Abstract
Hepatocellular carcinoma (HCC) has high morbidity and mortality rates. It is therefore imperative to study the underlying mechanism of HCC to identify potential prognostic biomarkers and therapeutic targets. Recently, GINS2 has been identified to be a cancer-promoting gene in different cancer types. Nevertheless, the exact mechanism of GINS2 in HCC remains to be elucidated. To systematically explore the significance of GINS2, we first assessed the relative expression of GINS2 in pan-cancers based on data obtained from the HCCDB, TIMER, and TCGA databases. Then, we explored the clinical significance of GINS2 in HCC through Kaplan-Meier method as well as univariate and multivariate cox regression analysis. Additionally, functional enrichment analysis of GINS2 was done through GO, KEGG, PPI network, and immune cell infiltration analyses. Functional experiments were also conducted to investigate the biological significance of GINS2 in HCC cell lines. Our research revealed that GINS2 is involved in HCC progression and highlighted its potential value as a crucial diagnostic and therapeutic target for HCC.
Keywords: Hepatocellular carcinoma, E2F1, GINS2, PI3K/AKT/mTOR
Introduction
Hepatocellular carcinoma (HCC) is a morphologically heterogeneous cancer type with several histologic subtypes and different structural growth patterns [1]. Due to the high prevalence of hepatitis B, the incidence rate of HCC has been rising in China [2]. Most HCC cases are diagnosed at advanced stages that lack curative therapies and have low resectability rates [3-5]. Meanwhile, the development of new therapies for advanced HCC has been hampered by an insufficient understanding of the underlying molecular mechanisms [6]. As a result, it is critical to identify possible genes and signaling pathways that regulate the progression of HCC.
The GINS complex (GINS) consists of PSF1, PSF2, PSF3, and SLD5 [7,8], and maintains DNA replication forks by interacting with the MCM2-MCM7 complex [9]. It is involved in regulating cell cycle as well as cell proliferation and apoptosis [10]. GINS complex subunit 2 (GINS2, also named PSF2), encoded by the GINS2 gene, is located on human chromosome 16q24 [11]. Previous studies have indicated aberrant expression of GINS2 in multiple tumors. For example, the overexpression of GINS2 promotes EMT via activating the ERK/MAPK signal pathway in pancreatic cancer [12]. Upregulated GINS2 promotes tumor progression in NSCLC [13]. And knockdown of GINS2 inhibits cell proliferation in human gliomas [14]. It has also been shown that GINS2 is upregulated in HCC tumor tissues, suggesting the potential of GINS2 as a therapeutic target [15-17]. Nevertheless, how GINS2 functions in the progression of HCC is still unclear. We describe in this report our investigation of the role of GINS2 in HCC progression and strive to shed light on its underlying mechanism.
Materials and methods
PPI network construction and functional annotation analysis
The PPI (protein-protein interaction) network of GINS2-related proteins was constructed through GeneMANIA database (http://genemania.org/). Gene ontology analysis (GO) has broad applications in the functional annotation of large-scale genomic data. The KEGG database contains information about biological pathways. GO and KEGG analyses were completed with the Cluster Profiler in R package.
Analysis of tumor-infiltrating immune cells
The tumor-infiltrating immune cell analysis was performed using TIMER2.0 (https://cistrome.shinyapps.io/timer/). Additionally, single sample Gene Enrichment Analysis (ssGSEA) was used in the determination of correlation between GINS2 and 24 subtypes of immune cells using the R package.
HCCDB database analysis
We examined the mRNA expression level of GINS2 through the HCCDB database (http://lifeome.net/database/hccdb), which consists of 15 datasets and nearly 4,000 clinical samples from the TCGA database and Gene Expression Omnibus (GEO) [18].
Kaplan-Meier survival curve analysis
The OS (overall survival), PFS (progression-free survival), DSS (disease-free survival), and RFS (recurrence-free survival) curves of HCC patients were obtained from the Kaplan-Meier database.
Single-cell analysis
CancerSEA (http://biocc.hrbmu.edu.cn/CancerSEA/home.jsp), a cancer single-cell database, was applied to reveal the correlation between GINS2 and 14 functional states in different tumor types.
University of Alabama Cancer Database (UALCAN) analysis
The online tool for gene expression and clinical data analysis UALCAN (http://ualcan.path.uab.edu/) was employed to explore the correlation between GINS2 expression and clinicopathological characteristics of HCC, including weight, nodal metastasis status, TP53 mutation status, tumor grade, histological subtype, cancer stage, gender, age, and race.
Multiple models on prognosis analysis
The ROC curve and time-dependent curve analyses were performed to investigate the performance of the prognostic classifier in predicting HCC patient outcome by estimating the AUC (area under the ROC curve) value. We also constructed univariate and multivariate cox regression models to explore the effect of various prognostic factors on overall survival (OS) of HCC patients.
Tissue specimens and cell lines
The HCC cell lines (HCC-LM3, SMMC-7721, MHCC-97L, and MHCC-97H) and immortalized non-cancerous hepatic LO2 cells were obtained from the Chinese Academy of Sciences (Shanghai, China). HUVECs were purchased from ATCC. HCC cells were maintained in DMEM (Gibco, USA) supplemented with 10% fetal bovine serum (FBS) (Gibco, USA). 45 paired (HCC and adjacent normal) tissues were obtained from patients undergoing hepatic resection at The First Affiliated Hospital of Nanjing Medical University. We obtained written informed consent from all the patients before the surgery. This research received ethics approval from the Ethics Committee of the First Affiliated Hospital of Nanjing Medical University. RNAlater solution (Thermo, USA) was used to save the tissue samples immediately after resection.
RT-qPCR analysis
Total RNA was isolated after cell lysis in TRIzol reagent (Invitrogen, USA). cDNA was obtained with the High Capacity cDNA Kit (Applied Biosystems, Germany). GAPDH served as an internal reference and the standard 2-ΔΔCT method was applied. The specific primers are shown in Table S1.
Cell treatment
Commercially available human LV-NC and LV-sh-GINS2 lentivirus were purchased from GenePharma (Shanghai, China). Transfected cells were selected by puromycin to generate stable cells. siRNAs targeting E2F1 were purchased from Tsingke Biological Technology (Beijing, China). cDNAs encoding E2F1 and GINS2 were cloned into the pcDNA3.1 empty vector to consistently express E2F1 (pcDNA3.1-E2F1) or GINS2 (pcDNA3.1-GINS2). Lipofectamine 2000 reagent (Invitrogen, USA) was used in cell transfections according to the manufacturer’s recommendations.
5-Ethynyl-2’-deoxyuridine (Edu) incorporation assay
The treated cells were plated into 24-well plates. The EdU assay kit (Ribobio) was conducted to assess cell proliferative ability. Briefly, after 2 h incubation with EdU, cells were stained with Apollo Dye Solution to label proliferating cells. Then, the cells were stained with DAPI. Images were captured with a fluorescence microscope.
CCK-8 assay and colony formation assay
1 × 103 treated cells were plated in each well of 96-well plates. Then, 10 μL of the CCK-8 (Doindo, Japan) solution was added to each well every 24 h. After 2 h incubation, optical density (OD) values at 450 nm were detected with a spectrophotometer. The treated cells were plated into each well of 6-well plates for colony-formation assay. Two weeks later, cells were fixed in 1% crystal violet for 1 h. Individual colonies were visualized and counted by the naked eye.
Flow cytometry analysis
Processed cells were fixed in pre-cooled ethanol under -20°C overnight. After washing three times to remove residual ethanol, cells were resuspended in 500 uL PI staining solution of the Cell Cycle Kit (MultiScience, China). After 30 min incubation, we used flow cytometry (FCM) to analyze the cell cycle.
Scratch-wound healing assay
The 8 × 105 stably transfected cells were plated into each well of the 6-well plates. A 200 µL sterile tip was used for wound scratching. Cells that have migrated were measured with phase-contrast microscopy at 0 h and 48 h.
Cell migration and invasion assays
For cell migration assays, the treated cells (2 × 104 cells/well) were seeded into the upper insert while the complete culture medium mixed with 20% FBS was added to the lower chambers. 24 h later, we washed the transwell membrane with PBS and stained the membrane with crystal violet solution. The migrated cells were imaged with a microscope. Similarly, cell invasion assays were performed using the Matrigel matrix-coated chamber, and cells were incubated for 48 h before staining.
Tube formation assay
Stably transfected 97H and LM3 cells were cultured with serum-free DMEM for 48 h and cell supernatants were collected and stored at -80°C. Then, 10 μL of Matrigel was pipetted into each well of 15-well plates (Ibidi, Germany) and polymerized for 30 min. Finally, a total of 50 μL conditional medium containing 1 × 104 HUVECs were planted into each well. After 4 h, the capillary-like structures were imaged using a microscope, and the number of capillary tubes was counted with ImageJ software.
Immunohistochemistry (IHC)
The collected subcutaneous and xenograft tumor tissues were fixed and then cut into sections. Each slide was incubated with antibodies specific for GINS2 or Ki-67 overnight at 4°C, followed by 1 h incubation with HRP-conjugated secondary antibodies. After that, a DAB kit was used to stain the slides. Subsequently, hematoxylin was used as a light nuclear counterstain. IHC staining signals were scored independently by both a pathologist and the author.
Western blot
The cell lysis buffer was used to extract the total proteins from HCC tissues and cells. The samples were then mixed with loading buffer, resolved by SDS-PAGE gel, and transferred onto PVDF membranes (Millipore, USA). Subsequently, the membranes were probed with appropriate primary antibodies and visualized by the ECL detection system (Millipore, USA). The primary antibodies used in this study is provided in Table S2.
Chromatin immunoprecipitation assay (ChIP)
The ChIP assay was conducted with the Magna ChIP assay kit (Millipore, USA). 97H and LM3 cells were fixed with 1% formaldehyde to crosslink proteins and DNA. Cell lysates were then subjected to immunoprecipitation with either the negative control IgG or anti-E2F1 antibodies at 4°C overnight with constant rotation. Finally, the amount of co-precipitated DNA was detected by qRT-PCR. Primer sequences for the ChIP assay are shown in Table S3.
In vivo tumorigenesis and metastasis assays
For the subcutaneous xenograft study, ten mice (5-week-old, female, BALB/c) were randomly grouped into two groups. Then, HCC cell lines LM3 that inhibited expression of GINS2 were inoculated into the left flank of nude mice (2 × 106 cells/200 μl). We recorded and assessed the xenograft using the formula: volume = 0.5 × (length × width2). Four weeks after injection, the xenografts were harvested, weighed, and fixed.
For in vivo tumor metastasis assays, twelve mice were randomly grouped into two groups. A total of 2 × 106 cells were injected into the tail veins of nude mice. After eight weeks, each mouse was injected with 100 mg/kg D-luciferin (Yisheng, China) and evaluated using the Berthold Imaging System (Berthold, Germany). Then, the mice were sacrificed and their lungs analyzed by HE staining. All experiments and animal handling were conducted following the guidelines of Nanjing Medical University Institutional Animal Care Committee.
Dual-luciferase reporter assay
Cells were transfected with appropriate plasmids using Lipofectamine 2000 (Thermo Fisher Scientific, USA). After 48 h, luciferase activities were detected with the Dual Luciferase Reporter Assay system (Beyotime).
Statistical analyses
Data were analyzed using GraphPad Prism5 (GraphPad Software) and presented as mean ± S.D. Each experiment was conducted with at least three independent biological replicates. A p-value < 0.05 was considered statistically significant.
Results
GINS2 was upregulated in HCC tissues
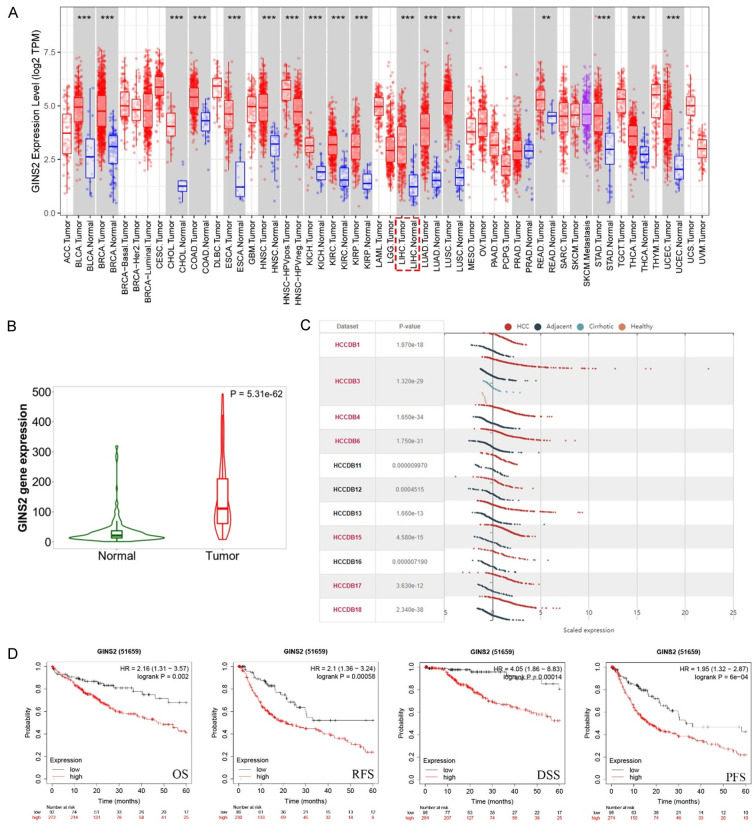
To investigate whether GINS2 was dysregulated in HCC tissues, we first examined its expression using the TIMER online database. GINS2 was upregulated in 17 cancer types including HCC (Figure 1A). Then, TCGA and HCCDB databases were examined to further explore the GINS2 expression in HCC. As shown in Figure 1B and 1C, GINS2 was significantly elevated in HCC tumor tissues. The results of RT-qPCR, western blot, and IHC analysis further confirmed the high expression of GINS2 in HCC tissues. (Figure 6A, 6D, and 6E). Moreover, we also demonstrated GINS2 expression was higher in HCC cells (SMMC-7721, MHCC-97L, MHCC-LM3, and Huh7) compared to LO2 liver cells, particularly in 97H and LM3 cells (Figure 6B). Subsequent survival analysis by the Kaplan-Meier plotter database illustrated that higher GINS2 expression correlated with poor OS, PFS, DSS, and RFS of HCC patients (Figure 1D).
Figure 1.
GINS2 expression levels in HCC from TIMER and HCCDB databases. A. Expression levels of GINS2 in pan-cancers from the TIMER database. B. Violin plot of GINS2 expression between HCC tissues and adjacent healthy tissues according to the TCGA database. C. Expression levels of GINS2 in HCC tissues and adjacent healthy tissues according to the HCCDB database. D. Kaplan-Meier survival curve analysis of the prognostic significance of GINS2 expression based on the Kaplan-Meier database. *P < 0.05; **P < 0.01; ***P < 0.001. (ACC, BLCA, CESC, CHOL, COAD, DLBC, ESCA, GBM, HNSC, KICH, KIRC, KIRP, LAML, LGG, LIHC, LUAD, LUSC, MESO, OV, PAAD, PCPG, PRAD, READ, SARC, SKCM, SKCM, TGCT, HCA, THYM, UCEC, UCS, UVM).
Figure 6.
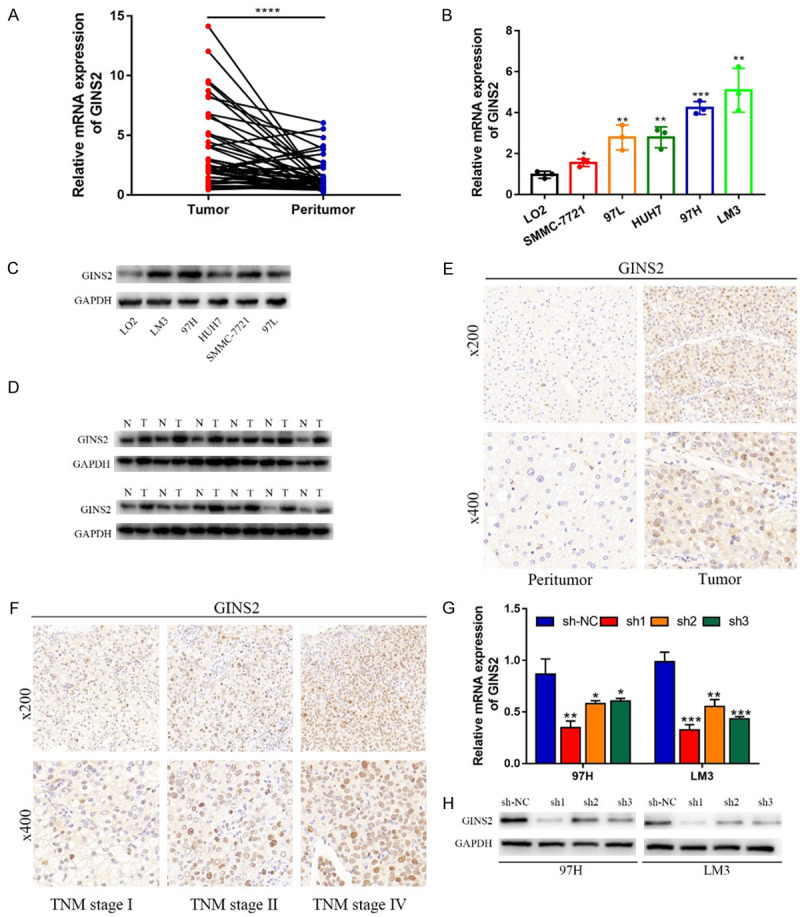
GINS2 expression decreased in HCC tissue samples and cell lines. A. qRT-PCR was used to examine the expression of GINS2 in 45 pairs of HCC tissues and corresponding peritumor tissues. B. The mRNA expression levels of GINS2 in HCC cell lines and normal LO2 cells. C, D. Western blot showing that GINS2 was upregulated in HCC tissues and HCC cell lines compared with adjacent healthy tissues and LO2 cells. E, F. The protein level of GINS2 in HCC specimens and adjacent normal tissues was detected by IHC. G, H. GINS2 knockdown efficiency was analyzed by qRT-PCR and western blot. All data are represented as mean ± S.E.M. *P < 0.05; **P < 0.01; ***P < 0.001.
GINS2 expression and clinicopathologic features of HCC patients
To explore the correlation between GINS2 and clinicopathologic features in HCC tissues, we downloaded the clinical data of LIHC in TCGA database. As illustrated in Table 1, GINS2 expression was associated with the Pathologic stage, T stage, Histologic grade, and AFP level of HCC patients. The outcome of logistic regression analysis also indicated that upregulated GINS2 expression was significantly associated with T stage, Pathologic stage, Tumor status, and Histologic grade in HCC (Table S4). Furthermore, the UALCAN database was employed to explore GINS2 expression in subgroups of patients based on different clinical features. For tumor grade, High GINS2 expression was observed in HCC patients in grade 1, 2, 3, and 4 (Figure 2A). In terms of individual cancer stages, GINS2 expression was significantly upregulated in stages 1, 2, and 3 (Figure 2B). Analysis of the node metastasis status showed that GINS2 expression was significantly elevated in the N0 group of HCC patients. Notably, GINS2 expression in N0 and N1 groups was not statistically significant due to the smaller sample in the N1 group (Figure 2C). TNMplot was then employed to further assess the association between GINS2 expression and metastasis status of HCC patients [19], and the results are presented in Figure S1. Interestingly, we found GINS2 expression was significantly increased in TP53-mutant and wild-type HCC patients (Figure 2D). Further analysis indicated that upregulated GINS2 expression was independent of patients’ gender, age, race, weight, or histological subtypes (Figure S1).
Table 1.
Association between GINS2 expression and clinicopathologic features in HCC samples from the TCGA database
| Characteristic | Low expression of GINS2 | High expression of GINS2 | P-value |
|---|---|---|---|
| n | 185 | 186 | |
| Age, n (%) | 0.405 | ||
| ≤ 60 | 84 (22.7%) | 93 (25.1%) | |
| > 60 | 101 (27.3%) | 92 (24.9%) | |
| Gender, n (%) | 0.530 | ||
| Female | 57 (15.4%) | 64 (17.3%) | |
| Male | 128 (34.5%) | 122 (32.9%) | |
| Pathologic stage, n (%) | 0.012 | ||
| Stage I | 97 (28%) | 74 (21.3%) | |
| Stage II | 40 (11.5%) | 46 (13.3%) | |
| Stage III | 32 (9.2%) | 53 (15.3%) | |
| Stage IV | 4 (1.2%) | 1 (0.3%) | |
| T stage, n (%) | 0.027 | ||
| T1 | 104 (28.3%) | 77 (20.9%) | |
| T2 | 42 (11.4%) | 52 (14.1%) | |
| T3 | 31 (8.4%) | 49 (13.3%) | |
| T4 | 6 (1.6%) | 7 (1.9%) | |
| N stage, n (%) | 1.000 | ||
| N0 | 122 (47.7%) | 130 (50.8%) | |
| N1 | 2 (0.8%) | 2 (0.8%) | |
| M stage, n (%) | 0.361 | ||
| M0 | 129 (47.8%) | 137 (50.7%) | |
| M1 | 3 (1.1%) | 1 (0.4%) | |
| Histologic grade, n (%) | < 0.001 | ||
| G1 | 34 (9.3%) | 21 (5.7%) | |
| G2 | 100 (27.3%) | 77 (21%) | |
| G3 | 44 (12%) | 78 (21.3%) | |
| G4 | 4 (1.1%) | 8 (2.2%) | |
| AFP (ng/ml), n (%) | 0.014 | ||
| ≤ 400 | 118 (42.4%) | 95 (34.2%) | |
| > 400 | 24 (8.6%) | 41 (14.7%) | |
| Child-Pugh grade, n (%) | 0.573 | ||
| A | 110 (46%) | 107 (44.8%) | |
| B | 12 (5%) | 9 (3.8%) | |
| C | 0 (0%) | 1 (0.4%) | |
| Vascular invasion, n (%) | 0.249 | ||
| No | 110 (34.9%) | 96 (30.5%) | |
| Yes | 50 (15.9%) | 59 (18.7%) |
Figure 2.
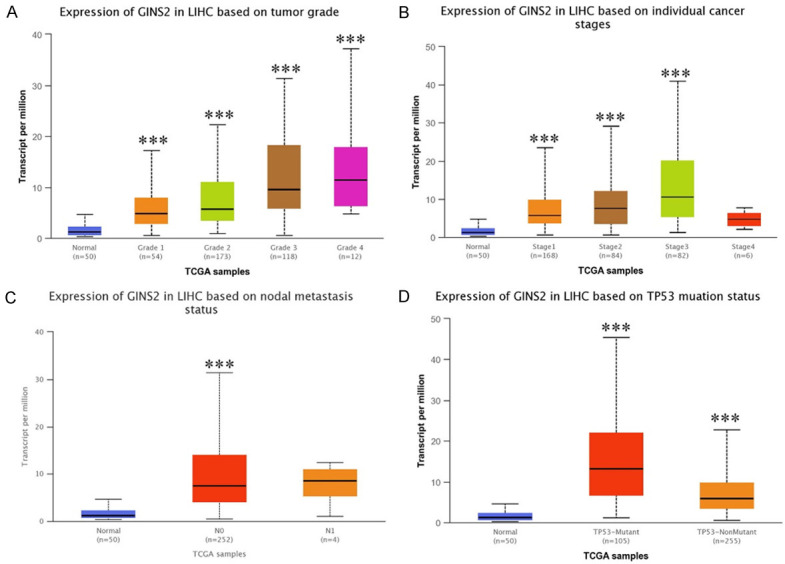
Box plot evaluation of GINS2 expression in subgroups of LIHC samples (UALCAN). Upregulated mRNA of GINS2 is independent of patients’ tumor grade (A), cancer stage (B), nodal metastasis status (C), and TP53 mutation status (D). The t-test was used to estimate the significance of differences in gene expression levels between groups. *P < 0.05; **P < 0.01; ***P < 0.001.
The prognostic value of GINS2 in HCC
We found the area under curves (AUC) of receiver operator characteristic (ROC) curve was 0.909, suggesting the ability of using GINS2 expression to accurately distinguish tumors from normal ones (Figure 3A). The time-dependent ROC curve analysis of GINS2 was also performed to predict overall survival of HCC. As illustrated in Figure 3B, AUC values of one-, three-, and five-year overall survival were above 0.6, which is considered suitable for prediction. In addition, the results of univariate regression analysis illustrated that T stage, Pathologic stage, Tumor status, and GINS2 expression all significantly correlated with HCC patient OS (Figure 3C and Table S5). Finally, GINS2 expression and Tumor status were selected as independent prognostic factors for patients with HCC according to the multivariate regression analysis (Figure 3D and Table S5). These results helped shed light on the prognostic value of GINS2.
Figure 3.
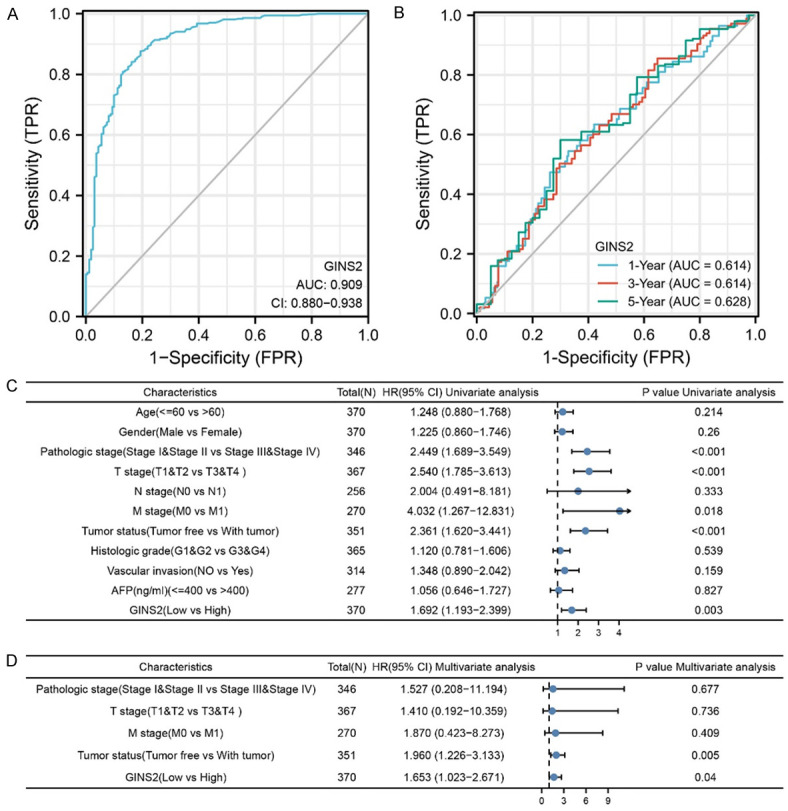
GINS2 expression and other clinicopathologic factors with OS in LIHC were calculated via univariate and multivariate regression analysis. A. The ROC curve of diagnosis to distinguish tumor from normal tissue. The AUC was 0.909. B. Time-dependent survival ROC curve analysis to predict 1-, 3-, and 5-year survival rates. C. The forest plot of univariate regression analysis. Among the factors, T stage, Pathologic stage, Tumor status, and GINS2 expression were identified as statistically significantly associated with the likelihood for OS univariate analysis. D. The forest plot of multivariate regression analysis.
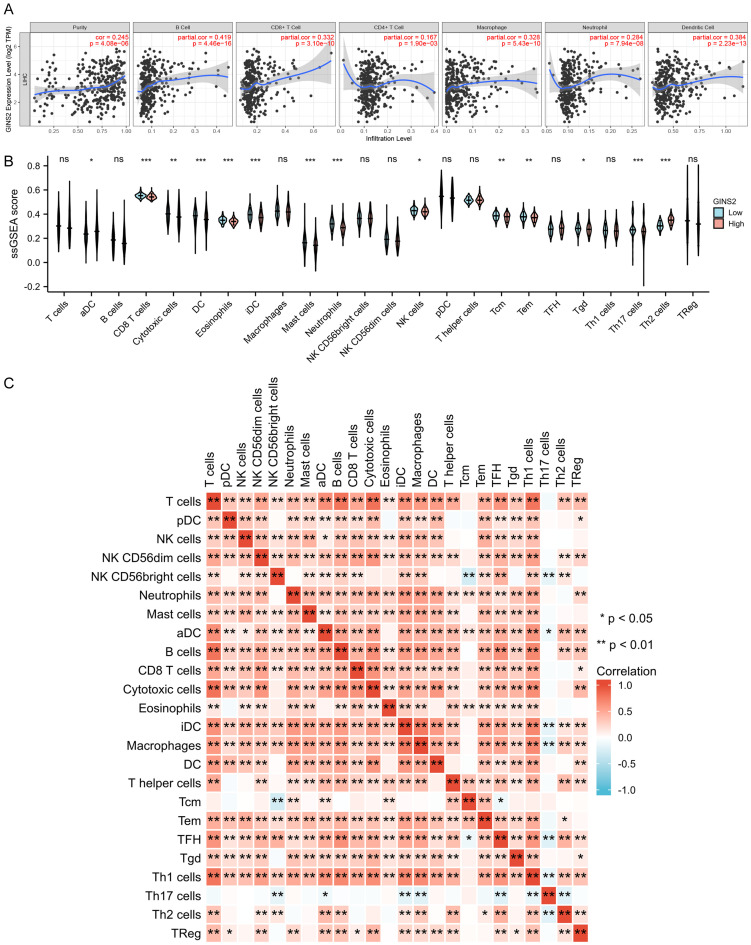
The correlation between GINS2 expression and immune cell infiltration
Recently, numerous studies have indicated the importance of immune cell infiltration in HCC occurrence and development [20-22]. Therefore, we assessed the potential correlation between GINS2 expression and immune cell markers in HCC using the TIMER2.0 database. As shown in Figure 4A, GINS2 expression was positively correlated with the infiltration of B cells, CD8+ T cells, macrophages, and dendritic cells in HCC tissues. Then, we carried out ssGSEA to further explore the immune cell infiltration landscape of HCC (recorded as ssGSEA score) according to the expression of GINS2. The results indicated a close relationship between GINS2 expression and immune cell infiltration (Figure 4B). Besides, the heat map was plotted to show the correlation between different tumor-infiltrating immune cells (Figure 4C).
Figure 4.
Correlations between GINS2 expression and immune infiltration level in HCC. A. The expression of GINS2 was significantly correlated with infiltrating levels of B cell, CD8+ T cells, CD4+ T cells, macrophages, neutrophils, and dendritic cells in HCC. B. The varied proportions of 24 subtypes of immune cells in high and low GINS2 expression groups in tumor samples. C. Heatmap of 24 immune infiltration cells in tumor samples. *P < 0.05; **P < 0.01; ***P < 0.001.
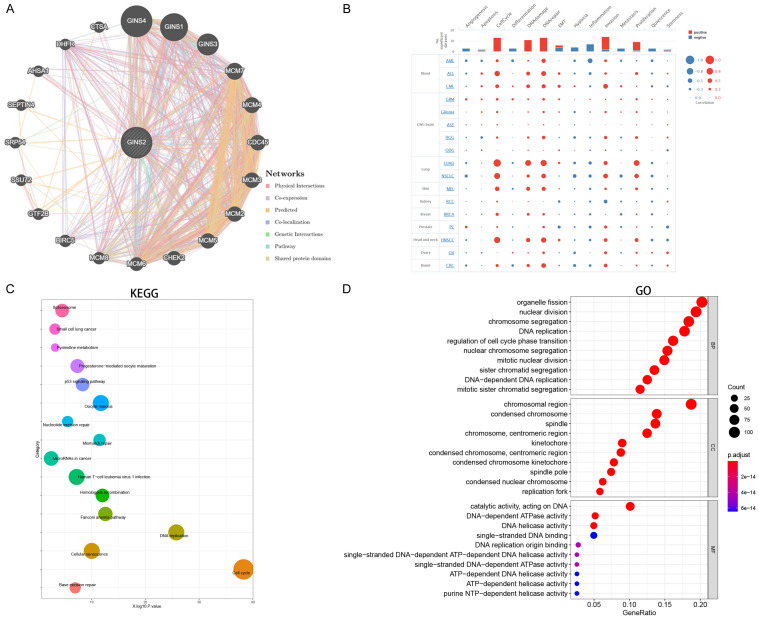
PPI, GO, and KEGG analysis of GINS2
A comprehensive protein-protein interaction network of GINS2 was constructed with the GeneMANIA database (Figure 5A), which revealed interactions between GINS1, GINS2, GINS3, GINS4, and MCM2-7. This is consistent with previously published studies [8,9]. Then, we conducted single-cell analysis to examine the association between GINS2 and 14 functional states in various cancer types. As illustrated in Figure 5B, GINS2 expression was positively associated with invasion, cell cycle, DNA repair, DNA damage, and proliferation. GO and KEGG analysis was next performed with genes co-expressed with GINS2 according to the LinkedOmics online tool. The enriched KEGG pathways illustrated that these co-expressed genes were mainly associated with cell cycle and DNA replication (Figure 5C). Meanwhile, GO function annotation was mainly enriched in nuclear division, DNA replication, and regulation of cell cycle phase transition (Figure 5D).
Figure 5.
PPI, GO and KEGG analysis of the genes co-expressed with GINS2 in HCC. A. The protein-protein interaction network of GINS2 was constructed using GeneMANIA. B. Single-cell analysis displayed the correlation between GINS2 and 14 functional states (including angiogenesis, apoptosis, cell cycle, differentiation, DNA damage, DNA repair, EMT, hypoxia, inflammation, invasion, metastasis, proliferation, stemness, and quiescence) in different tumor types. C, D. Significantly enriched KEGG pathways and GO annotations of the genes co-expressed with GINS2 in HCC based on the LinkedOmics database.
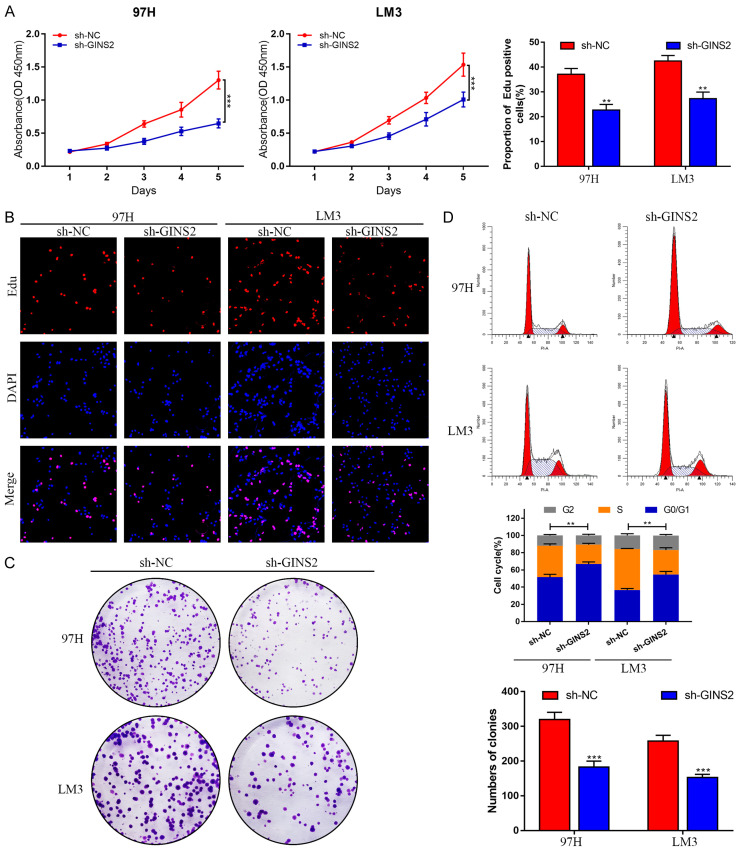
Knockdown of GINS2 induced cell proliferation restriction and cell cycle arrest of HCC cells
Stably transfected 97H and LM3 cell lines were generated using Lenti-sh-GINS2 or Lenti-sh-NC constructs. RT-qPCR and western blot experiments were performed to confirm the knockdown efficiency. As illustrated in Figure 6G and 6H, the expression of GINS2 decreased the most in the sh1 group. CCK-8, Edu, and colony formation assays were subsequently conducted to assess the effect of GINS2 on cell proliferation. Data from these experiments revealed that downregulating GINS2 suppressed the proliferative ability of 97H and LM3 cells (Figure 7A-C; Figure S2A). Moreover, cell cycle analysis demonstrated that depleting GINS2 resulted in a substantial increase in the GO/G1 phase (Figure 7D). Collectively, these findings demonstrated that silencing GINS2 could significantly suppress cell viability and induce G1 phase cell cycle arrest in HCC cells.
Figure 7.
Knockdown of GINS2 suppresses cell proliferation and induces cell cycle arrest of HCC cells in vitro. A. CCK-8 assay was performed in 97H and LM3 cells transfected with sh-NC or sh-GINS2. B. Cell proliferation was further examined in sh-NC and sh-GINS2 cells by EdU assay. C. The effect of GINS2 on colony-forming ability was detected by colony formation assay in 97H and LM3 cells after transfection. D. Flow cytometry results show the cell cycle distribution of GINS2 silenced 97H and LM3 cells. All data are represented as mean ± S.E.M. *P < 0.05; **P < 0.01; ***P < 0.001.
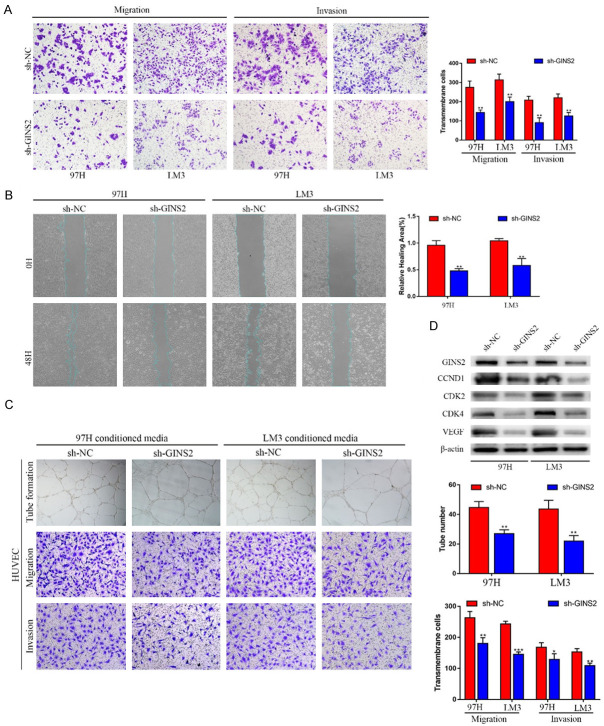
Knockdown of GINS2 inhibited migration and invasion of HCC cells
Given the positive correlation between GINS2 expression and the metastasis status in HCC patients, we hypothesized that GINS2 might facilitate the migration and invasion of HCC cells. Both wound healing assays and transwell assays showed the suppressed invasion and migration of 97H and LM3 cells, following GINS2 knockdown. (Figure 8A, 8B and Figure S2B). Additionally, we found that co-culturing with GINS2-silenced cells significantly suppressed the migration, invasion, and tube formation of HUVECs (Figure 8C). Furthermore, a significant decrease was observed in the proteins associated with angiogenesis (VEGF) and cell cycle (CCND1, CDK2, and CDK4) in GINS2-silenced cells (Figure 8D). Taken together, our results illustrated that GINS2 could increase cell migration and invasion capabilities and affect the angiogenesis, invasion, and migration of HUVECs, which may ultimately lead to enhanced metastasis of HCC.
Figure 8.
Knockdown of GINS2 suppresses invasion and migration of HCC cells in vitro. A. Invasive or migrated cells were measured by transwell assays with or without Matrigel. B. Wound-healing migration assays detected the effects of silencing GINS2 on cell migration. C. Tube formation, invasion, and migration of HUVECs in sh-GINS2 cell culture medium. D. Western blot analysis of cell angiogenesis and cell cycle-related proteins in 97H or LM3 cells with or without GINS2 knockdown. All data are represented as mean ± S.E.M. *P < 0.05; **P < 0.01; ***P < 0.001.
GINS2 accelerated the proliferation and metastasis of HCC cells in mouse models
Xenograft tumors were generated in nude mice to evaluate GINS2 in vivo. Compared with the control group, tumors from GINS2-silenced cells showed a marked decrease in tumor growth (Figure 9A-C). Immunohistochemistry staining with an anti-Ki67 antibody in xenograft tumors showed similar results (Figure 9D). A metastatic xenograft model was next generated by lateral tail-vein injection of LM3 cells. Mice were anesthetized after eight weeks to obtain lung tissues. The tissues were then subjected to HE staining, which showed decreased metastatic nodules in the GINS2-silencing group compared to the control group (Figure 9E).
Figure 9.
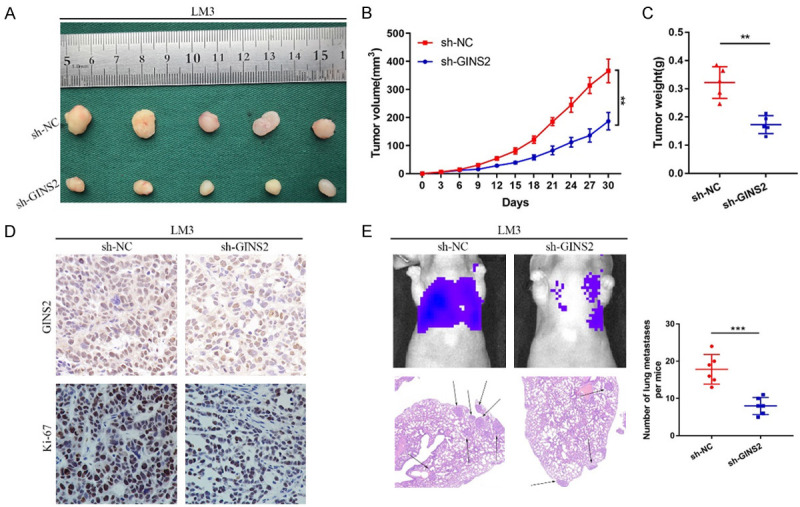
GINS2 promotes HCC progression in vivo. LM3 cells infected by sh-GINS2 or negative control sh-NC lentiviruses were implanted into nude mice and tumor growth was recorded. A-C. Images of tumors from different groups of nude mice. Tumor volume and average weight were assessed. D. IHC analysis of GINS2 and Ki-67 in the tumors derived from mice. E. GINS2 stable knockdown LM3 cells were injected into the tail vein of nude mice to induce lung metastasis. The relative luciferase activity of HCC tumor in nude mice from the sh-NC or sh-GINS2 group were determined using a live imaging system. Liver metastatic nodules were subjected to HE staining as indicated. All data are presented as mean ± S.E.M. *P < 0.05; **P < 0.01; ***P < 0.001.
GINS2 was transcriptionally activated by E2F1 in HCC cells
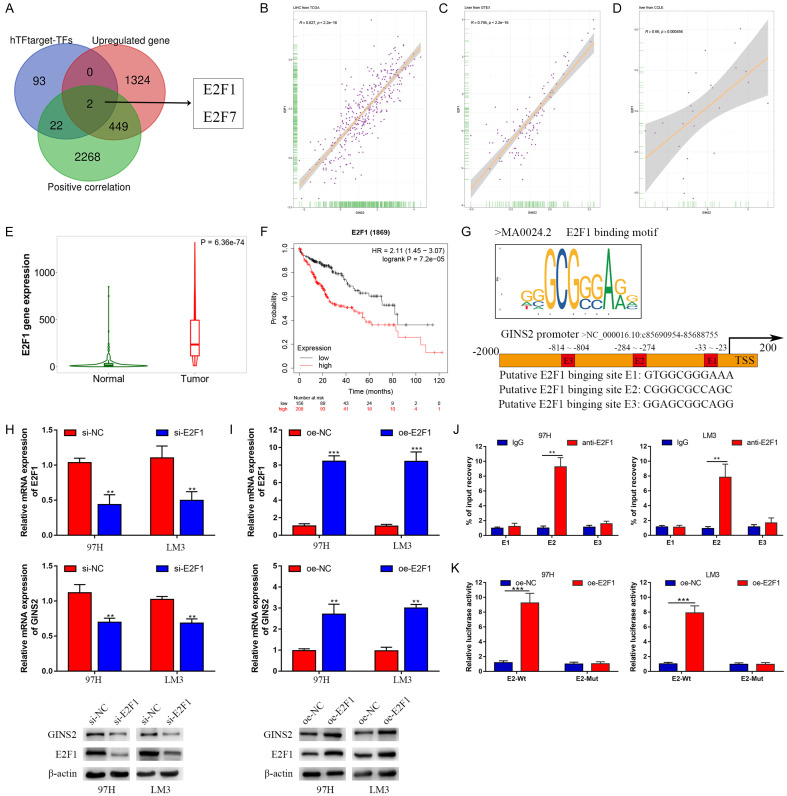
Previous studies have defined transcription factors (TFs) as key regulators of gene expression [23,24]. The hTFtarget database was used to predict potential TFs that could regulate GINS2 expression. E2F1 and E2F7 were selected for further study by plotting Venn diagrams of three gene lists: TFs predicted by the hTFtarget database, upregulated genes in LIHC according to TCGA data, and genes positively correlated with GINS2 expression in HCC according to the LinkedOmics database (Figure 10A). The analysis indicated a strong correlation between E2F1 and GINS2 in the TCGA, GTEx, and CCLE databases (Figure 10B-D and Figure S3). Moreover, E2F1 expression in HCC tissues was higher than in non-tumor tissues, where higher levels of E2F1 indicated poorer outcome in HCC patients (Figure 10E and 10F). Therefore, we chose E2F1 for further research.
Figure 10.
GINS2 was transcriptionally activated by E2F1. A. Venn diagrams of three gene lists: transcription factors predicted by the hTFtarget database, upregulated genes in LIHC according to the data from TCGA database, genes positively correlated with GINS2 expression in HCC according to the LinkedOmics database. B-D. Correlation between E2F1 and GINS2 in the TCGA, GTEx, and CCLE databases. E. Violin plot of E2F1 expression between HCC tissues and adjacent healthy tissues according to data from the TCGA database. F. Kaplan-Meier survival curve analysis of the prognostic significance of E2F1 expression based on the Kaplan-Meier database. G. E2F1-binding motifs and predicted E2F1-binding sites (E1, E2, and E3) on the promoter region of GINS2 were obtained from the JASPAR database. H, I. Expression of E2F1 and GINS2 was confirmed by qRT-PCR or western blot in 97H and LM3 cells transfected with si-E2F1, si-NC, pcDNA3.1, or pcDNA3.1-E2F1. J. qRT-PCR analysis of ChIP products validated the binding capacity of E2F1 to the GINS2 promoter. K. The luciferase reporter assay further confirmed direct binding of E2F1 to the GINS2 promoter in 97H and LM3 cells transfected with pcDNA3.1 or pcDNA3.1-E2F1. All data are presented as mean ± S.E.M. *P < 0.05; **P < 0.01; ***P < 0.001.
Using the JASPAR database, we acquired the first three E2F1-binding sequences from the promoter regions of GINS2 (Figure 10G and Figure S4). In addition, we demonstrated a potential regulatory relationship between E2F1 and GINS2 by knocking down or overexpressing E2F1 in 97H and LM3 cells, the efficiencies of which were further verified by qRT-PCR and western blot (Figure 10H and 10I). Chromatin immunoprecipitation (ChIP) assays also confirmed the predicted binding sites. Collectively, the data showed that E2 was the binding site of E2F1 on the GINS2 promoter region (Figure 10J). Subsequently, E2F1 overexpression could significantly drive the luciferase activity of GINS2 promoter-E2-Wt (Figure 10K). The above findings indicate that E2F1 could activate the transcription of GINS2 via binding to the GINS2 promoter region in HCC cells.
GINS2 could promote proliferative, migratory, and invasive capabilities of HCC cells through regulating the PI3K/AKT/mTOR signaling pathway
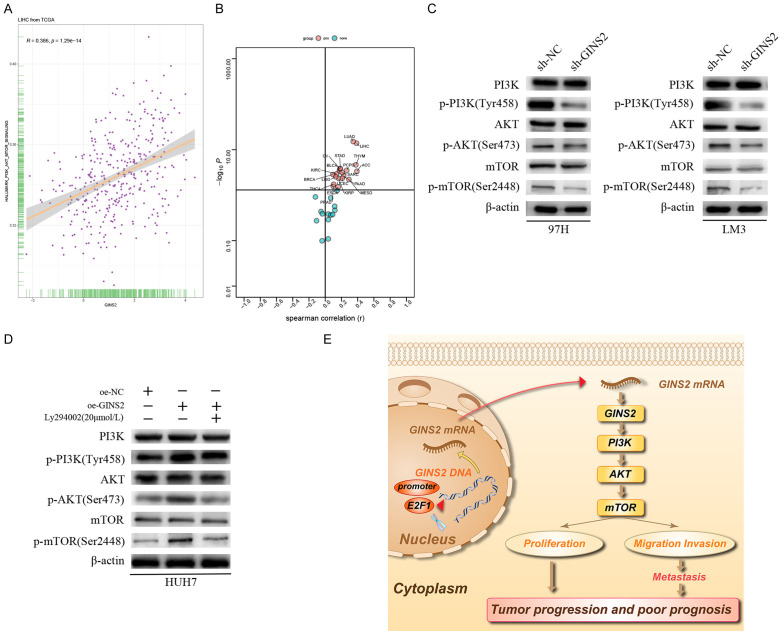
To find out the cellular mechanism of GINS2’s role in HCC progression, we analyzed the correlation between GINS2 and 50 signaling pathways in hallmark gene sets downloaded from the MsigDB database, which demonstrated that GINS2 expression was positively associated with HALLMARK_PI3K_AKT_MTOR_SIGNALING in most cancers types including HCC (Figure 11A and 11B). Therefore, we surmised that GINS2 could activate the PI3K/AKT/mTOR pathway. Western blot analysis indicated that knockdown of GINS2 inhibited levels of phosphorylated PI3K (Tyr458), AKT (Ser473), and mTOR (Ser2448). Conversely, a significant increase in the phosphorylation of these proteins was observed in GINS2-overexpression cells (Figure 11C and 11D). However, no change of PI3K, AKT, and mTOR was observed in 97H and LM3 cells. Addition of the PI3K inhibitor Ly294002 could partially reverse the altered protein levels caused by overexpressing GINS2 (Figure 11D). In summary, our findings support the notion that GINS2 could promote HCC progression through the PI3K/AKT/mTOR pathway.
Figure 11.
GINS2 promotes HCC cell proliferation, migration, and invasion by regulating the PI3K/AKT/mTOR signaling pathway. A. Correlation between HALLMARK_PI3K_AKT_MTOR_SIGNALING and GINS2 in LIHC according to data from the TCGA database. B. Correlation between HALLMARK_PI3K_AKT_MTOR_SIGNALING and GINS2 in pan-cancers according to data from the TCGA database. C. Expression levels of PI3K/p-PI3K (Tyr458), AKT/p-AKT (Ser473), and mTOR/p-mTOR (Ser2448) in 97H and LM3 cells transfected with sh-NC or sh-GINS2 were examined by western blot. D. HUH7 cells transfected with pcDNA3.1 or pcDNA3.1-GINS2 were treated with or without the indicated concentrations of Ly294002. Expression levels of PI3K/p-PI3K (Tyr458), AKT/p-AKT (Ser473), and mTOR/p-mTOR (Ser2448) were examined by western blot. E. The model of the E2F1/GINS2 axis mediating aggressive behaviors in HCC through activating the PI3K/AKT/mTOR signaling pathway.
Discussion
Despite remarkable advances in the technology of detection and treatment, hepatocellular carcinoma (HCC) is still one of the most common and deadliest malignancies worldwide [6,25]. Most HCC cases are diagnosed at an advanced stage and thus miss the best time window for surgery [26]. Hence, the identification of early detection biomarkers is crucial for the improvement of long-term survival of HCC patients. Various articles have reported that GINS2, as a novel oncogene, was upregulated in several malignant tumors, including breast cancer, lung cancer, pancreatic cancer, and thyroid cancer [12,27-29]. However, the biological significance and precise mechanism of GINS2 in HCC remain to be fully elucidated.
In our research, we first assessed the possibility of GINS2 as a HCC biomarker through a series of bioinformatic analyses. The results revealed that GINS2 was aberrantly high in HCC tissues in comparison with normal ones. Kaplan-Meier curves also illustrated that higher GINS2 expression correlated with poorer OS, PFS, DSS, and RFS of HCC patients (Figure 1). Multivariate regression analysis similarly illustrated that GINS2 was an independent prognostic marker for HCC patients, highlighting its potential as a novel biomarker (Figure 3). Recent research has indicated the importance of immune cell infiltration in HCC occurrence and development [20,22]. Besides, we found that GINS2 expression was closely involved in the infiltration of B cells, CD8+ T cells, macrophages, and dendritic cells, which indicated the importance of GINS2 in regulating HCC tumor immunity (Figure 4).
Functional experiments showed that the inhibition of GINS2 suppressed the proliferative, migratory, and invasive capabilities of HCC cells (Figures 7, 8 and 9). Transcription factors (TFs) are DNA-binding proteins that activate (and less frequently, inhibit) gene transcription [30]. Therefore, we speculated that the expression of GINS2 may be regulated by TFs. The hTFtarget database was then employed for the prediction of TFs that might regulate GINS2 expression. Additionally, the results of ChIP-qPCR and dual-luciferase reporter assays uncovered that E2F1 could bind to the promoter region of GINS2 and up-regulate its expression (Figure 10). Numerous studies have demonstrated the transcription factor E2F1 was involved in regulating cell cycle, apoptosis, metabolism, and metastasis [31,32]. Here, our study indicated that aberrant activation of E2F1 might upregulate the transcription of GINS2 and promote HCC progression.
The PI3K/AKT/mTOR pathway plays a crucial role in tumorigenesis [33-35]. An early study has established that KIF11 could regulate cell proliferative ability via the PI3K/AKT pathway in gallbladder cancer [36]. Another finding showed that Mex3a promotes lung adenocarcinoma metastasis via the PI3K/AKT pathway [37]. Our previous study also demonstrated that STK39 could promote the development of cholangiocarcinoma via the PI3K/AKT pathway [38]. Here, our data have added additional support to the notion that GINS2 can participate in regulating the PI3K/AKT/mTOR pathway. Knockdown of GINS2 suppressed the PI3K/AKT/mTOR pathway and vice versa. Additionally, we found that the effect of GINS2 overexpression on PI3K/AKT/mTOR signaling could be rescued with the PI3K inhibitor LY294002.
Conclusions
Collectively, increased GINS2 expression resulted in poorer outcome of HCC patients. And further experiments revealed that GINS2 could promote HCC cell proliferative, migratory, and invasive abilities by activating the PI3K/AKT/mTOR signaling pathway. Our experiments provide important preliminary evidence that GINS2 could be a promising therapeutic biomarker for HCC. Further experiments are required to elucidate the mechanism of action of GINS2 in HCC progression.
Acknowledgements
This work was supported by grants from the National Natural Science Foundation of China (81972768 and 81870488) and the Major Program of the National Natural Science Foundation of China (81530048 and 31930020).
Disclosure of conflict of interest
None.
Supporting Information
References
- 1.Vyas M, Zhang X. Hepatocellular carcinoma: role of pathology in the era of precision medicine. Clin Liver Dis. 2020;24:591–610. doi: 10.1016/j.cld.2020.07.010. [DOI] [PubMed] [Google Scholar]
- 2.Maluccio M, Covey A. Recent progress in understanding, diagnosing, and treating hepatocellular carcinoma. CA Cancer J Clin. 2012;62:394–399. doi: 10.3322/caac.21161. [DOI] [PubMed] [Google Scholar]
- 3.Lencioni R, Chen XP, Dagher L, Venook AP. Treatment of intermediate/advanced hepatocellular carcinoma in the clinic: how can outcomes be improved? Oncologist. 2010;15(Suppl 4):42–52. doi: 10.1634/theoncologist.2010-S4-42. [DOI] [PubMed] [Google Scholar]
- 4.Rimola J, Forner A, Tremosini S, Reig M, Vilana R, Bianchi L, Rodriguez-Lope C, Sole M, Ayuso C, Bruix J. Non-invasive diagnosis of hepatocellular carcinoma </= 2 cm in cirrhosis. Diagnostic accuracy assessing fat, capsule and signal intensity at dynamic MRI. J Hepatol. 2012;56:1317–1323. doi: 10.1016/j.jhep.2012.01.004. [DOI] [PubMed] [Google Scholar]
- 5.Akamatsu N, Cillo U, Cucchetti A, Donadon M, Pinna AD, Torzilli G, Kokudo N. Surgery and hepatocellular carcinoma. Liver Cancer. 2016;6:44–50. doi: 10.1159/000449344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 7.Chang YP, Wang G, Bermudez V, Hurwitz J, Chen XS. Crystal structure of the GINS complex and functional insights into its role in DNA replication. Proc Natl Acad Sci U S A. 2007;104:12685–12690. doi: 10.1073/pnas.0705558104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carroni M, De March M, Medagli B, Krastanova I, Taylor IA, Amenitsch H, Araki H, Pisani FM, Patwardhan A, Onesti S. New insights into the GINS complex explain the controversy between existing structural models. Sci Rep. 2017;7:40188. doi: 10.1038/srep40188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gambus A, Jones RC, Sanchez-Diaz A, Kanemaki M, van Deursen F, Edmondson RD, Labib K. GINS maintains association of Cdc45 with MCM in replisome progression complexes at eukaryotic DNA replication forks. Nat Cell Biol. 2006;8:358–366. doi: 10.1038/ncb1382. [DOI] [PubMed] [Google Scholar]
- 10.Zhang X, Zhong L, Liu BZ, Gao YJ, Gao YM, Hu XX. Effect of GINS2 on proliferation and apoptosis in leukemic cell line. Int J Med Sci. 2013;10:1795–1804. doi: 10.7150/ijms.7025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.MacNeill SA. Structure and function of the GINS complex, a key component of the eukaryotic replisome. Biochem J. 2010;425:489–500. doi: 10.1042/BJ20091531. [DOI] [PubMed] [Google Scholar]
- 12.Huang L, Chen S, Fan H, Ji D, Chen C, Sheng W. GINS2 promotes EMT in pancreatic cancer via specifically stimulating ERK/MAPK signaling. Cancer Gene Ther. 2021;28:839–849. doi: 10.1038/s41417-020-0206-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu X, Sun L, Zhang S, Zhang S, Li W. GINS2 facilitates epithelial-to-mesenchymal transition in non-small-cell lung cancer through modulating PI3K/Akt and MEK/ERK signaling. J Cell Physiol. 2020;235:7747–7756. doi: 10.1002/jcp.29381. [DOI] [PubMed] [Google Scholar]
- 14.Shen YL, Li HZ, Hu YW, Zheng L, Wang Q. Loss of GINS2 inhibits cell proliferation and tumorigenesis in human gliomas. CNS Neurosci Ther. 2019;25:273–287. doi: 10.1111/cns.13064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lian YF, Li SS, Huang YL, Wei H, Chen DM, Wang JL, Huang YH. Up-regulated and interrelated expressions of GINS subunits predict poor prognosis in hepatocellular carcinoma. Biosci Rep. 2018;38:BSR20181178. doi: 10.1042/BSR20181178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu C, Zhou Y, Wang M, Dai G, Liu X, Lai L, Tang S. Bioinformatics analysis explores potential hub genes in nonalcoholic fatty liver disease. Front Genet. 2021;12:772487. doi: 10.3389/fgene.2021.772487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang D, Liu J, Xie T, Jiang Q, Ding L, Zhu J, Ye Q. Oleate acid-stimulated HMMR expression by CEBPalpha is associated with nonalcoholic steatohepatitis and hepatocellular carcinoma. Int J Biol Sci. 2020;16:2812–2827. doi: 10.7150/ijbs.49785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lian Q, Wang S, Zhang G, Wang D, Luo G, Tang J, Chen L, Gu J. HCCDB: a database of hepatocellular carcinoma expression atlas. Genomics Proteomics Bioinformatics. 2018;16:269–275. doi: 10.1016/j.gpb.2018.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bartha A, Gyorffy B. TNMplot. com: a web tool for the comparison of gene expression in normal, tumor and metastatic tissues. Int J Mol Sci. 2021;22:2622. doi: 10.3390/ijms22052622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen S, Cao Q, Wen W, Wang H. Targeted therapy for hepatocellular carcinoma: challenges and opportunities. Cancer Lett. 2019;460:1–9. doi: 10.1016/j.canlet.2019.114428. [DOI] [PubMed] [Google Scholar]
- 21.Xu F, Jin T, Zhu Y, Dai C. Immune checkpoint therapy in liver cancer. J Exp Clin Cancer Res. 2018;37:110. doi: 10.1186/s13046-018-0777-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Prieto J, Melero I, Sangro B. Immunological landscape and immunotherapy of hepatocellular carcinoma. Nat Rev Gastroenterol Hepatol. 2015;12:681–700. doi: 10.1038/nrgastro.2015.173. [DOI] [PubMed] [Google Scholar]
- 23.Vaquerizas JM, Kummerfeld SK, Teichmann SA, Luscombe NM. A census of human transcription factors: function, expression and evolution. Nat Rev Genet. 2009;10:252–263. doi: 10.1038/nrg2538. [DOI] [PubMed] [Google Scholar]
- 24.Brouwer I, Lenstra TL. Visualizing transcription: key to understanding gene expression dynamics. Curr Opin Chem Biol. 2019;51:122–129. doi: 10.1016/j.cbpa.2019.05.031. [DOI] [PubMed] [Google Scholar]
- 25.Reghupaty SC, Sarkar D. Current status of gene therapy in hepatocellular carcinoma. Cancers (Basel) 2019;11:1265. doi: 10.3390/cancers11091265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Forner A, Reig M, Bruix J. Hepatocellular carcinoma. Lancet. 2018;391:1301–1314. doi: 10.1016/S0140-6736(18)30010-2. [DOI] [PubMed] [Google Scholar]
- 27.Sun D, Zong Y, Cheng J, Li Z, Xing L, Yu J. GINS2 attenuates the development of lung cancer by inhibiting the STAT signaling pathway. J Cancer. 2021;12:99–110. doi: 10.7150/jca.46744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yu S, Zhu L, Xie P, Jiang S, Wang K, Liu Y, He J, Ren Y. Mining the prognostic significance of the GINS2 gene in human breast cancer using bioinformatics analysis. Oncol Lett. 2020;20:1300–1310. doi: 10.3892/ol.2020.11651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ye Y, Song YN, He SF, Zhuang JH, Wang GY, Xia W. GINS2 promotes cell proliferation and inhibits cell apoptosis in thyroid cancer by regulating CITED2 and LOXL2. Cancer Gene Ther. 2019;26:103–113. doi: 10.1038/s41417-018-0045-y. [DOI] [PubMed] [Google Scholar]
- 30.Pennacchio LA, Bickmore W, Dean A, Nobrega MA, Bejerano G. Enhancers: five essential questions. Nat Rev Genet. 2013;14:288–295. doi: 10.1038/nrg3458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chun JN, Cho M, Park S, So I, Jeon JH. The conflicting role of E2F1 in prostate cancer: a matter of cell context or interpretational flexibility? Biochim Biophys Acta Rev Cancer. 2020;1873:188336. doi: 10.1016/j.bbcan.2019.188336. [DOI] [PubMed] [Google Scholar]
- 32.Tian W, Cui F, Esteban MA. E2F1 in renal cancer: Mr Hyde disguised as Dr Jekyll? J Pathol. 2013;231:143–146. doi: 10.1002/path.4238. [DOI] [PubMed] [Google Scholar]
- 33.Lien EC, Dibble CC, Toker A. PI3K signaling in cancer: beyond AKT. Curr Opin Cell Biol. 2017;45:62–71. doi: 10.1016/j.ceb.2017.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bertacchini J, Heidari N, Mediani L, Capitani S, Shahjahani M, Ahmadzadeh A, Saki N. Targeting PI3K/AKT/mTOR network for treatment of leukemia. Cell Mol Life Sci. 2015;72:2337–2347. doi: 10.1007/s00018-015-1867-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wu Y, Zhang Y, Qin X, Geng H, Zuo D, Zhao Q. PI3K/AKT/mTOR pathway-related long non-coding RNAs: roles and mechanisms in hepatocellular carcinoma. Pharmacol Res. 2020;160:105195. doi: 10.1016/j.phrs.2020.105195. [DOI] [PubMed] [Google Scholar]
- 36.Wei D, Rui B, Qingquan F, Chen C, Ping HY, Xiaoling S, Hao W, Jun G. KIF11 promotes cell proliferation via ERBB2/PI3K/AKT signaling pathway in gallbladder cancer. Int J Biol Sci. 2021;17:514–526. doi: 10.7150/ijbs.54074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liang J, Li H, Han J, Jiang J, Wang J, Li Y, Feng Z, Zhao R, Sun Z, Lv B, Tian H. Mex3a interacts with LAMA2 to promote lung adenocarcinoma metastasis via PI3K/AKT pathway. Cell Death Dis. 2020;11:614. doi: 10.1038/s41419-020-02858-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hao X, Zhang Y, Lu Y, Han G, Rong D, Sun G, Sun G, Tang W, Wu J, Wang X. STK39 enhances the progression of Cholangiocarcinoma via PI3K/AKT pathway. iScience. 2021;24:103223. doi: 10.1016/j.isci.2021.103223. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.