteins Proteom 1865, 664–673. 10.1016/j.bbapap.2017.03.010. [DOI] [PubMed] [Google Scholar]
- Kowal J, Arras G, Colombo M, Jouve M, Morath JP, Primdal-Bengtson B, Dingli F, Loew D, Tkach M, and Thery C (2016). Proteomic comparison defines novel markers to characterize heterogeneous populations of extracellular vesicle subtypes. Proc Natl Acad Sci U S A 113, E968–977. 10.1073/pnas.1521230113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai CP, Kim EY, Badr CE, Weissleder R, Mempel TR, Tannous BA, and Breakefield XO (2015). Visualization and tracking of tumour extracellular vesicle delivery and RNA translation using multiplexed reporters. Nat Commun 6, 7029. 10.1038/ncomms8029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lasser C, Shelke GV, Yeri A, Kim DK, Crescitelli R, Raimondo S, Sjostrand M, Gho YS, Van Keuren Jensen K, and Lotvall J (2017). Two distinct extracellular RNA signatures released by a single cell type identified by microarray and next-generation sequencing. RNA Biol 14, 58–72. 10.1080/15476286.2016.1249092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H, Li C, Zhang Y, Zhang D, Otterbein LE, and Jin Y (2019). Caveolin-1 selectively regulates microRNA sorting into microvesicles after noxious stimuli. J Exp Med 216, 2202–2220. 10.1084/jem.20182313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JE, Cathey PI, Wu H, Parker R, and Voeltz GK (2020). Endoplasmic reticulum contact sites regulate the dynamics of membraneless organelles. Science 367. 10.1126/science.aay7108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leidal AM, Huang HH, Marsh T, Solvik T, Zhang D, Ye J, Kai F, Goldsmith J, Liu JY, Huang YH, et al. (2020). The LC3-conjugation machinery specifies the loading of RNA-binding proteins into extracellular vesicles. Nat Cell Biol 22, 187–199. 10.1038/s41556-019-0450-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li K, Wong DK, Hong KY, and Raffai RL (2018). Cushioned-Density Gradient Ultracentrifugation (C-DGUC): A Refined and High Performance Method for the Isolation, Characterization, and Use of Exosomes. Methods Mol Biol 1740, 69–83. 10.1007/978-1-4939-7652-2_7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Zhou X, Wei M, Gao X, Zhao L, Shi R, Sun W, Duan Y, Yang G, and Yuan L (2019). In Vitro and in Vivo RNA Inhibition by CD9-HuR Functionalized Exosomes Encapsulated with miRNA or CRISPR/dCas9. Nano Lett 19, 19–28. 10.1021/acs.nanolett.8b02689. [DOI] [PubMed] [Google Scholar]
- Lin F, Zeng Z, Song Y, Li L, Wu Z, Zhang X, Li Z, Ke X, and Hu X (2019). YBX-1 mediated sorting of miR-133 into hypoxia/reoxygenation-induced EPC-derived exosomes to increase fibroblast angiogenesis and MEndoT. Stem Cell Res Ther 10, 263. 10.1186/s13287-019-1377-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, and Anders S (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15, 550. 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu P, Li H, Li N, Singh RN, Bishop CE, Chen X, and Lu B (2017). MEX3C interacts with adaptor-related protein complex 2 and involves in miR-451a exosomal sorting. PLoS One 12, e0185992. 10.1371/journal.pone.0185992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucero R, Zappulli V, Sammarco A, Murillo OD, Cheah PS, Srinivasan S, Tai E, Ting DT, Wei Z, Roth ME, et al. (2020). Glioma-Derived miRNA-Containing Extracellular Vesicles Induce Angiogenesis by Reprogramming Brain Endothelial Cells. Cell Rep 30, 2065–2074 e2064. 10.1016/j.celrep.2020.01.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma W, and Mayr C (2018). A Membraneless Organelle Associated with the Endoplasmic Reticulum Enables 3'UTR-Mediated Protein-Protein Interactions. Cell 175, 1492–1506 e1419. 10.1016/j.cell.2018.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maas SL, Breakefield XO, and Weaver AM (2017). Extracellular Vesicles: Unique Intercellular Delivery Vehicles. Trends in cell biology 27, 172–188. 10.1016/j.tcb.2016.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mantel PY, Hjelmqvist D, Walch M, Kharoubi-Hess S, Nilsson S, Ravel D, Ribeiro M, Gruring C, Ma S, Padmanabhan P, et al. (2016). Infected erythrocyte-derived extracellular vesicles alter vascular function via regulatory Ago2-miRNA complexes in malaria. Nat Commun 7, 12727. 10.1038/ncomms12727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maroney PA, Yu Y, Fisher J, and Nilsen TW (2006). Evidence that microRNAs are associated with translating messenger RNAs in human cells. Nat Struct Mol Biol 13, 1102–1107. 10.1038/nsmb1174. [DOI] [PubMed] [Google Scholar]
- Mateescu B, Kowal EJ, van Balkom BW, Bartel S, Bhattacharyya SN, Buzas EI, Buck AH, de Candia P, Chow FW, Das S, et al. (2017). Obstacles and opportunities in the functional analysis of extracellular vesicle RNA - an ISEV position paper. J Extracell Vesicles 6, 1286095. 10.1080/20013078.2017.1286095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto K, Tanaka KJ, and Tsujimoto M (2005). An acidic protein, YBAP1, mediates the release of YB-1 from mRNA and relieves the translational repression activity of YB-1. Mol Cell Biol 25, 1779–1792. 10.1128/MCB.25.5.1779-1792.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenzie AJ, Hoshino D, Hong NH, Cha DJ, Franklin JL, Coffey RJ, Patton JG, and Weaver AM (2016). KRAS-MEK Signaling Controls Ago2 Sorting into Exosomes. Cell Rep 15, 978–987. 10.1016/j.celrep.2016.03.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melo SA, Sugimoto H, O'Connell JT, Kato N, Villanueva A, Vidal A, Qiu L, Vitkin E, Perelman LT, Melo CA, et al. (2014). Cancer exosomes perform cell-independent microRNA biogenesis and promote tumorigenesis. Cancer Cell 26, 707–721. 10.1016/j.ccell.2014.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercier V, Larios J, Molinard G, Goujon A, Matile S, Gruenberg J, and Roux A (2020). Endosomal membrane tension regulates ESCRT-III-dependent intra-lumenal vesicle formation. Nat Cell Biol 22, 947–959. 10.1038/s41556-020-0546-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mesmin B, Bigay J, Moser von Filseck J, Lacas-Gervais S, Drin G, and Antonny B (2013). A four-step cycle driven by PI(4)P hydrolysis directs sterol/PI(4)P exchange by the ER-Golgi tether OSBP. Cell 155, 830–843. 10.1016/j.cell.2013.09.056. [DOI] [PubMed] [Google Scholar]
- Mukherjee K, Ghoshal B, Ghosh S, Chakrabarty Y, Shwetha S, Das S, and Bhattacharyya SN (2016). Reversible HuR-microRNA binding controls extracellular export of miR-122 and augments stress response. EMBO Rep 17, 1184–1203. 10.15252/embr.201541930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neefjes J, and Cabukusta B (2021). What the VAP: The Expanded VAP Family of Proteins Interacting With FFAT and FFAT-Related Motifs for Interorganellar Contact. Contact (Thousand Oaks) 4, 25152564211012246. 10.1177/25152564211012246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura AL, Mitne-Neto M, Silva HC, Richieri-Costa A, Middleton S, Cascio D, Kok F, Oliveira JR, Gillingwater T, Webb J, et al. (2004). A mutation in the vesicle-trafficking protein VAPB causes late-onset spinal muscular atrophy and amyotrophic lateral sclerosis. Am J Hum Genet 75, 822–831. 10.1086/425287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolte-'t Hoen E, Cremer T, Gallo RC, and Margolis LB (2016). Extracellular vesicles and viruses: Are they close relatives? Proc Natl Acad Sci U S A 113, 9155–9161. 10.1073/pnas.1605146113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nottrott S, Simard MJ, and Richter JD (2006). Human let-7a miRNA blocks protein production on actively translating polyribosomes. Nat Struct Mol Biol 13, 1108–1114. 10.1038/nsmb1173. [DOI] [PubMed] [Google Scholar]
- O'Brien K, Breyne K, Ughetto S, Laurent LC, and Breakefield XO (2020). RNA delivery by extracellular vesicles in mammalian cells and its applications. Nat Rev Mol Cell Biol. 10.1038/s41580-020-0251-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogretmen B (2018). Sphingolipid metabolism in cancer signalling and therapy. Nat Rev Cancer 18, 33–50. 10.1038/nrc.2017.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel MR, and Weaver AM (2021). Astrocyte-derived small extracellular vesicles promote synapse formation via fibulin-2-mediated TGF-beta signaling. Cell Rep 34, 108829. 10.1016/j.celrep.2021.108829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pathan M, Fonseka P, Chitti SV, Kang T, Sanwlani R, Van Deun J, Hendrix A, and Mathivanan S (2019). Vesiclepedia 2019: a compendium of RNA, proteins, lipids and metabolites in extracellular vesicles. Nucleic Acids Res 47, D516–D519. 10.1093/nar/gky1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peretti D, Dahan N, Shimoni E, Hirschberg K, and Lev S (2008). Coordinated lipid transfer between the endoplasmic reticulum and the Golgi complex requires the VAP proteins and is essential for Golgi-mediated transport. Mol Biol Cell 19, 3871–3884. 10.1091/mbc.E08-05-0498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry RJ, and Ridgway ND (2006). Oxysterol-binding protein and vesicle-associated membrane protein-associated protein are required for sterol-dependent activation of the ceramide transport protein. Mol Biol Cell 17, 2604–2616. 10.1091/mbc.e06-01-0060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips MJ, and Voeltz GK (2016). Structure and function of ER membrane contact sites with other organelles. Nat Rev Mol Cell Biol 17, 69–82. 10.1038/nrm.2015.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quaresma AJ, Bressan GC, Gava LM, Lanza DC, Ramos CH, and Kobarg J (2009). Human hnRNP Q re-localizes to cytoplasmic granules upon PMA, thapsigargin, arsenite and heat-shock treatments. Exp Cell Res 315, 968–980. 10.1016/j.yexcr.2009.01.012. [DOI] [PubMed] [Google Scholar]
- Raposo G, and Stoorvogel W (2013). Extracellular vesicles: exosomes, microvesicles, and friends. J Cell Biol 200, 373–383. 10.1083/jcb.201211138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, and Smyth GK (2015). limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 43, e47. 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts RL, Barbieri MA, Pryse KM, Chua M, Morisaki JH, and Stahl PD (1999). Endosome fusion in living cells overexpressing GFP-rab5. J Cell Sci 112 (Pt 21), 3667–3675. [DOI] [PubMed] [Google Scholar]
- Rocha N, Kuijl C, van der Kant R, Janssen L, Houben D, Janssen H, Zwart W, and Neefjes J (2009). Cholesterol sensor ORP1L contacts the ER protein VAP to control Rab7-RILP-p150 Glued and late endosome positioning. J Cell Biol 185, 1209–1225. 10.1083/jcb.200811005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero-Brey I, and Bartenschlager R (2016). Endoplasmic Reticulum: The Favorite Intracellular Niche for Viral Replication and Assembly. Viruses 8. 10.3390/v8060160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santangelo L, Giurato G, Cicchini C, Montaldo C, Mancone C, Tarallo R, Battistelli C, Alonzi T, Weisz A, and Tripodi M (2016). The RNA-Binding Protein SYNCRIP Is a Component of the Hepatocyte Exosomal Machinery Controlling MicroRNA Sorting. Cell Rep 17, 799–808. 10.1016/j.celrep.2016.09.031. [DOI] [PubMed] [Google Scholar]
- Schmittgen TD, and Livak KJ (2008). Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc 3, 1101–1108. 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- Shen M, Dong C, Ruan X, Yan W, Cao M, Pizzo D, Wu X, Yang L, Liu L, Ren X, and Wang SE (2019). Chemotherapy-Induced Extracellular Vesicle miRNAs Promote Breast Cancer Stemness by Targeting ONECUT2. Cancer Res 79, 3608–3621. 10.1158/0008-5472.CAN-18-4055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirasawa S, Furuse M, Yokoyama N, and Sasazuki T (1993). Altered growth of human colon cancer cell lines disrupted at activated Ki-ras. Science 260, 85–88. 10.1126/science.8465203. [DOI] [PubMed] [Google Scholar]
- Shurtleff MJ, Temoche-Diaz MM, Karfilis KV, Ri S, and Schekman R (2016). Y-box protein 1 is required to sort microRNAs into exosomes in cells and in a cell-free reaction. Elife 5. 10.7554/eLife.19276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shurtleff MJ, Yao J, Qin Y, Nottingham RM, Temoche-Diaz MM, Schekman R, and Lambowitz AM (2017). Broad role for YBX1 in defining the small noncoding RNA composition of exosomes. Proc Natl Acad Sci U S A 114, E8987–E8995. 10.1073/pnas.1712108114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinha S, Hoshino D, Hong NH, Kirkbride KC, Grega-Larson NE, Seiki M, Tyska MJ, and Weaver AM (2016). Cortactin promotes exosome secretion by controlling branched actin dynamics. J Cell Biol 214, 197–213. 10.1083/jcb.201601025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skog J, Wurdinger T, van Rijn S, Meijer DH, Gainche L, Sena-Esteves M, Curry WT Jr., Carter BS, Krichevsky AM, and Breakefield XO (2008). Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat Cell Biol 10, 1470–1476. 10.1038/ncb1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stalder L, Heusermann W, Sokol L, Trojer D, Wirz J, Hean J, Fritzsche A, Aeschimann F, Pfanzagl V, Basselet P, et al. (2013). The rough endoplasmatic reticulum is a central nucleation site of siRNA-mediated RNA silencing. EMBO J 32, 1115–1127. 10.1038/emboj.2013.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenmark H, Parton RG, Steele-Mortimer O, Lutcke A, Gruenberg J, and Zerial M (1994). Inhibition of rab5 GTPase activity stimulates membrane fusion in endocytosis. EMBO J 13, 1287–1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung BH, and Weaver AM (2017). Exosome secretion promotes chemotaxis of cancer cells. Cell Adh Migr 11, 187–195. 10.1080/19336918.2016.1273307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Temoche-Diaz MM, Shurtleff MJ, Nottingham RM, Yao J, Fadadu RP, Lambowitz AM, and Schekman R (2019). Distinct mechanisms of microRNA sorting into cancer cell-derived extracellular vesicle subtypes. Elife 8. 10.7554/eLife.47544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thery C, Witwer KW, Aikawa E, Alcaraz MJ, Anderson JD, Andriantsitohaina R, Antoniou A, Arab T, Archer F, Atkin-Smith GK, et al. (2018). Minimal information for studies of extracellular vesicles 2018 (MISEV2018): a position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J Extracell Vesicles 7, 1535750. 10.1080/20013078.2018.1535750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thul PJ, Akesson L, Wiking M, Mahdessian D, Geladaki A, Ait Blal H, Alm T, Asplund A, Bjork L, Breckels LM, et al. (2017). A subcellular map of the human proteome. Science 356. 10.1126/science.aal3321. [DOI] [PubMed] [Google Scholar]
- Trajkovic K, Hsu C, Chiantia S, Rajendran L, Wenzel D, Wieland F, Schwille P, Brugger B, and Simons M (2008). Ceramide triggers budding of exosome vesicles into multivesicular endosomes. Science 319, 1244–1247. 10.1126/science.1153124. [DOI] [PubMed] [Google Scholar]
- Tsugawa H, Ikeda K, Takahashi M, Satoh A, Mori Y, Uchino H, Okahashi N, Yamada Y, Tada I, Bonini P, et al. (2020). A lipidome atlas in MS-DIAL 4. Nat Biotechnol 38, 1159–1163. 10.1038/s41587-020-0531-2. [DOI] [PubMed] [Google Scholar]
- Valadi H, Ekstrom K, Bossios A, Sjostrand M, Lee JJ, and Lotvall JO (2007). Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol 9, 654–659. 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- van Niel G, D'Angelo G, and Raposo G (2018). Shedding light on the cell biology of extracellular vesicles. Nat Rev Mol Cell Biol 19, 213–228. 10.1038/nrm.2017.125. [DOI] [PubMed] [Google Scholar]
- Villarroya-Beltri C, Gutierrez-Vazquez C, Sanchez-Cabo F, Perez-Hernandez D, Vazquez J, Martin-Cofreces N, Martinez-Herrera DJ, Pascual-Montano A, Mittelbrunn M, and Sanchez-Madrid F (2013). Sumoylated hnRNPA2B1 controls the sorting of miRNAs into exosomes through binding to specific motifs. Nat Commun 4, 2980. 10.1038/ncomms3980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber-Boyvat M, Kentala H, Lilja J, Vihervaara T, Hanninen R, Zhou Y, Peranen J, Nyman TA, Ivaska J, and Olkkonen VM (2015). OSBP-related protein 3 (ORP3) coupling with VAMP-associated protein A regulates R-Ras activity. Exp Cell Res 331, 278–291. 10.1016/j.yexcr.2014.10.019. [DOI] [PubMed] [Google Scholar]
- Wegner CS, Malerod L, Pedersen NM, Progida C, Bakke O, Stenmark H, and Brech A (2010). Ultrastructural characterization of giant endosomes induced by GTPase-deficient Rab5. Histochem Cell Biol 133, 41–55. 10.1007/s00418-009-0643-8. [DOI] [PubMed] [Google Scholar]
- Wozniak AL, Adams A, King KE, Dunn W, Christenson LK, Hung WT, and Weinman SA (2020). The RNA binding protein FMR1 controls selective exosomal miRNA cargo loading during inflammation. J Cell Biol 219. 10.1083/jcb.201912074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Carvalho P, and Voeltz GK (2018). Here, there, and everywhere: The importance of ER membrane contact sites. Science 361. 10.1126/science.aan5835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying W, Riopel M, Bandyopadhyay G, Dong Y, Birmingham A, Seo JB, Ofrecio JM, Wollam J, Hernandez-Carretero A, Fu W, et al. (2017). Adipose Tissue Macrophage-Derived Exosomal miRNAs Can Modulate In Vivo and In Vitro Insulin Sensitivity. Cell 171, 372–384 e312. 10.1016/j.cell.2017.08.035. [DOI] [PubMed] [Google Scholar]
- Yoshida T, Akatsuka T, and Imanaka-Yoshida K (2015). Tenascin-C and integrins in cancer. Cell Adh Migr 9, 96–104. 10.1080/19336918.2015.1008332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Freitas D, Kim HS, Fabijanic K, Li Z, Chen H, Mark MT, Molina H, Martin AB, Bojmar L, et al. (2018). Identification of distinct nanoparticles and subsets of extracellular vesicles by asymmetric flow field-flow fractionation. Nat Cell Biol 20, 332–343. 10.1038/s41556-018-0040-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao YG, and Zhang H (2019). Autophagosome maturation: An epic journey from the ER to lysosomes. J Cell Biol 218, 757–770. 10.1083/jcb.201810099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zietzer A, Hosen MR, Wang H, Goody PR, Sylvester M, Latz E, Nickenig G, Werner N, and Jansen F (2020). The RNA-binding protein hnRNPU regulates the sorting of microRNA-30c-5p into large extracellular vesicles. J Extracell Vesicles 9, 1786967. 10.1080/20013078.2020.1786967. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data S1: microRNA data from small RNA-Seq data for control, VAP-A KD1, VAP-A KD2 cells, small EVs and large EVs. Related to Figure 2.
Data S2: snoRNA data from small RNA-Seq data for control, VAP-A KD1, VAP-A KD2 cells, small EVs and large EVs. Related to Figure 2 and Figure S3.
Data S3: tRNA data from small RNA-Seq data for control, VAP-A KD1, VAP-A KD2 cells, small EVs and large EVs. Related to Figure 2 and Figure S3.
Data S4: Untargeted lipid mass spectrometry data for control, VAP-A KD1, VAP-A KD2 cells, small EVs and large EVs. Related to Figure 6.
Table S1: Curated data for RNA binding proteins (RBPs). Related to Figure 1.
List of 61 RBPs shown in Figure 1A overlapped with endoplasmic reticulum and EV proteomes; 22 RBPs shown in Figure 1B overlapped with endoplasmic reticulum proteome.
Video S1. Intraluminal vesicle formation at an ER membrane contact site. Related to Figure 1.
Video shows a tomographic reconstruction of an MVB/ER contact site observed in a DKs-8 (Sc) cell (see also Figure 1H). Three-dimensional segmentations of organelles depict MVB (light green), intraluminal vesicles (ILVs) (dark blue), and ER tubules (purple). Note the presence of an ILV still connected to the MVB limiting membrane at the ER contact site.
Video S2. VAP-A-positive ER connects nSMase2-positive vesicles to an MVB. Related to Figure 7.
GFP-VAP-A- and mCherry-Rab5Q79L-cotransfected DKs-8 cell with nSMase2 immunofluorescence staining was deconvolved. Zooming effect is shown at 250 nm Z-step (see Figure 7J). The selected area is converted to a 3D structure reconstructed with 650 nm thickness. An arrow indicates GFP-VAP-A penetration into the lumen of mCherry-Rab5Q79L-labeled MVB. An arrowhead indicates the presence of nSMase2 at the ER-MVB contact site.