Abstract
Maternal and fetal pregnancy outcomes related to placental function vary based on fetal sex, which may be due to sexually dimorphic epigenetic regulation of RNA expression. We identified sexually dimorphic miRNA expression throughout gestation in human placentae. Next-generation sequencing identified miRNA expression profiles in first and third trimester uncomplicated pregnancies using tissue obtained at chorionic villous sampling (n = 113) and parturition (n = 47). Sequencing analysis identified 986 expressed mature miRNAs from female and male placentae at first and third trimester (baseMean>10). Of these, 11 sexually dimorphic (FDR < 0.05) miRNAs were identified in the first and 4 in the third trimester, all upregulated in females, including miR-361-5p, significant in both trimesters. Sex-specific analyses across gestation identified 677 differentially expressed (DE) miRNAs at FDR < 0.05 and baseMean>10, with 508 DE miRNAs in common between female-specific and male-specific analysis (269 upregulated in first trimester, 239 upregulated in third trimester). Of those, miR-4483 had the highest fold changes across gestation. There were 62.5% more female exclusive differences with fold change>2 across gestation than male exclusive (52 miRNAs vs 32 miRNAs), indicating miRNA expression across human gestation is sexually dimorphic. Pathway enrichment analysis identified significant pathways that were differentially regulated in first and third trimester as well as across gestation. This work provides the normative sex dimorphic miRNA atlas in first and third trimester, as well as the sex-independent and sex-specific placenta miRNA atlas across gestation, which may be used to identify biomarkers of placental function and direct functional studies investigating placental sex differences.
Keywords: stable miRNAs, developmental epigenetics, sexually dimorphic normative miRNA atlas, human transcriptome, miRNome, pregnancy, chorionic villous sampling, placenta sex differences, microRNA
Sex dimorphism in miRNA expression is more pronounced in first compared to third trimester placenta, and there are 62.5% more female exclusive gestational differences, indicating miRNA abundance across human gestation is also sexually dimorphic.
Introduction
The effects of fetal sex on neonatal and pregnancy outcomes under varied conditions have been examined for decades [1–4]. Fetal growth is influenced by fetal sex, with males heavier at birth [4, 5]. Sexually dimorphic fetal outcomes more common in males than females include macrosomia, shoulder dystocia, cord dysfunctions and low Apgar scores [4, 5]. Maternal outcomes more common with male fetuses include gestational diabetes, placental abruption, dysfunctional labor, prematurity and assisted or cesarean deliveries [1, 4]. Some studies have shown an increased risk of preeclampsia and hyperemesis gravidarum with male fetuses [1, 6], although this has not been found consistently [7, 8]. The pathophysiologic mechanism driving sexually dimorphic outcomes remain poorly understood. As the primary route of communication between the fetomaternal unit, the placenta may drive many of these outcomes. The placental cellular structure, and thus genome derives from the fetus [9]. Therefore sex-specific outcomes may derive from sexually dimorphic gene expression in the placenta, which has been identified in the placenta [10, 11].
Epigenetic modifications, including post-transcriptional regulation, control overall gene expression and phenotype. MicroRNAs (miRNAs) are small, single-stranded, noncoding RNA molecules, on average 22 nucleotides in length [12]. Of known miRNAs, approximately half originate from within introns of the protein coding genes they regulate, and can be co-transcribed [12]. miRNAs generally cause target messenger RNA to be degraded, or prevent translation to protein [13]; small changes in miRNA expression can result in multi-fold changes in gene expression [14]. In the placenta, miRNAs are important as early as trophectoderm development and implantation, some through export in exosomes [15–18], but these miRNA signatures change in the placenta throughout gestation during normal development [19–20]. In addition, there are two large miRNA clusters enriched in placenta, the chromosome 14 miRNA cluster (C14MC) and the chromosome 19 miRNA cluster (C19MC) [21, 22]. C14MC is a large, imprinted, maternally expressed miRNA cluster, with several members predominantly expressed in placenta and epithelial tissues [23]. C19MC is a large, imprinted, paternally expressed miRNA cluster whose members have highest expression in placenta and cancer, with relatively weak expression in other tissue [22–27]. In addition, two small clusters on chromosome 13 were recently identified to be uniquely present in the first trimester placenta [19, 28]. miRNAs have been identified in several pregnancy-related diseases, including pre-eclampsia [29–34], fetal growth [36–37] and gestational diabetes [38, 39], all sexually dimorphic pregnancy complications. However, sex differences and sex-specific miRNA signatures in normal healthy gestations have not been defined.
Therefore, to identify potential biomarkers of placental function, it is imperative to first determine if sex differences in miRNA signatures exist in the first and third trimester placenta of normal healthy gestations. Furthermore, it is critical to identify miRNA signatures that are sex independent and sex specific to develop normative miRNA signatures. Therefore, we performed next-generation sequencing (NGS) and expression analysis to identify and compare sexually dimorphic miRNA expression in first and third trimester placentae of healthy pregnancies resulting in delivery. Furthermore, we identified miRNAs that were sex specific across gestation in either females or males, including common differentially expressed (DE) miRNAs significant in both, as well as sex exclusive miRNAs significant in only females or males. These findings will aid future development of biomarkers for placental diseases that manifest early in gestation that are sex specific as well as sex independent.
Materials and methods
Study population
The study population consisted of 157 singleton pregnancies between 2009 and 2018, including 113 with leftover chorionic villus sampling (CVS) tissue (58 female and 55 male) and 47 with third trimester placenta collected at delivery (19 female and 28 male). These numbers include three patients with matched first and third trimester samples, for 160 samples in total. All subjects were enrolled with informed written consent under IRB approved protocols (Pro00006806, Pro00008600). The cohorts of male and female fetuses were matched for parental age, maternal medical conditions, and fetal race and ethnicity at the time of enrollment, to minimize confounding factors that could impact outcomes. All pregnancies had a normal karyotype and resulted in the delivery of a viable infant.
Collection of placental samples
Samples from the first trimester of pregnancy were collected at 10.0–14.5 weeks gestation during CVS procedures done for prenatal diagnosis. Samples used for research consisted of tissue, which is normally discarded once enough tissue is obtained for prenatal diagnosis. Fetal-derived chorionic villi were cleaned and separated from any maternally derived decidua. Samples from the third trimester of pregnancy were collected between 36.3 and 41.4 weeks gestation, after delivery of a viable neonate. Samples used for research consisted of tissue, which would have otherwise been discarded. One-centimeter cubed placental tissue samples were obtained immediately after delivery from the fetal side of the placenta near the site of cord insertion beneath the amnion, then separated into smaller pieces. Tissue samples were kept on ice and submerged in RNAlater RNA stabilization reagent (QIAGEN, Hilden, Germany) within 30 minutes of collection, and stored in −80°C.
Analysis of demographic data
For demographic analyses, three patients with matched first and third trimester samples were omitted, resulting in N = 110 (57 female and 53 male) first trimester and N = 44 (18 female and 26 male) third trimester samples for this analysis. Demographic data were collected including parental ages, races, and ethnicities, maternal pre-pregnancy body mass index (BMI), fetal sex, maternal medical history and medication use, pregnancy complications, mode of delivery, gestational age at delivery, and birth weight. Means and standard deviations were reported for continuous variables. T-test was used for normally distributed continuous variables, and the Wilcoxon rank-sum test for non-parametric data. Fisher’s exact test was also used as appropriate. Chi-square test was used for comparison of categorical variables.
RNA extraction from chorionic villi
RNA extraction was performed from the 113 first trimester placental samples utilizing a method optimized for extracting DNA and total RNA including small RNAs [10]. Briefly, tissue samples were thawed on ice with 600 μl of RLT Plus lysis buffer (QIAGEN) and 1% β-mercaptoethanol added to each sample, then homogenized by passing the tissue through progressively thinner needles (22G, 25G and 27G) attached to an RNase-free syringe. Homogenates were loaded onto AllPrep spin columns and the remainder of sample processing was performed following manufacturer instructions using the AllPrep DNA/RNA/miRNA Universal Kit (QIAGEN). RNA was eluted with 30–45 μl of RNase-free water at room temperature and the elution was passed through the column twice to improve yields, as previously described [10, 40]. Equal numbers of male and female samples were processed each time to reduce batch effects. The average RNA integrity number (RIN) for sequenced samples was 8.87.
RNA extraction from third trimester placenta
RNA extraction was performed from the 47 third trimester placenta samples utilizing a method similar to that used for the RNA extraction from chorionic villi. However, to homogenize third trimester placental tissue, the tissue was sonicated in ice-cold RLT + 1% beta-mercaptoethanol buffer, using 5-s pulses on a low setting (#2) until tissue fragments were small enough to complete homogenization with RNase-free needles. The average RIN for sequenced samples was 8.84.
Library preparation and miRNA sequencing
All 160 samples were sequenced. A miRNA sequencing library was prepared using the QIASeq miRNA Library Kit (QIAGEN, Hilden, Germany) from total RNA. A pre-adenylated DNA adapter was ligated to the 3′ ends of miRNAs, followed by ligation of an RNA adapter to the 5′ end. A reverse-transcription primer containing an integrated Unique Molecular Index (UMI) was used to convert the 3′/5′ ligated miRNAs into cDNA. After cDNA cleanup, indexed sequencing libraries were generated via sample indexing during library amplification, followed by library cleanup. Libraries were sequenced on a NextSeq 500 (Illumina, San Diego, CA) with a 1 × 75 bp read length and an average sequencing depth of 10.64 million reads per sample.
Differential expression analysis of miRNAs
The demultiplexed raw reads were uploaded to GeneGlobe Data Analysis Center (QIAGEN) at https://www.qiagen.com/us/resources/geneglobe/ for quality control, alignment and expression quantification. Briefly, 3′ adapter and low-quality bases were trimmed off from reads first using cutadapt v1.13 [41] with default settings, then reads with less than 16 bp insert sequences or with less than 10 bp UMI sequences were discarded. The remaining reads were collapsed to UMI counts and aligned sequentially to miRBase v21 mature, hairpin and piRNA (piRNABank) databases using Bowtie v1.2 [42, 43]. The UMI counts of each miRNA category were quantified, and then normalized by a size factor-based method in the R package DESeq2 version 1.22.2 (Bioconductor) [44]. Four separate differential expression analyses were performed: female vs male expression in first trimester, female vs male expression in the third trimester, first vs third trimester expression in all female samples and first vs third trimester expression in all male samples. In each analysis, data were averaged across all samples in each comparison group for each miRNA and were reported as baseMean, with miRNAs with baseMean = 0 removed from the analyses. Next, the R package FactoMineR release 1.41 was used to conduct principal components analysis (PCA), which was used to investigate clustering and potential outliers. Each miRNA was fitted into a negative binomial generalized linear model, and the Wald test was applied to assess the differential expressions between two sample groups. Benjamini and Hochberg procedure was applied to adjust for multiple hypothesis testing, and significantly DE miRNA candidates were selected as those with a false discovery rate (FDR) less than 0.05. The chromosomal location of DE miRNAs was ascertained as previously described using miRBase v21 and biomaRt v2.45.8 R package with Ensembl release 91 [45–47]. For miRNAs derived from more than one chromosome, each miRNA precursor was counted separately in order to capture all chromosome sources (e.g. in barplots, miR-514a-3p is encoded by three different precursors on chromosome X, and thus was counted three times). To avoid variable copy number regions, precursor duplications on the same chromosome were not included. For scatter plots, each mature miRNA was plotted only once per chromosome. Venn diagrams were created with the R package VennDiagram v1.6.20. Finally, the R package ggplot2 v3.3.5 was used to plot chromosome distributions, all bar plots, mean expression against fold change (FC) and FDR distributions.
Thresholds for expression
A threshold of mean baseMean>10 (reached by either comparison group) was selected to define “all expressed” miRNAs and applied throughout. A threshold of mean baseMean>2000 (either group) was selected for “highly expressed” miRNAs.
Predictive analysis of RNA targets for miRNAs with highest expression
Ingenuity Pathways Analysis (IPA) software’s microRNA Target Filter application (QIAGEN, Redwood City, CA, USA, http://www.qiagenbioinformatics.com/IPA) was used to generate a list of predicted target mRNAs based on sequence and experimental confirmation. Each sex and trimester was analyzed individually. For all RNA target lists except the cluster miRNAs, RNA targets were only included if biochemically confirmed using human tissue or non-species-specific methods (sourced from QIAGEN’s curated Ingenuity Knowledge Base [48], or the publicly available miRecords [49] or TarBase [50]), based on the TargetScan algorithm previously described [51]. For the cluster miRNAs, RNA targets included both experimentally confirmed and high confidence targets (based on sequence) to increase target numbers to IPA-recommended levels for analysis. IPA’s Core Analysis function was then used to identify enriched canonical pathways, as previously described. The Comparison Analysis function was used to generate ranked lists of Canonical Pathways from IPA’s Core Analysis output. The procedure described above was applied for highly expressed female and male miRNAs of both trimesters (baseMean>2000), sexually dimorphic first and third trimester miRNAs (FDR < 0.05, baseMean>10 in either sex), female-specific DE miRNAs across gestation (FDR < 1 × 10−5, baseMean>100 in either trimester, and FC > 2 in either direction), male-specific DE miRNAs across gestation (FDR < 1 × 10−5, baseMean>100 in either trimester and FC > 2 in either direction), and sex-specific DE miRNAs in placenta miRNA clusters across gestation (FDR < 0.05, baseMean>10 in each trimester), with each sex and trimester analyzed separately.
Heatmaps
Heatmaps with dendrograms of miRNAs vs samples were generated with matrices of log2(baseMean) data scaled and centered by rows. The plots were then generated with hierarchical clustering using the R package gplots v3.1.1. Heatmaps of gene enrichment data were generated using the R package pheatmap v1.0.12 with matrices of −log10(P) output from IPA Core Enrichment Analysis. For comparisons of −log10(P) values across rows in gene enrichment heatmaps of experimentally confirmed targets only, a difference of 2 or greater was considered the threshold at which visually distinct results were achieved, and was thus defined as “different.” A P-value difference of less than two orders of magnitude (−log10P difference less than 2) was defined as “similar.” For the placenta miRNA clusters in which highly predicted data targets were added, due to the different scales of the data, a −log10P value difference of 0.3 or greater was defined as “different,” and less than 0.3 was defined as “similar.”
Validation with qRT-PCR
Expression of 11 selected miRNAs was quantitatively re-analyzed in a subset of 32 Caucasian sequencing samples with remaining RNA (n = 8/group) by qRT-PCR using the miRCURY LNA miRNA PCR system (QIAGEN). The 11 mRNAs were selected (miR-361-5p, miR-9-5p, miR-1-3p, miR-143-3p, miR-20b-5p, miR-486-5p, miR-934, miR-125b-5p, miR-218-5p, miR-144-5p and miR-451a) based on high expression and significant differences (FDR < 0.05, baseMean>1000) in the sequencing cohorts. Two internal reference miRNAs were selected for each type of comparison after using sequencing data to identify stably and highly expressed miRNAs (baseMean>1000, P-value>0.95). References for female vs male analysis were miR-532-5p and miR-146a-5p. References for first vs third trimester analysis were miR-532-5p and miR-130a-3p. Power calculations were performed using sequencing data and methods described by Cohen [52] to determine sample size requirements, using power = 0.8 and alpha = 0.05. Total RNA was used to synthesize cDNA with universal primers in the miRCURY LNA RT Kit (QIAGEN). Expression was quantified by qRT-PCR using the miRCURY LNA SYBR Green PCR Kit (QIAGEN) and a BioRad MyIQ instrument. Analysis was performed using the ΔΔCt method and Wilcoxon rank-sum tests were performed on the ΔCt values to determine significance.
Results
Cohort demographics and birth outcomes
Female and male singleton pregnancies in the first (N = 113, 58 female and 55 male) and third trimester (N = 47, 19 female and 28 male fetuses) were matched for maternal age, race, ethnicity, and pre-existing medical conditions. Principal component analysis (PCA) of placenta miRNA expression shows no distinct segregation by sex, although PCA by trimester shows a distinct segregation by first and third trimester placenta divided by principal component 1 (Supplemental File 1). Parental and fetal race and ethnicity were not significantly different between sexes in either first or third trimester (Table 1A). Maternal pre-pregnancy BMI was not significantly different between the sexes in either trimester. All pregnancies resulted in live births and there were no significantly different pregnancy complications between the sexes in either trimester cohort. Males were larger than females at birth in both sequencing cohorts, though only significant in the first trimester cohort.
Table 1.
Demographics of study patient cohorts.
| (A) Comparison of male and female fetuses in each gestation | ||||||
|---|---|---|---|---|---|---|
| First trimester (N = 110) | Third trimester (N = 44) | |||||
| Female | Male | P | Female | Male | P | |
| N | 57 | 53 | 18 | 26 | ||
| Maternal age, years | 37.8 (2.9) | 37.5 (3.1) | 0.61 | 37.2 (2.5) | 37.3 (3.4) | 0.45 |
| Paternal age, years | 39.5 (4.6) | 39.4 (5.1) | 0.96 | 39.4 (4.8) | 38.0 (4.8) | 0.51 |
| Maternal race/ethnicity | ||||||
| Caucasian | 53 (93.0%) | 53 (100%) | 0.12 | 13 (72.2%) | 22 (84.6%) | 0.45 |
| Non-Hispanic | 55 (96.5%) | 52 (98.1%) | 1 | 14 (77.8%) | 21 (80.8%) | 1 |
| Paternal race/ethnicity | ||||||
| Caucasian | 54 (94.7%) | 50 (94.3%) | 1 | 15 (83.3%) | 22 (84.6%) | 1 |
| Non-Hispanic | 56 (98.3%) | 51 (96.2%) | 0.61 | 15 (83.3%) | 21 (80.8%) | 1 |
| Fetal race/ethnicity | ||||||
| Caucasian | 53 (93.0%) | 50 (94.3%) | 1 | 12 (66.7%) | 20 (76.9%) | 0.51 |
| Non-Hispanic | 55 (96.5%) | 51 (96.2%) | 1 | 13 (72.2%) | 20 (76.9%) | 0.74 |
| Maternal pre-pregnancy BMI, kg/m2 | 22.0 (3.4) | 21.8 (3.4) | 0.54 | 23.9 (4.6) | 24.0 (4.8) | 0.98 |
| Maternal preexisting medical conditions | ||||||
| Hypertension | 0 (0%) | 0 (0%) | - | 0 (0%) | 0 (0%) | - |
| Diabetes | 0 (0%) | 0 (0%) | - | 0 (0%) | 0 (0%) | - |
| Pregnancy complications | ||||||
| Hypertension | 0 (0%) | 0 (0%) | - | 3 (16.7%) | 5 (19.2%) | 1 |
| Diabetes | 1 (1.8%) | 2 (3.8%) | 0.42 | 0 (0%) | 1 (3.9%) | 1 |
| Placenta previa | 0 (0%) | 0 (0%) | - | 0 (0%) | 1 (3.9%) | 1 |
| Placental abruption | 0 (0%) | 0 (0%) | - | 0 (0%) | 0 (0%) | - |
| Hypertension management in pregnancy | ||||||
| Anti-hypertensives | 0 (0%) | 0 (0%) | - | 1 (5.6%) | 2 (7.7%) | 1 |
| Any magnesium use (ante- or postpartum) | 0 (0%) | 0 (0%) | - | 2 (11.1%) | 4 (15.4%) | 1 |
| Mode of delivery- Cesarean section | 19 (33.3%) | 14 (26.4%) | 0.45 (chi2) | 7 (38.9%) | 8 (30.8%) | 0.59 (chi2) |
| Gestational age at delivery, days | 276.2 (7.0) | 276.3 (7.0) | 0.95 | 278.8 (5.2) | 275.0 (9.3) | 0.13 |
| Birthweight, g | 3342.0 (430.1) | 3536.0 (481.3) | 0.03*, (ttest) | 3397.6 (409.5) | 3525.5 (504.1) | 0.48 |
| (B) Comparison of sex-specific first and third trimester samples | ||||||
| Female (N = 75) | Male (N = 79) | |||||
| First | Third | P value | First | Third | P value | |
| N | 57 | 18 | 53 | 26 | ||
| Maternal age, years | 37.8 (2.9) | 37.2 (2.5) | 0.34 | 37.5 (3.1) | 37.3 (3.4) | 0.87 |
| Paternal age, years | 39.5 (4.6) | 39.4 (4.8) | 0.79 | 39.4 (5.1) | 38.0 (4.8) | 0.27 |
| Maternal race/ethnicity | ||||||
| Caucasian | 53 (93.0%) | 13 (72.2%) | 0.03* | 53 (100%) | 22 (84.6%) | 0.01* |
| Non-Hispanic | 55 (96.5%) | 14 (77.8%) | 0.03* | 52 (98.1%) | 21 (80.8%) | 0.01* |
| Paternal race/ethnicity | ||||||
| Caucasian | 54 (94.7%) | 15 (83.3%) | 0.15 | 50 (94.3%) | 22 (84.6%) | 0.21 |
| Non-Hispanic | 56 (98.3%) | 15 (83.3%) | 0.04* | 51 (96.2%) | 21 (80.8%) | 0.04* |
| Fetal race/ethnicity | ||||||
| Caucasian | 53 (93.0%) | 12 (66.7%) | 0.01* | 50 (94.3%) | 20 (76.9%) | 0.053 |
| Non-Hispanic | 55 (96.5%) | 13 (72.2%) | 0.007* | 2 (3.8%) | 6 (23.1%) | 0.014* |
| Maternal pre-pregnancy BMI, kg/m2 | 22.0 (3.4) | 23.9 (4.6) | 0.11 | 21.8 (3.4) | 24.0 (4.8) | 0.03* |
| Maternal preexisting medical conditions | ||||||
| Hypertension | 0 (0%) | 0 (0%) | - | 0 (0%) | 0 (0%) | - |
| Diabetes | 0 (0%) | 0 (0%) | - | 0 (0%) | 0 (0%) | - |
| Thyroid disorder | 1 (1.8%) | 2 (11.1%) | 0.14 | 2 (3.8%) | 4 (15.4%) | 0.087 |
| Pregnancy complications | ||||||
| Hypertension | 0 (0%) | 3 (16.7%) | 0.012* | 0 (0%) | 5 (19.2%) | 0.003* |
| Diabetes | 1 (1.8%) | 0 (0%) | 1 | 2 (3.8%) | 1 (3.9%) | 1 |
| Placenta previa | 0 (0%) | 0 (0%) | - | 0 (0%) | 1 (3.9%) | 0.55 |
| Placental abruption | 0 (0%) | 0 (0%) | - | 0 (0%) | 0 (0%) | - |
| Hypertension management in pregnancy | ||||||
| Anti-hypertensives | 0 (0%) | 1 (5.6%) | 0.24 | 0 (0%) | 2 (7.7%) | 0.11 |
| Any magnesium use (ante- or Postpartum) | 0 (0%) | 2 (11.1%) | 0.055 | 0 (0%) | 4 (15.4%) | 0.003* |
| Mode of delivery- Cesarean section | 19 (33.3%) | 7 (38.9%) | 0.67 (chi2) | 14 (26.4%) | 8 (30.8%) | 0.72 (chi2) |
| Gestational age at delivery, days | 276.2 (7.0) | 278.8 (5.2) | 0.099 | 276.3 (7.0) | 275.0 (9.3) | 0.84 |
| Birthweight, g | 3342.0 (430.1) | 3397.6 (409.5) | 0.77 | 3536.0 (481.3) | 3525.5 (504.1) | 0.93 |
When comparing sexes across gestation, there were no significant differences in parental ages but there were significant differences in parental and fetal race and ethnicity (Table 1B). Pre-pregnancy BMI was greater in mothers of males in the third trimester cohort compared to the first trimester cohort. Maternal pre-existing medical conditions were not significantly different among the groups but there were three mothers who developed hypertension in the third trimester female cohort (some of which required magnesium) and five mothers in the third trimester male cohort, compared to none in the first trimester cohorts, which was significantly different.
MiRNAs expressed by male and female placentae throughout gestation
Human placenta expressed 986 mature miRNAs combined from females and males across gestation (baseMean>10) (Supplemental File 2AB). In first trimester, female placenta expressed 863 mature miRNAs derived from 958 precursors (baseMean>10), and male placenta expressed 864 mature miRNAs derived from 958 precursors (baseMean>10), from all 22 autosomes and the X chromosome (Figure 1Ai, Supplemental File 2A). No annotated miRNAs from the Y chromosome were identified in miRBase release 21. The majority of expressed miRNAs in both sexes originate with similar distribution from chromosome 19 (female, 123/958 [12.84%]; male, 123/958 [12.84%]), chromosome 14 (female, 103/958 [10.75%]; male, 104/958 [10.86%]), chromosome X (female, 93/958 [9.71%]; male, 91/958 [9.50%]) and chromosome 1 (female, 67/958 [6.99%]; male, 64/958 [6.68%]) (Figure 1Ai). Among the most highly expressed miRNAs (baseMean > 2000), females expressed 159 mature miRNAs from 189 precursors, and males expressed 155 mature miRNAs from 185 precursors, both also predominantly from chromosome 19 (female, 61/189 [32.28%]; male, 61/185 [32.97%]) (Figure 1Aii, Supplemental File 2A).
Figure 2.
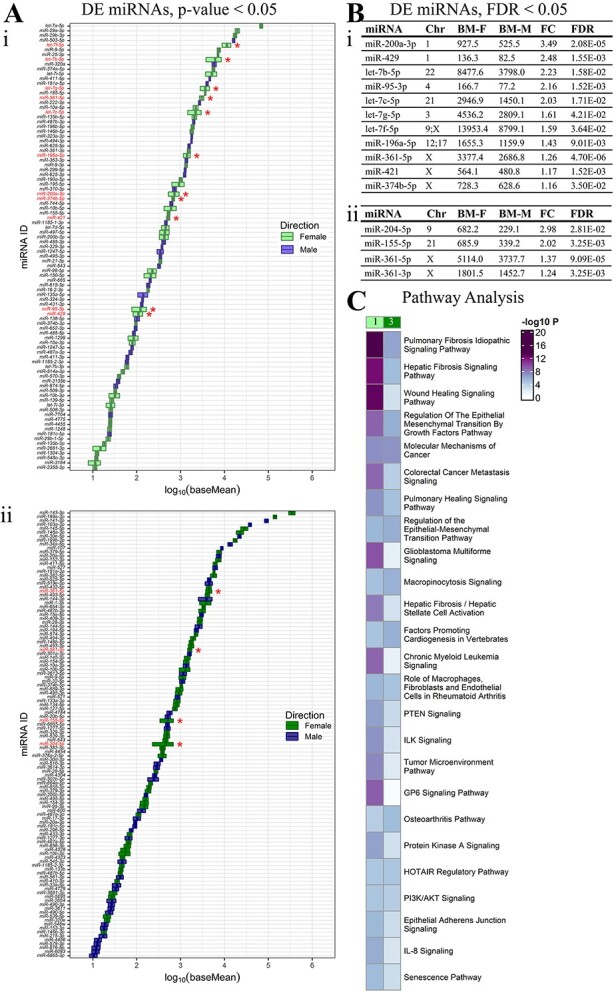
Sexually dimorphic miRNAs in first and third trimester. (A) 94 differentially expressed (DE) miRNAs (58 up in female and 36 in male, baseMean>10, P-value<0.05) listed based on increasing expression (log10 baseMean). Red stars and red labels: DE miRNAs that remain significant after multiple comparisons (FDR < 0.05) in (i) first and (ii) third trimesters. (B) Significant miRNAs at FDR < 0.05 in (i) first and (ii) third trimesters are tabulated with chromosome (Chr) location(s), average baseMeans of female (BM-F) and male (BM-M) samples, fold change (FC, female:male) and FDR. (C) Comparison of the pathways targeted by differentially expressed (FDR < 0.05) and female-upregulated miRNAs reveals sexually dimorphic differences between first (1) and third (3) trimesters.
Figure 1.
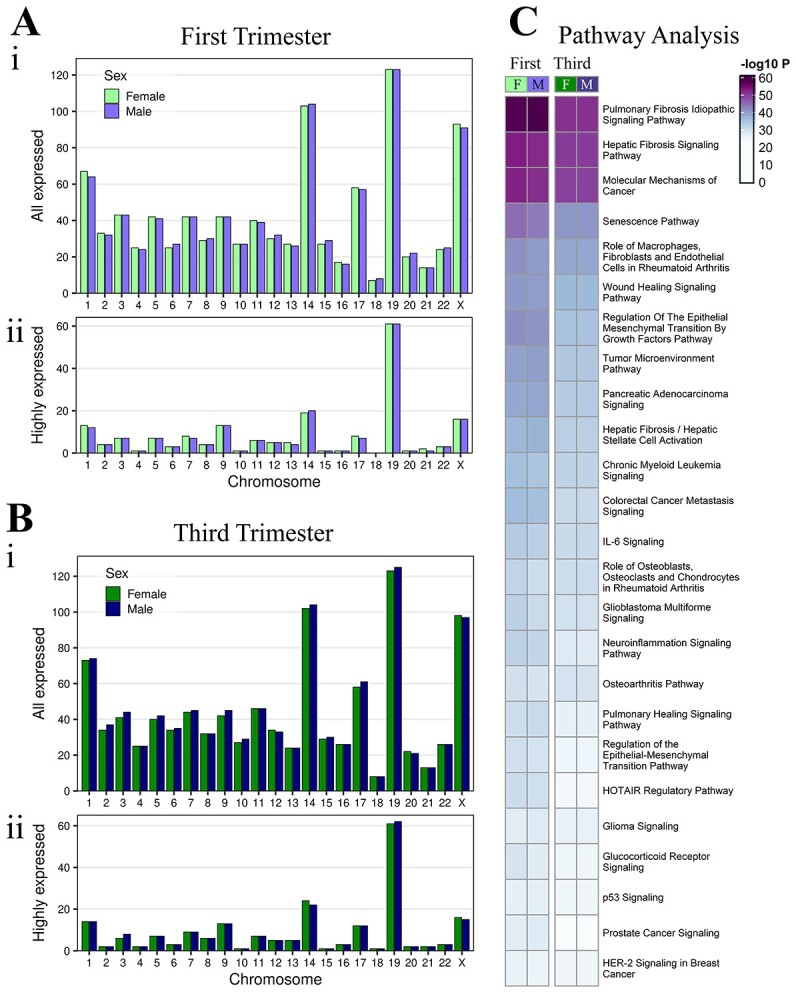
Expressed miRNAs in male and female placentae. (A) Chromosome frequency of precursor miRNAs in male and female placentae at first trimester, (i) all expressed miRNAs (baseMean>10) and (ii) the highest expressed miRNAs (baseMean>2000). Similarly, (B) chromosome frequency at third trimester. (C) Comparison of the pathways targeted by highly expressed miRNAs (baseMean>2000) in first and third trimester female and male placenta.
The number of expressed miRNAs in third trimester was similar to first with 898 mature miRNAs expressed in the third trimester female placenta derived from 1001 precursors (baseMean > 10), and 918 mature miRNAs expressed in the third trimester male placenta derived from 1022 precursors (baseMean > 10) (Figure 1Bi, Supplemental File 2B) from all 22 autosomes and the chromosome X but not Y. Among the expressed miRNAs, the majority originate with similar sex distribution from chromosome 19 (female, 123/1001 [12.29%]; male, 125/1022 [12.23%]), chromosome 14 (female, 102/1001 [10.19%]; male, 104/1022 [10.18%]), chromosome X (female, 98/1001 [9.79%]; male, 97/1022 [9.49%]), and chromosome 1 (female, 73/1001 [7.29%]; male, 74/1022 [7.24%]). Among the most highly expressed miRNAs (baseMean>2000), females and males each expressed 173 mature miRNAs encoded by 205 precursors, predominantly from chromosome 19 (female, 61/205 [29.76%]; male, 62/205 [30.24%]) (Figure 1Bii, Supplemental File 2B).
Pathway enrichment analysis was performed using experimentally confirmed targets of the most highly expressed miRNAs. Of the overall most significantly enriched pathways, there were no observable differences between females and males in either trimester (-log10P value differences of <2) (Figure 1C, Supplemental File 3). Of the top 10 pathways, all were more significantly enriched in the first compared to the third trimester in females (-log10P value differences of ≥2). In males, the majority were also more significantly enriched in the first compared to the third trimester, except “hepatic fibrosis signaling pathway” and “role of macrophages, fibroblasts and endothelial cells in rheumatoid arthritis,” which were similarly enriched in the first and third trimester. Among the top 30 pathways, immune function-related pathways were more enriched in the first vs third trimester: “wound healing signaling pathway” and “neuroinflammation signaling pathway” in both sexes. “IL-6 signaling” was significantly enriched in first vs third trimester in females only. Nine different cancer signaling pathways were also more significantly enriched in the first vs third trimester in both sexes among the top 30 pathways (Figure 1C, Supplemental File 3A).
Differentially expressed miRNAs between male and female placentae
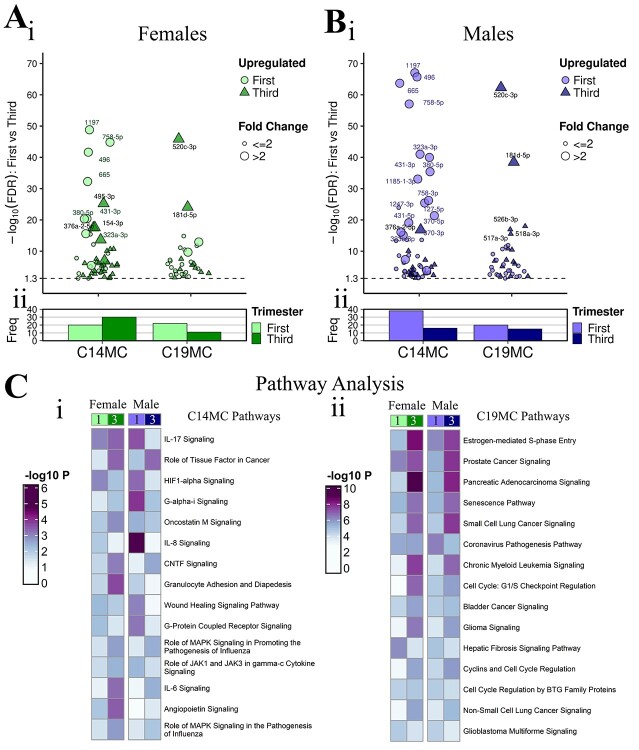
Differences in individual miRNAs between female and male placentae were identified despite lack of subject clustering on PCA plots and heatmaps of all expressed miRNAs (Supplemental Files 1, 4). In the first trimester, there were 94 differentially expressed miRNAs, with 58 upregulated in females and 36 upregulated in males (P-value<0.05, baseMean>10); however, only 11 miRNAs were differentially expressed following adjustment for multiple comparisons (FDR < 0.05, baseMean>10), all upregulated in females (Figure 2Ai). Of the 11 differentially expressed miRNAs (FDR < 0.05), the majority had high baseMeans>500, except miR-429 (female 136.3; male 82.5 baseMeans) and miR-95-3p (female 166.7; male 77.2 baseMeans). The most sexually dimorphic miRNA was miR-200a-3p with an FC of 3.49 and baseMeans of 927.5 in females and 525.5 in males. Four of eleven miRNAs were expressed on chromosome X (36.4%), two on chromosome 1 (18.2%) and one each on five other autosomes (9.1% each) (Figure 2Bi, Supplemental File 2A).
Figure 3.
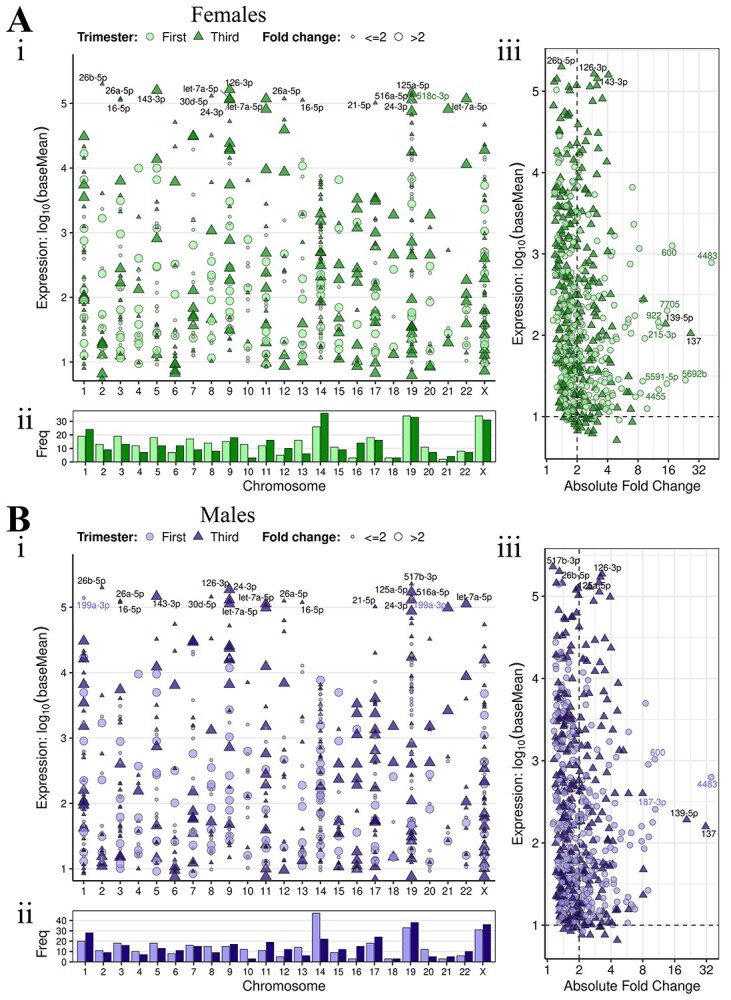
Sex specific expression differences in miRNAs across gestation. DE miRNAs in (A) females and (B) males. (i) Scatter plot of expression (log10 baseMean) distribution across chromosomes for all DE miRNAs at FDR <0.05, baseMean>10. (ii) Chromosome frequency of precursor miRNAs encoding the upregulated mature miRNAs for each trimester at FDR <0.05, baseMean>10. (iii) Scatter plots of expression (log10 baseMean) vs Absolute Fold Change for DE miRNAs (FDR <0.05). Dotted lines are at baseMean = 10 (horizontal) and FC = 2 (vertical). For all scatter plots, point shape indicates direction of upregulation.
In the third trimester, there were 114 DE miRNAs, with 64 upregulated in females and 50 upregulated in males (P-value<0.05, baseMean>10); however, only 4 miRNAs were differentially expressed following adjustment for multiple comparisons (FDR < 0.05, baseMean>10), all upregulated in females (Figure 2Aii). Of the four DE miRNAs (FDR < 0.05), the most sexually dimorphic miRNA was miR-204-5p with 2.98-fold higher expression in females (682.2 female; 229.1 male baseMean). The DE miRNAs in the third trimester originated from chromosome X (2 miRNAs), 9 (1 miRNA) and 21 (1 miRNA) (Figure 2Bii). The miRNA, miR-361-5p was sexually dimorphic and upregulated in females in both trimesters (Supplemental File 2B).
Pathway enrichment analysis was performed using experimentally confirmed targets of the 11 and 4 sexually dimorphic miRNAs in the first and third trimester, respectively. Among the top 10 significant pathways, these were more significantly enriched by sexually dimorphic miRNAs in the first trimester compared to the third trimester (−log10P-value differences of ≥2): “pulmonary fibrosis idiopathic signaling pathway,” “hepatic fibrosis signaling pathway,” “wound healing signaling pathway,” “regulation of the epithelial mesenchymal transition by growth factors pathway,” “colorectal cancer metastasis signaling,” “pulmonary healing signaling pathway” and “glioblastoma multiforme signaling.” The following pathways were enriched by sexually dimorphic miRNAs, with similar significance (−log10P value differences of <2) in first and third trimester: “molecular mechanisms of cancer,” “regulation of the epithelial-mesenchymal transition pathway” and “macropinocytosis signaling.” The “GP6 signaling pathway” was enriched by sexually dimorphic miRNAs only in the first trimester (Figure 2C, Supplemental File 3B).
Sex-specific differential miRNAs expression across gestation
In order to identify sex-specific differences throughout gestation, differential expression analysis of first vs third trimester placenta was performed separately with the female and male placentae. There was sample separation by trimester in both sexes on PCA plots and heatmaps of all expressed miRNAs (Supplemental Files 1, 4). In female placentae, 580 mature miRNAs were significantly differentially expressed between first and third trimester placentae (FDR < 0.05, baseMean>10), with 308 miRNAs upregulated in the first trimester and 272 miRNAs upregulated in the third trimester placentae (Figure 3Ai-ii, Supplemental File 2C). Of the 580 miRNAs derived from 637 precursors in female placenta throughout gestation, the majority originate from chromosomes 19 (first trimester, 34/330 [10.30%]; third trimester, 33/307 [10.75%]), 14 (first trimester, 26/330 [7.88%]; third trimester, 36/307 [11.73%]), and X (first trimester, 34/330 [10.30%]; third trimester, 31/307 [10.10%]) (Figure 3Aii). Of the differentially expressed miRNAs in females across gestation, those with the greatest FC (>8) had lower to moderate expression (baseMean 16.18–1643.77) and were primarily elevated in the first trimester, whereas those with the highest expression (baseMean>10,000) had lower fold changes (FC 1.17–4.08) and were predominantly elevated in the third trimester. The DE miRNA with greatest fold change was miR-4483, with 42.5-fold higher expression in first trimester placenta (FDR = 4.32 × 10−166) and a base mean decrease from 1030.93 to 24.24 in first to third trimester placentae (Figure 3Aiii).
Figure 4.
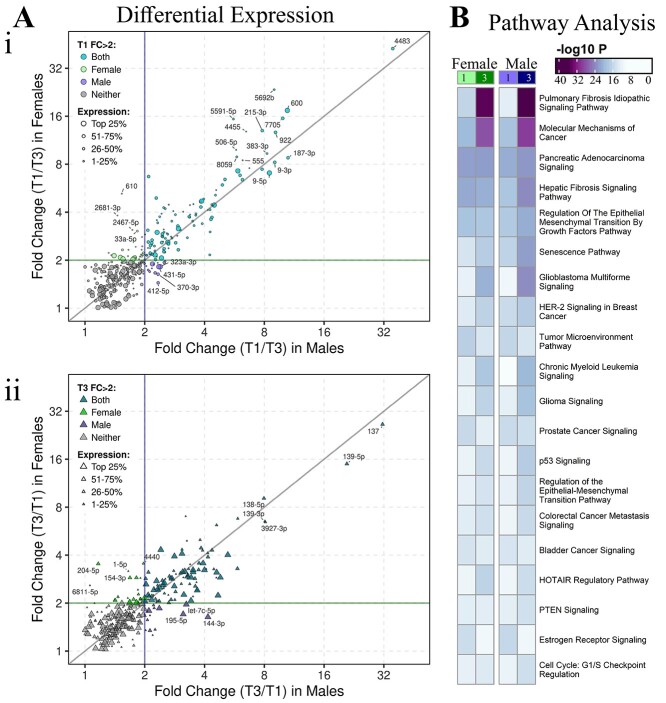
Common and sex-specific miRNA expression changes across gestation. (A) Scatter plots of first vs third trimester DE miRNA fold changes in females compared to males, highlighting miRNAs upregulated in (i) first and (ii) third trimester (FDR < 0.05, baseMean>10, FC > 2). Colors: common DE miRNAs, which reach FC > 2 in both sexes (aqua/turquoise), female exclusive DE miRNAs (green), male exclusive DE miRNAs (purple), or neither (gray). (B) Comparison of the pathways targeted by the most upregulated miRNAs (FDR < 1 × 10−5, baseMean>100 in either group, FC > 2) for each sex cohort in the first vs third trimester analysis.
In the male cohort, 605 mature miRNAs were significantly differentially expressed between first and third trimester placentae (FDR < 0.05, baseMean>10), with 315 miRNAs upregulated in the first trimester and 290 miRNAs upregulated in the third trimester placentae (Figure 3Bi-ii, Supplemental File 2D). Chromosomal distribution was similar for males across gestation with 605 miRNAs derived from 673 precursors. The majority of male miRNAs originate from chromosomes 19 (first trimester, 33/338 [9.76%]; third trimester, 38/335 [11.34%]), 14 (first trimester, 47/338 [13.91%]; third trimester, 22/335 [6.57%]), and X (first trimester, 31/338 [9.17%]; third trimester, 36/335 [10.75%]) (Figure 3Bii). Of the DE miRNAs in males across gestation, those with the greatest FC (>8) ranged from low to high expression (baseMean 37.90–7090.23) and were primarily elevated in the first trimester, whereas those with the highest expression (baseMean>10,000) had lower FC (1.13–4.68) and were predominantly elevated in the third trimester. Similar to females, the DE miRNA with greatest FC was also miR-4483, with 35.5-fold higher expression in first trimester placenta (FDR = 1.22 × 10−182) and a baseMean decrease from 936.26 to 26.33 in first to third trimester placentae (Figure 3Biii). Of the 580 DE miRNAs in females and 605 DE miRNAs in males, there were also 508 miRNAs that overlapped between females and males (common DE) across gestation. Of these common DE miRNAs, 269 were upregulated in first trimester and 239 were upregulated in third trimester (Supplemental Files 2CD, 5A).
Comparison of sex-specific differentially expressed miRNAs
There were sex exclusive DE miRNAs in first vs third trimester analyses, defined as sex-specific DE miRNAs with FC >2 in first vs third trimester analyses either in females or males only, suggesting sexually dimorphic miRNA regulation across gestation. Of the sex-specific DE miRNAs (Figure 3), the female cohort had more DE miRNAs across gestation (FC >2) that were not present in the male analysis (Figure 4A, Supplemental File 5B). There were 52 miRNAs DE across gestation only in females, including 28 upregulated in first trimester and 24 upregulated in third trimester (FDR < 0.05, FC > 2, baseMean>10 in either trimester), whereas there were 32 DE only in males, including 16 upregulated in first trimester and 16 upregulated in third trimester (FDR < 0.05, FC > 2, baseMean>10 in either trimester) (Figure 4A, Supplemental File 5B). Of the sex exclusive DE miRNAs, female exclusive DE miRNAs upregulated in the first trimester had greater FC than male exclusive DE miRNAs upregulated in the first trimester (Figure 4A). Among female exclusive DE miRNAs which had FC < 2 in males, miR-610 had the greatest FC across gestation, 5.22-fold higher in first trimester (FDR = 4.21 × 10−4) with a baseMean decrease of 11.38–2.21 from first to third trimester (Supplemental File 2E). Among male exclusive DE miRNAs, which had FC < 2 in females, miR-144-3p had the greatest FC across gestation with 4.16-fold higher expression in third trimester placenta (FDR = 3.75 × 10−16) and a baseMean increase of 1142.11–4752.80 from first to third trimester placentae (Supplemental File 2E).
Figure 5.
Expression differences in C14MC and C19MC clusters. Scatter plots for (Ai) female and (Bi) male cohorts show mature DE miRNAs (FDR < 0.05, baseMean>10) that are upregulated in first or third trimester (color/shape) and highly expressed (size) for each cluster. (Aii-Bii) Corresponding frequency plots count the upregulated miRNAs (FDR < 0.05, baseMean>10) for each trimester (color). (Ci-ii) Comparison of the pathways targeted by these upregulated miRNAs (FDR < 0.05, baseMean>10), by cluster.
Among the 252 DE miRNAs in females and 232 DE miRNAs in males that reached FC >2, there were 200 common DE miRNAs including 105 upregulated in first trimester and 95 upregulated in third trimester (Figure 4A, Supplemental File 5B). Common DE miRNAs in both sexes with high FC (>8 in both sexes) include miR-4483, miR-600, miR-7705, miR-5692b, miR-922, miR-187-3p, miR-383-3p and miR-9-3p upregulated in first trimester, and miR-137 and miR-139-5p upregulated in third trimester. The common DE miRNA most upregulated in first trimester placenta (compared to third trimester) was miR-4483 with a 42.52-fold difference in female placenta and a 35.47-fold difference in male placenta (Figure 4Ai). The common DE miRNA most upregulated in third trimester placenta (compared to first trimester) was miR-137 with a 26.49-fold difference in female placenta and a 31.52-fold difference in male placenta (Figure 4Aii, Supplemental File 2E).
Pathway enrichment analysis on experimentally confirmed targets of the sex-specific DE miRNAs across gestation identified “pulmonary fibrosis idiopathic signaling pathway” to be the most significantly enriched in the third trimester for both sexes, and in males the difference in significance of this pathway across gestation was greater (−log10P value difference of ≥2). Of the top 10 pathways, the majority of pathways were more significant in the third trimester in both sexes with “hepatic fibrosis signaling,” “regulation of the epithelial mesenchymal transition by growth factors pathway,” “senescence pathway” and “glioblastoma multiforme signaling,” the most significantly enriched in the third trimester in males (Figure 4B, Supplemental File 3C).
Sex differences in the placenta-specific C14MC and C19MC miRNAs
The placenta-specific miRNA clusters on chromosomes 14 and 19 were evaluated. There were 83 mature miRNAs expressed from C14MC and 62 mature miRNAs expressed from C19MC total from females and males across gestation (baseMean>10 any group) (Supplemental File 2FG). Female-specific first vs third trimester analysis (FDR < 0.05, baseMean>10) identified 50 DE mature miRNAs from C14MC (20 upregulated in first and 30 upregulated in third trimester) and 33 DE miRNAs from C19MC (22 upregulated in first and 11 upregulated in third trimester) (Figure 5A, Supplemental File 2F). Male-specific first vs third trimester placenta analysis (FDR < 0.05, baseMean>10) identified 54 DE mature miRNAs from C14MC (38 upregulated in first and 16 upregulated in third trimester) and 35 DE miRNAs from C19MC (20 upregulated in first and 15 upregulated in third trimester) (Figure 5B, Supplemental File 2G). From C14MC, miR-1197 was the most significant DE miRNA across gestation in both sexes and over 6-fold upregulated in the first trimester (FDR = 1.52 × 10−49 females, 9.16 × 10−68 males). From C19MC, miR-520c-3p was the most significant DE miRNA across gestation in both sexes and over 2.7-fold upregulated in the third trimester (FDR = 1.32 × 10−46 females, 4.83 × 10−63 males) (Figure 5A-B, Supplemental File 2FG). There were few sex exclusive DE miRNAs across gestation (FDR < 0.05, baseMean>10, FC > 2 only in females or only in males), 12 from C14MC and only 2 from C19MC (Supplemental File 2). From C14MC, there were 4 female exclusive DE miRNAs all upregulated in third trimester (miR-154-3p, miR-485-3p, miR-495-3p) and 8 male exclusive DE miRNAs all upregulated in first trimester (miR-323a-3p, miR-323b-3p, miR-370-3p, miR-370-5p, miR-412-5p, miR-431-5p, miR-1185-1-3p and miR-1247-3p). From C19MC, there were two female exclusive DE miRNAs both upregulated in first trimester (miR-520d-5p, miR-520 g-5p) and no male exclusive DE miRNAs.
Pathways enrichment analysis was performed using both the experimentally confirmed and highly predicted targets of the sex-specific cluster miRNAs (FDR < 0.05, baseMean>10). C14MC analysis identified top pathways “IL-17 signaling” “role of tissue factor in cancer” and “HIF1α signaling.” “IL-17 signaling” was more significantly enriched in third trimester in females whereas it was more significantly enriched in first trimester in males (-log10P value differences of ≥0.3). Other top immune related pathways that were differentially regulated across gestation in a sex-specific manner include “Gαi signaling,” “oncostatin M signaling,” “granulocyte adhesion and diapedesis” and “role of JAK1 and JAK3 in γc cytokine signaling,” all more significantly enriched in third trimester in females whereas they were more enriched in first trimester in males. The “IL-8 signaling” pathway was the most highly enriched pathway in the first trimester in males. The “wound healing signaling pathway” and “G-protein coupled receptor signaling” pathway were more highly enriched in first trimester females but only enriched in the first trimester in males. The “angiopoietin signaling” pathway was also only enriched in females, predominantly in the third trimester. The C19MC pathways were less sexually dimorphic across gestation with most of the pathways more significant in third versus first trimester, including top pathways “estrogen-mediated S-phase entry,” “prostate cancer signaling” and “pancreatic adenocarcinoma signaling.” Of the top 20, few pathways were highly significant in first trimester compared to third, with “coronavirus pathogenesis pathway” and “cell cycle regulation by BTG family proteins” enriched in the first trimester only in males. “Hepatic fibrosis signaling pathway” and “glioblastoma multiforme signaling” were more significantly enriched in the first trimester in both sexes (Figure 5C, Supplemental File 3DE).
Validation of miRNAs
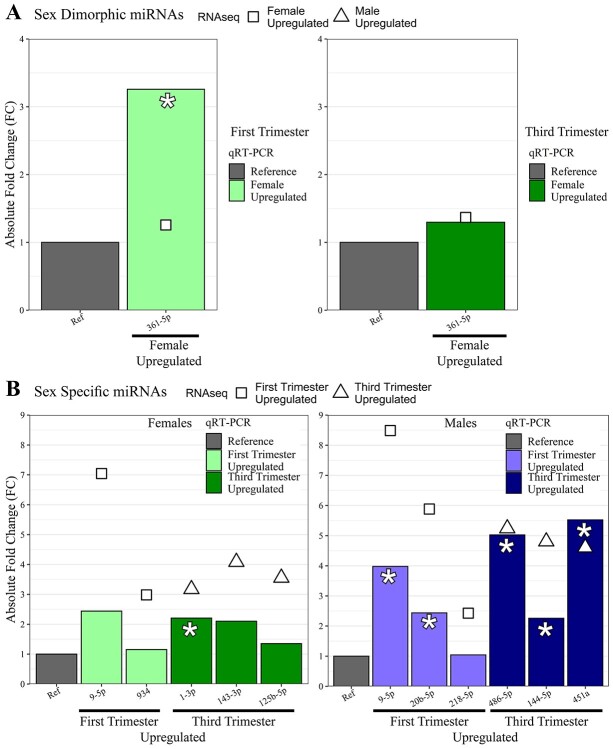
qRT-PCR was performed to validate miRNA-seq results. In the sex differences qRT-PCR comparison, one miRNA was tested at each trimester. Expression of miR-361-5p was higher in females than males at both trimesters, consistent with our sequencing data, however it was only statistically significant in the first trimester (3.26-fold higher, P-value = 0.034) (Figure 6A). In the female-specific DE across gestation, two miRNAs upregulated in the first trimester and three miRNAs upregulated in the third trimester were selected. All miRNAs were upregulated in the direction consistent with our sequencing data; however, only miR-1-3p was statistically significant (2.21-fold higher in third trimester, P-value = 0.022). In the male-specific DE across gestation, three miRNAs upregulated in the first trimester and three upregulated in the third were selected. All miRNAs were upregulated in the direction consistent with our sequencing data. MiR-20b-5p (2.44-fold higher in first trimester, P-value = 0.029), miR-486-5p (5.03-fold higher in third trimester, P-value = 0.021), miR-144-5p (2.26-fold higher in third trimester, P-value = 0.029) and miR-451a (5.53-fold higher in third trimester, P-value = 0.021) were statistically significant (Figure 6B).
Figure 6.
miRNA validation data for all cohorts. Bar plots of absolute fold change (FC) data from RNAseq (points) compared to qPCR (bars) for miRNAs tested in validation for (A.) Sexually dimorphic miRNAs, in which the Reference is the average of miR-532-5p and miR-146a-5p. (B.) Sex-specific miRNAs, in which the Reference is the average of miR-532-5p and miR-130a-3p. Point shape in each: RNAseq absolute FC direction. Bar color in each: qPCR absolute FC direction. Asterisks: denote those miRNAs that achieved significance (P-value<0.05) inqPCR.
Discussion
We identified the sex-specific miRNA signature of the first and third trimester placenta in pregnancies resulting in live births. To our knowledge, this is the largest normative sex dimorphic and sex-specific placenta miRNA atlas across gestation. There were similar numbers of miRNAs expressed in both sexes in each trimester, with similar chromosome distributions and peaks at chromosomes 1, 14, 19 and X as previously described [47]. Overall, the majority of miRNAs were not sexually dimorphic at FDR < 0.05, with 11 DE miRNAs in first and 4 in third trimester placentae, all upregulated in females. However, greater differences were seen in sex-specific differential expression analyses between first and third trimester, which may better explain pregnancy outcomes affected by fetal sex. Chromosomal distributions were sexually dimorphic in chromosomes 14 and 19, with chromosome 14 more representative in the third trimester in females but more representative in the first trimester in males, likely due to C14MC and C19MC. There were first vs third trimester DE miRNAs common to females and males, including miR-4483 upregulated in the first trimester and miR-137 and miR-139-5p upregulated in third trimester. There were 62.5% more female exclusive differences across gestation as male exclusive (52 miRNAs vs 32 miRNAs), indicating that gestational changes of miRNA expression in human placenta are sexually dimorphic. A subset of DE miRNAs was validated using qRT-PCR, all results were consistent with miRNA-seq results, with 7 of 13 miRNAs tested to be statistically significant.
Sex differences in the first and third trimester miRNAs were small, but differences were more prevalent in the first trimester placenta. Greater sexual dimorphism in first trimester may also account for more significant upregulated pathways identified in the first compared to the third trimester placenta, suggesting miRNAs have an impact on placental development early in gestation in a sex-specific manner, a result consistent with previous studies [10, 53–55]. The only miRNA that was sexually dimorphic across both trimesters examined was miR-361-5p. This miRNA has been minimally studied, with no references to placental development; however, Tsamou et al. report that this miRNA was also sexually dimorphic in newborns, with higher expression in girls [56].
Although other studies have identified sexually dimorphic miRNA expression in the placenta, these studies were small, which may lead to subject variability [54], or utilized microarrays [56] and most of them were in placenta disease states where differences may be attributed to development of placental dysfunction possibly due to sexually dimorphic miRNA regulation of these sexually dimorphic disease states early in gestation [10, 57]. Recently, sexually dimorphic miRNA expression was described by Eaves in second trimester placenta of extremely low gestational age newborns. Of those identified by Eaves, miR-361-5p, miR-196a-5p and miR-374b-5p were also sexually dimorphic in the first trimester in our study [20]. These miRNAs may ultimately be sex-specific early markers of extremely low gestational age. Another recent study by Guo et al. of a small number of third trimester placentae also found miR-361-3p and miR-361-5p upregulated in females vs males, in common with our third trimester results [54], suggesting these may be specific markers of normal gestational age. In addition, other miRNAs defined in our study have been previously shown to be associated with preterm labor, such as miR-421, miR-374b-5p and miR-155-5p [58]. Other gestational disease states have been previously studied as well, including recurrent spontaneous abortion, associated with miR-196a-5p [59] and pre-eclampsia, associated with miR-374b-5p [29], and may ultimately become sex-specific markers of these sex dimorphic placental pathologic states.
There were 508 miRNAs with differential expression across gestation common in both sexes, with 200 that reached a FC > 2. These are potentially critical miRNAs, particularly those that are expressed early in gestation, that can be used for future biomarker development, since they are independent of fetal sex. We previously identified miR-4483 as strongly downregulated in third trimester compared to first trimester placenta in an analysis adjusted for fetal sex [47], and now here show that miR-4483 is sex independent. There were also sex-specific miRNAs that had differential expression across gestation, including those that were sex exclusive, suggesting miRNA regulation of placental development throughout gestation is sexually dimorphic. These miRNAs are important for sex-specific normal placental development and may provide a foundation for miRNA expression changes leading to placental pathology in a sex-specific fashion.
None of the miRNAs that were sexually dimorphic in the first or third trimester were from the placenta-specific C14MC and C19MC [21, 22]. However, there were sex-specific differences across gestation in C14MC and C19MC. In females, there were more DE miRNAs that were upregulated in the third trimester from C14MC compared to the first trimester, whereas there were more DE miRNAs that were upregulated from C19MC in the first trimester, suggesting these clusters may play different roles throughout gestation in females, with C14MC potentially having a greater regulatory role in the third and C19MC in the first trimester. In contrast, in males, there were more differentially expressed miRNAs that were upregulated from C14MC and C19MC in the first compared to the third trimester, suggesting a greater regulatory role of C14MC and C19MC in males in the first trimester. There were six female exclusive DE miRNAs across gestation, including 4 from C14MC and 2 miR-520 family members from C19MC. There were eight male exclusive DE miRNAs across gestation from C14MC including two miR-323 family members, but no male exclusive DE miRNAs from C19MC. All of the sex exclusive DE miRNAs identified from the clusters have been associated with cancer including reproductive cancers [60–62], which may be suggestive that these miRNAs are under sex hormone control, which is fetal sex specific early in gestation during fetal sex differentiation [63, 64]. The sex-specific differences across gestation in C14MC and C19MC may account for known sex differences in placenta development, as these clusters are placenta specific. There were significant sex differences in immune-related pathways targeted by the C14MC cluster miRNAs, while there was less sexual dimorphism in C19MC overall.
Previous studies have identified miRNAs that are DE in first versus third trimester in both C14MC and C19MC clusters that play a role in immune suppressive, anti-inflammatory responses, as well as innate/adaptive immune responses [20]. We identified miRNAs in the C14MC cluster to regulate immune function in a sexually dimorphic fashion. miRNAs have been identified in regulating immune function, which may be sexually dimorphic [65] that may also translate in the placenta. These sexually dimorphic immune mediated pathways may impact placental function including malperfusion and subsequent sequela [66]. Furthermore, this altered intrauterine environment may have long term impact, including deficient development of the immune system into childhood [67–69], predisposition to inflammatory and immune disease states that develop in adulthood [69–71], including metabolic syndrome and cardiovascular disease [72–76], which are sexually dimorphic disease states.
The major strength of this study is the cohort size of normal healthy pregnancies resulting in delivery and the availability of detailed demographic information including birth outcomes. Additionally, the use of high-throughput sequencing, as opposed to other techniques such as arrays, allows for greater confidence regarding differential expression, since all known miRNA species previously annotated in the human genome are considered. Although other studies have identified sexually dimorphic miRNA expression in the placenta, these studies were small, and most of them were in placenta disease states where differences may be attributed to development of placental dysfunction, which may be the result of sexually dimorphic miRNA regulation of these sexually dimorphic disease states [56, 57].
Our study has some limitations. Female and male singleton pregnancies in the first and third trimester were matched for maternal age, race, ethnicity and pre-existing medical conditions. However, when comparing sexes across gestation, differences in parental and fetal race and ethnicity although small were identified. Maternal pre-existing medical conditions were not significantly different among the groups, but there were a few mothers that developed hypertension in the third trimester cohorts. However, we did not control for outcomes since subjects were enrolled in the first trimester. Pre-pregnancy BMI was greater in mothers of males in the third trimester cohort compared to the first trimester cohort. Although sexual dimorphism in miR-210 expression has been identified in the placenta associated with maternal obesity [77], our cohorts were not obese and BMI differences were only identified in the male-specific cohorts. As a result, miR-210 was not identified to be different among the groups. During predictive target enrichment analysis, most of the mRNA targets used were experimentally validated, but due to few experimentally validated targets for the C14MC DE miRNAs, we included additional predicted targets for both cluster analyses to improve the pathway enrichment predictions, at the cost of some confidence. DE expressed miRNAs were validated using qRT-PCR, with results consistent with miRNA-seq. However, not all were statistically significant. This may be due to the smaller sample size of the validation cohort.
Overall, our goals were: first, to identify the normative sex dimorphic miRNA signatures in first and third trimester and second, identify and compare the normative sex stable and sex-specific normative miRNA signature across gestation in a healthy population. With the identification of these normative signatures, future studies on placental pathologic disease states can be performed, controlling for variability due to fetal sex and gestational age. Furthermore, due to their small size and stability, these miRNAs may become potential biomarkers of disease early in gestation, particularly miRNAs from plasma exosomes [78–80], and utilized as additional metadata for machine learning models of placental disease [81].
Supplementary Material
Acknowledgements
The authors would like to acknowledge the members within the department of OB/GYN who assisted with subject recruitment, and most of all we are grateful for the subjects who are willing to participate in our studies.
Footnotes
† Grant Support: This work was supported by the National Institute of Health grants: R01 HD091773, R01 HD074368, T32 DK007770, U01 EB026421 and R01 AI154535. The funding agency was not involved in the design, analysis or interpretation of the data reported. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Contributor Information
Amy E Flowers, Department of Obstetrics and Gynecology, Division of Reproductive Endocrinology and Infertility, Cedars-Sinai Medical Center, Los Angeles, CA, USA.
Tania L Gonzalez, Department of Obstetrics and Gynecology, Division of Reproductive Endocrinology and Infertility, Cedars-Sinai Medical Center, Los Angeles, CA, USA.
Nikhil V Joshi, Department of Obstetrics and Gynecology, Division of Reproductive Endocrinology and Infertility, Cedars-Sinai Medical Center, Los Angeles, CA, USA; David Geffen School of Medicine, University of California, Los Angeles, CA, USA.
Laura E Eisman, Department of Obstetrics and Gynecology, Division of Reproductive Endocrinology and Infertility, Cedars-Sinai Medical Center, Los Angeles, CA, USA.
Ekaterina L Clark, Department of Obstetrics and Gynecology, Division of Reproductive Endocrinology and Infertility, Cedars-Sinai Medical Center, Los Angeles, CA, USA.
Rae A Buttle, Department of Obstetrics and Gynecology, Division of Reproductive Endocrinology and Infertility, Cedars-Sinai Medical Center, Los Angeles, CA, USA.
Erica Sauro, Department of Obstetrics and Gynecology, Division of Reproductive Endocrinology and Infertility, Cedars-Sinai Medical Center, Los Angeles, CA, USA.
Rosemarie DiPentino, Department of Obstetrics and Gynecology, Division of Reproductive Endocrinology and Infertility, Cedars-Sinai Medical Center, Los Angeles, CA, USA.
Yayu Lin, Department of Obstetrics and Gynecology, Division of Reproductive Endocrinology and Infertility, Cedars-Sinai Medical Center, Los Angeles, CA, USA.
Di Wu, Genomics Core, Cedars-Sinai Medical Center, Los Angeles, CA, USA.
Yizhou Wang, Genomics Core, Cedars-Sinai Medical Center, Los Angeles, CA, USA.
Chintda Santiskulvong, CS Cancer Applied Genomics Shared Resource, CS Cancer, Cedars-Sinai Medical Center, Los Angeles, CA, USA.
Jie Tang, Department of Biomedical Sciences, Cedars-Sinai Medical Center, Los Angeles, CA, USA.
Bora Lee, Department of Obstetrics and Gynecology, Division of Reproductive Endocrinology and Infertility, Cedars-Sinai Medical Center, Los Angeles, CA, USA.
Tianyanxin Sun, Department of Obstetrics and Gynecology, Division of Reproductive Endocrinology and Infertility, Cedars-Sinai Medical Center, Los Angeles, CA, USA.
Jessica L Chan, Department of Obstetrics and Gynecology, Division of Reproductive Endocrinology and Infertility, Cedars-Sinai Medical Center, Los Angeles, CA, USA; David Geffen School of Medicine, University of California, Los Angeles, CA, USA.
Erica T Wang, Department of Obstetrics and Gynecology, Division of Reproductive Endocrinology and Infertility, Cedars-Sinai Medical Center, Los Angeles, CA, USA; David Geffen School of Medicine, University of California, Los Angeles, CA, USA.
Caroline Jefferies, Department of Biomedical Sciences, Cedars-Sinai Medical Center, Los Angeles, CA, USA.
Kate Lawrenson, Department of Biomedical Sciences, Cedars-Sinai Medical Center, Los Angeles, CA, USA; Department of Obstetrics and Gynecology, Cedars-Sinai Medical Center, Los Angeles, CA, USA.
Yazhen Zhu, California NanoSystems Institute, Crump Institute for Molecular Imaging, Department of Molecular and Medical Pharmacology, University of California, Los Angeles, Los Angeles, CA, USA.
Yalda Afshar, David Geffen School of Medicine, University of California, Los Angeles, CA, USA.
Hsian-Rong Tseng, California NanoSystems Institute, Crump Institute for Molecular Imaging, Department of Molecular and Medical Pharmacology, University of California, Los Angeles, Los Angeles, CA, USA.
John Williams, III, David Geffen School of Medicine, University of California, Los Angeles, CA, USA; Department of Obstetrics and Gynecology, Cedars-Sinai Medical Center, Los Angeles, CA, USA.
Margareta D Pisarska, Department of Obstetrics and Gynecology, Division of Reproductive Endocrinology and Infertility, Cedars-Sinai Medical Center, Los Angeles, CA, USA; David Geffen School of Medicine, University of California, Los Angeles, CA, USA; Department of Biomedical Sciences, Cedars-Sinai Medical Center, Los Angeles, CA, USA.
Author contributions
Author(s) contributed to conceptualization and design of work (AEF, TLG, NVJ, LEE, KL, CJ, YZ, YA, HRT, JW III, MDP), acquisition of samples (ELC, RAB, ES, RD, JW III), sample processing (TLG, NVJ, ELC, RD, YL, CS, BL, TS), analysis (AEF, TLG, NVJ, DW, YW, JT, JLC, ETW, MDP) and interpretation of data (AEF, TLG, NVJ, DW, YW, MDP). The original draft was written by AEF, TLG, NVJ, LEE andMDP.
Conflict of interest
None to declare.
References
- 1. Broere-Brown ZA, Adank MC, Benschop L, Tielemans M, Muka T, Gonçalves R, Bramer WM, Schoufour JD, Voortman T, Steegers EAP, Franco OH, Schalekamp-Timmermans S. Fetal sex and maternal pregnancy outcomes: a systematic review and meta-analysis. Biology of Sex Differences 2020; 11:26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Lorente-Pozo S, Parra-Llorca A, Torres B, Torres-Cuevas I, Nuñez-Ramiro A, Cernada M, García-Robles A, Vento M. Influence of sex on gestational complications, Fetal-to-neonatal transition, and postnatal adaptation. Frontiers in Pediatrics 2018; 6:63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Traccis F, Frau R, Melis M. Gender differences in the outcome of offspring prenatally exposed to drugs of abuse. Frontiers in Behavioral Neuroscience 2020; 14:72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Sheiner E. The relationship between Fetal gender and pregnancy outcomes. Archives of Gynecology and Obstetrics 2007; 275:317–319. [DOI] [PubMed] [Google Scholar]
- 5. Kiserud T, Benachi A, Hecher K, Perez RG, Carvalho J, Piaggio G, Platt LD. The World Health Organization Fetal growth charts: concept, findings, interpretation, and application. American Journal of Obstetrics and Gynecology 2018; 218:S619–S629. [DOI] [PubMed] [Google Scholar]
- 6. Basso O, Olsen J. Sex ratio and twinning in women with hyperemesis or pre-Eclampsia. Epidemiology 2001; 12:747–749. [DOI] [PubMed] [Google Scholar]
- 7. Kuru O, Sen S, Akbayir O, Goksedef BPC, Ozsurmeli M, Attar E, Saygili H. Outcomes of pregnancies complicated by hyperemesis Gravidarum. Archives of Gynecology and Obstetrics 2011; 285:1517–1521. [DOI] [PubMed] [Google Scholar]
- 8. Nurmi M, Rautava P, Gissler M, Vahlberg T, Polo-Kantola P. Recurrence patterns of hyperemesis Gravidarum. American Journal of Obstetrics and Gynecology 2018; 219:e1–e10. [DOI] [PubMed] [Google Scholar]
- 9. Boss AL, Chamley LW, James JL. Placental formation in early pregnancy: how is the Center of the Placenta Made. Human Reproduction Update 2018; 24:750–760. [DOI] [PubMed] [Google Scholar]
- 10. Gonzalez TL, Sun T, Koeppel AF, Lee B, Wang ET, Farber CR, Rich SS, Sundheimer LW, Buttle RA, Chen Y-DI, Rotter JI, Turner SD et al. Sex differences in the late first trimester human placenta transcriptome. Biology of Sex Differences 2018; 9:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Buckberry S, Bianco-Miotto T, Bent SJ, Dekker GA, Roberts CT. Integrative transcriptome meta-analysis reveals widespread sex-biased gene expression at the human Fetal-maternal Interface. Molecular Human Reproduction 2014; 20:810–819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hayder H, O'Brien J, Nadeem U, Peng C. MicroRNAs: crucial regulators of placental development. Reproduction 2018; 155:R259–R271. [DOI] [PubMed] [Google Scholar]
- 13. Mouillet J-F, Ouyang Y, Coyne C, Sadovsky Y. MicroRNAs in placental health and disease. American Journal of Obstetrics and Gynecology 2015; 213:S163–S172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. de Sousa MC, Gjorgjieva M, Dolicka D, Sobolewski C, Foti M. Deciphering miRNAs' action through miRNA editing. International Journal of Molecular Sciences 2019; 20:6249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ali A, Bouma GJ, Anthony RV, Winger QA. The role of LIN28-let-7-ARID3B pathway in placental development. International Journal of Molecular Sciences 2020; 21:3637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zhang WM, PC LX, Zhang Y, Liu Z, Yao N, Ma Y-Y. Effect of miR-133 on apoptosis of trophoblasts in human placenta tissues via rho/ROCK signaling pathway. European Review for Medical and Pharmacological Sciences 2019; 23:10600–10608. [DOI] [PubMed] [Google Scholar]
- 17. Zhang WM, Liu X-L, Yang Y, Ayi N, Li L-H. Highly expressed microRNA-940 promotes early abortion by regulating placenta implantation by targeting ZNF672. European Review for Medical and Pharmacological Sciences 2019; 23:2693–2700. [DOI] [PubMed] [Google Scholar]
- 18. Burnett LA, Nowak RA. Exosomes mediate embryo and maternal interactions at implantation and during pregnancy. Frontiers in Bioscience (Scholar Edition) 2016; 8:79–96. [DOI] [PubMed] [Google Scholar]
- 19. Eaves LA, Phookphan P, Rager JE, Bangma J, Santos HP Jr, Smeester L, O'Shea TM, Fry RC. A role for microRNAs in the epigenetic control of sexually dimorphic gene expression in the human placenta. Epigenomics 2020; 17:1543–1558.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gu Y, Sun J, Groome L, Wang Y. Differential miRNA expression profiles between the first and third trimester human placentas. American Journal of Physiology: Endocrinology and Metabolism 2013; 304:E836–E843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Bentwich I, Avniel A, Karov Y, Aharonov R, Gilad S, Barad O, Barzilai A, Einat P, Einav U, Meiri E, Sharon E, Spector Y et al. Identification of hundreds of conserved and nonconserved human microRNAs. Nat Genet 2005; 37:766–770. [DOI] [PubMed] [Google Scholar]
- 22. Morales-Prieto DM, Ospina-Prieto S, Chaiwangyen W, Schoenleben M, Markert UR. Pregnancy-associated miRNA-clusters. J Reprod Immunol 2013; 97:51–61. [DOI] [PubMed] [Google Scholar]
- 23. Liang Y, Ridzon D, Wong L, Chen C. Characterization of microRNA expression profiles in normal human tissues. BMC Genomics 2007; 8:166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Jinesh GG, Flores ER, Brohl AS. Chromosome 19 miRNA cluster and CEBPB expression specifically mark and potentially drive triple negative breast cancers. PLoS One 2018; 13:e0206008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Nguyen PN, Huang CJ, Sugii S, Cheong SK, Choo KB. Selective activation of miRNAs of the primate-specific chromosome 19 miRNA cluster (C19MC) in cancer and stem cells and possible contribution to regulation of apoptosis. J Biomed Sci 2017; 24:20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Radovich M, Solzak JP, Hancock BA, Conces ML, Atale R, Porter RF, Zhu J, Glasscock J, Kesler KA, Badve SS, Schneider BP, Loehrer PJ. A large microRNA cluster on chromosome 19 is a transcriptional hallmark of WHO type a and AB thymomas. Br J Cancer 2016; 114:477–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Noguer-Dance M, Abu-Amero S, Al-Khtib M, Lefevre A, Coullin P, Moore GE, Cavaille J. The primate-specific microRNA gene cluster (C19MC) is imprinted in the placenta. Hum Mol Genet 2010; 19:3566–3582. [DOI] [PubMed] [Google Scholar]
- 28. Dunham A, Matthews LH, Burton J, Ashurst JL, Howe KL, Ashcroft KJ, Beare DM, Burford DC, Hunt SE, Griffiths-Jones S, Jones MC, Keenan SJ et al. The DNA sequence and analysis of human chromosome 13. Nature 2004; 428:522–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Zhu X-M, Han T, Sargent IL, Yin G-W, Yao Y-Q. Differential expression profile of microRNAs in human placentas from preeclamptic pregnancies vs normal pregnancies. American Journal of Obstetrics and Gynecology 2009; 200:661.e661–661.e667. [DOI] [PubMed] [Google Scholar]
- 30. Ishibashi O, Ohkuchi A, Ali M, Kurashina R, Luo S-S, Ishikawa T, Takizawa T, Hirashima C, Takahashi K, Migita M, Ishikawa G, Yoneyama K et al. Hydroxysteroid (17-β) dehydrogenase 1 is dysregulated by Mir-210 and Mir-518c that are aberrantly expressed in preeclamptic placentas: a novel marker for predicting preeclampsia. Hypertension 2012; 59:265–273. [DOI] [PubMed] [Google Scholar]
- 31. Yang S, Li H, Ge Q, Guo L, Chen P. Deregulated microRNA species in the plasma and placenta of patients with preeclampsia. Molecular Medicine Reports 2015; 12:527–534. [DOI] [PubMed] [Google Scholar]
- 32. Anton L, Olaerin-George AO, Hogenesch JB, Elovitz MA. Placental expression of miR-517a/b and miR-517c contributes to trophoblast dysfunction and preeclampsia. PLoS One 2015; 10:e0122707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Vashukova ES, Glotov AS, Fedotov PV, Efimova OA, Pakin VS, Mozgavaya EV, Pendina AA, Tikhonov AV, Koltsova AS, Baranov VS. Placental microRNA expression in pregnancies complicated by superimposed pre-eclampsia on chronic hypertension. Molecular Medicine Reports 2016; 14:22–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hromadnikova I, Kotlabova K, Ondrackova M, Pirkova P, Kestlerova A, Novotna V, Hympanova L, Krofta L. Expression profile of C19MC microRNAs in placental tissue in pregnancy-related complications. DNA and Cell Biology 2015; 34:437–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Sõber S, Rull K, Reiman M, Ilisson P, Mattila P, Laan M. RNA sequencing of chorionic villi from recurrent pregnancy loss patients reveals impaired function of basic nuclear and cellular machinery. Scientific Reports 2016; 6:38439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Higashijima A, Miura K, Mishima H, Kinoshita A, Jo O, Abe S, Hasegawa S, Miura S, Yamasaki K, Yashida A, Yashiura K-I, Masuzaki H. Characterization of placenta-specific microRNAs in fetal growth restriction pregnancy. Prenatal Diagnosis 2013; 33:214–222. [DOI] [PubMed] [Google Scholar]
- 37. Herbert JF, Millar JA, Raghavan R, Romney A, Podrabsky JE, Rennie MY, Felker AM, O'Tierney-Ginn P, Morita M, DuPriest EA, Morgan TK. Male fetal sex affects uteroplacental angiogenesis in growth restriction mouse model. Biology of Reproduction 2021; 104:924–934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Li J, Song L, Zhou L, Wu J, Sheng C, Chen H, Liu Y, Gao S, Huang W. A microRNA signature in gestational diabetes mellitus associated with risk of macrosomia. Cellular Physiology and Biochemistry 2016; 37:243–252. [DOI] [PubMed] [Google Scholar]
- 39. Ding R, Guo F, Zhang Y, Liu X-M, Xiang Y-Q, Zhang C, Liu Z-W, Sheng J-Z, Huang H-F, Zhang J-Y, Fan J-X. Integrated transcriptome sequencing analysis reveals role of miR-138-5p/TBL1X in placenta from gestational diabetes mellitus. Cellular Physiology and Biochemistry 2018; 51:630–946. [DOI] [PubMed] [Google Scholar]
- 40. Pisarska MD, Akhlaghpaur M, Lee B, Barlow GM, Xu N, Wang ET, Mackey AJ, Farber CR, Rich SS, Rotter JI, Chen YI, Goodorzi MO et al. Optimization of techniques for multiple platform testing in small precious samples such as human chorionic villus sampling. Prenatal Diagnosis 2016; 36:1061–1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet Journal 2011; 17:3. [Google Scholar]
- 42. Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biology 2009; 10:R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kozomara A, Griffiths-Jones S. miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Research 2014; 42:D68–D73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biology 2014; 15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Durinck S, Moreau Y, Kasprzyk A, Davis S, De Moor B, Brazma A, Huber W. BioMart and Bioconductor: a powerful link between biological databases and microarray data analysis. Bioinformatics 2005; 21:3439–3440. [DOI] [PubMed] [Google Scholar]
- 46. Durinck S, Spellman PT, Birney E, Huber W. Mapping identifiers for the integration of genomic datasets with the R/Bioconductor package biomaRt. Nature Protocols 2009; 4:1184–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Gonzalez TL, Eisman LE, Joshi NV, Flowers AE, Wu D, Wang Y, Santiskulvong C, Tang J, Buttle RA, Sauro E, Clark EL, DiPentino R et al. High-throughput miRNA sequencing of the human placenta: expression throughout gestation. Epigenomics 2021; 13:995–1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Krämer A, Green J, Pollard JJ, Tugendreich S. Causal analysis approaches in ingenuity pathway analysis. Bioinformatics 2014; 30:523–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Xiao F, Zuo Z, Cai G, Kang S, Gao X, Li T. miRecords: an integrated resource for microRNA-target interactions. Nucleic Acids Research 2009; 37:D105–D110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Karagkouni D, Paraskevopoulou MD, Chatzopoulos S, Vlachos IS, Tastsoglou S, Kanellos I, Papadimitriou D, Kavakiotis I, Maniou S, Skoufous G, Vergoulis T, Dalamagas T et al. DIANA-TarBase v8: a decade-long collection of experimentally supported miRNA-gene interactions. Nucleic Acids Research 2018; 46:D239–D246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Lewis BP, Jones-Rhoades MW, Bartel DP, Burge CP. Prediction of mammalian microRNA targets. Cell 2003; 115:787–798. [DOI] [PubMed] [Google Scholar]
- 52. Cohen J. Statistical power analysis for the Behavioral sciences. New York: Routledge 1988:19–66. [Google Scholar]
- 53. Sun T, Gonzalez TL, Deng N, DiPentino R, Clark EL, Lee B, Tang J, Wang Y, Stripp BR, Yao C, Tseng H-R, Karumanchi SA et al. Sexually dimorphic crosstalk at the maternal-fetal interface. Journal of Clinical Endocrinology and Metabolism 2020; 105:e4831–e4847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Guo S, Huang S, Jiang X, Hu H, Han D, Moreno CS, Fairbrother GL, Hughes DA, Stoneking M, Khaitovich P. Variation of microRNA expression in the human placenta driven by population identity and sex of the newborn. BMC Genomics 2021; 22:286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Howe CG, F HB, Kennedy EM, Eckel SP, Chavez TA, Faham D, Grubbs BH, Al-Marayati L, Lerner D, Suglia S, Bastain TM, Marsit CJ et al. Extracellular vesicle microRNA in early versus late pregnancy with birth outcomes in the MADRES study. Epigenetics 2021. 10.1080/15592294.2021.1899887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Tsamou M, Vrijens K, Wang C, Winckelmans E, Neven KY, Madhloum N, de Kok TM, Nawrot TS. Genome-wide microRNA expression analysis in human placenta reveals sex-specific patterns: an ENVIRONAGE birth cohort study. Epigenetics 2020; 16:373–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Saha B, Ganguly A, Home P, Bhattacharya B, Ray S, Ghosh A, Rumi MAK, Marsh C, French VA, Gunewardena S, Paul S. TEAD4 ensures postimplantation development by promoting trophoblast self-renewal: an implication in early human pregnancy loss. Proceedings of the National Academy of Sciences of the U.S.A. 2020; 117:17864–17875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Fallen S, Baxter D, Wu X, Kim T-K, Shynlova O, Lee MY, Scherler K, Lye S, Hood L, Wang K. Extracellular vesicle RNAs reflect placenta dysfunction and are a biomarker source for preterm labor. Journal of Cellular and Molecular Medicine 2018; 22:2760–2773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Jeon YJ, Choi YS, Rah H, Kim SY, Choi DH, Cha SH, Shin JE, Shim SH, Lee WS, Kim NK. Association study of microRNA polymorphisms with risk of idiopathic recurrent spontaneous abortion in Korean women. Gene 2012; 494:168–173. [DOI] [PubMed] [Google Scholar]
- 60. Pourteimoor V, Paryan M, Mohammadi-Yeganeh S. microRNA as a systemic intervention in the specific breast cancer subtypes with C-MYC impacts; introducing subtype-based appraisal tool. Journal of Cellular Physiology 2018; 233:5655–5669. [DOI] [PubMed] [Google Scholar]
- 61. Gao Q, Yao X, Zheng J. MiR-323 inhibits prostate cancer vascularization through adiponectin receptor. Cell Physiology and Biochemistry 2015; 36:1491–1498. [DOI] [PubMed] [Google Scholar]
- 62. Bian X, Shen Y, Zhang G, Gu C, Cai Y, Wang C, Zhu Y, Zhang H, Dai B, Ye D. Expression of dicer and its related miRNAs in the progression of prostate cancer. PLoS One 2015; 10:e0120159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Conte FA, Grumbach MM. Disorders of sex determination and differentiation. In: Gardner DG, Shoback DM, Greenspan FS (eds.), Greenspan's basic & clinical endocrinology. New York: McGraw-Hill Medical; 2007: 479–572. [Google Scholar]
- 64. Grumbach MM, Gluckman PD. The human fetal hypothalamus and pituitary gland: the maturation of neuroendocrine mechanisms controlling the secretion of pituitary growth hormone, prolactin, gonadotropins, adrenocorticotropin-related peptides, and thyrotropin. In: Tulchinsky D, Little AB (eds.), Maternal fetal endocrinology, 2nd ed. Philadelphia: Saunders; 1994: 193–262. [Google Scholar]
- 65. Dai R, Ahmed SA. Sexual dimorphism of miRNA expression: a new perspective in understanding sex bias of autoimmune diseases. Therapeutics and Clinical Risk Management 2014; 10:151–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Eloundou SN, Lee J, Wu D, Lei J, Feller MC, Ozen M, Zhu Y, Hwang M, Jia B, Zie H, Clemens JL, McLane MW et al. Placental malperfusion in response to intrauterine inflammation and its connection to fetal sequelae. PLoS One 2019; 14:e0214951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Ege MJ, Bieli C, Frei R, van Strien RT, Riedler J, Ublagger E, Schram-Bijkerk D, Brunekreef B, van Hage M, Scheynius A, Pershagen G, Benz MR et al. Prenatal farm exposure is related to the expression of receptors of the innate immunity and to atopic sensitization in school-age children. J Allergy Clin Immunol 2006; 117:817–823. [DOI] [PubMed] [Google Scholar]
- 68. Schaub B, Liu J, Hoppler S, Schleich I, Huehn J, Olek S, Wieczorek G, Illi S, von Mutius E. Maternal farm exposure modulates neonatal immune mechanisms through regulatory T cells. J Allergy Clin Immunol 2009; 123:774–782.e775. [DOI] [PubMed] [Google Scholar]
- 69. Chen T, Liu HX, Yan HY, Wu DM, Ping J. Developmental origins of inflammatory and immune diseases. Mol Hum Reprod 2016; 22:858–865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Huizink AC, Mulder EJ, Buitelaar JK. Prenatal stress and risk for psychopathology: specific effects or induction of general susceptibility? Psychol Bull 2004; 130:115–142. [DOI] [PubMed] [Google Scholar]
- 71. Fowden AL, Giussani DA, Forhead AJ. Intrauterine programming of physiological systems: causes and consequences. Physiology (Bethesda) 2006; 21:29–37. [DOI] [PubMed] [Google Scholar]
- 72. Barker DJ, Eriksson JG, Forsen T, Osmond C. Fetal origins of adult disease: strength of effects and biological basis. Int J Epidemiol 2002; 31:1235–1239. [DOI] [PubMed] [Google Scholar]
- 73. Barker DJ, Osmond C, Law CM. The intrauterine and early postnatal origins of cardiovascular disease and chronic bronchitis. J Epidemiol Community Health 1989; 43:237–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Barker DJ, Winter PD, Osmond C, Margetts B, Simmonds SJ. Weight in infancy and death from ischaemic heart disease. Lancet 1989; 2:577–580. [DOI] [PubMed] [Google Scholar]
- 75. Barker DJ, Hales CN, Fall CH, Osmond C, Phipps K, Clark PM. Type 2 (non-insulin-dependent) diabetes mellitus hypertension and hyperlipidaemia (syndrome X) relation to reduced fetal growth. Diabetologia 1993; 36:62–67. [DOI] [PubMed] [Google Scholar]
- 76. Vaughan OR, Rosario FJ, Powell TL, Jansson T. Regulation of placental amino acid transport and Fetal growth. Prog Mol Biol Transl Sci 2017; 145:217–251. [DOI] [PubMed] [Google Scholar]
- 77. Muralimanoharan S, Guo C, Myatt L, Maloyan A. Sexual dimorphism in miR-210 expression and mitochondrial dysfunction in the placenta with maternal obesity. International Journal of Obesity 2015; 39:1274–1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Li H, Ouyang Y, Sadovsky E, Parks WT, Chu T, Sadovsky Y. Unique microRNA signals in plasma exosomes from pregnancies complicated by preeclampsia. Hypertension 2020; 75:762–771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Srinivasan S, Treacy R, Herrero T, Olsen R, Leonardo TR, Zhang X, DeHoff P, To C, Poling LG, Fernando A, Leon-Garcia S, Knepper K et al. Discovery and verification of extracellular miRNA biomarkers for non-invasive prediction of pre-eclampsia in asymptomatic women. Cell Rep Med 2020; 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Gillet V, Ouellet A, Stepanov Y, Rodosthenous RS, Croft EK, Brennan K, Abdelouahab N, Baccarelli A, Takser L. miRNA profiles in extracellular vesicles from serum early in pregnancies complicated by gestational diabetes mellitus. J Clin Endocrinol Metab 2019; 104:5157–5169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Sufriyana H, Wu YW, Su EC. Prediction of preeclampsia and intrauterine growth restriction: development of machine learning models on a prospective cohort. JMIR Medical Informatics 2020; 8:e15411. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.