Abstract
We established a split nanoluciferase complementation assay to rapidly screen for inhibitors that interfere with binding of the receptor binding domain (RBD) of the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) spike glycoprotein with its target receptor, angiotensin-converting enzyme 2 (ACE2). After a screen of 1,200 US Food and Drug Administration (FDA)-approved compounds, we identified bifonazole, an imidazole-based antifungal agent, as a competitive inhibitor of RBD-ACE2 binding. Mechanistically, bifonazole binds ACE2 around residue K353, which prevents association with the RBD, affecting entry and replication of spike-pseudotyped viruses as well as native SARS-CoV-2 and its variants of concern (VOCs). Intranasal administration of bifonazole reduces lethality in K18-hACE2 mice challenged with vesicular stomatitis virus (VSV)-spike by 40%, with a similar benefit after live SARS-CoV-2 challenge. Our screen identified an antiviral agent that is effective against SARS-CoV-2 and VOCs such as Omicron that employ the same receptor to infect cells and therefore has high potential to be repurposed to control, treat, or prevent coronavirus disease 2019 (COVID-19).
Keywords: COVID-19, SARS-CoV-2, high-throughput screening, drug discovery, nanoluciferase bioreporter, bifonazole
Graphical abstract
Taha et al. screened more than 1,200 FDA-approved compounds for their ability to block SARS-CoV-2 from attaching to the host receptor, ACE2. Bifonazole was identified as the top hit and demonstrated a robust ability to impede SARS-CoV2 infection of the host in vitro and in vivo.
Introduction
Coronaviruses (CoVs) are enveloped, positive-sense, single-stranded RNA viruses. Severe acute respiratory syndrome (SARS)-CoV and SARS-CoV-2 infect cells through interaction of the receptor binding domain (RBD) of the viral spike protein with the host receptor, angiotensin-converting enzyme 2 (ACE2).1 Since the SARS outbreak in 2002/2003, the current pandemic caused by a genetically related virus has acutely highlighted a missed opportunity and urgent need for development of antiviral agents against SARS-like viruses. Systematic reviews have concluded that no approved antiviral regimens are currently effective against SARS-CoV or SARS-CoV-2.2 Remdesivir (Veklury) has been granted approval or emergency authorization in approximately 50 countries worldwide, but because of controversial clinical effects it remains conditionally recommended by the World Health Organization (WHO). Molnupiravir, a nucleoside analog like remdesivir, has recently become the first oral antiviral agent for the treatment of coronavirus disease 2019 (COVID-19) to be approved because of its effect of reducing hospitalization or death by 50%. Paxlovid, consisting of the protease inhibitors nirmatrelvir and ritonavir, was more recently authorized in December 2021 as an oral treatment for mild to moderate COVID-19 in individuals over 12 years of age at high risk of severe disease progression. Paxlovid has been shown to reduce hospitalization and death by 88% in a clinical trial but has multiple drug interactions. Monoclonal antibody (mAb) treatments such as bamlanivimab have shown promise in individuals at high risk for progression to severe disease. However, mAbs can lead to increased chances of viral escape, are harder to make, require intravenous delivery with medical supervision, are relatively bulky, and may not be broadly effective or capable of dislodging pre-formed virus-receptor interactions. Taking into consideration our current options, there still remains an urgent need for new and effective antiviral drugs to treat and prevent COVID-19.
The specific molecular interaction between SARS-CoV-2 and its receptor is a key therapeutic target for the identification of effective antiviral agents that could have a direct contribution to our global response to controlling COVID-19 spread. Small molecules are more versatile in this regard, and their use to prevent viral entry is an approved strategy in the context of HIV (e.g., maraviroc).3,4 Identification of efficient agents from existing drugs is the most rapid approach for development of a therapy given their proven safety, and it is anticipated that a number of existing small-molecule therapeutic agents could be repurposed to treat COVID-19. Because crystal structures are now available, in silico approaches or artificial intelligence (AI) tools can be implemented to identify potential drug candidates, and some have already used this approach to identify compounds that may block the ACE2-RBD interaction.2,5,6 However, high-throughput screening can better identify allosteric and non-competitive drugs and remains a gold standard for antiviral drug discovery. We established a split nanoluciferase complementation assay, easily amenable to high-throughput applications, where the RBD-ACE2 interaction facilitates activation of a strong luminescent nanoluciferase signal. We showed that the resulting signal is specific and requires interaction between the spike protein and ACE2.7 Our assay is based on NanoLuc binary technology (NanoBiT), which dissects nanoluciferase into two components, small BiT (SmBiT) and large BiT (LgBiT), that display strong conformational stability, creating an ideal split reporter for investigation of protein-protein interactions.7, 8, 9, 10, 11, 12, 13, 14, 15 We hypothesized that impairing the interactions between the spike RBD and host ACE2 using small-molecule drugs can disrupt viral attachment and prevent subsequent viral entry, ultimately precluding viral infection and spread. In this study, we adapted our split-luciferase assay for high-throughput screening to identify inhibitors of the RBD-ACE2 interaction from a collection of approved drugs.
Results
A nanoluciferase complementation bioreporter screen identifies potential SARS-CoV-2 blocking agents
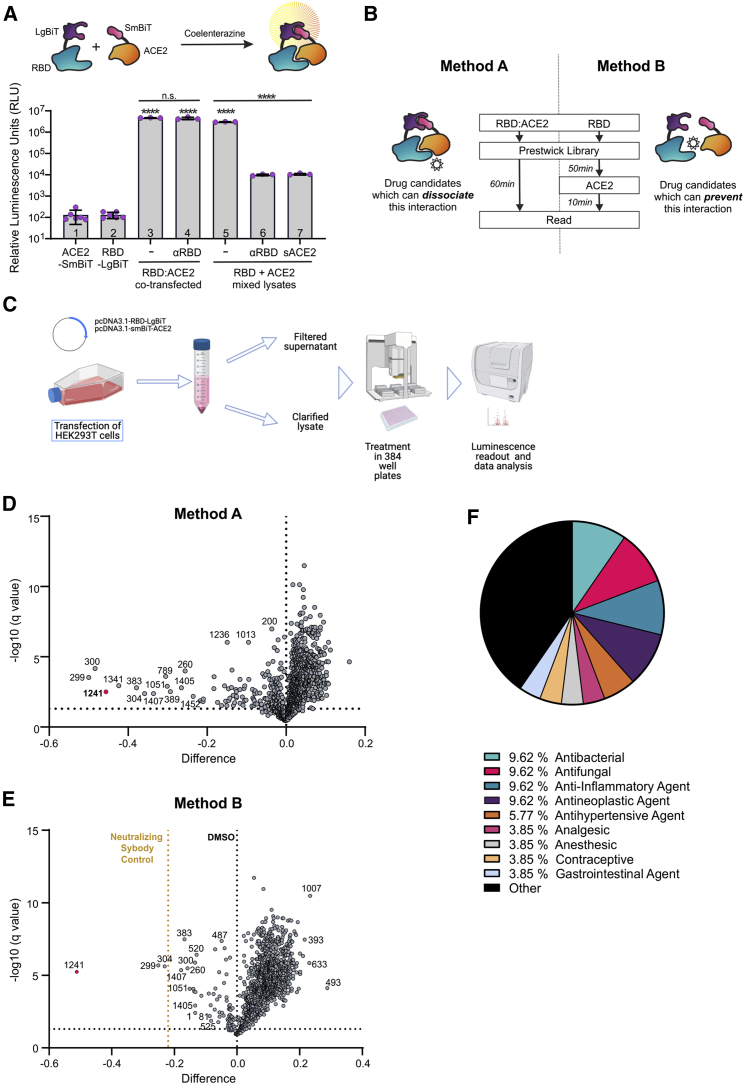
The components of our bioreporter have been described previously,7 LgBiT or SmBiT, fused to the SARS-CoV-2 spike RBD or the extracellular domain of human ACE2, respectively (Figure 1A). These constructs were cloned into plasmids and transfected into HEK293 cells for their expression in whole-cell lysates or supernatants. We confirmed that the combination of lysates from transfected cells expressing the SmBiT-ACE2 and LgBiT-RBD could lead to complementation of the nanoluciferase by producing a strong and stable luminescence signal upon addition of the substrate native coelenterazine. We demonstrate that treatment of LgBiT-RBD with an antibody against the spike RBD or addition of soluble ACE2 (sACE2) to SmBiT-ACE2 could compete with its counterpart and impair the luminescence signal. However, this effect only occurred under conditions where each construct was transfected separately and the resulting lysates were mixed, in contrast with when the constructs were co-transfected and, thus, the complex pre-formed (Figure 1A, lanes 6 and 7). We therefore employed our bioreporter to screen a library consisting of 1,200 US Food and Drug Administration (FDA)-approved compounds. Two parallel methods were employed (Figure 1B). For method A, library compounds were added to the pre-formed SmBiT-ACE2/LgBiT-RBD complex to identify compounds that could dissociate this complex. Method B involved addition of the compound library to LgBiT-RBD, followed by addition of an equal quantity of SmBiT-ACE2 to identify compounds that could prevent this interaction. Method B also included an RBD-neutralizing sybody as a positive control.16 Both methods were found to retain high signal-to-noise ratios (Figure S1A) and appropriate Z′ factors (Figure S1B), confirming suitability for high-throughput screening (HTS; Figures S1C and S1D).
Figure 1.
A bioreporter screen identifies a subset of an FDA-approved library as potential SARS-CoV-2 blocking agents
(A) SmBiT-ACE2 and LgBiT-RBD bioreporter constructs were transfected separately (columns 1–4) or co-transfected (columns 5–7) into HEK293 cells, and luminescence was measured from lysates. Bars 4, 6, and 7: a luciferase assay was performed on lysates in the presence of anti-RBD or sACE2, as indicated, for 15 min. n = 3–6, mean ± SEM, one-way ANOVA compared with mock with Tukey’s multiple corrections test; asterisks directly above columns are compared with control columns 1 or 2 (RBD or ACE2 alone). (B) The bioreporter screen was performed in two ways. Method A (left) involved addition of the library to the pre-formed RBD:ACE2 complex to identify compounds that could disrupt this interaction. Method B (right) involved addition of the compounds to LgBiT-RBD, followed by addition of an equal quantity of SmBiT-ACE2 to identify compounds that could prevent this interaction. Method B also included an RBD-neutralizing sybody-positive control. (C) Flowchart of the biosensor screen, depicting plasmid transfection, harvest of clarified lysates, treatment of 384-well plates with the Prestwick library (n = 3 replicates per drug on separate plates, 4 μM final concentration), followed by substrate (CTZ) addition and luminescence readout. (D and E) Volcano plots depicting the difference in average luminescence signal for each of the 1,200 compounds over q value at a 5% FDR cutoff for method A (D) and method B (E). (F) Pie chart of the top 45 hits from method A and method B, grouped by therapeutic class.
Methods A and B were used to screen the Prestwick library at 4 μM final concentration. Following 1-h incubation with the compounds, coelenterazine (CTZ) was added to each well, and luminescence was measured (n = 3; Figures 1B and 1C). Controls consisting of vehicle (DMSO) alone or RBD alone were added to each 384-well plate (n = 32/plate, 2 per row).
A small subset of the library compounds led to a statistically significant reduction in the luminescence signal from the bioreporter using method A or method B (5% false discovery rate [FDR] cutoff; Figures 1D and 1E).Volcano plots and Z scores (Figure S1E) were used to select approximately 45 hit compounds for further validation. Some of these hits were common to method A and method B, whereas some were unique to either method (Figure S1F). Because of the larger number of hits obtained from method A, we eliminated hits affecting bioreporter luminescence by 15% or less to enrich for the most potent compounds. Compound Prestw-1241 (bifonazole) consistently ranked the highest in terms of its ability to disrupt the biosensor through both methods and yielded more than twice the reduction in bioreporter signal using method A (−46%) and B (−51%) compared with the anti-RBD positive control (sybody, −22%). Pharmacological classification of our top 45 hits reveal an equal number (9.62%) of antifungal, antibacterial, anti-inflammatory, and anti-neoplastic agents, which represent 14.3%, 3.03%, 5.21%, and 8.06%, respectively, of each of these classes in the Prestwick library, ultimately revealing an enrichment in antifungal agents such as bifonazole (Figure 1F).
Screen validation identifies bifonazole as a top SARS-CoV-2 blocking candidate
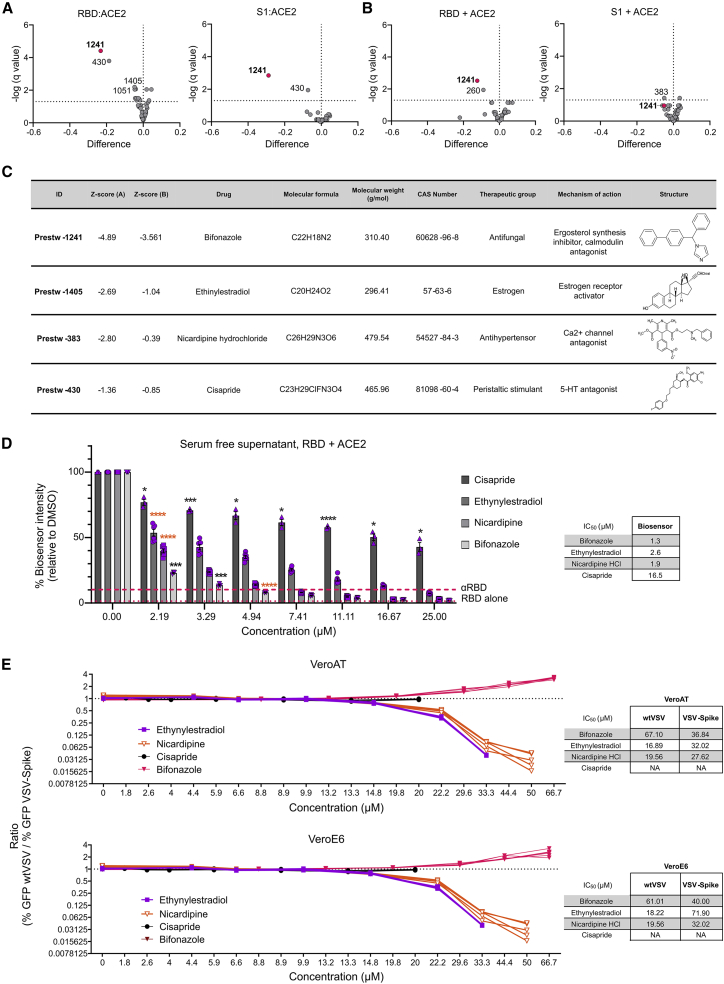
To validate the hits from our initial screen, we treated our bioreporter with the top 45 candidates identified at 4 μM final concentration in quadruplicate to confirm their inhibitory activity on the SARS-CoV-2 RBD-ACE2 interaction by methods A and B. In addition, we examined the effect of our top hits on a bioreporter designed to detect the interaction of ACE2 with the larger S1 domain of SARS-CoV-2 spike. As negative controls, we also included an unrelated control bioreporter (the large tumor suppressor kinase or LATS biosensor)9 as well as wild-type nanoluciferase to eliminate compounds that could non-specifically inhibit the enzyme (Figures S2A, S2B, and S2E). Our results confirmed that candidate Prestw-1241 (bifonazole) led to a significant reduction in binding between ACE2 and the RBD with method A or B (Figures 2A and 2B, left, and Z scores in Figure 2C). Similar results were obtained with ACE2 and the S1 domain of spike, where an ∼30% reduction was observed using method A (co-transfected; Figures 2A and 2B, right). The reduction in SARS-CoV-2 bioreporter luminescence generated by some of the other candidates was much lower than Prestw-1241 (<10% inhibition). Prestw-1405 (ethynylestradiol) and Prestw-383 (nicardipine) yielded an 4%–6% reduction in luminescence specific to the RBD/spike:ACE2 interaction. Although Prestw-430 (cisapride) led to a slightly more pronounced (6%–19%) effect (Figure S2E), it also led to a reduction in luminescence with the LATS biosensor (−13%; Figure S2A) but not nanoluciferase on its own, suggesting some non-specific ability to disrupt the molecular complex.
Figure 2.
Screen validation identifies bifonazole as the top SARS-CoV-2 blocking candidate
(A and B) Validation (n = 4) of the top 45 candidates using the RBD or the S1 domain of SARS-CoV-2 spike protein following method A (A) or method B (B). (C) Table describing the top 4 candidates selected for subsequent validation, including Z scores obtained following method A or method B. (D) SARS-CoV-2 RBD lysates were incubated as in method B with a dose range (0–25 μM) of ethinylestradiol, nicardipine, cisapride, and bifonazole, followed by addition of ACE2 lysates, and luminescence was measured. Anti-RBD was added as a positive control (dashed line), and the RBD alone (LgBiT, dotted line) was used as a negative control (n = 3–6, two-way ANOVA with Dunnett’s multiple corrections test compared with mock for each drug; orange asterisks signify that all subsequent values have the same level of significance). (E) VeroE6 or Vero-hACE2-TMPRSS2 (VeroAT) cells were treated with non-toxic concentrations of the indicated drugs for 15 min, followed by infection of cells with SARS-CoV-2-spike-pseudotyped VSV (VSV-GFP-Spike) or WT VSV-GFP (MOI 1.0), followed by high-content fluorescence imaging. The ratio of GFP foci obtained for WT VSV-GFP- over VSV-Spike-GFP-infected cells was graphed across the concentrations tested for all four compounds in both cell lines. Tables on the right indicate concentrations that are able to reduce GFP focus counts from WT VSV- or VSV-Spike-infected cells by 50% (n = 5 per condition; see Figures S2G–S2I for raw data).
Given that the 2002 SARS-CoV also utilizes ACE2 as a receptor for viral entry, we examined the potential effect of bifonazole on this CoV. Alongside the above validation, we tested the top 45 hits from our bioreporter screen with constructs encoding the RBD of SARS-CoV and ACE2. Our data clearly demonstrate a significant reduction in luminescence with 4 μM bifonazole using method A (45% inhibition) and method B (22% inhibition), suggesting that this compound also has the potential to impede SARS-CoV viral entry (Figures S2C and S2D). Ethynylestradiol and nicardipine led to a less pronounced but significant reduction in bioreporter signal using both methods (3%–6% inhibition; Figure S2E).
To compare head to head the potency of these lead compounds in impairing our SARS-CoV-2 bioreporter, we treated lysates with a broader range of concentrations (2–25 μM) of newly purchased drugs using method B (Figures 2D and S2F). Although we observed a dose-dependent reduction in bioluminescence with all four compounds, bifonazole led to an 80% signal inhibition at 3.29 μM with half maximal inhibitory concentration (IC50) of 1.3 μM. This inhibition was on par with the positive control anti-RBD antibody. Nicardipine-HCl yielded a similar level of bioreporter impairment as bifonazole (IC50 = 1.9 μM), followed by ethynylestradiol (IC50 = 2.6 μM) and cisapride (IC50 = 16.5 μM), which was noticeably the least potent with minimal effect on the bioreporter.
To assess the effectiveness of these compounds in a biological system, we treated VeroE6 cells, which express detectable levels of ACE2, or VeroE6 cells stably overexpressing human ACE2 and TMPRSS2 (VeroAT; Figure S2G) cells with non-toxic concentrations (∼2–50 μM) of bifonazole, nicardipine-HCl, or cisapride for 15 min. Treated cells were then infected with SARS-CoV-2-Spike pseudotyped vesicular stomatitis virus (VSV) expressing green fluorescent protein (VSV-GFP-Spike)17 or wild-type VSV-GFP (multiplicity of infection [MOI] 1.0). 12–24 h after infection, high-content fluorescence imaging was performed, and cell viability was assessed 48 h after infection. Our results demonstrate that bifonazole leads to a significant reduction in pseudotyped VSV-Spike-GFP-infected cells (IC50 = 30–40 μM in both model cell lines without any overt toxicity up to 100 μM; Figures 2E and S2E–S2I). Bifonazole also led to a reduction in wild-type (WT) VSV-GFP-infected cells but at much higher concentrations (IC50 = 60–70 μM). In contrast, ethynylestradiol and nicardipine were able to inhibit WT VSV-GFP (IC50 = ∼15–20 μM) more efficiently than VSV-Spike-GFP (IC50 = ∼25–70 μM). Cisapride did not affect VSV infection at all concentrations tested and was toxic at doses of more than 20 μM. To better illustrate the specificity toward spike, the ratio of percent GFP foci obtained for WT VSV over VSV-Spike was graphed across all concentrations for all four compounds in both cell lines. Although the curve for bifonazole trends upward from ∼20–67 μM, indicating greater inhibitory potential toward VSV-Spike, the curves for ethynylestradiol and nicardipine trend downward demonstrating increased inhibition of WT VSV viral growth and less specificity toward spike.
Validation attempts using our bioreporter as well as biological assays with spike-pseudotyped VSV identify bifonazole as a promising antiviral candidate for SARS-CoV-2 because of its ability to specifically block the RBD-ACE2 interaction in model cell systems.
Imidazole antifungals can impair interaction between ACE2 and the SARS-CoV-2 RBD
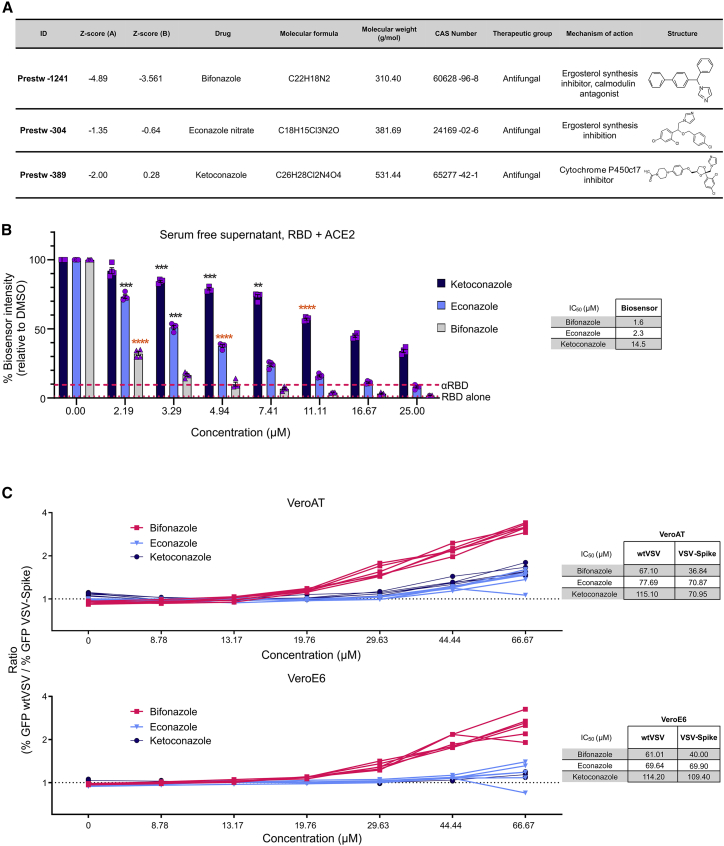
Further analysis of the screening data revealed that many other imidazole-category antifungal agents, like bifonazole, have the capacity to block the RBD-ACE2 interaction. These include econazole nitrate (Prestw-304, −36% method A and −23% method B), ketoconazole (Prestw-389, −29% method A), clotrimazole (Prestw-267, −21% method A) and oxiconazole nitrate (Prestw-304, −4% method B) (Figures 3A, S1E, and S1F). Prestw-1019 (tribenoside) is an anti-inflammatory agent also classified as an imidazole with inhibitory potential (−21% method A and −9% method B). The identification of multiple compounds containing an imidazole pharmacophore suggests a potential mechanistic or structural relevance for RBD-ACE2 interaction blockade observed with the biosensor. For these reasons, we further investigated and compared the antiviral potential of the top three imidazole compounds: bifonazole, econazole, and ketoconazole.
Figure 3.
Bifonazole outperforms other imidazole antifungal agents at impairing the interaction between ACE2 and the SARS-CoV-2 RBD
(A) Table describing the top 3 imidazole antifungal agents identified in the Prestwick library screen, including Z scores for method A and method B. (B) SARS-CoV-2 RBD and ACE2 lysates were treated using method B, as done previously, using bifonazole, econazole, or ketoconazole (n = 3–6, two-way ANOVA with Dunnett’s multiple corrections test compared with mock for each drug; orange asterisks signify that all subsequent values have the same level of significance). (C) VeroE6 or VeroAT cells were treated with non-toxic concentrations of the indicated drugs for 15 min, followed by infection of cells with VSV-Spike or WT VSV-GFP (MOI 1.0) and high-content fluorescence imaging. The ratio of GFP foci obtained for WT VSV- over VSV-Spike-GFP-infected cells was graphed across the concentrations tested for all compounds in both cell lines. Tables on the right indicate concentrations that are able to reduce GFP focus counts from WT VSV- or VSV-Spike-infected cells by 50% (n = 5 per condition; see Figures S3B–S3D for raw data).
As shown in Figure 2D, we treated our bioreporter lysates with a broader range of concentrations of newly purchased compounds using method B. Reflecting initial screen results, we observed superior performance by bifonazole (IC50 = 1.6 μM), followed by econazole (IC50 = 2.3 μM) and ketoconazole (IC50 = 14.5 μM) (Figures 3B and S3A).
To biologically assess antiviral potency, we subsequently treated VeroE6 and VeroAT cells with increasing concentrations of the three imidazoles up to 100 μM, as shown in Figure 2. Fluorescence imaging was performed 12–24 h after infection, and viability was assessed by resazurin 48 h after infection. The ratio of GFP foci obtained for WT VSV over VSV-Spike was graphed across concentrations tested for all four compounds in both cell lines. Bifonazole led to a spike-specific reduction in viral growth at concentrations of ∼35 μM, whereas econazole and ketoconazole demonstrated similar effects but at higher concentrations of ∼60 and 80 μM, respectively (Figures 3C and S3B–S3D).
Our screening data and subsequent validation reveal that members of the imidazole antifungal class of compounds exhibit antiviral activity toward SARS-CoV-2, with bifonazole outperforming all in terms of potency and specificity for disrupting the RBD-ACE2 interaction.
Bifonazole blocks the RBD binding site of SARS-CoV-2 on ACE2
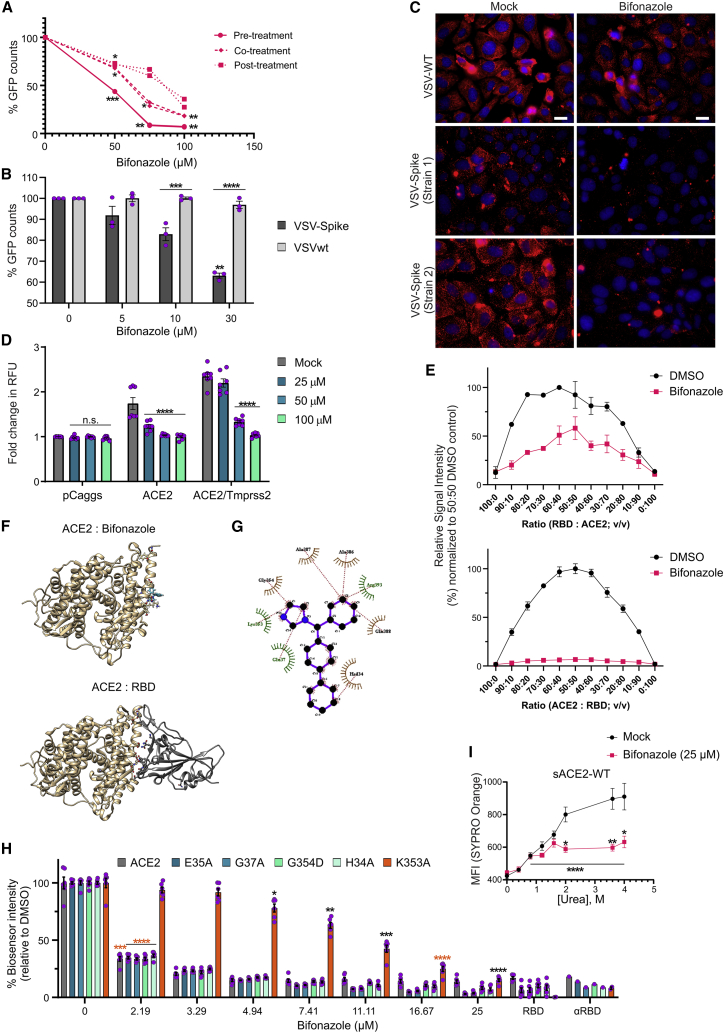
To investigate the mechanism of action of bifonazole, we first evaluated the effect of the timing of bifonazole addition on spike:ACE2 inhibition via infection of cells with two different strains of VSV-Spike (strain 1, VSV-Spike-GFP; strain 2, VSV-Spike) at MOI 1.0. Briefly, we pre-treated VeroAT cells with bifonazole for 15 min (“pre-treatment”) and added the drug at the same time as infection (“co-treatment”) or 60 min after infection (“post-treatment”). VSV-Spike spread was quantified by GFP imaging (strain 1; Figure 4A), and viral output was measured from supernatants 48 h after infection by standard plaque assay (Figure S4A). We found that treatment of ACE2-expressing VeroAT cells for as little as 15 min prior to infection with both strains of VSV-Spike led to the greatest increase in the magnitude of viral blockade by bifonazole.
Figure 4.
Bifonazole impairs viral entry and interacts with ACE2 to block the RBD binding site of SARS-CoV-2
(A) Bifonazole was added to VeroAT cells prior to infection with VSV-Spike-GFP (pre-treatment), at the time of infection (co-treatment), or after infection (post treatment), and GFP counts were quantified 24 h later (n = 2, two-way ANOVA with Dunnett’s multiple comparisons test over mock treated). (B) Vero cells were pre-treated with bifonazole and infected with the indicated viruses at MOI 1, and GFP counts were determined 24 hpi (n = 3, two-way ANOVA with Dunnett’s multiple comparison test over mock treated and Sidak’s test for counts within concentrations). (C) VeroAT cells were pre-treated with bifonazole, followed by infection with the indicated viruses and immunofluorescence staining (magnification, 63×; scale bar, 20 μm). (D) HEK293T cells expressing hACE2 or hACE2/TMPRSS2 and Zip-Venus-1 were treated with bifonazole for 30 min, and then HEK293T cells expressing SARS-CoV-2 spike and Zip-Venus-2 were added to wells. Cells were imaged for fusion and Venus signal. Data show fold change in relative fluorescent units (RFU) from 3 independent experiments (n = 7–11, two-way ANOVA with Dunnett’s multiple comparisons test over mock treated for each condition). (E) Top: bifonazole was added to the RBD, followed by ACE2. Bottom: bifonazole was added to ACE2, followed by the RBD (n = 3 for each). (F) The preferred binding pocket of bifonazole on hACE2 was determined using an automated in silico molecular docking approach (top). The most favorable models are clustered at a pocket of ACE2 that overlaps with RBD binding (bottom). (G) Interacting residues were identified using LigPlot. (H) The RBD was incubated with bifonazole, followed by addition of hACE2 or selected mutants (αRBD, positive control; RBD alone, negative control; n = 6; two-way ANOVA with Dunnett’s multiple comparisons test over mock treated). (I) sACE2 was treated with DMSO or bifonazole, and urea was added at the indicated concentrations, followed by SYPRO orange, and kinetic endpoint fluorescence was measured (n = 4; a bar indicates two-way ANOVA with Dunnett’s multiple comparison test over mock treated for both groups; Sidak’s test was used to compare means within the same concentration).
We next compared the effect of bifonazole on entry and spread of VSV-Spike versus WT VSV. To assess entry, we treated VeroAT cells with increasing concentrations of bifonazole (0, 5, 10, and 30 μM) for 15 min prior to infection with VSV-Spike-GFP or Vero76 cells with bifonazole followed by WT VSV-GFP, with infection occurring for 1 h at MOI 1. The virus and drug were then removed, and cells were imaged for fluorescence 24 h after infection. GFP quantification revealed that bifonazole treatment led to significant impairment in viral entry after brief high-MOI infection with pseudotyped VSV-Spike, whereas WT VSV infection was largely unaffected (Figure 4B). Upon infection at a low MOI (0.01), bifonazole also significantly affected the spread of VSV-Spike and WT VSV, with the effect on WT VSV occurring earlier, at 24 hours post-infection (hpi), because of more rapid growth kinetics (Figure S4B). We also observed a reduction in VSV-Spike plaque size, with no effect on WT VSV, after pre-treatment of cells with 100 μM bifonazole and infection for 1 h (Figure S4C).
As a second approach to assess viral entry, VeroAT cells were pre-treated with 100 μM bifonazole, followed by infection with WT VSV or both strains of pseudotyped VSV-Spike. After an incubation of 4 h, followed by fixation and immunofluorescent staining for VSV, we observed complete abrogation of VSV staining when using either of the spike-pseudotyped VSV strains, in sharp contrast with WT VSV (Figure 4C).
To more conclusively determine bifonazole’s effect on VSV-Spike viral entry, we interrogated its effect on fusion between spike and ACE2 using a bimolecular fluorescence complementation (BiFC) assay.18 Briefly, HEK293T cells expressing human ACE2 (hACE2), hACE2/TMPRSS2, or pCaggs, and Zip-Venus-1 (target cells) were seeded in dishes overnight and then pre-treated with 0, 25, 50, or 100 μM bifonazole. Next, HEK293T cells expressing spike and Zip-Venus-2 (effector cells) were added to the wells. Cells were imaged for the next 3–6 h for fusion morphology and Venus complementation through Zip dimerization as a result of fusion between target and effector cells (Figures 4D and S4D). Consistent with a mechanism involving blocking of the spike:ACE2 interaction, we observed a dose-dependent reduction in fusion of ACE2- and ACE2/TMPRSS2-expressing cells with spike-expressing HEK293T cells after treatment with bifonazole. A similar effect was obtained when using spike from SARS-CoV (Figure S4E).
To better understand whether bifonazole’s effect on viral entry was dependent on binding to the RBD or ACE2, we titrated one component of our bioreporter against its bifonazole-pre-treated counterpart at 4 μM final concentration (Figure 4E). Pre-treatment of ACE2, followed by addition of RBD, led to complete abrogation of bioreporter complementation and luminescence, whereas the opposite order of events (as performed with method B of our screen) led to a peak inhibition of only 50%. These findings suggest a preferential effect of bifonazole on ACE2 rather than the RBD. Given this observation, we next found that the impairment in ACE2:spike interaction by bifonazole is not a result of a reduction in ACE2 protein levels in our model cell lines (VeroE6 and VeroAT; Figure S4F) and does not involve inhibition of ACE2 enzymatic activity (Figure S4G). Extensive testing eliminated the possibility of non-specific effects mediated by micellar formations of bifonazole. Briefly, if the inhibitory effects of bifonazole were a result of non-specific biological effects resulting from micelle formations or aggregates, disrupting these aggregates would nullify the effect of bifonazole on ACE2:spike, including solubilizing serum protein (Figure S5A) or introduction of micelle-disrupting 0.01% Triton X-100 (Figure S5B). We found no evidence of precipitation by means of spectral scans of our solutions of bifonazole (Figure S5C) and no evidence of aggregation by means of an SYPRO orange aggregation assay (Figure S5D). These data support the capacity of bifonazole to specifically disrupt the ACE2:RBD interaction, likely by binding to ACE2.
To assess whether bifonazole may affect ACE2 through direct binding, we first performed in silico molecular docking using human ACE2 and bifonazole (Figure 4F). An automated in silico molecular docking approach that uses the SwissDock web server19 was employed to predict the preferred binding pocket of bifonazole on human ACE2. The clusters were visualized with University of California San Francisco (UCSF) Chimera, and the best predicted models were selected based on ΔG values. The most favorable binding models are clustered at a specified pocket of ACE2. The best binding model of bifonazole (ΔG, −7.03 kcal/mol) to ACE2 was selected for further analysis by LigPlot to identify interacting residues. Molecular docking predictions suggest that binding of bifonazole does not occur within the peptidase catalytic active site and, therefore, does not likely affect the enzymatic activity of ACE2, as demonstrated experimentally (Figure S4G). The prediction suggests that bifonazole binds within the ACE2 N-terminal small lobe and that the interaction involves amino acid residues crucial for maintaining stable ACE2-SARS-CoV-2 RBD attachment.20 Amino acid residues forming the predicted bifonazole binding pocket of ACE2 include H34, E37, K353, G354, A386, A387, Q388, and R393 (Figure 4G).
We have previously determined critical residues within ACE2 that enable RBD binding using our bioreporter through mutational analysis.11 Based on our predicted binding model of bifonazole to ACE2, we performed a bioreporter assay where the RBD was incubated with 2–25 μM bifonazole followed by addition of WT human ACE2 as before or selected ACE2 mutants: H34A, E35A, G37A, K353A, or G354D. Anti-RBD was added as a positive control to block the interaction of the bioreporter, and the RBD alone was used as a negative control. Although bifonazole was able to potently inhibit WT ACE2 binding to the RBD to similar levels as αRBD with concentrations as low as 3.29 μM, we found that only the K353A mutant was less sensitive to the inhibitory activity of bifonazole and required ∼25 μM to achieve similar signal quenching as αRBD. Bifonazole was able to reduce the bioreporter signal with the other ACE2 mutants as efficiently as WT ACE2 (Figures 4H and S4H). These data suggest that bifonazole selectively binds ACE2 in an area involving residue K353, blocking its interaction with spike/RBD.
Finally, to confirm whether bifonazole physically engages with ACE2, we treated recombinant sACE2 with increasing concentrations of urea in the presence of 25 μM bifonazole to induce denaturation and unfolding. SYPRO orange was used to determine changes in hydrophobic surfaces as a surrogate for unfolding. Our data show that bifonazole stabilizes sACE2, leading to a reduction of unfolding with up to 4 M of urea (Figure 4I). These findings suggest that ACE2 is the primary binding site for bifonazole required to exert its SARS-CoV-2 blocking effects; more specifically, the area involving K353.
Bifonazole impairs live SARS-CoV-2 and variants of concern (VOCs) in vitro
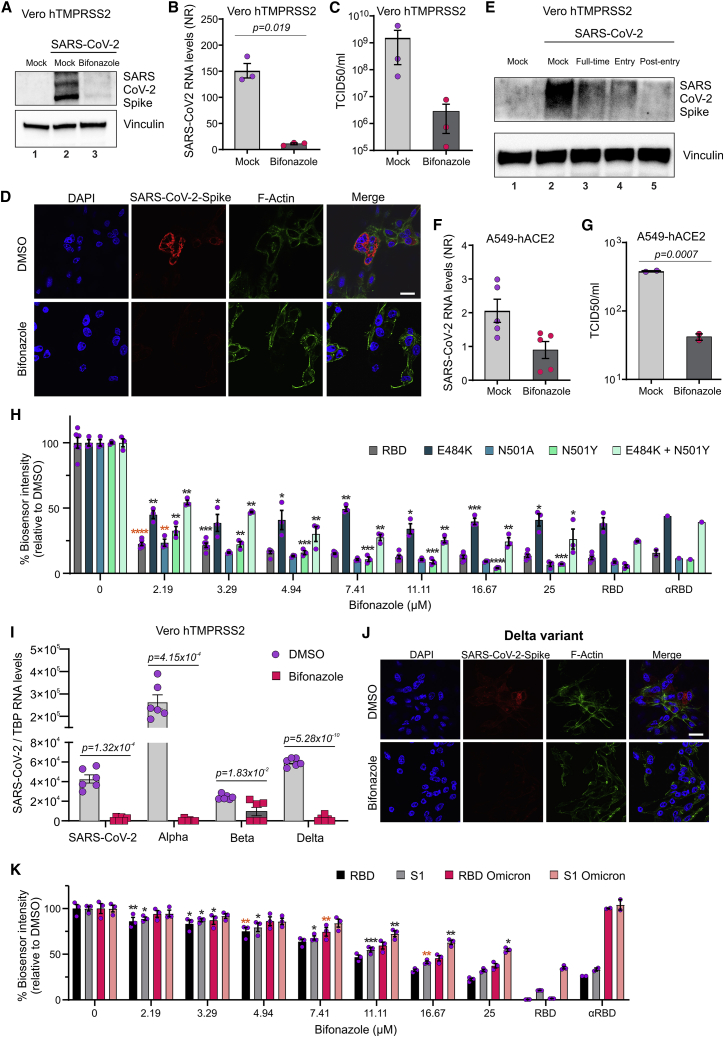
Motivated by these results, we next assessed the potential of bifonazole to affect native SARS-CoV-2 (strain 291.3 FR-4286). VeroE6 cells expressing human TMPRSS2 were treated with vehicle or 100 μM bifonazole for 15 min and infected with SARS-CoV-2 (MOI 0.1). 48 h later, cell lysates were collected and subject to immunoblotting for spike protein. Bifonazole led to a dramatic reduction in spike protein levels in infected VeroE6 cells (Figure 5A). 100 μM bifonazole also led to a significant reduction in viral mRNA levels 48 h after infection with SARS-CoV-2 (Figure 5B). The viral burden was also reduced in the presence of bifonazole by median tissue culture infectious dose (TCID50) assay (Figure 5C). We observed a similar reduction upon immunofluorescence staining for SARS-CoV-2 spike protein in bifonazole-treated cells challenged with SARS-CoV-2 (Figure 5D). Cytotoxicity assays performed in VeroE6-hTMPRSS2 cells confirmed that bifonazole has no effect on viability at the antiviral dose tested (Figure S6A). To evaluate the effect of our drug on viral kinetics, VeroE6-hTMPRSS2 cells were treated with bifonazole 15 min before infection (pre-entry), at the time of infection (entry), or throughout the whole experiment (full time) before challenge with SARS-CoV-2, and infection was assessed by immunoblotting of spike protein within cell lysates. A reduction in spike levels by bifonazole was evident under all conditions tested (Figure 5E). A549 human lung epithelial cells expressing human ACE2 were similarly pre-treated with 100 μM bifonazole and challenged with SARS-CoV-2 at MOI 0.1. In this context, bifonazole also led to a reduction in viral mRNA as well as infectious particles, as measured by TCID50 assay (Figures 5F and 5G), with no effect on cell viability at the dose tested (Figure S6B).
Figure 5.
Bifonazole restricts infection and replication of native SARS-CoV-2 and VOCs
(A–D) Vero-hTMPRSS2 cells were pre-treated with bifonazole (100 μM) and infected with SARS-CoV-2. Spike levels were detected 48 hpi by immunoblotting (A), viral RNA levels were evaluated by qPCR (B), virus replication assessed by TCID50 assay (C), and SARS-CoV-2 spike protein was visualized by immunostaining, with nuclei stained using DAPI and F-actin with Phalloidin (scale bar, 20 μm) (D). Data are the means ± SEM from 3 experiments with Student’s t test (B and C). (E) Vero hTMPRSS2 cells were treated with bifonazole before infection (pre-entry), at the time of infection (entry), or throughout the experiment (full time) before challenge with SARS-CoV-2, and lysates were immunoblotted for spike protein. (F and G) A549 cells expressing hACE2 were pre-treated with bifonazole (100 μM) and challenged with SARS-CoV-2. 48 hpi, viral RNA levels were evaluated by qPCR (F), and replication was assessed by TCID50 assay (G). Data are the means ± SEM from 2–5 experiments with Student’s t test. (H) Mutant or native RBD bioreporters were incubated with bifonazole, followed by ACE2, and luminescence was measured (n = 6, two-way ANOVA with Dunnett’s multiple comparison test over mock treated; orange asterisks signify that all subsequent values have the same level of significance). (I) Vero-hTMPRSS2 cells were pre-treated with bifonazole before infection with the original SARS-CoV-2 or the different VOCs (MOI 0.1). qPCR was performed 48 h after infection, and the data represent means ± SEM of three independent experiments performed in biological duplicates with Student’s t test. (J) Vero hTMPRSS2 cells were pre-treated with bifonazole and infected with the Delta variant. The SARS-CoV-2 spike protein was visualized by immunostaining, and nuclei were stained using DAPI and F-actin with Phalloidin (scale bar, 20 μm). (K) The ACE2 bioreporter was incubated with bifonazole, followed by addition of Omicron-mutant RBD or S1 bioreporters, the original RBD, or S1, and luminescence was measured (n = 3, two-way ANOVA with Dunnett’s multiple comparison test over mock treated; orange asterisks signify that all subsequent values have the same level of significance).
To evaluate the potential effect of bifonazole on VOCs, we first evaluated a panel of relevant mutant RBD bioreporters. The single mutants E484K (Alpha and Gamma lineages), N501Y (Alpha, Beta, and Gamma lineages), and N501A and the double mutant E484K/N501Y, or RBD as a control, were treated with increasing doses of bifonazole, followed by addition of ACE2. We demonstrated that all four bioreporters led to significant inhibition in luminescence, similar to the WT RBD reporter (>50% inhibition), suggesting that bifonazole operates comparably with these RBD mutants representative of VOCs that utilize ACE2 as their receptor (Figures 5H and S6C).
To confirm this observation, VeroE6-hTMPRSS2 cells were pre-treated with bifonazole before infection with the original SARS-CoV-2 or VOCs, including the Alpha (B1.1.7), Beta, and Delta variants, or cluster5 at MOI 0.1. Viral RNA levels obtained 48 h after infection revealed a significant reduction in output of the VOCs in the presence of bifonazole (Figures 5I and S6D). Confocal imaging of DMSO or bifonazole-treated cells challenged with VOCs demonstrate a reduction in staining for the viral spike protein (Figures 5J and S6E). Most notably, we demonstrate significantly reduced binding of bioreporters consisting of the Omicron variant RBD or S1 domains to hACE2 in the presence of bifonazole, whereas αRBD had no inhibitory effect on this variant. These results confirm that bifonazole can significantly reduce infection with native SARS-CoV-2 and multiple VOCs in cultured cells.
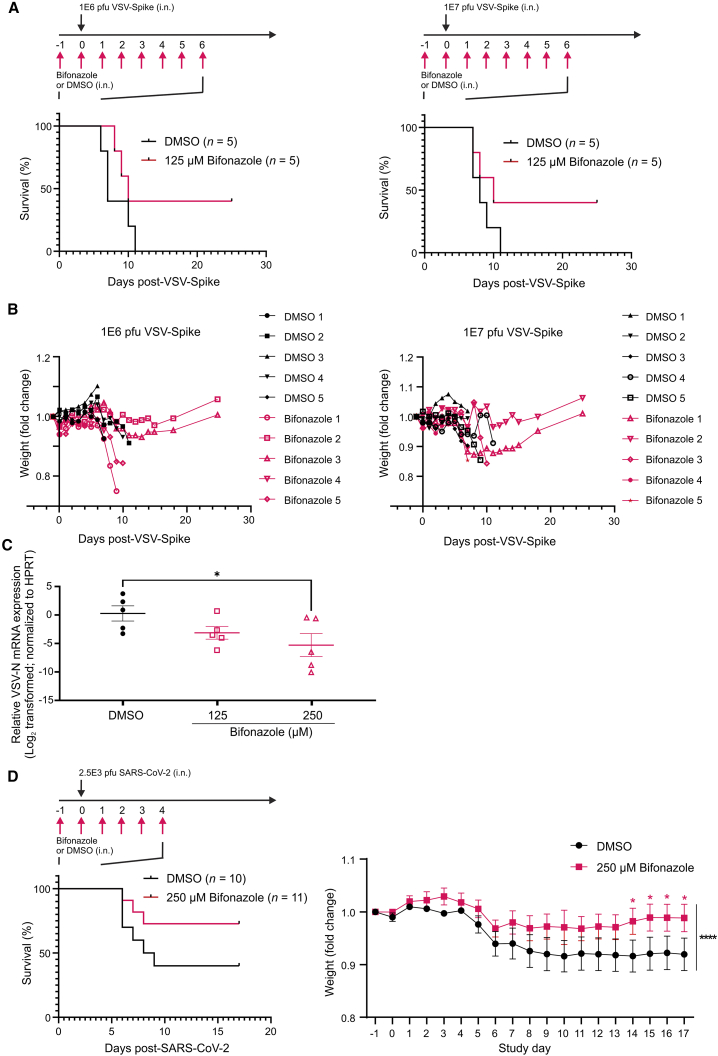
Bifonazole reduces the viral burden in mouse models of COVID-19
To evaluate the therapeutic potential of bifonazole, we assessed the effect of this drug in K18-hACE2 mice following challenge with VSV-Spike. The VSV-Spike used (strain 2) was ultimately lethal after intranasal delivery in this strain of mice (K.G.P., unpublished data). 9-week old mice were administered vehicle or 125 μM bifonazole intranasally based on a previous maximal tolerated dose (MTD) study (Figure S6F). The next day, mice were given the same treatments followed by VSV-Spike at a lethal dose of 1E6 or 1E7 plaque-forming units (PFU) intranasally in 20 μL PBS. Drug treatments were continued for another 6 days with weights recorded and mice assessed for wellness. Although all mice succumbed in the control groups, bifonazole treatment led to survival of 2 of 5 mice in both groups challenged with 1E6 or 1E7 PFU VSV-Spike (Figures 6A and 6B). Surviving mice gained weight, and lungs were normal upon autopsy performed 25 days after treatment. A similar experiment was performed where mice were given vehicle or 125 or 250 μM bifonazole 24 h prior to VSV-Spike (1E6 PFU). Mice were treated with vehicle or bifonazole daily as in Figure 6A but euthanized 4 days after VSV-Spike administration, and right lungs were homogenized and subject to qRT-PCR. A significant reduction in viral load, as detected by qRT-PCR for VSV-M protein, was observed in mice given 250 μM bifonazole compared with vehicle (Figure 6C). Left lungs were fixed in formalin, paraffin embedded, and stained for tyrosine phosphorylated (pY705)-Stat3 as a marker of inflammation. Quantification of positive staining revealed a statistically significant, ∼50% reduction in pY705-Stat3 in the lungs of mice treated with 125 μM bifonazole and administered VSV-Spike compared with those treated with DMSO. The pY705-Stat3 levels observed following bifonazole treatment were comparable with those observed in the lungs of healthy, uninfected mice (dotted line, Figure S6G).
Figure 6.
Bifonazole suppresses VSV-Spike and SARS-CoV-2 infection in K18-hACE2 mice
(A) Treatment schematic and survival study results. 5 K18-hACE2 mice per group were anesthetized daily and treated intranasally with 0 or 125 μM bifonazole in 10 μL containing 3.25% DMSO. 20 min after the second daily administration of drug, mice were given a lethal dose of 1E6 (left) or 1E7 PFU (right) of VSV-Spike (strain 2). Daily drug administration was terminated 6 days after VSV-Spike administration. (B) Weights were recorded daily, and mice were assessed for wellness. (C) Mice were treated as above with DMSO or 125 or 250 μM bifonazole, and lungs were collected 4 days after VSV-Spike (1E6 PFU) administration (n = 5 per group). qRT-PCR was performed, and VSV-N mRNA levels were quantified and normalized to HPRT. (D) Schematic of treatment. K18-hACE2 mice (10–11 mice/group) were treated intranasally with DMSO or 250 μM bifonazole as prepared in (A). 20 min after the second daily administration of drug, mice were anesthetized and given 2.5E3 PFU SARS-CoV-2. Drug administration was stopped 4 days after virus administration. Weights were recorded daily (n = 10–11 mice, mean ± SEM; black asterisks indicate that two-tailed paired-ratio t test was used to compare the DMSO and 250 μM bifonazole groups; red asterisks indicate two-way ANOVA with Fisher’s LSD test at each time point).
To further test the effect of bifonazole in a relevant preclinical context, K18-hACE2 mice were treated with vehicle or 250 μM bifonazole prior to intranasal challenge with live SARS-CoV-2 (2.5E3 PFU; Figure 6D). Drug treatments were continued intranasally after virus administration, weight progression was recorded, and mice were closely monitored. Similar to our observations with VSV-Spike, bifonazole treatment led to survival of 8 of 11 mice compared with 4 of 10 in the vehicle control group, and the treated group lost less weight after infection (Figure 6D, right). Our data demonstrate the safety and efficacy of intranasal bifonazole in 2 different mouse models of COVID-19, coupled with a reduction in viral burden in the lungs of treated animals.
Discussion
Although the current COVID-19 pandemic has been somewhat tamed by deployment of multiple vaccines globally, there still remains an urgent need for additional medical countermeasures. This is especially important because we do not fully understand SARS-CoV-2 vaccine longevity or its effectiveness against VOCs. For these reasons, we set out to find approved drugs that could be rapidly deployed as COVID-19 antiviral agents. The ACE2-RBD biosensor we developed previously was successfully adapted for HTS and led us to identify bifonazole as the most potent SARS-CoV-2 blocking agent from a collection of 1,200 FDA-approved compounds. Our results suggest that bifonazole binds ACE2 to disrupt its interaction with the SARS-CoV-2 RBD and that it can be effective at blocking SARS-CoV-2 infection after intranasal administration. Bifonazole appears to be more potent and specific to the spike-ACE2 interaction than atovaquone, a drug that, as we recently demonstrated, could reduce bioreporter luminescence and is currently in clinical trials.21
Mechanistic testing guided by a series of in silico molecular docking experiments suggests that the K353 residue of ACE2 is critical for the effect of bifonazole. K353 is also crucial for binding of the SARS-CoV-2 RBD to ACE2.11,20,22 Located at the center of the β-hairpin loop, it is involved in stabilizing intermolecular interactions with the RBD. Based on our model, we speculate that binding of bifonazole to E37, K353, G354, or R393 may impair formation of hydrogen bonds and hydrophobic interactions with Y505, Y491, G496, and G502 of SARS-CoV-2.20 The Glu37, K353, Gly354, and D355 cluster is one of two main hotspot regions between ACE2 and the viral spike protein.23 The impairment of the listed interactions by bifonazole has the potential to significantly affect the affinity of SARS-CoV-2 for ACE2 and offers a valid explanation for our findings demonstrating an effect on viral entry (Figure 4).
Identification of bifonazole as a binding partner of ACE2, specifically at the RBD binding site, has several advantages. By interacting with the host receptor rather than the viral RBD, the action of bifonazole in blocking viral entry may be applicable to VOCs, SARS-CoV, and potentially other new and emerging strains of betacoronaviruses that utilize the same RBD binding pocket within ACE2. Our data demonstrate that bifonazole can impair the interaction between ACE2 and the RBD or S1 domain from SARS-CoV as well as RBD mutants representing VOCs, including an Omicron variant-based bioreporter where αRBD has no effect. To validate these findings we demonstrate that bifonazole treatment of VeroE6-hTMPRSS2 cells can significantly reduce viral replication of SARS-CoV-2 and isolated Alpha, Beta, Delta, and cluster 5 variants in vitro (Figure 5). We found that bifonazole does not impair the catalytic activity of ACE2 (Figure S4G), likely reducing the likelihood of vascular side effects or adverse interactions with ACE inhibitor antihypertensive medications. Initial dose escalation studies revealed that bifonazole was well tolerated in mice (up to 500 μM × 9 daily doses) with no significant changes in weight or wellness (Figure S6F). Given that COVID-19 is a disease of the lungs, we opted for intranasal administration of bifonazole to allow greater exposure to virus binding sites. In our challenge studies, we observed 40% survival of challenged mice with daily bifonazole treatments when VSV-Spike was administered at lethal doses. A similar protective effect was observed upon challenge with live SARS-CoV-2.
Known for its antifungal properties, bifonazole is typically used as a topical agent to treat infections such as tinea pedis. A 1983 in-human study showed that systemic administration of 0.016 mg/kg 14C bifonazole could lead to rapid tissue uptake and metabolism, with only 30% of the initial dose remaining unaltered after 30 min.24 Elimination was also rapid and biphasic (half-life or t1/2 of 7 h and 42 h), with 45% of the dose excreted renally and 39% in the feces within 5 days. Although preclinical safety data suggest that bifonazole is not genotoxic after acute dosing, reproduction toxicology studies caution that oral doses in excess of 30 mg/kg in rabbits or 100 mg/kg in rats could result in embryotoxicity and retarded skeletal development, respectively. However, this effectively equates to between 1.5 × 103 and 5 × 103 times in excess of the amounts used in this study. Adaptation of bifonazole as a formulation for use in a nebulizer or inhaler would be attractive for human application and may further improve drug delivery to the upper and lower respiratory tract. Further studies will be needed to determine the best route of administration and dosing regimens for clinical application, including understanding the effect of treatment with increasing time after exposure.
We identified a number of other candidates belonging to the same imidazole antifungal pharmacological class that exhibited similar but inferior activity. Imidazole-related drugs are known to have high therapeutic properties.25 A few studies have identified itraconazole and fluconazole to have some in vitro activity against SARS-CoV-2 by interfering with cholesterol homeostasis or interacting with the viral protease,26,27 but a clinical study with itraconazole administered orally revealed no efficacy in hospitalized individuals.26 Given bifonazole’s superiority over the other imidazole anti-fungal agents identified in our screen, its activity in mouse models of COVID-19 and the possibility of an intranasal formulation, bifonazole should be considered a valid candidate for further clinical testing. Bifonazole has the potential to complement emerging antiviral agents, such as remdesivir and Paxlovid,28 as well as antibody therapeutic agents toward SARS-CoV-2 and variant lineages, especially given that individual neutralizing mAbs show reduced potency against some VOCs.29
We demonstrate proof of concept that our bioreporter is a reliable and effective HTS tool and identify a potent, existing drug that can be repurposed as an antiviral agent, warranting future work on formulation and dosing strategies prior to clinical testing for its effect on COVID-19 progression. Our characterized biosensor platform can be easily adapted to mimic other host-pathogen interactions to rapidly identify potential therapeutic agents that could be effective in controlling future epidemics.
Materials and methods
Cell lines
Vero cells (Vero76, African Green Monkey kidney cells, CCL-81) were obtained from the American Type Culture Collection (ATCC) and maintained in Dulbecco’s modified Eagle’s medium (DMEM, HyClone, Waltham, MA or Corning, Manassas, VA) with 10% fetal bovine serum (FBS; VWR, Mississauga, ON, Canada). pLenti cytomegalovirus (CMV) Hygro TMPRSS2 and pLenti CMV Puro ACE2 (a kind gift from Dr. A.C. Gingras, University of Toronto, Toronto, ON, Canada)30 were used to generate lentivirus via second-generation pSPAX2 and pMD2.G (encoding VSV-G). VeroE6 cells (ATCC, CRL-1586) were co-transduced with ACE2 and TMPRSS2 lentivirus to generate Vero hACE2-hTMPRSS2 and selected with 3 μg/mL puromycin and 200 μg/mL hygromycin.
Human lung adenocarcinoma epithelial A549 cells expressing hACE2 (kindly provided by Brad Rosenberg, Icahn School of Medicine at Mount Sinai, New York, NY, USA) were grown as a monolayer in DMEM supplemented with 10% heat-inactivated fetal calf serum (hiFCS), 2 mM L-glutamine, 100 U/mL penicillin, and 100 μg/mL streptomycin. For experiments involving native SARS-CoV-2, VeroE6 cells expressing TMPRSS2 (VeroE6-hTMPRSS2, kindly provided by Prof. Stefan Pöhlmann, University of Göttingen),31 were grown in DMEM supplemented with 5% hiFCS, 2 mM L-glutamine, 100 U/mL penicillin, and 100 μg/mL streptomycin with 10μg/mL blasticidin (InvivoGen) to maintain TMPRSS2 expression.
All cells were incubated at 37°C in a 5% CO2 humidified incubator, routinely tested for mycoplasma contamination by Hoechst staining and PCR (Diamed, Mississauga, ON, Canada, catalog number ABMG238) and used within 3–10 passages after thawing.
Viruses
WT VSV (Indiana serotype) encoding the SARS-CoV-2 S protein in place of the native envelope spike glycoprotein (VSV-EGFP-Spike, strain #1) was obtained from Dr. S.P.J. Whelan.17 WT VSV-Spike (strain #2) was obtained from Dr. D. Mahoney and was constructed by replacing the native VSV glycoprotein sequence with the Wuhan-Hu-1 isolate (GenBank: MN908947.3) of the SARS-CoV-2 spike sequence that was codon optimized by GenScript (Piscataway, NJ, USA) for human expression from VSV and contained a 21-amino-acid deletion in the cytoplasmic tail (K.G.P., unpublished data).
WT VSV-Spike and VSV-EGFP-Spike were propagated by inoculating 95% confluent Vero-hACE2-hTMPRSS2 cells at MOI 0.01 in roller bottles. Cells were incubated at 34°C with 5% CO2 for 48 h. Supernatants were collected and pelleted at 780 × g to clear heavy debris. Virus contained within the cleared supernatant was subsequently subject to 0.22 μm membrane filtration and then purified using a 5%–50% Optiprep gradient (Sigma-Aldrich, Oakville, ON, Canada, catalog number D1556).
WT VSV was propagated on Vero76 cells as described previously.32 Live SARS-CoV-2 strains were isolated and propagated as described previously.21 All virus suspensions were aliquoted and frozen at −80°C. All viruses were quantified by standard plaque assay on Vero32 or VeroE6 cells.17
Plasmids
Plasmids have been described previously;7 codon-optimized inserts were ordered from GenScript. Plasmids encoding synthetic nanobodies were purchased from Addgene (Sb#68-152527, Sb#45-152526, Sb#42-152525, and Sb#16-152525), purified as described previously,16 and pooled prior to addition. Plasmids encoding His-tagged sACE2 were purchased from Addgene (pcDNA3-sACE2WT-8his, catalog number #149268).
Biosensor HTS
The Prestwick Chemical Library (Prestwick Chemical, Illkirch, France) of approved pharmaceutical compounds was transferred to 384-well plates and diluted to 100 μM in 10% DMSO using the Bravo liquid handler (Agilent, CA, USA).33,34
HEK293T cells were transfected with the expression plasmids LgBiT-RBD or SmBiT-ACE2 using PolyJet transfection reagent (SignaGen Laboratories). Cells were lysed in nanoluciferase-compatible passive lysis buffer (Promega, catalog number E1910).
The effect of our library on SARS receptor binding was assessed in two ways: (1) with the compounds added to SmBiT-ACE2 for 50 min, followed by addition of an equal amount of LgBiT-RBD or LgBiT-S1 for another 10 min (“mixed lysates”), or (2) with the compounds added to the pre-formed SmBiT-ACE2+LgBiT-RBD/S1 complex for 1 h (“co-transfected”). After incubation, the nanoluciferase substrate native CTZ (3.33 mM final concentration; Nanolight Technologies – Prolume, Pinetop, AZ, USA) was added, and luminescence was measured using the Synergy microplate reader (BioTek Instruments, Winooski, VT, USA).
For validation purposes, we tested the effect of cherry-picked compounds in the drug library (n = 3–4) on SARS-CoV binding to ACE2 using a variety of plasmid constructs, notably the RBD or S1 domain from SARS-CoV-2, the RBD domain from SARS-CoV, or human ACE2, as described previously.7 For validation, we also generated a doxycycline-inducible, stable HEK293T cell line to separately produce components of our bioreporter, which we later harvested from clarified cell supernatants 72 h after induction.
Compounds
Bifonazole (B3563, Sigma-Aldrich), cisapride (21657, Cayman Chemicals), 17a-ethynylestradiol (E4876, Sigma-Aldrich), nicardipine HCl (N7510, Sigma-Aldrich), ketoconazole (K1003, Sigma-Aldrich), and econazole nitrate salt (E4632, Sigma-Aldrich) were purchased and reconstituted to 50 or 100 mM in DMSO and used as indicated for validation experiments.
Pseudotyped virus assays
For evaluation of the antiviral potential of candidate small molecules, VeroE6 or VeroAT cells were seeded at 2.5E4 cells/well in 96-well dishes and treated with various concentrations of drug using the Agilent Bravo and incubated for 15 min. Cells were infected at MOI 1.0 with WT VSV-Spike-GFP or WTVSV-GFP. Eleven hours after infection, cells were imaged, and GFP counts were obtained using the Arrayscan HCS Platform (Thermo Scientific Cellomics). 48 h after infection, viability was determined using resazurin sodium salt (Sigma-Aldrich).
To assess viral spread, a plaque expansion assay was used as described previously.35 VeroAT cells were treated with bifonazole (100 μM) or DMSO for 1 h at 37°C. Cells were subsequently infected with VSV-Spike (strain 1 or 2, MOI 0.01) or WT VSV (MOI 0.001) and incubated for 1 h at 37°C. Viral supernatants were then washed off, and a 1% agarose-DMEM overlay was applied. WT VSV plaques were fixed at 24 hpi using 3:1 methanol:acetic acid fixative, followed by staining with Coomassie blue solution. VSV-Spike plaques were fixed and stained 48 h after infection.
SARS-CoV-2 infection experiments and limiting dilution or TCID50 assay
VeroE6-TMPRSS2 cells and A549 cells expressing the human ACE2 receptor were seeded at 1.5 × 105 cells per well in complete DMEM in a 24-well plate. The following day, cells were pre-treated with different concentrations of bifonazole for 15 min prior to viral infection. SARS-CoV-2 (the original Freiburg strain or the different VOCs) was added at MOI 0.1 for 1 h in the presence of the drug. After 1 h, the virus-containing medium was removed and replaced with fresh complete DMEM containing bifonazole for 48 h. Cells were then rinsed in PBS and lysed in 400 μL of lysis buffer (Roche, 11828665001), followed by RNA extraction and gene expression analysis. For the experiments investigating the mode of action of bifonazole, the drug treatment was added full time, as described above, during pre-treatment only (entry) or only after infection (post entry). To quantify infectious virus, a limiting dilution assay (TCID50) was performed as described previously.21
Immunofluorescence and confocal microscopy
For experiments involving native SARS-CoV-2, Vero cells expressing human TMPRSS2 were seeded onto glass coverslips placed on the bottom of 12-well plates and treated with DMSO or bifonazole (100 μM) for 15 min. Cells were then incubated with SARS-CoV-2 MOI 1.0 for 48 h.
For spike-pseudotyped VSV experiments, VeroAT or VeroE6 cells were seeded onto glass coverslips (number 1.5) in 6-well plates and incubated overnight. The next day, cells were treated with DMSO or 100 μM bifonazole for 4 h. Cells were then infected with WT VSV or WT VSV-Spike at MOI 1.0 for 1 h at 37°C. Viral supernatant was then washed off thoroughly, and cells were incubated at 37°C for 4–6 h.
After incubation, all coverslips were fixed with 4% paraformaldehyde, quenched with 100 mM glycine in PBS∗ (with 1 mM CaCl2 and 0.5 mM MgCl2), permeabilized with 0.1% Triton X-100, and then blocked in 5% BSA in PBS∗ for 2 h at room temperature. Coverslips were incubated overnight at 4°C with anti-SARS-CoV-2 spike mAb (1:200, GeneTex) or polyclonal anti-VSV rabbit serum (a gift from Dr. Earl Brown, 1:5,000), in 2% BSA-PBS∗, followed by goat anti-mouse immunoglobulin G (IgG) Alexa Fluor 555 (Invitrogen, ON, Canada, 1:400), Phalloidin-Alexa Fluor 647 (Invitrogen, 1:400), PureBlu DAPI nuclear stain (Bio-Rad, 1:100), or goat anti-rabbit IgG Alexa Fluor 594 for 1 h at room temperature in the dark. Coverslips were washed and mounted using ProLong Gold Antifade with DAPI (Invitrogen, catalog number P36930) and stored at 4°C. Images were taken using the AxioCam HRm camera (Carl Zeiss, Toronto, ON, Canada) mounted on a Zeiss Axioscope Imager M1 microscope or Zeiss LSM 710 inverted confocal microscope with the corresponding Zeiss Zen software.
Immunohistochemistry
Following collection of lung tissue, all samples were fixed in 10% neutral buffered formalin and processed for paraffin embedding. Blocks were serially sectioned and stained with H&E or for pY705-Stat3 (1:200 dilution) (Cell Signaling Technology, catalog number 9145S) and developed using the ImmPRESS HRP Horse Anti-rabbit IgG Kit (Vector Laboratories, catalog number MP-7401-15). Slides were imaged using an AxioScan.Z1. For quantification of pY705-Stat3 3,3′-Diaminobenzidine (DAB) staining, mean pY705-Stat3 pixel intensity per nucleus was calculated by determining DAB mean nuclear pixel intensity per section and dividing it by the total number of nuclei in the same section. The Color Deconvolution 2 ImageJ plugin was used for immunohistochemistry (IHC) quantification.36
Quantitative PCR
For experiments involving VSV-Spike, cells were collected, and RNA extraction was performed using QiaShredder columns and the RNeasy kit (QIAGEN), according to the manufacturer’s protocol. RNA (2 μg) was converted to cDNA with the RevertAid First Strand cDNA Synthesis Kit (Thermo Scientific, catalog number K1621). Real-time PCR reactions were performed with PowerUp SYBR Green Master Mix (Applied Biosystems, catalog number A25741) on an ABI 7500 Fast system (Applied Biosciences). Melt curves for each primer exhibited a single peak, indicating specific amplification. Cycle threshold (Ct) values were determined using the ABI 7500 Fast PCR System software at the optimal threshold determined previously for each gene. Fold induction was calculated relative to the mock-treated control for each gene.37 Primers were designed using Primer 3 v.4.0, and sequences are as follows for hypoxanthine phosphoribosyltransferase (HPRT) or VSV-N :
mHPRT Forward: 5′-TGAAGAGCTACTGTAATGATCAGTCAA-3′,
mHPRT Reverse: 5′-AGCAAGCTTGCAACCTTAACCA-3′,
VSV-N Forward: 5′-ATGTCTGTTACAGTCAAGAGAATC-3′, and
VSV-N Reverse: 5′-TCATTTGTCAAATTCTGACTTAGCATA-3′.
For experiments involving native SARS-CoV-2, gene expression was determined by real-time quantitative PCR using TaqMan detection systems (Applied Biosciences) as described previously.21 For the SARS-CoV-2 gene, primers and probe sequences were provided by the CDC and purchased from Eurofins:
Forward primer: 5′-AAATTTTGGGGACCAGGAAC-3′,
Reverse primer: 5′-TGGCACCTGTGTAGGTCAAC-3′, and
Probe: 5′-FAM-ATGTCGCGCATTGGCATGGA-BHQ-3′.
Aggregation testing
Serum protein solubilization testing
HEK293T cells stably expressing doxycycline-inducible secreted versions of LgBiT-RBD or SmBiT-ACE2 were induced using 500 ng/mL doxycycline for 48 h in serum-free DMEM or in 10% FBS-DMEM. Supernatants were separately collected and centrifuged at 800 × g for 10 min to remove large cellular debris. Clarified supernatants were combined in the presence of a dose-range of bifonazole (0–25 μM), followed by addition of CTZ and reading of the luminescence signal.
Detergent disruption
HEK293T cells were transfected with LgBiT-RBD or SmBiT-ACE2. 48 h after transfection, whole-cell lysates were harvested using passive lysis buffer (PLB; Promega) or micelle-disrupting 0.01% Triton X-100 lysis buffer. Cell lysates were mixed in the presence or absence of 5 μM bifonazole, followed by addition of CTZ and reading of the luminescence signal.
Spectral scanning of bifonazole solutions
Bifonazole was dissolved to biologically relevant concentrations in 0.16% DMSO-DMEM (final DMSO concentration of the in vivo intranasal formulation), 0.25% DMSO-DMEM, or 0.5% DMSO-DMEM (final DMSO concentration for in vitro testing). Solutions were analyzed using the BioTek Synergy H1 plate reader under the absorbance spectral scan protocol. Noticeable shifts in absorbance spectra were noted at concentrations of 500 μM or greater.
ACE2 ligand binding assay
To assess ACE2 binding, we purified recombinant sACE2 (pcDNA3-sACE2(WT)-8his, a gift from Erik Procko; Addgene plasmid 149268; http://n2t.net/addgene:149268; RRID: Addgene_149268)38 by nickel column purification. After validation and quantification of protein concentration, 10 μg of sACE2 was dispensed into black clear-bottom 96-well plates, followed by addition of urea up to 4.25 M. Samples were incubated at 37°C for 15 min and then treated with warmed 25 μM bifonazole for 30 min at 37°C. 1× SYPRO™ orange dye (Invitrogen, catalog number S6650) was added and incubated for 5 min at 37°C, and fluorescence was read using the BioTek Synergy H1 plate reader (Ex470 nm/Ex570 nm).
ACE2 enzymatic assay
ACE2 enzymatic activity was assessed using the Angiotensin II Converting Enzyme (ACE2) Activity Assay Kit (Fluorometric) (Abcam, catalog number ab273297). Recombinant ACE2 enzymatic activity was assessed at 37°C over 3 h (Ex320 nm/Em420 nm kinetic read; BioTek Synergy H1) in the presence or absence of treatment with 25 μM bifonazole. Similarly, the effect of bifonazole on inhibition of ACE2 enzymatic activity by a defined ACE2 inhibitor was assessed. To confirm the enzymatic activity of ACE2 in the model cell line VeroAT, cells were harvested according to the manufacturer’s protocol, and cellular extracts were assayed at 37°C over 3 h.
Quantification of cell-to-cell fusion using biFC
HEK293T cells were seeded in a 12-well microplate (500,000 cells/well) in complete medium for 24 h. The original BiFC constructs GCN4 leucine zipper-Venus1 (ZipV1) and GCN4 leucine zipper-Venus2 (ZipV2) were sourced from Stephen W. Michnick.39 Transient transfections were performed using JetPRIME (Polyplus Transfection, France) according to the manufacturer’s instructions. Target cells were transfected with ZipV1 (0.5 μg), pCEP4(myc-ACE2) (0.05 μg), and ZipV1 (0.5 μg). Effector cell populations were transfected with ZipV2 (0.5 μg) and SARS-CoV-2-S (0.125 μg). Total DNA was normalized to 1 μg using the empty pCAGGS vector DNA. After transfection, cells were incubated at 37°C for 24 h. Then cells were rinsed with PBS and detached with Versene (PBS, 0.53 mM EDTA) and counted. 40,000 cells/well of both populations were co-seeded in DMEM without serum in a 384-well black plate with an optical clear bottom and incubated for 3 h at 37°C and 5% CO2. A biFC signal was acquired using the BioTek Synergy Neo2 plate reader using the monochromator set to excitation/emission of 500 and 542 nm, respectively.
Molecular docking
Docking analysis was carried out by Swiss-Dock software from the Swiss Institute of Bioinformatics (http://www.swissdock.ch/). The crystal structure of native human ACE2 (PDB: 1R42)40 was chosen as a target using the target selection tab in Swiss-Dock, and bifonazole and econazole were uploaded using the ligand molecular selection tab in Swiss-Dock. The binding affinities of binding clusters were evaluated by deltaG (kcal/mol). The resulting docking prediction models were viewed and analyzed using UCSF Chimera (https://www.cgl.ucsf.edu/chimera/). LigPlot (https://www.ebi.ac.uk/thornton-srv/software/LIGPLOT/)41 was applied for analysis of the interaction of residues between drugs and ACE2.
Western blotting
Immunoblotting was performed as described previously.42,43 All membranes were probed overnight at 4°C with any of the following specific primary antibodies in Tris-buffered saline (TBS) Tween (TBST) 0.05%: anti-spike SARS-CoV-2 (GeneTex, 1:1,000), anti-hACE2 (R&D Systems, catalog number AF933, 1:1,000), anti-Vinculin (Sigma-Aldrich, 1:10,000), anti-β-actin (CST, catalog number 4970, 1:1,000), and mouse anti-hemagglutinin (HA; 1:1,000, Invitrogen, catalog number 26183). After three washes in TBST, secondary antibodies, horseradish peroxidase (HRP)-conjugated F(ab)2 donkey anti-mouse IgG (heavy and light/H + L, 1:10,000), F(ab)2 donkey anti-rabbit IgG (H + L, 1:10,000, Jackson ImmunoResearch Laboratories), donkey anti-Goat IgG (H + L, Invitrogen, catalog number A16005, 1:10,000), and goat anti-rabbit IgG (H + L, CST, catalog number 7074, 1:10,000) were added to the membrane in TBST 1% milk for 1 h at room temperature. All membranes were exposed using the Bio-Rad ChemiDoc imaging system and the SuperSignal West Pico PLUS or Bio-Rad Clarity western blotting enhanced chemiluminescence (ECL) substrates.
Cytotoxicity assays
The metabolic activity of VeroE6 and VeroAT cells was assessed using resazurin sodium salt (Sigma-Aldrich) according to the manufacturer’s protocol. Treated and/or infected cells were administered 10% (v/v, final) resazurin in each well and incubated for 2–4 h, depending on the cell line. Fluorescence was measured at 590 nm upon excitation at 530 nm using the Fluoroskan Ascent FL (Thermo Labsystems, Beverly, MA) or the BioTek microplate reader.
For experiments involving native SARS-CoV-2, cell death was measured in Vero-hTMPRSS2 cells by lactate dehydrogenase (LDH) release using the Pierce LDH Cytotoxicity Assay Kit (Thermo Fisher Scientific) in sextuplicate according to the manufacturer’s instructions. Untreated cells were used as a negative control, whereas cells lysed with the provided lysis buffer served as a positive control. LDH activity was measured at 490 nm and 680 nm absorbance using a BioTek microplate reader. LDH activity was determined by subtracting the background 680-nm absorbance value from the 490-nm absorbance value. The percentage of cytotoxicity was calculated according to the manufacturer’s protocol.
Animal models
K18-hACE2 transgenic mice express the human receptor for SARS and SARS-CoV-2 in the airway and other epithelia under control of the human cytokeratin 18 (K18) promoter (strain B6.Cg-Tg(K18-hACE2)2Prlmn/J).44
For studies with VSV-Spike, 9-week-old female K18-hACE2 mice were obtained from The Jackson Laboratory and administered 0 or 125 μM bifonazole in 10 μL PBS containing 3.25% DMSO under general anesthesia. The next day, drug was administered again, and 20 min later, mice were anesthetized and given 1E6 or 1E7 PFU VSV-Spike in 20 μL with PBS. Daily drug administration was halted 6 days after VSV-Spike. Weights were recorded daily, and wellness was assessed. Mice were euthanized according to the institutional guideline review board for animal care. Surviving mice were kept for 25 days after VSV-Spike administration, until they reached initial body weight. For assessment of viral load, 8-week old mice were treated as above with 125 or 250 μM bifonazole daily starting 1 day prior to 1E6 PFU VSV-Spike, all given intranasally. Drug administration was halted 4 days after VSV-Spike administration, when all mice were culled. Lungs were collected and portions snap frozen and subjected to qRT-PCR. In vivo experiments were performed via protocol OHRI-3340, which is in good standing with the Animal Care Committee, and care and treatment of animals was in accordance with the ethical standards of the Canadian Council on Animal Care and with the Animals for Research Act. Each treatment group (DMSO, VSV-Spike, and VSV-Spike + bifonazole) comprised 5 mice. Studies were not formally blinded, but mouse assessments and injections were carried out by an animal technician who was not aware of the treatment groups.
For studies with SARS-CoV-2, age-matched K18-hACE2 male and female mice were randomized, grouped, fed standard chow diets, and housed in a pathogen-free facility. Animals were anesthetized with isoflurane and administered 2.5E3 PFU SARS-CoV-2 (FR-4286 strain), vehicle, or 250 μM bifonazole via intranasal administration. The mice received DMSO or bifonazole on day −1 until day 4 after infection. Mice were weighed every day at the same time until day 19 after infection, and the animals were killed at 20% weight loss or when they reached the humane endpoint. This study was approved in advance by the Animal Ethics Committee at the Danish Veterinary and Food Administration (Glostrup, Denmark) and was carried out in accordance with the Danish Animal Welfare Act for the Care and Use of Animals for Scientific Purposes. All aspects of this study were approved by the office of the Danish Working Environment Authority (Copenhagen, Denmark). Work with SARS-CoV-2 was performed in a biosafety level 2+ laboratory by personnel equipped with powered air-purifying respirators.
Statistics and reproducibility
All graphs and statistical analyses were performed using Excel or GraphPad Prism v.9. Means of two groups were compared using two-tailed unpaired Student’s t test. Means of more than two groups were compared by one-way ANOVA with Dunnett’s or Tukey’s multiple correction test or Fisher’s least significant difference (LSD) test. Two-way ANOVA with Sidak’s or Dunnett’s multiple correction test was applied when groups were split on two independent variables. Normal distribution of the data was assessed using D’Agostino and Pearson omnibus and Shapiro-Wilk normality tests. Alpha levels for all tests were 0.05 (confidence levels of 95%). Biological replicates are indicated by a number n and defined according to NIH guidelines, and error was calculated as the standard error of the mean (SEM). Measurements were taken from distinct samples. For all analyses, ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, and ∗∗∗∗p < 0.0001; n.s., not significant. Data were reproduced by at least two different operators.
Acknowledgments
J.-S.D. is supported by the Canadian Institutes of Health Research New Investigator Award – Infection and Immunity (INI-147824). J.-S.D. is also the recipient of an Ontario Research Fund – COVID-19 Rapid Research Fund grant (project C-134-2426131-DIALLO). The views expressed in the publication are the views of the recipient and do not necessarily reflect those of the Province. D.O. is supported by the Lundbeck Foundation (R335-2019-2138), a young talented cancer researcher grant from Kræftens Bekæmpelse (R279-A16218), the Brødrene Hartman Fond, the Hørslev Fond, the fabrikant Einer Willumsens mindelegat, and the Eva og Henry Fraenkels Mindefond. This research was also supported by a COVID-19 rapid research grant from the Canadian Institutes of Health Research (OV3 170632 to M.C. and P.G.). M.C. is a Canada Research Chair in Molecular Virology and Antiviral Therapeutics and a recipient of an Ontario Early Researcher Award. Z.T. is funded by NSERC Alexander Graham Bell Canada Graduate Scholarship, Ontario Graduate Scholarship, and Mitacs CanPRIME Accelerate fellowship. N.K.’s salary is covered by a scholarship from the Graduate School of Health at Aarhus University. S.R.P. is funded by The Independent Research Fund Denmark (0214-00001B), the European Research Council (ERC-AdG ENVISION, 786602), and the Novo Nordisk Foundation (NNF18OC0030274). E.E.F.B. is funded by a CIHR Frederick Banting and Charles Best Canada graduate scholarship and an OICR Lebovic fellowship. N.A. is funded by a scholarship from King Faisal Specialist Hospital and Research Centre represented by the Saudi Arabian Cultural Bureau in Canada. M.J.F.C. is the recipient of a Mitacs CanPRIME Accelerate and Taggart-Parkes fellowship. R.S. is funded by a CIHR postdoctoral fellowship. Figure 1C was generated using BioRender, licensed to Boaz Wong (OHRI) EC22Z6360S.
Author contributions
Z.T., R.A., T. Azad, J.-S.D., and J.C.B. conceived the project. T. Azad and R.R. designed the bioreporter constructs. Z.T., R.A., and G.M. performed and analyzed the library screens and the validation experiments. R.S. and E.E.F.B. generated the Vero cells expressing ACE2 and Tmprss2. M.I. expanded the SARS-CoV-2 clinical isolates. M.E.C.-T., N.K., and D.O. performed in vitro experiments with SARS-CoV-2 and variants. E.G. performed molecular modeling and data analysis. S.B., M.J.F.C., A.A., A.J., and N.A. assisted with experiments. G.L., A.P., P.G., and M.C. designed and performed bimolecular fluorescence complementation (BiFC) experiments. Z.T., R.A., A.C., and J.P. performed animal experiments with Spike-pseudotyped VSV. L.S.R. performed animal studies with live SARS-CoV-2. K.G.P. and H.T. generated the VSV-Spike (strain #2) recombinant virus. C.J. characterized the susceptibility of K18-hACE2 mice to VSV-Spike-mediated toxicity (strain #2). Z.T., R.A., T. Azad, E.G., S.B., M.J.F.C., R.S., J.-S.D., D.O., D.J.M., C.S.I., M.C., T. Alain, and S.R.P. contributed to study design. Z.T., R.A., E.G., and J.-S.D drafted the manuscript with input from all authors.
Declaration of interests
The authors declare no competing interests.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.ymthe.2022.04.025.
Supplemental information
Data and materials availability
All relevant data are in the main text or the supplemental information and are available from the authors upon request to the corresponding author.
References
- 1.Hoffmann M., Kleine-Weber H., Schroeder S., Kruger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.H., Nitsche A., et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280.e8. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mei M., Tan X. Current strategies of antiviral drug discovery for COVID-19. Front. Mol. Biosci. 2021;8:671263. doi: 10.3389/fmolb.2021.671263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Qian K., Morris-Natschke S.L., Lee K.H. HIV entry inhibitors and their potential in HIV therapy. Med. Res. Rev. 2009;29:369–393. doi: 10.1002/med.20138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dorr P., Westby M., Dobbs S., Griffin P., Irvine B., Macartney M., Mori J., Rickett G., Smith-Burchnell C., Napier C., et al. Maraviroc (UK-427,857), a potent, orally bioavailable, and selective small-molecule inhibitor of chemokine receptor CCR5 with broad-spectrum anti-human immunodeficiency virus type 1 activity. Antimicrob. Agents Chemother. 2005;49:4721–4732. doi: 10.1128/AAC.49.11.4721-4732.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shweta C., Yashpal S.M., Shailly T. Identification of SARS-CoV-2 cell entry inhibitors by drug repurposing using in silico structure-based virtual screening approach. ChemRxiv. 2020 doi: 10.26434/chemrxiv.12005988.v1. Preprint at. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kaushal K., Sarma P., Rana S.V., Medhi B., Naithani M. Emerging role of artificial intelligence in therapeutics for COVID-19: a systematic review. J. Biomol. Struct. Dyn. 2020:1–16. doi: 10.1080/07391102.2020.1855250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Azad T., Singaravelu R., Brown E.E.F., Taha Z., Rezaei R., Arulanandam R., Boulton S., Diallo J.S., Ilkow C.S., Bell J.C. SARS-CoV-2 S1 NanoBiT: a nanoluciferase complementation-based biosensor to rapidly probe SARS-CoV-2 receptor recognition. Biosens. Bioelectron. 2021;180:113122. doi: 10.1016/j.bios.2021.113122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dixon A.S., Schwinn M.K., Hall M.P., Zimmerman K., Otto P., Lubben T.H., Butler B.L., Binkowski B.F., Machleidt T., Kirkland T.A., et al. NanoLuc complementation reporter optimized for accurate measurement of protein interactions in cells. ACS Chem. Biol. 2016;11:400–408. doi: 10.1021/acschembio.5b00753. [DOI] [PubMed] [Google Scholar]
- 9.Azad T., Janse van Rensburg H.J., Lightbody E.D., Neveu B., Champagne A., Ghaffari A., Kay V.R., Hao Y., Shen H., Yeung B., et al. A LATS biosensor screen identifies VEGFR as a regulator of the Hippo pathway in angiogenesis. Nat. Commun. 2018;9:1061. doi: 10.1038/s41467-018-03278-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Azad T., Singaravelu R., Taha Z., Jamieson T.R., Boulton S., Crupi M.J.F., Martin N.T., Brown E.E.F., Poutou J., Ghahremani M., et al. Nanoluciferase complementation-based bioreporter reveals the importance of N-linked glycosylation of SARS-CoV-2 S for viral entry. Mol. Ther. 2021;29:1984–2000. doi: 10.1016/j.ymthe.2021.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brown E.E.F., Rezaei R., Jamieson T.R., Dave J., Martin N.T., Singaravelu R., Crupi M.J.F., Boulton S., Tucker S., Duong J., et al. Characterization of critical determinants of ACE2-SARS CoV-2 RBD interaction. Int. J. Mol. Sci. 2021;22:2268. doi: 10.3390/ijms22052268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Azad T., Rezaei R., Singaravelu R., Jamieson T.R., Crupi M.J.F., Surendran A., Poutou J., Taklifi P., Cowan J., Cameron D.W., Ilkow C.S. A high-throughput NanoBiT-based serological assay detects SARS-CoV-2 seroconversion. Nanomaterials. 2021;11:807. doi: 10.3390/nano11030807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jiang X., Dahlin A., Weiss S.T., Tantisira K., Lu Q. A high-throughput chemical screen identifies novel inhibitors and enhancers of anti-inflammatory functions of the glucocorticoid receptor. Sci. Rep. 2017;7:7405. doi: 10.1038/s41598-017-07565-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Moasses Ghafary S., Soriano-Teruel P.M., Lotfollahzadeh S., Sancho M., Serrano-Candelas E., Karami F., Barigye S.J., Fernandez-Perez I., Gozalbes R., Nikkhah M., et al. Identification of NLRP3(PYD) homo-oligomerization inhibitors with anti-inflammatory activity. Int. J. Mol. Sci. 2022;23:1651. doi: 10.3390/ijms23031651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Azad T., Tashakor A., Rahmati F., Hemmati R., Hosseinkhani S. Oscillation of apoptosome formation through assembly of truncated Apaf-1. Eur. J. Pharmacol. 2015;760:64–71. doi: 10.1016/j.ejphar.2015.04.008. [DOI] [PubMed] [Google Scholar]
- 16.Walter J.D., Hutter C.A.J., Zimmermann I., Wyss M., Egloff P., Sorgenfrei M., Hürlimann L.M., Gonda I., Meier G., Remm S., et al. Sybodies targeting the SARS-CoV-2 receptor-binding domain. bioRxiv. 2020 doi: 10.1101/2020.04.16.045419. Preprint at. [DOI] [Google Scholar]
- 17.Case J.B., Rothlauf P.W., Chen R.E., Kafai N.M., Fox J.M., Smith B.K., Shrihari S., McCune B.T., Harvey I.B., Keeler S.P., et al. Replication-competent vesicular stomatitis virus vaccine vector protects against SARS-CoV-2-mediated pathogenesis in mice. Cell Host Microbe. 2020;28:465–474.e4. doi: 10.1016/j.chom.2020.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rocheleau L., Laroche G., Fu K., Stewart C.M., Mohamud A.O., Côté M., Giguère P.M., Langlois M.-A., Pelchat M. Identification of a high-frequency intra-host SARS-CoV-2 spike variant with enhanced cytopathic and fusogenic effect. bioRxiv. 2021 doi: 10.1101/2020.12.03.409714. Preprint at. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Grosdidier A., Zoete V., Michielin O. SwissDock, a protein-small molecule docking web service based on EADock DSS. Nucleic Acids Res. 2011;39:W270–W277. doi: 10.1093/nar/gkr366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lan J., Ge J., Yu J., Shan S., Zhou H., Fan S., Zhang Q., Shi X., Wang Q., Zhang L., et al. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020;581:215–220. doi: 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- 21.Carter-Timofte M.E., Arulanandam R., Kurmasheva N., Fu K., Laroche G., Taha Z., van der Horst D., Cassin L., van der Sluis R.M., Palermo E., et al. Antiviral potential of the antimicrobial drug Atovaquone against SARS-CoV-2 and emerging variants of concern. ACS Infect. Dis. 2021;7:3034–3051. doi: 10.1021/acsinfecdis.1c00278. [DOI] [PubMed] [Google Scholar]
- 22.Wang Q., Zhang Y., Wu L., Niu S., Song C., Zhang Z., Lu G., Qiao C., Hu Y., Yuen K.Y., et al. Structural and functional basis of SARS-CoV-2 entry by using human ACE2. Cell. 2020;181:894–904.e9. doi: 10.1016/j.cell.2020.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lim H., Baek A., Kim J., Kim M.S., Liu J., Nam K.Y., Yoon J., No K.T. Hot spot profiles of SARS-CoV-2 and human ACE2 receptor protein protein interaction obtained by density functional tight binding fragment molecular orbital method. Sci. Rep. 2020;10:16862. doi: 10.1038/s41598-020-73820-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Patzschke K., Ritter W., Siefert H.M., Weber H., Wegner L.A. Pharmacokinetic studies following systemic and topical administration of [14C]bifonazole in man. Arzneimittelforschung. 1983;33:745–750. [PubMed] [Google Scholar]
- 25.Sharma D., Narasimhan B., Kumar P., Judge V., Narang R., De Clercq E., Balzarini J. Synthesis, antimicrobial and antiviral evaluation of substituted imidazole derivatives. Eur. J. Med. Chem. 2009;44:2347–2353. doi: 10.1016/j.ejmech.2008.08.010. [DOI] [PubMed] [Google Scholar]
- 26.Van Damme E., De Meyer S., Bojkova D., Ciesek S., Cinatl J., De Jonghe S., Jochmans D., Leyssen P., Buyck C., Neyts J., et al. In vitro activity of itraconazole against SARS-CoV-2. bioRxiv. 2020 doi: 10.1101/2020.11.13.381194. Preprint at. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Das S., Sarmah S., Lyndem S., Singha Roy A. An investigation into the identification of potential inhibitors of SARS-CoV-2 main protease using molecular docking study. J. Biomol. Struct. Dyn. 2020;39:3347–3357. doi: 10.1080/07391102.2020.1763201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fischer W., Eron J.J., Holman W., Cohen M.S., Fang L., Szewczyk L.J., Sheahan T.P., Baric R., Mollan K.R., Wolfe C.R., et al. Molnupiravir, an oral antiviral treatment for COVID-19. medRxiv. 2021 doi: 10.1101/2021.06.17.21258639. Preprint at. [DOI] [Google Scholar]
- 29.Chen R.E., Zhang X., Case J.B., Winkler E.S., Liu Y., VanBlargan L.A., Liu J., Errico J.M., Xie X., Suryadevara N., et al. Resistance of SARS-CoV-2 variants to neutralization by monoclonal and serum-derived polyclonal antibodies. Nat. Med. 2021;27:717–726. doi: 10.1038/s41591-021-01294-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Abe K.T., Li Z., Samson R., Samavarchi-Tehrani P., Valcourt E.J., Wood H., Budylowski P., Dupuis A.P., II Girardin R.C., Rathod B., et al. A simple protein-based surrogate neutralization assay for SARS-CoV-2. JCI Insight. 2020;5:e142362. doi: 10.1172/jci.insight.142362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dittmar M., Lee J.S., Whig K., Segrist E., Li M., Kamalia B., Castellana L., Ayyanathan K., Cardenas-Diaz F.L., Morrisey E.E., et al. Drug repurposing screens reveal cell-type-specific entry pathways and FDA-approved drugs active against SARS-Cov-2. Cell Rep. 2021;35:108959. doi: 10.1016/j.celrep.2021.108959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Diallo J.S., Vaha-Koskela M., Le Boeuf F., Bell J. Propagation, purification, and in vivo testing of oncolytic vesicular stomatitis virus strains. Methods Mol. Biol. 2012;797:127–140. doi: 10.1007/978-1-61779-340-0_10. [DOI] [PubMed] [Google Scholar]
- 33.Diallo J.S., Boeuf F.L., Lai F., Cox J., Vaha-Koskela M., Abdelbary H., MacTavish H., Waite K., Falls T., Wang J., et al. A high-throughput pharmacoviral approach identifies novel oncolytic virus sensitizers. Mol. Ther. 2010;18:1123–1129. doi: 10.1038/mt.2010.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Keller B.A., Laight B.J., Varette O., Broom A., Wedge M.E., McSweeney B., Cemeus C., Petryk J., Lo B., Burns B., et al. Personalized oncology and BRAF(K601N) melanoma: model development, drug discovery, and clinical correlation. J. Cancer Res. Clin. Oncol. 2021;147:1365–1378. doi: 10.1007/s00432-021-03545-2. [DOI] [PubMed] [Google Scholar]
- 35.Arulanandam R., Taha Z., Garcia V., Selman M., Chen A., Varette O., Jirovec A., Sutherland K., Macdonald E., Tzelepis F., et al. The strategic combination of trastuzumab emtansine with oncolytic rhabdoviruses leads to therapeutic synergy. Commun. Biol. 2020;3:254. doi: 10.1038/s42003-020-0972-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Landini G., Martinelli G., Piccinini F. Colour deconvolution: stain unmixing in histological imaging. Bioinformatics. 2020;37:1485–1487. doi: 10.1093/bioinformatics/btaa847. [DOI] [PubMed] [Google Scholar]
- 37.Pfaffl M.W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chan K.K., Dorosky D., Sharma P., Abbasi S.A., Dye J.M., Kranz D.M., Herbert A.S., Procko E. Engineering human ACE2 to optimize binding to the spike protein of SARS coronavirus 2. Science. 2020;369:1261–1265. doi: 10.1126/science.abc0870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Michnick S.W., Ear P.H., Landry C., Malleshaiah M.K., Messier V. A toolkit of protein-fragment complementation assays for studying and dissecting large-scale and dynamic protein-protein interactions in living cells. Methods Enzymol. 2010;470:335–368. doi: 10.1016/S0076-6879(10)70014-8. [DOI] [PubMed] [Google Scholar]
- 40.Towler P., Staker B., Prasad S.G., Menon S., Tang J., Parsons T., Ryan D., Fisher M., Williams D., Dales N.A., et al. ACE2 X-ray structures reveal a large hinge-bending motion important for inhibitor binding and catalysis. J. Biol. Chem. 2004;279:17996–18007. doi: 10.1074/jbc.M311191200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wallace A.C., Laskowski R.A., Thornton J.M. LIGPLOT: a program to generate schematic diagrams of protein-ligand interactions. Protein Eng. 1995;8:127–134. doi: 10.1093/protein/8.2.127. [DOI] [PubMed] [Google Scholar]
- 42.Olagnier D., Brandtoft A.M., Gunderstofte C., Villadsen N.L., Krapp C., Thielke A.L., Laustsen A., Peri S., Hansen A.L., Bonefeld L., et al. Nrf2 negatively regulates STING indicating a link between antiviral sensing and metabolic reprogramming. Nat. Commun. 2018;9:3506. doi: 10.1038/s41467-018-05861-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Olagnier D., Farahani E., Thyrsted J., Blay-Cadanet J., Herengt A., Idorn M., Hait A., Hernaez B., Knudsen A., Iversen M.B., et al. SARS-CoV2-mediated suppression of NRF2-signaling reveals potent antiviral and anti-inflammatory activity of 4-octyl-itaconate and dimethyl fumarate. Nat. Commun. 2020;11:4938. doi: 10.1038/s41467-020-18764-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McCray P.B., Jr., Pewe L., Wohlford-Lenane C., Hickey M., Manzel L., Shi L., Netland J., Jia H.P., Halabi C., Sigmund C.D., et al. Lethal infection of K18-hACE2 mice infected with severe acute respiratory syndrome coronavirus. J. Virol. 2007;81:813–821. doi: 10.1128/JVI.02012-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All relevant data are in the main text or the supplemental information and are available from the authors upon request to the corresponding author.