Abstract
Thermostable nanoparticles have numerous applications in catalysis and in the oil/gas industry. However, synthesizing these nanoparticles requires expensive polymers. Here, a novel thermostable ferritin named PcFn, originally from the hyperthermophilic archaeon Pyrococcus yayanosii CH1, was overexpressed in Escherichia coli, purified and characterized, which could successfully direct the synthesis of thermostable magnetoferritins (M-PcFn) with monodispersed iron oxide nanoparticles in one step. Transmission electron microscopy and magnetic measurements show that the cores of the M-PcFn have an average diameter of 4.7 nm, are well-crystalline and superparamagnetic. Both the PcFn and M-PcFn can resist temperatures up to 110 °C, which is significantly higher than for human H-chain ferritin (HFn) and M-HFn, and comparable to temperatures previously reported for Pyrococcus furiosus ferritin (PfFn) and M-PfFn. After heating at 110 °C for 30 minutes, PcFn and M-PcFn maintained their secondary structures and PcFn retained 87.4% of its iron uptake activity. This remarkable thermostability of PcFn and M-PcFn suggests potential applications in elevated temperature environments.
A novel thermostable ferritin from Pyrococcus yayanosii CH1 directs the synthesis of thermostable magnetoferritins (M-PcFn5000) in one step.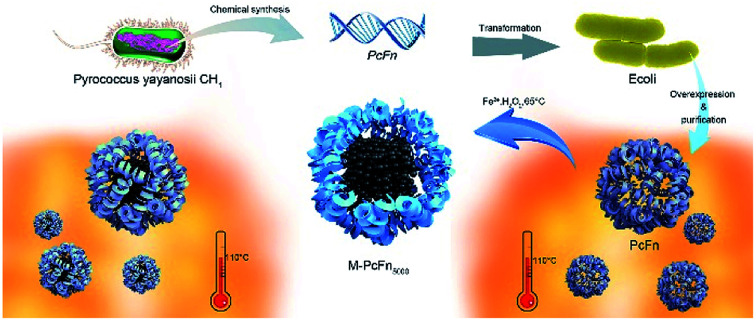
Introduction
Magnetic nanoparticles (MNPs) consisting of magnetite (Fe3O4) or maghemite (γ-Fe2O3) have been studied for applications at elevated temperatures, including in subsurface reservoirs1–4 and in catalysis.5,6 For example, superparamagnetic iron oxide nanoparticle dispersions can be applied as imaging agents for visualization of oil in underground oil reservoirs.7 These nanoparticles can be induced to move to the oil-water or air–water interface by a magnetic field, and the motion of magnetic nanoparticles at the interface can be detected by an external magnetic field which generates acoustic waves.8 In many cases, MNPs need to be coated with functional polymers to obtain optimal dispersibility and thermal stability, and limit magnetic interactions in aqueous mediums; examples include poly (2-methyl-2-acrylamidopropanesulfonate-co-acrylic acid),1 alkanedisulfamic acid5 and tetraethyl orthosilicate.6 However, these functional MNPs were fabricated using expensive polymer stabilizers, limiting their large-scale production and practical applications.
Ferritin is an ubiquitous iron storage protein consisting of 24 subunits, which self-assembles into a spherical nanocage with an exterior diameter of 12 nm and an internal diameter of 8 nm.9 Thus, it has been exploited as an excellent nanocontainer for synthesis of a variety of uniform-sized nanoparticles, including Fe,10 Fe3O4,11 Co(O)OH,12 Mn3O4,13 CaCO3,14 CdSe,15 CoPt,13 ZnSe,16 and Au.17 Magnetoferritin is a synthetic derivate of ferritin, consisting of an apoferritin shell and a magnetic iron oxide core (Fe3O4, γ-Fe2O3). The protein shell endows magnetoferritin with good dispersity and biocompatibility, and the magnetic core provides good superparamagnetism. This allows magnetoferritin to be monodispersed without extra modification, reducing its cost of production and simplifying the production procedures. Therefore, magnetoferritin serves as an excellent vehicle for biomedical applications, such as targeted drug delivery, delivery of contrast agents for early diagnosis of tumors, and cancer hyperthermia.18–21 From perspective of application in subsurface, the small size (about 12 nm) and superparamagnetism of magnetoferritin may make it easily detected with minimal retention through the porous mediums. In addition, ferritin has relatively high thermostability compared to most proteins. Most mesophilic ferritin cages, such as recombinant human H chain ferritin (HFn), can resist temperatures as high as 77 °C;22 indeed, this temperature is commonly used for ferritin purification. However, under some elevated temperature conditions, including underground reservoirs where the temperature is always above 80 °C,23 these mesophilic ferritins become unstable. To improve the thermostability of ferritin-based materials, several surface chemical modifications and gene mutation methods have been implemented. Yang et al. increased the Tm (denaturation temperature) of apo-red bean seed ferritin (OFN) from 73.12 °C to 78.16 °C with oligochitosan modification.24 Kim et al. reported an N-terminal fusion mutant of human L-chain ferritin maintaining about 60% concentration after heating at 72 °C for 30 min at both high (1.3 mg mL−1) and low (0.2 mg mL−1) protein concentrations.25 Nevertheless, these methods only slightly increased the thermostability of ferritins.
The discovery of hyperthermophilic organisms in extreme environments has paved the way for obtaining highly thermostable ferritins and magnetoferritins. Pyrococcus furiosus, is one of the most-studied hyperthermophilic archaeon, which was isolated from geothermally heated marine sediments with optimal growth temperature of 100 °C.26 Biologists have created a number of highly thermostable biomaterials from this organism, including Pfu-DNA polymerase,27 which is widely used in high-fidelity PCR amplification. Recombinant ferritin from P. furiosus (PfFn) is an extremely thermostable protein, with no melting occurring (by differential scanning calorimetry) up to 120 °C.28,29 Mackenzie et al. used PfFn as a template for the synthesis of magnetic nanoparticles by improving the synthesis temperature, and obtained particles with significant enhancement of magnetic hysteresis compared with HFn-mineral composites.30 Moreover, the ferritin AfFn from the hyperthermophilic archaeon Archaeoglobus fulgidus, has also been studied31 and used for enzyme encapsulation.32 These hyperthermophilic ferritins have remarkably elevated thermal tolerance for ferritin cages. However, there are currently no reports in the literature showing the thermostability of ferritins from new hyperthermophilic organisms (aside from PfFn), indicating that hyperthermophilic ferritins need to be further investigated.
In this study, we report a novel ferritin (PcFn) from the hyperthermophile Pyrococcus yayanosii CH1, which was isolated from a vent field at a depth of 4100 m and characterized as the first obligate piezophilic hyperthermophilic microorganism (optima of 52 MPa and 98 °C).33–35 According to these extremely harsh living conditions, we hypothesized that the ferritin and magnetoferritin from P. yayanosii CH1 might be resistant to high temperatures. To test this hypothesis, we cloned and overexpressed the PcFn gene from hyperthermophile P. yayanosii CH1 in Escherichia coli (E. coli), and successfully used the purified PcFn protein for biomimetic synthesis of magnetoferritin (M-PcFn5000), with a loading factor of 5000 Fe/cage. In addition, two other ferritins, HFn and PfFn, were also cloned, overexpressed in E. coli, and purified in order to synthesize the magnetoferritins M-HFn5000 and M-PfFn5000 for comparative study with PcFn and M-PcFn5000. To our knowledge, this is the first systematic study of the differences in thermostability between these three ferritin cages and their magnetoferritins. Furthermore, the crystalline structures and magnetic properties were also investigated between these different magnetoferritins, which has not previously been shown.
Experimental
Materials
Ammonium ferrous sulfate ((NH4)2Fe(SO4)2·6H2O) was purchased from Sigma-Aldrich (USA). Sodium chloride, Tris and N,N,N′,N′-tetramethylenebis(acrylamide) (TEMED) were obtained from Sangon Biotech (China). All experiments were performed using ultrapure water (MilliPore). Bovine serum albumin (BSA) was obtained from Thermo Scientific (USA).
Preparation of recombinant ferritins
Recombinant HFn was prepared as previously described.36 The gene sequences of two thermophilic ferritins, PcFn and PfFn, were identified and downloaded from GenBank, having respective gene IDs of 10837266 and 1468595. The obtained genes were modified for codon preference of E coli and synthesized. Subsequently, the expression vector pET-22b, containing either PcFn or PfFn, was transformed into E. coli BL21 (DE3). We cultured the E. coli at 37 °C to an OD600 of 0.6 in ampicillin-containing liquid Luria-Bertani (LB) medium, and induced expression with 0.5 mM isopropyl-β-d-thiogalactoside (IPTG) overnight at 30 °C. The cells were harvested by centrifugation at 8000 rpm for 8 min and the pellet was washed once and re-suspended in Tris–HCl buffer (0.025 M Tris, 0.1 M NaCl, pH 8.5). The cells were then incubated in lysis buffer (1 mM EDTA, 50 μg mL−1 lysozyme, 0.025 M Tris, 0.1 M NaCl, pH 8.5) for 2 h at 37 °C, and then heated at 75 °C (PcFn) or 100 °C (PFn) for 20 min. The preliminarily purified proteins were obtained by collecting the supernatant after centrifugation at 10 000 rpm for 30 min at 4 °C. Further purification was conducted by size exclusion chromatography (Sepharose 6B, GE Healthcare). Finally, purity of the ferritins was analyzed by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE). The Pierce™ BCA protein assay kit was used to determine protein concentrations.
Synthesis and transmission electron microscope (TEM) characterization of ferritin cages and magnetoferritins
The magnetoferritins were synthesized according to the method of our previously studies.36,37 All solutions were degassed and transferred into an anaerobic chamber. Taking M-PcFn5000 for example, the solution of 40 mL PcFn (1 mg mL−1) in 0.1 M NaCl was transferred into a reaction vessel, and the reaction temperature was kept at 65 °C and the pH was stabilized at 8.5 using 50 mM NaOH solution with a pH stat titrator. 8.3 mL of 50 mM (NH4)2Fe(SO4)2·6H2O was added at a rate of 50 Fe/(protein min) using a dosing device (800 Dosino). Simultaneously, stoichiometric equivalents (1 : 3H2O2 : Fe2+) of freshly prepared 8.3 mL H2O2 (16.7 mM) were added as an oxidant. The magnetite-forming reaction can be expressed as3Fe2+ + H2O2 + 2H2O → Fe3O4 + 6H+.
After adding theoretical 5000 iron atoms per protein cage, 1 mL of 0.3 M sodium citrate was added to chelate any free iron species. Centrifugation (10 000g for 30 min) was required to remove impurities suspended in the solution. The three of magnetoferritin nanoparticles synthesized were named M-HFn5000, M-PcFn5000, and M-PFn5000. After desalting, 3 μL of diluted samples (0.2 mg mL−1) were dropped onto a plasma-cleaned carbon-coated copper grid and dried at room temperature overnight. The morphology and crystallography of M-HFn5000, M-PcFn5000, and M-PFn5000 were then analyzed by TEM with an accelerating voltage of 200 kV. The size distribution of the three magnetoferritins was measured over 500 particles and crystallographic orientation of the core was examined by high-resolution TEM (HR-TEM). For negative-staining TEM observation, apoferritin and magnetoferritin samples (3 μL, 0.2 mg mL−1) were embedded in a Plasma Cleaner HPDC32G treated copper grid and stained with 1% uranyl acetate for 1 min, then imaged with a JEM-1400 100 kV TEM (JEOL, Japan).
Thermogravimetric analysis (TGA) and ICP Optical Emission Spectrometer (ICP-OES) analysis
TGA was used to obtain the proportion of ferritin cage of entire magnetoferritin nanoparticles by using TGA/DSC 3 STARe thermogravimetric analyzer (Mettler Toledo). The desalted and dried samples were heated from 30 °C to 700 °C at 5 °C min−1 under a N2 flow at 50 mL min−1. N2 flow is used to prevent the further oxidation of Fe3O4. ICP-OES (Avio 200 ICP Optical Emission Spectrometer, PerkinElmer) was used to determine the iron content of magnetoferritins. The iron standard sample was diluted into 0 ppm, 20 ppm, 40 ppm, 60 ppm, 80 ppm and 100 ppm, and the samples were dissolved by 5% (v/v %) hydrochloric acid and prepared with the concentration lower than 100 ppm before measured.
Magnetic measurements of magnetoferritins
Magnetic measurements were conducted with a Magnetic Property Measurement System (MPMS-5XL, Quantum Design, Inc.). Magnetoferritin samples (M-HFn5000, M-PcFn5000, and M-PFn5000) were desalted and dried before magnetic measurements were taken. The isothermal remanent magnetization (IRM) acquisition and demagnetization remanence (DCD) curves were measured at 5 K within ±1000 mT for calculation of magnetostatic interactions. Hysteresis loops were measured in field range of −3 T to 3 T at 5 K and 300 K, respectively.
Temperature gradient treatment of the ferritin cages and magnetoferritins
All samples (1 mL of a 0.5 mg mL−1 solution in 0.1 M NaCl, pH 8.5) were prepared in 1.5 mL centrifuge tubes and heated for 30 min in a heat block (at 70 °C, 80 °C, 90 °C, and 100 °C) or oil bath (110 °C and 120 °C) with a temperature gradient. The centrifuge tubes were wrapped with sealing film to prevent evaporation. After heating, all samples were centrifuged (14 500 rpm) for 10 min and stabilized at 4 °C overnight.
Dynamic light scattering (DLS) analysis of ferritin cages and magnetoferritins
The hydrodynamic size of apoferritins and magnetoferritins were determined by dynamic light scattering (DLS) (DynaPro NanoStar, Wyatt Technology Corporation. USA) at 25 °C with a scattering angle of 90 °C. Diluted samples (0.5 mg mL−1 in 0.1 M NaCl solution) were filtered with a 0.22 μm filter membrane and 10 μL of sample was used for DLS measurements.
Circular dichroism (CD) measurements of ferritin cages and magnetoferritins
CD spectra were taken on a Chirascan™-Plus CD spectrometer (Applied Photophysics, Leatherhead, UK) at 200–260 nm (bandwidth: 1 nm) at protein concentrations ∼0.1 mg mL−1. An attached software CDNN was used to calculate the secondary structure content and composition of all data by auto-deconvolution.
Differential scanning calorimetry (DSC) analysis
DSC was performed in a Nano DSC Microcalorimeter (TA instruments, Waters LLC, USA). Before DSC measurements were taken, 1 mg mL−1 of each sample in 0.1 M NaCl (pH 8.5) was filtered through a 0.22 μm filter, degassed for 15 min, and loaded into the Nano DSC. The experiments were carried out under pressure of 303 kPa to avoid degassing during heating. The protein was heated in the Nano DSC at a scan rate of 1 °C min−1 over a range of 60 to 130 °C. After buffer subtraction and concentration normalization, a plot of excess heat capacity versus temperature was obtained, and baseline was determined by manually adjusting the left and right linear line segments. Each peak was then set to the median temperature (Tm), the curve was fit with a non-two-state model, and the calorimetric enthalpy (ΔH) was calculated as the integral of the fitting curve area.
Iron uptake assay of recombinant ferritins
Iron incorporation experiment was conducted at 25 °C and monitored spectrophotometrically at 315 nm for 20 min. Specifically, 10 mM ammonium ferrous sulfate solution with 4.95 mg mL−1 thiourea was prepared as Fe2+ reagent and prevented the autoxidation of Fe2+. HEPES (0.1 M, pH 7.0) was prepared as reaction buffer. After adding ferritins to HEPES reaction buffer at a final concentration of 0.025 mg mL−1, 20 μL Fe2+ reagent was rapidly mixed and scanned in a UV/visible spectrophotometer.
Results and discussion
Sequence alignment and structure analysis
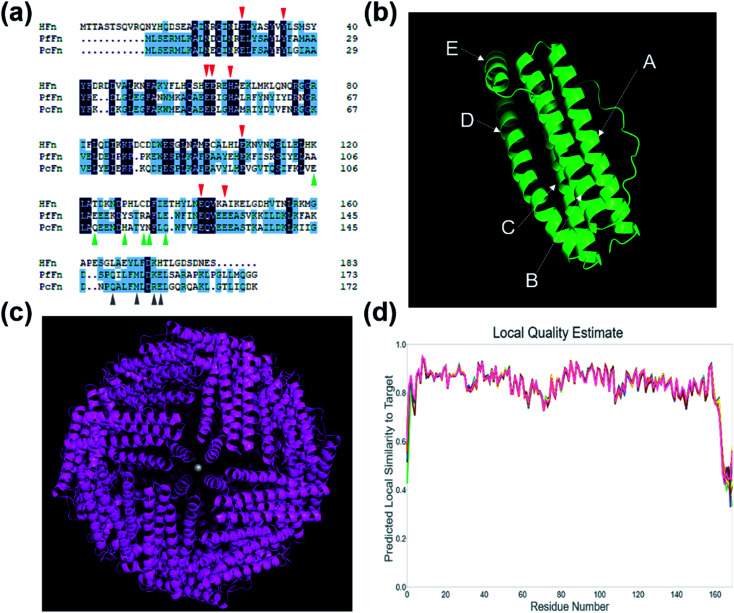
Fig. 1a shows the comparison of amino acid sequence of PcFn with HFn (36% identity, 55% similarity, 10% gaps) and PfFn (63% identity, 80% similarity, 0% gaps). The ferroxidase center of these three ferritins are highlighted in red triangle. Similar to PfFn, the residues E17, Y24, E50, and H53 of PcFn are characterized as motif EXXH, and Q127 and E130 arranged as the motif QXXE,28 which are highly conserved. The threefold channels sites of PcFn: E106, Q109, H114, Y117, N118, and Q121 are totally different from that of PfFn and HFn (shown in green triangle). However, the fourfold channels sites of PcFn: Q149, M153, R156, E157 are identical to that of PfFn (gray triangle),29 but different from HFn. In addition, the tertiary structure and quaternary structure of PcFn were predicted by using a template ferritin (PDB ID: 2X17) on the website (https://swissmodel.expasy.org). The subunit structure of PcFn is similar to HFn (PDB ID: 2FHA) and PfFn (PDB ID: 2X17), which is composed of five α-helix: A, B, C, D, and E (Fig. 1b). The predicted quaternary structure of PcFn is also cage-like protein with 24 subunits and consistent with the HFn and PfFn (Fig. 1c).
Fig. 1. Amino acid sequence analysis and protein structure prediction. (a) Amino acid sequence analysis of HFn, PfFn and PcFn. The red triangle shows the ferroxidase sites, the green triangle shows the threefold channels sites, and the gray triangle shows the fourfold channel sites. (b) The predicted subunit of PcFn with five α-helix: A, B, C, D and E. (c) The predicted 24-mer of PcFn templated by ferritin (PDB ID: 2X17). (d) The local quality estimate of predicted model.
Synthesis and characteristics of recombinant ferritin cages
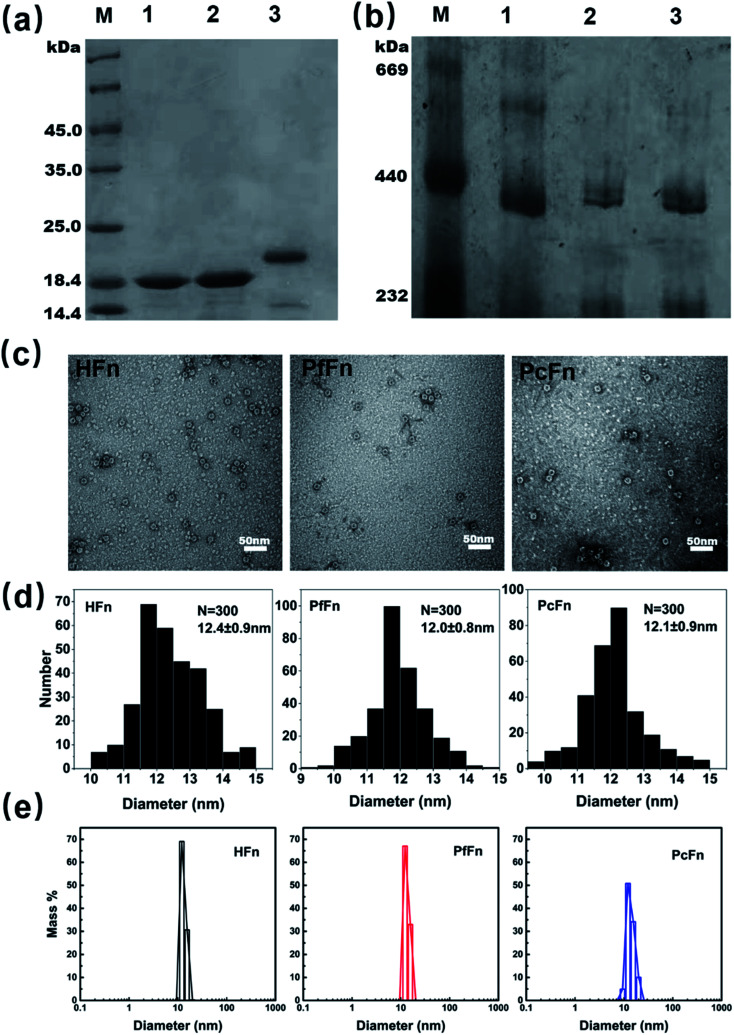
The recombinant ferritins (HFn, PfFn, and PcFn) were overexpressed in E. coli BL21 cells. After purification, the proteins were analyzed by SDS-PAGE and native PAGE. According to the amino acid sequence, the predicted molecular weights (MW) of HFn, PfFn and PcFn subunit are 21.2 kDa, 20.3 kDa, 20.1 kDa, respectively. The MW of the native HFn, PfFn, and PcFn subunits in the SDS-PAGE result were determined to be ∼20 kDa, ∼19 kDa, and ∼19 kDa, with only one protein band observed for each, indicating high purity of each protein (Fig. 2a). In the native PAGE gel, we can see that all three proteins had an apparent band at approximately 440 kDa, suggesting that the full protein PcFn may be composed of 24 subunits (Fig. 2b), which is accordant with the predict structure in Fig. 1c. To further investigate the size and protein architecture of PcFn, TEM and DLS were performed. Bright-field TEM images of negative stained apo-ferritins (Fig. 2c) showed that all the protein cages were monodispersed and intact, and the outer diameter of HFn, PfFn, and PcFn is 12.4 ± 0.9 nm, 12.0 ± 0.8 nm and 12.1 ± 0.9 nm, respectively (Fig. 2d). DLS results confirmed that the native HFn, PfFn, and PcFn protein cages were monodispersed, with average outer diameters of 12.1, 13.3, and 13.9 nm in solution (0.1 M NaCl), respectively (Fig. 2e), which is accordance with the TEM results and previously reported ferritin sizes.38
Fig. 2. Characteristics of recombinant ferritin cages. (a) SDS-PAGE and (b) native-PAGE analyses of recombinant ferritins. Lane M: protein markers and their corresponding molecular masses, lane 1: PcFn, lane 2: PfFn, lane 3: HFn. (c) Negative-stained TEM images of ferritin cages, scale bar is 50 nm. (d) Histograms of size distributions of ferritin cages. (e) DLS analysis of HFn, PfFn, and PcFn nanocages, with diameters in solution of 12.1, 13.3, and 13.9 nm, respectively.
Synthesis and characteristics of magnetoferritins
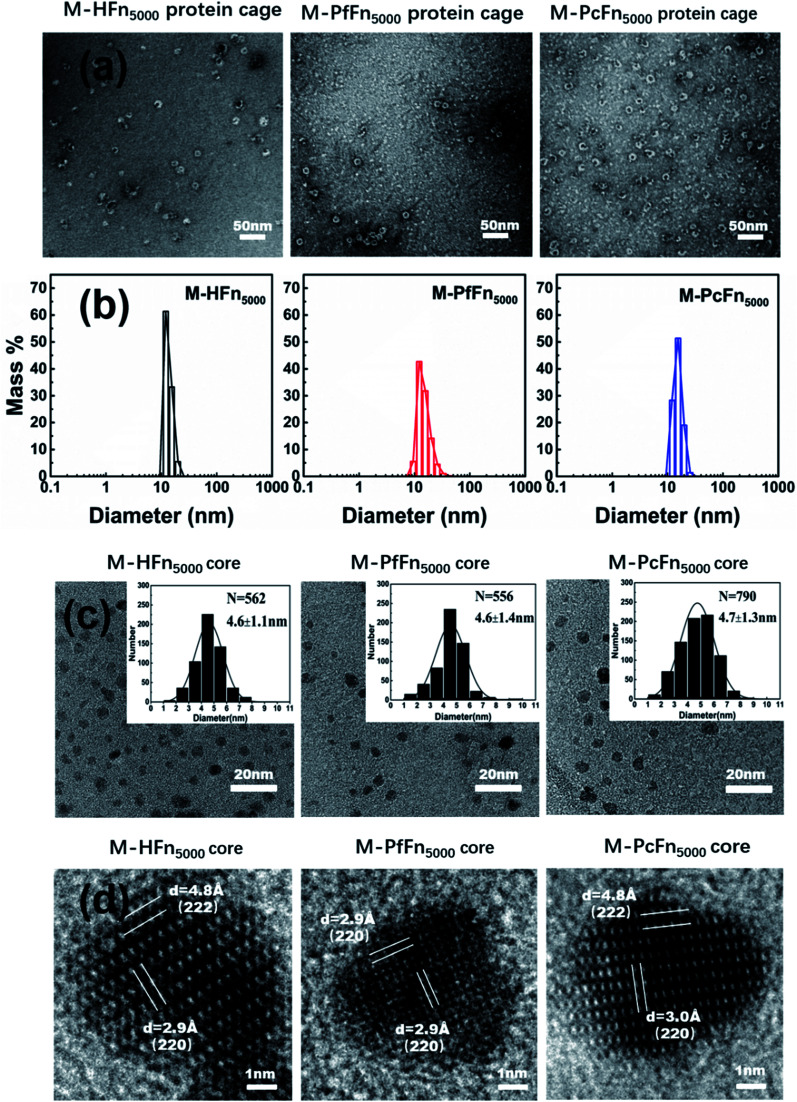
The magnetoferritins (M-HFn5000, M-PfFn5000, and M-PcFn5000) were synthesized by step-wise loading of iron oxide nanoparticles into the HFn, PfFn, and PcFn nanocages. After synthesis and purification by centrifugation and size exclusion chromatography, the protein cages could be seen clearly in negative-stained TEM images, indicating no disruption of the protein cages during synthesis (Fig. 3a). DLS data for the magnetoferritins shows hydrodynamic diameters (HD) of 13.6, 15.3, and 15.5 nm for M-HFn5000, M-PfFn5000, and M-PcFn5000 (Fig. 3b), respectively, which were only slightly larger than the original protein cages. This also confirms that the iron oxide nanoparticles were encapsulated by protein cages, because the aggregation of iron oxide nanoparticles always causes little changes in HD. TEM images show all the synthesized cores of magnetoferritins are monodispersed and sphere-like nanoparticles with mean diameters of 4.6 ± 1.1 nm, 4.6 ± 1.4 nm, and 4.7 ± 1.3 nm for M-HFn5000, M-PfFn5000, and M-PcFn5000, respectively (Fig. 3c). The size of M-HFn5000 is in accordance with previously published data,36,39 while the size distributions of M-PfFn5000 and M-PcFn5000 are slightly broader than for M-HFn5000. High-resolution TEM images (Fig. 3d) indicate that the cores of all three magnetoferritins are well-crystalline with clear lattice fringes. The measured lattice planes (220), (222) indicate that the mineral structure is magnetite (Fe3O4) or maghemite (γ-Fe2O3). Despite the fact that PcFn shares only 36% amino-acid sequence identity with HFn, and 63% identity with PfFn, the protein cages are very similar in shape and size. All of these ferritins can be used as good templates for controlled synthesis of magnetic nanoparticles with uniform shape and size distributions. In addition, these protein shells can greatly improve their dispersibility and help maintain lower magnetic interactions of the inner magnetite cores.
Fig. 3. Characteristics of magnetoferritins. (a) TEM images of negative-stained magnetoferritins (M-HFn5000, M-PfFn5000, and M-PcFn5000). Scale bar is 50 nm. (b) DLS analysis of native M-HFn5000, M-PfFn5000, M-PcFn5000 nanocages, with diameters in solution of 13.6, 15.3, 15.5 nm, respectively. (c) TEM images of M-HFn5000, M-PfFn5000, and M-PcFn5000 nanoparticles, and histograms of size distributions of magnetoferritins. Scale bar is 20 nm. (d) High-resolution TEM images showing lattice fringes of the magnetite cores. Scale bar is 1 nm.
In order to determine the ferritin cage and iron content of magnetoferritins, TGA combined with ICP-OES was conducted. TGA results indicate that organic content (ferritin cage) of M-HFn5000, M-PfFn5000, and M-PcFn5000 were 42.6%, 47.8%, and 56.9% (Fig. S1†), and Fe% of these particles measured ICP-MOS are listed in Table 1. According to the data, we calculated that each ferritin cage of M-HFn5000, M-PfFn5000, and M-PcFn5000 contain 4244, 4330, and 4132 Fe atoms, respectively. The iron atoms number inside protein cages are lower than theoretical 5000 iron atoms per protein cage and the loss of iron may be due to aggregation of Fe outside the ferritin cage, which were chelated by citrate and removed by centrifugation after reaction.
Ferritin cage and iron content of synthesized M-HFn5000, M-PfFn5000, and MPcFn5000.
| Sample | Ferritin cage (wt%) | Fe (wt%) | Number (Fe atom/ferritin cage) |
|---|---|---|---|
| M-HFn5000 | 42.6 | 19.9 | 4244 |
| M-PfFn5000 | 47.8 | 28.6 | 4132 |
| M-PcFn5000 | 56.9 | 22.7 | 4330 |
Magnetic properties of magnetoferritin nanoparticles
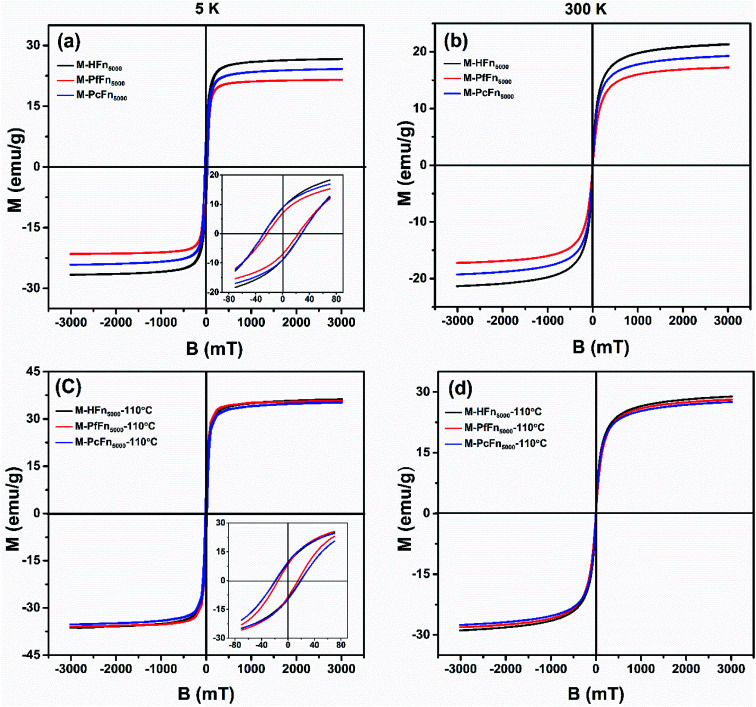
The magnetic properties of magnetoferritins (before and after heating at 110 °C for 30 min) were measured and summarized in Table 2 and Fig. 4. The 5 K coercivity (Hc) of M-HFn5000, M-PfFn5000, and M-PcFn5000 were 28.9 mT, 23.1 mT, and 27.8 mT, respectively, which are in agreement with the coercivity of magnetite or maghemite among 20–30 mT.40 At 5 K, the shape of the hysteresis loops among these samples are slightly constricted (wasp-waisted), meaning that some ultrasmall iron oxide nanoparticles are not unblocking at 5 K, and superparamagnetic and single-domain magnetite nanoparticles coexist in the samples. The ratios of saturation remanence to saturation magnetization (Mrs/Ms) at 5 K were 0.32, 0.35, and 0.37 for M-HFn5000, M-PfFn5000, and M-PcFn5000, respectively, indicating that all these samples are dominated by uniaxial anisotropy.41 Based on the Wohlfarth–Cisowski test for randomly oriented noninteracting single-domain (SD) particles, the isothermal remanent magnetization acquisition curve (IRM) and direct current demagnetization curve (DCD) cross R = 0.5.42 In this study, the R values of M-HFn5000, M-PfFn5000, and M-PcFn5000 were 0.42, 0.44, and 0.43, respectively, indicating very weak magnetic interactions in all these samples, likely because the magnetite nanoparticles were well-dispersed by protein cages (Table 2 and Fig. S2†). At 300 K, these three particle types showed no Mrs (Fig. 4b). Moreover, after heating at 110 °C, the value of Mrs, Ms at 5 K and Ms at 300 K increases but the value of Hc decreases, which could be explained by that some surface atoms rearranged to align dominant axial orientation after heating and reduced the surface spin disorder thickness of the magnetoferritin core. The R value of each sample is nearly the same after heating at 110 °C, indicating weak magnetic interaction among each magnetic nanoparticle and no disruption of ferritin cages under high temperature.
Magnetic parameters of magnetoferritinsa.
| Samples | Mean size (nm) | M rs (5 K) (emu g−1) | M s (5 K) (emu g−1) | M rs/Ms (5 K) | H c (5 K) (mT) | M s (300 K) (emu g−1) | R |
|---|---|---|---|---|---|---|---|
| M-HFn5000 | 4.6 | 8.7 | 26.7 | 0.33 | 28.9 | 21.3 | 0.42 |
| M-HFn5000-110 °C | 4.6 | 9.1 | 36.3 | 0.25 | 18.8 | 28.9 | 0.42 |
| M-PfFn5000 | 4.6 | 6.9 | 21.5 | 0.32 | 23.1 | 17.2 | 0.44 |
| M-PfFn5000-110 °C | 4.6 | 8.5 | 35.9 | 0.24 | 14.7 | 28.1 | 0.43 |
| M-PcFn5000 | 4.7 | 8.8 | 24.1 | 0.37 | 27.8 | 19.3 | 0.43 |
| M-PcFn5000-110 °C | 4.7 | 9.6 | 35.2 | 0.27 | 19.3 | 27.5 | 0.42 |
M rs, saturation remanence; Ms, saturation magnetization; Hc, coercivity; R value was determined by Wohlfarth–Cisowski test, as shown in Fig. S2. R value of 0.5 suggesting no magnetostatic interaction.
Fig. 4. Hysteresis loops of the desalted and dried magnetoferritins before and after heating at 110 °C for 30 min measured at 5 K (a and c) and 300 K (b and d).
Thermostability of recombinant ferritins
The protein concentration of each sample heated at different temperatures (25 °C, 70 °C, 80 °C, 90 °C, 100 °C, 110 °C, and 120 °C) for 30 min was determined by BSA method after centrifugation (14 500 rpm) for 10 min and stabilized at 4 °C overnight. It was clear that the concentration of HFn decreased when the heating temperature reached 90 °C. When the heating temperature was increased to 110 °C, only 18.7% of the original protein was preserved, while most of the HFn aggregated and precipitated (Fig. S3†). In contrast, most of the PfFn and PcFn were still soluble when the temperature reached 110 °C, and maintained 78.3% and 76.2% of the protein concentration, respectively.
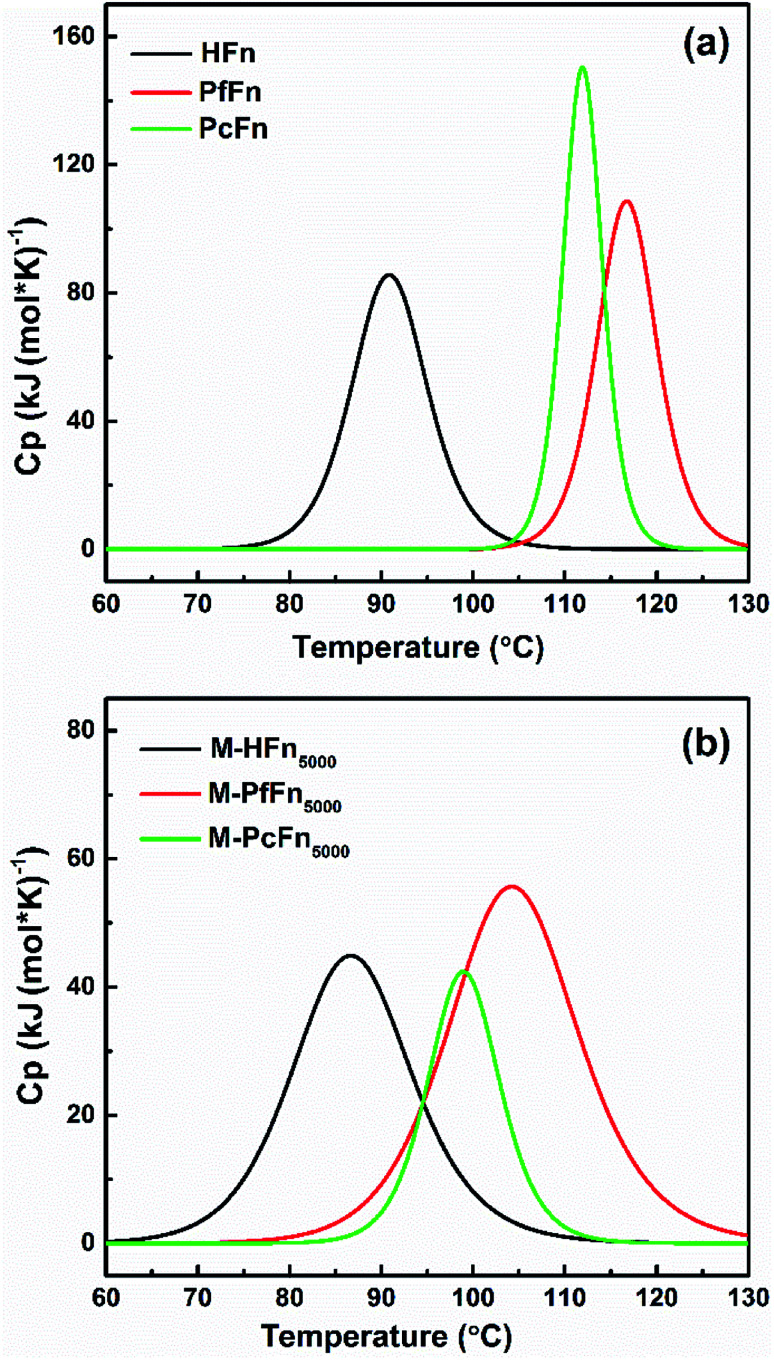
DSC is a technology that can characterize protein stability by measuring the thermal denaturation of samples while heating at a constant rate. The excess heat capacity (Cp) temperature curves and fitting curves are shown in Fig. 5. Tm value and the calorimetric enthalpy (ΔH) were acquired according to the fitting curve. It is shown that the Tm values for HFn, PfFn, and PcFn were 90.9 °C, 116.8 °C and 112.0 °C, respectively. The Tm values for PfFn and PcFn were significantly higher than for HFn, reflecting that the structures of PcFn and PfFn in solution are more stable than that of HFn (Table 3). The ΔH values of HFn, PfFn, and PcFn were 402.8, 580.1, 861.0 kJ mol−1, respectively. The differences in ΔH values are probably related to the different processes of protein unfolding during heating. It should be noted that a previous study reported the DSC results of PfFn with no Tm from 20 °C to 120 °C,28 which is different from our results of a Tm for PfFn of 116.8 °C. There might be two explanations for this: (1) ferritin has been demonstrated to be relatively thermolabile at high protein concentrations,43 and the DSC experiments were performed at a relatively higher concentration of PfFn in this study (1 mg mL−1) than that of previous study (∼0.15 mg mL−1); (2) we used 0.1 M NaCl to stabilize the ferritins which has a lower ionic strength than the buffer of PfFn previously used (50 mM HEPES, 250 mM NaCl solution, pH 7 or 1× PBS solution, pH 7.4).
Fig. 5. DSC profiles of ferritins (HFn, PfFn, and PcFn) and magnetoferritins (M-HFn5000, M-PfFn5000, and M-PcFn5000). Measurements were carried out in 0.1 M NaCl at pH 8.5. The scan rate was 1 °C min−1.
Thermodynamic parameters for thermal denaturation of ferritin cages and magnetoferritins.
| Sample (1 mg mL−1) | T m (°C) | ΔH (kJ mol−1) |
|---|---|---|
| HFn | 90.9 | 402.8 |
| PFn | 116.8 | 580.1 |
| PcFn | 112.0 | 861.0 |
| M-HFn5000 | 86.9 | 248.4 |
| M-PFn5000 | 104.5 | 250.5 |
| M-PcFn5000 | 100.1 | 441.9 |
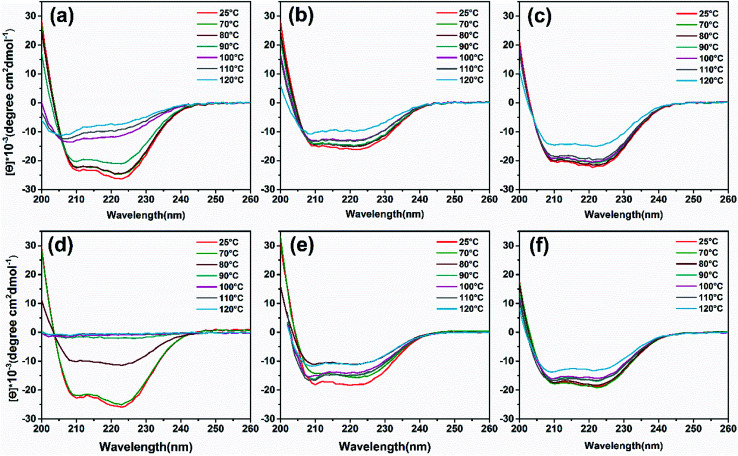
To analyze heat-induced structural changes in the α-helical structure of ferritin, CD spectra were performed at protein concentrations of 0.1 mg mL−1. As shown in Fig. 6a–c, the curves of HFn obviously shift upward with increased temperature. However, the curves of PcFn and PfFn began to shift until the heating temperature reached 120 °C. To quantify the proportion of secondary structure in the samples, the original data was imported into a software CDNN to calculate the α-helical, β-sheet, and random coil contents. The α-helical content of HFn, PcFn, and PfFn heated to 110 °C decreased by 45.3%, 16.3%, and 16.5%, respectively (Fig. S4†), while the α-helical content of PcFn and PfFn heated to 120 °C was reduced to 34.5% and 34%, respectively, indicating that the structures of PcFn and PfFn changed irreversibly at 120 °C. This result is consistent with the DSC and protein concentration experiements.
Fig. 6. CD wavelength scans of ferritins and magnetoferritins with temperature gradient treated (control group, 70 °C, 80 °C, 90 °C, 100 °C, 110 °C, 120 °C) and composition of α-helix. (a) HFn, (b) PfFn, (c) PcFn. (d) M-HFn5000, (e) M-PfFn5000, (f) M-PcFn5000.
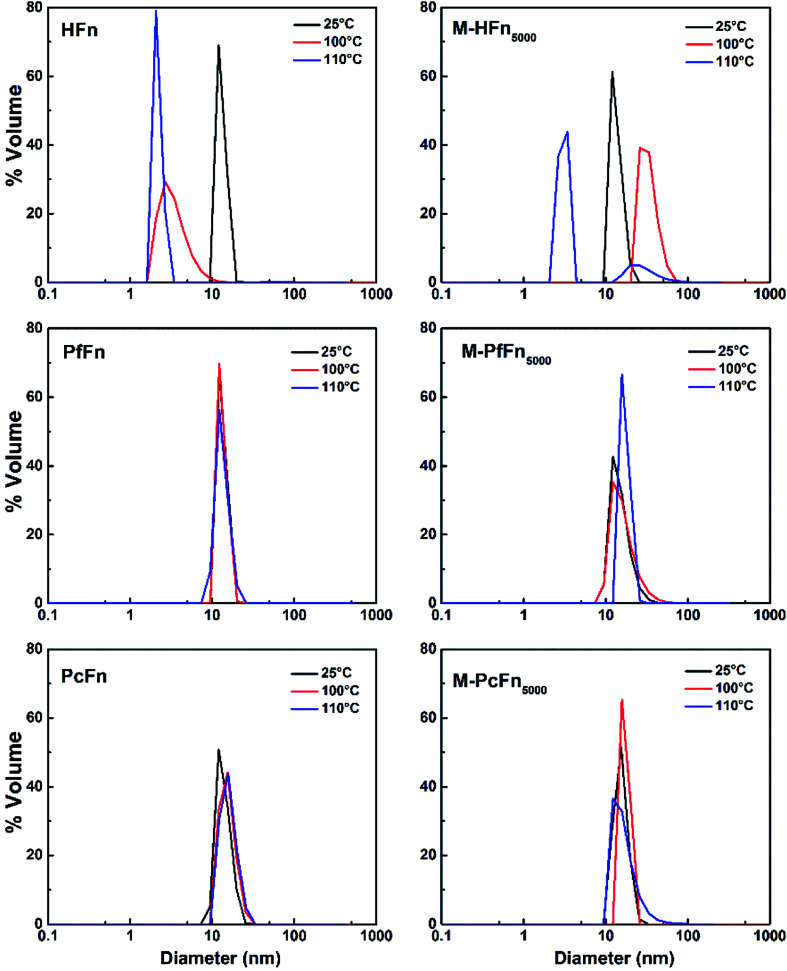
DLS was performed to detect the hydrodynamic size distribution of ferritins in aqueous solution.44 As shown in Fig. 7 and Table 4, the average hydrodynamic sizes of HFn, PfFn, and PcFn in 0.1 M NaCl before heating were 12.1 nm, 13.3 nm, and 13.9 nm, respectively. However, the size distribution of HFn was markedly altered toward smaller particles, with hydrodynamic sizes of 3.5 nm and 2.2 nm observed after heating at 100 °C and 110 °C, respectively, for 30 min. The sudden change in the hydrodynamic size of HFn is attributed to protein disassembly. In contrast, the hydrodynamic size of PfFn had no significant change before and after heating. And the hydrodynamic size of PcFn turned slight larger, to 15.5 nm and 16.0 nm, after heating at 100 °C and 110 °C, respectively.
Fig. 7. DLS analysis of ferritins (HFn, PfFn, and PcFn) and magnetoferritins heated at 100 °C (red plot) and 110 °C (blue plot) for 30 min with a control group (25 °C) (black plot).
The average hydrodynamic size of apoferritins and magnetoferritins (0.5 mg mL−1) in 0.1 M NaCl after heating (control group, 100 °C, and 110 °C) for 30 mina.
| Sample (0.5 mg mL−1) | HD before heating (nm) | HD after heating at 100 °C (nm) | HD after heating at 110 °C (nm) |
|---|---|---|---|
| HFn | 12.1 | 3.5 | 2.2 |
| PfFn | 13.3 | 13.3 | 13.4 |
| PcFn | 13.9 | 15.5 | 16.0 |
| M-HFn5000 | 13.6 | 3.1/29.5 | 2.6/39.8 |
| M-PfFn5000 | 15.3 | 16.8 | 17.3 |
| M-PcFn5000 | 15.5 | 17.3 | 17.3 |
HD: hydrodynamic diameter.
The thermostability of a protein is determined by various factors, including hydrogen bonds,45–47 salt bridges,48 and amino acid properties.49,50 By comparing the amino acid composition of mesophilic ferritin (HFn) with thermophilic ferritins (PcFn and PfFn) (Fig. S5†), we found that the prevalence of fully charged residues (Glu, Arg, and Lys)51 is much higher in PcFn (24.3%) and PfFn (24.1%) than in HFn (16.4%). It may be that the formation of extended networks of ion pairs by the charged groups represents a major stabilizing factor associated with adaptation to extreme temperatures.52,53 Additionally, we calculated the hydrogen bonds of the three ferritins with HBPLUS,54 and found that the total hydrogen bonds in PcFn (2683) and PfFn (2788) are significantly more than that of HFn (2373), despite HFn have more residues than other two thermophiles ferritins. This may explain why PfFn and PcFn have much higher thermostability than HFn, and why the PfFn is slightly more stable than PcFn as the DSC and CD data shown. As shown in Table 5, the default program divides the hydrogen bonds results into three classes: between main-chain atoms only (MM); between main-chain and side-chain atoms (MS/SM); and between side-chain atoms only (SS). The MM hydrogen bonds are a dominant portion of total hydrogen bonds, which mainly influence the thermostability of protein. MS/SM hydrogen bonds may not be an important factor of thermostability because the PfFn has the lowest of it but with highest thermostability. Furthermore, a previous study reported the preferentially replacement of mesophilic protein residues: Gly to Ala, Ser to Ala, Ser to Thr, Lys to Arg, and Asp to Glu in a thermophilic protein, respectively, strengthening the internal and weakened external hydrophobicity, and also facilitated helix stabilizing in α-helical proteins.55
Hydrogen bonds in 24-mer of HFn, PcFn, and PfFn, calculated with HBPLUSa.
| PDB ID | Sample | Total H-bondsb | MM | MS/SM | SS |
|---|---|---|---|---|---|
| 2FHA | HFn | 2373 | 1309 | 894 | 415 |
| —c | PcFn | 2683 | 1648 | 958 | 690 |
| 2JD6 | PfFn | 2788 | 2047 | 273 | 468 |
MM: H-bonds between main-chain atoms only; MS/SM: H-bonds between main-chain and side-chain atoms; SS: H-bonds between side-chain atoms only.
Calculated using code: “HBPLUS-d 3.0”.
The PDB format file of predicted PcFn obtained from (https://swissmodel.expasy.org/interactive/SEENpX/models/).
Thermostability of magnetoferritins
Similar to the thermostability of ferritin cages above, mineralized M-PcFn5000 and M-PfFn5000 have much higher thermostability than M-HFn5000. DSC curves show that Tm values for M-PcFn5000 (100.1 °C) and M-PfFn5000 (104.5 °C) are much higher than for M-HFn5000 (86.9 °C) (Fig. 5 and Table 3), suggesting that M-PcFn5000 and M-PfFn5000 remained the strong thermostability of protein cages.
CD spectra (Fig. 6d and e) and α-helical content results (Fig. S4b†) showed that the α-helical content of M-PfFn5000 decreased drastically with a temperature increased to 80 °C, while the secondary structure of M-PfFn5000 and M-PcFn5000 were able to resist temperatures of 110 °C. However, after heating at 120 °C for 30 minutes, the α-helical content of M-PfFn5000 and M-PcFn5000 had declined to 22.3% and 21.7% of their original α-helical content, respectively.
As shown in Fig. 7 and Table 4, the hydrodynamic diameters of M-HFn5000 increased from 13.6 nm before heating to 29.5 nm and 39.5 nm after heating at 100 °C and 110 °C, respectively, due to disassembly of HFn protein cages and aggregation of proteins and the magnetite/maghemite cores. But the hydrodynamic diameters of M-PfFn5000 and M-PcFn5000 experienced no significant changes after heating to 100 °C and 110 °C, indicating high thermostability of these two nanoparticles.
Moreover, we found that the Tm values for all the magnetoferritins (M-HFn5000, M-PfFn5000, and M-PcFn5000) declined (4 °C, 12.3 °C, and 11.9 °C, respectively) compared with their apoferritins (HFn, PfFn, and PcFn), which has not been previously reported. An explanation might be that the inorganic inner core (Fe3O4 or γ-Fe2O3) influenced the original connection of subunits of protein cage.
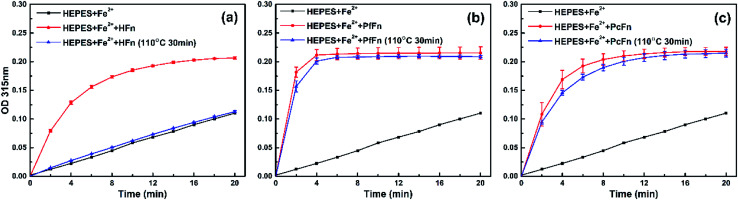
Iron incorporation of ferritin cages
To further investigate whether apo-ferritins retain iron-incorporating ability after heating at high temperature, we performed iron incorporation experiments with each apoferritin before and after heating at 110 °C, including a negative control that lacked apoferritins. A ferrihydrite core would be generated in the ferritin cage with the oxidation of Fe2+, and the absorbance obtained at 315 nm was used to quantify ferrihydrite.56 As expected, the HFn heated at 110 °C was unable to incorporate any Fe2+ due to disassembly of the protein cage. However, both of PcFn and PfFn exhibited the typical ability of iron incorporation after heating at 110 °C. We calculated the velocity of the first two minutes V0→2min (micromol per L per min) of ferrihydrite generation by determining the slope of each curve over the first two minutes. The PfFn and PcFn remained 86.3% and 87.4% of V0→2min, respectively (Fig. 8).
Fig. 8. A comparison of the iron uptake activity of ferritins: (a) HFn, (b) PfFn, and (c) PcFn before (red plot) and after (blue plot) heated at 110 °C for 30 min. Black plot is control group without adding ferritin. The reactions were monitored in time scan mode at 315 nm for 20 min.
Conclusion
We have overexpressed a novel ferritin, PcFn, from hyperthermophilic archaeon Pyrococcus CH1 in E. coli BL21 (DE3), and successfully used PcFn as a template for synthesis of M-PcFn5000 nanoparticles in vitro. To explore the structure, magnetic parameters, and thermostability of PcFn, we chose HFn and PfFn for comparisons, as the former is one of the most-studied mesophilic ferritins, and the latter is the most thermostable ferritin known to come from hyperthermophilic archaeon. TEM images and magnetic measurement results demonstrated that M-PcFn5000 particles are superparamagnetic, monodispersed, and well-crystalline magnetite/maghemite nanoparticles with uniform shape and size distribution. In comparison with HFn/M-HFn5000 and PfFn/M-PfFn5000, the thermostability of both PcFn and M-PcFn5000 was as high as 110 °C. After heating of the PcFn at 110 °C for 30 min, it still maintained 87.4% of V0→2min for iron incorporation ability. These encouraging findings open up the possibility of using PcFn as a new nanoplatform for controlled synthesis of thermostable nanoparticles for application in elevated temperature systems.
Conflicts of interest
There are no conflicts of interest to declare.
Supplementary Material
Acknowledgments
This work was supported by National Natural Science Foundation of China (NSFC) grants (41574062, 41774076, 41621004), Key Program of Chinese Academy of Sciences (QYZDJ-SSW-DQC024). We are grateful to Xu Tang from the Institute of Geology and Geophysics, Chinese Academy of Sciences for helping capture the high resolution TEM images, and we are grateful to B. X. Huangfu from the Institute of Biophysics, Chinese Academy of Sciences for helping with preparation of the negative-stained samples. We thank professor Xiang Xiao from Shanghai Jiaotong University for kindly sharing the gene sequence of PcFn with us. We also thank Yan Ma from Institute of Geology, China Earthquake Administration for help in the iron content measurement by ICP-OES.
Electronic supplementary information (ESI) available: Additional figures. See DOI: 10.1039/c9ra07397c
References
- Bagaria H. G. Yoon K. Y. Neilson B. M. Cheng V. Lee J. H. Worthen A. J. Xue Z. Huh C. Bryant S. L. Bielawski C. W. Johnston K. P. Langmuir. 2013;29:3195–3206. doi: 10.1021/la304496a. [DOI] [PubMed] [Google Scholar]
- Kotsmar C. Yoon K. Y. Yu H. Ryoo S. Y. Barth J. Shao S. Prodanović M. a. Milner T. E. Bryant S. L. Huh C. Ind. Eng. Chem. Res. 2010;49:12435–12443. doi: 10.1021/ie1010965. [DOI] [Google Scholar]
- Iqbal M. Lyon B. A. Ureña-Benavides E. E. Moaseri E. Fei Y. McFadden C. Javier K. J. Ellison C. J. Pennell K. D. Johnston K. P. Colloids Surf., A. 2017;520:257–267. doi: 10.1016/j.colsurfa.2017.01.080. [DOI] [Google Scholar]
- Rahmani A. R. Bryant S. Huh C. Athey A. Ahmadian M. Chen J. Wilt M. SPE J. 2015;20:1067–1082. doi: 10.2118/166140-PA. [DOI] [Google Scholar]
- Karimi A. Eftekhari Z. Karimi M. Dalirnasab Z. Synthesis. 2014;46:3180–3184. doi: 10.1055/s-0034-1378667. [DOI] [Google Scholar]
- Li W. Zhang B. Li X. Zhang H. Zhang Q. Appl. Catal., A. 2013;459:65–72. doi: 10.1016/j.apcata.2013.04.010. [DOI] [Google Scholar]
- Yoon K. Y. Kotsmar C. Ingram D. R. Huh C. Bryant S. L. Milner T. E. Johnston K. P. Langmuir. 2011;27:10962–10969. doi: 10.1021/la2006327. [DOI] [PubMed] [Google Scholar]
- Ryoo S. Rahmani A. R. Yoon K. Y. Prodanović M. Kotsmar C. Milner T. E. Johnston K. P. Bryant S. L. Huh C. J. Pet. Sci. Eng. 2012;81:129–144. doi: 10.1016/j.petrol.2011.11.008. [DOI] [Google Scholar]
- Arosio P. Ingrassia R. Cavadini P. Biochim. Biophys. Acta, Gen. Subj. 2009;1790:589–599. doi: 10.1016/j.bbagen.2008.09.004. [DOI] [PubMed] [Google Scholar]
- Hosein H.-A. Strongin D. R. Allen M. Douglas T. Langmuir. 2004;20:10283–10287. doi: 10.1021/la0491100. [DOI] [PubMed] [Google Scholar]
- Meldrum F. C. Heywood B. R. Mann S. Science. 1992;257:522–523. doi: 10.1126/science.1636086. [DOI] [PubMed] [Google Scholar]
- Douglas T. Stark V. T. Inorg. Chem. 2000;39:1828–1830. doi: 10.1021/ic991269q. [DOI] [PubMed] [Google Scholar]
- Jolley C. C. Uchida M. Reichhardt C. Harrington R. Kang S. Klem M. T. Parise J. B. Douglas T. Chem. Mater. 2010;22:4612–4618. doi: 10.1021/cm100657w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukano H. Takahashi T. Aizawa M. Yoshimura H. Inorg. Chem. 2011;50:6526–6532. doi: 10.1021/ic200117x. [DOI] [PubMed] [Google Scholar]
- Yamashita I. Hayashi J. Hara M. Chem. Lett. 2004;33:1158–1159. doi: 10.1246/cl.2004.1158. [DOI] [Google Scholar]
- Li S. Lee A. R. Kim J. H. Park S. J. Sci. Adv. Mater. 2015;7:2743–2746. doi: 10.1166/sam.2015.2599. [DOI] [Google Scholar]
- Pulsipher K. W. Honig S. Deng S. Dmochowski I. J. J. Inorg. Biochem. 2017;174:169–176. doi: 10.1016/j.jinorgbio.2017.06.012. [DOI] [PubMed] [Google Scholar]
- Fan K. Cao C. Pan Y. Lu D. Yang D. Feng J. Song L. Liang M. Yan X. Nat. Nanotechnol. 2012;7:459–464. doi: 10.1038/nnano.2012.90. [DOI] [PubMed] [Google Scholar]
- Cao C. Wang X. Cai Y. Sun L. Tian L. Wu H. He X. Lei H. Liu W. Chen G. Zhu R. Pan Y. Adv. Mater. 2014;26:2566–2571. doi: 10.1002/adma.201304544. [DOI] [PubMed] [Google Scholar]
- Cai Y. Wang Y. Xu H. Cao C. Zhu R. Tang X. Zhang T. Pan Y. Nanoscale. 2019;11:2644–2654. doi: 10.1039/C8NR06812G. [DOI] [PubMed] [Google Scholar]
- Fantechi E. Innocenti C. Zanardelli M. Fittipaldi M. Falvo E. Carbo M. Shullani V. Mannelli L. D. Ghelardini C. Ferretti A. M. Ponti A. Sangregorio C. Ceci P. ACS Nano. 2014;8:4705–4719. doi: 10.1021/nn500454n. [DOI] [PubMed] [Google Scholar]
- Stefanini S. Cavallo S. Wang C.-Q. Tataseo P. Vecchini P. Giartosio A. Chiancone E. Arch. Biochem. Biophys. 1996;325:58–64. doi: 10.1006/abbi.1996.0007. [DOI] [PubMed] [Google Scholar]
- Zhang F. She Y. H. Li H. M. Zhang X. T. Shu F. C. Wang Z. L. Yu L. J. Hou D. J. Appl. Microbiol. Biotechnol. 2012;95:811–821. doi: 10.1007/s00253-011-3717-1. [DOI] [PubMed] [Google Scholar]
- Yang R. Zuo P. Zhang M. Meng D. Wang B. Zhen T. Food Hydrocolloids. 2019;94:500–509. doi: 10.1016/j.foodhyd.2019.03.049. [DOI] [Google Scholar]
- Kim S.-W. Kim Y.-H. Lee J. Biochem. Biophys. Res. Commun. 2001;289:125–129. doi: 10.1006/bbrc.2001.5931. [DOI] [PubMed] [Google Scholar]
- Fiala G. Stetter K. O. Arch. Microbiol. 1986;145:56–61. doi: 10.1007/BF00413027. [DOI] [Google Scholar]
- Lundberg K. S. Shoemaker D. D. Adams M. W. W. Short J. M. Sorge J. A. Mathur E. J. Gene. 1991;108:1–6. doi: 10.1016/0378-1119(91)90480-Y. [DOI] [PubMed] [Google Scholar]
- Tatur J. Hagedoorn P. L. Overeijnder M. L. Hagen W. R. Extremophiles. 2006;10:139–148. doi: 10.1007/s00792-005-0484-x. [DOI] [PubMed] [Google Scholar]
- Tatur J. Hagen W. R. Matias P. M. J. Biol. Inorg Chem. 2007;12:615–630. doi: 10.1007/s00775-007-0212-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker M. J. Allen M. A. Ramsay B. Klem M. T. Young M. Douglas T. Chem. Mater. 2008;20:1541–1547. doi: 10.1021/cm702732x. [DOI] [Google Scholar]
- Johnson E. Cascio D. Sawaya M. R. Gingery M. Schroder I. Structure. 2005;13:637–648. doi: 10.1016/j.str.2005.01.019. [DOI] [PubMed] [Google Scholar]
- Tetter S. Hilvert D. Angew. Chem., Int. Ed. Engl. 2017;56:14933–14936. doi: 10.1002/anie.201708530. [DOI] [PubMed] [Google Scholar]
- Zeng X. Birrien J. L. Fouquet Y. Cherkashov G. Jebbar M. Querellou J. Oger P. Cambon-Bonavita M. A. Xiao X. Prieur D. ISME J. 2009;3:873–876. doi: 10.1038/ismej.2009.21. [DOI] [PubMed] [Google Scholar]
- Michoud G. Jebbar M. Sci. Rep. 2016;6:27289. doi: 10.1038/srep27289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jun X. Lupeng L. Minjuan X. Oger P. Fengping W. Jebbar M. Xiang X. J. Bacteriol. 2011;193:4297–4298. doi: 10.1128/JB.05345-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Y. Cao C. He X. Yang C. Tian L. Zhu R. Pan Y. Int. J. Nanomed. 2015;10:2619–2634. doi: 10.2147/IJN.S80025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T. Cao C. Tang X. Cai Y. Yang C. Pan Y. Nanotechnology. 2017;28:045704. doi: 10.1088/1361-6528/28/4/045704. [DOI] [PubMed] [Google Scholar]
- Jiang B. Yan L. Zhang J. Zhou M. Shi G. Tian X. Fan K. Hao C. Yan X. ACS Appl. Mater. Interfaces. 2019;11:9747–9755. doi: 10.1021/acsami.8b20942. [DOI] [PubMed] [Google Scholar]
- Yang C. Cao C. Cai Y. Xu H. Zhang T. Pan Y. J. Nanopart. Res. 2017;19:101. doi: 10.1007/s11051-017-3805-y. [DOI] [Google Scholar]
- Zhang T. Pan Y. Earth, Planets Space. 2018;70:206. doi: 10.1186/s40623-018-0978-2. [DOI] [Google Scholar]
- Cao C. Tian L. Liu Q. Liu W. Chen G. Pan Y. J. Geophys. Res. 2010;115:B07103. [Google Scholar]
- Wohlfarth E. P. J. Appl. Phys. 1958;29:595–596. doi: 10.1063/1.1723232. [DOI] [Google Scholar]
- Kim S. W. Kim Y. H. Lee J. Biochem. Biophys. Res. Commun. 2001;289:125–129. doi: 10.1006/bbrc.2001.5931. [DOI] [PubMed] [Google Scholar]
- Yang R. Chen L. Zhang T. Yang S. Leng X. Zhao G. Chem. Commun. 2014;50:481–483. doi: 10.1039/C3CC47847E. [DOI] [PubMed] [Google Scholar]
- Myers J. K. Pace C. N. Biophys. J. 1996;71:2033–2039. doi: 10.1016/S0006-3495(96)79401-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogt G. Woell S. Argos P. J. Mol. Biol. 1997;269:631–643. doi: 10.1006/jmbi.1997.1042. [DOI] [PubMed] [Google Scholar]
- Bleicher L. Prates E. T. Gomes T. C. Silveira R. L. Nascimento A. S. Rojas A. L. Golubev A. Martínez L. Skaf M. S. Polikarpov I. J. Phys. Chem. B. 2011;115:7940–7949. doi: 10.1021/jp200330z. [DOI] [PubMed] [Google Scholar]
- Strop P. Mayo S. L. Biochemistry. 2000;39:1251–1255. doi: 10.1021/bi992257j. [DOI] [PubMed] [Google Scholar]
- Gromiha M. M. Oobatake M. Kono H. Uedaira H. Sarai A. J. Protein Chem. 1999;18:565–578. doi: 10.1023/A:1020603401001. [DOI] [PubMed] [Google Scholar]
- Gromiha M. M. Oobatake M. Sarai A. Biophys. Chem. 1999;82:51–67. doi: 10.1016/S0301-4622(99)00103-9. [DOI] [PubMed] [Google Scholar]
- Tompa D. R. Gromiha M. M. Saraboji K. J. Mol. Graphics Modell. 2016;64:85–93. doi: 10.1016/j.jmgm.2016.01.001. [DOI] [PubMed] [Google Scholar]
- Yip K. Stillman T. Britton K. Artymiuk P. Baker P. Sedelnikova S. Engel P. Pasquo A. Chiaraluce R. Consalvi V. Structure. 1995;3:1147–1158. doi: 10.1016/S0969-2126(01)00251-9. [DOI] [PubMed] [Google Scholar]
- Haney P. J. Badger J. H. Buldak G. L. Reich C. I. Woese C. R. Olsen G. J. Proc. Natl. Acad. Sci. U. S. A. 1999;96:3578–3583. doi: 10.1073/pnas.96.7.3578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald I. K. Thornton J. M. J. Mol. Biol. 1994;238:777–793. doi: 10.1006/jmbi.1994.1334. [DOI] [PubMed] [Google Scholar]
- Argos P. Rossmann M. G. Grau U. M. Zuber H. Frank G. Tratschin J. D. Biochemistry. 1979;18:5698–5703. doi: 10.1021/bi00592a028. [DOI] [PubMed] [Google Scholar]
- Bonomi F. Kurtz Jr D. M. Cui X. JBIC, J. Biol. Inorg. Chem. 1996;1:67–72. doi: 10.1007/s007750050024. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.