Abstract
Background
Accurate detection of arrhythmic events in the intensive care units (ICU) is of paramount significance in providing timely care. However, traditional ICU monitors generate a high rate of false alarms causing alarm fatigue. In this work, we develop an algorithm to improve life threatening arrhythmia detection in the ICUs using a deep learning approach.
Methods and Results
This study involves a total of 953 independent life‐threatening arrhythmia alarms generated from the ICU bedside monitors of 410 patients. Specifically, we used the ECG (4 channels), arterial blood pressure, and photoplethysmograph signals to accurately detect the onset and offset of various arrhythmias, without prior knowledge of the alarm type. We used a hybrid convolutional neural network based classifier that fuses traditional handcrafted features with features automatically learned using convolutional neural networks. Further, the proposed architecture remains flexible to be adapted to various arrhythmic conditions as well as multiple physiological signals. Our hybrid‐ convolutional neural network approach achieved superior performance compared with methods which only used convolutional neural network. We evaluated our algorithm using 5‐fold cross‐validation for 5 times and obtained an accuracy of 87.5%±0.5%, and a score of 81%±0.9%. Independent evaluation of our algorithm on the publicly available PhysioNet 2015 Challenge database resulted in overall classification accuracy and score of 93.9% and 84.3%, respectively, indicating its efficacy and generalizability.
Conclusions
Our method accurately detects multiple arrhythmic conditions. Suitable translation of our algorithm may significantly improve the quality of care in ICUs by reducing the burden of false alarms.
Keywords: convolutional neural networks, false alarms, intensive care unit monitors, machine learning, multi‐class classification
Subject Categories: Arrhythmias, Quality and Outcomes
Nonstandard Abbreviations and Acronyms
- CNN
convolutional neural network
- EB
extreme bradycardia
- ET
extreme tachycardia
- FA
false alarm
- SR
sinus rhythm
Clinical Perspective
What Is New?
We proposed a novel arrhythmia detection algorithm that uses handcrafted features along with features learned from a machine learning algorithm.
While conventional intensive care unit monitors use single physiological signals to raise an alarm, our algorithm uses information from multiple physiological signals simultaneously to reduce the incidence of false alarms in the intensive care unit.
What Are the Clinical Implications?
Deployment of the proposed method could reduce the number of false intensive care unit alarms, making the organization, integration, and interpretation of the enormous amount of intensive care unit data less time‐consuming and more efficient.
Intensive care units (ICUs) are generally equipped with physiological monitoring systems to alert the caregivers about the onset of an adverse condition. However, the majority of such alarms are triggered because of innocuous conditions such as motion artifacts and electrode problems. Although such a system generally does not miss many true alarms, it generates false alarms (FAs) at rates as high as 88.8%. 1 , 2 Frequent FAs can cause delirium in patients and also gradually impact the responsiveness of the staff to alarms. Indeed, reducing the harm associated with clinical alarm systems has been consistently listed as a National Patient Safety Goal from 2012 to 2020. 3
Various algorithms have been developed to address FAs in ICUs. Early attempts used only ECG signals to alert the onset of an arrhythmic condition. 4 , 5 Recent algorithms that included the arterial blood pressure (BP) signal with the ECG signals have reported significant reduction in FA burden. 6 , 7 , 8 , 9 Algorithms based on wavelet transform, data mining, and machine learning approaches have been reported to further reduce FAs. 10 , 11 , 12 , 13 To foster the development of such algorithms, the “PhysioNet/Computing in Cardiology Challenge 2015: Reducing False Arrhythmia Alarms in the ICU” was introduced, especially for the scenario where prior knowledge of the alarm event is available. In response to the challenge, many time domain and frequency domain techniques were proposed. 14 , 15 , 16 , 17 , 18 Although such methods achieve impressive performance when the alarm type is known, detecting the presence of an unknown arrhythmia remains challenging. Consequently, such methods remain as an add‐on to existing monitoring systems to filter FAs. However, it has been observed that the alarm type on the monitor and the underlying arrhythmia may sometimes mismatch and hence alarm suppression based on a mismatched arrhythmia type can result in catastrophic consequences.
In the same vein, the “China Physiological Signal Challenge 2018” 19 and the recent “PhysioNet/Computing in Cardiology Challenge 2020: Classification of 12‐lead ECGs”, provided a large repository of 12‐lead ECG recordings from various databases, with the goal of identifying the clinical diagnosis. In such databases, although an annotation is provided for the entire record, the actual onset and offset of the arrhythmia remain unavailable. Furthermore, the aforementioned databases include only the ECG signals for analysis. It is important to note that certain life‐threatening conditions including ventricular tachycardia/ventricular fibrillation (VT/VF) can be better diagnosed if other vital‐sign signals such as arterial BP and photoplethysmogram are included.
To date, most reported methods have relied in processing handpicked features that are tailored to the signals and the arrhythmia at hand. Recently developed algorithms have reported significant improvement in performance by using automated feature learning with deep learning methods. 20 , 21 , 22 A method proposed by Hannun et al 23 used a deep convolutional neural network (CNN) architecture to detect twelve arrhythmia types from a single ECG recorded from an ambulatory device. Methods that combine CNNs with recurrent neural networks have been reported to achieve improved performance on ECG classification. 24 , 25 , 26 However, these methods do not classify certain life‐threatening arrhythmias including extreme bradycardia (EB), extreme tachycardia (ET), asystole and VF.
Given that deep learning methods, in particular CNNs have been proven effective and the preferred tools for various classification tasks, 27 in this study we propose to use a hybrid‐CNN technique that fuses conventional handcrafted features with the features learned from CNN. Such a network, when appropriately trained, is expected to enjoy the benefits of automated learning as well as traditional features. Furthermore, the proposed approach is expected to be suitable for different arrhythmias, without requiring major architectural changes.
Methods
Data Availability
The training data set will be available to any investigator upon request.
Code Availability
The code will be available to any investigator upon request.
Ethical Approval and Consent to Participate
The study was approved by the Institutional Review Board of Massachusetts General Hospital. We also used data from the “PhysioNet/Computing in Cardiology Challenge 2015: Reducing False Arrhythmia Alarms in the ICU”. The data used are open source and are available at https://physionet.org/content/challenge‐2015/1.0.0/.
Data Set
The study was approved by the Institutional Review Board of Massachusetts General Hospital. Adhering to the institutional review board guidelines, we obtained de‐identified data from the bedside monitors of the ICUs of Massachusetts General Hospital. The data consist of the ECG (4 channels), arterial BP, and photoplethysmogram waveforms recorded using 2 different device manufacturers. We used streaming data with the information corresponding to the time and type of the alarm recorded by the monitoring system.
The Association for the Advancement of Medical Instrumentation standards require that the alarm be raised within 10 seconds from the start of the event. Accordingly, we created a running 5‐second window buffer for the 15 seconds of data before the onset of the alarm. We considered only alarms corresponding to asystole, extreme tachycardia (ET), extreme bradycardia (EB), ventricular tachycardia (VT), ventricular fibrillation (VF). and atrial fibrillation (AF). The definitions/criteria for each arrhythmia were taken from the PhysioNet 2015 challenge, 16 and are listed in Table 1. Further, we used the open source data from the “PhysioNet/Computing in Cardiology Challenge 2015: Reducing False Arrhythmia Alarms in the ICU”, 16 as an independent data set to evaluate the proposed algorithm. The data are available at https://physionet.org/content/challenge‐2015/1.0.0/. The data consist of 300 seconds long records of 2‐lead ECG, BP, and photoplethysmogram signals before an alarm, as acquired by the bedside ICU monitors. The data also contain the corresponding annotations for each alarm, namely the alarm type and whether the alarm is true or false. We considered ≥15 second windows from each record and annotated it to mark the onset and offset of each arrhythmia (please, see Data S1 for details).
Table 1.
Definitions of the 7 Classes
| Class | Definition |
|---|---|
| Aystole | No heartbeats at all for a period of 4 s or more |
| EB | Heart rate is lower than 40 beats per minute; fewer than 5 beats occur within a period of 6 s |
| ET | Heart rate is higher than 140 beats per min; more than 17 beats occur within a period of 6.85 s |
| VF | A rapid fibrillatory, flutter, or oscillatory waveform for at least 4 s |
| VT | Five or more consecutive ventricular beats within a period of 2.4 s (a heart rate of 100 beats per min) |
| SR | Heart rate between 40 and 100 beats per min, for 8 s |
| AF | Tachyarrhythmia characterized by predominantly uncoordinated atrial activation with consequent deterioration of atrial mechanical function |
AF indicates atrial fibrillation; EB, extreme bradycardia; ET, extreme tachycardia; SR, sinus rhythm; VF, ventricular fibrillation; and VT, ventricular tachycardia.
Human Annotations: Ground Truth and Arrhythmia Onset and Offset
We built a custom‐made user interface to mark the onset and offset of noise portions and those of arrhythmia, corresponding to aystole, EB, ET, VF, and VT in each channel of ECG, BP, and photoplethysmogram signals. Based on the arrhythmia definitions/criteria listed in Table 1, the onset and offset of each arrhythmia record has been manually annotated by 2 experts. For complex cases, a third expert independently reviewed the record, and the majority view was used as the final annotation. Finally, we annotated noise and sinus rhythm (SR) portions in each of the channels of ECG, BP, and photoplethysmogram waveforms for further training and validations of our algorithms. Specifically, we annotated 953 independent alarms from 410 critical care subjects with diverse medical conditions. It has been observed that the alarm type indicated by the monitor may not correspond with the true alarm type. Table 2 provides the number of alarms raised for each event and their correspondence to the true underlying condition. About 50% of VF alarms that required immediate attention corresponded with a different arrhythmia type. Favorably, 80% of these mismatched alarms corresponded with VT. However, surprisingly, 22.6% of the EB alarms that require immediate attention corresponded with ET. Therefore, it becomes imperative to develop standalone arrhythmia detectors without considering the monitor alarm type.
Table 2.
Number of Records in Each Alarm Type and Their Correspondence to the Gold Standard
| Alarm annotation by clinicians | Total records | PPV | % of mismatched true alarms | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Aystole | EB | ET | VF | VT | AF | SR | |||||
| Alarm type from monitor | Aystole | 19 | 2 | 2 | 0 | 4 | 18 | 123 | 168 | 11.31 | 29.63 |
| EB | 0 | 60 | 7 | 0 | 0 | 66 | 132 | 265 | 22.64 | 10.45 | |
| ET | 0 | 1 | 39 | 0 | 3 | 96 | 89 | 228 | 17.11 | 9.3 | |
| VF | 0 | 0 | 2 | 10 | 8 | 5 | 21 | 46 | 21.74 | 50 | |
| VT | 0 | 1 | 5 | 3 | 132 | 26 | 79 | 246 | 53.66 | 6.38 | |
| Total records | 19 | 64 | 55 | 13 | 147 | 211 | 444 | 953 | |||
AF indicates atrial fibrillation; EB, extreme bradycardia; ET, extreme tachycardia; PPV, positive predictive value; SR, sinus rhythm; VF, ventricular fibrillation; and VT, ventricular tachycardia.
Based on the gold standard annotations indicating the onset and offset of each arrhythmia, we derived the ground truth labels of the 4‐ and 8‐second windows, as follows: A 4‐second window is marked as VT if there are at least 5 beats meeting the VT criteria. In particular, the onset of VT is determined by the approximate midpoint between the first ventricular beat and the previous sinus beat. In the same vein, the offset is determined by the approximate midpoint between the last ventricular beat and the next sinus beat or the offset is considered as the end of the record if the arrhythmia persists until the end of the record. Similarly, the window is marked as VF only if the entire 4‐second duration is marked as VF by the clinician. An 8‐second window is considered as EB if the heart rate is <40 beats per minute in the given window. While the window is considered as ET, if at least 17 beats occur in the window with heart rate >140 beats per minute. Finally the window is considered as AF or SR if the entire window falls under the AF and SR criteria, respectively.
Performance Metrics
To report the performance of the classifier, along with the overall accuracy, we report the percent score (inspired from PhysioNet/Computing in Cardiology 2015 challenge) that heavily penalizes false negatives, to reduce or eliminate the lack of emergency care during a life‐threatening event. The classifier’s accuracy and score values are calculated as:
where, |TP|, |TN|, |FP|, and |FN| refer to the counts of true positives, true negatives, false positives, and false negatives, respectively. Contrary to accuracy, the score value weighs the FNs, 5 times more than the FPs. We also report the classifier’s sensitivity and positive predictive value , in each window (4 or 8 seconds), respectively.
To estimate the performance of the proposed method on unseen data, we performed k‐fold cross‐validation (with stratified random sampling) on the available data. We assessed the 5‐fold cross‐validation performance of our method, and then took the sum performance across all the folds to represent the classifier performance on the entire data. Further, to account for the randomness associated with model training and stochastic selection of k‐folds, we repeated the 5‐fold cross validation, 5 times, and reported the mean and SD of the classifier performance.
Data Preprocessing
Baseline‐wander was corrected using median filters of window sizes 200 and 600 milliseconds, respectively. 28 Next, R‐wave peak locations were identified using an R‐wave detector that fuses information from multiple channels of ECG. 12 We excluded, across all channels, portions of records of ECG signals with amplitude <0.5 mV as being of low quality (normally, an alarm should be raised that would require lead repositioning to improve contact), resulting to a total of 1262.2 seconds of data being removed from the analysis. After data exclusion, the remaining 13032.8 seconds of data were used in the analysis. The proportion of data within each class were, SR=44.82%, AF=23.27%, VT=16.57%, VF=1.63%, EB=6.2%, ET=6.57%, and asystole=0.93%.
Block Schematic for Arrhythmia Detectors
In general, the majority of false alarms are attributed to noise, artifacts, or connection problems in ≥1 channels/signals. Accordingly, the first step is to identify such conditions. To this end, we first pass each signal through a noise detector and identify the signals that are corrupted with noise.
Next, depending on the arrhythmia type, we use either a window length of 4 or 8 seconds, respectively. Specifically, arrhythmias such as asystole, VT, and VF can be detected based only on 4 seconds of data, while EB, ET, AF, and SR require 8 seconds of data. As shown in Figure 1, we use a multi‐tiered approach for arrhythmia detection. In particular, noise in each channel of ECG, BP, and photoplethysmogram is identified in Tier‐0, and only those channels that are non‐noisy are passed to the later stages. VT and VF are identified in Tier‐1, ET and EB are identified in Tier‐2, and finally Tier‐3 distinguishes between AF and SR signals. At each tier, we use a classifier that is specific to the arrhythmia under consideration. In general, such classifiers are usually based on the handpicked arrhythmia dependent futures. As suggested earlier, we propose a hybrid‐CNN based generalizable building block that can be trained to serve as a classifier at different tiers.
Figure 1. Block schematic of the proposed classifier.
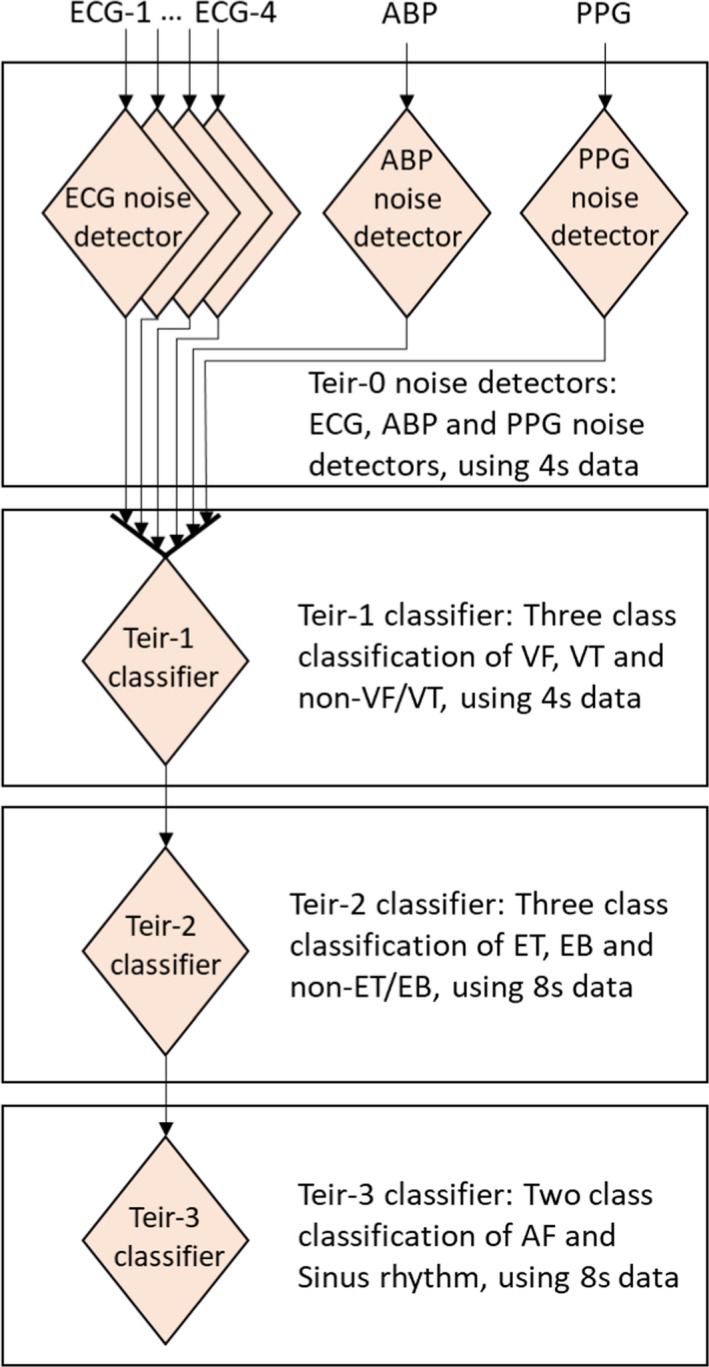
ABP indicates arterial blood pressure; AF, atrial fibrillation; EB, extreme bradycardia; ET, extreme tachycardia; PPG, photoplethysmogram; VF, ventricular fibrillation; and VT, ventricular tachycardia.
Hybrid Architecture as a Building Block
We consider a hybrid‐CNN architecture comprising multiple convolutional layers and a fully connected layer with handcrafted features augmented before the output layer, as shown in Figure 2.
Figure 2. Hybrid‐ convolutional neural network architecture that fuses the information from learned and handcrafted features.
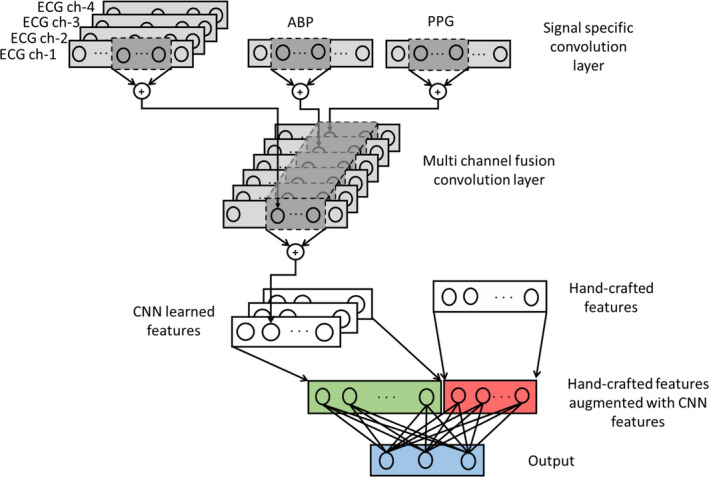
ABP indicates arterial blood pressure; CNN, convolutional neural network; and PPG, photoplethysmogram.
Handcrafted Features
We extracted a set of signal specific features from each ECG channel, as well as the BP and photoplethysmogram signals, to determine if the corresponding signal is noisy. Furthermore, a set of arrhythmia‐specific features, which characterize each arrhythmia class, were also extracted. 12 Details of these features are made available in the Data S1 (Tables S1 through S4). Missing ECG feature values were replaced with the average feature value from non‐noisy channels of ECG. Whereas for BP and photoplethysmogram features, NaN values in the training and test set are replaced with the median value of the corresponding non‐NaN feature values in the training set.
Hybrid‐CNN Architecture
The input consists of a convolution layer with a single filter for all ECG channels and a different filter for each of the photoplethysmogram and BP channels. Each filter operates on the corresponding signal and an output is obtained by convolving the signal with the filter weights. Further, each filter component is assumed to have the same length. The convolved output is passed through a rectified linear unit and is pooled to reduce the dimension by a factor of 2. Next the output from the ECG, BP, and photoplethysmogram filters are concatenated and passed to the later stages. The filters of the subsequent layer fuse the information from all the channels into a single vector followed by rectified linear unit activation and pooling. The convolution, non‐linear activation and pooling are treated as a single layer, which is repeated until penultimate layer (the second to last layer). The handcrafted features are augmented to the flattened convolution features, before fully connecting to the output layer.
Choice of Network Parameters
We used CNNs as building blocks to achieve the desired classification at each tier of the overall classifier. While training the CNNs, we used the binary cross entropy and the categorical cross entropy cost functions for binary and multi class classification tasks respectively. We also optimized the classifier performance by varying the number of filters, filter length, and pooling operations, to determine the effect of the convolution filter length as well as the network capacity, in terms of the number of trainable parameters.
Results
Noise Detector Performance Evaluation
Before proceeding to the evaluation of the overall algorithm performance, we first evaluated the performance of the noise detector using 5‐fold cross‐validation. Based on the manual annotations corresponding with the onset and offset of the noise segment, we extracted 4 seconds of clean and noisy data windows from each record in the training and test folds. We developed signal specific noise detectors for ECG, BP, and photoplethysmogram signals using (1) a fully connected network with handcrafted features, (2) only CNN and (3) hybrid‐CNN.
Using each classifier, we obtained the probability of determining whether the test window is a clean or noisy signal. Using this approach, we compared various classifiers using mean receiver operating characteristic curves, over 5‐fold. The overall optimal operating point is provided by the hybrid‐CNN classifier with a sensitivity and specificity of 94.0% and 91.9%, respectively.
The performance of the ECG noise classifier is shown in Figure S1. The desired operating point is obtained by computing the point on the receiver operating characteristic curve that is closest to the ideal classifier, ie, sensitivity=1 and specificity=1. The area under the curve of the ECG noise classifier for feature‐based, only CNN, and hybrid CNN classifiers were 93.56%, 96.97%, and 97.17%, respectively. While the hybrid CNN and only CNN classifiers achieved similar performance, both classifiers achieved significant performance improvement compared with the only feature‐based noise detector, perhaps because the features extracted from a short (4 seconds) window may not adequately capture the noise characteristics. The hybrid‐CNN BP noise classifier provided a sensitivity and specificity of 88.6% and 90.9% respectively, and the hybrid‐CNN photoplethysmogram noise classifier provided a sensitivity and specificity of 98.5% and 94.9%, respectively.
The proposed noise detectors appeared to be robust to the morphological changes in the signals during arrhythmic events, and have not classified any arrhythmia records as noise.
Algorithm Performance Evaluation
The data are processed sequentially in windows of 4 and 8 seconds while striding with 0.5‐second steps. Next, each 4‐second window of data is passed through the noise detector, and the noisy channels of ECG, BP, and photoplethysmogram signals are masked with zeros. If all ECG channels are noisy, the corresponding 4‐second window is considered noisy, and disregarded. Otherwise, the window is passed through Tier‐1 classifier to detect the presence of VT or VF. An 8‐second window that does not contain a 4‐second window of either noise, VT or VF, is passed on to the Tier‐2 classifier to detect EB or ET; if EB/ET arrhythmias are not found, the 8‐second window is marked as AF or SR based on the Tier‐3 classifier output.
We developed 3 classifiers, with: (1) handcrafted features alone, (2) CNN, and (3) hybrid‐CNN. We optimized the performance of only CNN and hybrid‐CNN by varying the number of filters and the filter length. The performance for each hyper‐parameter configuration is presented in Table 3. In terms of network capacity, for a Tier‐1 classifier, the minimum and maximum number of trainable parameters in a hybrid‐CNN were 15 457 (filter size=5, number of filters=4) and 3 134 669 (filter size=500, number of filters=32), respectively. Similarly, for only CNN classifier the minimum and maximum number of trainable parameters were 1015 (filter size=5, number of filters=4) and 3 120 227 (filter size=500, number of filters=32), respectively. It should be noted that the network capacity is increased with the filter size and the number of filters; also, the capacity of the hybrid network is greater than the equivalent only CNN classifier, because of the inclusion of the handcrafted features.
Table 3.
Performance of Various Network Architectures
| Filter size | No. of filters | Pooling | Hybrid‐CNN classifier | Only‐CNN classifier | ||
|---|---|---|---|---|---|---|
| Overall accuracy (%) | Score (%) | Overall accuracy (%) | Score (%) | |||
| 5 | 4 | Max pooling | 83.71 | 79.05 | 72.81 | 61.39 |
| 5 | 8 | Max pooling | 86.37 | 81.11 | 82.45 | 68.10 |
| 25 | 8 | Max pooling | 86.93 | 80.14 | 85.75 | 72.92 |
| 50 | 8 | Max pooling | 87.64 | 81.44 | 81.40 | 64.81 |
| 75 | 8 | Max pooling | 87.53 | 81.40 | 79.91 | 64.33 |
| 50 | 8 | Average pooling | 77.14 | 57.20 | 72.67 | 52.00 |
| 50 | 32 | Max pooling | 69.33 | 56.46 | 84.25 | 66.86 |
| 100 | 8 | Max pooling | 76.82 | 62.39 | 78.23 | 64.33 |
| 500 | 8 | Max pooling | 62.33 | 56.41 | 74.16 | 61.98 |
| 500 | 32 | Max pooling | 52.83 | 52.83 | 76.09 | 60.80 |
| Only feature‐based classifier | 86.72 | 80.48 | ||||
CNN indicates convolutional neural network.
Interestingly, an increased network capacity does not translate to improved performance. It is observed that the hybrid‐CNN achieved a slightly improved performance over only CNN classifier. Also, the performance increased with filter size, reached a peak, and then decreased. Intuitively, a small convolution filter operates on a short temporal window within the signal, and may not fully exploit the temporal dependency efficiently. On the other hand, a long filter also achieves low performance because, while observing for a significant duration, CNN filters might fuse and encode the information from the entire beat into few filter outputs. A similar observation is reported in earlier work, 26 and the choice of filter dimension plays a crucial role in determining the classifier performance. For hybrid‐CNN, a filter dimension of 50, 8 filters, and maximum pooling operation, among all classifiers, achieved the highest accuracy and score, of 87.6% and 81.4%, respectively.
To account for the stochasticity in model convergence as well as the randomness in the choice of data in each fold, we performed 5 times 5‐fold cross‐validation and reported the mean and SD of the classifier performance, for optimal hyper‐parameters. Our hybrid‐CNN model, only‐CNN model, and only feature‐based classifiers achieved a mean (±SD) accuracy of 87.5% (±0.48), 81.2% (±0.94) and 84.3% (±1.35) and a score of 81.0% (±0.89), 64.6% (±4.1) and 80.7% (±0.48), respectively. Favorably, the low SD indicates that our method generalizes well to new data. Further, we verified the statistical similarity between hybrid‐CNN and only feature‐based classifier using McNemar test, 29 , 30 and we observed that the Tier‐1, Tier‐2, and Tier‐3 classifiers are significantly different (P<0.001). The classification performance in terms of sensitivity, PPV, and accuracy, specific to each rhythm are presented in Table 4 and Table S5. For asystole classification, the SD is zero as it gets detected through a deterministic process, before passing through the classifier. The majority of EB and ET misclassifications are borderline cases in which the heart rate is close to 40 and 140 beats per minute, respectively. Such misclassifications can be corrected setting a hard‐threshold on the heart rate, although such minor misclassifications are unlikely to have a meaningful clinical impact. Finally, although the present work appears to have a poor sensitivity for VF detection, all the misclassified VF signals are assigned as VT and in practice, alarms will be raised for 100% of the VF signals. Furthermore, clinical differentiation of VT from VF is somewhat arbitrary and therefore, binning the alarms together serves to maximize clinical utility.
Table 4.
Sensitivity, PPV, Accuracy and Score for Each Rhythm Following 5‐Times 5‐fold Cross‐Validation (Highlighted in Grey), as Well as the Positive Predictive Value Observed by Bedside Monitors
| Rhythm | Sensitivity (%) | PPV (%) | Accuracy (%) | Score (%) | PPV (%) Current study (clinical annotations) | PPV (%) Physionet 2015 Challenge | PPV (%) MIMIC II study | PPV (%) UCSF study |
|---|---|---|---|---|---|---|---|---|
| Aystole | 100.00±0.00 | 61.58±0.00 | 99.42±0.00 | 99.42±0.00 | 11.31 | 16.67 | 9.33 | 32.83 |
| EB | 99.29±0.78 | 82.64±3.04 | 98.65±0.27 | 98.65±0.26 | 22.64 | 50 | 70.71 | NA |
| ET | 91.55±0.85 | 96.11±0.96 | 99.20±0.02 | 98.64±0.11 | 17.11 | 94.92 | 76.93 | NA |
| VF | 79.45±9.84 | 78.83±12.51 | 99.29±0.33 | 99.29±0.33 | 21.74 | 10.34 | 20.33 | 67.72 |
| VT | 97.33±0.60 | 95.13±0.99 | 98.73±0.21 | 98.22±0.27 | 53.66 | 26.23 | 53.42 | 13.00 |
| AF | 89.99±1.13 | 74.41±1.44 | 90.48±0.42 | 84.61±0.89 | NA | NA | NA | NA |
| SR | 80.33±1.41 | 94.74±0.81 | 89.16±0.45 | NA | NA | NA | NA | NA |
AF indicates atrial fibrillation; EB, extreme bradycardia; ET, extreme tachycardia; MIMIC, Medical Information Mart for Intensive Care; NA, not applicable; PPV, positive predictive value; SR, sinus rhythm; USCF, University of California, San Francisco; VF, ventricular fibrillation; and VT, ventricular tachycardia.
Next, we attempted to understand the improvement in the performance of hybrid CNN compared with only CNN approach and feature‐based approach. To this end, we used a Gradient‐Weighted Class Activation Map 31 to visualize the discriminatory parts of the signal that generated the output of a CNN‐based classifier (please, see Figure S2). Also, we used permutation‐importance 32 to depict the importance of handcrafted features in a feature‐based classifier (please, see Figures S3A through S3C). It appears, that since each arrhythmia is characterized by an associated morphology and well defined features, capturing these features using a hand engineered feature extraction approach helps in classification; although CNNs attempt to characterize these features, a hybrid approach would combine the benefits of both approaches.
Algorithm Performance Comparison
Thereafter, we compared the performance of the present method with the existing bedside monitoring systems (Table 4). Although the existing monitors continuously process data to detect arrhythmias, the exact onset and offset locations of arrhythmias remain unavailable. In this setting, assuming that the existing systems do not miss any arrhythmic events, we compared the PPV of our proposed system to those of the bedside monitors used in the PhysioNet 2015 challenge, 16 MIMIC (Medical Information Mart for Intensive Care) II study, 8 and the alarm fatigue study from University of California, San Francisco 1 (Table 4). Specifically, we computed the PPV as the ratio of the reported true alarms to the total alarms raised corresponding to that particular arrhythmia. We aim to maximize the PPV while maintaining high sensitivity.
For 5 life‐threatening arrhythmias, our method achieved an average PPV and sensitivity of 82.9% and 93.5%, respectively. The monitor‐based average PPV of the current study (based on clinician’s annotations), PhysioNet 2015 challenge and MIMIC II study are 25.29, 39.63, and 46.14, respectively. Among the common arrhythmias used in the present and the University of California, San Francisco studies (asystole, VT, and VF), we achieved a PPV of 78.57 with a sensitivity of 92.2% while the University of California, San Francisco study exhibited a PPV of 37.9%.
Independent Data Set Validation
We used data from PhysioNet 2015 challenge, 16 as an independent data set to validate the proposed algorithm. Record‐wise annotations indicating signal quality, rhythm type, and the onset and offset of each arrhythmia are presented in Table S6. In particular, we used 2‐channel ECG, BP, and photoplethysmogram signals as input and processed 4‐ and 8‐second long data, while striding with 0.5‐second steps. Although the proposed network is trained with 4‐channel ECG, BP, and photoplethysmogram, it remains adaptable and effective even with 2 missing channels of ECG. In Table 5, we report the confusion matrix of the proposed classifier with an overall accuracy and score of 93.93% and 84.32%, respectively. While VF arrhythmia achieved a lesser sensitivity and PPV, all the VF misclassifications correspond to VT and in practice an alarm would have been raised for each VF incidence. The score of the proposed hybrid CNN is close to the top scoring entry of the challenge, 33 with a score of 85.04%. Favorably, the proposed algorithm in this article is trained to detect AF in addition to the arrhythmia considered in the challenge and is flexible to be extended to other arrhythmias. Only CNN based classifier achieved an overall accuracy and score of 88.18% and 68.96% (please, see the confusion matrix in the Table 6), respectively, and only feature based classifier achieved an overall accuracy and score of 88.4% and 72.48%, respectively (please, see the confusion matrix in the Table 7).
Table 5.
Performance of Hybrid CNN Classifier on an Independent Validation Data Set From the PhysioNet 2015 Challenge
| Aystole | EB | ET | VF | VT | SR | Sensitivity (%) | |
|---|---|---|---|---|---|---|---|
| Aystole | 28 | 0 | 0 | 0 | 0 | 0 | 100 |
| EB | 0 | 227 | 0 | 0 | 0 | 15 | 93.80 |
| ET | 0 | 0 | 932 | 0 | 35 | 186 | 80.83 |
| VF | 0 | 0 | 0 | 10 | 12 | 0 | 45.45 |
| VT | 0 | 0 | 0 | 17 | 142 | 9 | 84.52 |
| SR | 17 | 25 | 69 | 0 | 62 | 5584 | 96.99 |
| PPV (%) | 62.22 | 90.08 | 93.11 | 37.04 | 56.57 | 96.38 |
Accuracy = 93.93% Score = 84.32% |
CNN indicates convolutional neural network; EB, extreme bradycardia; ET, extreme tachycardia; PPV, positive predictive value; SR, sinus rhythm; VF, ventricular fibrillation; and VT, ventricular tachycardia.
Table 6.
Performance of Only CNN Classifier on an Independent Validation Data Set From the PhysioNet 2015 Challenge
| Aystole | EB | ET | VF | VT | SR | Sensitivity (%) | |
|---|---|---|---|---|---|---|---|
| Aystole | 28 | 0 | 0 | 0 | 0 | 0 | 100.00 |
| EB | 0 | 184 | 0 | 0 | 0 | 51 | 78.30 |
| ET | 0 | 0 | 675 | 23 | 11 | 409 | 60.38 |
| VF | 0 | 0 | 0 | 18 | 2 | 2 | 81.82 |
| VT | 0 | 0 | 0 | 12 | 100 | 40 | 65.79 |
| SR | 4 | 68 | 64 | 55 | 110 | 5346 | 94.67 |
| PPV (%) | 87.50 | 73.02 | 91.34 | 55.56 | 11.92 | 91.42 |
Accuracy = 88.18% Score = 68.96% |
CNN indicates convolutional neural network; EB, extreme bradycardia; ET, extreme tachycardia; PPV, positive predictive value; SR, sinus rhythm; VF, ventricular fibrillation; and VT, ventricular tachycardia.
Table 7.
Performance of Only Feature‐Based Classifier on an Independent Validation Data Set from the PhysioNet 2015 Challenge
| Aystole | EB | ET | VF | VT | SR | Sensitivity (%) | |
|---|---|---|---|---|---|---|---|
| Aystole | 28 | 0 | 0 | 0 | 0 | 0 | 100.00 |
| EB | 0 | 203 | 0 | 0 | 0 | 32 | 86.38 |
| ET | 0 | 0 | 572 | 2 | 149 | 316 | 55.05 |
| VF | 0 | 0 | 0 | 15 | 5 | 2 | 68.18 |
| VT | 0 | 0 | 0 | 18 | 107 | 44 | 63.31 |
| SR | 4 | 62 | 54 | 14 | 130 | 5418 | 95.35 |
| PPV (%) | 87.50 | 76.60 | 91.37 | 30.61 | 27.37 | 93.22 |
Accuracy = 88.40% Score = 72.48 |
EB indicates extreme bradycardia; ET, extreme tachycardia; PPV, positive predictive value; SR, sinus rhythm; VF, ventricular fibrillation; and VT, ventricular tachycardia.
Discussion
The majority of ICU abnormal heart rhythm triggered monitor alarms are found to be false, 1 primarily attributable to noise and artifacts in the physiological signals, which often result from patient motion or loose electrodes. Excessive numbers of false alarms create a noisy environment and cause alarm desensitization among caregivers. In the present study, we observed that about 74.7% of the critical ECG arrhythmia alerts are false alarms. In particular, individual arrhythmia rates vary between 46.3% and 88.6%, which are observations consistent with the reported ICU FA rates ranging between 40% and 90%. 5 , 8
Various attempts have been made to address the issue of FAs. Recent algorithms that used machine learning techniques and prior information with respect to the alarm type have reported significant improvement in FA suppression. 17 , 18 , 34 , 35 However in practice it has been observed that, although some of the critical arrhythmia alarms are true, the condition indicated on the monitor may not indicate the true underlying arrhythmia. For instance, in our database, 22.6% of EB alarms that require attention correspond to ET. Therefore, it is imperative to develop arrhythmia detectors without prior knowledge of the alarm type. In this report, we present a standalone arrhythmia alerting system that identified life‐threatening arrhythmias without prior knowledge of the arrhythmia type. Several conclusions can be drawn from this study: first, a hybrid‐CNN approach performs better than either only CNN or only feature‐based approaches; second, the proposed method can be adapted to use multiple modules such as signal‐specific noise detectors and arrhythmia detectors; third, our algorithm is flexible to operate on different duration signals and can be extended to other arrhythmias with suitable training; fourth, the proposed hybrid‐CNN algorithm would have suppressed 77.05% of the FA generated by an existing monitoring system, without prior knowledge of the underlying alarm, which is superior to any other algorithm 8 , 9 , 36 ; fifth, our classifier generalizes well with an excellent performance when validated in an independent data set.
Early attempts in heart rhythm classification have extensively analyzed the signal morphology in single and multiple channels of ECG signals. 4 , 5 Recent algorithms have determined that BP and photoplethysmogram improve the arrhythmia detection performance, 6 , 7 , 8 , 9 considering, for example, that arrhythmias such as VT and VF are accompanied with a drop in BP. Although various algorithms consider the information from BP and photoplethysmogram signals, features from each physiological signal are extracted independently. In contrast, our proposed solution fuses the information from multiple physiological signals through convolution filters and learns suitable features to achieve the desired classification goals. In addition, we also augment the learned features with handcrafted features to fully exploit the benefits of feature engineering and feature learning. Further, the proposed algorithm is flexible and effective when independently validated using PhysioNet 2015 challenge data, even when 2 ECG channels are missing. For 5 life‐threatening arrhythmias (asystole, ET, EB, VT, and VF) recorded from the bedside monitors, the monitor‐based average PPV (based on clinician’s annotations) was 25.29%, resulting in 74.71% of false alarms. Using the same data, the proposed method achieved an average PPV of 82.9% resulting in 17.1% of false alarms. In practice, our proposal would have suppressed 77.05% of the false alarms generated by an existing monitoring system. Further, the proposed method facilitates real‐time operation by raising an alarm as soon as the arrhythmia criteria are met.
AF is the most frequent arrhythmia in the ICU across all populations, with an incidence ranging from 47.4% to 61%. 37 Although the majority of ICU physiologic monitors (ie, the GE EK‐Pro), are capable of detecting and alarming when AF is present (albeit they do not include classification of atrial flutter or atrioventricular block), the AF alarm is often kept inaudible, because AF is considered a non‐critical alarm. However, because of the high incidence of AF in the ICU and its impact on electrocardiographic features, it is imperative to include AF within an arrhythmia detection algorithm.
With the ever‐increasing use of wearable and mobile‐based devices for ambulatory patient monitoring, multiple physiological signals including ECG, BP, and photoplethysmogram can be monitored from the comfort of the home, thus providing abundant information for the accurate detection of abnormal heart rhythms such as those described in this study.
Conclusions
In this study, we proposed a method for detecting critical arrhythmias encountered in ICUs using a hybrid‐CNN based approach. In particular, we sought to accurately identify different life‐threatening arrhythmias and reduce the burden of FA fatigue. In the process, we developed a generalizable hybrid‐CNN architecture that fuses the hand‐picked features with those learned by the CNN. Although only CNN based classifiers learn suitable features from the data to optimize the classification performance, CNNs augmented by hand engineered features that characterize various arrhythmic conditions resulted in improved overall arrhythmia detection accuracy. The hybrid approach gave superior performance to both traditional handcrafted feature‐based methods and CNN based methods. While the proposed method is developed to process 4 and 8 seconds of data, the method remains generic to a variable processing window duration. Further, our method also retains the flexibility of including new arrhythmia detectors and novel handcrafted arrhythmia specific features, if needed, by simply reconfiguring and training the hybrid‐CNN modules appropriately.
Study Limitations
ICUs encompass patient groups with diverse medical conditions and a wide variety of abnormal heart rhythms and generated alarms. Ideally all monitor alarms should be considered for analysis, however, because of the resource‐intensive nature of the manual data annotations, this study was confined to the presented critical alarms. In addition, similarly to other studies, because of the limited available labeled data, the trained models are prone to overfit to the training data. To this end, we have used early stopping of classifier training based on the validation accuracy and, evaluated the model’s generalizability on an external database. In summary, while the proposed method is developed to suppress only false critical alarms, the framework remains generic and can be extended to other conditions via suitable training and network modifications.
Sources of Funding
The work was supported by a Grant‐in‐Aid (#15GRNT23070001) from the American Heart Association, the Institute of Precision Medicine (17UNPG33840017) from the American Heart Association, the RICBAC Foundation, National Institutes of Health grants 1 R01 HL135335‐01, 1 R21 HL137870‐01, 1 R21EB026164‐01, and 3R21EB026164‐02S1, the Founders Affiliate Postdoctoral Fellowships (award numbers 19POST34450149 and 834897) from the American Heart Association. This work was conducted with support from Harvard Catalyst, The Harvard Clinical and Translational Science Center (National Center for Research Resources, and the National Center for Advancing Translational Sciences, National Institutes of Health Award 8UL1TR000170‐05, and financial contributions from Harvard University and its affiliated academic healthcare centers). The content is solely the responsibility of the authors and does not necessarily represent the official views of Harvard Catalyst, Harvard University, and its affiliated academic healthcare centers, or the National Institutes of Health.
Disclosures
None.
Supporting information
Data S1
Tables S1–S6
Figures S1–S3
Acknowledgments
Author contributions: Sandeep Chandra Bollepalli participated in the development of the algorithms, data analysis, and writing the article; Rahul K. Sevakula participated in the development of the algorithms and writing the article; Wan‐Tai M. Au‐Yeung participated in the development of the algorithms and writing the article; Mohamad B. Kassab participated in the data analysis and writing the article; Faisal M. Merchant participated in the data analysis and writing the article; George Bazoukis participated in the data analysis and writing the article; Richard Boyer participated in the data analysis and writing the article; Eric M. Isselbacher participated in the conception of the study and writing the article; Antonis A. Armoundas participated in the conception of the study, the development of the algorithms, data analysis, and writing the article.
Supplementary Material for this article is available at https://www.ahajournals.org/doi/suppl/10.1161/JAHA.121.023222
For Sources of Funding and Disclosures, see page 11.
References
- 1. Drew BJ, Harris P, Zègre‐Hemsey JK, Mammone T, Schindler D, Salas‐Boni R, Bai Y, Tinoco A, Ding Q, Hu X. Insights into the problem of alarm fatigue with physiologic monitor devices: a comprehensive observational study of consecutive intensive care unit patients. PLoS One. 2014;9:e110274. doi: 10.1371/journal.pone.0110274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Tsien CL, Fackler JC. Poor prognosis for existing monitors in the intensive care unit. Crit Care Med. 1997;25:614–619. doi: 10.1097/00003246-199704000-00010 [DOI] [PubMed] [Google Scholar]
- 3. Keller JP. Clinical alarm hazards: a “top ten” health technology safety concern. J Electrocardiol. 2012;45:588–591. doi: 10.1016/j.jelectrocard.2012.08.050 [DOI] [PubMed] [Google Scholar]
- 4. Mäkivirta A, Koski E, Kari A, Sukuvaara T. The median filter as a preprocessor for a patient monitor limit alarm system in intensive care. Comput Methods Programs Biomed. 1991;34:139–144. doi: 10.1016/0169-2607(91)90039-V [DOI] [PubMed] [Google Scholar]
- 5. Sittig DF, Factor M. Physiologic trend detection and artifact rejection: a parallel implementation of a multi‐state Kalman filtering algorithm. Comput Methods Programs Biomed. 1990;31:1–10. doi: 10.1016/0169-2607(90)90026-6 [DOI] [PubMed] [Google Scholar]
- 6. Orphanidou C, Bonnici T, Charlton P, Clifton D, Vallance D, Tarassenko L. Signal‐quality indices for the electrocardiogram and photoplethysmogram: derivation and applications to wireless monitoring. IEEE J Biomed Health Inform. 2015;19:832–838. [DOI] [PubMed] [Google Scholar]
- 7. Zong W, Moody GB, Mark RG. Reduction of false arterial blood pressure alarms using signal quality assessment and relationships between the electrocardiogram and arterial blood pressure. Med Biol Eng Comput. 2004;42:698–706. doi: 10.1007/BF02347553 [DOI] [PubMed] [Google Scholar]
- 8. Aboukhalil A, Nielsen L, Saeed M, Mark RG, Clifford GD. Reducing false alarm rates for critical arrhythmias using the arterial blood pressure waveform. J Biomed Inform. 2008;41:442–451. doi: 10.1016/j.jbi.2008.03.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Li Q, Clifford GD. Signal quality and data fusion for false alarm reduction in the intensive care unit. J Electrocardiol. 2012;45:596–603. doi: 10.1016/j.jelectrocard.2012.07.015 [DOI] [PubMed] [Google Scholar]
- 10. Salas‐Boni R, Bai Y, Harris PR, Drew BJ, Hu X. False ventricular tachycardia alarm suppression in the ICU based on the discrete wavelet transform in the ECG signal. J Electrocardiol. 2014;47:775–780. doi: 10.1016/j.jelectrocard.2014.07.016 [DOI] [PubMed] [Google Scholar]
- 11. Tanantong T, Nantajeewarawat E, Thiemjarus S. False alarm reduction in BSN‐based cardiac monitoring using signal quality and activity type information. Sensors. 2015;15:3952–3974. doi: 10.3390/s150203952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Au‐Yeung W‐TM, Sahani AK, Isselbacher EM, Armoundas AA. Reduction of false alarms in the intensive care unit using an optimized machine learning based approach. NPJ Digit Med. 2019;2:86. doi: 10.1038/s41746-019-0160-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Au‐Yeung W‐TM, Sevakula RK, Sahani AK, Kassab M, Boyer R, Isselbacher EM, Armoundas AA. Real‐time machine learning‐based intensive care unit alarm classification without prior knowledge of the underlying rhythm. Eur Heart J ‐ Digital Health. 2021;2:437–445. doi: 10.1093/ehjdh/ztab058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ansari S, Belle A, Ghanbari H, Salamango M, Najarian K. Suppression of false arrhythmia alarms in the ICU: a machine learning approach. Physiol Meas. 2016;37:1186–1203. doi: 10.1088/0967-3334/37/8/1186 [DOI] [PubMed] [Google Scholar]
- 15. Antink CH, Leonhardt S, Walter M. Reducing false alarms in the ICU by quantifying self‐similarity of multimodal biosignals. Physiol Meas. 2016;37:1233–1252. doi: 10.1088/0967-3334/37/8/1233 [DOI] [PubMed] [Google Scholar]
- 16. Clifford GD, Silva I, Moody B, Li Q, Kella D, Chahin A, Kooistra T, Perry D, Mark RG. False alarm reduction in critical care. Physiol Meas. 2016;37:E5–E23. doi: 10.1088/0967-3334/37/8/E5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Eerikäinen LM, Vanschoren J, Rooijakkers MJ, Vullings R, Aarts RM. Reduction of false arrhythmia alarms using signal selection and machine learning. Physiol Meas. 2016;37:1204–1216. doi: 10.1088/0967-3334/37/8/1204 [DOI] [PubMed] [Google Scholar]
- 18. Plesinger F, Klimes P, Halamek J, Jurak P. Taming of the monitors: reducing false alarms in intensive care units. Physiol Meas. 2016;37:1313–1325. doi: 10.1088/0967-3334/37/8/1313 [DOI] [PubMed] [Google Scholar]
- 19. Liu F, Liu C, Zhao L, Zhang X, Wu X, Xu X, Liu Y, Ma C, Wei S, He Z, et al. An open access database for evaluating the algorithms of electrocardiogram rhythm and morphology abnormality detection. J Med Imaging Health Inform. 2018;8:1368–1373. doi: 10.1166/jmihi.2018.2442 [DOI] [Google Scholar]
- 20. Bazoukis G, Stavrakis S, Zhou J, Bollepalli SC, Tse G, Zhang Q, Singh JP, Armoundas AA. Machine learning versus conventional clinical methods in guiding management of heart failure patients‐a systematic review. Heart Fail Rev. 2021;26:23–34. doi: 10.1007/s10741-020-10007-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Sevakula RK, Au‐Yeung WM, Singh JP, Heist EK, Isselbacher EM, Armoundas AA. State‐of‐the‐art machine learning techniques aiming to improve patient outcomes pertaining to the cardiovascular system. J Am Heart Assoc. 2020;9:e013924. doi: 10.1161/JAHA.119.013924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Au‐Yeung W‐TM, Sevakula RK, Singh JP, Heist EK, Isselbacher EM, Armoundas AA. State of the art in artificial intelligence and machine learning techniques for improving patient outcomes pertaining to the cardiovascular and respiratory systems. In: Efimov IR, Ng FS, Laughner JI, eds. Cardiac bioelectric therapy: mechanisms and practical implications. Cham: Springer International Publishing; 2021:335–352. [Google Scholar]
- 23. Hannun AY, Rajpurkar P, Haghpanahi M, Tison GH, Bourn C, Turakhia MP, Ng AY. Cardiologist‐level arrhythmia detection and classification in ambulatory electrocardiograms using a deep neural network. Nat Med. 2019;25:65–69. doi: 10.1038/s41591-018-0268-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Xu X, Jeong S, Li J. Interpretation of electrocardiogram (ECG) rhythm by combined CNN and BILSTM. IEEE Access. 2020;8:125380–125388. doi: 10.1109/ACCESS.2020.3006707 [DOI] [Google Scholar]
- 25. Yildirim O, Talo M, Ciaccio EJ, Tan RS, Acharya UR. Accurate deep neural network model to detect cardiac arrhythmia on more than 10,000 individual subject ECG records. Comput Methods Programs Biomed. 2020;197:105740. doi: 10.1016/j.cmpb.2020.105740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Chandra BS, Sastry CS, Jana S. Robust heartbeat detection from multimodal data via CNN‐based generalizable information fusion. IEEE Trans Biomed Eng. 2018;66:710–717. doi: 10.1109/TBME.2018.2854899 [DOI] [PubMed] [Google Scholar]
- 27. Quer G, Arnaout R, Henne M, Arnaout R. Machine learning and the future of cardiovascular care. J Am Coll Cardiol. 2021;77:300–313. doi: 10.1016/j.jacc.2020.11.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. de Philip C, Dwyer MO, Reilly RB. Automatic classification of heartbeats using ECG morphology and heartbeat interval features. IEEE Trans Biomed Eng. 2004;51:1196–1206. doi: 10.1109/TBME.2004.827359 [DOI] [PubMed] [Google Scholar]
- 29. McNemar Q. Note on the sampling error of the difference between correlated proportions or percentages. Psychometrika. 1947;12:153–157. doi: 10.1007/BF02295996 [DOI] [PubMed] [Google Scholar]
- 30. Dietterich TG. Approximate statistical tests for comparing supervised classification learning algorithms. Neural Comput. 1998;10:1895–1923. doi: 10.1162/089976698300017197 [DOI] [PubMed] [Google Scholar]
- 31. Selvaraju RR, Cogswell M, Das A, Vedantam R, Parikh D, Batra D. Grad‐cam: visual explanations from deep networks via gradient‐based localization. IEEE Int Conf Comput Vis. 2017;2017:618–626. [Google Scholar]
- 32. Altmann A, Toloşi L, Sander O, Lengauer T. Permutation importance: a corrected feature importance measure. Bioinformatics. 2010;26:1340–1347. doi: 10.1093/bioinformatics/btq134 [DOI] [PubMed] [Google Scholar]
- 33. Fallet S, Yazdani S, Vesin JM. False arrhythmia alarms reduction in the intensive care unit: a multimodal approach. Physiol Meas. 2016;37:1217–1232. doi: 10.1088/0967-3334/37/8/1217 [DOI] [PubMed] [Google Scholar]
- 34. Kalidas V, Tamil LS. Cardiac arrhythmia classification using multi‐modal signal analysis. Physiol Meas. 2016;37:1253–1272. doi: 10.1088/0967-3334/37/8/1253 [DOI] [PubMed] [Google Scholar]
- 35. Fernandes CO, Miles S, De Lucena CJP, Cowan D. Artificial intelligence technologies for coping with alarm fatigue in hospital environments because of sensory overload: algorithm development and validation. J Med Internet Res. 2019;21:e15406. doi: 10.2196/15406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Sayadi O, Shamsollahi MB. Life‐threatening arrhythmia verification in ICU patients using the joint cardiovascular dynamical model and a Bayesian filter. IEEE Trans Biomed Eng. 2011;58:2748–2757. doi: 10.1109/TBME.2010.2093898 [DOI] [PubMed] [Google Scholar]
- 37. Heinz G. Atrial fibrillation in the intensive care unit. Intensive Care Med. 2006;32:345–348. doi: 10.1007/s00134-005-0033-1 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data S1
Tables S1–S6
Figures S1–S3
Data Availability Statement
The training data set will be available to any investigator upon request.


