Abstract
A comprehensive investigation of the neoantigen spectrum and immune infiltration in patients with hepatocellular carcinoma (HCC) is lacking. This study aimed to examine the molecular features correlating with better prognoses in HCC patients. 27 paired tumor and normal tissues from 27 HCC patients were collected and performed with whole-exome sequencing. The most frequently mutated gene in 27 HCC patients was TP53 (16/27, 59.26%). Based on the whole median disease-free survival (DFS), all patients were divided into ‘long-term’ (n = 14, median DFS = 318 weeks) and ‘short-term’ (n = 13, median DFS = 11 weeks) groups. RNA-seq was performed to compare differentially expressed genes, immune infiltration, and neoantigens. Immunohistochemistry was performed to evaluate the immune infiltration. There were no significant differences in tumor mutation burden, immune score, cytolytic activity score, or neoantigen load between two groups. Compared with the long-term group, significantly increased B lineage (P = 0.0463), myeloid dendritic cells (P = 0.0152), and fibroblast (P = 0.0244) infiltration levels were observed in the short-term group, in which genes involved in ribosome, proteasome, and ECM-receptor interaction pathways were also overexpressed. Additionally, 16 patients with tumor thrombus were explored to identify specific biomarkers for prognosis. We found that patients with tumor thrombus carrying TP53/ARID2 neoantigens had significantly longer DFS. In conclusion, higher B lineage, myeloid dendritic cells, and fibroblast infiltration levels might cause poor prognosis in the short-term group, which also showed higher expression of genes involved in ribosome, proteasome, and ECM-receptor interaction pathways. In patients with tumor thrombus, specific TP53/ARID2 neoantigens may be used as biomarkers toward personalized immunotherapy.
Keywords: Hepatocellular carcinoma (HCC), neoantigen, integrated multiomics analysis, immune
Introduction
Hepatocellular carcinoma (HCC) is one of the most common malignancies causing cancer-related deaths worldwide, with very limited therapeutic options [1]. Although most HCC cases can be attributed to hepatitis B virus (HBV) infection, other etiological factors include hepatitis C virus (HCV) infection, alcohol, aflatoxins, and nonalcoholic fatty liver disease, NAFLD, etc. [2]. In China, HCC patients carry a relatively low 5-year overall survival rate (11.8%-13.3%) [3], and long-term survival is uncommon, particularly in advanced stages. According to previous studies, factors including gene abnormalities [4], immune infiltration [5], and tumor stage [6], have been generally considered as predictors of prognosis in cancers. However, it is also worth exploring novel prognostic biomarker for HCC patients with different DFS.
Mutations inside the tumor tissue are caused by genome instability, which could stimulate tumor-specific mutated neoantigens accumulation, minimizing central T cell immune tolerance [7]. Therefore, neoantigens are an attractive target to increase antitumor immunity and personalized cancer immunotherapy [7]. With advances in next-generation sequencing (NGS) technologies, neoantigen discovery has become possible. Immune cells could contribute a complicated antitumor immune process of suppression or promotion by activating cytotoxic T cells or expressing immunosuppressive factors, respectively [8]. It has been observed that neoantigens strongly correlate with tumor mutation burden (TMB) as well as the clinical benefit in several cancers treated with immune checkpoint inhibitors [9]. Additionally, the efficacy of neoantigen-based personal vaccines has been verified in melanoma and glioblastoma, and it enhances a supplement for neoantigen as a promising target in cancer immunotherapy [10,11]. Several studies have descripted the neoantigen and/or immunogenomic features in HCC, which were focused on western population and heterogeneous immunogenomic landscapes for multifocal HCC [12,13]. Despite the above-mentioned recent advances in cancer treatment and neoantigen research, specific neoantigen biomarker predicted for a better prognosis has not been comprehensively discovered in patients with HCC in China.
Here, we sought to examine the molecular features correlating with a better prognosis of HCC patients, hoping to find biomarkers for prognosis prediction and optimization of personalized treatments. For that, we collected tissues from 27 HCC patients. Subsequently, we performed whole-exome sequencing (WES) and RNA sequencing, and molecular features were characterized, including somatic gene mutation, TMB, neoantigen, immune infiltration, and differentially expressed genes (DEGs).
Materials and methods
Patients and sample characteristics
27 paired formalin-fixed paraffin-embedded (FFPE) samples (tumor and matched adjacent non-cancerous tissue) were collected from Chinese HCC patients undergoing surgery from June 2012 to April 2016 at the Fifth Medical Center of Chinese PLA General Hospital. The enrollment criteria of patients is: (a) histologically proven HCC, (b) 18~70 years old, (c) no gender limitation, (d) enough tissue samples are available for sequencing, (e) no serious comorbidities of other organs, (f) regular follow-up after surgery. The diagnosis of all HCC cases (stages ranging from T1NM to T3NM) was confirmed by two independent hepatic pathologists. All the clinical characteristics were shown in Supplementary Table 1. All the patients did not receive immunotherapy treatment before or after surgery. We collected tumor tissues from primary HCC at the time of surgery, and the tissues had been confirmed by two pathologists before sequenced. We have obtained the informed consent from all patients. This study was approved by the ethical committee of the Fifth Medical Center of Chinese PLA General Hospital and was performed in accordance with the ethical standards of the World Medical Association Declaration of Helsinki. To better identify differences in gene mutation, gene expression, and immune infiltration of the 27 HCCs, the cohort was divided into ‘short-term’ and ‘long-term’ survivors based on the median DFS (78 weeks). The ‘long-’ and ‘short-term’ group included patients with DFS ≥ 78 weeks from surgery and those with DFS < 78 weeks from surgery, respectively.
WES and somatic mutation analysis
DNA samples were collected from tumor and matched normal FFPE tissues with the GeneRead DNA FFPE Kit. SureSelect Kit (v.4, Agilent) was used to construct DNA libraries, and WES was performed using HiSeq X10 platform (Illumina Inc., San Diego, CA, USA). The WES depth of tumor tissues is > 300× and the WES depth of normal tissues is > 100×. Sequencing reads containing many Ns (> 5%), low-quality bases (> 15% bases with quality ≤ 19) and adaptor sequences were discarded. Then paired-end reads with high-quality were subjected to mutation analysis and mapped to the human genome (hg19) using Burrows-Wheeler Aligner (BWA version 0.7.15, default parameters, BWA-MEM algorithm). Software GATK MuTect2 (version 4.1, default parameters) was used to call somatic SNVs and InDels.
RNA-seq and gene expression
RNA from FFPE tissues was extracted with RNeasy FFPE Kit (Qiagen Inc., Germantown, USA). RNA library was constructed with TruSeq RNA Exome kit (Illumina, Inc., San Diego, CA, USA) and then was sequenced on HiSeq X10 platform (Illumina Inc., San Diego, CA, USA). Sequencing reads containing many Ns (> 5%), low-quality bases (> 15% bases with quality ≤ 19) and adaptor sequences were discarded. Then paired-end reads with high-quality were retained for further data analysis. High-quality reads were mapped to the reference genome hg19 via Bowtie software (version 2.2.4) with default parameters. Gene expression was calculated as reads per kilobase per million reads (FPKM) with Cufflinks software (version 2.2.1, default parameters).
Identification of DEGs and KEGG pathway enrichment
DEGs were identified with Cuffdiff (version 2.2.1, default parameters). Significant DEGs met with the following criteria: the absolute value of log2 (fold change) ≥ 1, and P < 0.05. KEGG (http://www.genome.jp/kegg/) function enrichment analysis was carried out with KOBAS (version 2.1.1, default parameters). The enriched pathway with P < 0.05 were considered statistically significant.
Gene set enrichment analysis (GSEA)
The GSEA method (version 4.0.3, default parameters) was used to identify the significant pathways of the two groups. The KEGG subset of CP in C2 curated gene sets of MSigDB (http://software.broadinstitute.org/gsea/msigdb) served as the reference gene sets. The permutation analysis (1000 permutations) was performed to determine the thresholds for significance. Enrichment results with |NES| > 1, nominal P-value < 0.05, and false discovery rate (FDR) q-value < 0.25 were considered statistically significant.
Neoantigen predictions
OptiType (version 1.3.1) or seq2HLA (version 2.2) software was used to get the HLA I or HLA II typing from normal DNA with default parameters. Neoantigens were identified with pVACseq [14] (version 4.0.10, default parameters). NetMHC (version 2.22.1) or PickPocket (version 2.22.1), NetMHCIIpan (version 2.22.1) software was used to predict the affinity between peptide and MHC with half-maximal inhibitory concentration (IC50) with default parameters. In general, IC50 < 500 nM is considered as a basic binder threshold, and IC50 ≤ 150 nM as a strong binder [15]. Mutated peptides with IC50 < 500 nM was deemed as candidate neoantigens, and peptides with IC50 ≤ 150 nM were considered as strong-quality neoantigens [11]. Moreover, gene expression of mutated peptides should be at least 1 in FPKM [6].
Immune infiltration estimation
Software ESTIMATE (version 1.0.13, default parameters) was used to calculate the fraction of stromal and immune cells in tumor samples based on FPKM, which characterized the infiltration of immune cells with ‘Immune score’. The absolute cell abundance in the tumor was estimated with MCP-counter software (version 1.1.0, default parameters), including 8 immune cells and other 2 stromal cells. Additionally, CYT score was calculated as the geometric mean [16] based on the expression (expressed in TPM) of two genes (GZMA, PRF1).
Immunohistochemistry (IHC)
We selected four gene markers (CD8, CD45RO, CD3 and FOXP3) to perform IHC, and antibodies from Abcam (USA) were used. Four 5-μm thick sections were prepared from patient and dewaxed. Then slides were treated with 1 mM EDTA pH 8.0 at boiling condition, followed by 15 min at sub-boiling temperature. Slides were treated with 3% hydrogen peroxide for 15 min, washed 3 times with PBS for 5 min each, and then blocked with 10% goat serum for 10 min. Diluent primary antibodies used were as follows: CD8 (1:100, ab4055), CD45RO (1:150, ab23), CD3 (1:100, ab16669) and FOXP3 (1:200, ab20034). After incubation overnight at 4°C, the sections were treated with biotinylated secondary antibody for 30 min. Slides were stained with DAB and following washed by water. Then hematoxylin was used for counterstaining 90 seconds. Finally, slides were rinsed by water and mounted for observation of antibody-antigen binding complex.
Clinical risk factors associated with DFS in HCC
To assess the risk factors for DFS in this HCC cohort, hazard ratios (HR) were calculated to evaluate the clinical features at cancer diagnosis in association with survival using univariate and multivariate Cox regression analyses. Variables with a P-value < 0.0500 in the univariate Cox model were used in the multivariate Cox regression analysis.
Statistical analysis
Statistical analysis was performed using GraphPad Prism 8.0 software (GraphPad Software, San Diego, USA). Comparisons between groups were performed with two-sided Fisher’s exact test for categorical variables and two-tailed unpaired t test for numerical variables. The Log-rank test was used to calculate the difference in survival. P < 0.0500 was deemed as statistically significant. Figures were plotted with GraphPad Prism 8.0 software (GraphPad Software, San Diego, USA) or R (https://cran.r-project.org/).
Results
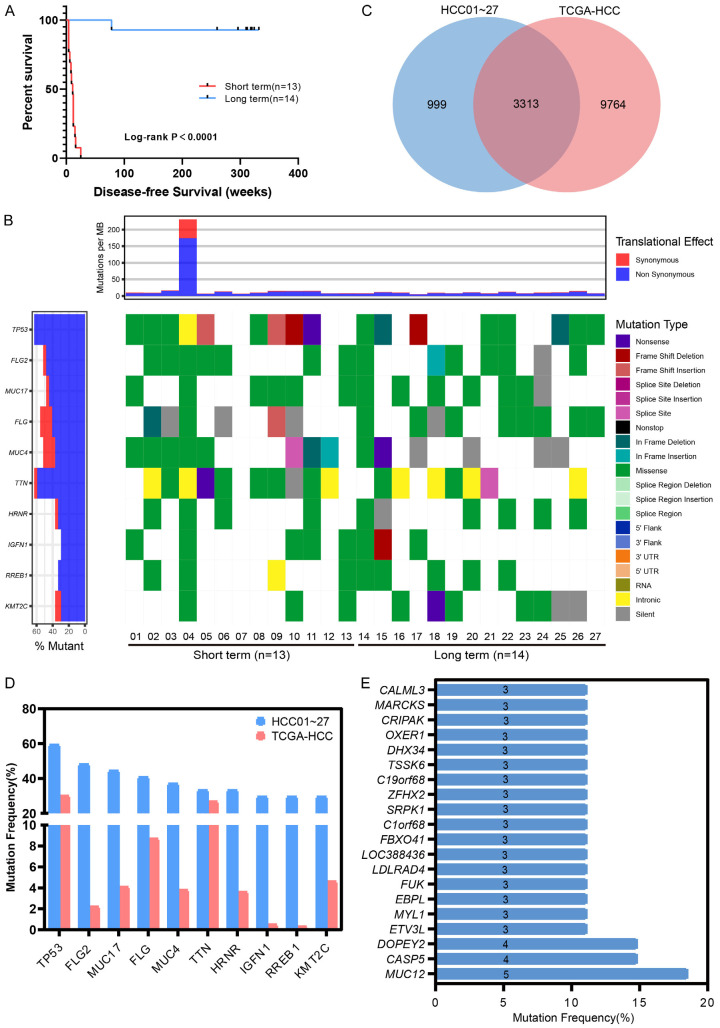
Disease-free survival (DFS) comparison and gene mutation landscape
Twenty-seven HCC patients (median DFS, 78 weeks) undergoing treatment at Fifth Medical Center of Chinese PLA General Hospital were enrolled in this study. The ‘long-term’ (n = 14, median DFS, 318 weeks) and ‘short-term’ (n = 13, median DFS, 11 weeks) groups significantly differed in terms of DFS with P < 0.0001 (Figure 1A).
Figure 1.
Disease-free survival comparison and gene mutation landscape in HCC patients. A. DFS of 27 HCC cohort patients. B. Gene mutation landscape of 27 HCC cohort patients. C. Mutated gene comparison between HCC01-27 and TCGA-HCC. D. Mutation frequency comparison between TCGA-HCC and top 10 mutated genes in HCC01-27. E. Gene mutation frequency (specific mutations in the 27 HCC) in HCC01-27. Genes mutated in at least 3 samples are shown.
All the FFPE tissues were performed with WES. A median of 149 somatic mutations (57-2283) was observed in each patient. Among all these mutations, only genes mutated in at least two patients were selected for further analysis. A total of 4312 gene mutation were obtained, and 20.80% of them (897/4312) were mutated in at least two patients. TP53 was the most frequently mutated gene (16/27, 59.26%), followed by FLG2 (48.15%), MUC17 (44.44%), FLG (40.74%), MUC4 (37.04%), TTN and HRNR (33.33%), IGFN1 (29.63%), RREB1 and KMT2C (29.63%) (Figure 1B). Compared with gene mutation from HCC cases in the TCGA database, we identified 3313 common genes (Figure 1C). Top 10 frequently mutated genes in this study were all in TCGA-HCC, although they showed a higher mutation frequency than in the database (Figure 1D), which may have been caused by our limited number of patients or ethnicity. In addition, we found 999 previously unreported mutated genes, 126 of which were found in at least 2 samples, and 20 in at least 3 samples (Figure 1E).
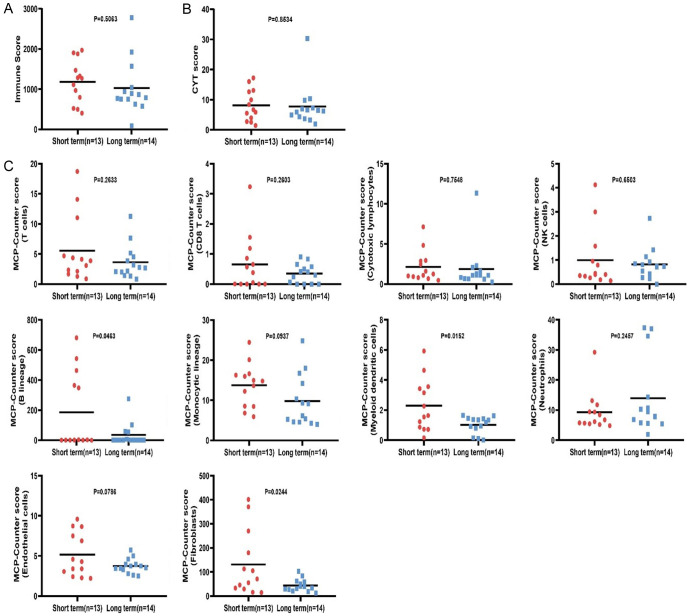
Immune infiltration comparison between long/short-term groups
RNA-seq data were used to calculate immune infiltration with three scores, Immune, CYT, and MCP-Counter scores. Immune infiltration was compared between the two groups. The immune and CYT scores showed no statistically significant differences between groups (Figure 2A, 2B). Significantly increased myeloid dendritic cells infiltration was observed in the short-term group compared with the long-term group, and the similar trend was witnessed in B lineage and fibroblast infiltration but with a relatively large variance (Figure 2C). While other cell infiltration showed no statistically significant differences (Figure 2C).
Figure 2.
Immune infiltration differences between the long- and short-term groups. A. Immune score. B. CYT score. C. MCP-Counter score (10 kinds of cells).
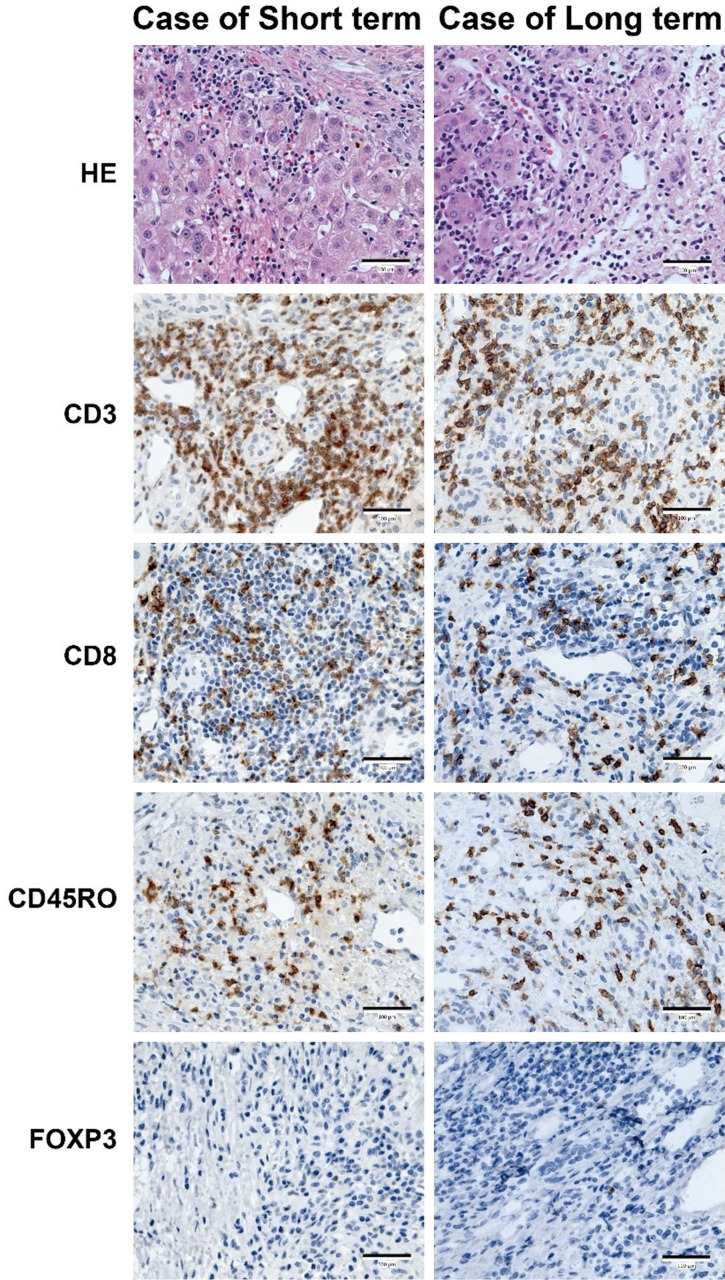
Immune infiltration evaluation of the long- and short-term groups via IHC
To further understand the immune infiltration differences between the long- and short-term groups, CD3, CD8, CD45RO, and FOXP3 were chosen to perform IHC in tumor samples. As shown in Figure 3, a tendency toward higher CD3+ T cells was observed in the short-term group (consistent with Figure 2C, but no statistical significance). No obvious difference of CD8+, CD45RO+, and FOXP3+ were witnessed between two groups.
Figure 3.
HE (400×) and immunohistochemistry (400×) images from patients belonging to the long- and short-term groups. Bar, 100 μm.
Different gene expression spectrum and pathway enrichment in the long- and short-term groups
To identify the DEGs and key pathways, the transcriptomes of samples from the long- and short-term groups were compared. In comparison with ‘long-term’ samples, a total of 951 DEGs were identified, including 386 upregulated and 565 downregulated genes in the ‘short-term’ group based on the criteria of P-value < 0.05 and |log2(Fold Change)| ≥ 1 (Figure 4A). Further, we conducted a KEGG pathway enrichment analysis to understand the functional roles of these DEGs and identify key pathways in our HCC patients. There were 33 pathways enriched based on the criteria of P-value < 0.05, including 9 for upregulated genes and 24 for downregulated genes. The top 20 statistically significant pathways are shown in Figure 4B based on the ranked P-value. Pathways, including protein digestion and absorption, and ECM-receptor interaction, were significantly enriched in the short-term group (Figure 4B). Similarly, a higher fibroblast infiltration exists in short-term group (shown in Figure 2C), which indicates that fibroblasts could construct new extracellular matrix (ECM) necessary to support cell ingrowth and potentially are involved in matrix remodeling. Drug metabolism-cytochrome P450, metabolism of xenobiotics by cytochrome P450, and chemical carcinogenesis were significantly enriched in the long-term group (Figure 4B). Here, we only focused on those pathways most possibly relevant to phenotype in this study.
Figure 4.
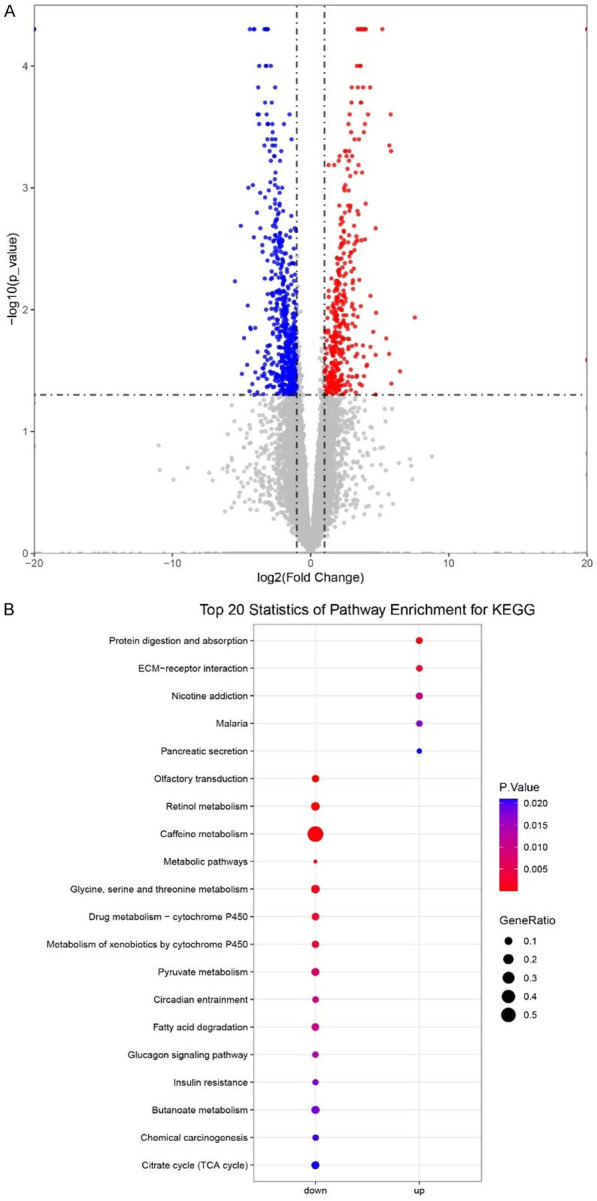
Gene expression comparison between the long- and short-term groups. A. Volcano plot of gene expression. Red dot, significantly higher expressed genes in the short-term group. Blue dot, significantly higher expressed genes in the long-term group. Gray dot, non-significant expressed genes. B. Top 20 significant pathways of DEGs.
GSEA of long- vs. short-term groups
To characterize the functional signaling pathway differences, GSEA was additionally utilized to identify the pathways enriched in the two groups. The results demonstrated that 20 pathways were enriched in the short-term group (NES < 0). Detailed information on several enriched pathways is shown in Supplementary Table 2. It suggested a higher expression of genes involved in the ribosome, proteasome, ECM-receptor interaction, etc., in the short-term group (Supplementary Table 2 and Figure 5), which is similar with KEGG pathway enrichment in Figure 4B.
Figure 5.
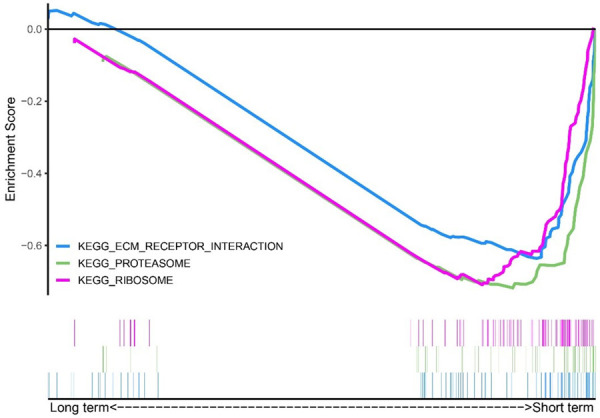
Enrichment score of several significantly enriched pathways in the short-term group.
TMB or neoantigen load has no relation to patient survival
As reported in the previous study, TMB and neoantigen load are considered to be associated with cancer prognosis [17,18]. Therefore, we analyzed the relationship between these two factors with prognosis in this study. TMB value ranged from 0.87 to 35.12 (median ≈ 2.30) in these HCC patients. The number of neoantigen (IC50 < 500 nM) ranged from 1 to 562 (median = 119) with. The number of neoantigen with high-quality (IC50 ≤ 150 nM) ranged from 0 to 205 (median = 34).
We compared TMB and neoantigen loads between the two groups. However, there were no significant differences (Supplementary Figure 1A-D). Then, we regrouped all patients into new groups based on the median value of TMB and neoantigen load. There was no significant correlation between TMB, neoantigen load, and DFS in HCC patients (Supplementary Figure 2A-C). Therefore, high mutation and neoantigen load may not contribute to DFS in HCC patients who do not undergo immunotherapy.
Clinical risk factors associated with DFS
A total of 35 clinical features were included in the univariate Cox regression analysis, and the results are summarized in Supplementary Table 3. Nine variables (including tumor thrombus, AST, DBIL, ALT, ALP, TBIL, AFP, CHE, and virus) with P < 0.0500 were included in the multivariate Cox analysis (variables in bold of Supplementary Table 3). According to our analyses, tumor thrombus was an independent risk factor for DFS in HCC (Figure 6, HR = 9.553, 95% CI = 1.624-56.191, P = 0.0126). In conclusion, the results demonstrate that carrying a tumor thrombus at diagnosis increases the risk of relapse or death in HCC patients.
Figure 6.
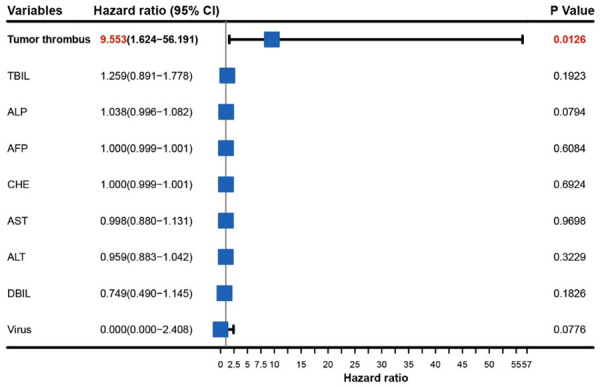
The association between 9 variables and relapse or death risk using multivariate Cox regression analysis. Note: AST, Aspartate Aminotransferase; DBIL, Direct bilirubin; ALT, Alanine transaminase; ALP, Alkaline phosphatase; TBIL, Total bilirubin; AFP, Alpha-fetoprotein; CHE, Cholinesterase.
The TP53/ARID2 neoantigens predict better prognosis in HCC patients with tumor thrombus
HCC tends to involve vascular structures in the liver, such as the portal veins and the hepatic veins. The prognosis of patients with HCC accompanied by portal vein tumor thrombus is generally poor if left untreated. However, we noted that in this project, several untreated patients also had a long DFS even with tumor thrombus. Thus, we analyzed neoantigens among 16 patients with tumor thrombus and explored whether specific neoantigens contributed to their survival. The 16 patients were grouped according to the median DFS. As shown in Figure 7A, the number of patients carrying the TP53/ARID2 neoantigens in the two groups was significantly different (Fisher’s exact test, P = 0.0338), and a tendency toward higher frequency was observed in the long-term DFS group. Patients were then regrouped based on the TP53/ARID2 neoantigens, suggesting that patients carrying the TP53/ARID2 neoantigens had a longer DFS (Figure 7B, P = 0.0127).
Figure 7.
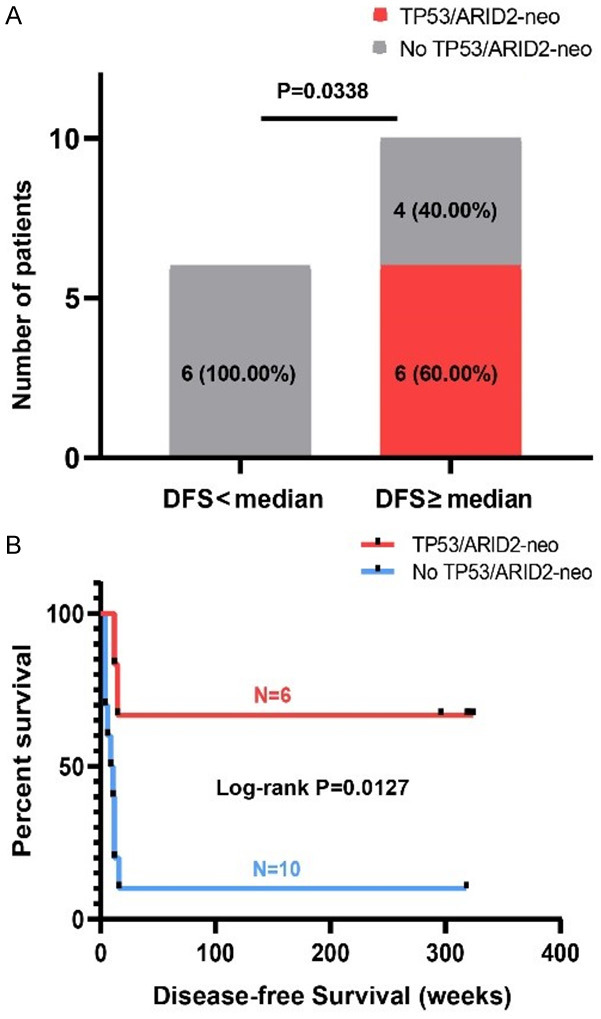
Identification of specific neoantigen markers among 16 HCC patients with tumor thrombus. A. Comparison of the number of patients (grouped according to the median DFS) carrying the TP53/ARID2 neoantigen. B. DFS comparison of 16 HCC patients with tumor thrombus.
Discussion
A better understanding of the molecular mechanisms and relative pathways that play a key role in prolonging patient survival may potentially provide new clinical therapeutic strategies. Here, we describe the mutation, immune, and neoantigen landscape of 27 HCC patients in a Chinese population. For the top mutated genes, a higher mutation frequency was observed in this project than in public mutations from HCC patients in TCGA. Additionally, 999 genes were only mutated in this project, which may have been due to ethnicity differences or our limited number of patients.
It was reported that higher TMB and neoantigen loads were related to worse prognosis in head and neck squamous cell carcinoma, NSCLC, adenoid cystic carcinoma, and myeloma [6,19]. However, controversial result was obtained in clear cell renal cell carcinoma [17], which showed TMB or neoantigen load had no association with prognosis. In our study, we observed the same result that DFS was not correlated with neoantigen or TMB load. The specfic reasons for such inconsistency are indistinct, which may be attributed to cancer histologic type, treatment, or sample sizes.
We found no difference in neoantigen or TMB load between two groups. Besides, all patients had a less favorable mutational profile for immunotherapy, with very low TMB (range 0.87-4.44), except one patient (HCC03, TMB ≈ 35.12). Therefore, we concluded that immunotherapy, such as checkpoint inhibitors, is unlikely to achieve high response rates in patients with HCC. No differences in immune and CYT scores were observed between the long- and short-term groups. Interestingly, we observed significantly increased B lineage, myeloid dendritic cells, and fibroblast infiltration in the short-term group. In this study, higher expression of transforming growth factor beta (TGFβ) was observed in short-term group (data not shown) and PI3K-AKT signaling pathway was significantly enriched in KEGG enrichment analysis in short-term group (data not shown). Therefore, we speculate the possible molecular mechanism that induces the increased infiltration of these cells may be attributed to increased TGFβ expression and PI3K-AKT signaling activation. TGFβ has been suggested to play an important role in shaping the cancer-associated fibroblasts (CAF) landscape, which contributes to CAF proliferation and differentiation [20-22]. Also, TGFβ could involve in the generation of dendritic cells by protecting progenitor cells from apoptosis [23]. In previous studies, researchers found that activation of PI3K-Akt signaling pathway would increase the proliferation of fibroblasts and suppress the autophagy and apoptosis [24,25].
There remains considerable controversy over the influence of tumor-infiltrating B lymphocytes. Although positive results have shown that high tumor-infiltrating B cells predicts a better prognosis in HCC [26], several studies have shown that high infiltration of B cells is associated with shorter survival and tumor progression in melanoma [27], renal cell cancer [28], and lung adenocarcinoma [29], as it is likely that patients with malfunctioning B cells and less efficient B-cell subsets have a higher infiltration of B TILs [30]. B cells exerted pro-tumoral functions and caused poor prognosis due to numerous mechanisms, including immune regulatory cytokines, production of autoantibodies and complement conjugation [31].
For myeloid dendritic cells, despite the common belief that myeloid dendritic cells provide specific stimulatory functions, this concept has been overturned in many reports [32,33]. Dendritic cells might be polarized into immunosuppressive or tolerogenic regulatory cells, limiting the activity of effector T cells and supporting tumor growth and progression. This means that myeloid dendritic cells may differentiate into different phenotypes and exhibit differential anti-tumoral or pro-tumoral functions. CAFs are components of the tumor microenvironment that promote the proliferation and invasion of cancer cells by secreting various growth factors and cytokines, which has been shown to correlate with poor prognosis tumor progression in cancers, including HCC [34]. Activated fibroblasts could secrete a lot of extracellular matrix (ECM), which can cause liver fibrosis. In summary, poor prognosis in the short-term group is likely attributable to stronger immunosuppression.
In this study, genes involved in ribosome, proteasome, and ECM-receptor interaction was significantly higher expressed in the short-term group, which might be caused by higher expression of TGFβ and antigen processing and presentation signaling. TGFβ could promote CAF to secrete ECM-cell-adhesion proteins including collagen, fibronectin, and integrins [35,36] and stimulate fibroblast to evolve into myofibroblast. Ribosomes are cytoplasmic granules containing ribosomal RNA (rRNAs) and ribosomal proteins (RPs) in which protein synthesis occurs. Over the decades, accumulative studies have shown that many genes with tumorigenic extra-ribosomal functions involved in translation [37,38] and played important roles in cell cycle regulation and apoptosis [39]. Also, researchers have found that elevated expression of ribosomal genes was associated with a poor prognosis of patients with solid tumors [40-43], including HCC. Proteasome is a multifunctional proteinase account for the degradation of intracellular proteins, like proteins vital for cell cycle regulation and cell apoptosis. For example, several studies have shown that overexpression of PSMD14 gene (the regulatory particle subunit of proteasome) could promote the development of cancer and cause a poor prognosis in HCC by: (a) deubiquitinating and stabilizing E2F1 [44], (b) deubiquitinating GRB2 and inducing continuous activation of PI3K/AKT signaling [45], (c) deubiquitinating P53/Bim to prevent cell apoptosis [46]. ECM could control homeostasis and tissue development, and its dysregulation might lead to neoplastic progression. ECM plays an important role in biochemical process that contributes to cell growth, differentiation and migration. ECM molecules could stimulate signaling pathways by bind to cell surface receptors. Sustaining proliferative signals from ERK and PI3K could be induced by ECM adhesion and its receptors [47]. Also, cell-ECM interactions can inhibit growth suppressors to subvert cell cycle arrest by regulating TGFβ signaling [48]. Besides, a stiffened ECM could promote TGFβ to induce EMT and stimulate cancer metastasis [49]. Higher expression of ITGA6 and laminin predicts a poor prognosis for HCC patients [50,51].
Tumor thrombus plays a vital role in the prognosis among patients with HCC [52]. Interestingly, among the 16 patients with tumor thrombus, those carrying the TP53/ARID2 neoantigens showed a better prognosis. Although TP53 and ARID2 are important tumor suppressor genes [53], we found a positive relationship between TP53/ARID2 neoantigens and better prognosis. It is in line with the previous study that MUC16 neoantigen, which facilitates a lasting neoantigen-specific immunity, had an association with a long-term survival in patients with pancreatic cancer [15]. It is probable that the ‘TP53/ARID2 neoantigens’ stimulate the specific antitumor immune response in long-term HCC in this study. Thus, the TP53/ARID2 neoantigens may be used for predicting prognosis or offer new treatment options among HCC cohort with tumor thrombus in the future. Moreover, it is worthwhile to perform more research to further explore the potential molecular mechanisms of ‘TP53/ARID2 neoantigen’ via other techniques, like T cell receptor (TCR) sequencing or single-cell sequencing.
Conclusions
In summary, this study provided insights into the mutational spectrum, immune infiltration, and potential mechanisms responsible for different clinical prognoses. Additionally, we provide new insight that patients carrying the specific TP53/ARID2 neoantigens showed a longer DFS with tumor thrombus, which might help guide future investigations of developing personalized immunotherapeutic approaches. More studies are also required to thoroughly explore the molecular mechanisms and factors that can affect or predict better prognosis in HCC patients.
Acknowledgements
This work was supported by grants from the National Natural Science Foundation of China (No. 81971720).
Disclosure of conflict of interest
None.
Abbreviations
- FFPE
Formalin-fixed paraffin-embedded
- HCC
Hepatocellular carcinoma
- TMB
Tumor mutation burden
- WES
Whole-exome sequencing
- PRF1
Perforin 1
- GZMA
Granzyme A
- CYT
Cytolytic activity
- IHC
Immunohistochemistry
- MHC
Histocompatibility complex
- NSCLC
Non-small cell lung cancer
- GSEA
Gene set enrichment analysis
- AST
Aspartate Aminotransferase
- DBIL
Direct bilirubin
- ALT
Alanine transaminase
- ALP
Alkaline phosphatase
- TBIL
Total bilirubin
- AFP
Alpha-fetoprotein
- CHE
Cholinesterase
- TCR
T cell receptor
Supplementary Tables 1, 3 and Supplementary Figures 1, 2
Supplementary Table 2
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Yang JD, Hainaut P, Gores GJ, Amadou A, Plymoth A, Roberts LR. A global view of hepatocellular carcinoma: trends, risk, prevention and management. Nat Rev Gastroenterol Hepatol. 2019;16:589–604. doi: 10.1038/s41575-019-0186-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Qiu WQ, Shi JF, Guo LW, Mao AY, Huang HY, Hu GY, Dong P, Bai FZ, Yan XL, Liao XZ, Liu GX, Bai YN, Ren JS, Sun XJ, Zhu XY, Zhou JY, Gong JY, Zhu L, Mai L, Du LB, Zhou Q, Xing XJ, Song BB, Liu YQ, Lou PA, Sun XH, Wu SL, Cao R, Qi X, Lan L, Ren Y, Zhang K, He J, Qu C, Dai M. Medical expenditure for liver cancer in urban China: a 10-year multicenter retrospective survey (2002-2011) J Cancer Res Ther. 2018;14:163–170. doi: 10.4103/jcrt.JCRT_709_16. [DOI] [PubMed] [Google Scholar]
- 4.He X, Wei Q, Sun M, Fu X, Fan S, Li Y. LS-CAP: an algorithm for identifying cytogenetic aberrations in hepatocellular carcinoma using microarray data. Front Biosci. 2006;11:1311–1322. doi: 10.2741/1885. [DOI] [PubMed] [Google Scholar]
- 5.Zhou C, Weng J, Liu C, Zhou Q, Chen W, Hsu JL, Sun J, Atyah M, Xu Y, Shi Y, Shen Y, Dong Q, Hung MC, Ren N. High RPS3A expression correlates with low tumor immune cell infiltration and unfavorable prognosis in hepatocellular carcinoma patients. Am J Cancer Res. 2020;10:2768–2784. [PMC free article] [PubMed] [Google Scholar]
- 6.Miller A, Asmann Y, Cattaneo L, Braggio E, Keats J, Auclair D, Lonial S, Russell S, Stewart A. High somatic mutation and neoantigen burden are correlated with decreased progression-free survival in multiple myeloma. Blood Cancer J. 2017;7:e612. doi: 10.1038/bcj.2017.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yarchoan M, Johnson BA, Lutz ER, Laheru DA, Jaffee EM. Targeting neoantigens to augment antitumour immunity. Nat Rev Cancer. 2017;17:569. doi: 10.1038/nrc.2017.74. [DOI] [PubMed] [Google Scholar]
- 8.Klink M. Interaction of immune and cancer cells. Anticancer Res. 2014;34:203–206. [Google Scholar]
- 9.Li L, Goedegebuure SP, Gillanders WE. Preclinical and clinical development of neoantigen vaccines. Ann Oncol. 2017;28(Suppl):xii11–xii17. doi: 10.1093/annonc/mdx681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sahin U, Derhovanessian E, Miller M, Kloke BP, Simon P, Löwer M, Bukur V, Tadmor AD, Luxemburger U, Schrörs B, Omokoko T, Vormehr M, Albrecht C, Paruzynski A, Kuhn AN, Buck J, Heesch S, Schreeb KH, Müller F, Ortseifer I, Vogler I, Godehardt E, Attig S, Rae R, Breitkreuz A, Tolliver C, Suchan M, Martic G, Hohberger A, Sorn P, Diekmann J, Ciesla J, Waksmann O, Brück AK, Witt M, Zillgen M, Rothermel A, Kasemann B, Langer D, Bolte S, Diken M, Kreiter S, Nemecek R, Gebhardt C, Grabbe S, Höller C, Utikal J, Huber C, Loquai C, Türeci Ö. Personalized RNA mutanome vaccines mobilize poly-specific therapeutic immunity against cancer. Nature. 2017;547:222–226. doi: 10.1038/nature23003. [DOI] [PubMed] [Google Scholar]
- 11.Keskin DB, Anandappa AJ, Sun J, Tirosh I, Mathewson ND, Li S, Oliveira G, Giobbie-Hurder A, Felt K, Gjini E, Shukla SA, Hu Z, Li L, Le PM, Allesøe RL, Richman AR, Kowalczyk MS, Abdelrahman S, Geduldig JE, Charbonneau S, Pelton K, Iorgulescu JB, Elagina L, Zhang W, Olive O, McCluskey C, Olsen LR, Stevens J, Lane WJ, Salazar AM, Daley H, Wen PY, Chiocca EA, Harden M, Lennon NJ, Gabriel S, Getz G, Lander ES, Regev A, Ritz J, Neuberg D, Rodig SJ, Ligon KL, Suvà ML, Wucherpfennig KW, Hacohen N, Fritsch EF, Livak KJ, Ott PA, Wu CJ, Reardon DA. Neoantigen vaccine generates intratumoral T cell responses in phase Ib glioblastoma trial. Nature. 2019;565:234–239. doi: 10.1038/s41586-018-0792-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Löffl er MW, Mohr C, Bichmann L, Freudenmann LK, Walzer M, Schroeder CM, Trautwein N, Hilke FJ, Zinser RS, Mühlenbruch L. Multi-omics discovery of exome-derived neoantigens in hepatocellular carcinoma. Genome Med. 2019;11:1–16. doi: 10.1186/s13073-019-0636-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dong LQ, Peng LH, Ma LJ, Liu DB, Zhang S, Luo SZ, Rao JH, Zhu HW, Yang SX, Xi SJ. Heterogeneous immunogenomic features and distinct escape mechanisms in multifocal hepatocellular carcinoma. J Hepatol. 2020;72:896–908. doi: 10.1016/j.jhep.2019.12.014. [DOI] [PubMed] [Google Scholar]
- 14.Hundal J, Carreno BM, Petti AA, Linette GP, Griffith OL, Mardis ER, Griffith M. pVAC-Seq: a genome-guided in silico approach to identifying tumor neoantigens. Genome Med. 2016;8:11. doi: 10.1186/s13073-016-0264-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Balachandran VP, Łuksza M, Zhao JN, Makarov V, Moral JA, Remark R, Herbst B, Askan G, Bhanot U, Senbabaoglu Y, Wells DK, Cary CIO, Grbovic-Huezo O, Attiyeh M, Medina B, Zhang J, Loo J, Saglimbeni J, Abu-Akeel M, Zappasodi R, Riaz N, Smoragiewicz M, Kelley ZL, Basturk O Australian Pancreatic Cancer Genome Initiative; Garvan Institute of Medical Research; Prince of Wales Hospital; Royal North Shore Hospital; University of Glasgow; St Vincent’s Hospital; QIMR Berghofer Medical Research Institute; University of Melbourne, Centre for Cancer Research; University of Queensland, Institute for Molecular Bioscience; Bankstown Hospital; Liverpool Hospital; Royal Prince Alfred Hospital, Chris O’Brien Lifehouse; Westmead Hospital; Fremantle Hospital; St John of God Healthcare; Royal Adelaide Hospital; Flinders Medical Centre; Envoi Pathology; Princess Alexandria Hospital; Austin Hospital; Johns Hopkins Medical Institutes; ARC-Net Centre for Applied Research on Cancer. Gönen M, Levine AJ, Allen PJ, Fearon DT, Merad M, Gnjatic S, Iacobuzio-Donahue CA, Wolchok JD, DeMatteo RP, Chan TA, Greenbaum BD, Merghoub T, Leach SD. Identification of unique neoantigen qualities in long-term survivors of pancreatic cancer. Nature. 2017;551:512–516. doi: 10.1038/nature24462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rooney MS, Shukla SA, Wu CJ, Getz G, Hacohen N. Molecular and genetic properties of tumors associated with local immune cytolytic activity. Cell. 2015;160:48–61. doi: 10.1016/j.cell.2014.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Matsushita H, Sato Y, Karasaki T, Nakagawa T, Kume H, Ogawa S, Homma Y, Kakimi K. Neoantigen load, antigen presentation machinery, and immune signatures determine prognosis in clear cell renal cell carcinoma. Cancer Immunol Res. 2016;4:463–471. doi: 10.1158/2326-6066.CIR-15-0225. [DOI] [PubMed] [Google Scholar]
- 18.Devarakonda S, Rotolo F, Tsao MS, Lanc I, Brambilla E, Masood A, Olaussen KA, Fulton R, Sakashita S, McLeer-Florin A, Ding K, Le Teuff G, Shepherd FA, Pignon JP, Graziano SL, Kratzke R, Soria JC, Seymour L, Govindan R, Michiels S. Tumor mutation burden as a biomarker in resected non-small-cell lung cancer. J. Clin. Oncol. 2018;36:2995–3006. doi: 10.1200/JCO.2018.78.1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Owada-Ozaki Y, Muto S, Takagi H, Inoue T, Watanabe Y, Fukuhara M, Yamaura T, Okabe N, Matsumura Y, Hasegawa T, Ohsugi J, Hoshino M, Shio Y, Nanamiya H, Imai JI, Isogai T, Watanabe S, Suzuki H. Prognostic impact of tumor mutation burden in patients with completely resected non-small cell lung cancer: brief report. J Thorac Oncol. 2018;13:1217–1221. doi: 10.1016/j.jtho.2018.04.003. [DOI] [PubMed] [Google Scholar]
- 20.Grauel AL, Nguyen B, Ruddy D, Laszewski T, Schwartz S, Chang J, Chen J, Piquet M, Pelletier M, Yan Z. TGFβ-blockade uncovers stromal plasticity in tumors by revealing the existence of a subset of interferon-licensed fibroblasts. Nat Commun. 2020;11:1–17. doi: 10.1038/s41467-020-19920-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Biffi G, Oni TE, Spielman B, Hao Y, Elyada E, Park Y, Preall J, Tuveson DA. IL1-induced JAK/STAT signaling is antagonized by TGFβ to shape CAF heterogeneity in pancreatic ductal adenocarcinoma. Cancer Discov. 2019;9:282–301. doi: 10.1158/2159-8290.CD-18-0710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Morikawa M, Derynck R, Miyazono K. TGF-β and the TGF-β family: context-dependent roles in cell and tissue physiology. Cold Spring Harb Perspect Biol. 2016;8:a021873. doi: 10.1101/cshperspect.a021873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Riedl E, Strobl H, Majdic O, Knapp W. TGF-beta 1 promotes in vitro generation of dendritic cells by protecting progenitor cells from apoptosis. J Immunol. 1997;158:1591–1597. [PubMed] [Google Scholar]
- 24.Dai J, Sun Y, Chen D, Zhang Y, Yan L, Li X, Wang J. Negative regulation of PI3K/AKT/mTOR axis regulates fibroblast proliferation, apoptosis and autophagy play a vital role in triptolide-induced epidural fibrosis reduction. Eur J Pharmacol. 2019;864:172724. doi: 10.1016/j.ejphar.2019.172724. [DOI] [PubMed] [Google Scholar]
- 25.Xie T, Xu Q, Wan H, Xing S, Shang C, Gao Y, He Z. Lipopolysaccharide promotes lung fibroblast proliferation through autophagy inhibition via activation of the PI3K-Akt-mTOR pathway. Lab Invest. 2019;99:625–633. doi: 10.1038/s41374-018-0160-2. [DOI] [PubMed] [Google Scholar]
- 26.Garnelo M, Tan A, Her Z, Yeong J, Lim CJ, Chen J, Lim KH, Weber A, Chow P, Chung A, Ooi LL, Toh HC, Heikenwalder M, Ng IO, Nardin A, Chen Q, Abastado JP, Chew V. Interaction between tumour-infiltrating B cells and T cells controls the progression of hepatocellular carcinoma. Gut. 2017;66:342–351. doi: 10.1136/gutjnl-2015-310814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Martinez-Rodriguez M, Thompson AK, Monteagudo C. A significant percentage of CD20-positive TILs correlates with poor prognosis in patients with primary cutaneous malignant melanoma. Histopathology. 2014;65:726–728. doi: 10.1111/his.12437. [DOI] [PubMed] [Google Scholar]
- 28.Sjöberg E, Frödin M, Lövrot J, Mezheyeuski A, Johansson M, Harmenberg U, Egevad L, Sandström P, Östman A. A minority-group of renal cell cancer patients with high infiltration of CD20+B-cells is associated with poor prognosis. Br J Cancer. 2018;119:840–846. doi: 10.1038/s41416-018-0266-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kurebayashi Y, Emoto K, Hayashi Y, Kamiyama I, Ohtsuka T, Asamura H, Sakamoto M. Comprehensive immune profiling of lung adenocarcinomas reveals four immunosubtypes with plasma cell subtype a negative indicator. Cancer Immunol Res. 2016;4:234–247. doi: 10.1158/2326-6066.CIR-15-0214. [DOI] [PubMed] [Google Scholar]
- 30.Fremd C, Schuetz F, Sohn C, Beckhove P, Domschke C. B cell-regulated immune responses in tumor models and cancer patients. Oncoimmunology. 2013;2:e25443. doi: 10.4161/onci.25443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gunderson AJ, Coussens LM. B cells and their mediators as targets for therapy in solid tumors. Exp Cell Res. 2013;319:1644–1649. doi: 10.1016/j.yexcr.2013.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lutz MB, Schuler G. Immature, semi-mature and fully mature dendritic cells: which signals induce tolerance or immunity? Trends Immunol. 2002;23:445–449. doi: 10.1016/s1471-4906(02)02281-0. [DOI] [PubMed] [Google Scholar]
- 33.Mahnke K, Schmitt E, Bonifaz L, Enk AH, Jonuleit H. Immature, but not inactive: the tolerogenic function of immature dendritic cells. Immunol Cell Biol. 2002;80:477–483. doi: 10.1046/j.1440-1711.2002.01115.x. [DOI] [PubMed] [Google Scholar]
- 34.Baglieri J, Brenner DA, Kisseleva T. The role of fibrosis and liver-associated fibroblasts in the pathogenesis of hepatocellular carcinoma. Int J Mol Sci. 2019;20:1723. doi: 10.3390/ijms20071723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Massague J. The transforming growth factor-beta family. Annu Rev Cell Biol. 1990;6:597–641. doi: 10.1146/annurev.cb.06.110190.003121. [DOI] [PubMed] [Google Scholar]
- 36.Pickup M, Novitskiy S, Moses HL. The roles of TGFβ in the tumour microenvironment. Nat Rev Cancer. 2013;13:788–799. doi: 10.1038/nrc3603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kim JH, You KR, Kim IH, Cho BH, Kim CY, Kim DG. Over-expression of the ribosomal protein L36a gene is associated with cellular proliferation in hepatocellular carcinoma. Hepatology. 2004;39:129–138. doi: 10.1002/hep.20017. [DOI] [PubMed] [Google Scholar]
- 38.Wang C, Cigliano A, Jiang L, Li X, Fan B, Pilo MG, Liu Y, Gui B, Sini M, Smith JW. 4EBP1/eIF4E and p70S6K/RPS6 axes play critical and distinct roles in hepatocarcinogenesis driven by AKT and N-Ras proto-oncogenes in mice. Hepatology. 2015;61:200–213. doi: 10.1002/hep.27396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stumpf CR, Moreno MV, Olshen AB, Taylor BS, Ruggero D. The translational landscape of the mammalian cell cycle. Mol Cell. 2013;52:574–582. doi: 10.1016/j.molcel.2013.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Grinchuk OV, Yenamandra SP, Iyer R, Singh M, Lee HK, Lim KH, Chow PK, Kuznetsov VA. Tumor-adjacent tissue co-expression profile analysis reveals pro-oncogenic ribosomal gene signature for prognosis of resectable hepatocellular carcinoma. Mol Oncol. 2018;12:89–113. doi: 10.1002/1878-0261.12153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang S, Hu B, Lv X, Chen S, Liu W, Shao Z. The prognostic role of ribosomal protein S6 kinase 1 pathway in patients with solid tumors: a meta-analysis. Front Oncol. 2019;9:390. doi: 10.3389/fonc.2019.00390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhou C, Sun J, Zheng Z, Weng J, Atyah M, Zhou Q, Chen W, Zhang Y, Huang J, Yin Y. High RPS11 level in hepatocellular carcinoma associates with poor prognosis after curative resection. Ann Trans Med. 2020;8:466. doi: 10.21037/atm.2020.03.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ji H, Zhang X. RPL38 regulates the proliferation and apoptosis of gastric cancer via miR-374b-5p/VEGF signal pathway. Onco Targets Ther. 2020;13:6131–6141. doi: 10.2147/OTT.S252045. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 44.Wang B, Ma A, Zhang L, Jin WL, Qian Y, Xu G, Qiu B, Yang Z, Liu Y, Xia Q. POH1 deubiquitylates and stabilizes E2F1 to promote tumour formation. Nat Commun. 2015;6:1–15. doi: 10.1038/ncomms9704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lv J, Zhang S, Wu H, Lu J, Lu Y, Wang F, Zhao W, Zhan P, Lu J, Fang Q. Deubiquitinase PSMD14 enhances hepatocellular carcinoma growth and metastasis by stabilizing GRB2. Cancer Lett. 2020;469:22–34. doi: 10.1016/j.canlet.2019.10.025. [DOI] [PubMed] [Google Scholar]
- 46.Wang CH, Lu SX, Liu LL, Li Y, Yang X, He YF, Chen SL, Cai SH, Wang H, Yun JP. POH1 knockdown induces cancer cell apoptosis via p53 and Bim. Neoplasia. 2018;20:411–424. doi: 10.1016/j.neo.2018.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pylayeva Y, Gillen KM, Gerald W, Beggs HE, Reichardt LF, Giancotti FG. Ras- and PI3K-dependent breast tumorigenesis in mice and humans requires focal adhesion kinase signaling. J Clin Invest. 2009;119:252–266. doi: 10.1172/JCI37160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kim W, Seok Kang Y, Soo Kim J, Shin NY, Hanks SK, Song WK. The integrin-coupled signaling adaptor p130Cas suppresses Smad3 function in transforming growth factor-β signaling. Mol Biol Cell. 2008;19:2135–2146. doi: 10.1091/mbc.E07-10-0991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Leight JL, Wozniak MA, Chen S, Lynch ML, Chen CS. Matrix rigidity regulates a switch between TGF-β1-induced apoptosis and epithelial-mesenchymal transition. Mol Biol Cell. 2012;23:781–791. doi: 10.1091/mbc.E11-06-0537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ke AW, Shi GM, Zhou J, Huang XY, Shi YH, Ding ZB, Wang XY, Devbhandari RP, Fan J. CD151 amplifies signaling by integrin α6β1 to PI3K and induces the epithelial-mesenchymal transition in HCC cells. Gastroenterology. 2011;140:1629–1641. doi: 10.1053/j.gastro.2011.02.008. [DOI] [PubMed] [Google Scholar]
- 51.Giannelli G, Fransvea E, Bergamini C, Marinosci F, Antonaci S. Laminin-5 chains are expressed differentially in metastatic and nonmetastatic hepatocellular carcinoma. Clin Cancer Res. 2003;9:3684–3691. [PubMed] [Google Scholar]
- 52.Sun J, Shi J, Li N, Guo W, Wu M, Lau W, Cheng S. Portal vein tumor thrombus is a bottleneck in the treatment of hepatocellular carcinoma. Cancer Biol Med. 2016;13:452–458. doi: 10.20892/j.issn.2095-3941.2016.0059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Khemlina G, Ikeda S, Kurzrock R. The biology of hepatocellular carcinoma: implications for genomic and immune therapies. Mol Cancer. 2017;16:149. doi: 10.1186/s12943-017-0712-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.