Abstract
The glycosylated receptor-binding domain (glycoRBD) of SARS-CoV-2 can induce protective neutralizing antibodies to function as a vaccine. However, it is unclear whether vaccines using non-glycosylated RBD (non-glycoRBD) can induce protective immunity. Here, we report the efficacy of a SARS-CoV-2 non-glycoRBD vaccine produced by prokaryotic system in mice. The recombinant non-glycoRBD protein was overexpressed in Escherichia coli in the form of inclusion bodies, and was obtained after renaturation and three-step purification. From HPLC analysis, the purity of the RBD was 99%. Additionally, angiotensin converting enzyme 2 (ACE2)-binding assays revealed that E.coli-derived non-glycoRBD had binding activity consistent with glycoRBD. The RBD was formulated with CpG ODN and Al(OH)3 adjuvants and the obtained RBD candidate vaccine elicited potent antibody responses and neutralized SARS-CoV-2 wild-type, Delta, and Omicron pseudoviruses. In summary, our data showed that a non-glycoRBD candidate vaccine produced by E.coli provided a robust immune response and had pseudovirus neutralizing activity, making it a solid candidate vaccine for protection against SARS-CoV-2.
Keywords: SARS-CoV-2, Escherichia coli, Receptor-binding domain (RBD), Neutralizing antibody
1. Introduction
Since December 2019, the coronavirus disease 2019 (COVID-19) outbreak has had a worldwide impact. Globally, as of March 12, 2022, there have been more than 45.22 million confirmed cases of COVID-19, including 6,029,852 deaths. The World Health Organization has declared this novel coronavirus outbreak a global pandemic (www.who.int, 2020). The SARS-CoV-2 spike protein is a homotrimeric transmembrane glycoprotein, which is composed of S1 and S2 subunits (Sternberg et al., 2020; Huang et al., 2020). The spike protein receptor-binding domain (RBD), which binds to the ACE2 receptor and initiates the membrane fusion between the virus and host cell (Felice et al., 2020; Lai et al., 2020), is the immunodominant part of the spike protein and is the main target of most of the neutralizing activity in immune serum. Therefore, the RBD is an ideal target for vaccines (Luan et al., 2020; Shi et al., 2020).
Ongoing improvements in SARS-CoV-2 vaccines are targeted toward developing stable, cost-effective, and convenient candidates for worldwide use, particularly in developing countries (Gupta et al., 2020). Subunit vaccines are the safest vaccines because of the lack of replication, precise composition, and high homogeneity. Spike and RBD-based subunit vaccines are currently produced by mammalian (Liang et al., 2021), insect (Goepfert et al., 2021), or glycoengineered yeast cells (Bo et al., 2021) because of the glycosylation modification of the RBD. A RBD tandem repeat (RBD dimer) subunit vaccine (ZF2001) (Yang et al., 2021) has been produced using CHO cells by the Institute of Microbiology, Chinese Academy of Sciences. Clinical data have shown that this vaccine had good safety and immunogenicity. Novavax has used an insect expression system to develop a recombinant nanoparticle vaccine based on the full-length S protein, which induced Th1-biased immunity (Zhang et al., 2015). In addition, a RBD vaccine candidate derived from glycoengineered yeast elicited potent RBD specific antibody responses in mice. The persistence of pseudovirus neutralizing antibody titers was more than 6 months (Bo et al., 2021). However, whether non-glycoRBD vaccines can induce specific and neutralizing antibodies is unknown.
Among the platforms used for producing recombinant antigens, those using E. coli have simple culture conditions, short culture periods, and are suitable for large-scale fermentation (Tripathi, 2009; Xu et al., 1999; Losen et al., 2004). E. coli is the preferred expression system for biological product production (Liang et al., 2018; Chen et al., 2010). However, prokaryotic cells lack the ability to produce post-translational modifications that require the presence of organelles and enzymes. In the present study, a non-glycoRBD recombinant subunit vaccine of SARS-CoV-2 was produced using E. coli and its immunogenicity was explored.
2. Materials and methods
2.1. Experimental materials
The prokaryotic expression vector pET28a was preserved in our laboratory. Competent E. coli. Trans1 T1 and BL21 (DE3) cells were purchased from Beijing TransGen Biotech Co., Ltd. (China). Anti-SARS-CoV RBD antibody was obtained from Sino Biological Inc. Anti-Rabbit IgG (H + L) X-Adsorbed-horseradish peroxidase (HRP) (Sigma-Aldrich Lot: R133652). Goat pAb to Ms. IgG1 (HRP) (Lot: GR3320187–9), goat pAb to Ms. IgG2a (HRP) (Lot: GR3324477–8), goat pAb to Ms. IgG2b (HRP) (Lot: GR3342724–4), and goat pAb to Ms. IgG3 (HRP) (Lot: GR3285205–10) were obtained from Abcam (USA). The molecular weight protein markers were purchased from Solarbio (China). The DNA extraction kit was purchased from Thermo Fisher Scientific (USA). SARS-CoV-2-Fluc wild type (WT), Delta, and Omicron were purchased from Vazyme Biotech Co., Ltd. (China).
2.2. Construction of the prokaryotic expression vector
The coding sequence of RBD domain (R319-V534) of Wuhan-Hu-1 (MN908947.3) was codon-optimized, synthesized by Tsingke Biotechnology Co., Ltd. (China). The recombinant expression plasmid pET28a-RBD was constructed by inserting the SARS-CoV-2 RBD gene clone into the pET28a vector. Selected positive clones were screened and sent to BGI Genomics Co., Ltd. (China) for sequencing.
2.3. Expression of recombinant protein
E. coli BL21 (DE3) cells (TransGen) were transformed with the RBD construct. The cells were plated on LB agar containing kanamycin (50 μg mL−1) and were grown overnight at 37 °C. The strains were incubated at 37 °C in liquid LB medium with 50 μg mL−1 kanamycin, with shaking until the OD600 values were 0.6–0.8. Isopropyl β-D-1 galactopyranoside (IPTG) was added to 0.5 mmol L−1 and the strains were further induced for 6 h. The expression of the recombinant RBD fusion proteins was evaluated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and western blotting.
2.4. Purification and renaturation of inclusion bodies
Bacterial bodies were harvested through centrifugation of the LB medium and ultrasonication for 30 min on ice. Inclusion bodies were isolated by centrifugation (8000 rpm) for 20 min at 4 °C and were washed once with 20 mmol L−1 Tris-HCl pH 8.0, with 0.5% Tween 20. The inclusion bodies were dissolved by stirring at room temperature in denaturing buffer (20 mmol L−1 Tris-HCl pH 8.0, 5 mmol L−1 dithiothreitol, and 8 mol L−1 urea). The supernatant was collected after centrifugation at 8000 rpm for 20 min at room temperature. Strong anion exchange columns (Source 30Q; GE Healthcare) were then used to purify the RBD in the supernatant using an AKTA pure 25 protein purification system (GE Healthcare).
After preliminary purification, refolding was carried out by dilution into a buffer containing 20 mmol L−1 Tris pH 8.5, 0.8 mmol L−1 reduced glutathione, and 0.2 mmol L−1 oxidized glutathione and renaturing was continued for 12 h at 4 °C. Then, purification using strong cation exchange (Source 30S; GE Healthcare) was performed. The protein was concentrated via ultrafiltration (Millipore; molecular mass cutoff, 10 kDa) and then purified by gel filtration chromatography using Superdex G75. The protein was stored at −20 °C until further use. To analyze the glycosylation modifications, the recombinant RBD was treated with peptide-N-asparaginase (PNGase F) at 37 °C.
2.5. Measurement of binding affinity between RBD and ACE2
The binding affinity of the E. coli-produced RBD to ACE2 was analyzed by ELISA. The antigen [non-glycoRBD, glycoRBD (expressed in glycoengineered Pichia pastoris Bo et al., 2021), human serum albumin (HSA)] (4 μg mL−1) was packaged in ELISA plates and left at 4 °C overnight, washed with PBST (0.5% Tween-20), blocked with 5% skim milk powder, and incubated for 1 h at 37 °C in an incubator. ACE2-His was incubated at 37 °C for 1 h after a twofold dilution from 8 μg·mL−1. The wells were rinsed with PBST buffer and 100 μL of HRP-labeled anti-His antibody diluted 1:2500 was added, and the wells were incubated for 1 h at 37 °C in an incubator, then rinsed with PBST buffer followed by 3,3′,5,5′-tetramethylbenzidine color development for 3 min. Sulfuric acid (2 mol/L) was added to stop the reaction.
2.6. Purity analysis of the recombinant protein
The purity of the target proteins was analyzed by size exclusion chromatography-high performance liquid chromatography (SEC-HPLC) and reversed-phase high-performance liquid chromatography (RP-HPLC); the absorbance values at 280 nm were recorded. SEC-HPLC (TSK gel G2000SWXL, 5 μm, Ф7.8 × 300 mm) was performed using a mobile phase of 20 mmol·L−1 PB pH 7.0 containing 150 mmol·L−1 NaCl at a flow rate of 0.5 mL·min−1. RP-HPLC was performed using an Agilent ZORBAX 300SB-C8 column, 5 μm, Ф4.6 × 250 mm for analysis. The mobile phase A was 1‰ aqueous trifluoroacetic acid (TFA), and mobile phase B was1‰TFA in 95% acetonitrile at a flow rate of 1 mL·min−1, and the elution conditions were 0%–80% B over 40 min.
2.7. Animal immunization
BALB/c mice were purchased from Beijing Vital River Animal Technology Co., Ltd. (China). The experiments were accredited by the Institutional Animal Care and Use Committee (IACUC) of the Beijing Institute of Biotechnology and the IACUC approved experimental animal welfare ethics number was DWZX-2020-039. The experimenter had obtained a certificate as a laboratory animal practitioner and completed the experiments according to the experimental guidelines.
Four experimental groups (n = 5) were set up using 7-week-old female BALB/c mice: E. coli-expressed non-glycoRBD 10 μg-dose group; the protein termed glycoRBD 10 μg-dose group; an adjuvant group; and a saline group. Except for the saline group, all groups were supplemented with 100 μg of Al(OH)3 and 50 μg of CpG ODN adjuvant, and each mouse was immunized with 100 μL of vaccine intramuscularly. Mice were immunized at days 0, 14, and 28 by intramuscular injection, and blood was collected 2 weeks after each immunization and centrifuged at 5000 rpm for 15 min at 4 °C. Sera were collected and stored at −20 °C.
2.8. Detection of antibody titer using ELISA
ELISA plates were coated with RBD antigen at 2 μg·mL−1 per well, incubated overnight at 4 °C, then blocked with 5% skim milk in a 37 °C incubator for 1 h, mouse serum from each group diluted with 5% skim milk was added, and the plates were incubated in a 37 °C incubator for 1 h. After washing four times, 100 μL of HRP-labeled sheep anti-mouse antibody (15,000 dilution) was added to each well and the wells were incubated at 37 °C for 1 h. The plate was washed three times with PBST, and the reaction was performed using 3,3′,5,5′-tetramethylbenzidine for 3 min, and the response was terminated by addition of 2 mmol·L−1 sulfuric acid. The antibody titers were defined as >2.1 times the background absorbance.
2.9. Neutralization of pseudovirus
Serum neutralizing antibodies were detected using pseudoviruses that did not require BSL-3 biosafety conditions. Serum samples were inactivated at 56 °C for 30 min prior to use. The commercial SARS-CoV-2-Fluc wild-type (WT), Delta, and Omicron with serum were serially diluted threefold and incubated at 37 °C for 1 h. The 293T-ACE2 cells were cultured overnight in 96-well plates, and then the virus-serum mixture was transferred to the cells. 8 h later DMEM containing 10% FBS was added. After 48-h incubation at 37 °C in a 5% CO2 incubator, luciferase detection substrate (100 μL) (Promega) was added. The plate was shaken for 2 min and incubated at room temperature for 5 min, and the fluorescent signal was detected by a multi-mode plate reader. Median effective dose (ED50) values were calculated according to the pseudovirus protocal.
2.10. Statistical analysis
The obtained data were plotted by GrapHPad Prism®8 software and statistically analyzed using a t-test. P < 0.05 indicated a statistically meaningful difference [P > 0.05, not significant (ns); P ≤ 0.05, *; P ≤ 0.01, **; P ≤ 0.001, ***].
3. Results
3.1. A prokaryotic expression system can produce recombinant RBD protein
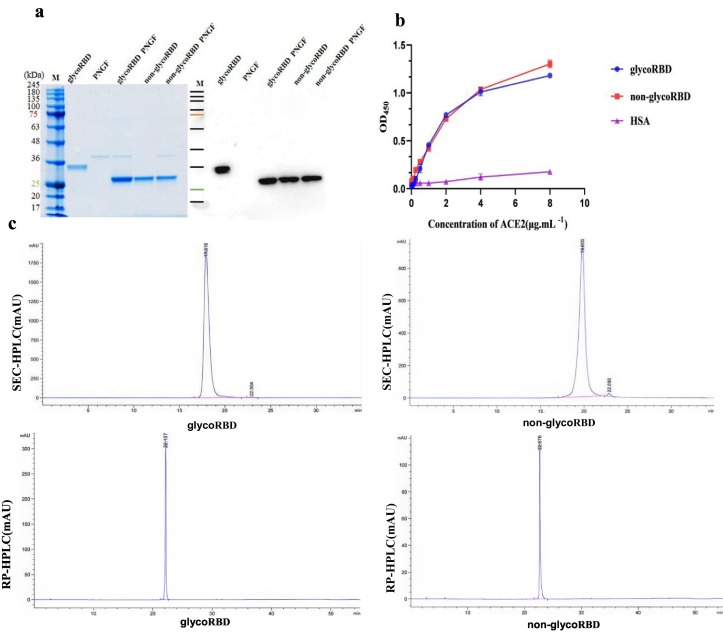
The cell lysates of E. coli were analyzed by SDS-PAGE electrophoresis. Compared with before IPTG induction [Fig. 1(a), lanes 1 and 2], a specific band appeared after the induction [Fig. 1(a), lane 4]. The observed molecular weight of approximately 26 kDa was consistent with the theoretical molecular weight. Western blotting indicated that the recombinant protein could be recognized specifically by SARS-CoV-2 antibodies [Fig. 1(a)]. In addition, the target protein was mainly located in the broken bacteria precipitate, indicating that the protein existed in the form of inclusion bodies.
Fig. 1.
Expression and purification of the RBD protein of SARS-CoV-2. M: marker; 1: bacteriolytic supernatant before induction with IPTG; 2: bacteriolytic precipitate before induction with IPTG; 3: bacterial supernatant after induction with IPTG; 4: bacterial precipitate after induction with IPTG; 5: Source 30Q purification; 6: Source 30S purification; 7: G75 purification.
We established a tag-free RBD protein purification process. The recombinant RBD was purified by anion chromatography [Fig. 1(a), lane 5], and the target protein was not bound to the column. After dilution and renaturation, the RBD protein was enriched by cation chromatography and eluted in 0.5 mmol·L−1 NaCl. Finally, the protein was obtained using gel filtration chromatography (Fig. 1, lanes 6 and 7). The results from SDS-PAGE and western blot analysis indicated that E. coli could produce recombinant RBD protein, and the purified RBD ran as a single band.
3.2. Non-glycoRBD had similar properties to glycoRBD
The molecular mass of non-glycoRBD remained unchanged before and after PNGase F digestion [Fig. 1(a), lanes 4 and 5], and the glycoRBD protein treated with PNGase F had the same molecular weight as non-glycoRBD [Fig. 2(a), lane 3]. These results suggested that the RBD expressed by E. coli was non-glycosylated.
Fig. 2.
Analysis of RBD protein. (a) Analysis of glycosylation modification results by RBD digestes with PNGase F. (b) Detection of RBD and ACE2 binding activity. (c) Analysis of glycoRBD and non-glycoRBD proteins by SEC-HPLC and RP-HPRC.
The binding ability of glycoRBD and non-glycoRBD to human ACE2 was measured by ELISA. No significant difference was found between the binding of the non-glycoRBD group and that of the glycoRBD control group (P = 0.9932) [Fig. 2(b)]. These results suggest that the purified non-glycoRBD had the correct spatial structure.
The HPLC results showed that the RBD protein presented a single, precise profile [Fig. 2(c)]. The purity of the non-glycoRBD protein expressed and purified by the prokaryotic system was >99% [Fig. 2(c)]. The non-glycoRBD retention time in SEC-HPLC was 19.806 min and the glycoRBD retention time was 17.918 min, which was because of the smaller molecular weight of the non-glycoRBD. In addition, the non-glycoRBD retention time in RP-HPLC was 22.676 min and the glycoRBD retention time was 22.167 min. The stronger hydrophobicity of the non-glycoRBD may have caused this result.
3.3. Non-glycoRBD can induce specific and neutralizing antibodies
To evaluate the potential of the non-glycoRBD as a vaccine, mice were immunized with the recombinant RBD protein, and 2 weeks after each immunization, blood was collected from the mice, and the serum was isolated for the detection of specific antibodies. The mice showed an overall trend of weight gain between the groups [Fig. 3(b)]. The antibody titer of non-glycoRBD was 3.98 × 103 at 14 days after the initial immunization and 8.51 × 105 at 14 days after the prime-boost immunization [Fig. 3(c)]. The results showed that 10 μg of non-glycoRBD formulated with aluminum and CpG ODN induced high titers of RBD-specific IgG titers. The different IgG antibody typing was determined by ELISA. Non-glycoRBD formulated with aluminum and CpG ODN induced high antibody IgG titers. The specific IgG1, IgG2a, IgG2b, and IgG3 antibody titers induced by 10 μg of non-glycoRBD 2 weeks after the prime boost were 4.89 × 105, 2.95 × 105, 1.48 × 105, and 1.99 × 104, respectively [Fig. 3(d)].
Fig. 3.
IgG antibody titers after immunization. (a) Immunization scheme. (b)Body weight of mice after immunization. (c) IgG antibody titers after immunizations. (d) Detection of IgG antibody typing titers.
The most successful vaccines currently available protect the body from infection by inducing effective neutralizing antibodies. Therefore, detection of serum neutralizing antibody levels is important to assess the immune efficacy of vaccines. Sera from mice 2 weeks after prime-boost vaccination were tested for neutralizing activity against SARS-CoV-2 wild type (WT), Delta, and Omicron pseudoviruses. Non-glycoRBD (10 μg) neutralized SARS-CoV-2 WT with an antibody titer of 1:1122 and the neutralization titers against Delta and Omicron pseudoviruses were 1:182 and 1:112, respectively. GlycoRBD (10 μg) neutralized SARS-CoV-2 WT, Delta, and Omicron pseudoviruses with antibody titers of 1:891, 1:281, and 1:269, respectively [Fig. 4(a)]. These results suggest that non-glycoRBD can induce cross-neutralizing antibodies in mice.
Fig. 4.
Neutralizing activity. (a) Neutralizing activity against the SARS-CoV-2 WT, Delta, and Omicron pseudoviruses. ED50: median effective dose. (b) Mutation sites on the RBD of the SARS-CoV-2 Delta and Omicron mutants.
4. Discussion
To determine whether non-glycoRBD could induce neutralizing antibody production, we prepared recombinant RBD using a prokaryotic expression system. After the inclusion bodies were diluted and denatured, recombinant RBD protein was obtained after a three-step purification. The prepared recombinant SARS-CoV-2 RBD protein did not contain a tag and had a high homogeneity of ACE2 binding capacity. The non-glycoRBD was used to immunize BALB/c mice and in combination with an Al(OH)3 and CpG ODN double adjuvant induced a specific immune response against the RBD.
According to the WHO, the SARS-CoV-2 Delta and Omicron mutants are variants of current concern with increased transmissibility and increased virulence characteristics. Three mutations in the SARS-CoV-2 Delta spike protein play a key role in the transmission properties of Delta, including the L452R mutation in the RBD of the spike protein, which enhances the ability of the virus to invade cells, and the P681R mutation, which enables the virus to enter cells more efficiently. In addition, Delta also carries a T478K mutation, which may directly enhance the interaction between the RBD and ACE2 and thus, assist the virus to evade immune system surveillance (Cherian et al., 2021; Mlcochova et al., 2021). The SARS-CoV-2 Omicron mutant strain has 15 mutant loci in the RBD region; the Omicron spike-in trimer has a sixfold to ninefold increase in affinity for ACE2 binding (Yin et al., 2022). We used sera from non-glycoRBD-immunized mice to detect neutralizing activity against Delta and Omicron pseudoviruses and showed that cross-neutralizing antibodies against SARS-CoV-2 Delta and Omicron mutant strains could be induced. Non-glycoRBD provided a robust immune response in mice, making it a potential vaccine candidate.
The RBD protein of SARS-CoV-2 has four disulfide bonds and two N-glycosylation sites (Zhou et al., 2021). The protein was efficiently expressed using a prokaryotic expression system, but the expressed product exists in the form of inclusion bodies with no biological activity. After protein denaturation, all the hydrogen and hydrophobic bonds are broken, and the hydrophobic side chains are fully exposed, but the primary structure and covalent bonds are intact. Recombinant proteins are automatically folded into the correct active conformation by dilution complexation (Hespenheide et al., 2002; Dill, 1990). However, the yield of recombinant RBD protein after renaturation is not ideal. Approximately 0.38 mg of RBD can be obtained by renaturation per liter of cultured bacteria. To solve this problem, we investigated the use of E. coli-secreted expression to provide the RBD, to improve the yield of the recombinant protein.
Owing to the short culture period of E. coli, a RBD candidate vaccine can be expected to be stably produced, from the construction of the vector to the acquisition of the target protein, within 1 week. In addition, E. coli platforms are inexpensive and easy to operate. With the emergence of SARS-CoV-2 mutant strains (Tang et al., 2021; Covid and Team, 2021), the use of prokaryotic expression systems may be necessary to prepare corresponding vaccines rapidly. Our results provide relevant data on non-glycoRBD. This study provides a foundation for basic research in the preparation of recombinant RBD subunit vaccines.
Funding
This work was supported by the National Key Research and Development Program of China (2020YFC0841400–008).
Author contributions
Bo Liu and Jun Wu conceived this study; Qian Ke, Peng Sun, Tiantian Wang, Huifang Xu, and Taotao Mi carried out the experiments and performed data analysis; Qian Ke drafted the manuscript; Qian Ke, Peng Sun, Huifang Xu wrote, edited, and reviewed the manuscript; Bo Liu and Jun Wu provided resources; Bo Liu and Jun Wu supervised the work. All authors read and approved the final manuscript.
Ethics approval
This work was supported by the National Key Research and Development Program of China (2020YFC0841400–008).
Declaration of Competing Interest
The authors have no relevant financial or non-financial interests to disclose.
Data availability
Data will be made available on request.
References
- Bo L.A., Ying Y.A., Ylb C., et al. A vaccine based on the receptor-binding domain of the spike protein expressed in glycoengineered Pichia pastoris targeting SARS-CoV-2 stimulates neutralizing and protective antibody responses. Engineering. 2021 doi: 10.1016/j.eng.2021.06.012. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen A.Q., Liu X.J., Cui X.D., et al. Construction of prokaryotic expression vector of β-synuclein and expression in E. coli BL21. Acta Lab. Anim. Sci. Sinica. 2010;18(6):495–497. [Google Scholar]
- Cherian S., Potdar V., Jadhav S., et al. SARS-CoV-2 spike mutations, L452R, T478K, E484Q and P681R, in the second wave of COVID-19 in Maharashtra, India. Microorganisms. 2021;9(7):1542. doi: 10.3390/microorganisms9071542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Covid C.D.C., Team R. SARS-CoV-2 B. 1.1. 529 (Omicron) variant—United States, December 1–8, 2021. Morbid. Mortal. Week. Rep. 2021;70(50):1731. doi: 10.15585/mmwr.mm7050e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dill K.A. Dominant forces in protein folding. Biochemistry. 1990;29(31):7133–7155. doi: 10.1021/bi00483a001. [DOI] [PubMed] [Google Scholar]
- Felice F., Tovar-Moll F., Moll J., et al. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and the central nervous system. Trends Neurosci. 2020;43(6) doi: 10.1016/j.tins.2020.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goepfert P.A., Fu B., Chabanon A.L., et al. Safety and immunogenicity of SARS-CoV-2 recombinant protein vaccine formulations in healthy adults:interim results of a randomised, placebo-controlled, phase 1-2, dose-ranging study. Lancet Infect. Dis. 2021;21(9):1257–1270. doi: 10.1016/S1473-3099(21)00147-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta E., Mishra R.K., Niraj R.R.K., et al. Identification of potential vaccine candidates against SARS-CoV-2 to fight COVID-19: reverse vaccinology approach. JMIR Bioinformatics and Biotechnology. 2020;3(1):e320401. doi: 10.2196/32401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hespenheide B.M., Rader A.J., Thorpe M.F., et al. Identifying protein folding cores from the evolution of flexible regions during unfolding. J. Mol. Graph. Model. 2002;21(3):195–207. doi: 10.1016/s1093-3263(02)00146-8. [DOI] [PubMed] [Google Scholar]
- Huang Y., Yang C., Xu X., et al. Structural and functional properties of SARS-CoV-2 spike protein: potential antivirus drug development for COVID-19. Acta Pharmacol. Sin. 2020;41(9):1141–1149. doi: 10.1038/s41401-020-0485-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai C.C., Shih T.P., Ko W.C., et al. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and coronavirus disease-2019 (COVID-19): the epidemic and the challenges. Int. J. Antimicrob. Agents. 2020;55(3) doi: 10.1016/j.ijantimicag.2020.105924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang H., Yang R., Liu Z., et al. Recombinant Zika virus envelope protein elicited protective immunity against Zika virus in immunocompetent mice. PLoS One. 2018;13:0194860. doi: 10.1371/journal.pone.0194860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang J.G., Su D., Song T.Z., et al. S-Trimer, a COVID-19 subunit vaccine candidate, induces protective immunity in non-human primates. Nat. Commun. 2021;12(1):1346. doi: 10.1038/s41467-021-21634-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Losen M., Frölich B., Pohl M., Büchs J. Effect of oxygen limitation and medium composition on Escherichia coli fermentation in shake-flask cultures. Biotechnol. Prog. 2004;20(4):1062–1068. doi: 10.1021/bp034282t. [DOI] [PubMed] [Google Scholar]
- Luan R.S., Wang X., Sun X., et al. Epidemiology, treatment, and epidemic prevention and control of the coronavirus disease 2019: a review. Sichuan da xue xue bao. Yi Xue ban J. Sichuan Univ. Med. Sci. Edn. 2020;51(2):131–138. doi: 10.12182/20200360505. [DOI] [PubMed] [Google Scholar]
- Mlcochova P., Kemp S.A., et al., Dhar M.S., et al. SARS-CoV-2 B. 1.617. 2 Delta variant replication and immune evasion. Nature. 2021;599(7883):114–119. doi: 10.1038/s41586-021-03944-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi R., Shan C., Duan X., et al. A human neutralizing antibody targets the receptor-binding site of sars-cov-2. Nature. 2020;584:120–124. doi: 10.1038/s41586-020-2381-y. [DOI] [PubMed] [Google Scholar]
- Sternberg A., Naujokat C., et al. Structural features of coronavirus SARS-CoV-2 spike protein: targets for vaccination. Life Sci. 2020;118056 doi: 10.1016/j.lfs.2020.118056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang J.W., Tambyah P.A., Hui D.S.C. Emergence of a new SARS-CoV-2 variant in the UK. J. Infect. 2021;82(4):27–28. doi: 10.1016/j.jinf.2020.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripathi N.K. High yield production of heterologous proteins with Escherichia coli. Def. Sci. J. 2009;59(2):137. [Google Scholar]
- www.who.int [Internet]. Geneva: World Health Organization. 2020. https://covid19.who.int/region/euro/country/tj [cited 2022 March 12]. Available from:
- Xu B., Jahic M., Blomsten G., Enfors S.O. Glucose overflow metabolism and mixed-acid fermentation in aerobic large-scale fed-batch processes with Escherichia coli. Appl. Microbiol. Biotechnol. 1999;51(5):564–571. doi: 10.1007/s002530051433. [DOI] [PubMed] [Google Scholar]
- Yang S., et al. Safety and immunogenicity of a recombinant tandem-repeat dimeric RBD-based protein subunit vaccine (ZF2001) against COVID-19 in adults: two randomised, double-blind, placebo-controlled, phase 1 and 2 trials. Lancet Infect. Dis. 2021;21(8):1107–1119. doi: 10.1016/S1473-3099(21)00127-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin W., Xu Y., Xu P., et al. Structures of the omicron spike trimer with ACE2 and an anti-omicron antibody. Science. 2022;375(6584):1048–1053. doi: 10.1126/science.abn8863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Zhang X.F., Huang S.J., et al. Long-term efficacy of a hepatitis E vaccine. N. Engl. J. Med. 2015;372(10):914–922. doi: 10.1056/NEJMoa1406011. [DOI] [PubMed] [Google Scholar]
- Zhou D., Tian X., Qi R., et al. Identification of 22 N-glycosites on spike glycoprotein of SARS-CoV-2 and accessible surface glycopeptide motifs: implications for vaccination and antibody therapeutics. Glycobiology. 2021;31(1):69–80. doi: 10.1093/glycob/cwaa052. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data will be made available on request.