Abstract
Tumour‐associated angiogenesis plays a critical role in metastasis, the main cause of malignancy‐related death. Extracellular vesicles (EVs) can regulate angiogenesis to participate in tumour metastasis. Our previous study showed that EVs rich in HAX1 are associated with in metastasis of nasopharyngeal carcinoma (NPC). However, the mechanism by which HAX1 of EVs promotes metastasis and angiogenesis is unclear. In this study, we demonstrated that EVs rich in HAX1 promote angiogenesis phenotype by activating the FAK pathway in endothelial cells (ECs) by increasing expression level of ITGB6. The expression level of HAX1 is markedly correlated with microvessel density (MVDs) in NPC and head and neck cancers based on an analysis of IHC. In addition to a series of in vitro cellular analyses, in vivo models revealed that HAX1 was correlated with migration and blood vessel formation of ECs, and metastasis of NPC. Using ribosome profiling, we found that HAX1 regulates the FAK pathway to influence microvessel formation and promote NPC metastasis by enhancing the translation efficiency of ITGB6. Our findings demonstrate that HAX1 can be used as an important biomarker for NPC metastasis, providing a novel basis for antiangiogenesis therapy in clinical settings.
Keywords: angiogenesis, extracellular vesicles, FAK pathway, HAX1, ITGB6, nasopharyngeal carcinoma, translation
1. INTRODUCTION
Nasopharyngeal carcinoma (NPC), an epithelial carcinoma arising from the nasopharyngeal mucosal lining, is the most common malignancy among head and neck squamous cell carcinomas (HNSCCs) and is highly invasive and metastatic (Zhu et al., 2021). It is common in southern China and Southeast Asia (Chen et al., 2019). Early detection and timely therapy can help patients achieve a better prognosis (Agulnik et al., 2017). However, tumour metastases typically occur in the terminal stage (Li et al., 2015; Zhang et al., 2021), and the benefits of treatment for advanced patients are usually limited.
Angiogenesis, the process of new vessel formation, is an original hallmark of cancer (Hanahan & Weinberg, 2011). This process requires ‘tip’ cells and ‘stalk’ cells, which follow tip cells and proliferate to provide new ECs for growing sprouts (Kim et al., 2019; Watson et al., 2016). Angiogenesis is regulated by a balance between angiogenic and angiostatic factors and is involved in cancer (Carmeliet & Jain, 2000; Pralhad et al., 2003). In 1971, the late Dr. Judah Folkman first proposed the concept that tumour growth and metastasis require pathological angiogenesis (Carmeliet & Jain, 2000). Subsequent studies have shown that tumour‐associated vessels transport oxygen and nutrients and produce factors that can support the cancer microenvironment thus promoting tumour growth, progression and metastasis (Stuelten et al., 2018). Currently, vascular endothelial growth factor A (VEGFA) is considered the most predominant angiogenic factor, which initiates this process by engaging VEGF receptor 2 (VEGFR2) (Stratman et al., 2020; Wu et al., 2017). Therefore, VEGF is the most important therapeutic target for cancer angiogenesis (Lin et al., 2017). However, the currently available antiangiogenic therapies for malignant solid tumours are disappointing (Zhou et al., 2020). Therefore, understanding a distinct mechanism is an urgent requirement.
HS1‐related protein X‐1 (HAX1) is a 35 kDa ubiquitously expressed protein that is mainly localised in the mitochondria, but can also be found in the endoplasmic reticulum and nuclear membrane (Jeyaraju et al., 2009; Skokowa et al., 2012). HAX1 is involved in various important physiological processes including the regulation of apoptosis, migration, endocytosis and calcium homeostasis (Fadeel & Grzybowska, 2009). HAX1 plays complex biological roles by directly interacting with various proteins as well as the 3′untranslated region (3′‐UTR) of mRNA (Simmen, 2011). HAX‐1 expression is significantly increased several tumours including esophageal squamous cell carcinoma, colorectal cancer, oral squamous cell carcinoma, lung cancer, lymphoma, melanoma, leukaemia, myeloma, breast cancer and hepatoma (Farajifard et al., 2018; Karapınar et al., 2017; Liang et al., 2020; Sheng & Ni, 2015; Trebinska‐Stryjewska et al., 2019; Wu et al., 2017). HAX1 overexpression is also implicated in the poor prognosis of patients with tumours. Our previous studies revealed that EVs rich in HAX1 affect the prognosis of NPC by promoting vessel formation (You et al., 2016). However, the molecular mechanism by which HAX1 promotes angiogenesis in NPC has not been elucidated. A study on the complex function of HAX has indicated that it can act as a post‐transcriptional regulator by interacting with the 3′ UTR of a specific mRNA (Simmen, 2011). However, the biological functions and effects of this binding remain unclear. Therefore, it is necessary to explore the possibility that HAX1 promotes angiogenesis by interacting with the 3′ UTR of vasculogenic mRNA.
The 3′‐UTR is considered to be involved in regulating RNA transcription (Zhao et al., 2017). However, Solomon et al. found that the 3‐UTR can also be a target for regulating protein translation (Backlund et al., 2009; Solomon et al., 2017). Translation deregulation is an important hallmark of most cancers (Fagan et al., 2017; Howard et al., 2019). Regulation of translation is largely dependent on the eukaryotic initiation factor 4F (eukaryotic initiation factor 4F, eIF4F) complex, which scans the 5′ end of mRNA to recognise the initiation codon (Ho & Lee, 2016). Many oncogenic signalling pathways are known to modulate protein synthesis (Fagan et al., 2017; Howard et al., 2019). For example, the PI3K/AKT/mTOR pathway affects eIF4F complex assembly to promote cap‐dependent mRNA translation (Gandin et al., 2016; Guo et al., 2020; Thoreen, 2013). eIF4F is a heterotrimeric complex comprising the 5 cap binding protein eIF4E, the scaffold protein eIF4G, and an ATP‐dependent helicase eIF4A, and its function is modulated by eIF4E‐binding proteins (4E‐BPs) (Boussemart et al., 2014). Previously, scientists studied the effect of uncontrolled protein synthesis in the tumour itself. However, the effect of tumour exosomes on protein synthesis in ECs remains to be elucidated.
EVs are membrane‐enclosed vesicles that are released by prokaryotic and eukaryotic cells, including exosomes, ectosomes, microvesicles, microparticles, apoptotic bodies and other EV subsets (Adamiak et al., 2018). Currently, EVs play an indispensable role in intercellular communication by carrying a rich cargo, including packaged proteins, DNA, RNA and lipids (Madeo et al., 2018). Increasing evidence indicates that EVs are involved in the progression of various human disorders through several mechanisms, including tumour metastasis, recurrence and drug resistance (Geng et al., 2020; Simeoli et al., 2017). In tumour microenvironment studies, EVs derived from cancer cells have been implicated in premetastatic niche formation by accelerating angiogenesis (Geng et al., 2020; Wu et al., 2019). In our previous study, we found that EVs secreted by NPC promote angiogenesis and tumour metastasis (You et al., 2016). However, many important details of this mechanism are yet unclear.
In the present study, we explored the molecular mechanisms by which EVs HAX1 affects angiogenesis in NPC. We confirmed the role of HAX1 playing in angiogenesis by animal and cell models. Furthermore, using ribosome profiling, we identified the downstream protein modulated though translation by HAX1, and test it by rescue experiments.
2. MATERIALS AND METHODS
2.1. Human NPC specimens
With approval from the ethics committee, serum samples and tumour tissues were collected from 51 patients with pathologically confirmed NPC at the Affiliated Hospital of Nantong University. Blood samples were collected in 10 ml Vacutech Vacuum Blood Collection Tubes K2 EDTA purchased from Zhejiang Kangshi Medical Equipment Co., Ltd. (Zhejiang, China) and stored at 4°C. And the volume of serum samples collected is 5 ml per person. Tumour tissues samples were stored at –80°C prior to use. All included patients provided informed consent and did not receive any cancer treatment before the biopsy. Immunohistochemistry was performed to detect HAX1 and CD31 expression. Kaplan–Meier analysis was used to evaluate the prognostic significance of HAX1.
2.2. Cell culture
The human NPC cell lines cells were cultured in RPMI 1640 (Biological Industries Israel Beit‐Haemek, 01‐100‐1ACS) containing 10% FBS (Biological Industries Israel Beit‐Haemek, 1304‐001‐1ACS), and HUVECs cells were cultured in DMEM/F‐12(HAM)1:1 (Biological Industries Israel Beit‐Haemek, 01‐19 72‐1ACS) containing 10% FBS. Cells were grown at 37°C with 5% CO2.
2.3. Immunohistochemistry (IHC)
IHC for HAX1 and CD31 were performed as previously described. The staining intensity score was divided into four grades: 1 (negative), 2 (weakly positive), 3 (moderately positive) and 4 (strongly positive) and the staining area score was divided into :1 (0%–25%), 2 (26%–50%), 3 (51%–75%) and 4 (>75%). the final staining score was defined as the product of the two scores, with scores of 0–8 defined as HAX1 low expression and scores of 9–16 defined as HAX1 high expression.
2.4. Transfection with plasmids and lentiviral vectors
All transfection experiments were performed as described previously. Plasmids, lentiviral vectors and their negative controls were constructed and produced by GeneChem (Shanghai, China). The short hairpin RNA (shRNA) sequences are listed below:
shHAX1: ccAGAGGCCATTTCATAGGTT
shITGB6: gcCTCCAAACATTCCCATGAT
HUVECs and CNE2 cells were transfected with LV‐HAX1‐GFP or LV‐ITGB6 (GV218; GeneChem) for 12 h.
2.5. Zebrafish experiments
Zebrafish were kept at 26.5°C and bred under standard conditions. Tg(fli:egfp) and Tg(LR57:GFP) transgenic zebrafish lines were used as previously described (Krueger et al., 2011). Morpholino antisense oligomers (MOs; Gene Tools) were prepared at a stock concentration of 1 mM according to the manufacturer's protocol. The MOs or EVs were injected into the yolk of one‐cell‐stage embryos. After 24 h, zebrafish were anesthetised with 0.16 mg/ml tricaine and embedded in 1% agarose. Using a 25× objective, confocal stack images of the trunk and head region were obtained in time intervals of 20 min. For zebrafish tumour models, 300 tumour cells in 5 nl RPMI 1640 medium stained with 2 g/ml Dil (Beyotime, C1036) were injected into the common cardinal of zebrafish embryos at 48 h post fertilisation.
2.6. EVs isolation and uptake
EVs were collected essentially as previously described (Théry et al., 2018). To isolate EVs from cell culture supernatants, the culture supernatant of CNE2 cells was subjected to differential centrifugation at 500 × g for 10 min, 3000 × g for 60 min, and 10,000 × g for 60 min at 4°C. After filtering through a 0.22 m filter, the supernatant was transferred to an ultracentrifuge tube. Next, the supernatant was centrifuged at 100,000 × g (XPN‐100, Beckman Coulter) for 90 min at 4°C. CNE2 cells were cultured in RPMI 1640 (Biological Industries Israel Beit‐Haemek, 01‐100‐1ACS) without FBS for 24 h prior to collect the culture supernatant. The isolated EVs were resuspended in phosphate‐buffered saline (PBS). To detect the uptake of EVs by recipient cells, we used a PKH‐26 labeling kit (Sigma–Aldrich, MINI26‐1KT) to label EVs and then cocultured them with HUVECs for 2 h. Finally, the HUVECs ‐nuclei were stained with Hoechst. Images were acquired using a confocal microscope (Axio Observer, Zeiss, Göttingen, Germany,). Further, to detect the uptake of HAX1 of EVs by HUVECs, we used an upper and lower chamber cocultivation system. After 48 h, the HUVECs in the lower cavity were stained with Hoechst stain and imaged.
2.7. BALB/c nude mice models
To assess the role of HAX1 in NPC lung metastasis in vivo, CNE2 cells were transfected with lentivirus. Next, 2 × 106 luciferase‐labeled CNE2 cells in 200 µl RPMI 1640 medium were injected into the tail veins of 5‐to 6‐week‐old male nude mice. To confirm successful injection, the photon flux from the whole body of mice was measured weekly using IVIS Lumina Series III (Caliper Life Sciences, Mountain View, CA, USA). Over the course of 6 weeks, bioluminescence imaging (BLI) analysis of each mouse was performed to monitor lung metastasis. To identify the mechanism by which HAX1 is involved in metastasis, HAX1‐overexpressing CNE2 cells were transfected with or without lentiviral shITGB6 or were treated with PF573228. The resultant cells were then injected into the tail veins of nude mice at 2 × 106 cancer cells per mouse. BLI was then performed after 6 weeks. All BALB/c mice were obtained from the Laboratory Animal Center of Nantong University
2.8. Transmission electron microscopy
First, 20 µl of fresh EVs were dried on a copper grid for 15 min. The samples were then fixed in 2.5% glutaraldehyde for 1 h and stained with 3% phosphotungstic acid for 1 min. The EVs were then immediately observed using a transmission electron microscope (JEOL Ltd., Tokyo, Japan, JEM‐1230).
2.9. Western blot assays
Immunoblotting was performed using whole‐cell lysates. Protein content was quantified using a BCA kit (Thermo Fisher Scientific, 23327). Protein samples were separated using polyacrylamide gel electrophoresis and transferred to nitrocellulose membranes. After blocking with blocking buffer for 1 h, the cells were incubated with the appropriate antibodies (indicated in the table below) at 4°C overnight.
2.10. Transwell assay
Migration assays were performed using Transwell inserts (Corning, 3422) with a pore size of 8 µm. In total, 5 × 104 cells suspended in serum‐free medium were added to the upper chambers, and medium containing 10% fetal bovine serum was added to the lower chambers. After 16 h of incubation, the cells attached to the upper side were removed, and the cells attached to the underside of the membrane were fixed and stained with crystal violet. Digital images were obtained from the membranes, and five random fields were counted per chamber. Furthermore, we measured the number of cell by ImageJ.
2.11. Tube formation assay
Tube formation assays were performed according to a published protocol. Fifty microlitres BD Matrigel (Corning, 354234) was coated onto each well of a 96‐well plate. Then, the plate was incubated at 37°C for 30 min. Next, HUVECs (2 × 104 cells) were seeded into the wells of the plate and incubated for 6 h. Images were taken with a microscope (Zeiss, Göttingen, German, Axio Obse) at ×100 magnification. The tube formation ability was quantified using Image J (Wayne Rasband, National Institute of Health, USA) and Angiogenesis Analyzer plugin (Gilles Carpentier, Universite Paris Est Creteil Val de Marne, France). Each experiment was performed in triplicate.
2.12. Chorioallantoic membrane assay
Chicken chorioallantoic membrane (CAM) assays were performed according to a published protocol with slight modifications. Fertilised chicken eggs were cleaned with 70% ethanol and incubated at 37–38°C under 50%–60% humidity. After 3 days, a window (10 × 10 mm) was cut in the shell and a sterilised gelatin sponge treated with EVs was placed on the chorioallantoic membrane. After 10 days, the vessels surrounding each sponge were counted using a microscope.
2.13. Immunoprecipitation and mass spectrometry
Preparation of cell extracts followed by coimmunoprecipitation (coIP) was performed using the Co‐Immunoprecipitation Kit (Thermo Scientific, 14321D), according to manufacturer's instructions. Briefly, for each IP reaction, 1–1.5 mg of cell extracts and 15 µg of rabbit HAX1 antibody or 15 µg of mouse eIF4E antibody were combined in a final volume of 500 µl of the IP buffer and incubated overnight at 4°C, followed by four rapid washes with coIP buffer. Finally, proteins were eluted using elution buffer. The IP samples are used to perform Western blot ting and LC‐MS/MS experiments
LC‐MS/MS experiments were performed on a Q Exactive mass spectrometer that was coupled to Easy nLC (Thermo Scientific). The MS data was analysed using MaxQuant software version 1.6.0.16. MS data was searched against the UniProtKB Rattus norvegicus database. The database search results were filtered and exported with <1% false discovery rate (FDR) at peptide‐spectrum‐matched level, and protein level, respectively. Finally, KEGG analysis was performed using KEGG Mapper.
2.14. Immunofluorescence assay
Cells were seeded in 15‐mm glass bottom cell culture dishes (Corning, 354469) at a density of 5 × 104 cells. After a day, the cells were washed three times with PBS and fixed with 4% paraformaldehyde for 20 min. The cells were then washed three times with PBS, and permeabilised with 0.2% Triton X‐100 (Beyotime, ST795) at room temperature for 10 min. After washing three times with PBS, the cells were incubated overnight at 4°C with the primary antibodies antieIF4E and antieIF4G antibodies. After washing three times with PBS, and incubating with a fluorescent dye‐labelled secondary antibody for 1 h in the dark, an antifluorescence attenuator containing Hoechst stain (Thermo Fisher Scientific, 62249) was then added followed by imaging.
2.15. Polysomal fractionation experiment
Sucrose density gradient centrifugation was performed to separate the subpolysomal and polysomal ribosome fractions. Fifteen minutes before collection, the cells were incubated with 100 µg/ml cycloheximide at 37°C. Next, the cells were washed, scraped into ice‐cold PBS supplemented with 100 µg/ml cycloheximide, and centrifuged at 3000 rpm for 5 min; next, 400 µl of LSB buffer (20 mM Tris, pH 7.4, 100 mM NaCl, 3 mM MgCl2, 0.5 M sucrose, 2% Triton X‐100, 1 mM DTT, 100 U/ml RNAsin, and 100 µg/ml cycloheximide) was added after homogenisation, and was supplemented with 0.2% Triton X‐100 and 0.25 M sucrose. The sample was centrifuged at 12,000 × g for 10 min at 4°C. The resulting supernatant was adjusted with 5 M NaCl and 1 M MgCl2. The lysate was then loaded onto a 10%–45% sucrose density gradient and centrifuged using an SW41 rotor at 38,000 rpm at 4°C for 2 h. A gradient grading system (Isco) was used to monitor and collect the polysomal fractions.
2.16. Translation efficiency analysis
HUVECs in the NC and shHAX1 groups were lysed following the vendor's recommended protocol (lc‐bio). Ribosome profiling, RNA sequencing, and preliminary data analysis were performed by lc‐bio Co. Translation efficiency was calculated by comparing transcriptome and ribosome profiling data. Genes with a log2(FC) greater than 1 (p < 0.05) were considered significant, allowing us to identify the groups of genes regulated only at the translational or transcriptional levels. Genes that were downregulated at the translational level under HAX1 knockdown conditions were further studied in subsequent experiments.
2.17. Statistical analysis
Calculations were performed using GraphPad Prism Software. Results are presented as means ± S.D. of at least three independent experiments unless otherwise specified. Analysis of correlation was performed in R using the Pearson correlation test. Kaplan–Meier curves were used to analyse survival. Datasets with Gaussian distributions were compared using One‐way ANOVA and two‐tailed Student's t‐tests. p < 0.05 was considered statistically significant.
Antibodies used for Western blot and immunohistochemistry
| Antibodies | Manufacturer | Catalogue numbers | Dilution (WB) | Dilution (IHC) | Dilution (IF) |
| HAX1 | Abclonal | A5551 | 1:500 | 1:200 | |
| ACTB | Proteintech | 20536‐1‐AP | 1:1000 | ||
| eIF4A1 | Abcam | Ab31217 | 1:1000 | ||
| eIF4E | Sangon Biotech | sc‐271480 | 1:500 | 1:100 | |
| 4EBP1 | Abclonal | A19045 | 1:500 | ||
| p‐ 4EBP1 (T70) | Proteintech | A0031 | 1:500 | ||
| p‐ 4EBP1 (S65) | Proteintech | AP0032 | 1:500 | ||
| eIF4G | Proteintech | 15704‐1‐AP | 1:100 | ||
| ITGB6 | Abclonal | A16904 | 1:500 | ||
| FAK | Abclonal | A11195 | 1:1000 | ||
| p‐FAK (Tyr397) | Cell Signaling Technology | 8556 | 1:1000 |
3. RESULTS
3.1. HAX1 upregulation promotes NPC metastasis and angiogenesis
To examine the role of HAX1 on NPC progression, we analysed the expression of HAX1 in 51 NPC tissues by immunohistochemistry (IHC) (Figure 1a). High expression of HAX1 was associated with the late clinical stage (Figure 1b). Further, data from 521 patients with HNSCC from The Cancer Genome Atlas (TCGA) showed that patients with high HAX1 expression had a significantly poorer prognosis (Figure 1c, p = 0.011). Next, we calculated the microvessel densities (MVDs) in NPC tissues using CD31 IHC. Patients in the late clinical stage showed high MVD in the NPC tissues (Figure 1d,e). Furthermore, MVDs and HAX1 expression were positively correlated (Figure 1f,g).
FIGURE 1.
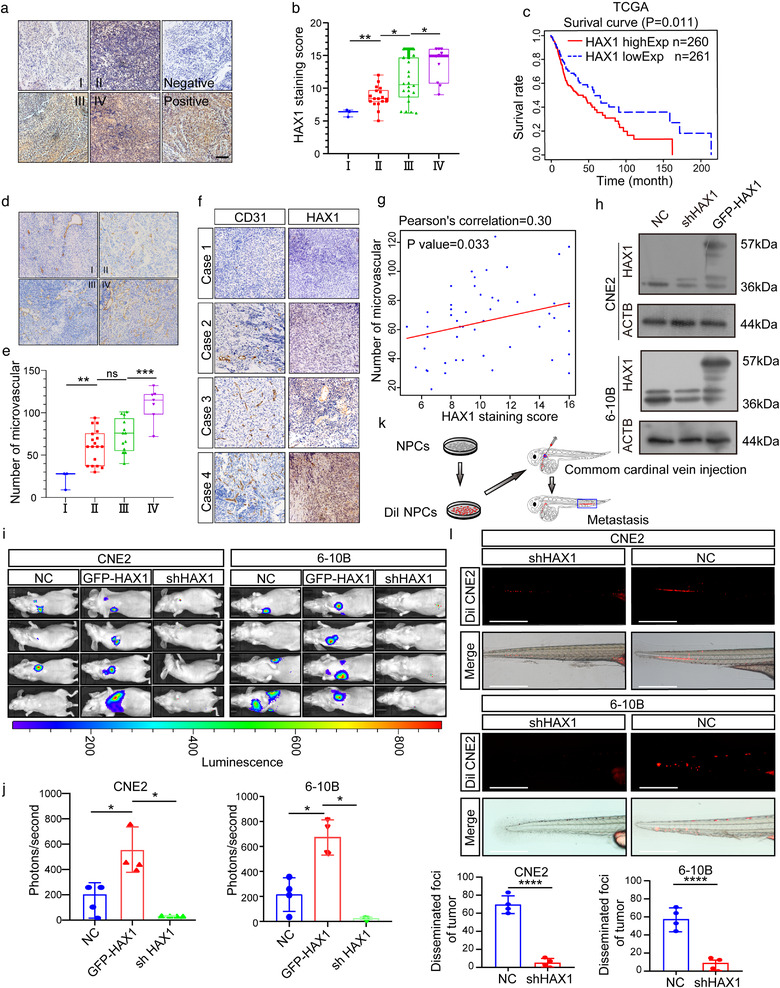
HAX1 overexpression is related to NPC metastasis and angiogenesis. (a) Representative immunohistochemical analysis (IHC) images of HAX1 staining in tumour tissues of different clinical grades from patients with NPC, bar: 100 µm. (b) Statistical comparison of HAX1 scores in different clinical grades. (*p < 0.05, **p < 0.01, one‐way ANOVA). (c) Kaplan–Meier curves showing the correlation between HAX1 expression and overall survival by the log‐rank test (Data from TCGA‐HNSCC). (d) Tumours were analysed by IHC for microvessel densities (MVDs), bar: 100 µm. (e) Statistical comparison of number of MVDs in different clinical grades (**p < 0.01, ***p < 0.001, one‐way ANOVA). (f) Representative IHC images of MVDs and HAX1 staining. (g) Spearman correlation between HAX1 score and MVDs in NPC. Pearson correlation coefficient (r2) and p‐values are shown. (h) Up: the HAX1 levels of CNE2‐NC, CNE2‐shHAX1 and CNE2‐GFP‐HAX1 were measured by Western blotting. Down: The HAX1 levels of 6‐10B‐NC, 6‐10B‐shHAX1 and 6‐10B‐GFP‐HAX1 were measured by Western blotting. (i) Visualisation of lung metastasis after intravenous injection of transfected NPC cells in BALB/c mice. (j) Mean fluorescence intensity (*p < 0.05, one‐way ANOVA). (k) Schematic diagram of common cardinal vein injection in zebrafish. (l) NPC cells (red) were injected into the common cardinal vein of zebrafish at 48 hpf, and photographed using a confocal microscope (representative of three embryos per group, bar: 200 µm (m) Quantification of migratory cell numbers. (****p < 0.0001, Student's t‐test)
To further investigate the correlation of HAX1 and NPC metastasis, we examined the expression of HAX1 in six NPC cell lines and a normal nasopharyngeal epithelial cell line. The highest expressing cells (CNE2) and a cell line with low to moderate expression levels (6‐10B) were selected for further study (Data not shown). Zebrafish and BALB/c mouse models were used to further determine the relationship between HAX1 expression and NPC metastasis. First, CNE2 cells and 6‐10B cells expressing shHAX1 or GFP‐HAX1 were generated using lentiviral transduction (Figure 1h). We then injected luciferase‐labelled transduced CNE2 cells and 6‐10B cells into nude mice via tail vein injection, and monitored their tumour growth by measuring the resulting bioluminescence (BLI). We found that HAX1 knockdown significantly inhibited the metastatic ability of CNE2 cells and 6‐10B cells (Figure 1i,j).
We then established a zebrafish tumour model for NPC metastasis research. We stained CNE2 cells with DIL and injected them into the common cardinal vein of zebrafish (Figure 1k). On the third day after the injection, we found that the number of metastatic tumour cells in the zebrafish tail was lower in the shHAX1 group compared to those in the NC group (Figure 1l,m). Overall, HAX1 expression in NPC was found to affect NPC progression and angiogenesis.
3.2. EVs rich in HAX1 promote angiogenesis in NPC
To determine the role of HAX1 in promoting angiogenesis, we transfected NPC cells with shHAX1 lentivirus or the vector virus, cocultured them with HUVECs, and then proceeded to subsequent experiments in vitro (Figure 2a). Compared with the NC group, formation of vessel structures and migration capacity of HUVECs cocultured with CNE2‐shHAX1 and 6‐10B‐shHAX1 was substantially attenuated (Figure 2b–g). Tumours can promote angiogenesis by secreting proangiogenic factors directly or by transferring proteins via EVs. To identify the mode of angiogenesis induced by NPC, GW4869, an inhibitor of neutral sphingomyelinase that effectively reduces EVs release (Catalano & O'driscoll, 2020), was used in the coculture system. GW4869 treatment significantly inhibited the ability of CNE2‐NC to promote angiogenesis (Figure 2h–j). These findings, we investigated whether HAX1 could be taken up by HUVECs in the form of EVs. We thus transduced CNE2 cells with GFP‐HAX1 and separated the EVs from CNE2 cell culture medium. These EVs were characterised by electron microscopy and marker expression (Figure 2k,l). We cocultured HUVECs with isolated EVs for 24 h, and found green fluorescence in HUVECs by confocal microscopy, indicating that HAX1 could be taken up by HUVECs from CNE2 cells (Figure 2m).
FIGURE 2.
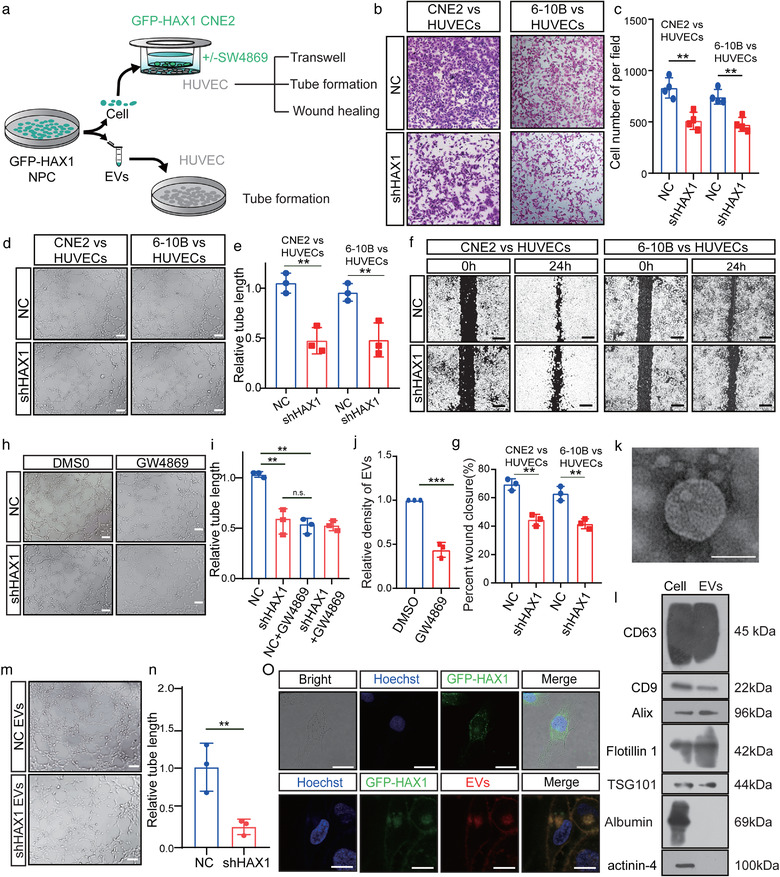
HAX1‐rich EVs promote angiogenesis in vitro. (a) Schematic diagram of HUVECs processing. (b–g) Co‐culture of HUVECs with NPC‐NC or NPC‐shHAX1 (b) The migration potential of HUVECs was measured using the Transwell assay, bar: 100 µm. (c) Statistical comparison of the cell number. (**p < 0.01, Student's t‐test). (d, e) Tube formation assays were performed to measure the tube forming ability of HUVECs, bar: 100 µm. (**p < 0.01, Student's t‐test). (f, g) The migration potential of HUVECs was measured using the wound closure assay, bar 200 µm. (**p < 0.01, Student's t‐test). (h, i) After 4 h of treatment with DMSO or GW4869, the tube forming ability of HUVECs co‐cultured with two groups of CNE2 was measured. (n.s. p > 0.05 and **p < 0.01, Student's t‐test). (j) CNE2 cells treated with DMSO or GW4869 for 4 h, and measured the density of EVs in CNE2 cells supernatants (k) Representative transmission electron micrograph of EVs derived from CNE2 cells, bar 100 nm. (l) Western blotting analysis for CD63, CD9, Alix, Flotillin 1, TSG101, Albumin and actinin‐4 in CNE2 cells and CNE2 cells EVs. (m, n) Tube formation assays were performed to measure the tube forming ability of HUVECs treated with two groups of EVs, bar: 100 µm. (**p < 0.01, Student's t‐test). (o) Top, HUVECs were cocultured with CNE2 cells transfected with fluorescent GFP‐HAX1 lentivirus in a chamber. Down, uptake of EVs released by CNE2 cells in HUVECs. Images were obtained by confocal microscopy, blue: Hoechst staining; red: PKH26‐labelled EVs; green: GFP‐HAX1, bar: 25 µm
Therefore, we investigated whether HAX1 of EVs affected angiogenesis and metastasis in subsequent studies. We collected the cell culture supernatants of CNE2‐NC and CNE2‐shHAX1 cells at 48 h and isolated the respective EVs. Compared to the coculture with NC‐EVs, HUVECs cocultured with shHAX1‐EVs showed inhibition of tubule formation (Figure 2i). Next, we injected these two groups of EVs into zebrafish embryos or placed them on a chick chorioallantoic membrane (CAM). As observed in the in vitro experiments, shHAX1‐EVs significantly inhibited the sprouting of CAM and zebrafish embryonic vessels (Figure 3a–c). Angiogenic sprouting behaviour was not affected initially (Figure 3c, 30 and 35 hpf). However, at later stages, compared with PBS and shHAX1‐EVs, the intersegmental vessels of zebrafish embryos treated with NC‐EVs formed more vascular sprouts. (Figure 3c, 40 hpf). Notably, that new vessels may provide ducts for tumour metastasis. Therefore, to determine whether HAX1 of EVs can promote metastasis, we injected luciferase‐labelled CNE2 cells and 6–10B cells into the tail vein of nude mice, and then injected NC‐EVs, shHAX1‐EVs or GFP‐HAX1‐EVs into the mice, once every 3 days. We found that compared with the NC‐EVs group, the shHAX1‐EVs group had fewer distant metastases, whereas the GFP‐HAX1‐EVs group had more distant metastases (Figure 3d–f). To determine whether the function of EVs depends on the number of EVs or the HAX1 content in EVs, we determined the abundance of EVs in the serum of patients with NPC (n = 170). We found no significant statistical correlation between the density of serum EVs and disease progression and patient prognosis (Table 1, Figure S1A–F).
FIGURE 3.
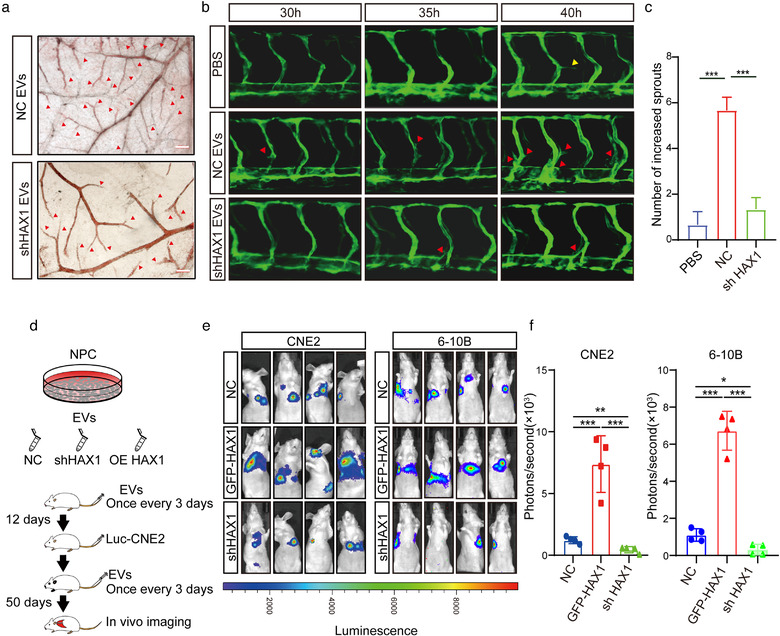
EVs rich in HAX1 promote angiogenesis in vivo.(a) The blood vessel formation ability treatment with EVs derived from CNE2‐NC or CNE2‐shHAX1 was measured using the CAM assay. Red arrows: blood vessel sprouts, bar 200 µm. (b, c) Injection of two groups of EVs affected the branching of zebrafish intersegmental vessels. Red arrows: sprouts of intersegmental vessel, bar: 200 µm. (c) The number of sprouts of intersegmental vessels at 40 hpf. (***p < 0.001, one‐way ANOVA). (d) Schematic diagram of tail vein injection protocol of EVs and CNE2 cells. (e, f) Visualization of lung metastasis in BALB/c mice. (*p < 0.05, ***p < 0.001, one‐way ANOVA)
TABLE 1.
Correlation of the serum EVs density with and disease progression and prognosis
| Factors | Density of EVs | p | |
|---|---|---|---|
| Low | High | ||
| Survival | Overall survival | 0.377 | |
| T stage | |||
| T1–T2 | 24 | 30 | 0.2429 |
| T3–T4 | 28 | 36 | |
| N stage | |||
| N0–N1 | 35 | 39 | 0.4207 |
| N2–N3 | 17 | 27 | |
| M stage | |||
| M0 | 50 | 64 | 0.5034 |
| M1 | 2 | 2 | |
| Grade | |||
| I‐II | 18 | 16 | 0.0937 |
| III‐IV | 42 | 42 | |
3.3. HAX1 affects angiogenesis both in vitro and vivo
To further identify the effect of HAX1 on angiogenesis, we knocked down HAX1 in HUVECs using lentiviral transduction (Figure 4a). In vitro experiments showed that HAX1 knockdown inhibited the migration and tube formation capability of HUVECs (Figure 4b–d). Next, we injected HUVECs mixed with Matrigel into the skin of nude mice. Compared with the NC group, Matrigel plugs in the shHAX1 group exhibited decreased angiogenesis (Figure 4e–g).
FIGURE 4.
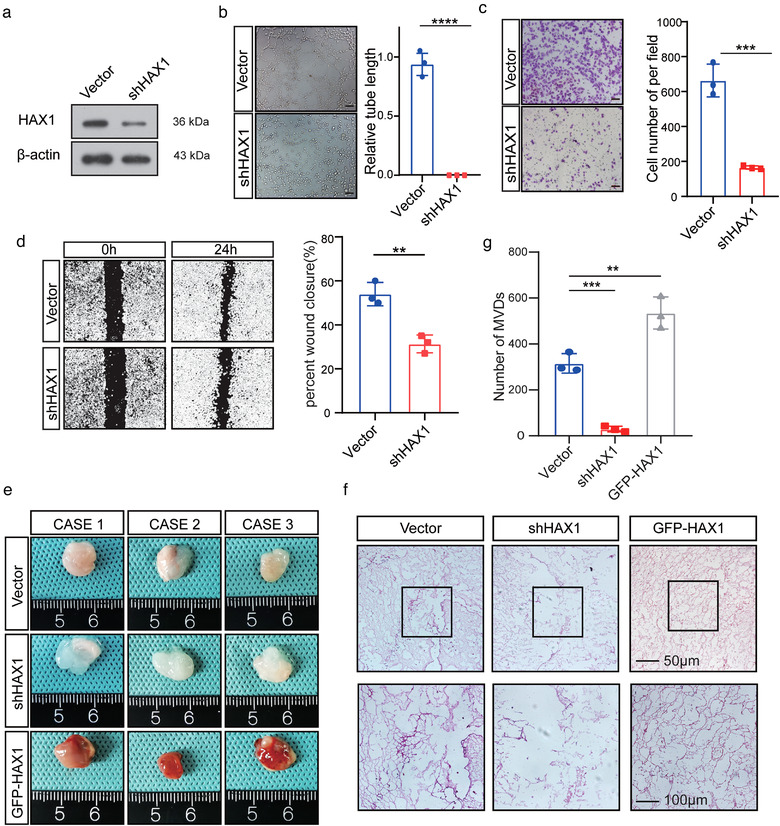
HAX1 expression affects the tube formation ability of HUVECs. (a) HUVECs transfected with control (CON) or shHAX‐1 were analysed by Western blotting. (b–d) HUVECs were transfected with the control (CON) or shHAX‐1 lentivirus, followed by angiogenesis‐related experiments. (b) Tube formation assays were performed to measure the tube forming ability of two groups of HUVECs, bar: 100µm. (****p < 0.0001, Student's t‐test). (c) The migration potential of HUVECs was measured using the Transwell assay, bar: 100 µm. (**p < 0.01, Student's t‐test) (d) The migration potential of HUVECs was measured using the wound closure assay, bar: 200 µm. (***p < 0.001, Student's t‐test). (e, f) HUVECs were mixed with Matrigel for subcutaneous injection. (e) Left, gross‐observation of HAX1‐modulated angiogenesis in Matrigel plugs. Right, H&E was performed to observe blood vessel formation. Scale bar in 200× images: 100 µm. Scale bar in 400× images: 50 µm. (f) Quantitative evaluation of angiogenesis based on vessel area (**p < 0.01, ***p < 0.001, one‐way ANOVA)
To study whether HAX1 expression is vessel‐specific, we analysed its expression in various tissues and cell lines using the Human Protein Atlas (HPA) database. Although HAX1 expression in the cardiovascular system and ECs was not prominent (Figure S2), HAX1 was highly expressed in some tissues and cells with strong regenerative abilities, such as the liver, rectum and spermatocytes (Figure S2). As the expression of HAX1 provided by the database was for the mature cardiovascular system, we conjecture that HAX1 expression in immature vessels may be high.
As zebrafish embryos are transparent, they provide a great advantage in studying vessel formation. In toto in situ hybridisation of zebrafish embryos showed that HAX1 was expressed in the brain, cardinal vein and developing vessels before 24 hpf. As the zebrafish matured, HAX1 expression in the vessels of zebrafish gradually decreased (Figure 5a). This result suggests that HAX1 may play an important role in angiogenesis.
FIGURE 5.
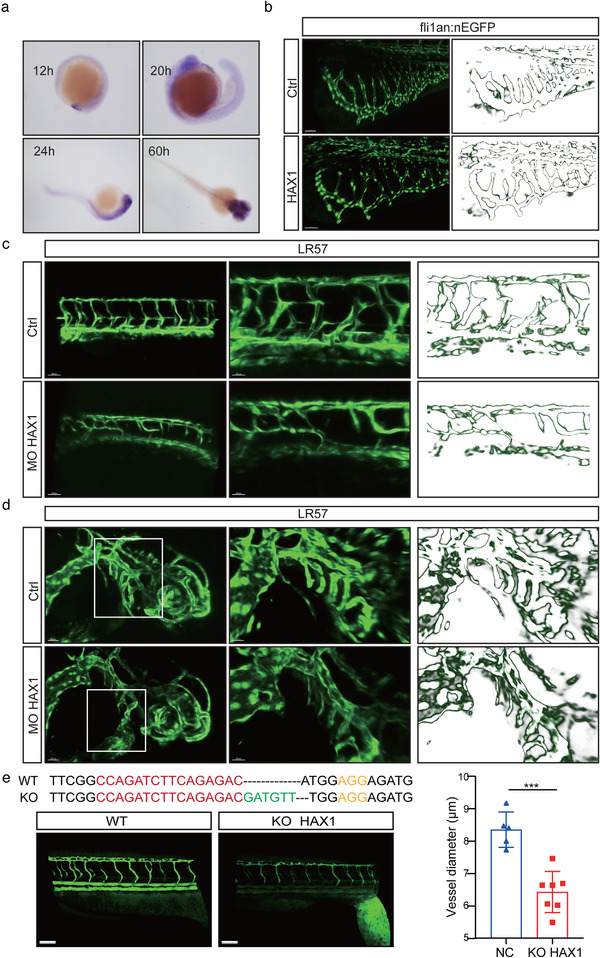
HAX1 regulates vascular morphogenesis in zebrafish. (a) In situ hybridization was performed on zebrafish embryos at 12 hpf, 20 hpf, 24 hpf and 60 hpf to observe the HAX1 level. (b) HAX1 mRNA was injected in the embryo. Left, representative pictures of the sub‐intestinal vessel using confocal microscopy. Right, stroke outlines of the sub‐intestinal vessel. (c, d) Reduced HAX1 level. Left, representative pictures of (c) the intersegmental vessel and (d) head vessel. Right, stroke outlines of the intersegmental vessel. (e) Top: CRISPR/Cas9 targeting sequences. Down, representative pictures of the intersegmental vessel, bar: 20 µm. (***p < 0.001, Student's t‐test)
To verify the role of HAX1 in vascular development in vivo, we performed experiments by injecting HAX1 mRNA or HAX1 ATG‐targeting morphants (Figure 5b–d). Compared with the Ctrl group, the HAX1 group showed excessive branching of subintestinal vessels (Figure 5b). For ATG‐blocking morphants, we observed a striking absence in the intersegmental and head vessel branching of HAX1 morphants (Figure 5c,d). Furthermore, as previously described (Torraca & Mostowy, 2018), we knocked out HAX1 in zebrafish using CRISPR/Cas9 (Figure 5e). Consistent with the effect of HAX1 morphants, vessel development was delayed after HAX1 knockout, and the intersegmental vessel diameter was significantly smaller than that in the wild type (Figure 5e).
3.4. HAX1 promotes protein translation in HUVECs
The cancer angiogenesis‐promoting role of HAX1 has been studied extensively, but the underlying mechanism has not been elucidated yet. Therefore, we performed whole‐transcriptome sequencing of wild‐type and HAX1‐KO zebrafish. HAX1 knockout resulted in 11,703 differentially expressed genes (Figure S3A). We then performed a pathway enrichment analysis of these differentially expressed genes (Figure S3B). Unfortunately, it was difficult to identify a molecular mechanism to explain the angiogenesis promotion. Next, we examined the correlation between HAX1 mRNA expression and that of angiogenesis‐related molecules in TCGA database (Figure S3C). The results showed no significant correlation between HAX1 and the expression of CD34, VEGFA, PDGFA, CD31, LYVE1 and PROX1, while HAX1 was negatively correlated with VEGFC and PDPN (Figure S3C). These results are inconsistent with the effects of HAX1 on angiogenesis.
We speculate that there may be other mechanisms that regulate angiogenesis, such as regulation of protein translation. Therefore, we measured the nascent protein synthesis levels. Compared to the NC group, these levels were significantly decreased in shHAX1 HUVECs (Figure 6a). Likewise, polysome analysis showed that HAX1 downregulation is not conducive to polysome formation (Figure 6b). The activity of eIF4F is considered a key determinant of translational efficiency. Thus, we examined the expression of eIF4F components, 4EBP1, and the phosphorylation level of 4EBP1. HAX1 knockdown did not change the expression of eIF4A1 and 4EBP1, whereas the expression of eIF4E and 4EBP1 phosphorylation level decreased significantly (Figure 6c,d). In our previous study, we confirmed that HAX1 can regulate the PI3K pathway, and the PI3K pathway can affect the phosphorylation of 4EBP1. This is consistent with our phosphorylation change of 4EBP1. However, as transcriptome sequencing shown in Figure S3A, HAX1 unable to affect the transcription level of eIF4E. Previous studies have shown that HAX1 can bind to many proteins to produce a variety of effects. We speculated that HAX1 may affect protein translation in the way of protein interaction. To test this conjecture, immunoprecipitation‐mass spectrometry (IP‐MS) was performed. Enrichment analysis of the identified proteins revealed that HAX1 is highly correlated with ribosome formation (Figure 6e). Mass spectrometry data revealed that HAX1 binds to various components of eIF4F. Among them eIF4E is predicted domains are present in combination with HAX1 (Figure 6f). The result of co‐IP between HAX1 and eIF4E is consistent with IP‐MS. (Figure 6g). Using immunofluorescence, we found that the binding between eIF4E and eIF4 decreased with HAX1 knockdown, indicating that the formation of eIF4F is reduced. (Figure 6h). 4EGI can suppress the formation of the 4F initiation complex by destroying the eIF4E/eIF4G interaction. To verify that the vascular phenotype was due to the reduction in translation, 4EGI was used in HAX1 overexpressing cell (Figure S4). Under the treatment of 4EGI, the ability of HAX1 to promote HUVEC migration and tube formation is significantly inhibited (Figure S4A–F).
FIGURE 6.
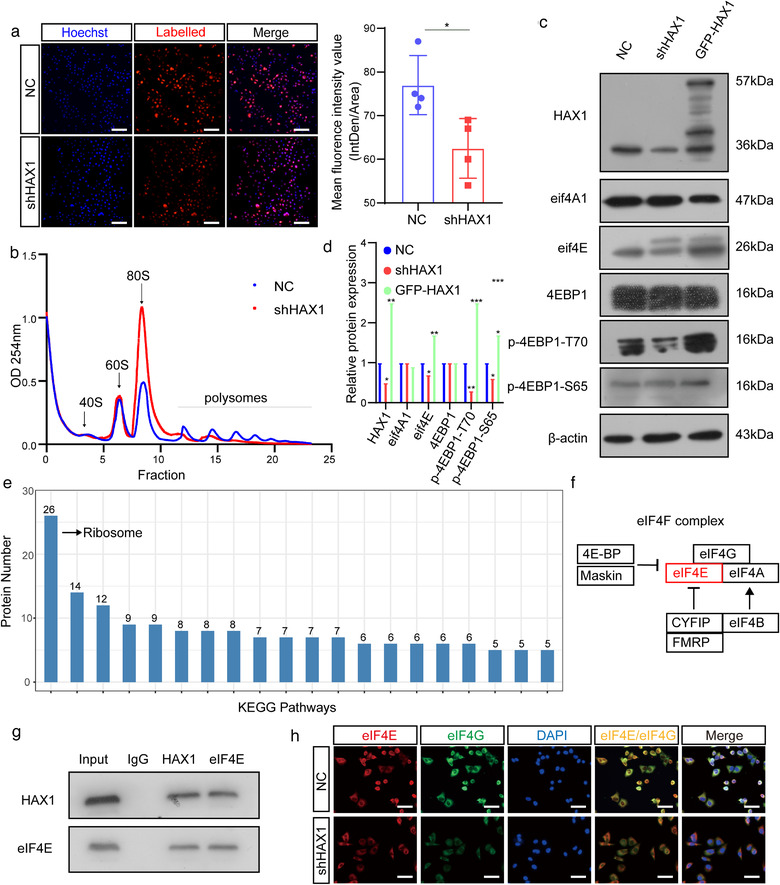
HAX1 promotes protein translation in HUVECs. (a) Analyses of nascent protein synthesis labeled with L‐Homopropargyl glycine incorporation, detected in HUVECs‐NC and HUVECs‐shHAX1 by fluorescence microscopy. Quantify nascent protein expression by the quantification of fluorescence intensity. Blue: nuclei; Red: nascent protein, bar: 100 µm. (*p < 0.05, Student's t‐test). (b) Polysome profiling of HUVECs‐NC and HUVECs‐shHAX1. Absorbance at 254 nm. (c, d) Western blot analysis of the protein levels in HUVECs‐NC and HUVECs‐shHAX1 (*p < 0.05, **p < 0.01, p < 0.001, Student's t‐test). (e) KEGG pathway enrichment analysis of the mass spectrometry results. (f) Schematic diagram of HAX1 affecting eIF4F. (g) Co‐immunoprecipitation analysis between HAX1 and eIF4E in HUVECs. (h) Immunofluorescence of eIF4G and eIF4E in HUVECs, bar: 50 µm
3.5. HAX1 regulates the translation of ITGB6 mRNA in HUVECs
Based on the results above, we hypothesised that HAX1 might first improve the translation of mRNAs encoding proteins promoting angiogenesis. To obtain a global view of the HAX1‐dependent translational effects of HUVECs, we performed ribosome profiling on HUVECs that knockdown HAX1 or control. By comparing transcriptional and translational landscapes, we found that 6035 mRNAs are transcriptionally significantly regulated by HAX1, while their translation is regulated oppositely, therefore there may be no difference in protein expression, and 4872 mRNAs are only translationally regulated (Figure 7a–c). By comparing control cells, we observed 2497 down‐ and 2375 up‐regulated transcripts in ribosome footprints under knockdown HAX1 (Figure 7b, d). Of note, a subset of eukaryotic translation initiation factors can be regulated in their translational efficiency (Figure 7e). Furthermore, we observed that the TE of eIF4E was dramatically decreasing (Figure 7f).
FIGURE 7.
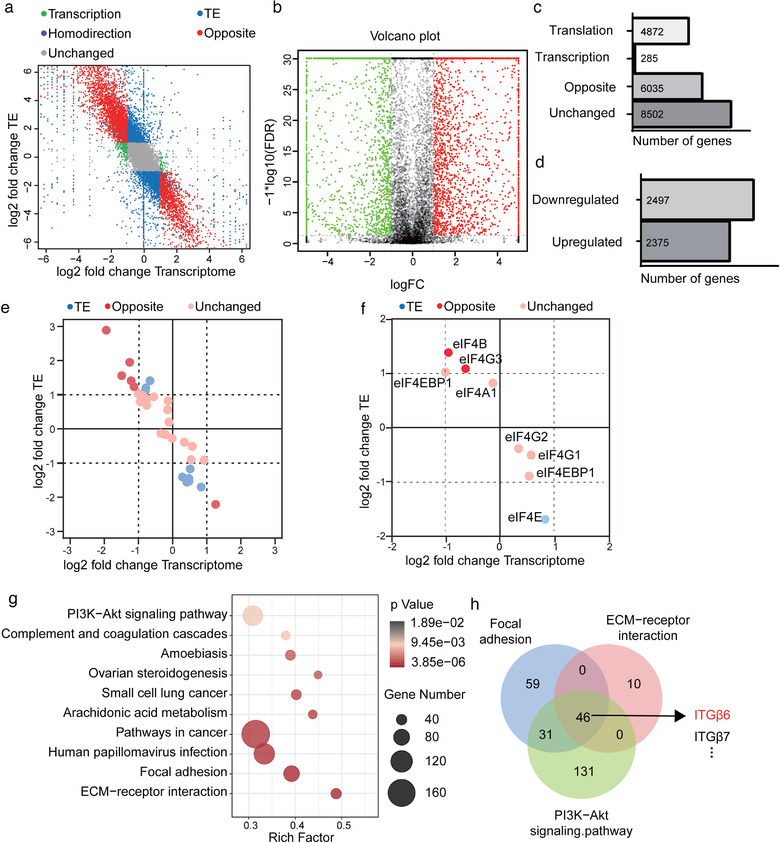
HAX1 regulates global gene translation in HUVECs. (a) Genome‐wide distribution of ribosome profiling and RNA abundance between HUVECs‐NC and HUVECs‐shHAX1. Green: transcription; Blue: translation; Purple: homodirection; Red: opposite. (b) The volcano plot reflects changes in translation efficiency. (c, d) Stacked bar charts indicate (c) the absolute numbers of transcription, translation, homodirection, opposite genes (d) and the number of genes whose translation efficiency is increased or decreased. (e, f) Ribosome profiling of (e) translation initiation factor and (f) the components of eIF4F. (g) KEGG pathways enrichment analysis for genes upregulated in translation efficiency. (h) The Venn diagram shows the overlap of genes in the PI3K signalling pathway, focal adhesion, and ECM‐receptor interaction
To further investigate possible translational regulation by HAX1, pathway enrichment analyses were performed on the 2497 transcripts that significantly down‐regulated ribosome footprints in knockdown HAX1 HUVECs compared with control. This revealed several significant KEGG pathways, the most significant among them including PI3K signalling pathway, focal adhesion and ECM‐receptor interaction, etc. (Figure 7g). Further research on genes enriched in these three pathways revealed that Integrin family is enriched in all those pathways (Figure 7h). Integrins can mediate cell migration and are thought to play a key role in EC function and angiogenesis. Among these integrins, ITGB6 was the most differentially regulated, and was seemed particularly relevant to cell migration and angiogenesis because of its well‐known role as an activator of FAK. We confirmed that HAX1 knockdown inhibited the expression of ITGB6 in HUVECs and inhibited FAK phosphorylation without affecting the total level of FAK (Figure 8a).
FIGURE 8.
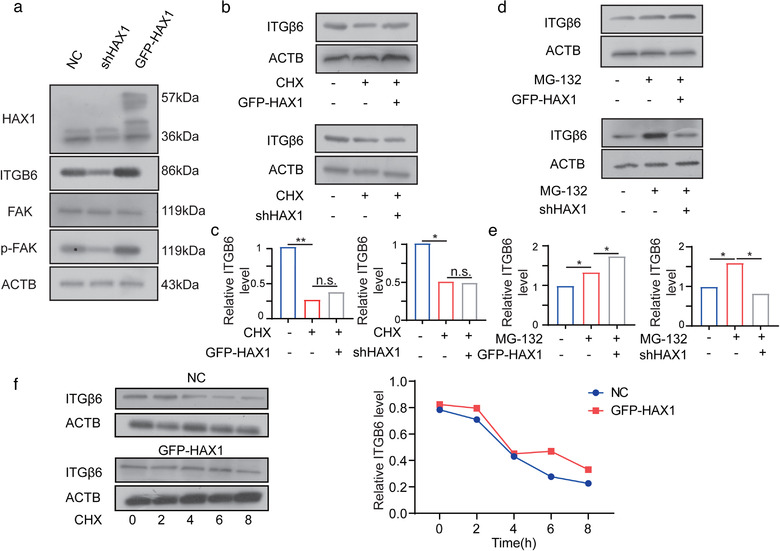
HAX1 enhances ITGB6 translation. (a) Western blot analysis of the protein levels in HUVECs‐NC and HUVECs‐shHAX1. (b, c) After 24 h treatment with CHX, cells were used for Western blot analysis (d, e) After 24 h treatment with MG132, cells were used for Western blot analysis. (f) HUVECs‐NC and HUVECs‐GFP‐HAX1 were treated with CHX (100 µg/ml) and collected at different time points for Western blot analysis
Next, we treated HUVECs with cycloheximide (CHX), a protein synthesis inhibitor, and determined that HAX1 could not regulate ITGB6 expression by affecting its half‐life (Figure 8b,d). Moreover, treatment with MG132, a proteasome inhibitor, further confirmed that HAX1 affected the translational efficiency of ITGB6 (Figure 8c).
3.6. HAX1 promotes the angiogenesis and metastasis of NPC via the FAK pathway
To verify the contribution of the FAK pathway to HAX1‐mediated angiogenesis and NPC metastasis, we performed rescue experiments and found that PF573228 (a kinase inhibitor of FAK) treatment and ITGB6 knockdown blocked the FAK phosphorylation induced by HAX1 (Figure 9a,b). Further, ITGB6 overexpression rescued cellular migration and angiogenesis in vitro, which was suppressed by HAX1 knockdown; in contrast, PF573228 treatment and ITGB6 knockdown blocked the phenotype induced by HAX1 overexpression (Figure 9c–f). The results of the in vivo experiments were consistent with those of the in vitro experiments as PF573228 treatment and ITGB6 knockdown significantly inhibited the NPC metastasis promoted by HAX1, whereas ITGB6 overexpression rescued the phenotype that was suppressed by HAX1 knockdown (Figure 9g).
FIGURE 9.
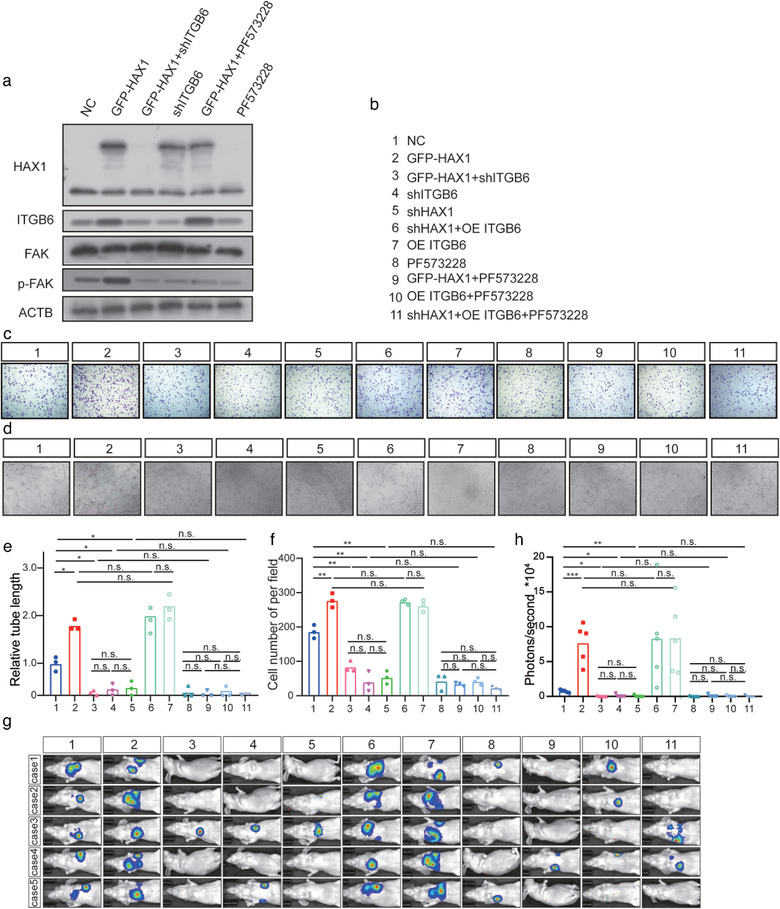
HAX1 promotes angiogenesis and metastasis of NPC via the FAK pathway. (a) Western blot analysis of HAX1, ITGB6, FAK and p‐FAK. (b–f) Tube forming and migration ability of HUVECs measured using the (c, e) Transwell assay and (d, f) tube formation assay, bar: 100 µm. (n.s. p > 0.05, *p < 0.05, **p < 0.001, one‐way ANOVA). (g, h) Visualisation of lung metastasis in BALB/c mice. (n.s. p > 0.05, ***p < 0.001, ****p < 0.05 by one‐way ANOVA)
4. DISCUSSION
Over the past decade, angiogenesis has been established to play a key role in tumour growth and metastasis by providing tumour cells with sufficient oxygen and nutrients (Zhang et al., 2021). Therefore, antiangiogenic drugs are considered an effective antitumour treatment and are a widely accepted clinical treatment (Jayson et al., 2018). Tumours can enhance angiogenesis by secreting proteins and EVs, generating a hypoxic environment, and through the infiltration of other cells (Carmeliet & Jain, 2000; Pralhad et al., 2003). In response to these processes, various antiangiogenic drugs have been developed. Currently, many protein molecules are considered therapeutic antiangiogenic targets including VEGF, VEGFR1, VEGFR2 and PDGF. Many antiangiogenic small‐molecule compounds including aflibercept, sorafenib, vanucizumab, cabozantinib and bevacizumab have been approved by the FDA (Fukumura et al., 2018). However, these drugs may only be effective against some tumour types. Most antiangiogenic drugs offer a limited survival benefit only in the early stages, but hardly alter tumour progression towing to incomplete inhibition of angiogenesis (Fukumura et al., 2018). Furthermore, VEGF‐targeted therapies have multiple adverse effects, including hypertension, proteinuria, impaired wound healing, gastrointestinal perforation, haemorrhage, thrombosis, reversible posterior leukoencephalopathy, cardiac toxicity and endocrine dysfunction (Vasudev & Reynolds, 2014). Therefore, new antiangiogenesis treatment targets are urgently needed to improve patient prognosis. HAX1 is known to be associated with the progression and recurrence of various tumours. Our previous research also found that HAX1 expression accelerates NPC tumour growth and angiogenesis. Based on histological and bioinformatics analyses, we confirmed that HAX1 expression was significantly higher in patients with late‐stage disease and that HAX1 levels were significantly correlated with patient prognosis. Notably, in NPC tissue samples, MVD and HAX1 levels were found to be positively correlated. These findings suggest HAX1 upregulation affect NPC prognosis by promoting angiogenesis. Subsequently, in a nude mouse/zebrafish xenograft model, an increase in HAX1 expression was found to enhance NPC metastasis. Similar tendencies were observed in nude mice treated with HAX1‐enriched EVs. Data from the Human Protein Atlas Database suggest that HAX1 is only highly expressed in tissues with active proliferation, but not in general tissues. Our results of in situ hybridisation in zebrafish suggest a stage of high HAX1 expression during vascular development. Thus, we concluded that HAX1 plays a key role in tumour angiogenesis. Overall, we report that HAX1 mediates angiogenesis by promoting ITGB6 expression to activate the FAK pathway, as ITGB6 knockdown or inhibition of FAK pathway activation completely blocked this effect.
The role of translation initiation factors and protein translation in tumourigenesis and immune escape has been examined in melanoma (Boussemart et al., 2014; Guo et al., 2020; Oblinger et al., 2018). Currently, owing to advances in ribosome profiling (Alexander et al., 2021; Zlotorynski, 2016) technology, scientists have again focused on the regulation of translation. However, the effect of translational control on angiogenesis remains largely unknown. Based on our ribosome‐profiling data, we found that HAX1 is involved in regulating angiogenesis, by promoting the translation of ITGB6. Importantly, ITGB6 has been shown to activate the FAK pathway to affect cell migration. Consistently, using cell models and various animal models, we found that HAX1 mainly affects angiogenesis by enhancing the migration ability of ECs. Considering the current lack of effective translation inhibitors, we only used FAK inhibitors for our in vivo experiments and obtained positive results. Our ribosome profiling data suggest that besides HAX1, several other proteins involved in angiogenesis are regulated by HAX1 overexpression. Considering the role of protein translation in the tumour, we believe that the combination of translation inhibitors and FAK inhibitors may have the potential to achieve beneficial outcomes.
Currently, the efficacy of treatments in patients with metastasis or on NPC recurrence remains unclear. Our data provide an important advance in understanding the importance and mechanism of HAX1 in regulating angiogenesis in NPC. Thus, our results provide a new prognostic indicator for patients with NPC and open novel avenues for therapeutic intervention.
CONFLICT OF INTEREST
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
AUTHOR CONTRIBUTIONS
Bo You: Data curation; Formal analysis; Investigation; Methodology; Resources; Software; Supervision; Validation; Visualisation; Writing – original draft; Writing – review & editing. Si Pan: Data curation; Formal analysis; Investigation; Methodology; Resources; Software; Supervision; Validation; Visualisation; Writing – original draft; Writing – review & editing. Miao Gu: Investigation; Validation. Kaiwen Zhang: Investigation; Software. Tian Xia: Investigation; Visualisation. Siyu Zhang: Investigation; Validation. Wenhui Chen: Investigation. Haijing Xie: Investigation. Yue Fan: Investigation. Hui Yao: Investigation. Tianyi Cheng: Investigation. Panpan Zhang: Investigation. Dong Liu: Conceptualisation; Investigation; Methodology. Yiwen You: Conceptualisation; Investigation; Methodology; Project administration
ETHICS STATEMENT
This study received ethics approval from the Affiliated Hospital of Nantong University (IRB number: 2016‐099). In vivo studies were approved by the committee on the Ethics of Animal Experiments of Nantong University (RDD number: 20180227‐008). All mouse experiments followed NIH Guidelines and were approved by the Administration Committee of Experimental Animals, Jiangsu Province, China (Approval ID: SYXK (SU) 2017‐0046).
Supporting information
Supporting information.
ACKNOWLEDGEMENTS
This work was supported by grants from the National Natural Science Foundation of China (82173288, 81972554, 81672682 and 81602385), the Natural Science Foundation of Jiangsu (No. BK20201208), Clinical Frontier Technology of Jiangsu (BE2017680), CSCO Clinical Oncology Research Foundation of Beijing (Y‐HS2017‐074, Y‐HS2017‐074), Innovative Research Project for Postgraduate Students in Jiangsu Province (SJCX19_0871, SJCX19_0872, SJCX20_1164), Scientific Research Project of Nantong Municipal Health Commission (QA2019060).
You, B. , Pan, S. , Gu, M. , Zhang, K. , Xia, T. , Zhang, S. , Chen, W. , Xie, H. , Fan, Y. , Yao, H. , Cheng, T. , Zhang, P. , Liu, D. , & You, Y. (2022). Extracellular vesicles rich in HAX1 promote angiogenesis by modulating ITGB6 translation. Journal of Extracellular Vesicles, 11, e12221. 10.1002/jev2.12221
Bo You and Si Pan contributed equally.
Contributor Information
Dong Liu, Email: liudongtom@gmail.com.
Yiwen You, Email: youyiwen_nantong@163.com.
REFERENCES
- Adamiak, M. , Cheng, G. , Bobis‐Wozowicz, S. , Zhao, L. , Kedracka‐Krok, S. , Samanta, A. , Karnas, E. , Xuan, Yu‐T. , Skupien‐Rabian, B. , Chen, X. , Jankowska, U. , Girgis, M. , Sekula, M. , Davani, A. , Lasota, S. , Vincent, R. J. , Sarna, M. , Newell, K. L. , Wang, O.‐L. , … Zuba‐Surma, E. K. (2018). Induced Pluripotent Stem Cell (iPSC)‐derived extracellular vesicles are safer and more effective for cardiac repair than iPSCs. Circulation Research, 122(2), 296–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agulnik, M. , Costa, R. L. B. , Milhem, M. , Rademaker, A. W. , Prunder, B. C. , Daniels, D. , Rhodes, B. T. , Humphreys, C. , Abbinanti, S. , Nye, L. , Cehic, R. , Polish, A. , Vintilescu, C. , Mcfarland, T. , Skubitz, K. , Robinson, S. , Okuno, S. , & Van Tine, B. A. (2017). A phase II study of tivozanib in patients with metastatic and nonresectable soft‐tissue sarcomas. Annals of Oncology, 28(1), 121–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander, M. R. , Brice, A. M. , Jansen Van Vuren, P. , Rootes, C. L. , Tribolet, L. , Cowled, C. , Bean, A. G. D. , & Stewart, C. R. (2021). Ribosome‐profiling reveals restricted post transcriptional expression of antiviral cytokines and transcription factors during SARS‐CoV‐2 infection. International Journal of Molecular Sciences, 22(7), 3392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Backlund, M. , Paukku, K. , Daviet, L. , De Boer, R. A. , Valo, E. , Hautaniemi, S. , Kalkkinen, N. , Ehsan, A. , Kontula, K. K. , & Lehtonen, J. Y. A. (2009). Posttranscriptional regulation of angiotensin II type 1 receptor expression by glyceraldehyde 3‐phosphate dehydrogenase. Nucleic Acids Research, 37(7), 2346–2358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boussemart, L. , Malka‐Mahieu, H. , Girault, I. , Allard, D. , Hemmingsson, O. , Tomasic, G. , Thomas, M. , Basmadjian, C. , Ribeiro, N. , Thuaud, F. , Mateus, C. , Routier, E. , Kamsu‐Kom, N. , Agoussi, S. , Eggermont, A. M. , Désaubry, L. , Robert, C. , & Vagner, S. (2014). eIF4F is a nexus of resistance to anti‐BRAF and anti‐MEK cancer therapies. Nature, 513(7516), 105–109. [DOI] [PubMed] [Google Scholar]
- Carmeliet, P. , & Jain, R. K. (2000). Angiogenesis in cancer and other diseases. Nature, 407(6801), 249–257. [DOI] [PubMed] [Google Scholar]
- Catalano, M. , & O'driscoll, L. (2020). Inhibiting extracellular vesicles formation and release: A review of EV inhibitors. Journal of Extracellular Vesicles, 9(1), 1703244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, Yu‐P. , Chan, A. T. C. , Le, Q. T. , Blanchard, P. , Sun, Y. , & Ma, J. (2019). Nasopharyngeal carcinoma. The Lancet, 394(10192), 64–80. [DOI] [PubMed] [Google Scholar]
- Fadeel, B. , & Grzybowska, E. (2009). HAX‐1: A multifunctional protein with emerging roles in human disease. Biochimica Et Biophysica Acta, 1790(10), 1139–1148. [DOI] [PubMed] [Google Scholar]
- Fagan, D. H. , Fettig, L. M. , Avdulov, S. , Beckwith, H. , Peterson, M. S. , Ho, Y.‐Y. , Wang, F. , Polunovsky, V. A. , & Yee, D. (2017). Acquired tamoxifen resistance in MCF‐7 breast cancer cells requires hyperactivation of eIF4F‐mediated translation. Hormones and Cancer, 8(4), 219–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farajifard, H. , Zavvar, M. , Rajaei, T. , Noorbakhsh, F. , Nikougoftar‐Zarif, M. , Azadmanesh, K. , Kompani, F. , & Rezaei, N. (2018). In vitro study of HAX1 gene therapy by retro viral transduction as a therapeutic target in severe congenital neutropenia. European Cytokine Network, 29(4), 146–152. [DOI] [PubMed] [Google Scholar]
- Fukumura, D. , Kloepper, J. , Amoozgar, Z. , Duda, D. G. , & Jain, R. K. (2018). Enhancing cancer immunotherapy using antiangiogenics: Opportunities and challenges. Nature Reviews Clinical Oncology, 15(5), 325–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gandin, V. , Masvidal, L. , Cargnello, M. , Gyenis, L. , Mclaughlan, S. , Cai, Y. , Tenkerian, C. , Morita, M. , Balanathan, P. , Jean‐Jean, O. , Stambolic, V. , Trost, M. , Furic, L. , Larose, L. , Koromilas, A. E. , Asano, K. , Litchfield, D. , Larsson, O. , & Topisirovic, I. (2016). mTORC1 and CK2 coordinate ternary and eIF4F complex assembly. Nature Communication, 7, 11127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geng, T. , Song, Z. Y. , Xing, J. X. , Wang, B. X. , Dai, S. P. , & Xu, Z.‐S. (2020). Exosome derived from coronary serum of patients with myocardial infarction promotes angiogenesis through the miRNA‐143/IGF‐IR pathway. International Journal of Nanomedicine, 15, 2647–2658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo, C. , Hou, Y. , Yu, X. , Zhang, F. , Wu, X. , Wu, X. , & Wang, L. (2020). The ERK‐MNK‐eIF4F signaling pathway mediates TPDHT‐induced A549 cell death in vitro and in vivo. Food and Chemical Toxicology, 137, 111158. [DOI] [PubMed] [Google Scholar]
- Hanahan, D. , & Weinberg, R A. (2011). Hallmarks of cancer: The next generation. Cell, 144(5), 646–674. [DOI] [PubMed] [Google Scholar]
- Ho, J. J. D. , & Lee, S. (2016). A cap for every occasion: Alternative eIF4F complexes. Trends in Biochemical Sciences, 41(10), 821–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard, C. M. , Bearss, N. , Subramaniyan, B. , Tilley, A. , Sridharan, S. , Villa, N. , Fraser, C. S. , & Raman, D. (2019). The CXCR4‐LASP1‐eIF4F Axis promotes translation of oncogenic proteins in triple‐negative breast cancer cells. Frontiers in Oncology, 9, 284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayson, G. C. , Zhou, C. , Backen, A. , Horsley, L. , Marti‐Marti, K. , Shaw, D. , Mescallado, N. , Clamp, A. , Saunders, M. P. , Valle, J. W. , Mullamitha, S. , Braun, M. , Hasan, J. , Mcentee, D. , Simpson, K. , Little, R. A. , Watson, Y. , Cheung, S. , Roberts, C. , … Dive, C. (2018). Plasma Tie2 is a tumor vascular response biomarker for VEGF inhibitors in metastatic colorectal cancer. Nature Communication, 9(1), 4672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeyaraju, D. V. , Cisbani, G. , De Brito, O. M. , Koonin, E. V. , & Pellegrini, L. (2009). Hax1 lacks BH modules and is peripherally associated to heavy membranes: Implications for Omi/HtrA2 and PARL activity in the regulation of mitochondrial stress and apoptosis. Cell Death and Differentiation, 16(12), 1622–1629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karapınar, T. H. , Yılmaz Karapinar, D. , Oymak, Y. , Ay, Y. , Demirağ, B. , Aykut, A. , Onay, H. , Hazan, F. , Aydınok, Y. , Özkınay, F. , & Vergin, C. (2017). HAX1 mutation positive children presenting with haemophagocytic lymphohistiocytosis. British Journal of Haematology, 177(4), 597–600. [DOI] [PubMed] [Google Scholar]
- Kim, Y. H. , Choi, J. , Yang, M. J. , Hong, S. P. , Lee, C. K. , Kubota, Y. , Lim, D. S. , & Koh, G. Y. (2019). A MST1‐FOXO1 cascade establishes endothelial tip cell polarity and facilitates sprouting angiogenesis. Nature Communication, 10(1), 838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krueger, J. , Liu, D. , Scholz, K. , Zimmer, A. , Shi, Y. , Klein, C. , Siekmann, A. , Schulte‐Merker, S. , Cudmore, M. , Ahmed, A. , & Le Noble, F. (2011). Flt1 acts as a negative regulator of tip cell formation and branching morphogenesis in the zebrafish embryo. Development (Cambridge, England), 138(10), 2111–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Ze‐L. , Ye, S. B. , Ouyang, Li‐Y. , Zhang, H. , Chen, Y.‐S. , He, J. , Chen, Q. Y. , Qian, C. N. , Zhang, X. S. , Cui, J. , Zeng, Yi‐X. , & Li, J. (2015). COX‐2 promotes metastasis in nasopharyngeal carcinoma by mediating interactions between cancer cells and myeloid‐derived suppressor cells. Oncoimmunology, 4(11), e1044712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang, Z. , Zhong, Y. , Meng, L. , Chen, Y. , Liu, Y. , Wu, A. , Li, X. , & Wang, M. (2020). HAX1 enhances the survival and metastasis of non‐small cell lung cancer through the AKT/mTOR and MDM2/p53 signaling pathway. Thorac Cancer, 11(11), 3155–3167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, S. , Negulescu, A. , Bulusu, S. , Gibert, B. , Delcros, J. G. , Ducarouge, B. , Rama, N. , Gadot, N. , Treilleux, I. , Saintigny, P. , Meurette, O. , & Mehlen, P. (2017). Non‐canonical NOTCH3 signalling limits tumour angiogenesis. Nature Communication, 8, 16074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madeo, M. , Colbert, P. L. , Vermeer, D. W. , Lucido, C. T. , Cain, J. T. , Vichaya, E. G. , Grossberg, A. J. , Muirhead, D. , Rickel, A. P. , Hong, Z. , Zhao, J. , Weimer, J. M. , Spanos, W. C. , Lee, J. H. , Dantzer, R. , & Vermeer, P. D. (2018). Cancer exosomes induce tumor innervation. Nature Communication, 9(1), 4284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oblinger, J L. , Burns, S S. , Huang, J. , Pan, L. , Ren, Y. , Shen, R. , Kinghorn, A. D , Welling, D. B , & Chang, L. S. (2018). Overexpression of eIF4F components in meningiomas and suppression of meningioma cell growth by inhibiting translation initiation. Experimental Neurology, 299(Pt B), 299–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pralhad, T. , Madhusudan, S. , & Rajendrakumar, K. (2003). Concept, mechanisms and therapeutics of angiogenesis in cancer and other diseases. Journal of Pharmacy and Pharmacology, 55(8), 1045–1053. [DOI] [PubMed] [Google Scholar]
- Sheng, C. , & Ni, Q. (2015). Expression of HAX1 and Ki‐67 in breast cancer and its correlations with patient's clinicopathological characteristics and prognosis. International Journal of Clinical and Experimental Medicine, 8(11), 20904–20910. [PMC free article] [PubMed] [Google Scholar]
- Simeoli, R. , Montague, K. , Jones, H. R. , Castaldi, L. , Chambers, D. , Kelleher, J. H. , Vacca, V. , Pitcher, T. , Grist, J. , Al‐Ahdal, H. , Wong, L.‐F. , Perretti, M. , Lai, J. , Mouritzen, P. , Heppenstall, P. , & Malcangio, M. (2017). Exosomal cargo including microRNA regulates sensory neuron to macrophage communication after nerve trauma. Nature Communication, 8(1), 1778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmen, T. (2011). Hax‐1: A regulator of calcium signaling and apoptosis progression with multiple roles in human disease. Expert Opinion on Therapeutic Targets, 15(6), 741–751. [DOI] [PubMed] [Google Scholar]
- Skokowa, J. , Klimiankou, M. , Klimenkova, O. , Lan, D. , Gupta, K. , Hussein, K. , Carrizosa, E. , Kusnetsova, I. , Li, Z. , Sustmann, C. , Ganser, A. , Zeidler, C. , Kreipe, H.‐H. , Burkhardt, J. , Grosschedl, R. , & Welte, K. (2012). Interactions among HCLS1, HAX1 and LEF‐1 proteins are essential for G‐CSF‐triggered granulopoiesis. Nature Medicine, 18(10), 1550–1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solomon, O. , Di Segni, A. , Cesarkas, K. , Porath, H. T. , Marcu‐Malina, V. , Mizrahi, O. , Stern‐Ginossar, N. , Kol, N. , Farage‐Barhom, S. , Glick‐Saar, E. , Lerenthal, Y. , Levanon, E. Y. , Amariglio, N. , Unger, R. , Goldstein, I. , Eyal, E. , & Rechavi, G. (2017). RNA editing by ADAR1 leads to context‐dependent transcriptome‐wide changes in RNA secondary structure. Nature Communication, 8(1), 1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stratman, A. N. , Farrelly, O. M. , Mikelis, C. M. , Miller, M. F. , Wang, Z. , Pham, V. N. , Davis, A. E. , Burns, M. C. , Pezoa, S. A. , Castranova, D. , Yano, J. J. , Kilts, T. M. , Davis, G. E. , Gutkind, J. S. , & Weinstein, B. M. (2020). Anti‐angiogenic effects of VEGF stimulation on endothelium deficient in phosphoinositide recycling. Nature Communication, 11(1), 1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuelten, C. H. , Parent, C. A. , & Montell, D. J. (2018). Cell motility in cancer invasion and metastasis: insights from simple model organisms. Nature Reviews Cancer, 18(5), 296–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Théry, C. , Witwer, K. W. , Aikawa, E. , Alcaraz, M. J. , Anderson, J. D. , Andriantsitohaina, R. , Antoniou, A. , Arab, T. , Archer, F. , Atkin‐Smith, G. K. , Ayre, D. C. , Bach, J. M. , Bachurski, D. , Baharvand, H. , Balaj, L. , Baldacchino, S. , Bauer, N. N. , Baxter, A. A. , Bebawy, M. , … Zuba‐Surma, E. K. (2018). Minimal information for studies of extracellular vesicles 2018 (MISEV2018): A position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. Journal of Extracellular Vesicles, 7(1), 1535750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoreen, C. C. (2013). Many roads from mTOR to eIF4F. Biochemical Society Transactions, 41(4), 913–916. [DOI] [PubMed] [Google Scholar]
- Torraca, V. , & Mostowy, S. (2018). Zebrafish Infection: From Pathogenesis to Cell Biology. Trends in Cell Biology, 28(2), 143–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trebinska‐Stryjewska, A. , Wakula, M. , Tabor, S. , Sienkiewicz, R. , Owczarek, J. , Balcerak, A. , Felisiak‐Golabek, A. , & Grzybowska, E. A. (2019). Cytoplasmic HAX1 is an independent risk factor for breast cancer metastasis. Journal of Oncology, 2019, 6375025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasudev, N. S. , & Reynolds, A. R. (2014). Anti‐angiogenic therapy for cancer: Current progress, unresolved questions and future directions. Angiogenesis, 17(3), 471–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson, E. C. , Whitehead, L. , Adams, R. H. , Dewson, G. , & Coultas, L. (2016). Endothelial cell survival during angiogenesis requires the pro‐survival protein MCL1. Cell Death and Differentiation, 23(8), 1371–1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, F. , Chen, W. , Gillis, B. , Fischbach, C. , Estroff, L. A. , & Gourdon, D. (2017). Protein‐crystal interface mediates cell adhesion and proangiogenic secretion. Biomaterials, 116, 174–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, H. , Chen, J. , Wang, Q. , Yin, Y. , Da, P. , Le, H. , Zhang, Z. , & Qiu, X. (2017). Abnormal expression of HAX1 is associated with cellular proliferation and migration in human hypopharyngeal squamous cell carcinoma. Molecular Medicine Reports, 16(4), 4664–4670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, X. G. , Zhou, C. F. , Zhang, Y. M. , Yan, R. M. , Wei, W. F. , Chen, X. J. , Yi, H. Y. , Liang, L. J. , Fan, L. S. , Liang, L. , Wu, S. , & Wang, W. (2019). Cancer‐derived exosomal miR‐221‐3p promotes angiogenesis by targeting THBS2 in cervical squamous cell carcinoma. Angiogenesis, 22(3), 397–410. [DOI] [PubMed] [Google Scholar]
- You, B. , Cao, X. , Shao, X. , Ni, H. , Shi, S. , Shan, Y. , Gu, Z. , & You, Y. (2016). Clinical and biological significance of HAX‐1 overexpression in nasopharyngeal carcinoma. Oncotarget, 7(11), 12505–12524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, K. , Liu, D. , Zhao, J. , Shi, S. , He, X. , Da, P. , You, Y. , & You, B. (2021). Nuclear exosome HMGB3 secreted by nasopharyngeal carcinoma cells promotes tumour metastasis by inducing angiogenesis. Cell Death & Disease, 12(6), 554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, W. , Siegel, D. , Biton, A. , Tonqueze, O. L. e. , Zaitlen, N. , Ahituv, N. , & Erle, D. J. (2017). CRISPR‐Cas9‐mediated functional dissection of 3'‐UTRs. Nucleic Acids Research, 45(18), 10800–10810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, Q. , Perakis, S. O. , Ulz, P. , Mohan, S. , Riedl, J. M. , Talakic, E. , Lax, S. , Tötsch, M. , Hoefler, G. , Bauernhofer, T. , Pichler, M. , Gerger, A. , Geigl, J. B. , Heitzer, E. , & Speicher, M. R. (2020). Cell‐free DNA analysis reveals POLR1D‐mediated resistance to bevacizumab in colorectal cancer. Genome Medicine, 12(1), 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, Q. , Zhang, Q. , Gu, M. , Zhang, K. , Xia, T. , Zhang, S. , Chen, W. , Yin, H. , Yao, H. , Fan, Y. , Pan, S. , Xie, H. , Liu, H. , Cheng, T. , Zhang, P. , Zhang, T. , You, B. , & You, Y. (2021). MIR106A‐5p upregulation suppresses autophagy and accelerates malignant phenotype in nasopharyngeal carcinoma. Autophagy, 17(7), 1667–1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zlotorynski, E. (2016). Translation: Profiling ribosome dynamics. Nature Reviews Molecular Cell Biology, 17(9), 535. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting information.


