Abstract
According to the EPOC study, chemotherapy could improve 5-year disease-free survival of stage IV colon cancer patients by 8.1%. However, more molecular biomarkers are required to identify patients who need neoadjuvant chemotherapy. DENND2D expression was evaluated by immunohistochemistry in 181 stage IV colon cancer patients. The prognosis was better for patients with DENND2D expression than patients without DENND2D expression (5-year overall survival [OS]: 42% vs. 12%, p = 0.038; 5-year disease-free survival: 20% vs. 10%, p = 0.001). Subgroup analysis of the DENND2D-negative group showed that patients treated with neoadjuvant chemotherapy achieved longer OS than patients without neoadjuvant chemotherapy (RR = 0.179; 95% CI = 0.054–0.598; p = 0.003). DENND2D suppressed CRC proliferation in vitro and in vivo. Downregulation of DENND2D also promoted metastasis to distant organs in vivo. Mechanistically, DENND2D suppressed the MAPK pathway in CRC. Colon cancer patients who were DENND2D negative always showed a worse prognosis and were more likely to benefit from neoadjuvant chemotherapy. DENND2D may be a new prognostic factor and a predictor of the need for neoadjuvant chemotherapy in stage IV colon cancer.
Subject terms: Colon cancer, Tumour biomarkers
Introduction
Colon cancer is the leading cause of cancer-related mortality worldwide [1]. Surgical resection with or without adjuvant chemotherapy remains the standard treatment for nonmetastatic colorectal cancer (CRC) [2]. The survival of nonmetastatic CRC patients has improved with the development of multiple therapeutic strategies, including surgery, chemotherapy, and radiotherapy. However, improving the outcome of stage IV CRC remains a challenge for oncologists. Approximately 50–60% of patients diagnosed with CRC develop distant metastases [3]. Based on the report that almost half of colon cancer patients have liver metastasis at autopsy, liver metastases are the leading cause of death in CRC patients [4].
The median 5-year survival rate of colon cancer patients with liver metastasis (CCLM) could reach 38% if R0 resection could be performed [5]. The 5-year overall survival (OS) was as high as 71% following surgical resection for patients with a single lesion of liver metastasis [6–8]. According to the results of the EPOC study, systemic chemotherapy could improve disease-free survival (DFS) by 8.1% in CRC patients with liver metastasis, and chemotherapy was recommended for all patients with liver metastasis [9]. Whether CCLM should receive neoadjuvant chemotherapy remains debatable. CCLM patients with a poor prognosis could benefit more from neoadjuvant chemotherapy because micrometastatic disease could be treated earlier, and palliative surgery could be avoided for those with early disease progression [10]. However, no standard approach is available for these groups of patients. Especially for initially resectable CCLM patients, there is still controversy whether neoadjuvant chemotherapy should be administered. Most medical centers use the clinical risk score (CRS) to guide clinical practice [11]. For patients with high risk (CRS ≥3), neoadjuvant chemotherapy will be administered before surgical resection [11]. Patients with low risk (CRS ≤2) could receive surgical resection before adjuvant chemotherapy. However, CRS was created almost 20 years ago, and all variables are clinical parameters. Presently, more solid biomarkers are needed to help identify patients who might benefit from neoadjuvant chemotherapy.
Many studies have tried to molecularly characterize tumors [12–14]. Some studies have reported that RAS mutation is a negative prognostic factor for stage IV colon cancer patients [15–18]. Other studies have shown that RAS mutation status has no relationship with survival [18–21]. Therefore, the relationship between RAS mutation and survival remains unclear. BRAF mutation has a strong relationship with worse survival for stage IV colon cancer [21–23]. MSI-H colon cancer is a special subtype of colon cancer comprising a special subgroup of patients who could respond to immunotherapy [24]. Whether MSI could be a prognostic factor for stage IV colon cancer remains debatable [25, 26]. TP53 and PIK3CA gene mutations did not affect long-term outcomes [27, 28]. Many other potential molecular prognostic markers have been described, including POLE, POLD, HER2, NTRK, ALK, and ROS1 [29–32]; however, none are currently incorporated into routine clinical practice.
As a member of the DENND2 family, DENND2D consists of four subregions: the full DENN domain, the upstream DENN, the core DENN, and the downstream DENN. DENN/MADD (DENND) proteins represent a newly recognized class of membrane trafficking proteins that regulate Rab GTPases [33, 34]. DENND proteins play an important role as guanine nucleotide exchange factors for this GTPase and interact with Rab35 [35, 36]. However, the function of DENND2D in malignant tumors is still unclear. Previous studies investigated the function of DENND2D as a tumor-suppressor gene in gastric cancer (GC) [37, 38], lung cancer [39], hepatocellular carcinoma (HCC) [40], and bladder cancer [41]. DENND2D was reported to activate Rab pathways and function in intracellular signaling pathways [33, 34]. To our knowledge, no study has reported the relationship and regulatory mechanisms of DENND2D in CRC. In this study, we investigated the relationship among DENND2D expression, prognosis, and response to neoadjuvant chemotherapy in stage IV colon cancer patients.
Materials and methods
Patients and tissue samples
One hundred eighty-one samples were obtained from patients at Sun Yat-Sen University Cancer Center from May 1, 2003, to May 1, 2016. Informed consent was obtained from all the patients before tissue collection. All patients were diagnosed with initially resectable CRLM and received R0 or R1 surgical resection. All the patients were pathologically confirmed to have colon adenocarcinoma. And all the baseline information of patients was collected blindly, without knowing the expression level of DENND2D. Studies involving human tissue samples were carried out in accordance with the guidelines approved by the Ethics Committees of Sun Yat-Sen University Cancer Center (IRB NO. B2021–192–01).
Cell culture
HCT116, HT29, and SW620 CRC cell lines were purchased from American Type Culture Collection (ATCC, Manassas, VA, USA) and cultured according to ATCC guidelines. HCT116 and HT29 cells were cultivated in McCoy’s 5A medium (KGM4892N; KeyGEN BioTECH), and SW620 cells were cultured in Leibovitz’s L-15 medium (KGM41300N; KeyGEN BioTECH) supplemented with 10% fetal bovine serum (P30–2302, Pan Biotech) and 1% penicillin-streptomycin (P4333; Sigma-Aldrich). The cells were maintained at 37 °C in 5% CO2. All the cell lines were authenticated by short tandem repeat analysis at the China Center for Type Culture Collection (Wuhan, China), and the absence of mycoplasma contamination was verified using a PCR detection kit (Shanghai Biothrive Sci. & Tech. Ltd.). The cells were frozen in liquid nitrogen and used for experiments in passages 3–10.
SiRNA knockdown
Knockdown experiments were performed by transfecting 25 nM DENND2D siRNA (Sigma-Aldrich) into cells with Lipofectamine 3000 (L300075; Invitrogen). The RNA sequences were as follows: DENND2D: si-1, GGATGATTACGAGCCTATAAT; si-2, CCATTATGCTTCCTATATCAA; si-3, TTGGATCCCTGGTATTGATTT. Si-Ctrl is the Mission shRNA nontargeting Pool from Sigma-Aldrich. The cells were transfected with siRNAs for 72 h; siRNA suppression of target protein was validated by western blot analysis.
Generation of stable cell lines
The full-length cDNA of human DENND2D was cloned into the pCDH-EF1α-MCS-T2A-Puro Cloning and Expression Lentivector (CD526A-1; System Bioscience) tagged with 3×Flag. The shRNA sequences targeting DENND2D (sh1 and sh2) and negative control shRNA (shNC) were inserted into the pLko.1 Cloning and Expression Lentivector (SHC001; Sigma-Aldrich), and the sequences were as follows: DENND2D: sh1, CCGGTTGGATCCCTGGTATTGATTTCTCGAGAAATCAATACCAGGGATCCAATTTTTG; sh2, CCGGCCATTATGCTTCCTATATCAACTCGAGTTGATATAGGAAGCATAATGGTTTTTTG. HEK293T cells were transfected using Lipofectamine 3000 with pSPAX2, pMD2. G and these recombinant plasmids. Stable overexpression (-vec, -DENND2D) or knockdown (-shNC, -sh1/2) cell lines were used after selection with puromycin (1–2 µg/ml; BS080A; Biosharp) for 7 days and verification by western blot analysis.
Western blotting (WB)
Cells were lysed in lysis buffer (9803; Cell Signaling Technology) supplemented with a protease and phosphatase inhibitor cocktail (78442; Thermo Fisher Scientific), and the protein concentration was quantified using a BCA protein assay kit (KGP903; KeyGen Biotech). The protein samples were denatured at 95 °C for 10 min and separated by SDS-PAGE. PageRuler Prestained Protein Ladder (26616) (Thermo Fisher Scientific) was used as a size standard to indicate the molecular weight, and the proteins were transferred onto PVDF membranes (03010040001; Sigma-Aldrich), which were blocked with 5% milk in TBS and incubated overnight at 4 °C with specific antibodies against the following proteins: DENND2N (ab184799; Abcam); GAPDH (D16H11), phosphor-MEK1/2 (Ser221) (166F8), MEK1/2 (L38C12), phosphor-ERK1/2 (Thr202/Tyr204) (#9101), and ERK1/2 (20G11). Subsequently, the membranes were incubated with HRP-conjugated goat anti-rabbit IgG (#7074) or anti-mouse IgG (#7076P2) (CST). The signals were visualized using an enhanced chemiluminescence reagent (WP20005; Thermo Fisher Scientific) for immunoblotting. The proteins were analyzed using the ChemiDoc XRS system (Bio-Rad).
Clonogenic assay
Cells were seeded into six-well plates at a density of 1 × 103 cells per well and cultured for 10–14 days at 37 °C to allow colony formation. The colonies (containing more than 50 cells) were stained with 0.5% crystal violet and counted.
CCK-8 assay
Cell viability was determined using a CCK-8 kit (HY-K0301; MCE). Briefly, cells (2500/well) were cultured in 96-well plates for 72 h. Ten microliter of CCK-8 (Dojindo Laboratories) was added to each well, and the cells were incubated for an additional 3 h at 37 °C. The absorbance was then measured at 450 nm using a scanning multiwell spectrophotometer (Thermo Scientific). Cells were treated with various concentrations of 5FU for 72 h. The 50% inhibitory concentration (IC50) value was calculated.
Quantitative reverse transcription-PCR
Total RNA was isolated from cells and tissues using the TRIzolTM RNA Purification Kit (12183555; Invitrogen). cDNA was obtained by reverse transcription using the M-MLV Kit (M1705; Promega) for qRT-PCR according to the manufacturer’s instructions. iTaq SYBR Green Mix (720001564; Bio-Rad) was used for qRT-PCR gene expression analysis, which was performed on the Bio-Rad CFX platform. The relative DENND2D mRNA levels were normalized to the actin mRNA levels. The primer sequences were as follows:
DENND2D
forward: 5’-CACTGCTCTACCCCTTCAGC-3’;
reverse: 5’-TTTTTCATCACCAACCGACA-3’.
Actin
Forward: 5’-CGTGAAAAGATGACCCAGATCA-3’;
Reverse: 5’-CACAGCCTGGATGGCTACGT-3’.
Nude mouse xenograft model, liver metastasis model, and in vivo analysis
The animal experiments were approved by the SYSUCC Institutional Animal Care and Usage Committee in accordance with the Animal Welfare and Rights in China. Female BALB/c nude mice (4–5 weeks; 15–18 g; SLRC Laboratory Animal Co., Shanghai, China) were divided into five groups with five mice each randomly and used to generate xenograft models by injecting HCT116 cells (shNC; DENND2D-knockdown-sh1/sh2 or Vector; DENND2D-overexpression) into the right flanks of the mice. Starting on the 7th day post transplantation, mouse xenografts were monitored every 3 days for tumor formation, and mice were sacrificed when the largest diameter reached 20 mm, and the tumors were resected and weighed. The tumor volume was calculated using the following formula: tumor volume = 0.52 × width2 × length. To study liver metastasis, we established a mouse liver metastasis model by injecting stably modified HCT116 cell lines into the spleen. We injected 5 × 106 cells from each clone into the spleens of BALB/c nude mice. Three weeks later, all mice were sacrificed to examine the livers for metastases [42].
Tissue microarrays and immunohistochemistry
We used a tissue array instrument (personal tissue arrayer, Beeche, USA) to convert the paraffin-embedded specimens of individual tumor and normal mucosa into construction [43]. Immunohistochemistry (IHC) was performed as previously reported [40]. Briefly, paraffin-embedded specimens were serially cut into three 4-μm-thick sections. One section was used for routine hematoxylin and eosin staining, while the other two sections were used for staining using the streptavidin peroxidase (SP) IHC method. The procedures were performed according to the manufacturer’s instructions for each reagent kit. After deparaffinization and rehydration, sections were washed three times in PBS and boiled in a high-pressure cooker for 2.5 min in EDTA buffer (pH 8.0) for antigen retrieval. Non-specific binding was blocked using 5% BSA, after which the sections were consecutively incubated with the primary antibody, secondary antibody, and enzyme-labeled SP. Finally, the sections were developed using 3,3’-diaminobenzidine and counterstained with hematoxylin. The stained sections were cleared, mounted, and examined under a microscope.
The primary antibody solution consisted of a rabbit anti-human DENND2D polyclonal antibody (1:200 dilution; HPA048642; Sigma-Aldrich, USA), p-ERK (1:200 dilution, #4370, Cell Signaling Technology, Inc.) in blocking buffer, or ki67 (Working Concentration; ZM-0167, Zhongshan Golden Bridge Bio-technology, Beijing, China), and it was incubated with the sections at 4 °C overnight in a humidified chamber. Blocking buffer without the primary antibody was used as a negative control.
Each slide was evaluated using the IHC scoring system used in our previous study [44]. If the conclusions of the two pathologists differed, a third pathologist independently evaluated each case and decided the final score.
Gene expression data and statistical analyses
Gene expression data of each cancer were downloaded from the TCGA data portal (https://portal.gdc.cancer.gov/) (Supplementary Table 1). A total of 20,531 genes (protein coding and noncoding) were included in the TCGA Illumina HiSeq RNASeq V2 data. We used level 3 gene expression data, which were derived from the reads per kilobase of transcript per million reads mapped (RPKM). The gene expression values were logarithmically transformed (base 2) prior to further analysis. Gene expression was visualized with box plots by the R package ggplot2, version 3.3.5.
The clinical and follow-up data were analyzed using SPSS v19.0 and R language. χ2, continuity correction χ2, and Fisher’s exact tests were used to assess the patients’ baseline variables. The significance of the variables was tested using Kaplan–Meier, multivariate Cox regression, and logistic regression models. The variation of each group had been estimated properly and met the basic assumptions of the tests. OS was defined as the interval between surgical resection and death or the end of follow-up. DFS was defined as the interval between surgical resection and recurrence, metastasis, or the end of follow-up. Values of p < 0.05 indicated significant differences. All data in our study have been recorded at Sun Yat-Sen University Cancer Center for future reference (Number RDDB2021001606).
Results
DENND2D expression in TCGA database and TNMplot web tool
Different gene expression data of each cancer were downloaded from TCGA data portal (https://portal.gdc.cancer.gov/) (Supplementary Table). A total of 20,531 genes (protein coding and noncoding) were included in the TCGA Illumina HiSeq RNASeq V2 data (Supplementary Table). DENND2D was expressed at higher levels in normal tissue than in the tumor tissue in colon cancer (p = 0.003). Next, we confirmed our results using the TNMplot web tool [45] (Fig. 1B).
Fig. 1. DENND2D is more highly expressed in adjacent normal tissues than colorectal cancer tissues, confirmed by online database and our clinical samples.
A Expression level of DENND2D across 31 TCGA cancer types, in comparison with their normal controls if available. The middle line in the box is the median, the bottom and top of the box are the first and third quartiles, the whiskers extend to 1.5 IQR of the lower quartile and the upper quartile respectively, and the black solid circles represent outliers. P values were derived from two-sided Wilcoxon’s rank-sum test. B DENND2D gene expression among normal, tumor, and metastatic tissues in colorectal cancer patients obtained from the TNMplot.com web tool. C Protein expression of DENND2D in colon cancer tissues from 181 stage IV CRC patients. The level of DENND2D was classified as negative, weak, moderate, and strong. Protein expression of DENND2D in normal tissue (×40 and ×200). D Protein expression of DENND2D in normal tissues. E, F Western blot analysis of six pairs of tumor tissues and normal tissues from six colon cancer patients.
General characteristics of the patients
From May 1, 2003, to May 1, 2016, 181 patients who were pathologically confirmed to have stage IV colon cancer were recruited from the Sun Yat-Sen University Cancer Center. All patients were initially deemed resectable as determined by two hepatic surgeons. In the end, 123 patients (67.9%) received R0 resection, and 39 patients (32.1%) received R1 resection. Among all the patients in this study, samples from 150 patients (82.1%) were DENND2D positive, and 31 (17.9%) were DENND2D negative. Forty-eight patients (26.5%) received neoadjuvant chemotherapy and 133 patients (73.5%) did not.
DENND2D expression in colorectal cancer tissue and normal tissue
Compared with normal tissue, DENND2D expression was lower (Fig. 1A). DENND2D expression was examined in the tissues of 181 colon cancer patients and in normal tissues by IHC (Fig. 1C, D). The expression level of DENND2D was defined as negative, weak, moderate, and strong. According to the percentage of positive cells, the results were classified as follows: negative (negative was defined as no DENND2D expression) and positive (weak, moderate, and strong expressions were defined as positive). The classification of IHC expression levels was performed as previously described [46]. Six pairs of tumor tissue and normal colon tissue from six colon cancer patients were also used to evaluate DENND2D expression by WB (p = 0.01, Fig. 1E, F). We confirmed that DENND2D expression was stronger in normal tissue than in tumor tissue.
DENND2D expression and survival
In total, 181 patients were followed up until December 1, 2020, and were included in the survival analysis. No other difference, in terms of characteristics including sex, age, and tumor location, was detected between the DENND2D-positive and DENND2D-negative groups (more details are provided in Table 1). The median follow-up period was 27 months. At the end of the follow-up time, 9 patients (29.0%) in the DENND2D-negative group were alive, while 69 patients (43.1%) in the DENND2D-positive group were alive.
Table 1.
Clinicopathologic characteristics of stage IV CRC patients.
| Characteristic | DENND2D negative (n = 31) | DENND2D positive (n = 150) | p value |
|---|---|---|---|
| Sex | 0.222 | ||
| Male | 17 (54.8%) | 100 (66.7) | |
| Female | 14 (45.2%) | 50 (33.3%) | |
| Age | 55.55 (50.04–61.06) | 55.93 (53.78–60.66) | 0.888 |
| Tumor location | 0.676 | ||
| Ascending colon | 10 (32.3%) | 43 (28.7%) | |
| Transverse colon | 1 (3.2%) | 6 (4.0%) | |
| Descending colon | 4 (12.9%) | 12 (8.0%) | |
| Sigmoid colon | 9 (29.0%) | 63 (42.0%) | |
| Rectum | 7 (22.6%) | 26 (17.3%) | |
| Surgery resection | 0.526 | ||
| R0 | 23 (74.2%) | 119 (79.3%) | |
| R1 | 8 (25.8%) | 31 (20.7%) | |
| CEA | 9.78 (0.96–2586) | 13.81 (0.35–4935) | 0.181 |
| CA199 | 13.99 (0.60–10,426.40) | 26.77 (0.60–20,000) | 0.176 |
| Surgery procedure | 0.584 | ||
| Right hemicolectomy | 12 (38.7%) | 42 (28.0%) | |
| Transverse colectomy | 0 (0%) | 5 (3.3%) | |
| Left colectomy | 3 (9.7%) | 15 (10.0%) | |
| Sigmoidectomy | 6 (19.4%) | 35 (23.3%) | |
| Anterior resection | 9 (29.0%) | 51 (34.0%) | |
| Abdominal perineal resection | 0 (0%) | 1 (0.7%) | |
| Hartmann | 1 (3.2%) | 1 (0.7%) | |
| Neoadjuvant chemotherapy | 0.728 | ||
| None | 9 (29.0%) | 39 (26.0%) | |
| Yes | 22 (71.0%) | 111 (74.0%) | |
| Neoadjuvant chemotherapy | 0.069 | ||
| None | 9 (29.0%) | 39 (26.0%) | |
| FOLFIRI | 2 (6.5%) | 14 (9.3%) | |
| FOLFOX | 6 (19.4%) | 29 (19.3%) | |
| XELOX | 12 (38.7%) | 58 (38.7%) | |
| XELODA | 0 (0%) | 7 (4.7%) | |
| XELIRI | 1 (3.1%) | 0 (0%) | |
| XELOX + XELODA | 0 (0%) | 2 (1.3%) | |
| 5FU/CF | 1 (3.2%) | 1 (0.7%) |
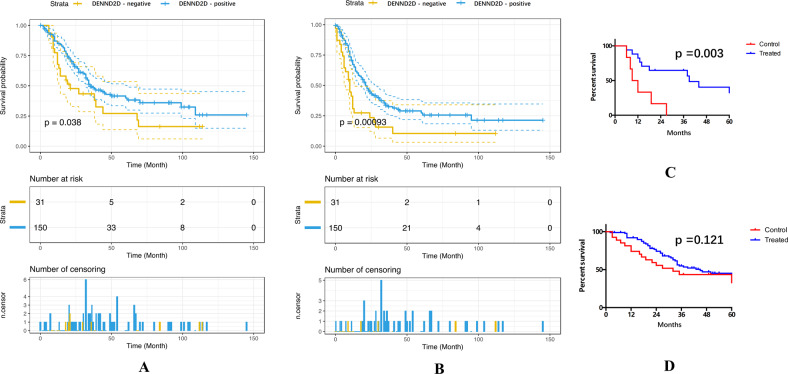
Univariate analysis revealed that DENND2D expression (RR = 0.611; 95% CI: 0.381–0.981; p = 0.038), CEA level (RR = 1.003; 95% CI: 1.000–1.004; p = 0.001), and neoadjuvant chemotherapy (RR = 0.615; 95% CI: 0.403–0.938; p = 0.024), R0 resection (RR = 0.542; 95% CI: 0.335–0.876; p = 0.012), N stage (RR = 0.419; 95% CI: 0.240–0.732; p = 0.001), and number of lymph nodes with metastasis (RR = 0.970; 95% CI: 0.944–0.997; p = 0.032) were strongly related to OS among colon cancer patients (Fig. 2A). In addition, DENND2D expression (RR = 0.493; 95% CI: 0.319–0.761; p = 0.00093), the CEA level (RR = 1.00; 95% CI: 1.000–1.001; p = 0.015), the CA19–9 level (RR = 1.00; 95% CI: 1.000–1.001; p = 0.001), number of lymph nodes with metastasis (RR = 0.967; 95% CI: 0.944–0.991; p = 0.008), and N stage (RR = 0.595; 95% CI: 0.369–0.961; p = 0.034) affected DFS (Fig. 2B). Other variables showed no relationship with the survival of colon cancer patients (additional details are presented in Table 2).
Fig. 2. DENND2D-negative patients with R0 resction benefit more from neoadjuvant chemotherapy.
A Overall survival of 181 stage IV colon cancer patients with 95% confidence intervals. B Disease-free survival of 181 stage IV colon cancer patients with 95% confidence intervals. C Comparison of the survival rate between patients who had received R0 resection with or without neoadjuvant chemotherapy in the DENND2D-negative group. D Comparison of the survival rate between patients who had received R0 resection with or without neoadjuvant chemotherapy in the DENND2D-positive group.
Table 2.
Univariate and multivariate analyses of prognostic factors for disease-free survival and overall survival in 181 stage IV colorectal cancer patients.
| DFS | OS | |||||||
|---|---|---|---|---|---|---|---|---|
| Univariate | Multivariate | Univariate | Multivariate | |||||
| Variable | RR (95% CI) | p | RR (95% CI) | p | RR (95% CI) | p | RR (95% CI) | p |
| Sex | 0.790 (0.549–1.138) | 0.206 | 0.843 (0.564–1.262) | 0.408 | ||||
| Age | 0.998 (0.985–1.010) | 0.723 | 1.003 (0.988–1.017) | 0.713 | ||||
| T stage | 0.912 (0.656–1.266) | 0.580 | 1.415 (0.194–10.346) | 0.732 | ||||
| N stage | 0.595 (0.369–0.961) | 0.034 | 0.448 (0.269–0.747) | 0.002 | 0.419 (0.240–0.732) | 0.001 | 0.291 (0.160–0.528) | 0.001 |
| LN | 0.967 (0.944–0.991) | 0.008 | 0.970 (0.944–0.997) | 0.032 | ||||
| Ki67 | 1.005 (0.994–1.015) | 0.379 | 1.005 (0.099–1.017) | 0.369 | ||||
| Positive LN | 1.029 (0.981–1.079) | 0.241 | 1.031 (0.979–1.085) | 0.247 | ||||
| R0 resection | 0.766 (0.494–1.188) | 0.233 | 0.542 (0.335–0.876) | 0.012 | ||||
| Tumor size | 0.932 (0.833–1.043) | 0.216 | 0.919 (0.806–1.049) | 0.210 | ||||
| Pathology | 1.249 (1.009–1.545) | 0.051 | 4.782 (1.293–17.68) | 0.019 | 0.713 (0.224–2.268) | 0.566 | 5.499 (0.582–51.953) | 0.001 |
| DENND2D expression | 0.493 (0.319–0.761) | 0.001 | 0.353 (0.210–0.595) | 0.001 | 0.611 (0.381–0.981) | 0.041 | 0.327 (0.207–0.517) | 0.001 |
| CEA | 1.00 (1.000–1.001) | 0.015 | 1.003 (1.000–1.004) | 0.001 | ||||
| CA 19–9 | 1.00 (1.000–1.001) | 0.001 | 1.000 (1.000–1.001) | 0.001 | ||||
| Neoadjuvant chemotherapy | 0.933 (0.627–1.387) | 0.730 | 0.615 (0.403–0.938) | 0.024 | ||||
| Tumor location | 0.935 (0.835–1.049) | 0.251 | 1.399 (0.798–2.453) | 0.241 | ||||
Bold values indicate statistically significant p values.
Cox multivariate analysis revealed that the significant prognostic factors for DFS were DENND2D expression (RR = 0.353; 95% CI = 0.210–0.595; p = 0.001), N stage (RR = 0.448; 95% CI = 0.269–0.747; p = 0.002) and pathology types (RR = 4.782; 95% CI = 1.293–17.68; p = 0.019). DENND2D expression (RR = 0.327; 95% CI = 0.207–0.517; p = 0.001), pathology types (RR = 5.499; 95% CI = 0.582–51.953; p = 0.001), N stage (RR = 0.291; 95% CI = 0.160–0.528; p = 0.001) were significant prognostic factors for OS (additional details are presented in Table 2).
DENND2D was associated with the efficacy of neoadjuvant chemotherapy in the R0 resection group
Kaplan–Meier analysis was used to examine the relationship between neoadjuvant chemotherapy, DENND2D expression, and long-term survival in 123 patients who had received R0 surgical resection. In the DENND2D-negative group (including 23 patients), the patients who had received neoadjuvant chemotherapy achieved a longer median survival time than those without neoadjuvant chemotherapy (RR = 0.179; 95% CI = 0.054–0.598; p = 0.003; Fig. 2C). Regarding DENND2D-positive patients, neoadjuvant chemotherapy did not improve long-term survival significantly compared with those without neoadjuvant chemotherapy (RR = 0.659; 95% CI = 0.093–4.262; p = 0.121; Fig. 2D). For the patients who only received R1 resection, neoadjuvant therapy did not improve long-term survival regardless of whether DENND2D expression was positive or negative. The most significant difference was found in the DENND2D-positive group with neoadjuvant chemotherapy and the DENND2D-negative group without neoadjuvant chemotherapy (Supplementary Fig. 1).
DENND2D suppressed CRC cell proliferation and metastasis in vitro and in vivo
Compared with normal tissue, DENND2D was expressed at a lower level in CRC tissues, and patients with lower DENND2D expression were more likely to show a poor prognosis than those with higher DENND2D expression. To determine whether DENND2D is a tumor-suppressor gene for CRC, we investigated the relationship among DENND2D expression, CRC tumorigenesis, and progression. First, we knocked down DENND2D expression in the colon cancer cell lines HCT116, HT29, and SW480 (Fig. 3A) by siRNA and confirmed the results by western blot assays (Fig. 3B). To explore the function of DENND2D in CRC cell lines, colony formation, CCK-8, and migration assays were performed. DENND2D knockdown significantly promoted CRC cell proliferation and migration (Fig. 3C–F, G, I). According to the DENND2D expression level in common CRC cell lines (Supplementary Fig. 2A), we used shRNA to knock down DENND2D expression and established stable colon cell lines, which were confirmed by qPCR, WB, and IHC (Fig. 3J, K). Next, CCK-8, colony formation, and migration assays were performed. DENND2D knockdown significantly promoted CRC cell proliferation and migration (Fig. 3L–N). The same results were also obtained in the HT29 cell line (Supplementary Fig. 2B–F). We also overexpressed DENND2D in the HCT116 cell line, which was confirmed at both the transcriptional and protein levels (Fig. 3O, P). The CCK-8, colony formation, and migration assay results showed that DENND2D overexpression significantly suppressed CRC cell proliferation and migration (Fig. 3Q–S). The same results were also obtained in the RKO cell line (Supplementary Fig. 3A–E). By calculating the 50% inhibitory concentration (IC50) value, we noticed that HCT116 cells were less sensitive to 5FU after DENND2D knockdown, and DENND2D-overexpressing HCT116 cells were more sensitive to 5FU (Fig. 3T, U). In addition, the results were confirmed at both levels in HT29 and RKO cell lines (Supplementary Figs. 2G and 3F). Next, a xenograft model was used to test whether DENND2D could influence the growth of CRC in nude mice. DENND2D-silenced or DENND2D-overexpressing HCT116 cells were subcutaneously injected into the flanks of nude mice to establish a xenograft model (5 mice in each group). The volumes of xenografts derived from HCT116-sh1/sh2 cells were significantly larger than those from HCT116-shNC cells (Fig. 4A–C), whereas DENND2D overexpression significantly suppressed tumor growth (Fig. 4D–F). IHC of animal tumor samples confirmed that DENND2D was knocked down or overexpressed in stable cell lines (Fig. 4G, H). We established a mouse liver metastasis model by injecting HCT116 cell lines stably expressing shRNA or overexpressing DENND2D into the spleen. DENND2D-silenced HCT116 cells produced more metastatic lesions in the liver, and the DENND2D-overexpressing HCT116 group had fewer liver metastatic lesions (Fig. 4I–K). Collectively, these findings indicate that DENND2D suppresses CRC cell proliferation and metastasis in vitro and in vivo, regulating CRC tumorigenesis.
Fig. 3. DENND2D suppressed tumor proliferation, migration and colony formation in vitro.
A, B DENND2D was knockdown by siRNA in HCT116, HT29 and SW620 confirmed by qPCR and western blot. C, D CRC cell proliferation was promoted by DENND2D knockdown. E, F CRC cell migration was promoted by DENND2D knockdown. G–I CRC cell proliferation was promoted by DENND2D knockdown. J, K DENND2D was knocked down by shRNA in HCT116 confirmed by qPCR, western blot, and IHC. L, M CRC cell proliferation was promoted by DENND2D knockdown. N CRC cell migration was promoted by DENND2D knockdown. O, P DENND2D was overexpressed in CRC cells by shRNA confirmed by qPCR, western blot, and IHC. Q, R CRC cell proliferation was suppressed by DENND2D overexpression. S CRC cell migration was suppressed by DENND2D overexpression. T, U CRC cells are more sensitive to 5FU after DENND2D knockdown.
Fig. 4. DENND2D suppressed CRC cell proliferation and metastasis in vivo.
A–C Volumes of xenografts generated from HCT116-sh1/sh2 and HCT-shNC cells. D–F The volumes of xenografts generated from HCT116 and HCT overexpressing cells. G, H DENND2D and Ki67 expression in xenografts generated from HCT116-sh1/sh2, HCT-shNC, HCT116, and DENND2D-overexpressing cells. I–K Liver metastasis model generated by spleen injection of HCT116-sh2, HCT-shNC, HCT116, and DENND2D-overexpressing cells.
DENND2D functions by suppressing the MAPK pathway in CRC tumorigenesis
To elucidate the molecular mechanisms by which DENND2D contributes to CRC tumorigenesis, we examined the expression of different possible pathway-related proteins between shNC HCT116 and sh2 HCT116 cells to identify possible signaling pathways involved. MEK was significantly upregulated after DENND2D was knocked down (Fig. 5A). We also confirmed the correlation of five related genes in the TCGA database (Fig. 5B). Therefore, we hypothesized that DENND2D functions by suppressing the MAPK pathway. WB assays revealed that DENND2D knockdown increased the levels of p-MEK1/2 (Ser221) and p-ERK1/2 (Thr202/Tyr204), whereas DENND2D overexpression resulted in decreased levels of p-MEK1/2 and p-ERK1/2 in HCT116 cells (Fig. 5C). We also confirmed our results in animal tumor samples. IHC demonstrated that DENND2D overexpression decreased the levels of p-ERK1/2 (Thr202/Tyr204), whereas DENND2D knockdown resulted in increased levels of p-ERK1/2 in HCT116 cells (Fig. 5D, E). To confirm the results, we restored DENND2D expression in DENND2D-silenced CRC cells by transiently transfecting a DENND2D expression plasmid and found that the re-expression of DENND2D decreased the activity of the MAPK pathway (Fig. 5F–H). In summary, DENND2D functions by suppressing the MAPK pathway in CRC tumorigenesis.
Fig. 5. DENND2D suppressed CRC cell proliferation and metastasis in vitro by MAPK pathway.
A Differential pathway-related protein expression between shNC and sh2 DENND2D cell lines. B The relationship between six key molecules using Pearson correlation analysis of the data from the TCGA database. C DENND2D overexpression or knockdown changed the levels of p-MET1/2 (Ser221) and p-ERK1/2 (Thr202/Tyr204) in HCT116 cells, as determined by WB assay. D, E DENND2D and pERK were expressed in xenografts generated from HCT116-sh1/sh2, HCT-shNC, HCT116, and DENND2D-overexpressing cells. F–H Restoring DENND2D expression in DENND2D-silenced CRC cells rescued the activity of the MAPK pathway.
Discussion
DENND2D, a regulator of Rab GTPases, is a member of the DENND2 family [36]. DENND2D plays a crucial role in cancer proliferation and metastasis. However, little is known concerning the relationship between DENND2D expression and colon cancer. In a non-small cell lung cancer cell line, DENND2D was identified as a tumor-suppressor gene. The same phenomenon was also observed in HCC [40], esophageal squamous cell carcinoma [47], and GC [48]. In our study, DENND2D acted as a tumor-suppressor gene. DENND2D expression was downregulated in CRC tissues compared with that in normal tissues in colon cancer samples from six stage IV patients. By knocking down DENND2D expression in HCT116, HT29, and SW480 cells, CRC cell proliferation was significantly increased. The same phenomenon was observed in CRC xenografts in a nude mouse model, where we subcutaneously injected DENND2D-silenced or DENND2D-overexpressing HCT116 cells into the flanks of nude mice. A total of 181 stage IV colon cancer patients who had undergone surgical resection were enrolled in our study, and DENND2D-positive patients had achieved better DFS and OS than DENND2D-negative patients (DENND2D-positive group vs. DENND2D-negative group; OS: HR = 0.611, 95% CI = 0.381–0.981, p = 0.038; DFS: HR = 0.493, 95% CI = 0.319–0.761; p = 0.001). By comparing HCT116 cells expressing different levels of DENND2D mRNA by overexpression and shRNA-mediated knockdown, we observed that DENND2D suppressed the MAPK pathway. Our findings may have uncovered a novel regulatory mechanism for the DENND2D-mediated MAPK pathway in CRC.
Whether resectable CCLM should receive neoadjuvant chemotherapy remains debatable. CRS is still used in clinical practice; for patients with low risk (CRS ≤2), surgical resection is recommended. However, even low-risk patients still face a high risk of distant metastasis. In our study, of the 181 patients, 103 (56.9%) died, and 78 (43.15%) were still alive. In addition, 78 (43.1%) did not receive neoadjuvant chemotherapy, and 103 (56.8%) received neoadjuvant chemotherapy. Patients without DENND2D expression could benefit more from neoadjuvant chemotherapy. However, for patients with DENND2D expression, the benefit of neoadjuvant chemotherapy was not significant. Neoadjuvant chemotherapy could improve DENND2D-negative patient survival. DENND2D-negative patients who did not receive neoadjuvant chemotherapy experienced the worst survival.
Many oncologists have tried to identify a biomarker to identify who could benefit more from neoadjuvant chemotherapy, particularly those with low-risk CRS. MSI or MMR has been proposed as a potential biomarker. MSI-H or dMMR status has been associated with a favorable prognosis in stage II colon cancer patients. However, only a few studies have reported the relationship between the MMR status and prognosis in stage IV colon cancer [49]. A study of an Australian registry reported that MSI-H patients experienced a shorter OS than MSS patients [50]. Another study from the Mayo Clinic also reported a similar result: a shorter OS in patients with an MSI-H versus MSS status [50, 51]. However, less than 5% of stage IV colon cancer patients are MSI-H; thus, MMR status is not a good biomarker for most stage IV patients [52]. In a report based on the SEER database, KRAS mutations were found in approximately 23% of all stage IV colon cancers [52]. Of all patients who had been evaluated for KRAS mutation, no survival differences were found between KRAS-mutant patients and KRAS-WT patients [53]. Other studies reported that KRAS mutation is associated with a poor prognosis in stage IV colon cancer [54]. These discrepancies demonstrate that, although we have assumed until recently that KRAS status is a predictor for the use of EGFR inhibitors, more data are needed to support that KRAS mutation is a poor prognosis.
For the past few years, the Immunoscore® has been used to help doctors and oncologists predict the prognosis of CRC patients [55]. However, all previous studies were focused on stage I to III CRC, and whether the Immunoscore® could be used in stage IV colon cancer patients remains unclear [54]. In addition, the Immunoscore® was also used as a predictor for chemotherapy. Chemotherapy is recommended for all stage III CC patients. We still do not have data to show the relationship between stage IV colon cancer and the response to chemotherapy. Another factor that could potentially predict the effect of adjuvant chemotherapy is circulating tumor DNA (ctDNA). Typically, ctDNA constitutes only a small proportion of total circulating free DNA [56]. An increasing number of oncologists believe ctDNA is a reliable tool that can be used as a prognostic factor in the follow-up of CRC patients because assay techniques are improving and providing better sensitivity to detect ctDNA [57]. The data related to stage IV colon cancer remain limited.
Ki-67 antigen expression is one of the most widely used markers to evaluate the proliferation of tumor cells, except for quiescent (G0 and early G1 phases) cells [58]. Whether Ki-67 expression has a relationship with the prognosis of CRC patients is unclear [59, 60]. Some studies have reported that CRC patients with high Ki-67 expression were more likely to show a poor OS [61, 62], whereas several studies have reported that high Ki-67 expression was correlated with a favorable OS [63–65]. Most studies have used a combined model including the expression of Ki-67 and other pathological parameters to predict the prognosis of CRC without distant metastasis. In our study, Ki-67 did not show a relationship with the prognosis in survival analysis. The reason is likely that the value of Ki-67 as a predictor in stage IV colon cancer was not as good as that in nonstage IV CRC.
Our data showed that stage IV colon cancer patients with DENND2D expression had better DFS and OS than those without DENND2D expression. Based on subgroup analysis, we also indicated that stage IV colon cancer patients without DENND2D expression had a worse prognosis and benefited more from neoadjuvant chemotherapy, although these results were not statistically significant. DENND2D-positive patients had a better prognosis and did not seem to benefit from neoadjuvant chemotherapy.
Based on the results of our study, DENND2D could help make decisions for stage IV colon cancer patients regarding whether they should receive neoadjuvant chemotherapy. Nine DENND2D-positive patients and 39 DENND2D-negative patients did not receive chemotherapy. Most patients in both groups (38.7% in both groups) had received XELOX as neoadjuvant chemotherapy. Based on the results of the NEW Epoc study [66], initial resectable CCLM could not benefit from EGFR inhibitor therapy. VEGF inhibitors (bevacizumab) are associated with a risk for major complications [67], and VEGF inhibitors are also not recommended by the NCCN guidelines. In our study, no patient had received targeted therapy as neoadjuvant chemotherapy. The limitations of this study are its retrospective nature and limited number of enrolled patients. More patients are needed to confirm our results. Further study is currently underway.
In summary, stage IV colon cancer patients without DENND2D expression consistently showed a worse prognosis and were more likely to benefit from neoadjuvant chemotherapy. Downregulation of DENND2D promoted CRC cell proliferation and progression in vitro and in vivo by activating the MAPK pathway. DENND2D may not only be a prognostic factor but also a predictor of sensitivity to neoadjuvant chemotherapy for stage IV colon cancer. All biopsy tissue, which was obtained by colonoscopy, should be detected for the expression of DENND2D routinely for every stage IV patient. Based on these findings, neoadjuvant chemotherapy should be strongly recommended for DENND2D-negative CCLM, even if they can achieve R0 resection for the first time. For those with DENND2D-positive expression, surgical resection could be the first choice.
Supplementary information
Acknowledgements
We would like to thank Dr Di Cao, Miao-qing Wu, and Shanshan Sun for collecting the clinical samples.
Author contributions
All authors helped to perform the study. W-JM and YC wrote the manuscript and performed the procedures and data analysis. J-HP, CT, LZ, and ML performed the experiments and data analysis. Z-ZP wrote the manuscript and analyzed the data. GC collected the data and designed the study. HNX, SH, and HT analyzed the data and designed the study. Z-GZ wrote the manuscript and conceived the study. R-XZ designed the study and analyzed the data.
Funding
The work was supported by grants from the National Natural Science Foundation of China (Grant No. 82002628), Natural Science Foundation of Guangdong Province (Grant No. 2021A1515010096, 2018A030313426), China Postdoctoral Science Foundation (Grant No. 2019M660227), Chinese Society of Clinical Oncology Foundation (Grant Nos. Y-HR2018–319, Y-L2017–002, and Y-JS2019–009), Sun Yat-Sen University Basic Research Fund (Grant No. 19ykpy180), and open research funds from the Sixth Affiliated Hospital of Guangzhou Medical University, Qingyuan People’s Hospital (202011–103).
Data availability
The online datasets analyzed during the current study are available in the TCGA repository (https://portal.gdc.cancer.gov/) and other experimental and clinical data are included in this article.
Competing interests
The authors declare no competing interests.
Ethics approval
This study was carried out according to the guidelines of the Declaration of Helsinki and approved guidelines by the Ethics Committees of Sun Yat-Sen University Cancer Center.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Wen-juan Ma, Yukun Chen, Jian-hong Peng, Chaoming Tang.
Contributor Information
Gong Chen, Email: chengong@sysucc.org.cn.
Zhong-guo Zhou, Email: zhouzhg@sysucc.org.cn.
Rong-xin Zhang, Email: zhangrx@sysucc.org.cn.
Supplementary information
The online version contains supplementary material available at 10.1038/s41419-022-04885-8.
References
- 1.Siegel RL, Miller KD, Fedewa SA, Ahnen DJ, Meester RG, Barzi A, et al. Colorectal cancer statistics, 2017. CA Cancer J Clin. 2017;67:177–93. doi: 10.3322/caac.21395. [DOI] [PubMed] [Google Scholar]
- 2.Sobon L, Wittekind C. UICC TNM classification of malignant tumors. New York: Wiley; 2002.
- 3.Van Cutsem E, Nordlinger B, Adam R, Köhne C-H, Pozzo C, Poston G, et al. Towards a pan-European consensus on the treatment of patients with colorectal liver metastases. Eur J Cancer. 2006;42:2212–21. doi: 10.1016/j.ejca.2006.04.012. [DOI] [PubMed] [Google Scholar]
- 4.Foster JH. Treatment of metastatic disease of the liver: a skeptic's view. Semin Liver Dis. 1984;4:170–9. [DOI] [PubMed]
- 5.Kanas GP, Taylor A, Primrose JN, Langeberg WJ, Kelsh MA, Mowat FS, et al. Survival after liver resection in metastatic colorectal cancer: review and meta-analysis of prognostic factors. Clin Epidemiol. 2012;4:283. doi: 10.2147/CLEP.S34285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Aloia TA, Vauthey J-N, Loyer EM, Ribero D, Pawlik TM, Wei SH, et al. Solitary colorectal liver metastasis: resection determines outcome. Arch Surg. 2006;141:460–7. doi: 10.1001/archsurg.141.5.460. [DOI] [PubMed] [Google Scholar]
- 7.Hur H, Ko YT, Min BS, Kim KS, Choi JS, Sohn SK, et al. Comparative study of resection and radiofrequency ablation in the treatment of solitary colorectal liver metastases. Am J Surg. 2009;197:728–36. doi: 10.1016/j.amjsurg.2008.04.013. [DOI] [PubMed] [Google Scholar]
- 8.Lee W-S, Yun SH, Chun H-K, Lee WY, Kim S-J, Choi S-H, et al. Clinical outcomes of hepatic resection and radiofrequency ablation in patients with solitary colorectal liver metastasis. J Clin Gastroenterol. 2008;42:945–9. doi: 10.1097/MCG.0b013e318064e752. [DOI] [PubMed] [Google Scholar]
- 9.Nordlinger B, Sorbye H, Glimelius B, Poston GJ, Schlag PM, Rougier P, et al. Perioperative chemotherapy with FOLFOX4 and surgery versus surgery alone for resectable liver metastases from colorectal cancer (EORTC Intergroup trial 40983): a randomised controlled trial. Lancet. 2008;371:1007–16. doi: 10.1016/S0140-6736(08)60455-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Leonard GD, Brenner B, Kemeny NE. Neoadjuvant chemotherapy before liver resection for patients with unresectable liver metastases from colorectal carcinoma. J Clin Oncol. 2005;23:2038–48. doi: 10.1200/JCO.2005.00.349. [DOI] [PubMed] [Google Scholar]
- 11.Fong Y, Fortner J, Sun RL, Brennan MF, Blumgart LH. Clinical score for predicting recurrence after hepatic resection for metastatic colorectal cancer: analysis of 1001 consecutive cases. Ann Surg. 1999;230:309. doi: 10.1097/00000658-199909000-00004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gertler R, Rosenberg R, Stricker D, Friederichs J, Hoos A, Werner M, et al. Telomere length and human telomerase reverse transcriptase expression as markers for progression and prognosis of colorectal carcinoma. J Clin Oncol. 2004;22:1807–14. doi: 10.1200/JCO.2004.09.160. [DOI] [PubMed] [Google Scholar]
- 13.Diep CB, Thorstensen L, Meling GI, Skovlund E, Rognum TO, Lothe RA. Genetic tumor markers with prognostic impact in Dukes’ stages B and C colorectal cancer patients. J Clin Oncol. 2003;21:820–9. doi: 10.1200/JCO.2003.05.190. [DOI] [PubMed] [Google Scholar]
- 14.Allegra CJ, Paik S, Colangelo LH, Parr AL, Kirsch I, Kim G, et al. Prognostic value of thymidylate synthase, Ki-67, and p53 in patients with Dukes’ B and C colon cancer: a National Cancer Institute–National Surgical Adjuvant Breast and Bowel Project Collaborative Study. J Clin Oncol. 2003;21:241–50.. doi: 10.1200/JCO.2003.05.044. [DOI] [PubMed] [Google Scholar]
- 15.Karagkounis G, Torbenson MS, Daniel HD, Azad NS, Diaz LA, Jr, Donehower RC, et al. Incidence and prognostic impact of KRAS and BRAF mutation in patients undergoing liver surgery for colorectal metastases. Cancer. 2013;119:4137–44. doi: 10.1002/cncr.28347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Amikura K, Akagi K, Ogura T, Takahashi A, Sakamoto H. The RAS mutation status predicts survival in patients undergoing hepatic resection for colorectal liver metastases: the results from a genetic analysis of all-RAS. J Surg Oncol. 2018;117:745–55.. doi: 10.1002/jso.24910. [DOI] [PubMed] [Google Scholar]
- 17.Denbo JW, Yamashita S, Passot G, Egger M, Chun YS, Kopetz SE, et al. RAS mutation is associated with decreased survival in patients undergoing repeat hepatectomy for colorectal liver metastases. J Gastrointest Surg. 2017;21:68–77. doi: 10.1007/s11605-016-3189-9. [DOI] [PubMed] [Google Scholar]
- 18.Kemeny NE, Chou JF, Capanu M, Gewirtz AN, Cercek A, Kingham TP, et al. KRAS mutation influences recurrence patterns in patients undergoing hepatic resection of colorectal metastases. Cancer. 2014;120:3965–71. doi: 10.1002/cncr.28954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lin Q, Ye Q, Zhu D, Wei Y, Ren L, Ye L, et al. Determinants of long-term outcome in patients undergoing simultaneous resection of synchronous colorectal liver metastases. PLoS One. 2014;9:e105747. doi: 10.1371/journal.pone.0105747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Russo A, Migliavacca M, Bazan V, Maturi N, Morello V, Dardanoni G, et al. Prognostic significance of proliferative activity, DNA‐ploidy, p53 and Ki-ras point mutations in colorectal liver metastases. Cell Prolif. 1998;31:139–53. doi: 10.1111/j.1365-2184.1998.tb01192.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schirripa M, Bergamo F, Cremolini C, Casagrande M, Lonardi S, Aprile G, et al. BRAF and RAS mutations as prognostic factors in metastatic colorectal cancer patients undergoing liver resection. Br J Cancer. 2015;112:1921–8. doi: 10.1038/bjc.2015.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Teng HW, Huang YC, Lin JK, Chen WS, Lin TC, Jiang JK, et al. BRAF mutation is a prognostic biomarker for colorectal liver metastasectomy. J Surg Oncol. 2012;106:123–9. doi: 10.1002/jso.23063. [DOI] [PubMed] [Google Scholar]
- 23.Umeda Y, Nagasaka T, Mori Y, Sadamori H, Sun D-S, Shinoura S, et al. Poor prognosis of KRAS or BRAF mutant colorectal liver metastasis without microsatellite instability. J Hepatobiliary Pancreat Sci. 2013;20:223–33. doi: 10.1007/s00534-012-0531-9. [DOI] [PubMed] [Google Scholar]
- 24.Jácome AA, Johnson B, editors. More than FOLFOX and FOLFIRI: the management of metastatic colorectal cancer in the era of precision oncology. EMJ Oncol. 2021;9:43–52.
- 25.Uhlig J, Cecchini M, Sheth A, Stein S, Lacy J, Kim HS. Microsatellite instability and KRAS mutation in stage IV colorectal cancer: prevalence, geographic discrepancies, and outcomes from the national cancer database. J Natl Compr Cancer Netw. 2021;19:307–18.. doi: 10.6004/jnccn.2020.7619. [DOI] [PubMed] [Google Scholar]
- 26.Bläker H, Alwers E, Arnold A, Herpel E, Tagscherer KE, Roth W, et al. The association between mutations in BRAF and colorectal cancer–specific survival depends on microsatellite status and tumor stage. Clin Gastroenterol Hepatol. 2019;17:455–62.e6. doi: 10.1016/j.cgh.2018.04.015. [DOI] [PubMed] [Google Scholar]
- 27.Khan ZA, Jonas SK, Feldmann KA, Patel H, Wharton RQ, Tarragona A, et al. P53 mutation and response to hepatic arterial floxuridine in patients with colorectal liver metastases. J Cancer Res Clin Oncol. 2001;127:675–80. doi: 10.1007/s004320100248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Benhattar J, Cerottini JP, Saraga E, Metthez G, Givel JC. p53 mutations as a possible predictor of response to chemotherapy in metastatic colorectal carcinomas. Int J Cancer. 1996;69:190–2. doi: 10.1002/(SICI)1097-0215(19960621)69:3<190::AID-IJC7>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- 29.Domingo E, Freeman-Mills L, Rayner E, Glaire M, Briggs S, Vermeulen L, et al. Somatic POLE proofreading domain mutation, immune response, and prognosis in colorectal cancer: a retrospective, pooled biomarker study. Lancet Gastroenterol Hepatol. 2016;1:207–16.. doi: 10.1016/S2468-1253(16)30014-0. [DOI] [PubMed] [Google Scholar]
- 30.Hino H, Shiomi A, Kusuhara M, Kagawa H, Yamakawa Y, Hatakeyama K, et al. Clinicopathological and mutational analyses of colorectal cancer with mutations in the POLE gene. Cancer Med. 2019;8:4587–97.. doi: 10.1002/cam4.2344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pietrantonio F, Di Nicolantonio F, Schrock AB, Lee J, Tejpar S, Sartore-Bianchi A, et al. ALK, ROS1, and NTRK rearrangements in metastatic colorectal cancer. J Natl Cancer Inst. 2017;109. [DOI] [PubMed]
- 32.Yakirevich E, Resnick MB, Mangray S, Wheeler M, Jackson CL, Lombardo KA, et al. Oncogenic ALK fusion in rare and aggressive subtype of colorectal adenocarcinoma as a potential therapeutic target. Clin Cancer Res. 2016;22:3831–40. doi: 10.1158/1078-0432.CCR-15-3000. [DOI] [PubMed] [Google Scholar]
- 33.Imai A, Ishida M, Fukuda M, Nashida T, Shimomura H. MADD/DENN/Rab3GEP functions as a guanine nucleotide exchange factor for Rab27 during granule exocytosis of rat parotid acinar cells. Arch Biochem Biophys. 2013;536:31–7. doi: 10.1016/j.abb.2013.05.002. [DOI] [PubMed] [Google Scholar]
- 34.Marat AL, Dokainish H, McPherson PS. DENN domain proteins: regulators of Rab GTPases. J Biol Chem. 2011;286:13791–800. doi: 10.1074/jbc.R110.217067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Marat AL, McPherson PS. The connecdenn family, Rab35 guanine nucleotide exchange factors interfacing with the clathrin machinery. J Biol Chem. 2010;285:10627–37. doi: 10.1074/jbc.M109.050930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yoshimura S-i, Gerondopoulos A, Linford A, Rigden DJ, Barr FA. Family-wide characterization of the DENN domain Rab GDP-GTP exchange factors. J Cell Biol. 2010;191:367–81. doi: 10.1083/jcb.201008051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Subramani D, Alahari SK. Integrin-mediated function of Rab GTPases in cancer progression. Mol Cancer. 2010;9:312. doi: 10.1186/1476-4598-9-312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cheng KW, Lahad JP, Gray JW, Mills GB. Emerging role of RAB GTPases in cancer and human disease. Cancer Res. 2005;65:2516–9. doi: 10.1158/0008-5472.CAN-05-0573. [DOI] [PubMed] [Google Scholar]
- 39.Ling B, Zheng H, Fu G, Yuan J, Shi T, Chen S, et al. Suppression of non-small cell lung cancer proliferation and tumorigenicity by DENND2D. Lung Cancer. 2013;79:104–10. doi: 10.1016/j.lungcan.2012.10.012. [DOI] [PubMed] [Google Scholar]
- 40.Kanda M, Nomoto S, Oya H, Takami H, Hibino S, Hishida M, et al. Downregulation of DENND2D by promoter hypermethylation is associated with early recurrence of hepatocellular carcinoma. Int J Oncol. 2014;44:44–52. doi: 10.3892/ijo.2013.2165. [DOI] [PubMed] [Google Scholar]
- 41.He Y, Wu Y, Liu Z, Li B, Jiang N, Xu P, et al. Identification of signature genes associated with invasiveness and the construction of a prognostic model that predicts the overall survival of bladder cancer. Front Genet. 2021;12:694777. doi: 10.3389/fgene.2021.694777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gavert N, Sheffer M, Raveh S, Spaderna S, Shtutman M, Brabletz T, et al. Expression of L1-CAM and ADAM10 in human colon cancer cells induces metastasis. Cancer Res. 2007;67:7703–12. doi: 10.1158/0008-5472.CAN-07-0991. [DOI] [PubMed] [Google Scholar]
- 43.Peng J, Ou Q, Wu X, Zhang R, Zhao Q, Jiang W, et al. Expression of voltage-gated sodium channel Nav1. 5 in non-metastatic colon cancer and its associations with estrogen receptor (ER)-β expression and clinical outcomes. Chin J Cancer. 2017;36:1–10.. doi: 10.1186/s40880-016-0161-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Peng J-H, Fang Y-J, Li C-X, Ou Q-J, Jiang W, Lu S-X, et al. A scoring system based on artificial neural network for predicting 10-year survival in stage II A colon cancer patients after radical surgery. Oncotarget. 2016;7:22939. doi: 10.18632/oncotarget.8217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bartha A, Gyorffy B. TNMplot.com: a web tool for the comparison of gene expression in normal, tumor and metastatic tissues. Int J Mol Sci. 2021;22:2622. [DOI] [PMC free article] [PubMed]
- 46.Zhang R-X, Zhou Z-G, Lu S-X, Lu Z-H, Wan D-S, Pan Z-Z, et al. Pim-3 as a potential predictor of chemoradiotherapy resistance in locally advanced rectal cancer patients. Sci Rep. 2017;7:1–10. doi: 10.1038/s41598-016-0028-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hibino S, Kanda M, Oya H, Takami H, Shimizu D, Nomoto S, et al. Reduced expression of DENND2D through promoter hypermethylation is an adverse prognostic factor in squamous cell carcinoma of the esophagus. Oncol Rep. 2014;31:693–700. doi: 10.3892/or.2013.2901. [DOI] [PubMed] [Google Scholar]
- 48.Kanda M, Shimizu D, Nomoto S, Takami H, Hibino S, Oya H, et al. Prognostic impact of expression and methylation status of DENN/MADD domain-containing protein 2D in gastric cancer. Gastric Cancer. 2015;18:288–96. doi: 10.1007/s10120-014-0372-0. [DOI] [PubMed] [Google Scholar]
- 49.Hutchins G, Southward K, Handley K, Magill L, Beaumont C, Stahlschmidt J, et al. Value of mismatch repair, KRAS, and BRAF mutations in predicting recurrence and benefits from chemotherapy in colorectal cancer. J Clin Oncol. 2011;29:1261–70. doi: 10.1200/JCO.2010.30.1366. [DOI] [PubMed] [Google Scholar]
- 50.Price TJ, Karapetis CS, Joanne Y, Roy A, Padbury R, Maddern G, et al. Outcomes for metastatic colorectal cancer (mCRC) based on microsatellite instability. J Clin Oncol. 2018;36:759.
- 51.Jin Z, Sanhueza CT, Johnson B, Nagorney DM, Larson DW, Mara KC, et al. Outcome of mismatch repair‐deficient metastatic colorectal cancer: The Mayo Clinic Experience. Oncologist. 2018;23:1083. doi: 10.1634/theoncologist.2017-0289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fujiyoshi K, Yamamoto G, Takenoya T, Takahashi A, Arai Y, Yamada M, et al. Metastatic pattern of stage IV colorectal cancer with high-frequency microsatellite instability as a prognostic factor. Anticancer Res. 2017;37:239–47.. doi: 10.21873/anticanres.11313. [DOI] [PubMed] [Google Scholar]
- 53.Charlton ME, Karlitz JJ, Schlichting JA, Chen VW, Lynch CF. Factors associated with guideline-recommended KRAS testing in colorectal cancer patients: a population-based study. Am J Clin Oncol. 2017;40:498. doi: 10.1097/COC.0000000000000191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sinicrope FA, Mahoney MR, Smyrk TC, Thibodeau SN, Warren RS, Bertagnolli MM, et al. Prognostic impact of deficient DNA mismatch repair in patients with stage III colon cancer from a randomized trial of FOLFOX-based adjuvant chemotherapy. J Clin Oncol. 2013;31:3664. doi: 10.1200/JCO.2013.48.9591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Prall F, Dührkop T, Weirich V, Ostwald C, Lenz P, Nizze H, et al. Prognostic role of CD8+ tumor-infiltrating lymphocytes in stage III colorectal cancer with and without microsatellite instability. Hum Pathol. 2004;35:808–16. doi: 10.1016/j.humpath.2004.01.022. [DOI] [PubMed] [Google Scholar]
- 56.Holdhoff M, Schmidt K, Donehower R, Diaz LA. Analysis of circulating tumor DNA to confirm somatic KRAS mutations. J Natl Cancer Inst. 2009;101:1284–5. doi: 10.1093/jnci/djp240. [DOI] [PubMed] [Google Scholar]
- 57.Diehl F, Schmidt K, Choti MA, Romans K, Goodman S, Li M, et al. Circulating mutant DNA to assess tumor dynamics. Nat Med. 2008;14:985–90. doi: 10.1038/nm.1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schlüter C, Duchrow M, Wohlenberg C, Becker M, Key G, Flad H-D, et al. The cell proliferation-associated antigen of antibody Ki-67: a very large, ubiquitous nuclear protein with numerous repeated elements, representing a new kind of cell cycle-maintaining proteins. J Cell Biol. 1993;123:513–22. doi: 10.1083/jcb.123.3.513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Meyer A, Merkel S, Brückl W, Schellerer V, Schildberg C, Campean V, et al. Cdc2 as prognostic marker in stage UICC II colon carcinomas. Eur J Cancer. 2009;45:1466–73. doi: 10.1016/j.ejca.2009.01.010. [DOI] [PubMed] [Google Scholar]
- 60.Shin IY, Sung NY, Lee YS, Kwon TS, Si Y, Lee YS, et al. The expression of multiple proteins as prognostic factors in colorectal cancer: cathepsin D, p53, COX-2, epidermal growth factor receptor, C-erbB-2, and Ki-67. Gut Liver. 2014;8:13. doi: 10.5009/gnl.2014.8.1.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Fernández-Cebrián JM, Nevado Santos M, Vorwald Kuborn P, Pardo de Lama M, Martin-Cavanna J, Pacheco Martínez P, et al. Can the clinical outcome in state II colon carcinomas be predicted by determination of molecular marker expression? Clin Transl Oncol. 2007;9:663–70. doi: 10.1007/s12094-007-0119-z. [DOI] [PubMed] [Google Scholar]
- 62.Furudoi A, Tanaka S, Haruma K, Yoshihara M, Sumii K, Kajiyama G, et al. Clinical significance of human erythrocyte glucose transporter 1 expression at the deepest invasive site of advanced colorectal carcinoma. Oncology. 2001;60:162–9. doi: 10.1159/000055314. [DOI] [PubMed] [Google Scholar]
- 63.Salminen E, Palmu S, Vahlberg T, Roberts PJ, Söderström K-O. Increased proliferation activity measured by immunoreactive Ki67 is associated with survival improvement in rectal/recto sigmoid cancer. World J Gastroenterol. 2005;11:3245. doi: 10.3748/wjg.v11.i21.3245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Xi H-Q, Zhao P. Clinicopathological significance and prognostic value of EphA3 and CD133 expression in colorectal carcinoma. J Clin Pathol. 2011;64:498–503. doi: 10.1136/jcp.2010.087213. [DOI] [PubMed] [Google Scholar]
- 65.Ivanecz A, Kavalar R, Palfy M, Pivec V, Sremec M, Horvat M, et al. Can we improve the clinical risk score? The prognostic value of p53, Ki-67 and thymidylate synthase in patients undergoing radical resection of colorectal liver metastases. HPB. 2014;16:235–42. doi: 10.1111/hpb.12089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Primrose J, Falk S, Finch-Jones M, Valle J, O’Reilly D, Siriwardena A, et al. Systemic chemotherapy with or without cetuximab in patients with resectable colorectal liver metastasis: the New EPOC randomised controlled trial. Lancet Oncol. 2014;15:601–11. doi: 10.1016/S1470-2045(14)70105-6. [DOI] [PubMed] [Google Scholar]
- 67.Nasti G, Piccirillo M, Izzo F, Ottaiano A, Albino V, Delrio P, et al. Neoadjuvant FOLFIRI+ bevacizumab in patients with resectable liver metastases from colorectal cancer: a phase 2 trial. Br J Cancer. 2013;108:1566–70. doi: 10.1038/bjc.2013.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The online datasets analyzed during the current study are available in the TCGA repository (https://portal.gdc.cancer.gov/) and other experimental and clinical data are included in this article.