Abstract
Parvalbumin-positive (PV+) interneurons play a critical role in maintaining circuit rhythm in the brain, and their reduction is implicated in autism spectrum disorders. Animal studies demonstrate that maternal immune activation (MIA) leads to reduced PV+ interneurons in the somatosensory cortex and autism-like behaviors. However, the underlying molecular mechanisms remain largely unknown. Here, we show that MIA down-regulates microglial Gpr56 expression in fetal brains in an interleukin-17a–dependent manner and that conditional deletion of microglial Gpr56 [Gpr56 conditional knockout (cKO)] mimics MIA-induced PV+ interneuron defects and autism-like behaviors in offspring. We further demonstrate that elevated microglial tumor necrosis factor–α expression is the underlying mechanism by which MIA and Gpr56 cKO impair interneuron generation. Genetically restoring Gpr56 expression in microglia ameliorates PV+ interneuron deficits and autism-like behaviors in MIA offspring. Together, our study demonstrates that microglial GPR56 plays an important role in PV+ interneuron development and serves as a salient target of MIA-induced neurodevelopmental disorders.
Maternal inflammation causes autism-like behaviors by down-regulating microglial GPR56 expression in the fetal brain.
INTRODUCTION
In the adult neocortices of humans and mice, about 20% of cortical neurons are γ-aminobutyric acid–containing inhibitory interneurons (1, 2). Cerebral interneurons provide the main cortical inhibitory input and are essential for the functional rhythms and activity of neural circuits in the cortex (3). Disrupting interneuron number and function causes neurological and psychiatric disorders, including autism spectrum disorder (ASD) (4, 5). Reduction or dysfunction of one main type of interneurons, parvalbumin-positive (PV+) interneurons, leads to ASD in humans and autism-like behaviors in rodents (4, 6). Moreover, maternal immune activation (MIA) has been linked to abnormal neurodevelopmental disorders in both humans and mice (7, 8), with a decreased density of cortical PV+ interneurons as a common underlying mechanism (6, 9, 10).
Most, if not all, cortical interneurons are derived from three progenitor regions in the embryonic subpallium, the medial ganglionic eminence (MGE), the caudal ganglionic eminence (CGE), and the preoptic area (POA) (1). MGE development—regulated by transcription factors like Nkx2.1, Lhx6, Lhx8, and Sox6—gives rise to most PV+ and somatostatin-positive (SST+) cortical interneurons. CGE and POA generate reelin-, calretinin-, and vasoactive intestinal peptide–positive cortical interneurons (1). MGE neurogenesis occurs in a temporal manner, producing more SST+ interneurons in early stage [embryonic days 10 to 13 (E10 to E13)] and more PV+ interneurons in mid- (E13 to E15) and late stage (E15 to E18) (11, 12). However, it is unclear how MIA adversely affects PV+ interneuron development.
Microglia, the brain-resident tissue macrophages, are the primary innate immune cells in the brain parenchyma. They are yolk sac–derived myeloid cells that enter the brain as early as E8.5/E9.5 in mice (13). In addition to their immune functions, which include immune surveillance and pathogen response (14), microglia regulate brain developmental processes such as myelination (15), neurogenesis (16), synaptogenesis (17), neuronal survival (18), and synaptic pruning (19).
We reasoned that fetal microglia could be the cellular targets of MIA during cortical interneuron development. First, microglia populate the fetal brain at the onset of mouse brain development (20). Second, rodent MIA models demonstrate that MIA induces a proinflammatory response in fetal microglia, including increased cytokine expression and less ramified morphology (21, 22). Third, microglial activation contributes to neurodevelopmental disorders, such as ASD (23). Last, microglia regulate cortical interneuron development, as ablation of microglia impairs cortical positioning of interneuron subsets in the mouse cortex (24).
G Protein-Coupled Receptor 56 (GPR56), also known as adhesion G protein-coupled receptor G1 (ADGRG1), is an adhesion G protein–coupled receptor. It is one of the critical genes that define yolk sac–derived microglia: Gpr56 is only expressed in yolk sac–derived microglia, but not in fetal liver- and bone marrow–derived microglia-like cells, even after long-term adaptation in the central nervous system in vivo (25). In mice, Gpr56 transcripts are detected in microglia as early as E10.5 and markedly increase at E14.5 to eventually reach adult levels during the early postnatal stage (19, 26). The expression of microglial Gpr56 in adult mice down-regulates in response to immune challenge (27, 28). However, it is unknown whether fetal microglial Gpr56 is down-regulated by immune activation.
In this study, we tested the hypothesis that microglial Gpr56 is mechanistically involved in MIA-induced PV+ interneuron reduction and autism-like behaviors. Using a mouse MIA model, conditional deletion and restoration of Gpr56 in microglia, ex vivo embryonic brain slice culture model, and behavior assays, we provided evidence supporting our hypothesis and further demonstrated that elevated tumor necrosis factor–α underlines MIA- and microglial Gpr56 deletion-induced PV+ interneuron impairment.
RESULTS
MIA down-regulates fetal microglial Gpr56 expression in an IL-17a–dependent manner
MIA causes neurodevelopmental abnormalities in offspring through maternal interleukin-17a (IL-17a) (7, 29). However, the underlying cellular and molecular targets in the fetal brain remain unknown. Encoded by one of the genes that define yolk sac–derived microglia (25, 30), the investigation into the function of GPR56 in microglia in the developing brain is gaining interest (19). RNA sequencing data from late B. Barres laboratory show that lipopolysaccharide-induced neuroinflammation down-regulates Gpr56 expression in adult microglia (27). We hypothesize that microglial GPR56 is the molecular target of MIA in the fetal brain by acting downstream of IL-17a.
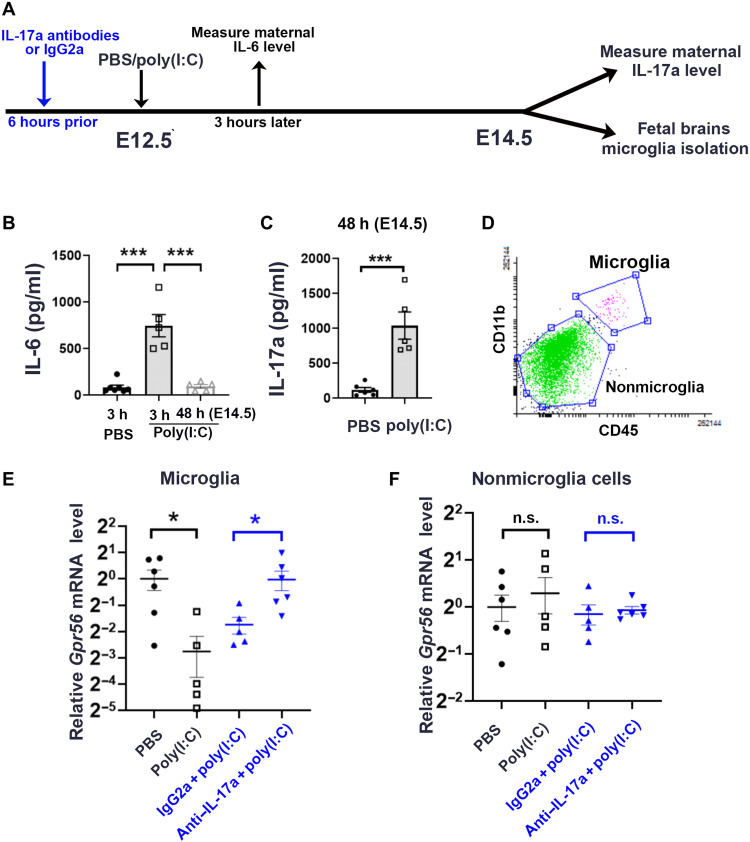
To test this hypothesis, we first investigated whether MIA down-regulates microglial Gpr56 expression in the fetal brain by induction of MIA at E12.5, a developmental stage when maternal inflammation leads to neurodevelopmental abnormalities, including interneuron deficits and autistic behaviors in offspring (6, 7, 29). We intraperitoneally injected synthetic double-strand RNA, polyinosinic:polycytidylic acid [poly(I:C)], which mimics viral infection, to pregnant mice at E12.5 to induce MIA (Fig. 1A). We observed a significant elevation of maternal serum IL-6 concentration at 3 hours and IL-17a at 48 hours (E14.5) after poly(I:C) injection compared to those injected with phosphate-buffered saline (PBS) (Fig. 1, B and C), confirming that we had successfully induced MIA in our mouse model (7). We then examined Gpr56 mRNA expression in microglia isolated from E14.5 fetal brains of control and MIA animals (Fig. 1D). As expected, we saw a significant down-regulation of Gpr56 in microglia isolated from MIA fetuses compared to controls (Fig. 1E), while the mRNA expression of Gpr56 in nonmicroglial fetal brain cells including neural progenitor cells and postmitotic neurons was comparable between the two groups (Fig. 1F).
Fig. 1. MIA down-regulates microglial Gpr56 expression in fetal brains in an IL-17a–dependent manner.
(A) A schematic timeline for MIA induction. (B) Serum concentration of IL-6 was significantly up-regulated at 3 hours after poly(I:C) injection and went back to baseline 48 hours later at E14.5 (n = 5). (C) Serum concentration of IL-17a was significantly up-regulated at E14.5 48 hours after poly(I:C) injection (n = 5). (D) Fluorescence-activated cell sorting (FACS) strategy to collect CD11b+CD45medium microglia and CD11b−CD45− cells as nonmicroglial cells. (E) Relative levels of Gpr56 mRNA in microglia isolated from E14.5 fetal brains whose mothers were treated with PBS (n = 6), poly(I:C) (n = 5), IgG2a isotype control antibodies and poly(I:C) (n = 5), and IL-17a antibodies and poly(I:C) (n = 6). (F) Relative levels of Gpr56 mRNA in nonmicroglia cells isolated from E14.5 fetal brains. PBS versus poly(I:C): P = 0.55; IgG2a + poly(I:C) versus IL-17a antibodies + poly(I:C): P = 0.69. Unpaired Student’s t test. n.s., not significant. *P < 0.05 and ***P < 0.001. Data presented as means ± SEM.
Next, we examined whether MIA-induced microglial Gpr56 down-regulation is mediated by maternal IL-17a by injecting anti–IL-17a–neutralizing antibodies 6 hours before poly(I:C) injection (Fig. 1A). We saw a significantly attenuated Gpr56 down-regulation in E14.5 microglia isolated from MIA fetuses whose mothers were pretreated with IL-17a–neutralizing antibodies compared to those who received immunoglobulin G2a (IgG2a) isotype control antibodies (Fig. 1E). Together, our results support the hypothesis that microglial Gpr56 serves as a molecular target of MIA by acting downstream of IL-17a.
Deleting microglial Gpr56 mimics MIA-induced PV+ interneuron reduction in the neocortex
To further establish the role of microglial GPR56 in interneuron development, we investigated the brain phenotype of microglial Gpr56 knockout mice. We chose Cx3cr1Cre/+ mice as the microglia-specific Cre driver based on the following reasoning: (i) Cx3cr1Cre/+ induces Cre recombination specifically in myeloid cells (19), and (ii) Gpr56 is restrictively expressed in microglia, but not in macrophages (25, 30). We used Gpr56f/f; Cx3cr1Cre/+ as conditional knockout (cKO) and Gpr56+/+; Cx3cr1Cre/+ as controls.
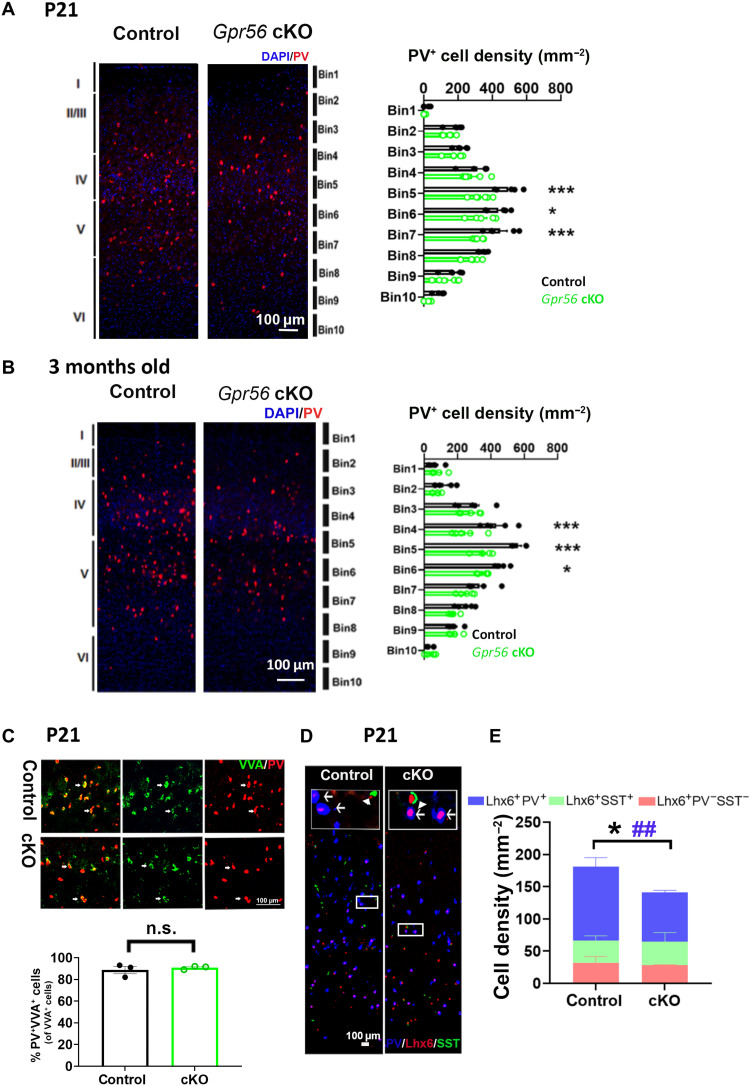
We detected a significant decrease in PV+ interneurons in the somatosensory cortex (SSC) of both juvenile [postnatal day 21 (P21)] and adult (3 months old) Gpr56 cKO mice compared to controls (Fig. 2, A and B), similar to the phenotype reported in MIA offspring (6, 9, 10). The decrease in PV+ interneurons was also detected in the primary auditory cortices of P21 Gpr56 cKO mice (fig. S1A). We did not observe any PV+ interneuron deficits in other brain regions, including the hippocampus, striatum, and cingulate cortex (fig. S1, B to D), nor in other types of interneurons, including SST+, reelin+, and calretinin+ interneurons (fig. S2, A to C). Next, we examined the distribution and density of special AT-rich sequence-binding protein 2 (SatB2)–positive excitatory neurons in the SSC and saw no difference in their densities and distribution between Gpr56 cKO and control mice (fig. S2D). Together, we concluded that deleting microglial Gpr56 recapitulates PV+ interneuron deficits associated with MIA (6).
Fig. 2. Deleting microglial Gpr56 mimics MIA-induced PV+ interneuron reduction in the SSC.
(A and B) The density of PV+ interneurons is significantly decreased in the primary SSC of microglial Gpr56 cKO mice at P21 and 3 months old [(A) n = 4 in control and n = 5 in cKO; (B) n = 5]. DAPI, 4′,6-diamidino-2-phenylindole. (C) Representative images of VVA and PV double labeling. The percentage of PV and VVA double-positive cells are comparable in the SSC of control and microglial Gpr56 cKO mice (86% versus 89%; n = 3, P = 0.50). (D) Representative images of Lhx6, PV, and SST immunostaining. Arrows show Lhx6+PV+ cells, and arrowheads indicate Lhx6+SST+ cells. (E) Bar graph depicting the composition of Lhx6+PV−SST−, Lhx6+PV+, and Lhx6+SST+ cells in the SSC. The densities of Lhx6+PV−SST− and Lhx6+SST+ cells are comparable between control and Gpr56 cKO mice (P = 0.53 and P = 0.80). The densities of Lhx6+ cells and Lhx6+PV+ cells are significantly decreased in Gpr56 cKO mice compared to controls. n = 4 in control and n = 3 in cKO. Two-way analysis of variance (ANOVA) and post hoc Bonferroni’s test for (A) and (B). *P < 0.05 and ***P < 0.001. Unpaired t test for (C) and (E). CR versus cKO for (E) in the density of Lhx6+ cells (*P < 0.05) and Lhx6+PV+ cells (##P < 0.01). Data are presented as means ± SEM.
Our above observation begs the question whether the decreased PV+ interneuron density is a result of impaired PV protein expression. To answer this question, we performed staining with fluorescein-conjugated Vicia Villosa agglutinin (VVA), a lectin labeling the perineural network around most cortical PV+ interneurons independent of PV expression (31). We detected a comparable percentage of PV+ VVA+ interneurons in Gpr56 cKO mice and controls (Fig. 2C), suggesting a decrease in the cell number other than PV expression in the setting of microglial Gpr56 deletion.
Both cortical PV+ and SST+ interneurons express LIM homeobox 6–positive (Lhx6+) into adulthood (32). We performed triple immunohistochemistry (IHC) of Lhx6, PV, and SST. We detected a significant reduction in the number of Lhx6+ cells and Lhx6+PV+ cells in the SSC of Gpr56 cKO mice compared to the controls (Fig. 2, D and E). On the other hand, Lhx6+SST+ and Lhx6+SST−PV− cell numbers were comparable between Gpr56 cKO mice and controls (Fig. 2, D and E). In summary, our study results support the idea that deleting microglial Gpr56 leads to a decrease in the number of PV+ interneurons.
Deleting microglial Gpr56 decreases PV+ interneurons born at E14.5 and E15.5
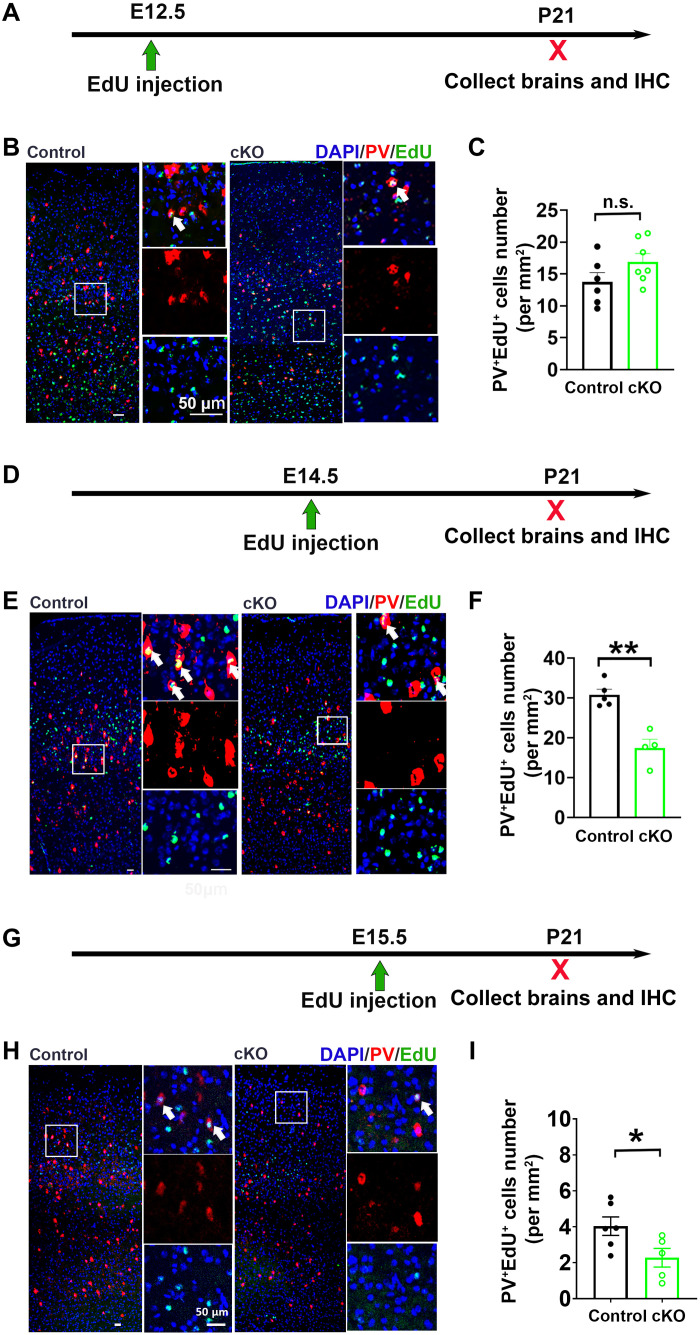
To study whether a temporal factor is involved in Gpr56 cKO–induced PV+ interneuron reduction, we performed a fate-mapping experiment by injecting 5-ethynyl-2′-deoxyuridine (EdU) at either E12.5, E14.5, or E15.5 followed by quantification of PV+EdU+ interneurons in the SSC at P21 (Fig. 3). We found that EdU-labeled PV+ interneurons were mostly distributed in layer V/VI of the SSC of control mice when injected at E12.5, in layer IV/V when injected at E14.5, and in layer II/III and upper layer IV when injected at E15.5 (Fig. 3, B, E, and H), consistent with previous reports (12, 33). The density of PV+EdU+ interneurons in the SSC was comparable between controls and Gpr56 cKO mice when EdU was injected at E12.5 (Fig. 3, B and C) but significantly reduced in Gpr56 cKO mice when EdU was injected at E14.5 and E15.5 (Fig. 3, E, F, H, and I). Together, our study results indicate that microglial Gpr56 deletion affects PV+ interneuron development in a temporal manner.
Fig. 3. Deleting microglial Gpr56 impairs mid- and late-born PV+ interneurons in the MGE.
(A) The experimental design to investigate the PV+ interneuron lineage born at E12.5, the early-stage neurogenesis. (B) Representative images of EdU and PV staining in the SSC of Gpr56 cKO mice and controls. (C) A bar graph showing no significant difference in the number of PV+EdU+ cells between control and Gpr56 cKO mice (P = 0.13); n = 6 in control and n = 7 in cKO. (D and G) The experimental design to evaluate the PV+ interneuron lineage born at E14.5 and E15.5, the mid- and late-stage neurogenesis. (E and H) Representative images of EdU and PV staining in the SSC of Gpr56 cKO mice and controls. (F and I) Bar graphs showing the number of PV+EdU+ cells is significantly decreased in the SSC of microglial Gpr56 cKO mice when EdU was injected at E14.5 or E15.5. [n = 5 and 4 in (D) to (F); n = 6 and 5 in (G) to (I)]. White arrows indicate PV+EdU+ cells. Unpaired t test. *P < 0.05 and **P < 0.01. Data are presented as means ± SEM.
Microglial Gpr56 cKO affects mid- and late-stage progenitor proliferation in the MGE
To investigate the cellular mechanism underlying PV+ interneuron deficits, we first examined the potential role of microglial Gpr56 in cell death. We used cleaved caspase 3 to label apoptotic cells at P1, known as the starting time point of interneuron apoptosis, and at P10, a time point with high interneuron apoptosis (34), in the SSC region. We observed a comparable number of apoptotic cells between control and microglial Gpr56 cKO mice at both time points (fig. S3).
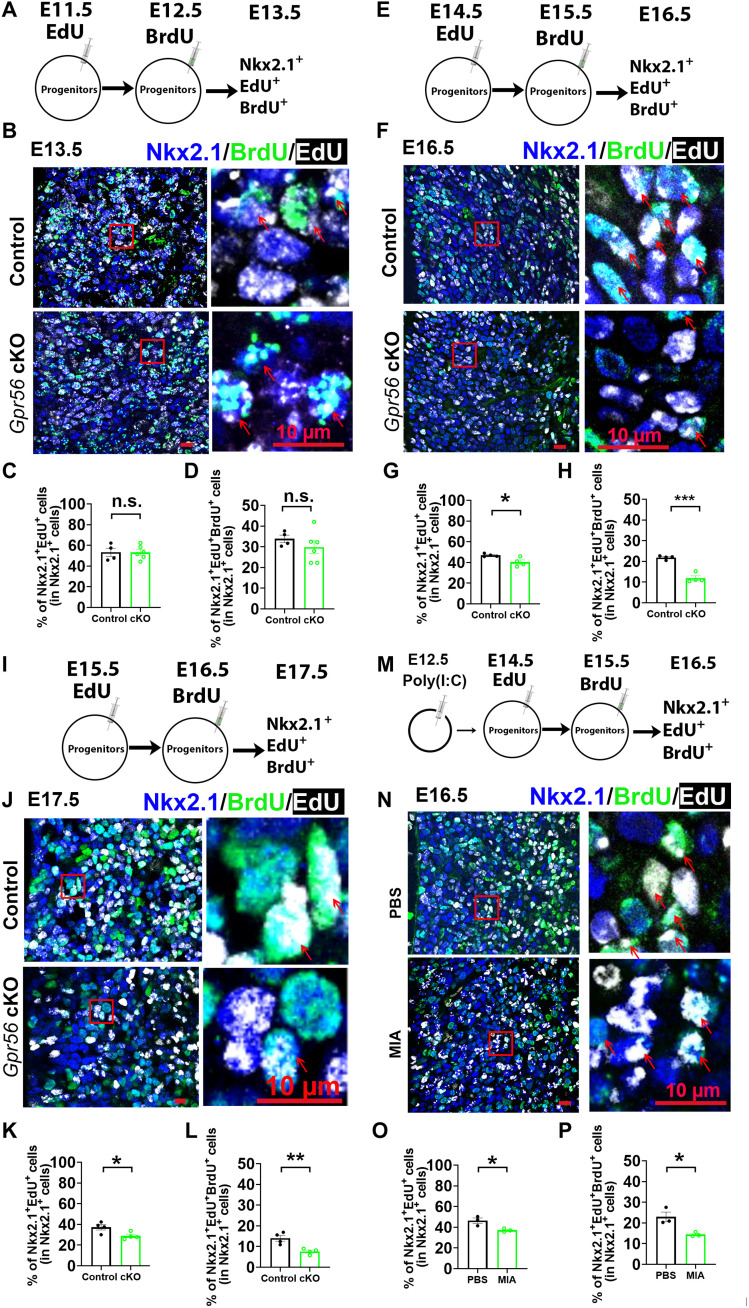
Next, we investigated the role of microglial Gpr56 in MGE progenitor proliferation by sequential labeling with EdU and 5-bromo-2′-deoxyuridine (BrdU). We examined early neurogenesis by intraperitoneally injecting EdU to pregnant dams at E11.5 and BrdU at E12.5. The fetal brains were collected at E13.5 and subjected to triple IHC with EdU, BrdU, and the MGE marker NK2 homeobox 1 (Nkx2.1) (Fig. 4A). The percentage of both EdU-labeled and EdU/BrdU double-labeled Nkx2.1 cells was comparable between control and cKO mice, suggesting no difference in MGE proliferation between control and Gpr56 cKO mice during E11.5 to E13.5 (Fig. 4, B to D). To investigate the role of microglial GPR56 in mid-stage neurogenesis in the MGE, we examined MGE proliferation from E14.5 to E16.5 by injecting EdU at E14.5 and BrdU at E15.5 and collecting the fetal brains at E16.5 (Fig. 4E), while in late-stage neurogenesis, we examined MGE proliferation from E15.5 to E17.5 by injecting EdU at E15.5 and BrdU at E16.5 and collecting at E17.5 (Fig. 4I). Excitingly, we found a significantly decreased percentage of Nkx2.1+EdU+ cells and Nkx2.1+EdU+BrdU+ cells in Gpr56 cKO mice at both time points (Fig. 4, F to H and J to L), suggesting that microglial Gpr56 deletion impairs progenitor proliferation during mid- and late-stage MGE neurogenesis.
Fig. 4. Both microglial Gpr56 deletion and MIA impairs mid-stage neurogenesis in MGE.
(A) A schematic drawing of early neurogenesis evaluation. (B) Representative images of triple staining of EdU, BrdU, and Nkx2.1 in the E13.5 MGE with EdU injected at E11.5 and BrdU injected at E12.5. (C and D) Bar graphs showing no significant differences between control and Gpr56 cKO mice in the percentages of Nkx2.1+EdU+ cells (C; P = 0.98) and Nkx2.1+EdU+BrdU+ cells (D; P = 0.35). n = 4 in control and n = 6 in cKO. (E to H) Triple staining of EdU, BrdU, and Nkx2.1 to evaluate mid-stage MGE neurogenesis ranging from E14.5 to E16.5. Bar graphs show significantly decreased percentages of Nkx2.1+EdU+ cells (G) and Nkx2.1+EdU+BrdU+ cells (H) in the MGE of Gpr56 cKO mice compared to controls. n = 4. (I to L) Triple staining of EdU, BrdU, and Nkx2.1 to evaluate late-stage MGE neurogenesis ranging from E15.5 to E17.5. Bar graphs showing significantly decreased percentages of Nkx2.1+EdU+ cells (K) and Nkx2.1+EdU+BrdU+ cells (L) in the MGE of Gpr56 cKO mice compared to controls. n = 4. (M to P) Triple staining of EdU, BrdU, and Nkx2.1 to evaluate mid-stage MGE neurogenesis ranging from E14.5 to E16.5 in MIA fetal brains. Bar graphs show a significantly decreased percentage of Nkx2.1+EdU+ cells (O) and Nkx2.1+EdU+BrdU+ cells (P) in the MGE of MIA offspring. n = 3. Red arrows show examples of Nkx2.1+EdU+BrdU+ cells. Unpaired t test. *P < 0.05, **P < 0.01, and ***P < 0.001. Data are presented as means ± SEM.
We questioned whether MIA has a similar impact on neurogenesis in the MGE. To address this question, we induced MIA by injecting poly(I:C) at E12.5, followed by sequential EdU and BrdU labeling at E14.5 and E15.5, as described above (Fig. 4M). We saw a significant decrease in MGE progenitor proliferation in MIA fetal brains, consistent with our results in Gpr56 cKO mice (Fig. 4, N to P), indicating that MIA impairs neurogenesis in the MGE of offspring in a similar manner as deleting microglial Gpr56.
Deleting Gpr56 in microglia leads to decreased microglia density and ramification in the MGE and elevates microglial TNFα mRNA expression
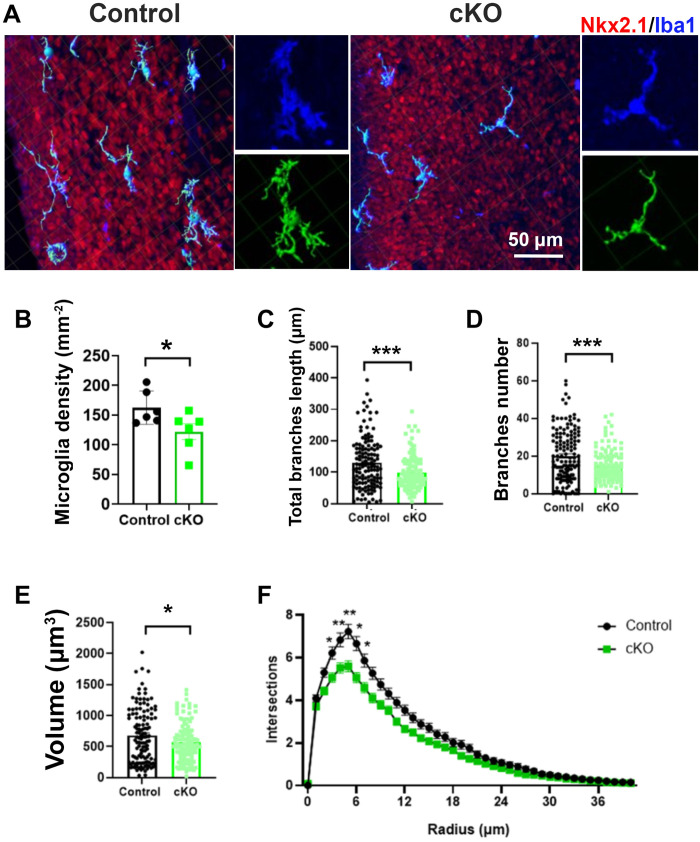
Our study results thus far indicate that microglial GPR56 is important for the development of PV+ interneurons. To test the role of GPR56 in microglial function, we first examined microglial cell number and morphology in the E16.5 MGE of Gpr56 cKO and control mice with IHC of ionized calcium-binding adaptor molecule 1 (Iba1), which labels microglia and macrophages and provides excellent visualization of microglial morphology (35). We observed a significant decrease in the density of microglia in the MGE of Gpr56 cKO mice (Fig. 5, A and B). Three-dimensional reconstruction of microglia showed that the length and number of branches, as well as the cell volume, were significantly reduced in microglia in the MGE of Gpr56 cKO mice compared to controls (Fig. 5, A and C to E). Furthermore, Sholl analysis confirmed less ramified microglial morphology in the MGE of Gpr56 cKO mice compared to controls (Fig. 5, A and F).
Fig. 5. Deleting microglial Gpr56 alters microglial density and morphology in the MGE.
(A) IHC and three-dimensional reconstructions (green) of microglia in the E16.5 MGE of control and Gpr56 cKO brains. (B) Microglia density is significantly reduced in the E16.5 MGE of Gpr56 cKO brains compared to controls. n = 6. (C to E) The number and length of branches, as well as cell volume, were significantly decreased in Gpr56 cKO microglia in the MGE compared to controls. Cell numbers: 132 in control and 121 in cKO. (F) Sholl analysis indicates a reduction in the ramification of microglia in the MGE of Gpr56 cKO mice compared to controls. Unpaired t test for (B) to (E) and two-way ANOVA and post hoc Bonferroni’s test for (F). *P < 0.05, **P < 0.01, and ***P < 0.001. Data are presented as means ± SEM.
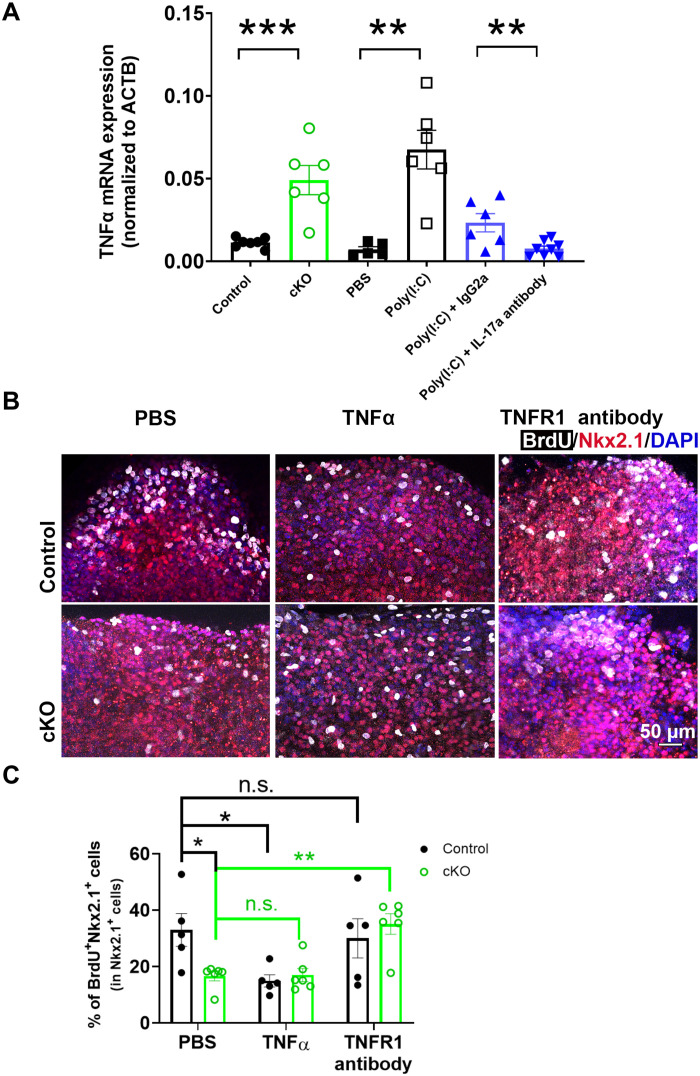
Cytokines and growth factors are key modulators in neurogenesis (36). Notably, microglia-specific cKO of Ras homolog family member A (RhoA) increases TNFα expression in microglia (37). Furthermore, elevated TNFα inhibits neurogenesis (38). Since GPR56 activates the RhoA pathway (39, 40), we reasoned that microglial GPR56 might function through the TNFα pathway. To test this hypothesis, we first examined TNFα mRNA levels in purified E14.5 microglia. We detected a significantly higher level of TNFα transcripts in microglia isolated from Gpr56 cKO brains compared to controls (Fig. 6A). We saw a similar elevation of TNFα transcripts in E14.5 microglia isolated from MIA fetal brains in an IL-17a–dependent manner (Fig. 6A).
Fig. 6. Elevated TNFα mediates Gpr56 cKO–induced MGE proliferation deficits.
(A) TNFα mRNA was significantly elevated in E14.5 microglia isolated from Gpr56 cKO and MIA fetal brains. IL-17a–neutralizing antibodies significantly attenuated microglial TNFα mRNA elevation in MIA-treated fetal brains. n = 5 to 8. ACTB, actin beta. (B) Representative images of BrdU and Nkx2.1 in ex vivo culture slices. (C) The percentage of BrdU+ cells in the MGE were significantly decreased in Gpr56 cKO slices compared to control slices under PBS treatment. In control slices, TNFα treatment significantly decreased the percentage of BrdU+ progenitors in the MGE, while treatment with TNFR1-neutralizing antibodies had no effect (P > 0.99). In Gpr56 cKO slices, TNFR1-neutralizing antibody treatment significantly increased MGE proliferation, while TNFα treatment did not alter MGE proliferation (P > 0.99). Control: n = 5; cKO: n = 6. Unpaired t test for (A) and two-way ANOVA and Bonferroni’s post hoc test for (C). *P < 0.05, **P < 0.01, and ***P < 0.001. Data are presented as means ± SEM.
To further examine whether TNFα plays a role in MGE neurogenesis, we performed ex vivo embryonic brain slice cultures. We treated the brain slices with carrier solution PBS, TNFα, or TNF receptor 1 (TNFR1)–neutralizing antibodies for 48 hours and pulsed with BrdU for the last 12 hours (Fig. 6B). Excitingly, the percentage of BrdU+Nkx2.1+ cells in the MGE was significantly decreased in control slices treated with TNFα compared to those treated with PBS (Fig. 6, B and C), indicating that TNFα impairs MGE proliferation. To explore whether elevated TNFα is the molecular mechanism underlying Gpr56 cKO–induced MGE proliferation impairment, we treated Gpr56 cKO brain slices with carrier solution PBS, TNFα, or TNFR1-neutralizing antibodies. We observed a significant decrease in MGE proliferation in Gpr56 cKO slices compared to controls in the PBS-treated group, confirming that deleting microglial Gpr56 impairs MGE proliferation (Fig. 6, B and C). Furthermore, TNFR1-neutralizing antibody treatment significantly increased MGE proliferation, whereas TNFα treatment did not affect MGE proliferation in Gpr56 cKO slices compared to controls (Fig. 6, B and C). Together, our study results support the idea that raised TNFα expression mediates Gpr56 cKO–induced MGE proliferation deficits.
Restoring microglial Gpr56 expression corrects PV+ interneuron deficits in Gpr56 global KOs and MIA offspring
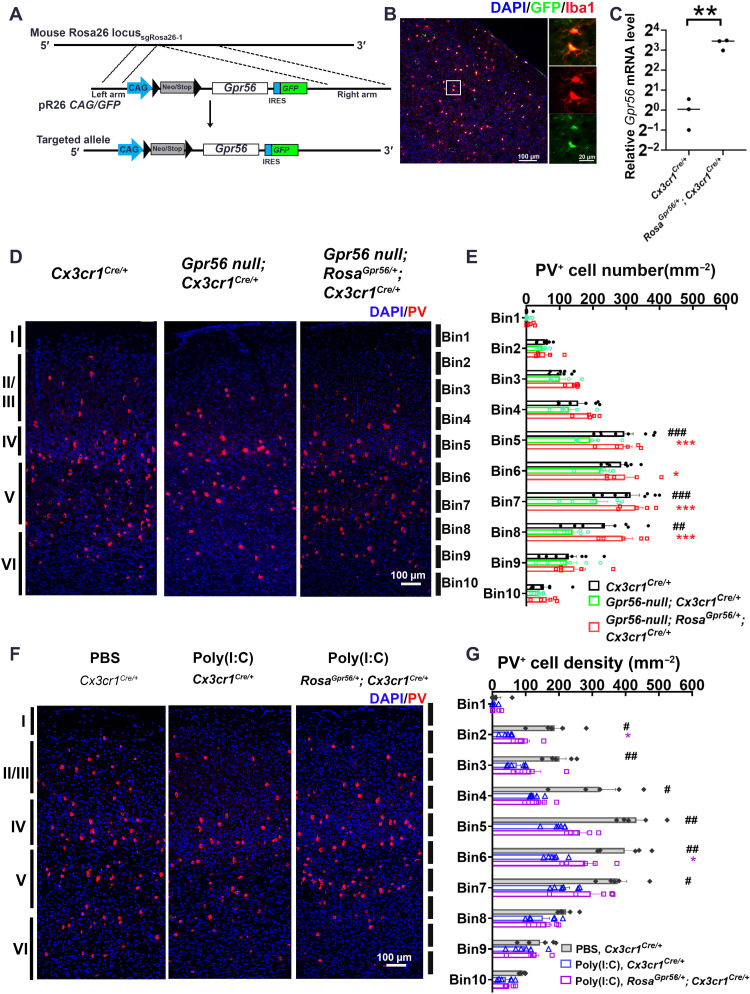
To unambiguously establish the role of microglial Gpr56 in interneuron development, we engineered a new transgenic mouse line, RosaGpr56, that drives Gpr56 expression in a cell-specific manner (Fig. 7A). Upon crossing with Cx3cr1-Cre mice, we detected specific cre-loxP recombination in microglia (Fig. 7B) and a high level of Gpr56 expression in purified microglia from E14.5 RosaGpr56/+; Cx3cr1Cre/+ mouse brains (Fig. 7C).
Fig. 7. Restoring microglial Gpr56 expression corrects PV+ interneuron deficits in both global Gpr56 KO mice and MIA offspring.
(A) Strategy for the generation of the RosaGpr56 mouse line. IRES, internal ribosomal entry site. (B) IHC staining showing reporter green fluorescent protein (GFP) is colocalized with Iba1 in the RosaGpr56/+; Cx3cr1Cre/+ mouse brain. (C) Reverse transcription qPCR (RT-qPCR) showing Gpr56 mRNA expression is largely increased in FACS-isolated microglia from RosaGpr56/+; Cx3cr1Cre/+ mice (n = 3). (D) Representative images of PV+ interneurons in the posterior SSC. (E) A bar graph showing PV+ interneuron density in the posterior SSC is decreased in global Gpr56 KO mice compared to control mice, and restoring microglial Gpr56 expression corrects the PV+ interneuron deficits. n = 8 (Cx3cr1Cre/+), n = 6 (Gpr56 null; Cx3cr1Cre/+), and n = 5 (Gpr56 null; RosaGpr56/+; Cx3cr1Cre/+). (F) Representative images of PV+ interneurons in the SSC of control mice (PBS, Cx3cr1Cre/+), MIA offspring [poly(I:C), Cx3cr1Cre/+], and MIA offspring with restored expression of microglial Gpr56 [poly(I:C), RosaGpr56/+; Cx3cr1Cre/+]. (G) A bar graph showing PV+ interneuron density is decreased in MIA offspring compared to controls. The density of PV+ interneurons is increased in MIA offspring with restored expression of microglial Gpr56 compared to control MIA offspring. n = 5 (PBS, Cx3cr1Cre/+), n = 6 [poly(I:C), Cx3cr1Cre/+], and n = 6 [poly(I:C), RosaGpr56/+; Cx3cr1Cre/+]. Unpaired t test for (C): **P < 0.01; two-way ANOVA and post hoc Bonferroni’s test for (E): ##P < 0.01 and ###P < 0.001 between Cx3cr1Cre/+ and Gpr56 null; Cx3cr1Cre/+; *P < 0.05, **P < 0.01, and ***P < 0.001 between Gpr56 null; Cx3cr1Cre/+ and Gpr56 null; RosaGpr56/+; Cx3cr1Cre/+. Two-way ANOVA and post hoc Bonferroni’s test for (G): #P < 0.05 and ##P < 0.01 between PBS, Cx3cr1Cre/+ and poly(I:C), and Cx3cr1Cre/+; *P < 0.05 between poly(I:C), Cx3cr1Cre/+ and poly(I:C), and RosaGpr56/+; Cx3cr1Cre/+. Data are presented as means ± SEM.
We first sought to rescue PV+ interneuron phenotypes associated with Gpr56 deletion by crossing RosaGpr56/+; Cx3cr1Cre/+ with Gpr56 null mice, in which Gpr56 is removed globally (19). As deleting Gpr56 leads to cortical lamination defects in the frontal cortex (41), we quantified the distribution of PV+ interneurons in the posterior part of the SSC to stay away from regions with cortical ectopias (19, 41). We detected a decreased PV+ interneuron density in the posterior SSC of Gpr56 null mice (Gpr56null; Cx3cr1Cre/+) compared to controls (Cx3cr1Cre/+) at P21 (Fig. 7, D and E), a similar phenotype seen in microglial Gpr56 cKO mice. The density of PV+ interneurons in Gpr56null; RosaGpr56/+; Cx3cr1Cre/+ mice are at a similar level as in Cx3cr1Cre/+ control mice, supporting a cell-autonomous function of microglial Gpr56 in PV+ interneuron development.
To confirm the involvement of Gpr56 in MIA-induced PV+ interneuron reduction, we investigated whether RosaGpr56/+; Cx3cr1Cre/+ mice would resist MIA-induced PV+ interneuron deficits. We observed that MIA induction at E12.5 significantly decreased the density of PV+ interneurons in layers II to V of the SSC in P21 Cx3cr1Cre/+ mice (Fig. 7, F and G). As expected, PV+ interneuron density was significantly improved in layers II (Bin2) and V (Bin6) of the SSC from MIA-treated RosaGpr56/+; Cx3cr1Cre/+ offspring compared to MIA-treated Cx3cr1Cre/+ offspring (Fig. 7, F and G), indicating that microglial Gpr56 down-regulation is a critical link between MIA and PV+ interneuron reduction. The density of PV+ interneurons in MIA offspring with restored expression of microglial Gpr56 did not fully reach the levels of PBS-treated controls, suggesting that other pathways may also be involved in the effect of MIA on interneuron development.
Gpr56 cKO mice exhibit autism-like behaviors, and restoring microglial Gpr56 expression rescues behavior abnormalities in MIA offspring
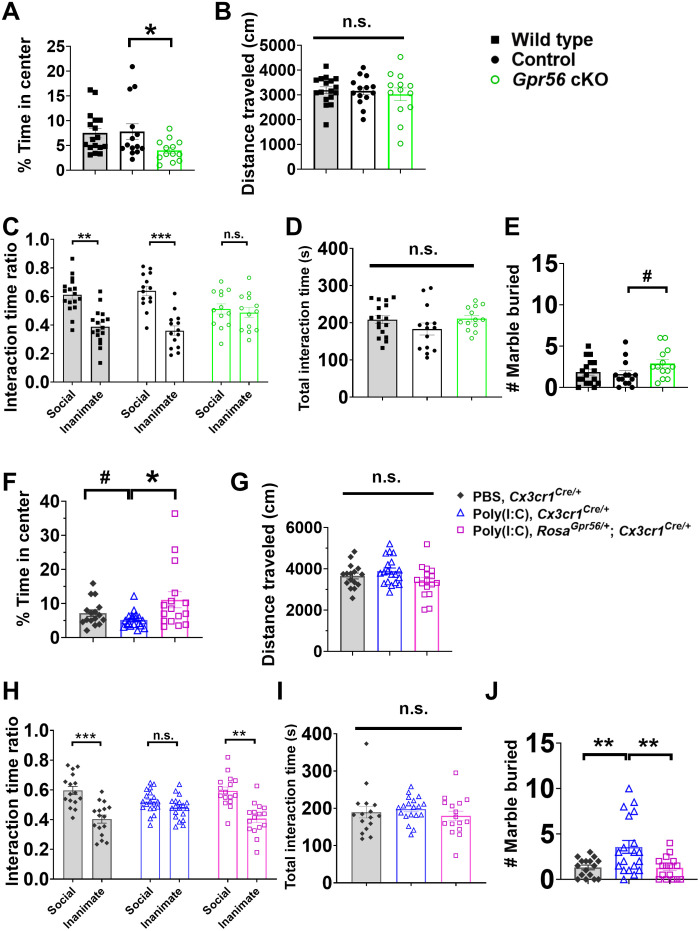
We investigated the functional relevance of microglial Gpr56 in respect to mouse behaviors. First, we evaluated mouse anxiety levels using an open-field test (7). While there was no difference in the time spent in the center of the open field between wild-type and Cx3cr1Cre/+ mice, we found Gpr56 cKO mice spent significantly less time in the center, indicating that Gpr56 cKO mice had an elevated anxiety level (Fig. 8A). Second, we tested social preference in our experimental mice with a three-chamber test (7). Mice in both wild-type and control groups showed a preference for a social animal, while Gpr56 cKO mice spent similar time between a social animal and an inanimate object (Fig. 8C). Third, we conducted a marble-burying test in our experimental mice to evaluate repetitive behaviors, which is another key feature of ASD (42). Our results showed a trend that Gpr56 cKO mice buried more marbles compared to controls (P = 0.058; Fig. 8E). Given that the total distances traveled in the open field and the total interaction time in the three-chamber test were comparable among different groups, all differences detected in these behavior tests were not due to activity or arousal level differences among different groups of mice (Fig. 8, B and D).
Fig. 8. GPR56 expression status in microglia affects mouse behaviors.
(A) The percentage of time spent in the center of an open field, (B) total distance traveled in an open field (P = 0.80), (C) interaction time ratio in the three-chamber test, (D) total interaction time in the three-chamber test (P = 0.23), and (E) number of marbles buried in marble-burying test in wild-type (n = 17), control (Cx3cr1Cre/+; n = 14), and Gpr56 cKO mice (Gpr56f/f; Cx3cr1Cre/+; n = 13). (F) The percentage of time spent in the center of an open field, (G) total distance traveled in an open field (P = 0.09), (H) interaction time ratio in the three-chamber test, (I) total interaction time in the three-chamber test (P = 0.55), and (J) number of marbles buried in marble-burying test in control mice (PBS, Cx3cr1Cre/+; n = 16), MIA offspring [poly(I:C), Cx3cr1Cre/+; n = 19], and MIA offspring with restored expression of Gpr56 in microglia [poly(I:C), RosaGpr56/+; Cx3cr1Cre/+; n = 16). Two independent experiments. Unpaired t test for (A), (E), (F), and (J); one-way ANOVA test for (B), (D), (G), and (I); and two-way ANOVA and post hoc Bonferroni’s test for (C) and (H). #P < 0.1, *P < 0.05, **P < 0.01, and ***P < 0.001. Data are presented as means ± SEM.
Next, we investigated whether sustained microglial Gpr56 expression corrects autism-like behaviors in MIA offspring. We observed that MIA offspring [poly(I:C), Cx3cr1Cre/+] exhibited social disability in the three-chamber test (Fig. 8H) and significantly elevated repetitive behaviors in the marble-burying test (Fig. 8J), consistent with literature (7, 29). Restoring microglial Gpr56 expression (Poly(I:C), RosaGpr56/+; Cx3cr1Cre/+) aborted those autism-like behaviors in MIA offspring (Fig. 8, F to J).
DISCUSSION
Microglia play an important role in MGE proliferation during mid-stage neurogenesis
Despite increasing evidence indicating the role of microglia in regulating excitatory (43) and adult neurogenesis (16, 44), our understanding of microglial function in interneuron development is sparse. Squarzoni et al. (24) reported that early microglia depletion affects interneuron positioning and numbers in the mouse cortex. Our results ascertain the role of microglia in MGE proliferation through a GPR56/TNFα pathway. Gpr56 cKO specifically impairs PV+, but not SST+, interneurons in the neocortex. The lack of reduction of calretinin+ and reelin+ interneurons suggests that CGE-derived interneurons are not affected by Gpr56 cKO (1). Selective alternations in PV+ interneurons have been reported in multiple psychiatric disorders with suspected neurodevelopmental etiology. For example, studies indicate specific PV+ interneurons deficits in MIA offspring and ASD mouse models (4, 10). It has also been found that PV+ interneurons were selectively decreased, or PV expression is reduced in brain tissues from patients with schizophrenia (45, 46).
The temporal effect of microglial Gpr56 cKO on MGE progenitor proliferation might explain the selective reduction of PV+ interneurons in cKO mice. Our fate-mapping experiments show that most PV+EdU+ cells were distributed in layer IV/V when EdU was injected at E14.5 (Fig. 3E). On the other hand, PV+EdU+ cells were more concentrated in layer V/VI when EdU was injected at E12.5 (Fig. 3B), which is consistent with previous observations that interneurons have an “inside-out” relationship between birthdate and laminar locations (47, 48). The reduction of PV+ interneuron density is mainly seen in layer IV/V of the Gpr56 cKO mouse SSC (Fig. 2, A and B), further supporting the time of action for microglial Gpr56 at mid-stage neurogenesis. Of note, we did not see a significant reduction in PV+ interneuron density in layer II/III of the SSC of Gpr56 cKO mice (Fig. 2, A and B) despite a reduced proliferation in MGE during E15.5 to E16.5 (Fig. 4, J to L). One possible explanation could be lower contribution of PV+ interneuron from late-stage neurogenesis relative to early- and mid-stage MGE neurogenesis. Our fate-mapping experiments showed a marked reduction of PV+ interneurons derived from E15.5 compared to E14.5—from ~30 PV+EdU+ cells/mm2 when EdU was injected at E14.5 to ~4 cells/mm2 at E15.5 (Fig 3, E, F, H, and I). Another possibility could be an unknown compensatory effect that is selective for the superficial cortical layer. Together, our data support that the temporal factor dictates the effect of microglial GPR56 function in MGE interneuron development. Future studies are needed to delineate whether spatial distribution (33) and intrinsic properties (49) of MGE progenitors also influence the impact of microglial GPR56.
This time-specific function of microglial GPR56 is consistent with the stepwise developmental program of microglia in the mouse brain (26). During early MGE neurogenesis, “early microglia” before E14.5 are enriched in genes associated with proliferation and the cell cycle, similar to myeloid progenitors in the yolk sac, while “premicroglia” from E14 to P9 show high heterogeneity and express distinct genes related to neural migration, neurogenesis, and cytokine release (26). Microglial Gpr56 transcripts were increased ~8-fold from E12.5 to E14.5, followed by a gradual elevation to an adult level at early postnatal age (19, 26). Therefore, a potential explanation for the time-specific function of microglial GPR56 in MGE neurogenesis is that only those “premicroglia,” which have high expression of Gpr56 and other genes related to neurogenesis and cytokine release, are able to influence MGE progenitor proliferation.
Microglial GPR56 functions as a key regulator in neuroinflammation
Microglial GPR56 is down-regulated in the setting of inflammation. Down-regulation of microglial Gpr56 was observed in both embryonic brains in response to MIA (Fig. 1) and adult brains upon lipopolysaccharide-induced neuroinflammation in mice (27). In humans, GPR56 expression was significantly reduced in microglia isolated from the white matter of individuals with multiple sclerosis, a disease characterized by neuroinflammation and demyelination (50). Moreover, Gosselin et al. (51) showed that cultured primary human microglia expressed high level of genes related to neuroinflammation, while the expression of GPR56, together with other homeostatic genes such as P2RY12 and CX3CR1, was markedly down-regulated. Together, literatures and our results support the idea that GPR56 down-regulation in microglia is indicative of neuroinflammation.
Deleting microglial GPR56 results in less ramification and elevated TNFα expression in microglia (Figs. 5 and 6A). The role of microglial GPR56 in microglial morphology and TNFα expression may be mediated by the RhoA pathway. First, GPR56 couples to Gα12/13 and activates the RhoA pathway in multiple types of cells (15, 39, 40, 52). Second, the RhoA pathway is a key modulator of cytoskeleton dynamics (53). Last, the RhoA pathway plays a critical role in TNFα production, as microglial RhoA cKO mice showed an elevated expression of TNFα in microglia (37). It would be interesting to further investigate whether RhoA is the downstream signaling pathway of GPR56 in microglia.
Our research did not rule out the involvement of other pathways in MIA-induced interneuron deficits. In addition to TNFα, other cytokines have also been known to play a role in the regulation of neurogenesis (36, 54). For example, IL-1β has been reported to be regulated by the RhoA pathway (55) and inhibit neurogenesis (54). It would be worthwhile to investigate the comprehensive effect of GPR56 on interneuron development through modulation of cytokine release.
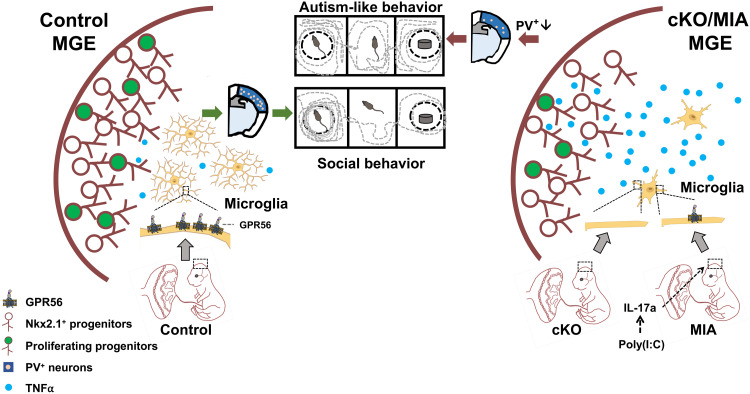
Microglial GPR56 is a molecular target of MIA-induced neurodevelopmental disorder
Epidemiological studies link maternal inflammation to an increased risk of ASD in offspring (8, 56–58). Animal studies recapitulate this finding and reveal PV+ interneuron deficits as a major mechanism (6, 9). However, little is known about the underlying molecular mechanism in the fetal brain. Here, we show that fetal microglial Gpr56 down-regulation, either in response to MIA or in our Gpr56 cKO model, impairs MGE proliferation at late-stage neurogenesis and results in PV+ interneuron deficits in the neocortex and autism-like behaviors in adult mice through elevated TNFα expression (Fig. 9). We did not detect any significant change in Gpr56 expression in nonmicroglial cells isolated from MIA offspring. Restoration of microglial Gpr56 expression in Gpr56 global knockout mice ameliorates PV+ interneuron reduction in the SSC, supporting a cell-autonomous role of microglial Gpr56 in PV+ interneuron development. Moreover, sustained microglial Gpr56 expression attenuates MIA-induced PV+ interneuron deficits and autism-like behaviors. Together, our study results support the idea that microglial Gpr56 serves as a molecular target of MIA-associated neurodevelopmental disorders. We demonstrated that MIA down-regulates fetal microglial Gpr56 expression via elevated maternal IL-17a levels. Further epidemiological studies are needed to test whether an elevated maternal IL-17a level is associated with increased risk of developing autism in offspring. If so, one could potentially treat pregnant people with a short course of IL-17a–neutralizing antibodies in the setting of MIA. In this regard, secukinumab, an anti–IL-17a monoclonal antibody, is a Food and Drug Administration–approved therapy for adult autoimmune disorders (59).
Fig. 9. Microglial GPR56 function.
In normal conditions, microglial GPR56 regulates microglia density, ramification, and cytokine release. Deletion or MIA-induced down-regulation of microglial Gpr56 leads to elevated microglial TNFα, which results in MGE proliferation deficits at mid-/late-stage neurogenesis, PV+ interneuron reduction in the neocortex, and autism-like behaviors in adult mice.
MATERIALS AND METHODS
Animals
All mice were handled according to the guidelines of the Institutional Animal Care and Use Committee at the University of California, San Francisco. For all experiments, animals were independently randomized to different experimental groups. Gpr56f/f mice were generated by our laboratory and have been previously reported (15, 52). Cx3cr1Cre/+ mice were purchased from the Jackson Laboratory (strain no. 025524). Gpr56f/f; Cx3cr1Cre/+ mice were named as Gpr56 cKO mice and Gpr56+/+; Cx3cr1cre/+ mice as control mice. Both male and female mice were used for experiments and quantifications other than behavior tests. For the behavioral assay, only males aged between 2 and 3 months old were used as experimental mice according to previous research (6, 29). Gpr56 global knockout mice, Gpr56 null, were generated by crossing Gpr56f/f mice with CMVCre mice (the Jackson Laboratory, no. 006054), and CMVCre was removed by further crossing with C57/B6 mice (19). RosaGpr56 mice were generated via CRISPR-Cas9 as previously reported (60). Briefly, wild-type murine Gpr56 complementary DNA was inserted into the mouse Rosa26 locus under the CAG promoter and floxed by Neo/Stop cassette through modifying the targeting vector Rosa26 CAG/GFP Asc, a gift from the laboratory of M. Kühn. Modified Cas9 mRNA and single-guide RNA targeting the intronic Xba I site of Rosa26 were microinjected into C57/B6 zygotes together with targeting vectors. The transgenic RosaGpr56 mice were screened from survived offspring by three different polymerase chain reaction (PCR) reactions, indicating correct insertion of the transgene (60).
For timed pregnancy, males and females were mated together overnight and separated on the second morning. After the mating night, females were placed back in their home cages. The noon of the day showing vaginal plugs was noted as E0.5. In the BrdU or EdU labeling experiment, a single dose of BrdU (100 mg/kg; Abcam, ab142567) or EdU (10 mg/kg; provided in Click-it EdU Alexa Fluor 647 Imaging Kit from Invitrogen, #C10340) was injected intraperitoneally into pregnant mice at the designed time points. Wild-type mice used for MIA induction were C57/B6 mice purchased originally from Taconic Biosciences and bred in the laboratory. In the MIA rescue experiment, all mice were cohoused with the mice ordered from Taconic Biosciences for 2 to 3 weeks. All female mice used for MIA had been tested to have segmented filamentous bacteria according to previously reported methods (29). For the MIA induction study, pregnant females were intraperitoneally injected with poly(I:C) (20 mg/kg; Sigma-Aldrich, P9582) or PBS. Successful induction of MIA was further confirmed by elevated IL-6 serum concentration in 3 hours after poly(I:C) injection and elevated IL-17a serum concentration at E14.5. In the antibody rescue experiment, IL-17a–neutralizing antibodies (10 g/kg; R&D Systems, MAB421) and IgG2a isotype controls (10 g/kg; R&D Systems, IC006V) were intraperitoneally injected 6 hours before poly(I:C) injection.
Enzyme-linked immunosorbent assay
Blood was collected from lateral tail veins of pregnant dams 3 and 48 hours after poly(I:C) or PBS injection. Fresh blood was left undisturbed at room temperature for 30 min to clot. Serum was isolated from clotted blood by centrifuging for 15 min at 1000 rpm at 4°C (Eppendorf Centrifuge 5415D) and was diluted 10 times for interleukin enzyme-linked immunosorbent assay (ELISA) tests. IL-6 (Invitrogen, #88-7064) and IL-17A ELISA kits (BioLegend, #432501) were used to test the concentration of IL-6 and IL-17A in the serum following the manufacturer’s instructions. Briefly, standard IL-6 and IL-17A provided in the kits were diluted sequentially and applied to make a standard curve. Absorbances at 450 nm (A450) were read, and the concentrations of IL-6 and IL-17A of samples were calculated according to A450 and the standard curve. The original concentration of plasma was calculated accordingly.
Immunohistochemistry
We followed the IHC protocol as previously reported (19). All postnatal mouse brains were collected after transcardial perfusion with ice-cold PBS followed by 4% paraformaldehyde (PFA; Thermo Fisher Scientific, J19943-K2) and were post-fixed in 4% PFA overnight. Embryonic brains were collected without perfusion and fixed directly in 4% PFA overnight. All samples were moved to 30% sucrose in PBS overnight for cryostat protection followed by O.C.T. (SCIgen, #4586) embedding. Brain sections of 12 or 40 μm were used for IHC.
Antigen retrieval was performed for 12 min at 95° to 99°C with antigen retrieval buffer (BD Pharmingen, #550524). After antigen retrieval, brain slices were blocked in blocking buffer [5% serum, 1% bovine serum albumin (BSA), and 0.025% Triton X-100 in PBS] for 1.5 hours before proceeding to primary and secondary antibody incubation at room temperature. For VVA staining, fluorescein-conjugated VVA (Vector Laboratories, FL-1231) was diluted at a 1:200 ratio and applied to mouse brain slices together with PV primary antibodies to incubate overnight. For slices from the embryonic stage, TrueBlack Lipofuscin Autofluorescence Quencher (Biotium, #23007) at a dilution ratio of 1:20 in 70% alcohol was applied to slices for 30 s to block autofluorescence. For BrdU staining, brain slices were treated with 1 M HCl for 1 hour at 37°C and neutralized with 0.1 M sodium borate (Sigma-Aldrich, SX0355) before antigen retrieval. For EdU staining, EdU working solution was applied to slices after IHC following the manufacturer’s instructions. All slides were counterstained with 4′,6-diamidino-2-phenylindole (1:1000) before mounting (VWR International Inc., H-5000-60) and microscope imaging. For microglia morphology analysis, the IMARIS software (Oxford Instruments) was used for three-dimensional reconstruction of microglia.
The antibodies used in this study are as follows: mouse anti-SatB2 (Abcam, ab51502), guinea pig anti-Iba1 (Synaptic system, catalog no. 234004), rabbit anti–cleaved caspase 3 (Cell Signaling Technology, catalog no. 9661S), rabbit anti-PV (Swant, PV27), rat anti-SST (EMD, MAB354), rat anti-BrdU (Abcam, ab6326), rabbit anti-calretinin (Abcam, ab702), mouse anti-calbindin (ab82812), rabbit anti-reelin (Abcam, ab230820), rabbit anti-Nkx2.1 (Abcam, ab76013), and mouse anti-Lhx6 (Santa Cruz Biotechnology, sc-271433).
Microglia isolation
E14.5 embryonic brains were collected without meninges and homogenized in ice-cold DPBS (Dulbecco’s Phosphate-Buffered Saline) with a 2-ml pestle-mortar system on ice. For MIA induction, two to three brains from the same litter were pooled and homogenized together. For RosaGpr56 mice, each brain was individually homogenized, and two to three brain homogenates with the same genotypes from the same litter were pooled after homogenization. Brain homogenates were centrifuged at 300g for 10 min at 4°C to produce cell pellets. Cell pellets were washed and resuspended in fluorescence-activated cell sorting (FACS) buffer (Hanks’ balanced salt solution + 0.5% BSA) as a single-cell suspension. The single-cell suspension was then blocked in rat anti-mouse CD16/32 (1:100) for 20 min and further stained with fluorescence-conjugated rat anti-mouse CD45 (1:50), rat anti-mouse CD11b (1:50), and LIVE/DEAD marker (DraQ7 far red, 1:100). BD FACS Aria2 SORP Cell Sorters with 70-μm nozzles were used for microglia sorting. DraQ7−CD45mediumCD11b+ cells were collected as microglia, and DraQ7−CD45−CD11b− cells were collected as nonmicroglial cells. The cells were sorted directly into RLT lysis buffer from the RNeasy Mini Kit (QIAGEN, 74104) and stored at −80°C for later RNA extraction.
RNA extraction and reverse transcription quantitative PCR
RNA was extracted from isolated microglia and nonmicroglial cells with the RNeasy Mini Kit (QIAGEN, 74104), following the manufacturer’s instructions. Reverse transcription was conducted using the iScript Reverse Transcription Supermix (Bio-Rad, 1708840). Quantitative PCR (qPCR) was conducted with the iTaq Universal SYBR Green Supermix (Bio-Rad, 1725121) in a Roche LightCycler 480 Instrument II. Gpr56 mRNA expression level was tested with primers: 5′- TTGCTGCCTACCTCTGCTCC-3′ and 5′-AGCAGGAAGACAGCGGACAG-3′. ΤΝFα mRNA expression was tested with primers: 5′-GACGTGGAACTGGCAGAAGAG-3′ and 5′-TGCCACAAGCAGGAATGAGA-3′. The expression of Gpr56 mRNA and TNFα tested in the reverse transcription qPCR (RT-qPCR) experiments was normalized to the widely used housekeeping reference gene β-actin (Actb). Actb mRNA was tested with primers: 5′-CAGCAAGCAGGAGTACGATGAGTC-3′ and 5′-CAGTAACAGTCCGCCTAGAAGCAC-3′.
Ex vivo slice culture
Embryonic brains were isolated at E13.5 and embedded with 4% low-melt agarose (Apex, CAS no.: 9012-36-6). Brain slices (200 μm) with the MGE were collected with a Leica Vibratome (Leica VT 1000s) and placed onto inserts (0.4 μm; Millicell, Millipore) in Neurobasal medium (Gibco, #21103-049) supplemented with 2% B27 (Gibco, #17504-044), 2 mM GlutaMAX (Gibco, #35050061), and 1% penicillin-streptomycin (Gibco, #15070063). After 24 hours, the culture mediums were changed to new medium with PBS (1 μl per 1 ml medium), TNFα (100 ng/ml; Abcam, ab259411), or ΤΝFR1-neutralizing antibody (5 μg/ml; R&D systems, MAB430). Thirty-six hours later, the culture media in each condition were changed to a new medium with the same treatment plus 10 μM BrdU. All slices were fixed with 4% PFA for 20 min after 12 hours of incubation. In this way, the total treatment time for TNFα and TNFR1 antibody is 48 hours and 12 hours for BrdU.
For IHC, cultured slices were treated with 1 M HCl for 1 hour at 37°C for DNA hydrolysis and permeabilized with 10% Triton X-100 in PBS for 20 min. After neutralizing and washing, slices were blocked and incubated with primary antibodies over two nights. The following steps are the same as IHC mentioned above.
Mouse behavior assay
All experimental mice in behavior tests were 2- to 3-month-old males and housed at two to five mice per cage at all times unless otherwise specified. The genotyping and treatment conditions of all mice were blinded to experimental conductors and data analyzers. Arenas and contents were thoroughly cleaned between testing sessions with 70% ethanol.
Open-field test
All experimental mice were habituated in the experimental room for 1 hour before the experiment. During the experiment, mice were placed in one corner of an open testing arena (arena size, 40 cm by 40 cm; Kinder Scientific) with heads facing the corner and were allowed to explore the whole arena freely for 10 min. The center of the arena was defined as a 20 cm by 20 cm square in the center. Infrared photobeam breaks were recorded, and movement metrics were tracked and analyzed by ActiTrack tracking software (ActiTrack v2.7.13, Panlab). The percentages of time spent in the center and total distance were analyzed by the tracking software.
Three-chamber social tests
One week after the open-field test, experimental mice were conducted a 2-day, three-chamber sociability test. On day 1, experimental mice were single-housed for 1 hour in separate clean holding cages in the experimental room for habituation and then placed into a three-chamber arena (23.5 inches by 17.5 inches) with two empty object/mouse holders located in the left and right chamber. Experimental mice were allowed to explore the arena and holders for 10 min and then placed back in their home cages. On day 2, one holder contained an unfamiliar C57BL/6 mouse (social animal) at a similar age as the experimental mouse, and the other holder contained a plastic mouse toy of a similar size (inanimate object). Experimental mice were placed in the center chamber without access to the left and right chamber and were only allowed to explore the center. Five minutes later, gates to the lateral chambers were lifted, and mice were allowed to explore the whole arena freely for 10 min. The interaction was recognized when experimental mice were approaching either object holder within 2 cm with attention at the object holder. The sessions were video-recorded and analyzed with the SMART Video Tracking System (Smart v3.0, Panlab). Each social mouse was used no more than four times.
Marble-burying test
One week after the three-chamber social test, experimental mice were single-housed for 0.5 to 1 hour in separate clean holding cages in the experimental room for habituation. Mice were placed in a new home cage with deep bedding (arena size: 12.5 inches by 6.5 inches by 6 inches; bedding depth: 2 inches) containing 20 glass marbles in an array of 4 × 5 for 15 min. After this exploration period, mice were gently removed and put back in their home cage. The testing cages were recorded and manually counted: 1 for marbles covered more than 50%, 0.5 for ~50% covered, and 0 for anything less.
Statistical analysis
Statistical analyses were performed using the GraphPad Prism Version 8 software (GraphPad Software Inc., San Diego, CA). The results were analyzed with one-way analysis of variance (ANOVA) tests, two-way ANOVA followed by Bonferroni’s post hoc tests, or unpaired Student’s t tests as described in figure legends. Sample size n is defined in figure legends as well as individual dots in bar graphs. Each embryonic experiment was conducted with at least two litters in each condition. All average data are shown as means ± SE in bar graph [not significant (n.s.)]. * or #P < 0.05, ** or ##P < 0.01, and *** or ###P < 0.001 unless specified.
Acknowledgments
We thank R. Kühn of the Max-Delbrück-Center for Molecular Medicine for the gift of Rosa26 CAG/GFP Asc vector, M. Bhaumik of the Mouse Gene Manipulation Core at Boston Children’s Hospital for technical support in the generation of RosaGpr56 mice, and J. Huh for thoughtful comments on the manuscript.
Funding: This research is supported in part by the NIH/National Research Service Award Fellowships F32 F32MH118785 (to G.P.) and NIH/National Institute of Neurological Disorders and Stroke grants P01 NS083513 (to X.P.), R21 NS108312 (to X.P.), R01 NS094164 (to X.P.), and R01 NS108446 (to X.P.).
Author contributions: D.Y. and X.P. conceived the study, analyzed the data, and wrote the manuscript. D.Y. performed most of the experiments. T.L., J.-C.D., P.K., T.K., B.Z., G.P., and T.J.N. contributed to experiments and data analysis. K.J.B.P. and S.A.V. assisted in mouse behavioral assay. R.L. generated RosaGpr56 mouse line. All authors read and edited the manuscript.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. The Gpr56-floxed mice can be provided by X.P. pending scientific review and a completed material transfer agreement. Requests for the Gpr56-floxed mice should be submitted to X.P. (xianhua.piao@ucsf.edu).
Supplementary Materials
This PDF file includes:
Figs. S1 to S3
REFERENCES AND NOTES
- 1.Xu Q., Cobos I., De La Cruz E., Rubenstein J. L., Anderson S. A., Origins of cortical interneuron subtypes. J. Neurosci. 24, 2612–2622 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.D. M. Gelman, O. Marin, J. L. R. Rubenstein, in Jasper’s Basic Mechanisms of the Epilepsies, J. L. Noebels, M. Avoli, M. A. Rogawski, R. W. Olsen, A. V. Delgado-Escueta, Eds. (National Center for Biotechnology Information, 2012). [PubMed] [Google Scholar]
- 3.Cho K. K. A., Hoch R., Lee A. T., Patel T., Rubenstein J. L. R., Sohal V. S., Gamma rhythms link prefrontal interneuron dysfunction with cognitive inflexibility in Dlx5/6+/− mice. Neuron 85, 1332–1343 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lauber E., Filice F., Schwaller B., Parvalbumin neurons as a hub in autism spectrum disorders. J. Neurosci. Res. 96, 360–361 (2018). [DOI] [PubMed] [Google Scholar]
- 5.Sohal V. S., Rubenstein J. L. R., Excitation-inhibition balance as a framework for investigating mechanisms in neuropsychiatric disorders. Mol. Psychiatry 24, 1248–1257 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shin Yim Y., Park A., Berrios J., Lafourcade M., Pascual L. M., Soares N., Yeon Kim J., Kim S., Kim H., Waisman A., Littman D. R., Wickersham I. R., Harnett M. T., Huh J. R., Choi G. B., Reversing behavioural abnormalities in mice exposed to maternal inflammation. Nature 549, 482–487 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Choi G. B., Yim Y. S., Wong H., Kim S., Kim H., Kim S. V., Hoeffer C. A., Littman D. R., Huh J. R., The maternal interleukin-17a pathway in mice promotes autism-like phenotypes in offspring. Science 351, 933–939 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee B. K., Magnusson C., Gardner R. M., Blomström Å., Newschaffer C. J., Burstyn I., Karlsson H., Dalman C., Maternal hospitalization with infection during pregnancy and risk of autism spectrum disorders. Brain Behav. Immun. 44, 100–105 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vasistha N. A., Pardo-Navarro M., Gasthaus J., Weijers D., Müller M. K., García-González D., Malwade S., Korshunova I., Pfisterer U., von Engelhardt J., Hougaard K. S., Khodosevich K., Maternal inflammation has a profound effect on cortical interneuron development in a stage and subtype-specific manner. Mol. Psychiatry 25, 2313–2329 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Canetta S., Bolkan S., Padilla-Coreano N., Song L. J., Sahn R., Harrison N. L., Gordon J. A., Brown A., Kellendonk C., Maternal immune activation leads to selective functional deficits in offspring parvalbumin interneurons. Mol. Psychiatry 21, 956–968 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hu J. S., Vogt D., Sandberg M., Rubenstein J. L., Cortical interneuron development: A tale of time and space. Development 144, 3867–3878 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Inan M., Welagen J., Anderson S. A., Spatial and temporal bias in the mitotic origins of somatostatin- and parvalbumin-expressing interneuron subgroups and the chandelier subtype in the medial ganglionic eminence. Cereb. Cortex 22, 820–827 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gomez Perdiguero E., Klapproth K., Schulz C., Busch K., Azzoni E., Crozet L., Garner H., Trouillet C., de Bruijn M. F., Geissmann F., Rodewald H.-R., Tissue-resident macrophages originate from yolk-sac-derived erythro-myeloid progenitors. Nature 518, 547–551 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bernier L. P., York E. M., Kamyabi A., Choi H. B., Weilinger N. L., MacVicar B. A., Microglial metabolic flexibility supports immune surveillance of the brain parenchyma. Nat. Commun. 11, 1559 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Giera S., Luo R., Ying Y., Ackerman S. D., Jeong S.-J., Stoveken H. M., Folts C. J., Welsh C. A., Tall G. G., Stevens B., Monk K. R., Piao X., Microglial transglutaminase-2 drives myelination and myelin repair via GPR56/ADGRG1 in oligodendrocyte precursor cells. eLife 7, e33385 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ribeiro Xavier A. L., Kress B. T., Goldman S. A., Lacerda de Menezes J. R., Nedergaard M., A distinct population of microglia supports adult neurogenesis in the subventricular zone. J. Neurosci. 35, 11848–11861 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Parkhurst C. N., Yang G., Ninan I., Savas J. N., Yates J. R. III, Lafaille J. J., Hempstead B. L., Littman D. R., Gan W. B., Microglia promote learning-dependent synapse formation through brain-derived neurotrophic factor. Cell 155, 1596–1609 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Seo Y., Kim H.-S., Shin Y., Kang I., Choi S. W., Yu K.-R., Seo K.-W., Kang K.-S., Excessive microglial activation aggravates olfactory dysfunction by impeding the survival of newborn neurons in the olfactory bulb of Niemann-Pick disease type C1 mice. Biochim. Biophys. Acta 1842, 2193–2203 (2014). [DOI] [PubMed] [Google Scholar]
- 19.Li T., Chiou B., Gilman C. K., Luo R., Koshi T., Yu D., Oak H. C., Giera S., Johnson-Venkatesh E., Muthukumar A. K., Stevens B., Umemori H., Piao X., A splicing isoform of GPR56 mediates microglial synaptic refinement via phosphatidylserine binding. EMBO J. 39, e104136 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.De S., Van Deren D., Peden E., Hockin M., Boulet A., Titen S., Capecchi M. R., Two distinct ontogenies confer heterogeneity to mouse brain microglia. Development 145, dev152306 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schaafsma W., Basterra L. B., Jacobs S., Brouwer N., Meerlo P., Schaafsma A., Boddeke E. W. G. M., Eggen B. J. L., Maternal inflammation induces immune activation of fetal microglia and leads to disrupted microglia immune responses, behavior, and learning performance in adulthood. Neurobiol. Dis. 106, 291–300 (2017). [DOI] [PubMed] [Google Scholar]
- 22.Li X., Tian X., Lv L., Hei G., Huang X., Fan X., Zhang J., Zhang J., Pang L., Song X., Microglia activation in the offspring of prenatal Poly I: C exposed rats: A PET imaging and immunohistochemistry study. Gen. Psychiatr. 31, e000006 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pardo C. A., Vargas D. L., Zimmerman A. W., Immunity, neuroglia and neuroinflammation in autism. Int. Rev. Psychiatry 17, 485–495 (2005). [DOI] [PubMed] [Google Scholar]
- 24.Squarzoni P., Oller G., Hoeffel G., Pont-Lezica L., Rostaing P., Low D., Bessis A., Ginhoux F., Garel S., Microglia modulate wiring of the embryonic forebrain. Cell Rep. 8, 1271–1279 (2014). [DOI] [PubMed] [Google Scholar]
- 25.Bennett F. C., Bennett M. L., Yaqoob F., Mulinyawe S. B., Grant G. A., Gephart M. H., Plowey E. D., Barres B. A., A combination of ontogeny and CNS environment establishes microglial identity. Neuron 98, 1170–1183.e8 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Matcovitch-Natan O., Winter D. R., Giladi A., Aguilar S. V., Spinrad A., Sarrazin S., Ben-Yehuda H., David E., González F. Z., Perrin P., Keren-Shaul H., Gury M., Lara-Astaiso D., Thaiss C. A., Cohen M., Halpern K. B., Baruch K., Deczkowska A., Lorenzo-Vivas E., Itzkovitz S., Elinav E., Sieweke M. H., Schwartz M., Amit I., Microglia development follows a stepwise program to regulate brain homeostasis. Science 353, aad8670 (2016). [DOI] [PubMed] [Google Scholar]
- 27.Bennett M. L., Bennett F. C., Liddelow S. A., Ajami B., Zamanian J. L., Fernhoff N. B., Mulinyawe S. B., Bohlen C. J., Adil A., Tucker A., Weissman I. L., Chang E. F., Li G., Grant G. A., Hayden Gephart M. G., Barres B. A., New tools for studying microglia in the mouse and human CNS. Proc. Natl. Acad. Sci. U.S.A. 113, E1738–E1746 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang Y., Chen K., Sloan S. A., Bennett M. L., Scholze A. R., O’Keeffe S., Phatnani H. P., Guarnieri P., Caneda C., Ruderisch N., Deng S., Liddelow S. A., Zhang C., Daneman R., Maniatis T., Barres B. A., Wu J. Q., An RNA-sequencing transcriptome and splicing database of glia, neurons, and vascular cells of the cerebral cortex. J. Neurosci. 34, 11929–11947 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim S., Kim H., Yim Y. S., Ha S., Atarashi K., Tan T. G., Longman R. S., Honda K., Littman D. R., Choi G. B., Huh J. R., Maternal gut bacteria promote neurodevelopmental abnormalities in mouse offspring. Nature 549, 528–532 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gosselin D., Link V. M., Romanoski C. E., Fonseca G. J., Eichenfield D. Z., Spann N. J., Stender J. D., Chun H. B., Garner H., Geissmann F., Glass C. K., Environment drives selection and function of enhancers controlling tissue-specific macrophage identities. Cell 159, 1327–1340 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Filice F., Vorckel K. J., Sungur A. O., Wohr M., Schwaller B., Reduction in parvalbumin expression not loss of the parvalbumin-expressing GABA interneuron subpopulation in genetic parvalbumin and shank mouse models of autism. Mol. Brain 9, 10 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liodis P., Denaxa M., Grigoriou M., Akufo-Addo C., Yanagawa Y., Pachnis V., Lhx6 activity is required for the normal migration and specification of cortical interneuron subtypes. J. Neurosci. 27, 3078–3089 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xu Q., Guo L., Moore H., Waclaw R. R., Campbell K., Anderson S. A., Sonic hedgehog signaling confers ventral telencephalic progenitors with distinct cortical interneuron fates. Neuron 65, 328–340 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Southwell D. G., Paredes M. F., Galvao R. P., Jones D. L., Froemke R. C., Sebe J. Y., Alfaro-Cervello C., Tang Y., Garcia-Verdugo J. M., Rubenstein J. L., Baraban S. C., Alvarez-Buylla A., Intrinsically determined cell death of developing cortical interneurons. Nature 491, 109–113 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shapiro L. A., Perez Z. D., Foresti M. L., Arisi G. M., Ribak C. E., Morphological and ultrastructural features of Iba1-immunolabeled microglial cells in the hippocampal dentate gyrus. Brain Res. 1266, 29–36 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Borsini A., Zunszain P. A., Thuret S., Pariante C. M., The role of inflammatory cytokines as key modulators of neurogenesis. Trends Neurosci. 38, 145–157 (2015). [DOI] [PubMed] [Google Scholar]
- 37.Socodato R., Portugal C. C., Canedo T., Rodrigues A., Almeida T. O., Henriques J. F., Vaz S. H., Magalhães J., Silva C. M., Baptista F. I., Alves R. L., Coelho-Santos V., Silva A. P., Paes-de-Carvalho R., Magalhães A., Brakebusch C., Sebastião A. M., Summavielle T., Ambrósio A. F., Relvas J. B., Microglia dysfunction caused by the loss of RhoA disrupts neuronal physiology and leads to neurodegeneration. Cell Rep. 31, 107796 (2020). [DOI] [PubMed] [Google Scholar]
- 38.Peng H., Whitney N., Wu Y., Tian C., Dou H., Zhou Y., Zheng J., HIV-1-infected and/or immune-activated macrophage-secreted TNF-alpha affects human fetal cortical neural progenitor cell proliferation and differentiation. Glia 56, 903–916 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Luo R., Jeong S.-J., Jin Z., Strokes N., Li S., Piao X., G protein-coupled receptor 56 and collagen III, a receptor-ligand pair, regulates cortical development and lamination. Proc. Natl. Acad. Sci. U.S.A. 108, 12925–12930 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhu B., Luo R., Jin P., Li T., Oak H. C., Giera S., Monk K. R., Lak P., Shoichet B. K., Piao X., GAIN domain-mediated cleavage is required for activation of G protein-coupled receptor 56 (GPR56) by its natural ligands and a small-molecule agonist. J. Biol. Chem. 294, 19246–19254 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li S., Jin Z., Koirala S., Bu L., Xu L., Hynes R. O., Walsh C. A., Corfas G., Piao X., GPR56 regulates pial basement membrane integrity and cortical lamination. J. Neurosci. 28, 5817–5826 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Baribeau D. A., Vigod S., Pullenayegum E., Kerns C. M., Mirenda P., Smith I. M., Vaillancourt T., Volden J., Waddell C., Zwaigenbaum L., Bennett T., Duku E., Elsabbagh M., Georgiades S., Ungar W. J., Zaidman-Zait A., Szatmari P., Repetitive behavior severity as an early indicator of risk for elevated anxiety symptoms in autism spectrum disorder. J. Am. Acad. Child Adolesc. Psychiatry 59, 890–899.e3 (2020). [DOI] [PubMed] [Google Scholar]
- 43.Cunningham C. L., Martinez-Cerdeno V., Noctor S. C., Microglia regulate the number of neural precursor cells in the developing cerebral cortex. J. Neurosci. 33, 4216–4233 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nikolakopoulou A. M., Dutta R., Chen Z., Miller R. H., Trapp B. D., Activated microglia enhance neurogenesis via trypsinogen secretion. Proc. Natl. Acad. Sci. U.S.A. 110, 8714–8719 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dienel S. J., Lewis D. A., Alterations in cortical interneurons and cognitive function in schizophrenia. Neurobiol. Dis. 131, 104208 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kaar S. J., Angelescu I., Marques T. R., Howes O. D., Pre-frontal parvalbumin interneurons in schizophrenia: A meta-analysis of post-mortem studies. J. Neural Transm. (Vienna) 126, 1637–1651 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Miller M. W., Cogeneration of retrogradely labeled corticocortical projection and GABA-immunoreactive local circuit neurons in cerebral cortex. Brain Res. 355, 187–192 (1985). [DOI] [PubMed] [Google Scholar]
- 48.Rymar V. V., Sadikot A. F., Laminar fate of cortical GABAergic interneurons is dependent on both birthdate and phenotype. J. Comp. Neurol. 501, 369–380 (2007). [DOI] [PubMed] [Google Scholar]
- 49.Petros T. J., Bultje R. S., Ross M. E., Fishell G., Anderson S. A., Apical versus basal neurogenesis directs cortical interneuron subclass fate. Cell Rep. 13, 1090–1095 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.van der Poel M., Ulas T., Mizee M. R., Hsiao C.-C., Miedema S. S. M., Adelia, Schuurman K. G., Helder B., Tas S. W., Schultze J. L., Hamann J., Huitinga I., Transcriptional profiling of human microglia reveals grey-white matter heterogeneity and multiple sclerosis-associated changes. Nat. Commun. 10, 1139 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gosselin D., Skola D., Coufal N. G., Holtman I. R., Schlachetzki J. C. M., Sajti E., Jaeger B. N., O’Connor C., Fitzpatrick C., Pasillas M. P., Pena M., Adair A., Gonda D. D., Levy M. L., Ransohoff R. M., Gage F. H., Glass C. K., An environment-dependent transcriptional network specifies human microglia identity. Science 356, eaal3222 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Giera S., Deng Y., Luo R., Ackerman S. D., Mogha A., Monk K. R., Ying Y., Jeong S.-J., Makinodan M., Bialas A. R., Chang B. S., Stevens B., Corfas G., Piao X., The adhesion G protein-coupled receptor GPR56 is a cell-autonomous regulator of oligodendrocyte development. Nat. Commun. 6, 6121 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bustelo X. R., Sauzeau V., Berenjeno I. M., GTP-binding proteins of the Rho/Rac family: Regulation, effectors and functions in vivo. Bioessays 29, 356–370 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Crampton S. J., Collins L. M., Toulouse A., Nolan Y. M., O’Keeffe G. W., Exposure of foetal neural progenitor cells to IL-1β impairs their proliferation and alters their differentiation - A role for maternal inflammation? J. Neurochem. 120, 964–973 (2012). [DOI] [PubMed] [Google Scholar]
- 55.Wong S. S. C., Lee U. M., Wang X. M., Chung S. K., Cheung C. W., Role of DLC2 and RhoA/ROCK pathway in formalin induced inflammatory pain in mice. Neurosci. Lett. 709, 134379 (2019). [DOI] [PubMed] [Google Scholar]
- 56.Estes M. L., McAllister A. K., Maternal immune activation: Implications for neuropsychiatric disorders. Science 353, 772–777 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Brown A. S., Sourander A., Hinkka-Yli-Salomäki S., McKeague I. W., Sundvall J., Surcel H.-M., Elevated maternal C-reactive protein and autism in a national birth cohort. Mol. Psychiatry 19, 259–264 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Atladóttir H. O., Thorsen P., Østergaard L., Schendel D. E., Lemcke S., Abdallah M., Parner E. T., Maternal infection requiring hospitalization during pregnancy and autism spectrum disorders. J. Autism Dev. Disord. 40, 1423–1430 (2010). [DOI] [PubMed] [Google Scholar]
- 59.Fala L., Cosentyx (secukinumab): First IL-17A antagonist receives FDA approval for moderate-to-severe plaque psoriasis. Am. Health Drug Benefits 9, 60–63 (2016). [PMC free article] [PubMed] [Google Scholar]
- 60.Chu V. T., Weber T., Graf R., Sommermann T., Petsch K., Sack U., Volchkov P., Rajewsky K., Kühn R., Efficient generation of Rosa26 knock-in mice using CRISPR/Cas9 in C57BL/6 zygotes. BMC Biotechnol. 16, 4 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S3