Abstract
Ubiquitylation is an essential post-translational modification that regulates intracellular signalling networks by triggering proteasomal substrate degradation, changing the activity of substrates or mediating changes in proteins that interact with substrates. Hundreds of enzymes participate in reversible ubiquitylation of proteins, some acting globally and others targeting specific proteins. Ubiquitylation is essential for innate immune responses, as it facilitates rapid regulation of inflammatory pathways, thereby ensuring sufficient but not excessive responses. A growing number of inborn errors of immunity are attributed to dysregulated ubiquitylation. These genetic disorders exhibit broad clinical manifestations, ranging from susceptibility to infection to autoinflammatory and/or autoimmune features, lymphoproliferation and propensity to malignancy. Many autoinflammatory disorders result from disruption of components of the ubiquitylation machinery and lead to overactivation of innate immune cells. An understanding of the disorders of ubiquitylation in autoinflammatory diseases could enable the development of novel management strategies.
Subject terms: Rheumatic diseases, Pathogenesis, Health care, Disease genetics
Ubiquitylation is involved in the regulation of most cellular systems, including the innate immune system. Here, the authors describe the molecular pathogenesis of disorders of ubiquitylation that result in innate immune overactivation and systemic autoinflammatory disease.
Key points
Autoinflammatory diseases are disorders of overactivation of the innate immune system, and some of these conditions are caused by pathogenic mutations affecting components of the ubiquitylation pathways.
Ubiquitylation is a reversible post-translational modification that is essential for regulation of cellular responses to immune signals, in particular NF-κB and interferon pathways, as well as the unfolded protein response.
Dysregulation of ubiquitylation leads to autoinflammation through both disruption of specific signalling networks and induction of generalized stress pathways.
Mutations affecting ubiquitylation pathway components that regulate large sets of substrates (UBA1 and the proteasome) and components regulating specific substrates (E3 ligases and deubiquitylases) can result in overlapping phenotypes.
Consideration of ubiquitylation disorders as a group could provide additional insights into these diseases and suggest novel treatment avenues.
Introduction
Since the identification in the early 1980s of the ubiquitin pathway as the major component of intracellular proteolysis, it has emerged as an essential regulator of most cellular systems1–5. Ubiquitylation is a post-translational modification (PTM) that temporally, spatially and specifically regulates substrate stability and signalling, and is therefore fundamental to health and disease6. Ubiquitylation is central to many dynamically regulated processes such as cell-cycle progression, DNA-damage response and inflammatory and immune signalling pathways. Not surprisingly, dysregulation of the ubiquitylation cascade is associated with many disease states, from infantile neurodevelopmental disorders to inborn errors of immunity7,8.
In the past decade, evidence has linked monogenic autoinflammatory diseases, which result from the hyperactivation of the innate immune system, to alterations in the ubiquitin–proteasome machinery. Although these disorders are rare, they provide important insights into the complex mechanisms regulating innate immune responses9. A broader understanding of the multifactorial role of ubiquitylation in chronic inflammatory disease could enable the development of new therapeutic modalities for both rare and common rheumatological diseases. In this Review, we outline the known disorders of ubiquitylation that lead to excessive innate immune activation, describe the molecular pathogenesis of these conditions, and propose that disruption of the ubiquitin pathway is a key contributor to inflammation through both dysregulation of specific signalling networks and induction of generalized stress pathways.
Ubiquitin, inflammation and disease
Ubiquitin machinery
Ubiquitin is a 76-amino acid protein that is covalently attached to substrates through the concerted activities of three different sets of enzymes (Box 1). First, structurally related ubiquitin E1 enzymes (ubiquitin-like modifier-activating enzymes UBA1 and UBA6) utilize ATP to activate ubiquitin10. Second, the activated ubiquitin moiety is transferred to the cysteine residue in the active site of one of around 40 human ubiquitin E2 carrier enzymes. In the final step, ubiquitin-charged E2 enzymes co-operate with ubiquitin E3 ligases (a protein family with around 600 members in humans), which catalyse ubiquitin transfer to specific substrate proteins by forming an isopeptide bond between the C-terminal carboxyl group of ubiquitin and the ε-amino group of a lysine residue of the target protein10. Ubiquitin can also be conjugated via a peptide bond to the N-terminal amino group of a substrate, or via esterification to a threonine or serine residue of a protein substrate, and ubiquitin can even be joined to non-proteinaceous substrates11–13. Typically, one E3 ligase can modify several substrates, and conversely one protein can be ubiquitylated by several E2/E3 combinations, resulting in a diversity of ubiquitylation sites and patterns14.
The ubiquitylation cascade is organized in a hierarchical manner (Box 1). Ubiquitin activation by E1 enzymes is essential for all cellular ubiquitylation, whereas activity of E2 enzymes that bind specific sets of E3 ligases is required for ubiquitylation of many substrates and contributes to generation of particular ubiquitylation patterns. The nature of the ubiquitin modification determines the fate of a particular substrate: ubiquitin can be attached as a monomer to one or several sites within a substrate (monoubiquitylation or multi-monoubiquitylation), or it can be attached in the form of ubiquitin chains (polyubiquitylation). Polyubiquitin chains can be interconnected by ubiquitin conjugation to one or more of the seven lysine residues or the N-terminal methionine (K6, K11, K27, K29, K33, K48, K63, M1) of any ubiquitin monomer, resulting in a staggering complexity of homotypic and heterotypic architectures, each of which has the biochemical capacity to elicit distinct downstream functional consequences15 (Box 1). Notable ubiquitin modifications are the homotypic K48-linked ubiquitin chains that mediate degradation by the proteasome16, and the homotypic and heterotypic M1–K63-linked ubiquitin chains that assemble multi-protein signalling platforms during immune responses17,18. Different types of ubiquitylation are recognized by a variety of proteins with ubiquitin-binding domains (ubiquitin receptors) that translate specific ubiquitin linkages into cellular responses19,20. Ubiquitylation is reversed or edited by deubiquitylases (a family of around 100 proteins in humans), which catalyse either the removal of ubiquitin from substrates or the trimming of polymeric modifications21. In addition, there are many ubiquitin-like modifications that use analogous biochemical pathways for activation and conjugation, including SUMO (small ubiquitin-like modifier), FAT10 (human leukocyte antigen-F adjacent transcript, also known as ubiquitin D), ISG15 (interferon-stimulated gene 15), UFM1 (ubiquitin fold modifier 1), URM1 (ubiquitin-related modifier 1) and NEDD8 (neuronal precursor cell-expressed developmentally down-regulated protein 8)22.
The consequences of the ubiquitylation pathways even within a single cell are hard to fathom, with thousands of proteins undergoing modification at a time, each at a particular combination of target residues, with varying chain types and lengths, to achieve precisely coordinated functions. In addition, cell-specific and temporally dynamic patterns of ubiquitylation indicate the importance of ubiquitylation as a regulatory mechanism for interpretation of signalling changes and changes in the cellular environment. These precise functions are demonstrated by the subtle perturbations that can happen in human disease following the loss of a specific enzyme23,24. In many ways, this requirement for proper ubiquitylation to maintain continuity of physiological functions is an elegant example of “destruction for the sake of construction”25.
Box 1 Overview of the ubiquitin conjugation and deconjugation machinery and ubiquitin signals.
Ubiquitin is conjugated by a hierarchical, three-step enzymatic cascade involving ubiquitin activation and thioester formation by an E1 activating enzyme, ubiquitin transthiolation to an E2-conjugating enzyme and E3 ligase-assisted attachment of ubiquitin to substrates. Depending on the class of E3 ligase, ubiquitin conjugation to substrates either involves transthiolation to the E3 ligase (for the classes ‘homologous to E6AP C-terminus’ (HECT) and ‘RING between RING’ (RBR)) or occurs directly from the E2 enzyme (for the class of ‘really interesting new gene’ (RING) E3 ligases). Ubiquitin can be deconjugated from substrates via deubiquitylases (DUBs). The concerted action of E1, E2 and E3 enzymes and DUBs generates specific ubiquitylation patterns. Substrates can be mono-ubiquitylated or multi-ubiquitylated, or ubiquitin can be further ubiquitylated on one of its seven lysine residues (K6, K11, K27, K29, K33, K48 and K63) or its N-terminus (M1, for linear ubiquitylation). Polyubiquitin chains can either contain one linkage type (homotypic), several linkage types (heterotypic), or they can be branched, if at least one ubiquitin in the chain is subject to modification at multiple sites. These different types of ubiquitin modification encode diverse messages and are interpreted by ubiquitin-binding proteins that distinguish between different signals and initiate the correct cellular responses. For example, homotypic K48-linked ubiquitin chains mediate recognition and degradation of substrates by the 26S proteasome.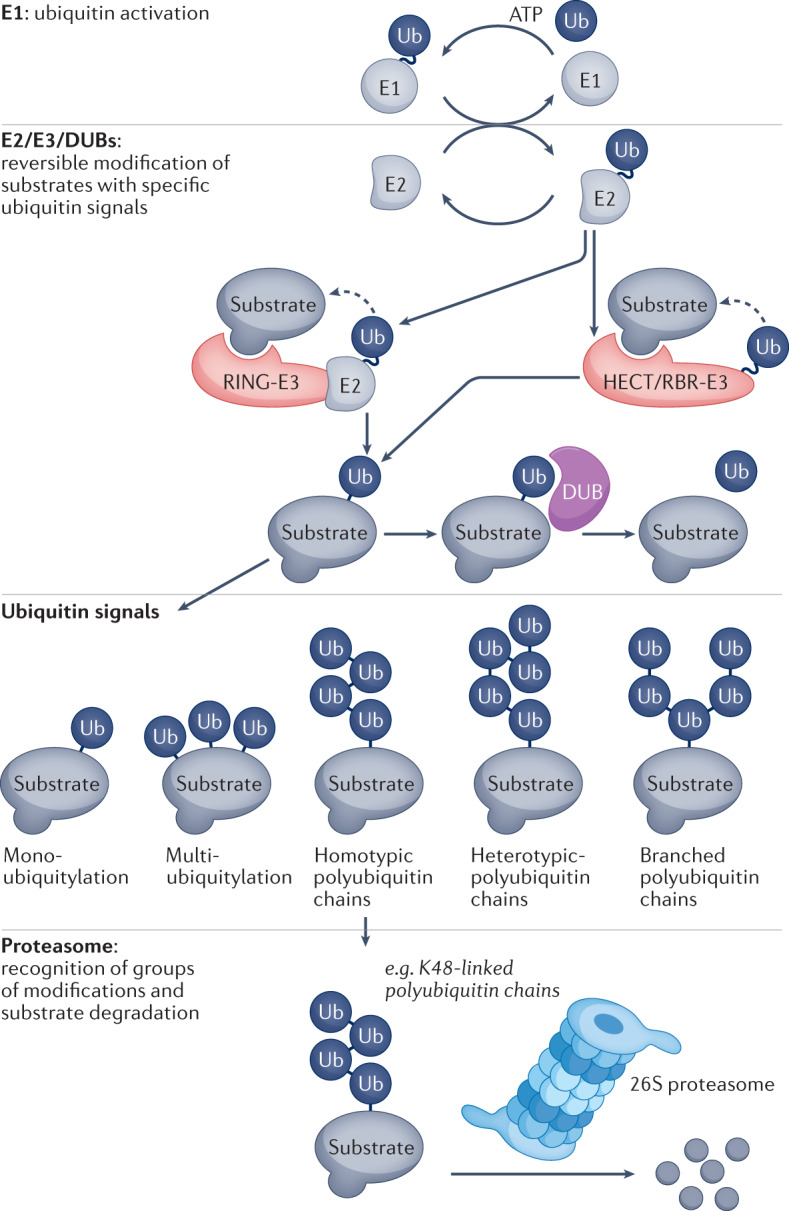
Autoinflammation and ubiquitylation
Dysregulation of ubiquitylation is linked to many distinct disorders of recurrent systemic inflammation that are collectively known as systemic autoinflammatory diseases (SAIDs)26. Studies of patients with SAIDs and of animal models that phenocopy these conditions have revealed basic mechanisms related to innate immunity that are widely applicable across many different disease states9. SAIDs are caused by genetic mutations that lead to enhanced or constitutive inflammation, either gain-of-function mutations in activators or sensors of immune responses (such as NLRP3, mutation of which results in cryopyrin-associated periodic syndrome)27,28, or loss-of-function mutations in inflammatory repressors (such as IL1RN, mutation of which results in deficiency of IL-1 receptor antagonist)29,30. Identification of the molecular causes of such diseases is important to enable correct clinical diagnosis and selection of targeted therapies.
SAIDs can be categorized according to a number of parameters, such as clinical manifestation31, genetics32 and molecular pathogenesis33; however, in this Review, we focus on the biochemical pathways of ubiquitylation in autoinflammatory diseases as a defining feature. Depending on which step in the ubiquitylation hierarchy is compromised, dysregulation of ubiquitylation can directly affect a particular signalling pathway (for example, through mutations affecting E3 ligases and deubiquitylases) that will ultimately result in activation of cellular stress responses, or it can cause general cellular stress as an immediate consequence (for example, through mutations affecting E1 enzymes or the proteasome). We focus here on existing data from human genetic diseases and supporting evidence from relevant model organisms to review the current understanding of the pathology and molecular basis of associated phenotypes. Strikingly, defects in general components of the ubiquitin–proteasome system (UPS) and in highly substrate-specific ubiquitin ligation and deconjugation enzymes can lead to overlapping clinical presentations, suggesting commonalities that are not apparent at the level of known molecular aetiology. Given the highly targetable nature of the ubiquitin pathway34, conceptually connecting these diseases may be important for designing rational therapeutics across diagnoses.
In the following sections, we describe multiple specific inflammatory pathways that are known to be dysregulated in inflammatory disease, organized by the type of ubiquitylation enzyme that is affected in human disease, with information on clinical manifestations, genetics and molecular signalling (Table 1). Importantly, given the pleiotropic role of many of these regulatory enzymes, we briefly highlight supporting evidence relating to other genes where possible. In many cases, ubiquitylation coordinates multiple signalling pathways during immune responses, and mutations that affect ubiquitylation enzymes often alter various cellular processes that contribute to disease manifestations.
Table 1.
Disorders of ubiquitylation: genetics, mechanisms and phenotypes
| Gene | Genomic location | Protein name | Inheritance | Disease mechanism | Phenotype | OMIM disease nomenclature | OMIM identifier |
|---|---|---|---|---|---|---|---|
| OTULIN | 5p15.2 | Ubiquitin thioesterase otulin | AR | Loss-of-function | Early-onset recurrent fever, erythematous rash with nodules, joint swelling, lipodystrophy | Autoinflammation, panniculitis, and dermatosis syndrome; otulipenia; ORAS | 617099 |
| POMP | 13q12.3 | Proteasome maturation protein | AD | Dominant negative | Early-onset autoinflammatory disease with erythematous plaques, lipodystrophy, fever and immunodeficiency | Proteasome-associated autoinflammatory syndrome 2 | 618048 |
| PSMA3 | 14q23.1 | Proteasome subunit alpha type 3 | Digenic | Hypomorphic | Early-onset autoinflammatory disease with erythematous plaques, lipodystrophy and fever | Proteasome-associated autoinflammatory syndrome | NA |
| PSMB4 | 1q21.3 | Proteasome subunit beta type 4 | AR, digenic | Hypomorphic | Early-onset autoinflammatory disease with erythematous plaques, lipodystrophy and fever | Proteasome-associated autoinflammatory syndrome 3 | 617591 |
| PSMB8 | 6p21.32 | Proteasome subunit beta type 8 | AR, digenic | Hypomorphic | Early-onset autoinflammatory disease with erythematous plaques, lipodystrophy and fever | Proteasome-associated autoinflammatory syndrome 1 | 256040 |
| PSMB9 | 6p21.2 | Proteasome subunit beta type 9 | AR, digenic, heterozygous de novo | Hypomorphic | Early-onset autoinflammatory disease with erythematous plaques, lipodystrophy, fever and immunodeficiency | Proteasome-associated autoinflammatory syndrome 3 | 617591 |
| PSMB10 | 16q22.1 | Proteasome subunit beta type 10 | AR | Hypomorphic | Early-onset autoinflammatory disease with erythematous plaques, lipodystrophy and fever | Proteasome-associated autoinflammatory syndrome 5 | 619175 |
| PSMD12 | 17q24.2 | 26 S proteasome non-ATPase regulatory subunit 12 | AD | Haploinsufficiency | Rash, arthritis, uveitis, neurodevelopmental, various congenital defects | Stankiewicz–Isidor syndrome | 617516 |
| PSMG2 | 18p11.21 | Proteasome assembly chaperone 2 | AR | Hypomorphic | Early-onset autoinflammatory disease with erythematous plaques, lipodystrophy and fever | Proteasome-associated autoinflammatory syndrome 4 | 619183 |
| RBCK1 | 20p13 | RanBP-type and C3HC4-type zinc finger-containing protein 1 | AR | Loss-of-function | Progressive proximal muscle weakness, cardiomyopathy, severe immunodeficiency, autoinflammation | Polyglucosan body myopathy 1 with or without immunodeficiency | 615895 |
| RNF31 | 14q21 | E3 ubiquitin-protein ligase RNF31/HOIL-1-interacting protein | AR | Loss-of-function | Severe immunodeficiency, autoinflammation, myopathy | HOIP deficiency | NA |
| TNFAIP3 | 6q23.3 | Tumour necrosis factor alpha-induced protein 3 | AD | Haplo-insufficiency, aberrant phosphorylation | Early-onset systemic inflammation, arthralgia, oral and genital ulcers, uveitis, autoimmunity, immunodeficiency | Familial Behçet-like autoinflammatory syndrome; HA20 | 616744 |
| UBA1 | Xp11.23 | Ubiquitin activating enzyme 1 | Somatic | Hypomorphic | Late-onset systemic, skin, lung and cartilage inflammation, also associated with haematological abnormalities | Vacuoles, E1 activating, X-linked, Autoinflammatory, somatic syndrome; VEXAS | 301054 |
| XIAP | Xq25 | E3 ubiquitin-protein ligase XIAP | XLR | Hypomorphic, loss-of-function | Haemophagocytic lymphohistiocytosis, splenomegaly and inflammatory bowel disease | Lymphoproliferative syndrome, X-linked, 2 | 300635 |
AD, autosomal-dominant; AR, autosomal-recessive; HA20, haploinsufficiency of A20; HOIP, HOIL-1-interacting protein; NA, not available; OMIM, Online Mendelian Inheritance in Man; ORAS, OTULIN-related autoinflammatory syndrome; XLR, X-linked recessive.
UBA1 E1 enzyme, mutations and disease
VEXAS syndrome
Vacuoles, E1 activating enzyme, X-linked autoinflammatory somatic (VEXAS) syndrome is the first example of an autoinflammatory disease arising exclusively from somatic mutations, and one of an emerging class of acquired errors of immunity. Although this disorder was only first reported in 2020, hundreds of patients have already been identified35. VEXAS syndrome is caused by pathogenic variants in UBA1, a gene located on the X chromosome that encodes ubiquitin-like modifier activating enzyme 1 (UBA1); these mutations are restricted to myeloid-lineage cells35 (Fig. 1). UBA1 is the E1 enzyme that is required for initiation of most cellular ubiquitylation, and it has two well-characterized isoforms, UBA1a and UBA1b36,10. UBA1a is a long isoform that is localized to the nucleus, whereas UBA1b is a shorter isoform, without a nuclear localization signal. Individuals with VEXAS have acquired missense mutations that abrogate expression of UBA1b and initiate translation of a new catalytically impaired isoform in mutant cells. These loss-of-function mutations are primarily found at, or around, the start codon for UBA1b (p.Met41), leading to a reduction in protein isoform production35. Individual variants at p.Met41 confer particular phenotypes and levels of disease severity through differential UBA1b translation37 (D. Beck, personal communication). Several other deleterious mutations that affect the sequence of the N-terminal domain can also reduce UBA1b protein expression or activity38–40. Nearly all instances of VEXAS syndrome are identified in men, but it can also occur in women with inherited or acquired monosomy of the X chromosome41–45. UBA1 function is essential in model organisms, and pathogenic mutations are only physiologically tolerated in somatic form in haematopoietic stem cells and myeloid lineages, whereas they are lethal in other tissues35,46.
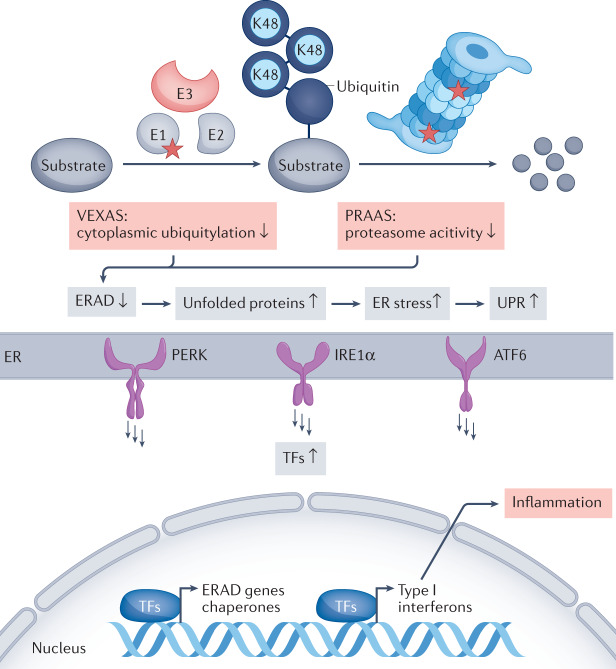
Fig. 1. Mutations affecting the major ubiquitin activating enzyme UBA1 and proteasome subunits result in type I interferon production and inflammation.
Reduction of cytoplasmic ubiquitylation by mutations affecting ubiquitin-like modifier-activating enzyme UBA1 (resulting in vacuoles, E1 activating enzyme, X-linked autoinflammatory somatic (VEXAS) syndrome) or of proteasome activity by mutations affecting particular proteasomal subunits (resulting in proteasome-associated autoinflammatory syndromes (PRAAS)) decreases the efficiency of endoplasmic reticulum (ER)-associated degradation (ERAD). This alteration results in imbalances in cellular proteostasis and accumulation of unfolded and unnecessary proteins, which activate the three different sensors of the unfolded protein response (UPR), PERK, IRE1α and ATF6. Sensor activation initiates signalling cascades that culminate in the production of specific transcription factors (TFs) that mediate upregulation of expression of ERAD components and chaperones as well as type I interferons, which drive the autoinflammation in VEXAS syndrome and PRAAS. Red stars denote proteins affected by mutation. Blue circles denote K48 linked polyubiquitin chains destined for proteasomal degradation.
Patients with VEXAS syndrome have multisystem inflammation involving primarily the skin, cartilage and lungs (although the clinical spectrum also includes many other disease manifestations), with onset in middle age or later47–49. VEXAS syndrome can present in a similar manner to a variety of haematological and inflammatory conditions50, and patients with VEXAS syndrome are likely to meet diagnostic criteria for relapsing polychondritis, systemic lupus erythematosus, rheumatoid arthritis, polyarteritis nodosa and Sweet’s syndrome, along with haematological diseases including multiple myeloma and myelodysplastic syndrome49. However, VEXAS syndrome can be distinguished by the presence of the distinct genetic mutation, and by unique clinical characteristics including histopathological vacuoles and late-onset treatment-refractory disease.
The mechanism by which mutation in UBA1 causes disease has remained elusive, although preliminary evidence suggests activation of the unfolded protein response (UPR) and of multiple inflammatory pathways, including the IFNα and IFNβ pathways. Activation of the UPR can lead to a type I interferon response, which is a feature of proteasomopathies (Fig. 1). The proteasome acts at the completion of the degradative ubiquitylation pathway, and, as with the initiation of the cascade by UBA1, it represents a bottleneck in the regulation of the vast majority of ubiquitylated proteins, so that loss of either step disrupts protein homeostasis (proteostasis)51. Therefore, it is no surprise that similar clinical manifestations occur in patients with different proteasomopathies (such as proteasome maturation protein (POMP)-related autoinflammation and immune dysregulation, a disease with prominent neutrophilic dermatosis), interferonopathies (such as STING-associated vasculopathy with onset in infancy or COPA syndrome, inherited disorders with prominent interstitial lung disease)52,53 and VEXAS syndrome. Finally, there is emerging evidence that blocking Janus kinases (JAKs), which are downstream effectors in interferon signalling, might be an important approach to treatment for VEXAS syndrome39. However, as is seen with other disorders of constitutive inflammation, and depending on the disease stage in individual patients, allogenic haematopoietic stem cell transplantation (HSCT) could end up being the only therapy with sustained efficacy43,45,54.
UBA1 and spinal muscular atrophy
In contrast to VEXAS syndrome, germline missense and synonymous mutations affecting the C-terminal portion of UBA1 occur in patients with a neuromuscular disease that resembles spinal muscular atrophy (SMA)55. The reported mutations cluster within the active adenylation domain that is required for catalytic activity in UBA1a and UBA1b, although not all of the mutations have a demonstrable effect on enzyme activity56. Loss of Uba1 in zebrafish causes impairment of movement, and exogenous expression of UBA1 can improve symptoms related to classic SMA in mouse models with pathogenic hypomorphic variants in the spliceosome biogenesis factor survival motor neuron protein (Smn1)57,58. The precise molecular connection between UBA1-dependent ubiquitylation and SMN1-driven spliceosome assembly remains elusive. However, given the manifold mechanisms by which ubiquitylation regulates spliceosome function59, it is likely that an essential ubiquitin-dependent pathway coordinates splicing during neurodevelopment.
Predisposition to phenotypically varied disorders as a result of mutations affecting differentially expressed protein isoforms (such as UBA1) also occurs in other diseases such as CDC42-associated or POMP-associated phenotypes60–63. For VEXAS syndrome and SMA, the relationship between UBA1 functions in neurodevelopment and innate immunity is still unclear and it remains to be seen whether UBA1-related SMA is effected by severe inflammation. Further characterization of these disparate UBA1-related conditions may provide a better understanding of the cell-specific and tissue-specific functions of this protein.
Proteasome, mutations and disease
Proteasome-associated SAIDs
The proteasome is a multisubunit complex that is required for degradation of ubiquitylated substrates, enabling the removal of misfolded and unnecessary proteins64. The proteasome consists of a 20 S core particle with proteolytic activity (caspase-like, trypsin-like and chymotrypsin-like)65 and 19 S regulatory particles that can associate with either or both ends of the core particle. The regulatory particle consists of ‘base’ and ‘lid’ sub-complexes and serves to recruit ubiquitylated substrates. The proteasome is constitutively expressed, but the core particle can also contain multiple tissue-specific subunits, generating immune-specific, thymus-specific and testis-specific proteasomes66.
Biallelic loss-of-function mutations affecting components of the proteasome lead to a class of SAIDs called proteasome-associated autoinflammatory syndromes (PRAAS), which are also known as Nakajo–Nishimura syndrome, CANDLE (chronic atypical neutrophilic dermatosis and lipodystrophy and elevated temperature) or joint contractures, muscular atrophy, microcytic anaemia and panniculitis-induced lipodystrophy67. Patients with these conditions present with symptoms that include early-onset fevers, skin manifestations, intracranial calcifications, neurological impairment, arthritis and lipomuscular dystrophy68.
Most of the identified causative genetic changes affect subunits that constitute the immunoproteasome (such as PSMB8, PSMB9 and PSMB10), although some affect core proteasome subunits (such as PSMA3 and PSMB4) or regulatory proteins (such as POMP, PSMG2 and PSMD12)60,69,70. This observation suggests the involvement of tissue-specific expression of proteasome core and/or regulatory subunits, and/or a high demand for proteasome activity in specific cells71, which could explain why, even though proteasome activity is essential for most cellular pathways, loss of proteasome function can lead to inflammation without other broad pathological conditions.
Initial clues to the pathophysiological mechanism in PRAAS came from the demonstration that defects in the immunoproteasome, both in patient-derived cells and in cells from healthy individuals with proteasome knockdown or inhibition, lead to production of type I interferons69 (Fig. 1). Further studies of POMP-related autoinflammation and immune dysregulation, with heterozygous dominant-negative mutations in POMP, demonstrated that the UPR is activated in cells from affected patients, thereby providing further evidence for a link between proteasome-dependent quality control and interferon production, which is consistent with the observation of clinical response to JAK inhibition in patients with PRAAS60. Furthermore, evidence indicates that protein kinase R (PKR) functions as an innate immune sensor for proteotoxic stress triggered by proteasome dysfunction72. Interestingly, knockout of the catalytic β5i/LMP7 subunit of the immunoproteasome in mice does not result in spontaneous inflammation, limiting efforts to further model these diseases73.
Neurodevelopmental disease
Mutations in PSMD12, which encodes one of the 19 S regulatory proteasomal subunits, are linked to neurodevelopmental disease, with heterozygous loss-of-function alleles identified in patients with intellectual disability, congenital malformations and autistic features74. Interpretation of the effects of these mutations is complicated by the variability of the severity of disease presentation among affected family members75,76. Reduced penetrance of pathogenic variants is a common feature of many primary interferonopathies77. In the same way that distinct UBA1 mutations can lead to either VEXAS syndrome or SMA, there may be a link between the neurodevelopmental and inflammatory phenotypes associated with PSMD12 (ref.70). Proteasomal regulatory proteins, such as PSMD12, might also have previously unrecognized cell-specific or tissue-specific functions, which are now being discovered by human genetics studies. Whether inflammation associated with the haploinsufficiency of PSMD12 contributes to the neurodevelopmental phenotype has yet to be identified but might be an important consideration for optimization of treatment strategies.
E3 ligases and deubiquitylases
Ensuring a proper innate immune response requires rapid and highly regulated remodelling of signalling networks, for which immune cells often utilize PTMs. In particular, ubiquitylation has emerged as a critical means of fine-tuning the strength and duration of immune signals. In this respect, it is not surprising that mutations that affect the enzymatic machinery catalysing ubiquitylation (E3 ligases) and deubiquitylation (deubiquitylases) can lead to alteration of many cytokine pathways.
NF-κB signalling pathway
NF-κB is a transcription factor that lies at the centre of multiple innate and adaptive immune pathways78. NF-κB has both canonical and non-canonical pathways that regulate activation of immune responses79. In general, the canonical NF-κB pathway utilizes an abundance of cell-specific and stimulus-specific regulators to activate immune responses, including pathogen-associated molecular patterns (PAMPs) and cytokines such as TNF and IL-1β, whereas the non-canonical pathway responds mainly to ligands of the TNF superfamily. Although the non-canonical pathway has important roles in inflammation, it is canonical signalling that is essential for rapid regulation of host responses. Inhibitors of the pathways (IκB proteins) directly bind to NF-κB family members and prevent activation of signalling80. Activity of NF-κB pathways is extensively regulated by a variety of PTMs, including phosphorylation and ubiquitylation. The central importance of NF-κB to human disease is supported by an abundance of evidence from in vitro experiments, cellular systems, model organisms and observations in human disease81. Pathogenic variants of proteins that are involved in NF-κB pathways can behave either as gain-of-function or as loss-of-function alleles and, depending on the affected domain and the function of the altered protein, can cause either immunodeficiency or systemic inflammation. Nevertheless, all the diseases resulting from disruption of NF-κB pathways present with a continuum of clinical features, including immunodeficiency, autoinflammation, autoimmunity and atopy78. A strong additional piece of evidence comes from identification of an autoinflammatory disease caused by haploinsufficiency of RELA (encoding a subunit of NF-κB)82, which has overlapping clinical features with the disease caused by dysregulated ubiquitylation, termed haploinsufficiency of A20 (HA20; A20 is encoded by TNFAIP3). This condition is discussed in greater detail in the following section.
TNFAIP3-associated diseases
A20 is a bi-functional ubiquitin-modulating enzyme with both ubiquitin E3 ligase and deubiquitylase activities, which can also act as a linear ubiquitin effector83,84 (Fig. 2a). Because A20 functions as an inhibitor of inflammation, mutations that result in loss of function of this protein are linked to autoimmune and autoinflammatory phenotypes. A20 has deubiquitylase activity towards K63-linked ubiquitin chains on various proteins, including RIPK1 (receptor-interacting serine/threonine protein kinase 1) and NEMO (NF-κB essential modulator), and this activity results in disassembly of inflammatory complexes and suppression of immune responses26. Similarly, A20 has ubiquitin ligase activity, and modifies RIPK1 and other substrates with proteasome-targeting K48 chains, again halting NF-κB signalling. These contrasting, yet synergistic, functions of A20 are carried out by specific protein domains. The ovarian tumour (OTU) domain harbours deubiquitylase activity, whereas C-terminal zinc finger (ZnF) domains have ubiquitin-binding functions, with the ZnF4 domain binding K63 and K48 chains, and the ZnF7 domain binding M1 ubiquitin chains85. Although both the deubiquitylase and ubiquitin-ligase domains of A20 are implicated in regulatory functions, results from in vitro experiments and transgenic knock-in mice deficient in one or other of these activities fail to recapitulate the spontaneous inflammatory phenotype of Tnfaip3 knockout mice or of patients with TNFAIP3 loss-of-function mutations86. Results from studies of mouse models indicate that the anti-inflammatory role of A20 is also the result of its non-catalytic function in protecting cells from cell death86. Specifically, the activity of ZnF7 is essential for the inhibitory function of A20 by preventing TNF-induced necroptosis via inflammasome activation84,87. Although these models do not completely recapitulate the knockout mouse, they clearly demonstrate the importance of linear ubiquitin binding in A20 function in vivo. Moreover, only the combined inactivation of ZnF4 and ZnF7 phenocopied the perinatal lethality and severe multiorgan inflammation of A20-deficient mice87. In addition, A20 phosphorylation is critical for its anti-inflammatory function88. Hypomorphic missense mutations that diminish A20 phosphorylation can be evolutionarily advantageous to humans because they induce a subtle hyperinflammatory phenotype in leukocytes. The ancient Denisovan A20 haplotype carrying p.Thr108Ala and p.Ile207Leu variants, which are common in populations in Southeast Asia and Oceania, are associated with greater NF-κB response and heightened immune responsiveness89.
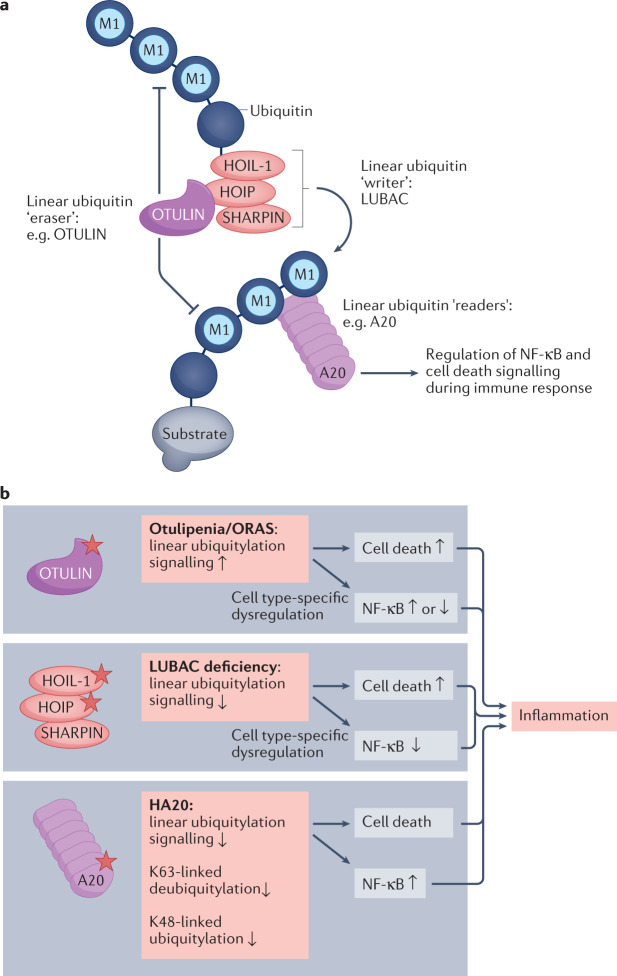
Fig. 2. Mutations affecting substrate-specific E3 ligases and deubiquitylases lead to dysregulation of immune signalling networks and inflammation.
a | Simplified model of how the linear-ubiquitylation pathway regulates NF-κB and cell-death signalling during responses to immune stimuli. Upon immune stimulation, the linear-ubiquitin assembly complex (LUBAC), which consists of HOIL-1 (haem-oxidized IRP2 ubiquitin ligase 1), HOIP (HOIL-1-interacting protein) and SHARPIN (SHANK-associated RH domain interactor), assembles linear ubiquitin chains on specific substrates. This is counteracted by the linear-ubiquitin-specific deubiquitylase OTULIN, which also reverses autoubiquitylation and thus stabilizes LUBAC in a cell-type-specific manner. Linear ubiquitin chains are recognized and interpreted by dedicated binding proteins that initiate NF-κB or cell-death signalling pathways. One such linear-ubiquitin reader is the hybrid deubiquitylase–E3 ligase enzyme A20. In this model, LUBAC is a ubiquitin ‘writer’, OTULIN is a ubiquitin ‘eraser’ and A20 is a ubiquitin ‘reader’. b | Loss-of-function mutations affecting subunits of the linear-ubiquitin writer LUBAC, the linear-ubiquitin eraser OTULIN and the linear-ubiquitin effector protein A20 result in autoinflammatory diseases. HA20, haploinsufficiency of A20; ORAS, OTULIN-related autoinflammatory syndrome.
TNFAIP3 coding and non-coding genetic variants are implicated in many different human diseases90, at the level of both common, low-penetrance (single-nucleotide polymorphisms (SNPs)), and rare, high-penetrance variants. TNFAIP3 SNPs are associated with conditions including systemic lupus erythematosus91, rheumatoid arthritis92 and psoriatic arthritis93. Some of these SNPs result in downregulation of A20 protein expression. Rare nonsense and frameshift variants are associated with a severe dominantly inherited autoinflammatory disease, HA20 (ref.94); somatic TNFAIP3 variants are also found in up to 30% of B cell lymphomas95. Patients with HA20 present with childhood-onset systemic disease manifesting with oral and genital ulcers, musculoskeletal, gastrointestinal and integumentary (skin) inflammation, and features of autoimmunity96. Immune profiling of these patients has identified high concentrations of numerous cytokines, many of which result directly from upregulation of NF-κB signalling94 (Fig. 2b). Patients with HA20 tend to respond to cytokine inhibitors, although severe, refractory cases may require more extensive combined therapies97 or HSCT. What is abundantly clear is that A20 acts as a central regulator of many pathways with various functions in particular cell types, and its perturbations underlie a spectrum of rheumatological and haematological diseases. In summary, although ‘mild’ hypomorphic A20 variants can be beneficial to human health by enhancing immune responses, more-detrimental variants can overactivate inflammatory signalling, resulting in a spectrum of autoinflammatory and autoimmune phenotypes87.
Diseases linked to dysregulation of linear ubiquitylation
Ubiquitylation frequently involves the addition of protein chains, with the most proximal ubiquitin attached to the substrate, typically to a lysine reside. An important chain subtype is the linear ubiquitin chain, in which the N-terminal α-amino group of Met1 of the growing chain is used for linkage to the C terminus of the previous ubiquitin moiety. Linear (M1) ubiquitin chains are critical for innate immune signalling and are conjugated by the E3 ligase complex LUBAC (linear ubiquitin chain activating complex) and removed by the deubiquitylase OTULIN (ubiquitin thioesterase otulin), which exclusively hydrolyses M1 ubiquitin chains, and CYLD (ubiquitin carboxyl-terminal hydrolase), which is less specific and can hydrolyse K63-linked chains and other ubiquitin linkages98 (Fig. 2a). SPATA2 (spermatogenesis-associated protein 2) acts as activator for the K63 and Met1 deubiquitylase function of CYLD in the TNFR1 signalling pathway99. Intriguingly, deficiencies of either LUBAC or OTULIN are implicated in monogenic autoinflammatory diseases98. LUBAC is composed of three proteins, HOIL-1 (haem-oxidized IRP2 ubiquitin ligase 1), HOIP (HOIL-1-interacting protein) and SHARPIN (SHANK-associated RH domain interactor), and mutations affecting HOIL-1 and HOIP lead to an autosomal-recessive immunodeficiency with concomitant autoinflammation100,101. Autosomal-recessive mutation resulting in the loss of OTULIN expression causes a rare, potentially fatal disease manifesting with constitutive severe systemic inflammation102–105. Notably, although LUBAC and OTULIN have opposing functions, their deficiencies lead to overlapping clinical manifestations, possibly (at least partly) as a result of the role of OTULIN in regulation of LUBAC activity. In the mouse model, OTULIN was shown to prevent autoubiquitylation of LUBAC, thereby promoting its E3 ligase function106 (Fig. 2a). Many components of TNF receptor (TNFR) signalling cascades are modified with, or can directly bind to, linear ubiquitin chains, so linear ubiquitylation is an important regulator of many aspects of innate and adaptive immunity. M1-linked ubiquitin chains are not only essential for TNFR-dependent NF-κB signalling but are also required for NOD2 (nucleotide-binding oligomerization domain-containing protein 2)-dependent NF-κB signalling and proteasome function107–109. Patients with otulipenia present with severe inflammation of the skin, adipose tissue and joints, whereas immunodeficiency is a more prominent feature of LUBAC deficiencies98. These differences reflect cell-specific and tissue-specific functions of individual components of the linear ubiquitin system. In general, patients with OTULIN deficiency respond to therapy with cytokine inhibitors, provided that treatment is initiated early in life. Alternatively, a substantial resolution of systemic inflammation and/or immunodeficiency is possible using HSCT110.
As described above, because the binding of A20 to linear ubiquitin chains is important for its anti-inflammatory function, we hypothesize that HA20 manifestations might in part result from dysregulation of linear ubiquitylation (Fig. 2).
Disease linked to dysregulation of the E3 ligase XIAP
E3 ubiquitin-protein ligase XIAP (also known as X-linked inhibitor of apoptosis) regulates cell-death pathways and NF-κB-dependent immune signalling events111. The multiple functions of XIAP are mediated by its modular domain architecture, which harbours three zinc-binding baculovirus IAP repeat (BIR) domains, a ubiquitin-associated (UBA) domain and a C-terminal RING ubiquitin E3 ligase domain. The BIR2 and BIR3 domains bind to and inhibit caspases, enabling XIAP to exert anti-apoptotic effects that are important during expansion of adaptive immune cells in the setting of viral infection112. The UBA domain of XIAP binds directly to K63-linked ubiquitin chains on NEMO, to activate NF-κB signalling113. Through its ubiquitin E3 ligase activity, XIAP prevents TNF-mediated and RIPK3-dependent cell death (via ubiquitylation of RIPK1)114 and promotes NOD2 pro-inflammatory signalling by mediating K63-linked polyubiquitylation of RIPK2 within the NOD2 signalling complex, to recruit LUBAC and to enhance NF-κB-dependent and MAPK-dependent cytokine production107,115. Thus, XIAP is important for the clearance of pathogens and for broad regulation of inflammatory and immune responses.
The XIAP gene is located on the X chromosome, and loss-of-function mutations (including point mutations affecting the RING domain) in males lead to a broad spectrum of immune dysregulation characterized by cytopenia, hypogammaglobulinaemia and/or recurrent infection, splenomegaly, colitis and a propensity to haemophagocytic lymphohistiocytosis, often triggered by Epstein–Barr virus infection112. The inflammatory phenotypes are in part attributed to inflammasome activation and secretion of various pro-inflammatory cytokines and chemokines. Disease onset and presentation is highly variable, and some men with loss-of-function mutations in XIAP remain asymptomatic, suggesting that environmental factors, such as exposure to infection, contribute to the disease expressivity. Elucidation of these factors will be important, as HCST is the only curative treatment described to date, and it has poor outcomes116.
Interferon signalling pathway
Type I interferons are cytokines that mediate host responses to pathogens, in particular viruses. These cytokines are produced in response to stimulation of host pathogen sensors by binding of specific PAMPs, primarily through nucleic acid sensing117,118. PAMP recognition initiates a cascade of signalling, through JAK–signal transducer and activator of transcription (STAT) pathways that lead to activation of ISGs, which is a hallmark of type I interferon-mediated inflammation119. This transcriptional programme leads to antiviral activities including apoptosis of infected cells, and maturation and proliferation of lymphocytes as a coordinated means of clearing pathogens. Furthermore, secretion of type I interferons leads to autocrine and paracrine signalling, which enhances anti-viral effects. In this pathway, inherited genetic defects or acquired defects in the form of anti-interferon autoantibodies result in a compromised responsiveness to infections, including SARS-CoV-2 (refs120–122). Notably, some viruses have evolved strategies whereby, upon infection, they attenuate interferon responses by use of specific enzymes that can cleave the ubiquitin-like modifier ISG15 from proteins such as IRF3 (ref.123).
Type I interferonopathies
Type I interferonopathies are a class of monogenic autoinflammatory diseases characterized by mutations that lead to constitutive activation of the type I interferon pathway. These disorders have substantial overlap of phenotypes, including arthritis, necrotizing vasculitis, intracranial calcifications, interstitial lung disease and lipodystrophy, with similar molecular signatures at the transcriptional and cytokine level124. Evidence of the importance of this signalling pathway is abundant, and derives from both inherited diseases and acquired conditions that lead to hyperinflamed or immunodeficient states125. Similar to the activation in NF-κB signalling, mutations in ubiquitin E3 ligases and deubiquitylases can cause constitutive activation of the type I interferon pathway126.
The OTU domain-containing protein 1 (OTUD1) deubiquitylase was implicated as a potential disease-causing gene in multiple systemic inflammatory diseases in a small-scale case–control study127. OTUD1 enzymatically controls inflammation by inhibiting IRF3-dependent transcriptional activity and activation of RIG-I (antiviral innate immune response receptor RIG-I). In experiments in vitro and in vivo in mice, OTUD1 repressed type I interferon-mediated disease128. Other ubiquitylation enzymes identified in mice as being critical to regulation of interferon signalling are not yet known to be associated with human disease129. Upregulation of type I interferon pathways also occurs in OTULIN-deficient humans and mice106, and in patients with HA20 (ref.97).
Diseases linked to dysregulation of the ubiquitin-like modifier ISG15
ISG15 is expressed in response to type I interferon signalling and has important roles in anti-pathogenic responses98,130. In a similar manner to ubiquitylation, ISG15 undergoes cycles of conjugation and deconjugation to substrates, and there is thought to be considerable crosstalk between ISG15 and ubiquitylation pathways131. Historically, ISG15 was thought to function as a pro-inflammatory antiviral effector. Results from studies in patients who lack ISG15 showed that, depending on the circumstances and target substrates, ISG15 can either suppress or activate the interferon pathway. ISG15 loss by recessively inherited mutations leads to a disease manifesting with basal ganglia calcifications and sporadic seizures that is similar to Aicardi–Goutières Syndrome (AGS), as a result of upregulation of the expression of ISGs132. Interestingly, patients with ISG15 deficiency also have susceptibility to mycobacterial disease, which is likely to be explained by loss of the extracellular function of ISG15 to induce IFNγ production in lymphocytes133–135. Thus, patients have manifestations of both autoinflammation and immunodeficiency. Similarly, autosomal-recessive loss-of-function mutations affecting the ISG15-specific deconjugating enzyme ubiquitin specific peptidase 18 (USP18) also result in primary interferonopathy. Loss of USP18 is primarily thought to cause disease in a manner that does not rely on its catalytic activity but rather on its ability to bind and inhibit signalling through the IFNα receptor (IFNAR)136. The binding between free intracellular ISG15 and USP18 increases the stability of USP18 by preventing its degradation. Although loss of ISG15 is compatible with life, USP18 deficiency is a potentially lethal disease with a phenotype similar to that of severe AGS as a result of brain calcification. Both USP18-deficient and ISG15-deficient patients have a strong interferon gene-expression signature in their peripheral blood leukocytes and cerebrospinal fluid, which can be effectively ameliorated with JAK inhibitors137. Interestingly, deficiency of ISG15 results in specific immunodeficiency in humans but not in mice, and as there is minimal conservation between species of the gene and pathway, ISG15-deficient patients are potentially a critical resource for further studies of this pathway138.
Stress and cell death in immune cells
In contrast to the many specific pathways described above, there are a few examples of human diseases that are caused by dysregulated ubiquitylation leading to disruption of general cellular functions. These general pathways are probably not causing inflammation because of aberrant levels of a single downstream effector but instead through activation of stress or cell-death pathways with many potential triggers (Fig. 3). These factors might also have a role in some of the specific pathways, as a downstream consequence of inflammation.
Fig. 3. Disorders of ubiquitylation lead to systemic inflammation and have potential for therapeutic intervention.
Mutations that affect enzymes involved in ubiquitylation pathways (indicated here by red stars) can lead to activation of cell death, unfolded protein response and inflammatory pathways that result in inflammatory disease. Therapeutic intervention might be possible at the levels of enzyme-specific activation and inhibition, pathway-specific inhibition and cytokine inhibition. LUBAC, linear-ubiquitin assembly complex; OTULIN, ubiquitin thioesterase otulin; UBA1, ubiquitin-like modifier-activating enzyme 1.
The UPR and autoinflammation
The UPR is a cellular stress pathway that regulates immune responses139,140. The UPR is activated when protein-folding requirements exceed the processing capacity of the endoplasmic reticulum (ER), and misfolded and unfolded proteins accumulate. The UPR is composed of three major pathways that use inositol-requiring enzyme 1α (IRE1α), PRKR-like ER kinase (PERK) and activating transcription factor 6α (ATF6α) as sensors, all of which are activated by misfolded proteins139,140. Stimulation of these pathways can directly result in activation of multiple innate immune responses, including NF-κB pathways, with reciprocal activation of the UPR as a consequence of pathogen-receptor activation. Studies of autoimmune and autoinflammatory diseases such as inflammatory bowel disease have implicated the UPR and ER integrity in disease mechanisms141,142. Dysregulation of both ubiquitin activation (in VEXAS) and processing of degradative ubiquitylation signals (in PRAAS) lead to activation of the UPR. As further evidence of the importance of the UPR in inflammation, mutations in APS13 (ref.143) and COPA144, which encode proteins that maintain Golgi–ER trafficking of TLR3 and STING proteins, respectively, can lead to monogenic autoinflammatory diseases with distinct features, likely through altered proteostasis. How these proteins directly contribute to specific forms of inflammation (such as pustular psoriasis in APS13 deficiency) remains unclear, but it might involve inappropriate processing of a key factor, such as STING145, or regulation of a number of other cell-specific proteins.
Cell death and autoinflammation
Cell death is an essential process that is required for proper tissue homeostasis and development. Many forms of programmed cell death exist, including apoptosis, necroptosis, pyroptosis and NETosis, and extensive reviews of these processes are available elsewhere146,147. Instead of death bringing finality, it has manifold roles, depending on the context of cell type and stimuli, in preventing, causing and perpetuating inflammation146. Programmed cell death can result directly in the release of cytokines, as is the case for pyroptosis, or it can result in the presentation of damage-associated molecular patterns (DAMPs), which then promote inflammation. In particular, apoptosis and necroptosis integrate external signals, such as binding of death-receptor ligands (including TNF, FASL and TRAIL), either directly or indirectly resulting in a pro-inflammatory milieu. These mechanisms can involve cell-intrinsic and cell-extrinsic processes.
A number of inborn errors of immunity are caused by mutations in genes encoding key regulators of cell death, including CASP8 (ref.148) and RIPK1 (refs149–151), or receptor–ligand combinations, such as FAS and FASLG (also known as TNFSF6)152. These disorders can present with a combination of immunodeficiency and/or autoinflammation, similar to LUBAC-related disorders. FAS and TNFR1 directly mediate immune signalling upon specific cytokine binding and activate cell-death pathways, and dysregulation of either activity contributes to disease pathogenesis. These diseases directly involve cell death, whether through lack of apoptosis in autoimmune lymphoproliferative syndrome caused by FAS mutations152,153, or via increased cell death in RIPK1-related disorders. In addition, many of the downstream ubiquitylation enzymes and ubiquitylated substrates in programmed cell death are important for regulation of inflammation as they mediate the balance between TNFR1-induced apoptosis complex 1 (membrane-bound TNFR1 complex) and complex 2 (cytoplasmic RIPK1-associated complex)9. Notably, because of the overlap between the signals that modulate inflammation and cell death, many factors have pleiotropic effects on inflammation.
PTMs such as ubiquitylation are important downstream changes that enable dynamic modulation of cell-death processes, and alteration of PTM machinery has varied physiological consequences, particularly in immunity. For example, loss of the E3 ligase LUBAC leads to autoinflammation, although the reduction of NF-κB signalling is a major feature of disease in patients with HOIL-1 loss of function100, which seems paradoxical, given that increased NF-κB signalling would be expected in autoinflammation. Nevertheless, the downregulation in NF-kB pro-survival signalling can shift the balance towards RIPK1-mediated pro-death pathways, resulting in increased sensitivity of LUBAC-deficient cells to TNF-induced cell death. Evidence from mouse models indicates that programmed cell death, whether apoptosis or necroptosis, is pro-inflammatory in these LUBAC deficiencies146. LUBAC prevents cell death in response to TRAIL, FAS, TLR and TNF signalling, and A20, in addition to its inhibitory role in TNF-dependent inflammation, is implicated in dysregulation of cell death86. Another deubiquitylase, CYLD, has a role in an autoinflammatory disease that is caused by deficiency of TANK binding kinase 1 (TBK1)154. Although TBK1 is known to regulate the type I interferon pathway, TBK1-deficient individuals maintain hypomorphic but sufficient interferon induction via the RIG-I–MAVS pathway, and present with severe systemic inflammation resulting from dysregulation of TNF-induced RIPK1-mediated cell death154. Fibroblasts derived from patients with TBK1 deficiency display heightened sensitivity to necroptosis that results from the lack of inhibitory phosphorylation on CYLD, which increases its deubiquitylation activity towards RIPK1. CYLD is phosphorylated by TBK1 to restrain the pro-death propensity of RIPK1154. A similar mechanism may operate in patients with HA20. The contribution of dysregulated cell death to broader classes of inflammatory disease remains poorly understood, but it is likely to be an under-recognized effector of pathogenesis that could also act in VEXAS syndrome and PRAAS as a secondary effect. In the future, it will be important to study the contribution of these pathways in a cell-specific context and in human cells.
Conclusions
Innate immune sensing and activation requires the concerted ability to detect pathogens or stimuli and rapidly trigger production of cytokines or mediators of inflammation. In turn, efficient inactivation is critical to prevent prolonged inflammation once the threat is neutralized. In many cases, activation is accomplished by sensors or receptors that bind specific molecules and trigger highly targeted transcriptional, translational or post-translational programmes to promote clearance of pathogens155. In SAIDs, and rheumatological disorders as a whole, these pathways can be inappropriately co-opted and lead to recurrent or continued inflammation.
Genetic discovery of Mendelian disorders of autoinflammation has revealed new insights into the molecular mechanisms that are important for fine tuning of the innate immune system. Many disorders are linked to enzymes that regulate ubiquitylation, which is a pleiotropic set of processes that regulate many different aspects of immunity. Despite being linked to specific pathways of inflammation, ubiquitylation-related disorders cannot be easily treated with single cytokine-targeting therapies. Notably, the most proximal enzyme (UBA1) and some of the most distal enzymes (A20 and OTULIN) of ubiquitylation, when affected by mutation, can have similar effects in disease manifestation (resulting in VEXAS syndrome, HA20 and otulipenia) (Fig. 3). This observation suggests that shared mechanisms of disease exist for disorders of ubiquitylation, beyond systemic cytokine production, although the nature of these shared mechanisms is not yet fully known. They could involve specific shared substrates or pathways, many different substrates with similar effects, global shifts in cytoplasmic ubiquitin machinery, or combinations of these factors. In particular, the contribution of dysregulation of cell-death pathways warrants further investigation. Studying diseases caused by ubiquitylation defects consistently in humans and in animal models will help to further our understanding of potential commonalities, and could lead to more general therapies than are available at present (Fig. 3). For example, we propose that developing small-molecule activators of UBA1 or the proteasome that counterbalance dysregulation in protein turnover might not only provide a treatment option for PRAAS and VEXAS syndromes, but these drugs might also be beneficial for amelioration of autoinflammatory phenotypes caused by mutations affecting substrate-specific ubiquitin-system components. Other general treatment strategies could involve the use of inhibitors of the UPR and cell-death pathways that are frequently overactivated in SAIDs.
The genes that are currently implicated in disorders of ubiquitylation are likely to be just the first of many, and associations of further elements of the ubiquitylation machinery with human immunological and non-immunological disorders might soon be identified. Given the identification of disease-causing variants in E1 enzymes, E3 ligases and deubiquitylases, there could also be phenotypes that are associated with variants in E2 enzymes, ubiquitin sensors or effector proteins, and other cell-specific deubiquitylases. In addition, given the disease manifestations related to ISG15 and UPS18, we predict that variants in the ISG15 conjugation or ubiquitin-like machineries could also contribute to human immunological diseases. Similarly, other forms of protein degradation, such as autophagy, are likely to be implicated in the pathogenesis of autoinflammation156. Although we have focused here on germline causes, additional, both germline and somatic, pathogenic mutations causing aberrant ubiquitylation will likely be uncovered in the future.
Acknowledgements
We thank Dr Hirotsugu Oda for thoughtful comments on the manuscript. This work was supported by the intramural research program of the NIH including the National Human Genome Research Institute and the National Institute of Dental and Craniofacial Research. D.B.B. was supported in part by funding from the NIH (R00AR078205).
Author contributions
All authors contributed equally to all aspects of the article.
Peer review
Peer review information
Nature Reviews Rheumatology thanks G. van Loo; M. A. Lee-Kirsch; and M. McDermott, who co-reviewed with L. Pieper Pournara, for their contribution to the peer review of this work.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.No Author Listed. A prize for protein degradation. Nat. Cell Biol. 2004;6:1011. doi: 10.1038/ncb1104-1011. [DOI] [PubMed] [Google Scholar]
- 2.Wilkinson KD. The discovery of ubiquitin-dependent proteolysis. Proc. Natl Acad. Sci. USA. 2005;102:15280–15282. doi: 10.1073/pnas.0504842102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ciechanover A, Heller H, Elias S, Haas AL, Hershko A. ATP-dependent conjugation of reticulocyte proteins with the polypeptide required for protein degradation. Proc. Natl Acad. Sci. USA. 1980;77:1365–1368. doi: 10.1073/pnas.77.3.1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hershko A, Ciechanover A, Heller H, Haas AL, Rose IA. Proposed role of ATP in protein breakdown: conjugation of protein with multiple chains of the polypeptide of ATP-dependent proteolysis. Proc. Natl Acad. Sci. USA. 1980;77:1783–1786. doi: 10.1073/pnas.77.4.1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Goldstein G, et al. Isolation of a polypeptide that has lymphocyte-differentiating properties and is probably represented universally in living cells. Proc. Natl Acad. Sci. USA. 1975;72:11–15. doi: 10.1073/pnas.72.1.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Komander D, Rape M. The ubiquitin code. Annu. Rev. Biochem. 2012;81:203–229. doi: 10.1146/annurev-biochem-060310-170328. [DOI] [PubMed] [Google Scholar]
- 7.Rape M. Ubiquitylation at the crossroads of development and disease. Nat. Rev. Mol. Cell Biol. 2018;19:59–70. doi: 10.1038/nrm.2017.83. [DOI] [PubMed] [Google Scholar]
- 8.Basar MA, Beck DB, Werner A. Deubiquitylases in developmental ubiquitin signaling and congenital diseases. Cell Death Differ. 2021;28:538–556. doi: 10.1038/s41418-020-00697-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Manthiram K, Zhou Q, Aksentijevich I, Kastner DL. The monogenic autoinflammatory diseases define new pathways in human innate immunity and inflammation. Nat. Immunol. 2017;18:832–842. doi: 10.1038/ni.3777. [DOI] [PubMed] [Google Scholar]
- 10.Schulman BA, Harper JW. Ubiquitin-like protein activation by E1 enzymes: the apex for downstream signalling pathways. Nat. Rev. Mol. Cell Biol. 2009;10:319–331. doi: 10.1038/nrm2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Otten EG, et al. Ubiquitylation of lipopolysaccharide by RNF213 during bacterial infection. Nature. 2021;594:111–116. doi: 10.1038/s41586-021-03566-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kelsall IR, Zhang J, Knebel A, Arthur JSC, Cohen P. The E3 ligase HOIL-1 catalyses ester bond formation between ubiquitin and components of the Myddosome in mammalian cells. Proc. Natl Acad. Sci. USA. 2019;116:13293–13298. doi: 10.1073/pnas.1905873116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pao KC, et al. Activity-based E3 ligase profiling uncovers an E3 ligase with esterification activity. Nature. 2018;556:381–385. doi: 10.1038/s41586-018-0026-1. [DOI] [PubMed] [Google Scholar]
- 14.Metzger MB, Pruneda JN, Klevit RE, Weissman AM. RING-type E3 ligases: master manipulators of E2 ubiquitin-conjugating enzymes and ubiquitination. Biochim. Biophys. Acta. 2014;1843:47–60. doi: 10.1016/j.bbamcr.2013.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Islam AB, Dave M, Amin S, Jensen RV, Amin AR. Genomic, lipidomic and metabolomic analysis of cyclooxygenase-null cells: eicosanoid storm, cross talk, and compensation by COX-1. Genomics Proteom. Bioinforma. 2016;14:81–93. doi: 10.1016/j.gpb.2014.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pickart CM. Targeting of substrates to the 26S proteasome. FASEB J. 1997;11:1055–1066. doi: 10.1096/fasebj.11.13.9367341. [DOI] [PubMed] [Google Scholar]
- 17.Iwai K. Diverse ubiquitin signaling in NF-κB activation. Trends Cell Biol. 2012;22:355–364. doi: 10.1016/j.tcb.2012.04.001. [DOI] [PubMed] [Google Scholar]
- 18.French ME, Koehler CF, Hunter T. Emerging functions of branched ubiquitin chains. Cell Discov. 2021;7:6. doi: 10.1038/s41421-020-00237-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Randles L, Walters KJ. Ubiquitin and its binding domains. Front. Biosci. 2012;17:2140–2157. doi: 10.2741/4042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Husnjak K, Dikic I. Ubiquitin-binding proteins: decoders of ubiquitin-mediated cellular functions. Annu. Rev. Biochem. 2012;81:291–322. doi: 10.1146/annurev-biochem-051810-094654. [DOI] [PubMed] [Google Scholar]
- 21.Clague MJ, Urbe S, Komander D. Breaking the chains: deubiquitylating enzyme specificity begets function. Nat. Rev. Mol. Cell Biol. 2019;20:338–352. doi: 10.1038/s41580-019-0099-1. [DOI] [PubMed] [Google Scholar]
- 22.Cappadocia L, Lima CD. Ubiquitin-like protein conjugation: structures, chemistry, and mechanism. Chem. Rev. 2018;118:889–918. doi: 10.1021/acs.chemrev.6b00737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Boyden LM, et al. Mutations in kelch-like 3 and cullin 3 cause hypertension and electrolyte abnormalities. Nature. 2012;482:98–102. doi: 10.1038/nature10814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Choi J, et al. Loss of KLHL6 promotes diffuse large B-cell lymphoma growth and survival by stabilizing the mRNA decay factor roquin2. Nat. Cell Biol. 2018;20:586–596. doi: 10.1038/s41556-018-0084-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Glickman MH, Ciechanover A. The ubiquitin-proteasome proteolytic pathway: destruction for the sake of construction. Physiol. Rev. 2002;82:373–428. doi: 10.1152/physrev.00027.2001. [DOI] [PubMed] [Google Scholar]
- 26.Aksentijevich I, Zhou Q. NF-κB pathway in autoinflammatory diseases: dysregulation of protein modifications by ubiquitin defines a new category of autoinflammatory diseases. Front. Immunol. 2017;8:399. doi: 10.3389/fimmu.2017.00399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hoffman HM, Mueller JL, Broide DH, Wanderer AA, Kolodner RD. Mutation of a new gene encoding a putative pyrin-like protein causes familial cold autoinflammatory syndrome and Muckle-Wells syndrome. Nat. Genet. 2001;29:301–305. doi: 10.1038/ng756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Swanson KV, Deng M, Ting JP. The NLRP3 inflammasome: molecular activation and regulation to therapeutics. Nat. Rev. Immunol. 2019;19:477–489. doi: 10.1038/s41577-019-0165-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Aksentijevich I, et al. An autoinflammatory disease with deficiency of the interleukin-1-receptor antagonist. N. Engl. J. Med. 2009;360:2426–2437. doi: 10.1056/NEJMoa0807865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stone DL, et al. The systemic autoinflammatory diseases: coming of age with the human genome. J. Allergy Clin. Immunol. 2020;146:997–1001. doi: 10.1016/j.jaci.2020.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McGonagle D, McDermott MF. A proposed classification of the immunological diseases. PLoS Med. 2006;3:e297. doi: 10.1371/journal.pmed.0030297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ben-Chetrit E, et al. Consensus proposal for taxonomy and definition of the autoinflammatory diseases (AIDs): a Delphi study. Ann. Rheum. Dis. 2018;77:1558–1565. doi: 10.1136/annrheumdis-2017-212515. [DOI] [PubMed] [Google Scholar]
- 33.Savic S, Caseley EA, McDermott MF. Moving towards a systems-based classification of innate immune-mediated diseases. Nat. Rev. Rheumatol. 2020;16:222–237. doi: 10.1038/s41584-020-0377-5. [DOI] [PubMed] [Google Scholar]
- 34.Wertz IE, Wang X. From discovery to bedside: targeting the ubiquitin system. Cell Chem. Biol. 2019;26:156–177. doi: 10.1016/j.chembiol.2018.10.022. [DOI] [PubMed] [Google Scholar]
- 35.Beck DB, et al. Somatic mutations in UBA1 and severe adult-onset autoinflammatory disease. N. Engl. J. Med. 2020 doi: 10.1056/NEJMoa2026834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hyer ML, et al. A small-molecule inhibitor of the ubiquitin activating enzyme for cancer treatment. Nat. Med. 2018;24:186–193. doi: 10.1038/nm.4474. [DOI] [PubMed] [Google Scholar]
- 37.Georgin-Lavialle S, et al. Further characterization of clinical and laboratory features in VEXAS syndrome: large-scale analysis of a multicentre case series of 116 French patients. Br. J. Dermatol. 2021 doi: 10.1111/bjd.20805. [DOI] [PubMed] [Google Scholar]
- 38.Poulter JA, et al. Novel somatic mutations in UBA1 as a cause of VEXAS syndrome. Blood. 2021;137:3676–3681. doi: 10.1182/blood.2020010286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bourbon E, et al. Therapeutic options in VEXAS syndrome: insights from a retrospective series. Blood. 2021;137:3682–3684. doi: 10.1182/blood.2020010177. [DOI] [PubMed] [Google Scholar]
- 40.Staels F, et al. Case report: VEXAS syndrome: from mild symptoms to life-threatening macrophage activation syndrome. Front. Immunol. 2021;12:678927. doi: 10.3389/fimmu.2021.678927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Barba T, et al. VEXAS syndrome in a woman. Rheumatology. 2021 doi: 10.1093/rheumatology/keab392. [DOI] [PubMed] [Google Scholar]
- 42.Arlet JB, Terrier B, Kosmider O. Mutant UBA1 and severe adult-onset autoinflammatory disease. N. Engl. J. Med. 2021;384:2163. doi: 10.1056/NEJMc2102124. [DOI] [PubMed] [Google Scholar]
- 43.Beck DB, Grayson PC, Kastner DL. Mutant UBA1 and severe adult-onset autoinflammatory disease. N. Engl. J. Med. 2021;384:2164–2165. doi: 10.1056/NEJMc2102124. [DOI] [PubMed] [Google Scholar]
- 44.Luzzatto L, Risitano AM, Notaro R. Mutant UBA1 and severe adult-onset autoinflammatory disease. N. Engl. J. Med. 2021;384:2164. doi: 10.1056/NEJMc2102124. [DOI] [PubMed] [Google Scholar]
- 45.Diarra A, Duployez N, Terriou L. Mutant UBA1 and severe adult-onset autoinflammatory disease. N. Engl. J. Med. 2021;384:2163–2164. doi: 10.1056/NEJMc2102124. [DOI] [PubMed] [Google Scholar]
- 46.Pfleger CM, Harvey KF, Yan H, Hariharan IK. Mutation of the gene encoding the ubiquitin activating enzyme uba1 causes tissue overgrowth in Drosophila. Fly. 2007;1:95–105. doi: 10.4161/fly.4285. [DOI] [PubMed] [Google Scholar]
- 47.Lacombe V, Kosmider O, Prevost M, Lavigne C, Urbanski G. Severe joint involvement in VEXAS syndrome: a case report. Ann. Intern. Med. 2021;174:1025–1027. doi: 10.7326/L21-0023. [DOI] [PubMed] [Google Scholar]
- 48.Grey A, et al. A case of VEXAS syndrome complicated by hemophagocytic lymphohistiocytosis. J. Clin. Immunol. 2021 doi: 10.1007/s10875-021-01070-y. [DOI] [PubMed] [Google Scholar]
- 49.Magnol M, et al. VEXAS syndrome in a patient with previous spondyloarthritis with favorable response to intravenous immunoglobulin anti-IL17 therapy. Rheumatology. 2021 doi: 10.1093/rheumatology/keab211. [DOI] [PubMed] [Google Scholar]
- 50.Grayson PC, Patel BA, Young NS. VEXAS syndrome. Blood. 2021;137:3591–3594. doi: 10.1182/blood.2021011455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Goetzke CC, Ebstein F, Kallinich T. Role of proteasomes in inflammation. J. Clin. Med. 2021 doi: 10.3390/jcm10081783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fremond ML, Crow YJ. STING-Mediated Lung Inflammation and Beyond. J. Clin. Immunol. 2021;41:501–514. doi: 10.1007/s10875-021-00974-z. [DOI] [PubMed] [Google Scholar]
- 53.Liu Y, et al. Activated STING in a vascular and pulmonary syndrome. N. Engl. J. Med. 2014;371:507–518. doi: 10.1056/NEJMoa1312625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Diarra A, et al. Successful allogeneic hematopoietic stem cell transplantation in patients with VEXAS syndrome: a two center experience. Blood Adv. 2021 doi: 10.1182/bloodadvances.2021004749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ramser J, et al. Rare missense and synonymous variants in UBE1 are associated with X-linked infantile spinal muscular atrophy. Am. J. Hum. Genet. 2008;82:188–193. doi: 10.1016/j.ajhg.2007.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Balak CD, et al. Functional characterizations of rare UBA1 variants in X-linked Spinal Muscular Atrophy. F1000Res. 2017;6:1636. doi: 10.12688/f1000research.11878.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Groen EJN, Gillingwater TH. UBA1:at the crossroads of ubiquitin homeostasis and neurodegeneration. Trends Mol. Med. 2015;21:622–632. doi: 10.1016/j.molmed.2015.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Powis RA, et al. Systemic restoration of UBA1 ameliorates disease in spinal muscular atrophy. JCI Insight. 2016;1:e87908. doi: 10.1172/jci.insight.87908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chanarat S, Mishra SK. Emerging roles of ubiquitin-like proteins in pre-mRNA splicing. Trends Biochem. Sci. 2018;43:896–907. doi: 10.1016/j.tibs.2018.09.001. [DOI] [PubMed] [Google Scholar]
- 60.Poli MC, et al. Heterozygous truncating variants in POMP escape nonsense-mediated decay and cause a unique immune dysregulatory syndrome. Am. J. Hum. Genet. 2018;102:1126–1142. doi: 10.1016/j.ajhg.2018.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lam MT, et al. A novel disorder involving dyshematopoiesis, inflammation, and HLH due to aberrant CDC42 function. J. Exp. Med. 2019;216:2778–2799. doi: 10.1084/jem.20190147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Endo M, Druso JE, Cerione RA. The two splice variant forms of Cdc42 exert distinct and essential functions in neurogenesis. J. Biol. Chem. 2020;295:4498–4512. doi: 10.1074/jbc.RA119.011837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Dahlqvist J, et al. A single-nucleotide deletion in the POMP 5′ UTR causes a transcriptional switch and altered epidermal proteasome distribution in KLICK genodermatosis. Am. J. Hum. Genet. 2010;86:596–603. doi: 10.1016/j.ajhg.2010.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bard JAM, et al. Structure and function of the 26S proteasome. Annu. Rev. Biochem. 2018;87:697–724. doi: 10.1146/annurev-biochem-062917-011931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rousseau A, Bertolotti A. Regulation of proteasome assembly and activity in health and disease. Nat. Rev. Mol. Cell Biol. 2018;19:697–712. doi: 10.1038/s41580-018-0040-z. [DOI] [PubMed] [Google Scholar]
- 66.Murata S, Takahama Y, Kasahara M, Tanaka K. The immunoproteasome and thymoproteasome: functions, evolution and human disease. Nat. Immunol. 2018;19:923–931. doi: 10.1038/s41590-018-0186-z. [DOI] [PubMed] [Google Scholar]
- 67.Brehm A, Kruger E. Dysfunction in protein clearance by the proteasome: impact on autoinflammatory diseases. Semin. Immunopathol. 2015;37:323–333. doi: 10.1007/s00281-015-0486-4. [DOI] [PubMed] [Google Scholar]
- 68.Ohmura K. Nakajo-Nishimura syndrome and related proteasome-associated autoinflammatory syndromes. J. Inflamm. Res. 2019;12:259–265. doi: 10.2147/JIR.S194098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Brehm A, et al. Additive loss-of-function proteasome subunit mutations in CANDLE/PRAAS patients promote type I IFN production. J. Clin. Invest. 2015;125:4196–4211. doi: 10.1172/JCI81260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yan K, et al. Haploinsufficiency of PSMD12 causes proteasome dysfunction and subclinical autoinflammation. Arthritis Rheumatol. 2022 doi: 10.1002/art.42070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Aksentijevich I, Schnappauf O. Molecular mechanisms of phenotypic variability in monogenic autoinflammatory diseases. Nat. Rev. Rheumatol. 2021;17:405–425. doi: 10.1038/s41584-021-00614-1. [DOI] [PubMed] [Google Scholar]
- 72.Davidson S, et al. Protein kinase R is an innate immune sensor of proteotoxic stress via accumulation of cytoplasmic IL-24. Sci. Immunol. 2022;7:eabi6763. doi: 10.1126/sciimmunol.abi6763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Opitz E, et al. Impairment of immunoproteasome function by beta5i/LMP7 subunit deficiency results in severe enterovirus myocarditis. PLoS Pathog. 2011;7:e1002233. doi: 10.1371/journal.ppat.1002233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kury S, et al. De novo disruption of the proteasome regulatory subunit PSMD12 causes a syndromic neurodevelopmental disorder. Am. J. Hum. Genet. 2017;100:352–363. doi: 10.1016/j.ajhg.2017.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Isidor B, et al. Stankiewicz-Isidor syndrome: expanding the clinical and molecular phenotype. Genet. Med. 2022;24:179–191. doi: 10.1016/j.gim.2021.09.005. [DOI] [PubMed] [Google Scholar]
- 76.Khalil R, et al. PSMD12 haploinsufficiency in a neurodevelopmental disorder with autistic features. Am. J. Med. Genet. B Neuropsychiatr. Genet. 2018;177:736–745. doi: 10.1002/ajmg.b.32688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Crow YJ, Manel N. Aicardi-Goutieres syndrome and the type I interferonopathies. Nat. Rev. Immunol. 2015;15:429–440. doi: 10.1038/nri3850. [DOI] [PubMed] [Google Scholar]
- 78.Schnappauf O, Aksentijevich I. Mendelian diseases of dysregulated canonical NF-κB signaling: From immunodeficiency to inflammation. J. Leukoc. Biol. 2020;108:573–589. doi: 10.1002/JLB.2MR0520-166R. [DOI] [PubMed] [Google Scholar]
- 79.Sun SC. The non-canonical NF-κB pathway in immunity and inflammation. Nat. Rev. Immunol. 2017;17:545–558. doi: 10.1038/nri.2017.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hayden MS, Ghosh S. NF-κB, the first quarter-century: remarkable progress and outstanding questions. Genes Dev. 2012;26:203–234. doi: 10.1101/gad.183434.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zhang Q, Lenardo MJ, Baltimore D. 30 years of NF-κB: a blossoming of relevance to human pathobiology. Cell. 2017;168:37–57. doi: 10.1016/j.cell.2016.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Badran YR, et al. Human RELA haploinsufficiency results in autosomal-dominant chronic mucocutaneous ulceration. J. Exp. Med. 2017;214:1937–1947. doi: 10.1084/jem.20160724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wertz IE, et al. De-ubiquitination and ubiquitin ligase domains of A2κκ0 downregulate NF-κB signalling. Nature. 2004;430:694–699. doi: 10.1038/nature02794. [DOI] [PubMed] [Google Scholar]
- 84.Polykratis A, et al. A20 prevents inflammasome-dependent arthritis by inhibiting macrophage necroptosis through its ZnF7 ubiquitin-binding domain. Nat. Cell Biol. 2019;21:731–742. doi: 10.1038/s41556-019-0324-3. [DOI] [PubMed] [Google Scholar]
- 85.Tokunaga F, et al. Specific recognition of linear polyubiquitin by A20 zinc finger 7 is involved in NF-κB regulation. EMBO J. 2012;31:3856–3870. doi: 10.1038/emboj.2012.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Priem D, van Loo G, Bertrand MJM. A20 and cell death-driven inflammation. Trends Immunol. 2020;41:421–435. doi: 10.1016/j.it.2020.03.001. [DOI] [PubMed] [Google Scholar]
- 87.Martens A, et al. Two distinct ubiquitin-binding motifs in A20 mediate its anti-inflammatory and cell-protective activities. Nat. Immunol. 2020;21:381–387. doi: 10.1038/s41590-020-0621-9. [DOI] [PubMed] [Google Scholar]
- 88.Hutti JE, et al. IκB kinase beta phosphorylates the K63 deubiquitinase A20 to cause feedback inhibition of the NF-κB pathway. Mol. Cell Biol. 2007;27:7451–7461. doi: 10.1128/MCB.01101-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Zammit NW, et al. Denisovan, modern human and mouse TNFAIP3 alleles tune A20 phosphorylation and immunity. Nat. Immunol. 2019;20:1299–1310. doi: 10.1038/s41590-019-0492-0. [DOI] [PubMed] [Google Scholar]
- 90.Vereecke L, Beyaert R, van Loo G. Genetic relationships between A20/TNFAIP3, chronic inflammation and autoimmune disease. Biochem. Soc. Trans. 2011;39:1086–1091. doi: 10.1042/BST0391086. [DOI] [PubMed] [Google Scholar]
- 91.Adrianto I, et al. Association of a functional variant downstream of TNFAIP3 with systemic lupus erythematosus. Nat. Genet. 2011;43:253–258. doi: 10.1038/ng.766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Westra HJ, et al. Fine-mapping and functional studies highlight potential causal variants for rheumatoid arthritis and type 1 diabetes. Nat. Genet. 2018;50:1366–1374. doi: 10.1038/s41588-018-0216-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Stuart PE, et al. Genome-wide association analysis of psoriatic arthritis and cutaneous psoriasis reveals differences in their genetic architecture. Am. J. Hum. Genet. 2015;97:816–836. doi: 10.1016/j.ajhg.2015.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Zhou Q, et al. Loss-of-function mutations in TNFAIP3 leading to A20 haploinsufficiency cause an early-onset autoinflammatory disease. Nat. Genet. 2016;48:67–73. doi: 10.1038/ng.3459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Kato M, et al. Frequent inactivation of A20 in B-cell lymphomas. Nature. 2009;459:712–716. doi: 10.1038/nature07969. [DOI] [PubMed] [Google Scholar]
- 96.Aeschlimann FA, et al. A20 haploinsufficiency (HA20): clinical phenotypes and disease course of patients with a newly recognised NF-kB-mediated autoinflammatory disease. Ann. Rheum. Dis. 2018;77:728–735. doi: 10.1136/annrheumdis-2017-212403. [DOI] [PubMed] [Google Scholar]
- 97.Schwartz DM, et al. Type I interferon signature predicts response to JAK inhibition in haploinsufficiency of A20. Ann. Rheum. Dis. 2020;79:429–431. doi: 10.1136/annrheumdis-2019-215918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Jahan AS, Elbaek CR, Damgaard RB. Met1-linked ubiquitin signalling in health and disease: inflammation, immunity, cancer, and beyond. Cell Death Differ. 2021;28:473–492. doi: 10.1038/s41418-020-00676-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Schlicher L, et al. SPATA2 promotes CYLD activity and regulates TNF-induced NF-κB signaling and cell death. EMBO Rep. 2016;17:1485–1497. doi: 10.15252/embr.201642592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Boisson B, et al. Immunodeficiency, autoinflammation and amylopectinosis in humans with inherited HOIL-1 and LUBAC deficiency. Nat. Immunol. 2012;13:1178–1186. doi: 10.1038/ni.2457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Boisson B, et al. Human HOIP and LUBAC deficiency underlies autoinflammation, immunodeficiency, amylopectinosis, and lymphangiectasia. J. Exp. Med. 2015;212:939–951. doi: 10.1084/jem.20141130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Zinngrebe J, et al. Compound heterozygous variants in OTULIN are associated with fulminant atypical late-onset ORAS. EMBO Mol. Med. 2022;14:e14901. doi: 10.15252/emmm.202114901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Damgaard RB, et al. The deubiquitinase OTULIN is an essential negative regulator of inflammation and autoimmunity. Cell. 2016;166:1215–1230.e20. doi: 10.1016/j.cell.2016.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Verboom L, Hoste E, van Loo G. OTULIN in NF-κB signaling, cell death, and disease. Trends Immunol. 2021;42:590–603. doi: 10.1016/j.it.2021.05.003. [DOI] [PubMed] [Google Scholar]
- 105.Zhou Q, et al. Biallelic hypomorphic mutations in a linear deubiquitinase define otulipenia, an early-onset autoinflammatory disease. Proc. Natl Acad. Sci. USA. 2016;113:10127–10132. doi: 10.1073/pnas.1612594113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Heger K, et al. OTULIN limits cell death and inflammation by deubiquitinating LUBAC. Nature. 2018;559:120–124. doi: 10.1038/s41586-018-0256-2. [DOI] [PubMed] [Google Scholar]
- 107.Damgaard RB, et al. The ubiquitin ligase XIAP recruits LUBAC for NOD2 signaling in inflammation and innate immunity. Mol. Cell. 2012;46:746–758. doi: 10.1016/j.molcel.2012.04.014. [DOI] [PubMed] [Google Scholar]
- 108.Gerlach B, et al. Linear ubiquitination prevents inflammation and regulates immune signalling. Nature. 2011;471:591–596. doi: 10.1038/nature09816. [DOI] [PubMed] [Google Scholar]
- 109.Tao P, et al. Deubiquitination of proteasome subunits by OTULIN regulates type I IFN production. Sci. Adv. 2021;7:eabi6794. doi: 10.1126/sciadv.abi6794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Damgaard RB, et al. OTULIN deficiency in ORAS causes cell type-specific LUBAC degradation, dysregulated TNF signalling and cell death. EMBO Mol. Med. 2019 doi: 10.15252/emmm.201809324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Jost PJ, Vucic D. Regulation of cell death and immunity by XIAP. Cold Spring Harb. Perspect. Biol. 2020 doi: 10.1101/cshperspect.a036426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Mudde ACA, Booth C, Marsh RA. Evolution of our understanding of XIAP deficiency. Front. Pediatr. 2021;9:660520. doi: 10.3389/fped.2021.660520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Gyrd-Hansen M, et al. IAPs contain an evolutionarily conserved ubiquitin-binding domain that regulates NF-κB as well as cell survival and oncogenesis. Nat. Cell Biol. 2008;10:1309–1317. doi: 10.1038/ncb1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Lawlor KE, et al. RIPK3 promotes cell death and NLRP3 inflammasome activation in the absence of MLKL. Nat. Commun. 2015;6:6282. doi: 10.1038/ncomms7282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Goncharov T, et al. Disruption of XIAP-RIP2 association blocks NOD2-mediated inflammatory signaling. Mol. Cell. 2018;69:551–565 e557. doi: 10.1016/j.molcel.2018.01.016. [DOI] [PubMed] [Google Scholar]
- 116.Marsh RA, et al. Allogeneic hematopoietic cell transplantation for XIAP deficiency: an international survey reveals poor outcomes. Blood. 2013;121:877–883. doi: 10.1182/blood-2012-06-432500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Lee-Kirsch MA. The type I interferonopathies. Annu. Rev. Med. 2017;68:297–315. doi: 10.1146/annurev-med-050715-104506. [DOI] [PubMed] [Google Scholar]
- 118.Rodero MP, Crow YJ. Type I interferon-mediated monogenic autoinflammation: the type I interferonopathies, a conceptual overview. J. Exp. Med. 2016;213:2527–2538. doi: 10.1084/jem.20161596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Liu C, Kieltyka J, Fleischmann R, Gadina M, O’Shea JJ. A decade of jakinibs: what have we learned and what may be the future. Arthritis Rheumatol. 2021 doi: 10.1002/art.41906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Zhang Q, et al. Inborn errors of type I IFN immunity in patients with life-threatening COVID-19. Science. 2020 doi: 10.1126/science.abd4570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Bastard P, et al. Autoantibodies against type I IFNs in patients with life-threatening COVID-19. Science. 2020 doi: 10.1126/science.abd4585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Beck DB, Aksentijevich I. Susceptibility to severe COVID-19. Science. 2020;370:404–405. doi: 10.1126/science.abe7591. [DOI] [PubMed] [Google Scholar]
- 123.Shin D, et al. Papain-like protease regulates SARS-CoV-2 viral spread and innate immunity. Nature. 2020;587:657–662. doi: 10.1038/s41586-020-2601-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.d’Angelo DM, Di Filippo P, Breda L, Chiarelli F. Type I interferonopathies in children: an overview. Front. Pediatr. 2021;9:631329. doi: 10.3389/fped.2021.631329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Crow YJ, Stetson DB. The type I interferonopathies: 10 years on. Nat. Rev. Immunol. 2021 doi: 10.1038/s41577-021-00633-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Wang HT, Hur S. Substrate recognition by TRIM and TRIM-like proteins in innate immunity. Semin. Cell Dev. Biol. 2021;111:76–85. doi: 10.1016/j.semcdb.2020.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Lu D, et al. Mutations of deubiquitinase OTUD1 are associated with autoimmune disorders. J. Autoimmun. 2018;94:156–165. doi: 10.1016/j.jaut.2018.07.019. [DOI] [PubMed] [Google Scholar]
- 128.Zhang Z, Wang D, Wang P, Zhao Y, You F. OTUD1 negatively regulates type I IFN induction by disrupting noncanonical ubiquitination of IRF3. J. Immunol. 2020;204:1904–1918. doi: 10.4049/jimmunol.1900305. [DOI] [PubMed] [Google Scholar]
- 129.Fuchs SY. Ubiquitination-mediated regulation of interferon responses. Growth Factors. 2012;30:141–148. doi: 10.3109/08977194.2012.669382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.D’Cunha J, Knight E, Jr, Haas AL, Truitt RL, Borden EC. Immunoregulatory properties of ISG15, an interferon-induced cytokine. Proc. Natl Acad. Sci. USA. 1996;93:211–215. doi: 10.1073/pnas.93.1.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Fan JB, et al. Identification and characterization of a novel ISG15-ubiquitin mixed chain and its role in regulating protein homeostasis. Sci. Rep. 2015;5:12704. doi: 10.1038/srep12704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Zhang X, et al. Human intracellular ISG15 prevents interferon-alpha/beta over-amplification and auto-inflammation. Nature. 2015;517:89–93. doi: 10.1038/nature13801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Swaim CD, et al. Modulation of extracellular ISG15 signaling by pathogens and viral effector proteins. Cell Rep. 2020;31:107772. doi: 10.1016/j.celrep.2020.107772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Bogunovic D, et al. Mycobacterial disease and impaired IFN-gamma immunity in humans with inherited ISG15 deficiency. Science. 2012;337:1684–1688. doi: 10.1126/science.1224026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Meuwissen ME, et al. Human USP18 deficiency underlies type 1 interferonopathy leading to severe pseudo-TORCH syndrome. J. Exp. Med. 2016;213:1163–1174. doi: 10.1084/jem.20151529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Basters A, Knobeloch KP, Fritz G. USP18 — a multifunctional component in the interferon response. Biosci. Rep. 2018 doi: 10.1042/BSR20180250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Strong WB. Cholesterol screening in children: a consensus statement — finally. J. Med. Assoc. Ga. 1991;80:451–452. [PubMed] [Google Scholar]
- 138.Speer SD, et al. ISG15 deficiency and increased viral resistance in humans but not mice. Nat. Commun. 2016;7:11496. doi: 10.1038/ncomms11496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Metcalf MG, Higuchi-Sanabria R, Garcia G, Tsui CK, Dillin A. Beyond the cell factory: homeostatic regulation of and by the UPR(ER) Sci. Adv. 2020;6:eabb9614. doi: 10.1126/sciadv.abb9614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Grootjans J, Kaser A, Kaufman RJ, Blumberg RS. The unfolded protein response in immunity and inflammation. Nat. Rev. Immunol. 2016;16:469–484. doi: 10.1038/nri.2016.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Henckaerts L, et al. Genetic variation in the autophagy gene ULK1 and risk of Crohn’s disease. Inflamm. Bowel Dis. 2011;17:1392–1397. doi: 10.1002/ibd.21486. [DOI] [PubMed] [Google Scholar]
- 142.Larabi A, Barnich N, Nguyen HTT. New insights into the interplay between autophagy, gut microbiota and inflammatory responses in IBD. Autophagy. 2020;16:38–51. doi: 10.1080/15548627.2019.1635384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Setta-Kaffetzi N, et al. AP1S3 mutations are associated with pustular psoriasis and impaired Toll-like receptor 3 trafficking. Am. J. Hum. Genet. 2014;94:790–797. doi: 10.1016/j.ajhg.2014.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Watkin LB, et al. COPA mutations impair ER-Golgi transport and cause hereditary autoimmune-mediated lung disease and arthritis. Nat. Genet. 2015;47:654–660. doi: 10.1038/ng.3279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Deng Z, et al. A defect in COPI-mediated transport of STING causes immune dysregulation in COPA syndrome. J. Exp. Med. 2020 doi: 10.1084/jem.20201045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Anderton H, Wicks IP, Silke J. Cell death in chronic inflammation: breaking the cycle to treat rheumatic disease. Nat. Rev. Rheumatol. 2020;16:496–513. doi: 10.1038/s41584-020-0455-8. [DOI] [PubMed] [Google Scholar]
- 147.Samir P, Malireddi RKS, Kanneganti TD. The PANoptosome: a deadly protein complex driving pyroptosis, apoptosis, and necroptosis (PANoptosis) Front. Cell Infect. Microbiol. 2020;10:238. doi: 10.3389/fcimb.2020.00238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Chun HJ, et al. Pleiotropic defects in lymphocyte activation caused by caspase-8 mutations lead to human immunodeficiency. Nature. 2002;419:395–399. doi: 10.1038/nature01063. [DOI] [PubMed] [Google Scholar]
- 149.Lalaoui N, et al. Mutations that prevent caspase cleavage of RIPK1 cause autoinflammatory disease. Nature. 2020;577:103–108. doi: 10.1038/s41586-019-1828-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Tao P, et al. A dominant autoinflammatory disease caused by non-cleavable variants of RIPK1. Nature. 2020;577:109–114. doi: 10.1038/s41586-019-1830-y. [DOI] [PubMed] [Google Scholar]
- 151.Cuchet-Lourenco D, et al. Biallelic RIPK1 mutations in humans cause severe immunodeficiency, arthritis, and intestinal inflammation. Science. 2018;361:810–813. doi: 10.1126/science.aar2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.McDermott MF, et al. Germline mutations in the extracellular domains of the 55 kDa TNF receptor, TNFR1, define a family of dominantly inherited autoinflammatory syndromes. Cell. 1999;97:133–144. doi: 10.1016/S0092-8674(00)80721-7. [DOI] [PubMed] [Google Scholar]
- 153.Molnar E, et al. Key diagnostic markers for autoimmune lymphoproliferative syndrome with molecular genetic diagnosis. Blood. 2020;136:1933–1945. doi: 10.1182/blood.2020005486. [DOI] [PubMed] [Google Scholar]
- 154.Taft J, et al. Human TBK1 deficiency leads to autoinflammation driven by TNF-induced cell death. Cell. 2021;184:4447–4463.e20. doi: 10.1016/j.cell.2021.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Broz P, Dixit VM. Inflammasomes: mechanism of assembly, regulation and signalling. Nat. Rev. Immunol. 2016;16:407–420. doi: 10.1038/nri.2016.58. [DOI] [PubMed] [Google Scholar]
- 156.Grumati P, Dikic I. Ubiquitin signaling and autophagy. J. Biol. Chem. 2018;293:5404–5413. doi: 10.1074/jbc.TM117.000117. [DOI] [PMC free article] [PubMed] [Google Scholar]