Abstract
Background:
B1a and B1b lymphocytes produce IgM that inactivates oxidation-specific epitopes (IgMOSE) on low-density lipoprotein and protects against atherosclerosis. Loss of inhibitor of differentiation 3 (ID3) in B cells selectively promotes B1b but not B1a cell numbers leading to higher IgMOSE production and reduction in atherosclerotic plaque formation. Yet, the mechanism underlying this regulation remains unexplored.
Methods:
Bulk RNA sequencing was utilized to identify differentially expressed genes (DEGs) in B1a and B1b cells from Id3KO and Id3WT mice. CRISPR/Cas9 and lentiviral genome editing coupled with adoptive transfer were used to identify key Id3-dependent signaling pathways regulating B1b cell proliferation and the impact on atherosclerosis. Biospecimens from humans with advanced coronary artery disease (CAD) imaging were analyzed to translate murine findings to human subjects with CAD.
Results:
Through RNAseq, P62 was found to be enriched in Id3KO B1b cells. Further in vitro characterization reveals an novel role for P62 in mediating BAFF induced B1b cell proliferation through interacting with TRAF6 and activating NFκB leading to subsequent C-MYC upregulation. Promoter-reporter assays reveal that Id3 inhibits the E2A protein from activating the P62 promoter. Mice adoptively transferred with B1 cells overexpressing P62 exhibited an increase in B1b cell number, IgMOSE levels, and were protected against atherosclerosis. Consistent with murine mechanistic findings, P62 expression in human B1 cells was significantly higher in subjects harboring a function-impairing SNP (rs11574) in the ID3 gene and directly correlated with plasma IgMOSE levels.
Conclusions:
This study unveils a novel role for P62 in driving BAFF induced B1b cell proliferation and IgMOSE production to attenuate diet-induced atherosclerosis. Results identify a direct role for Id3 in antagonizing E2A from activating the p62 promoter. Moreover, analysis of putative human B1 cells also implicates these pathways in CAD subjects, suggesting P62 as a new immunomodulatory target for treating atherosclerosis.
Keywords: Atherosclerosis, Basic Science, Research, Cell Signaling/Signal Transduction, Mechanisms, Vascular Disease
Graphical Abstract
Introduction
Cardiovascular disease (CVD) is the leading cause of death worldwide and atherosclerosis is the major underlying pathology of most CVD1. Atherosclerosis is characterized by accumulation and subsequent oxidative modification of lipoproteins within the artery wall, leading to inflammatory cell infiltration and lesion formation2. Innate and adaptive immunity contribute to atherosclerosis and B cells have emerged as important modulators of both pro- and anti-inflammatory effects in atherosclerosis. Murine B cells are divided into 2 subsets: B-2 cells, which include conventional follicular and marginal zone B cells; and B-1 cells, which are fetal liver derived and persist throughout life through self-renewal3. A wealth of evidence, both murine mechanistic and human associative studies, has implicated an atheroprotective role for B1 cells, mainly through T cell-independent production of IgMs that inactivate oxidation specific epitopes (OSE) on LDL, and prevent lipid uptake and inflammatory cytokine production by macrophages4,5,6,7. In mice, B1 cells can be subtyped into B1a and B1b subsets based on their expression of CD58. Both B-1a and B-1b cells produce IgM in vivo, however, adoptive transfer studies in ApoE−/−Rag1−/− mice revealed that B1b cells produce significantly higher levels of IgM in vivo than B1a cells 9. As such, identifying the molecular and cellular mechanisms that promote increased B-1b cell numbers may have important implications for preventing atherosclerosis and its progression.
ID3 is a member of the helix-loop-helix (HLH) family of transcription factors. The HLH transcription factor family contains members with a basic DNA-binding domain (bHLH) such as E12 and E47 10,11 and those without the DNA-binding domain, such as the ID proteins, which can antagonize E12 and E47 function by dimerizing with them, inhibiting their DNA binding12,13. There are 4 ID proteins (ID1–4) that have both unique and redundant properties12. ID3 has emerged as an important factor in atherosclerosis10. Specific deletion of Id3 in B cells (Id3BKO) in ApoE−/− mice selectively increased B-1b cell numbers in the peritoneal cavity (PerC), bone marrow (BM) and spleen, increased plasma atheroprotective IgM levels and attenuated diet-induced atherosclerosis9. Yet, the molecular mechanisms mediating the selective increase in B1b cell number in Id3BKO mice are unknown.
The present study demonstrated that loss of Id3 increased proliferation in B-1b but not B-1a cells. Bulk RNA sequencing on sorted PerC B1a and B1b cells from Id3WT and Id3KO mice identified only 20 differentially expressed genes (DEGs) in B-1a cells compared to 1903 DEGs in B-1b cells. While genes involved in B cell proliferation were among the DEGs in B-1b cells from Id3KO mice, P62, a scaffold and ubiquitin binding protein, was found to be one of the most highly enriched genes. Further pathway analysis also suggested that P62 might regulate B1b cell proliferation through B cell activating factor (BAFF) and NFkB signaling. Imaging flow cytometry and proliferation assays provided evidence that BAFF stimulation recruited P62 to TRAF6 and further activated NFKB signaling to drive B1b proliferation. E12 and E47 gain of function through overexpression or loss of ID3 resulted in increased P62 promoter activation and protein expression. Increased expression of P62 in B1b cells enhanced proliferation, increased atheroprotective IgM in plasma and reduced atherosclerosis formation in mice. Moreover, humans harboring a SNP in ID3 at rs11574, shown to attenuate Id3 function10 had higher expression of P62 expression on their B1 cells. Higher P62 expression on human B1 cells was significantly associated with the % of B1 cells in circulation and plasma levels of IgM to MDA. This study provides insights into mechanisms that mediate ID3 effects on B-1b proliferation and is the first to identify a role for P62 in B1 cell-mediated atheroprotection, both novel findings with important therapeutic implications.
Methods
Data Availability
Human Subjects
Coronoary artery disease subjects were recruited for study through the Cardiac Catheterization laboratory at the University of Virginia. All participants provided written informed consent before enrollment, and the study was approved by the Human Institutional Review Board. Peripheral blood was obtained from these participants prior to catherization. In addition, peripheral blood from healthy volunteers were also obtained after providing with written informed consent.
Mice
All animal protocols were approved by the Animal Care and Use Committee at the University of Virginia. Id3fl/fl mice were a generous gift from Dr. Yuan Zhuang at Duke University. CD19Cre/+ mice were provided by Dr. Bender (University of Virginia). C57BL/6J mice were purchased from Jackson Laboratory. These mice were fed a standard chow diet or a Western diet. All the mice utilized in all the experiments were 8–10 weeks old female mice.
Cell Preparations for Murine and Human flow cytometry
Peritoneal cavity cells were processed for flow cytometry or fluorescence activated cell (FAC) sorting as previously described9,14. Isolation of human PBMCs was performed as previously described9,14. Clone and fluorophore information as well as isotype control for flow cytometry antibodies were provided in supplementary table 3.
Samples preparation of Bulk RNA sequencing
Sort-purified peritoneal B1a and B1b obtained from Id3KO and Id3WT C57BL/6 mice were RNA extracted by using Qiagen RNeasy Plus kit. The purified RNAs were stored at −80°C and sent to a Novogene to perform sequencing.
Differentially expressed genes and pathway analysis
RNA sequences in raw FASTQ data files were obtained from Novogene and HISAT2/Stringtie/Ballgown pipeline was utilized for RNASeq analysis following the previously published protocol15 Volcano plots of differentially expressed genes were visualized by using the python bioinfokit package. Ingenuity Pathway Analysis was performed on all the annotated RNA to analyze for differentially regulated cellular processes, canonical pathways and network analysis.
In Vitro and In Vivo Cell Proliferation Assay
Peritoneal cavity cells were enriched for B cells using the EasySep Mouse Pan-B cell enrichment kit (STEMCELL), and then labeled with 5uM Celltrace-violet for 20 minutes at 37oC, and then washed with FACS buffer (1% FBS, 0.1%NaN3 in PBS). Labeled cells were cultured in B cell culture media for 3 days with 20ng/mL recombinant BAFF or PBS control to measure in vitro proliferation. For in vivo proliferation, sort-purified p62-gRNA-ATTO+ Celltrace-violet+ B1 cells were intraperitoneally transferred into C57BL/6 mice.
Lentiviral Production and P62 Overexpression on mouse B cells.
P62-eGFP lentivirus was generated using pLV-eGFP vector (purchased from Addgene) and mouse P62 was subcloned into the vector. The eGFP or P62-eGFP lentiviruses were generated using four plasmid system (pLV-eGFP or pLV-p62-eGFP, pLP1, pLP2, and pVSV-G) into 293T cells. The eGFP ctrl and P62-eGFP lentiviruses were used to transduce enriched peritoneal mouse B cells. Transduced murine B cells were harvested 48 hrs post transduction and FAC-sorted for GFP+ B1 cells to be used for adoptive transfer.
p62 CRISPR/sgRNA Knockout and PP62-GFP transfection on Mouse B cells.
crRNA targeting p62 (IDT) was conjugated with tracRNA-ATTO (IDT). Pp62-GFP plasmid was generated by subcloning 1000bp of p62 promoter to replace CMV promoter in CMV-GFP (Addgene). Murine peritoneal cells stimulated with 50ng/mL LPS overnight. The stimulated B cells were then nucleotransfected with Cas9, and pre-conjugated P62 crRNA-tracRNA ATTO, Pp62-GFP, or CMV-GFP ctrl plasmid using the P3 Primary cells Nucleofection Kit (Lonza). B cells were collected for analysis and sorting 24 hrs after nucleofection.
Nucleofection of Engineered Plasmids into Human B cells.
Heatlhy donors’ PBMCs were thawed and washed with RPMI +5% FBS. B cells were enriched using EasySep human pan-B cell enrichment kit (STEMCELL). CMV-E12-Flag, CMV-E47-Flag were generated using CMV-Flag plasmid to subclone human E12 and E47 into. These plasmids were nucleotransfected into enriched human B cells t using the P3 Primary cells Nucleofection Kit (Lonza). B cells were collected for analysis and sorting 24 hrs after nucleofection.
Quantification of markers colocalization by using imaging flow cytometry
Murine peritoneal cells obtained from Id3KO and Id3WT mice were enriched for B cells. The enriched B cells were incubated with 20ng/mL murine recombinant BAFF or PBS control for 12 hrs in B cell media. Prior to running on the Imagestream imaging flow cytometry machine, cells were stained using previously developed surface staining protocol9,14 following with permeabilization and intracellular staining using FIX & PERM Cell Permeabilization Kit (Invitrogen). Images of 1000 Id3KO or Id3WT B1b cells from both unstimulated, BAFF stimulated conditions were collected, and quantitative co-localization analysis was performed using Amnis Imagestream colocalization software.
ELISA for Quantification of Total and Anti-OSE IgM or IgG Isotypes in Mice and Humans
Total IgM subtypes in mouse plasma were measured using colorimetric ELISA as described previously9. Levels of IgM and IgG against the MDA mimotope16 in human plasma were measured by chemiluminescent ELISA as previously described16.
Adoptive Transfer
Celltrace violet+ or eGFP+ (eGFP-ctrl or P62-eGFP) B1 (CD3-CD19+B220-) cells were sort-purified and adoptively transferred into C57BL/6 mice intraperitoneally. Mice adoptively transferred with Celltrace-violet+ cells were fed a chow diet for 2 weeks. Mice adoptively transferred with eGFP-ctrl or P62-eGFP B1 cells were fed with Western diet for 8 weeks and harvested for peritoneal cells, hearts and aortas.
Analysis of Atherosclerotic Lesions
Hearts and aortas were harvested as previously described10. Plaque areas were assessed using Image-Pro Plus software (Media Cybernetics). The analysis was done by the lab specialist without knowing the group allocation.
Statistics
Statistics were performed using GraphPad Prism Version 7.0a (GraphPad Software, Inc), Python 3.0, R 3.6.1, or SAS 9.4. Results from all replicated experiments are displayed, and bar graphs display Mean ± SD.
Results
Loss of ID3 promoted BAFF-P62-NFκB expression and proliferation in B1b but not B1a cells.
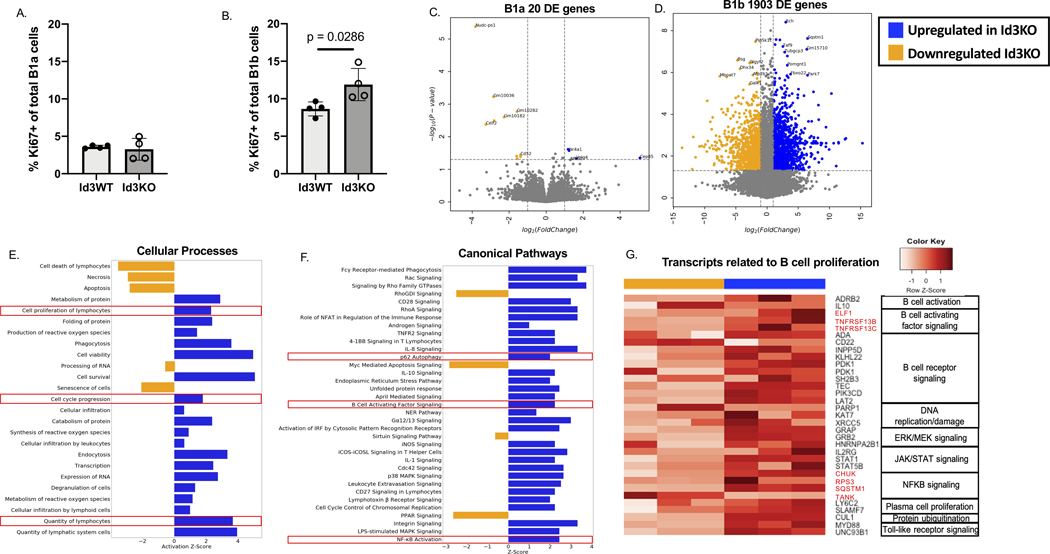
To determine potential mechanisms mediating the selective increase in B-1b cell number in Id3BKO mice we measured Ki67 expression in B1a and B1b cells obtained from the peritoneal cavity of C57BL/6 mice null for Id3 (Id3KO) and littermate controls (Id3WT). Results showed an average of 3.59% of B-1a and 8.64% of B-1b cells in the PerC expressed Ki67 at homeostasis and that B-1b but not B-1a cells had a significant increase in the %Ki67 positivity with loss of Id3 (Figure 1A-B, gating strategy shown in Supplementary Figure. 1). To identify genes and pathways whereby Id3 may regulate B1b cell number, bulk RNA sequencing was performed on PerC B-1a and B-1b cells obtained from Id3KO and Id3WT mice. Differentially expressed gene (DEG) analysis revealed only 20 DEG (FDR ≤ 0.05) out of 14933 aligned genes in B1a cells between Id3WT and Id3KO. In contrast, there were 1903 DEG (FDR ≤ 0.05) out of 14399 aligned genes in B1b cells (Figure 1C-D). Notably, Sqstm1 or p62 was markedly overexpressed in Id3KO B1b cells compared to Id3WT B1b cells with a 90.8 folds increase and a p-value of 8.7×10−5 (supplementary table 1). Sqstm1/p62 was not differentially expressed in B1a cells (supplementary table 2).
Figure 1: Loss of Id3 promoted BAFF-P62-NFKB expression and proliferation in B1b but not B1a cells.
(A-B) Percentage of Ki67+ of total B1a (A) and B1b (B) obtained from Id3WT (n = 4) and Id3KO (n =4) peritoneal cells (C-D) Volcano plots compared between Id3WT (yellow, n = 3) vs Id3KO (blue, n = 3) B1a (C) and B1b (D) cells obtained from peritoneal cavity. (E-F) Cellular processes (E) and canonical (F) pathway analysis determining pathways downregulated in Id3KO (yellow) and upregulated in Id3KO (blue) B1b cells. (G) Transcripts related to B cell proliferation categorized by canonical pathways downregulated in Id3KO (yellow) and upregulated in Id3KO (blue) in B1b cells. Results were represented with Mean ± SD with unpaired Mann-Whitney t-test.
Consistent with the increase in the %Ki67+ B-1b cells in Id3KO mice, pathway analysis of cellular processes indicated upregulation of cell proliferation, cell cycle progression, as well as cell survival/viability in Id3KO B1b cells compared to Id3WT B1b cells (Figure 1E). Canonical pathway analysis demonstrated an enrichment of cell survival/proliferation related pathways such as P62 autophagy, B cell activating factor (BAFF), and NFκB signaling in Id3KO B1b cells compared to Id3WT (Figure 1F). Consistent with the increase in proliferation and survival pathways in Id3KO B1b cells, genes involving in B cell activation, BCR activation, DNA replication and repair, ERK/MEK, JAK/STAT, BAFF, NFκB, TLR signaling, plasma cell proliferation, and protein ubiquitination were enriched in Id3KO B-1b cells as shown in Figure 1G. CD22 (a negative regulator of BCR activation), PARP1 (Poly ADP-ribose polymerase 1 which regulates DNA protein interaction during DNA repair), and TANK (a negative regulator of TRAF2 mediated NFκB activation) were downregulated.
Loss of Id3 increased BAFFR expression and BAFF-induced B-1b proliferation in a P62-dependent manner
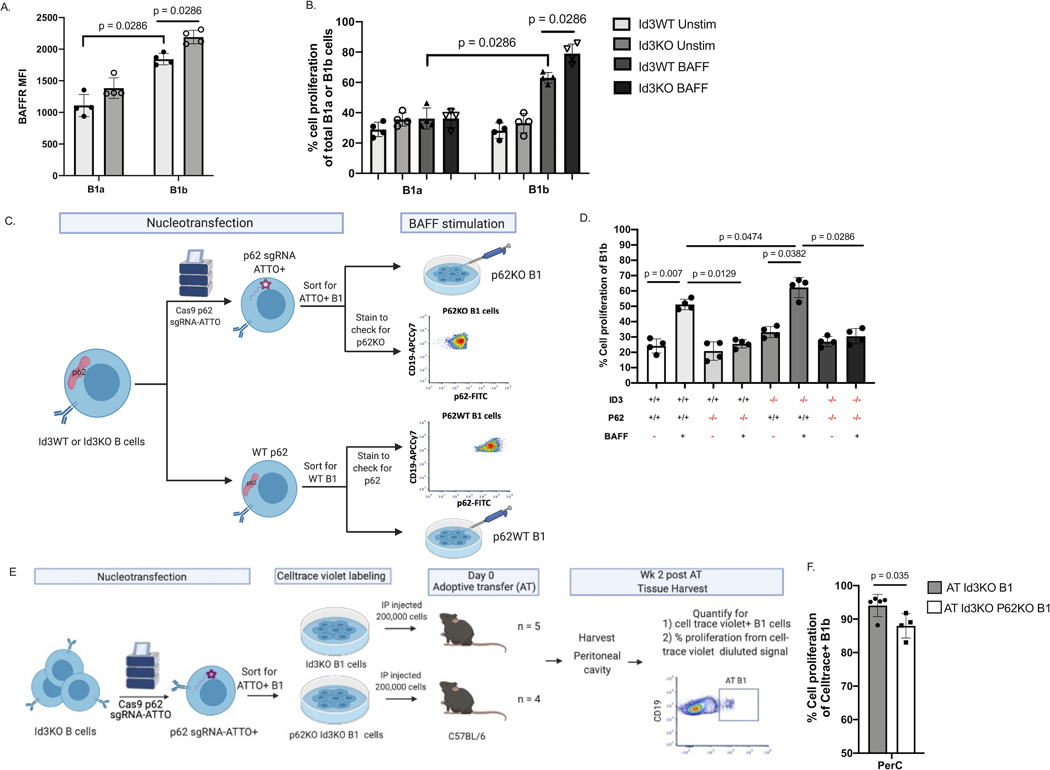
BAFF is a key factor that promotes B cell proliferation and survival17 and DEG analysis from bulk RNAseq indicated an enrichment in Baffr/Tnfrsf13c gene in B-1b cells from Id3KO compared to Id3WT (Figure 1D and supplementary table 2). We investigated BAFFR protein expression level on B-1a and B-1b cells from Id3KO and Id3WT mice by flow cytometry. BAFFR is more highly expressed on B1b compared to B1a cells and expression was significantly increased in B1b cells but not in B1a cells from Id3KO compared to Id3WT mice (Figure 2A). Consistent with these findings, BAFF stimulation of B1a and B1b cells obtained from Id3WT and Id3KO mice revealed a significant increase in cell proliferation in B-1b but not B-1a cells, that was further augmented in the Id3KO mice (Figure 2B).
Figure 2: Loss of P62 significantly reduces B1b cell proliferation in vitro and in vivo.
(A) MFI of BAFFR in Id3WT and Id3KO B1a and B1b cells (n = 4 per each group). (B) percentage of cell proliferation measured by Celltrace-violet on Id3WT and Id3KO B1a and B1b cells under unstimulated and BAFF stimulated conditions (n = 4 per each group). (C) Schematics showing nucleofection of Cas9 p62 targeted gRNA labeled with ATTO fluorophore ribonucleoprotein complex (p62-gRNA-RNP) into murine Id3WT and Id3KO B cells following with sorting for ATTO+ B1 cells to select for p62KO B1 cells. (D) Percentage of cell proliferation of Id3WT and Id3KO B1b cells with conditional p62KO and BAFF stimulation measured by Celltrace-violet (n = 4 per each group). (E) Schematics showing nucleofection of p62-gRNA-RNP into Id3KO B cells and sorting for ATTO+ B1 cells to select for p62KO B1 cells, following with AT of Id3KOp62KO or Id3KO B1 cells into C57BL/6 to quantify proliferation of AT cells in various tissue compartment 2 wks post AT. (F) Percentage of cell proliferation of AT Id3KO (grey, n = 5) and Id3KOp62KO (white, n = 3) B1b cells 2 wk post transfer measured by Celltrace-violet. Results were represented with Mean ± SD with unpaired Mann-Whitney t-test.
As Sqstm1/p62 was one of the most highly enriched transcripts in Id3KO compared to Id3WT B1b cells, yet a role for P62 in B cell proliferation has not been reported, we sought to determine whether P62 was critical for B-1b proliferation. Nucleofection of Cas9 and p62 targeted gRNA was utilized to knockout P62 in B cells (Figure 2C). Consistent with the prior finding that loss of Id3 in B1b cells led to an increase in BAFFR level (Figure 2A), BAFF induced B1b cell proliferation was significantly higher in Id3KO than Id3WT B1b cells (Figure 2D). In addition, this result also demonstrated a significant reduction of BAFF-induced B1b cell proliferation with knockout of p62 in both Id3WTand Id3KO mice (Figure 2D). These results suggested a crucial role of P62 in mediating BAFF induced B1b cell proliferation.
To determine if P62 promoted B1b cell proliferation under loss of Id3 in vivo, p62 was knocked out in Id3KO B1 cells (Figure 2E). Equal numbers of B1 cells with double knockout of p62 and Id3 and B1 cells with only single knockout of Id3 were adoptively transferred (AT) through intraperitoneal injection into WT C57BL/6 mice. Mice were harvested 2 weeks post AT and cell trace violet was used to track the transferred cells as well as measured their proliferation (Figure 2E). Results demonstrated that transferred Id3KO-p62KO B1b cells had significantly lower proliferation rates when compared to Id3KO B1b cells (Figure 2F) despite the comparable level of BAFF in the host mice (Figure S2).
Loss of Id3 and increased expression of E12 and E47 promote P62 promoter activation and protein expression
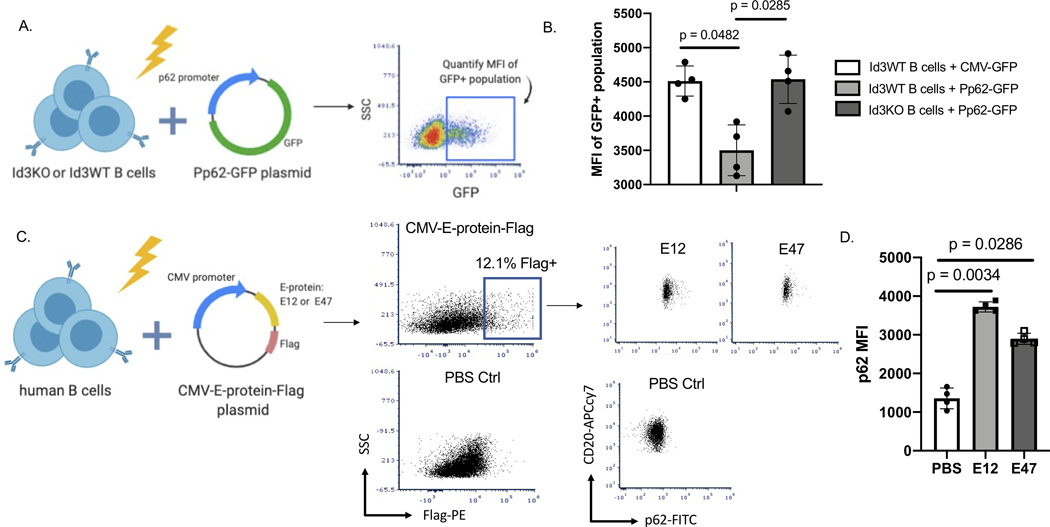
To determine potential mechanisms whereby ID3 may regulate P62 expression, we analyzed the P62’s promoter. ID3 is a transcription regulator that dimerizes with bHLH factors, particularly the E-proteins E12 and E4710,18,19, to inhibit their ability to regulate promoter activation. Indeed, analysis of the p62 promoter region revealed multiple consensus sequences for E-protein binding (Ebox: CANNTG) (Supplementary Figure. 3). To determine the impact of loss of Id3 on the p62 promoter, we cloned a 1000 bp region of the p62 promoter into a plasmid upstream of the GFP coding region. CMV-GFP (control) and the p62 promoter-GFP plasmid (Pp62-GFP) were nucleo-transfected into Id3WT and Id3KO murine B cells. We confirmed the ability of the constructs to produce eGFP protein using flow cytometry (Figure 3A). Results demonstrated that B cells from Id3KO mice had significantly greater p62 promoter activation compared to Id3WT (Figure 3B). To determine whether increased expression of E12 and E47 impacted P62 protein expression in human B cells, we cloned C-terminal FLAG-tagged E12 or E47 cDNA downstream of the CMV promoter in an expression plasmid and performed nucleofection with PBS or these constructs into sort-purified human B cells (Figure 3C). Flow cytometry of the FLAG+ population for intracellular staining of P62 revealed significantly more P62 protein in the cells with E12 and E47 overexpressed compared to PBS control (Figure 3D).
Figure 3: Loss of Id3 and increased expression of E12 and E47 promote p62 promoter activation.
(A) Schematics showing nucleofection of p62 promoter GFP plasmid (Pp62-GFP) or CMV-GFP plasmid control in murine Id3WT or Id3KO B cells to quantify GFP expression. (B) MFI of GFP+ B cells to compare GFP expression between Id3WT B cells transfected with CMV-GFP control (n = 4), Id3WT B cells transfected with Pp62-GFP (n = 4) and Id3KO B cells transfected with Pp62-GFP (n = 4). (C) Schematics demonstrating nucleo-transfection CMV-E12-Flag and CMV-E47-Flag plasmids into human B cells to measure P62 expression driven by overexpression of E-protein. (D) MFI of P62 compared across PBS control (n = 4), CMV-E12-Flag (n = 4) and CMV-E47-Flag (n = 4) transfection in human B cells. Results were represented with Mean ± SD. Statistical analyses were performed using Kruskal-Wallis test with Dunn’s correction for multiple comparison.
BAFF-induced binding of P62 to TRAF6 in B1b cells, but not B1a cells, led to NFκB activation, upregulation of c-myc expression and cell proliferation
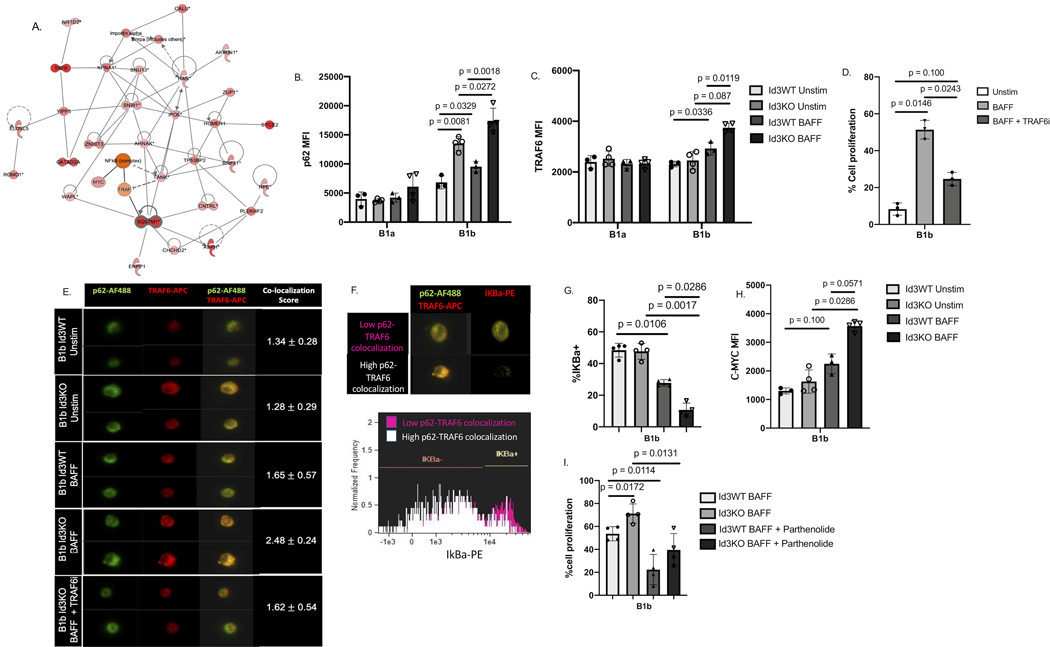
While SQSTM1/P62 has been implicated in cellular proliferation in cancer cells, it’s role in BAFF-induced B cell proliferation is unknown. To identify potential P62 interacting proteins in B cells that may mediate P62-dependent BAFF-induced proliferation, a protein-protein interaction network using DEGs obtained from our RNAseq data was performed. Through interaction network analysis, SQSTM1/P62 was shown to interact with TRAF and TANK which are proteins involved in NFκB activation and cell proliferation (Figure 4A).
Figure 4: BAFF induced binding of P62 to TRAF6 in B-1b, but not B-1a cells, leading to NFKB activation and upregulation of c-myc driven cell proliferation.
(A) protein interaction analysis indicated SQSTM1/P62 interaction with proteins related to cell proliferation and cell cycle. (B-C) MFI of P62 (B) and TRAF6 (C) on Id3WT (n = 4) and Id3KO (n = 4) B1a and B1b cells under unstimulated and BAFF stimulated conditions. (D) percentage of in vitro cell proliferation of total B1b (n = 4) measured by Celltrace -violet under unstimulated, BAFF stimulated, and BAFF + TRAF6 inhibitor conditions. (E) Co-localization between P62 and TRAF6 on B1b cells (n = 3, 1,000 B1b cells per mouse) under unstimulated, BAFF stimulated, and BAFF + TRAF6 inhibitor conditions quantified by imaging flow cytometry. (G) Representative images of IKBa degradation with low and high P62 and TRAF6 co-localization. (G) percentage of IKBa+ on Id3WT (n = 4) and Id3KO (n = 4) B1b cells under unstimulated and BAFF stimulated conditions. (H) MFI of C-MYC on Id3WT (n = 4) and Id3KO (n = 4) B1b cells under unstimulated and BAFF stimulated conditions. (I) percentage of in vitro cell proliferation of total Id3WT (n = 4) and Id3KO (n = 4) B1b measured by Celltrace -violet under BAFF stimulated, and BAFF + Parthenolide inhibitor conditions. Results were represented with Mean ± SD. Statistical analyses were performed using Kruskal-Wallis test with Dunn’s correction for multiple comparison.
Flow cytometry demonstrated increased P62 protein in response to BAFF stimulation in B-1b but not B-1a cells from Id3KO and Id3WT mice. Notably, the BAFF-stimulated P62 protein expression was significantly greater in B-1b cells from Id3KO compared to Id3WT mice (Figure 4B). Moreover, consistent with the RNAseq results, no difference in TRAF6 expression was observed between Id3KO and Id3WT mice in either B1a or B1b cells under unstimulated condition (Figure 4C). However, BAFF stimulation increased TRAF6 expression in B-1b but not B-1a cells and this increase was further augmented in Id3KO B1b cells (Figure 4C), suggesting that TRAF6 is an important mediator of BAFF-induced proliferation in B-1b cells. To test this hypothesis, we measured cell proliferation of B1b cells stimulated with BAFF in the presence and absence of the TRAF6 inhibitor (TRAF6i, C17H17NO)20. BAFF-induced B1b cell proliferation was significantly reduced by the TRAF6i (Figure 4D).
To determine if P62 and TRAF6 interact in B-1b cells and if BAFF stimulation promotes this interaction, imaging flow cytometry was performed (Figure 4E). Under unstimulated conditions, colocalization of P62 and TRAF6 was observed in Id3WT B1b cells (score = 1.34 ± 0.28). Consistent with our flow cytometry findings (Figure 4B), higher P62 expression was observed in Id3KO B1b cells, however the co-localization score (score = 1.28 ± 0.29) was unchanged. In contrast, BAFF stimulation increased P62-TRAF6 colocalization particularly in B-1b cells from Id3KO mice (score = 2.48 ±0.24). The addition of the TRAF6i to the BAFF-stimulated Id3KO B-1b cells resulted in abrogation of BAFF-induced increase in P62-TRAF6 co-localization (score = 1.62 ±0.54). In addition, we utilized confocal microscopy to visualize P62 and TRAF6 colocalization (Figure S4A) as well as proximity ligation assay (PLA) to visualize co-localization of P62 and TRAF6 (Figure S4B) and found that BAFF stimulation enhanced P62 and TRAF6 co-localization.To determine if P62-TRAF6 interaction impacts downstream NFκB signaling, we measured the endogenous NFκB inhibitor, IκBa, in B-1b cells with low (pink, co-localization score < 1.5) and high (white, co-localization score > 2) P62-TRAF6 colocalization (Figure 4F). IκBa was observed at higher level in cells with low P62-TRAF6 co-localization (Figure 4G). Consistent with this finding, IκBa levels in BAFF stimulated B-1b cells were significantly reduced compared to unstimulated B-1b cells, and Id3KO B-1b cells demonstrated a further decrement compared to Id3WT (Figure 4G). BAFF stimulation increased C-MYC expression in B-1b cells at a trending significance, an effect further increased in Id3KO mice (Figure 4H). To further validate that BAFF-P62-TRAF6 driven B-1b cell proliferation is NFκB dependent, we utilized parthenolide (NFκB inhibitor)21 to perform in vitro proliferation assay. Results indicated a significant reduction of BAFF-induced cell proliferation in parthenolide treated B1b cells from both Id3WT and Id3KO mice (Figure 4I), implicating a crucial role of NFκB in mediating BAFF induced B1b cell proliferation.
In addition, we also investigated the impact of BAFF stimulation as well as loss of Id3 on phosphorylation of mTOR (p-mTOR), another potential pathway driving cell proliferation20. The result demonstrated that BAFF induced mTOR phosphorylation in both Id3WT and Id3KO B1b cells with a trend toward higher phosphorylation in the absence of Id3 (Figure S5A).
As NRF2 is a known transcription factor involving in P62-mediated autophagy22,23 and autophagy can be a potential mechanism underlining an increase in B1b cell number, we assessed expression level of NRF2 as well as cell viability in Id3WT and Id3KO B1b cells with and without BAFF stimulation. The results demonstrated that loss of Id3 increased NRF2 expression in B1b cells in a BAFF-independent manner (Figure S5B). In addition, both BAFF stimulation and loss of Id3 were shown to increase B1b cell viability (Figure S5C).
Overexpression of P62 increased B-1b cell number and plasma IgM levels and reduced diet induced atherosclerosis
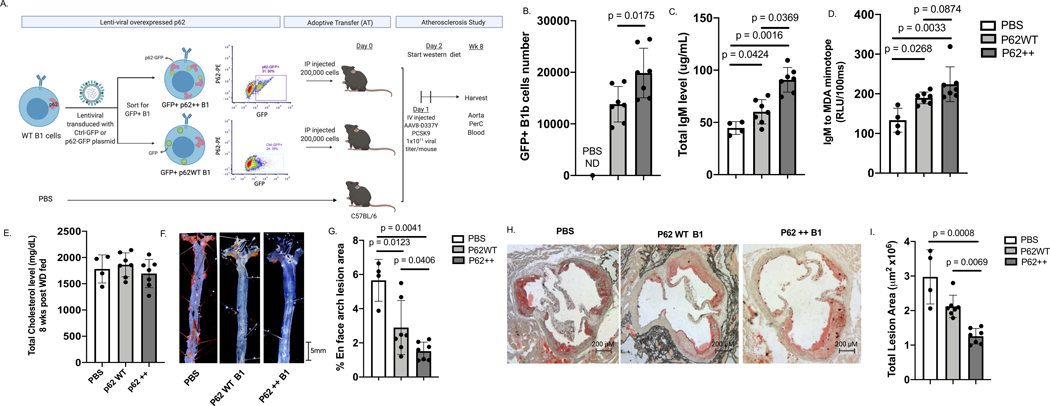
We utilized a lentiviral system to overexpress GFP-P62 and GFP control in murine B1 cells. Equal numbers of the engineered cells or PBS were intraperitoneal injected into C57BL/6 WT mice (Figure 5A). Hyperlipemia was induced by tail vein injections of AAV8 D337Y PCSK924 and Western diet (WD) feeding (Figure 5A). After 8 weeks of WD, the number of GFP+ B1b cells in the PerC of mice injected with GFP-P62-transduced cells was significantly higher than those injected with equal numbers of GFP control-transduced cells and undetectable in PBS injected mice (Figure 5B), while there was no difference in the number of GFP+ B1a cells (Figure S6). Additionally, levels of plasma total IgM and IgM specific to MDA-mimotope16 were higher in mice adoptively transferred with GFP+ B1 cells and GFP+ P62++ B1 cells compared to PBS control (Figure 5C+5D). More importantly, plasma levels of total IgM was higher at statistical significance level and IgM to MDA-mimotope was higher at a trending significance level in mice receiving GFP+ P62++ B1 cells than mice receiving GFP+ B1 cells (Figure 5C+5D), while no difference in IgG to MDA-mimotope post AT (Figure S7A) nor differences in IgM and IgG to MDA-mimotope pre AT were observed (Figure S7B+7C). However, one particular mouse in P62++ B1 cells group had a value of IgM to MDA-mimotope > 2SD above the average. Excluding this mouse from the analysis, IgM to MDA-mimotope in mice receiving P62++ B1 cells was not higher than mice receiving P62WT B1 cells (p = 0.138). Additionally, plasma IgM level was significantly lower when P62 was knocked out of B1b cells from Id3KO mice (Figure S8). There was no difference in plasma total cholesterol levels (p = 0.47) among PBS, P62++ and P62 WT B1 groups (Figure 5E). Despite similar level of cholesterol, Sudan IV en face staining revealed less atherosclerosis in mice adoptively transferred with GFP+ B1 cells and GFP+ P62++ B1 cells than the PBS control group and the P62++ B1 group had lower atherosclerosis level when compared to the P62WT B1 control group (Figure 5F+5G). To confirm and extend these findings, cross-sectional analysis of the ascending aorta was employed. Consistent with en face analysis, mice adoptively transferred with GFP+P62++ B1 cells had significantly smaller total atherosclerotic plaque area than mice with adoptively transferred GFP+P62WT B1 cells (Figure 5H+5I).
Figure 5: Overexpression of P62 increased B-1b cell number and plasma IgM levels and reduced diet induced atherosclerosis.
(A) Schematics depicting lentiviral overexpression of P62-eGFP plasmid and GFP plasmid control in murine B cells following FAC sorting for eGFP+ B1 cells and adoptive transfer of P62-eGFP overexpressed B1 (P62++), eGFP overexpressed B1 (P62WT), or PBS into C57BL/6 host mice to perform diet induced atherosclerosis study. (B) Number of recovered GFP+ B1b post atherosclerosis study compared among PBS (n = 4), P62++ (n = 7) and P62 WT groups (n = 6). (C) Plasma total IgM level 8 weeks post-AT compared among PBS (n = 4), P62++ (n = 7) and P62 WT groups (n = 6 ) measured by ELISA. (D) Plasma IgM to MDA mimotope level 8 weeks post-AT compared compared among PBS (n = 4), P62++ (n = 7) and P62 WT groups (n = 6) measured by ELISA. (E) Plasma total cholesterol level compared compared among PBS (n = 4), P62++ (n = 7) and P62 WT groups (n = 6) after 8 weeks western diet fed. (F) Representative images of en face stained aortic arch regions in P62++ B1 and P62WT B1 AT groups at the end of 8 weeks western diet fed. (G) Quantitative analysis of en face arch lesion area compared among PBS (n = 4), P62++ (n = 7) and P62 WT groups (n = 6). (H) Representative images of cross sections of the matched region of the aortas obtained from PBS control, P62++ B1 and P62WT B1 AT groups at the end of 8 weeks western diet fed. (I) Quantitative analysis of atheroscleorotic plaque areas compared among PBS (n = 4), P62++ (n = 7) and P62 WT groups (n = 6). Results were represented with Mean ± SD. Statistical analyses were performed using Kruskal-Wallis test with Dunn’s correction for multiple comparison.
Human B1 cells from subjects with an ID3 SNP at rs11574, known to attenuate Id3 function, have increased P62 expression; a finding associated with B1 frequency and plasma IgM to MDA-mimotope levels.
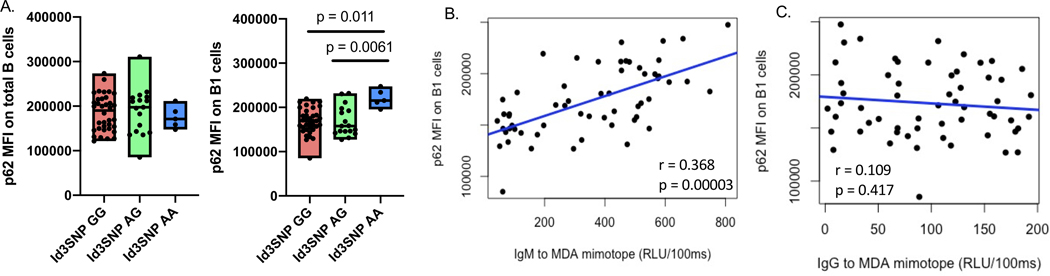
Human missense polymorphism of ID3 rs11574 (A105T) has a reduced ability in dimerizing with its binding partners10. This human ID3 rs11574 SNP was also previously shown to be associated with an increased human B1 cell (CD20+CD27+CD43+) frequency and higher level of plasma IgM to MDA-LDL9. Here, we measured expression (MFI) of P62 in 58 CAD subjects with the homozygous major (GG, n = 36), heterozygous (AG, n = 17), and homozygous minor allele of ID3 rs11574 SNPs (AA, n = 5) otherwise matched for age, sex, and total cholesterol. Result demonstrated significantly higher P62 expression in human B1 cells (CD20+CD27+CD43+) in subjects with ID3 rs11574 homozygous minor allele when compared to the heterozygous, and homozygous major allele (Figure 6A). In addition, P62 expression directly correlated with the frequency of human B1 cells (Figure S9) and plasma level of atheroprotective IgM specific to MDA-mimotope (Figure 6C) with no correlation with IgG to MDA-mimotope (Figure 6D) in these 58 CAD subjects.
Figure 6: Human B1 cells from subjects with an ID3 SNP at rs11574, known to attenuate Id3 function, have increased P62 expression; a finding associated with B1 frequency and plasma IgM to MDA-LDL levels.
(A) MFI of P62 within P62+ population in human total B and human B1 (CD27+CD43+) cells across major (n = 36), heterozygous (n = 17), and minor allele (n = 5) ID3 rs11574 SNP otherwise age and sex matched. (B-C) correlations between IgM specific to MDL mimotope (B) with P62 expression (n = 58), and IgG specific to MDL mimotope with P62 expression (n = 58) (C). Results were represented with Mean ± SD. Statistical analyses were performed using Kruskal-Wallis test with Dunn’s correction for multiple comparison and correlation analysis was performed by using Spearman correlation.
Discussion
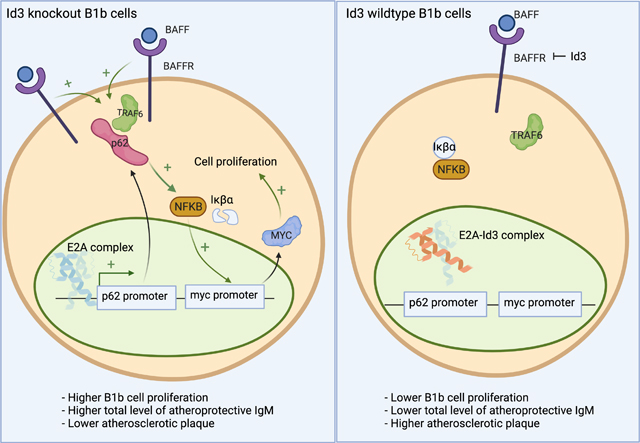
The B1b subtype of B1 cells has been shown to have unique features compared to B1a cells, allowing for prolonged protection from infection25 and increased in vivo production of IgM that can block inflammation, protecting from diet-induced atherosclerosis6,26–29. A previous study by Rosenfeld et al9 has demonstrated that loss of helix-loop-helix factor, Id3 in B cells results in an increase in B1b but not in B1a cell number and a reduction in atherosclerosis formation. Yet, the molecular and cellular mechanisms that allow Id3 to uniquely regulate B1b but not B1a cells are poorly understood. The present study provides the first evidence that Id3 is a key regulator of B1b cell self-renewal through a novel BAFF-P62-NFκB signaling pathway.
BAFF signaling is known to regulate self-renewal and development of B cells through activation of NFκB30,31. Yet, whether BAFF promoting survival and proliferation of B cells is subtype dependent still remains controversial. Schiemannn et al have shown that BAFFRWT and BAFFRKO mice possess similar number of peritoneal B1 cells as well as B1a/B1b ratio32, while others have demonstrated that BAFF might regulate B1 cells self-renewal through coupling with toll-like receptor activation33,34. Additionally, Sage et al have utilized bone marrow transplant technique to show that the LDLRKO host mice with BAFFRKO bone marrow reconstitution has a decreased frequency of B1b and not B1a when compared to the wildtype transplant control, suggesting that BAFF mediating B1 cell proliferation might be B1b subtype specific35. Here, we show that BAFFR is significantly expressed at higher level in B1b compared to B1a cells and BAFF stimulation augments in vitro cell proliferation of B1b but not B1a cells. Interestingly, loss of Id3 also leads to a significant increase in BAFFR expression in B1b cells resulting in higher BAFF driven B1b cell proliferation in Id3KO compared to Id3WT. However, future studies are needed to further explore how loss of Id3 leads to an increase in BAFFR expression.
Additionally, our results also suggest that BAFF driven B1b cell proliferation is P62 dependent. P62 is a scaffold and ubiquitination protein known in its role for selective autophagy and cancer cell proliferation36,37. Here, we are the first to show that BAFF induces binding between P62 and TRAF6 in B1b cells to activate NFκB signaling. NFκB signaling in B cells is known to drive expression of C-MYC and subsequent proliferation38. Consistent with this prior discovery, we found that C-MYC expression pattern in B1b cells is similar to NFκB activation pattern measured by the decrease of IκBa level. These findings altogether suggest that P62 drives B1b cell proliferation through binding with TRAF6, activating NFκB, and enhancing pro-proliferative C-MYC gene expression. Even though P62 is crucial in mediating BAFF induced B1b cell proliferation, BAFF is not the only ligand driving B1 cell proliferation. Prior literature has demonstrated that IL-5 as well as CD40 are also important molecules in promoting B1 cell survival and expansion39,40. This is consistent with our results showing a significant albeit modest statistical difference of B1b in vivo cell proliferation once P62 was knocked out while a greater reduction of B1b cell proliferation was observed with the knockout of P62 in the BAFF induced in vitro proliferation assay.
Current literature underscores the complex and ubiquitous roles of P62 in mediating both proliferation and autophagy41,42. Therefore, there might be other mechanisms by which ID3 and BAFF regulate B1b cell numbers. We found that BAFF stimulates the phosphorylation of mTOR and promotes B1b cell proliferation, an effect that appears to be augmented by loss of Id3. However, whether P62-TRAF6 interaction is crucial in mediating this effect still requires further study. As mentioned earlier, P62 has been known as a scaffold protein promoting autophagy and autophagy was shown in cancer cells to increase cell survival42. Increased NRF2 and cell viability in Id3KO B1b cells raises the interesting possibility that P62-NRF2 mediated autophagy might be another putative mechanism underlying an increase in B1b cell number in Id3KO mice.
In addition to exploring a novel role of P62 in BAFF driven B1b cell proliferation, we also investigate a regulatory function of Id3 in mediating P62 expression. P62 expression has been previously shown to be induced by a bHLH-zipper family transcription factor, TFEB43,44, although no prior studies of Id3 and the bHLH proteins, E12 and E47 has been reported. Our results demonstrate that the bHLH proteins, E12 and E47, drive P62 expression. This discovery unveils a mechanism underlining a relationship between loss of Id3 and an increase in B1b cell number observed by Rosenfeld et al9.
Furthermore, we explore the role of P62 in mediating atherosclerosis. Sergin et al have previously shown that P62 facilitates clearance of inclusion bodies from the macrophage, lowering transformation of macrophage into foam cells, resulting in reduction in plaque burden in mice45. Yet, the role of P62 in B cell mediated atherosclerosis has never been investigated. Here, we provide the first evidence that overexpression of P62 in B1 cells drives proliferation of B1b cells, increases plasma level of total IgM and IgM to OSE, and reduces atherosclerosis progression in hyperlipidemic mice. Since P62 activates NFκB pathway, there is a possibility that B1b cell proliferation might not be the only pathway leading to an augmentation of IgM production. Previous literatures provides evidence that activation of NFκB can also enhance antibody secretion as well as plasma cell differentiation46,47. Therefore, P62 might potentially mediate B1b cell differentiation into antibody secreting cells resulting in an increase in IgM production. However, validation and investigation of the detailed mechanism is still awaited.
To translate our murine findings to human, we investigate P62 expression in CD20+CD27+CD43+ human B1 cells48 obtained from CAD subjects. Consistent with our murine study, higher expression of P62 in circulating human B1 cells is found in subjects homozygous for a SNP in the ID3 gene at rs11574, which is known to attenuate the function of the Id3 protein. Moreover, that higher P62 expression on human B1 cells associated with higher frequency of human B1 cells and higher levels of IgM to MDA-mimotope, underscores the clinical relevance of this finding. These findings also raise a possibility that CD20+CD27+CD43+ human B1 cells might behave similar to murine B1b cells underscoring the importance of deeper characterization of these cells.
In summary, this report provides novel insight into the mechanism whereby BAFF and P62 regulate cell proliferation in B1b cells as well as unfolds a regulatory function of Id3 in mediating P62 expression. Atheroprotective roles of P62 in both B1 cells and macrophages highlight P62 as a promising immunomodulatory target for atherosclerosis treatment. Our translational studies provide potential human relevance to this pathway and underscore the need for larger and more in depth human studies.
Supplementary Material
Novelty and Significance.
What is known?
B1 cells spontaneously produce natural antibody IgM to oxidation specific epitopes (OSEs) which confer atheroprotection.
Loss of ID3 (Inhibitor of differentiation 3) in B1 cells leads to an increase in B1b cell number. Yet, mechanisms whereby loss of Id3 leads to greater B1b cell number are unknown.
P62 or SQSTM1, a scaffold and ubiquitin binding protein, plays major roles in regulating autophagy, survival, and proliferation of various cancer cells.
What New Information Does This Article Contribute?
P62 promotes B-cell activating factor (BAFF)-induced B1b self-renewal leading to increased levels of IgM and atheroprotection.
ID3 has a regulatory function in inhibiting the E2A protein from activating P62 promoter.
Human subjects with a function-attenuating SNP in the human ID3 gene have higher expression of P62 in B1 cells, a finding significantly associated with plasma levels of IgMOSE.
B cells have emerged as important immune cells in modulating atherosclerosis with B1 subtypes of B cells protecting from diet-induced atherosclerosis through production of IgM which neutralizes OSEs on LDL. Yet, the molecular mechanisms that regulate B1 cell numbers are poorly understood. Here, we provide the first evidence that the helix-loop helix factor, ID3 regulates multiple genes in the BAFF-induced prolifation pathway in B1b cells, most notably P62. We identify a novel role for P62 in driving BAFF-induced B1b cell self-renewal resulting in higher IgM production and atheroprotection. These findings were translated to humans linking P62 on B1 cells with a function-attenuating SNP in human ID3 and plasma levels of atheroprotective IgMOSE. Results provide novel potential targets for bolstering B1-mediated immune protection to limit CAD.
Acknowledgments
We thank Mike Solga, Claude Chew, Alex Wendling, and Taylor Harper from the UVa flow cytometry core for their excellent technical assistance and Fabrizio Drago for statistical assistance.
Source of Funding
This work was supported by National Institute of Health R01HL136098 and HL148109 (C.A.M).
Nonstandard Abbreviations and Acronyms
- Id3/ID3
Inhibitor of differentiation 3
- KO
knockout
- WT
Wild type
- OSE
Oxidation specific epitopes
- MDA
Malondialdehyde
- PerC
Peritoneal cavity
- BM
Bone marrow
- AT
Adoptive transfer
Footnotes
Disclosures: None.
References:
- 1.Benjamin EJ, Virani SS, Callaway CW, Chamberlain AM, Chang AR, Cheng S, Chiuve SE, Cushman M, Delling FN, Deo R, De Ferranti SD, Ferguson JF, Fornage M, Gillespie C, Isasi CR, Jiménez MC, Jordan LC, Judd SE, Lackland D, et al. Heart Disease and Stroke Statistics - 2018 Update: A Report from the American Heart Association. Vol 137.; 2018. doi: 10.1161/CIR.0000000000000558 [DOI] [PubMed] [Google Scholar]
- 2.Libby P. Inflammation in atherosclerosis. Arterioscler Thromb Vasc Biol. 2012;32(9):2045–2051. doi: 10.1161/ATVBAHA.108.179705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pattarabanjird T, Li C, McNamara C. B Cells in Atherosclerosis: Mechanisms and Potential Clinical Applications. JACC Basic to Transl Sci. 2021;6(6):546–563. doi: 10.1016/J.JACBTS.2021.01.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alugupalli KR, Gerstein RM. Divide and conquer: Division of labor by B-1 B cells. Immunity. 2005;23(1):1–2. doi: 10.1016/j.immuni.2005.07.001 [DOI] [PubMed] [Google Scholar]
- 5.Baumgarth N. B-1 cell heterogeneity and the regulation of natural and antigen-induced IgM production. Front Immunol. 2016;7(SEP):324. doi: 10.3389/fimmu.2016.00324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Prasad A, Clopton P, Ayers C, Khera A, De Lemos JA, Witztum JL, Tsimikas S. Relationship of Autoantibodies to MDA-LDL and ApoB-Immune Complexes to Sex, Ethnicity, Subclinical Atherosclerosis, and Cardiovascular Events. Arterioscler Thromb Vasc Biol. 2017;37(6):1213–1221. doi: 10.1161/ATVBAHA.117.309101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tsimikas S, Willeit P, Willeit J, Santer P, Mayr M, Xu Q, Mayr A, Witztum JL, Kiechl S. Oxidation-specific biomarkers, prospective 15-year cardiovascular and stroke outcomes, and net reclassification of cardiovascular events. J Am Coll Cardiol. 2012;60(21):2218–2229. doi: 10.1016/j.jacc.2012.08.979 [DOI] [PubMed] [Google Scholar]
- 8.Berland R, Wortis HH. Origins and functions of B-1 cells with notes on the role of CD5. Annu Rev Immunol. 2002;20:253–300. doi: 10.1146/annurev.immunol.20.100301.064833 [DOI] [PubMed] [Google Scholar]
- 9.Rosenfeld SM, Perry HM, Gonen A, Prohaska TA, Srikakulapu P, Grewal S, Das D, McSkimming C, Taylor AM, Tsimikas S, Bender TP, Witztum JL, McNamara CA. B-1b Cells Secrete Atheroprotective IgM and Attenuate Atherosclerosis. Circ Res. 2015;117(3):e28–39. doi: 10.1161/CIRCRESAHA.117.306044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Doran AC, Lehtinen AB, Meller N, Lipinski MJ, Slayton RP, Oldham SN, Skaflen MD, Yeboah J, Rich SS, Bowden DW, McNamara CA. Id3 Is a novel atheroprotective factor containing a functionally significant single-nucleotide polymorphism associated with intima-media thickness in humans. Circ Res. 2010;106(7):1303–1311. doi: 10.1161/CIRCRESAHA.109.210294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Doran AC, Meller N, Cutchins A, Deliri H, Slayton RP, Oldham SN, Kim JB, Keller SR, McNamara CA. The Helix–Loop–Helix Factors Id3 and E47 Are Novel Regulators of Adiponectin. Circ Res. 2008;103(6):624–634. doi: 10.1161/CIRCRESAHA.108.175893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Engel I, Murre C. The function of E- and id proteins in lymphocyte development. Nat Rev Immunol. 2001;1(3):193–199. doi: 10.1038/35105060 [DOI] [PubMed] [Google Scholar]
- 13.Roschger C, Cabrele C. The Id-protein family in developmental and cancer-associated pathways. Cell Commun Signal. 2017;15(1):7. doi: 10.1186/s12964-016-0161-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Upadhye A, Srikakulapu P, Gonen A, Hendrikx S, Perry HM, Nguyen A, McSkimming C, Marshall MA, Garmey JC, Taylor AM, Bender TP, Tsimikas S, Holodick NE, Rothstein TL, Witztum JL, McNamara CA. Diversification and CXCR4-Dependent Establishment of the Bone Marrow B-1a Cell Pool Governs Atheroprotective IgM Production Linked to Human Coronary Atherosclerosis. Circ Res. 2019;125(10):e55–e70. doi: 10.1161/CIRCRESAHA.119.315786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pertea M, Kim D, Pertea GM, Leek JT, Salzberg SL. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat Protoc. 2016;11(9):1650–1667. doi: 10.1038/nprot.2016.095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Amir S, Hartvigsen K, Gonen A, Leibundgut G, Que X, Jensen-Jarolim E, Wagner O, Tsimikas S, Witztum JL, Binder CJ. Peptide mimotopes of malondialdehyde epitopes for clinical applications in cardiovascular disease. J Lipid Res. 2012;53(7):1316–1326. doi: 10.1194/jlr.M025445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schneider P, MacKay F, Steiner V, Hofmann K, Bodmer JL, Holler N, Ambrose C, Lawton P, Bixler S, Acha-Orbea H, Valmori D, Romero P, Werner-Favre C, Zubler RH, Browning JL, Tschopp J. BAFF, a novel ligand of the tumor necrosis factor family, stimulates B cell growth. J Exp Med. 1999;189(11):1747–1756. doi: 10.1084/jem.189.11.1747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Doran AC, Lipinski MJ, Oldham SN, Garmey JC, Campbell KA. B cell Aortic Homing and Atheroprotection Depend on Id3. 110(1):e1–e12. doi: 10.1371/journal.pone.0178059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lipinski MJ, Campbell KA, Duong SQ, Welch TJ, Garmey JC, Doran AC, Skaflen MD, Oldham SN, Kelly KA, McNamara CA. Loss of Id3 increases VCAM-1 expression, macrophage accumulation, and atherogenesis in Ldlr−/− mice. Arterioscler Thromb Vasc Biol. 2012;32(12):2855–2861. doi: 10.1161/ATVBAHA.112.300352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Seijkens TTP, van Tiel CM, Kusters PJH, Atzler D, Soehnlein O, Zarzycka B, Aarts SABM, Lameijer M, Gijbels MJ, Beckers L, den Toom M, Slütter B, Kuiper J, Duchene J, Aslani M, Megens RTA, van ‘t Veer C, Kooij G, Schrijver R, et al. Targeting CD40-Induced TRAF6 Signaling in Macrophages Reduces Atherosclerosis. J Am Coll Cardiol. 2018;71(5):527–542. doi: 10.1016/j.jacc.2017.11.055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.López-Franco O, Hernández-Vargas P, Ortiz-Muñoz G, Sanjuán G, Suzuki Y, Ortega L, Blanco J, Egido J, Gómez-Guerrero C. Parthenolide Modulates the NF-\&\#x3ba;B\&\#x2013;Mediated Inflammatory Responses in Experimental Atherosclerosis. Arterioscler Thromb Vasc Biol. 2006;26(8):1864–1870. doi: 10.1161/01.ATV.0000229659.94020.53 [DOI] [PubMed] [Google Scholar]
- 22.Komatsu M, Kurokawa H, Waguri S, Taguchi K, Kobayashi A, Ichimura Y, Sou YS, Ueno I, Sakamoto A, Tong KI, Kim M, Nishito Y, Iemura S ichiro, Natsume T, Ueno T, Kominami E, Motohashi H, Tanaka K, Yamamoto M. The selective autophagy substrate P62 activates the stress responsive transcription factor Nrf2 through inactivation of Keap1. Nat Cell Biol. 2010;12(3):213–223. doi: 10.1038/ncb2021 [DOI] [PubMed] [Google Scholar]
- 23.Jain A, Lamark T, Sjøttem E, Larsen KB, Awuh JA, Øvervatn A, McMahon M, Hayes JD, Johansen T. P62/SQSTM1 is a target gene for transcription factor NRF2 and creates a positive feedback loop by inducing antioxidant response element-driven gene transcription. J Biol Chem. 2010;285(29):22576–22591. doi: 10.1074/jbc.M110.118976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vozenilek AE, Blackburn CMR, Schilke RM, Chandran S, Castore R, Klein RL, Woolard MD. AAV8-mediated overexpression of mPCSK9 in liver differs between male and female mice. Atherosclerosis. 2018;278:66–72. doi: 10.1016/j.atherosclerosis.2018.09.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Haas KM, Poe JC, Steeber DA, Tedder TF. B-1a and B-1b Cells Exhibit Distinct Developmental Requirements and Have Unique Functional Roles in Innate and Adaptive Immunity to S. pneumoniae. Immunity. 2005;23(1):7–18. doi: 10.1016/J.IMMUNI.2005.04.011 [DOI] [PubMed] [Google Scholar]
- 26.Shaw PX, Goodyear CS, Chang MK, Witztum JL, Silverman GJ. The autoreactivity of anti-phosphorylcholine antibodies for atherosclerosis-associated neo-antigens and apoptotic cells. J Immunol. 2003;170(12):6151–6157. doi: 10.4049/jimmunol.170.12.6151 [DOI] [PubMed] [Google Scholar]
- 27.Choi SH, Sviridov D, Miller YI. Oxidized cholesteryl esters and inflammation. Biochim Biophys Acta - Mol Cell Biol Lipids. 2017;1862(4):393–397. doi: 10.1016/j.bbalip.2016.06.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gonen A, Hansen LF, Turner WW, Montano EN, Que X, Rafia A, Chou MY, Wiesner P, Tsiantoulas D, Corr M, VanNieuwenhze MS, Tsimikas S, Binder CJ, Witztum JL, Hartvigsen K. Atheroprotective immunization with malondialdehyde-modified LDL is hapten specific and dependent on advanced MDA adducts: Implications for development of an atheroprotective vaccine. J Lipid Res. 2014;55(10):2137–2155. doi: 10.1194/jlr.M053256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Srikakulapu P, McNamara CA. B cells and atherosclerosis. Am J Physiol - Hear Circ Physiol. 2017;312(5):H1060–H1067. doi: 10.1152/ajpheart.00859.2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yoshimura LC, Tamayo AT, Lin-Lee YC, Pham LV., Fu L, Ford RJ. BAFF-R promotes cell proliferation and survival through interaction with IKKβ and NF-κB/c-Rel in the nucleus of normal and neoplastic B-lymphoid cells. Blood. 2009;113(19):4627–4636. doi: 10.1182/blood-2008-10-183467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Smulski CR, Eibel H. BAFF and BAFF-Receptor in B Cell Selection and Survival. Front Immunol. 2018;9(OCT):2285. doi: 10.3389/FIMMU.2018.02285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schiemann B, Gommerman JL, Vora K, Cachero TG, Shulga-Morskaya S, Dobles M, Frew E, Scott ML. An Essential Role for BAFF in the Normal Development of B Cells Through a BCMA-Independent Pathway. Science (80-). 2001;293(5537):2111–2114. doi: 10.1126/SCIENCE.1061964 [DOI] [PubMed] [Google Scholar]
- 33.Ng LG, Ng CH, Woehl B, Sutherland APR, Huo J, Xu S, Mackay F, Lam KP. BAFF costimulation of Toll-like receptor-activated B-1 cells. Eur J Immunol. 2006;36(7):1837–1846. doi: 10.1002/eji.200635956 [DOI] [PubMed] [Google Scholar]
- 34.Amezcua Vesely MC, Schwartz M, Bermejo DA, Montes CL, Cautivo KM, Kalergis AM, Rawlings DJ, Acosta-Rodríguez EV, Gruppi A. FcγRIIb and BAFF Differentially Regulate Peritoneal B1 Cell Survival. J Immunol. 2012;188(10):4792 LP–4800. doi: 10.4049/jimmunol.1102070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sage AP, Tsiantoulas D, Baker L, Harrison J, Masters L, Murphy D, Loinard C, Binder CJ, Mallat Z. BAFF receptor deficiency reduces the development of atherosclerosis in mice--brief report. Arterioscler Thromb Vasc Biol. 2012;32(7):1573–1576. doi: 10.1161/ATVBAHA.111.244731 [DOI] [PubMed] [Google Scholar]
- 36.Umemura A, He F, Taniguchi K, Nakagawa H, Yamachika S, Font-Burgada J, Zhong Z, Subramaniam S, Raghunandan S, Duran A, Linares JF, Reina-Campos M, Umemura S, Valasek MA, Seki E, Yamaguchi K, Koike K, Itoh Y, Diaz-Meco MT, et al. P62, Upregulated during Preneoplasia, Induces Hepatocellular Carcinogenesis by Maintaining Survival of Stressed HCC-Initiating Cells. Cancer Cell. 2016;29(6):935–948. doi: 10.1016/j.ccell.2016.04.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Valencia T, Kim JY, Abu-Baker S, Moscat-Pardos J, Ahn CS, Reina-Campos M, Duran A, Castilla EA, Metallo CM, Diaz-Meco MT, Moscat J. Metabolic Reprogramming of Stromal Fibroblasts through P62-mTORC1 Signaling Promotes Inflammation and Tumorigenesis. Cancer Cell. 2014;26(1):121–135. doi: 10.1016/j.ccr.2014.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Duyao MP, Buckler AJ, Sonenshein GE. Interaction of an NF-kappa B-like factor with a site upstream of the c-myc promoter. Proc Natl Acad Sci U S A. 1990;87(12):4727–4731. doi: 10.1073/pnas.87.12.4727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rush JS, Hodgkin PD. B cells activated via CD40 and IL-4 undergo a division burst but require continued stimulation to maintain division, survival and differentiation. Eur J Immunol. 2001;31(4):1150–1159. doi: [DOI] [PubMed] [Google Scholar]
- 40.Moon B gon, Takaki S, Miyake K, Takatsu K The Role of IL-5 for Mature B-1 Cells in Homeostatic Proliferation, Cell Survival, and Ig Production. J Immunol. 2004;172(10):6020–6029. doi: 10.4049/jimmunol.172.10.6020 [DOI] [PubMed] [Google Scholar]
- 41.Linares JF, Duran A, Yajima T, Pasparakis M, Moscat J, Diaz-Meco MT. K63 polyubiquitination and activation of mTOR by the P62-TRAF6 complex in nutrient-activated cells. Mol Cell. 2013;51(3):283–296. doi: 10.1016/j.molcel.2013.06.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Moscat J, Karin M, Diaz-Meco MT. Leading Edge Minireview P62 in Cancer: Signaling Adaptor Beyond Autophagy. Cell. 2016;167:606–609. doi: 10.1016/j.cell.2016.09.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sergin I, Evans TD, Zhang X, Bhattacharya S, Stokes CJ, Song E, Ali S, Dehestani B, Holloway KB, Micevych PS, Javaheri A, Crowley JR, Ballabio A, Schilling JD, Epelman S, Weihl CC, Diwan A, Fan D, Zayed MA, et al. Exploiting macrophage autophagy-lysosomal biogenesis as a therapy for atherosclerosis. Nat Commun. 2017;8. doi: 10.1038/ncomms15750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pan B, Li J, Parajuli N, Tian Z, Wu P, Lewno MT, Zou J, Wang W, Bedford L, Mayer RJ, Fang J, Liu J, Cui T, Su H, Wang X. The Calcineurin-TFEB-P62 Pathway Mediates the Activation of Cardiac Macroautophagy by Proteasomal Malfunction. Circ Res. 2020;127(4):502–518. doi: 10.1161/CIRCRESAHA.119.316007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sergin I, Bhattacharya S, Emanuel R, Esen E, Stokes CJ, Evans TD, Arif B, Curci JA, Razani B, Mo L. atherosclerosis. 2016;9(409). doi: 10.1126/scisignal.aad5614.P62-enriched [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hosseini H, Li Y, Kanellakis P, Tay C, Cao A, Liu E, Peter K, Tipping P, Toh BH, Bobik A, Kyaw T. Toll-Like Receptor (TLR)4 and MyD88 are Essential for Atheroprotection by Peritoneal B1a B Cells. J Am Heart Assoc. 2016;5(11). doi: 10.1161/JAHA.115.002947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Genestier L, Taillardet M, Mondiere P, Gheit H, Bella C, Defrance T. TLR Agonists Selectively Promote Terminal Plasma Cell Differentiation of B Cell Subsets Specialized in Thymus-Independent Responses. J Immunol. 2007;178(12):7779–7786. doi: 10.4049/jimmunol.178.12.7779 [DOI] [PubMed] [Google Scholar]
- 48.Griffin DO, Rothstein TL. Human B1 Cell Frequency: Isolation and Analysis of Human B1 Cells. Front Immunol. 2012;3(MAY):122. doi: 10.3389/fimmu.2012.00122 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Human Subjects
Coronoary artery disease subjects were recruited for study through the Cardiac Catheterization laboratory at the University of Virginia. All participants provided written informed consent before enrollment, and the study was approved by the Human Institutional Review Board. Peripheral blood was obtained from these participants prior to catherization. In addition, peripheral blood from healthy volunteers were also obtained after providing with written informed consent.
Mice
All animal protocols were approved by the Animal Care and Use Committee at the University of Virginia. Id3fl/fl mice were a generous gift from Dr. Yuan Zhuang at Duke University. CD19Cre/+ mice were provided by Dr. Bender (University of Virginia). C57BL/6J mice were purchased from Jackson Laboratory. These mice were fed a standard chow diet or a Western diet. All the mice utilized in all the experiments were 8–10 weeks old female mice.
Cell Preparations for Murine and Human flow cytometry
Peritoneal cavity cells were processed for flow cytometry or fluorescence activated cell (FAC) sorting as previously described9,14. Isolation of human PBMCs was performed as previously described9,14. Clone and fluorophore information as well as isotype control for flow cytometry antibodies were provided in supplementary table 3.
Samples preparation of Bulk RNA sequencing
Sort-purified peritoneal B1a and B1b obtained from Id3KO and Id3WT C57BL/6 mice were RNA extracted by using Qiagen RNeasy Plus kit. The purified RNAs were stored at −80°C and sent to a Novogene to perform sequencing.
Differentially expressed genes and pathway analysis
RNA sequences in raw FASTQ data files were obtained from Novogene and HISAT2/Stringtie/Ballgown pipeline was utilized for RNASeq analysis following the previously published protocol15 Volcano plots of differentially expressed genes were visualized by using the python bioinfokit package. Ingenuity Pathway Analysis was performed on all the annotated RNA to analyze for differentially regulated cellular processes, canonical pathways and network analysis.
In Vitro and In Vivo Cell Proliferation Assay
Peritoneal cavity cells were enriched for B cells using the EasySep Mouse Pan-B cell enrichment kit (STEMCELL), and then labeled with 5uM Celltrace-violet for 20 minutes at 37oC, and then washed with FACS buffer (1% FBS, 0.1%NaN3 in PBS). Labeled cells were cultured in B cell culture media for 3 days with 20ng/mL recombinant BAFF or PBS control to measure in vitro proliferation. For in vivo proliferation, sort-purified p62-gRNA-ATTO+ Celltrace-violet+ B1 cells were intraperitoneally transferred into C57BL/6 mice.
Lentiviral Production and P62 Overexpression on mouse B cells.
P62-eGFP lentivirus was generated using pLV-eGFP vector (purchased from Addgene) and mouse P62 was subcloned into the vector. The eGFP or P62-eGFP lentiviruses were generated using four plasmid system (pLV-eGFP or pLV-p62-eGFP, pLP1, pLP2, and pVSV-G) into 293T cells. The eGFP ctrl and P62-eGFP lentiviruses were used to transduce enriched peritoneal mouse B cells. Transduced murine B cells were harvested 48 hrs post transduction and FAC-sorted for GFP+ B1 cells to be used for adoptive transfer.
p62 CRISPR/sgRNA Knockout and PP62-GFP transfection on Mouse B cells.
crRNA targeting p62 (IDT) was conjugated with tracRNA-ATTO (IDT). Pp62-GFP plasmid was generated by subcloning 1000bp of p62 promoter to replace CMV promoter in CMV-GFP (Addgene). Murine peritoneal cells stimulated with 50ng/mL LPS overnight. The stimulated B cells were then nucleotransfected with Cas9, and pre-conjugated P62 crRNA-tracRNA ATTO, Pp62-GFP, or CMV-GFP ctrl plasmid using the P3 Primary cells Nucleofection Kit (Lonza). B cells were collected for analysis and sorting 24 hrs after nucleofection.
Nucleofection of Engineered Plasmids into Human B cells.
Heatlhy donors’ PBMCs were thawed and washed with RPMI +5% FBS. B cells were enriched using EasySep human pan-B cell enrichment kit (STEMCELL). CMV-E12-Flag, CMV-E47-Flag were generated using CMV-Flag plasmid to subclone human E12 and E47 into. These plasmids were nucleotransfected into enriched human B cells t using the P3 Primary cells Nucleofection Kit (Lonza). B cells were collected for analysis and sorting 24 hrs after nucleofection.
Quantification of markers colocalization by using imaging flow cytometry
Murine peritoneal cells obtained from Id3KO and Id3WT mice were enriched for B cells. The enriched B cells were incubated with 20ng/mL murine recombinant BAFF or PBS control for 12 hrs in B cell media. Prior to running on the Imagestream imaging flow cytometry machine, cells were stained using previously developed surface staining protocol9,14 following with permeabilization and intracellular staining using FIX & PERM Cell Permeabilization Kit (Invitrogen). Images of 1000 Id3KO or Id3WT B1b cells from both unstimulated, BAFF stimulated conditions were collected, and quantitative co-localization analysis was performed using Amnis Imagestream colocalization software.
ELISA for Quantification of Total and Anti-OSE IgM or IgG Isotypes in Mice and Humans
Total IgM subtypes in mouse plasma were measured using colorimetric ELISA as described previously9. Levels of IgM and IgG against the MDA mimotope16 in human plasma were measured by chemiluminescent ELISA as previously described16.
Adoptive Transfer
Celltrace violet+ or eGFP+ (eGFP-ctrl or P62-eGFP) B1 (CD3-CD19+B220-) cells were sort-purified and adoptively transferred into C57BL/6 mice intraperitoneally. Mice adoptively transferred with Celltrace-violet+ cells were fed a chow diet for 2 weeks. Mice adoptively transferred with eGFP-ctrl or P62-eGFP B1 cells were fed with Western diet for 8 weeks and harvested for peritoneal cells, hearts and aortas.
Analysis of Atherosclerotic Lesions
Hearts and aortas were harvested as previously described10. Plaque areas were assessed using Image-Pro Plus software (Media Cybernetics). The analysis was done by the lab specialist without knowing the group allocation.
Statistics
Statistics were performed using GraphPad Prism Version 7.0a (GraphPad Software, Inc), Python 3.0, R 3.6.1, or SAS 9.4. Results from all replicated experiments are displayed, and bar graphs display Mean ± SD.