Abstract
Several knowledgebases are manually curated to support clinical interpretations of thousands of hotspot somatic mutations in cancer. However, discrepancies or even conflicting interpretations are observed among these databases. Furthermore, many previously undocumented mutations may have clinical or functional impacts on cancer but are not systematically interpreted by existing knowledgebases. To address these challenges, we developed CancerVar to facilitate automated and standardized interpretations for 13 million somatic mutations based on the AMP/ASCO/CAP 2017 guidelines. We further introduced a deep learning framework to predict oncogenicity for these variants using both functional and clinical features. CancerVar achieved satisfactory performance when compared to several independent knowledgebases and, using clinically curated datasets, demonstrated practical utility in classifying somatic variants. In summary, by integrating clinical guidelines with a deep learning framework, CancerVar facilitates clinical interpretation of somatic variants, reduces manual work, improves consistency in variant classification, and promotes implementation of the guidelines.
CancerVar is an automated tool for somatic variant interpretation based on the AMP/ASCO/CAP 2017 guidelines and deep learning.
INTRODUCTION
A large number of somatic variants have been identified by next-generation sequencing (NGS) during clinical oncology practice to facilitate precision medicine (1, 2). To better understand the clinical impacts of somatic variants in cancer, several knowledgebases have been curated, including OncoKB (1), My Cancer Genome (3), CIViC (4), Precision Medicine Knowledge Base (5), JAX Clinical Knowledgebase (CKB) (6), and Cancer Genome Interpreter (7). Although clinically relevant, interpretation of somatic variants is not a standardized practice, and different clinical groups often generate different or even conflicting results. To standardize clinical interpretation of somatic variants in cancer and support clinical decision-making, the Association for Molecular Pathology (AMP), American Society of Clinical Oncology (ASCO), and College of American Pathologists (CAP) jointly proposed standards and guidelines for the interpretation and reporting of somatic variants, classifying somatic variants into four tiers: strong clinical significance (tier I), potential clinical significance (tier II), uncertain significance (tier III), and benign (tier IV) (8). These AMP/ASCO/CAP 2017 guidelines incorporate 12 pieces of evidence, including diagnostic, prognostic, and therapeutic clinical evidence; mutation type; variant allele fraction [mosaic variant frequency (likely somatic) and nonmosaic variant frequency (potential germline)]; population databases; germline databases; somatic databases; predictive results of different computational algorithms; pathway involvement; and publications (8, 9).
However, as the AMP/ASCO/CAP classification scheme heavily relies on published clinical evidence for a given variant, ambiguous assignments among human curators frequently occur when using the same evidence for a given variant. For example, Sirohi et al. (10) compared human classifications for 51 variants by 20 randomly selected molecular pathologists from 10 institutions. The original overall observed agreement was only 58%; when providing the same evidential data for variants to the pathologists, the agreement rate of reclassification increased to 70%. The reasons for such discordance are as follows: (i) gathering information/evidence is complicated and may not be reproducible by the same interpreter at different time points; (ii) different researchers may use different algorithms, cutoffs, and parameters, rendering interpretation less reproducible; and (iii) newly published evidence for certain variants may not be incorporated into the evaluation system instantly and systematically, which is especially relevant for variants of unknown significance (VUSs).
To standardize interpretation of somatic variants across multiple knowledgebases, a more recently published knowledgebase, MetaKB from The Variant Interpretation for Cancer Consortium, has aggregated evidence based on the AMP/ASCO/CAP 2017 guidelines (11). However, this MetaKB knowledgebase also has the following limitations: (i) it only focuses on consensus interpretations for a limited number of known hotspot mutations, such that a large number of variants are now classified as unknown clinical significance, but they may be oncogenic through “loss of function” or “gain of function” in cancer; (ii) it only provides a summarized classification for each variant, without demonstrating itemized evidence in detail when mapping to the 12 criteria of the AMP/ASCO/CAP 2017 guidelines, and therefore, users cannot conduct customized evaluations based on their own protocols, experiences, and updated clinical knowledge; and (iii) it uses a simple scoring system to rank driver mutations without considering heterogeneity of functional consequences (such as predictions of deleteriousness) of variants, especially for newly identified variants reported in the literature.
In clinical practice, when a somatic mutation is considered to have strong confidence in causing a functional impact on protein changes, clinicians likely interpret it with clinical significance or likely clinical significance (12, 13). Although a number of useful software tools (2, 14–22), especially sorting intolerant from tolerant (SIFT) (23), PolyPhen-2 (24), and functional analysis through hidden Markov models (FATHMM) (21, 25), have been developed to predict functional impacts, disagreements regarding certain mutations are consistently observed. Some of these tools use similar background information based on alignment, evolutionary conservation, and homology, such as MutationAssessor (20), FATHMM (21, 25), cancer-specific high-throughput annotation of somatic mutations (CHASM) based on random forest (RF) classifier (15), and CanDrA based on support vector machine (SVM) (16); others use consensus information by integrating multiple sources of information from many computational tools, such as combined tool adjusted total CTAT-cancer (2). Although some meta-analysis tools, such as deleterious annotation of genetic variants using neural networks (DANN) (26) and DriverPower (27), were later developed to prioritize functionally important variants using more comprehensive functional scoring features as input, they have limitations in jointly modeling clinical impact features based on the AMP/ASCO/CAP guidelines. Because the guidelines tend to be conservative (“negative diagnosis” is preferred to “wrong diagnosis”), more variants than expected were misinterpreted as VUSs (28–36). In addition, the AMP/ASCO/CAP guidelines only designate seven functional impact prediction tools, such as MutationAssessor (20), as the official recommended tools, and only the variant from majority voting (more than four from seven tools) can be considered “with clinical significance,” which oversimplifies the heterogeneity of predictions of functional consequence in cancer progression. Although using existing tools may be useful in the prediction of the overall impacts of cancer driver genes, it may not be optimal for prioritizing novel mutations in these genes. In addition to the in silico predictive methods above, interactome network approaches have attracted much attention for identifying oncogenic variants in cancer through genotype-phenotype studies (37, 38). In interactome networks, certain perturbed mutations (network nodes) can disrupt certain signaling pathways and protein-protein interactions (PPIs), resulting in similar cancer phenotypes in different patients. These perturbed mutations, termed “edgetic” mutations, are functionally important but are understudied with existing cancer variant interpretation tools (39, 40). To address these challenges and improve automated clinical interpretations of somatic variants in cancer, there is a strong need for reliable and accurate computational methods using both clinical evidence and functional impact score features.
We previously developed the standalone software VIC written in Java, which was among the first tools for interpreting clinical impacts of somatic variants using a rule-based scoring system based on 12 criteria of the AMP/ASCO/CAP 2017 guidelines (9). In the current study, we developed an improved somatic variant interpretation tool called CancerVar (cancer variant interpretation) implemented in Python with an accompanying web server. Compared to VIC, CancerVar is a markedly improved tool providing more options to users: (i) Python implementation provides more flexibility to incorporate CancerVar into custom command-line workflows; (ii) CancerVar involves a user-friendly web server with precomputed clinical evidence for 13 million variants from 1911 cancer census genes through literature mining and database aggregations; (iii) we use a flexible AMP/ASCO/CAP rule-based score system and a deep learning–based scoring system that allows for improved interpretations; and (iv) RESTful application programming interface (API) is used to enable developers to freely access complied knowledge. CancerVar allows users to query clinical interpretations for variants using the chromosome position, cDNA change, or protein change and interactively fine-tune weights of scoring features based on prior knowledge or additional user-specified criteria. The CancerVar web server generates automated text with summarized descriptive interpretations, such as diagnostic, prognostic, targeted drug responses, and clinical trial information for many hotspot mutations, significantly reducing the workload of human reviewers in drafting clinical reports in the practice of precision oncology.
RESULTS
Summary of the functionality of the CancerVar web server
The CancerVar web server provides multiple query options at variant, gene, and copy number alteration (CNA) levels across 30 cancer types and two versions of reference genomes: hg19 (GRCh37) and hg38 (GRCh38). With user-supplied input, CancerVar generates an output web page, with information organized as cards, including interpretation summary, gene overview, mutation information, evidence overview, pathways, clinical publications, protein domains, in silico predictions, and exchangeable information from other knowledgebases. The CancerVar web server provides full details for variants, including all automatically generated criteria, most of the supporting evidence, and predictive scores for clinical significance. In the CancerVar webserver, we compiled a variant database from a list of 1911 cancer censuses or driver genes with 13 million exonic variants from seven existing cancer knowledge-bases, including catalogue of somatic mutations in cancer (COSMIC), CIViC, and OncoKB, and two datasets collected from the literature about driver gene predictions (table S1). CancerVar can be accessed at https://cancervar.wglab.org, and the Python command-line program can be downloaded at https://github.com/WGLab/CancerVar.
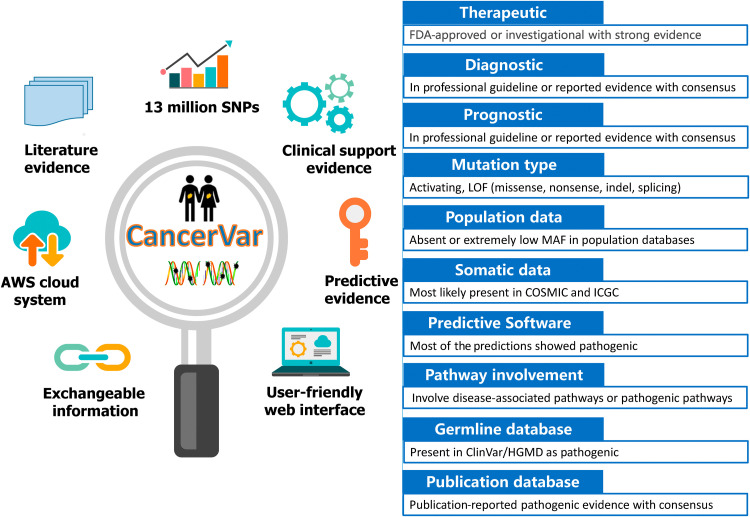
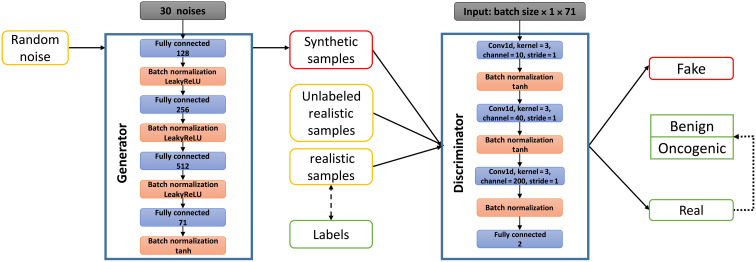
By implementing a rule-based approach, users have the ability to manually adjust the criteria and perform reinterpretation based on their prior knowledge or experience. If the user already knows the information of each of the scoring criteria for the variant (possibly inferred by using other software tools), they can alternatively compute the clinical significance of the variant from the “Interpret by Criteria” service. Each variant is provided with a prediction score and clinical classification categories, which are strong clinical significance, potential clinical significance, uncertain significance, and likely benign/benign, based on the 12 criteria of the AMP/ASCO/CAP 2017 guidelines. Figure 1 provides descriptions of 12 types of evidence and summarizes the functionality of the CancerVar server. Descriptions of the complete scoring system for each piece of evidence can be found in table S2. In addition, we developed a scoring method for oncogenic prioritization by artificial intelligence (OPAI). Using a deep learning–based approach, CancerVar provides a probability score predicted by OPAI to determine the oncogenicity of a variant using 12 evidence features from the AMP/ASCO/CAP guidelines and 23 functional features with scoring metrics predicted by various computational tools. However, the absolute value of the predictive score cannot provide meaningful information, and the classification greatly depends on the cutoff of the predictive score. Unlike typical deep learning algorithms that train a predictive model from a set of positive and negative training samples, OPAI is based on a semisupervised generative adversarial network that includes observed but unclassified somatic mutations from real-world sequencing data for patients with cancer. Figure 2 illustrates the workflow and architecture of the generator and discriminator/classifier used in the generative adversarial network of OPAI. The OPAI’s functionality and user instructions are already integrated within CancerVar software, allowing users to predict their own variants in the future.
Fig. 1. Summary of the functionality of CancerVar and descriptions of 12 types of evidence.
AWS, Amazon Web Services; LOF, Loss of Function; MAF, Minor allele frequency; HGMD, Human Gene Mutation Database.
Fig. 2. Workflow and architecture of the generator and discriminator/classifier used in OPAI.
The generator contains three linear layers with batch normalization, LeakyReLu as the activation layer, and a 60% dropout rate in each layer. The final layer is a linear layer with batch normalization and tanh as the activation layer. For the discriminator we implemented three Convolutional Neural Network (CNN) layers with tanh as the activation layer.
Comparative evaluations of CancerVar with human interpreters
Sirohi et al. (10) measured the reliability of the 2017 AMP/ASCO/CAP guidelines using 51 variants [31 single-nucleotide variants (SNVs), 14 insertions and deletions (indels), 5 CNAs, and 1 fusion] based on a literature review. Among these variants, we selected 43, including all 31 SNVs and 12 indel variants (we did not find alternative allele information for two indels in the CHEK1 and MET genes). CancerVar interpreted the clinical impacts of these 43 variants with respect to the specified cancer types. As these 43 variants do not have solid/consistent clinical interpretation, we compared 20 pathologists’ opinions from 10 institutions with CancerVar’s predictions. As shown in Table 1, CancerVar assigned 21 variants as tier I/II (strong or potential clinical significance). Among these 21 variants, the pathologists classified 17 variants (17 of 21, approximately 81%) as tier I/II, in agreement with CancerVar. Moreover, CancerVar assigned 21 variants as VUS; among these 21 variants, 9 variants (9 of 21, approximately 43%) were also classified as VUS by pathologists. In total, the clinical significance of 26 variants (approximately 61%) matched between human interpreters and CancerVar.
Table 1. Comparison of classification of 43 variants between 20 pathologists and CancerVar.
| Annotators | Classifications | 20 Pathologists | |||
| I/II* | III | IV | Total | ||
| CancerVar | I/II | 17 | 4 | 0 | 21 |
| III | 12 | 9 | 0 | 21 | |
| IV | 0 | 1 | 0 | 1 | |
| Total | 29 | 14 | 0 | 43 | |
*Tier I, strong clinical significance; tier II, potential clinical significance; tier III, variants of unknown clinical significance (VUS); and tier IV, benign/likely benign.
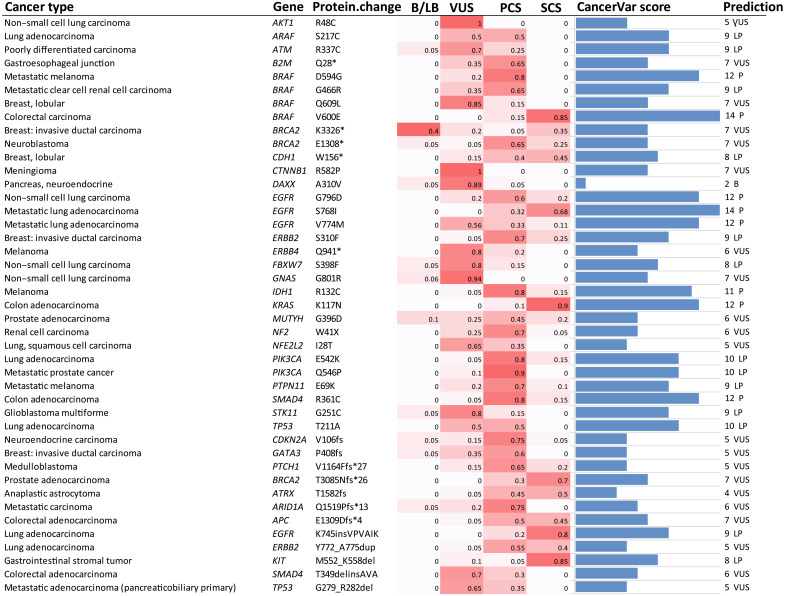
Details regarding the interpretation of these 43 variants can be found in table S3 and Fig. 3. Compared to human interpreters, the advantage of CancerVar is clear, in that it can automatically generate clinical interpretations with standardized, consistent, and reproducible workflows, with evidence-based support for each of the 12 mentioned criteria. Therefore, CancerVar can greatly reduce the workload of human reviewers and facilitate the generation of precise and reproducible clinical interpretations.
Fig. 3. Comparison of the interpretation of 43 variants between 20 pathologists and CancerVar.
The heatmap shows the ratio of 20 pathologists voting for the four tiers: tier I, strong clinical significance (SCS); tier II, potential clinical significance (PCS); tier III, variant of uncertain clinical significance (VUS); and tier IV, benign/likely benign (B/LB). The last two columns are CancerVar-predicted scores and classifications. CancerVar showed an 81% (17 of 21) agreement rate with pathologists’ majority voting for tier I/II and a 60.5% (26 of 43) agreement rate for all tiers. This agreement rate is comparable to the 58% agreement rate among the 20 pathologists, but CancerVar can automate the interpretation process. P, Pathogenic/strong clinical significance; LP:Likely Pathogenic/potential clinical significance; B:(Likely)Benign.
Benchmark studies on OncoKB annotations
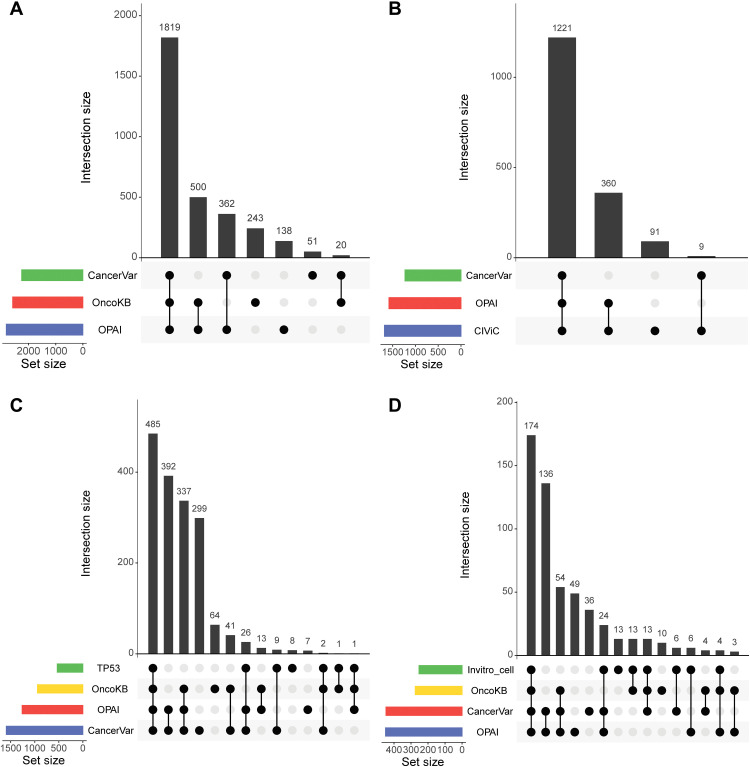
OncoKB (1), a manually curated database of somatic mutations with oncogenic effects, is widely used in the cancer research community. OncoKB provides an evidence-based classification system to interpret somatic variants and classifies them as inconclusive, likely neutral, predicted oncogenic, likely oncogenic, or oncogenic. In total, 3455 SNVs involving 245 genes were downloaded from the OncoKB annotation database (downloaded 1 March 2020). This version contains 2582 oncogenic/likely oncogenic (O/LO) mutations, 587 likely neutral mutations, and 286 mutations annotated as inconclusive for this study. CancerVar evidence-based and deep learning–based prediction methods were applied to classify the mutations and compared them with OncoKB classifications. For the O/LO group in OncoKB, the CancerVar rule-based method classified 1839 (1839 of 2582, 71.2% consistent with OncoKB classification) variants as having strong or potential clinical significance; the CancerVar deep learning–based method (OPAI) classified 2319 variants (2319 of 2582, 90% consistent with OncoKB classification). The details are given in Table 2, Fig. 4A, and fig. S1. An UpSet plot shows the intersections of predictions between OncoKB, the CancerVar rule-based method, and the deep learning–based method (OPAI). Although most variants were consistently interpreted by the three methods, OPAI appears to be more concordant with OncoKB than rule-based interpretations.
Table 2. Summary of CancerVar prediction on OncoKB mutations.
| OncoKB | Model-based CancerVar (OPAI) | Rule-based CancerVar | |||
| Oncogenic | Neutral | I/II (S/P*) | III (VUS*) | IV (Benign) | |
| Oncogenic/likely oncogenic |
2319 | 263 | 1839 | 690 | 53 |
| Neutral/likely neutral | 348 | 239 | 281 | 279 | 27 |
| Inconclusive | 152 | 134 | 132 | 150 | 4 |
| Total | 2819 | 636 | 2252 | 1119 | 84 |
*S, strong clinical significance (tier I); P, potential clinical significance (tier II); VUS, variant of unknown clinical significance (tier III).
Fig. 4. UpSet plot highlighting the intersection of multiple methods with oncogenic prediction from different datasets.
(A) Mutations were taken from the OncoKB dataset. (B) Mutations were taken from CIViC. (C) Mutations were taken from the IARC TP53 transactivation dataset. (D) Mutations were taken from in vitro cell viability by Ng et al. (42).
Benchmark studies on CIViC annotations
CIViC is a crowdsourced and expert-moderated public resource for somatic variants in cancer (4) with five evidence levels to differentiate reported mutations, namely, A: validated, B: clinical, C: case study, D: preclinical, and E: inferential. In total, 1681 unique SNVs/indels of 113 unique genes were retrieved from the CIViC website (https://civicdb.org/releases, accessed 1 May 2020) and assessed by CancerVar. The CancerVar rule-based method predicted 1230 (1230 of 1681, 73.2% consistent with CIViC classification) variants as having strong or potential clinical significance, whereas the CancerVar deep learning–based method OPAI predicted 1581 (94.1% consistent with CIViC classification) variants. Table 3 and Fig. 4B show the details of the CancerVar predictions. Similar to what we report above, OPAI appears to be more concordant with CIViC than rule-based interpretations.
Table 3. Summary of CancerVar prediction of CIViC mutations.
| CIViC | Model-based CancerVar (OPAI) | Rule-based CancerVar | ||||
| Oncogenic | Neutral | I (Strong) | II (Potential) | III (VUS) | IV (Benign) | |
| A: Validated | 17 | 2 | 2 | 7 | 0 | 10 |
| B: Clinical | 259 | 40 | 111 | 109 | 61 | 18 |
| C: Case study | 792 | 30 | 178 | 439 | 198 | 7 |
| D: Preclinical | 466 | 22 | 91 | 277 | 119 | 1 |
| E: Inferential | 47 | 6 | 5 | 11 | 33 | 4 |
| Total | 1581 | 100 | 387 | 843 | 411 | 40 |
Benchmark studies on IARC TP53 transactivation mutations
TP53 is the most frequently mutated gene in human cancer, and many of its mutants have been functionally assessed based on median transactivation levels and compiled in the International Agency for Research on Cancer (IARC) TP53 database (41). On the basis of the median of eight different yeast functional assays (WAF1, MDM2, BAX, h1433s, AIP1, GADD45, noxa, and P53R2), TP53 mutations can be classified as oncogenic, resulting in lower transactivation (a median transactivation level ≤ 25% wild type), or neutral, resulting in higher transactivation (level ≥ 25% wild type). We retrieved 1915 missense mutations (532 mutations used as oncogenic cases and 1383 mutations used as neutral cases) from the IARC TP53 database. For 532 oncogenic mutations, the CancerVar rule-based method predicted 522 [true-positive rate (TPR) = 98%] variants, and the model-based method predicted 512 (TPR = 96.2%) variants as having strong/potential clinical significance. Compared to OncoKB, which predicted 489 (489 of 532 = 91.9%) oncogenic variants, CancerVar rule-based and deep learning–based methods have a higher TPR. The details of CancerVar and OncoKB prediction are shown in Fig. 4C, table S4, and fig. S2.
Benchmark studies on cell viability in vitro assays
The oncogenic effects of somatic mutations can be directly assessed by preferential growth or survival advantage to cells using cellular assays. Ng et al. (42) recently developed a medium-throughput in vitro system to assess the functional effects of mutations using two growth factor–dependent cell lines: Ba/F3 (a sensitive leukemia cell line frequently used in drug screening) and MCF10A (a breast epithelial cell line). Cell viability data of mutations in these two cell lines were used to generate consensus functional annotation to interpret the functional impacts of somatic mutations. The mutations were considered oncogenic when cell viability was labeled as activating and neutral when labeled as neutral from the consensus functional annotation. Although we recognize that results from in vitro assays cannot be directly translated to the clinic, they offer a comprehensive and unbiased way of assessing the potential functional significance of somatic variants. From the published study, we retrieved 717 missense mutations (253 oncogenic and 464 neutral) in 44 genes. For 253 oncogenic variants, the CancerVar rule-based method predicted 217 (TP = 85.7%) and the deep learning–based method predicted 208 (82.2%) as having strong/potential clinical significance; OncoKB predicted 204 (TP = 80.6%) variants as oncogenic, likely, or predicted oncogenic (Fig. 4D, table S5, and fig. S3). Therefore, CancerVar rule-based and deep learning–based methods perform slightly better than OncoKB.
Performance of OPAI in predicting oncogenic variants
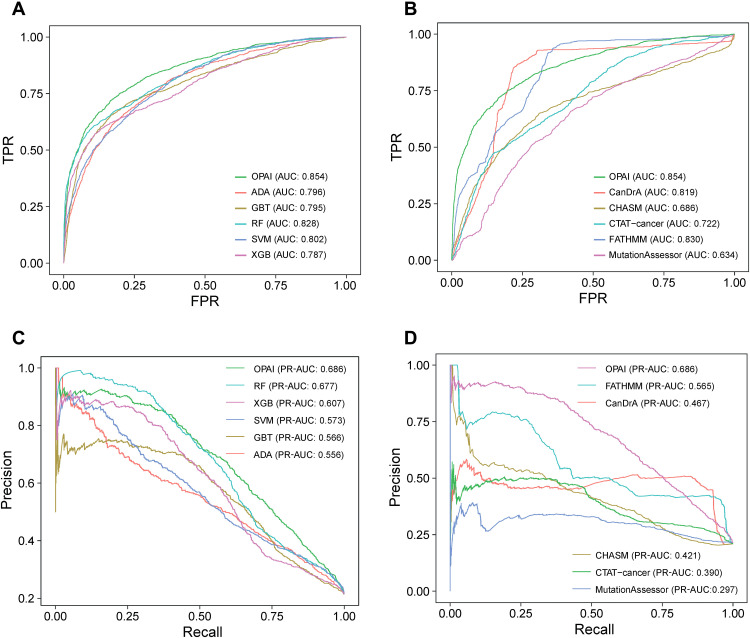
One unique feature of CancerVar is the inclusion of the OPAI approach, which allows prediction of oncogenic variants that have never been reported in public databases. Unlike conventional models trained on labeled data, OPAI can learn the hidden distribution of unlabeled mutations collected from clinical data. In the current study, training the OPAI models required ~100 hours with 1000 epochs on an Nvidia Tesla M40 GPU. Next, we assessed the ability to predict novel oncogenic variants. OPAI was compared with five other machine learning algorithms, including gradient boosting tree, SVM, AdaBoost, RF, and XGBoost, using the Python package scikit-learn (43). We further compared performance with the other five cancer-specific driver mutation analysis tools, including CanDrA, CHASM, CTAT-cancer, and MutationAssessor, using the area under the curve (AUC) score from receiver operating characteristic (ROC) plots and the true-negative rate (or specificity) as performance measurements. To evaluate imbalanced classes, we also calculated the area under precision-recall curve (AUPRC) to assess the predictive performance of various models. On the basis of an independent testing set of 6226 somatic variants, Fig. 5 (A and B) shows that when using the AUC measure, the OPAI method in CancerVar (AUC-ROC = 0.854) performed the best compared to cancer-specific driver predicting methods and any individual functional prediction tool. Figure 5 (C and D) highlights that OPAI performed the best (AUPRC = 0.686) among these methods. Because of the imbalanced classes, that is, the fraction of positives (oncogenic) was less than that of negatives (benign), it was expected that OPAI’s AUPRC value would be lower than the AUC-ROC value.
Fig. 5. Performance comparisons.
(A and B) Receiver operating characteristic (ROC) curves for performance comparison between OPAI and five other machine learning algorithms, including gradient boosting tree (GBT), support vector machine (SVM), AdaBoost (ADA), random forest (RF), and XGBoost (XGB), and five other in silico predictive tools using 6226 somatic mutations as the testing set. (C and D) Area under the precision-recall curve (AUPRC) comparison between OPAI and five other machine learning tools and in silico predictive tools. OPAI outperformed any individual tool in the prediction of somatic driver mutations in cancer. TPR, true-positive rate; FPR, false-positive rate.
Evaluation of FDA-approved or -recognized cancer biomarkers for therapeutic, diagnostic, and prognostic purposes
To further evaluate the performance and reliability of CancerVar in clinical applications, we collected 22 cancer biomarkers specified by the U.S. Food and Drug Administration (FDA) and interpreted these biomarkers and predicted their oncogenicity. Among them, 9 were classified as tier I strong clinical significance when using only the evidence-based method; the other 13 biomarkers were classified as tier II potential clinical significance. For the OPAI model, most of the biomarkers (19 of 22) were predicted to have scores ≥0.95, suggesting a very high probability of having clinical significance. The interpretation of these biomarkers is shown in Table 4.
Table 4. FDA-approved or -recognized biomarkers (therapeutic, diagnostic, and prognostic) from the rule-based model and the deep learning model (OPAI) in CancerVar.
| Gene | Alternation | Cancers | Levels | Rule-based CancerVar | OPAI |
| ABL1 | T315I | B-lymphoblastic leukemia/lymphoma/ chronic myelogenous leukemia |
Therapeutic (ponatinib) | Tier II (score 8) | 0.93 |
| AKT1 | E17K | Breast cancer/ovarian cancer/endometrial cancer |
Therapeutic (AZD5363) | Tier II (score 10) | 0.98 |
| BRAF | V600E | Melanoma/non–small cell lung cancer |
Therapeutic (dabrafenib + trametinib; vemurafenib) |
Tier I (score 11) | 0.98 |
| EGFR | T790M | Non–small cell lung cancer |
Therapeutic (osimertinib) |
Tier II (score 9) | 0.98 |
| EGFR | L861Q | Non–small cell lung cancer |
Therapeutic (afatinib) | Tier II (score 10) | 0.99 |
| EGFR | S768I | Non–small cell lung cancer |
Therapeutic (afatinib) | Tier I (score 11) | 0.99 |
| EZH2 | Y646F | Follicular lymphoma | Therapeutic (tazemetostat) |
Tier I (score 11) | 0.99 |
| Y646H | Tier I (score 11) | 0.99 | |||
| Y646N | Tier I (score 11) | 0.99 | |||
| Y646S | Tier I (score 11) | 0.99 | |||
| FGFR3 | R248C | Bladder cancer | Therapeutic (erdafitinib) |
Tier II (score 9) | 0.99 |
| S249C | Tier II (score 9) | 0.99 | |||
| Y373C | Tier II (score 10) | 0.99 | |||
| JAK2 | V617F | Primary myelofibrosis | Prognostic | Tier I (score 11) | 0.87 |
| KIT | A829P | Gastrointestinal stromal tumor |
Therapeutic (imatinib, regorafenib, ripretinib, and sunitinib) |
Tier II (score 9) | 0.83 |
| T670I | Tier II (score 9) | 0.98 | |||
| V654A | Tier I (score 11) | 0.99 | |||
| Y823D | Tier II (score 10) | 0.99 | |||
| D816V | Systemic mastocytosis | Diagnostic | Tier II (score 9) | 0.99 | |
| KRAS | G12C | Non–small cell lung cancer |
Therapeutic (AMG-510) | Tier I (score 11) | 0.99 |
| PDGFRA | D842V | Gastrointestinal stromal tumor |
Therapeutic (avapritinib) |
Tier II (score 9) | 0.97 |
| D842Y | Tier II (score 10) | 0.99 |
Oncogenicity prediction of edgetic mutations
Gene products can interact with each other in PPI networks or cellular signaling networks. Network rewiring is crucial for understanding the complex genotype-phenotype relationships in cancer (37, 39). Some cancer mutations have been found to broadly or specifically affect these interactome networks or pathways via perturbation (38). These mutations in cancer are called edgetic mutations, and they may cause loss of molecular function, representing novel oncogenic candidates (44).
Li et al. (45) built an e-MutPath (edgetic mutation–mediated pathway perturbations) database to report cancer somatic edgetic mutations based on genome-wide somatic mutation profiles with gene expression and PPI networks. We downloaded 2541 edgetic mutations (P < 0.05) related to non–small cell lung cancer from the e-MutPath database and predicted their oncogenicity using CancerVar. Among these 2541 variants, CancerVar predicted 987 (39%) variants to be oncogenic using clinical features only; it predicted 2047 (81%) variants to be oncogenic using ensemble features, including both clinical and functional evidence. This result is consistent with the expectation that the functional impact of edgetic variants contributes more weight than clinical features in the prediction model for oncogenicity.
Enrichment analysis for CancerVar-predicted driver mutations in well-known ONCs and TSGs
We also performed enrichment analysis on CancerVar-predicted driver mutations in 10 well-known tumor suppressor genes (TSGs) and 10 oncogenes (ONCs) reported in Bayley’s driver gene list (46). In total, ~223,000 exome mutations (147,141 in TSGs and 76,362 in ONCs) were observed. CancerVar predicted 1641 driver variants in TSGs (approximately 1641 of 147,141 = 1.1%) and 875 in ONCs (approximately 875 of 76,362 = 1.1%). According to the theoretical estimation of Bozic et al. (47), on average, ~114 driver mutations are expected in one TSG, and ~14 driver mutations are expected in one ONC. CancerVar predicted ~164 (1641/10) driver mutations in each TSG and ~88 (875/10) in each ONC, greater than the theoretical expectations. This observation may be partially attributed to the fact that although ONCs tend to have hotspot (recurring) somatic mutations as gain-of-function mutations, this is not taken into account in the prediction model. The numbers and frequencies of predicted oncogenic variants are shown in table S6.
For comparison, we used CancerVar to predict oncogenic variants in 10 randomly selected nonessential genes (“human knockout” genes) from two studies [Narasimhan et al. (48) and Saleheen et al. (49)] with 250 overlapping genes. We scanned 86,094 possible exome mutations in these 10 genes, including AKR1E2, BROX, BTN3A3, EFCAB13, KLHL25, LRRC69, MGST1, MROH2A, TTLL2, and ZSCAN16. CancerVar did not detect any oncogenic variants (0%) in these genes but predicted 84,445 (98%) as benign and 1649 (2%) as VUSs. These results further suggest that oncogenic variants predicted by CancerVar are enriched (1.1%) in putative ONCs and TSGs compared to the lack of findings (0%) for the human knockout genes.
Use case: Comprehensive interpretation of FOXA1 somatic mutations in prostate cancer
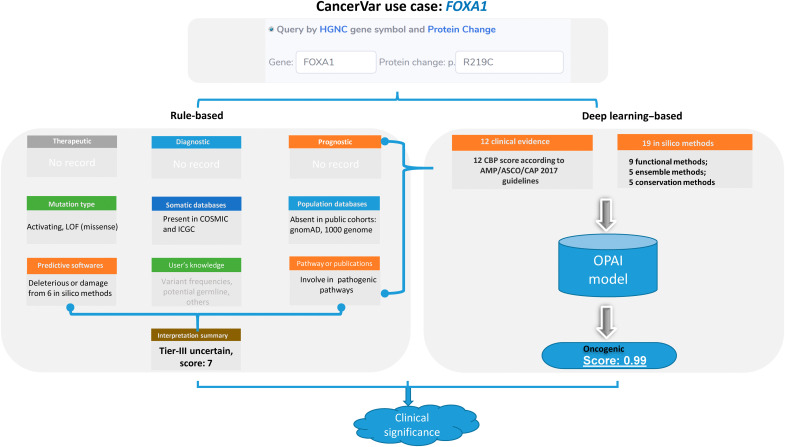
In this use case, we show the clinical interpretation of the FOXA1’s mutation in prostate cancer (Fig. 6) from rule-based and deep learning–based models in CancerVar. Prostate cancer is the most commonly diagnosed cancer in men worldwide (50). The FOXA1 protein (forkhead Box A1, previously known as HNF3a) is essential for normal development of the prostate (51). FOXA1 somatic mutations are frequently observed in prostate cancer (52) and associated with poor outcomes. In 2019, two studies demonstrated that FOXA1 acts as an ONC in prostate cancer (53, 54), whereby the hotspot mutation at R219 (R219S and R219C) drives a proluminal phenotype in prostate cancer exclusive to other fusions or mutations (53, 54). We interpreted these two mutations, but, here, we only illustrate the clinical interpretation of R219C, as that of R219S is very similar. We searched for the missense mutation R219C using the protein change and gene name “FOXA1” in the CancerVar web server. CancerVar did not find any therapeutic, diagnostic, or prognostic evidence for this mutation. Because this mutation has recently been incorporated into somatic databases, including COSMIC (ID: COSM3738526) and international cancer genome consortium (ICGC) (ID: MU67448716), CBP_9 was applied as moderate evidence. Recently, two publications reported its biological functions in prostate cancer, and CBP_12 was applied. According to CBP_7, this mutation is absent or has an extremely low minor allele frequency in the public allele frequency database. As all seven in silico methods predicted this mutation as (likely) pathogenic, CBP_10 was applied. According to the AMP/ASCO/CAP/CGC guidelines, this variant falls into the class “tier III uncertain significance,” with a score of 7, but very close to the class “tier II potential.” On the other hand, on the basis of the OPAI model, the score on this variant is 0.99, suggesting that it is very likely oncogenic. This use case demonstrates that a semiautomated interpretation approach can greatly improve prediction accuracy for each variant given existing knowledge and domain expertise. In addition, this user case shows that a model-based approach involving machine intelligence can be applied as additional evidence to support rule-based methods.
Fig. 6. A use case of using rule-based and deep learning-based models in CancerVar for interpretation of FOXA1 variants.
– We queried the FOXA1 mutation R219C in prostate cancer. The rule-based prediction of this variant was tier III (uncertain significance), with a score of 7, which is very close to tier II. However, the OPAI model predicted this variant to be oncogenic, with a score of 0.99. On the basis of a manual review of the results, we suggest that this variant has clinical significance.
DISCUSSION
Clinical interpretation of cancer somatic variants is important for clinicians and researchers working in the field of precision oncology, especially given the transition from panel sequencing to whole-exome/genome sequencing in cancer genomics studies. To build a standardized, rapid, and user-friendly interpretation tool, we developed command-line software tools together with a web server to assess the clinical impacts of somatic variants using the AMP/ASCO/CAP 2017 guidelines. CancerVar is an enhanced version of the cancer variant knowledgebase incorporated from our previously developed tools for variant annotations and prioritizations, including ANNOVAR (55), InterVar (56), VIC (9), and iCAGES (57), as well as assembling existing variant annotation databases such as CIViC (4), CKB (6), and OncoKB (1). We stress here that CancerVar will not replace human acumen in clinical interpretation but rather generate evidence to help human reviewers by providing standardized, reproducible, and precise output for interpreting somatic variants.
With CancerVar, we did not reconcile the well-known “conflicting interpretation” issues across knowledgebases; instead, we documented and harmonized all types of clinical evidence (i.e., drug information and publications) for both hotspot and non-hotspot mutations in detail to allow users to make informed clinical decisions based on their own domain knowledge and expertise. Compared to existing knowledgebases such as OncoKB, CIViC, and metaKB, CancerVar provides an improved platform in four areas: (i) comprehensive, evidence-based annotations with rigorous quality control for ~13 million somatic variants, which is not limited to the small number of known hotspot mutations; (ii) a flexible scoring system allowing users to fine-tune the importance of clinical evidence criteria according to their own domain knowledge; (iii) improved prioritization for cancer driver mutations using the novel semisupervised deep learning method OPAI; and (iv) automatically summarized interpretation text such that users do not need to compile evidence from multiple knowledgebases manually. We expect CancerVar to become a useful web service for the interpretation of somatic variants in clinical cancer research.
We also need to acknowledge several limitations in CancerVar. First, the scoring weight system is not very robust and may be considered ad hoc by some users. We note that existing clinical guidelines did not provide recommendations for weighting different evidence types, and therefore, we treat all weights as equal by default. Nevertheless, with increasing amounts of clinical knowledge regarding somatic mutations, we expect that we may build a weighted model in the future to enhance prediction accuracy. Second, a small number of CNAs (similar to hotspot mutations) have emerged as important biomarkers for disease characterization and therapeutic decision-making; however, there is a lack of a specific database for clinically actionable somatic CNAs. Although AMP/ASCO/CAP recently published a CNA guideline, CNAs are very heterogeneous in size, and their significance is much harder to score in practice. Therefore, in the future, we will design and implement the scoring system for CNAs based on the platform used to discover CNAs (which determines the resolution of calls), the reliability of the CNA calls, the genes covered by the CNAs, and additional cancer type–specific information from existing databases (given that different cancer types have different CNA profiles). Third, CancerVar currently cannot interpret inversions or gene fusions and cannot interpret gene expression alterations, although these genomic alterations may play important roles in cancer development/progression. Before a specific guideline for these types of mutations becomes available, we suggest that users treat them as CNAs (gene inversions/fusions as deletions and gene expression down-regulation or up-regulation as deletions or duplications). Last, CancerVar web servers cannot process indels because there are many more possible indels than SNVs in cancer to be precomputed en masse. However, users have the flexibility to use the command-line version to predict the clinical significance of indels, including those in genes that are not included in the cancer gene census.
Overall, accurate interpretation of clinical significance depends greatly on evidence harmonization, which should be precisely derived and standardized from multiple databases and annotations. Compared to existing knowledgebases that document a limited number of hotspot mutations, CancerVar provides polished, comprehensive, and semiautomated clinical interpretations for somatic variants with clinical evidence, and it greatly facilitates human reviewers’ drafts of clinical reports for panel sequencing, exome sequencing, or whole-genome sequencing in cancer. Although some commercial software tools also use the AMP/ASCO/CAP rules to standardize variant interpretation, they require a high license fee that many academic researchers may not afford. In addition to interpretation based on the AMP/ASCO/CAP rules specified by consensus from human experts, the CancerVar deep learning–based approach jointly models both rule-based clinical features and functional prediction features to support oncogenic predictions for novel unreported mutations. We believe that CancerVar allows for comprehensive clinical interpretations and prioritizations for both hotspot and non-hotspot variants, which may facilitate the implementation of precision oncology.
In summary, CancerVar is both a web server and a command-line software tool that provides polished and semiautomated clinical interpretations for somatic variants in cancer. Moreover, CancerVar facilitates drafting clinical reports semiautomatically for panel sequencing, exome sequencing, or genome sequencing in cancer. We expect to continuously improve CancerVar and incorporate new functionalities in the future, similar to the wInterVar server (56) and wANNOVAR server (58).
MATERIALS AND METHODS
Overview of mapping of clinical evidence to the AMP/ASCO/CAP 2017 guidelines
According to the AMP/ASCO/CAP 2017 guidelines, there are a total of 12 types of clinical-based evidence to predict the clinical significance of somatic variants, including therapies, mutation types, variant allele fraction [mosaic variant frequency (likely somatic) and nonmosaic variant frequency (potential germline)], population databases, germline databases, somatic databases, predictive results of different computational algorithms, pathway involvement, and publications (8, 9). As shown in Fig. 1, CancerVar contains all the above 12 pieces of evidence, among which 10 are automatically generated; the other two, including the variant allele fraction and potential germline, require user input [possibly included in variant call format (VCF) files] for manual adjustment.
Cancer variant collection and preprocessing
The cancer gene census list and potential driver gene list are essential for somatic variant annotation. We curated a list of 1911 cancer census or driver genes with 13 million exonic variants from seven existing cancer knowledgebases, including COSMIC, CIViC, and OncoKB, and two datasets collected from the literature on driver gene predictions (table S1). For each exon position in these 1911 genes, we generated all three possible nucleotide changes. Unlike other knowledgebases, which only compile variants reported or documented previously, we precomputed annotations for all possible variants for CancerVar interpretation. We compiled clinical evidence based on the AMP/ASCO/CAP 2017 guidelines, which makes the variant searching in the CancerVar web server very fast. In CancerVar, we document all types of clinical evidence, such as in silico prediction, drug information, and publications, in detail to help users adjust criteria and make their own decisions using prior knowledge.
The evidence-based scoring method to prioritize the clinical significance of somatic variants
CancerVar evaluates each set of evidence and then scores each clinical-based prediction (CBP). The variant evidence receives 2 points for strong clinical significance or oncogenic, 1 point for supporting clinical significance or oncogenic, 0 for no support, and −1 for benign or neutral. The CancerVar score is the sum of all evidence scores. The complete scoring system for each CBP can be found in table S2. Let CBP[i] be the ith evidence score, and let weight [i] be the score for the ith evidence; the CancerVar score is calculated using Eq. 1. The weight is 1 by default, but users can adjust it based on its importance from prior knowledge. On the basis of the score range in Eq. 2, we classify each variant into one of the four tiers: strong clinical significance, potential clinical significance, variants of unknown clinical significance (VUS), and benign/likely benign (neutral)
| (1) |
| (2) |
The deep learning–based scoring method to predict oncogenic variants
We developed a scoring method for OPAI using 12 clinical evidence prediction scores and 23 precomputed scores predicted by other computational tools, such as SIFT (23), DANN (26), and SiPhy (59), by improving a semisupervised generative adversarial network model that we previously built. As the score ranges are diverse among the predictive tools, we used their categorical outputs as prediction features. Some variants have missing values on certain features, and we thus excluded variants with more than two missing features. After filtering, 12.9 million variants were used for downstream analysis.
OPAI uses one type of deep learning architecture, the semisupervised generative adversarial network method, to predict the probability of oncogenic variants. As depicted in Fig. 2, this architecture consists of two parts: the generator (G) and the discriminator (D). The generator generates synthetic samples (fake) by random noise from the normal distribution, and the discriminator differentiates realistic samples and synthetic data. In our model, the input data consist of labeled samples, unlabeled samples, and random noise from a normal distribution as the synthetic data. Then, the discriminator/classifier classifies the sample into three classes: (i) neutral, (ii) nonneutral (oncogenic mutations), and (iii) fake synthetic data, in which the unlabeled real samples can be identified as 1 or 2 and the synthetic samples are 3. Therefore, loss function L can be written as two parts
| (3) |
where pdata is the underlying distribution of real samples and pG is the distribution of the output from the generator. For the loss of the generator, we used feature matching as our loss function: .
OPAI training and testing
Labeled data were obtained from expert curation on diagnostic reports of patients with cancer in our in-house database; we used 4000 variants (1000 positive) as the training set and 1234 variants (669 positive) as the validation set. We tested our model on 6226 variants (1335 positive), which were compiled from a literature review. For unlabeled data, we randomly selected 60,000 variants from 12.9 million samples with non-missing features and repeated this process multiple times.
For synthetic samples, the generator produces random noise from a standard normal distribution in each batch step and outputs the synthetic samples. In each minibatch, the model calculates 2000 labeled samples, 10,000 unlabeled samples, and 10,000 synthetic samples from the generator. The discriminator/classifier is trained by calculating the loss from supervised learning and unsupervised training separately. Then, the generator is trained by minimizing the feature matching in each batch.
To improve analysis interpretability and evaluate the feature contribution in our study, RF analysis was used to evaluate the importance of 35 features (23 in silico functional features and 12 clinical evidence scores) in the above expert manually labeled variant database. The importance score of each feature was calculated and ranked, as provided in the Supplementary Materials (fig. S4).
Pan-cancer benchmarks from public datasets
Complementary and comprehensive benchmark datasets are needed for systematic evaluation of the performance of CancerVar. In the current study, we used several different benchmark datasets with references: (i) a multi-institutional evaluation study with 51 variants from Sirohi et al. (10); (ii) a literature annotation database from OncoKB (1) and CIViC (4); (iii) TP53 mutations and their target transcription activity from the IARC database (41); and (iv) functional annotation based on in vitro cell viability assays from the study of Ng et al. (42).
High-quality expert-labeled variants from in-house clinical reports
In addition to datasets from public resources, we also have an in-house dataset with 7967 somatic mutations from deidentified patients at the Children’s Hospital of Philadelphia (CHOP). Each variant has been manually annotated and classified by human experts in the diagnostic labs. Using the AMP/ASCO/CAP guidelines, the four-tier classification assignment for each somatic mutation was agreed upon by at least two cancer experts from the Division of Genomic Diagnostics Laboratory at CHOP. Furthermore, to train the deep learning model, we used variants from strong clinical significance (tier I) and potential clinical significance (tier II) categories as positive samples and used benign/likely benign variants (tier IV) as negative samples. There were 5234 variants, 1668 positive samples and 3566 negative samples, for training and validation.
CancerVar and OPAI software accessibility
Users can access CancerVar and OPAI in three ways, including a web server that is free and open to all users without login requirements (https://cancervar.wglab.org), a command-line software written in Python that is freely available from GitHub (https://github.com/wglab/CancerVar) for noncommercial users, and a RESTful API service to facilitate other web developers in accessing our precomputed evidence and OPAI scores for ~13 million coding variants. Users can use the command-line version to predict the clinical significance of indels, including indels in genes not included in the cancer gene list precomputed by CancerVar.
Acknowledgments
We would like to thank S. N. Martins-Filho and N.-A. Pham for constructive comments. We also thank members of the Wang laboratory for helpful comments on the user interface of the CancerVar web server and for testing the CancerVar web server. Q.L. dedicates this study to the memory of his beloved father, Hong Li.
Funding: This work was supported by the National Institutes of Health (NIH)/National Library of Medicine (NLM)/National Human Genome Research Institute (NHGRI) (grant number LM012895), NIH/National Institute of General Medical Sciences (NIGMS) (grant number GM120609 and GM132713), CHOP Pathology Diagnostic Innovation Award, and CHOP Research Institute.
Author contributions: Q.L., Y.Z., and K.W. conceived the study. Q.L. and Z.R. performed the coding and generated and interpreted the results. K.C. and M.M.L. provided manual annotation data and advised on the study. Q.L., Y.Z., and K.W. wrote the original manuscript with input from all authors. All authors reviewed and approved the final manuscript.
Competing interests: K.W. indirectly owned shares in PierianDx. All other authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.
Supplementary Materials
This PDF file includes:
Figs. S1 to S4
Tables S1 and S2, S4 to S6
Other Supplementary Material for this manuscript includes the following:
Table S3
REFERENCES AND NOTES
- 1.Chakravarty D., Gao J., Phillips S. M., Kundra R., Zhang H., Wang J., Rudolph J. E., Yaeger R., Soumerai T., Nissan M. H., Chang M. T., Chandarlapaty S., Traina T. A., Paik P. K., Ho A. L., Hantash F. M., Grupe A., Baxi S. S., Callahan M. K., Snyder A., Chi P., Danila D., Gounder M., Harding J. J., Hellmann M. D., Iyer G., Janjigian Y., Kaley T., Levine D. A., Lowery M., Omuro A., Postow M. A., Rathkopf D., Shoushtari A. N., Shukla N., Voss M., Paraiso E., Zehir A., Berger M. F., Taylor B. S., Saltz L. B., Riely G. J., Ladanyi M., Hyman D. M., Baselga J., Sabbatini P., Solit D. B., Schultz N., OncoKB: A precision oncology knowledge base. JCO Precis. Oncol. 2017, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bailey M. H., Tokheim C., Porta-Pardo E., Sengupta S., Bertrand D., Weerasinghe A., Colaprico A., Wendl M. C., Kim J., Reardon B., Ng P. K.-S., Jeong K. J., Cao S., Wang Z., Gao J., Gao Q., Wang F., Liu E. M., Mularoni L., Rubio-Perez C., Nagarajan N., Cortes-Ciriano I., Zhou D. C., Liang W. W., Hess J. M., Yellapantula V. D., Tamborero D., Gonzalez-Perez A., Suphavilai C., Ko J. Y., Khurana E., Park P. J., Van Allen E. M., Liang H.; MC3 Working Group; Cancer Genome Atlas Research Network, Lawrence M. S., Lawrence M. S., Godzik A., Lopez-Bigas N., Stuart J., Wheeler D., Getz G., Chen K., Lazar A. J., Mills G. B., Karchin R., Ding L., Comprehensive characterization of cancer driver genes and mutations. Cell 174, 1034–1035 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Micheel C. M., Sweeney S. M., LeNoue-Newton M. L., Andre F., Bedard P. L., Guinney J., Meijer G. A., Rollins B. J., Sawyers C. L., Schultz N., Shaw K. R. M., Velculescu V. E., Levy M. A.; AACR Project GENIE Consortium , American Association for Cancer Research Project Genomics Evidence Neoplasia Information Exchange: From inception to first data release and beyond-lessons learned and member institutions’ perspectives. JCO Clin. Cancer Inform. 2, 1–14 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Griffith M., Spies N. C., Krysiak K., McMichael J. F., Coffman A. C., Danos A. M., Ainscough B. J., Ramirez C. A., Rieke D. T., Kujan L., Barnell E. K., Wagner A. H., Skidmore Z. L., Wollam A., Liu C. J., Jones M. R., Bilski R. L., Lesurf R., Feng Y. Y., Shah N. M., Bonakdar M., Trani L., Matlock M., Ramu A., Campbell K. M., Spies G. C., Graubert A. P., Gangavarapu K., Eldred J. M., Larson D. E., Walker J. R., Good B. M., Wu C., Su A. I., Dienstmann R., Margolin A. A., Tamborero D., Lopez-Bigas N., Jones S. J., Bose R., Spencer D. H., Wartman L. D., Wilson R. K., Mardis E. R., Griffith O. L., CIViC is a community knowledgebase for expert crowdsourcing the clinical interpretation of variants in cancer. Nat. Genet. 49, 170–174 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Huang L., Fernandes H., Zia H., Tavassoli P., Rennert H., Pisapia D., Imielinski M., Sboner A., Rubin M. A., Kluk M., Elemento O., The cancer precision medicine knowledge base for structured clinical-grade mutations and interpretations. J. Am. Med. Inform. Assoc. 24, 513–519 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Patterson S. E., Liu R., Statz C. M., Durkin D., Lakshminarayana A., Mockus S. M., The clinical trial landscape in oncology and connectivity of somatic mutational profiles to targeted therapies. Hum. Genomics 10, 4 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tamborero D., Rubio-Perez C., Deu-Pons J., Schroeder M. P., Vivancos A., Rovira A., Tusquets I., Albanell J., Rodon J., Tabernero J., de Torres C., Dienstmann R., Gonzalez-Perez A., Lopez-Bigas N., Cancer Genome Interpreter annotates the biological and clinical relevance of tumor alterations. Genome Med. 10, 25 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li M. M., Datto M., Duncavage E. J., Kulkarni S., Lindeman N. I., Roy S., Tsimberidou A. M., Vnencak-Jones C. L., Wolff D. J., Younes A., Nikiforova M. N., Standards and guidelines for the interpretation and reporting of sequence variants in cancer: A joint consensus recommendation of the Association for Molecular Pathology, American Society of Clinical Oncology, and College of American Pathologists. J. Mol. Diagn. 19, 4–23 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.He M. M., Li Q., Yan M., Cao H., Hu Y., He K. Y., Cao K., Li M. M., Wang K., Variant Interpretation for Cancer (VIC): A computational tool for assessing clinical impacts of somatic variants. Genome Med. 11, 53 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sirohi D., Schmidt R. L., Aisner D. L., Behdad A., Betz B. L., Brown N., Coleman J. F., Corless C. L., Deftereos G., Ewalt M. D., Fernandes H., Hsiao S. J., Mansukhani M. M., Murray S. S., Niu N., Ritterhouse L. L., Suarez C. J., Tafe L. J., Thorson J. A., Segal J. P., Furtado L. V., Multi-institutional evaluation of interrater agreement of variant classification based on the 2017 Association for Molecular Pathology, American Society of Clinical Oncology, and College of American Pathologists standards and guidelines for the interpretation and reporting of sequence variants in cancer. J. Mol. Diagn. 22, 284–293 (2020). [DOI] [PubMed] [Google Scholar]
- 11.Wagner A. H., Walsh B., Mayfield G., Tamborero D., Sonkin D., Krysiak K., Deu-Pons J., Duren R. P., Gao J., McMurry J., A harmonized meta-knowledgebase of clinical interpretations of somatic genomic variants in cancer. Nat. Genet. 52, 448–457 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kalia S. S., Adelman K., Bale S. J., Chung W. K., Eng C., Evans J. P., Herman G. E., Hufnagel S. B., Klein T. E., Korf B. R., Recommendations for reporting of secondary findings in clinical exome and genome sequencing, 2016 update (ACMG SF v2. 0): A policy statement of the American College of Medical Genetics and Genomics. Genet. Med. 19, 249–255 (2017). [DOI] [PubMed] [Google Scholar]
- 13.Harrison S. M., Rehm H. L., Is ‘likely pathogenic’ really 90% likely? reclassification data in ClinVar. Genome Med. 11, 72 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bertrand D., Chng K. R., Sherbaf F. G., Kiesel A., Chia B. K., Sia Y. Y., Huang S. K., Hoon D. S., Liu E. T., Hillmer A., Nagarajan N., Patient-specific driver gene prediction and risk assessment through integrated network analysis of cancer omics profiles. Nucleic Acids Res. 43, e44 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Carter H., Chen S., Isik L., Tyekucheva S., Velculescu V. E., Kinzler K. W., Vogelstein B., Karchin R., Cancer-specific high-throughput annotation of somatic mutations: Computational prediction of driver missense mutations. Cancer Res. 69, 6660–6667 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mao Y., Chen H., Liang H., Meric-Bernstam F., Mills G. B., Chen K., CanDrA: Cancer-specific driver missense mutation annotation with optimized features. PLOS ONE 8, e77945 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mularoni L., Sabarinathan R., Deu-Pons J., Gonzalez-Perez A., Lopez-Bigas N., OncodriveFML: A general framework to identify coding and non-coding regions with cancer driver mutations. Genome Biol. 17, 128 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Porta-Pardo E., Godzik A., e-Driver: A novel method to identify protein regions driving cancer. Bioinformatics 30, 3109–3114 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Reimand J., Bader G. D., Systematic analysis of somatic mutations in phosphorylation signaling predicts novel cancer drivers. Mol. Syst. Biol. 9, 637 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Reva B., Antipin Y., Sander C., Predicting the functional impact of protein mutations: Application to cancer genomics. Nucleic Acids Res. 39, e118 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shihab H. A., Gough J., Cooper D. N., Day I. N., Gaunt T. R., Predicting the functional consequences of cancer-associated amino acid substitutions. Bioinformatics 29, 1504–1510 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tamborero D., Gonzalez-Perez A., Lopez-Bigas N., OncodriveCLUST: Exploiting the positional clustering of somatic mutations to identify cancer genes. Bioinformatics 29, 2238–2244 (2013). [DOI] [PubMed] [Google Scholar]
- 23.Ng P. C., Henikoff S., SIFT: Predicting amino acid changes that affect protein function. Nucleic Acids Res. 31, 3812–3814 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Adzhubei I., Jordan D. M., Sunyaev S. R., Predicting functional effect of human missense mutations using PolyPhen-2. Curr. Protoc. Hum. Genet. 76, Chapter 7:Unit7.20, (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shihab H. A., Gough J., Cooper D. N., Stenson P. D., Barker G. L., Edwards K. J., Day I. N., Gaunt T. R., Predicting the functional, molecular, and phenotypic consequences of amino acid substitutions using hidden Markov models. Hum. Mutat. 34, 57–65 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Quang D., Chen Y., Xie X., DANN: A deep learning approach for annotating the pathogenicity of genetic variants. Bioinformatics 31, 761–763 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shuai S., Gallinger S., Stein L., Combined burden and functional impact tests for cancer driver discovery using DriverPower. Nat. Commun. 11, 1–12 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Posey J. E., Rosenfeld J. A., James R. A., Bainbridge M., Niu Z., Wang X., Dhar S., Wiszniewski W., Akdemir Z. H., Gambin T., Xia F., Person R. E., Walkiewicz M., Shaw C. A., Sutton V. R., Beaudet A. L., Muzny D., Eng C. M., Yang Y., Gibbs R. A., Lupski J. R., Boerwinkle E., Plon S. E., Molecular diagnostic experience of whole-exome sequencing in adult patients. Genet. Med. 18, 678–685 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang Y., Muzny D. M., Reid J. G., Bainbridge M. N., Willis A., Ward P. A., Braxton A., Beuten J., Xia F., Niu Z., Hardison M., Person R., Bekheirnia M. R., Leduc M. S., Kirby A., Pham P., Scull J., Wang M., Ding Y., Plon S. E., Lupski J. R., Beaudet A. L., Gibbs R. A., Eng C. M., Clinical whole-exome sequencing for the diagnosis of mendelian disorders. N. Engl. J. Med. 369, 1502–1511 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Retterer K., Juusola J., Cho M. T., Vitazka P., Millan F., Gibellini F., Vertino-Bell A., Smaoui N., Neidich J., Monaghan K. G., McKnight D., Bai R., Suchy S., Friedman B., Tahiliani J., Pineda-Alvarez D., Richard G., Brandt T., Haverfield E., Chung W. K., Bale S., Clinical application of whole-exome sequencing across clinical indications. Genet. Med. 18, 696–704 (2016). [DOI] [PubMed] [Google Scholar]
- 31.Trujillano D., Bertoli-Avella A. M., Kumar Kandaswamy K., Weiss M. E., Koster J., Marais A., Paknia O., Schroder R., Garcia-Aznar J. M., Werber M., Brandau O., Calvo Del Castillo M., Baldi C., Wessel K., Kishore S., Nahavandi N., Eyaid W., Al Rifai M. T., Al-Rumayyan A., Al-Twaijri W., Alothaim A., Alhashem A., Al-Sannaa N., Al-Balwi M., Alfadhel M., Rolfs A., Abou Jamra R., Clinical exome sequencing: Results from 2819 samples reflecting 1000 families. Eur. J. Hum. Genet. 25, 176–182 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee H., Deignan J. L., Dorrani N., Strom S. P., Kantarci S., Quintero-Rivera F., Das K., Toy T., Harry B., Yourshaw M., Fox M., Fogel B. L., Martinez-Agosto J. A., Wong D. A., Chang V. Y., Shieh P. B., Palmer C. G., Dipple K. M., Grody W. W., Vilain E., Nelson S. F., Clinical exome sequencing for genetic identification of rare Mendelian disorders. JAMA 312, 1880–1887 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Clark M. M., Stark Z., Farnaes L., Tan T. Y., White S. M., Dimmock D., Kingsmore S. F., Meta-analysis of the diagnostic and clinical utility of genome and exome sequencing and chromosomal microarray in children with suspected genetic diseases. NPJ Genom. Med. 3, 16 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Salfati E. L., Spencer E. G., Topol S. E., Muse E. D., Rueda M., Lucas J. R., Wagner G. N., Campman S., Topol E. J., Torkamani A., Re-analysis of whole-exome sequencing data uncovers novel diagnostic variants and improves molecular diagnostic yields for sudden death and idiopathic diseases. Genome Med. 11, 83 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ramchand J., Wallis M., Macciocca I., Lynch E., Farouque O., Martyn M., Phelan D., Chong B., Lockwood S., Weintraub R., Thompson T., Trainer A., Zentner D., Vohra J., Chetrit M., Hare D. L., James P., Prospective evaluation of the utility of whole exome sequencing in dilated cardiomyopathy. J. Am. Heart Assoc. 9, e013346 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dong X., Liu B., Yang L., Wang H., Wu B., Liu R., Chen H., Chen X., Yu S., Chen B., Wang S., Xu X., Zhou W., Lu Y., Clinical exome sequencing as the first-tier test for diagnosing developmental disorders covering both CNV and SNV: A Chinese cohort. J. Med. Genet. 57, 558–566 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yi S., Lin S., Li Y., Zhao W., Mills G. B., Sahni N., Functional variomics and network perturbation: Connecting genotype to phenotype in cancer. Nat. Rev. Genet. 18, 395–410 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bouhaddou M., Eckhardt M., Chi Naing Z. Z., Kim M., Ideker T., Krogan N. J., Mapping the protein-protein and genetic interactions of cancer to guide precision medicine. Curr. Opin. Genet. Dev. 54, 110–117 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sahni N., Yi S., Zhong Q., Jailkhani N., Charloteaux B., Cusick M. E., Vidal M., Edgotype: A fundamental link between genotype and phenotype. Curr. Opin. Genet. Dev. 23, 649–657 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kuenzi B. M., Ideker T., A census of pathway maps in cancer systems biology. Nat. Rev. Cancer 20, 233–246 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bouaoun L., Sonkin D., Ardin M., Hollstein M., Byrnes G., Zavadil J., Olivier M., TP53 variations in human cancers: New lessons from the IARC TP53 database and genomics data. Hum. Mutat. 37, 865–876 (2016). [DOI] [PubMed] [Google Scholar]
- 42.Ng P. K., Li J., Jeong K. J., Shao S., Chen H., Tsang Y. H., Sengupta S., Wang Z., Bhavana V. H., Tran R., Soewito S., Minussi D. C., Moreno D., Kong K., Dogruluk T., Lu H., Gao J., Tokheim C., Zhou D. C., Johnson A. M., Zeng J., Ip C. K. M., Ju Z., Wester M., Yu S., Li Y., Vellano C. P., Schultz N., Karchin R., Ding L., Lu Y., Cheung L. W. T., Chen K., Shaw K. R., Meric-Bernstam F., Scott K. L., Yi S., Sahni N., Liang H., Mills G. B., Systematic functional annotation of somatic mutations in cancer. Cancer Cell 33, 450–462.e10 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pedregosa F., Varoquaux G., Gramfort A., Michel V., Thirion B., Grisel O., Blondel M., Prettenhofer P., Weiss R., Dubourg V., Scikit-learn: Machine learning in Python. J. Mach. Learn. Res. 12, 2825–2830 (2011). [Google Scholar]
- 44.Sahni N., Yi S., Taipale M., Bass J. I. F., Coulombe-Huntington J., Yang F., Peng J., Weile J., Karras G. I., Wang Y., Kovacs I. A., Kamburov A., Krykbaeva I., Lam M. H., Tucker G., Khurana V., Sharma A., Liu Y. Y., Yachie N., Zhong Q., Shen Y., Palagi A., San-Miguel A., Fan C., Balcha D., Dricot A., Jordan D. M., Walsh J. M., Shah A. A., Yang X., Stoyanova A. K., Leighton A., Calderwood M. A., Jacob Y., Cusick M. E., Salehi-Ashtiani K., Whitesell L. J., Sunyaev S., Berger B., Barabasi A. L., Charloteaux B., Hill D. E., Hao T., Roth F. P., Xia Y., Walhout A. J. M., Lindquist S., Vidal M., Widespread macromolecular interaction perturbations in human genetic disorders. Cell 161, 647–660 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li Y., Burgman B., Khatri I. S., Pentaparthi S. R., Su Z., McGrail D. J., Li Y., Wu E., Eckhardt S. G., Sahni N., Yi S. S., e-MutPath: Computational modeling reveals the functional landscape of genetic mutations rewiring interactome networks. Nucleic Acids Res. 49, e2 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bailey M. H., Tokheim C., Porta-Pardo E., Sengupta S., Bertrand D., Weerasinghe A., Colaprico A., Wendl M. C., Kim J., Reardon B., Comprehensive characterization of cancer driver genes and mutations. Cell 173, 371–385.e18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bozic I., Antal T., Ohtsuki H., Carter H., Kim D., Chen S., Karchin R., Kinzler K. W., Vogelstein B., Nowak M. A., Accumulation of driver and passenger mutations during tumor progression. Proc. Natl. Acad. Sci. 107, 18545–18550 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Narasimhan V. M., Hunt K. A., Mason D., Baker C. L., Karczewski K. J., Barnes M. R., Barnett A. H., Bates C., Bellary S., Bockett N. A., Health and population effects of rare gene knockouts in adult humans with related parents. Science 352, 474–477 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Saleheen D., Natarajan P., Armean I. M., Zhao W., Rasheed A., Khetarpal S. A., Won H.-H., Karczewski K. J., O’Donnell-Luria A. H., Samocha K. E., Human knockouts and phenotypic analysis in a cohort with a high rate of consanguinity. Nature 544, 235–239 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bray F., Ferlay J., Soerjomataram I., Siegel R. L., Torre L. A., Jemal A., Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 68, 394–424 (2018). [DOI] [PubMed] [Google Scholar]
- 51.Gao N., Zhang J., Rao M. A., Case T. C., Mirosevich J., Wang Y., Jin R., Gupta A., Rennie P. S., Matusik R. J., The role of hepatocyte nuclear factor-3 alpha (Forkhead Box A1) and androgen receptor in transcriptional regulation of prostatic genes. Mol. Endocrinol. 17, 1484–1507 (2003). [DOI] [PubMed] [Google Scholar]
- 52.Cancer Genome Atlas Research Network , The molecular taxonomy of primary prostate cancer. Cell 163, 1011–1025 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Adams E. J., Karthaus W. R., Hoover E., Liu D., Gruet A., Zhang Z., Cho H., DiLoreto R., Chhangawala S., Liu Y., Watson P. A., Davicioni E., Sboner A., Barbieri C. E., Bose R., Leslie C. S., Sawyers C. L., FOXA1 mutations alter pioneering activity, differentiation and prostate cancer phenotypes. Nature 571, 408–412 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Parolia A., Cieslik M., Chu S. C., Xiao L., Ouchi T., Zhang Y., Wang X., Vats P., Cao X., Pitchiaya S., Su F., Wang R., Feng F. Y., Wu Y. M., Lonigro R. J., Robinson D. R., Chinnaiyan A. M., Distinct structural classes of activating FOXA1 alterations in advanced prostate cancer. Nature 571, 413–418 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang K., Li M., Hakonarson H., ANNOVAR: Functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 38, e164 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Li Q., Wang K., InterVar: Clinical interpretation of genetic variants by the 2015 ACMG-AMP guidelines. Am. J. Hum. Genet. 100, 267–280 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dong C., Guo Y., Yang H., He Z., Liu X., Wang K., iCAGES: Integrated CAncer GEnome Score for comprehensively prioritizing driver genes in personal cancer genomes. Genome Med. 8, 135 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chang X., Wang K., wANNOVAR: Annotating genetic variants for personal genomes via the web. J. Med. Genet. 49, 433–436 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Garber M., Guttman M., Clamp M., Zody M. C., Friedman N., Xie X., Identifying novel constrained elements by exploiting biased substitution patterns. Bioinformatics 25, i54–i62 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S4
Tables S1 and S2, S4 to S6
Table S3