Abstract
From whole-cell DNA of a clinical isolate of the enterobacterial species Rahnella aquatilis, a β-lactamase gene was cloned that encoded a chromosomally encoded Ambler class A enzyme, RAHN-1. RAHN-1, with a pI of 7.2, shares 76, 73, and 71% amino acid identity with the extended-spectrum β-lactamase of chromosomal origin from Serratia fonticola and with the plasmid-mediated β-lactamases CTX-M-2 and CTX-M-1, respectively. The hydrolysis spectrum of the clavulanic acid-inhibited RAHN-1 was expanded to cephalosporins such as cefuroxime, cefotaxime, and ceftriaxone, but not to ceftazidime. Its expression was not inducible.
Rahnella aquatilis is an enterobacterial species (11) that is widely distributed in water and soils as well as in foods (26). It is an opportunistic pathogen that predominantly causes bacteremia and septicemia, as well as endocarditis, urinary tract infections, and wound infections, mostly in immunocompromised patients (7, 14, 17, 26).
A recent study investigated the antibiotic susceptibility patterns of 72 R. aquatilis strains of clinical and environmental origins (26). It showed that R. aquatilis is resistant to amoxicillin, ticarcillin, cefuroxime, and cephalothin; is of intermediate susceptibility to cefotaxime and ceftriaxone; and is susceptible to ceftazidime and imipenem. R. aquatilis is susceptible to β-lactam combinations such as amoxicillin-clavulanic acid and piperacillin-tazobactam. This β-lactam susceptibility pattern suggested the presence of a clavulanic acid-inhibited Ambler class A β-lactamase (26).
In this work, we report on characterization of a novel class A β-lactamase with an extended-spectrum hydrolysis profile from R. aquatilis.
Bacterial strains, plasmid analysis, and conjugation.
R. aquatilis clinical strain DON-1 was isolated from a central venous catheter of a 48-year-old patient hospitalized in the neurosurgery unit of the Hôpital Bicêtre (France) in September 2000. It was identified by the API 20-E system (bioMérieux, Marcy-L'Etoile, France). R. aquatilis reference strain CIP 103.904 was from the Institut Pasteur strain collection (Paris). Plasmid DNA extractions, performed as described previously (24), failed to identify plasmids in R. aquatilis DON-1. Direct transfer of an amoxicillin resistance marker from R. aquatilis DON-1 to rifampin-resistant Escherichia coli JM109 also failed (24).
Cloning and sequence analysis of the β-lactamase gene from R. aquatilis DON-1.
Whole-cell DNA of R. aquatilis DON-1 was extracted as described previously (23). It was partially digested with Sau3AI and ligated into the BamHI site of the pBK-CMV phagemid (Stratagene, Amsterdam, The Netherlands) as described previously (23). The ligation products were electroporated into electrocompetent E. coli DH10B (Bio-Rad, Ivry-sur-Seine, France). E. coli DH10B cells harboring recombinant plasmids were selected on kanamycin (30 μg/ml)- and amoxicillin (20 μg/ml)-containing Trypticase soy agar plates. The sizes of the inserts were determined by double restriction analysis (25).
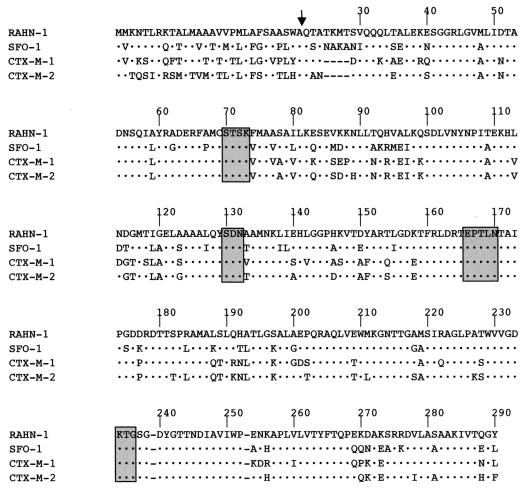
Fifteen E. coli DH10B recombinant clones were obtained. One of the recombinant plasmids that had the shortest insert was retained for further analysis (pRAHN-1). The 5.6-kb DNA insert of recombinant plasmid pRAHN-1 was sequenced on both strands by using an ABI 377 sequencer (Applied Biosystems, Foster City, Calif.). The nucleotide and the deduced protein sequences were analyzed with software available over the internet at the National Center for Biotechnology Information web site (http://www.ncbi.nlm.nih.gov). An open reading frame (ORF) of 888 bp was identified (data not shown). The G+C content of this ORF was 55.7%, which lies within the G+C ratio of enterobacterial genes. Within the deduced protein of this ORF (295 amino acids), named RAHN-1, a serine-threonine-phenylalanine-lysine tetrad was found in amino acid positions 70 to 73 (Fig. 1) according to the designation of Ambler et al. (1). This tetrad included the conserved serine and lysine amino acid residues characteristic of β-lactamases possessing a serine active site or penicillin-binding proteins (13). Three other structural elements were also found: serine-aspartic acid-asparagine (S-D-N) in positions 130 to 132, glutamate-X-X-leucine-asparagine (E-X-X-L-N) in position 166 to 170, which is part of the so-called omega loop of class A β-lactamases, and lysine-threonine-glycine (K-T-G) in positions 234 to 236 (Fig. 1).
FIG. 1.
Alignment of the RAHN-1 amino acid sequence with those of the most closely related enzymes, SFO-1, CTX-M-1, and CTX-M-2. Numbering is according to that of Ambler et al. (1). Dots indicate identical amino acid residues, and dashes indicate gaps introduced to optimize the alignment. The vertical arrow is the putative cleavage site of the leader peptide of RAHN-1 β-lactamase. Four structural elements characteristic of class A β-lactamases are boxed in gray.
RAHN-1 is related to class A β-lactamases. The highest percentages of identity were found with the plasmid-mediated extended-spectrum β-lactamase (ESBL) SFO-1 (75.9%), which originated from Serratia fonticola (18, 20), and with the plasmid-mediated extended-spectrum β-lactamases CTX-M-2 (73.2%) and CTX-M-1 (71.5%) (3, 4). CTX-M-2 β-lactamase has been recently reported as being related to the chromosomally encoded class A β-lactamase from Kluyvera ascorbata (GenBank accession no. AJ 272538). RAHN-1 shared 69, 65, and 62% amino acid identity with the chromosomally encoded class A β-lactamases from Klebsiella oxytoca (2), Citrobacter diversus (22), and Proteus vulgaris (21), respectively.
Upstream of the ORF coding for RAHN-1, no putative LysR-type regulator gene was identified, while such chromosome-encoded AmpR regulators were divergently transcribed usptream of the β-lactamase genes coding for the class A β-lactamases of C. diversus, P. vulgaris, and S. fonticola (9, 12, 18). However, 126 bp upstream of blaRAHN-1, a stop codon of a 1,664-bp ORF was identified that encoded a putative protein sharing 50% identity with a methyl-accepting chemotaxis protein of E. coli (5). Further upstream, another 996-bp ORF was identified that encoded a putative protein that shared 36% amino acid identity with an ATP-binding protein of E. coli (10). Downstream of blaRAHN-1, no ORF could have been identified, since the end of the 5.6-kb insert of pRAHN-1 was located 2 bp downstream of the stop codon of blaRAHN-1. A Southern transfer (25) was performed with a PCR-obtained 861-bp internal fragment of blaRAHN-1 as a labeled probe (23) and whole-cell DNAs of R. aquatilis DON-1 and the CIP 103.904 reference strain. In both cases, a positive signal was detected at the chromosome migration position, further indicating the chromosomal origin of blaRAHN-1 (data not shown).
Susceptibility testing.
MICs of selected β-lactams were determined by an agar dilution technique on Mueller-Hinton plates as described previously (24) and interpreted according to the guidelines of the National Committee for Clinical Laboratory Standards (19). R. aquatilis DON-1 was resistant to amoxicillin, ticarcillin, cephalothin, and cefuroxime (Table 1). It was of intermediate susceptibility to piperacillin, cefotaxime, ceftriaxone, cefepime, and aztreonam and remained susceptible to ceftazidime and imipenem (Table 1). This pattern of β-lactam susceptibility corresponded to that described previously for R. aquatilis strains and R. aquatilis reference strain CIP 103.904 (Table 1) (26). Once cloned in pBK-CMV (pRAHN-1) and expressed in E. coli DH10B (Table 1), the same antibiotic resistance pattern was obtained, thus showing the contribution of RAHN-1 to the natural β-lactam resistance phenotype of R. aquatilis DON-1. Addition of clavulanic acid and tazobactam decreased the MICs of β-lactams, especially those of cefotaxime and ceftriaxone, thus showing that RAHN-1 is a clavulanic acid-inhibited ESBL.
TABLE 1.
MICs of β-lactams for R. aquatilis clinical isolate DON-1 and reference strain CIP 103.904 and E. coli DH10B harboring recombinant plasmid pRAHN-1 and reference strain DH10B
| β-Lactam(s)a | MIC (μg/ml) for strain:
|
|||
|---|---|---|---|---|
|
R. aquatilis
|
E. coli
|
|||
| DON-1 | CIP 103.904 | DH10B(pRAHN-1) | DH10B | |
| Amoxicillin | 256 | 256 | >512 | 2 |
| Amoxicillin + CLA | 2 | 4 | 4 | 2 |
| Ticarcillin | >512 | >512 | >512 | 1 |
| Ticarcillin + CLA | 4 | 4 | 8 | 1 |
| Piperacillin | 8 | 16 | 16 | 1 |
| Piperacillin + TZB | <1 | <1 | 1 | 1 |
| Cephalothin | 512 | 512 | 256 | 2 |
| Cefuroxime | 128 | 128 | 256 | 2 |
| Cefotaxime | 4 | 4 | 4 | <0.06 |
| Cefotaxime + CLA | <0.06 | 0.06 | <0.06 | <0.06 |
| Cefotaxime + TZB | <0.06 | <0.06 | <0.06 | <0.06 |
| Ceftazidime | 0.25 | 0.5 | 0.25 | 0.12 |
| Ceftazidime + CLA | 0.12 | 0.12 | 0.12 | <0.06 |
| Ceftazidime + TZB | 0.12 | 0.12 | <0.06 | <0.06 |
| Ceftriaxone | 4 | 4 | 4 | <0.06 |
| Cefepime | 1 | 0.5 | 0.25 | <0.06 |
| Cefepime + CLA | <0.06 | 0.06 | <0.06 | <0.06 |
| Cefepime + TZB | <0.06 | <0.06 | <0.06 | <0.06 |
| Cefpirome | 1 | 4 | 4 | <0.06 |
| Cefpirome + CLA | <0.06 | 0.06 | <0.06 | <0.06 |
| Cefpirome + TZB | <0.06 | <0.06 | <0.06 | <0.06 |
| Cefoxitin | 4 | 8 | 4 | 2 |
| Moxalactam | 0.25 | 0.5 | 0.25 | 0.12 |
| Aztreonam | 1 | 1 | 1 | 0.06 |
| Meropenem | 0.06 | 0.06 | <0.06 | <0.06 |
| Imipenem | 0.5 | 0.5 | 0.5 | 0.06 |
CLA, clavulanic acid at a fixed concentration of 2 μg/ml; TZB, tazobactam at a fixed concentration of 4 μg/ml.
The β-lactam resistance pattern of R. aquatilis was similar to that of S. fonticola (20). Moreover, R. aquatilis strains, like C. diversus and P. vulgaris strains, are of intermediate susceptibility or resistance to cefuroxime. In contrast to K. oxytoca, which overproduces its chromosomally encoded class A β-lactamase (8), R. aquatilis was not resistant to aztreonam.
Biochemical properties of β-lactamase RAHN-1.
Cultures of E. coli DH10B(pRAHN-1) were grown overnight at 37°C in 4 liters of Trypticase soy broth containing amoxicillin (30 μg/ml) and kanamycin (30 μg/ml). The β-lactamase extract was obtained after sonification as described previously (24). It was loaded onto a preequilibrated Q-Sepharose column (Amersham Pharmacia Biotech). The enzyme was eluted with a linear NaCl gradient (0 to 1 M) in Tris-HCl buffer (pH 9.0). The β-lactamase was eluted at a concentration of 100 to 150 mM NaCl. Fractions containing β-lactamase activity were loaded onto the same preequilibrated Q-Sepharose column in the same buffer and eluted with a linear NaCl gradient (0 to 200 mM) in the same Tris-HCl buffer at a concentration of 100 mM NaCl. Fractions containing β-lactamase activity were pooled and dialyzed overnight against 100 mM phosphate buffer (pH 7.0) and then concentrated 10-fold with Centrisart-C30 spin column (Sartorius, Goettingen, Germany). The specific activity of the purified RAHN-1 enzyme was 15,100 mU mg of protein−1, determined with 100 μM cephalothin as a substrate with a 35-fold purification. Its purity was estimated to be 80% by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (25).
Purified RAHN-1 β-lactamase was used for kinetic measurements performed at 30°C in 100 mM sodium phosphate (pH 7.0). The initial rates of hydrolysis were determined with an ULTROSPEC 2000 UV spectrophotometer (Amersham Pharmacia Biotech) as described previously (24). The 50% inhibitory concentrations (IC50s) were determined for clavulanic acid and tazobactam as reported previously (24).
RAHN-1 had a strong activity against benzylpenicillin, piperacillin, cephalothin, cefuroxime, and ceftriaxone (Table 2). A significant hydrolytic activity against cefotaxime was also observed, while no activity against ceftazidime was detectable. The activity against cefuroxime is shared by the β-lactamases of C. diversus, P. vulgaris, and S. fonticola (9, 15, 16, 20, 21). RAHN-1 affinity for cefepime, cefpirome, and aztreonam was low (high Km values) (Table 2). Inhibition studies showed that the IC50s of clavulanic acid and tazobactam (0.4 and 0.06 μM, respectively) were similar to those of TEM-1 β-lactamase. These results showed that the RAHN-1 enzyme is a β-lactamase that may belong to the group 2be β-lactamases of the Bush-Jacoby-Medeiros classification (6).
TABLE 2.
Kinetic parameters of purified β-lactamase RAHN-1a
| Substrate | kcat (s−1) | Km (μM) | kcat/Km (mM s−1) |
|---|---|---|---|
| Benzylpenicillin | 2 | 10 | 240 |
| Amoxicillin | 0.5 | 15 | 40 |
| Ticarcillin | 0.2 | 5 | 40 |
| Piperacillin | 1 | 10 | 100 |
| Cephalothin | 11 | 70 | 160 |
| Cephaloridine | 6 | 130 | 45 |
| Cefuroxime | 1 | 10 | 100 |
| Cefoxitin | <0.01 | NDb | ND |
| Cefotaxime | 0.5 | 10 | 50 |
| Ceftazidime | <0.01 | ND | ND |
| Ceftriaxone | 0.5 | 5 | 120 |
| Cefpirome | 7 | 650 | 10 |
| Cefepime | 5 | 1400 | 5 |
| Aztreonam | 0.5 | 340 | 1 |
| Imipenem | <0.01 | ND | ND |
Standard deviations of the kinetic parameters were within 15%.
ND, not determined.
Analytic isoelectric focusing, performed as previously reported (24), showed that cultures of R. aquatilis DON-1 and E. coli DH10B(pRAHN-1) gave a β-lactamase with a pI of 7.2. The expected cleavage site of the peptide leader was between an alanine residue and a glutamine residue (between the S-W-A and Q-T motifs [Fig. 1]). The same pI was found for culture of R. aquatilis reference strain CIP 103.904. The relative molecular mass of RAHN-1, determined with purified enzyme by SDS-PAGE as described previously (24), was ca. 29 kDa (data not shown). This value corresponds to those reported for class A β-lactamases (6).
Induction studies with 0.5 μg of cefoxitin per ml as a β-lactam inducer (23) and 100 μM benzylpenicillin as a substrate failed to detect induction of β-lactamase with cultures of R. aquatilis DON-1 and CIP 103.904. These results, although different from those reported previously (14), are consistent with the lack of a LysR-type regulator gene located just upstream of blaRAHN-1. The chromosomally encoded class A β-lactamase of R. aquatilis is not inducible, as opposed to the naturally occurring class A enzymes of P. vulgaris, S. fonticola, and C. diversus.
Conclusion.
Comparison of the RAHN-1 sequence to those of other class A ESBLs identified several amino acid residues that may be involved in its extended spectrum of hydrolysis (15). This is the case for the serine residue in position 237 found, for example, in β-lactamases of S. fonticola, P. vulgaris, and CTX-M-type enzymes (20, 21, 27, 28). The hydrolysis spectrum of RAHN-1 is similar to that of CTX-M-type enzymes that do not compromise ceftazidime.
This work identified a novel chromosomally encoded class A ESBL. The identication of this ESBL should be taken into account for the choice of β-lactams in treating R. aquatilis infections, since failure of a cefotaxime-containing regimen has been reported (17). Finally, β-lactamases from this enterobacterial species, R. aquatilis, may be progenitors of plasmid-encoded ESBLs, as recently described for the plasmid-encoded β-lactamases SFO-1 and CTX-M-2 related to the chromosomally encoded β-lactamases of S. fonticola and K. ascorbata, respectively.
Nucleotide sequence accession number.
The nucleotide sequences of the RAHN-1 gene and the sequence of the 5.6-kb insert of pRAHN-1 have been assigned to the GenBank nucleotide database under accession no. AF338038.
Acknowledgments
This work was funded by a grant from the Ministères de l'Education Nationale et de la Recherche (UPRES, JE 2227), Université Paris XI, Faculté de Médecine Paris-Sud, Le Kremlin Bicêtre, France.
REFERENCES
- 1.Ambler R P, Coulson A F W, Frère J-M, Ghuysen J M, Joris B, Forsman M, Lévesque R C, Tiraby G, Waley S G. A standard numbering scheme for the class A β-lactamases. Biochem J. 1991;276:269–270. doi: 10.1042/bj2760269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arakawa Y, Ohta M, Kido N, Mori M, Ito H, Komatsu T, Fujii Y, Kato N. Chromosomal β-lactamase of Klebsiella oxytoca, a new class A enzyme that hydrolyzes broad-spectrum β-lactam antibiotics. Antimicrob Agents Chemother. 1989;33:63–70. doi: 10.1128/aac.33.1.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barthélémy M, Péduzzi J, Bernard H, Tancrède C, Labia R. Close amino acid sequence relationship between the new plasmid-mediated extended-spectrum β-lactamase MEN-1 and chromosomally encoded enzymes of Klebsiella oxytoca. Biochim Biophys Acta. 1992;1122:15–22. doi: 10.1016/0167-4838(92)90121-s. [DOI] [PubMed] [Google Scholar]
- 4.Bauernfeind A, Stemplinger I, Jungwirth R, Ernst S, Casellas J M. Sequences of β-lactamase genes encoding CTX-M-1 (MEN-1) and CTX-M-2 and relationship of their amino acid sequences with those of other β-lactamases. Antimicrob Agents Chemother. 1996;40:509–513. doi: 10.1128/aac.40.2.509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boyd A, Kendall K, Simon M I. Structure of the serine chemoreceptor in Escherichia coli. Nature. 1983;301:623–626. doi: 10.1038/301623a0. [DOI] [PubMed] [Google Scholar]
- 6.Bush K, Jacoby G A, Medeiros A A. A functional classification scheme for β-lactamases and its correlation with molecular structure. Antimicrob Agents Chemother. 1995;39:1211–1233. doi: 10.1128/aac.39.6.1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Caroff N, Chamoux C, Le Gallou F, Espaze E, Gavini F, Gautreau D, Richet H, Reynaud A. Two epidemiologically related cases of Rahnella aquatilis bacteremia. Eur J Clin Microbiol Infect Dis. 1998;17:349–352. doi: 10.1007/BF01709459. [DOI] [PubMed] [Google Scholar]
- 8.Fournier B, Lagrange P H, Philippon A. In-vitro susceptibility of Klebsiella oxytoca strains to 13 β-lactams in the presence and absence of β-lactamase inhibitors. J Antimicrob Chemother. 1996;37:931–942. doi: 10.1093/jac/37.5.931. [DOI] [PubMed] [Google Scholar]
- 9.Ishiguro K, Sugimoto K. Purification and characterization of the Proteus vulgaris BlaA protein, the activator of the β-lactamase gene. J Biochem. 1996;120:98–103. doi: 10.1093/oxfordjournals.jbchem.a021399. [DOI] [PubMed] [Google Scholar]
- 10.Itoh T, Aiba H, Baba T, Hayashi H, Inada T, Isono K, Kasai H, Kimura S, Kitakawa M, Makino K, Miki T, Mizobuchi K, Mori H, Mori T, Motomura K, Nakade S, Nakamura Y, Nashimoto H, Nishio Y, Oshima T, Saito N, Sampei G, Seki Y, Horiuchi T. A 460-kb DNA sequence of the Escherichia coli K-12 genome corresponding to the 40.1–50.0 min region on the linkage map. DNA Res. 1996;3:379–392. doi: 10.1093/dnares/3.6.379. [DOI] [PubMed] [Google Scholar]
- 11.Izard D, Gavini F, Trinel P A, Leclerc H. Rahnella aquatilis, nouveau membre de la famille des Enterobacteriaceae. Ann Microbiol (Paris) 1979;130A:163–177. [PubMed] [Google Scholar]
- 12.Jones M E, Bennett P M. Inducible expression of the chromosomal cdiA from Citrobacter diversus NF85, encoding an Ambler class A β-lactamase, is under similar genetic control to the chromosomal ampC, encoding an Ambler class C enzyme from Citrobacter diversus OS60. Microb Drug Resist. 1995;1:285–291. doi: 10.1089/mdr.1995.1.285. [DOI] [PubMed] [Google Scholar]
- 13.Joris B, Ledent P, Dideberg O, Fonze E, Lamotte-Brasseur J, Kelly J A, Ghuysen J M, Frère J-M. Comparison of the sequences of class A β-lactamases and of the secondary structure elements of penicillin-recognizing proteins. Antimicrob Agents Chemother. 1991;35:2294–2301. doi: 10.1128/aac.35.11.2294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Maraki S, Samonis G, Marnelakis E, Tselentis Y. Surgical wound infection caused by Rahnella aquatilis. J Clin Microbiol. 1994;32:2706–2708. doi: 10.1128/jcm.32.11.2706-2708.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Matagne A, Lamotte-Brasseur J, Frère J-M. Catalytic properties of class A β-lactamases: efficiency and diversity. Biochem J. 1998;330:581–598. doi: 10.1042/bj3300581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Matsubara N, Yotsuji A, Kumano K, Inoue M, Mitsuhashi S. Purification and some properties of a cephalosporinase from Proteus vulgaris. Antimicrob Agents Chemother. 1981;19:185–187. doi: 10.1128/aac.19.1.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Matsukura H, Katayama K, Kitano N, Kobayashi K, Kamegane C, Higuchi A, Kyotami S. Infective endocarditis caused by an unusual gram-negative rod, Rahnella aquatilis. Pediatr Cardiol. 1996;17:108–111. doi: 10.1007/BF02505093. [DOI] [PubMed] [Google Scholar]
- 18.Matsumoto Y, Inoue M. Characterization of SFO-1, a plasmid-mediated inducible class A β-lactamase from Enterobacter cloacae. Antimicrob Agents Chemother. 1999;43:307–313. doi: 10.1128/aac.43.2.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.National Committee for Clinical Laboratory Standards. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. 5th ed. Approved standard M7–A4. Wayne, Pa: National Committee for Clinical Laboratory Standards; 2000. [Google Scholar]
- 20.Péduzzi J, Farzaneh S, Reynaud A, Barthélémy M, Labia R. Characterization and amino acid sequence analysis of a new oxyimino cephalosporin-hydrolyzing class A β-lactamase from Serratia fonticola CUV. Biochim Biophys Acta. 1997;1341:58–70. doi: 10.1016/s0167-4838(97)00020-4. [DOI] [PubMed] [Google Scholar]
- 21.Péduzzi J, Reynaud A, Baron P, Barthélémy M, Labia R. Chromosomally encoded cephalosporin-hydrolyzing β-lactamase of Proteus vulgaris RO104 belongs to Ambler's class A. Biochim Biophys Acta. 1994;1207:31–39. doi: 10.1016/0167-4838(94)90048-5. [DOI] [PubMed] [Google Scholar]
- 22.Perilli M, Franceschini N, Segatore B, Amicosante G, Oratore A, Duez C, Joris B, Frère J-M. Cloning and nucleotide sequencing of the gene encoding the β-lactamase from Citrobacter diversus. FEMS Microbiol Lett. 1991;67:79–84. doi: 10.1016/0378-1097(91)90448-j. [DOI] [PubMed] [Google Scholar]
- 23.Poirel L, Guibert M, Girlich D, Naas T, Nordmann P. Cloning, sequence analyses, expression, and distribution of ampC-ampR from Morganella morganii clinical isolates. Antimicrob Agents Chemother. 1999;43:769–776. doi: 10.1128/aac.43.4.769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Poirel L, Le Thomas I, Naas T, Karim A, Nordmann P. Biochemical sequence analyses of GES-1, a novel class A extended-spectrum β-lactamase, and the class 1 integron In52 from Klebsiella pneumoniae. Antimicrob Agents Chemother. 2000;44:622–632. doi: 10.1128/aac.44.3.622-632.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 26.Stock I, Grüger T, Wiedemann B. Natural antibiotic susceptibility of Rahnella aquatilis and R. aquatilis-related strains. J Chemother. 2000;12:30–39. doi: 10.1179/joc.2000.12.1.30. [DOI] [PubMed] [Google Scholar]
- 27.Tamaki M, Nukaga M, Sawai T. Replacement of serine 237 in class A β-lactamase of Proteus vulgaris modifies its unique substrate specificity. Biochemistry. 1994;33:10200–10206. doi: 10.1021/bi00199a049. [DOI] [PubMed] [Google Scholar]
- 28.Tzouvelekis L S, Tzelepi E, Tassios P T, Legakis N J. CTX-M-type β-lactamases: an emerging group of extended-spectrum enzymes. Int J Antimicrob Agents. 2000;14:137–142. doi: 10.1016/s0924-8579(99)00165-x. [DOI] [PubMed] [Google Scholar]