Abstract
Climate change will have numerous impacts on crop production worldwide necessitating a broadening of the germplasm base required to source and incorporate novel traits. Major variation exists in crop progenitor species for seasonal adaptation, photosynthetic characteristics, and root system architecture. Wheat is crucial for securing future food and nutrition security and its evolutionary history and progenitor diversity offer opportunities to mine favourable functional variation in the primary gene pool. Here we provide a review of the status of characterisation of wheat progenitor variation and the potential to use this knowledge to inform the use of variation in other cereal crops. Although significant knowledge of progenitor variation has been generated, we make recommendations for further work required to systematically characterise underlying genetics and physiological mechanisms and propose steps for effective use in breeding. This will enable targeted exploitation of useful variation, supported by the growing portfolio of genomics and accelerated breeding approaches. The knowledge and approaches generated are also likely to be useful across wider crop improvement.
Subject terms: Natural variation in plants, Plant hybridization
Introduction
Modern crop breeding involving targeted crossing and selection has led to the development of elite, high yielding cultivars. The genetic components of yield have been improved through constant selection for desirable traits, initially in landraces and early varieties and then through trait driven plant breeding (Fradgley et al. 2019). In wheat, the positive impact of this is exemplified by the introduction of semi-dwarfing genes contributing to large increases in yield potential during the so-called Green Revolution (Borlaug 1968). In addition to genetic improvement, agronomic potential is strongly influenced by the environment. Environmental adaptation, through direct breeding and selection, allows for optimisation of yield within the seasonal constraints of a given region (Worland and Snape 2001), control of biotic stresses including pests and diseases (either via crop management or the deployment of disease resistance genes) and targeting of abiotic response, for example to available water (Reynolds et al. 2007), applied fertiliser (Swarbreck et al. 2019) and other production-limiting stresses. The quest to optimise both genetic potential and environmental response for a range of crop production regions around the world is being enhanced by the array of genetic and bioinformatics tools now available (Adamski et al. 2020).
Climate is the driver of environmental change with an impact for crop production capacity (Rosenzweig et al. 2008). Global climate change creates an urgency for the development of cultivars with enhanced resilience to environmental changes in order to secure future food security.
Expanding the wheat gene pool
Hexaploid wheat (Triticum aestivum) arose through a limited number of hybridisation events between a domesticated form of the tetraploid wild emmer wheat, Triticum turgidum ssp dicoccoides (AABB) and Aegilops tauschii (DD) around 10,000 years ago (McFadden and Sears 1946; Cox 1997; Petersen et al. 2006). An intermediate, hulled hexaploid is proposed by Kerber and Rowland (1974) though this is not supported by the archaeological record (Feldman 2001). As bread wheat spread, the crop became adapted to local conditions through selection and the resulting distinct, locally adapted wheats are known as landraces (Camacho Villa et al. 2005; Jones et al. 2012). Landraces of hexaploid wheat have long been used for wheat improvement and are a reservoir of readily available diversity that can be introduced into breeding programmes with relative ease (Wingen et al. 2014). Domestication and subsequent selection have created bottlenecks, reducing genetic diversity in all cultivated wheat species derived from wild emmer wheat including pasta or durum wheat (T. turgidum ssp. durum) and bread wheat (Tanksley and McCouch 1997; Lopes et al. 2015). Some of this diversity may be reintroduced to bread wheat by interrogating progenitor species for functional variation in target traits.
Tetraploid (AABB) wild emmer wheat has a modern-day range that spans the western Fertile Crescent, southeastern Turkey, and the mountainous regions of eastern Iraq and western Iran. Tetraploid wheats related to wild emmer include emmer wheat (T. turgidum ssp. dicoccum), a domesticated tetraploid wheat that was widely cultivated prior to the adoption of hexaploid wheat (Salamini et al. 2002), and durum wheat which is widely cultivated, predominantly within the Mediterranean Rim (Martínez-Moreno et al. 2020). Tetraploid wheats are readily crossable with hexaploid wheat and allelic diversity from tetraploid donors or ‘tetraploid derived alleles’ can be introgressed via direct crossing and backcrossing (Ullah et al. 2018).
The diploid progenitor species Ae. tauschii (DD) is part of the large Aegilops genus (van Slageren 1994) that includes at least 10 diploid and 12 polyploid species (Matsuoka et al. 2015). Many (up to 14; reviewed by Schneider et al. 2008) Aegilops species have been used in wheat crossing programmes although most species in the genus are challenging to introgress due to issues with chromosome pairing. This limitation does not exist with Ae. tauschii that is characterised as the specific wheat D-genome donor (Kihara 1944, McFadden and Sears 1946) and it has been frequently used for introgression into hexaploid wheat because there is little inhibition of meiotic chromosome pairing between D-genome chromosomes (Kishii 2019). The distribution of Ae. tauschii centres on a region to the south of the Caspian Sea and into Azerbaijan. The species range spreads eastward, to Pakistan and western China, via the Kopet Dag Mountains of Turkmenistan, and westward, to central Syria, via the valleys of southeastern Turkey (van Slageren 1994). Although the genus and specific species have a wide geographical range, the genetic diversity of hexaploid wheat’s D-genome is severely limited because of the small number of polyploidisation events that gave rise to it (Giles and Brown 2006). Collections and populations of Ae. tauschii have been used to identify useful genes for specific traits, many of which are disease-related (recently reviewed by Kishii 2019) including resistance genes for foliar pathogens and insect pests (Gaurav et al. 2021).
Whilst direct hexaploid × Ae. tauschii crossing has been documented, Ae. tauschii is predominantly captured via the creation of synthetic allohexaploids made by chromosome doubling of triploid hybrids from an inter-specific AABB × DD cross (also called synthetic wheats or synthetic hexaploid wheats (SHW); Dreisigacker et al. 2008; Mujeeb-Kazi et al. 2013). These can be used to introduce diversity from either or both the tetraploid or diploid donor. Synthetic wheats have been used for breeding to increase diversity (Dreisigacker et al. 2008; Li et al. 2014a), for adaption (Li et al. 2014a), disease resistance (Ogbonnaya et al. 2008) and yield improvement (Jafarzadeh et al. 2016). The creation of octoploid (AABBDDDD) synthetics has also been reported (Chèvre et al. 1989), as have synthetic amphiploids created using introgressions with other wheat species such as Ae. crassa, Ae. cylindrica and Ae. ventricosa (Mirzaghaderi et al. 2020). These however have not typically been used for downstream breeding applications due to the complexities of ploidy, recombination and tracking introgression segments.
Ancestral wheat species such as Triticum urartu (the AA genome donor of bread wheat) and members of the Aegilops tribe including Ae. speltioides (a relative of the BB genome donor) offer a wealth of diversity in agronomically important traits such as disease resistance (Rowland and Kerber 1974). Many of these species do not cross readily with bread wheat due to the presence of Ph1 genes preventing recombination between chromosomes (Sears 1977). Instead, a wheat line carrying a mutant allele of ph1 may be used to induce bread wheat and ancestor homoeologous recombination (Rey et al. 2017). The resulting lines carry large introgressions and development of high-throughput single-nucleotide polymorphism (SNP)-based marker systems designed to screen wild relative species has facilitated rapid validation and tracking of these introgressions (Przewieslik-Allen et al. 2019; King et al. 2017, 2019). Such marker systems are likely to facilitate enhanced and targeted deployment of diversity from wild relatives in breeding programmes in the future.
Climate change is predicted to increase the frequency and intensity of abiotic stress events and their impacts on wheat productivity (Lopes et al. 2015). Here we review the potential for further detailed interrogation of adaptive and physiological variation in wheat’s progenitor species. Work to date has focussed primarily on biotic stresses but there is evidence to support the usefulness of progenitor species for introducing targeted variation for optimising responses to changing climates. Our review demonstrates that there is a gap in the systematic characterisation of progenitor variation specifically for responses to abiotic stress including seasonal adaptation, physiological response, and root system architecture (RSA). Further understanding the genetic and physiological basis of these responses will support future targeted use of progenitor variation for mobilisation into wheat breeding.
Progenitor species provide additional variation for flowering time and adaptive response
If heat or drought stress occurs during grain filling, abortion of tillers and/or lower kernel weight reduces wheat yield (reviewed by Fleury et al. 2010 and Ni et al. 2018). The manipulation of flowering time can shift grain production away from risk periods, thereby providing an escape strategy. Research undertaken in both Arabidopsis and agronomically important grasses (maize, rice and wheat) over the past 20 years has revealed that floral transition is controlled by complex overlapping genetic pathways (reviewed by Cockram et al. 2007; Colasanti and Coneva 2009). Wheat is a long-day species in which floral initiation is accelerated by exposure to lengthening days. Although the underlying genetics of flowering are complex (reviewed by Hyles et al. 2020), manipulation of the major vernalisation and photoperiod response genes are widely used in wheat breeding programmes to provide adaption to agroeconomic environments (Bentley et al. 2011).
Adaption in terms of phenology is a powerful tool, particularly in marginal environments. Since the 1990s, 25% of reported global wheat yield improvement has come from wheat grown in marginal environments due to breeding for wide adaption (Lantican et al. 2005). Marginal environments and the necessity to mitigate climate-based yield impacts are likely to become more prominent under a climate change scenario. Climate change is also likely to have impacts on crop production in temperate and cold regions of the world where flowering is a function of both winter cold and spring heat (Yu et al. 2010). Temperate cereals grow across a wide range of semi-arid environments but show marked reductions in productivity (Reynolds et al. 2010) and yield (Lobell and Field 2007) at high temperatures. Increased temperatures in winter may delay fulfilment of vernalisation requirement (a prolonged period of cold, non-freezing, temperatures required for subsequent competence to flower) resulting in later flowering, although increased spring temperatures could mask or offset this (Yu et al. 2010). In areas of high latitude and altitude the effect could be exacerbated, as plants in these regions are particularly sensitive to temperature cues. Vernalisation in wheat is controlled by the major Vrn-1 locus (Dubcovsky et al. 1998) with the additional Vrn-2 and Vrn-3 loci also contributing to variation (Yoshida et al. 2010). Hexaploid wheat has three homoeologous Vrn-1 loci (denoted -A1, -B1 and -D1) located on group 5 chromosomes. Dominant alleles confer a spring growth habit meaning that a cold period is not required for induction of flowering.
Natural plant populations often have wide flowering time variation (Grazzani et al. 2003) and therefore progenitor species offer potential functional genetic variation for fine-tuning adaptive response. In hexaploid wheat, the photoperiod response Ppd-1 loci are a homoallelic series on group 2 chromosomes (Worland and Snape 2001; Beales et al. 2007; Bentley et al. 2011). In tetraploid (AABB) wheat Wilhelm et al. (2009) described two mutations of the Ppd-A1 gene leading to photoperiod insensitivity (PI) and early flowering. These effects have also been confirmed in hexaploid and SHW (Bentley et al. 2011). However, screening of ancestral tetraploids (T. dicoccoides (n = 122) and T. dicoccum (n = 276)) for these mutant Ppd-A1a alleles revealed no variation, suggesting that these are photoperiod sensitive species, and that insensitivity arose post-domestication, being first observed in T. durum landrace accessions as well as in collections from southern Europe (Italy, Spain, France), North Africa and North America (Bentley et al. 2011).
Diversity in flowering time has been further characterised by several studies in tetraploids wheats (Nishimura et al. 2018; Wright et al. 2020; Würschum et al. 2019). Alleles of the Ppd-A1 associated with early flowering (but distinct from the Ppd-A1a alleles described by Wilhelm et al. 2009) were detected in emmer wheat by Nishimura et al. (2018) who also identified an early heading date QTL associated with Vrn-A3. This QTL was found to be a cis-element GATA box in Vrn-A3 (located on chromosome 7AS), which suppressed the late-flowering (photoperiod sensitive) Ppd-A1b allele (Nishimura et al. 2018). A QTL controlling flowering time was also reported on 7B linked to Vrn-B3 in an emmer mapping population (Wright et al. 2020). Takenaka and Kawahara (2012) identified novel loss of function alleles in tetraploid Ppd-A1 in emmer wheat that do not confer PI but may induce small variations in flowering time.
Compared with work in tetraploid progenitors, little is currently known about the diversity of flowering time response in the diploid wheat progenitor Ae. tauschii. Matsouka et al. (2008) assessed natural flowering time variation in a collection of 200 accessions representing the latitudinal range (30°N–45°N) of the species. Flowering time phenotypes could be divided into early-, intermediate- and late-flowering groups that enabled detection of geographical patterns: with early-flowering lines being dominant in southern regions compared to late-flowering lines in northern regions. However, the impacts of environmental differences varied between the western and eastern parts of the species range preventing a clear attribution of genetic effects (Matsouka et al. 2008).
Range expansion occurs when species adapt beyond native habitats and has been documented for Ae. tauschii associated with shifts in phenology and seed production ability (Matsuoka et al. 2015). Of the species within the Aegilops genus, Ae. tauschii is the only diploid species to have expanded its range east and Matsuoka et al. (2015) suggest that early flowering at least partially explains range expansion into Asia. Further work by Koyama et al. (2018) used a F2-based QTL mapping approach to determine genetic differences between photoperiod sensitive and insensitive lines. This allowed for mapping of a QTL locus on 5DL for heading under short days, proximal to the Vrn-D1 locus, along with three QTLs (one on 4D, two on 7D) for flowering under field conditions. Quantitative variation for vernalisation was also observed in Ae. tauschii accessions (Koyama et al. 2018).
Golovnina et al. (2010) identified spring variants of Ae. tauschii including a recessive Vrn-D1 allele. Vernalisation-insensitive accessions of the species have been previously described in germplasm originating from Pakistan and Afghanistan (Tanaka and Yamashita 1957; Tsunewaki 1966) but there is little evidence for the use of derived alleles in breeding. Takumi et al. (2011) used 211 accessions collected across the Ae. tauschii habitat range to assess flowering in the absence of vernalisation. Sequencing of the Vrn-D1 locus and haplotype analysis revealed distinct variation in Ae. tauschii, including a large deletion leading to a loss of vernalisation requirement (Takumi et al. 2011). The authors however conclude that this deletion is discreet from mutations in Vrn-D1 dominant alleles in hexaploid wheat, indicating that the loss of vernalisation requirement in the progenitor and domesticated forms of wheat occurred separately, but followed a similar mutational event (Takumi et al. 2011). Understanding the vernalisation response and the interactions between Vrn-1 and other genes (e.g., the floral repressors Vrn-2), particularly at high temperatures will be important for future resilience breeding.
Dixon et al. (2019) demonstrate that diverse material can provide variants of many of these genes and that understanding their interactions can potentially facilitate their use for incorporating resilience to temperature fluctuations. Overall, although significant variation has been reported for adaptive response in wheat progenitor species, gaps exist in deployment into breeding. We propose that this is due to two main factors: the lack of resolution available for genetic trait dissection in wild progenitors and the confounding effects of genotype × environment. Many of the alleles or QTLs described from progenitor species to date have not been genetically resolved and many co-locate in forward genetic studies. The availability of sequenced progenitor collections (e.g., Gaurav et al. 2021) is likely to improve the resolution of novel alleles from progenitors, thereby enabling their rapid extraction and validation. This will also likely address the other current limitation in separating the confounding effects of environment and masking effects of interacting loci. Overlapping flowering time pathways introduce functional redundancy, particularly in hexaploid wheat, and they are influenced by multiple environmental factors. Therefore, the priority requirement for extraction of useful functional adaptive trait variation from progenitors is rapid and accurate assaying, extraction and validation of variants to enable quantification of phenotypic effects independent of genetic background and environmental effects.
Novel physiological traits can potentially be mined from progenitor species
Cultivars bred for high yield potential under optimal conditions typically maintain performance in moderately stressful environments (Richards et al. 2002; Foulkes and Reynolds 2015; Voss-Fels et al. 2019). The yield potential of a crop can be simplified to a function of light interception (LI), harvest index (HI) and radiation use efficiency (RUE, Reynolds et al. 2009). Progression in crop breeding has brought HI and LI close to theoretical maximum (Long et al. 2006) indicating that selection for improved RUE may be the most rewarding opportunity for breeders to increase yield potential. RUE is effectively the slope of correlation between dry matter content at harvest and total intercepted radiation (Murchie et al. 2009). Optimising RUE is key to utilising available resources when breeding for variable or resource-limited environments. Thus, enhancing crop canopy photosynthesis is an important breeding target and progenitor species may offer novel physiological variation that can be exploited in breeding.
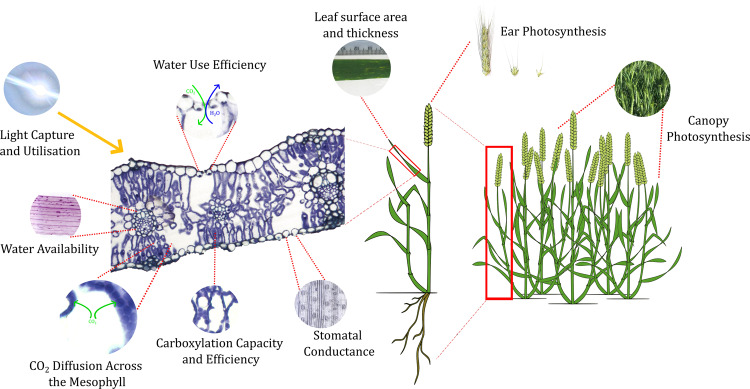
Crop photosynthesis is a complex process, consisting of dynamic networks from the molecular to canopy level (Fig. 1). When considering CO2 assimilation expressed on a standardised leaf area basis (A), there are numerous morphological and biochemical traits underpinning performance. Past experiments have highlighted that wheat progenitors harbour higher A than hexaploid wheat cultivars (Evans and Dunstone 1970; Austin et al. 1982). Since domestication, due to selection programmes for other agronomic traits, there has been limited historic selection pressure from breeders on leaf photosynthetic capacity (Driever et al. 2014). If progenitor diversity can be captured to target a single aspect of the process of photosynthesis giving a moderate increase in flag leaf A of modern wheat then, as the canopy carbon fixation is an integrated process multiplied over the entire growing season there could be consequential overall improvements in RUE and yield (Parry et al. 2011). To harness diversity from wild relatives, components driving high A need to be identified to facilitate their use in targeted genetic dissection, direct use in pre-breeding and future application in wheat breeding using marker- and phenotypic-based screening methods.
Fig. 1. A schematic showing key targets for photosynthetic improvement where diversity from wild relatives could be utilised to increase productivity or stress tolerance in modern wheat.
The flag leaf cross-section highlights important traits underpinning CO2 assimilation on a standardised leaf area basis. When considering photosynthesis on a plant or canopy basis, other targets for improvement include organ size, ear photosynthesis and CO2 assimilation across the whole canopy.
The determinants of A (Fig. 1), and thus potential targets for improvement, include components that govern the rate of delivery of CO2 to the sites of carboxylation; the availability of products from photochemical reactions; and downstream enzyme-regulated mechanisms of the Calvin–Benson cycle. Within these components, superior characteristics found in progenitors can be targeted to improve either photosynthetic productivity or tolerance under environmental stress in modern cultivars (e.g., Merchuk-Ovnat et al. 2016a, b).
The delivery of CO2 to the sites of carboxylation is governed by several diffusive boundaries, particularly those imposed by the leaf stomata. When stomata are closed, water loss is minimal, but the closed pores act as the sole limitation to carbon fixation (Farquhar et al. 1982). Therefore, there is a fundamental trade-off between the flux of CO2 entering the leaf and flux of H2O exiting (Lawson and Blatt 2014). The proportion of CO2 gained in relation to H2O transpired is termed instantaneous water use efficiency (WUE) (Farquhar and Richards 1984). Wheat progenitors have been shown to maintain higher instantaneous WUE in drought-prone conditions compared to hexaploid wheat (Li et al. 2017). Furthermore, Merchuk-Ovnat et al. (2016a) found that introgressions from T. dicocciodes into hexaploid wheat were linked to greater grain yield under drought. Plants originating from drier climates, such as wild relatives, would require increased hydraulic supply to the leaves to maintain photosynthesis under increased evaporative loss (Scoffoni et al. 2016). Austin et al. (1982) found higher stomatal and vein densities in tetraploid wheat flag leaves compared to hexaploid varieties, which could reflect a strategy for maintaining A in drought-prone environments. An alternative strategy could aim to reduce stomal density to minimise water loss and improve drought tolerance (Hughes et al. 2017). As variation in leaf stomatal density and size has been observed across wheat ploidy levels (Dunstone et al. 1973; Khazaei et al. 2009), wild relatives could be a genetic reserve for optimising the balance between CO2 and water loss depending on the targeted breeding environment.
In rice, a distinct group of landraces known as aus-type rice (McNally et al. 2009) evolved and were cultivated under environmental stress conditions in India and Bangladesh. Aus-type rice has been shown to be a valuable source of novel diversity; varieties developed from this material have been shown to be highly tolerant of drought (Henry et al. 2011) and heat stress (Li et al. 2015). A range of physiologies underpins such adaptation including increased rooting depth and lateral root formation resulting in increased water uptake, thus reduced canopy temperature prevented stomatal closure and prolonged photosynthetic activity in the drought-tolerant rice lines (Henry et al. 2011). In combination, heat and drought stress have a negative additive effect on many aspects of wheat plant physiology (reviewed by Tricker et al. 2018), and identifying a suite of tolerance traits pertaining to a fine balance of gas exchange, WUE and assimilation from wild relatives is a breeding target in order to maintain yield under combined stresses.
Another diffusive boundary that acts as a limitation to A is the diffusion of CO2 across the mesophyll (gm, Fig. 1). This boundary is governed by mesophyll anatomical or biochemical features (Evans et al. 2009; Flexas et al. 2012). There has been limited investigation of how gm varies across wheat ploidy levels. The grasses are generally considered to have comparatively high gm (Flexas et al. 2012), which may have decreased through the domestication process, as negative correlations have been observed with gm and potentially desirable traits such as leaf mass area (Gu et al. 2012). Mesophyll cell size is thought to have increased across wheat ploidy, with ancestral species possessing smaller cells (Dunstone and Evans 1974; Wilson et al. 2021). Smaller mesophyll cells may facilitate higher gm due to an increased surface area for gas exchange (Lundgren and Fleming 2020). Further work is required to establish if the comparatively high rates of A found within progenitor species are driven by higher gm.
Improved photochemistry is another trait targeted for improvement (Fig. 1). In a large wheat wild relative comparison using 41 species, McAusland et al. (2020) identified accessions that outperformed modern varieties in traits linked to photochemistry, including T. dicoccoides and lines from the Amblyopyrum and Aegilops genera that demonstrated high Photosystem II (PSII) operating efficiency or electron transport. They hypothesised that high rates of maximum electron transport and carboxylation resulted in high photosynthetic capacity in some wild relative accessions. An introgression from wild emmer into bread wheat has also been linked to improved electron transport rate during booting (Merchuk-Ovnat et al. 2016b). Under high light intensity, not all captured energy is utilised in photochemistry and plants have developed mechanisms for dissipating possibly detrimental excess energy through photoprotection (Demmig-Adams and Adams 1992). This protection process is termed nonphotochemical quenching (NPQ). When the leaf is returned to lower light intensities, the time required for the relaxation of NPQ is a limit to crop productivity (Kromdijk et al. 2016). Improved RUE and photosynthetic efficiency have been achieved through manipulation of the NPQ process through genetic engineering (see: Kromdijk et al. 2016; Hubbart et al. 2018). However, while there has been promising diversity observed in NPQ kinetics in diverse relatives of wheat (McAusland et al. 2020), the degree to which natural variation could be exploited from wild ancestors still needs to be determined.
The determinants, and limitations, imposed at the sites of carboxylation relate to the enzyme-regulated mechanisms of the Calvin–Benson cycle (Fig. 1). Johnson et al. (1987) concluded that a higher capacity for mesophyll photosynthesis may be linked to variation in CO2 assimilation across wheat ploidy. Demand for CO2 is restricted by the carboxylation and oxygenation activities of the enzyme Rubisco (Farquhar et al. 1980), the capacity and efficiency of this enzyme is a major bottleneck in raising wheat yields (Parry et al. 2011). Prins et al. (2016) demonstrated the superior Rubisco catalytic properties of several wheat genotypes (including progenitors) compared to the modern wheat variety Cadenza when assessed across different temperatures. Scafaro et al. (2012) found a wild relative of rice maintained a higher activation state of Rubisco under higher temperatures compared to domesticated rice, which was linked to the high heat tolerance of the wild relative. Progenitors of wheat, originating from warmer climates, may also possess superior Rubisco kinetics, which could be utilised in breeding for marginal environments; this requires further systematic characterisation. Rubisco is also responsible for catalysing the oxygenation of ribulose 1,5-bisphosphate. Photorespiration is the energetically expensive process of converting the by-products of the oxygenation reaction and is a significant constraint on wheat productivity (Long et al. 2006; Parry et al. 2011).
In tobacco, South et al. (2019) showed that genetic engineering of pathways linked to photorespiration produced promising improvements to biomass production and photosynthetic efficiency. Yield penalties linked to photorespiration may lesson under future predicted climates but will still remain important (Walker et al. 2016). While the most promising gains in improving photorespiration losses may be through genetic engineering routes, targeting superior Rubisco characteristics from relatives could hold promise, such as selection for the Rubisco specificity factor that may be higher in plants from drier environments (Galmés et al. 2005). Natural sources of variation in Rubisco kinetics (e.g., Prins et al. 2016) may be a more readily available tool for breeders to utilise in ongoing selection programmes compared to genetic engineering routes.
Targeting photosynthetic improvement should also be considered on both a leaf and canopy basis (Fig. 1). When considering CO2 capture on a per leaf basis, the total organ surface area and thickness are key components. The smaller leaf area typical of progenitor species (Evans and Dunstone 1970) may have more concentrated photosynthetic capacity (Long et al. 2006). McAusland et al. (2020) found that the thicker and narrower leaves found in wild relatives underpinned a higher maximum carboxylation rate, which was also supported by the observed negative relationship between specific leaf area and photosynthetic capacity. Furthermore, leaf surface area and A are typically negatively correlated (Evans and Dunstone 1970; Austin et al. 1982) and a major challenge in utilising photosynthetic diversity from wild relatives will be transferring high A found in progenitor lines into a larger flag leaf typical of modern wheat. This could be addressed by using existing pre-breeding material (derived from progenitors) to screen for genotypes that show a deviation from the negative correlation between leaf area and A. As a first step, segregating pre-breeding material could be used to extract extreme individuals based on leaf area and used either for reciprocal recurrent selection or for bulk segregant analysis to move forwards the genetic understanding of the link between A and leaf area.
Long et al. (2006) outlined that the efficiency of the canopy to intercept light is controlled by canopy characteristics linked to size, architecture, longevity, speed of development and closure. Successful breeding efforts across recent decades have limited opportunities for improvements in canopy LI efficiency (Zhu et al. 2010). Furthermore, canopy architecture has been optimised through domestication (Li et al. 2014b), suggesting wild ancestors may not be a useful source of variation. Canopy conditions are very heterogeneous, particularly in terms of light distribution (Horton 2000). A crop canopy that responds quickly to these changes will be more efficient in maximising resource capture (Taylor and Long 2017). Fast photosynthetic and photoprotection induction has been observed in wild rice accessions (Acevedo‐Siaca et al. 2021) and in wild wheat relatives (McAusland et al. 2020), respectively. Incorporating these faster light transition responses into modern wheat could be an objective for improving resource capture. Targeting earlier photosynthetic improvement before canopy closure is another potential route for improvement, as pre-anthesis photosynthesis is known to correlate with grain yield (Gaju et al. 2016; Carmo-Silva et al. 2017). Gaju et al. (2016) found at a pre-anthesis growth stage (during the onset of stem extension) a synthetic-derived hexaploid genotype maintained higher A than its recurrent hexaploid parent.
Taken together, there is good evidence to support the need for further characterisation of the component traits underpinning photosynthesis in wheat progenitors. Although much of the trait variation described is likely to be quantitatively controlled, there is an opportunity to identify specific progenitor accessions for direct use as donors in physiological pre-breeding. In addition, the development of protocols and tools for rapid screening of these physiological traits will enhance future genetic dissection. At present most methods require detailed experimentation and specialist equipment so the development of predictive phenotyping tools also offers promise to enable accurate forward genetic studies to discover trait-linked markers, and the selection of favourable variants in marker-assisted breeding. This is likely to yield significant benefits for breeding offering new potential to transfer higher WUE for drought tolerance or increased A from progenitor species.
Another strand of potential variation for further investigation towards application is the photosynthetic potential of reproductive tissues in progenitor species (Fig. 1). Ear photosynthesis is heritable, varies across different wheat genotypes and is an important determinant of grain yield (Molero and Reynolds 2020), highlighting the importance of ear photosynthesis as a breeding target. Li et al. (2017) found that ears of T. dicoccoides maintained higher CO2 assimilation during grain-filling when compared to hexaploid wheat, along with higher WUE under drought stress. Progenitor wheat species, particularly tetraploids, typically have a larger awn surface area than hexaploid wheat (Blum 1985). As a photosynthetic organ, awns have been reported to have high instantaneous WUE (Blum 1985; Weyhrich et al. 1995) potentially explaining why in a drought-prone environment, the presence of awns is reported to be beneficial to grain yield (Evans et al. 1972). Other components of the ear may also harbour useful stress tolerance characteristics, Araus et al. (1993) found that WUE was 33% higher in the ear bracts compared to the leaf blade using carbon isotope analysis, linking the higher efficiency to a lower gs and the xeromorphic features of the ear bracts (glumes, paleas and lemnas). Under heat stress, positive correlations have been observed between grain yield and the contribution of ear photosynthesis to grain yield (Molero and Reynolds 2020). Progenitors originating from drier and hotter environments may possess strategies, such as high ear CO2 fixation linked to the preservation of photosynthesis under unfavourable conditions and these could become increasingly useful for adapting modern wheat to more marginal environments.
Although little data exist on the quantitative differences in ear photosynthesis in wheat progenitors, and their relative contributions under stress, further work is warranted. As breeders seek to incorporate additional diversity into their programmes, the selection of progenitor donors with high ear CO2 fixation could be prioritised. Further evidence is required to confirm the consistency of photosynthetic contributions from the presence of awns. If consistently higher photosynthetic capacity can be demonstrated without reducing photosynthetic activity in other parts of the plant, then this trait can be readily incorporated as a breeding target due to the additional benefit and ease of phenotypic and genotypic selection. In many regions, awned wheat varieties predominate making it likely this benefit is already present and fixed, but it could also be applied where awned varieties are not widespread, and/or to prioritise selections within segregating pre-breeding material derived from progenitors.
Progenitor species are a source of new root system architecture ideotypes
RSA plays a pivotal role in drought tolerance and nutrient acquisition and enhancing root systems is a target for improving climate resilience (recently reviewed by Ober et al. 2021). Deeper roots can extract more water from subsoils, particularly during late developmental stages and grain fill, thereby improving yield in water limiting environments (Manschadi et al. 2010). However, the characterisation of mature RSA in wheat can be time consuming making it difficult to use as a selection target in breeding (Richard et al. 2015). Techniques that use early rooting traits (seminal root angle and seminal root number, e.g., the clear pot system developed by Richard et al. 2015) or root crowns extracted from the field at maturity (e.g., using the shovelomics method adapted from maize (Trachsel et al. 2011)) can be used to infer wheat RSA (Fradgley et al. 2020). A ‘pasta strainer’ technique described by El Hassouni et al. (2018) allows characterisation of the mature root system when grown within a perforated basket submerged in the field. All these tools allow RSA of genotypes to be characterised into wide or narrow/deep rooting types.
Wheat progenitor species may be used to augment the diversity in RSA that exists in the bread wheat gene pool. Tetraploid wheats have been shown to offer RSA diversity; using recombinant inbred lines of durum × wild emmer, QTLs for drought resistance and related traits were mapped (Peleg et al. 2009). Marker-assisted selection (MAS) enabled the QTL regions to be introgressed into both durum and hexaploid wheat (Merchuk-Ovnat et al. 2016a; b). This produced one hexaploid wheat isogenic line with introgression of a QTL from chromosome 7A of the wild emmer donor showing greater productivity (biomass, flag leaf area and grain yield) and photosynthetic capacity than the recurrent parent when grown under water limiting conditions. RSA was found to differ in this line, with greater development of deep roots and associated root tips whilst under drought stress (Merchuk-Ovnat et al. 2016a). This RSA enhanced the plant’s ability to access water at a greater soil depth and conferred greater drought tolerance as subsoil water levels are generally more stable than those in the upper layers of the soil.
Iannucci et al. (2017) identified 17 QTLs relating to root and shoot morphology in a durum × emmer wheat population, three of which were previously undescribed (two for the number of root tips and one for rooting depth). Root morphology QTL co-segregated with the height reducing Rht-B1 gene on chromosome 4B, indicating these alleles are involved in the control of both root and shoot traits, with tall plants having longer and larger root systems in this population. However, Christopher et al. (2013) found no co-segregation of root traits with dwarfing genes and most studies agree that root and shoot development are under the control of different sets of loci (Iannucci et al. 2017). QTL clusters for root morphology traits have also been reported to coincide with those for thousand grain weight and yield (Maccaferri et al. 2008; Iannucci et al. 2017) but further work is required to resolve these interactions. El Hassouni et al. (2018) found that in trials with low water availability, durum accessions with deep roots achieved a 37–38% yield increase but suffered a 20–40% yield penalty in irrigated environments.
Previous work has shown yield and biomass increases in synthetic-derived wheat lines can be attributed to a greater proportion of deep roots (Reynolds et al. 2007). Becker et al. (2016) also demonstrated that increased rooting depth and fine root mass allowed for the maintenance of plant growth under drought stress in two synthetic wheat lines, thus maintaining yields. However, a third synthetic line lacked deep roots but tolerated drought stress through increased stomatal density and reduced stomatal aperture (Becker et al. 2016). Recently Liu et al. (2020) detected eight QTL associated with drought tolerance in a SHW × commercial wheat F2 population with most of the positive alleles attributable to the Ae. tauschii (four QTLs) or tetraploid (durum; two QTLs) components of the synthetic. Ober et al. (2021) reviewed the range of wheat trait variation reported in wheat as well as summarised available evidence linking deeper roots to access to soil moisture.
Understanding the RSA diversity available in the wheat gene pool will allow the selection of targeted root types to suit environmental conditions such as drought or waterlogging, and nutrient availability. This remains a medium- to long-term breeding objective as there is still relatively little known about the heritability, environmental and management independence of RSA in elite cultivars (Fradgley et al. 2020). As highlighted by Ober et al. (2021) many upstream research questions remain including the mechanisms by which architectural traits impact water and nutrient acquisition. In addition, there remains a gap in understanding the linkage and direction of interactions between root and agronomic/crop production traits, and their environmental dependencies. Progenitor species typically have a wide eco-geographical adaption range, and it is proposed that this is likely to confer functional RSA variation. Whilst surveying large collections of progenitors for RSA variation is possible, more rapid progress is likely through the identification of pre-breeding material (capturing progenitor variation) with contrasting root types and comprehensive analysis of the linkages between trait variation and root functions. As for photosynthetic traits, high-throughput screening methods that can be scaled and applied for forward genetic screens and MAS are likely to accelerate progress in exploiting progenitor variation for RSA.
Prospects for climate-responsive breeding across crop species
Major and minor crops worldwide are likely to face both new limitations and opportunities for maintaining and increasing productivity due to changing climates. The identification of useful variation as described for wheat progenitor species and the successful application of approaches to mobilise it into cultivated wheat can serve as an exemplar for other crops. In addition to supporting productivity, this will also incentivise the search for useful variation in their progenitors and wild relatives. Exploration of progenitors or crop wild relatives has already begun in a variety of crop species (e.g., legumes (Porch et al. 2013; Coyne et al. 2020) apples (Volk et al. 2015) and numerous others (reviewed in (Hajjar and Hodgkin 2007; Dempewolf et al. 2017)) to identify genomic regions linked to phenotypes of interest for both biotic and abiotic stresses. For major cereals such as rice and barley, there are already examples of the successful introgression of traits linked to climate change adaptions such as drought tolerance (Talame et al. 2004; Zhang et al. 2006) and flowering traits (Ishimaru et al. 2010; Wiegmann et al. 2019). For minor cereal grain crops there are few confirmed examples to date, e.g., sorghum (reviewed in (Ananda et al. 2020)), pearl millet (reviewed in (Sharma et al. 2020)), finger millet (blast resistance (Akech et al. 2016)), oats (reviewed in (Ociepa 2019)) and rye (plant height and yield (Falke et al. 2009)), indicating that allocating resources to the exploration of diversity within progenitors and wild relatives would reveal further useful adaptations that could improve the resilience of these crops to changing climates. Examples of monocot crops, their progenitor species and breeding priorities linked to changing climates are shown in Table 1.
Table 1.
Monocot crops and their progenitor species or wild relatives that offer genetic diversity for targeted crop improvement.
| Crop | Progenitors | Breeding priorities linked to climate stresses | References |
|---|---|---|---|
| Maize (Zea mays) | Teosinte (Z. mays ssp. parviglumis) | Drought, heat, waterlogging | Mano and Omori 2013; Challinor et al. 2016 |
| Rice (Oryza sativa) | O. rufipogon | Drought, heat, flooding, salinity, C4 photosynthesis | Zhang et al. 2006; Ishimaru et al. 2010; Covshoff and Hibberd 2012; Singh et al. 2021 |
| Wheat (Triticum aestivum) | T. turgidum ssp dicoccoides and Aegilops tauschii | Drought, heat, C4 photosynthesis | Covshoff and Hibberd 2012; Lopes et al. 2015 |
| Barley (Hordeum vulgare) | H. vulgare ssp. spontaneum | Drought, heat, waterlogging, C4 photosynthesis | Setter and Waters 2003; Talame et al. 2004; Covshoff and Hibberd 2012; Weigmann et al. 2019 |
| Sorghum (Sorghum bicolor) | S. bicolor subsp. verticilliflorum | Cold, drought, heat | Ananda et al. 2020 |
| Pearl millet (Pennisetum glaucum) | P. glaucum subsp. monodii | Drought and heat | Sharma et al. 2020 |
| Oats (Avena sativa) | A. ventricosa, A. longiglumis, A. insularis, A. canariensis and A. agadiriana | Cold, drought and heat, C4 photosynthesis | Covshoff and Hibberd 2012; Ociepa 2019 |
| Rye (Secale cereale) | S. cereale subsp. vavilovii | Drought and heat, C4 photosynthesis | Covshoff and Hibberd 2012; Miedaner and Laidig 2019 |
| Finger millet (Eleusine coracana) | E. coracana subsp. africana. | Drought and salinity | Mirza and Marla 2019 |
Opportunities also exist to transfer desirable characteristics from minor to major cereal grain crops. An avenue that holds much promise, along with numerous technical challenges, is the incorporation of the C4 photosynthetic pathway, a characteristic of a C4 crop (e.g., sorghum or millet), into a C3 crop (e.g., rice or wheat). The C4 pathway utilises a carbon concentrating mechanism to diminish photorespiration, a process that takes place at the sites of carboxylation that limits productivity in C3 crops. The C4 pathway evolved due to increased abiotic stress, including heat and drought, which are conditions that can enhance photorespiration (Sage 2004). There is scope for breeding photosynthetic improvements within C4 crop species (von Caemmerer and Furbank 2016). However, major cereal crops are still cultivated in climates that favour photorespiration, meaning the enhanced water and nitrogen use efficiency characteristics of the C4 pathway is an attractive breeding target for C3 crops (Mitchell and Sheehy 2006). Climate change could exacerbate this need further, which has contributed to a concerted effort to incorporate the C4 pathway into C3 crops, in particular rice (e.g., www.c4rice.com). Challenges still need to be overcome before these improvements are available to the breeding community and C3 wild progenitors may provide a more accessible source of improvement for major C3 crops.
Opportunities exist to use genomics to accelerate the use of progenitors in crop breeding
Whilst traditional breeding approaches have been successfully used to cross cultivated materials with their wild relatives to introduce traits of interest, the success rate varies between species and becomes increasingly difficult with more distantly related species. There also remain barriers to using genomics-based advances to accelerate the uptake of novel alleles. Linkage drag is traditionally one of the major barriers to incorporating diversity from progenitors. Here, unwanted genes are introgressed simultaneously with a targeted region from a donor into the desired background. Backcross breeding is typically used to increase the recurrent parent (background) genotype and reduce unwanted genes. This strategy can be complemented by MAS, allowing the selection of a specific trait based on a linked genetic marker. MAS can be employed to facilitate more accurate introgression from a progenitor donor and reduce linkage drag from a wild background (Tanksley et al. 1989). This has been used successfully to make introgressions from several wild relatives into domesticated wheat (Nevo and Chen 2010; Merchuk-Ovnat et al. 2016a; King et al. 2017). Beyond linkage drag, other factors can pose issues to capturing wild diversity. The merging of genomes across wheat species can lead to intergenomic gene suppression (Feldman and Levy 2012). This phenomenon leads to the silencing of homoeologous genes and is reported to be common in hexaploid bread wheat (Bottley et al. 2006). This poses a potential problem for utilising newly synthesised wheats in pre-breeding programmes. However, the establishment of homoeologs does not necessarily result in functional silencing or suppression through dominance; phenotypes can be influenced by an additive dosage effect or complex interactions linked to the homoeologs (Borrill et al. 2015). Another potential roadblock is the genomic instability and radical changes which can occur because of allopolyploidization (Kraitshtein et al. 2010). However, there is evidence to suggest the severity of these changes may be of little consequence to the overall development of the plant (Zhao et al. 2011). Recent advancements in next-generation sequencing provide an opportunity for increasing our understanding of the functional genomics that underpin relationships across homoeologs (reviewed in Borrill et al. 2015). These tools could contribute to providing an improved understanding of the functional genetics of newly formed pre-breeding resources such as synthetic wheats incorporating progenitor diversity.
Advancements in sequencing technologies have facilitated the discovery of large numbers of DNA markers in crop species. In wheat (Winfield et al. 2012), this has led to the development of numerous genotyping platforms (Adamski et al. 2020) that have aided the application of QTL mapping and have enhanced the accessibility of diversity in progenitors and related species (Winfield et al. 2016, 2018; Wingen et al. 2017). SNPs are very effective markers in high-throughput genotyping due to their abundance across the wheat genome (Rimbert et al. 2018). Specific platforms have been developed to characterise wheat progenitors and wild relatives, including the Axiom® HD Wheat Genotyping Array (Winfield et al. 2016) and the Axiom® Wheat-Relative Genotyping Array (Przewieslik-Allen et al. 2019) in addition to arrays developed for elite varieties (e.g., Axiom® Wheat Breeder’s Genotyping Array; Allen et al. 2017). The wheat-relative array has been used to aid the introgression of the diploid wheat-relative Ambylopyrum muticum into a hexaploid wheat background through MAS (King et al. 2017). Furthermore, the Wheat Breeders’ array has been used in several studies for identifying QTLs in tetraploid wheat (Lucas et al. 2017; Wright et al. 2020). Low-cost genotyping platforms designed to demonstrate potential genetic variability between progenitor species and elite varieties are a tool of growing importance in exploring and harnessing diversity and have been deployed in many crops such as barley (Bayer et al. 2017), rice (Chen et al. 2013) and maize (Xu et al. 2017).
The availability of sequenced genomes from crop species, for example, the annotated reference genome assembly of the wheat cultivar Chinese Spring (International Wheat Genome Sequencing Consortium et al. 2018) augmented by the multiple genome assembly of Walkowiak et al. (2020) improve our understanding of the size and context of targeted introgressions through knowledge of the physical chromosome location of markers used for selection. In addition, the resources can improve our understanding of synteny with ancestral genomes (Grewal et al. 2018). Introgression fragments can be queried to identify the genes and any potentially favourable alleles present (Cheng et al. 2019). Due to the reducing expense of sequencing technologies (Jia et al. 2018), the number of cultivars sequenced is increasing, including many important elite wheat varieties (e.g., the 10+ genomes project: www.10wheatgenomes.com; Montenegro et al. 2017; Walkowiak et al. 2020). Increasing the number of modern wheat varieties sequenced, or genotyped through high-density marker arrays, will help characterise the haplotype diversity within the modern wheat gene pool. Haplotypes present in low diversity may reflect regions that have been under past selection (Fradgley et al. 2019) or where variation has been lost due to the domestication bottleneck (Haudry et al. 2007). Regardless, using this knowledge, targeted comparisons can then be made with extended progenitor gene pools to capture novel haplotypes (Uauy 2017). This comparison is being accelerated in wheat by the availability of increasing numbers of progenitors sequenced, including Ae. tauschii (Luo et al. 2017), T. urartu (Ling et al. 2013), T. dicoccoides (Avni et al. 2017) and T. durum (Maccaferri et al. 2019). A recent study by Cheng et al. (2019) compared re-sequenced genome data from a mixture of cultivated and progenitor wheat accessions, flagging regions of past introgression and identifying haplotype blocks that are nearly completely fixed in cultivated varieties. These regions of low diversity highlight the potential for identifying regions to target for improving genetic diversity from progenitor species.
In addition to the characterisation of haplotype diversity, enhanced sequencing resources will also support genetic mapping, cloning and functional characterisation from progenitor species. Kishii (2019) summarised the progress in generating genetic and physical mapping resources for Ae. tauschii documenting the progression from the early use of restriction fragment length polymorphism mapping in Ae. tauschii mapping populations (Gill et al. 1991) through to single sequence repeat genotyping (Nishijima et al. 2018). This supported the production of a 10 K Ae. tauschii Infinium SNP array by Luo et al. (2013) and the draft sequence of Ae. tauschii (Luo et al. 2017). The availability of reference genomes supports the use of data-driven approaches to selections, including linking phenotype to gene expression as demonstrated by Gálvez et al. (2019) for drought tolerance. This highlights the potential impact of understanding gene networks underpinning traits, and how genomics may identify novel breeding targets (Gálvez et al. 2019).
Resources supporting reverse genetics have also been developed in progenitor species with Targeting Induced Local Lesions in Genomes populations available in the wheat tetraploid (durum wheat Kronos; Krasileva et al. 2017) and diploid species (Ae. tauschii; Rawat et al. 2018), as well as being available for hexaploid wheat (cultivar Cadenza; Krasileva et al. 2017). A wheat exome capture was developed to focus sequencing efforts on exons, thereby reducing sequencing costs (Winfield et al. 2012). Along with genome sequences, these provide a useful resource for allele mining and gene discovery and could be used in future to support gene identification and cloning directly from the progenitor species. Direct cloning of favourable genes from progenitor species has been demonstrated using a combination of association genetics and resistance gene enrichment and sequencing (AgRenSeq; Arora et al. 2019). This method has been used to both discover and clone functional stem rust resistance genes in a panel of diverse Ae. tauschii accessions (Arora et al. 2019). Molecular breeding technologies provide the potential to directly introduce useful variation discovered in one crop into another, either by the introduction of the gene via genetic transformation or gene editing to introduce variation within homoeologous genes. The efficiency of the approaches discussed in this review remains to be seen for different genes and crops and can be impacted by the genetic background of particular varieties, but identifying a set of variants that already exist in nature and that can be used to introduce variation within genes of interest is an exciting prospect for the future.
Summary
There is a wealth of variation present in crop progenitor species for traits of relevance to plant breeding including flowering time, physiological response and RSA. Although initial characterisation demonstrates that functional variation exists, there remains a significant opportunity to systematically characterise this variation in order to make it accessible for use in breeding. In particular, more work is required to fully understand the genetic and physiological basis of progenitor trait variation in order to accurately inform future breeding strategies. The growing availability of sequencing and genomics tools offers great potential for targeted and accelerated progress in the systematic use of functional progenitor variation. The advances in use of wheat progenitors and the techniques developed for the capture of novel diversity may be applicable for the improvement of other cereal crop species.
Acknowledgements
We acknowledge support from the Biotechnology and Biological Sciences Research Council (BBSRC) Cross-Institute Strategic Programme ‘Designing Future Wheat’ BB/P016855/1. We thank Lorna McAusland (University of Nottingham) for the image of a cleared flag leaf (Fig. 1), Bethany Love (NIAB) for the image of the wheat ear (Fig. 1) and Howard Griffiths (University of Cambridge) for insightful discussions on physiological variation. This paper is dedicated to the memory of our friend Shigeo Takumi from Kobe University, Japan who passed away on 4 June 2020. We are grateful for his kindness and for all that he contributed to the understanding of variation in Aegilops tauschii.
Author contributions
FJL, TICW, RAH and ARB developed and designed the review content and format. SD was responsible for broadening the review scope to include further cereal crop species and progenitors. All authors contributed to writing and reviewing the manuscript.
Competing interests
The authors declare no competing interests.
Footnotes
Associate editor: Frank Hailer.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Acevedo‐Siaca LG, Dionora J, Laza R, Paul Quick W, Long SP (2021) Dynamics of photosynthetic induction and relaxation within the canopy of rice and two wild relatives. Food Energy Secur 10:e286 [DOI] [PMC free article] [PubMed]
- Adamski NM, Borrill P, Brinton J, Harrington SA, Marchal C, Bentley AR, et al. A roadmap for gene functional characterisation in crops with large genomes: lessons from polyploid wheat. eLife. 2020;9:e55646. doi: 10.7554/eLife.55646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akech V, Ojulong H, Okori P, Biruma M. Resistance to blast in interspecific finger millet progenies. Cape Town, South Africa 14: Fifth RUFORUM Bienn Reg Conf 17–21 Oct 2016; 2016. pp. 725–730. [Google Scholar]
- Allen AM, Winfield MO, Burridge AJ, Downie RC, Benbow HR, Barker GLA, et al. Characterization of a Wheat Breeders’ Array suitable for high-throughput SNP genotyping of global accessions of hexaploid bread wheat (Triticum aestivum) Plant Biotechnol J. 2017;15:390–401. doi: 10.1111/pbi.12635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ananda GKS, Myrans H, Norton SL, Gleadow R, Furtado A, Henry RJ. Wild sorghum as a promising resource for crop improvement. Front Plant Sci. 2020;11:1–14. doi: 10.3389/fpls.2020.01108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araus JL, Brown HR, Febrero A, Bort J, Serret MD. Ear photosynthesis, carbon isotope discrimination and the contribution of respiratory CO2 to differences in grain mass in durum wheat. Plant, Cell Environ. 1993;16:383–392. doi: 10.1111/j.1365-3040.1993.tb00884.x. [DOI] [Google Scholar]
- Arora S, Steuernagel B, Gaurav K, Chandramohan S, Long Y, Matny O et al. (2019) Resistance gene cloning from a wild crop relative by sequence capture and association genetics. Nat Biotechnol 37:139–143. 10.1038/s41587-018-0007-9 [DOI] [PubMed]
- Austin R, Morgan C, Ford M, Bhagwat S (1982) Flag leaf photosynthesis of Triticum aestivum and related diploid and tetraploid species. Ann Bot 49:177–189
- Avni R, Nave M, Barad O, Baruch K, Twardziok SO, Gundlach H, et al. Wild emmer genome architecture and diversity elucidate wheat evolution and domestication. Science. 2017;97:93–97. doi: 10.1126/science.aan0032. [DOI] [PubMed] [Google Scholar]
- Bayer MM, Rapazote-Flores P, Ganal M, Hedley PE, Macaulay M, Plieske J, et al. Development and evaluation of a barley 50k iSelect SNP array. Front Plant Sci. 2017;8:1792. doi: 10.3389/fpls.2017.01792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beales J, Turner A, Griffiths S, Snape JW, Laurie DA. A Pseudo-Response Regulator is misexpressed in the photoperiod insensitive Ppd-D1a mutant of wheat (Triticum aestivum L.) Theor Appl Genet. 2007;115:721–733. doi: 10.1007/s00122-007-0603-4. [DOI] [PubMed] [Google Scholar]
- Becker SR, Byrne PF, Reid SD, Bauerle WL, McKay JK, Haley SD. Root traits contributing to drought tolerance of synthetic hexaploid wheat in a greenhouse study. Euphytica. 2016;207:213–224. doi: 10.1007/s10681-015-1574-1. [DOI] [Google Scholar]
- Bentley AR, Turner AS, Gosman N, Leigh F, Maccaferri M, Dreisigacker S, et al. The frequency of photoperiod insensitive Ppd-A1a alleles in tetraploid, hexaploid and synthetic hexaploid wheat germplasm. Plant Breed. 2011;130:10–15. doi: 10.1111/j.1439-0523.2010.01802.x. [DOI] [Google Scholar]
- Blum A. Photosynthesis and transpiration in leaves and ears of wheat and barley varieties. J Exp Bot. 1985;36:432–440. doi: 10.1093/jxb/36.3.432. [DOI] [Google Scholar]
- Borlaug NE (1968) Wheat breeding and its impact on world food supply. In: Third International Wheat Genetics Symposium, Mexico: CIMMYT, 1–36
- Borrill P, Adamski N, Uauy C. Genomics as the key to unlocking the polyploid potential of wheat. N Phytologist. 2015;208:1008–1022. doi: 10.1111/nph.13533. [DOI] [PubMed] [Google Scholar]
- Bottley A, Xia GM, Koebner RMD. Homoeologous gene silencing in hexaploid wheat. Plant J. 2006;47:897–906. doi: 10.1111/j.1365-313X.2006.02841.x. [DOI] [PubMed] [Google Scholar]
- Camacho Villa TC, Maxted N, Scholten M, Ford-Lloyd B. Defining and identifying crop landraces. Plant Genet Resour. 2005;3:373–384. doi: 10.1079/PGR200591. [DOI] [Google Scholar]
- Carmo-Silva E, Andralojc PJ, Scales JC, Driever SM, Mead A, Lawson T, et al. Phenotyping of field-grown wheat in the UK highlights contribution of light response of photosynthesis and flag leaf longevity to grain yield. J Exp Bot. 2017;68:3473–3486. doi: 10.1093/jxb/erx169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Challinor A, Koehler AK, Ramirez-Villegas J, Whitfield S, Das B. Current warming will reduce yields unless maize breeding and seed systems adapt immediately. Nat Clim Change. 2016;6:954–958. doi: 10.1038/nclimate3061. [DOI] [Google Scholar]
- Chen H, Xie W, He H, Yu H, Chen W, Li J, et al. A high-density SNP genotyping array for rice biology and molecular breeding molecular. Plant. 2013;7:541–553. doi: 10.1093/mp/sst135. [DOI] [PubMed] [Google Scholar]
- Cheng H, Liu J, Wen J, Nie X, Xu L, Chen N, et al. Frequent intra- and inter-species introgression shapes the landscape of genetic variation in bread wheat. Genome Biol. 2019;20:1–16. doi: 10.1186/s13059-019-1744-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chèvre AM, Jahier J, Trottet M. Expression of disease resistance genes in amphiploid wheats-Triticum tauschii (Coss.) Schmal. Cer Res Comm. 1989;17:23–29. [Google Scholar]
- Christopher J, Christopher M, Jennings R, Jones S, Fletcher S, Borrell A, et al. QTL for root angle and number in a population developed from bread wheats (Triticum aestivum) with contrasting adaptation to water-limited environments. Theor Appl Genet. 2013;126:1563–1574. doi: 10.1007/s00122-013-2074-0. [DOI] [PubMed] [Google Scholar]
- Cockram J, Jones H, Leigh FJ, O’Sullivan D, Powell W, Laurie D, et al. Control of flowering time in temperate cereals: Genes, domestication, and sustainable productivity. J Exp Bot. 2007;58:1231–1244. doi: 10.1093/jxb/erm042. [DOI] [PubMed] [Google Scholar]
- Colasanti J, Coneva V. Mechanisms of floral induction in grasses: something borrowed, something new. Plant Physiol. 2009;149:56–62. doi: 10.1104/pp.108.130500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Covshoff S, Hibberd JM. Integrating C4 photosynthesis into C3 crops to increase yield potential. Curr Opin Biotechnol. 2012;23:209–214. doi: 10.1016/j.copbio.2011.12.011. [DOI] [PubMed] [Google Scholar]
- Cox TS. Deepening the wheat gene pool. J Crop Prod. 1997;1:1–25. doi: 10.1300/J144v01n01_01. [DOI] [Google Scholar]
- Coyne CJ, Kumar S, Wettberg EJB, Marques E, Berger JD, Redden RJ et al. (2020) Potential and limits of exploitation of crop wild relatives for pea, lentil, and chickpea improvement. Legum Sci 2:e36 10.1002/leg3.36
- Demmig-Adams B, Adams WW. Photoprotection and other responses of plants to high light stress. Annu Rev Plant Physiol Plant Mol Biol. 1992;43:599–626. doi: 10.1146/annurev.pp.43.060192.003123. [DOI] [Google Scholar]
- Dempewolf H, Baute G, Anderson J, Kilian B, Smith C, Guarino L. Past and future use of wild relatives in crop breeding. Crop Sci. 2017;57:1070–1082. doi: 10.2135/cropsci2016.10.0885. [DOI] [Google Scholar]
- Dixon LE, Karsai I, Kiss T, Adamski NM, Liu Z, Ding Y et al. (2019) VERNALIZATION1 controls developmental responses of winter wheat under high ambient temperatures. Development 146:dev172684. 10.1242/dev.172684 [DOI] [PMC free article] [PubMed]
- Dreisigacker S, Kishii M, Lage J, Warburton M. Use of synthetic hexaploid wheat to increase diversity for CIMMYT bread wheat improvement. Crop Pasture Sci. 2008;59:413–420. doi: 10.1071/AR07225. [DOI] [Google Scholar]
- Driever SM, Lawson T, Andralojc PJ, Raines CA, Parry MAJ. Natural variation in photosynthetic capacity, growth, and yield in 64 field-grown wheat genotypes. J Exp Bot. 2014;65:4959–4973. doi: 10.1093/jxb/eru253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubcovsky J, Lijavetzky D, Appendino L, Tranquilli G. Comparative RFLP mapping of Triticum monococcum genes controlling vernalization requirement. Theor Appl Genet. 1998;97:968–975. doi: 10.1007/s001220050978. [DOI] [Google Scholar]
- Dunstone RL, Evans LT. Role of changes in cell size in the evolution of wheat. Aust J Plant Physiol. 1974;1:157–165. [Google Scholar]
- Dunstone RL, Gifford RM, Evans LT. Photosynthetic characteristics of modern and primitive wheat species in relation to ontogeny and adaption to light. Aust J Biol Sci. 1973;26:295–307. doi: 10.1071/BI9730295. [DOI] [Google Scholar]
- El Hassouni K, Alahmad S, Belkadi B, Filali-Maltouf A, Hickey LT, Bassi FM. Root system architecture and its association with yield under different water regimes in durum wheat. Crop Sci. 2018;58:2331–2346. doi: 10.2135/cropsci2018.01.0076. [DOI] [Google Scholar]
- Evans LT, Dunstone RL. Some physiological aspects of evolution in wheat. Aust J Biol Sci. 1970;23:725–742. doi: 10.1071/BI9700725. [DOI] [Google Scholar]
- Evans LT, Bingham J, Jackson P, Sutherland J. Effect of awns and drought on the supply of photosynthate and its distribution within wheat ears. Ann Appl Biol. 1972;70:67–76. doi: 10.1111/j.1744-7348.1972.tb04689.x. [DOI] [Google Scholar]
- Evans JR, Kaldenhoff R, Genty B, Terashima I. Resistances along the CO2 diffusion pathway inside leaves. J Exp Bot. 2009;60:2235–2248. doi: 10.1093/jxb/erp117. [DOI] [PubMed] [Google Scholar]
- Falke KC, Sušić Z, Wilde P, Wortmann H, Möhring J, Piepho HP, et al. Testcross performance of rye introgression lines developed by marker-assisted backcrossing using an Iranian accession as donor. Theor Appl Genet. 2009;118:1225–1238. doi: 10.1007/s00122-009-0976-7. [DOI] [PubMed] [Google Scholar]
- Farquhar GD, von Caemmerer S, Berry J. A biochemical model of photosynthetic CO2 assimilation in leaves of C3 species. Planta. 1980;149:78–90. doi: 10.1007/BF00386231. [DOI] [PubMed] [Google Scholar]
- Farquhar G, O’Leary M, Berry J. On the relationship between carbon isotope discrimination and the intercellular carbon dioxide concentration in leaves. Aust J Plant Physiol. 1982;9:121. [Google Scholar]
- Farquhar GD, Richards RA. Isotopic composition of plant carbon correlates with water-use efficiency of wheat genotypes. Funct Plant Biol. 1984;11:539–552. doi: 10.1071/PP9840539. [DOI] [Google Scholar]
- Feldman M (2001) Origin of cultivated wheat. In: Bonjean AP, Angus WJ (eds) The world wheat book: a history of wheat breeding. Intercept Ltd., London. p. 3–56
- Feldman M, Levy AA. Genome evolution due to allopolyploidization in wheat. Genetics. 2012;192:763–774. doi: 10.1534/genetics.112.146316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleury D, Jefferies S, Kuchl H, Langridge P. Genetic and genomic tools to improve drought tolerance in wheat. J Exp Bot. 2010;61:3211–22. doi: 10.1093/jxb/erq152. [DOI] [PubMed] [Google Scholar]
- Flexas J, Barbour MM, Brendel O, Cabrera HM, Carriqui M, Diaz-Espejo A. Mesophyll diffusion conductance to CO2: an unappreciated central player in photosynthesis. Plant Sci. 2012;193–194:70–84. doi: 10.1016/j.plantsci.2012.05.009. [DOI] [PubMed] [Google Scholar]
- Foulkes MJ, Reynolds MP (2015) Breeding challenge: improving yield potential, 2nd edn. Elsevier Inc
- Fradgley N, Evans G, Biernaskie JM, Cockram J, Marr EC, Oliver AG, et al. Effects of breeding history and crop management on the root architecture of wheat. Plant Soil. 2020;452:587–600. doi: 10.1007/s11104-020-04585-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fradgley N, Gardner KA, Cockram J, Elderfield J, Hickey JM, Howell P, et al. A large-scale pedigree resource of wheat reveals evidence for adaptation and selection by breeders. PLoS Biol. 2019;17(2):e3000071. doi: 10.1371/journal.pbio.3000071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaju O, DeSilva J, Carvalho P, Hawkesford MJ, Griffiths S, Greenland A, et al. Leaf photosynthesis and associations with grain yield, biomass and nitrogen-use efficiency in landraces, synthetic-derived lines and cultivars in wheat. Field Crops Res. 2016;193:1–15. doi: 10.1016/j.fcr.2016.04.018. [DOI] [Google Scholar]
- Galmés J, Flexas J, Keys AJ, Cifre J, Mitchell RAC, Madgwick PJ, et al. Rubisco specificity factor tends to be larger in plant species from drier habitats and in species with persistent leaves. Plant Cell Environ. 2005;28:571–579. doi: 10.1111/j.1365-3040.2005.01300.x. [DOI] [Google Scholar]
- Gálvez S, Mérida-García R, Camino C, Borrill P, Abrouk M, Raminez-Gonzalez RH, et al. Hotspots in the genomic architecture of field drought responses in wheat as breeding targets. Funct Integr Genomics. 2019;19:295–309. doi: 10.1007/s10142-018-0639-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaurav K, Arora S, Silva P, Sánchez-Martín J, Horsnell R, Gao L et al. (2021) Population genomic analysis of Aegilops tauschii identifies targets for bread wheat improvement. Nat Biotechnol 40:422–431 10.1038/s41587-021-01058-4 [DOI] [PMC free article] [PubMed]
- Giles RJ, Brown TA. GluDy allele variations in Aegilops tauschii and Triticum aestivum: implications for the origins of hexaploid wheats. Theor Appl Genet. 2006;112:1563–1572. doi: 10.1007/s00122-006-0259-5. [DOI] [PubMed] [Google Scholar]
- Gill KS, Lubbers EL, Gill BS, Raupp WJ, Cox TS. A genetic linkage map of Triticum tauschii (DD) and its relationship to the D-genome of bread wheat (AABBDD) Genome. 1991;14:362–374. doi: 10.1139/g91-058. [DOI] [Google Scholar]
- Golovnina KA, Kondratenko EY, Blinov AG, Goncharov NP. Molecular characterization of vernalization loci VRN1 in wild and cultivated wheats. BMC Plant Biol. 2010;10:168. doi: 10.1186/1471-2229-10-168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grazzani S, Gendall AR, Lister C, Dean C. Analysis of the molecular basis of flowering time variation in Arabidopsis accessions. Plant Physiol. 2003;132:1107–14. doi: 10.1104/pp.103.021212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grewal S, Hubbart-Edwards S, Yang C, Scholefield D, Ashling S, Burridge A, et al. Detection of T. urartu introgressions in wheat and development of a panel of interspecific introgression lines. Front Plant Sci. 2018;871:1–10. doi: 10.3389/fpls.2018.01565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu J, Yin X, Stomph TJ, Wang H, Struik PC. Physiological basis of genetic variation in leaf photosynthesis among rice (Oryza sativa L.) introgression lines under drought and well-watered conditions. J Exp Bot. 2012;63:5137–5153. doi: 10.1093/jxb/ers170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajjar R, Hodgkin T. The use of wild relatives in crop improvement: a survey of developments over the last 20 years. Euphytica. 2007;156:1–13. doi: 10.1007/s10681-007-9363-0. [DOI] [Google Scholar]
- Haudry A, Cenci A, Ravel C, Bataillon T, Brunel D, Poncet C, et al. Grinding up wheat: a massive loss of nucleotide diversity since domestication. Mol Biol Evol. 2007;24:1506–1517. doi: 10.1093/molbev/msm077. [DOI] [PubMed] [Google Scholar]
- Henry A, Gowda VRP, Torres RO, McNally KL, Serraj R. Variation in root system architecture and drought response in rice (Oryza sativa): phenotyping of the OryzaSNP panel in rainfed lowland fields. Field Crop Res. 2011;120:205–214. doi: 10.1016/j.fcr.2010.10.003. [DOI] [Google Scholar]
- Horton P. Prospects for crop improvement through the genetic manipulation of photosynthesis: morphological and biochemical aspects of light capture. J Exp Bot. 2000;51:475–485. doi: 10.1093/jexbot/51.suppl_1.475. [DOI] [PubMed] [Google Scholar]
- Hubbart S, Smillie IRA, Heatley M, Swarup R, Foo CC, Zhao L, et al. Enhanced thylakoid photoprotection can increase yield and canopy radiation use efficiency in rice. Commun Biol. 2018;1:22. doi: 10.1038/s42003-018-0026-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes J, Hepworth C, Dutton C, Dunn JA, Hunt L, Stephens J, et al. Reducing stomatal density in barley improves drought tolerance without impacting on yield. Plant Physiol. 2017;174:776–787. doi: 10.1104/pp.16.01844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyles J, Bloomfield MT, Hunt JR, Threthowan RM, Trevaskis B (2020) Phenology and related traits for wheat adaptation. Heredity 125:417–430 10.1038/s41437-020-0320-1 [DOI] [PMC free article] [PubMed]
- Iannucci A, Marone D, Russo MA, De Vita P, Miullo V, Ferragonio P et al. (2017) Mapping QTL for root and shoot morphological traits in a durum wheat × T. dicoccum segregating population at seedling stage. Int J Genomics 2017;6876393. 10.1155/2017/6876393 [DOI] [PMC free article] [PubMed]
- Appels A, Eversole K, Feuillet C, Keller B, et al. Shifting the limits in wheat research and breeding using a fully annotated reference genome. Science. 2018;361:eaar7191. doi: 10.1126/science.aar7191. [DOI] [PubMed] [Google Scholar]
- Ishimaru T, Hirabayashi H, Ida M, Takai T, San-Oh YA, Yoshinaga S, et al. A genetic resource for early-morning flowering trait of wild rice Oryza officinalis to mitigate high temperature-induced spikelet sterility at anthesis. Ann Bot. 2010;106:515–520. doi: 10.1093/aob/mcq124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jafarzadeh J, Bonnett D, Jannink J-L, Akdemir D, Dreisigacker S, Sorrells ME. Breeding value of primary synthetic wheat genotypes for grain yield. PLoS One. 2016;11:e0162860. doi: 10.1371/journal.pone.0162860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia M, Guan J, Zhai Z, Geng S, Zhang X, Mao L, et al. Wheat functional genomics in the era of next generation sequencing: an update. Crop J. 2018;6:7–14. doi: 10.1016/j.cj.2017.09.003. [DOI] [Google Scholar]
- Johnson RC, Kebede H, Mornhinweg DW, Carver BF, Rayburn AL, Nguyen HT. Photosynthetic differences among Triticum accessions at tillering. Crop Sci. 1987;27:1046–1050. doi: 10.2135/cropsci1987.0011183X002700050044x. [DOI] [Google Scholar]
- Jones G, Jones H, Charles MP, Jones MK, College S, Leigh FJ, et al. Phylogeographic analysis of barley DNA as evidence for the spread of Neolithic agriculture through Europe. J Archaeol. Science. 2012;39:3230–3238. [Google Scholar]
- Kerber R, Rowland GG (1974) Origin of the free threshing character in hexaploid wheat. Can J Genet Cytol 16:145–154. 10.1139/g74-014
- Khazaei H, Monneveux P, Hongbo S, Mohammady S. Variation for stomatal characteristics and water use efficiency among diploid, tetraploid and hexaploid Iranian wheat landraces. Genet Resour Crop Evol. 2009;57:307–314. doi: 10.1007/s10722-009-9471-x. [DOI] [Google Scholar]
- Kihara H. Discovery of the DD-analyser, one of the ancestors of Triticum vulgare (Japanese) Agric Hort (Tokyo) 1944;19:13–14. [Google Scholar]
- King J, Grewal S, Yang CY, Hubbart S, Scholefield D, Ashling S, et al. A step change in the transfer of interspecific variation into wheat from Amblyopyrum muticum. Plant Biotechnol J. 2017;15:217–226. doi: 10.1111/pbi.12606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King J, Newell C, Grewal S, Hubbart-Edwards S, Yang C-Y, Scholefield D et al. (2019) Development of stable homozygous wheat/Amblyopyrum muticum (Aegilops mutica) introgression lines and their cytogenetic and molecular characterization. Front Plant Sci 10:34 10.3389/fpls.2019.00034 [DOI] [PMC free article] [PubMed]
- Kishii M (2019) An update of recent use of Aegilops species in wheat breeding. Front Plant Sci 10:585. 10.3389/fpls.2019.00585 [DOI] [PMC free article] [PubMed]
- Koyama K, Okumura Y, Okamoto E, Nishijima R, Takumi S. Natural variation in photoperiodic flowering pathway and identification of photoperiod-insensitive accessions in wild wheat, Aegilops tauschii. Euphytica. 2018;214:3. doi: 10.1007/s10681-017-2089-8. [DOI] [Google Scholar]
- Krasileva KV, Vasquez-Gross HA, Howell T, Bailey P, Paraiso F, Clissold L, et al. Uncovering hidden variation in polyploid wheat. Proc Natl Acad Sci USA. 2017;114:E913–E921. doi: 10.1073/pnas.1619268114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraitshtein Z, Yaakov B, Khasdan V, Kashkush K. Genetic and epigenetic dynamics of a retrotransposon after allopolyploidization of wheat. Genetics. 2010;186:801–812. doi: 10.1534/genetics.110.120790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kromdijk J, Głowacka K, Leonelli L, Gabilly ST, Iwai M, Niyogi KK, et al. Improving photosynthesis and crop productivity by accelerating recovery from photoprotection. Science. 2016;354:857–861. doi: 10.1126/science.aai8878. [DOI] [PubMed] [Google Scholar]
- Lantican MA, Pingali PL, Rajaram S. Is research on marginal lands catching up? The case of unfavourable wheat growing environments. Agric Econ. 2005;29:353–361. doi: 10.1111/j.1574-0862.2003.tb00171.x. [DOI] [Google Scholar]
- Lawson T, Blatt MR. Stomatal size, speed, and responsiveness impact on photosynthesis and water use efficiency. Plant Physiol. 2014;164:1556–1570. doi: 10.1104/pp.114.237107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Wan HS, Yang WY. Synthetic hexaploid wheat enhances variation and adaptive evolution of bread wheat in breeding processes. J Syst Evol. 2014;52:735–742. doi: 10.1111/jse.12110. [DOI] [Google Scholar]
- Li P-F, Cheng Z-G, Ma B-L, Palta JA, Kong H-Y, Mo F, et al. Dryland wheat domestication changed the development of aboveground architecture for a well-structured canopy. PLoS One. 2014;9:e95825. doi: 10.1371/journal.pone.0095825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Lawas LMF, Malo R, Glaubitz U, Erban A, Mauleon R, et al. Metabolic and transcriptomic signatures of rice floral organs reveal sugar starvation as a factor in reproductive failure under heat and drought stress. Plant Cell Environ. 2015;38:2171–2192. doi: 10.1111/pce.12545. [DOI] [PubMed] [Google Scholar]
- Li YP, Li YY, Li DY, Wang SW, Zhang SQ. Photosynthetic response of tetraploid and hexaploid wheat to water stress. Photosynthetica. 2017;55:1–13. doi: 10.1007/s11099-016-0659-y. [DOI] [Google Scholar]
- Ling H-Q, Zhao S, Liu D, Wang J, Sun H, Zhang C. Draft genome of the wheat A-genome progenitor Triticum urartu. Nature. 2013;496:87–90. doi: 10.1038/nature11997. [DOI] [PubMed] [Google Scholar]
- Liu R, Wu F, Yi X, Lin Y, Wang Z, Liu S, et al. Quantitative trait loci analysis for root traits in synthetic hexaploid wheat under drought stress conditions. J Integr Agric. 2020;19:1947–1960. doi: 10.1016/S2095-3119(19)62825-X. [DOI] [Google Scholar]
- Lobell DB, Field CB. Global scale climate-crop yield relationships and the impacts of recent warming. Environ Res Lett. 2007;2:014002. doi: 10.1088/1748-9326/2/1/014002. [DOI] [Google Scholar]
- Long SP, Zhu X-G, Naidu SL, Ort DR. Can improvement in photosynthesis increase crop yields? Plant, Cell Environ. 2006;29:315–330. doi: 10.1111/j.1365-3040.2005.01493.x. [DOI] [PubMed] [Google Scholar]
- Lopes MS, El-Basyoni I, Baenziger PS, Singh S, Royo C, Ozbek K, et al. Exploiting genetic diversity from landraces in wheat breeding for adaptation to climate change. J Exp Bo. 2015;66:3477–3486. doi: 10.1093/jxb/erv122. [DOI] [PubMed] [Google Scholar]
- Lucas SJ, Salantur A, Yazar S, Budak H. High-throughput SNP genotyping of modern and wild emmer wheat for yield and root morphology using a combined association and linkage analysis. Funct Integr Genomics. 2017;17:667–685. doi: 10.1007/s10142-017-0563-y. [DOI] [PubMed] [Google Scholar]
- Lundgren MR, Fleming AJ. Cellular perspectives for improving mesophyll conductance. Plant J. 2020;101:845–857. doi: 10.1111/tpj.14656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo MC, Gu YQ, You FM, Deal KR, Ma Y, Hu Y, et al. A 4-gigabase physical map unlocks the structure and evolution of the complex genome of Aegilops tauschii, the wheat D-genome progenitor. Proc Natl Acad Sci USA. 2013;110:7940–7945. doi: 10.1073/pnas.1219082110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo MC, Gu YQ, Puiu D, Wang H, Twardziok SO, Deal KR, et al. Genome sequence of the progenitor of the wheat D genome Aegilops tauschii. Nature. 2017;551:498–502. doi: 10.1038/nature24486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNally KL, Childs KL, Bohnert R, Davidson RM, Zhao K, Ulat VJ, et al. Genomewide SNP variation reveals relationships among landraces and modern varieties of rice. Proc Natl Acad Sci USA. 2009;106:12273–12278. doi: 10.1073/pnas.0900992106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maccaferri M, Sanguineti MC, Corneti S, Ortega JLA, Ben Salem M, Bort J, et al. Quantitative trait loci for grain yield and adaptation of durum wheat (Triticum durum Desf.) across a wide range of water availability. Genetics. 2008;178:489–511. doi: 10.1534/genetics.107.077297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maccaferri M, Harris NS, Twardziok SO, Pasam RK, Gundlach H, Spannagl B, et al. Durum wheat genome highlights past domestication signatures and future improvement targets. Nat Genet. 2019;51:885–895. doi: 10.1038/s41588-019-0381-3. [DOI] [PubMed] [Google Scholar]
- Mano Y, Omori F. Flooding tolerance in interspecific introgression lines containing chromosome segments from teosinte (Zea nicaraguensis) in maize (Zea mays subsp. mays) Ann Bot. 2013;112:1125–1139. doi: 10.1093/aob/mct160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manschadi AM, Christopher JT, Hammer GL, Devoil P. Experimental and modelling studies of drought‐adaptive root architectural traits in wheat (Triticum aestivum L.) Plant Biosyst. 2010;144:458–462. doi: 10.1080/11263501003731805. [DOI] [Google Scholar]
- Martínez-Moreno F, Solís I, Nogueoro D, Blanco A, Özberk İ, Nsarellah N, et al. Durum wheat in the Mediterranean Rim: historical evolution and genetic resources. Genet Resour Crop Evol. 2020;67:1415–1436. doi: 10.1007/s10722-020-00913-8. [DOI] [Google Scholar]
- Matsouka Y, Takumi S, Kawahara T. Flowering time diversification and dispersal in central Eurasian wild wheat Aegilops tauschii Coss.: genealogical and ecological framework. PLoS One. 2008;3:e3138. doi: 10.1371/journal.pone.0003138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuoka Y, Kawahara T, Takumi S. Intraspecific lineage divergence and its association with reproductive trait change during species range expansion in central Eurasian wild wheat Aegilops tauschii Coss. (Poaceae) BMC Evol Biol. 2015;15:213. doi: 10.1186/s12862-015-0496-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAusland L, Vialet-Chabrand S, Jauregui I, Burridge A, Hubbart-Edwards S, Fryer MJ, et al. Variation in key leaf photosynthetic traits across wheat wild relatives is accession dependent not species dependent. N Phytol. 2020;228:1767–1780. doi: 10.1111/nph.16832. [DOI] [PubMed] [Google Scholar]
- McFadden E, Sears E. The origin of Triticum spelta and its free-threshing hexaploid relatives. J Hered. 1946;37:81–89. doi: 10.1093/oxfordjournals.jhered.a105590. [DOI] [PubMed] [Google Scholar]
- Merchuk-Ovnat L, Barak V, Fahima T, Ordon F, Lidzbarsky GA, Krugman T, et al. Ancestral QTL alleles from wild emmer wheat improve drought resistance and productivity in modern wheat cultivars. Front Plant Sci. 2016;7:452. doi: 10.3389/fpls.2016.00452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merchuk-Ovnat L, Fahima T, Krugman T, Saranga Y. Ancestral QTL alleles from wild emmer wheat improve grain yield, biomass and photosynthesis across environments in modern wheat. Plant Sci. 2016;251:23–34. doi: 10.1016/j.plantsci.2016.05.003. [DOI] [PubMed] [Google Scholar]
- Miedaner T, Laidig F (2019) Hybrid breeding in rye (Secale cereale L.). In: Al-Khayri JM, Jain SM, Johnson DV (eds) Advances in plant breeding strategies: cereals. Springer, Cham. pp 343–372. 10.1007/978-3-030-23108-8_9
- Mirzaghaderi G, Abdolmalaki Z, Ebrahimzadegan R, Bahmani F, Orooji F, Majdi M, et al. Production of synthetic wheat lines to exploit the genetic diversity of emmer wheat and D genome containing Aegilops species in wheat breeding. Sci Rep. 2020;10:19698. doi: 10.1038/s41598-020-76475-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirza N, Marla S (2019) Finger millet (Eleusine coracana L. Gartn.) breeding. In: Al-Khayri JM, Jain SM, Johnson DV (eds) Advances in plant breeding strategies: cereals. Springer, Cham. p 83–132. 10.1007/978-3-030-23108-8_3
- Mitchell PL, Sheehy JE. Supercharging rice photosynthesis to increase yield. N Phytol. 2006;171:688–693. doi: 10.1111/j.1469-8137.2006.01855.x. [DOI] [PubMed] [Google Scholar]
- Molero G, Reynolds MP. Spike photosynthesis measured at high throughput indicates genetic variation independent of flag leaf photosynthesis. Field Crops Res. 2020;255:107866. doi: 10.1016/j.fcr.2020.107866. [DOI] [Google Scholar]
- Montenegro JD, Golicz AA, Bayer PE, Hurgobin B, Lee HT, Chan CKK, et al. The pangenome of hexaploid bread wheat. Plant J. 2017;90:1007–1013. doi: 10.1111/tpj.13515. [DOI] [PubMed] [Google Scholar]
- Mujeeb-Kazi A, Kaz, AG, Dundas I, Farrakh S (2013) Genetic diversity for wheat improvement as a conduit to food security. Advances in agronomy, 1st edn, vol 122. Elsevier Inc
- Murchie EH, Pinto M, Horton P. Agriculture and the new challenges for photosynthesis research. N Phytol. 2009;181:532–552. doi: 10.1111/j.1469-8137.2008.02705.x. [DOI] [PubMed] [Google Scholar]
- Nevo E, Chen G. Drought and salt tolerances in wild relatives for wheat and barley improvement. Plant Cell Environ. 2010;33:670–685. doi: 10.1111/j.1365-3040.2009.02107.x. [DOI] [PubMed] [Google Scholar]
- Ni Z, Li H, Zhao Y, Peng H, Hu Z, Xin M, et al. Genetic improvement of heat tolerance in wheat: recent progress in understanding the underlying molecular mechanisms. Crop J. 2018;6:32–41. doi: 10.1016/j.cj.2017.09.005. [DOI] [Google Scholar]
- Nishijima R, Ikeda TM, Takumi S. Genetic mapping reveals a dominant awn-inhibiting gene related to differentiation of the variety anathera in the wild diploid wheat Aegilops tauschii. Genetica. 2018;146:75–84. doi: 10.1007/s10709-017-9998-2. [DOI] [PubMed] [Google Scholar]
- Nishimura K, Moriyama R, Katsura K, Saito H, Takisawa R, Kitajima A, et al. The early flowering trait of an emmer wheat accession (Triticum turgidum L. ssp. dicoccum) is associated with the cis‑element of the Vrn-A3 locus. Theor Appl Genet. 2018;131:2037–2053. doi: 10.1007/s00122-018-3131-5. [DOI] [PubMed] [Google Scholar]
- Ober ES, Alahmad S, Cockram J, Forestan C, Hickey LT, Kant J, et al. Wheat root systems as a breeding target for climate resilience. Theor Appl Genet. 2021;134:1645–1662. doi: 10.1007/s00122-021-03819-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ociepa T. The oat gene pools – review about the use of wild species in improving cultivated oat. J Cent Eur Agric. 2019;20:251–261. doi: 10.5513/JCEA01/20.1.2044. [DOI] [Google Scholar]
- Ogbonnaya FC, Imtiaz M, Bariana HS, McLean M, Shankar MM, Hollaway GJ. Mining synthetic hexaploids for multiple disease resistance to improve bread wheat. Aust J Agric Res. 2008;59:421–431. doi: 10.1071/AR07227. [DOI] [Google Scholar]
- Parry MAJ, Reynolds M, Salvucci ME, Raines C, Andralojc PJ, Zhu X-G, et al. Raising yield potential of wheat. II. Increasing photosynthetic capacity and efficiency. J Exp Bot. 2011;62:453–467. doi: 10.1093/jxb/erq304. [DOI] [PubMed] [Google Scholar]
- Peleg Z, Fahima T, Krugman T, Abbo S, Yakir D, Korol AB. Genomic dissection of drought resistance in durum wheat × wild emmer wheat recombinant inbred line population. Plant Cell Environ. 2009;32:758–779. doi: 10.1111/j.1365-3040.2009.01956.x. [DOI] [PubMed] [Google Scholar]
- Petersen G, Seberg O, Yde M, Berthelsen K. Phylogenetic relationships of Triticum and Aegilops and evidence for the origin of the A, B, and D genomes of common wheat (Triticum aestivum) Mol Phylogenet Evol. 2006;30:70–82. doi: 10.1016/j.ympev.2006.01.023. [DOI] [PubMed] [Google Scholar]
- Porch T, Beaver J, Debouck D, Jackson S, Kelly J, Dempewolf H. Use of wild relatives and closely related species to adapt common bean to climate change. Agronomy. 2013;3:433–461. doi: 10.3390/agronomy3020433. [DOI] [Google Scholar]
- Prins A, Orr DJ, Andralojc PJ, Reynolds MP, Carmo-Silva E, Parry MAJ. Rubisco catalytic properties of wild and domesticated relatives provide scope for improving wheat photosynthesis. J Exp Bot. 2016;67:1827–1838. doi: 10.1093/jxb/erv574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Przewieslik-Allen AM, Burridge AJ, Wilkinson PA, Winfield MO, Shaw DS, McAusland L et al. (2019) Developing a high-throughput SNP-based marker system to facilitate the introgression of traits from Aegilops species into bread wheat (Triticum aestivum). Front Plant Sci 9:1993. 10.3389/fpls.2018.01993 [DOI] [PMC free article] [PubMed]
- Rawat N, Schoen A, Singh L, Mahlandt A, Wilson DL, Liu S, et al. TILL-D: An Aegilops tauschii TILLING resource for wheat improvement. Front Plant Sci. 2018;9:1665. doi: 10.3389/fpls.2018.01665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rey MD, Martín AC, Higgins J, Swarbreck D, Uauy C, Shaw P et al. (2017) Exploiting the ZIP4 homologue within the wheat Ph1 locus has identified two lines exhibiting homoeologous crossover in wheat-wild relative hybrids. Mol Breed 37:95. 10.1007/s11032-017-0700-2 [DOI] [PMC free article] [PubMed]
- Reynolds M, Dreccer F, Trethowan R. Drought-adaptive traits derived from wheat wild relatives and landraces. J Exp Bot. 2007;58:177–186. doi: 10.1093/jxb/erl250. [DOI] [PubMed] [Google Scholar]
- Reynolds M, Foulkes MJ, Slafer GA, Berry P, Parry MAJ, Snape JW, et al. Raising yield potential in wheat. J Exp Bot. 2009;60:1899–1918. doi: 10.1093/jxb/erp016. [DOI] [PubMed] [Google Scholar]
- Reynolds MP, Hays D, Chapman S. Breeding for adaptation to heat and drought stress. In: Reynolds MP, editor. Climate change and crop production. Wallingford, Oxfordshire: CABI; 2010. pp. 71–91. [Google Scholar]
- Richard CA, Hickey LT, Fletcher S, Jennings R, Chenu K, Christopher JT. High-throughput phenotyping of seminal root traits in wheat. Plant Methods. 2015;11:13. doi: 10.1186/s13007-015-0055-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards RA, Rebetzke GJ, Condon AG, van Herwaarden AF. Breeding opportunities for increasing the efficiency of water use and crop yield in temperate Cereals. Crop Sci. 2002;42:111. doi: 10.2135/cropsci2002.0111. [DOI] [PubMed] [Google Scholar]
- Rimbert H, Darrier B, Navarro J, Kitt J, Choulet F, Leveugle M, et al. High throughput SNP discovery and genotyping in hexaploid wheat. PLoS One. 2018;13:1–19. doi: 10.1371/journal.pone.0186329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenzweig CD, Vicarelli KM, Neofotis P, Wu Q, Casassa G, Menzel A, et al. Attributing physical and biological impacts to anthropogenic climate change. Nature. 2008;453:353–357. doi: 10.1038/nature06937. [DOI] [PubMed] [Google Scholar]
- Rowland GG, Kerber ER (1974) Telocentric mapping in hexaploid wheat of genes for leaf rust resistance and other characters derived from Aegilops squarrosa. Can J Genet Cytol 16:137–144 10.1139/g74-013
- Sage RF. The evolution of C4 photosynthesis. N Phytol. 2004;161:341–370. doi: 10.1111/j.1469-8137.2004.00974.x. [DOI] [PubMed] [Google Scholar]
- Salamini F, Özkan H, Brandolini A, Schäfer-Pregl R, Martin W. Genetics and geography of wild cereal domestication in the near east. Nat Rev Genet. 2002;3:429–441. doi: 10.1038/nrg817. [DOI] [PubMed] [Google Scholar]
- Scafaro AP, Yamori W, Carmo-Silva AE, Salvucci ME, von Caemmerer S, Atwell BJ. Rubisco activity is associated with photosynthetic thermotolerance in a wild rice (Oryza meridionalis) Physiol Plant. 2012;146:99–109. doi: 10.1111/j.1399-3054.2012.01597.x. [DOI] [PubMed] [Google Scholar]
- Schneider A, Molnár I, Molnár-Láng M. Utilisation of Aegilops (goatgrass) species to widen the genetic diversity of cultivated wheat. Euphytica. 2008;163:1–19. doi: 10.1007/s10681-007-9624-y. [DOI] [Google Scholar]
- Scoffoni C, Chatelet DS, Pasquet-kok J, Rawls M, Donoghue MJ, Edwards EJ, et al. Hydraulic basis for the evolution of photosynthetic productivity. Nat Plants. 2016;2:16072. doi: 10.1038/nplants.2016.72. [DOI] [PubMed] [Google Scholar]
- Sears ER (1977) Genetics Society Of Canada Award Of Excellence Lecture: an induced mutant with homoeologous pairing in common wheat. Can J Genet Cytol 19:4. 10.1139/g77-063
- Setter T, Waters I. Review of prospects for germplasm improvement for waterlogging tolerance in wheat, barley and oats. Plant Soil. 2003;253:1–34. doi: 10.1023/A:1024573305997. [DOI] [Google Scholar]
- Sharma S, Sharma R, Govindaraj M, Mahala RS, Satyavathi CT, Srivastava RK et al. (2020) Harnessing wild relatives of pearl millet (Pennisetum glaucum L. R. Br) for germplasm enhancement: challenges and opportunities. Crop Sci 61:177–200. 10.1002/csc2.20343
- Singh VJ, Vinod KK, Krishnan SG, Singh AK (2021) Rice adaptation to climate change: opportunities and priorities in molecular breeding. In: Hossain MA, Hassan L, Ifterkharuddaula KM, Kumar A, Henry R (eds) Molecular breeding for rice abiotic stress tolerance and nutritional quality. John Wiley & Sons, Ltd. pp 1–25 10.1002/9781119633174.ch1
- South PF, Cavanagh AP, Liu HW, Ort DR (2019). Synthetic glycolate metabolism pathways stimulate crop growth and productivity in the field. Science 363:eaat9077. 10.1126/science.aat9077 [DOI] [PMC free article] [PubMed]
- Swarbreck SM, Wang M, Wang Y, Kindred D, Sylvester-Bradley R, Shi W et al. (2019) A roadmap for lowering crop N requirement. Trends Plant Sci 24:892–904 10.1016/j.tplants.2019.06.006. [DOI] [PubMed]
- Takenaka S, Kawahara T. Evolution and dispersal of emmer wheat (Triticum sp.) from novel haplotypes of Ppd-1 (photoperiod response) genes and their surrounding DNA sequences. Theor Appl Genet. 2012;125:999–1014. doi: 10.1007/s00122-012-1890-y. [DOI] [PubMed] [Google Scholar]
- Takumi S, Koyama K, Fujiwara K, Kobayashi F (2011) Identification of a large deletion in the first intron of the Vrn-D1 locus, associated with loss of vernalization requirement in wild wheat progenitor Aegilops tauschii Coss. Genes Genet Syst 86:183–195 [DOI] [PubMed]
- Talame V, Sanguineti MC, Chiapparino E, Bahri H, Salem M, Forster BP, et al. Identification of Hordeum spontaneum QTL alleles improving field performance of barley grown under rainfed conditions. Ann Appl Biol. 2004;144:309–319. doi: 10.1111/j.1744-7348.2004.tb00346.x. [DOI] [Google Scholar]
- Tanaka M, Yamashita K. Growth habit of Aegilops squarrosa strains collected in Pakistan, Afghanistan and Iran. Wheat Inform Serv. 1957;6:16–18. [Google Scholar]
- Tanksley SD, McCouch SR. Seed banks and molecular maps: unlocking genetic potential from the wild. Science. 1997;5329:1063–1066. doi: 10.1126/science.277.5329.1063. [DOI] [PubMed] [Google Scholar]
- Tanksley SD, Young ND, Paterson AH, Bonierbale MW. RFLP mapping in plant breeding: new tools for an old science. Bio/Technol. 1989;7:257–264. doi: 10.1038/nbt0389-257. [DOI] [Google Scholar]
- Taylor SH, Long SP. Slow induction of photosynthesis on shade to sun transitions in wheat may cost at least 21% of productivity. Philos Trans R Soc B: Biol Sci. 2017;372:20160543. doi: 10.1098/rstb.2016.0543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trachsel S, Kaeppler SM, Brown KM, Lynch JP. Shovelomics: high throughput phenotyping of maize (Zea mays L.) root architecture in the field. Plant Soil. 2011;341:75–87. doi: 10.1007/s11104-010-0623-8. [DOI] [Google Scholar]
- Tricker PJ, ElHabti A, Schmidt J, Fleury D. The physiological and genetic basis of combined drought and heat tolerance in wheat. J Exp Bot. 2018;69:3195–3210. doi: 10.1093/jxb/ery081. [DOI] [PubMed] [Google Scholar]
- Tsunewaki K. Comparative gene analysis of common wheat and its ancestral species. II. Waxiness, growth habit and awnedness. Jpn J Bot. 1966;19:175–229. doi: 10.1093/genetics/53.2.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uauy C. Wheat genomics comes of age. Curr Opin Plant Biol. 2017;36:142–148. doi: 10.1016/j.pbi.2017.01.007. [DOI] [PubMed] [Google Scholar]
- Ullah S, Bramley H, Daetwyler H, He S, Mahmood T, Thistlethwaite R, et al. Genetic contribution of emmer wheat (Triticum dicoccon Schrank) to heat tolerance of bread wheat. Front Plant Sci. 2018;9:1529. doi: 10.3389/fpls.2018.01529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Slageren MW (1994) Wild wheats: a monograph of Aegilops L. and Amblyopyrum (Jaub. and Spach) Eig (Poaceae). Wageningen Agricultural University, 1994.
- Volk GM, Chao CT, Norelli J, Brown SK, Fazio G, Peace C, et al. The vulnerability of US apple (Malus) genetic resources. Genet Resour Crop Evol. 2015;62:765–794. doi: 10.1007/s10722-014-0194-2. [DOI] [Google Scholar]
- von Caemmerer S, Furbank RT. Strategies for improving C4 photosynthesis. Curr Opin Plant Biol. 2016;31:125–134. doi: 10.1016/j.pbi.2016.04.003. [DOI] [PubMed] [Google Scholar]
- Voss-Fels KP, Stahl A, Wittkop B, Lichthardt C, Nagler S, Rose T. Breeding improves wheat productivity under contrasting agrochemical input levels. Nat Plants. 2019;5:706–714. doi: 10.1038/s41477-019-0445-5. [DOI] [PubMed] [Google Scholar]
- Walker BJ, VanLoocke A, Bernacchi CJ, Ort DR. The costs of photorespiration to food production now and in the future. Annu Rev Plant Biol. 2016;67:107–129. doi: 10.1146/annurev-arplant-043015-111709. [DOI] [PubMed] [Google Scholar]
- Walkowiak S, Gao L, Monat C, Haberer G, Kassa MT, Brinton J. Multiple wheat genomes reveal global variation in modern breeding. Nature. 2020;588:277–283. doi: 10.1038/s41586-020-2961-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weyhrich RA, Carver BF, Martin BC. Photosynthesis and water-use efficiency of awned and awnletted near-isogenic lines of hard red winter wheat. Crop Sci. 1995;35:172–176. doi: 10.2135/cropsci1995.0011183X003500010032x. [DOI] [Google Scholar]
- Weigmann M, Maurer A, Pham A, March TJ, Al-Abdallat A, Thomas WTB, Bull HJ et al. (2019) Barley yield formation under abiotic stress depends on the interplay between flowering time genes and environmental cues. Sci Rep 9:1–16. 10.1038/s41598-019-42673-1 [DOI] [PMC free article] [PubMed]
- Wilhelm EP, Turners AS, Laurie DA. Photoperiod insensitive Ppd-A1a mutations in tetraploid wheat (Triticum durum Desf.) Theor Appl Genet. 2009;118:285–294. doi: 10.1007/s00122-008-0898-9. [DOI] [PubMed] [Google Scholar]
- Wilson MJ, Fradera‐Soler M, Summers R, Sturrock CJ, Fleming AJ. Ploidy influences wheat mesophyll cell geometry, packing and leaf function. Plant Direct. 2021;5:e00314. doi: 10.1002/pld3.314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winfield MO, Wilkinson PA, Allen AM, Barker GLA, Coghill JA, Burridge A, et al. Targeted re-sequencing of the allohexaploid wheat exome. Plant Biotechnol J. 2012;10:733–742. doi: 10.1111/j.1467-7652.2012.00713.x. [DOI] [PubMed] [Google Scholar]
- Winfield MO, Allen AM, Burridge AJ, Barker GLA, Benbow HR, Wilkinson PA, et al. High-density SNP genotyping array for hexaploid wheat and its secondary and tertiary gene pool. Plant Biotechnol J. 2016;14:1195–1206. doi: 10.1111/pbi.12485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winfield MO, Allen AM, Wilkinson PA, Burridge AJ, Barker GLA, Coghill JA, et al. High-density genotyping of the A.E. Watkins Collection of hexaploid landraces identifies a large molecular diversity compared to elite bread wheat. Plant Biotechnol J. 2018;16:165–175. doi: 10.1111/pbi.12757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wingen LU, Orford S, Goram R, Leverington-Waite M, Bilham L, Patsiou TS, et al. Establishing the A. E. Watkins landrace cultivar collection as a resource for systematic gene discovery in bread wheat. Theor Appl Genet. 2014;127:1831–1842. doi: 10.1007/s00122-014-2344-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wingen LU, West C, Leverington M, Collier S, Orford S, Goram R, et al. Wheat landrace genome diversity. Genetics. 2017;205:1657–1676. doi: 10.1534/genetics.116.194688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worland AJ, Snape JW. Genetic basis of worldwide wheat varietal improvement. In: Bonjean AP, Angus WP, editors. World wheat book – a history of wheat breeding. Paris, France: Lavoisier Publishing; 2001. pp. 59–100. [Google Scholar]
- Wright TIC, Burnett AC, Griffiths H, Kadner M, Powell JS, Oliveira HR, et al. Identification of quantitative trait loci relating to flowering time, flag leaf and awn characteristics in a novel Triticum dicoccum mapping population. Plants. 2020;9:829. doi: 10.3390/plants9070829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Würschum T, Rapp M, Miedaner T, Longin CFH, Leiser WL. Copy number variation of Ppd-B1 is the major determinant of heading time in durum wheat. BMC Genet. 2019;20:1–8. doi: 10.1186/s12863-019-0768-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu C, Ren Y, Jian Y, Guo Z, Zhang Y, Xie C, et al. Development of a maize 55 K SNP array with improved genome coverage for molecular breeding. Mol Breed. 2017;37:20. doi: 10.1007/s11032-017-0622-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida T, Nishida H, Zhu J, Nitcher R, Distelfield A, Akashi Y, et al. Vrn-D4 is a vernalisation gene located on the centromeric region of chromosome 5D in hexaploid wheat. Theor Appl Genet. 2010;120:543–552. doi: 10.1007/s00122-009-1174-3. [DOI] [PubMed] [Google Scholar]
- Yu H, Luedeling E, Xu J. Winter and spring warming result in delayed spring phenology on the Tibetan Plateau. Proc Natl Acad Sci USA. 2010;107:22151–22156. doi: 10.1073/pnas.1012490107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao N, Zhu B, Li M, Wang L, Xu L, Zhang H, et al. Extensive and heritable epigenetic remodeling and genetic stability accompany allohexaploidization of wheat. Genetics. 2011;188:499–510. doi: 10.1534/genetics.111.127688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Zhou S, Fu Y, Su Z, Wang X, Sun C. Identification of a drought tolerant introgression line derived from Dongxiang common wild rice (O. rufipogon Griff.) Plant Mol Biol. 2006;62:247–259. doi: 10.1007/s11103-006-9018-x. [DOI] [PubMed] [Google Scholar]
- Zhu X-G, Long SP, Ort DR. Improving photosynthetic efficiency for greater yield. Annu Rev Plant Biol. 2010;61:235–261. doi: 10.1146/annurev-arplant-042809-112206. [DOI] [PubMed] [Google Scholar]