Abstract
We describe two mutants of Mycoplasma hominis PG-21 which show resistance to 16-membered macrolides but susceptibility to lincosamides, obtained by in vitro exposure to increasing doses of josamycin. The 23S rRNA gene showed that each had a mutation (A2062G and A2062T) corresponding to nucleotide 2062 in Escherichia coli, which was associated with the acquired phenotype.
The genetic bases for macrolide-lincosamide-streptogramin B group resistance have been extensively studied for many bacteria (2, 4–6, 16, 18, 27, 29, 30, 32–34) and mycoplasmas (8, 14). In Mycoplasma pneumoniae (14, 22), as well as in other microorganisms, resistance to erythromycin has been associated with point mutations (A-to-G transition) in the loop of domain V of the 23S rRNA (2, 4–6, 8, 16, 18, 27, 29, 30, 32). Specific residues within domain V of 23S rRNA are involved in the action of macrolide-lincosamide-streptogramin antibiotics and chloramphenicol (2–6, 8, 11, 14–18, 20, 27–30, 32). It is well known that Mycoplasma hominis and many other mycoplasmas are resistant to erythromycin but susceptible to 16-membered macrolides (josamycin and miocamycin) (1, 7–9, 19, 23) and lincosamides (23). Recently the natural resistance of M. hominis to erythromycin has been associated with a guanine-to-adenine transition in position 2057 (Escherichia coli numbering), located in the central loop of the 23S rRNA domain V (8), and such a transition is present in Mycoplasma flocculare and Mycoplasma hyopneumoniae, which are also resistant to erythromycin (24).
These features encouraged us to investigate the ability of josamycin to select for resistance in M. hominis strains, in order to see whether the acquired resistance pattern includes either macrolides only or both macrolides and lincosamides and to establish a possible relationship between the resistant phenotype and the appearance of specific point mutations in the region coding for the peptidyl transferase loop in the 23S rRNA gene.
The type strain M. hominis PG-21 was chosen for our study. The strain was grown in SP-4 broth (pH 7.0) (7, 8, 21, 25, 26, 31). The MIC was determined by a broth microdilution assay as previously described (7–9, 23).
For multistep selection for resistance, SP-4 broth medium containing doubling concentrations of josamycin was inoculated with strain PG-21, incubated at 37°C, and examined for growth each day. To test for the development of resistance, 0.1 ml of the culture was withdrawn from each tube every day and spread on an SP-4 agar plate containing 1 μg of josamycin/ml. All the colonies growing on josamycin-agar were challenged with increasing concentrations of the drug in broth (from 16 to 128 μg/ml). All selected mutants were single colony purified on SP-4 agar-josamycin and drug-free SP-4 agar. Resistance to josamycin was assayed after the microrganisms were repeatedly transferred to antibiotic-free media.
Two resistant clones of the M. hominis PG-21 strain were obtained by the selection procedure outlined above and were found to be stably resistant. The resistance/susceptibility patterns for the two strains, called PG-21/JR and PG-21/JR2, are shown in Table 1 (23).
TABLE 1.
The resistance/susceptibility patterns of M. hominis PG-21/JR and PG-21/JR2 compared to that of M. hominis PG-21
DNA was extracted and purified by standard methods. Oligonucleotide primers were designed upon alignment of 23S rRNAs from a number of closely related species (8). PCR amplification was performed by standard methods. The cycling programs were as follows: one cycle at 98°C for 10 min; 30 cycles of 95°C for 30 s, 53°C for 30 s, and 72°C for 30 s; and a final elongation step at 72°C for 5 min. The DNA sequences were determined with dye terminators.
Nucleotide sequence accession numbers.
The partial 23S rRNA sequences of the M. hominis strains PG-21/JR and PG-21/JR2 have been submitted to GenBank under accession numbers AF184237 and AF317663.
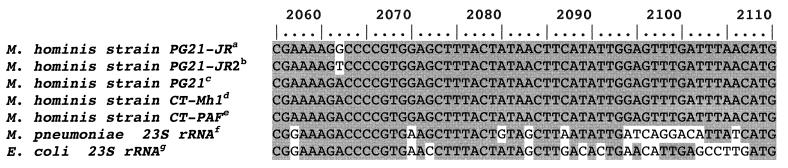
Since specific positions within the central loop of domain V of 23S rRNA have been associated with the development of erythromycin resistance in mycoplasma and in many other microrganisms, we examined the sequence of this domain by amplifying the corresponding ribosomal DNA gene of the two josamycin-resistant PG-21/JR strains. The sequences are shown in Fig. 1 aligned with the corresponding sequences from a number of related microorganisms and Escherichia coli J01695 in order to number the nucleotide positions. The G-to-A transition at position 2057, already described for M. hominis as a naturally occurring transition, helped further in establishing the nucleotide correspondence in domain V. In the two josamycin-resistant derivatives, two new mutations, an A2062G transition and an A2062T transversion, were observed in the region coding for the peptidyl transferase loop. These represent new mutations, so far not reported in any microorganism. In the original electropherogram, the first appeared as two coincident G and A peaks while the second appeared as a unique T peak (data not shown). In order to exclude a sequence artifact for the two coincident peaks seen, an allele-specific PCR experiment was performed (35). Two different primers were designed whose 3′ position matched the nucleotide at position 2062 either for the mutated operon (endG, 5′-CCGCATCTAGACGAAAAGG-3′; endT, 5′-CCGCATCTAGACGAAAAGT-3′) or for the wild type (endA: 5′-CCGCATCTAGACGAAAAGA-3′). These primers were used in separate experiments in conjunction with the reverse primer R1 (5′-CCTCCGTTACCTTTTAGGA-3′) or R2 (5′-GGTCCTCTCGTACTAGAAG-3′). KCl was replaced with (NH4)2SO4, and the annealing temperature was increased to 59°C to increase stringency, keeping unchanged the remaining PCR parameters.
FIG. 1.
Alignment (partial) of 23S rRNA genes from selected organisms corresponding to the loop of the peptidyl transferase (domain V). The nucleotides are numbered on the basis of the E. coli sequence. Raised letters denote the following: a, GenBank accession no. AF184237, M. hominis strain PG-21/JR 23S rRNA gene (partial sequence); b, AF317663, M. hominis 23S rRNA gene (partial sequence, strain PG-21/JR2); c, AF101242, M. hominis strain PG-21 23S rRNA gene (partial sequence); d, AF131860, M. hominis strain CT-Mh1 23S rRNA gene (partial sequence); e, AF131073, M. hominis strain CT-PAF 23S rRNA gene (partial sequence); f, X68422, M. pneumoniae gene for 23S rRNA; g, J01695, E. coli rRNA operon (rrnB).
As expected, the use of primers with different 3′ ends gave rise to the expected amplification product in the mutant strains, whereas only the wild-type-specific primer worked in the original PG-21 strain (data not shown). The heterozygous state of the A2062G transition in the mutant strain was thus definitively assessed, while in the case of transversion, an apparent homozygous state was evidenced by both the electropherogram and allele-specific PCR. Control experiments, done on pools of either uninduced or induced (resistant) strain cultures using oligonucleotides designed to match all possible mutations at position 2062, failed to detect any other mutation at that position (data not shown).
The involvement of domain V in the mechanisms of resistance has been demonstrated by chemical footprinting studies (2, 17), which showed that the reactivity of certain purines within domain V was specifically altered by macrolides (erythromycin and carbomycin, a 16-membered macrolide), lincosamides, vernamicin B, chloramphenicol, or azalides. Moazed and Noeller (17) incubated 70S ribosomes together with antibiotics and showed direct protection of both A2058 and A2059 by both erythromycin and carbomycin against derivatization by dimethyl sulfate; carbomycin additionally protected position A2062. Of the three protectable adenine residues, vernamycin B, a streptogramin, protected A2062 but not A2058 or A2059. Moreover, clindamycin protected both A2058 and A2059, whereas lincomycin protected only A2058 (4–5).
The role of the adenine residue at position 2062 is not fully defined. An A2062C transversion, associated with chloramphenicol and with linezolid resistance in Halobacterium halobium, was found at the same position (12, 15). More recently, an A2062C transversion has been associated with 16-membered macrolide and streptogramin resistance in Streptococcus pneumoniae (3). While no drug effects could be directly attributed to the A2062 site, raising the possibility that such an effect, if it exists, may be attributed to ribosomal protein (20), A2062 was shielded by virginiamycin M (a streptogramin A) in a mutant strain resistant to virginiamycin S (a streptogramin B) (28). The aforementioned features may explain why our mutant strains became resistant to 16-membered macrolides due to mutation at position 2062 and retained their susceptibility to lincomycin and clindamycin. An analysis of our mutants suggests that A2062 is incompatible with 16-membered macrolide resistance; however, since a role of A2062 in resistance has been questioned by some authors, as shown above, we hypothesize that A2062 incompatibility with macrolide resistance could be associated with species-specific sequence context, particularly with the naturally occurring G2057A transition. Further studies will be necessary to confirm this hypothesis.
Since there are two rRNA operons in M. hominis (10, 13), a new mutation in any operon should be present in a heterozygous state. Under selective pressure the mutant “allele” should behave as dominant, allowing the strain to overcome antibiotic inhibition. The effect of antibiotics on the rRNA stability in both wild-type and mutant strains should also be taken into account.
Acknowledgments
This work was supported by grants from University of Catania (1997 to 2000).
REFERENCES
- 1.Braun P, Klein J O, Kass E H. Susceptibility of genital mycoplasmas to antimicrobial agents. Appl Microbiol. 1970;19:62–70. doi: 10.1128/am.19.1.62-70.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cundliffe E. Recognition sites for antibiotics within rRNA. In: Hill W E, Dahlberg A, Garrett R A, Moore P B, Schlessinger D, Warner J R, editors. The ribosome: structure, functions, and evolution. Washington, D.C.: American Society for Microbiology; 1990. pp. 479–490. [Google Scholar]
- 3.Depardieu F, Courvalin P. Mutation in 23S rRNA responsible for resistance to 16-membered macrolides and streptogramins in Streptococcus pneumoniae. Antimicrob Agents Chemother. 2001;45:319–323. doi: 10.1128/AAC.45.1.319-323.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Douthwaite S. Functional interactions within 23S rRNA involving the peptidyltransferase center. J Bacteriol. 1992;174:1333–1338. doi: 10.1128/jb.174.4.1333-1338.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Douthwaite S, Aagaard C. Erythromycin binding is reduced in ribosomes with conformational alteration in the 23S rRNA peptidyl transferase loop. J Mol Biol. 1993;232:725–731. doi: 10.1006/jmbi.1993.1426. [DOI] [PubMed] [Google Scholar]
- 6.Ettayebi M, Prasad S M, Morgan E A. Chloramphenicol-erythromycin resistance mutations in 23S rRNA gene of Escherichia coli. J Bacteriol. 1985;162:551–557. doi: 10.1128/jb.162.2.551-557.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Furneri P M, Bisignano G, Cerniglia G, Tempera G, Nicoletti G. In vitro antimycoplasmal activity of flurithromycin. J Antimicrob Chemother. 1995;35:161–165. doi: 10.1093/jac/35.1.161. [DOI] [PubMed] [Google Scholar]
- 8.Furneri P M, Rappazzo G, Musumarra M P, Tempera G, Roccasalva L S. Genetic basis of natural resistance to erythromycin by Mycoplasma hominis. J Antimicrob Chemother. 2000;45:547–548. doi: 10.1093/jac/45.4.547. [DOI] [PubMed] [Google Scholar]
- 9.Furneri P M, Tempera G, Bisignano G, Cottarelli F V, Nicoletti G. Anti-mycoplasmal activity of a new macrolide: miocamycin. Chemioterapia. 1987;VI:341–345. [PubMed] [Google Scholar]
- 10.Gobel U, Butler G H, Stanbridge E J. Comparative analysis of Mycoplasma ribosomal RNA operons. Isr J Med Sci. 1984;20:762–764. [PubMed] [Google Scholar]
- 11.Hansen L H, Mauvais P, Douthwaite S. The macrolide-ketolide antibiotic binding site is formed by structures in domains II and V of 23S ribosomal RNA. Mol Microbiol. 1999;31:623–631. doi: 10.1046/j.1365-2958.1999.01202.x. [DOI] [PubMed] [Google Scholar]
- 12.Kloss P, Xiong L, Shinabarger D L, Mankin A S. Resistance mutations in 23S rRNA identify the site of action of the protein synthesis inhibitor linezolid in the ribosomal peptidyl transferase center. J Mol Biol. 1999;294:93–103. doi: 10.1006/jmbi.1999.3247. [DOI] [PubMed] [Google Scholar]
- 13.Ladefoged S A, Christiansen G. Physical and genetic mapping of the genomes of five Mycoplasma hominis strains by pulsed-field gel electrophoresis. J Bacteriol. 1992;174:2199–2207. doi: 10.1128/jb.174.7.2199-2207.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lucier T S, Heitzman K, Liu S-K, Hu P-C. Transition mutations in the 23S rRNA of erythromycin-resistant isolates of Mycoplasma pneumoniae. Antimicrob Agents Chemother. 1995;39:2270–2273. doi: 10.1128/aac.39.12.2770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mankin A S, Garret R A. Chloramphenicol resistance mutations in the single 23S rRNA gene of the archaeon Halobacterium halobium. J Bacteriol. 1991;173:3559–3563. doi: 10.1128/jb.173.11.3559-3563.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Meier A, Kirschner P, Springer B, Steingrube V A, Brown B A, Wallace R J, Jr, Böttger E C. Identification of mutations in 23S rRNA gene of clarithromycin-resistance Mycobacterium intracellulare. Antimicrob Agents Chemother. 1994;38:381–384. doi: 10.1128/aac.38.2.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Moazed D, Noller H F. Chloramphenicol, erythromycin, carbomycin and vernamycin B protect overlapping sites in the peptidyl transferase region of 23S ribosomal RNA. Biochimie. 1987;69:879–884. doi: 10.1016/0300-9084(87)90215-x. [DOI] [PubMed] [Google Scholar]
- 18.Nash K A, Inderlied C B. Genetic basis of macrolide resistance in Mycobacterium avium isolated from patients with disseminated disease. Antimicrob Agents Chemother. 1995;39:2625–2630. doi: 10.1128/aac.39.12.2625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Palù G, Valisena S, Sassella D, Furneri P, Meloni G A. Antimycoplasmal activity of roxithromycin. Br J Clin Pract. 1988;42(Suppl. 55):21–23. [Google Scholar]
- 20.Rodriguez-Fonseca C, Amils R, Garrett R A. Fine structure of the peptidyl transferase centre on 23 S-like rRNAs deduced from chemical probing of antibiotic-ribosome complexes. J Mol Biol. 1995;247:224–235. doi: 10.1006/jmbi.1994.0135. [DOI] [PubMed] [Google Scholar]
- 21.Senterfit L B. Laboratory diagnosis of mycoplasmal infections. Isr J Med Sci. 1984;20:905–907. [PubMed] [Google Scholar]
- 22.Stopler T, Bransky D. Resistance of Mycoplasma pneumoniae to macrolides, lincomycin and streptogramin B. J Antimicrob Chemother. 1986;18:359–364. doi: 10.1093/jac/18.3.359. [DOI] [PubMed] [Google Scholar]
- 23.Taylor-Robinson D, Bebear C. Antibiotic susceptibilities of mycoplasmas and treatment of mycoplasmal infections. J Antimicrob Chemother. 1998;40:622–630. doi: 10.1093/jac/40.5.622. [DOI] [PubMed] [Google Scholar]
- 24.ter Laak E A, Pijpers A, Noordergraaf J H, Schoevers E C, Verheijden J H M. Comparison of methods for in vitro testing of susceptibility of porcine Mycoplasma species to antimicrobial agents. Antimicrob Agents Chemother. 1991;35:228–233. doi: 10.1128/aac.35.2.228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tully J G, Rose D L, Whitcomb R F, Wenzel R P. Enhanced isolation of Mycoplasma pneumoniae from throat washings with a newly-modified culture medium. J Infect Dis. 1979;139:478–482. doi: 10.1093/infdis/139.4.478. [DOI] [PubMed] [Google Scholar]
- 26.Tully J G, Taylor-Robinson D, Rose D L, Furr P M, Hawkins D A. Evaluation of culture media for the recovery of Mycoplasma hominis from the urogenital tract. Sex Transm Dis. 1983;10(Suppl. 4):256–260. [PubMed] [Google Scholar]
- 27.Vannuffel P, Di Giambattista M, Cocito C. The role of rRNA bases in the interaction of peptidyl-transferase inhibitors with bacterial ribosomes. J Biol Chem. 1992;267:16114–16120. [PubMed] [Google Scholar]
- 28.Vannuffel P, Di Giambattista M, Cocito C. Chemical probing of a virginiamycin M-promoted conformational change of the peptidyl-transferase domain. Nucleic Acids Res. 1994;22:4449–4453. doi: 10.1093/nar/22.21.4449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Versalovic J, Shortridge D, Kibler K, Griffy M V, Beyer J, Flamm R K, Tanaka S K, Graham D Y, Go M F. Mutations in 23S rRNA are associated with clarithromycin resistance in Helicobacter pylor. Antimicrob Agents Chemother. 1996;40:477–480. doi: 10.1128/aac.40.2.477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vester B, Hansen L H, Douthwaite S. The conformation of 23S rRNA nucleotide A2058 determines its recognition by the ermE methyltransferase. RNA. 1995;1:501–509. [PMC free article] [PubMed] [Google Scholar]
- 31.Waites K B, Duffy L B, Schmid T, Crabb D, Pate M S, Cassell G H. In vitro susceptibilities of Mycoplasma pneumoniae, Mycoplasma hominis, and Ureaplasma urealyticum to sparfloxacin and PD 127391. Antimicrob Agents Chemother. 1991;35:1181–1185. doi: 10.1128/aac.35.6.1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Weisblum B. Erythromycin resistance by ribosome modification. Antimicrob Agents Chemother. 1995;39:577–585. doi: 10.1128/AAC.39.3.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Weisblum B. Insights into erythromycin action from studies of its activity as inducers of resistance. Antimicrob Agents Chemother. 1995;39:797–805. doi: 10.1128/aac.39.4.797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Weisblum B. Macrolide resistance. Drug Resist Updates. 1998;1:29–41. doi: 10.1016/s1368-7646(98)80212-4. [DOI] [PubMed] [Google Scholar]
- 35.Wu D Y, Ugozzoli L, Pol B K, Wallace R B. Allele-specific enzymatic amplification of β-globin genomic DNA for diagnosis of sickle cell anemia. Proc Nat Acad Sci USA. 1989;86:2757–2760. doi: 10.1073/pnas.86.8.2757. [DOI] [PMC free article] [PubMed] [Google Scholar]