Abstract
Kinase insert domain receptor (KDR) activation is associated with the immunosuppressive microenvironment. However, the efficacy of immunotherapy in patients with KDR mutations is still unclear. To investigate the relationship between KDR gene mutations and the prognosis of pan-cancer, and whether immune checkpoint inhibitors (ICIs) may improve the prognosis of patients with KDR mutations, we analyzed public cohorts of pan-cancer immunotherapeutic patients including genomic and clinical data.Further analysis was performed on an internal validation data set including 67 non-small cell lung cancer. Through bioinformatics analysis, potential mechanism was studied in TCGA data. We found better responses to ICIs in patients with KDR mutation from pan-cancer public datasets (objective response rate [ORR], 45.0% vs 25.1%, P=0.0058; progression-free survival [PFS], P=0.039, HR=0.586, 95% CI 0.353-0.973) and validation cohort (overall survival (OS), P=0.05, HR=0.62; 95% CI, 0.38-1.00). Our NSCLC cohort verified the value of KDR mutation in predicting better clinical outcomes, including ORR (70.0% vs 22.81%, P=0.0057) and PFS (HR=0.158; 95% CI, 0.045-0.773, P=0.007). KDR mutation was associated with tumor mutation burden high, neoantigen burden and immune cellular activities. Meanwhile, KDR mutation was indicative of an immune-hot status, characterized by higher expression of PD-L1 and abundance of cytotoxic lymphocytes. KDR mutations may be potential positive predictors for pan-cancer received ICIs.
Keywords: KDR, pan-cancer, immune checkpoint inhibitors, biomarker, NSCLC
Introduction
Immune Checkpoint Inhibitors (ICIs) are transforming the landscape of treatment in cancers [1]. With ICIs initially approved for advanced melanoma (MM) [2], immunotherapy has become an important treatment for cancers, including non-small cell lung cancer (NSCLC), breast cancer (BRC), clear cell renal cell carcinoma (ccRCC), head and neck squamous cell carcinoma (HNSCC), etc [3-5].
However, ICIs are not effective for all patients. Patients with different molecular, histological, or genetic characteristics may have different response rates to ICIs. The response rate in monotherapy of ICIs is just 15%-20% [6-9]. Although some biomarkers were proven to be significant positive predictors of ICIs treatment, such as microsatellite instability (MSI), PD-L1, and tumor mutation burden (TMB), nearly half of patients could not benefit from ICIs even if the biomarker was positive [10-12]. The development of biomarkers lacks a breakthrough. The expression of PD-L1 is the most widely accepted predictor of immunotherapy. It has been included in the majority of clinical trials of immunotherapy as an important observation indicator. It is generally believed that patients with PD-L1 positive have a better response rate and prognosis during immunotherapy [13,14]. There are still many technical limitations in PD-L1 expression detection [7,15]. Cytoplasmic PD-L1 protein seems to have an uncertain function in immunotherapy. The antibody used to detect of PD-L1 protein expression remains specific. Tumor specimens collected from bronchoscopy or percutaneous lung biopsy are not enough to get stable results. TMB is evolving as a potential biomarker of ICIs efficacy. It also has some limitations. Each tumor mutation was given equal weight, but not all mutations had the same efficacy to the ICIs. For example, RCC patients have promising efficacy with moderate TMB level [16,17]. In addition, some genes mutations, such as NORTH 1/2/3 mutation, were associated with good ICIs efficacy, while JAK1/2 mutations, MDM2/4 amplification, and EGFR mutations were associated with poor efficacy [18-20]. As a biomarker, these results were not imperfect. Therefore, new biomarkers are urgently needed to screen more patients suitable for ICIs treatment.
Kinase insert domain receptor (KDR) encodes a key receptor that mediates tumor angiogenesis/metastasis switches [21]. Several studies have reported that VEGF-VEGFR2 axis activation is associated with the immunosuppressive microenvironment. Activation of VEGF-VEGFR2 could induce the aggregation of immature dendritic cells, bone marrow-derived suppressor cells and regulatory T cells (Tregs), and inhibit T lymphocyte migration [22,23]. However, the clinical significance of KDR mutations for ICIs treated pan-cancer is unclear.
Methods
Clinical cohorts and TCGA cohort
Based on six published immunotherapy studies, survival and mutation dates were collected and integrated as discovery cohort for this study (Supplementary Figure 1) [24-29]. In the discovery cohort, samples were sequenced using whole-exome sequencing (WES) and the Memorial Sloan Kettering-Integrated Mutation Profiling of Actionable Cancer Targets (MSK-IMPACT) panel. Patients with a sample size of <10 (n=3), receiving other treatments (n=10), or unknown treatments (n=4) were excluded. 662 patients were finally included in the discovery cohort, including ccRCC (n=35), NSCLC (n=385), MM (n=205), HNSCC (n=10), and bladder cancer (BLCA) (n=27).
To verify the predictive power of KDR gene to ICIs treatment, we used an expanded pan-cancer cohort with overall survival (OS) and mutational data as the validation cohort [30]. MSK-IMPACT panel were used for the samples from this cohort. After filtering, 1333 patients were included from eight cancer types: renal cell carcinoma (RCC) (n=146), NSCLC (n=303), MM (n=302), HNSSC (n=133), esophagogastric cancer (ESCA) (n=104), colorectal cancer (CRC) (n=106), BRC (n=39), and BLCA (n=200, Figure 1B). All the clinical and mutational dates of the discovery and the validation cohort were obtained from cBioPortal.
Figure 1.
KDR-MUT was associated with the clinical benefit to ICIs in the discovery cohort. A. Progression-free survival curves comparing the KDR-WT and KDR-MUT groups in patients treated with ICIs therapy from the discovery cohort. B. Overall survival curves comparing the KDR-WT and KDR-MUT groups in patients treated with ICIs therapy from the discovery cohort. C. Bar graph showing proportions of ORR in KDR-MUT and KDR-WT patients. D. Bar graph showing proportions of DCB in KDR-MUT and KDR-WT patients (Fisher’s exact test). E. Overall survival curves comparing the KDR-WT and KDR-MUT groups in the TCGA cohort. F. Progression-free survival curves comparing the KDR-WT and KDR-MUT groups in the TCGA cohort.
Sixty-seven patients from our hospital who received at least two cycles of immunotherapy (camrelizumab, nivolumab, pembrolizumab toripalimab, or tislelizumab) were included in our NSCLC cohort (Figure 1C). All patients had genomic profiling of tumor tissue before anti-PD-1 treatment (288-gene panel).
The clinical data, somatic mutation data, and RNA-seq data of the TCGA cohort were retrieved from the UCSC Xena data portal.
Calculation of the TMB and NAL
The TMB was defined as the total number of non-synonymous mutations/exome size or normalized to the exonic coverage of MSK-IMPACT panels for the discovery or validation cohorts, respectively. The high TMB cutoff was the highest 20% of TMB in each cancer, as previously described [30]. NAL (neoantigen load) and TCR (T cell receptor) diversity scores were from pan-cancer immune landscape project [31].
Immune infiltration estimation, leukocyte, lymphocyte and TIL fraction analyses
Cell-type identification was performed by estimating relative subgroup of RNA transcripts (CIBERSORT) based on an online method. Gene profiles was used to describe cellular composition of complex tissues [32]. Hematopoietic subsets were counted using mixed mRNA mixtures from TCGA database.
By analyzing Thorsson’s data, we estimated TIL level using genomics and HE-stained in the TCGA pan-cancer cohort [31]. The leukocyte fraction data was derived from DNA methylation, and an aggregation of plasma cells, NK cells, CD8 T cells, gamma-delta T cells, Tregs, follicular helper T cells, CD4 T cells, and B cells estimated by CIBERSORT. TIL fraction analysis was based on the mappings of TILs for >5,000 HE-stained images on TCGA dataset [33].
Assessment of immune signatures
Gene set enrichment analysis (GSEA) was used to associate the gene signature with KDR-MUT and KDR-WT with the “Cluster Profiler” R package. The normalized enrichment score is the main statistical data to test results of gene set enrichment. In addition, in order to study the relationship between the antitumor immunity and KDR mutation, we obtained 29 classical immune signatures from a previous study [34]. GSEA was used to quantitatively analyze enrichment levels of 29 immune signatures in each sample.
Immunohistochemistry
The paraffin sections of NSCLC were taken from pathology department of our hospital. After dewaxing and rehydrating, the tumor sections were immersed into pH 6.0 citrate buffer for antigen extraction. Endogenous peroxidase was blocked with 3% H2O2, and 3% BSA was added to evenly cover the tissue at 37°C for 30 minutes, then blocking solution was gently removed. The primary antibody was added at 4°C overnight. Then, the HRP labeled secondary antibody from the corresponding species of the primary antibody was added and incubated at 37°C for 50 minutes. DAB staining, hematoxylin dying at 37°C for 3 minutes, dehydration and neutral resin sealing were performed. Anti-CD3 (Servicebio, GB111337), anti-CD31 (Servicebio, GB11063-1), anti-ICAM1 (Servicebio, GB11106) were used. Immunohistochemical staining was quantified by HALO image analysis software.
Statistical analysis
All statistical analyses were conducted with R software (version 3.6.3). Fisher’s exact test was used to assess the KDR status and the response. Log-rank test and Cox proportional hazards regression analysis were used to analyze the differences between KDR-MUT and KDR-WT in PFS and OS. The Cox proportional hazards model was used for subgroup survival analysis and adjustments were made for available confounding. Interactions between the KDR status and age, sex, cancer type, TMB level, and drug class were assessed in the validation cohort. Statistical analysis of comparisons between two groups was conducted using the Wilcoxon test. P values were two-tailed, and <0.05 were considered statistically significant.
Results
Association between VEGF signaling pathway and ICIs efficacy in the discovery cohort
Considering that KDR is a gene in the VEGF signaling pathway, we further explored the relationships between genes in the VEGF pathway and the efficacy of immunotherapy. In the discovery cohort, eight genes involved in the VEGF signaling pathway were investigated, including KDR, FLT1, FLT3, FLT4, VEGFC, VEGFA, VEGFD, and VEGFB. Among these genes, KDR mutation was the only one that significantly prolonged PFS and OS (Supplementary Figure 2A, 2B). In addition, only KDR mutation was gathered in patients with ORR and DCB (Supplementary Figure 2C), indicating that KDR mutation may be the best biomarker for predicting the efficacy of immunotherapy.
Association between KDR-MUT and ICIs efficacy in the discovery cohort
Six publicly available studies involving 5 cancer types were consolidated into the discovery cohort. The basic characteristics were summarized in Supplementary Table 1. Fifty-one patients were harboring KDR mutations (KDR-MUT), accounting for 7.7% of the discovery cohort, and 611 patients were KDR wild-type (KDR-WT). 3 (5.9%) patients were confirmed harboring p.S803Y mutations and 3 (5.9%) patients with p.R1032Q mutations (Supplementary Figure 3A). We found that patients with KDR-MUT had longer PFS than patients with KDR-WT (median PFS: 48.91 vs 6.63 months, log-rank test P=0.003, multivariable-adjusted P=0.039, multivariable-adjusted HR 0.586; 95% CI, 0.353-0.973, Figure 1A). Superior OS was also observed in KDR-MUT group (median OS: not reached vs 24.08 months, log-rank test P=0.028, multivariable-adjusted P=0.069, multivariable-adjusted HR=0.595; 95% CI, 0.340-1.040, Figure 1B). After adjusted for sex, age, cancer types, drug class, and TMB level, the significant difference remained in PFS (multivariable-adjusted P=0.039, multivariable-adjusted HR=0.586; 95% CI, 0.353-0.973), but only a numerically significant OS benefit (multivariable-adjusted P=0.069, multivariable-adjusted HR=0.595; 95% CI, 0.340-1.040). According to RECIST 1.1, the overall response of 594 patients was evaluable, including 40 KDR-MUT patients and 554 KDR-WT patients. As expected, the ORR in patients with KDR-MUT was almost twice as higher as in patients with KDR-WT (45.0% vs 25.1%, P=0.0058, Figure 1C). As for DCB, 56% of patients with KDR-MUT were from ICIs treatment, while 41.8% of patients with KDR-WT were from ICIs (P=0.0522, Figure 1D).
Further, to assess the potential prognostic value of KDR mutations, we performed survival analyses based on KDR status in the TCGA database. There was no significant difference in OS between the KDR-WT and KDR-MUT patients treated with standard treatment (Figure 1E), and the same results across multiple cancer types were presented in Supplementary Figure 4. Although significantly worse PFS was observed in KDR-MUT patients (median PFS 49.249 vs 70.125 months, log-rank test P=0.04, HR=1.170; 95% CI, 1.01-1.36), it was no longer significant after adjusting for sex, age, and cancer types (multivariable-adjusted P=0.36, multivariable-adjusted HR=0.93; 95% CI, 0.80-1.08, Figure 1F). Taken together, KDR-MUT may potentially predict the efficacy and favorable clinical outcomes of ICIs treatment.
Association between KDR-MUT and ICIs efficacy in the validation cohort
The basic characteristics of an expanded ICI-treated cohort (n=1333) were summarized in Supplementary Table 2. There were 73 cases of KDR-MUT, including 39 MM, 11 BLCA, 9 NSCLC, 5 HNSCC, 3 ESCA, 3 RCC, 2 CRC, and 1 BRC, accounting for 5.5% of the population. In the validation cohort, 73 of 1333 patients were confirmed to have KDR mutations (Supplementary Figure 2B). p.R1032Q (2.7%) was high frequency mutation site in KDR mutation, followed by p.P351S (2.7%), p.E469K (2.7%). Adjusted for confounding factors (sex, age, cancer types, drug class, and TMB level), KDR mutation remained an independent predictor for superior OS (median OS 46.0 vs 22.0 months, log-rank test P=0.0001, multivariable-adjusted P=0.05, multivariable-adjusted HR=0.62; 95% CI, 0.38-1.00, Figure 2A). Even compared with other oncogenes, KDR remained the most stable predictor for multiple cancers (Supplementary Table 3) [25,26,35-38]. In the stratification analysis of OS, KDR-MUT also had a survival advantage over KDR-WT in subgroups of age, sex, cancer type, TMB status, and drug class (Figure 2B, P>0.05).
Figure 2.
Validation of the predictive function of KDR-MUT. A. Overall survival curves comparing the KDR-WT and KDR-MUT groups in patients treated with ICIs therapy from the validation cohort. B. Stratification analysis of OS in the validation cohort. NSCLC, non-small cell lung cancer; SKCM, melanoma; HNSC, head and neck cancer; CRC, colorectal cancer; BLCA, bladder cancer; ESCA, esophagogastric cancer; RCC, renal cell carcinoma; BRCA, Breast invasive carcinoma. C. Overall survival curves comparing KDR-MUT&TMB high, KDR-MUT&TMB low, KDR-WT&TMB high, and KDR-WT&TMB low groups in the validation cohort.
Recently, TMB has been proven to be an effective immunotherapy biomarker [39]. Hence, to evaluate the predictive performances of TMB and KDR, patients were divided into four groups based on KDR status and TMB level. We found that KDR-MUT patients achieved the longest OS. Moreover, KDR mutation was associated with higher TMB (adjusted P=0.04, HR=0.40; 95% CI, 0.19-0.94, Figure 2C). We also explored the effect of the most frequent types of KDR mutations on TMB in discovery and validation cohort. As shown in Supplementary Figure 6, patients with specific KDR mutation sites had higher TMB than KDR-WT patients, but there was no difference between patients with different KDR mutation subtypes.
Association between KDR-MUT and ICIs efficacy in NSCLC cohort
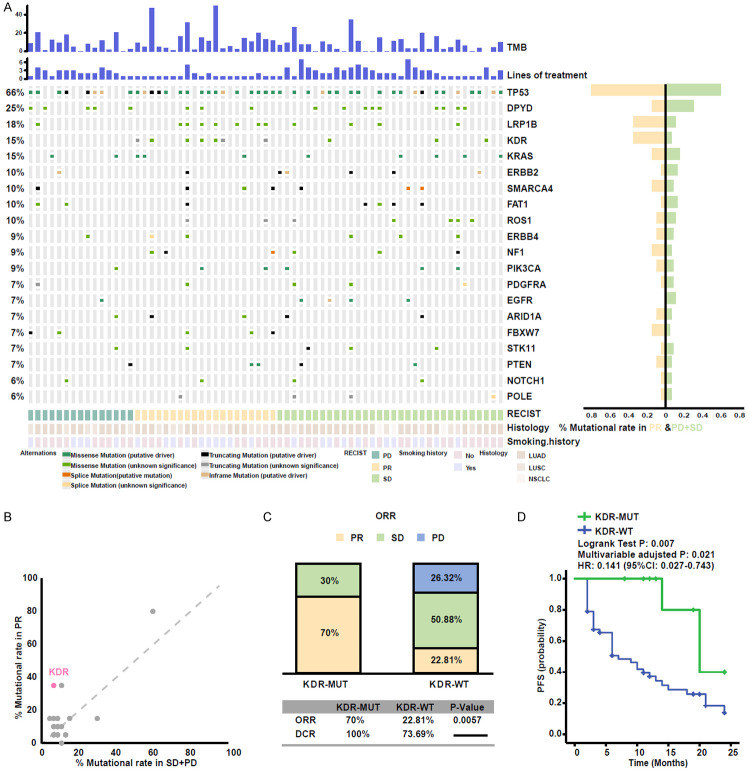
In our study, 67 NSCLC patients with genomic profiling were included (Supplementary Table 4). In NSCLC cohort, 54 of the 67 patients (80.6%) were male with a median age of 64-year-old (range, 32-80). The ORR was 29.8%. In the NSCLC cohort, the frequency of KDR mutations was 14.9%. And p.R1032Q mutation was not found in NSCLC with KDR mutation (Supplementary Figure 3C).
The genomic mutational landscape of 67 patients is displayed in Figure 3A. Consistent with other studies, the rate of TP53 and LRP1B mutations were higher in responders than non-responders [37,40]. Besides these, KDR mutation enrichment was discovered in responders as well (Figure 3B). KDR mutation was associated with higher ORR (70.0% vs 22.81%, P=0.0057, Figure 3C), longer PFS (median PFS 20 months vs 7 months, log-rank test P=0.007, HR=0.158; 95% CI, 0.045-0.773, Figure 3D) in NSCLC patients. These results further demonstrate the predictive function of KDR mutation to ICIs treatment.
Figure 3.
KDR-MUT was associated with a better response to ICIs in the NSCLC cohort. A. Stacked plots show mutational burden (histogram, top); lines of treatment (histogram); mutations in TP53, DPYD, LRP1B, KDR, KRAS, ERBB2, SMARCA4, FAT1, ROS1, ERBB4, NF1, PIK3CA, PDGFRA, EGFR, ARID1A, FBXW7, STK11, PTEN, NOTCH1, and POLE (tile plot, middle); their mutational rates in patients having achieved objective response or progressive disease (histogram, right); and mutational marks (bottom). B. Scatter diagram displaying the mutational rate in patients having achieved objective response or progressive disease. KDR is emphasized in red. C. Histogram depicting proportions of ORR and DCR in KDR-MUT and KDR-WT patients. D. Progression-free survival curves in the patients with or without KDR-MUT.
Association of KDR-MUT with enhanced immunogenicity and activated immune response
The mutational landscape of KDR and clinical characteristics were shown in Figure 4A, with MM patients (16.4%) having the highest levels of KDR mutations, followed by NSCLC (10.5%) and GBM (8.8%) (Figure 4B). Across all patients, the mutational frequencies of KDR was 5.0%. The somatic mutations of KDR were evenly distributed (Figure 4C, Supplementary Figure 4A), and there was no difference in PFS and OS with the common KDR mutation subtypes (Supplementary Figure 7).
Figure 4.
The pan-cancer mutational landscape of KDR in TCGA cohort. A. Association between KDR status and annotated clinical characteristics in TCGA cohort. (cancer type, sex, age, CNA, TMB, PFS, and OS were annotated. Samples were sorted by KDR status, while KDR-MUT and KDR-WT samples were separated by a gap. B. The proportion of KDR-MUT tumors identified for each cancer type with alteration frequency above 1%. C. Lollipop plot showing the loci distribution of mutations across the KDR altered patient cohorts from the TCGA database. Truncating mutations included nonsense, nonstop, splice site mutations, and frameshift insertion and deletion; Non truncating mutations included missense mutations and inframe insertion and deletion.
To further explore the potential mechanisms associated with KDR-MUT in predicting the efficacy of ICIs. We first analyzed the correlation between KDR and TMB and neoantigen burden. As shown, KDR mutations were associated with higher TMB in the TCGA, discovery and validation cohorts, as well as significantly higher predicted neoantigens in the TCGA cohort (Figure 5A; Supplementary Figure 3A), suggesting that the KDR positive patients may have a higher immune reaction to tumor neoantigens.
Figure 5.
KDR-MUT was associated with enhanced tumor immunogenicity and anti-tumor immunity. A. Comparison of the TMB and NAL levels between the KDR-MUT and KDR-WT tumors in the TCGA cohort. B. Significantly enriched pathways with GSEA between KDR-MUT and KDR-WT tumors in the TCGA dataset. C. Comparison of the MHC molecules and co-stimulators expression levels between KDR-WT and KDR-MUT tumors in the TCGA cohort. D. Comparison of the expression of TCR richness between KDR-WT and KDR-MUT tumors. E. Heatmap shows clustering of tumor types based on angiogenesis and APC correlated gene sets. The heatmap is colored by the normalized enrichment score of a gene set for a tumor type. F. Comparison of the adhesion molecules expression levels in KDR-MUT and KDR-WT groups. G. Quantitative immunohistochemistry (IHC) analysis of CD31 and ICAM1 protein expression in the KDR-MUT and KDR-WT groups (n=4/group). Representative micrographs of these samples are shown. Data represent the mean ± SD. (Mann-Whitney U test; ns, not significant; *P<0.05, **P<0.01, ***P<0.001, ****P<0.0001).
GSEA revealed the significant enrichment of antigen processing and presentation pathway in the KDR-MUT group, along with other immune activation-related pathways, including T-cell receptor signaling, NK cell-mediated immunity, CD8+ T cell activation, and IFN-gamma response (Figure 5B). In addition, KDR-MUT tumors had higher expression of MHC I- and II-associated antigen-presenting molecules than KDR-WT tumors (Figure 5C). Antigen-presenting cells (APCs) have long been known to present tumor-associated antigens to T cells, which elicited tumor-specific immune responses. Immune evasion associated with in antigen presentation to T lymphocytes defects is a common phenomenon and highlights the relevance of T cells in solid tumors rejection. These results attenuate this effect in KDR-MUT tumors, which may make tumor cells susceptible to ICIs.
Following antigen processing and presentation by APC, two signals are essential to activate the immune response against cancer cells: peptide-MHC complexes occupy TCRs and subsequent activate the costimulatory molecules [41]. The host immune system needs to maintain a diverse TCR repertoire to recognize multiple tumor neoantigens [42]. We found the TCR diversity and costimulatory molecules were significantly higher in KDR-MUT tumors than KDR-WT tumors (Figure 5C and 5D). Taken together, these results suggested that KDR-MUT was associated with enhanced immunogenicity and activation of tumor neoantigens immune response.
Notably, we also found that angiogenesis and VEGFR signaling was down-regulated in KDR-MUT tumors than KDR-WT tumors across multiple cancer types (Figure 5B and 5E). VEGFR2 activation promotes endothelial cell proliferation, migration. Consistently, significantly decreased mRNA expression levels of vascular epithelial cell migration and proliferation-related genes were observed (Supplementary Figure 5B). Abnormal angiogenesis was closely associated with immunosuppressive and is characterized by T cell infiltration overcoming higher interstitial fluid pressure, and down-regulated vasculature cell adhesion molecule-1 (VCAM-1) and intracellular cell adhesion molecule-1 (ICAM-1) to impair the T cell extravasation [43]. As shown in Figure 5E and 5F, vascular permeability and expression of ICAM1 were decreased and increased in KDR-MUT patients, respectively. Decreased tumor vessel density labeled by CD31 and increased expression of ICAM1 were further confirmed by immunohistochemistry (Figure 5G). These results implied that KDR-MUT was associated with the inhibition of angiogenesis, which could promote immune cell infiltration.
KDR-MUT was indicative of an immune-hot status
Hot tumor microenvironment (TME) characterized by the presence of tumor-infiltrating lymphocytes and PD-L1 expression is associated with increased response to anti-PD-1/L1 monotherapy [44,45]. By using CIBERSORT, we found that activated NK cells and CD8+ T cells were more abundant in KDR-MUT tumors. Besides, M0 and M1 macrophages infiltration increased, while the M2 macrophages infiltration decreased (Figure 6A). To cross-examine the above results with different immune cells assessment methods, we next analyzed the expression profiles of 29 immune signatures. As shown in Figure 6B, KDR-MUT tumors have rich immune signatures and microenvironment cell populations, such as dendritic cells (DCs) and cytotoxic lymphocytes. Then, using three different methods, we further verified the abundance of immune cells in KDR-MUT tumors. First, the larger leukocyte fraction in KDR-MUT tumors was assessed based on DNA methylation arrays (Figure 6C). Next, we obtained a similar high TIL results in KDR-MUT tumors based on TIL score estimated from HE-stained slides (Figure 6D) [33], which was consistent with the results of lymphocyte fraction estimated by the CIBERSORT method (Figure 6E). Importantly, CD3 immunohistochemical results further confirmed the presence of tumor-infiltrating T cells in KDR-MUT tumors (Figure 6F).
Figure 6.
KDR-MUT was indicative of an immune-hot status. A. Comparison of the 22 immune cells infiltration levels in KDR-MUT and KDR-WT tumors. CIBERSORT was used to calculate the infiltration degree of these immune cells. B. Volcano plots of 29 immune signatures estimated by the GSEA method. Immune signatures enriched in KDR-MUT tumors and KDR-WT tumors are marked in red and blue, respectively. C. Comparison of the leukocyte fractions between KDR-WT and KDR-MUT tumors. D. Comparison of the TIL regional fractions between KDR-WT and KDR-MUT tumors. E. Comparison of the lymphocyte fractions between KDR-WT and KDR-MUT tumors. F. Quantitative immunohistochemistry (IHC) analysis of CD3 protein expression in the KDR-MUT and KDR-WT groups (n=4/group). Representative micrographs of these samples are shown. Data represent the mean ± SD. G. Boxplot comparing the immune-related genes expression levels in KDR-MUT and KDR-WT groups (Mann-Whitney U test; ns, not significant; *P<0.05, **P<0.01, ***P <0.001, ****P<0.0001).
Some chemokines (such as CXCL9, CXCL10, and CCL5) were also up-regulated in the KDR-MUT tumors, and these chemokines have been shown to attract CD8 T cells and DCs [46,47]. Additionally, the expression of genes related with cytotoxic activity (such as GZMA, PRF1), the surrogate measures of cytotoxic T lymphocyte (CTL) activity, was higher in KDR-MUT tumors, indicating enhanced tumor-killing capacity. Moreover, PD-L1 (CD274) and CTLA4 were up-regulated in KDR-MUT tumors (Figure 6G). Both of these results demonstrate KDR-MUT was associated with a hot TME and enhanced the ICIs efficacy.
Discussion
In this work, KDR-MUT were identified for the first time as a positive factor for better clinical benefit in pan-cancer patients treated with ICIs, particularly NSCLC. However, patients receiving standard care did not receive the clinical benefits of OS. In the exploratory analyses, higher TMB, higher neoantigen burden, and reduced expression of genes associated with VEGFR pathway activation may be potential mechanism for predicting KDR mutation in pan-cancer (Supplementary Figure 8). GSEA revealed prominent enrichment of signatures related to antigen processing and presentation in patients with KDR-MUT. Using CIBERSORT, we found cytotoxic lymphocytes were more abundant in KDR-MUT tumors. GSEA showed that KDR-MUT tumors had rich immune signatures and microenvironment cell populations, further proving that KDR-MUT was associated with a hot tumor microenvironment. These results suggest that KDR-MUT may be a potentially positive predictor of pan-cancer patients treated with ICIs.
In recent years, more and more immunotherapy treatments has been applied to clinical practice. How to select patients who can benefit from immunotherapy has become a thorny clinical problem. Some specific mutations have been proved to influence the response to immunotherapy. It was reported that MMR, POLE, and POLD1 were related to the response to immunotherapy [27,28,37-40]. These genes seem to lack powerful clinical evidence. Our study demonstrated that KDR-MUT has the potential to become a biomarker for ICIs. We showed the association between KDR gene status and the responses of immunotherapy in three independent cohorts. Importantly, in our own NSCLC cohort, we excluded some confounding factors, such as the course of treatment which were not covered in the public cohort. It can reduce the statistical bias to some extent and make the result more reliable. KDR mutation is not so rare in tumors, accounting for about 5.0% of the pan-cancer cohort, with MM ranking first (16.4%) followed by NSCLC (10.5%) and GBM (8.8%). This means that KDR-MUT could serve more patients than other rare mutants. Unlike PD-L1 expression or TMB, KDR-MUT can be easily detected by next-generation sequencing. KDR-MUT is also included in most commercially available gene panels.
KDR is a type 2 vascular endothelial growth factor receptor that plays an important role in the regulation of angiogenesis and vascular integrity. However, a possible functional association between PD-1/PD-L1 and VEGF/VEGFR has been reported. In patients with ccRCC and classical Hodgkin lymphoma addition, PD-L1 expression was associated with VEGF and microvessel density [48,49]. Our study showed similar results, with significantly higher TMB values and PD-L1 expression associated with KDR-MUT compared to KDR-WT. It produced more neoantigens, which were processed by APCs and presented to T cells. Up-regulated MHC-related molecules and TCR diversity activated this process. Meanwhile, chemokines contributed to the invasion of CD8 T cells and DCs into the tumor tissues. CTL released GZMA and PRF1 and then enhanced tumor-killing capacity. This is one of the reasons why we believe that KDR nonsynonymous mutations are associated with good clinical outcomes in ICIs.
To our knowledge, this is the first time for exploring the relationship between KDR-MUT and ICIs in pan-cancer. Some limitations still existed in our study including these inherent to a retrospective design. The analysis was based on a universal carcinomatous public cohort that received WES or panel sequencing, which may have resulted in selection bias. However, our own independent cohort of NSCLC was included in the analysis as a strong complement. Due to the limited number of patients, the relationship between KDR-MUT and the efficacy of immunotherapy cannot be further explored, but the overall efficacy of KDR-MUT has been satisfactory. These results should be confirmed in large cohorts. In addition, the potential mechanisms by which KDR mutation enhance the efficacy of immunotherapy were explored only through bioinformatics analysis. Our conclusion is only that KDR mutation is related to the immune hot environment, but the specific mechanism how KDR-MUT caused the immune alterations is worthy of further experimental study.
In conclusion, our results suggested that KDR-MUT was associated with better PFS and OS in pan-cancer patients who received ICIs. KDR-MUT might be an important indicator of the immunogenetic landscape, and should be considered as a therapeutic biomarker for immunotherapy.
Acknowledgements
We are indebted and thankful to all participants for their valuable contributions. This study was supported by the National Natural Science Foundation of China (81972188) and the Medical Important Talents of Jiangsu Province (ZDRCA2016024).
Disclosure of conflict of interest
None.
Supporting Information
References
- 1.Johnson DB, Sullivan RJ, Menzies AM. Immune checkpoint inhibitors in challenging populations. Cancer. 2017;123:1904–1911. doi: 10.1002/cncr.30642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Billan S, Kaidar-Person O, Gil Z. Treatment after progression in the era of immunotherapy. Lancet Oncol. 2020;21:e463–e476. doi: 10.1016/S1470-2045(20)30328-4. [DOI] [PubMed] [Google Scholar]
- 3.Rassy EE, Khoury Abboud RM, Ibrahim N, Assi T, Aoun F, Kattan J. The current state of immune checkpoint inhibitors in the first-line treatment of renal cancer. Immunotherapy. 2018;10:1047–1052. doi: 10.2217/imt-2018-0017. [DOI] [PubMed] [Google Scholar]
- 4.Zhang Z, Xie T, Zhang X, Qi C, Shen L, Peng Z. Immune checkpoint inhibitors for treatment of advanced gastric or gastroesophageal junction cancer: current evidence and future perspectives. Chin J Cancer Res. 2020;32:287–302. doi: 10.21147/j.issn.1000-9604.2020.03.02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zam W, Ali L. Immune checkpoint inhibitors in the treatment of cancer. Curr Rev Clin Exp Pharmacol. 2022;17:103–113. doi: 10.2174/1574884716666210325095022. [DOI] [PubMed] [Google Scholar]
- 6.Borghaei H, Paz-Ares L, Horn L, Spigel DR, Steins M, Ready NE, Chow LQ, Vokes EE, Felip E, Holgado E, Barlesi F, Kohlhäufl M, Arrieta O, Burgio MA, Fayette J, Lena H, Poddubskaya E, Gerber DE, Gettinger SN, Rudin CM, Rizvi N, Crinò L, Blumenschein GR Jr, Antonia SJ, Dorange C, Harbison CT, Graf Finckenstein F, Brahmer JR. Nivolumab versus docetaxel in advanced nonsquamous non-small-cell lung cancer. N Engl J Med. 2015;373:1627–39. doi: 10.1056/NEJMoa1507643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ikeda S, Goodman AM, Cohen PR, Jensen TJ, Ellison CK, Frampton G, Miller V, Patel SP, Kurzrock R. Metastatic basal cell carcinoma with amplification of PD-L1: exceptional response to anti-PD1 therapy. NPJ Genom Med. 2016;1:16037. doi: 10.1038/npjgenmed.2016.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shitara K, Özgüroğlu M, Bang YJ, Di Bartolomeo M, Mandalà M, Ryu MH, Fornaro L, Olesiński T, Caglevic C, Chung HC, Muro K, Goekkurt E, Mansoor W, McDermott RS, Shacham-Shmueli E, Chen X, Mayo C, Kang SP, Ohtsu A, Fuchs CS KEYNOTE-061 investigators. Pembrolizumab versus paclitaxel for previously treated, advanced gastric or gastro-oesophageal junction cancer (KEYNOTE-061): a randomised, open-label, controlled, phase 3 trial. Lancet. 2018;392:123–133. doi: 10.1016/S0140-6736(18)31257-1. [DOI] [PubMed] [Google Scholar]
- 9.Jardim DL, Goodman A, de Melo Gagliato D, Kurzrock R. The challenges of tumor mutational burden as an immunotherapy biomarker. Cancer Cell. 2021;39:154–173. doi: 10.1016/j.ccell.2020.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mok TSK, Wu YL, Kudaba I, Kowalski DM, Cho BC, Turna HZ, Castro G Jr, Srimuninnimit V, Laktionov KK, Bondarenko I, Kubota K, Lubiniecki GM, Zhang J, Kush D, Lopes G KEYNOTE-042 Investigators. Pembrolizumab versus chemotherapy for previously untreated, PD-L1-expressing, locally advanced or metastatic non-small-cell lung cancer (KEYNOTE-042): a randomised, open-label, controlled, phase 3 trial. Lancet. 2019;393:1819–1830. doi: 10.1016/S0140-6736(18)32409-7. [DOI] [PubMed] [Google Scholar]
- 11.André T, Shiu KK, Kim TW, Jensen BV, Jensen LH, Punt C, Smith D, Garcia-Carbonero R, Benavides M, Gibbs P, de la Fouchardiere C, Rivera F, Elez E, Bendell J, Le DT, Yoshino T, Van Cutsem E, Yang P, Farooqui MZH, Marinello P, Diaz LA Jr KEYNOTE-177 Investigators. Pembrolizumab in microsatellite-instability-high advanced colorectal cancer. N Engl J Med. 2020;383:2207–2218. doi: 10.1056/NEJMoa2017699. [DOI] [PubMed] [Google Scholar]
- 12.Carbone DP, Reck M, Paz-Ares L, Creelan B, Horn L, Steins M, Felip E, van den Heuvel MM, Ciuleanu TE, Badin F, Ready N, Hiltermann TJN, Nair S, Juergens R, Peters S, Minenza E, Wrangle JM, Rodriguez-Abreu D, Borghaei H, Blumenschein GR Jr, Villaruz LC, Havel L, Krejci J, Corral Jaime J, Chang H, Geese WJ, Bhagavatheeswaran P, Chen AC, Socinski MA CheckMate 026 Investigators. First-line nivolumab in stage IV or recurrent non-small-cell lung cancer. N Engl J Med. 2017;376:2415–2426. doi: 10.1056/NEJMoa1613493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Reck M, Rodríguez-Abreu D, Robinson AG, Hui R, Csőszi T, Fülöp A, Gottfried M, Peled N, Tafreshi A, Cuffe S, O’Brien M, Rao S, Hotta K, Leiby MA, Lubiniecki GM, Shentu Y, Rangwala R, Brahmer JR KEYNOTE-024 Investigators. Pembrolizumab versus chemotherapy for PD-L1-positive non-small-cell lung cancer. N Engl J Med. 2016;375:1823–1833. doi: 10.1056/NEJMoa1606774. [DOI] [PubMed] [Google Scholar]
- 14.Balar AV, Castellano D, O’Donnell PH, Grivas P, Vuky J, Powles T, Plimack ER, Hahn NM, de Wit R, Pang L, Savage MJ, Perini RF, Keefe SM, Bajorin D, Bellmunt J. First-line pembrolizumab in cisplatin-ineligible patients with locally advanced and unresectable or metastatic urothelial cancer (KEYNOTE-052): a multicentre, single-arm, phase 2 study. Lancet Oncol. 2017;18:1483–1492. doi: 10.1016/S1470-2045(17)30616-2. [DOI] [PubMed] [Google Scholar]
- 15.Zhou ZQ, Zhao JJ, Pan QZ, Chen CL, Liu Y, Tang Y, Zhu Q, Weng DS, Xia JC. PD-L1 expression is a predictive biomarker for CIK cell-based immunotherapy in postoperative patients with breast cancer. J Immunother Cancer. 2019;7:228. doi: 10.1186/s40425-019-0696-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gunjur A. Nivolumab plus ipilimumab in advanced renal-cell carcinoma. Lancet Oncol. 2018;19:e232. doi: 10.1016/S1470-2045(18)30257-2. [DOI] [PubMed] [Google Scholar]
- 17.Motzer RJ, Escudier B, McDermott DF, George S, Hammers HJ, Srinivas S, Tykodi SS, Sosman JA, Procopio G, Plimack ER, Castellano D, Choueiri TK, Gurney H, Donskov F, Bono P, Wagstaff J, Gauler TC, Ueda T, Tomita Y, Schutz FA, Kollmannsberger C, Larkin J, Ravaud A, Simon JS, Xu LA, Waxman IM, Sharma P CheckMate 025 Investigators. Nivolumab versus everolimus in advanced renal-cell carcinoma. N Engl J Med. 2015;373:1803–13. doi: 10.1056/NEJMoa1510665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang K, Hong X, Song Z, Xu Y, Li C, Wang G, Zhang Y, Zhao X, Zhao Z, Zhao J, Huang M, Huang D, Qi C, Gao C, Cai S, Gu F, Hu Y, Xu C, Wang W, Lou Z, Zhang Y, Liu L. Identification of deleterious NOTCH mutation as novel predictor to efficacious immunotherapy in NSCLC. Clin Cancer Res. 2020;26:3649–3661. doi: 10.1158/1078-0432.CCR-19-3976. [DOI] [PubMed] [Google Scholar]
- 19.Shin DS, Zaretsky JM, Escuin-Ordinas H, Garcia-Diaz A, Hu-Lieskovan S, Kalbasi A, Grasso CS, Hugo W, Sandoval S, Torrejon DY, Palaskas N, Rodriguez GA, Parisi G, Azhdam A, Chmielowski B, Cherry G, Seja E, Berent-Maoz B, Shintaku IP, Le DT, Pardoll DM, Diaz LA Jr, Tumeh PC, Graeber TG, Lo RS, Comin-Anduix B, Ribas A. Primary resistance to PD-1 blockade mediated by JAK1/2 mutations. Cancer Discov. 2017;7:188–201. doi: 10.1158/2159-8290.CD-16-1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kato S, Goodman A, Walavalkar V, Barkauskas DA, Sharabi A, Kurzrock R. Hyperprogressors after immunotherapy: analysis of genomic alterations associated with accelerated growth rate. Clin Cancer Res. 2017;23:4242–4250. doi: 10.1158/1078-0432.CCR-16-3133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Peng FW, Liu DK, Zhang QW, Xu YG, Shi L. VEGFR-2 inhibitors and the therapeutic applications thereof: a patent review (2012-2016) Expert Opin Ther Pat. 2017;27:987–1004. doi: 10.1080/13543776.2017.1344215. [DOI] [PubMed] [Google Scholar]
- 22.Voron T, Colussi O, Marcheteau E, Pernot S, Nizard M, Pointet AL, Latreche S, Bergaya S, Benhamouda N, Tanchot C, Stockmann C, Combe P, Berger A, Zinzindohoue F, Yagita H, Tartour E, Taieb J, Terme M. VEGF-A modulates expression of inhibitory checkpoints on CD8+ T cells in tumors. J Exp Med. 2015;212:139–48. doi: 10.1084/jem.20140559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Terme M, Pernot S, Marcheteau E, Sandoval F, Benhamouda N, Colussi O, Dubreuil O, Carpentier AF, Tartour E, Taieb J. VEGFA-VEGFR pathway blockade inhibits tumor-induced regulatory T-cell proliferation in colorectal cancer. Cancer Res. 2013;73:539–49. doi: 10.1158/0008-5472.CAN-12-2325. [DOI] [PubMed] [Google Scholar]
- 24.Miao D, Margolis CA, Vokes NI, Liu D, Taylor-Weiner A, Wankowicz SM, Adeegbe D, Keliher D, Schilling B, Tracy A, Manos M, Chau NG, Hanna GJ, Polak P, Rodig SJ, Signoretti S, Sholl LM, Engelman JA, Getz G, Jänne PA, Haddad RI, Choueiri TK, Barbie DA, Haq R, Awad MM, Schadendorf D, Hodi FS, Bellmunt J, Wong KK, Hammerman P, Van Allen EM. Genomic correlates of response to immune checkpoint blockade in microsatellite-stable solid tumors. Nat Genet. 2018;50:1271–1281. doi: 10.1038/s41588-018-0200-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Miao D, Margolis CA, Gao W, Voss MH, Li W, Martini DJ, Norton C, Bossé D, Wankowicz SM, Cullen D, Horak C, Wind-Rotolo M, Tracy A, Giannakis M, Hodi FS, Drake CG, Ball MW, Allaf ME, Snyder A, Hellmann MD, Ho T, Motzer RJ, Signoretti S, Kaelin WG Jr, Choueiri TK, Van Allen EM. Genomic correlates of response to immune checkpoint therapies in clear cell renal cell carcinoma. Science. 2018;359:801–806. doi: 10.1126/science.aan5951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rizvi NA, Hellmann MD, Snyder A, Kvistborg P, Makarov V, Havel JJ, Lee W, Yuan J, Wong P, Ho TS, Miller ML, Rekhtman N, Moreira AL, Ibrahim F, Bruggeman C, Gasmi B, Zappasodi R, Maeda Y, Sander C, Garon EB, Merghoub T, Wolchok JD, Schumacher TN, Chan TA. Cancer immunology. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science. 2015;348:124–8. doi: 10.1126/science.aaa1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hellmann MD, Nathanson T, Rizvi H, Creelan BC, Sanchez-Vega F, Ahuja A, Ni A, Novik JB, Mangarin LMB, Abu-Akeel M, Liu C, Sauter JL, Rekhtman N, Chang E, Callahan MK, Chaft JE, Voss MH, Tenet M, Li XM, Covello K, Renninger A, Vitazka P, Geese WJ, Borghaei H, Rudin CM, Antonia SJ, Swanton C, Hammerbacher J, Merghoub T, McGranahan N, Snyder A, Wolchok JD. Genomic features of response to combination immunotherapy in patients with advanced non-small-cell lung cancer. Cancer Cell. 2018;33:843–852. doi: 10.1016/j.ccell.2018.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Snyder A, Makarov V, Merghoub T, Yuan J, Zaretsky JM, Desrichard A, Walsh LA, Postow MA, Wong P, Ho TS, Hollmann TJ, Bruggeman C, Kannan K, Li Y, Elipenahli C, Liu C, Harbison CT, Wang L, Ribas A, Wolchok JD, Chan TA. Genetic basis for clinical response to CTLA-4 blockade in melanoma. N Engl J Med. 2014;371:2189–2199. doi: 10.1056/NEJMoa1406498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rizvi H, Sanchez-Vega F, La K, Chatila W, Jonsson P, Halpenny D, Plodkowski A, Long N, Sauter JL, Rekhtman N, Hollmann T, Schalper KA, Gainor JF, Shen R, Ni A, Arbour KC, Merghoub T, Wolchok J, Snyder A, Chaft JE, Kris MG, Rudin CM, Socci ND, Berger MF, Taylor BS, Zehir A, Solit DB, Arcila ME, Ladanyi M, Riely GJ, Schultz N, Hellmann MD. Molecular determinants of response to anti-programmed cell death (PD)-1 and anti-programmed death-ligand 1 (PD-L1) blockade in patients with non-small-cell lung cancer profiled with targeted next-generation sequencing. J. Clin. Oncol. 2018;36:633–641. doi: 10.1200/JCO.2017.75.3384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Samstein RM, Lee CH, Shoushtari AN, Hellmann MD, Shen R, Janjigian YY, Barron DA, Zehir A, Jordan EJ, Omuro A, Kaley TJ, Kendall SM, Motzer RJ, Hakimi AA, Voss MH, Russo P, Rosenberg J, Iyer G, Bochner BH, Bajorin DF, Al-Ahmadie HA, Chaft JE, Rudin CM, Riely GJ, Baxi S, Ho AL, Wong RJ, Pfister DG, Wolchok JD, Barker CA, Gutin PH, Brennan CW, Tabar V, Mellinghoff IK, DeAngelis LM, Ariyan CE, Lee N, Tap WD, Gounder MM, D’Angelo SP, Saltz L, Stadler ZK, Scher HI, Baselga J, Razavi P, Klebanoff CA, Yaeger R, Segal NH, Ku GY, DeMatteo RP, Ladanyi M, Rizvi NA, Berger MF, Riaz N, Solit DB, Chan TA, Morris LGT. Tumor mutational load predicts survival after immunotherapy across multiple cancer types. Nat Genet. 2019;51:202–206. doi: 10.1038/s41588-018-0312-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Thorsson V, Gibbs DL, Brown SD, Wolf D, Bortone DS, Ou Yang TH, Porta-Pardo E, Gao GF, Plaisier CL, Eddy JA, Ziv E, Culhane AC, Paull EO, Sivakumar IKA, Gentles AJ, Malhotra R, Farshidfar F, Colaprico A, Parker JS, Mose LE, Vo NS, Liu J, Liu Y, Rader J, Dhankani V, Reynolds SM, Bowlby R, Califano A, Cherniack AD, Anastassiou D, Bedognetti D, Mokrab Y, Newman AM, Rao A, Chen K, Krasnitz A, Hu H, Malta TM, Noushmehr H, Pedamallu CS, Bullman S, Ojesina AI, Lamb A, Zhou W, Shen H, Choueiri TK, Weinstein JN, Guinney J, Saltz J, Holt RA, Rabkin CS Cancer Genome Atlas Research Network. Lazar AJ, Serody JS, Demicco EG, Disis ML, Vincent BG, Shmulevich I. The immune landscape of cancer. Immunity. 2018;48:812–830. doi: 10.1016/j.immuni.2018.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Newman AM, Liu CL, Green MR, Gentles AJ, Feng W, Xu Y, Hoang CD, Diehn M, Alizadeh AA. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods. 2015;12:453–7. doi: 10.1038/nmeth.3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Saltz J, Gupta R, Hou L, Kurc T, Singh P, Nguyen V, Samaras D, Shroyer KR, Zhao T, Batiste R, Van Arnam J Cancer Genome Atlas Research Network. Shmulevich I, Rao AUK, Lazar AJ, Sharma A, Thorsson V. Spatial organization and molecular correlation of tumor-infiltrating lymphocytes using deep learning on pathology images. Cell Rep. 2018;23:181–193. doi: 10.1016/j.celrep.2018.03.086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.He Y, Jiang Z, Chen C, Wang X. Classification of triple-negative breast cancers based on immunogenomic profiling. J Exp Clin Cancer Res. 2018;37:327. doi: 10.1186/s13046-018-1002-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Haradhvala NJ, Kim J, Maruvka YE, Polak P, Rosebrock D, Livitz D, Hess JM, Leshchiner I, Kamburov A, Mouw KW, Lawrence MS, Getz G. Distinct mutational signatures characterize concurrent loss of polymerase proofreading and mismatch repair. Nat Commun. 2018;9:1746. doi: 10.1038/s41467-018-04002-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mehnert JM, Panda A, Zhong H, Hirshfield K, Damare S, Lane K, Sokol L, Stein MN, Rodriguez-Rodriquez L, Kaufman HL, Ali S, Ross JS, Pavlick DC, Bhanot G, White EP, DiPaola RS, Lovell A, Cheng J, Ganesan S. Immune activation and response to pembrolizumab in POLE-mutant endometrial cancer. J Clin Invest. 2016;126:2334–40. doi: 10.1172/JCI84940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dong ZY, Zhong WZ, Zhang XC, Su J, Xie Z, Liu SY, Tu HY, Chen HJ, Sun YL, Zhou Q, Yang JJ, Yang XN, Lin JX, Yan HH, Zhai HR, Yan LX, Liao RQ, Wu SP, Wu YL. Potential predictive value of TP53 and KRAS mutation status for response to PD-1 blockade immunotherapy in lung adenocarcinoma. Clin Cancer Res. 2017;23:3012–3024. doi: 10.1158/1078-0432.CCR-16-2554. [DOI] [PubMed] [Google Scholar]
- 38.Wu YM, Cieślik M, Lonigro RJ, Vats P, Reimers MA, Cao X, Ning Y, Wang L, Kunju LP, de Sarkar N, Heath EI, Chou J, Feng FY, Nelson PS, de Bono JS, Zou W, Montgomery B, Alva A, Robinson DR, Chinnaiyan AM PCF/SU2C International Prostate Cancer Dream Team. Inactivation of CDK12 delineates a distinct immunogenic class of advanced prostate cancer. Cell. 2018;173:1770–1782. doi: 10.1016/j.cell.2018.04.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cui Y, Chen H, Xi R, Cui H, Zhao Y, Xu E, Yan T, Lu X, Huang F, Kong P, Li Y, Zhu X, Wang J, Zhu W, Wang J, Ma Y, Zhou Y, Guo S, Zhang L, Liu Y, Wang B, Xi Y, Sun R, Yu X, Zhai Y, Wang F, Yang J, Yang B, Cheng C, Liu J, Song B, Li H, Wang Y, Zhang Y, Cheng X, Zhan Q, Li Y, Liu Z. Whole-genome sequencing of 508 patients identifies key molecular features associated with poor prognosis in esophageal squamous cell carcinoma. Cell Res. 2020;30:902–913. doi: 10.1038/s41422-020-0333-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen H, Chong W, Wu Q, Yao Y, Mao M, Wang X. Association of LRP1B mutation with tumor mutation burden and outcomes in melanoma and non-small cell lung cancer patients treated with immune check-point blockades. Front Immunol. 2019;10:1113. doi: 10.3389/fimmu.2019.01113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pahari S, Negi S, Aqdas M, Arnett E, Schlesinger LS, Agrewala JN. Induction of autophagy through CLEC4E in combination with TLR4: an innovative strategy to restrict the survival of Mycobacterium tuberculosis. Autophagy. 2020;16:1021–1043. doi: 10.1080/15548627.2019.1658436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Riaz N, Havel JJ, Makarov V, Desrichard A, Urba WJ, Sims JS, Hodi FS, Martín-Algarra S, Mandal R, Sharfman WH, Bhatia S, Hwu WJ, Gajewski TF, Slingluff CL Jr, Chowell D, Kendall SM, Chang H, Shah R, Kuo F, Morris LGT, Sidhom JW, Schneck JP, Horak CE, Weinhold N, Chan TA. Tumor and microenvironment evolution during immunotherapy with nivolumab. Cell. 2017;171:934–949. doi: 10.1016/j.cell.2017.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Teng MW, Ngiow SF, Ribas A, Smyth MJ. Classifying cancers based on T-cell infiltration and PD-L1. Cancer Res. 2015;75:2139–45. doi: 10.1158/0008-5472.CAN-15-0255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Galon J, Bruni D. Approaches to treat immune hot, altered and cold tumours with combination immunotherapies. Nat Rev Drug Discov. 2019;18:197–218. doi: 10.1038/s41573-018-0007-y. [DOI] [PubMed] [Google Scholar]
- 45.Hegde PS, Karanikas V, Evers S. The where, the when, and the how of immune monitoring for cancer immunotherapies in the era of checkpoint inhibition. Clin Cancer Res. 2016;22:1865–74. doi: 10.1158/1078-0432.CCR-15-1507. [DOI] [PubMed] [Google Scholar]
- 46.Harlin H, Meng Y, Peterson AC, Zha Y, Tretiakova M, Slingluff C, McKee M, Gajewski TF. Chemokine expression in melanoma metastases associated with CD8+ T-cell recruitment. Cancer Res. 2009;69:3077–85. doi: 10.1158/0008-5472.CAN-08-2281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bergamaschi C, Pandit H, Nagy BA, Stellas D, Jensen SM, Bear J, Cam M, Valentin A, Fox BA, Felber BK, Pavlakis GN. Heterodimeric IL-15 delays tumor growth and promotes intratumoral CTL and dendritic cell accumulation by a cytokine network involving XCL1, IFN-γ, CXCL9 and CXCL10. J Immunother Cancer. 2020;8:e000599. doi: 10.1136/jitc-2020-000599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shin SJ, Jeon YK, Kim PJ, Cho YM, Koh J, Chung DH, Go H. Clinicopathologic analysis of PD-L1 and PD-L2 expression in renal cell carcinoma: association with oncogenic proteins status. Ann Surg Oncol. 2016;23:694–702. doi: 10.1245/s10434-015-4903-7. [DOI] [PubMed] [Google Scholar]
- 49.Koh YW, Han JH, Yoon DH, Suh C, Huh J. PD-L1 expression correlates with VEGF and microvessel density in patients with uniformly treated classical Hodgkin lymphoma. Ann Hematol. 2017;96:1883–1890. doi: 10.1007/s00277-017-3115-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.