Abstract
Pyroptosis plays a vital role in the development of cancers; however, its role in regulating immune cell infiltration in tumor microenvironment (TME) and pyroptosis-related molecular subtypes remain unclear. Herein, we comprehensively analyzed the molecular subtypes mediated by the pyroptosis-related genes (PRGs) in gastric cancer (GC). Three pyroptosis patterns were determined with distinct TME cell-infiltrating characteristics and prognosis. Principal component analysis was performed to establish the pyroptosis score. The high pyroptosis score group was featured by increased activated CD4+ T cell infiltration, better prognosis, elevated tumor mutation burden, higher immune and stromal scores, and enhanced response to immunotherapy. However, the low pyroptosis score group was characterized by poorer survival, decreased immune infiltration, and glycerolipid and histidine metabolism pathways. Additionally, high pyroptosis score was confirmed as an independent favorable prognostic factor for overall survival. Three cohorts designed to analyze the response to immunotherapy verified that patients with higher pyroptosis score showed treatment benefit. In summary, our study demonstrated that pyroptosis regulates the complex TME. Assessing the pyroptosis patterns will advance our understanding on TME features and tumor immunology and provide the rationale for designing personalized immunotherapy strategies.
Keywords: Pyroptosis, gastric cancer, immunotherapy, tumor microenvironment, prognosis
Introduction
Gastric cancer (GC) ranks as the fourth most common cancer worldwide and claims about 11,010 lives in the United States in 2020 [1]. Metastasis, local recurrences, and multidrug resistance are the major causes of death in patients with advanced GC [2]. Due to the lack of ideal diagnostic biomarkers, most GC patients are diagnosed at late stage, and radical surgery can’t be performed. Thus, chemotherapy plays a crucial role in the management of GC, especially for the advanced GC patients. Recently, immunotherapy is emerging as a promising treatment option for GC [3]. Immunotherapy uses patient’s own immune system to identify and eliminate the cancer cells. The most successful immunotherapy used so far is to target the immune checkpoint blockade such as programmed cell death-1/Ligand 1 (PD-1/L1) [4,5]. Nevertheless, only a small fraction of patients have benefited from the immunotherapy [6,7]. Therefore, it is in urgent need to identify the prognostic biomarkers and improve the effectiveness of immunotherapy. Previous studies have argued that genetic variation, epigenetic regulation, and environmental factors play crucial roles in the tumor initiation and progression [8,9]. However, increasing evidence demonstrates that the tumor microenvironment (TME) is also a key regulatory factor in tumor progression [10,11]. Tumor cells can directly or indirectly interact with the TME components, which, in turn promotes the malignant phenotypes of tumor cells [10,12,13]. Effective immunotherapy is based on the infiltration of immune cells in tumor sites, prediction of the response to immunotherapy based on the characterization of TME cell infiltration has become an important area of research, and novel immunotherapeutic drugs are currently being developed and tested [6,7]. Hence, TME is important for the management of tumor progression, the development of personalized immunotherapy, and the prediction of prognosis and the response to immunotherapy [11,14].
To personalize and optimize the treatments for patients with GC, an accurate prognosis prediction is also crucial for the GC management. To date, the tumor node metastasis (TNM) staging system is commonly adopted to predict the prognosis of cancer patients [15]. Nevertheless, increasing evidence showed the unsatisfactory discriminative capacity of staging system [16,17]. There is controversy over whether the risk factors other than TNM factors are important predictors of clinical outcomes [17]. Recently, increasing prognostic biomarkers have been identified for GC [2]. Nevertheless, the existing biomarkers may lack sufficient specificity and sensitivity in prognosis judgement [2]. A comprehensive understanding of the TME landscape and different tumor immune phenotypes as well as the biomarkers or molecular signatures will be helpful in guiding the development of cancer treatment and prognosis prediction, which would be a great complement to the TNM staging system.
Pyroptosis has been reported as a pro-inflammatory cell death characterized by cellular swelling, plasma membrane rupture, water influx, osmotic lysis, and the release of the intracellular pro-inflammatory contents [18]. Although it shares some features with apoptosis, including caspase dependence, nuclear condensation, and DNA damage [18], pyroptosis has unique features. For example, pyroptosis is morphologically different from apoptosis as it presents the features of lysis and cell swelling [19]. Pyroptosis can be triggered mainly by two pathways-canonical pathway and non-canonical pathway. The canonical pathway is activated when pathogens, pathogen associated molecular patterns (PAMPs), and damage associated molecular patterns (DAMPs) activates the inflammasome sensors such as nucleotide oligomerization domain (NOD)-like receptor family, pyrin domain-containing-1 and 3 (NLRP1, NLRP3), or absent in melanoma-2 (AIM2) and recruits caspase 1 (CASP1) to activate Gasdermin D (GSDMD), which then forms a pore in the plasma membrane [20,21]. As for the non-canonical pathway, lipopolysaccharide (LPS) and PAMPs directly activate CASP4, CASP5, and CASP11, causing pyroptosis via cleaving GSDMD. Subsequently, the activated GSDMD initiates pore formation in the cell membrane, finally inducing the cell death [20]. Pyroptosis is initially identified as an infection defending mechanism. Recent studies have revealed its important role in the development of various tumors [22,23]. For example, caspase-3/GSDME pathway is considered as a switch between apoptosis and pyroptosis in cancer [24]. Recent studies also reveal the significant roles of pyroptosis in regulating immunology and tumor immune microenvironment (TIME) [25]. GSDMD is reported to enhance the phagocytosis of tumor cells by tumor-associated macrophages, tumor-infiltrating natural-killer, and CD8+ T lymphocytes and promote neutrophil death [26]. Furthermore, PD-L1-regulated GSDMC expression can switch apoptosis to pyroptosis in cancer cells and facilitate tumor necrosis [27]. However, most of studies only explored the function of a single or several pyroptosis-related genes (PRGs), while comprehensive analysis on the correlation of PRGs and TME characterizations is scarce. In this study, we comprehensively assessed the correlation between the pyroptosis-related molecular patterns and the TME cell-infiltrating characteristics through analyzing the genomic data of 989 GC cases. Three distinct pyroptosis-related molecular patterns were identified with different prognosis and TME characteristics, indicating the importance of pyroptosis-related molecular clusters in modulating the characterizations of TME. Moreover, we established a pyroptosis score system to quantify the pyroptosis clusters for each GC patient. The pyroptosis score may be helpful in selecting the personalized immunotherapy and predicting the prognosis of GC patients.
Materials and methods
Materials
TRIzol reagent: Invitrogen, USA.
PrimeScriptTM RT Master Mix (Perfect Real Time) Kit: TaKaRa, Dalian, China.
SYBR® Premix Ex TaqTM II (Tli RNaseH Plus): TaKaRa, Dalian, China.
Anti-GSDMC antibody: 27630-1-AP; Proteintech Group, Inc., USA.
HRP-conjugated secondary antibodies: Sigma-Aldrich, USA.
DAB substrate liquid: Thermo Fisher Scientific, USA.
Dataset collection and preprocessing
Figure S1 presented the workflow of our study. Gene profiling data and full clinical information were searched in Gene-Expression Omnibus (GEO) (http://www.ncbi.nlm.nih.gov/geo/) and The Cancer Genome Atlas (TCGA) (https://portal.gdc.cancer.gov/) databases. Datasets without survival information were censored. Five eligible GC cohorts (GSE15459, GSE34942, GSE57303, GSE62254, and TCGA-Stomach Adenocarcinoma (STAD)) were selected in the study. GEO database are available in GPL570 platform (Affymetrix Human Genome U133 Plus 2.0 Array). The raw “CEL” files of these GEO datasets were downloaded, and “affy” and “simpleaffy” packages were used to perform the background adjustment and quantile normalization. For TCGA-STAD dataset, gene expression profiling of RNA seq data (FPKM value) was obtained from the Genomic Data Commons (GDC, oohttps://portal.gdc.cancer.gov/) website. Subsequently, the fragments per kilobase million (FPKM) values were transformed into transcripts per kilobase million (TPM). The “ComBat” algorithm of “sva” package was applied to correct the batch effects due to the non-biological technical bias. Table S1 presented the information of all enrolled datasets of GC patients. The genomic mutation data (copy number variation (CNV) and somatic mutation data) of TCGA-STAD were downloaded from the UCSC Xena database (https://gdc.xenahubs.net/). The CNV landscape of 33 PRGs in human chromosomes was plotted using the “Rcircos” package.
Unsupervised clustering for PRGs
Thirty-three PRGs were extracted from the prior reviews [28-31] (Table S2). These genes are known to be involved in the pyroptosis signaling, and encoding the core components of the pyroptosis machinery. Thus, they are collectively termed as PRGs. 30 overlapping PRGs were extracted from five integrated datasets to construct pyroptosis-related molecular patterns by unsupervised clustering analysis (Table S3). The optimal number of clusters and their stability were evaluated based on the consensus clustering algorithm. The “ConsensuClusterPlus” package was used to conduct the above steps and 1000 times repetitions were performed to guarantee the classification stability [32].
Gene set variation analysis (GSVA) and functional enrichment
GSVA enrichment was performed to decipher the biological process among the different pyroptosis clusters using “GSVA” R packages. The biological signatures (c2.cp.kegg.v7.2.symbols) were downloaded from the MSigDB database (http://www.gsea-msigdb.org/gsea/msigdb) for GSVA enrichment analysis. Adjusted P-value P<0.05 was regarded as statistically significant. Functional enrichment for PRGs was performed using the “clusterProfiler” R package with false discovery rate (FDR)-value cutoff below 0.05.
Estimation of immune cell infiltration
The single sample gene set enrichment analysis (ssGSEA) was performed to quantify the relative abundance of different immune cell types in the TME as reported [33]. Marker genes for each tumor-infiltrating immune cell type were selected based on previous publications [34]. The enrichment score was calculated by ssGSEA analysis and normalized to unity distribution from 0 to 1 to represent the relative abundance of each immune cell type.
Identification of pyroptosis molecular pattern-related differential expression genes (DEGs)
To identify the pyroptosis molecular pattern-related DEGs, we classified GC patients into three different pyroptosis molecular patterns based on the expression of 30 PRGs. The “limma” R package was utilized to screen the DEGs in GC patients in the distinct clusters [35]. The criteria for selecting the pyroptosis molecular pattern-related DEGs were set at adjusted p-value <0.001.
Estimation of STromal and Immune cells in MAlignant Tumour tissues using Expression data (ESTIMATE)
By performing the ESTIMATE algorithm, the ESTIMATE score, stromal score, and immune score were calculated to evaluate the levels of infiltrating stromal and immune cells [36]. The differences in the ESTIMATE score, stromal score, and immune score among the different groups was compared using Wilcoxon rank-sum test.
Development of the pyroptosis score
Pyroptosis scoring system was established to evaluate the pyroptosis-related molecular patterns of individual patient with GC by using principal components analysis (PCA) algorithm. Briefly, the overlapping DEGs screened from different pyroptosis clusters were used to select the overall survival (OS)-related DEGs by using the univariate Cox regression analysis. The OS-related DEGs were extracted for further analysis. The consensus clustering algorithm was used for identify the number of pyroptosis gene clusters as well as their stability using the OS-related DEGs. Then, PCA was performed to construct the pyroptosis-related gene signature (pyroptosis score). Both principal component 1 and 2 were extracted and served as the signature score. We subsequently used a formula similar to previous studies to define the pyroptosis score [37,38]: pyroptosis score=∑(PC1i+PC2i), where I represents the expression of final determined pyroptosis phenotype-related prognostic DEGs.
Analysis of somatic alteration data
Mutation data of GC patients in the TCGA-STAD cohort were downloaded from TCGA database (https://www.cancer.gov/tcga/). The total number of non-synonymous mutations (including frameshift mutation, inflame mutation, missense mutation, nonsense mutation, and splice site mutation) was counted to determine the mutational burden of GC [33]. The “maftool” package was used to identify and present the GC driver genes [39]. The distribution difference of somatic alterations between the low and high pyroptosis score group in TCGA-STAD cohort was also evaluated. The top 20 driver genes with the highest alteration frequency in these two groups were compared as reported [40].
Prediction of drug sensitivity for GC patients
The Genomics of Drug Sensitivity in Cancer (GDSC; https://www.cancerrxgene.org/) database was used to estimate the sensitivity of each patient to different chemotherapy drugs [41]. “pRRophetic” package was utilized to quantify the half-maximal inhibitory concentration (IC50) of drugs [42].
Identification of small molecule drugs for GC patients
The “limma” package was applied to screen the DEGs between the low and high pyroptosis score groups with p-value <0.05 and |logFC| >1.0 and visualized by volcano plot. “ClusterProfiler” package was used for Kyoto Encyclopedia of Genes and Genomes (KEGG) and Gene Ontology (GO) enrichment analyses. An adjusted p-value <0.05 was considered statistically significant. The upregulated and downregulated DEGs were then uploaded into the Connectivity map (CMap) database (http://portals.broadinstitute.org/cmap/) [43]. Small molecular drug candidates were identified by CMap mode-of-action (MoA) analysis.
Validation of pyroptosis score in predicting response to immunotherapy
The immunophenogram was established to predict the response to anti-PD-1/PD-L1 therapy in pan-cancer [34]. Immunophenoscore (IPS) was calculated by the immunophenogram among the four cancer subtypes. The clinical data containing IPS information of TCGA-STAD cohort were downloaded from The Cancer Immunome Atlas database (TCIA, https://tcia.at/home) [34]. We compared IPS (cytotoxic T lymphocyte-associated antigen-4 (CTLA4)_negative+PD-1_negative, CTLA4_positive+PD-1_negative, CTLA4_negative+PD-1_positive, CTLA4_positive+PD-1_positive) between the high and low pyroptosis score groups in each subtype. A high IPS in PD-1_positive/CTLA4_positive subtype predicted a better response to anti-PD-1/PD-L1/anti_CTLA4 therapy.
Three immunotherapy cohorts were enrolled in this study. The gene expression profiles and complete clinical information were downloaded from the GEO database. These three cohorts had 26 patients with metastatic melanoma treated with anti-PD-1 antibody (GSE78220 cohort) [44], 27 patients with advanced non-small cell lung carcinoma patients treated with anti-PD-1/PD-L1 antibody (GSE135222 cohort) [45,46], and 56 patients with advanced melanoma treated with anti-CTLA4 and ant-PD-1 (GSE91061 cohort) antibodies [47]. The gene expression profiles of GEO datasets were curated and converted to the TPM format for further analysis.
Tissues collection, RNA isolation, and quantitative reverse transcription PCR (RT-qPCR)
20 paired GC and adjacent normal tissues were collected from the GC patients who underwent radical resection at the Xijing Hospital of Digestive Diseases of Fourth Military Medical University during March 2017 to October 2019. Written informed consent was obtained from all patients. The patients didn’t receive other treatments prior to the surgery. After collection, the samples were immediately snap-frozen in liquid nitrogen and stored at -80°C. The study was approved by the Ethics Committee of the Xijing Hospital of Fourth Military Medical University.
TRIzol reagent was used for total RNA isolation. cDNA was synthetized using the PrimeScriptTM RT Master Mix (Perfect Real Time) Kit. RT-qPCR reaction was performed using SYBR® Premix Ex TaqTM II (Tli RNaseH Plus). mRNA expression was normalized with internal control GAPDH. Relative RNA expression was calculated by the 2-ΔΔCt method. The primer sequences used were: GAPDH Forward primer: 5’-GACAGTCAGCCGCATCTTCT-3’; Reverse primer: 5’-GCGCCCAATACGACCAAATC-3’. GSDMC Forward primer: 5’-CCTGGTGGTGCCATCCTAAA-3’; Reverse primer: 5’-GATGCTCCTTACCAGCTCCT-3’.
Immunohistochemistry (IHC) assay
IHC of GSDMC was performed on the tissue microarray (OD-CT-DgStm01-003) provided by Shanghai Outdo Biotech (Shanghai, China). Briefly, the slides were incubated with primary antibody and HRP-conjugated secondary antibody. The signal was visualized by DAB staining and scanned by 3D-histech scanner (3DHISTECH Ltd., Hungary) equipped with a Panoramic Viewer (3DHISTECH Ltd., Hungary) and a Caseviewer software (3DHISTECH Ltd., Hungary). The IHC results were independently evaluated and scored by two pathologists. The IHC signal was scored based on the total proportion of cells stained positively and the intensity of signal in cells. The intensity was scored as follows: 0, negative; 1, weak; 2, medium; or 3, strong. The proportion was scored as follows: 0, negative; 1, 1-25%; 2, 26-50%; 3, 51-75%; or 4, 76-100%. The IHC scores were obtained by multiplying the proportion score with the intensity score, which were in the range of 0-12.
Statistical analysis
All statistical analysis in this study was conducted using R 4.0.2 software. The Perl language and R 4.0.2 software were used to process the data. The “RCircos” package was used to plot the copy number variation landscape of 33 PRGs in human chromosomes. For two group comparison, Student’s t tests and Wilcoxon rank-sum test were used to estimate the statistical significance of normally distributed variables and non-normally distributed variables, respectively. One-way ANOVA and Kruskal-Wallis tests were conducted for multiple group comparison. The “survminer” package was utilized to identify the cut-off point of each subgroup. The survival curve for OS analysis was constructed using the Kaplan-Meier method and compared by the log-rank tests. Univariate and multivariate Cox regression analysis were conducted to determine the effect of clinicopathological factors and pyroptosis score on OS. Two-sided p-value less than 0.05 was considered statistically significant.
Results
Landscape of genetic variation of pyroptosis-related genes in GC
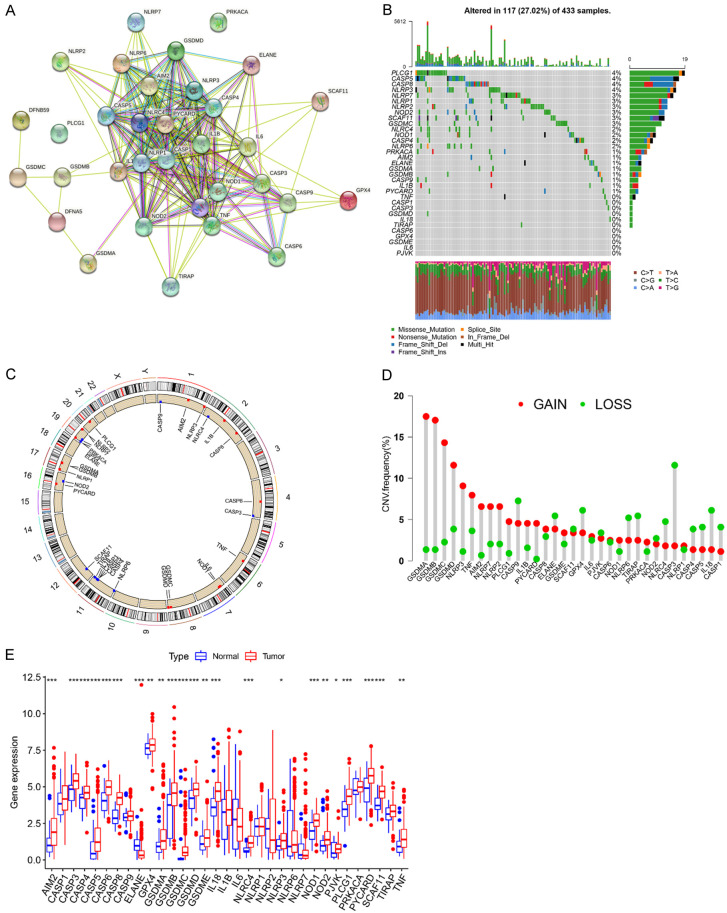
To explore the importance of PRGs in GC progression, we examined the genetic variation of PRGs in GC. Thirty-three PRGs that have been reported till now were included in our analysis [46] (Table S2). We used STRING (https://www.string-db.org/) to construct the protein-protein interaction (PPI) network [49]. As shown in Figure 1A, there were broad interactions among these genes. Somatic mutations were frequently observed in PRGs in GC patients. Out of 433 GC cases we examined, 117 samples (27.02%) had mutations in PRGs, including missense mutations, nonsense mutations, deep deletions, and etc. (Figure 1B). Among these PRGs, phospholipase cgamma 1 (PLCG1), CASP5, CASP8, and NLRP3 exhibited the highest mutation rates (4%), whereas tumor necrosis factor (TNF), CASP1, CASP3, GSDMD, interleukin 18 (IL18), TIR domain-containing adaptor protein (TIRAP), CASP6, glutathione peroxidase 4 (GPX4), GSDME, IL6, and pejvakin (PJVK) had the lowest mutation rates (0%). Figure 1C showed the location of CNV of these PRGs on chromosomes. GSDMA, GSDMB, GSDMC, and GSDMD genes had a relatively high frequency of amplification, while CASP3, CASP9, elastase, neutrophil expressed (ELANE), and GPX4 genes mainly had CNV deletions (Figure 1D). We also analyzed the mutation co-occurrence across all PRGs and identified several co-occurring mutation patterns in several PRGs, such as the co-occurring mutation between NLRP6 and CASP5, or NOD1 and NLRC4, as well as PLCG1 and CASP5 (Figure S2A). More importantly, our three-dimensional PCA (3D-PCA) showed that the 33 PRGs could completely distinguished GC patients from normal controls (Figure S2B). As shown in Figure 1E, most PRGs showed higher mRNA expression in GC tissues compared with that in the normal tissues. Particularly, the expression of PRGs with CNV amplification such as GSDMA, GSDMB, GSDMC, GSDMD, NLRP3, TNF and absent in melanoma 2 (AIM2), except for ELANE, was markedly increased in GC tissues compared to that in normal controls (Figure 1E). Moreover, we performed Spearman correlation analysis to explore the co-expression correlation among these PRGs. As shown in Figure S2C, CASP1 and CASP3 showed a significant positive correlation with most of other PRGs. KEGG pathway enrichment analysis of these PRGs was also performed, and we found signaling pathways such as NOD-like receptor-, TNF-, and IL17-signaling pathway were significantly enriched (Figure S2D), suggesting that these PRGs may play significant roles in the development and progression of GC. In addition, the expression levels of some PRGs were closely associated with The Cancer Genome Atlas (TCGA) molecular subtypes (Figure S3A), suggesting that the PRGs might contribute to the heterogeneity of GC. These results were further validated in the GSE62254 cohort (Figure S3B). Collectively, these findings indicated the high heterogeneity in the genetic alteration of PRGs in GC.
Figure 1.
The landscape of genetic and expression variation of PRGs in GC. A. PPI network of 33 PRGs. B. Genetic alterations of 33 PRGs with a frequency of 27.02% in 117 of 433 GC patients from TCGA-STAD cohort. Each column represents an individual patient. C. The location of CNV alteration of 33 PRGs on chromosomes using the data from TCGA-STAD cohort. D. The CNV mutation frequency of 33 PRGs in TCGA-STAD cohort. The column height represents the alteration frequency. The deletion frequency, green dot; The amplification frequency, red dot. E. The expression levels of 33 PRGs between the normal tissues and GC tissues. Normal tissues, blue; Tumor tissues, red. The asterisks represent the statistical p-value (*P<0.05; **P<0.01; ***P<0.001).
Prognostic role of PRGs and pyroptosis-related molecular patterns in GC patients
To further investigate the function of PRGs in GC development and progression, four independent GEO datasets (GSE15459, GSE34942, GSE57303, and GSE62254) and a TCGA-STAD dataset containing patient OS and clinical data were merged into one meta-cohort (Table S1). After analyzing the gene expression data, 30 overlapping PRGs were selected for further analysis (Table S3). First, the prognostic value of these PRGs was investigated using a univariate Cox regression model (Table S4), and PRGs related to OS of patients were summarized in Figure S4. The prognostic significance and the interactions of these PRGs were then depicted in a network plot (Figure 2A). We found that these PRGs showed a remarkably co-expression correlation. Since PLCG1, CASP5, CASP8, and NLRP3 genes exhibited a relatively higher mutation frequency, we analyzed the difference in the expression of PRGs between these wild type and mutants, respectively. We found a differential expression of PRGs, e.g., CASP3 was markedly upregulated in mutant as compared to wild type (Figures S5, S6, S7, S8). These results suggested that the crosstalk among the PRGs might play a significant role in the formation of pyroptosis-related molecular patterns.
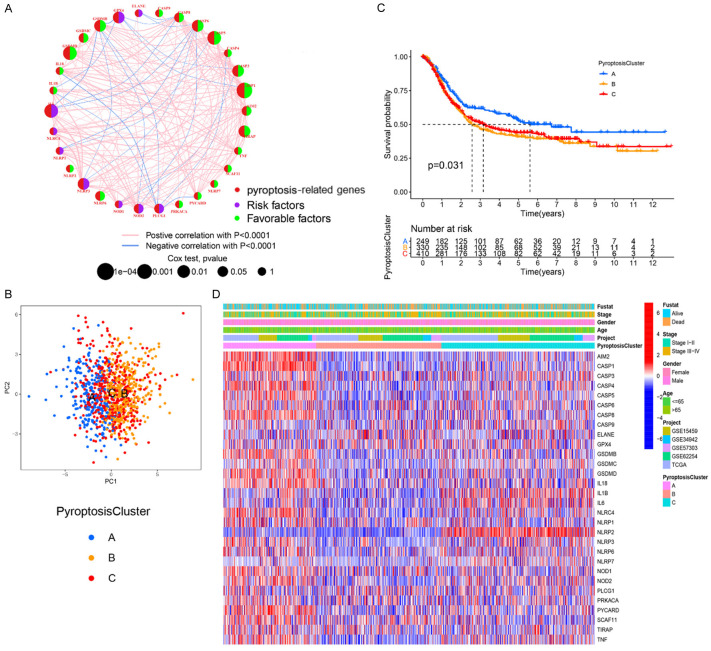
Figure 2.
Characteristics of pyroptosis-related molecular patterns. A. The interaction network of PRGs in GC. The size of circle represents the effects of PRGs on the clinical outcome of GC patients (P<0.0001, P<0.001, P<0.01, P<0.05 and P<1, Cox test). Green dots in the circle, favorable factors of prognosis. Purple dots in the circle, unfavorable factors of prognosis; B. PCA of the mRNA expression profiles of PRGs confirms the three pyroptosis clusters (blue, pyroptosis Cluster A; yellow, pyroptosis Cluster B; red, pyroptosis Cluster C). C. Survival analyses for the three pyroptosis-related molecular patterns based on 989 GC patients from five cohorts (GSE15459, GSE34942, GSE57303, GSE62254, and TCGA-STAD) including 249 patients in pyroptosis Cluster A, 330 patients in pyroptosis Cluster B, and 410 patients in pyroptosis Cluster C (Log-rank test, P=0.031). D. Heatmap presents the correlation between the three pyroptosis clusters and clinicopathological characteristics of GC patients.
Based on the above findings, we applied consensus clustering analysis to classify samples with qualitatively different pyroptosis-related molecular patterns by the mRNA expression profiles of overlapping PRGs, and we identified three pyroptosis-related molecular patterns using the R package of “ConsensusClusterPlus”, including 249 patients in pyroptosis Cluster-A, 330 cases in pyroptosis Cluster-B and 410 cases in pyroptosis Cluster-C (Figure S9A-D). PCA results showed that the three Clusters can be completely separated (Figure 2B). In addition, prognostic analysis for the three pyroptosis-related molecular patterns revealed that the pyroptosis Cluster-A pattern had survival advantage over the other two Clusters (Figure 2C). We observed a clear difference in the expression of PRGs between different pyroptosis-related molecular patterns. The expression of AIM2, CASP1, CASP4, CASP8, and IL18 was markedly upregulated in the pyroptosis Cluster-A subtype while ELANE and GPX4 was significantly increased in pyroptosis Cluster-B. IL1B, IL6 and NLRP2 levels were evidently elevated in pyroptosis Cluster-C (Figure 2D). The different clinicopathological characteristics of GC patients in these three Clusters were also shown in Figure 2D.
Immunological characteristics of distinct pyroptosis-related molecular patterns in GC
We further performed gene set variation analysis (GSVA) enrichment analysis to assess gene set enrichment and evaluate the pathway enrichment in these three pyroptosis-related molecular patterns. As shown in Figure S10A-C, pyroptosis Cluster-A was prominently enriched in the pathways related to the activation of immune system, including natural killer (NK) cell mediated cytotoxicity, NOD-like receptor signaling pathway, graft-versus-host disease, antigen processing and presentation, and allograft rejection. On the other hand, pathways related to basal cell carcinoma, propanoate metabolism, taurine and hypotaurine metabolism, cardiac muscle contraction and ribosome were enriched in Pyroptosis Cluster-B. Pyroptosis Cluster-C was mainly associated with biological processes related to steroid biosynthesis, terpenoid backbone biosynthesis, maturity onset diabetes of the young, nitrogen metabolism, protein export and several immune-related pathways such as NOD-like receptor signaling pathway, graft-versus-host disease, and allograft rejection. Based on the GSVA enrichment analysis, we found that these three pyroptosis-related molecular patterns had markedly distinct TME cell infiltration characteristics. Consistently, TME cell infiltration analyses showed that pyroptosis Cluster-C was mainly enriched in immune cell infiltration such as natural killer cell, immature dendritic cell, neutrophil, and plasmacytoid dendritic cell (Figure S11A). Nevertheless, GC patients in this subtype didn’t have survival advantage (Figure 2C). One possible explanation was that the immune cells were limited in the stroma of tumor microenvironment and couldn’t reach the tissue parenchyma, as a result, the anti-tumor immune response was inhibited by the stromal elements in the pyroptosis Cluster-C [50]. We also found a low level of major histocompatibility complex (MHC) class I and myeloid-derived suppressor cells (MDSCs) in pyroptosis Cluster-C whereas pyroptosis Cluster-B was associated with the absence of activated CD8+ T cells, low MHC class I expression, and fatty acid metabolism (Figure S11A, S11B). Based on these results, we categorized these three pyroptosis Clusters as immune-excluded phenotype (pyroptosis Cluster-C), immune-inflamed phenotype (pyroptosis Cluster-A), and immune-desert phenotype (pyroptosis Cluster-B) according to the previous published criteria (Figure S11A, S11B). Moreover, the correlation between the characteristics of tumor immune microenvironment (TIME) and pyroptosis-related molecular patterns were explored. As shown in Figure S11A, S11B, clear differences in the immune cell infiltration and immune functions were found among the three Clusters. Anti-tumor lymphocyte subpopulations, including activated B cells, activated CD4+ T cells, activated CD8+ T cells, and NK T cells were significantly enriched in the pyroptosis Cluster-A subtype as compared with other subtypes. Additionally, we found that patients in the pyroptosis Cluster-A pattern had markedly higher ESTIMATE score (P<0.001, Figure S11C), immune score (P<0.001, Figure S11D) and stromal score (P<0.01, Figure S11E) than pyroptosis Cluster-B or Cluster-C had, suggesting the critical roles of PRGs in the tumor immunology.
Similarly, Spearman’s correlation analyses were applied to analyze the specific correlation between PRGs and TME infiltration cell type. As shown in Figure S12A, GSDMC had a relatively higher correlation with the activated CD8+ T cells and activated CD4+ T cells, therefore, we focused our analysis on the role of GSDMC in tumor immunology. We first found that patients in GSDMC high expression group showed a relatively longer OS than those in GSDMC low expression group (Figures S4K, S12B and S12C). Second, gene set enrichment analysis (GSEA) suggested the enrichment of several immune-related pathways such antigen processing and presentation pathway, cytokine-cytokine receptor interaction pathway, janus kinase/signal transducer and activator of transcription (JAK-STAT) signaling pathway, NK cell mediated cytotoxicity pathway, and retinoic acid-inducible gene-I (RIG-I)-like receptor signal pathway in GSDMC high expression group (Figure S12D). Next, we detected the mRNA and protein expression levels of GSDMC in GC and the adjacent normal tissues. In consistent with the data in TCGA dataset, RT-qPCR result showed that the mRNA expression of GSDMC in GC tissues was significantly higher than that in the adjacent normal tissues (Figure S12E). However, the IHC results indicated that GSDMC protein level was much lower in the GC tissues (Figure S12F, S12G). We speculated that this discrepancy might result from the post-translational modifications, epigenetic modifications, and other modulations. Lastly, we investigated the difference in TME infiltrating immune cells between the high and low GSDMC expression groups. The results showed that patients with high GSDMC had markedly increased infiltration in resting memory CD4+ T cells, activated memory CD4+ T cells, regulatory T cells (TReg cells), resting NK cells, activated dendritic cell, and neutrophils than patients with low GSDMC (Figure S13A). To support this, we used ESTIMATE algorithm to evaluate the overall infiltration of immune cells between the patients with high and low GSDMC expression. Figure S13B showed the higher immune scores in GSDMC high group. The correlations between GSDMC expression and several immune cells were shown in Figure S13C.
Pyroptosis molecular pattern related DEGs and pyroptosis gene cluster in GC
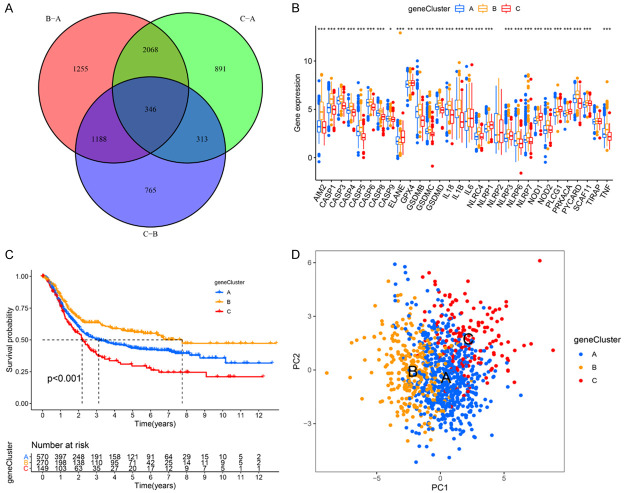
In order to further explore the underlying genetic alterations within each pyroptosis-related molecular pattern, we performed differential expression analysis among these three patterns using the “limma” package and identified 346 overlapping DEGs (Figure 3A; Table S5). We then conducted the KEGG pathway enrichment and GO enrichment analyses for these DEGs. The significant enrichment KEGG pathways and GO terms were summarized in Figure S14A and S14B, respectively. These DEGs showed the enrichment of pathways associated with immune response and inflammatory response, further suggesting the role of pyroptosis in the immunology regulation within the TME (Figure S14A, S14B). Then, the prognostic value of these DEGs in GC patients were explored via a univariate Cox regression analysis, and 143 OS-related DEGs were screened (Table S6). The unsupervised clustering analysis was performed, and the GC patients were clustered into three different genomic subtypes with different clinicopathologic characteristics (gene Clusters A-C; Figures S14C, S15A-D). The expression of PRGs and the OS in these three pyroptosis gene clusters were markedly different (Figure 3B, 3C). Patients in Cluster B had the best OS, while Cluster C exhibited the worst prognosis among these three clusters (P<0.001, Figure 3C). In consistent, GC patients with death status were markedly concentrated in the Cluster A and gene Cluster C, and clinical stage III-IV cases were also enriched in Cluster A (Figure S14C). These results suggested the potential prognostic value of the pyroptosis gene Clusters. We also observed that patients in Clusters A and C subtypes were enriched in the advanced age subtype (age >65). PCA analysis confirmed the separation of these clusters (Figure 3D).
Figure 3.
Characteristics of pyroptosis gene clusters. A. Venn diagram presents 346 DEGs among the three pyroptosis clusters. B. The expression levels of PRGs among the three pyroptosis gene clusters. *P<0.05; **P<0.01; ***P<0.001. C. Survival analyses for the pyroptosis gene clusters based on 989 GC patients including 570 patients in pyroptosis gene Cluster A, 270 patients in pyroptosis gene Cluster B, and 149 patients in pyroptosis gene Cluster C (Log-rank test, P<0.01). D. PCA of the mRNA expression profiles of PRGs confirms the pyroptosis gene clusters (blue, gene Cluster A; yellow, gene Cluster B; red, gene Cluster C).
Subsequently, we investigated whether these three pyroptosis gene Clusters exhibited different TME characteristics. As shown in Figure S16A, compared to pyroptosis gene Clusters A and C, Cluster B was particularly enriched in immune cell infiltration including active B cells, activated memory CD4+ T cells, activated memory CD8+ T cells, NK cells. Additionally, Cluster B was the highest in the scores of antigen presenting cell (APC) co-stimulation, chemokine receptors (CCR), check point, cytolytic activity, human leukocyte antigen (HLA), inflammation promoting, MHC class I, parainflammation, T cell co-stimulation and type I Interferon (IFN) response and the lowest in type II IFN response score (Figure S16B). Patients in Cluster-B had relative higher ESTIMATE score (P<0.001, Figure S16C), immune score (P<0.001, Figure S16D) and stromal score (P<0.01, Figure S16E) than patients in Cluster-A or Cluster-C. These data also confirmed our conclusion that there were indeed three distinct immune phenotype groups in GC with different clinicopathologic and TME features.
Construction of pyroptosis scoring system and its clinical significance
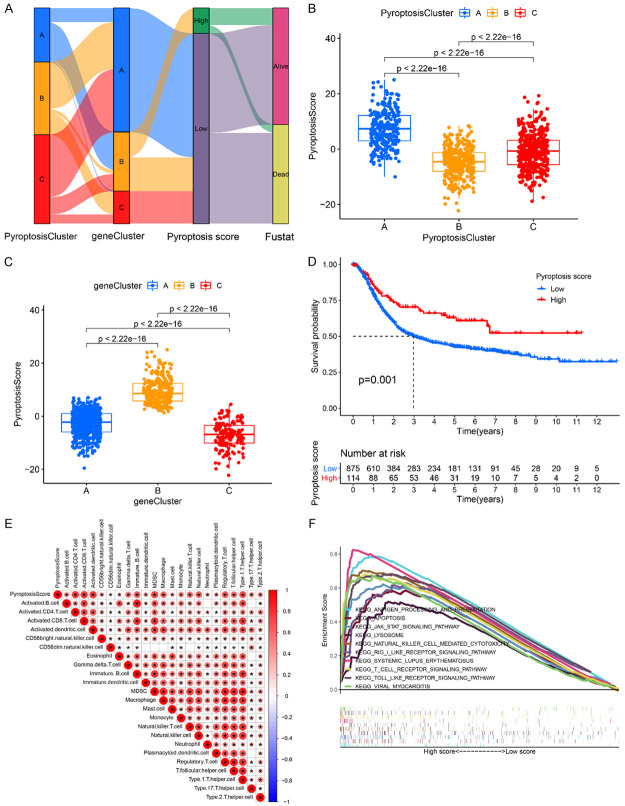
In order to quantify the pyroptosis-related molecular patterns of individual GC patient, we constructed a scoring system termed pyroptosis score based on the identified OS-related DEGs. The distribution of pyroptosis score and the survival status of GC patients in different gene Clusters and pyroptosis Clusters were shown in Figure 4A. Specifically, patients in pyroptosis Cluster A had the highest pyroptosis score among the three clusters (P<2.2e-16) (Figure 4B). At the same time, the highest pyroptosis score was also observed in pyroptosis gene Cluster B (P<2.2e-16, Figure 4C). Figure 4D showed that patients with high pyroptosis score usually exhibited a better outcome than those with low pyroptosis score (P=0.001). We further assessed the prognostic role of pyroptosis score in GC patients that were subgrouped by clinicopathological features (age, grade, and stage). As shown in Figure S17A-E, GC patients with higher pyroptosis score in subgroups of age ≤65, age >65, and male group had dramatically better OS than patients with lower pyroptosis score (P<0.05), suggesting that the pyroptosis score could also separate the patients with different OS. GC patients with stage I-II, stage III-IV, and female group also showed similar trend but it was not statistically significant (Figure S17C, S17E and S17F). The prognostic value of the pyroptosis score was further confirmed in the meta-GEO cohort (n=618), TCGA-STAD cohort (n=371), GSE62254 cohort (n=300), GSE57303 cohort (n=70), and GSE34942 cohort (n=56) (Figure S18A-E). The relationships between the pyroptosis score and the clinical features were also explored (Figure S19A-H). The results showed that high pyroptosis score group had higher percentage of alive patients during the follow-up (Figure S19A, S19B). However, no visible difference was observed in percent weight between the two groups considering the features of gender, age, and stage (Figure S19C-H). Furthermore, we explored the relationship between the pyroptosis score and TME infiltrating immune cells by Spearman analysis. The heatmap of the correlation matrix showed that the pyroptosis score was positively correlated with the activated CD4+ T cells, activated CD8+ T cells, activated dendritic cells, immature B cells, MDSC, macrophage, NK T cells, NK cells, T-helper cell type 1 (Th1) cells, T-regulatory (Treg) cells, and T follicular helper (Tfh) cells (Figure 4E). GSEA analysis showed that antigen processing and presentation pathway, apoptosis, JAK-STAT signaling pathway, lysosome, NK cell mediated cytotoxicity pathway, and RIG-I-like receptor signal pathway, systemic lupus erythematosus, T cell receptor signaling pathway, toll like receptor signal pathway and viral myocarditis were significantly enriched in the high pyroptosis score group (Figure 4F), while low pyroptosis score group was mainly enriched in pathways involved in cytochrome P450 of drug metabolism, glycerolipid and histidine metabolism, maturity-onset diabetes of the young, and reclamation of bicarbonate in the proximal tubule (Figure S20). Moreover, we also explored the correlation between the characteristics of TIME and pyroptosis score. As shown in Figure S21A, high pyroptosis score group was significantly enriched in immune cell infiltration including activated B cells, activated CD4+ T cells, activated CD8+ T cells, NK cells, etc. In addition, high pyroptosis score subtype showed higher APC co-stimulation, CCR, check point, cytolytic activity, HLA, inflammation promoting, MHC class I, parainflammation, T cell co-stimulation and type I IFN response scores and the lower type II IFN response score (Figure S21B). Importantly, we also observed that GC patients in the high pyroptosis score group had relative higher ESTIMATE score (P<2.22e-16; Figure S21C), immune score (P<2.22e-16; Figure S21D) and stromal score (P=0.00032; Figure S21E) as compared to those in low pyroptosis score group. In sum, we confirmed that the pyroptosis score was associated with TME and could be used to evaluate certain clinical features of GC patients.
Figure 4.
Construction of pyroptosis score system. A. Alluvial diagram shows the distribution of GC patients with different pyroptosis clusters, pyroptosis gene clusters, pyroptosis scores, and survival state. B. Differences in pyroptosis score among three pyroptosis clusters (P<0.001, Kruskal-Wallis test). C. Differences in pyroptosis score among three pyroptosis gene clusters (P<0.001, Kruskal-Wallis test). D. Kaplan-Meier curves for GC patient with high and low pyroptosis score. Log-rank test, P=0.001. E. Correlations between pyroptosis score and different types of immune cells using Spearman analysis. F. GSEA identified several immune-related pathways enriched in the high pyroptosis score group.
Correlation between the pyroptosis and TCGA molecular subtypes of GC
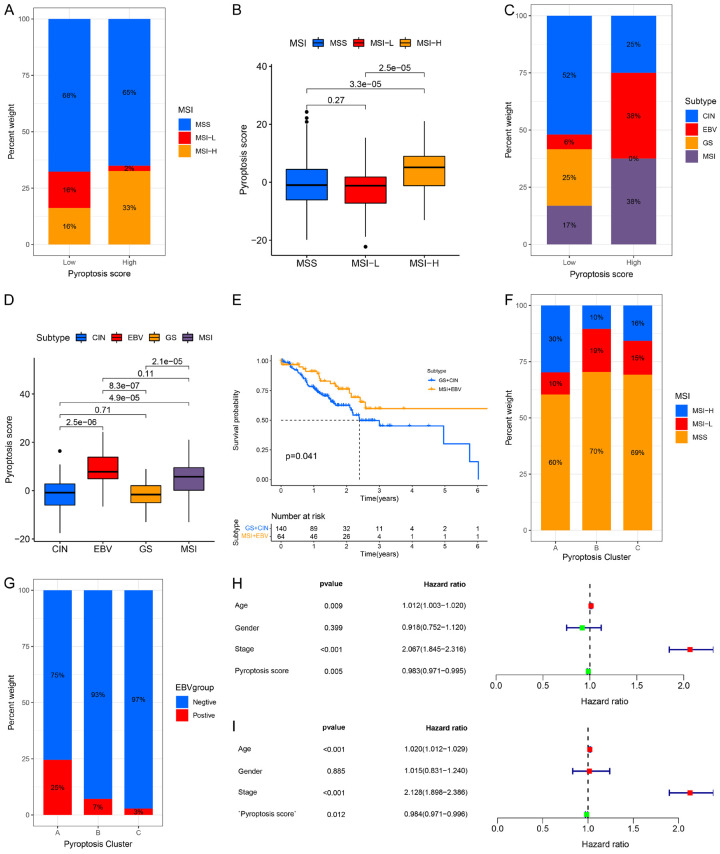
TCGA project classified GC into four molecular subtypes: Epstein-Barr virus (EBV) infection, chromosomal instability (CIN), genome stable (GS), and microsatellite instability (MSI) [51]. Studies also revealed that MSI patients or EBV-positive cases usually showed better response to immune checkpoint blockade treatment [52]. Thus, we assessed the correlation between our pyroptosis score and TCGA molecular subtypes. First, the expression levels of PRGs in different groups were examined. As shown in Figure S22A-D, AIM2, CASP1, CASP3, CASP4, CASP5, and IL18 were markedly elevated, while PLCG1 was significantly downregulated in the MSI group in both the TCGA cohort and GSE62254 cohort. The MSI-High subtype, featured by the better outcome, was associated with high pyroptosis score, whereas MSI-Low and MSS had a low pyroptosis score (Figure 5A, 5B). Moreover, the higher pyroptosis score was mainly concentrated on EBV infection and MSI subtype, which showed a better OS in GC patients (Figure 5C-E), The lower pyroptosis score was concentrated on the subtypes of CIN and GS group, which was associated with poorer OS (Figure 5E, P=0.041). Further analysis showed that patients in MSI-High subtype shared the features in pyroptosis Cluster-A, whilst cases with MSS subtype were like the pyroptosis Cluster-B and Cluster-C in TCGA cohort (Figure 5F). The levels of AIM2, CASP1, CASP4, GCDMB, GSDMC, GSDMD, IL18, NLRC4, PLCG1 and PYD and CARD domain containing (PYCARD) was much higher, whereas CASP9, ELANE, and NLRP6 level was lower in EBV-positive patients than in EBV-negative patients (Figure S22E). Moreover, pyroptosis Cluster-A was concentrated on the EBV positive subtype compared to pyroptosis Cluster-B and Cluster-C (Figure 5G). Univariate Cox regression analysis showed that age, tumor stage, and pyroptosis score were closely associated with OS (HR=1.012 (95% CI: 1.003-1.020), P=0.009; HR=2.067 (95% CI: 1.845-2.316), P<0.001; HR=0.983 (95% CI: 0.971-0.995), P=0.005, respectively) (Figure 5H). Multivariate Cox regression analysis indicated that age (HR=1.020 (95% CI: 1.012-1.029), P<0.001) and tumor stage (HR=2.128 (95% CI: 1.898-2.396), P<0.001) could be considered as independent risk factors for OS, while pyroptosis score acted as an independent favorable prognostic indicator for clinical outcome in GC (HR=0.984 (95% CI: 0.971-0.996), P<0.001) (Figure 5I).
Figure 5.
Characteristics of pyroptosis in TCGA molecular subtypes and identification of independent prognostic factors. A, B. Differences in pyroptosis score among different microsatellite subtypes. The Kruskal-Wallis test was used to compare the statistical difference between the three microsatellite subtypes. C, D. Differences in pyroptosis score among different TCGA-STAD molecular subtypes. The Kruskal-Wallis test was conducted to compare the statistical difference between the four TCGA-STAD molecular subtypes. E. Kaplan-Meier curves for GC patients in GS+CIN and MSI+EBV subtypes in the TCGA-STAD cohort. Log rank test, P=0.041. F. The proportion of three pyroptosis clusters in the MSS, MSI-High and MSI-Low subtypes. MSI, microsatellite instability; MSS, microsatellite stable. G. The proportion of three pyroptosis clusters in the EBV-positive and EBV-negative groups. H. Univariate Cox regression analysis of the clinicopathological factors and pyroptosis score in the TCGA-STAD cohort. I. Multivariate Cox regression analysis of the clinicopathological features and pyroptosis score in the TCGA-STAD cohort.
Correlation between the pyroptosis scores and somatic variants
It has been reported that high tumor mutation burden (TMB) tissues usually have an increased immune infiltration of CD8+ T cell [53,54]. Some studies have shown that elevated TMB level is associated with the prolonged survival time and improved response to PD-1-based immunotherapy [53,55,56]. These findings suggest the clinical significance of TMB in cancer treatment. We therefore sought to investigate the correlation between our pyroptosis scores and TMB. First, the TMB level was compared between the high and the low pyroptosis score of GC patients. We found that GC patients in high pyroptosis score group had a markedly higher TMB than patients in the low pyroptosis score group had (Figure 6A, P=0.011). Next, correlation analyses showed that the pyroptosis score was positively correlated with the TMB (R=0.13, P=0.011, Figure 6B). We further found that GC patients with high TMB showed longer OS than those with low TMB (P<0.001, Figure 6C). Additionally, the synergistic effect of TMB and pyroptosis score in predicting prognosis of GC patients was evaluated (Figure 6D). Stratified survival analysis showed that GC patients with both high TMB status and high pyroptosis score had the longest OS time as compared with patients in other groups. Different TMB status caused significant differences in OS in both high and low pyroptosis score subgroups (Figure 6D, P<0.001). Taken together, we concluded that the TMB status and pyroptosis score might act as promising prognostic markers in GC patients.
Figure 6.
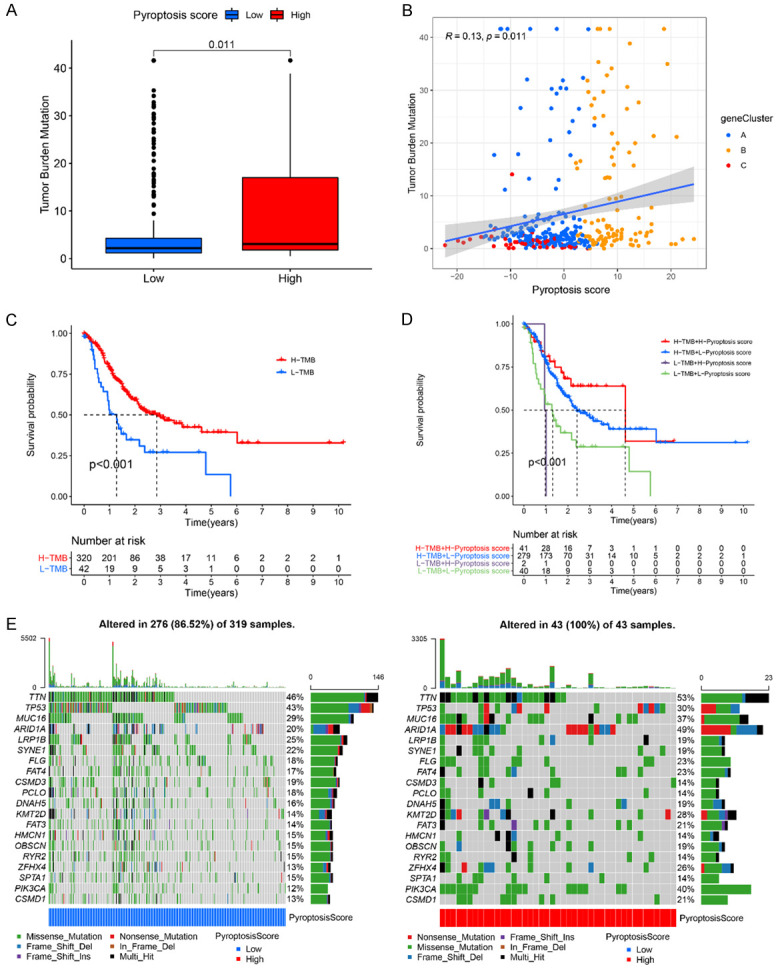
The correlation between the pyroptosis score and somatic variants. A. Difference of TMB level in the high and low pyroptosis score groups. Wilcoxon test, P=0.011. B. Scatterplots presents the positive correlation between pyroptosis score and mutation load in the TCGA-STAD cohort. Spearman correlation analysis, R=0.13, P=0.011. C. Kaplan-Meier curves for GC patients with high and low TMB in the TCGA-STAD cohort. Log rank test, P<0.001. D. Kaplan-Meier curves for GC patients with different TMB status and pyroptosis scores in the TCGA-STAD cohort. Log rank test, P<0.001. E. Mutational landscape of significantly mutated genes in TCGA-STAD cohort stratified by low (left panel, bule) and high pyroptosis score (right panel, red) subgroups. Each column represents an individual GC patient.
Moreover, the distribution of the somatic variants in GC driver genes between the low and high pyroptosis groups was analysis. The maftools was applied to evaluate the GC driver genes. The top 20 driver genes with the highest alteration frequency in the low and high pyroptosis groups were shown in Figure 6E. Among them, AT-rich interaction domain 1A (ARID1A) (P<0.001), lysine methyltransferase 2D (KMT2D) (P=0.002), zinc finger homeobox 4 (ZFHX4) (P=0.025), and phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha (PIK3CA) (P<0.001) showed the highest difference in alteration frequency between the low and high pyroptosis score groups, indicating the cross-talk between pyroptosis phenotypes and individual somatic mutations.
Chemo sensitivity screening for patients with GC based on the pyroptosis score
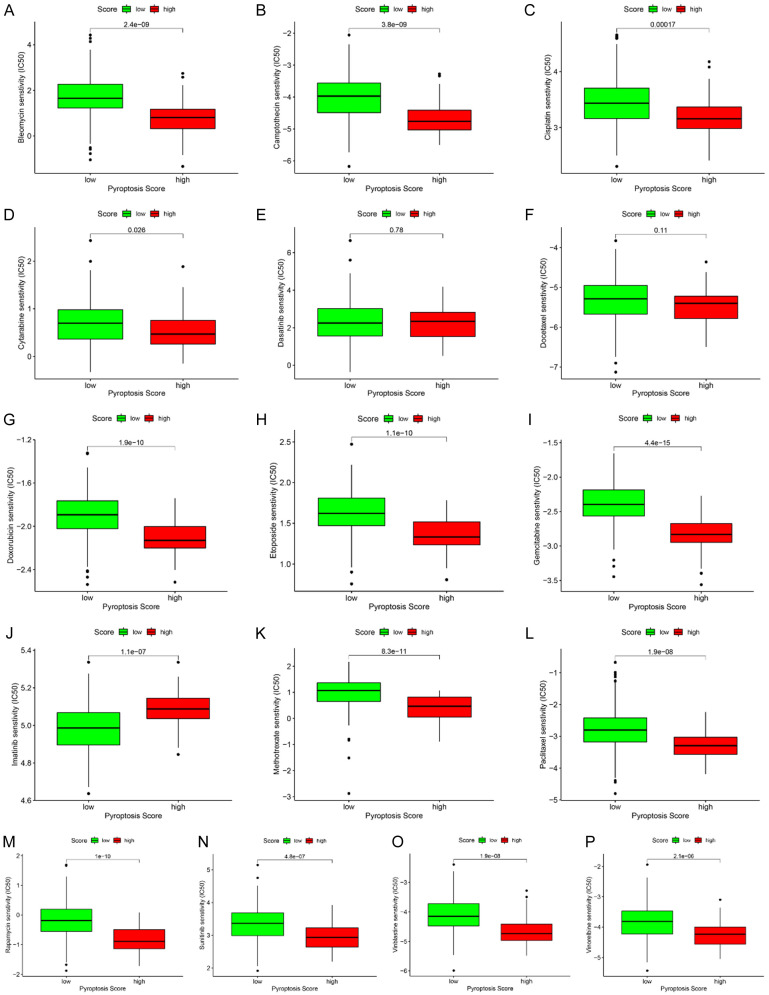
We evaluated the application of our pyroptosis score on predicting the response of GC patient to chemotherapeutic drugs. The estimated IC50 values were compared in different groups for 16 chemotherapy drugs: Bleomycin (Figure 7A), Camptothecin (Figure 7B), Cisplatin (Figure 7C), Cytarabine (Figure 7D), Dasatinib (Figure 7E), Docetaxel (Figure 7F), Doxorubicin (Figure 7G), Etoposide (Figure 7H), Gemcitabine (Figure 7I), Imatinib (Figure 7J), Methotrexate (Figure 7K), Paclitaxel (Figure 7L), Rapamycin (Figure 7M), Sunitinib (Figure 7N), Vinblastine (Figure 7O), and Vinorelbine (Figure 7P). We found that IC50 values of 13 chemotherapy drugs were higher in the low pyroptosis score group than that in the high pyroptosis score group (Figure 7A-D, G-I and K-P). On the contrary, the estimated IC50 value of lmatinib (P=1.1e-07, Figure 7J) was significantly lower in the low pyroptosis score group than that in the high pyroptosis score group, suggesting that low pyroptosis patients were more sensitive to lmatinib.
Figure 7.
Identification of sensitive chemotherapy drugs based on the pyroptosis score. Box plots depicts the differences in the estimated IC50 levels of Bleomycin (A), Camptothecin (B), Cisplatin (C), Cytarabine (D), Dasatinib (E), Docetaxel (F), Doxorubicin (G), Etoposide (H), Gemcitabine (I), Imatinib (J), Methotrexate (K), Paclitaxel (L), Rapamycin (M), Sunitinib (N), Vinblastine (O), and Vinorelbine (P) between the high and low pyroptosis score groups.
Identification of potential small molecule compounds for patients with GC based on the pyroptosis score
In order to identify the potential small molecule compounds for treating the GC patients, we screened the DEGs between the low and high pyroptosis score groups. A total of 309 DEGs were selected between the two groups with adjusted p value <0.05 and |logFC| >1.0, including 71 upregulated and 238 downregulated genes (Figure S23A; Table S7). GO enrichment analysis classified the DEGs into three functional subgroups: biological process (BP), cellular component (CC), and molecular function (MF). As shown in Figure S23B, the DEGs significantly enriched in the BP were related to regulation of innate immune response, T cell activation, positive regulation of innate immune response iron ion binding. In the CC group, the DEGs were mainly enriched in the tertiary granule membrane, secretory granule membrane, chromosomal region. DEGs related to cytokine receptor activity, helicase activity, tumor necrosis factor receptor superfamily binding was enriched in MF. KEGG analysis showed that the DEGs were obviously related to Epstein-Barr virus infection, influenza A, osteoclast differentiation, leishmaniasis, chemokine signaling pathway, Th1 and Th2 cell differentiation, measles, NOD-like receptor signaling pathway, allograft rejection, and NF-kappa B signaling pathway (Figure S23C). Finally, DEGs query in CMAP small molecule drug database identified 11 potential drugs, including caffeic acid, puromycin and alimemazine (Figure S23D), which might provide new clues for targeted therapy in GC patients in the future.
Predictive value of pyroptosis score in predicting response to immunotherapy
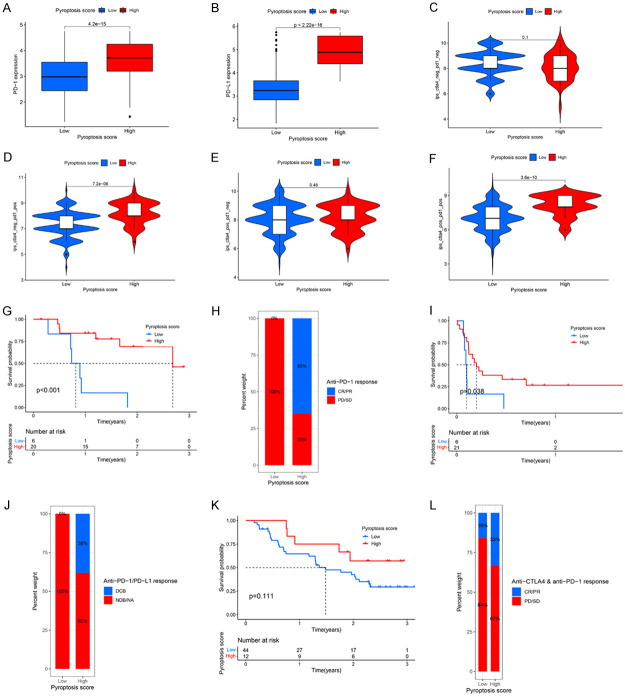
We first evaluated the expression of two crucial immune checkpoint inhibitors (PD-1 and PD-L1) in the two pyroptosis score groups. The results showed that high pyroptosis score group was characterized by a markedly higher PD-1 and PD-L1 expression levels compared to the low pyroptosis score group (Figure 8A, 8B). Since IPS has been reported to play important roles in predicting the response to immunotherapy [34], we assessed the predicting ability of pyroptosis score too. Immunophenogram analysis was used to explore the correlation between IPS and pyroptosis score in TCGA-STAD cohort (Figure 8C, 8F). Our results showed that in the CTLA4_negative+PD-1_positive and CTLA4_positive+PD-1_positive subtypes, the IPS in high pyroptosis group was significantly higher than that in low pyroptosis group (Figure 8D, 8F, both P<0.0001), suggesting that GC patients in high pyroptosis score group might benefit from treatment with anti-PD-1/PD-L1 antibodies alone or in combination with anti-CTLA4 blockers.
Figure 8.
Prediction values of pyroptosis score in immunotherapeutic benefits. (A, B) Difference in PD-1 (A) and PD-L1 (B) expression between high and low pyroptosis score groups (P<0.0001). (C-F) Comparison of IPS between the GC patients with high and low pyroptosis score groups in the CTLA4 negative/positive or PD-1 negative/positive groups. CTLA4_positive or PD1_positive represents anti-CTLA4 or anti-PD-1/PD-L1 therapy, respectively. (G, H) Kaplan-Meier curves (G) and clinical response (H) to anti-PD-1 therapy for patients in high and low pyroptosis score groups from the GSE78220 cohort. (I, J) Kaplan-Meier curves (G) and clinical response (H) to anti-PD-1/PD-L1 therapy for patients in high and low pyroptosis score groups from the GSE135222 cohort. (K, L) Kaplan-Meier curves (K) and clinical response (L) to anti-CTLA4 and anti-PD-1 therapy for patients in high and low pyroptosis score groups from the GSE91061 cohort. CR, Complete Response; PR, Partial Response; SD, Stable Disease; PD, Progressive Disease; DCB, Durable Clinical Benefit; NDB, Non-Durable Benefit; NA, Not Determined.
We further validated the correlation between pyroptosis score and the response to immunotherapy with another three independent cohorts (GSE78220, GSE135222, and GSE91061). The patients who received anti-PD-1 therapy in the GSE78220 cohort were divided into high and low pyroptosis score groups (Figure 8G). We found patients with high pyroptosis score had a better prognosis than patients with low pyroptosis score (GSE78220, P<0.001, Figure 8G). Moreover, the response rate to anti-PD-1 therapy in the high pyroptosis score group was higher than that in the low pyroptosis score group (Figure 8H). Similar results were also observed in the GSE135222 cohort (Figure 8I, 8J). Patients in high pyroptosis score group showed obvious clinical advantages and significantly prolonged OS (P=0.038, Figure 8I, 8J). Patients in GSE91061 cohort also showed similar trend, but it was not statistically significant (Figure 8K, 8L), partly due to the small sample size and tumor heterogeneity. Collectively, the above results strongly indicated that the pyroptosis score was closely related to the response to immunotherapy, and it might sever as a biomarker to help predict benefits of immunotherapy in GC patients.
Discussion
Numerous studies have shown that pyroptosis plays a significant role in inflammation, innate immunity, and cancer biology [57], while the overall characteristics of TME mediated by integrated PRGs remain poorly understood. Thus, exploring the functions of distinct pyroptosis-related molecular patterns in the TME will advance our understanding of the interactions of PRGs on anti-tumor immune response and assist doctors to make more effective immunotherapy strategies for GC patients.
In this study, we identified three distinct pyroptosis-related molecular patterns based on the mRNA expression profiles of pyroptosis genes. Distinct features in immune infiltrations, prognosis, and functions were observed among the three clusters. They can be classified as immune-inflamed phenotype (pyroptosis Cluster-A), immune-desert phenotype (pyroptosis Cluster-B), and immune-excluded phenotype (pyroptosis Cluster-C) according to the previously published studies [50]. Further, based on the OS-related DEGs that were correlated with the different pyroptosis clusters, we identified three pyroptosis gene clusters with distinct outcomes and functions and immune infiltrations for GC. By conducting PCA algorithms, we established the pyroptosis score to quantify the pyroptosis patterns in individual GC patient. The pyroptosis Cluster-A and pyroptosis gene Cluster B had the best clinical outcomes and had the highest pyroptosis score among three pyroptosis patterns and gene clusters. Importantly, GC patients with high pyroptosis scores usually exhibited longer OS time, suggesting that high pyroptosis score could contribute to better outcome for patients. Moreover, our results showed that pyroptosis score was an independent favorable indicator for predicting the prognosis of GC patients. Pyroptosis score was also closely associated with clinicopathological feature and molecular subtypes of GC patients. Recently, a pyroptosis-related signature has been developed for predicting the prognosis of GC patients, although this model didn’t perform well in terms of all-time survival stages in immunotherapy cohort [58]. Still, our findings and the results from other groups warrant further investigation on the role of pyroptosis in GC.
We also investigated the specific functions of individual pyroptosis gene in tumor immunity. The crucial role of pyroptosis in tumor immunity has been reported [57]. Recently, increasing evidence has also revealed the dysregulation and dysfunction of the GSDM family members in various cancers [60,61]. However, the diagnostic and prognostic roles of GSDM family members have not been clearly defined. Wei et al. reported that overexpression of GSDMC is a prognostic indicator for poor outcome in lung cancer [61]. Interestingly, our results showed that GSDMC was upregulated in GC tissues but related to better prognosis. There are several possible explanations for this contradiction. First, the effect of GSDMC expression might be tissue-specific or context-dependent. Furthermore, tumor immune cell infiltration is an important defense mechanism in tumorigenesis, and it may in turn modulate the functions of tumor-associated genes. Our data showed that higher expression of GSDMC was closely related to the activation of immune-related pathways, higher levels of activated CD4+ T cells and activated CD8+ T cells, and higher immune scores, suggesting that GSDMC mediated pyroptosis may be widely involved in the tumor immunity. The biological functions and predictive roles of GSDMC need to be further validated in larger sample sets and by experimental studies.
Increasing evidence has shown that pyroptosis is important in inflammation and immunity. Consistent with these findings, our GSEA results demonstrated that antigen processing and presentation pathway, JAK-STAT signaling pathway, NK cell mediated cytotoxicity pathway, and T cell receptor signaling pathway were significantly enriched in, suggesting that pyroptosis is crucial in the immune response. TME mainly contains immune cells and stromal cells [12]. Here, we used ESTIMATE algorithm to assess the immune and stromal scores for GC patients based on the pyroptosis patterns, gene clusters, and pyroptosis score groups. Patients in pyroptosis Cluster A, gene Cluster B, and high pyroptosis score group had significantly higher ESTIMATE score, higher immune score, and higher stromal scores, indicating that pyroptosis participated in the modulation of TME. Therefore, targeting pyroptosis might be a promising treatment strategy. It is well known that accumulated genetic mutations can lead to cancigenesis [62]. Higher TMB level has been reported to be related to better prognosis for cancer patients [63]. Consistently, our results verified that there was a significant difference in TMB level between the high and low pyroptosis score groups. In addition, patients with both high TMB status and high pyroptosis score had the longest OS time compared to other groups, suggesting TMB status and high pyroptosis score as the positive prognostic factors in GC patients.
Drug resistance remains the major challenge in GC treatment [2]. Therefore, it is of extreme importance to timely assess the drug resistance and identify more effective drug treatment options. Our study showed that patients with low pyroptosis were more sensitive to lmatinib, and inhibiting pyroptosis might alleviate the drug resistance in these patients. Moreover, we identified 11 potential small molecule compounds for treating GC patients based on the pyroptosis score, although the activity of these novel pharmacological and genetic inhibitors for pyroptosis needs to be further validated.
Immunotherapy is emerging as a new and effective approach in the treatment of GC [3]. Our results showed that higher pyroptosis score was associated with higher expression levels of PD-1 and PD-L1 in GC patients. Additionally, IPS, a newly identified predictor for immune response, was significantly higher in GC patients treated with PD-1 antibody alone or in combination with CTLA-4 in the high pyroptosis score group than that in the low pyroptosis score group, indicating that pyroptosis score hold the potential in predicting the response to immunotherapy. We also validated the prognostic values of pyroptosis score in another three independent immunotherapy cohorts. Due to the lack of datasets for immunotherapy in GC, immunotherapy cohorts of melanoma and lung cancer were selected. Using the GSE78220 and GSE135222 cohorts, the predictive value of pyroptosis score was evaluated for response to anti-PD-1/PD-L1 immunotherapy. GC Patients in high pyroptosis score group were more likely to benefit from treatment with anti-PD-1/PD-L1 antibodies. Patients in GSE91061 datasets showed similar trends, but it was not statistically significant, possibly due to the tumor heterogeneity and small sample size. Therefore, the current results need to be confirmed in a larger sample GC cohort.
Conclusion
In summary, we systematically studied the genetic and expression variation landscape of pyroptosis-related genes in GC, which advanced our understanding on the clinical features and implications of pyroptosis in GC. Three pyroptosis-related molecular patterns with distinct clinical prognosis and TME features were found in GC. Obvious differences in TME and the response to immunotherapy were also observed between the GC patients with high and low pyroptosis scores. Therefore, it is of great significance to comprehensively assess the pyroptosis scores for individual GC patient, which may provide novel insight on optimizing the immunotherapy strategies.
Acknowledgements
This study was supported in part by grant from the National Natural Science Foundation of China (No. 82073210), grant from the Scientific Foundation of Shaanxi Province (No. S2019ZDCXL01-02-01), grant from the National Clinical Research Center for Digestive Diseases (No. 2015BAI13B07). We appreciated STRING, GEO, TCGA, CMAP, UCSC Xena, MSigDB, TCIA, and GDSC databases for providing the platform or datasets.
Disclosure of conflict of interest
None.
Abbreviations
- GC
Gastric Cancer
- STAD
Stomach Adenocarcinoma
- TME
Tumor Microenvironment
- GEO
Gene-Expression Omnibus
- TCGA
The Cancer Genome Atlas
- PCA
Principal Component Analysis
- CNV
Copy Number Variation
- GSVA
Gene Set Variation Analysis
- ssGSEA
single sample Gene Set Enrichment Analysis
- DEGs
Differential Expression Genes
- FDR
False Discovery Rate
- OS
Overall Survival
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- GO
Gene Ontology
- BP
Biological Process
- CC
Cellular Component
- MF
Molecular Function
- CIN
Chromosomal Instability
- GS
Genome Stable
- MSI
Microsatellite Instability
- EBV
Epstein-Barr Virus
- MSS
Microsatellite Stability
- PRGs
Pyroptosis-Related Genes
- PAMPs
Pathogen-Associated Molecular Patterns
- DAMPs
Damage Associated Molecular Patterns
- AIM2
Absent In Melanoma-2
- GSDMD
Gasdermin D
- IPS
Immunophenoscore
- LPS
Lipopolysaccharide
- TIME
Tumor Immune Microenvironment
- ESTIMATE
Estimation of STromal and Immune cells in MAlignant Tumor tissues using Expression data
- TMB
Tumor Mutation Burden
- GDSC
Genomics of Drug Sensitivity in Cancer
- IC50
Half Maximal Inhibitory Concentration
- CMap
Connectivity Map
- MoA
Mode-of-Action
- DCs
Dendritic Cells
- Tfh
Follicular Helper T cells
- Th1
Type-1 T Helper Cells
- Th2
Type-2 T Helper Cells
- Treg
Regulatory T Cells
- aDCs
Activated Dendritic Cells
- HR
Hazard Ratio
- CI
Confidence Interval
- pDCs
plasmacytoid Dendritic Cells
- ZFHX4
Zinc Finger Homeobox 4
- JAK-STAT
Janus Kinase/Signal Transducer and Activator Of Transcription
- RIG-I
Retinoic Acid-Inducible Gene-I
- PYCARD
PYD and CARD Domain Containing
- NLRP1
Nucleotide Oligomerization Domain (NOD)-Like Receptor Family, Pyrin Domain-Containing-1
- APC
Antigen Presenting Cell
- CCR
Chemokine Receptors
- HLA
Human Leukocyte Antigen
- TNF
Tumor Necrosis Factor
- IL18
Interleukin 18
- TIRAP
TIR Domain-Containing Adaptor Protein
- GPX4
Glutathione Peroxidase 4
- PJVK
Pejvakin
- ELANE
Neutrophil Expressed
- IFN
Interferon
- PD-1/L1
Programmed Cell Death-1/Ligand 1
- PIK3CA
Phosphatidylinositol-4,5-Bisphosphate 3-Kinase Catalytic Subunit Alpha
- KMT2D
Lysine Methyltransferase 2D
- ARID1A
AT-Rich Interaction Domain 1A
- AIM2
Absent In Melanoma-2
- CASP1
Caspase 1
- CTLA4
Cytotoxic T Lymphocyte-Associated Antigen-4
Supporting Information
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin. 2020;70:7–30. doi: 10.3322/caac.21590. [DOI] [PubMed] [Google Scholar]
- 2.Yang W, Ma J, Zhou W, Cao B, Zhou X, Yang Z, Zhang H, Zhao Q, Fan D, Hong L. Molecular mechanisms and theranostic potential of miRNAs in drug resistance of gastric cancer. Expert Opin Ther Targets. 2017;21:1063–1075. doi: 10.1080/14728222.2017.1389900. [DOI] [PubMed] [Google Scholar]
- 3.Johnston FM, Beckman M. Updates on management of gastric cancer. Curr Oncol Rep. 2019;21:67. doi: 10.1007/s11912-019-0820-4. [DOI] [PubMed] [Google Scholar]
- 4.Helmy KY, Patel SA, Nahas GR, Rameshwar P. Cancer immunotherapy: accomplishments to date and future promise. Ther Deliv. 2013;4:1307–1320. doi: 10.4155/tde.13.88. [DOI] [PubMed] [Google Scholar]
- 5.Zhuo M, Chi Y, Wang Z. The adverse events associated with combination immunotherapy in cancers: challenges and chances. Asia Pac J Clin Oncol. 2020;16:e154–e159. doi: 10.1111/ajco.13365. [DOI] [PubMed] [Google Scholar]
- 6.Zhao Q, Cao L, Guan L, Bie L, Wang S, Xie B, Chen X, Shen X, Cao F. Immunotherapy for gastric cancer: dilemmas and prospect. Brief Funct Genomics. 2019;18:107–112. doi: 10.1093/bfgp/ely019. [DOI] [PubMed] [Google Scholar]
- 7.Coutzac C, Pernot S, Chaput N, Zaanan A. Immunotherapy in advanced gastric cancer, is it the future? Crit Rev Oncol Hematol. 2019;133:25–32. doi: 10.1016/j.critrevonc.2018.10.007. [DOI] [PubMed] [Google Scholar]
- 8.Spector LG, Pankratz N, Marcotte EL. Genetic and nongenetic risk factors for childhood cancer. Pediatr Clin North Am. 2015;62:11–25. doi: 10.1016/j.pcl.2014.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lewandowska AM, Rudzki M, Rudzki S, Lewandowski T, Laskowska B. Environmental risk factors for cancer-review paper. Ann Agric Environ Med. 2019;26:1–7. doi: 10.26444/aaem/94299. [DOI] [PubMed] [Google Scholar]
- 10.Hinshaw DC, Shevde LA. The tumor microenvironment innately modulates cancer progression. Cancer Res. 2019;79:4557–4566. doi: 10.1158/0008-5472.CAN-18-3962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wu T, Dai Y. Tumor microenvironment and therapeutic response. Cancer Lett. 2017;387:61–68. doi: 10.1016/j.canlet.2016.01.043. [DOI] [PubMed] [Google Scholar]
- 12.Quail DF, Joyce JA. Microenvironmental regulation of tumor progression and metastasis. Nat Med. 2013;19:1423–1437. doi: 10.1038/nm.3394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Maacha S, Bhat AA, Jimenez L, Raza A, Haris M, Uddin S, Grivel JC. Extracellular vesicles-mediated intercellular communication: roles in the tumor microenvironment and anti-cancer drug resistance. Mol Cancer. 2019;18:55. doi: 10.1186/s12943-019-0965-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhou Z, He H, Wang K, Shi X, Wang Y, Su Y, Wang Y, Li D, Liu W, Shen L, Han W, Shen L, Ding J, Shao F. Granzyme A from cytotoxic lymphocytes cleaves GSDMB to trigger pyroptosis in target cells. Science. 2020;368:eaaz7548. doi: 10.1126/science.aaz7548. [DOI] [PubMed] [Google Scholar]
- 15.Birla R, Gandea C, Hoara P, Caragui A, Marica C, Vasiliu E, Constantinoiu S. Clinical and therapeutic implications of the 8th edition TNM classification of adenocarcinomas of the esophagogastric junction. Chirurgia (Bucur) 2018;113:747–757. doi: 10.21614/chirurgia.113.6.747. [DOI] [PubMed] [Google Scholar]
- 16.Zheng Y, Fu S, He T, Yan Q, Di W, Wang J. Predicting prognosis in resected esophageal squamous cell carcinoma using a clinical nomogram and recursive partitioning analysis. Eur J Surg Oncol. 2018;44:1199–1204. doi: 10.1016/j.ejso.2018.04.011. [DOI] [PubMed] [Google Scholar]
- 17.Du F, Sun Z, Jia J, Yang Y, Yu J, Shi Y, Jia B, Zhao J, Zhang X. Development and validation of an individualized nomogram for predicting survival in patients with esophageal carcinoma after resection. J Cancer. 2020;11:4023–4029. doi: 10.7150/jca.40767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fang Y, Tian S, Pan Y, Li W, Wang Q, Tang Y, Yu T, Wu X, Shi Y, Ma P, Shu Y. Pyroptosis: a new frontier in cancer. Biomed Pharmacother. 2020;121:109595. doi: 10.1016/j.biopha.2019.109595. [DOI] [PubMed] [Google Scholar]
- 19.Schnappauf O, Chae JJ, Kastner DL, Aksentijevich I. The pyrin inflammasome in health and disease. Front Immunol. 2019;10:1745. doi: 10.3389/fimmu.2019.01745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shojaie L, Iorga A, Dara L. Cell death in liver diseases: a review. Int J Mol Sci. 2020;21:9682. doi: 10.3390/ijms21249682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Galluzzi L, Vitale I, Aaronson SA, Abrams JM, Adam D, Agostinis P, Alnemri ES, Altucci L, Amelio I, Andrews DW, Annicchiarico-Petruzzelli M, Antonov AV, Arama E, Baehrecke EH, Barlev NA, Bazan NG, Bernassola F, Bertrand MJM, Bianchi K, Blagosklonny MV, Blomgren K, Borner C, Boya P, Brenner C, Campanella M, Candi E, Carmona-Gutierrez D, Cecconi F, Chan FK, Chandel NS, Cheng EH, Chipuk JE, Cidlowski JA, Ciechanover A, Cohen GM, Conrad M, Cubillos-Ruiz JR, Czabotar PE, D’Angiolella V, Dawson TM, Dawson VL, De Laurenzi V, De Maria R, Debatin KM, DeBerardinis RJ, Deshmukh M, Di Daniele N, Di Virgilio F, Dixit VM, Dixon SJ, Duckett CS, Dynlacht BD, El-Deiry WS, Elrod JW, Fimia GM, Fulda S, García-Sáez AJ, Garg AD, Garrido C, Gavathiotis E, Golstein P, Gottlieb E, Green DR, Greene LA, Gronemeyer H, Gross A, Hajnoczky G, Hardwick JM, Harris IS, Hengartner MO, Hetz C, Ichijo H, Jäättelä M, Joseph B, Jost PJ, Juin PP, Kaiser WJ, Karin M, Kaufmann T, Kepp O, Kimchi A, Kitsis RN, Klionsky DJ, Knight RA, Kumar S, Lee SW, Lemasters JJ, Levine B, Linkermann A, Lipton SA, Lockshin RA, López-Otín C, Lowe SW, Luedde T, Lugli E, MacFarlane M, Madeo F, Malewicz M, Malorni W, Manic G, Marine JC, Martin SJ, Martinou JC, Medema JP, Mehlen P, Meier P, Melino S, Miao EA, Molkentin JD, Moll UM, Muñoz-Pinedo C, Nagata S, Nuñez G, Oberst A, Oren M, Overholtzer M, Pagano M, Panaretakis T, Pasparakis M, Penninger JM, Pereira DM, Pervaiz S, Peter ME, Piacentini M, Pinton P, Prehn JHM, Puthalakath H, Rabinovich GA, Rehm M, Rizzuto R, Rodrigues CMP, Rubinsztein DC, Rudel T, Ryan KM, Sayan E, Scorrano L, Shao F, Shi Y, Silke J, Simon HU, Sistigu A, Stockwell BR, Strasser A, Szabadkai G, Tait SWG, Tang D, Tavernarakis N, Thorburn A, Tsujimoto Y, Turk B, Vanden Berghe T, Vandenabeele P, Vander Heiden MG, Villunger A, Virgin HW, Vousden KH, Vucic D, Wagner EF, Walczak H, Wallach D, Wang Y, Wells JA, Wood W, Yuan J, Zakeri Z, Zhivotovsky B, Zitvogel L, Melino G, Kroemer G. Molecular mechanisms of cell death: recommendations of the Nomenclature Committee on Cell Death 2018. Cell Death Differ. 2018;25:486–541. doi: 10.1038/s41418-017-0012-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Man SM, Karki R, Kanneganti TD. Molecular mechanisms and functions of pyroptosis, inflammatory caspases and inflammasomes in infectious diseases. Immunol Rev. 2017;277:61–75. doi: 10.1111/imr.12534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu X, Zhang Z, Ruan J, Pan Y, Magupalli VG, Wu H, Lieberman J. Inflammasome-activated gasdermin D causes pyroptosis by forming membrane pores. Nature. 2016;535:153–158. doi: 10.1038/nature18629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jiang M, Qi L, Li L, Li Y. The caspase-3/GSDME signal pathway as a switch between apoptosis and pyroptosis in cancer. Cell Death Discov. 2020;6:112. doi: 10.1038/s41420-020-00349-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhao P, Wang M, Chen M, Chen Z, Peng X, Zhou F, Song J, Qu J. Programming cell pyroptosis with biomimetic nanoparticles for solid tumor immunotherapy. Biomaterials. 2020;254:120142. doi: 10.1016/j.biomaterials.2020.120142. [DOI] [PubMed] [Google Scholar]
- 26.Kambara H, Liu F, Zhang X, Liu P, Bajrami B, Teng Y, Zhao L, Zhou S, Yu H, Zhou W, Silberstein LE, Cheng T, Han M, Xu Y, Luo HR. Gasdermin D exerts anti-inflammatory effects by promoting neutrophil death. Cell Rep. 2018;22:2924–2936. doi: 10.1016/j.celrep.2018.02.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hou J, Zhao R, Xia W, Chang CW, You Y, Hsu JM, Nie L, Chen Y, Wang YC, Liu C, Wang WJ, Wu Y, Ke B, Hsu JL, Huang K, Ye Z, Yang Y, Xia X, Li Y, Li CW, Shao B, Tainer JA, Hung MC. PD-L1-mediated gasdermin C expression switches apoptosis to pyroptosis in cancer cells and facilitates tumour necrosis. Nat Cell Biol. 2020;22:1264–1275. doi: 10.1038/s41556-020-0575-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tan Y, Chen Q, Li X, Zeng Z, Xiong W, Li G, Li X, Yang J, Xiang B. Pyroptosis: a new paradigm of cell death for fighting against cancer. J Exp Clin Cancer Res. 2021;40:153. doi: 10.1186/s13046-021-01959-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kesavardhana S, Malireddi RKS, Kanneganti TD. Caspases in cell death, inflammation, and pyroptosis. Annu Rev Immunol. 2020;38:567–595. doi: 10.1146/annurev-immunol-073119-095439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Karki R, Kanneganti TD. Diverging inflammasome signals in tumorigenesis and potential targeting. Nat Rev Cancer. 2019;19:197–214. doi: 10.1038/s41568-019-0123-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xia X, Wang X, Cheng Z, Qin W, Lei L, Jiang J, Hu J. The role of pyroptosis in cancer: pro-cancer or pro- “host”? Cell Death Dis. 2019;10:650. doi: 10.1038/s41419-019-1883-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wilkerson MD, Hayes DN. ConsensusClusterPlus: a class discovery tool with confidence assessments and item tracking. Bioinformatics. 2010;26:1572–1573. doi: 10.1093/bioinformatics/btq170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chong W, Shang L, Liu J, Fang Z, Du F, Wu H, Liu Y, Wang Z, Chen Y, Jia S, Chen L, Li L, Chen H. m(6)A regulator-based methylation modification patterns characterized by distinct tumor microenvironment immune profiles in colon cancer. Theranostics. 2021;11:2201–2217. doi: 10.7150/thno.52717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Charoentong P, Finotello F, Angelova M, Mayer C, Efremova M, Rieder D, Hackl H, Trajanoski Z. Pan-cancer immunogenomic analyses reveal genotype-immunophenotype relationships and predictors of response to checkpoint blockade. Cell Rep. 2017;18:248–262. doi: 10.1016/j.celrep.2016.12.019. [DOI] [PubMed] [Google Scholar]
- 35.Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yoshihara K, Shahmoradgoli M, Martinez E, Vegesna R, Kim H, Torres-Garcia W, Trevino V, Shen H, Laird PW, Levine DA, Carter SL, Getz G, Stemke-Hale K, Mills GB, Verhaak RG. Inferring tumour purity and stromal and immune cell admixture from expression data. Nat Commun. 2013;4:2612. doi: 10.1038/ncomms3612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang B, Wu Q, Li B, Wang D, Wang L, Zhou YL. m(6)A regulator-mediated methylation modification patterns and tumor microenvironment infiltration characterization in gastric cancer. Mol Cancer. 2020;19:53. doi: 10.1186/s12943-020-01170-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zeng D, Li M, Zhou R, Zhang J, Sun H, Shi M, Bin J, Liao Y, Rao J, Liao W. Tumor microenvironment characterization in gastric cancer identifies prognostic and immunotherapeutically relevant gene signatures. Cancer Immunol Res. 2019;7:737–750. doi: 10.1158/2326-6066.CIR-18-0436. [DOI] [PubMed] [Google Scholar]
- 39.Mayakonda A, Lin DC, Assenov Y, Plass C, Koeffler HP. Maftools: efficient and comprehensive analysis of somatic variants in cancer. Genome Res. 2018;28:1747–1756. doi: 10.1101/gr.239244.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang X, Shi M, Chen T, Zhang B. Characterization of the immune cell infiltration landscape in head and neck squamous cell carcinoma to aid immunotherapy. Mol Ther Nucleic Acids. 2020;22:298–309. doi: 10.1016/j.omtn.2020.08.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yang W, Soares J, Greninger P, Edelman EJ, Lightfoot H, Forbes S, Bindal N, Beare D, Smith JA, Thompson IR, Ramaswamy S, Futreal PA, Haber DA, Stratton MR, Benes C, McDermott U, Garnett MJ. Genomics of Drug Sensitivity in Cancer (GDSC): a resource for therapeutic biomarker discovery in cancer cells. Nucleic Acids Res. 2013;41:D955–961. doi: 10.1093/nar/gks1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Geeleher P, Cox N, Huang RS. pRRophetic: an R package for prediction of clinical chemotherapeutic response from tumor gene expression levels. PLoS One. 2014;9:e107468. doi: 10.1371/journal.pone.0107468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lamb J, Crawford ED, Peck D, Modell JW, Blat IC, Wrobel MJ, Lerner J, Brunet JP, Subramanian A, Ross KN, Reich M, Hieronymus H, Wei G, Armstrong SA, Haggarty SJ, Clemons PA, Wei R, Carr SA, Lander ES, Golub TR. The connectivity map: using gene-expression signatures to connect small molecules, genes, and disease. Science. 2006;313:1929–1935. doi: 10.1126/science.1132939. [DOI] [PubMed] [Google Scholar]
- 44.Hugo W, Zaretsky JM, Sun L, Song C, Moreno BH, Hu-Lieskovan S, Berent-Maoz B, Pang J, Chmielowski B, Cherry G, Seja E, Lomeli S, Kong X, Kelley MC, Sosman JA, Johnson DB, Ribas A, Lo RS. Genomic and transcriptomic features of response to Anti-PD-1 therapy in metastatic melanoma. Cell. 2016;165:35–44. doi: 10.1016/j.cell.2016.02.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kim JY, Choi JK, Jung H. Genome-wide methylation patterns predict clinical benefit of immunotherapy in lung cancer. Clin Epigenetics. 2020;12:119. doi: 10.1186/s13148-020-00907-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jung H, Kim HS, Kim JY, Sun JM, Ahn JS, Ahn MJ, Park K, Esteller M, Lee SH, Choi JK. DNA methylation loss promotes immune evasion of tumours with high mutation and copy number load. Nat Commun. 2019;10:4278. doi: 10.1038/s41467-019-12159-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Riaz N, Havel JJ, Makarov V, Desrichard A, Urba WJ, Sims JS, Hodi FS, Martín-Algarra S, Mandal R, Sharfman WH, Bhatia S, Hwu WJ, Gajewski TF, Slingluff CL Jr, Chowell D, Kendall SM, Chang H, Shah R, Kuo F, Morris LGT, Sidhom JW, Schneck JP, Horak CE, Weinhold N, Chan TA. Tumor and microenvironment evolution during immunotherapy with nivolumab. Cell. 2017;171:934–949. doi: 10.1016/j.cell.2017.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ye Y, Dai Q, Qi H. A novel defined pyroptosis-related gene signature for predicting the prognosis of ovarian cancer. Cell Death Discov. 2021;7:71. doi: 10.1038/s41420-021-00451-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.von Mering C, Huynen M, Jaeggi D, Schmidt S, Bork P, Snel B. STRING: a database of predicted functional associations between proteins. Nucleic Acids Res. 2003;31:258–261. doi: 10.1093/nar/gkg034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chen DS, Mellman I. Elements of cancer immunity and the cancer-immune set point. Nature. 2017;541:321–330. doi: 10.1038/nature21349. [DOI] [PubMed] [Google Scholar]
- 51.Sohn BH, Hwang JE, Jang HJ, Lee HS, Oh SC, Shim JJ, Lee KW, Kim EH, Yim SY, Lee SH, Cheong JH, Jeong W, Cho JY, Kim J, Chae J, Lee J, Kang WK, Kim S, Noh SH, Ajani JA, Lee JS. Clinical significance of four molecular subtypes of gastric cancer identified by the cancer genome atlas project. Clin Cancer Res. 2017;23:4441–4449. doi: 10.1158/1078-0432.CCR-16-2211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kim TS, da Silva E, Coit DG, Tang LH. Intratumoral immune response to gastric cancer varies by molecular and histologic subtype. Am J Surg Pathol. 2019;43:851–860. doi: 10.1097/PAS.0000000000001253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rizvi NA, Hellmann MD, Snyder A, Kvistborg P, Makarov V, Havel JJ, Lee W, Yuan J, Wong P, Ho TS, Miller ML, Rekhtman N, Moreira AL, Ibrahim F, Bruggeman C, Gasmi B, Zappasodi R, Maeda Y, Sander C, Garon EB, Merghoub T, Wolchok JD, Schumacher TN, Chan TA. Cancer immunology. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science. 2015;348:124–128. doi: 10.1126/science.aaa1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Blaeschke F, Paul MC, Schuhmann MU, Rabsteyn A, Schroeder C, Casadei N, Matthes J, Mohr C, Lotfi R, Wagner B, Kaeuferle T, Feucht J, Willier S, Handgretinger R, StevanoviĆ S, Lang P, Feuchtinger T. Low mutational load in pediatric medulloblastoma still translates into neoantigens as targets for specific T-cell immunotherapy. Cytotherapy. 2019;21:973–986. doi: 10.1016/j.jcyt.2019.06.009. [DOI] [PubMed] [Google Scholar]
- 55.Braun DA, Hou Y, Bakouny Z, Ficial M, Sant’ Angelo M, Forman J, Ross-Macdonald P, Berger AC, Jegede OA, Elagina L, Steinharter J, Sun M, Wind-Rotolo M, Pignon JC, Cherniack AD, Lichtenstein L, Neuberg D, Catalano P, Freeman GJ, Sharpe AH, McDermott DF, Van Allen EM, Signoretti S, Wu CJ, Shukla SA, Choueiri TK. Interplay of somatic alterations and immune infiltration modulates response to PD-1 blockade in advanced clear cell renal cell carcinoma. Nat Med. 2020;26:909–918. doi: 10.1038/s41591-020-0839-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sabbatino F, Marra A, Liguori L, Scognamiglio G, Fusciello C, Botti G, Ferrone S, Pepe S. Resistance to anti-PD-1-based immunotherapy in basal cell carcinoma: a case report and review of the literature. J Immunother Cancer. 2018;6:126. doi: 10.1186/s40425-018-0439-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tang R, Xu J, Zhang B, Liu J, Liang C, Hua J, Meng Q, Yu X, Shi S. Ferroptosis, necroptosis, and pyroptosis in anticancer immunity. J Hematol Oncol. 2020;13:110. doi: 10.1186/s13045-020-00946-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shao W, Yang Z, Fu Y, Zheng L, Liu F, Chai L, Jia J. The pyroptosis-related signature predicts prognosis and indicates immune microenvironment infiltration in gastric cancer. Front Cell Dev Biol. 2021;9:676485. doi: 10.3389/fcell.2021.676485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Li L, Jiang M, Qi L, Wu Y, Song D, Gan J, Li Y. Pyroptosis, a new bridge to tumor immunity. Cancer Sci. 2021;112:3979–3994. doi: 10.1111/cas.15059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Berkel C, Cacan E. Differential expression and copy number variation of gasdermin (GSDM) family members, pore-forming proteins in pyroptosis, in normal and malignant serous ovarian tissue. Inflammation. 2021;44:2203–2216. doi: 10.1007/s10753-021-01493-0. [DOI] [PubMed] [Google Scholar]
- 61.Wei J, Xu Z, Chen X, Wang X, Zeng S, Qian L, Yang X, Ou C, Lin W, Gong Z, Yan Y. Overexpression of GSDMC is a prognostic factor for predicting a poor outcome in lung adenocarcinoma. Mol Med Rep. 2020;21:360–370. doi: 10.3892/mmr.2019.10837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Paul P, Malakar AK, Chakraborty S. The significance of gene mutations across eight major cancer types. Mutat Res Rev Mutat Res. 2019;781:88–99. doi: 10.1016/j.mrrev.2019.04.004. [DOI] [PubMed] [Google Scholar]
- 63.Lee DW, Han SW, Bae JM, Jang H, Han H, Kim H, Bang D, Jeong SY, Park KJ, Kang GH, Kim TY. Tumor mutation burden and prognosis in patients with colorectal cancer treated with adjuvant fluoropyrimidine and oxaliplatin. Clin Cancer Res. 2019;25:6141–6147. doi: 10.1158/1078-0432.CCR-19-1105. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.