Abstract
Background
Obstructive sleep apnea (OSA) has shown to be associated with an increased risk of atrial fibrillation in observational studies. Whether this association reflect causal effect is still unclear. The aim of this study was to evaluate the causal effect of OSA on atrial fibrillation.
Methods and Results
We used a 2‐sample Mendelian randomization (MR) method to evaluate the causal effect of OSA on atrial fibrillation. Summary data on genetic variant‐OSA association were obtained from a recently published genome‐wide association studies with up to 217 955 individuals and data on variant‐atrial fibrillation association from another genome‐wide association study with up to 1 030 836 individuals. Effect estimates were evaluated using inverse‐variance weighted method. Other MR analyses, including penalized inverse‐variance weighted, penalized robust inverse‐variance weighted, MR‐Egger, simple median, weighted median, weighted mode‐based estimate and Mendelian Randomization Pleiotropy Residual Sum and Outlier methods were performed in sensitivity analyses. The MR analyses in both the fixed‐effect and random‐effect inverse‐variance weighted models showed that genetically predicted OSA was associated with an increased risk of atrial fibrillation (odds ratio [OR], 1.21; 95% CI, 1.12–1.31, P<0.001; OR, 1.21; 95% CI, 1.11–1.32, P<0.001) using 5 single nucleotide polymorphisms as the instruments. MR‐Egger indicated no evidence of genetic pleiotropy (intercept, −0.014; 95% CI, −0.033 to 0.005, P=0.14). Results were robust using other MR methods in sensitivity analyses.
Conclusions
This MR analysis found that genetically predicted OSA had causal effect on an increased risk of atrial fibrillation.
Keywords: atrial fibrillation, causal association, genetics, Mendelian randomization, obstructive sleep apnea
Subject Categories: Genetic, Association Studies; Risk Factors
Nonstandard Abbreviations and Acronyms
- IVW
inverse‐variance weighted
- MR
Mendelian randomization
Clinical Perspective
What Is New?
Although accumulating observational evidence has demonstrated that obstructive sleep apnea was associated with development of atrial fibrillation, the causal effect of obstructive sleep apnea on atrial fibrillation is still unclear.
This study provides genetic evidence of causal effect of obstructive sleep apnea on atrial fibrillation.
What Are the Clinical Implications?
These findings support detection and treatment of obstructive sleep apnea for preventing atrial fibrillation.
Atrial fibrillation is the most common cardiac arrhythmia that affecting 1% to 4% of the population, and its prevalence is expected to increase globally over the next 30 years due to population growth and aging. 1 , 2 Previous studies showed that there were mechanistic links between cardiovascular risk factors and atrial fibrillation susceptibility; in particular, obstructive sleep apnea (OSA) was expected to contribute to development of atrial fibrillation. 3
Several studies have shown that OSA increased the risk of atrial fibrillation, potentially through hypoxia, oxidative stress, inflammation and atrial remodeling. 4 , 5 , 6 There were growing evidences of observational studies indicating that presence of OSA might increase the risk of atrial fibrillation in community population and post‐operative atrial fibrillation following cardiac surgery. 7 , 8 , 9 However, due to the potential confounding biases and reverse causation that exist in observational studies, 10 it is difficult to assess the causality of OSA on atrial fibrillation for the association observed in observational studies.
Mendelian randomization (MR) design is a method for evaluating causality of risk factors on disease by using genetic variants as instrumental variables for risk factors. Because the genetic variants are assigned randomly at conception, MR analysis can reduce potential unmeasured confounders and reverse causation, a major limitation of evidence from observational studies. 10 In this study, we aimed to evaluate the causal effect of OSA on atrial fibrillation using MR approaches.
METHODS
Study Design and Data Sources
The authors declare that all supporting data are available within the article. We used a 2‐sample MR model to evaluate the causal effect of OSA on atrial fibrillation (Figure 1). MR design is a method to test whether an exposure has a causal effect on the development of a disease, where genetic variations are deemed as instrumental variables. MR method can overcome unmeasured confounding and make stronger causal inferences. 10 MR design rests on 3 assumptions: (1) the genetic variants are robustly associated with the exposure; (2) the genetic variants are not associated with other confounders; (3) the genetic variants are associated with the outcome only through the investigated exposure. 11 Data on the associations of single nucleotide polymorphisms (SNPs) with OSA and atrial fibrillation were obtained from recently published genome‐wide association studies (GWAS). 12 , 13 Only summary data were used in this article. Appropriate ethical approval and patient informed consent were obtained in the original studies.
Figure 1. Mendelian randomization model of obstructive sleep apnea and risk of atrial fibrillation.
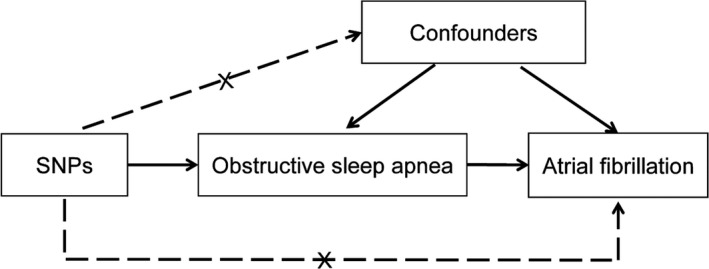
The design is under the assumption that the genetic variants are associated with obstructive sleep apnea, but not with confounders, and the genetic variants influence atrial fibrillation only through obstructive sleep apnea. SNP indicates single nucleotide polymorphism.
Instrument Variables
We used published genetic variants associated with OSA from a recent published GWAS from FinnGen Study. 12 This GWAS included 217 955 individuals with 16 761 OSA patients identified using Finland nationwide health registries. OSA was diagnosed according to International Classification of Diseases, Tenth Revision (ICD‐10) and Ninth Revision (ICD‐9) codes (ICD‐10: G47.3, ICD‐9: 3472A), which is based on subjective symptoms, clinical examination and sleep registration applying apnea‐hypopnea index ≥5/hour or respiratory event index ≥5/hour. This GWAS identified 5 loci associated with OSA (P<5.0×10−8). 12 These 5 locus index variants explained 1.6% of the variation in OSA (F statistic=709, indicating sufficient strength of the instruments). 12 All these 5 SNPs were in different genomic regions and not in linkage disequilibrium (r 2<0.20). 12 After a look‐up of the 5 SNPs in Phenoscanner, we found that the 1 SNP (rs9937053) was associated with body mass index another SNP (rs4837016) was associated with whole body fat‐free mass at genome‐wide significance level (P<5.0×10−8). 14 Potential pleiotropic effect was not found for other 3 SNPs. Considering causal effect of obesity on OSA and mediation effect of OSA in the obesity’s effect on atrial fibrillation, 12 we included these 2 SNPs in the main MR analysis but excluded them in sensitivity analysis. Table shows the characteristics and associations of the 5 included SNPs with OSA and atrial fibrillation.
Table 1.
Characteristics of the 5 Single Nucleotide Polymorphisms Associated With Obstructive Sleep Apnea
| SNP | Prioritized genes | Position (hg19) | Effect allele/other allele | Effect allele frequency | Effect on obstructive sleep apnea | Effect on atrial fibrillation | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| Beta | SE | P value | Beta | SE | P value | |||||
| rs9937053 | FTO | chr16:53799507 | A/G | 0.43 | 0.1044 | 0.0115 | 4.3×10−16 | 0.0279 | 0.0067 | 0.00003 |
| rs10507084 | RMST/NEDD1 | chr12:97753152 | T/C | 0.18 | 0.1044 | 0.0160 | 2.8×10−11 | 0.0108 | 0.0134 | 0.4194 |
| rs185932673 | CAMK1D | chr10:12698439 | T/C | 0.003 | 0.6259 | 0.1123 | 2.4×10−8 | 0.1610 | 0.0541 | 0.0029 |
| rs4837016 | GAPVD1 | chr9:128141809 | G/A | 0.53 | 0.0726 | 0.0110 | 1.5×10−8 | 0.0034 | 0.0067 | 0.6116 |
| rs10928560 | CXCR4 | chr2:136991807 | C/T | 0.80 | 0.0834 | 0.0139 | 2.8×10−8 | 0.0108 | 0.0089 | 0.2266 |
Outcomes
Summary data for the associations between the 5 OSA‐related SNPs and atrial fibrillation were obtained from the recently published GWAS. 13 In that study, associations between 34 740 186 genetic variants and atrial fibrillation were tested in a total of 60 620 cases and 970 216 controls from 6 contributing studies: HUNT (Nord‐Trøndelag Health Study), deCODE, MGI (Michigan Genomics Initiative), DiscovEHR, UK Biobank, and the AFGen (Atrial Fibrillation Genomics) Consortium. Atrial fibrillation was mainly diagnosed according to ICD‐9 or ICD‐10. The sample of this GWAS was nonoverlapping with the FinnGen Study and the majority (98.6%) of individuals were European ancestry. The associations between each OSA‐related SNP and atrial fibrillation are presented in Table.
Statistical Analysis
We used several 2‐sample MR approaches to estimate the effect of OSA on atrial fibrillation using summarized data of the SNP‐OSA and SNP‐atrial fibrillation associations. In primary analysis, we used both fixed‐effect and random‐effect inverse‐variance weighted (IVW) MR methods which assume that all SNPs are valid instruments and estimate the effect as the IVW average of ratio estimates of individual variants with first‐order weights. 15 In sensitivity analyses, we further performed several other MR methods that were more robust to the inclusion of pleiotropic and/or invalid instruments. The penalized IVW method improves the robustness by penalizing the weights of candidate instruments with heterogeneous causal estimates in the weighted regression model and the penalized robust IVW method further provides robustness both to outliers and to data points with high leverage through robust regression. 16 The MR‐Egger method uses weighted linear regression of SNP‐atrial fibrillation against SNP‐OSA estimates with the inverse‐variance of SNP‐atrial fibrillation estimate as weights. The MR‐Egger regression provides a valid effect estimate even when all the genetic variants are invalid instruments under the assumption that the association of each genetic variant with the exposure is independent of the pleiotropic effect of the variant (not via the exposure). 17 The intercept of MR‐Egger regression was adapted to test for bias from pleiotropy. It is a plausible assumption in this setting as no pleiotropic effect of the variant was observed after a look‐up of all the SNPs and the intercept of MR‐Egger regression was not significant. The median methods estimate the effect using the median of the empirical distribution function of individual SNP ratio estimates and may provide robust estimates even if up to 50% of genetic variants are invalid instruments. 18 The weighted mode‐based estimate method uses the mode of the IVW empirical density function as the effect estimate and is robust to horizontal pleiotropy. 19 The Mendelian Randomization Pleiotropy Residual Sum and Outlier approach is used to detect and correct for horizontal pleiotropic outliers through outlier removal in MR analysis with multi‐instruments. 20 The Q statistic and I2 index was applied to test the presence of heterogeneity between SNPs in the IVW analysis. 18 Evidence of pleiotropic effects were assessed using intercepts of the MR‐Egger regression. 17 An statistic was used to quantify the strength of violation of “NO Measurement Error” assumption (SNP‐exposure associations are measured without error) for MR‐Egger method and statistic <0.90 indicated potential violation of “NO Measurement Error” assumption. 21 To estimate the potential influence of outlying and/or pleiotropic SNPs, we further performed a leave‐one‐out analysis with fixed‐effect IVW MR method in which each SNP was omitted in turn. 22
The effect estimates of genetically predicted OSA on atrial fibrillation were presented as odds ratios (ORs) with their 95% CIs per 1‐unit‐higher log‐odds of OSA. The association of each SNP with OSA was further plotted against its effect on the risk of atrial fibrillation.
A power analysis was performed using a web‐based application (http://cnsgenomics.com/shiny/mRnd/). Based on the sample size of 217 955 and an alpha of 5%, our MR analysis have 80% power to detect an OR of 1.10 for atrial fibrillation per log odds of OSA.
An observed 2‐sided P<0.05 was considered as significant evidence for a causal effect. All analyses were conducted with R 4.0.3 (R Development Core Team).
RESULTS
The 2‐sample MR analyses showed a causal effect of genetically predicted OSA on the risk of atrial fibrillation using 5 SNPs as the instruments (Figure 2). Both the fixed‐effect and random‐effect IVW models showed that genetically predicted OSA was associated with an increased risk of atrial fibrillation (OR, 1.21; 95% CI, 1.12–1.31; P<0.001; OR, 1.21; 95% CI, 1.11–1.32; P<0.001). Similar results were observed using the penalized IVW, penalized robust IVW, MR‐Egger, weighted median, weighted mode‐based estimate, and Mendelian Randomization Pleiotropy Residual Sum and Outlier methods in sensitivity analyses. Associations of each variant with OSA and risk of atrial fibrillation are shown in Figure 3.
Figure 2. Risk of atrial fibrillation for genetically predicted obstructive sleep apnea.
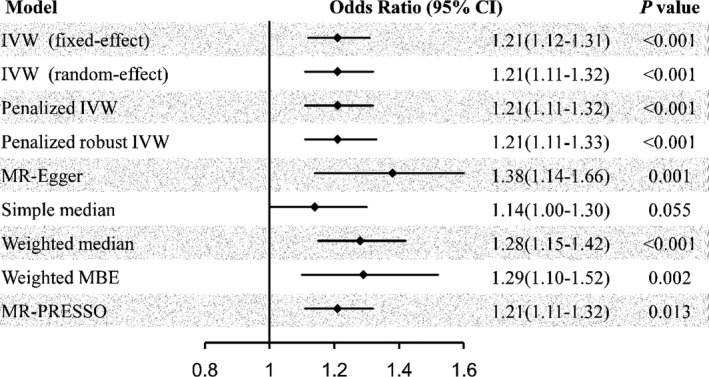
IVW indicates inverse‐variance weighted; MBE, mode‐based estimate; MR, Mendelian randomization; and MR‐PRESSO, Mendelian Randomization Pleiotropy Residual Sum and Outlier.
Figure 3. Associations of obstructive sleep apnea‐related variants with risk of atrial fibrillation.
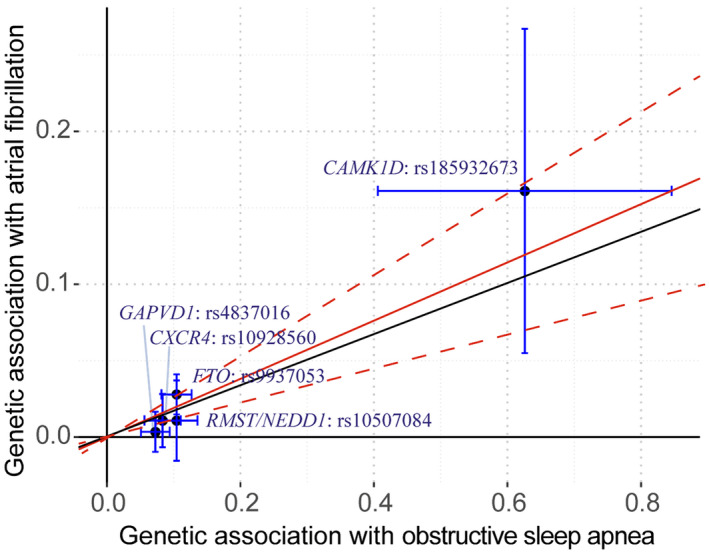
The solid red line indicates the estimate of effect using IVW method with the dashed red lines as the 95% CI. The solid black line indicates the estimate of effect using IVW method omitting of the SNP rs185932673. Circles indicate marginal genetic associations with obstructive sleep apnea and risk of atrial fibrillation for each variant. Error bars indicate 95% CIs.
There was no evidence of heterogeneity in the IVW analysis (Q=5.24, P=0.26; I2=23.6%). MR‐Egger regression showed no evidence of directional pleiotropic effect across the genetic variants (intercept, −0.014; 95% CI, −0.033 to 0.005; P=0.14). The MR‐Egger “NO Measurement Error” assumption was not fully satisfied ( statistic=72.8%). The leave‐one‐out sensitivity analysis showed that the association between OSA and atrial fibrillation was not substantially driven by any individual SNP (Figure 4).
Figure 4. MR leave‐one‐out sensitivity analysis for obstructive sleep apnea on atrial fibrillation.
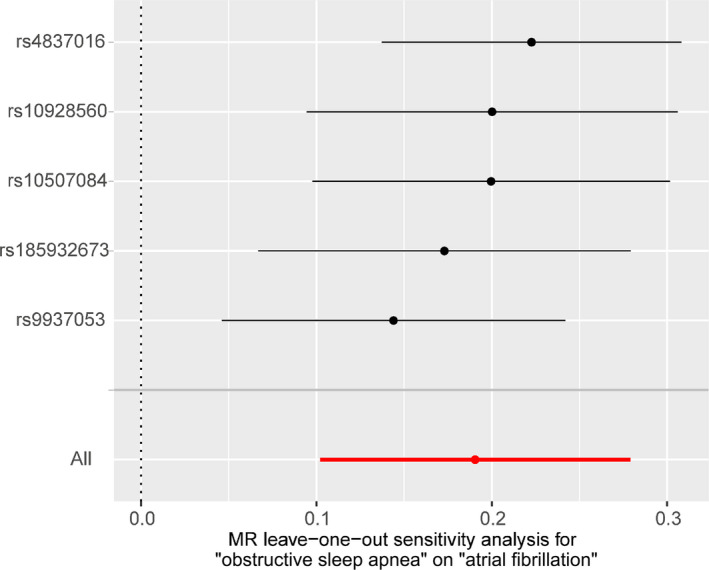
Circles indicate MR estimates for obstructive sleep apnea on atrial fibrillation using inverse‐variance weighted fixed‐effect method if each single nucleotide polymorphism was omitted in turn. The bars indicate the CI. MR indicates Mendelian randomization.
DISCUSSION
Results of this 2‐sample MR study based on data from large‐scale GWAS showed a consistent effect of genetically predicted obstructive sleep apnea on an increased risk of atrial fibrillation. The findings were robust in sensitivity analyses with different MR models.
OSA and atrial fibrillation are 2 common diseases among adults. The prevalence of OSA ranges from 3% to 49% in population‐based studies and highly prevalent among patients with atrial fibrillation ranging from 21% to 74%. 6 There were several assumed mechanisms underlying atrial fibrillation attributing to OSA. 23 , 24 OSA is characterized by a recurrent interruption in ventilation due to repetitive upper airway collapse, resulting in hypoxia, oxidative stress, inflammation, substantially negative intrathoracic pressure and hyperactivity of the cardiac autonomic nervous system which may lead to future development of atrial fibrillation. 23 , 24 Previous studies demonstrated that repetitive OSA may cause atrial structural and electrical remodeling characterized by atrial enlargement, reduction in voltage and conduction abnormalities which formed a substrate for atrial fibrillation vulnerability. 6 , 24 , 25
In recent decades, accumulating observational evidence has demonstrated that OSA was associated with development of atrial fibrillation. 7 , 26 , 27 , 28 , 29 Severity of OSA was found to be an independent predictor of incident atrial fibrillation hospitalization in a large sleep‐clinic cohort in Australia (n=6841). 7 The association between OSA and atrial fibrillation was observed in a retrospective cohort study of 3542 adults with a mean follow‐up of 4.7 years, 27 but not in another prospective, community‐based study of 2912 individuals with a mean follow‐up of 5.3 years. 28 In the REGARDS (Reasons for Geographic and Racial Differences in Stroke) study, high risk of OSA was associated with prevalent atrial fibrillation among Black participants but not White participants. 29 A recent meta‐analysis including 9 studies with 19 837 individuals showed that OSA was strongly associated with high risk of atrial fibrillation. 8 Other studies showed an increased risk of postoperative atrial fibrillation following cardiac surgery in patients with OSA. 9 , 30 However, these previous studies were most cross‐sectional, retrospective or prospective cohort studies, 8 and cannot overcome the influence of unmeasured confounders due to the nature of observational study. The present study found a causal effect of genetically predicted OSA on risk of atrial fibrillation using MR methods, which may control unmeasured confounders and reverse causation. 31 Previous studies have showed that continuous positive airway pressure therapy might reduce major adverse cardiovascular events and recurrence of atrial fibrillation in patients with OSA. 32 , 33 The present study supports more further research on strategies to detect and treat OSA and large‐scale randomized trials are warranted.
The strength of the study is the design of 2‐sample MR analysis based on OSA‐related SNPs and effects of SNP‐atrial fibrillation from large‐scale GWAS. With the 2‐sample MR design, we were able to investigate the effect of OSA on atrial fibrillation based on data with large sample sizes (16 761 OSA cases and 201 194 controls; 60 620 atrial fibrillation cases and 970 216 controls). Biases due to reverse causation or confounding are greatly reduced in MR design because the SNPs are randomly assigned at conception and are not associated with confounders. 15 Compared with observational study, MR methods can strengthen the evidence for causal inference.
Our study has several limitations. First, it is difficult to completely exclude potential horizontal pleiotropy that may lead to biased effect estimates. 17 This may also be caused by the obesity‐related SNPs (rs9937053 and rs4837016). We did not exclude this 2 SNPs in the main MR analysis, considering causal effect of obesity on OSA. 12 However, MR‐Egger regression showed no evidence of pleiotropic effect and similar results were observed in sensitivity analyses using several other robust models and leave‐one‐out analysis omitting this 2 SNPs. Second, risk of bias due to measurement error may exist for MR‐Egger regression. However, the MR‐Egger estimate of causal effect is biased towards the null when “NO Measurement Error” assumption is violated 21 ; whereas, the MR‐Egger estimate showed a positive result in our study. Finally, generalizability of our findings to other ethnics may be limited as the analysis population focused on subjects of European ancestry. However, the uniformity of participants ensures minimal risk of confounding by population admixture.
CONCLUSIONS
Our 2‐sample MR analysis provides genetic evidences of causal effect of obstructive sleep apnea on an increased risk of atrial fibrillation. Further research should investigate the strategies to detect and treat obstructive sleep apnea.
Sources of Funding
This work was supported by the National Natural Science Foundation of China (81971091, 81901177), Beijing Hospitals Authority Youth Programme (QML20190501, QML20200501), Young Elite Scientist Sponsorship Program from China Association for Science and Technology (2019QNRC001), Beijing Tiantan Hospital, Capital Medical University (2018‐YQN‐1, 2020MP01).
Disclosures
None.
For Sources of Funding and Disclosures, see page 6.
REFERENCES
- 1. Rahman F, Kwan GF, Benjamin EJ. Global epidemiology of atrial fibrillation. Nat Rev Cardiol. 2014;11:639–654. doi: 10.1038/nrcardio.2014.118 [DOI] [PubMed] [Google Scholar]
- 2. Lippi G, Sanchis‐Gomar F, Cervellin G. Global epidemiology of atrial fibrillation: an increasing epidemic and public health challenge. Int J Stroke. 2021;16:217–221. doi: 10.1177/1747493019897870 [DOI] [PubMed] [Google Scholar]
- 3. Lau DH, Nattel S, Kalman JM, Sanders P. Modifiable risk factors and atrial fibrillation. Circulation. 2017;136:583–596. doi: 10.1161/CIRCULATIONAHA.116.023163 [DOI] [PubMed] [Google Scholar]
- 4. Zhao J, Xu W, Yun F, Zhao H, Li W, Gong Y, Yuan Y, Yan S, Zhang S, Ding X, et al. Chronic obstructive sleep apnea causes atrial remodeling in canines: mechanisms and implications. Basic Res Cardiol. 2014;109:427. doi: 10.1007/s00395-014-0427-8 [DOI] [PubMed] [Google Scholar]
- 5. Linz B, Hohl M, Lang L, Wong DWL, Nickel AG, De La Torre C, Sticht C, Wirth K, Boor P, Maack C, et al. Repeated exposure to transient obstructive sleep apnea‐related conditions causes an atrial fibrillation substrate in a chronic rat model. Heart Rhythm. 2021;18:455–464. doi: 10.1016/j.hrthm.2020.10.011 [DOI] [PubMed] [Google Scholar]
- 6. Linz D, McEvoy RD, Cowie MR, Somers VK, Nattel S, Levy P, Kalman JM, Sanders P. Associations of obstructive sleep apnea with atrial fibrillation and continuous positive airway pressure treatment: a review. JAMA Cardiol. 2018;3:532–540. doi: 10.1001/jamacardio.2018.0095 [DOI] [PubMed] [Google Scholar]
- 7. Cadby G, McArdle N, Briffa T, Hillman DR, Simpson L, Knuiman M, Hung J. Severity of OSA is an independent predictor of incident atrial fibrillation hospitalization in a large sleep‐clinic cohort. Chest. 2015;148:945–952. doi: 10.1378/chest.15-0229 [DOI] [PubMed] [Google Scholar]
- 8. Youssef I, Kamran H, Yacoub M, Patel N, Goulbourne C, Kumar S, Kane J, Hoffner H, Salifu M, McFarlane SI. Obstructive sleep apnea as a risk factor for atrial fibrillation: a meta‐analysis. J Sleep Disord Ther. 2018;7:282. doi: 10.4172/2167-0277.1000282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Feng TR, White RS, Ma X, Askin G, Pryor KO. The effect of obstructive sleep apnea on readmissions and atrial fibrillation after cardiac surgery. J Clin Anesth. 2019;56:17–23. doi: 10.1016/j.jclinane.2019.01.011 [DOI] [PubMed] [Google Scholar]
- 10. Lawlor DA, Harbord RM, Sterne JA, Timpson N, Davey SG. Mendelian randomization: using genes as instruments for making causal inferences in epidemiology. Stat Med. 2008;27:1133–1163. doi: 10.1002/sim.3034 [DOI] [PubMed] [Google Scholar]
- 11. Emdin CA, Khera AV, Kathiresan S. Mendelian randomization. JAMA. 2017;318:1925–1926. doi: 10.1001/jama.2017.17219 [DOI] [PubMed] [Google Scholar]
- 12. Strausz S, Ruotsalainen S, Ollila HM, Karjalainen J, Kiiskinen T, Reeve M, Kurki M, Mars N, Havulinna AS, Luonsi E, et al.; FinnGen Research Group . Genetic analysis of obstructive sleep apnoea discovers a strong association with cardiometabolic health. Eur Respir J. 2021;57:2003091. [Epub ahead of print] doi: 10.1183/13993003.03091-2020 [DOI] [PubMed] [Google Scholar]
- 13. Nielsen JB, Thorolfsdottir RB, Fritsche LG, Zhou W, Skov MW, Graham SE, Herron TJ, McCarthy S, Schmidt EM, Sveinbjornsson G, et al. Biobank‐driven genomic discovery yields new insight into atrial fibrillation biology. Nat Genet. 2018;50:1234–1239. doi: 10.1038/s41588-018-0171-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Staley JR, Blackshaw J, Kamat MA, Ellis S, Surendran P, Sun BB, Paul DS, Freitag D, Burgess S, Danesh J, et al. PhenoScanner: a database of human genotype‐phenotype associations. Bioinformatics. 2016;32:3207–3209. doi: 10.1093/bioinformatics/btw373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Burgess S, Butterworth A, Thompson SG. Mendelian randomization analysis with multiple genetic variants using summarized data. Genet Epidemiol. 2013;37:658–665. doi: 10.1002/gepi.21758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Burgess S, Bowden J, Dudbridge F, Thompson SG. Robust instrumental variable methods using multiple candidate instruments with application to Mendelian randomization. arXiv. 2016:1606.03729. [Google Scholar]
- 17. Bowden J, Davey Smith G, Burgess S. Mendelian randomization with invalid instruments: effect estimation and bias detection through Egger regression. Int J Epidemiol. 2015;44:512–525. doi: 10.1093/ije/dyv080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Bowden J, Davey Smith G, Haycock PC, Burgess S. Consistent estimation in Mendelian randomization with some invalid instruments using a weighted median estimator. Genet Epidemiol. 2016;40:304–314. doi: 10.1002/gepi.21965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hartwig FP, Davey Smith G, Bowden J. Robust inference in summary data Mendelian randomization via the zero modal pleiotropy assumption. Int J Epidemiol. 2017;46:1985–1998. doi: 10.1093/ije/dyx102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Verbanck M, Chen CY, Neale B, Do R. Detection of widespread horizontal pleiotropy in causal relationships inferred from Mendelian randomization between complex traits and diseases. Nat Genet. 2018;50:693–698. doi: 10.1038/s41588-018-0099-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Bowden J, Del Greco MF, Minelli C, Davey Smith G, Sheehan NA, Thompson JR. Assessing the suitability of summary data for two‐sample Mendelian randomization analyses using MR‐Egger regression: the role of the I2 statistic. Int J Epidemiol. 2016;45:1961–1974. doi: 10.1093/ije/dyw220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Burgess S, Thompson SG. Interpreting findings from Mendelian randomization using the MR‐Egger method. Eur J Epidemiol. 2017;32:377–389. doi: 10.1007/s10654-017-0255-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Riaz S, Bhatti H, Sampat PJ, Dhamoon A. The converging pathologies of obstructive sleep apnea and atrial arrhythmias. Cureus. 2020;12:e9388. doi: 10.7759/cureus.9388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Zhang L, Hou Y, Po SS. Obstructive sleep apnoea and atrial fibrillation. Arrhythm Electrophysiol Rev. 2015;4:14–18. doi: 10.15420/aer.2015.4.1.14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Dimitri H, Ng M, Brooks AG, Kuklik P, Stiles MK, Lau DH, Antic N, Thornton A, Saint DA, McEvoy D, et al. Atrial remodeling in obstructive sleep apnea: implications for atrial fibrillation. Heart Rhythm. 2012;9:321–327. doi: 10.1016/j.hrthm.2011.10.017 [DOI] [PubMed] [Google Scholar]
- 26. Gami AS, Pressman G, Caples SM, Kanagala R, Gard JJ, Davison DE, Malouf JF, Ammash NM, Friedman PA, Somers VK. Association of atrial fibrillation and obstructive sleep apnea. Circulation. 2004;110:364–367. doi: 10.1161/01.CIR.0000136587.68725.8E [DOI] [PubMed] [Google Scholar]
- 27. Gami AS, Hodge DO, Herges RM, Olson EJ, Nykodym J, Kara T, Somers VK. Obstructive sleep apnea, obesity, and the risk of incident atrial fibrillation. J Am Coll Cardiol. 2007;49:565–571. doi: 10.1016/j.jacc.2006.08.060 [DOI] [PubMed] [Google Scholar]
- 28. Tung P, Levitzky YS, Wang R, Weng J, Quan SF, Gottlieb DJ, Rueschman M, Punjabi NM, Mehra R, Bertisch S, et al. Obstructive and central sleep apnea and the risk of incident atrial fibrillation in a community cohort of men and women. J Am Heart Assoc. 2017;6:e004500. doi: 10.1161/JAHA.116.004500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ghazi L, Bennett A, Petrov ME, Howard VJ, Safford MM, Soliman EZ, Glasser SP. Race, sex, age, and regional differences in the association of obstructive sleep apnea with atrial fibrillation: reasons for geographic and racial differences in stroke study. J Clin Sleep Med. 2018;14:1485–1493. doi: 10.5664/jcsm.7320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Karimi N, Kelava M, Kothari P, Zimmerman NM, Gillinov AM, Duncan AE. Patients at high risk for obstructive sleep apnea are at increased risk for atrial fibrillation after cardiac surgery: a cohort analysis. Anesth Analg. 2018;126:2025–2031. doi: 10.1213/ANE.0000000000002852 [DOI] [PubMed] [Google Scholar]
- 31. Allman PH, Aban IB, Tiwari HK, Cutter GR. An introduction to Mendelian randomization with applications in neurology. Mult Scler Relat Disord. 2018;24:72–78. doi: 10.1016/j.msard.2018.06.017 [DOI] [PubMed] [Google Scholar]
- 32. Khan SU, Duran CA, Rahman H, Lekkala M, Saleem MA, Kaluski E. A meta‐analysis of continuous positive airway pressure therapy in prevention of cardiovascular events in patients with obstructive sleep apnoea. Eur Heart J. 2018;39:2291–2297. doi: 10.1093/eurheartj/ehx597 [DOI] [PubMed] [Google Scholar]
- 33. Shukla A, Aizer A, Holmes D, Fowler S, Park DS, Bernstein S, Bernstein N, Chinitz L. Effect of obstructive sleep apnea treatment on atrial fibrillation recurrence: a meta‐analysis. JACC Clin Electrophysiol. 2015;1:41–51. doi: 10.1016/j.jacep.2015.02.014 [DOI] [PubMed] [Google Scholar]


