Abstract
Transposable element (TE) mobilization is a constant threat to genome integrity. Eukaryotic organisms have evolved robust defensive mechanisms to suppress their activity, yet TEs can escape suppression and proliferate, creating strong selective pressure for host defense to adapt. This genomic conflict fuels a never-ending arms race that drives the rapid evolution of TEs and recurrent positive selection of genes involved in host defense; the latter has been shown to contribute to postzygotic hybrid incompatibility. However, how TE proliferation impacts genome and regulatory divergence remains poorly understood. Here, we report the highly complete and contiguous (N50 = 33.8–38.0 Mb) genome assemblies of seven closely related Drosophila species that belong to the nasuta species group—a poorly studied group of flies that radiated in the last 2 My. We constructed a high-quality de novo TE library and gathered germline RNA-seq data, which allowed us to comprehensively annotate and compare TE insertion patterns between the species, and infer the evolutionary forces controlling their spread. We find a strong negative association between TE insertion frequency and expression of genes nearby; this likely reflects survivor bias from reduced fitness impact of TEs inserting near lowly expressed, nonessential genes, with limited TE-induced epigenetic silencing. Phylogenetic analyses of insertions of 147 TE families reveal that 53% of them show recent amplification in at least one species. The most highly amplified TE is a nonautonomous DNA element (Drosophila INterspersed Element; DINE) which has gone through multiple bouts of expansions with thousands of full-length copies littered throughout each genome. Across all TEs, we find that TEs expansions are significantly associated with high expression in the expanded species consistent with suppression escape. Thus, whereas horizontal transfer followed by the invasion of a naïve genome has been highlighted to explain the long-term survival of TEs, our analysis suggests that evasion of host suppression of resident TEs is a major strategy to persist over evolutionary times. Altogether, our results shed light on the heterogenous and context-dependent nature in which TEs affect gene regulation and the dynamics of rampant TE proliferation amidst a recently radiated species group.
Keywords: Drosophila, transposable elements, epigenetic suppression
Introduction
Eukaryotic genomes are littered with transposable elements (TEs). TEs are selfish genetic elements that self-replicate via copy and paste or cut and paste mechanisms. Despite their abundance and ubiquity in genomes (Kidwell 2002), they can be highly deleterious especially when active. When they transpose, TEs can create double-strand breaks and disrupt reading frames when inserted into genes (Hedges and Deininger 2007). Even when transpositionally inactive, they can induce nonallelic exchange due to sequence homology which can create devastating genome rearrangements (Athma and Peterson 1991; Xiao et al. 2000; Kidwell and Holyoake 2001; Zhang et al. 2011).
To combat their deleterious activity, eukaryotic genomes have evolved intricate defense pathways to inactivate TEs both transcriptionally and posttranscriptionally (for review see Ozata et al. 2019). Posttranscriptional silencing generally involves small RNA-targeted degradation of TE transcripts (for reviews see Czech et al. 2018; Ozata et al. 2019; Wang and Lin 2021). Transcriptional inactivation is achieved through compaction of the chromatin environment into a dense and inaccessible state, known as heterochromatin (for reviews see Richards and Elgin 2002; Elgin and Reuter 2013). Directed by complementary small RNAs, the formation of heterochromatin at TE insertions involves di- and trimethylation to the histone H3 tail at the 9th lysine (H3K9me2/3); this in turn recruits neighboring histones to be methylated, allowing heterochromatin to spread across broad domains (Bannister et al. 2001; Lachner et al. 2001; Nakayama et al. 2001; Hall et al. 2002). Interestingly, this spreading mechanism can also have the unintended effect of silencing genes nearby TE insertions (Choi and Lee 2020). Therefore, in addition to disrupting coding sequences, TE insertions can further impair gene function by disrupting gene expression (Hollister and Gaut 2009; Lee 2015; Lee and Karpen 2017).
However, even with strong repressive mechanisms, defense against TEs appears to be an uphill battle. TEs are among the most rapidly changing components of eukaryotic genomes. TE content can differ drastically even between closely related species and has been shown to be a key contributor to genome size disparities. In Drosophila, the P-element, a DNA transposon originating from D. willistoni, invaded both D. melanogaster (Anxolabéhère et al. 1988; Daniels et al. 1990) and subsequently D. simulans (Kofler et al. 2015). Both of these cross-species invasions occurred rapidly within the last century and resulted in world-wide sweeps of the P-element in wild populations. Previously suppressed TEs can also evolve to evade silencing; for example, the rice DNA transposon mPing emerged through a deletion in the Ping element and subsequently amplified to thousands of copies in some rice strains (Naito et al. 2006; Chen et al. 2019). Both horizontal transfer and suppression escape can lead to bursts of TE activity. TE mobilizations are accompanied by a reduction in host fertility and viability (Kidwell et al. 1977; Kidwell and Novy 1979; Schaefer et al. 1979), which in turn creates strong selective pressure for the host to reestablish silencing. This conflicting dynamic has been hypothesized to create an evolutionary arms race between host suppression mechanisms and TE suppression escape, driving recurrent adaptive evolution of many proteins involved in the TE silencing pathways (Kelleher and Barbash 2013; Simkin et al. 2013; Parhad and Theurkauf 2019; Luo et al. 2020). The rapid evolution of TEs and the repressive pathways have even been implicated in establishing postzygotic reproductive isolation between closely related Drosophila species (Kliman et al. 2000; Garrigan et al. 2012; Brand et al. 2013).
Beyond their deleterious potential, TEs can also be sources of novelty in the genome (Kidwell and Lisch 1997). TEs, or parts of their sequences, have been coopted for gene regulatory functions such as promoters and enhancers (Jacques et al. 2013; Merenciano et al. 2016; Sundaram and Wysocka 2020). Their recurrent transpositions across nascent sex chromosomes also mediated the evolution of dosage compensation chromosome wide (Ellison and Bachtrog 2013; Zhou et al. 2013). Insertions of TEs in the proximity of genes have also created functional chimeric retrogenes (Buzdin 2004; Xing et al. 2006). In mammals, Krüppel-associated box-zinc finger transcription factors have repeatedly coopted the transposase protein encoded by DNA transposons, allowing for the diversification of their binding targets (Cosby et al. 2021). Lastly, in flies, domesticated retrotransposons insert at chromosome ends for telomere extension, thus alleviating the need for telomerase to solve the end-replication problem (Traverse and Pardue 1988; Biessmann et al. 1990; Levis 1994). Therefore, TEs do not just force the host defense to adapt in order to suppress their activity, but they can also be beneficial drivers of genome evolution (Kidwell and Lisch 1997; Casacuberta and González 2013).
Whereas TEs can have multi-faceted influences on the genome and its evolution, the dynamics of TE amplification and suppression escape remain poorly understood, especially outside of select model species. This is in part due to the inherent challenge associated with studying highly repetitive sequences, an issue that became particularly problematic during the boom of short-read sequencing technologies in the last two decades. Most TE-derived short reads (typically <150 bps) cannot be uniquely assigned to a region of the genome, which causes errors in mapping and breakages in genome assemblies (Bourque et al. 2018; O’Neill et al. 2020). Numerous approaches have been devised that take advantage of different features of short-read sequencing platforms (e.g., paired sequencing) to call insertions (Linheiro and Bergman 2012; Cridland et al. 2013; Rahman et al. 2015; McGurk and Barbash 2018; Wei et al. 2020), but such methods are nevertheless limited by short-read lengths, often producing inconsistent results (Vendrell-Mir et al. 2019). With the advent of long-read (5 kb+) sequencing technologies from Oxford Nanopore and PacBio, many of these issues can finally be circumvented (Hotaling et al. 2021). The use of such technologies has already led to drastic improvements of genome assemblies across highly repetitive genomes in, for example, flies (Mahajan et al. 2018; Bracewell et al. 2019; Chakraborty et al. 2021), mosquitoes (Matthews et al. 2018), mammals (Bickhart et al. 2017), and humans (Nurk et al. 2022).
Highly contiguous genomes with well-represented repeat content permit comprehensive analyses of TE insertions across the genome. Multiple such high-quality genomes further enable analyses of the dynamics of TE proliferation through a comparative and phylogenomic framework. Therefore, to illuminate how TEs proliferate and potentially drive genome evolution and speciation, we used long-read technologies to generate high-quality genome assemblies of seven closely related Drosophila species (fig. 1A and B) in the nasuta species group. This species group radiated in the last 2 My (Kitagawa et al. 1982; Bachtrog 2006; Ranjini and Ramachandra 2013; Mai et al. 2020) and is widely distributed across Asia, with some populations found in eastern Africa, Oceania, and Hawaii (Wilson et al. 1969; Mai et al. 2020). Drosophila nasuta has recently been identified as an invasive species in Brazil that is spreading quickly in South America (Vilela and Goñib 2015; Silva et al. 2020). Whereas most of the species are geographically isolated, they have varying levels of reproductive isolation (Kitagawa et al. 1982); over half of interspecific crosses produce viable offspring. With these high-quality genomes, we sought to systematically understand how TE insertions around genes affect gene expression, and how frequently TEs escape repression and expand. To answer these questions, we generated a library of a common set of high-quality TE consensus sequences from de novo TE calls across the genome assemblies. With this library, we identified species-specific TE insertions and found that TEs frequently expand, likely due to suppression escape, with >50% of TEs showing evidence of lineage-specific expansion in at least one species. Species-specific TEs are disproportionately found near lowly expressing genes and only rarely have an impact on gene expression. Lastly, we show that the silencing of expanding TEs can lead to silencing of neighboring genes.
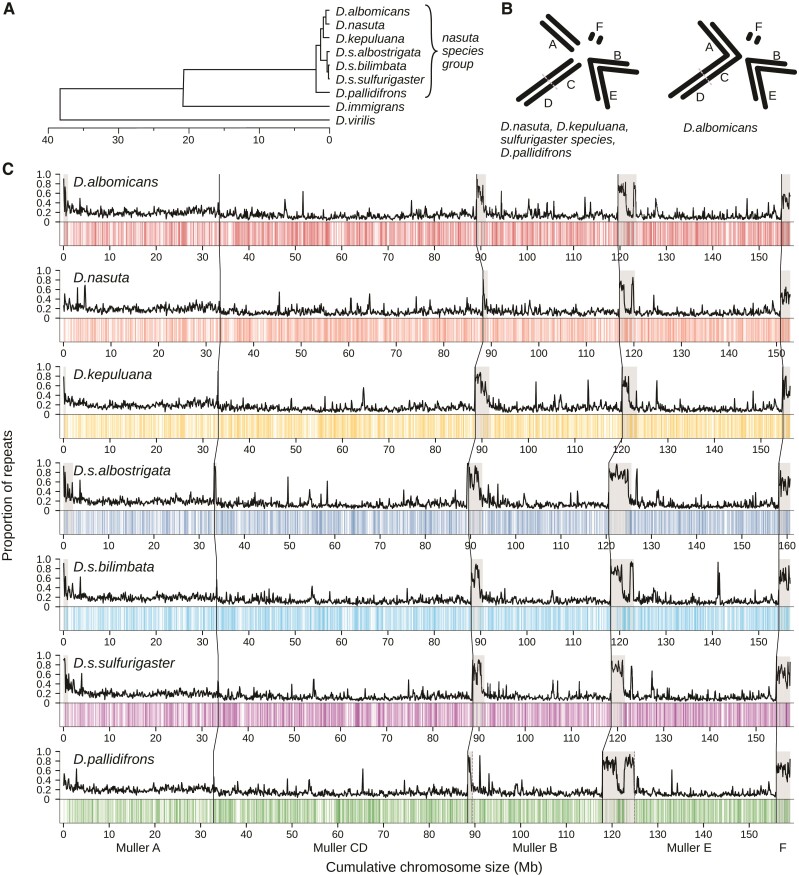
Fig. 1.
Genomes of the Drosophila nasuta species group. (A) Phylogeny of the nasuta species radiation within the Drosophila subgenus. Tree adapted from Mai et al. (2020) and Izumitani et al. (2016). (B) Karyotypes of the species group; chromosomes are oriented such that centromeres are pointed toward the center of circle. (C) Long-read-based genome assemblies of seven species. For each species, the top track depicts the repeat content estimated for 100 kb windows. Positions of annotate genes are represented on the bottom track as vertical lines. The centromeric ends are on the left side of each chromosome. Regions deemed as pericentromeric are highlighted in gray. Chromosomes are demarcated by black vertical lines. Unless otherwise stated, species are represented by colors used here: red (D. albomicans), orange (D. nasuta), yellow (D. kepuluana), navy (D. s. albostrigata), light blue (D. s. bilimbata), purple (D. s. sulfurigaster), and green (D. pallidifrons).
Results
High-Quality Genome Assemblies Across Seven Species
Genome assemblies for females of seven species in the nasuta clade—D. albomicans, D. nasuta, D. kepulauana, D. sulfurigaster albostrigata, D. sulfurigaster bilimbata, D. sulfurigaster sulfurigaster, and D. pallidifrons—were generated using Nanopore sequencing complemented by Hi-C scaffolding (table 1; supplementary figs. S1–S7, Supplementary Material online). The methodology for preparing reads and assembling genomes was adopted from Bracewell et al. (2019) and applied across all species: error-correct Nanopore reads with canu, generate contig assembly with wtdbg2 and flye, polish assembly with racon and pilon, remove contigs that belong to other organisms with BLAST, and stitch contig assemblies using Hi-C reads as input for Juicer and 3d-dna (Altschul et al. 1990; Walker et al. 2014; Durand et al. 2016; Dudchenko et al. 2017; Koren et al. 2017; Vaser et al. 2017; Bracewell et al. 2019; Kolmogorov et al. 2019; Ruan and Li 2020). However, there is no universal pipeline to generate ideal assemblies for each species, and the assembly pipeline for different flies underwent various adjustments for optimal results (see supplementary materials, Supplementary Material online). Overall, we generated consistent assemblies for each species using an average of 30.3× long-read coverage (std dev = 10; supplementary table S1, Supplementary Material online), resulting in a mean N50 of 35.9 Mb (std dev = 1.4 Mb; table 1), assembly size of 166.6 Mb (std dev = 2.59 Mb; table 1), and Benchmarking Universal Single-Copy Orthologs (BUSCO) score of 99.3% (std dev = 0.4%; supplementary table S2, Supplementary Material online).
Table 1.
Size of Genome Assemblies (and their Chromosomes) for Each Species and their Associated Summary Statistics.
| Chromosome | D. albomicans | D. nasuta | D. kepulauana | D. s. albostrigata | D. s. bilimbata | D. s. sulfurigaster | D. pallidifrons |
|---|---|---|---|---|---|---|---|
| size (bp) | size (bp) | size (bp) | size (bp) | size (bp) | size (bp) | size (bp) | |
| Muller A | 33,597,023 | 33,189,490 | 33,291,615 | 33,386,403 | 33,007,321 | 33,493,557 | 32,839,655 |
| Muller B | 30,469,903 | 28,690,150 | 31,604,248 | 30,983,057 | 30,102,783 | 29,954,194 | 29,503,240 |
| Muller CD | 55,495,487 | 55,283,848 | 55,283,860 | 56,209,854 | 54,959,191 | 55,186,638 | 55,584,481 |
| Muller E | 35,291,776 | 33,885,645 | 34,564,094 | 37,627,869 | 36,279,119 | 35,818,991 | 37,973,042 |
| Muller F | 1,839,965 | 2,061,818 | 1,552,407 | 2,495,194 | 2,423,155 | 2,918,623 | 3,067,770 |
| Chromosome total | 156,694,154 | 153,110,951 | 156,296,224 | 160,702,377 | 156,771,569 | 157,372,003 | 158,968,188 |
| Assembly total | 167,541,436 | 171,781,232 | 163,769,021 | 168,284,230 | 164,595,183 | 168,070,293 | 164,659,715 |
| N50 | 35,291,776 | 33,885,645 | 34,564,094 | 37,627,869 | 36,279,119 | 35,818,991 | 37,973,042 |
| Number of Scaffoldsa | 220 | 282 | 77 | 95 | 201 | 123 | 104 |
| BUSCOb | 99.62% | 99.16% | 99.72% | 98.50% | 99.72% | 98.87% | 99.62% |
| Repeat contentc | 20.71% | 23.26% | 18.75% | 21.27% | 19.79% | 19.60% | 20.10% |
| Annotated genes | 12,395 | 12,492 | 12,594 | 12,595 | 12,432 | 12,718 | 12,362 |
See supplementary figs. S1–S7, Supplementary Material online for Hi-C scaffolding of the chromosome arms in each species.
See supplementary table S2, Supplementary Material online for detailed BUSCO statistics.
See supplementary table S3, Supplementary Material online for repeat content and masking details.
We leveraged these chromosome level genome assemblies alongside RNA-seq datasets from D. albomicans and D. nasuta (Zhou and Bachtrog 2012) to annotate genes across all species (table 1). An average of 12,513 genes were annotated per species (std dev = 128.48), which is lower than the number of genes annotated in other Drosophila species (Drosophila 12 Genomes Consortium 2007). In order to analyze homologous genes, we clustered genes between species with OrthoDB and found 9,413 genes shared across all species (Kriventseva et al. 2019).
Generating a Curated De Novo TE Library
For each genome, we used RepeatModeler2 to generate a de novo TE library, which we then used to annotate the genome (Flynn et al. 2020). This resulted in 18.8–23.3% of the genomes being masked (supplementary table S3, Supplementary Material online). High repeat content near chromosome ends shows that these near-chromosome length scaffolds include some heterochromatin and pericentromeric regions. Expectedly, gene density and repeat density are negatively correlated (fig. 1C).
One major challenge with de novo TE identification using standard computational methods is that the resulting TE libraries are littered with redundant and fragmented entries. Furthermore, we find that secondary structures such as nested insertions or fragment duplications (supplementary fig. S8, Supplementary Material online) are frequently identified as unique TE entries in the libraries. To improve the de novo TE library and to generate a common set of TE consensus sequences across all the nasuta subgroup, we devised a pipeline that utilizes multiple steps and metrics (fig. 2A). After an initial de novo TE library call with RepeatModeler2 for each of the genomes, we demarcated the euchromatin/pericentromere boundaries (fig. 1C). Reasoning that recently active TEs are more likely to be intact and surrounded by unique sequences in the euchromatin, we then ran RepeatModeler2 for a second time on only the euchromatic portions of the genome assemblies. We note that although removing pericentric regions favors the identification of full-length TEs, this is at the expense of having the most complete repeat library and will remove TEs that exist only in the pericentric region (Bergman et al. 2006).
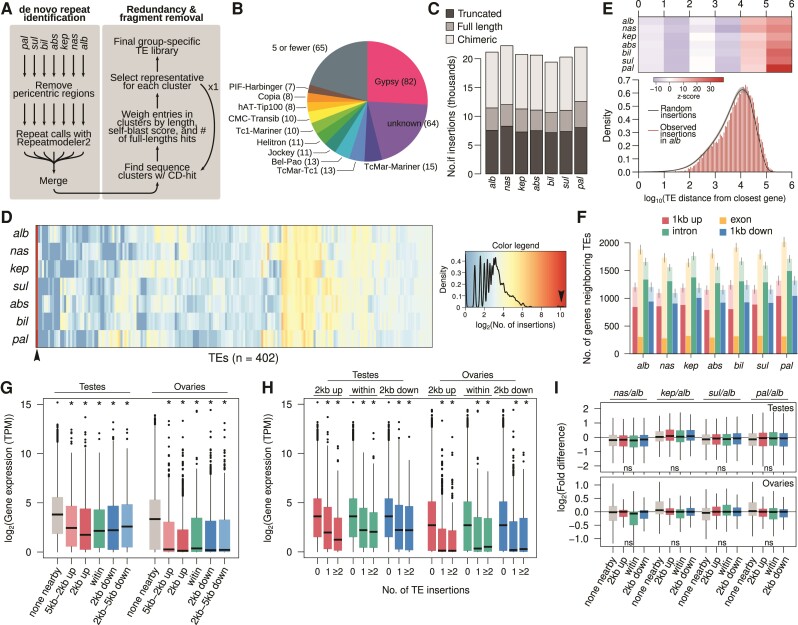
Fig. 2.
De novo identification and distribution of TE insertions across the genomes. (A) Pipeline to construct and refinea de novo TE reference library from genome assemblies. We used RepeatModeler2 to first identify repeats from the euchromatic regions of each species. The resulting repeat libraries are merged followed by sequence clustering with CD-HIT2. Multiple indexes were used to select the full-length representative TEs. (B) Breakdown of TE classes identified; for breakdown of the gray section see supplementary fig. S9, Supplementary Material online. (C) Number of full length and truncated insertions found in each genome. The chimeric class represents the merger of annotations that overlap or are contiguous. (D) Copy number of full-length insertions of 318 TE families across the seven genomes. (E) Distribution of the distance between genes and TEs in Drosophila albomicans (red histogram) compared with that from random TE insertions (black contour with 95% confidence interval denoted by gray). See supplementary fig. S10, Supplementary Material online for other species. The z-score between the random expectation and observed counts at different intervals are shown above for different size categories with lower and high z-score representing depletion and enrichment. Insertions within genes are not counted. See supplementary fig. S9, Supplementary Material online for distribution of intragenic insertions from exons. (F) Number of genes with TEs inserted in different regions of genes with and without insertions. Expectations from random distribution of insertions are shown in lighter bars with error bars demarcating 95% confidence intervals. (G) Transcript abundance of annotated genes in TPM, partitioned into different classes depending on where TE insertions are found. (H) Transcript abundance of genes with different numbers of TE insertions. (I) Fold-difference in transcript abundance of orthologous genes depending on different numbers of insertions in D. albomicans. For (G–I), “*” represents significant Wilcoxon rank sum tests (P < 0.00001) comparing categories with insertions to the 0-insertion categories. See supplementary fig. S10, Supplementary Material online, for comparisons using insertions in other species.
We merged all the TEs across species generating a library of 1,818 entries, and then used CD-HIT2 to group the entries into clusters based on sequence similarity (Fu et al. 2012). By default, CD-HIT2 outputs the longest sequence in each cluster. Whereas this means full-length entries will be favored over fragmented entries (when both exist in the library), entries with nested structures or chimeric TEs will be selected in favor of full length but shorter elements. Therefore, in addition to sequence length, we evaluate each TE in each cluster based on two additional metrics to preferentially select representative and full-length TE consensus sequences. Despite having higher entry lengths, chimeric structures are unlikely to have high copy number; we, therefore, blasted the TE entries to the genome and tallied the number of times hits cover 80% of the length of the entries. In addition, we blasted the TEs to themselves to determine internal redundancy; entries with internal duplications or nested insertions will have a high self-blast score. We then selected the representative sequence as the longest sequence with a high number of near-full-length blast hits and a low self-blast score. We then repeated this step one more time to further remove redundancies in the library. After these two rounds of clustering with CD-HIT2, the TE library size was reduced to 351 consensus sequences. Afterwards, we merged TE sequences that make up a larger, full-length element through patterns of cooccurrences in the genome. These series of steps progressively increased the size of TEs in the library and reduced the number of fragmented and chimeric annotations (supplementary table S4, Supplementary Material online). Overall, it resulted in a substantially reduced library with 318 TE entries.
The TEs generated from RepeatModeler2 are, by default, assigned to a TE category. To validate these assignments, we used ClassifyTE to reannotate the TE library (Panta et al. 2021). There is a 50% concordance between the annotations from RepeatModeler2 and ClassifyTE. Entries that were different between the two annotations were assigned the default category from RepeatModeler2. Gypsy elements make up the majority of the TE library, consisting of 82 entries (25.8%) followed by unknown families (57 entries, 17.9%; fig. 2B). All other TE families make up <5% of the TE library. The pattern of high number of Gypsy families is similar to that in other Drosophila species (Mérel et al. 2020).
TE Insertion Patterns Across the Genome
Using the refined nasuta group-specific TE library, we annotated TE insertions in each genome assembly using RepeatMasker (see Materials and Methods). We classified full-length insertions as annotations that cover at least 80% of the entry in the library; insertions covering <80% but are over 200 bp are classified as truncated TE insertions. In addition, we merged annotations that are contiguous or overlapping, which can be due to nested insertions or remaining redundancies in the repeat library. The number of full-length TEs range from 3,489 to 4,544 between species (fig. 2C) and the majority (73.6% on average) fall within euchromatic regions of the genomes (supplementary fig. S9C, Supplementary Material online), similar to previous reports (Biémont and Vieira 2005; Drosophila 12 Genomes Consortium 2007). Truncated insertions are nearly 2× as numerous (ranging between 7,164 and 8,273; fig. 2C). As expected given their mosaic nature, the merged annotations have the largest fractions fall within the heterochromatic regions (supplementary fig. S9, Supplementary Material online). With the exception of four TE families, all are found in low to intermediate copy numbers with fewer than 100 copies in any given genome, consistent with previous findings (fig. 2D). Interestingly, one TE stands out as having thousands of copies across all the genomes (fig. 2D, arrowhead, see Recurrent and Rampant Amplifications of Drosophila INterspersed Elements [DINEs]).
To evaluate if and how TEs impact gene function, we looked at TE insertion patterns with respect to neighboring genes (fig. 2E, supplementary fig. S9A, Supplementary Material online). On average, TE insertions are 18.5 kb away from the nearest genes; 41.5–46.9% of insertions are within 5 kb of genes (fig. 2E). Compared with the expectation of TEs inserting randomly across the genome, we see a significant depletion of TE insertions near genes, and an excess of TEs further away from genes (see Materials and Methods; fig. 2E and supplementary fig. S10, Supplementary Material online). Of the 12,362–12,718 genes annotated, only 3,276–3,889 have insertions within or nearby (<1 kb 5′ or 3′). While insertions are significantly depleted in or around genes (fig. 2F; P < 0.001 permutation test of random insertions, see Materials and Methods), they are most commonly found within introns (37.7–39.8%) and, expectedly, least likely in exons (8.0–9.5%). The nonrandom and severe depletion of TEs around genes affirms the notion that insertions near genes are deleterious (Bergman et al. 2006).
TE Insertions are Associated with Low Expression of Nearby Genes
To systematically examine the impact, if any, of TE insertions on gene expression, we generated ovarian and testes mRNA-seq for five of the seven species investigated (excluding D. s. bilimbata and D. s. albostrigata). Genes with TE insertions nearby or within are overrepresented for lowly expressed genes, in both testes and ovaries (P < 2.2e−16 Wilcoxon rank sum tests; fig. 2G, supplementary fig. S11, Supplementary Material online). Genes with insertions <2 kb upstream have the lowest expression in both the testes and ovaries (fig. 2G). Genes with TEs inserted further away (2–5 kb) also have significantly lower expression, though to a lesser extent (P < 2.2e−16 Wilcoxon rank sum tests; fig. 2G, supplementary fig. S11, Supplementary Material online). Moreover, we find that gene expression is inversely correlated with the number of TE insertions (fig. 2H). This negative relationship holds for insertions found within, upstream, and downstream of genes. Interestingly, ovarian expression appears to be more negatively associated with TE insertions, with no expression in ovaries of nearly half of the genes with TEs inserted nearby (fig. 2G and H).
Due to the spreading of heterochromatin, TE insertions can induce epigenetic silencing at neighboring genes (Choi and Lee 2020). Therefore, prima facie, these results are consistent with the epigenetic silencing of genes due to neighboring TE insertion. To further test this, we reasoned that if TE insertions are inducing down-regulation of surrounding genes, orthologous genes without insertions should be more highly expressed. To test this possibility, we compared the expression of orthologs when insertions are found in one species but not the other. Curiously, we do not find that expression between orthologs changes significantly depending on the presence of insertions nearby or within (fig. 2F, supplementary fig. S12, Supplementary Material online, all pairwise P-values > 0.1, Wilcoxon rank sum test). This suggests that insertions within/nearby genes are not systematically down-regulating expression. Instead, TEs appear to preferentially insert and/or accumulate around lowly expressed genes.
Survivor Bias Likely Drives Anticorrelation Between TE Insertion and Gene Expression
To elucidate the source of the negative association between gene expression and TE insertions nearby, we looked at all TE insertions found around/within the 9,413 genes with orthologs across all species. To ensure that only unique insertions are counted and ancestral insertions are counted only once, we removed insertions belonging to the same TE family that are within 100 bp of each other relative to the neighboring gene. Further, we removed pericentric genes from this analysis to avoid their high local TE counts driving correlations. For these gene orthologs, we indeed find a significant negative correlation between TE insertion counts and averaged gene expression in both testes and ovaries (fig. 3A and B; P < 2.2e−16). Similar correlations are also found when looking at the proportion of bases covered by TEs around and within genes (supplementary fig. S13, Supplementary Material online). Curiously, the negative correlation of ovarian expression is significantly stronger than that of testes expression (fig. 3A and B; P < 1e−8, Pearson and Filon’s z).
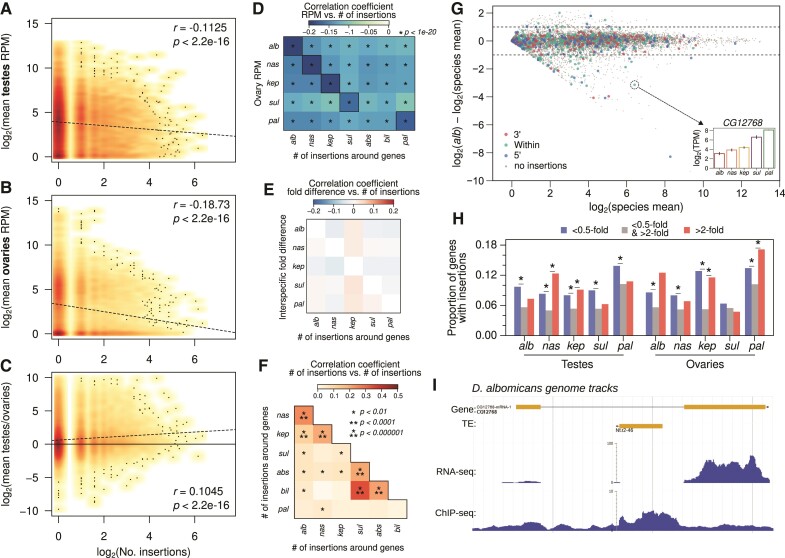
Fig. 3.
Negative association between TE insertions and genic expression. (A and B) Density scatterplots of number of unique (both full length and truncated) TE insertions around genes (±2 kb) across all the nasuta species genomes plotted against genic transcript abundances (averaged across the species) in the ovaries (A) and testes (B). Increased intensity of warm colors indicates higher density of points. Scattered black dots indicate positions of single points. Regression lines are depicted by dotted lines; the Pearson’s correlation coefficients and corresponding P-values are labeled in the top right. (C) Same as A and B, but with the fold difference of genic expression between testes and ovaries. (D) Pairwise correlation of TE insertion counts around genes in a particular species to the ovarian transcript abundance of the gene orthologs in another species. (E) Pairwise correlation of TE insertion counts around orthologous genes across species; genes with no insertions in either species are not used. (G) MA-plot of average gene expression (TPM) across species in the testes (x-axis) plotted against fold difference between the Drosophila albomicans expression and the average across species (y-axis). Colored points represent genes with TE insertions in different parts of the gene. Horizontal dotted line demarcates 0.5- and 2-fold differences. Inset shows the testes expression of the CG12768 gene across all five species. For MA-plots in ovaries and other species, see supplementary fig. S14, Supplementary Material online. (H) Proportions of genes with TE insertions grouped by expression levels; genes in each species are partitioned depending on their testis and ovary expression levels relative the species average (i.e., genes below or above the dotted lines in panel G), for each species and in ovaries and testes. (I) Genome browser shot of CG12768 showing tracks for gene structure, TE insertions, transcript abundance, and H3K9me3 enrichment. For genome browser shot of this gene in other species see supplementary fig. S15, Supplementary Material online.
We then looked at the extent of correlation between species-specific TE insertion counts to gene expression across species. If TEs insert independently at different genes and are down-regulating nearby genes in one species, we expect no cross-species correlations. Instead, significantly negative correlations are observed between all pairwise comparisons (fig. 3D), although the within-species correlations are significantly more negative than between-species correlations (fig. 3D, outlined boxes). Further, insertion-induced epigenetic down-regulation to neighboring genes is expected to increase expression divergence between species, since genes with insertions are expected to be more lowly expressed than their orthologs without insertions. We do not find any significant correlation between insertion counts around genes in one species and their expression fold differences when compared with orthologs without insertions (fig. 3E). However, when comparing the distribution of TEs between species, we find that the number of TE insertions at/near genes is correlated between many of the species (fig. 3F). Especially between more closely related species pairs, the correlation of insertions is highly significant, suggesting that TEs have a tendency to independently insert and/or accumulate near the same genes in different genomes. Thus, between-species correlations in TE counts versus gene expression (fig. 3D), and low interspecific expression divergence (fig. 3E) may in part be explained by the same genes being targeted by TEs in different species. Biased insertion counts near lowly expressed genes could be due to insertion bias or survival bias. The former can result from TEs preferentially targeting specific genomic features to insert such as promoters and accessible chromatin; the latter is likely the result of low fitness consequences due to insertions near lowly expressed genes.
TE Insertions Associated with Extreme Expression Changes in a Small Number of Genes
TE insertions do not appear to have pervasive silencing effects on neighboring genes (figs. 2G, 3B, C, and 4D). However, there are known cases where individual TE insertions modulate gene regulation of nearby genes. To identify such cases, we compared the expression of each gene in each species to the average expression across all species (fig. 3G, supplementary fig. S14, Supplementary Material online). For the vast majority of genes with/near insertions, their expression does not deviate from the cross-species average. However, interestingly, we notice multiple cases where insertions are associated with substantially lowered gene expression. Examining the small fraction of genes with expression less than half of the cross-species average, we find that there are between 55 and 167 genes in each species showing low expression and nearby/intronic insertions (supplementary table S5, Supplementary Material online). Consistent with TE-induced epigenetic silencing, these genes with reduced expression are significantly overrepresented for genes with TE insertions in almost every species, and in both ovaries and testes (fig. 3H).
To determine whether TE insertions can induce epigenetic silencing nearby in some cases, we selected one of the more significantly down-regulated genes, CG12768. This gene has a TE insertion in the first intron in D. albomicans (fig. 3I) and shows the lowest expression in D. albomicans testes (fig. 3G, inset). Accompanying its low expression in D. albomicans, we find elevated enrichment of H3K9me3 at the intronic insertion as well as across the gene body, exons and 5′ region (see below for ChIP analysis). Notably, this insertion did not appear to completely silence the gene, as abundant RNA-seq reads still map to the second exon, albeit at a substantially lower level than in other species (supplementary fig. S15, Supplementary Material online).
Interestingly, TE insertions are not just associated with highly down-regulated genes: we find that highly up-regulated genes in a species (>2-fold higher than species mean) can also be significantly overrepresented by genes with insertions. Although not significant in all species, up-regulated genes have proportionally more TE insertions in all comparisons (fig. 3G). For example, the gene Gyc88E in D. albomicans has an intronic insertion in the first exon and is the highest expressed ortholog in the testes comparing the different species (2.16-fold higher than the next highest; supplementary fig. S16, Supplementary Material online). Therefore, some TE insertions appear to be associated with increased expression divergence through both down- and up-regulation of nearby genes.
H3K9me3 Spreading Around TE Insertions Near Genes
To evaluate the extent to which epigenetic silencing of TEs can lead to reduction in expression of neighboring genes, we analyzed available ChIP-seq data for the repressive heterochromatic histone modification H3K9me3 in D. albomicans male 3rd instar larvae (Wei and Bachtrog 2019). We examined the extent of H3K9me3 spreading from TE insertions with different distances to the closest gene; to avoid TEs inside the pericentromeric or telomeric heterochromatin, we analyzed only those >5 Mb from the chromosome ends. TE insertions over 5 kb from genes show the highest H3K9me3 enrichment in neighboring regions (fig. 3A, top). TEs that are closer to genes (within 5 kb of genes), on the other hand, show lower levels of heterochromatin spreading. Less heterochromatin spreading from TE insertions nearby genes is consistent with opposing effects of heterochromatin formation and gene expression; transcriptionally active chromatin near genes may impede the spreading of silencing heterochromatin. Looking more closely, we find that high H3K9me3 enrichment is observed in the immediate vicinity up and downstream of the insertions and quickly drops off within 100 bp (fig. 4A, bottom). Interestingly, this rapid decline from highly elevated H3K9me3 enrichment is observed regardless of insertion distance. Therefore, despite a narrower spreading range of TEs close to genes, the silencing effect in the immediate vicinity is similar to those far from genes, and may explain the paucity of insertions within 100 bp of genes (fig. 2E) and exons (supplementary fig. S9A, Supplementary Material online).
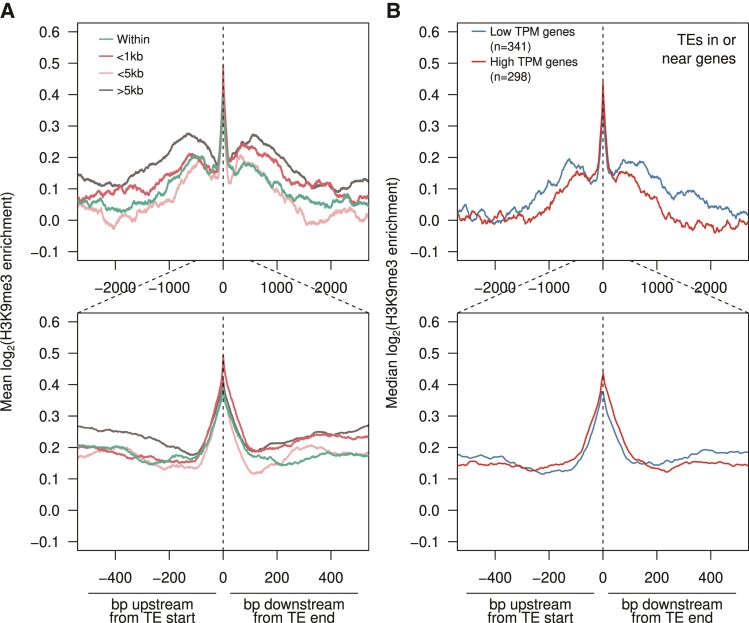
Fig.4.
Epigenetic silencing through H3K9me3 spreading around TE insertions. (A) Median H3K9me3 enrichment ± 5 kb upstream and downstream of TEs inserted at different distances to genes (enrichment across TE insertions not plotted). TE insertions within pericentric regions are removed from analyses. Zoomed in plot (±500 bp) is shown below. (B) As with A but with TEs inserted within genes or <2 kb around genes of different expression levels.
To address whether heterochromatin spreading from TEs reduces the expression of nearby genes, we evaluated the extent of H3K9me3 enrichment surrounding TE insertions that are nearby genes with different expression levels in testes. Insertions were partitioned by their proximity to genes with low (<8 transcripts per million [TPM]) and high expression (>8 TPM). Insertions around low TPM genes show a higher H3K9me3 enrichment and spreading than those around high TPM genes (fig. 4B, top). While these differences are consistent with epigenetic silencing of genes induced by neighboring TEs, they could also reflect high transcriptional activity opposing heterochromatin spreading from nearby TEs. Given the lack of systematic down-regulation between genes with insertions and their orthologs (fig. 2F), yet overrepresentation of TE insertions in genes that are down-regulated (fig. 3H), our data suggest that both forces are at play.
Recurrent and Rampant Amplifications of DINEs
The most abundant TE, accounting for 2.1–3.8 Mb across all the species, is a 770 bp repeat which shows homology to the DINE—a nonautonomous DNA transposon that is highly species-specific (Locke et al. 1999). DINE’s are widespread in the Drosophila genus, with hundreds to thousands of copies identified across a wide range of Drosophila species (Yang and Barbash 2008). They appear particularly abundant in the nasuta species complex, with 1,501–3,202 full length and 4,863–6,793 truncated DINE insertions identified across species.
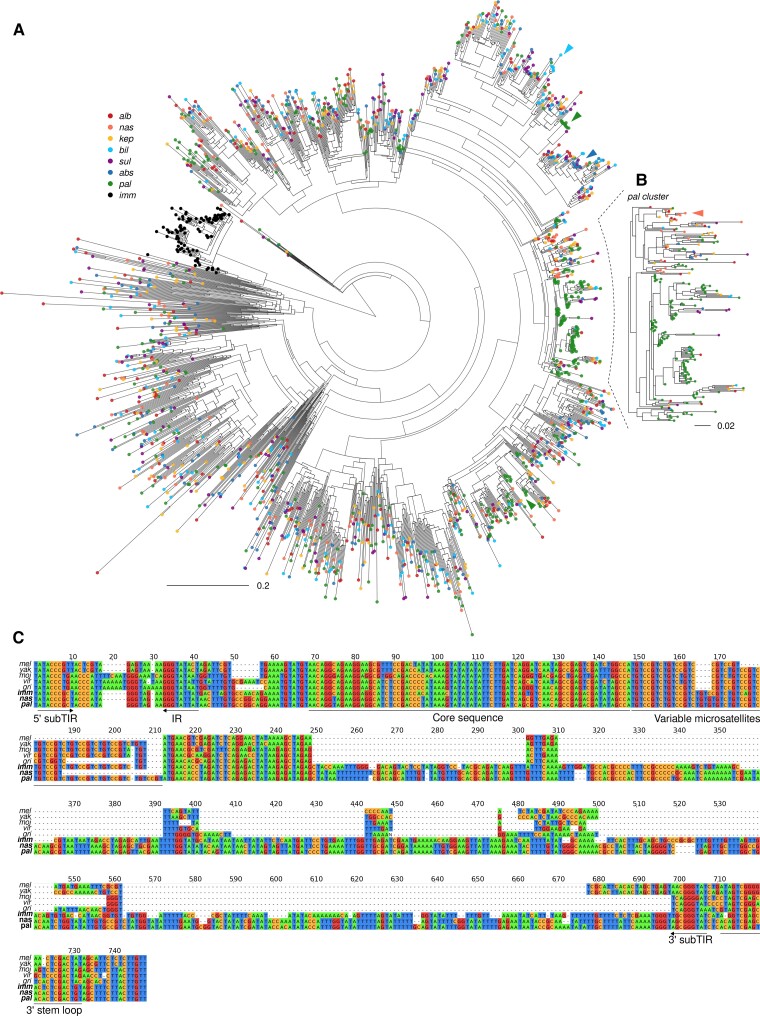
Phylogenetic analysis of individual TE insertions can reveal about their evolutionary history, including the timing of when a particular TE likely was transpositionally active. To study the explosion of DINE elements in the nasuta species group, we determined their phylogenetic relationship, using near-full-length copies with the addition of insertions found in the D. immigrans genome as the outgroup (fig. 5A). We find a complex phylogenetic tree where the majority of DINEs do not show species-specific clustering. Instead, insertions from different species in the nasuta subgroup are highly intermingled, indicating that the bulk of DINE amplification predated the radiation of this species complex (fig. 5A). Most of the elements are likely currently inactive given the lack of species-specific clusters and long terminal branches (supplementary fig. S17, Supplementary Material online).
Fig. 5.
Recurrent DINE expansions. (A) Radial tree of subsampled DINE insertions with the addition of Drosophila immigrans DINE elements as outgroup. Insertions from the same species have the same colored tips. Colored arrowheads point to small scale species-specific expansions on the tree. (B) Large cluster of D. pallidifrons DINE insertions indicate recent burst of species-specific activity. (C) Multiple sequence alignments of consensus DINE sequences of representative species. DINE-specific sequence features are annotated beneath the tracks.
Whereas most DINEs in the nasuta subgroup likely originated from old expansion events, we nevertheless identified multiple instances of species-specific clustering. First, we find that the D. immigrans DINEs form a monophyletic clade with short branch lengths, suggesting a relatively recent, immigrans-specific expansion of this element. Second, we identified multiple clusters of D. pallidifrons insertions throughout the tree, including one large branch containing 142 out of 400 (subsampled) DINE insertions. D. pallidifrons DINEs within this branch contain several distinct clusters with short branch lengths, suggesting that multiple copies of DINE are currently (or have been recently) amplifying in the genome (fig. 5B). Expansions of DINE in D. pallidifrons and D. immigrans are consistent with a small number of elements (if not a single copy) escaping silencing, which subsequently generated a large number of insertions. In contrast, horizontal transfer is expected to either create a deep monophyly of elements if transferred from unsampled, distant species, or nested expansion if transferred from one of the closely related nasuta species that had its own expansions.
Interestingly, multiple smaller clusters of D. pallidifrons DINE expansions (fig. 5, green arrows) are also found in distant branches across the phylogeny, suggesting that other DINE lineages may have reactivated (see Discussion). Lastly, though less obvious, smaller scale copy number increases of DINE can also be observed in other species, such as the large numbers of D. albomicans, D. nasuta, and D. kepulauana DINEs within the D. pallidifrons cluster that suggest both species-specific insertion events as well as older insertions events in their common ancestor. Similarly, small scale expansion events are also observed for the sulfurigaster species complex.
To better understand the sequence changes that may have precipitated the expansions, we first generated consensus sequences for DINEs in D. immigrans, across the nasuta subgroup, and in specifically the D. pallidifrons cluster (fig. 5B) from the D. pallidifrons genome. We then compared them to the previously reported consensus sequences from other Drosophila species (fig. 5C). Whereas DINEs are between 300 and 400 bps in the other species, they double to 695 and 726 bp in D. immigrans and the nasuta group, respectively. However, they still contain many of the main features such as the presence of subterminal inverted repeats, microsatellite regions consisting of variable lengths of simple repeats and 3′ stem loop. Conservation can be found across the core sequence near the 5′. Nearly all the sequence length increase can be found in the middle disordered region where alignment is poor even between D. virilis and D. melanogaster. We note that there are several indels and SNPs that differentiate between consensus from the nasuta group consensus and the pallidifrons cluster. However, many of these mutations are found in DINEs that are outside of the expanded clusters.
Frequent Expansion Likely Due to Suppression Escape
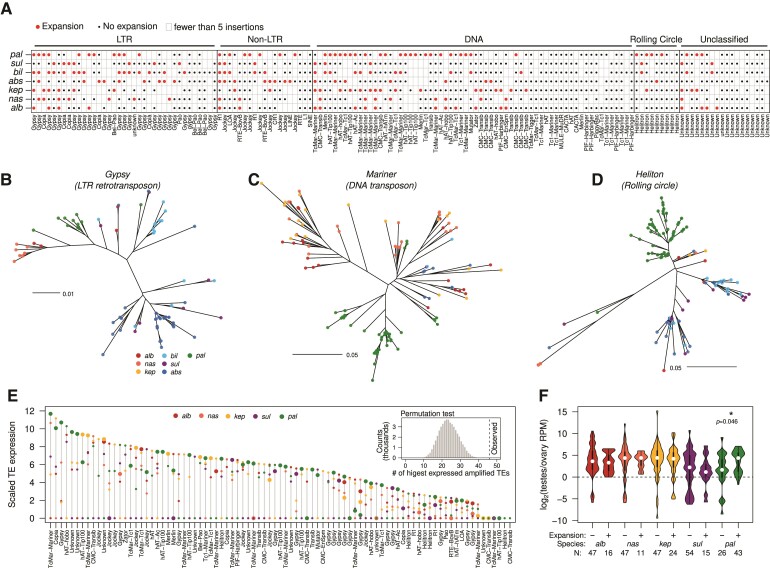
Given the pattern of proliferation of the DINEs, we were curious as to the frequency in which TEs can escape suppression and expand. We, therefore, generated phylogenetic trees of 147 TEs where we can find more than 20 copies summed across all seven species; expansions were identified as branches showing significant lineage and/or species-specific clustering (fig. 6A–D). We find that 78 TEs show significant species-specific clustering in at least one species, suggesting TE proliferation occurs frequently in different species (fig. 6A). In most cases, individual TE expansions do not reach beyond 50 copies. Expansion occurs across all types of elements although in different ways (fig. 6A). For example, for a variant of the Gypsy LTR retrotransposon, expansions are observed in four species as well as before the sulfurigaster semispecies split (fig. 6B). In contrast, for Merlin, a DNA transposon, expansions are observed in D. pallidifrons and D. nasuta and before the D. albomicans/D. nasuta/D. kepulauana species split (fig. 6C). Lastly, a rolling circle element expanded in D. pallidifrons and two of the sulfurigaster species (fig. 6D). Strikingly, there are 47 expanded TE families in D. pallidifrons which accounts for its higher repeat content compared with the other species (fig. 6C–E) and may suggest increased tolerance to TE load and/or reduced genomic defense.
Fig. 6.
Frequent lineage-specific amplifications and suppressions of TE families. (A) Species-specific expansion status of different TE families and types based on phylogenies of insertions. Red dots indicate amplification in a nasuta species, black dots indicate no amplification, and empty boxes indicate fewer than five insertions. (B–D) Unrooted trees of TE insertions of different types of TEs. Their positions on the table in (A) are marked by arrowheads. (E) Expression of expanded and unexpanded TE families in the testes of different species. For each TE family, the transcript abundance is scaled by the lowest expressed species, and the range of expression across the different species is plotted vertically as demarcated by the gray line. Along this line, the expression in the different species is positioned by colored circles. Large circles denote species-specific expansion. The observed positions of the expanded TEs along the expression ranges are tested against the null expectation using randomized permutation testing (top right inset). The null distribution is presented and the observed count is marked by the vertical dotted line. (F) Fold-difference in TE transcript abundance between testes and ovarian expression across species. TEs are subdivided into those that have species-specific expansions and those without.
To determine whether these expansions resulted from the escape of transcriptional and posttranscriptional silencing, we examined TE expression from the testes and ovaries in five species. Cross-species comparisons revealed that TEs frequently show elevated expression accompanying their expansion (fig. 6E). Out of those that have expanded, 46 TE families (58.9%) show the highest expression in the species in which the expansion occurred, significantly higher than the random expectation of 24 (fig. 6E; P < 0.00002, permutation testing, see Materials and Methods). However, this is not always the case; for example, whereas DINE shows recurrent and recent expansions in D. pallidifrons (fig. 5A), it is expressed at intermediate levels in this species (supplementary fig. S16, Supplementary Material online). Notably, even when adjusting the expression by the copy number of the TE found in each genome, TEs are significantly more elevated in expression in the species in which they expanded (supplementary fig. S18, Supplementary Material online). Interestingly, we also find at least 15 instances where the TE family is the most lowly expressed in the species in which it expanded; we suspect these may reflect successful suppression mechanisms that evolved after expansion. However, without germline piRNA sequencing, it remains unclear whether the expression differences between expanded TE families reflect the emergence of piRNA defense. Further, we note that posttranscriptional silencing of TEs does not always have to decrease transcript abundance; for example, the piRNA machinery induces alternative splicing of the P-element without affecting transcript levels (Teixeira et al. 2017).
In Drosophila, the activity of TEs and their silencing systems can both differ between the sexes (Chen et al. 2021). Across all species, TE expression is higher in testes than in ovaries, suggesting weaker silencing in the testes (fig. 6F). Curiously, the expression of expanded TEs in D. pallidifrons is on average 20.70-fold higher in testes compared with ovaries. This is significantly higher than unexpanded TEs which are only 3.16-fold higher expressed in the testes (P = 0.0464). This striking difference raises the interesting possibility that the numerous TEs that have expanded in D. pallidifrons may be exploiting the male germline for amplification. This hypothesis is consistent with our observation that insertions are found more frequently around genes with higher expression in testes compared with ovaries (fig. 3C). Further supporting this possibility, higher TE expression and transposition rates in males have been shown in other fly species to result from reduced suppression of Y-linked insertions in embryos and testes (Wei et al. 2020; Lawlor et al. 2021).
Epigenetic Silencing of Expanded TEs Moderately Reduces Expression in Neighboring Genes
Even though expanded TEs are typically highly expressed when compared with other species, several expanded TEs show low to no expression. We hypothesized that the lowly expressed expanded TEs may have been historically active elements that are now silenced. To evaluate this possibility, we looked at the expression of genes neighboring these expanded TEs, reasoning that silencing of TEs will likely lead to reduced expression of neighboring genes.
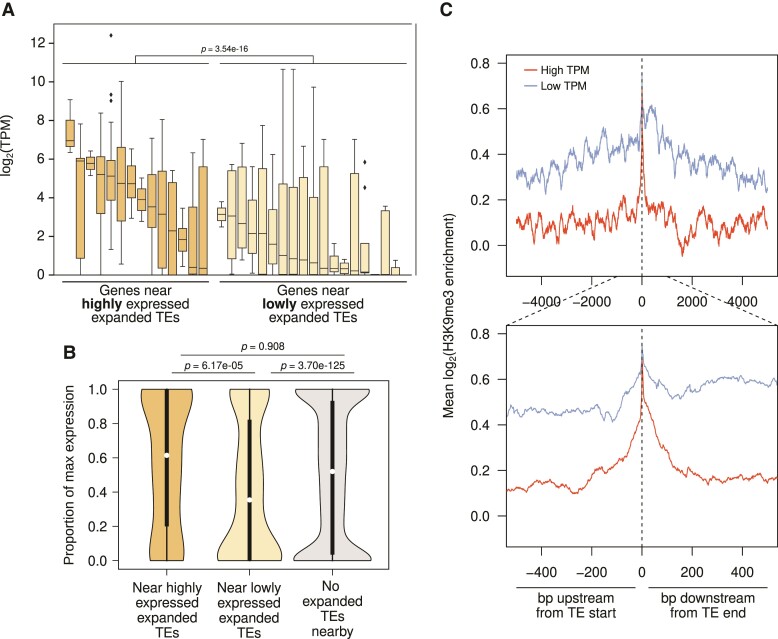
We identified genes with nearby TE insertions (internal or ±1 kb up- and downstream), and subdivided them into those with insertions of highly versus lowly expressed expanded TEs. We focused on D. pallidifrons as it has the highest number of expanded TEs, and identified 182 and 552 genes with expanded lowly expressed and expanded highly expressed nearby TEs, respectively. Interestingly, expression of genes nearby highly expressed expanded TEs is significantly higher than for genes nearby lowly expressed expanded TEs (fig. 7A, P-value < 3.5392 × 10−16, Wilcoxon rank sum test). This is consistent with the notion that silencing of expanded TEs is associated with lower expression of nearby genes.
Fig. 7.
Epigenetic silencing of expanded TEs down-regulates nearby genes. (A) Expanded TEs are categorized as either highly or lowly expressed depending on expression differences between species. Dark yellow boxes represent genes nearby highly expressed expanded TEs, whereas light yellow boxes represent genes nearby lowly expressed expanded TEs. Each box represents the distribution of transcript abundances (TPM) of genes with nearby insertions of a given expanded TE family. Genes (n = 785) near lowly expressed expanded TEs have significantly lower expression (Wilcoxon’s rank sum test, P < 3.54e−16). (B) Scaled expression of genes near highly (dark yellow, n = 82) and lowly expressed expanded TEs (light yellow, n = 552), as well as those with no expanded TEs nearby (gray, n = 8705). Genic expression is scaled by the TPM of the highest expressed orthologs across all species. Significance of pairwise comparisons of the three sets is labeled above the figure. (C) H3K9me3 enrichment around full-length TE insertions in Drosophila albomicans depending on whether the TE is highly expressed as compared with other species (red) versus lowly expressed (blue). Insertions within the pericentric regions are removed.
To differentiate between insertions/survival bias near lowly expressed genes versus bona fide spreading of epigenetic silencing into neighboring genes, we again compared the expression of the orthologs of these genes between species. To sensitively detect potential down-regulation, for each gene, we scaled the expression of the D. pallidifrons ortholog relative to the most highly expressed ortholog. For genes with no expanded TEs around them (fig. 7B, gray), the D. pallidifrons orthologs, expectedly, have a median relative expression of 0.50. Although not significantly different, genes with highly expressed expanded TEs nearby show a slightly higher median expression and are slightly skewed toward higher expression (fig. 7B, dark yellow). On the other hand, genes near lowly expressed expanded TEs (i.e., near those TEs that are putatively silenced) show a low relative expression of 0.37 (fig. 7B, light yellow). These genes show a clear skew toward low to no expression, and are significantly lower ranked than both the control set of genes (no expanded TEs nearby) and genes near highly expressed TEs (fig. 7B, light yellow; P = 3.70e−12 and 5.17e−05, Wilcoxon’s rank sum test). These results reveal that insertions of recently expanded TEs can cause a subtle but significant decrease in gene expression if inserted nearby, but only if the TEs are targeted for (presumably epigenetic) silencing. However, if a recently expanded TE is not being targeted for silencing, it may potentially induce higher expression of neighboring genes.
We used our H3K9me3 ChIP data in D. albomicans to further evaluate whether this effect is due to epigenetic silencing. We plotted H3K9me3 enrichment around TEs with elevated expression and TEs with low expression in D. albomicans, removing insertions in the pericentric regions (fig. 7C). Consistent with epigenetic spreading at putatively silenced TEs, we find that TEs with low expression show substantially higher H3K9me3 enrichment in surrounding regions, with both elevated and wider spreading of heterochromatin. More highly expressed TEs, in contrast, show substantially less enrichment and spreading of H3K9me3. Therefore, lowly expressed TEs are likely under stronger epigenetic silencing which leads to broader spreading of H3K9me3.
Discussion
Here, we generated highly contigous genomes of seven closely related Drosophila species, taking advantage of long-read sequencing technologies. Enabled by these high-quality genome assemblies, we systematically characterized the landscape of TE insertions and evaluated how their activities and regulation influence genome evolution. Specifically, we focused on two questions: how often do TEs impact gene regulation and how common do TEs escape silencing and expand in copy number?
The Regulatory Impact of TE Insertions on Gene Expression
There are numerous examples of TE insertions affecting the expression of neighboring genes, some of which even confer adaptive phenotypes (Casacuberta and González 2013; Mateo et al. 2014; Merenciano et al. 2016; Villanueva-Cañas et al. 2019). However, insertions around genes are primarily thought to be deleterious as they can induce epigenetic silencing of neighboring genes through heterochromatin spreading (Choi and Lee 2020). Here we comprehensively evaluate such an effect in a comparative genomics framework by combining high confidence TE insertion calls from de novo genome assemblies with gene expression data across a group of recently diverged species, the nasuta species group. While TE insertions are found more frequently near lowly expressed genes, TE-induced silencing does not appear to be a major cause of this negative association. The vast majority of genes with insertions around them do not show lower expression compared with other species lacking those insertions. Therefore, instead of TEs causing nearby down-regulation, it appears that they tend to accumulate and repeatedly insert near historically lowly expressed genes. The fact that independent insertion patterns are positively correlated between species suggests that two types of nonmutually exclusive biases could be at play. TEs may preferentially insert into specific regions, chromatin environments, or gene features, thereby resulting in similar insertion patterns between species. This alone is unlikely to fully account for the negative association between gene expression and insertion counts. We, therefore, suspect that the observed insertion landscape also reflects a survivorship bias; insertions with high fitness costs are unlikely to reach high population frequency, and most of the observable insertions in the genome will be those with low fitness impacts. Unlike highly expressed genes, such as housekeeping genes that are under strong purifying selection, lowly expressed genes may be more permissive to fluctuations in gene expression.
TEs are underrepresented near highly expressed genes, yet most TE insertions identified in our genomes do not appear to alter gene expression (figs. 2F and 3G). If observed TE insertions rarely influence gene expression, then how could they be more deleterious when inserted near highly expressed genes? One possible explanation for this apparent paradox may be that the regulatory effects of TEs becomes more substantial upon environmental perturbations (Capy et al. 2000). In plants, multiple classes of retrotransposons are activated upon stress (Wessler 1996; Grandbastien et al. 1997), and in flies and worms, TEs increase in activity during elevated temperature (Kidwell et al. 1977; Garza et al. 1991; Ratner et al. 1992; Kurhanewicz et al. 2020). The lack of expression change in genes with TEs inserted nearby may therefore be the product of maintaining stocks in stable lab conditions. But upon environmental perturbation, these genes might begin to show more drastic regulatory changes as TEs become active. In changing environmental conditions, insertions around highly expressed and functionally important genes may therefore be under strong purifying selection, accounting for the negative association between gene expression and TE insertions.
Context-Dependent Heterochromatin Spreading and Epigenetic Silencing
Despite no systematic support for widespread down-regulation of genes with TEs inserted nearby, we were able to find evidence of epigenetic silencing of genes due to TE insertions in some cases. In D. pallidifrons, insertions of recently expanded TEs can cause moderate down-regulation of gene expression, but only if the TEs have low expression—presumably due to epigenetic silencing (fig. 7A and B). Moreover, in every species, a few dozens of species-specific TE insertions appear to be associated with down-regulation of nearby genes (fig. 3G and H, supplementary table S4, Supplementary Material online). Notably, we also find cases where insertions are associated with large up-regulation in gene expression (fig. 3G and H, supplementary table S4, Supplementary Material online), but these cases are much rarer than those associated with down-regulation of nearby genes.
Although we do not have direct evidence of transcriptional or posttranscriptional silencing in most species, spreading of heterochromatin from TE insertions is observed in D. albomicans (fig. 4). TEs far from genes show the highest and broadest H3K9me3 enrichment, and TE insertions near lowly expressed genes also show more heterochromatin spreading. While consistent with epigenetic silencing of neighboring genes, these results are also consistent with the notion that active transcription antagonizes heterochromatin formation, and vice versa. Lower expression of genes near TEs that show higher levels of heterochromatin spreading could indicate that H3K9me3-inducing TEs are more tolerated near lowly expressed genes. Further, we find that insertions of TE families with low expression are associated with broader and stronger heterochromatin spreading to their surroundings. Indeed, lowly expressed and high copy number TEs are typically recently active and have robust small RNA targeting for posttranscriptional degradation and transcriptional silencing (Wei et al. 2021). Altogether, these results suggest that TE insertions can have multiple effects on gene expression and calls into question how pervasive TE-induced epigenetic silencing of neighboring genes is. The epigenetic effect TEs have on neighboring genes, if any, is likely dependent on multiple factors, such as the transcription rate of the gene, the local repeat density, and the 3D architecture of the genome.
Frequent and Recurrent TE Expansions and Silencing
A phylogenetic approach revealed that >50% of TE families show lineage and species-specific amplification. The most striking expansion is the DINE, which has exploded to thousands of copies across the nasuta species group. This expansion occurred once before the species radiation, and at least twice since, one in D. pallidifrons, and one in the related outgroup species D. immigrans (∼20 My diverged; Izumitani et al. 2016; O’Grady and DeSalle 2018) (fig. 5A). The repeated expansions suggest multiple bouts of suppression escape. Interestingly, we were unable to find unique mutations private to the D. pallidifrons expansion clade, which may be causal mutations allowing to avoid suppression. One possible explanation for the absence of such mutations is that gene conversion events have erased lineage-specific mutations (Fawcett and Innan 2019). Gene conversion among nonallelic TE insertions has previously been shown to allow for rapid adaptive changes at TE sequences coopted for X-chromosome dosage compensation (Ellison and Bachtrog 2015). Consistent with gene conversion, there are multiple smaller scale clusters of D. pallidifrons DINEs all across the tree which may represent elements that acquired the causal mutations for suppression escape, allowing for their own, albeit limited, proliferation. Previous analyses of DINEs across Drosophila have found their sequences to be species-specific (Yang and Barbash 2008), even for recently diverged species. This may be due to rapid homogenization of copies due to gene conversion events similar to what we are observing in D. pallidifrons.
In addition to DINEs, many other TEs also show lineage-specific expansions, though at much more limited scales. Most of these expanding TE families show elevated expression only in the species with the expansion, consistent with species-specific suppression escape and derepression allowing for expansion. Most strikingly, 32 families are currently or have been recently expanding in D. pallidifrons. This may in part reflect the fact that it is the least derived of our species and therefore has the longest terminal branches. However, we find high expression for many of these expanding TEs, indicating recent and perhaps on-going mobilizations. Why are so many TEs concurrently expanding in D. pallidifrons? P-element dysgenesis in D. melanogaster is caused by the absence of maternally deposited piRNAs against the P-element, yet derepression and mobilization of TEs may not be limited to P-elements (Engels 1984; Gerasimova et al. 1984; Petrov et al. 1995; Khurana et al. 2011). Therefore, the large numbers of expanding and highly expressed TEs may be reflecting an on-going sweep of a novel TE in the species. However, other studies argue that during dysgenesis, mobilization is restricted to only the P-element (Woodruff et al. 1987; Eggleston et al. 1988; Kofler et al. 2018). Thus, the concurrent expansions of TEs may instead raise the curious possibility of weakened genomic defense in D. pallidifrons. Interestingly, we also find that a fraction of these recently expanded TEs, paradoxically, have low expression, and genes around them show reduced expression. We suspect that these are recently active TEs that are now epigenetically silenced.
The importance of horizontal transfer to the long-term survival and expansion of TEs has been pointed out multiple times in the literature (Kidwell 1992; Silva et al. 2004; Loreto et al. 2008; Schaack et al. 2010; Zhang et al. 2020). Horizontal transfer can allow TEs to cross species boundaries and invade a naive genome that lacks suppressive mechanisms against this TE, where it can proliferate (Le Rouzic and Capy 2005). Once silencing mechanisms against a TE emerge, for example, targeting by small RNAs, mobilization of that TE is prevented (Khurana et al. 2011). Inactive TEs will accumulate mutations, and eventually all functional copies may die, and horizontal transfer to a new lineage would allow that TE to escape extinction. Our finding of species-specific escape from TE repression for a large fraction of TE families suggests a very dynamic evolution of host genomes and their TEs. Active TEs are temporarily silenced within a lineage, but over evolutionary timescales, some copies will escape silencing in different lineages, leading to species-specific bursts in TE activity. Thus, in addition to horizontal transfer, our data suggest that escape from host suppression seems to be an important strategy allowing for the long-term survival of TEs.
Long-Read Genome Assemblies Open New Doors for Studying TEs
In our study, high-quality genomes assembled via long reads have circumvented many of the previous challenges associated with studying TEs and repeats (Khost et al. 2017), and enabled high confidence annotation of TE insertions. Further, our approach of integrating phylogenetics, functional genomics, and comparative genomics has revealed a comprehensive picture of the dynamics of TE suppression escape and subsequent reestablishment of silencing and their effects on the rest of the genome. These high-quality genome assemblies will further facilitate the molecular dissection of the nucleotide changes in TEs causing suppression escape in future studies. With the rapidly decreasing cost and less input material in generating high-quality assemblies (Adams et al. 2020), it will become easier and cheaper to identify de novo insertions. Despite the rapid adoption of new sequencing technologies, TEs and repeats often remain understudied. Our study demonstrates that assembling repeats is among one of the greatest advantages to long-read sequencing, and allows for a comprehensive investigation of TEs in a phylogenetic context.
Materials and Methods
Fly Strains and Nanopore Sequencing
We extracted high molecular weight DNA from ∼50 females from D. nasuta 15112-1781.00, D. kepulauana 15112-1761.03, D. s. albostrigata 15112-1771.04, D. s. bilimbata 15112-1821.10, D. s. sulfurigaster 15112-1831.01, and D. pallidifrons PN175_E-19901 using the QIAGEN Gentra Puregene Tissue Kit. The D. kepulauana high molecular weight DNA was sequenced on PacBio RS II platform at UC Berkley QB3 genome sequencing center. The high molecular weight DNA of other species was sequenced on Nanopore MinIOn.
Genome Assemblies
We used a similar approach to obtain highly contiguous genome assemblies for each species. Pipelines differ slightly depending on the quality of the final genome. For details on assembly methods and statistics for each genome, see supplementary materials and table S2, Supplementary Material online. We used canu (Koren et al. 2017) to error-correct Nanopore or PacBio long reads, which were then used for an initial genome assembly using a combination of wtdbg2 (Ruan and Li 2020) and Flye (Kolmogorov et al. 2019). The assemblies were polished using Illumina paired end reads (from Mai et al. 2020), typically three times with minimap2 (Li 2018) and Racon (Vaser et al. 2017) followed by one round of Pilon (Walker et al. 2014). Bacterial contaminated contigs were identified by BLAST against the NCBI database and removed. The filtered genome was scaffolded using Hi-C data with the Juicer and 3d-dna pipeline (Dudchenko et al. 2017); strings of 50 N’s were placed between scaffolded contigs. To maximize assembly quality, we generated two genomes for the species D. kepulauana and D. s. albostrigata using both wtdbg2 and Flye, then used quickmerge (Chakraborty et al. 2016) to combine the assemblies. For D. albomicans, we used quickmerge to combine our new assembly with the previous Pacbio-based high-quality assembly to maximize the assembly quality. These strategies resulted in highly complete (BUSCO scores 98.5–99.7%) and highly contiguous assemblies (supplementary table S2, Supplementary Material online).
Gene Annotation and Clustering
We used MAKER to annotate genes in each species’ genome assembly (Campbell et al. 2014). To train MAKER’s gene inference model, we generated a transcriptome from D. albomicans and D. nasuta RNA seq data from Zhou and Bachtrog (2012). RNA seq data from D. albomicans and D. nasuta were aligned to the corresponding genome assemblies with HISAT2 under default settings (Kim et al. 2019). The alignments were then used to create transcriptomes using StringTie (Pertea et al. 2015). Additionally, satellites and repeats in the genome assemblies for each species were masked using RepeatMasker in preparation for gene annotations (Smit et al. 2013). Then, using both the D. albomicans and D. nasuta transcriptomes, we ran MAKER with default settings. We then took the annotations and determined gene homology between species with OrthoDB (Kriventseva et al. 2019).
TE Library Generation, Annotation, and Analyses
In order to lower the occurrence of nested TE structures, pericentromeric regions were removed from each assembly and the resulting sequences were separately used as input for RepeatModeler2 and the accompanying LTRharvest software with default options (Ellinghaus et al. 2008; Flynn et al. 2020). The resulting species-specific TE libraries were merged together. To remove redundancy from the merged library, we used CD-HIT2 to cluster TE entries with each other. However, instead of allowing CD-HIT2 to select the representative sequence of the cluster (which is usually the longest sequence), we evaluated the TEs within clusters based on three criteria: entry sequence length, self-identity, and probability of full-length insertions. For self-identity, we blasted each TE entry to itself and calculated the self-blast score as the proportion of the sequence showing alignment to another region of itself. For the probability of full-length insertions, we blasted each entry to the genome and calculated the proportion of near-full-length blast hits. We then weighed the three criteria to maximize length and probability of full-length insertion, whereas minimizing self-identity, in order to select the representative sequence per CD-HIT2 cluster. This procedure is done twice. We used both the Repeatmodeler2 TEs categorization as well as the program ClassifyTE (Panta et al. 2021). When the two disagreed with the TE classification, we used the assignment from RepeatModeler2. Note, even after two rounds of CD-HIT2, we found 10 redundant entries corresponding to variants of the DINE in the genome through manual NCBI BLASTn (Altschul et al. 1990). We removed entries with unique sequences flanking the DINEs and kept the longest entry.
TE insertions in genomes are annotated by RepeatMasking the final nasuta group-specific TE index to the respective species genomes. Because RepeatMasker can provide overlapping annotations, we used bedtools merge to merge overlapping annotations first, generating chimerics. We then blasted all the chimeric annotations to the repeat library and recategorized those where 90% of the sequence blasts to a specific TE. Full length and truncated elements are defined as annotations that are >80% length of the TE entries, or <80% length but >200 bp, respectively. Distances between full-length TE and the closest gene in each species were calculated using bedtools closest (Quinlan and Hall 2010), with species-specific TE and gene annotations as inputs.
To generate random TE insertions in the genome for permutation testing, we first removed all annotated TEs from the genome fasta files by masking their positions (bedtools maskfasta –mc M) followed by removal of the masked sites (sed “s/M//g”). Then for each entry in the TE annotation, we randomly reassigned a position on the same chromosome. Coordinates of gene annotations were adjusted according to the insertions/removal of TEs. We determined the distance between genes and TEs using bedtools closest, and overlap between genes and TEs with bedtools intersect.
Phylogenetic Analyses of TEs
We ran BLAST using the TE libraries as the query and the genome assemblies of each species as the database (Altschul et al. 1990). TE sequences from full-length BLAST alignments—defined as those in which the alignment length is at least 80% of the TE length—are extracted. We used Clustal Omega under default settings to perform a multiple sequence alignment for all sequences for each TE (Sievers et al. 2011); those with over 200 full-length copies across all species were subsampled down to 200 sequences. In order to maintain the different copy number in the different species, the subsampling procedure maintained the proportional difference of insertion counts across the species.
Phylogenies for TEs were then generated with RAxML using the command: raxmlHPC-PTHREADS-AVX -T 24 -f a -x 1255 -p 555 -# 100 -m GTRGAMMA -s input.MSA.fa -n input.MSA.tree > input.MSA.tree.stderr (Stamatakis 2014).
We tested for the presence of species-specific expansion of each TE by measuring the extent of clustering using the RRphylo R package (Serio et al. 2019). Tests were carried out for species where there were at least five sequences or 5% of the total sequences in the phylogeny. The resulting P-values from the analyses were adjusted for multiple testing using the Benjamini–Hochberg procedure. TEs from a particular species with P-values < 0.05 are considered to be expanded. We note that this program does not take into account the species relationships and therefore cannot capture lineage-specific expansion. Thus this approach underestimates the number of TEs that have recently expanded.
RNA Sample Collection and Sequencing
Two replicates of RNA sequencing libraries created from males and females of each species were generated and sequenced. Testes from five to eight males from live D. albomicans, D. nasuta, D. kepulauana, and D. s. sulfurigaster as well as frozen D. pallidifrons were dissected for each RNA sequencing library. Ovaries from three to five females from live D. albomicans, D. nasuta, D. kepulauana, and D. s. sulfurigaster as well as frozen D. pallidifrons were dissected for each RNA sequencing library. For each species, tissue samples were placed in Trizol for RNA extraction. RNA was extracted using the Trizol extraction method and enriched for ployA RNA using NEBNext Poly(A) mRNA Magnetic Isolation Module (E7490) as per manufacturer protocol. The RNA libraries were prepared as per NEBNext Ultra II Directional RNA kit (E7760S) and sequenced on Illumina NovSeq 6000 on SP flow cell for 150 PE reads.
RNA Transcript Abundance
Genes
Generated RNA sequencing data for D. albomicans, D. nasuta, D. kepulauana, D. s. sulfurigaster, and D. pallidifrons were aligned to their corresponding genome assembly. Using the alignment data and gene annotations, we used the featureCounts program from the Subread package to calculate the number of reads mapping to each gene. We then calculated gene TPM with the following formula:
Transposable Elements
Generated RNA sequencing data for D. albomicans, D. nasuta, D. kepulauana, D. s. sulfurigaster, and D. pallidifrons were aligned to the TE library. A custom script was used to count the number of reads mapping to each TE. The number of reads was then normalized by the TE length and then divided by the median of gene read counts that are normalized in the same way from the corresponding species.
Permutation Testing of TE Expression
The test statistic used for the permutation test is the number of times the highest expression for a particular TE comes from a species where that TE has expanded. We first calculate this test statistic from our data. We then randomly shuffle the species associated with each expansion event and calculate the test statistic 50,000 times. The P-value obtained is the proportion of tests with test statistics less than or equal to our original test statistic.
Cross-Species TE and Gene Expression Comparisons
With normalized testes and ovaries RNA-seq expression for five species, we determined the two species with the highest expression and lowest expression for each TE. We used the top two species instead of the highest species to increase the number of TEs for these comparisons. Then, to evaluate whether the expression of genes neighboring these TEs differ between species, we scaled the expression of each gene across the five species to span 0 and 1 where the species with the lowest and highest expression will have values of 0 and 1, respectively, using the formulae:
where 1 represents a pseudocount.
ChIP-seq Analyses
ChIP-seq analyses were slightly modified from methods in Wei et al. (2021). Briefly, larval H3K9me3 ChIP and input data (Wei and Bachtrog 2019) were aligned to the genome using bwa mem. The per base pair coverage was determined using bedtools coverageBed -d -ibam. Median autosomal coverage was estimated from 50 kb nonoverlapping sliding windows. We then inferred enrichment at every position as:
We averaged the enrichment across the three replicates. For H3K9me3 spreading around TE insertions, we lined up annotated TE insertions at either the 5′ or 3′, and averaged enrichment 5 kb upstream and downstream of the insertions, respectively.
Supplementary Material
Acknowledgments
We thank the three anonymous reviewers for their feedback. We also thank Carolus Chan for help generating the Hi-C libraries. This research was funded by NIH grants (R01 GM101255 and R56 AG057029) to D.B. Publication made possible in part by support from the Berkeley Research Impact Initiative (BRII) sponsored by the UC Berkeley Library.
Contributor Information
Kevin H.-C. Wei, Department of Integrative Biology, University of California Berkeley, Berkeley, CA 94720, USA
Dat Mai, Department of Integrative Biology, University of California Berkeley, Berkeley, CA 94720, USA.
Kamalakar Chatla, Department of Integrative Biology, University of California Berkeley, Berkeley, CA 94720, USA.
Doris Bachtrog, Department of Integrative Biology, University of California Berkeley, Berkeley, CA 94720, USA.
Supplementary Material
Supplementary data are available at Molecular Biology and Evolution online.
Author Contributions
D.B. and K.H.-C.W. conceived of the study. K.H.-C.W. and D.M. analyzed the data. K.C. and K.H.-C.W. collected the samples, prepared the libraries, and generated the sequencing. D.B., K.H.-C.W., and D.M. composed the manuscript. D.B. and K.H.-C.W. revised the manuscript and addressed reviewer comments.
Data Availability
All raw reads are deposited on the SRA under BioProject PRJNA736413. Genome assemblies, TE libraries, gene, and TE annotations can be found on Dryad at https://doi.org/10.6078/D11B01.
References
- Drosophila 12 Genomes Consortium . 2007. Evolution of genes and genomes on the Drosophila phylogeny. Nature 450:203–218. [DOI] [PubMed] [Google Scholar]
- Adams M, McBroome J, Maurer N, Pepper-Tunick E, Saremi NF, Green RE, Vollmers C, Corbett-Detig RB. 2020. One fly-one genome: chromosome-scale genome assembly of a single outbred Drosophila melanogaster. Nucleic Acids Res. 48:e75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol. 215:403–410. [DOI] [PubMed] [Google Scholar]
- Anxolabéhère D, Kidwell MG, Periquet G. 1988. Molecular characteristics of diverse populations are consistent with the hypothesis of a recent invasion of Drosophila melanogaster by mobile P elements. Mol Biol Evol. 5:252–269. [DOI] [PubMed] [Google Scholar]
- Athma P, Peterson T. 1991. Ac induces homologous recombination at the maize P locus. Genetics 128:163–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachtrog D. 2006. The speciation history of the Drosophila nasuta complex. Genet Res. 88:13–26. [DOI] [PubMed] [Google Scholar]
- Bannister AJ, Zegerman P, Partridge JF, Miska EA, Thomas JO, Allshire RC, Kouzarides T. 2001. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature 410:120–124. [DOI] [PubMed] [Google Scholar]
- Bergman CM, Quesneville H, Anxolabéhère D, Ashburner M. 2006. Recurrent insertion and duplication generate networks of transposable element sequences in the Drosophila melanogaster genome. Genome Biol. 7(11):R112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bickhart DM, Rosen BD, Koren S, Sayre BL, Hastie AR, Chan S, Lee J, Lam ET, Liachko I, Sullivan ST, et al. 2017. Single-molecule sequencing and chromatin conformation capture enable de novo reference assembly of the domestic goat genome. Nat Genet. 49:643–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biémont C, Vieira C. 2005. What transposable elements tell us about genome organization and evolution: the case of Drosophila. Cytogenet Genome Res. 110:25–34. [DOI] [PubMed] [Google Scholar]
- Biessmann H, Mason JM, Ferry K, d’Hulst M, Valgeirsdottir K, Traverse KL, Pardue ML. 1990. Addition of telomere-associated HeT DNA sequences “heals” broken chromosome ends in Drosophila. Cell 61:663–673. [DOI] [PubMed] [Google Scholar]
- Bourque G, Burns KH, Gehring M, Gorbunova V, Seluanov A, Hammell M, Imbeault M, Izsvák Z, Levin HL, Macfarlan TS, et al. 2018. Ten things you should know about transposable elements. Genome Biol. 19:199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bracewell R, Chatla K, Nalley MJ, Bachtrog D. 2019. Dynamic turnover of centromeres drives karyotype evolution in Drosophila. Elife 8:e49002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand CL, Kingan SB, Wu L, Garrigan D. 2013. A selective sweep across species boundaries in Drosophila. Mol Biol Evol. 30:2177–2186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buzdin AA. 2004. Retroelements and formation of chimeric retrogenes. Cell Mol Life Sci. 61:2046–2059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell MS, Holt C, Moore B, Yandell M. 2014. Genome annotation and curation using MAKER and MAKER-P. Curr Protoc Bioinform. 48:4.11.1–4.11.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capy P, Gasperi G, Biémont C, Bazin C. 2000. Stress and transposable elements: co-evolution or useful parasites? Heredity 85(Pt 2):101–106. [DOI] [PubMed] [Google Scholar]
- Casacuberta E, González J. 2013. The impact of transposable elements in environmental adaptation. Mol Ecol. 22:1503–1517. [DOI] [PubMed] [Google Scholar]
- Chakraborty M, Baldwin-Brown JG, Long AD, Emerson JJ. 2016. Contiguous and accurate de novo assembly of metazoan genomes with modest long read coverage. Nuc Acids Res. 44:e147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakraborty M, Chang C-H, Khost DE, Vedanayagam J, Adrion JR, Liao Y, Montooth KL, Meiklejohn CD, Larracuente AM, Emerson JJ. 2021. Evolution of genome structure in the Drosophila simulans species complex. Genome Res. 31:380–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen P, Kotov AA, Godneeva BK, Bazylev SS, Olenina LV, Aravin AA. 2021. piRNA-mediated gene regulation and adaptation to sex-specific transposon expression in D. melanogaster male germline. Genes Dev. 35:914–935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Lu L, Benjamin J, Diaz S, Hancock CN, Stajich JE, Wessler SR. 2019. Tracking the origin of two genetic components associated with transposable element bursts in domesticated rice. Nat Commun. 10(1):641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi JY, Lee YCG. 2020. Double-edged sword: the evolutionary consequences of the epigenetic silencing of transposable elements. PLoS Genet. 16:e1008872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosby RL, Judd J, Zhang R, Zhong A, Garry N, Pritham EJ, Feschotte C. 2021. Recurrent evolution of vertebrate transcription factors by transposase capture. Science 371:eabc6405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cridland JM, Macdonald SJ, Long AD, Thornton KR. 2013. Abundance and distribution of transposable elements in two Drosophila QTL mapping resources. Mol Biol Evol. 30:2311–2327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czech B, Munafò M, Ciabrelli F, Eastwood EL, Fabry MH, Kneuss E, Hannon GJ. 2018. piRNA-guided genome defense: from biogenesis to silencing. Annu Rev Genet. 52:131–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniels SB, Peterson KR, Strausbaugh LD, Kidwell MG, Chovnick A. 1990. Evidence for horizontal transmission of the P transposable element between Drosophila species. Genetics 124:339–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudchenko O, Batra SS, Omer AD, Nyquist SK, Hoeger M, Durand NC, Shamim MS, Machol I, Lander ES, Aiden AP, et al. 2017. De novo assembly of the genome using Hi-C yields chromosome-length scaffolds. Science 356:92–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durand NC, Shamim MS, Machol I, Rao SSP, Huntley MH, Lander ES, Aiden EL. 2016. Juicer provides a one-click system for analyzing loop-resolution Hi-C experiments. Cell Syst. 3:95–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eggleston WB, Johnson-Schlitz DM, Engels WR. 1988. P-M hybrid dysgenesis does not mobilize other transposable element families in D. melanogaster. Nature 331(6154):368–370. [DOI] [PubMed] [Google Scholar]
- Elgin SCR, Reuter G. 2013. Position-effect variegation, heterochromatin formation, and gene silencing in Drosophila. Cold Spring Harb Perspect Biol. 5:a017780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellinghaus D, Kurtz S, Willhoeft U. 2008. LTRharvest, an efficient and flexible software for de novo detection of LTR retrotransposons. BMC Bioinform. 9:18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellison CE, Bachtrog D. 2013. Dosage compensation via transposable element mediated rewiring of a regulatory network. Science 342:846–850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellison CE, Bachtrog D. 2015. Non-allelic gene conversion enables rapid evolutionary change at multiple regulatory sites encoded by transposable elements. Elife 4:e05899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engels WR. 1984. A trans-acting product needed for P factor transposition in Drosophila. Science (New York, N.Y.) 226(4679):1194–1196. [DOI] [PubMed] [Google Scholar]
- Fawcett JA, Innan H. 2019. The role of gene conversion between transposable elements in rewiring regulatory networks. Genome Biol Evol. 11:1723–1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flynn JM, Hubley R, Goubert C, Rosen J, Clark AG, Feschotte C, Smit AF. 2020. RepeatModeler2 for automated genomic discovery of transposable element families. Proc Natl Acad Sci U S A. 117:9451–9457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu L, Niu B, Zhu Z, Wu S, Li W. 2012. CD-HIT: accelerated for clustering the next-generation sequencing data. Bioinformatics 28:3150–3152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrigan D, Kingan SB, Geneva AJ, Andolfatto P, Clark AG, Thornton KR, Presgraves DC. 2012. Genome sequencing reveals complex speciation in the Drosophila simulans clade. Genome Res. 22:1499–1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garza D, Medhora M, Koga A, Hartl DL. 1991. Introduction of the transposable element mariner into the germline of Drosophila melanogaster. Genetics 128:303–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerasimova T, Mizrokhi L, Georgiev G. 1984. Transposition bursts in genetically unstable Drosophila melanogaster. Nature 309:714–716. [Google Scholar]
- Grandbastien MA, Lucas H, Morel JB, Mhiri C, Vernhettes S, Casacuberta JM. 1997. The expression of the tobacco Tnt1 retrotransposon is linked to plant defense responses. Genetica 100:241–252. [PubMed] [Google Scholar]
- Hall IM, Shankaranarayana GD, Noma K-I, Ayoub N, Cohen A, Grewal SIS. 2002. Establishment and maintenance of a heterochromatin domain. Science 297:2232–2237. [DOI] [PubMed] [Google Scholar]
- Hedges DJ, Deininger PL. 2007. Inviting instability: transposable elements, double-strand breaks, and the maintenance of genome integrity. Mutat Res. 616:46–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollister JD, Gaut BS. 2009. Epigenetic silencing of transposable elements: a trade-off between reduced transposition and deleterious effects on neighboring gene expression. Genome Res. 19:1419–1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hotaling S, Sproul JS, Heckenhauer J, Powell A, Larracuente AM, Pauls SU, Kelley JL, Frandsen PB. 2021. Long-reads are revolutionizing 20 years of insect genome sequencing. Genome Biol Evol. 13:evab138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izumitani HF, Kusaka Y, Koshikawa S, Toda MJ, Katoh T. 2016. Phylogeography of the subgenus Drosophila (Diptera: Drosophilidae): evolutionary history of faunal divergence between the old and the new worlds. PLoS One 11:e0160051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacques P-É, Jeyakani J, Bourque G. 2013. The majority of primate-specific regulatory sequences are derived from transposable elements. PLoS Genet 9:e1003504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelleher ES, Barbash DA. 2013. Analysis of piRNA-mediated silencing of active TEs in Drosophila melanogaster suggests limits on the evolution of host genome defense. Mol Biol Evol. 30:1816–1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khost DE, Eickbush DG, Larracuente AM. 2017. Single-molecule sequencing resolves the detailed structure of complex satellite DNA loci in Drosophila melanogaster. Genome Res. 27:709–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khurana JS, Wang J, Xu J, Koppetsch BS, Thomson TC, Nowosielska A, Li C, Zamore PD, Weng Z, Theurkauf WE. 2011. Adaptation to P element transposon invasion in Drosophila melanogaster. Cell 147:1551–1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidwell MG. 1992. Horizontal transfer of P elements and other short inverted repeat transposons. Genetica 86:275–286. [DOI] [PubMed] [Google Scholar]
- Kidwell MG. 2002. Transposable elements and the evolution of genome size in eukaryotes. Genetica 115:49–63. [DOI] [PubMed] [Google Scholar]
- Kidwell MG, Holyoake AJ. 2001. Transposon-induced hotspots for genomic instability. Genome Res. 11:1321–1322. [DOI] [PubMed] [Google Scholar]
- Kidwell MG, Kidwell JF, Sved JA. 1977. Hybrid dysgenesis in Drosophila melanogaster: a syndrome of aberrant traits including mutation, sterility and male recombination. Genetics 86:813–833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidwell MG, Lisch D. 1997. Transposable elements as sources of variation in animals and plants. Proc Natl Acad Sci U S A. 94:7704–7711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidwell MG, Novy JB. 1979. Hybrid dysgenesis in Drosophila melanogaster: sterility resulting from gonadal dysgenesis in the P–M system. Genetics 92:1127–1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Paggi JM, Park C, Bennett C, Salzberg SL. 2019. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat Biotechnol. 37:907–915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitagawa O, Wakahama K-I, Fuyama Y, Shimada Y, Takanashi E, Hatsumi M, Uwabo M, Mita Y. 1982. Genetic studies of the Drosophila nasuta subgroup, with notes on distribution and morphology. Jpn J Genet. 57:113–141. [Google Scholar]
- Kliman RM, Andolfatto P, Coyne JA, Depaulis F, Kreitman M, Berry AJ, McCarter J, Wakeley J, Hey J. 2000. The population genetics of the origin and divergence of the Drosophila simulans complex species. Genetics 156:1913–1931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kofler R, Hill T, Nolte V, Betancourt AJ, Schlötterer C. 2015. The recent invasion of natural Drosophila simulans populations by the P-element. Proc Natl Acad Sci USA. 112:6659–6663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kofler R, Senti KA, Nolte V, Tobler R, Schlötterer C. 2018. Molecular dissection of a natural transposable element invasion. Genome Res. 28(6):824–835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolmogorov M, Yuan J, Lin Y, Pevzner PA. 2019. Assembly of long, error-prone reads using repeat graphs. Nat Biotechnol. 37:540–546. [DOI] [PubMed] [Google Scholar]
- Koren S, Walenz BP, Berlin K, Miller JR, Bergman NH, Phillippy AM. 2017. Canu: scalable and accurate long-read assembly via adaptive -mer weighting and repeat separation. Genome Res. 27:722–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriventseva EV, Kuznetsov D, Tegenfeldt F, Manni M, Dias R, Simão FA, Zdobnov EM. 2019. OrthoDB v10: sampling the diversity of animal, plant, fungal, protist, bacterial and viral genomes for evolutionary and functional annotations of orthologs. Nucleic Acids Res. 47:D807–D811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurhanewicz NA, Dinwiddie D, Bush ZD, Libuda DE. 2020. Elevated temperatures cause transposon-associated DNA damage in C. elegans spermatocytes. Curr Biol. 30:5007–5017.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lachner M, O’Carroll D, Rea S, Mechtler K, Jenuwein T. 2001. Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature 410:116–120. [DOI] [PubMed] [Google Scholar]
- Lawlor MA, Cao W, Ellison CE. 2021. A transposon expression burst accompanies the activation of Y-chromosome fertility genes during Drosophila spermatogenesis. Nat Commun. 12(1):6854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YCG. 2015. The role of piRNA-mediated epigenetic silencing in the population dynamics of transposable elements in Drosophila melanogaster. PLoS Genet. 11:e1005269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YCG, Karpen GH. 2017. Pervasive epigenetic effects of Drosophila euchromatic transposable elements impact their evolution. Elife 6:e25762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Rouzic A, Capy P. 2005. The first steps of transposable elements invasion: parasitic strategy vs. genetic drift. Genetics 169:1033–1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levis RW. 1994. Transposons in place of telomeric repeats at a Drosophila telomere. Trends Genet. 10:77. [DOI] [PubMed] [Google Scholar]
- Li H. 2018. Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics (Oxford, England) 34(18):3094–3100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linheiro RS, Bergman CM. 2012. Whole genome resequencing reveals natural target site preferences of transposable elements in Drosophila melanogaster. PLoS One 7:e30008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locke J, Howard LT, Aippersbach N, Podemski L, Hodgetts RB. 1999. The characterization of DINE-1, a short, interspersed repetitive element present on chromosome and in the centric heterochromatin of Drosophila melanogaster. Chromosoma 108:356–366. [DOI] [PubMed] [Google Scholar]
- Loreto E, Carareto C, Capy P. 2008. Revisiting horizontal transfer of transposable elements in Drosophila. Heredity 100:545–554. [DOI] [PubMed] [Google Scholar]
- Luo S, Zhang H, Duan Y, Yao X, Clark AG, Lu J. 2020. The evolutionary arms race between transposable elements and piRNAs in Drosophila melanogaster. BMC Evol Biol. 20:14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahajan S, Wei KH-C, Nalley MJ, Gibilisco L, Bachtrog D. 2018. De novo assembly of a young Drosophila Y chromosome using single-molecule sequencing and chromatin conformation capture. PLoS Biol. 16:e2006348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mai D, Nalley MJ, Bachtrog D. 2020. Patterns of genomic differentiation in the Drosophila nasuta species complex. Mol Biol Evol. 37:208–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mateo L, Ullastres A, González J. 2014. A transposable element insertion confers xenobiotic resistance in Drosophila. PLoS Genet. 10:e1004560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews BJ, Dudchenko O, Kingan SB, Koren S, Antoshechkin I, Crawford JE, Glassford WJ, Herre M, Redmond SN, Rose NH, et al. 2018. Improved reference genome of Aedes aegypti informs arbovirus vector control. Nature 563:501–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGurk MP, Barbash DA. 2018. Double insertion of transposable elements provides a substrate for the evolution of satellite DNA. Genome Res. 28:714–725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mérel V, Boulesteix M, Fablet M, Vieira C. 2020. Transposable elements in Drosophila. Mobile DNA 11:23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merenciano M, Ullastres A, de Cara MAR, Barrón MG, González J. 2016. Multiple independent retroelement insertions in the promoter of a stress response gene have variable molecular and functional effects in Drosophila. PLoS Genet. 12:e1006249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naito K, Cho E, Yang G, Campbell MA, Yano K, Okumoto Y, Tanisaka T, Wessler SR. 2006. Dramatic amplification of a rice transposable element during recent domestication. Proc Natl Acad Sci U S A. 103(47):17620–17625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama J, Rice JC, Strahl BD, Allis CD, Grewal SI. 2001. Role of histone H3 lysine 9 methylation in epigenetic control of heterochromatin assembly. Science 292:110–113. [DOI] [PubMed] [Google Scholar]
- Nurk S, Koren S, Rhie A, Rautiainen M, Bzikadze AV, Mikheenko A, Vollger MR, Altemose N, Uralsky L, Gershman A, et al. 2022. The complete sequence of a human genome. Science 376:44–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Grady PM, DeSalle R. 2018. Phylogeny of the genus Drosophila. Genetics 209:1–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Neill K, Brocks D, Hammell MG. 2020. Mobile genomics: tools and techniques for tackling transposons. Philos Trans R Soc Lond B Biol Sci. 375:20190345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozata DM, Gainetdinov I, Zoch A, O’Carroll D, Zamore PD. 2019. PIWI-interacting RNAs: small RNAs with big functions. Nat Rev Genet. 20:89–108. [DOI] [PubMed] [Google Scholar]
- Panta M, Mishra A, Hoque MT, Atallah J. 2021. ClassifyTE: a stacking-based prediction of hierarchical classification of transposable elements. Bioinformatics. 37:2529–2536. [DOI] [PubMed] [Google Scholar]
- Parhad SS, Theurkauf WE. 2019. Rapid evolution and conserved function of the piRNA pathway. Open Biol. 9:180181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pertea M, Pertea GM, Antonescu CM, Chang T-C, Mendell JT, Salzberg SL. 2015. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat Biotechnol. 33:290–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrov DA, Schutzman JL, Hartl DL, Lozovskaya ER. 1995. Diverse transposable elements are mobilized in hybrid dysgenesis in Drosophila virilis. Proc Natl Acad Sci U S A. 92(17):8050–8054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM. 2010. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26:841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman R, Chirn G-W, Kanodia A, Sytnikova YA, Brembs B, Bergman CM, Lau NC. 2015. Unique transposon landscapes are pervasive across Drosophila melanogaster genomes. Nucleic Acids Res. 43:10655–10672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranjini MS, Ramachandra NB. 2013. Rapid evolution of a few members of nasuta-albomicans complex of Drosophila: study on two candidate genes, Sod1 and Rpd3. J Mol Evol. 76:311–323. [DOI] [PubMed] [Google Scholar]
- Ratner VA, Zabanov SA, Kolesnikova OV, Vasilyeva LA. 1992. Induction of the mobile genetic element Dm-412 transpositions in the Drosophila genome by heat shock treatment. Proc Natl Acad Sci U S A. 89:5650–5654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards EJ, Elgin SCR. 2002. Epigenetic codes for heterochromatin formation and silencing: rounding up the usual suspects. Cell 108:489–500. [DOI] [PubMed] [Google Scholar]
- Ruan J, Li H. 2020. Fast and accurate long-read assembly with wtdbg2. Nat Methods 17:155–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaack S, Gilbert C, Feschotte C. 2010. Promiscuous DNA: horizontal transfer of transposable elements and why it matters for eukaryotic evolution. Trends Ecol Evol. 25:537–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaefer RE, Kidwell MG, Fausto-Sterling A. 1979. Hybrid dysgenesis in Drosophila melanogaster: morphological and cytological studies of ovarian dysgenesis. Genetics 92:1141–1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serio C, Castiglione S, Tesone G, Piccolo M, Melchionna M, Mondanaro A, Di Febbraro M, Raia P. 2019. Macroevolution of toothed whales exceptional relative brain size. Evol Biol. 46:332–342. [Google Scholar]
- Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Söding J, et al. 2011. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol. 7:539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva JC, Loreto ELS, Clark JB. 2004. Factors that affect the horizontal transfer of transposable elements. Curr Issues Mol Biol. 6:57–71. [PubMed] [Google Scholar]
- Silva DG, Schmitz HJ, de Medeiros HF, Rohde C, Montes MA, Garcia ACL. 2020. Geographic expansion and dominance of the invading species Drosophila nasuta (Diptera, Drosophilidae) in Brazil. J Insect Conserv. 24:525–534. [Google Scholar]
- Simkin A, Wong A, Poh Y-P, Theurkauf WE, Jensen JD. 2013. Recurrent and recent selective sweeps in the piRNA pathway. Evolution 67:1081–1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smit AFA, Hubley R, Green P. 2013–2015. RepeatMasker Open-4.0. http://www.repeatmasker.org. (Accessed May 18, 2021).
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30:1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundaram V, Wysocka J. 2020. Transposable elements as a potent source of diverse cis-regulatory sequences in mammalian genomes. Philos Trans R Soc Lond B Biol Sci. 375:20190347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teixeira FK, Okuniewska M, Malone CD, Coux RX, Rio DC, Lehmann R. 2017. piRNA-mediated regulation of transposon alternative splicing in the soma and germ line. Nature 552(7684):268–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traverse KL, Pardue ML. 1988. A spontaneously opened ring chromosome of Drosophila melanogaster has acquired He-T DNA sequences at both new telomeres. Proc Natl Acad Sci U S A. 85:8116–8120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaser R, Sović I, Nagarajan N, Šikić M. 2017. Fast and accurate de novo genome assembly from long uncorrected reads. Genome Res. 27:737–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vendrell-Mir P, Barteri F, Merenciano M, González J, Casacuberta JM, Castanera R. 2019. A benchmark of transposon insertion detection tools using real data. Mob DNA 10:53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilela CR, Goñib B. 2015. Is Drosophila nasuta Lamb (Diptera, Drosophilidae) currently reaching the status of a cosmopolitan species? Rev Bras Entomol. 59:346–350. [Google Scholar]
- Villanueva-Cañas JL, Horvath V, Aguilera L, González J. 2019. Diverse families of transposable elements affect the transcriptional regulation of stress-response genes in Drosophila melanogaster. Nucleic Acids Res. 47:6842–6857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker BJ, Abeel T, Shea T, Priest M, Abouelliel A, Sakthikumar S, Cuomo CA, Zeng Q, Wortman J, Young SK, et al. 2014. Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS One 9:e112963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Lin H. 2021. Roles of piRNAs in transposon and pseudogene regulation of germline mRNAs and lncRNAs. Genome Biol. 22:27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei KH-C, Bachtrog D. 2019. Ancestral male recombination in Drosophila albomicans produced geographically restricted neo-Y chromosome haplotypes varying in age and onset of decay. PLoS Genet. 15:e1008502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei KH-C, Chan C, Bachtrog D. 2021. Establishment of H3K9me3-dependent heterochromatin during embryogenesis in Drosophila miranda. Elife 10:e55612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei KH-C, Gibilisco L, Bachtrog D. 2020. Epigenetic conflict on a degenerating Y chromosome increases mutational burden in Drosophila males. Nat Commun. 11:5537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wessler SR. 1996. Plant retrotransposons: turned on by stress. Curr Biol. 6:959–961. [DOI] [PubMed] [Google Scholar]
- Wilson FD, Wheeler MR, Harget M, Kambysellis M. 1969. Cytogenetic relations in the Drosophila nasuta subgroup of the immigrans group of species. In: Studies in Genetics V. Univ Texas Publ. p. 207–270. [Google Scholar]
- Woodruff RC, Blount JL, Thompson JN Jr. 1987. Hybrid dysgenesis in D. melanogaster is not a general release mechanism for DNA transpositions. Science (New York, N.Y.) 237(4819):1206–1218. [DOI] [PubMed] [Google Scholar]
- Xiao YL, Li X, Peterson T. 2000. Ac insertion site affects the frequency of transposon-induced homologous recombination at the maize p1 locus. Genetics 156:2007–2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing J, Wang H, Belancio VP, Cordaux R, Deininger PL, Batzer MA. 2006. Emergence of primate genes by retrotransposon-mediated sequence transduction. Proc Natl Acad Sci U S A. 103:17608–17613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H-P, Barbash DA. 2008. Abundant and species-specific DINE-1 transposable elements in 12 Drosophila genomes. Genome Biol. 9:R39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H-H, Peccoud J, Xu M-R-X, Zhang X-G, Gilbert C. 2020. Horizontal transfer and evolution of transposable elements in vertebrates. Nat Commun. 11:1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Yu C, Krishnaswamy L, Peterson T. 2011. Transposable elements as catalysts for chromosome rearrangements. Methods Mol Biol. 701:315–326. [DOI] [PubMed] [Google Scholar]
- Zhou Q, Bachtrog D. 2012. Chromosome-wide gene silencing initiates Y degeneration in Drosophila. Curr Biol. 22:522–525. [DOI] [PubMed] [Google Scholar]
- Zhou Q, Ellison CE, Kaiser VB, Alekseyenko AA, Gorchakov AA, Bachtrog D. 2013. The epigenome of evolving Drosophila neo-sex chromosomes: dosage compensation and heterochromatin formation. PLoS Biol. 11:e1001711. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All raw reads are deposited on the SRA under BioProject PRJNA736413. Genome assemblies, TE libraries, gene, and TE annotations can be found on Dryad at https://doi.org/10.6078/D11B01.