Abstract
Aims
To construct a polygenic risk score (PRS) for coronary artery disease (CAD) and comprehensively evaluate its potential in clinical utility for primary prevention in Chinese populations.
Methods and results
Using meta-analytic approach and large genome-wide association results for CAD and CAD-related traits in East Asians, a PRS comprising 540 genetic variants was developed in a training set of 2800 patients with CAD and 2055 controls, and was further assessed for risk stratification for CAD integrating with the guideline-recommended clinical risk score in large prospective cohorts comprising 41 271 individuals. During a mean follow-up of 13.0 years, 1303 incident CAD cases were identified. Individuals with high PRS (the highest 20%) had about three-fold higher risk of CAD than the lowest 20% (hazard ratio 2.91, 95% confidence interval 2.43–3.49), with the lifetime risk of 15.9 and 5.8%, respectively. The addition of PRS to the clinical risk score yielded a modest yet significant improvement in C-statistic (1%) and net reclassification improvement (3.5%). We observed significant gradients in both 10-year and lifetime risk of CAD according to the PRS within each clinical risk strata. Particularly, when integrating high PRS, intermediate clinical risk individuals with uncertain clinical decision for intervention would reach the risk levels (10-year of 4.6 vs. 4.8%, lifetime of 17.9 vs. 16.6%) of high clinical risk individuals with intermediate (20–80%) PRS.
Conclusion
The PRS could stratify individuals into different trajectories of CAD risk, and further refine risk stratification for CAD within each clinical risk strata, demonstrating a great potential to identify high-risk individuals for targeted intervention in clinical utility.
Keywords: Coronary artery disease, Polygenic risk score, Clinical risk score
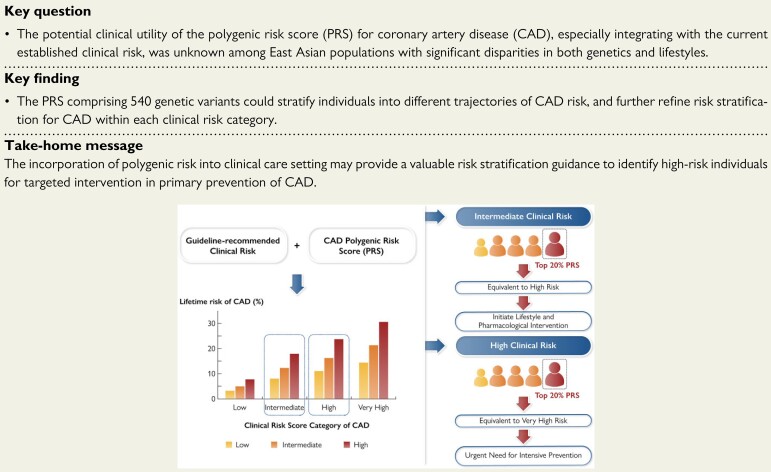
Structured Graphical Abstract
Structured Graphical Abstract.
The polygenic risk has a great potential to refine CAD risk stratification within each guideline-recommended clinical risk category and inform clinical decision making for primary prevention. Among individuals at intermediate clinical risk whose guideline-based recommendations are unclear, those with high polygenic risk should be recommended to initiate lifestyle and pharmacological intervention. Individuals with both high polygenic risk and high clinical risk urgently need intensive prevention. Combination of polygenic risk and clinical risk could promote precision prevention of CAD and reduce the disease burden, particularly considering inadequate primary prevention or statins and antihypertensive treatment in China.
See the editorial comment for this article ‘Polygenic risk score: a tool ready for clinical use?’, by M. Kavousi and H. Schunkert, https://doi.org/10.1093/eurheartj/ehab923.
Introduction
Cardiovascular disease (CVD), which is driven by both genetic susceptibility and environmental risk factors, is the leading cause of death and disease burden in China and worldwide.1 There has been a tremendous increase in the incidence of coronary artery disease (CAD) in China, due to ageing demography, unhealthy lifestyles, and environmental changes after several decades of rapid economic development.2 Cardiovascular risk assessment is recommended to identify those at high risk for lifestyle and pharmacological intervention by the current guidelines on the primary prevention of CVD issued by ESC and ACC/AHA.3–5 We have developed the Prediction for Atherosclerotic cardiovascular disease Risk in China (China-PAR) risk prediction equations, which were recommended by the ACC/AHA Guidelines to facilitate the primary prevention and management of CVD in clinical practice for populations of Chinese ethnicity.3,6
Genetic factors have also been recognized to make a substantial contribution to CVD risk. In the last decade, genome-wide association studies (GWAS) have successfully identified hundreds of genetic loci, which are robustly associated with CAD or CAD-related traits, such as blood lipid levels, blood pressure (BP), Type 2 diabetes (T2D), and body mass index (BMI).7 Genetic variants are attractive biomarkers because they are quantifiable at the time of birth, long time before the onset of clinical risk factors. Recently, polygenic risk scores (PRSs) for CAD by combining multiple risk alleles have been developed exclusively in European-descent populations and have shown promise in the prediction of CAD and risk stratification.8–15 However, the genetic prediction power was substantially lower in non-European populations due to the ancestry-specific differences in variant frequencies, effect sizes, and linkage disequilibrium patterns.16–19 Accordingly, it is of utmost importance to generate ancestry-specific PRS by performing the large-scale GWAS on non-European ancestries from which reference effect sizes are taken for PRS calculation. More importantly, the PRSs of CAD derived from European ancestry demonstrated modest improvements of predictive accuracy over existing clinical risk scores, such as pooled cohort equations (PCEs) or QRISK,20,21 while the clinical utility of PRS in risk prediction remains unclear.5 Therefore, it is imperative to evaluate the performance of PRS in a prospective setting among non-Europeans and the interplay of PRS and the clinical risk score in impacting the risk of CAD.
Here, we constructed a PRS for CAD by incorporating the large genome-wide association results for CAD and CAD-related traits in East Asians. Then we applied the PRS in large population-based prospective cohorts to assess how the polygenic risk affected lifetime trajectories of CAD risk and whether the polygenic score could refine risk stratification for CAD beyond the clinical risk prediction.
Materials and methods
Study design and population
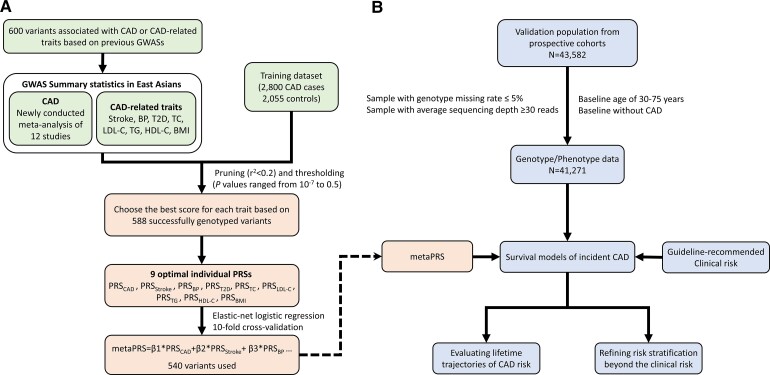
The study design is shown in Figure 1. A training set with 2800 cases of CAD and 2055 controls (see Supplementary material online, Table S1) was used to test the PRS performance. The CAD cases were enrolled from Fuwai Hospital, National Center for Cardiovascular Diseases, China. Diagnoses of cases with myocardial infarction (MI) followed strict diagnostic rules based on signs, symptoms, electrocardiograms, and the activity of cardiac enzymes.22 Individuals having >70% stenosis in one or more major epicardial vessel, or >50% stenosis for the left main coronary artery, were also diagnosed as CAD. The controls were randomly selected from individuals in the China-PAR project.
Figure 1.
Flow chart of the study. (A) Derivation of metaPRS for CAD in training dataset. (B) Validation of metaPRS in prospective cohorts. PRS, polygenic risk score; CAD, coronary artery disease; BP, blood pressure; BMI, body mass index; T2D, type 2 diabetes; TC, total cholesterol; LDL-C, LDL cholesterol; HDL-C, HDL cholesterol; TG, triglycerides.
The validation cohorts were derived from three cohorts in the China-PAR project, including the International Collaborative Study of Cardiovascular Disease in Asia (InterASIA), the China Multi-Center Collaborative Study of Cardiovascular Epidemiology (ChinaMUCA-1998), and Community Intervention of Metabolic Syndrome in China and Chinese Family Health Study (CIMIC). Details of the project’s design have been described elsewhere.6 Briefly, the ChinaMUCA-1998, InterASIA, and CIMIC were established in 1998, 2000–01, and 2007–08, respectively. According to a uniform protocol, both InterASIA and ChinaMUCA-1998 cohorts were first followed up during 2007–08, and all three cohorts were further followed up in 2012–15 and 2018–20. For CIMIC, we utilized a subset of 23 805 individuals based on the stratified sampling method according to survey sites. For this study, blood samples and the main covariate data were available for 43 582 participants, independent of individuals in the training set. We further excluded 561 individuals with high genotype missing rate (>5.0%) or low average sequencing depth (<30 reads), 1352 individuals aged <30 years or more than 75 years at baseline, and 398 participants with CAD at baseline. The final sample for analysis comprised 41 271 participants.
All studies were approved by the Institutional Review Board at Fuwai Hospital (Beijing, China). Written informed consent was obtained from each participant before data collection.
Data collection and outcomes
Information on baseline status and vital information during follow-up visits were collected by trained healthcare staff under strict quality control. A standard questionnaire was used to provide personal information, lifestyle information, disease conditions, and CAD family history. Participants also received a physical examination (weight, height, BP, etc.) and provided fasting blood samples to measure blood lipid and glucose levels.
To obtain disease outcomes and death information during follow-up duration, study participants or their proxies were identified and interviewed. Hospital records or death certificates were also collected. Two endpoint assessment committee members who were unaware of baseline information verified the events independently, and discrepancies were discussed to consensus with an additional committee member. Incident CAD was defined as the first occurrence of unstable angina, non-fatal acute MI, or CAD death. Unstable angina was identified as angina pectoris that changes or worsens. Acute MI was defined as changed biochemical markers of myocardial necrosis accompanied by any one of the following four characteristics: ischaemic symptoms, pathological Q waves, ST-segment elevation or depression, or coronary intervention. Fatal events resulting from MI or other coronary deaths were defined as CAD death. Person-years of follow-up for each participant were calculated as the interval between the date of the baseline examination and the date of occurrence of CAD, the date of death, or the last follow-up visit, whichever occurred first.
Variant selection and genotyping
We developed the PRS using the genetic variants that showed genome-wide significant associations (P < 5 × 10−8) with CAD and CAD-related traits based on the previous GWASs in East Asian ancestry (see Supplementary material online, Table S2). For CAD and stroke, we also included the variants identified by European populations. We finally selected 600 genetic variants associated with CAD or CAD-related traits, including stroke, BP, lipid, T2D, and obesity. The information of all variants was available in Supplementary material online, Table S3.
Individuals in the training set were genotyped with the Infinium Multi-Ethnic Genotyping Arrays covering ∼2 million markers. Subsequently, imputation to ∼47 million makers was carried out using the 1000 Genomes Phase 3 haplotype resource panel. In the validation set, we genotyped samples using multiplex PCR targeted amplicon sequencing technology. We designed multiplexed primers targeting the 600 genetic variants and amplified the target regions for high-throughput sequencing with Illumina Hiseq X Ten sequencer. After excluding 12 variants with a genotype call rate of <95% or being not available in the training dataset, 588 variants or their proxies remained for subsequent analyses, with 99.9% call rate and 982× median sequencing depth (see Supplementary material online, Figure S1). We performed strict quality control to assess genotyping reproducibility of genetic data and compared the genotyping consistency with Fludigm platform. A total of 1648 duplicate samples were genotyped, and the concordance rate was determined to be >99.4%. The principal component analysis showed minimal evidence for population stratification (see Supplementary material online, Figure S2).
Genome-wide association studies summary statistics and generation of trait-specific polygenic risk scores
Nine trait-specific PRSs (CAD, stroke, BP, T2D, TC, LDL-C, TG, HDL-C, and BMI) were separately constructed by summing the number of corresponding risk alleles (0, 1, or 2) for each individual, weighted by the effect size of variants on the corresponding trait (see Supplementary material online, Table S3). For CAD, to estimate the precise effect sizes of the selected variants in East Asian ancestry, we conducted a large GWAS meta-analysis in the East Asian population with a total sample size of 267 465 (51 531 CAD cases and 215 934 controls) from 12 cohorts, including BBJ, CAS, BAS.23,24 A detailed list of studies for CAD GWAS meta-analysis is available in Supplementary material online, Table S4. For CAD-related traits, the effect sizes were directly obtained from the large-scale GWASs for each trait conducted among East Asian populations, and those containing Chinese samples were selected in priority if more than one GWAS were reported. If a given variant was associated with an identical trait at GWAS significance in more than one GWAS, we selected the effect size with a minimal P-value. It was worth noting that each variant for BP PRS was weighted by the average effect size (β-coefficient) of systolic and diastolic BP.
To construct an optimized PRS for each trait, we generated a series of scores containing independent variants (r 2 < 0.2) at 12 different significance thresholds (P = 0.5, 0.4, 0.3, 0.2, 0.1, 0.05, 0.01, 10−3, 10−4, 10−5, 10−6, 10−7) based on trait-specific summary statistics from the large-scale GWAS in East Asian ancestry. The information on the variants and their weights used for trait-specific PRSs construction was provided in Supplementary material online, Table S3. The score with the largest magnitude of OR per PRS standard derivation (SD) for CAD in the training set was defined as the optimal PRS for each trait (see Supplementary material online, Figure S3). Each optimal PRS was standardized by calculating the z-score (zero mean, unit standard deviation).
MetaPRS calculation
A combined PRS (metaPRS) was then generated by integrating the nine optimal trait-specific PRSs. We conducted an elastic-net logistic regression with 10-fold cross-validation using the R package ‘glmnet’, a method having been used to construct a PRS for stroke with consideration of the correlation between distinct PRSs,25 to assess the association between the nine optimal PRSs and CAD in the training set, adjusting for age and sex. A series of models with different penalties were evaluated, and the model with the best performance, defined as the maximum cross-validated area under the receiver operator curve, was selected as the final model. Finally, the metaPRS for CAD was constructed by summing the standardized optimal trait-specific PRSs weighted by adjusted estimates β 1,…, β 9 derived from the final elastic-net model. The metaPRS can be calculated via a weighted sum by using variant-level genotype,
where m is the total number of variants, σ 1,…, σ 9 are the empirical standard deviations of each of the nine PRSs in the training set, α j1, …, α j9 are the variant effect sizes for the jth variant in each of the PRSs, respectively, and x ij is the genotype for the ith individual’s jth variant. A variant’s effect size α jk was considered to be zero for the kth score if the variant was not included in that score. All these procedures resulted in 540 variants for inclusion in the metaPRS. The weights of all variants for metaPRS were provided in Supplementary material online, Table S3.
Statistical analysis
Characteristics of study participants were described as mean (SD) for continuous variables and frequency (percentage) for categorical variables as appropriate. The participants in the validation cohorts were categorized into low (bottom quintile), intermediate (the second to the fourth quintile), and high (top quintile) polygenic risk categories according to quintiles of the metaPRS. The China-PAR equations for predicting 10-year atherosclerotic CVD (ASCVD) risk were developed from gender-specific Cox proportional hazards models, with variables in the equations including age, treated or untreated systolic BP, TC, HDL-C, current smoking, diabetes, waist circumference, geographic region, urbanization, and family history of ASCVD, as well as available interaction terms for age with risk factors that met predefined statistical criteria.6,26 The Chinese guideline on the assessment and management of cardiovascular risk recommended a scale of risk stratification for CVD prevention by classifying participants into low (<5%), intermediate (5–9.9%), and high (≥10%) clinical risk groups. For individuals with ASCVD risk ≥10%, drug therapy should be recommended in addition to lifestyle changes.27,28 The China-PAR model for ASCVD was recalibrated by estimating the baseline survival function and fitting the predicted log-hazard ratios (HRs) as covariates to predict CAD risk, as performed previously by Elliott et al.21 C-statistic and net reclassification improvement (NRI) for survival data were used to estimate the improvement in discrimination and reclassification after adding the metaPRS to the recalibrated China-PAR model.29,30 A risk threshold of 4.5% for 10-year CAD risk (equivalent to the ASCVD risk of 10%) was used to calculate NRI. We also obtained the 10-year and lifetime CAD risk in clinical risk categories (low: <2.5%, intermediate: 2.5–4.4%, high: 4.5–5.9%, and very high: ≥6%) and PRS categories using the recalibrated models, which were standardized to the mean of the predictor variables within each population.14 Fine and Gray’s proportional hazards model which accounted for competing risk of non-CAD deaths was used to evaluate the lifetime risk (up to 80 years of age) of incident CAD after adjustment of sex and the first four principal components with age as the time scale.31 A two-sided P-value of <0.05 is considered statistical significance. Statistical analysis was performed in R software, version 3.5.0 (R Foundation for Statistical Computing, Vienna, Austria) or SAS statistical package, version 9.4 (SAS Institute Inc., Cary, NC, USA).
Results
Study population
The mean age of disease onset was 51.59 years (SD, 7.36 years) for 2800 CAD cases in the training set while the age at entry into the study was 54.77 years (SD, 7.53 years) for 2055 controls (see Supplementary material online, Table S1). For 41 271 individuals in the validation cohorts, the mean age at baseline was 52.3 years (SD, 10.6 years), and 42.5% of them were men (Table 1). Men had a higher proportion of current smokers and higher clinical risk scores than women. During a total of 534 701 person-years (mean follow-up duration, 13.0 years), 1303 incident CAD cases occurred.
Table 1.
Baseline characteristics of the prospective cohorts
| Items | Total (n = 41 271) | Men (n = 17 560) | Women (n = 23 711) |
|---|---|---|---|
| Age at baseline, years | 52.3 (10.6) | 52.8 (10.8) | 51.9 (10.5) |
| Current smokers | 10 026 (24.4) | 9380 (53.5) | 646 (2.7) |
| Family history of CAD | 2255 (5.5) | 965 (5.5) | 1290 (5.4) |
| Body mass index, kg/m2 | 23.8 (3.6) | 23.4 (3.4) | 24.1 (3.8) |
| Systolic blood pressure, mmHg | 128.4 (21.9) | 129.1 (20.9) | 127.9 (22.6) |
| Diastolic blood pressure, mmHg | 79.4 (11.9) | 80.6 (12) | 78.5 (11.8) |
| Total cholesterol, mg/dL | 180.5 (36.3) | 177.9 (36) | 182.4 (36.5) |
| Blood glucose, mg/dL | 94.2 (27.2) | 93.2 (25.4) | 94.9 (28.4) |
| Hypertension | 14 038 (34) | 6187 (35.2) | 7851 (33.1) |
| Diabetes | 2705 (6.8) | 1012 (6) | 1693 (7.4) |
| Dyslipidaemia | 13 399 (33) | 6063 (35.2) | 7336 (31.5) |
| China-PAR scorea | |||
| Low (<5%) | 24 892 (62.2) | 9052 (53.2) | 15 840 (68.9) |
| Intermediate (5–9.9%) | 8342 (20.9) | 4057 (23.9) | 4285 (18.6) |
| High (≥10%) | 6768 (16.9) | 3898 (22.9) | 2870 (12.5) |
| Incident CAD events | 1303 (3.2) | 635 (3.6) | 668 (2.8) |
| Person-years of follow-up | 13.0 (4.8) | 12.9 (5.1) | 13.0 (4.6) |
Ten-year ASCVD risk score using the Prediction for Atherosclerotic cardiovascular disease Risk in China equations; values are presented as mean (standard deviations) or n (%); CAD, coronary artery disease.
Construction of polygenic risk score
To accurately assess the associations of 588 successfully genotyped variants with CAD, we carried out the large GWAS meta-analyses for CAD in the East Asian population comprising a total of 267 465 individuals from 12 cohorts (see Supplementary material online, Table S4). Among the 209 CAD variants, 140 showed nominal significance (P < 0.05) with CAD, whereas 91 variants showed significant associations even at a Bonferroni-corrected threshold (P-values ranged from 2.0 × 10−4 to 3.76 × 10−112 <0.05/209) (see Supplementary material online, Table S3 and Figure S4). As expected, the variants associated with CAD-related traits also displayed associations with CAD at different levels of significance.
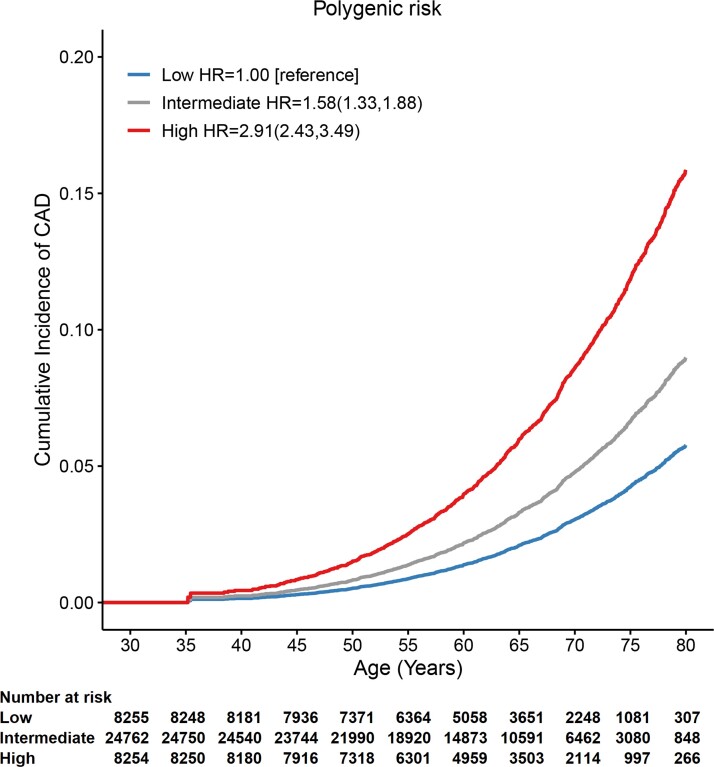
We compared the effect sizes of CAD with those observed in the UK Biobank, and 180 out of 209 (86%) CAD variants displayed associations with CAD in a consistent direction and the strengths of effect on CAD were moderately correlated (r 2 = 0.266) (see Supplementary material online, Figure S5A). We generated a set of CAD PRSs using effect size derived from our CAD GWAS at different significance thresholds and observed the strongest association of CAD PRS comprising 311 variants in the training set (see Supplementary material online, Figure S3 and Table S3). The ORs of CAD PRS would decay markedly when using effect sizes from the European population (see Supplementary material online, Figure S5B). Similarly, we obtained another eight optimal PRSs for specific traits using effect sizes from the East Asian population, which were correlated with each other to different degrees (see Supplementary material online, Figure S6). Then the metaPRS was generated by integrating the nine individual PRSs using elastic-net logistic regression with 10-fold cross-validation, with the estimate for each PRS being significantly adjusted (see Supplementary material online, Figure S7). As expected, the metaPRS had the greatest association with CAD risk than any other individual PRS in the validation cohorts (see Supplementary material online, Figure S8), with the HR of 1.44 [95% confidence interval (CI) 1.36–1.52] per SD increment in metaPRS (P = 2.84 × 10−39) and 1.41 (95% CI 1.33–1.49) per SD increment in CAD-only PRS. We also observed a more marked gradient of CAD risk across quintiles of metaPRS than CAD-only PRS. The metaPRS had a greater HR of 2.91 (95% CI 2.43–3.49) in the top quintile vs. the bottom quintile than the CAD-only PRS (HR: 2.59, 95% CI 2.17–3.10) (Figure 2 and Supplementary material online, Figure S9).
Figure 2.
Cumulative incidence curves for incident coronary artery disease across polygenic risk categories. Fine and Gray’s proportional hazards model accounting for competing risk was used to estimate the hazard ratios (95% confidence intervals) and the cumulative risk of coronary artery disease adjusted for sex and the first four principal components with age as the time scale. Polygenic risk categories: low (bottom quintile), intermediate (2nd–4th quintile), or high (top quintile) risk according to quintiles of the metaPRS. CAD, coronary artery disease; HR, hazard ratio; CI, confidence interval.
Polygenic risk and the lifetime trajectories of coronary artery disease risk
We then assessed how the metaPRS affected lifetime trajectories of CAD risk in 41 271 individuals. The cumulative risks by age of 80 for coronary events were 5.8% for individuals with low polygenic risk (bottom quintile of the metaPRS) and 15.9% among those with high polygenic risk (top quintile of the metaPRS), respectively (Figure 2). Similar results were observed in both sexes, with men having higher HR and cumulative risks (see Supplementary material online, Figure S10). These associations with the metaPRS were largely independent of the traditional risk factors, including CAD family history and the clinical risk score (see Supplementary material online, Table S5). The combination of CAD family history and PRS category would help further discriminate individuals. The lifetime risk of CAD was 5.6% among individuals with low polygenic risk and without a family history, whereas the presence of both could obtain the lifetime risk of CAD as high as 27.7%, conferring 5.66-fold increased risk of CAD (95% CI 3.98–8.04) (see Supplementary material online, Figure S11).
Predicting coronary artery disease risk by polygenic and clinical risk score
The recalibrated China-PAR model for CAD risk demonstrated good agreement between observed rate and expected rate (see Supplementary material online, Figure S12 and Table S6). We further evaluated the potential of PRS for CAD prediction beyond the recalibrated model. The addition of the metaPRS to a baseline model including age and sex increased the C-statistic from 0.705 to 0.728 (difference, 2.4%; P = 4.86 × 10−12). Adding the metaPRS to the recalibrated clinical risk model also significantly improved risk discrimination of incident CAD (C-statistic change, 1%; P = 7.72 × 10−7) (see Supplementary material online, Table S7). There was a significant reclassification improvement in a two-category risk assessment (<4.5%, ≥4.5%) (NRI 3.5%, 95% CI 1.2–6.0%; continuous NRI = 25.8%, 95% CI 18.5–32.5%) (Table 2).
Table 2.
Net reclassification improvement after adding metaPRS to clinical risk score
| Clinical risk score | MetaPRS + clinical risk score | ||
|---|---|---|---|
| <4.5% | ≥4.5% | Total | |
| CAD | |||
| <4.5% | 405 | 65 | 470 |
| ≥4.5% | 36 | 334 | 370 |
| Totals | 441 | 399 | 840 |
| Non-CAD | |||
| <4.5% | 28 154 | 870 | 29 024 |
| ≥4.5% | 866 | 2969 | 3835 |
| Totals | 29 020 | 3839 | 32 859 |
| Net reclassification improvement (NRI) | |||
| NRI for CAD (95% CI), % | 3.2 (0.9–5.8) | ||
| NRI for Non-CAD (95% CI), % | 0.3 (0.1–0.5) | ||
| NRI (95% CI), % | 3.5 (1.2–6.0) | ||
| Continuous NRI for CAD (95% CI), % | 15.7 (7.7–22.2) | ||
| Continuous NRI for Non-CAD (95% CI), % | 10.1 (9.1–11.1) | ||
| Continuous NRI, % | 25.8 (18.5–32.5) | ||
NRI, net reclassification improvement; CAD, coronary artery disease; CI, confidence interval; PRS, polygenic risk score.
The clinical risk score of CAD was obtained from the recalibrated 10-year China-PAR model. The risk of 4.5% for 10-year CAD risk is equivalent to the ASCVD risk of 10%.
We also performed sensitivity analysis to compare the performance of the PCE model. The recalibrated China-PAR model had better discrimination of CAD risk than the recalibrated PCE among Chinese population, regardless of adding metaPRS to the clinical risk models (see Supplementary material online, Table S7).
Polygenic and clinical risk stratification for coronary artery disease
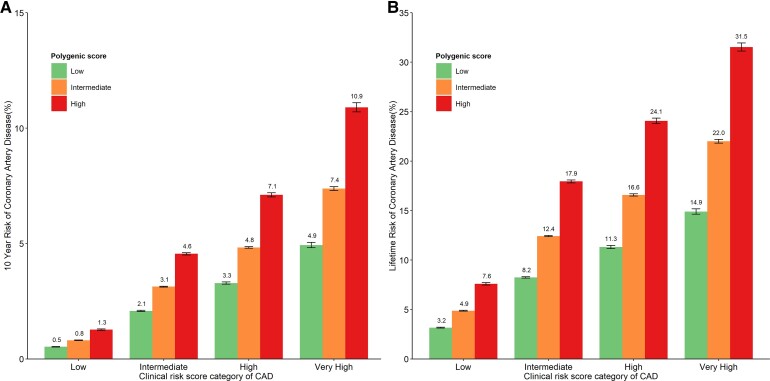
We further assessed the interplay of the metaPRS and the clinical risk in impacting the risk of CAD. We observed significant gradients in the 10-year and lifetime risk of incident CAD across PRS categories within each clinical risk strata (Figure 3). For example, among individuals with high clinical risk, the 10-year absolute CAD risk varied from 3.3% for those with the low genetic risk to 7.1% for those with the high genetic risk, and their corresponding lifetime absolute risk ranged from 11.3 to 24.1%. More interestingly, individuals at intermediate clinical risk with high genetic risk demonstrated the 10-year CAD risk of 4.6% over the threshold for high clinical risk of CAD (equating to the established treatment threshold of 10% ASCVD risk). The 10-year (4.6 vs. 4.8%) and lifetime risk (17.9 vs. 16.6%) of incident CAD for these 20% individuals at intermediate clinical risk also reached the risk levels of those at the high clinical risk group with intermediate (20–80%) genetic risk. We repeated the analysis and observed the similar patterns of the interplay between PCE score and metaPRS, although the absolute risk predicted by the recalibrated PCE seemed to be lower than the values predicted by the recalibrated China-PAR model (see Supplementary material online, Figures S13 and S14).
Figure 3.
Ten-year and lifetime risk of coronary artery disease according to clinical and polygenic risk categories. (A) Ten-year risk of coronary artery disease obtained from the recalibrated clinical risk and metaPRS model with follow-up time as the time scale. (B) Lifetime risk of coronary artery disease (till 80 years of age) obtained from the recalibrated clinical risk and metaPRS model accounting for competing risk with age as the time scale. Participants were stratified into low (<2.5%), intermediate (2.5–4.4%), high (4.5–5.9%), and very high (≥6%) 10-year risk of CAD categories, approximately equating to the atherosclerotic cardiovascular disease risk of <5, 5–9.9, 10–14.9, and ≥15%. According to the risk assessment guideline from China, the established treatment threshold for atherosclerotic cardiovascular disease is the 10-year risk of atherosclerotic cardiovascular disease of 10%. CAD, coronary artery disease; ASCVD, atherosclerotic cardiovascular disease; PRS, polygenic risk score.
We also observed marked variability in the 10-year and lifetime risk of incident CAD across PRS categories, regardless of the age and gender categories (see Supplementary material online, Figures S15 and S16). For example, among men aged 35–44 years and with a clinical risk score of 6% and above, 10-year absolute risk varied dramatically from 5.8 to 14.2%, which depended on the polygenic risk categories. The corresponding values of lifetime risk ranged from 15.8 to 35.0%. It is particularly noteworthy for individuals at intermediate clinical risk score (2.5–4.4%) due to their clinical uncertainty in primary prevention, the high genetic score made both the 10-year and lifetime risk of CAD be close to or exceed the values (20–80% of metaPRS) of high clinical risk score (4.5–6.0%) in all the sex and age-specific groups.
Discussion
We developed and evaluated a PRS for CAD incidence combined with the traditional clinical risk score in East Asian ancestry based on population-based prospective cohorts. We found that the polygenic score could enhance risk stratification for incident CAD beyond the clinical risk score, demonstrating substantial gradients in the 10-year and lifetime risk of CAD across the polygenic risk categories. In particular, the CAD risk for individuals at the intermediate clinical risk would reach the risk level of those at the high clinical risk when integrating high polygenic risk (Structured Graphical Abstract).
Polygenic scoring studies have primarily been conducted in European ancestry populations, but its predictive utility decreased largely in other ethnic groups.16,17 Due to the lack of comprehensively characterized and genotyped cohorts, few studies have evaluated the performance of PRS in a prospective setting among non-Europeans. The large-scale meta-analysis of GWAS of CAD we conducted allowed us precisely assess the effect sizes of the variants in East Asian population. The metaPRS we constructed by incorporating genetic information of CAD-related traits had the best performance compared with the trait-specific PRS.
We further compared the performance of our metaPRS with the reported genome-wide PRSs of CAD in our training dataset (see Supplementary material online, Table S8). As expected, our metaPRS comprising 540 genetic variants showed better performance than the two genome-wide PRSs (i.e. metaGRS and LDPred, 1.7M and 6.6M variants) derived from the European studies8,9 despite its limited number of variants, with ORs per SD-PRS of 1.83 (95% CI 1.70–1.96) for our metaPRS being higher than that for metaGRS (OR 1.69, 95% CI 1.57–1.81) and LDPred (OR: 1.52, 95% CI: 1.42, 1.63). We also constructed a set of genome-wide PRSs based on the summary statistics of our CAD GWAS and BBJ datasets by using the LDpred algorithm and pruning and thresholding strategies (see Supplementary material online, Table S9). The two best CAD PRSs (comprising 5.1M and 4.9M variants) obtained from our CAD meta-analysis and BBJ-only had ORs for per SD-PRS of 1.78 (95% CI 1.66–1.91) and 1.62 (95% CI 1.51–1.74), respectively. Of note, the trans-ancestry CAD PRS comprising 75 028 variants developed based on the trans-ancestry genome-wide meta-analysis (BBJ, C4D, and UKBB) had the largest OR of 1.97 (95% CI 1.83–2.12) in our training dataset. When using other two measurements, Nagelkerke’s pseudo-R 2 and C-statistic, our metaPRS also demonstrated comparable predictive performance to the genome-wide PRSs. Our metaPRS conferred an HR of 1.44 per SD increment of the metaPRS in the validation cohorts, which was larger than the scores derived by dozens to hundreds of variants,11,15,2–4 and was even comparable to the genome-wide polygenic scores derived from European population.9,14,20,21 Consistent with the genome-wide PRSs in European population, the metaPRS demonstrated a great potential to substantially stratify individual CAD risk trajectories.
The clinical utility of PRS in CAD risk reclassification was uncertain when combining the traditional clinical score. Several studies examined whether the genome-wide CAD PRSs improved risk prediction beyond the PCE in European ancestry populations.14,20,21 Adding the PRS to the PCE yielded an increment of about 0.02 in the C-statistic and a NRI of 4.0% at a risk threshold of 7.5% in the UK biobank. We also demonstrated that the metaPRS provided a statistically significant yet modest discrimination over China-PAR score. Of note, the NRI would be 0.165 in the UK biobank when assessed the reclassification in a four-category PCE risk (<5, 5–7.5, 7.5–20, and >20%).14 Accordingly, a striking gradient of longitudinal risk across CAD PRS categories was observed within each of the four PCE risk strata. Our metaPRS consistently displayed the ability to substantially stratify CAD risk trajectories within each China-PAR risk category. In particular, among individuals with clinical uncertainty, the absolute risk for those at the highest 20% of genetic risk would reach the level of high clinical risk group. Moreover, recent evidence from three US health care systems suggested that individuals at high polygenic risk for CAD were not well identified by current clinical risk estimators and the high CAD PRS were considered a risk-enhancing factor to up-classify risk in a guideline framework.35 Risk-enhancing factors are those independently associated with CAD and conferred a nearly two-fold risk of disease.36,37 In our analysis, we observed a comparable level of risk (HR 1.92, 95% CI 1.63–2.27) among those in the top 20% of the metaPRS compared with the remainder of the population, supporting that the metaPRS may serve as a risk-enhancing factor for CAD.
Age is the most important risk driver in the clinical risk equation, therefore resulting in overestimation or underestimation of risk, whereas genetic risk is age independent and can be determined early in life when few individuals express risk factors that exceed treatment thresholds established for older adults. Our findings highlight the concept that PRS may provide complementary information within guideline-supported frameworks to better stratify different trajectories of CAD risk and inform clinical decision-making for primary prevention, although not substantially influencing C-statistic.13,38 For individuals at intermediate clinical risk whose guideline-based recommendations are unclear, the addition of a polygenic risk estimate could clarify their risk and favour to decide against (low polygenic risk) or for (high polygenic risk) taking lifestyle and medical intervention action. For the individuals with high clinical risk, the incorporation of high genetic risk would facilitate capturing individuals who need intensive lifestyle changes and drug treatments, particularly when considering that treatment with statins and antihypertensive drugs in them was still inadequate.39 It has been demonstrated that high genetic risk of CAD may be mitigated by statin use and healthy lifestyle in both primary and secondary prevention and that individuals at high genetic risk were found to derive the greatest benefit from the therapeutic intervention.11,0–3 The randomized controlled trials focusing on individuals at intermediate or high clinical risk, especially for Chinese, are required to confirm the clinically meaningful benefit and the cost-effectiveness of polygenic risk stratification for CAD.
The major strengths of the current study include the large sample size of participants with up to 20 years’ follow-up, and rich baseline phenotyping according to a well-defined and standardized protocol, which enabled us to comprehensively evaluate the combination of polygenic risk and traditional clinical risk. Furthermore, to derive a PRS for CAD, we used the current large GWAS of CAD and CAD-related traits in East Asians. However, some limitations should also be noted. First, our metaPRS did not include all the variants that underlay CAD risk, which might result in underestimation of the true effect. Future studies that construct PRS capturing the full spectrum of genomic variants will likely provide additional gains in prediction and risk stratification. Second, it has been demonstrated that the performance of PRS varied across different ancestries, even populations of similar ethnic but different countries.44 Further external evaluation of our metaPRS in other populations of East Asian ancestry other than Chinese is warranted.
Conclusion
We developed a CAD polygenic score with good performance in risk stratification beyond the clinical risk score. The incorporation of polygenic risk into clinical care setting may provide a valuable risk stratification guidance to identify individuals who should be initiated or given intensive lifestyle changes and drug treatments.
Supplementary Material
Acknowledgements
The authors are grateful to Biobank Japan Project (BBJ) and Asian Genetic Epidemiology Network (AGEN) consortium for access to summary statistics data. The authors also thank Wenhao Chen and Haobin Wang (Boke Biotech Co., Wuxi, Jiangsu, China) for helping with the genotyping design and analysis.
Contributor Information
Xiangfeng Lu, Key Laboratory of Cardiovascular Epidemiology & Department of Epidemiology, State Key Laboratory of Cardiovascular Disease, Fuwai Hospital, National Center for Cardiovascular Diseases, Chinese Academy of Medical Sciences and Peking Union Medical College, Beijing 100037, China.
Zhongying Liu, Key Laboratory of Cardiovascular Epidemiology & Department of Epidemiology, State Key Laboratory of Cardiovascular Disease, Fuwai Hospital, National Center for Cardiovascular Diseases, Chinese Academy of Medical Sciences and Peking Union Medical College, Beijing 100037, China.
Qingmei Cui, Key Laboratory of Cardiovascular Epidemiology & Department of Epidemiology, State Key Laboratory of Cardiovascular Disease, Fuwai Hospital, National Center for Cardiovascular Diseases, Chinese Academy of Medical Sciences and Peking Union Medical College, Beijing 100037, China.
Fangchao Liu, Key Laboratory of Cardiovascular Epidemiology & Department of Epidemiology, State Key Laboratory of Cardiovascular Disease, Fuwai Hospital, National Center for Cardiovascular Diseases, Chinese Academy of Medical Sciences and Peking Union Medical College, Beijing 100037, China.
Jianxin Li, Key Laboratory of Cardiovascular Epidemiology & Department of Epidemiology, State Key Laboratory of Cardiovascular Disease, Fuwai Hospital, National Center for Cardiovascular Diseases, Chinese Academy of Medical Sciences and Peking Union Medical College, Beijing 100037, China.
Xiaoge Niu, Key Laboratory of Cardiovascular Epidemiology & Department of Epidemiology, State Key Laboratory of Cardiovascular Disease, Fuwai Hospital, National Center for Cardiovascular Diseases, Chinese Academy of Medical Sciences and Peking Union Medical College, Beijing 100037, China.
Chong Shen, Department of Epidemiology and Biostatistics, School of Public Health, Nanjing Medical University, Nanjing 211166, China.
Dongsheng Hu, Department of Epidemiology and Health Statistics, College of Public Health, Zhengzhou University, Zhengzhou 450001, China; Department of Biostatistics and Epidemiology, School of Public Health, Shenzhen University Health Science Center, Shenzhen 518071, China.
Keyong Huang, Key Laboratory of Cardiovascular Epidemiology & Department of Epidemiology, State Key Laboratory of Cardiovascular Disease, Fuwai Hospital, National Center for Cardiovascular Diseases, Chinese Academy of Medical Sciences and Peking Union Medical College, Beijing 100037, China.
Jichun Chen, Key Laboratory of Cardiovascular Epidemiology & Department of Epidemiology, State Key Laboratory of Cardiovascular Disease, Fuwai Hospital, National Center for Cardiovascular Diseases, Chinese Academy of Medical Sciences and Peking Union Medical College, Beijing 100037, China.
Xiaolong Xing, Key Laboratory of Cardiovascular Epidemiology & Department of Epidemiology, State Key Laboratory of Cardiovascular Disease, Fuwai Hospital, National Center for Cardiovascular Diseases, Chinese Academy of Medical Sciences and Peking Union Medical College, Beijing 100037, China.
Yingxin Zhao, Cardio-Cerebrovascular Control and Research Center, Institute of Basic Medicine, Shandong Academy of Medical Sciences, Jinan 250062, China.
Fanghong Lu, Cardio-Cerebrovascular Control and Research Center, Institute of Basic Medicine, Shandong Academy of Medical Sciences, Jinan 250062, China.
Xiaoqing Liu, Division of Epidemiology, Guangdong Provincial People’s Hospital and Cardiovascular Institute, Guangzhou 510080, China.
Jie Cao, Key Laboratory of Cardiovascular Epidemiology & Department of Epidemiology, State Key Laboratory of Cardiovascular Disease, Fuwai Hospital, National Center for Cardiovascular Diseases, Chinese Academy of Medical Sciences and Peking Union Medical College, Beijing 100037, China.
Shufeng Chen, Key Laboratory of Cardiovascular Epidemiology & Department of Epidemiology, State Key Laboratory of Cardiovascular Disease, Fuwai Hospital, National Center for Cardiovascular Diseases, Chinese Academy of Medical Sciences and Peking Union Medical College, Beijing 100037, China.
Hongxia Ma, Department of Epidemiology and Biostatistics, School of Public Health, Nanjing Medical University, Nanjing 211166, China.
Ling Yu, Department of Cardiology, Fujian Provincial People’s Hospital, Fuzhou 350014, China.
Xianping Wu, Sichuan Center for Disease Control and Prevention, Chengdu 610041, China.
Xigui Wu, Key Laboratory of Cardiovascular Epidemiology & Department of Epidemiology, State Key Laboratory of Cardiovascular Disease, Fuwai Hospital, National Center for Cardiovascular Diseases, Chinese Academy of Medical Sciences and Peking Union Medical College, Beijing 100037, China.
Ying Li, Key Laboratory of Cardiovascular Epidemiology & Department of Epidemiology, State Key Laboratory of Cardiovascular Disease, Fuwai Hospital, National Center for Cardiovascular Diseases, Chinese Academy of Medical Sciences and Peking Union Medical College, Beijing 100037, China.
Huan Zhang, Center for Genetic Epidemiology and Genomics, School of Public Health and Jiangsu Key Laboratory of Preventive and Translational Medicine for Geriatric Diseases, Medical College of Soochow University, Suzhou 215123, China.
Xingbo Mo, Center for Genetic Epidemiology and Genomics, School of Public Health and Jiangsu Key Laboratory of Preventive and Translational Medicine for Geriatric Diseases, Medical College of Soochow University, Suzhou 215123, China.
Liancheng Zhao, Key Laboratory of Cardiovascular Epidemiology & Department of Epidemiology, State Key Laboratory of Cardiovascular Disease, Fuwai Hospital, National Center for Cardiovascular Diseases, Chinese Academy of Medical Sciences and Peking Union Medical College, Beijing 100037, China.
Jianfeng Huang, Key Laboratory of Cardiovascular Epidemiology & Department of Epidemiology, State Key Laboratory of Cardiovascular Disease, Fuwai Hospital, National Center for Cardiovascular Diseases, Chinese Academy of Medical Sciences and Peking Union Medical College, Beijing 100037, China.
Laiyuan Wang, Key Laboratory of Cardiovascular Epidemiology & Department of Epidemiology, State Key Laboratory of Cardiovascular Disease, Fuwai Hospital, National Center for Cardiovascular Diseases, Chinese Academy of Medical Sciences and Peking Union Medical College, Beijing 100037, China.
Wanqing Wen, Department of Medicine, Vanderbilt University Medical Center, Nashville, TN, USA.
Xiao-Ou Shu, Department of Medicine, Vanderbilt University Medical Center, Nashville, TN, USA.
Fumihiko Takeuchi, Department of Gene Diagnostics and Therapeutics, Research Institute, National Center for Global Health and Medicine, Tokyo, Japan.
Woon-Puay Koh, Healthy Longevity Translational Research Programme, Yong Loo Lin School of Medicine, National University of Singapore, Singapore, Singapore.
E Shyong Tai, Saw Swee Hock School of Public Health, National University of Singapore and National University Health System, Singapore; Department of Medicine, Yong Loo Lin School of Medicine, National University Health System, Singapore.
Ching-Yu Cheng, Singapore Eye Research Institute, Singapore National Eye Centre, Singapore.
Tien yin Wong, Singapore Eye Research Institute, Singapore National Eye Centre, Singapore; Ophthalmology and Visual Sciences Academic Clinical Program, Duke-NUS, Medical School, Singapore.
Xuling Chang, Department of Paediatrics, Yong Loo Lin School of Medicine, National University of Singapore, Singapore; Khoo Teck Puat—National University Children’s Medical Institute, National University Health System, Singapore.
Mark Yan-Yee Chan, Department of Medicine, Yong Loo Lin School of Medicine, National University of Singapore, Singapore; National University Heart Centre, National University Health System, Singapore.
Wei Gao, Department of Cardiology, Institute of Vascular Medicine, Peking University Third Hospital, Beijing, China.
Hong Zheng, Department of Epidemiology and Biostatistics, Key Laboratory of Cancer Prevention and Therapy, Tianjin Key Laboratory of Breast Cancer Prevention and Therapy, Ministry of Education, National Clinical Research Center for Cancer, Tianjin Medical University Cancer Institute and Hospital, Tianjin, China.
Kexin Chen, Department of Epidemiology and Biostatistics, Key Laboratory of Cancer Prevention and Therapy, Tianjin Key Laboratory of Breast Cancer Prevention and Therapy, Ministry of Education, National Clinical Research Center for Cancer, Tianjin Medical University Cancer Institute and Hospital, Tianjin, China.
Jing Chen, Department of Medicine, Tulane University School of Medicine, and Tulane University Translational Science Institute, New Orleans, LA, USA.
Jiang He, Department of Epidemiology, Tulane University School of Public Health and Tropical Medicine, and Tulane University Translational Science Institute, New Orleans, LA, USA.
Clara Sze-man Tang, Department of Surgery, Li Ka Shing Faculty of Medicine, The University of Hong Kong, Hong Kong, China.
Karen Siu Ling Lam, Department of Medicine, Li Ka Shing Faculty of Medicine, The University of Hong Kong, Hong Kong, China.
Hung-fat Tse, Department of Medicine, Li Ka Shing Faculty of Medicine, The University of Hong Kong, Hong Kong, China.
Chloe Yu Yan Cheung, Department of Medicine, Li Ka Shing Faculty of Medicine, The University of Hong Kong, Hong Kong, China.
Atsushi Takahashi, Laboratory for Statistical and Translational Genetics, RIKEN Center for Integrative Medical Sciences, Yokohama, Japan; Department of Genomic Medicine, Research Institute, National Cerebral and Cardiovascular Center, Osaka, Japan.
Michiaki Kubo, Laboratory for Statistical and Translational Genetics, RIKEN Center for Integrative Medical Sciences, Yokohama, Japan.
Norihiro Kato, Department of Gene Diagnostics and Therapeutics, Research Institute, National Center for Global Health and Medicine, Tokyo, Japan.
Chikashi Terao, Laboratory for Statistical and Translational Genetics, RIKEN Center for Integrative Medical Sciences, Yokohama, Japan.
Yoichiro Kamatani, Laboratory for Statistical and Translational Genetics, RIKEN Center for Integrative Medical Sciences, Yokohama, Japan; Laboratory of Complex Trait Genomics, Graduate School of Frontier Sciences, The University of Tokyo, Tokyo, Japan.
Pak Chung Sham, Centre for PanorOmic Sciences, Li Ka Shing Faculty of Medicine, The University of Hong Kong, Hong Kong, China.
Chew-Kiat Heng, Department of Paediatrics, Yong Loo Lin School of Medicine, National University of Singapore, Singapore.
Zhibin Hu, Department of Epidemiology and Biostatistics, School of Public Health, Nanjing Medical University, Nanjing 211166, China.
Y Eugene Chen, Department of Internal Medicine, Division of Cardiovascular Medicine, University of Michigan, Ann Arbor, MI, USA.
Tangchun Wu, MOE Key Lab of Environment and Health, School of Public Health, Tongji Medical College, Huazhong University of Science & Technology, Wuhan 430030, China.
Hongbing Shen, Department of Epidemiology and Biostatistics, School of Public Health, Nanjing Medical University, Nanjing 211166, China.
Cristen J Willer, Department of Internal Medicine, Division of Cardiovascular Medicine, University of Michigan, Ann Arbor, MI, USA; Department of Human Genetics, University of Michigan, Ann Arbor, MI, USA.
Dongfeng Gu, Key Laboratory of Cardiovascular Epidemiology & Department of Epidemiology, State Key Laboratory of Cardiovascular Disease, Fuwai Hospital, National Center for Cardiovascular Diseases, Chinese Academy of Medical Sciences and Peking Union Medical College, Beijing 100037, China.
Supplementary material
Supplementary material is available at European Heart Journal online.
Funding
This work was supported by Chinese Academy of Medical Sciences (CAMS) Innovation Fund for Medical Sciences (2021-I2M-1-010, 2019-I2M-2-003, 2017-I2M-1-004), National Natural Science Foundation of China (82030102, 12126602, 91857118), and National Key Research and Development Program of China (2018YFE0115300, 2017YFC0211700).
Conflict of interest
The authors declare that there is no conflict of interest.
Data availability
Data are available upon reasonable request to the corresponding authors.
References
- 1. Wang H, Naghavi M, Allen C, Barber RM, Bhutta ZA, Carter A, et al. Global, regional, and national life expectancy, all-cause mortality, and cause-specific mortality for 249 causes of death, 1980–2015: a systematic analysis for the Global Burden of Disease Study 2015. Lancet 2016;388:1459–1544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. National Center for Cardiovascular Diseases . Report on cardiovascular diseases in China 2019 [Chinese] (Encyclopedia of China Publishing House, 2019).
- 3. Arnett DK, Blumenthal RS, Albert MA, Buroker AB, Goldberger ZD, Hahn EJ, et al. 2019 ACC/AHA Guideline on the Primary Prevention of Cardiovascular Disease: a report of the American College of Cardiology/American Heart Association Task Force on Clinical Practice Guidelines. Circulation 2019;140:e596–e646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Knuuti J, Wijns W, Saraste A, Capodanno D, Barbato E, Funck-Brentano C, et al. 2019 ESC Guidelines for the diagnosis and management of chronic coronary syndromes. Eur Heart J 2020;41:407–477. [DOI] [PubMed] [Google Scholar]
- 5. Visseren FLJ, Mach F, Smulders YM, Carballo D, Koskinas KC, Back M, et al. 2021 ESC Guidelines on cardiovascular disease prevention in clinical practice. Eur Heart J 2021;42:3227–3337. [DOI] [PubMed] [Google Scholar]
- 6. Yang X, Li J, Hu D, Chen J, Li Y, Huang J, et al. Predicting the 10-year risks of atherosclerotic cardiovascular disease in Chinese population: the China-PAR Project (Prediction for ASCVD Risk in China). Circulation 2016;134:1430–1440. [DOI] [PubMed] [Google Scholar]
- 7. Buniello A, MacArthur JAL, Cerezo M, Harris LW, Hayhurst J, Malangone C, et al. The NHGRI-EBI GWAS Catalog of published genome-wide association studies, targeted arrays and summary statistics 2019. Nucleic Acid Res 2019;47:D1005–D1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Khera AV, Chaffin M, Aragam KG, Haas ME, Roselli C, Choi SH, et al. Genome-wide polygenic scores for common diseases identify individuals with risk equivalent to monogenic mutations. Nat Genet 2018;50:1219–1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Inouye M, Abraham G, Nelson CP, Wood AM, Sweeting MJ, Dudbridge F, et al. Genomic risk prediction of coronary artery disease in 480,000 adults: implications for primary prevention. J Am Coll Cardiol 2018;72:1883–1893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Abraham G, Havulinna AS, Bhalala OG, Byars SG, De Livera AM, Yetukuri L, et al. Genomic prediction of coronary heart disease. Eur Heart J 2016;37:3267–3278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Khera AV, Emdin CA, Drake I, Natarajan P, Bick AG, Cook NR, et al. Genetic risk, adherence to a healthy lifestyle, and coronary disease. N Engl J Med 2016;375:2349–2358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Sun L, Pennells L, Kaptoge S, Nelson CP, Ritchie SC, Abraham G, et al. Polygenic risk scores in cardiovascular risk prediction: a cohort study and modelling analyses. PLoS Med 2021;18:e1003498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Aragam KG, Natarajan P. Polygenic scores to assess atherosclerotic cardiovascular disease risk: clinical perspectives and basic implications. Circ Res 2020;126:1159–1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hindy G, Aragam KG, Ng K, Chaffin M, Lotta LA, Baras A, et al. Genome-wide polygenic score, clinical risk factors, and long-term trajectories of coronary artery disease. Arterioscler Thromb Vasc Biol 2020;40:2738–2746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Said MA, Verweij N, van der Harst P. Associations of combined genetic and lifestyle risks with incident cardiovascular disease and diabetes in the UK biobank study. JAMA Cardiol 2018;3:693–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Duncan L, Shen H, Gelaye B, Meijsen J, Ressler K, Feldman M, et al. Analysis of polygenic risk score usage and performance in diverse human populations. Nat Commun 2019;10:3328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Martin AR, Gignoux CR, Walters RK, Wojcik GL, Neale BM, Gravel S, et al. Human demographic history impacts genetic risk prediction across diverse populations. Am J Hum Genet 2017;100:635–649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gurdasani D, Barroso I, Zeggini E, Sandhu MS. Genomics of disease risk in globally diverse populations. Nat Rev Genet 2019;20:520–535. [DOI] [PubMed] [Google Scholar]
- 19. Martin AR, Kanai M, Kamatani Y, Okada Y, Neale BM, Daly MJ. Clinical use of current polygenic risk scores may exacerbate health disparities. Nat Genet 2019;51:584–591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Mosley JD, Gupta DK, Tan J, Yao J, Wells QS, Shaffer CM, et al. Predictive accuracy of a polygenic risk score compared with a clinical risk score for incident coronary heart disease. JAMA 2020;323:627–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Elliott J, Bodinier B, Bond TA, Chadeau-Hyam M, Evangelou E, Moons KGM, et al. Predictive accuracy of a polygenic risk score-enhanced prediction model vs a clinical risk score for coronary artery disease. JAMA 2020;323:636–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Alpert JS, Thygesen K, Antman E, Bassand JP. Myocardial infarction redefined—a consensus document of The Joint European Society of Cardiology/American College of Cardiology Committee for the redefinition of myocardial infarction. J Am Coll Cardiol 2000;36:959–969. [DOI] [PubMed] [Google Scholar]
- 23. Lu X, Wang L, Chen S, He L, Yang X, Shi Y, et al. Genome-wide association study in Han Chinese identifies four new susceptibility loci for coronary artery disease. Nat Genet 2012;44:890–894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Koyama S, Ito K, Terao C, Akiyama M, Horikoshi M, Momozawa Y, et al. Population-specific and trans-ancestry genome-wide analyses identify distinct and shared genetic risk loci for coronary artery disease. Nat Genet 2020;52:1169–1177. [DOI] [PubMed] [Google Scholar]
- 25. Abraham G, Malik R, Yonova-Doing E, Salim A, Wang T, Danesh J, et al. Genomic risk score offers predictive performance comparable to clinical risk factors for ischaemic stroke. Nat Commun 2019;10:5819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Liu F, Li J, Chen J, Hu D, Li Y, Huang J, et al. Predicting lifetime risk for developing atherosclerotic cardiovascular disease in Chinese population: the China-PAR project. Sci Bull 2018;63:779–787. [DOI] [PubMed] [Google Scholar]
- 27. The Joint Task Force for Guideline on the Assessment and Management of Cardiovascular Risk in China . Guideline on the assessment and management of cardiovascular risk in China. Zhonghua Yu Fang Yi Xue Za Zhi 2019;53:13–35. [DOI] [PubMed] [Google Scholar]
- 28. Yang XL, Chen JC, Li JX, Cao J, Lu XF, Liu FC, et al. Risk stratification of atherosclerotic cardiovascular disease in Chinese adults. Chronic Dis Transl Med 2016;2:102–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Pencina MJ, D’Agostino R, Steyerberg EW. Extensions of net reclassification improvement calculations to measure usefulness of new biomarkers. Stat Med 2011;30:11–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Pencina MJ, D’Agostino RB. Overall C as a measure of discrimination in survival analysis: model specific population value and confidence interval estimation. Stat Med 2004;23:2109–2123. [DOI] [PubMed] [Google Scholar]
- 31. Fine JP, Gray RJ. A proportional hazards model for the subdistribution of a competing risk. J Am Stat Assoc 1999;94:496–509. [Google Scholar]
- 32. Thanassoulis G, Peloso GM, Pencina MJ, Hoffmann U, Fox CS, Cupples LA, et al. A genetic risk score is associated with incident cardiovascular disease and coronary artery calcium: the Framingham Heart Study. Circ Cardiovasc Genet 2012;5:113–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ripatti S, Tikkanen E, Orho-Melander M, Havulinna AS, Silander K, Sharma A, et al. A multilocus genetic risk score for coronary heart disease: case–control and prospective cohort analyses. Lancet 2010;376:1393–1400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Tikkanen E, Havulinna AS, Palotie A, Salomaa V, Ripatti S. Genetic risk prediction and a 2-stage risk screening strategy for coronary heart disease. Arterioscler Thromb Vasc Biol 2013;33:2261–2266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Aragam KG, Dobbyn A, Judy R, Chaffin M, Chaudhary K, Hindy G, et al. Limitations of contemporary guidelines for managing patients at high genetic risk of coronary artery disease. J Am Coll Cardiol 2020;75:2769–2780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Grundy SM, Stone NJ, Bailey AL, Beam C, Birtcher KK, Blumenthal RS, et al. 2018 AHA/ACC/AACVPR/AAPA/ABC/ACPM/ADA/AGS/APhA/ASPC/NLA/PCNA Guideline on the management of blood cholesterol: executive summary: a report of the American College of Cardiology/American Heart Association Task Force on Clinical Practice Guidelines. J Am Coll Cardiol 2019;73:3168–3209. [DOI] [PubMed] [Google Scholar]
- 37. Yeboah J, McClelland RL, Polonsky TS, Burke GL, Sibley CT, O'Leary D, et al. Comparison of novel risk markers for improvement in cardiovascular risk assessment in intermediate-risk individuals. JAMA 2012;308:788–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Torkamani A, Wineinger NE, Topol EJ. The personal and clinical utility of polygenic risk scores. Nat Rev Genet 2018;19:581–590. [DOI] [PubMed] [Google Scholar]
- 39. Lu J, Lu Y, Yang H, Bilige W, Li Y, Schulz W, et al. Characteristics of high cardiovascular risk in 1.7 million Chinese adults. Ann Intern Med 2019;170:298–308. [DOI] [PubMed] [Google Scholar]
- 40. Mega JL, Stitziel NO, Smith JG, Chasman DI, Caulfield MJ, Devlin JJ, et al. Genetic risk, coronary heart disease events, and the clinical benefit of statin therapy: an analysis of primary and secondary prevention trials. Lancet 2015;385:2264–2271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Natarajan P, Young R, Stitziel NO, Padmanabhan S, Baber U, Mehran R, et al. Polygenic risk score identifies subgroup with higher burden of atherosclerosis and greater relative benefit from statin therapy in the primary prevention setting. Circulation 2017;135:2091–2101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Damask A, Steg PG, Schwartz GG, Szarek M, Hagstrom E, Badimon L, et al. Patients with high genome-wide polygenic risk scores for coronary artery disease may receive greater clinical benefit from alirocumab treatment in the ODYSSEY OUTCOMES Trial. Circulation 2020;141:624–636. [DOI] [PubMed] [Google Scholar]
- 43. Marston NA, Kamanu FK, Nordio F, Gurmu Y, Roselli C, Sever PS, et al. Predicting benefit from evolocumab therapy in patients with atherosclerotic disease using a genetic risk score: results from the FOURIER Trial. Circulation 2020;141:616–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Gola D, Erdmann J, Lall K, Magi R, Muller-Myhsok B, Schunkert H, et al. Population bias in polygenic risk prediction models for coronary artery disease. Circ Genom Precis Med 2020;13:e002932. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data are available upon reasonable request to the corresponding authors.