Abstract
Research on tumour cell‐derived small extracellular vesicles (sEVs) that regulate tumour microenvironment (TME) has provided strategies for targeted therapy of head and neck squamous cell carcinoma (HNSCC). Herein, we demonstrated that sEVs derived from HNSCC cancer cells carried CD73 (sEVsCD73), which promoted malignant progression and mediated immune evasion. The sEVsCD73 phagocytosed by tumour‐associated macrophages (TAMs) in the TME induced immunosuppression. Higher CD73high TAMs infiltration levels in the HNSCC microenvironment were correlated with poorer prognosis, while sEVsCD73 activated the NF‐κB pathway in TAMs, thereby inhibiting immune function by increasing cytokines secretion such as IL‐6, IL‐10, TNF‐α, and TGF‐β1. The absence of sEVsCD73 enhanced the sensitivity of anti‐PD‐1 therapy through reversed immunosuppression. Moreover, circulating sEVsCD73 increased the risk of lymph node metastasis and worse prognosis. Taken together, our study suggests that sEVsCD73 derived from tumour cells contributes to immunosuppression and is a potential predictor of anti‐PD‐1 responses for immune checkpoint therapy in HNSCC.
Keywords: anti‐PD‐1 therapy, CD73, head and heck squamous cell carcinoma, macrophage, small extracellular vesicle
Abbreviations
- EVs
extracellular vesicles
- FDA
food and drug administration
- HNSCC
head and neck squamous cell carcinoma
- TAMs
tumour‐associated macrophages
- TME
tumour microenvironment
- sEVs
small extracellular vesicles
- sEVsCD73
CD73 carried on sEVs
- ICB
Immune checkpoint blockade
- Tregs
T regulatory cells
- NK
Natural killer cells
- DMEM
Dulbecco's modified Eagle's medium
- FBS
Foetal bovine serum
- THP‐1
Human monocyte leukemia cell line
- CM
Conditioned media
- CAFs
Cancer‐associated fibroblasts
- CR cells
Conditional reprograming cells
- DCs
Myeloid dendritic cells
- Mac
Macrophages
- M2
M2 Macrophages
- Machigh
Macrophages higher infiltration
- Maclow
Macrophages lower infiltration
- LAG3
Lymphocyte‐activation gene 3
- PD‐1
Programmed cell death protein 1
- PD‐L1
Programmed death ligand 1
- RNA‐seq
RNA sequencing assay
- PDTC
Pyrrolidine dithiocarbonate
- ELISA
Enzyme‐linked immunosorbent assay
- TNM
Tumours and the tumour node metastasis
- UICC
Union for International Cancer Control
- DAPI
4′,6‐ Diamidino‐2‐phenylindole
- TCGA
Cancer Genome Atlas dataset
- GSEA
Gene Set Enrichment Analysis
- Lat A
Latrunculin A
- DLNs
Draining lymph nodes
- H‐score
Histochemistry score
- ADO
Adenosine
- eADO
extracellular adenosine
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- AUC
Area under the curve
- E. coli
Escherichia coli
- GFP
Green fluorescent protein
1. INTRODUCTION
Immune checkpoint blockade (ICB) is an effective strategy to reverse immunosuppression in tumour therapy (Poggio et al., 2019). Anti‐programmed cell death protein 1 (PD‐1) therapy has been approved by the Food and Drug Administration (FDA) as a first‐line treatment of patients with metastatic or unresectable recurrent head and neck squamous cell carcinoma (HNSCC) (Cohen et al., 2019; H. H. Lee et al., 2019; H. Lee et al., 2019), an aggressive cancer (Y. Tang et al., 2021) with an overall 5‐year survival rate of approximately 50%, while more than 65% of patients show local recurrence or lymph node metastasis (Chow, 2020; Leemans et al., 2011; Lu et al., 2020). Although the overall treatment efficiency is improved with anti‐PD‐1 therapy, only 15%–20% of patients benefit from it (L. Zhou et al., 2020). Therefore, strategies for reactivating the immune response in patients insensitive to anti‐PD‐1 therapy are urgently needed.
The tumour microenvironment (TME) influences patients’ response to ICB. As a courier for substance transformation, extracellular vehicles (EVs) are important contributors to immune evasion and are valuable for guiding patient selection and therapy decisions (Chen et al., 2018). Small EVs (sEVs) have a diameter range of 30–150 nm and regulate cell function through autocrine or paracrine pathways (M. Li et al., 2021; Raposo & Stoorvogel, 2013; Théry et al., 2018). sEVs carrying abundant bioactive molecules such as proteins, nuclear acids, and lipids propagate signals to reshape TME and accelerate tumour progression (Brown et al., 2021; Qu et al., 2015; Xiao et al., 2019), which may hinder immunotherapy. For instance, activation of CD8+ T cells is inhibited by sEVs in the TME, resulting in insensitivity to anti‐programmed death‐ligand 1 (PD‐L1) therapy (Chen et al., 2018). Tumour‐associated macrophages (TAMs) play vital roles in the TME (Y. Zhang et al., 2018) and may be polarized by sEVs toward an immunosuppressive phenotype, which are associated with poor prognosis (Gu et al., 2016; Mantovani et al., 2017; Mei et al., 2016; Ngambenjawong et al., 2017; Quail & Joyce, 2017; Quail & Joyce, 2013; Ruffell et al., 2012). Accumulating evidence has shown that oncoproteins anchored on the tumour cell membranes are released into the TME and induce a series of downstream cascades, which involves sEVs, that change the function of immunocytes. Therefore, emerging treatment strategies beyond T cell coreceptors are focusing on TME remodellings, such as reappropriating TAMs or other transportation hubs, including sEVs, to promote ICB efficiency (Hume & Macdonald, 2012; Morrison et al., 2004; Poggio et al., 2019; Whiteside, 2016; Willingham et al., 2012). However, the complexity of the interactions between sEVs, immune cells, and the TME should be considered when developing anticancer treatment strategies.
In this study, we found that CD73 protein levels were significantly higher in HNSCC‐derived sEVs than in normal cell‐derived sEVs. CD73, encoded by NT5E, is an ectonucleotide present mainly in the membrane‐bound form (Yegutkin, 2008), overexpressed in various types of tumours, and is associated with adenosine (ADO) metabolism and poor prognosis (Meng et al., 2019). In the TME, CD73 was detected on the membranes of various cells, including tumour cells, T regulatory cells (Tregs), natural killer (NK) cells, and macrophages (Roh et al., 2020), thereby inducing immunosuppression and immune escape and promoting tumour progression (Neo et al., 2020). A recent study showed that CD73 was a specific immunotherapeutic target for improving antitumour immune responses to immune checkpoint therapy (Goswami et al., 2020). Clinical trials on anti‐CD73 agents remain in the early stages (B. Allard et al., 2020). We found that CD73 in sEVs (sEVsCD73) polarized TAMs toward an immunosuppressive phenotype and triggered tumour escape mechanisms. Moreover, our results suggest that sEVCD73 is a potential combined target for ICB in HNSCC and improves the response to ICB.
2. MATERIALS AND METHODS
2.1. Ethics statement
This study was approved by the Ethics Committee of the Ninth People's Hospital, Shanghai Jiao Tong University School of Medicine (Shanghai, China). Informed consent was obtained before starting the experiments. All animal studies were performed in accordance with the NIH Guide for the Care and Use of Laboratory Animals, with the approval of the Ninth People's Hospital, Shanghai Jiao Tong University School of Medicine Institute Animal Care and Use Committee.
2.2. Patients and specimens
All clinical samples were obtained from the Department of Oral and Maxillofacial‐Head & Neck Oncology, Ninth People's Hospital, Shanghai Jiao Tong University School of Medicine. Tumour tissues (n = 92) from patients with HNSCC were collected between September 2011 and June 2015 along with complete follow‐up data. As controls, 10 normal mucosa samples were obtained from patients who underwent periodontal surgery. Paired tumour and adjacent normal tissues for conditional reprogramming (CR) cell culture were obtained from patients diagnosed with HNSCC that underwent initial surgery between March 2018 and January 2021. Serum samples from patients with HNSCC (n = 54) were collected, and healthy controls were from 22 donors at the Ninth People's Hospital, from January 2021 to February 2021. Additionally, World Health Organization Classification of Tumours and the tumour node metastasis (TNM) staging system (8th edition) were used to determine the pathological differentiation and the clinical stage.
2.3. Cell culture
The HNSCC cell lines HN6, HN30, CAL27, SCC9, and SCC25 were used in this study. The SCC7 cell line is a murine HNSCC cell line, which is syngeneic to C3H mice (Ruan et al., 2020). Human bone marrow‐derived cells (hBMSCs) were kindly provided by Guangzhou ELGBIO Company (Guangzhou, China). Mouse BMSCs (mBMSCs) were extracted from the bone marrow of C3H mice. After 14 days of culture, mBMSCs were amplified for further experiments. All cells were cultured in Dulbecco's modified Eagle's medium (DMEM; Gibco, USA) with 10% foetal bovine serum (FBS; Gibco, USA), 100 U/mL penicillin, and 100 μg/mL streptomycin in a humidified 5% CO2 atmosphere at 37°C.
2.4. Macrophage culture and treatment
Macrophages were transformed from a human monocyte leukaemia cell line (THP‐1), kindly provided by Guangzhou ELGBIO Company (Guangzhou, China). Cells were cultured in RPMI 1640 medium (Gibco, USA) supplemented with 10% FBS at 37°C in a humidified 5% CO2 atmosphere. To transform the cells into macrophages, THP‐1 monocytes were cocultured with 100 ng/mL phorbol 12‐myristate 13‐acetate (PMA, Sigma, USA) in RPMI 1640 medium without FBS for 48 h (Park et al., 2007). Macrophages treated with 20 ng/ml interleukin 4 (IL‐4) (Sigma, USA) for 24 h were polarized into M2 phenotype.
2.5. CR cell culture
CR cells were prepared as previously described (Tjin et al., 2018). Tissues from patients with HNSCC were rinsed with phosphate‐buffered saline (PBS) and disinfected with 75% anhydrous alcohol. The samples were cut into pieces and digested with a mixture containing collagenase/hyaluronidase solution, dispase (Sigma, USA), and DMEM at 37°C for 2 h. The samples were centrifuged at 1,000×g for 8 min, and the supernatant was removed. The pellet was resuspended and treated with collagenase IV (1 mg/mL) at 37°C for 15 min. After adding 10 mL DMEM, the resuspended pellet was filtered through 70 μm cell strainers (SORFA, Zhejiang, China). After centrifugation for 10 min at 1,000×g, the cells were seeded into culture medium with processed Swiss 3T2 J2 as feeder cells to prevent the cells from ageing. According to the pathological state of the patients, the CR cells were classified into three groups: para‐cancerous epithelial cells, tumour cells without lymph node metastasis, and primary tumour cells with evidence of lymph node metastasis. To accumulate the supernatant for sEVs extraction, the feeder cells were digested, and CR cells were cultured in serum‐free DMEM medium for 48 h.
2.6. sEVs isolation and identification
CR cells without feeder cells, HNSCC cells (HN6, SCC25, CAL27, and SCC7) and h/mBMSC (2 × 106 cells) prepared for sEVs extraction were cultured in 10 mL DMEM without FBS or penicillin‐streptomycin. Conditioned media (CM) were collected after 48 h of cell culture via centrifugation at 300×g for 5 min, followed by 3,000×g for 15 min to remove cell debris. The CM was purified through a 0.22 μm polyvinylidene fluoride filter (Millipore, Billerica, MA, USA), followed by ultracentrifugation spins at 100,000×g for 70 min to pellet the sEVs using a Beckman Coulter Type 55.2 Ti Rotor (Brea, CA, USA). The pellets were washed with PBS and centrifuged at 100,000×g for 70 min to obtain purified sEVs. Finally, the sEVs were resuspended in PBS or other buffers as necessary. The sEVs were quantified using micro‐bicinchoninic acid. The initial cell number in the culture, expected doubling time, and collection frequency were the same in all groups to remove potential confounding factors.
For transmission electron microscopy, 5 μL of tumour‐derived sEVs suspension was dropped on a copper wire and incubated at room temperature for 5 min. Next, 2% uranyl acetate was added to the copper mesh and incubated at room temperature for 1 min. After incubation at room temperature for approximately 20 min, samples were observed using a Nano transmission electron microscope (Tecnai G2 Spirit BioTwin, FEI, Hillsboro, OR, USA).
For nanoparticle tracking analysis, isolated samples were appropriately diluted in 1×PBS buffer (Biological Industries, Beit‐Haemek, Israel) to measure the particle size and concentration. The nanoparticle tracking measurements were recorded and analyzed at 11 positions. The ZetaView system was calibrated using 110 nm polystyrene particles. The temperature was maintained at approximately 25°C. The particle size and concentration were measured using a ZetaView PMX 110 (Particle Metrix, Meerbusch, Germany) and corresponding software ZetaView 8.04.02. To isolate sEVs from the serum of patients with HNSCC, we used Minute™ Hi‐Efficiency Exosome Precipitation Reagent (Cat. No. EI‐027, Invent Biotechnologies, Plymouth, MN, USA), a non‐PEG‐based reagent. Briefly, 1 mL serum samples stored at ‐80°C were thawed and centrifuged at 2,000×g for 10 min to remove large debris. The supernatant was then mixed with two volumes of exosome precipitation reagent and incubated for 1 h. The sample was centrifuged at 10,000×g for 30 min at 4°C, washed with PBS, and recentrifuged at 100,000×g for an additional 70 min to obtain purified sEVs.
2.7. LC‐MS/MS‐based proteomics and proteogenomics
The sEVs were extracted from the supernatant of eighteen CR cultures obtained from six patients with nonmetastatic HNSCC, six patients with metastatic HNSCC, and six normal mucosa samples. The CR cells were divided into three categories: normal control cells from adjacent tissues, nonmetastatic HNSCC cells from patients without metastasis, and metastatic HNSCC cells from in situ tumours with lymph node metastasis. The sEVs from three different subjects in the same group were pooled to form the samples, which were isolated from the CR cell culture medium via ultracentrifugation.
After protein extraction, the samples were subjected to liquid chromatography‐tandem mass spectrometry (LC‐MS/MS)‐based proteomics and proteogenomics. The sEVs from HN6, SCC25, and CAL27 were evaluated in the same experiments. LC‐MS/MS analysis was performed on a Q Exactive mass spectrometer (Thermo Fisher Scientific, Waltham, MA, USA) coupled to an Easy nLC (Thermo Fisher Scientific). The MS data were analysed using MaxQuant software version 1.5.3.17 (Max Planck Institute of Biochemistry, Martinsried, Germany). MASCOT engine (Matrix Science, London, UK; version 2.2) was used to search MS/MS spectra embedded into Proteome Discoverer 1.4 (Thermo Electron, San Jose, CA.), against Uniprot Human database (133549 sequences, download on March 3rd, 2013) and the decoy database. The following options were used for protein identification. Peptide mass tolerance = 20 ppm, MS/MS tolerance = 0.1 Da, Missed cleavage = 2, Enzyme = Trypsin, Fixed modification: Carbamidomethyl (C), Variable modification: Oxidation(M), FDR ≤ 0.01.
2.8. CRISPR/Cas9 knockout system
The type II CRISPR system was selected according to the literature (Cong & Zhang, 2015). LentiCRISPRv2 was used as a vector. Three single‐guide RNAs (sgRNAs) were selected for human NT5E/mouse Nt5e and human RAB27A/mouse Rab27a. The sgRNAs were designed using the online Benchling CRISPR sgRNA design tool (http://www.benchling.com) and are listed in Table S1. The efficiency of depletion was evaluated using immunoblotting. After sequencing to screen for positive expression and plasmid extraction by endotoxin transfection, we selected one of the sequences showing high activity (human NT5E sgRNA: 5′‐CACCGGATCGAGCCACTCCTCAAAG‐3′, 5′‐AAACCTTTGAGGAGTGGCTCGATCC‐3′; mouse Nt5e sgRNA: 5′‐CACCGCAAGGTGCAGCAGATCCGCA‐3′, 5′‐AAACTGCGGATCTGCTGCACCTTGC‐3′), and knocked down NT5E. Then, single‐cell clones were prepared using these sequences. Similarly, the human RAB27A sgRNA (5′‐CACCGAATATGCATGCATCTGTAGC‐3′, 5′‐AAACGCTACAGATGCATGCATATTC‐3′) and mouse Rab27a sgRNA (5′‐ CACCGGTTTCCTCAATGTCCGAAAC‐3′, 5′‐AAACGTTTCGGACATTGAGGAAACC‐3′) showed the highest RAB27A/Rab27a knockout (KO) efficiency. A nontargeting control sgRNA was also used to rule out phenotypes resulting from nonspecific editing. A high concentration of puromycin (Yeasen, Shanghai, China) (HN6: 4 μg/mL, SCC25: 2 μg/mL, SCC7: 12 μg/mL) was used to select stably expressing cells: HN6 NT5E KO, SCC25 NT5E KO, SCC7 Nt5e KO, HN6 RAB27A KO, SCC25 RAB27A KO, SCC7 Rab27a KO.
2.9. NT5E/Nt5e overexpressing lentivirus construction and cell transfection
Lentivirus overexpressing NT5E‐GFP/Nt5e‐GFP (GPF, Green fluorescent protein) was purchased from Hanyin Biotechnology (Shanghai, China). CMV‐MCS‐ZsGreen1‐PGK‐Puro was used as a vector for human and mouse NT5E/Nt5e overexpression. The restriction enzyme sites were XhoI and EcoRI. The oligosaccharide single‐strand DNA sequence (5′ to 3′) for XhoI and EcoRI used in the experiments is listed in Table S1.
The HNSCC cells with NT5E/Nt5e knockout, including HN6 NT5E KO, SCC25 NT5E KO, SCC7 Nt5e KO, and BMSCs cells from human or mouse were transfected with lentivirus using polybrene (Yeasen, Shanghai, China). Overexpression of CD73 was confirmed via immunoblotting assay. A high concentration of puromycin (Yeasen) (HN6: 4 μg/mL, SCC25: 2 μg/mL, SCC7: 12 μg/mL, BMSC: 10 μg/mL) was used to select stably expressing cells: HN6 NT5E OE, SCC25 NT5E OE, SCC7 Nt5e OE (SCC7CD73‐GFP), hBMSC NT5E OE (hBMSCCD73‐GFP), mBMSC Nt5e OE (mBMSCCD73‐GFP).
2.10. Immunohistochemistry
For the specimens collected from 10 normal mucosa donors and 92 patients with HNSCC, all patients had clinical information available, including age, gender, tumour size, location, pathologic stage, number of lymph node metastases, and 5‐year follow‐up information. Paraffin‐embedded slides were deparaffinized using graded xylene. After blocking with 10% bovine serum albumin, the slides were incubated with primary antibodies at 4°C overnight. The secondary antibody (Yeasen, Shanghai, China) was incubated with the slides for 30 min at room temperature (25°C). Hematoxylin and dehydration were used to counterstain the nuclei. The antibodies used in these assays are listed in Table S2. The sections were scanned using a panoramic slice scanner (Pannoramic MIDI, 3DHISTECH, Budapest, Hungary) and analyzed using Aipathwell software (Servicebio Technology, Wuhan, China). We determined the histochemistry score (H‐score) for each slide to evaluate the relationship between the staining intensity of CD73 and prognosis of HNSCC: H‐score = ∑(pi × i) = (percentage of weak intensity area × 1) + (percentage of moderate intensity area × 2) + (percentage of strong intensity area × 3), where PI represents the percentage of the positive signal pixel area and I represents a positive grade. In addition, the staining intensity of CD73 varied in different samples and was classified as weakly positive, modestly positive, and strongly positive.
2.11. Immunofluorescence
Tumour tissues from patients with HNSCC or subcutaneous SCC7 tumours were prepared as paraffin‐embedded slides. The SCC7 tumour tissues and draining lymph nodes (DLNs) within sEVsCD73‐GFP were snap‐frozen in liquid nitrogen and stored at ‐80°C. After attaching and fixing the tissue sections on the slides, antigen retrieval was performed. The sections were sealed with 10% normal goat serum at room temperature for 1 h and then incubated with specific primary antibodies overnight at 4°C. For HNSCC patient's tumour tissues, CD73, CD68, CD4, CD8, Foxp3, and α‐SMA were analysed. CD73, CD4, CD8, F4/80, and Foxp3 were detected on subcutaneous SCC7 tumour tissues. The sample was incubated with a fluorescent secondary antibody. Cellular nuclei were stained with DAPI (CST, USA). Images were acquired using BioTek Cytation 5 (BioTek, Winooski, VT, USA).
For cellular immunofluorescence, macrophages were seeded into 35‐mm confocal dishes (Corning, USA). The sEVs (CD73‐GFP labelled or labelled with anti‐CD73‐FITC) were cocultured with macrophages for 1 h. After washing the cells three times, CellMask Plasma Membrane Stains (Thermo Fisher, USA) (5 μg/mL) were added and incubated for 1 h. The 4% paraformaldehyde was used to fix the macrophages. Images were taken using Leica Microsystems confocal fluorescence imaging microscope with LAS AF software (version 2.0; Wetzlar, Germany).
Macrophages were seeded in 24‐well plates and cocultured with sEVs for 3 h with or without 100 μM pyrrolidine dithiocarbonate (PDTC, Abmole, USA). The 4% paraformaldehyde was used for cell fixation. The samples were permeabilized with 0.2% Triton X‐100 and blocked with 10% goat serum. After incubating with specific primary antibodies (anti‐p65) overnight at 4°C, cells were incubated with a fluorescent secondary antibody. Images were taken using BioTek Cytation 5. The antibodies used are shown in Table S2.
2.12. Western blot analysis
The CR cells/HNSCC/BMSC cells‐derived sEVs and cell lysates from M2 were collected in sodium dodecyl sulfate lysis buffer (Beyotime, Shanghai, China) and stored at ‐80°C until use. The cell lysates (20 μg) and sEVs proteins (3 μg), were separated using sodium dodecyl sulfate‐polyacrylamide gel electrophoresis and then transferred onto 0.22‐μm polyvinylidene fluoride membranes (Merck Millipore, Billerica, MA, USA). Membranes were blocked with nonfat milk for 2 h at room temperature and incubated overnight at 4°C with primary antibodies. β‐Actin and GAPDH were used as loading controls. The membranes were treated with secondary antibodies and visualized using ECLUltra (New Cell and Molecular Biotech, Suzhou, China). The antibodies used are listed in Table S2.
2.13. Co‐culture assay
Macrophages were differentiated from THP‐1 cells for 48 h prior to the experiment. Macrophages (1 × 106 cells) were cocultured in a six‐well format with Transwell membranes (0.4‐μm pores, Merck Millipore, USA), whereas macrophages were placed below the membranes and the HNSCC cell lines HN6, HN6 NT5E KO, HN6 NT5E OE, HN6 RAB27A KO, SCC25, SCC25 NT5E KO, SCC25 NT5E OE, SCC25 RAB27A KO (1 × 105 cells) were placed on the permeable membranes. All cells were cultured with medium without FBS for 24 h and the macrophages were digested for flow cytometry analysis.
The sEVs were extracted from HN6 cells, HN6 NT5E KO cells, SCC25 cells, SCC25 NT5E KO cells, hBMSCs, and hBMSC NT5E OE. The sEVs (50 μg) were mixed into M2 macrophages (1 × 106 cells) and cultured for 24 h. All cells were cultured with medium without FBS.
2.14. RNA extraction and quantitative real‐time PCR
Total RNA was extracted from the cells using TRIzol reagent (Invitrogen, Carlsbad, CA, USA) and reverse‐transcribed into cDNA using a Prime Script RT reagent Kit (Takara, Shiga, Japan). A SYBR Premix Ex Taq reagent kit (Takara) was used to amplify the cDNA according to the manufacturer's protocol. The specific primers used for quantitative real‐time PCR are shown in Table S3. GAPDH (Sangon Biotech, Shanghai, China) was used as an internal control. The 2ΔΔct method was used to calculate the relative expressions of the genes (Biray Avci et al., 2020).
2.15. Cell proliferation assay
RAB27A in HNSCC cells (HN6, SCC25, and SCC7) was knocked out by transfection with LentiCRISPRv2. Three vectors carrying RAB27A sgRNA were used. The RAB27AKO cells or their wild‐type control cells were seeded into 96‐well plates (1 × 103 cells/well). CCK‐8 (NCM Biotech, Suzhou, China) was added to the plate, and the optical density value was measured at 450 nm for 3 days post‐seeding. All experiments were performed in triplicate.
2.16. ELISA
Macrophages (1 × 106 cells/well) were seeded into six‐well plates and cocultured with 50 μg sEVs derived from HN6, HN6 NT5E KO, SCC25, SCC25 NT5E KO, BMSC, and BMSC NT5E OE. The experiments were divided into seven groups: M0, M2, M2+sEVsHNSCC, M2+sEVsHNSCC‐ NT5E KO, M2+sEVsBMSC, M2+sEVsBMSC‐ NT5E OE, and M2+sEVsBMSC‐ NT5E OE + Lat A (30 μM, MedChemExpress, Monmouth Junction, NJ, USA). The CM was composed of RPMI1640 medium without FBS and was collected after incubation for 24 h. After centrifugation at 10,000×g for 10 min at 4°C, the coculture supernatant was added to a 96‐well plate to detect human IL‐6, IL‐10, TNF‐α, and TGF‐β1 using a human IL‐6 enzyme‐linked immunosorbent assay (ELISA) kit (R&D Systems, Minneapolis, MN, USA), human IL‐10 ELISA kit (R&D Systems), human TNF‐α ELISA kit (R&D Systems), and human TGF‐β1 ELISA kit (R&D Systems) according to the manufacturer's instructions.
The procedure for extracting sEVs from serum samples has been previously described in 2.6. A human CD73 ELISA kit (R&D Systems) and ELISA ancillary reagent kit (R&D Systems) were used to measure the CD73 level in sEVs from the serum.
2.17. Flow cytometry
The samples were processed into single‐cell suspensions and stained with antibodies for 20 min on ice. After washing with PBS and centrifugation for 5 min at 1,000×g, the pellet was suspended in 200 μL PBS and evaluated using a flow cytometer (FACS Calibur, BD Biosciences, Franklin Lakes, NJ, USA). CD68+ were used to label human macrophages, CD68+ CD163+ CD206+ to label human M2 macrophages. F4/80+ CD45+ or F4/80+ to label mouse macrophages, and CD8+ or CD45+ CD3+ CD8+ to gate mouse CD8+ T cells. In addition, CD4+ or CD45+ CD3+ CD4+ cells were used to gate mouse CD4+ T cells, whereas CD4+ Foxp3+ or CD45+ CD3+ CD4+ Foxp3+ cells were used to label Tregs. For mouse tissues, a fixable viability stain (BD Pharmingen, San Diego, CA, USA) was used. The expression of PD‐1, PD‐L1, and CD73 was also detected using flow cytometry. The antibodies used are listed in Table S1.
2.18. Phagocytosis analysis
Macrophages (1 × 106 cells/well) were seeded into six‐well plates. The sEVs (50 μg) from HNSCC cell lines (HN6 and SCC25) were added to macrophages and cultured for 24 h. Latrunculin A (Lat A, 30 μM) was co‐inoculated with sEVs into macrophages. For phagocytosis analysis, 1 mg/mL pHrodo™ Deep Red Escherichia coli BioParticles Conjugate (P35364, Invitrogen) was added to the macrophages (1 × 106 cells) and incubated for 2 h at 37°C according to the manufacturer's instructions. The macrophages were then rinsed with PBS and digested for flow cytometry analysis. Red fluorescence represents macrophages that took up the E. coli BioParticles.
2.19. In vivo sEVs injection and tumorigenicity assay
C3H/He mice aged 6–8 weeks were used for the animal experiments. The sEVs from SCC7 Nt5e OE‐GFP cells were injected into tumours from mice that underwent subcutaneous tumorigenesis. After 24 h, the tumours were dissected and frozen for immunofluorescence analysis.
In experiments involving sEVs foot pad injection, SCC7, SCC7CD73‐GFP, mBMSC, and mBMSCCD73‐GFP cells were prepared for sEVs extraction. First, sEVs derived from SCC7CD73‐GFP (10 ng‐50 μg) were injected into foot pads to determine the optimum concentration of sEVs injection and the DLNs were dissected after 24 h. GFP fluorescence was detected using flow cytometry. The sEVs derived from SCC7CD73‐GFP (10 μg) were injected, and the DLNs were harvested at 30 and 60 min. Immunofluorescence was performed to examine the presence of sEVs in the DLNs. The sEVs derived from SCC7CD73‐GFP (25 μg) were injected into the foot pad, and GFP fluorescence was detected using flow cytometry after 24 h. The sEVsSCC7 (25 μg) were injected, 24 h later, flow cytometry analysis was performed. The percentages of CD73+/PD‐1+ in CD4+ T cells, CD8+ T cells, macrophages, and Tregs were determined. Next, sEVsSCC7, CD73‐GFP+ sEVs from mouse bone marrow stromal cells (sEVsmBMSC), and sEVsmBMSC‐ Nt5e OE (25 μg) were injected every day, and DLNs were harvested at different time points for flow cytometry analysis. The percentages of CD4+ T cells, CD8+ T cells, macrophages, M2 macrophages, and Tregs were determined. In addition, subcutaneous tumorigenesis experiments using SCC7 and SCC7 Nt5e KO cells (2 × 105) were performed. The sEVs release inhibitor GW4869 (2 mg/kg) was administered intraperitoneally every 2 days. After 21 days, the DLNs were dissected for flow cytometry to analyse the infiltrations of macrophages and CD8+ T cells.
In the first subcutaneous tumorigenesis experiment, SCC7 and SCC7 Rab27a KO cells (8 × 105) were injected into the backs of mice. Meanwhile, the sEVs (50 μg) from SCC7, SCC7 Nt5e KO, and SCC7 Nt5e KO+ Nt5e OE (abbreviated as SCC7 Nt5e OE) were injected into the tumour every 2 days starting on day 3. Lat A (0.15 mg/kg) was injected intraperitoneally every 2 days starting on day 0. Tumour volumes were measured every 3 days after treatment. Mice were sacrificed at 15 days after injection and the tumours were removed for further analysis.
In the second subcutaneous tumorigenesis experiment, SCC7 and SCC7 Rab27a KO cells (8 × 105) were injected into the backs of mice. The sEVs (50 μg) from mBMSCs and mBMSC Nt5e OE cells were injected into the tumour every 2 days starting on day 3. Tumour volumes were measured every 3 days after treatment. Mice were sacrificed at 15 days after injection and the tumours were removed for further analysis.
Finally, in the third subcutaneous tumorigenesis experiment, SCC7 and SCC7 Rab27a KO cells (8 × 105) were injected into the backs of mice. The sEVs (50 μg) from SCC7 Nt5e KO and SCC7 Nt5e OE were injected into the tumour every 2 days starting on day 3. Anti‐PD‐1 drugs (10 mg/kg) (BioXcell, Lebanon, NH, USA) were injected into mice intraperitoneally every 2 days starting on day 0. Tumour volumes were measured every 3 days after treatment. Mice were sacrificed at 15 days after injection and the tumours were removed for further analysis.
2.20. Transcriptome analysis of NT5E based on TCGA datasets
The Timer2.0 platform was used for transcriptional analysis of CD73 (NT5E) across published datasets from The Cancer Genome Atlas (TCGA), and the Spearman correlation coefficient was determined to evaluate the NT5E expression level and TME cell compositions in the HNSCC cohort (T. Li et al., 2020). Transcriptome data of TCGA HNSCC were downloaded from the GDC portal via TCGA biolinks following a standard protocol (Mounir et al., 2019), and correlation analysis of the NT5E expression level and TME cell compositions was performed using the R package immunedeconv (Sturm et al., 2019). The TME matrix was calculated using the CIBERSORT algorithm (Newman et al., 2015). Kaplan–Meier and Cox regression analyses were performed to investigate the prognosis and clinical significance of NT5E expression levels, and the results are shown as survival curves.
2.21. Cell line transcriptome analysis
sEVs secreted by HNSCC cell lines (HN6, HN6 NT5E KO, SCC25, SCC25 NT5E KO, CAL27, and CAL27 NT5E KO) were harvested and cocultured with macrophages, and the total RNA was sequenced using the Illumina HiSeq 2500 platform (San Diego, CA, USA). Raw reads were processed into fragments per kilobase of exon model per million mapped reads (FPKM) values for further investigation. The expression levels of immune checkpoint markers were shown in a heatmap to determine the effect of incubation with sEVsHNSCC or sEVsHNSCC‐ NT5E KO. Genes upregulated by sEVsHNSCC and downregulated by sEVsHNSCC‐ NT5E KO were screened, and 143 genes were identified. Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis was performed using cluster profiler as previously described (Yu et al., 2012), and the enriched pathways and correlated genes are shown in a Circos‐gram. Regulatory factors were analyzed using the Metascape platform (Y. Zhou et al., 2019). The results indicated that NFκB1 was significantly enriched with the highest gene counts. Gene Set Enrichment Analysis was performed to validate the status of IκB phosphorylation and NIK NF‐κB signalling pathway. Changes in the downstream factors IL6, IL10, TNFA, TGFB1, CD274, CD279, and lymphocyte‐activation gene 3 (LAG3) were detected using quantitative real‐time PCR.
2.22. Validation of CD73 as a marker for immune checkpoint blockade therapy via published data
The Tumour Immune Dysfunction and Exclusion (TIDE; http://tide.dfci.harvard.edu) platform was used to evaluate the potential of CD73 as an ICB predictor (P. Jiang, Gu, et al., 2018), the data of provided clinical trials included Glioblastoma, Melanoma, HNSCC, NSCLC, Kidney cancer, Bladder cancer, Gastric cancer, while data from both pre and post ICB were available in some studies, the targeted immune check point or immunotherapy strategies are not limited to PD‐1, PD‐L1, CTLA4 and ACT (Adoptive immune Cell Therapy). The area under curve (AUC) values from different indices or markers were compared with CD73 (NT5E), including TIDE score, MSI.Score, TMB, CD274 CD8, IFNG, T. Clonality, B. Clonality, and Merck18. As described by Mariathasan et al. (2018), the R package IMvigor210CoreBiologies was downloaded to investigate the relationship between NT5E and the tumour response to PD‐L1 blockade, as well as response to therapy and immune phenotype.
2.23. Statistical methods
SPSS V.21.0 statistical software (SPSS, Inc., Chicago, IL, USA) was used to analyze the measurement data, and the mean ± standard deviation is shown in the figures. Student's t‐test and Mann–Whitney test were used to compare the means of two or more groups depending on whether the data was normally distributed. Spearman's correlation test was used for correlation analysis. Significance was determined as *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001. Statistical software R (The R Group for Statistical Computing, Vienna, Austria) and GraphPad Prism 9 (GraphPad, Inc., La Jolla, CA, USA) were used to visualize the data.
3. RESULTS
3.1. CD73 is overexpressed in sEVs derived from HNSCC tumour cells
Primary tumour and paired epithelial cells were obtained from HNSCC tumour tissues and adjacent tissues using a previously established CR cell system, which enabled epithelial cells from patients to rapidly generate and propagate indefinitely in vitro (X. Liu et al., 2012) (Figure 1a,b; Tjin et al., 2018). sEVs were isolated from the CR cell culture medium using ultracentrifugation. Transmission electron microscopy and particle size analysis confirmed the bilayer‐enclosed morphology of sEVs with an average diameter of 122.5 nm (Figure 1c,d). After validating the quantification of sEVs based on the positive expression of HSP90, CD9, CD63, HSP70, and Rab5 and negative expression of GRP94 (Greening et al., 2015) (Figure 1e; Witwer et al., 2013), we performed quantitative proteomic analysis of the sEVs from three different groups (NC sEVs from adjacent tissues, nonmetastatic HNSCC tumour cell‐derived sEVs, and sEVs from primary tumour cells with evidence of metastasis). The Venn diagram shows the identified proteins (Figure 1f and S1A,B). A total of 66 unique proteins were identified in both the nonmetastatic and metastatic HNSCC‐derived sEVs group, while 336 were found only in the metastatic sEVs group, in which CD73 was detected (Table S4). CD73 was found in all samples from the metastatic sEVs group and one sample from the nonmetastatic sEVs group, but not in sEVs from adjacent epithelial cells (Figure 1f,g). Western blot analysis showed that CD73 was overexpressed in sEVs from tumour tissues compared to in adjacent tissues (Figure 1h). In addition, both quantitative mass spectrometry and western blot analysis revealed that sEVs from HNSCC cell lines carried varying amounts of CD73, among which sEVs from HN6 and SCC25 cells showed higher expression of CD73 (Figure S1C–E).
FIGURE 1.
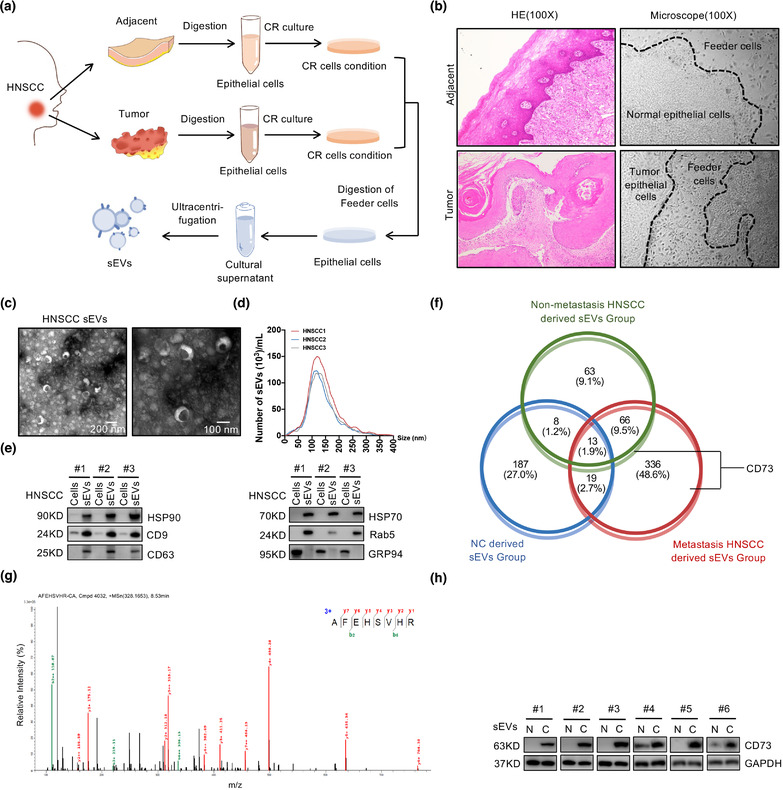
CD73 was overexpressed in the tumour cells derived sEVs of HNSCC patients. (a) Schematic of the progress of extracting sEVs from Conditional Reprogramming (CR) cells. (b) The histological and morphological characteristics of CR coculture of keratinocytes and feeder cells from HNSCC and adjacent normal tissues. (c) Electron microscopy images of sEVs from HNSCC. Scale bar, 100 and 200 nm. (d) Nanoparticle tracking analysis (NTA) of sEVs from HNSCC. (e) Immunoblotting for sEVs biomarkers. (f) Quantitative proteome results of cells released sEVs from HNSCC or adjacent normal tissues, shown by venn diagram. (g) The mass spectrum identified CD73 in sEVs derived from HNSCC cells. (h) Representative western blotting for CD73 in sEVs from six pairs of HNSCC samples
3.2. CD73 is internalized by macrophage through sEVs to promote HNSCC malignancy
Analysis of TCGA dataset showed that NT5E was highly expressed in different types of tumours, including HNSCC (Figure S2A). Moreover, higher NT5E levels were associated with a poorer overall survival rate of patients with HNSCC (p = 0.0039) (Figure S2B). Next, we investigated the correlation between CD73 protein levels in HNSCC tissues and sEVs in malignant tumour progression.
We first characterized CD73 expression via immunohistochemistry in 92 HNSCC tumour tissues and 10 oral epithelial tissues from healthy donors. The expression of CD73 varied across different HNSCC samples and was markedly increased compared to that in the normal oral mucosa (Figure 2a). Based on the staining intensity, analysis of CD73 expression with clinical information of patients with HNSCC revealed that subjects with higher levels of CD73 had an increased risk of lymph node metastasis, larger tumour size, poorer pathologic stage, and lower 5‐year overall survival rate (Figure 2b; Table 1). Taken together, these data indicate that CD73 is overexpressed in HNSCC tissues and is associated with a poor prognosis and high possibility of lymph node metastasis.
FIGURE 2.
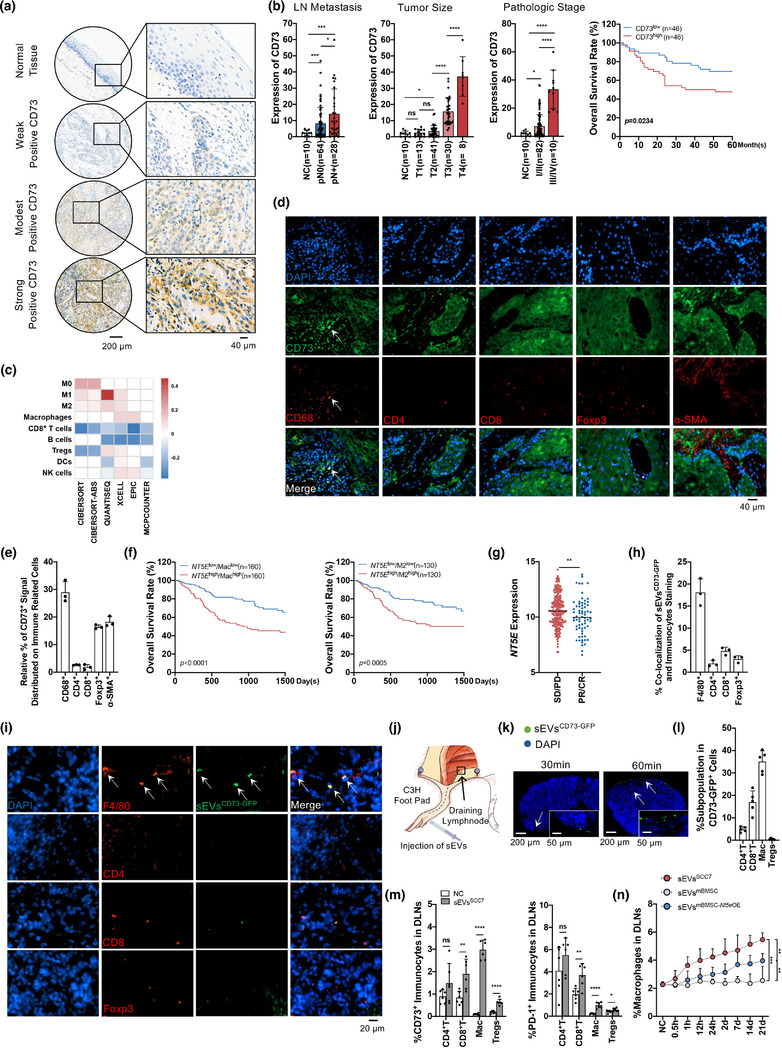
CD73 in sEVs was closely contributed to tumour associated macrophages and HNSCC malignant progress. (a) IHC analysis of CD73 expression levels in tissue microarrays, containing 10 normal tissues and 92 HNSCC tissues. Representative immunohistochemistry images of normal tissue, weak positive, modest positive, and strong positive CD73 staining were shown. Scale bar: 200 μm, 40 μm. (b) Statistical analysis about lymph node metastasis (LN metastasis), tumour stage, pathologic stage and overall survival rate with CD73 stain intensity in HNSCC tissues. (c) The correlation of NT5E expression with immune infiltration level in HNSCC investigated in TCGA database based on six deconvolution algorithms. (d) Immunofluorescence staining of CD73 distribution (green) and different resident immune‐associated cell types (red), including macrophages (CD68+), CD4+ T cells, CD8+ T cells, Tregs (Foxp3+) and CAFs(α‐SMA+) in tissues from HNSCC patients. Scale bar: 40 μm. (e) The percentage of costaining of CD73 with macrophages, CD4+ T cells, CD8+ T cells, Tregs and CAFs in HNSCC patient tumour samples. (f) NT5E expression (NT5E high, NT5E low) as a marker for prediction of overall survival rate in TCGA HNSCC cohort. Data were classified into low macrophage/low M2 macrophage signature (Maclow/M2low) and high macrophage/high M2 macrophage signature (Machigh/M2high). Log‐rank Mantel‐Cox test was used to assess significance. (g) The association between sensitivity of anti‐PD‐L1 treatment and NT5E expression were studied through the public data of IMvigor210CoreBiologies, lower NT5E group exhibited increased sensitivity to PD‐L1 blockade than higher NT5E group. (h) The percentage of SCC7 Nt5e OE‐GFP‐derived sEVsCD73‐GFP signal that distributed on macrophages (F4/80+), CD4+ T cells, CD8+ T cells, Tregs (Foxp3+) in SCC7 tumour‐bearing C3H mice. (i) The SCC7 Nt5e OE‐GFP‐derived sEVsCD73‐GFP were injected into tumours on C3H mice. After 24 h, fluorescence visualization identified the coexpression of CD73‐GFP in sEVs (green) with immunocytes(red) in tumours. Scale bar: 20 μm. (j) Schematic of sEVs injection through foot pad and its draining lymph node (DLNs). (k) Fluorescence microscopy showed sEVsCD73‐GFP (green) in whole DLNs imaging after 30 or 60 min of sEVsCD73‐GFP (10 μg) injection. Scale bar: 200 μm, 50 μm. (l) Flow cytometry analysis for subpopulation in CD73‐GFP+ cells of DLNs (sEVsCD73‐GFP: 25 μg, 24 h). (m) The sEVsSCC7 (25 μg) were injected into foot pads. After 24 h, flow cytometry analyzed the expression of CD73+ or PD‐1+ immunocytes in DLNs. (n) Flow cytometry analysis for percentage of macrophages in DLNs after injecting sEVs (25 μg) derived from SCC7 cells (sEVsSCC7), mBMSC cells (sEVsmBMSC) or mBMSC‐Nt5eOE cells (sEVsmBMSC‐ Nt5e OE) every day. DLNs were harvested at different time point. Lymph nodes from untreated mice were used as normal control. CD4+ T: CD4+ T cells, CD8+ T: CD8+ T cells, Mac: Macrophages. Data were analysed by Mann‐Whitney test. (ns, no significant difference, *: p < 0.05, **: p < 0.01, ***: p < 0.001, ****: p < 0.0001)
TABLE 1.
Relationship between CD73 level and clinicopathologic features in HNSCC tumour tissues (n = 92)
| Characteristics | Number of patients | % proportion | CD73 expression of tissues | p‐Value |
|---|---|---|---|---|
| Age (years) | 0.982 | |||
| ≥60 | 87 | 94.57 | 9.94 | |
| <60 | 5 | 5.43 | 9.82 | |
| Gender | 0.743 | |||
| Male | 48 | 52.17 | 10.50 | |
| Female | 44 | 47.83 | 9.67 | |
| Lymph node metastasis | 0.029 | |||
| Non‐metastasis | 64 | 69.57 | 8.26 | |
| Metastasis | 28 | 30.43 | 14.19 | |
| Tumour size | 0.018 | |||
| I | 13 | 14.13 | 3.87 | |
| II | 41 | 44.57 | 10.50 | |
| III | 30 | 32.61 | 11.06 | |
| IV | 8 | 8.69 | 27.37 | |
| Pathologic stage | <0.0001 | |||
| I | 29 | 31.52 | 9.37 | |
| II | 53 | 57.61 | 11.22 | |
| III | 10 | 10.87 | 33.39 | |
| IV | 0 | 0 | 0 |
By retrieving methods integrated with R package immunedeconv, the relationship between CD73 (NT5E) and tumour‐infiltrating immune cells in HNSCC was analyzed. Different deconvolution algorithms showed that NT5E expression was closely related to the macrophage composition (Figure 2c). The result suggests that macrophages had the highest sensitivity to NT5E than other immunocytes in HNSCC. Immunofluorescence staining of HNSCC tumour tissues confirmed that the CD73 signal was mostly overlapped with macrophages (Figure 2d,e), suggesting that macrophages were strongly correlated with CD73 in HNSCC.
In addition, HNSCC samples from TCGA database were distributed into groups based on the level of macrophage infiltration and NT5E expression level. Without considering the macrophage composition, the NT5E high group showed a lower overall survival rate (Figure S2B). The samples were then divided into high macrophage infiltration (Machigh) and low macrophage infiltration (Maclow) groups. Subjects with higher macrophage levels and higher NT5E expression exhibited the lowest overall survival rate (Figures 2f and S2C). In addition, through the analysis of data on tumour ICB in metastatic urothelial cancer (Mariathasan et al., 2018), we found that higher expression level of NT5E reduced the sensitivity of anti‐PD‐L1 treatment (Figure 2g), which illustrated the important role of NT5E in attenuating the effect of immune therapy. In the TME, macrophages tend to differentiate into M2 macrophages, which play a central role in tumour propagation (H. W. Wang & Joyce, 2010). The influence of M2 polarization and NT5E expression on clinical prognosis was similar to that of macrophages (Figures 2f and S2E,F). These data demonstrate that the levels of macrophages and NT5E have the potential to be used as biomarkers for predicting the survival rate and metastasis risk in HNSCC.
Studies have shown that the expression level of CD73 in immunocytes from patients with cancer is significantly higher and associated with poor prognosis (Neo et al., 2020). Since sEVs derived from HNSCC cells carried CD73, we determined whether sEVsCD73 could be further taken up by immunocytes and affect the tumour‐related immune response. To identify the immunocytes closely related to sEVsCD73, sEVsCD73‐GFP was injected into subcutaneous SCC7‐derived tumours in C3H mice. After 24 h, the tumours were removed, and immunofluorescence analysis was performed. Remarkably, sEVsCD73‐GFP were most colocalized with macrophages (Figure 2h,i), suggesting that sEVsCD73‐GFP was mainly phagocytosed by macrophages.
As sEVsCD73 are closely related to lymph node metastasis, SCC7‐derived sEVs were injected through the mouse foot pads to explore the effect of sEVsCD73 on DLNs (Figure 2j). At the lowest concentration of sEVsCD73 (10 μg), green fluorescence was observed in the DLNs at 30 min after sEVsCD73‐GFP injection (Figures 2k and S2G). Flow cytometry revealed that the CD73‐GFP signal was mostly distributed in the macrophages (Figure 2l). In addition, CD73 and PD‐1 were upregulated in CD4+ T cells, CD8+ T cells, macrophages, and Tregs at 24 h post injection (sEVsCD73, 25 μg) (Figure 2m). To detect the effects of CD73 in sEVs on immunocytes, we introduced engineered mBMSCCD73‐GFP. Compared with sEVmBMSC, sEVsmBMSC‐ Nt5e OE enhanced the infiltration of macrophages, CD4+ T cells, and Tregs, with fewer CD8+ T cells in the DLNs (Figures 2n and S2H,I). Next, subcutaneous tumour was performed in C3H mice for 21 days (Figure S2J,K), and DLNs were dissected to analyse the effect of tumour cell‐derived sEVsCD73 on lymph nodes microenvironment. GW4869 was introduced as an sEVs releasing inhibitor. In DLNs, HNSCC‐derived sEVsCD73 polarized macrophages toward an immunosuppressive phenotype and depleted CD8+ T cells, with increased expression of CD73 and PD‐1 (Figure S2L,M). These results indicate that tumour cell‐derived sEVs transform the microenvironment of DLNs into an immunocompromised niche to facilitate tumour metastasis, in a process that involved CD73 in sEVs.
3.3. HNSCC‐derived sEVs regulate macrophage‐mediated immunosuppression through CD73
sEVsCD73 was closely related to TAMs, hence we next evaluated the impact of CD73 in HNSCC‐derived sEVs on macrophages. We utilized CRISPR/Cas9‐mediated mutagenesis to knock out RAB27A from HNSCC cells (HN6, SCC25) to remove the sEVs (Figure S3A). Deletion of RAB27A did not affect cell proliferation (Figure S3B). Moreover, NT5E in HNSCC cell lines was knocked out using CRISPR/Cas9, with CD73 vanishing in sEVs (Figure S3C). NT5E was overexpressed in NT5EKO cells through lentivirus transfection, and the level of CD73 in sEVs was measured using western blotting (Figure S3D).
To investigate how HNSCC‐derived sEVsCD73 modulates TAMs, macrophages were cocultured with tumour cells (HN6, SCC25) under different conditions for 24 h (Figure 3a). Compared with NC cell medium, the tumour cell culture medium increased the level of CD73 in macrophages and had a stronger effect than the NT5EKO cell medium (p < 0.0001), whereas the percentage of CD73+ macrophages did not significantly differ between the NC cell medium and RAB27AKO cell medium (p = 0.093), suggesting that CD73 is mainly released through sEVs (Figures 3b and S3E,F). The rescue experiment between HNSCC cell medium and HNSCC NT5E OE cell medium showed no significant difference (p = 0.5837) (Figure 3b). The percentage of M2 macrophages showed a similar trend, suggesting that sEVsCD73 can transform macrophages into a malignant phenotype (Figure 3b).
FIGURE 3.
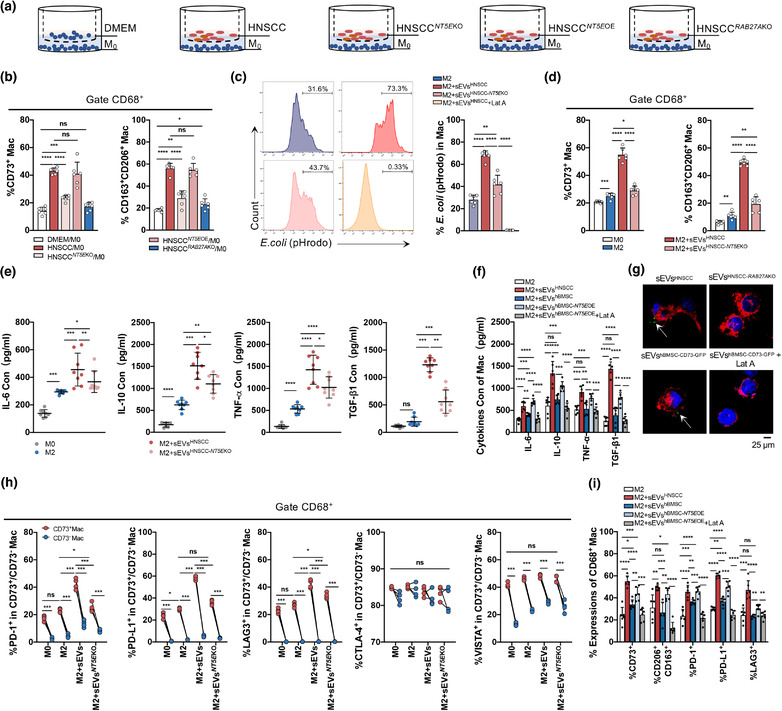
The effect of CD73 in sEVs derived from HNSCC cells on the function of macrophages. (a) Macrophages were cocultured with DMEM or two HNSCC lines (SCC25, HN6) with or without NT5E/RAB27AKO for 24 h. HNSCC NT5E OE, referred to overexpression of NT5E performed on HNSCC NT5E KO cells. (b) Flow cytometry analysis for percentage of CD73+ macrophages and M2 macrophages (CD163+CD206+) in coculture system. (c–i) The sEVs (50 μg) were cocultured with macrophages (1 × 106) for 24 h. The sEVsHNSCC derived from two HNSCC lines: SCC25 and HN6. The sEVshBMSC‐ NT5E OE derived from hBMSC cells with NT5E overexpression. Latrunculin A (Lat A, 30 μM) was used as the inhibitor of sEVs uptaken. (c) Compromised phagocytosis of M2 macrophages treated with or without sEVs or Lat A. The percentage of pHrodo dyes of M2 macrophages was analyzed by flow cytometry. (d) The percentage of CD73+ macrophages and M2 macrophages after macrophages cocultured with sEVs. (e, f) Concentration of IL‐6, IL‐10, TNF‐α, and TGF‐β1 levels in macrophages conditional medium after sEVs education by ELISA. (e)The sEVs were derived from HNSCC cells or HNSCC NT5E KO cells. (f) The sEVs were derived from HNSCC, hBMSC or hBMSC NT5E OE cells with or without Lat A (30 μM). (g) Macrophages were cocultured with anti‐CD73‐FITC labelled sEVs from HNSCC cell lines control or RAB27AKO, CD73‐GFP labelled sEVs from hBMSC treated with or without Lat A were cocultured with macrophages for 1 h, and Laser Scanning Confocal Microscopy was used to analyze the internalization of HNSCC‐derived sEVs into macrophages (Scale bar = 25 μm). (h) Flow cytometry analysis for differential expression of immune check point (PD‐1, PD‐L1, LAG3, CTLA‐4, VISTA) comparing M0 and M2 macrophages which were educated with sEVs from HNSCC cells (h) or hBMSC with or without Lat A (i). Data were analysed by Mann‐Whitney test (ns, no significant difference, *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001)
In the TME, TAMs account for the largest proportion of immunocytes and M2 macrophages have tumour‐promoting properties (W. Li et al., 2019). M2 macrophages are narrowly defined as TAMs (Laviron & Boissonnas, 2019); therefore, M2 macrophages were evaluated to assess the function of macrophages under various treatments. The level of CD73 in sEVs affected the proportion of CD73+ macrophages, which increased with the accumulation of sEVsCD73 in the medium and peaked after treatment with 50 μg sEVsHNSCC. At 100 μg/L sEVs, the ratio of CD73+ macrophages decreased, although not significantly (p = 0.1235) (Figure S3G), possibly because of a negative feedback regulation mechanism. Escherichia coli (E. coli) pHrodo, which shows red fluorescence, was added to macrophages 2 h before cell harvest to analyze phagocytosis of macrophages cocultured with purified sEVs (50 μg) for 24 h. Fluorescence‐activated cell sorting revealed that phagocytosis was significantly enhanced in the presence of sEVsCD73 (p < 0.0001), and downregulated by the phagocytosis inhibitor Lat A (30 μM). (Figure 3c). In addition, M2 macrophages had a higher level of CD73 than M0 macrophages, and sEVsCD73 increased the percentage of CD73+ macrophages and induced the production of more M2 macrophages (Figure 3d).
Given that CD73+ macrophages induce immunosuppression in some tumours, we analysed the secretion of macrophage‐related cytokines, including IL‐6, IL‐10, TNF‐α, and TGF‐β1, which indicate the malignant phenotype of TAMs (Q. Liu et al., 2020; Wan et al., 2014; Yang et al., 2021; H. Zhang et al., 2020). These inflammatory cytokines increased significantly in the supernatant of macrophages cocultured with sEVs containing CD73 (Figure 3e), indicating that macrophages with higher levels of CD73 have stronger tumour‐promoting effects compared to normal M2 macrophages. To examine the effect of CD73 in sEVs, sEVs derived from hBMSC NT5E OE was introduced. CD73 was present in hBMSC‐derived sEVs but showed lower expression than in sEVsHNSCC (Figure S3H). The expression of cytokines also increased when macrophages were cocultured with sEVs derived from hBMSCs overexpressing CD73, whereas this trend was reversed by Lat A (Figure 3f). These results demonstrate that engineered sEVs mimic the malignant effect on macrophages caused by sEVsHNSCC through CD73.
CD73 is a transmembrane protein, and immunofluorescence showed that sEVsCD73 could be taken up by macrophages and presented on the membrane and within vesicle‐like structures (Figure 3g). Similarly, sEVs derived from hBMSCs transfected with GFP‐labeled CD73 were phagocytosed, and green fluorescence was observed on the macrophage surface. However, these phenomena disappeared following knock out of RAB27A or treatment with Lat A (Figure 3g).
Next, we determined whether sEVsCD73 functions similarly to cell surface CD73 to influence classic immune checkpoints in macrophages. Flow cytometry showed that sEVsCD73 enhanced the expression of LAG3, PD‐1, and PD‐L1, but had little effect on VISTA and CTLA‐4 expression (Figure 3h). Macrophages were further classified into CD73high and CD73low groups based on their CD73 expression levels using flow cytometry analysis, with the negative control used as a cut‐off point. LAG3, PD‐1, PD‐L1, and VISTA were clearly upregulated in CD73high macrophages compared to in CD73low macrophages (Figure 3h), suggesting that macrophages can acquire CD73 from sEVsCD73 and facilitate the immunosuppressive function in tumours. When CD73 was overexpressed in engineered sEVs, the expression of CD73, LAG3, PD‐1, and PD‐L1 increased significantly, and more macrophages were differentiated into the M2 subtype, compared with sEVsBMSC group (Figure 3i). These results were consistent with tumour‐derived sEVsCD73 and indicate that sEVsCD73 is a key suppressor of the tumour immune microenvironment.
3.4. CD73‐deficient sEVs fail to trigger immune suppression and aggressive tumour growth in vivo
We next evaluated the effects of the absence of CD73 in sEVs versus that of Rab27a in tumorigenesis to determine whether sEVs function specifically through CD73 to mediate immunosuppression and tumour progression in vivo. Rab27a and Nt5e were stably knocked out in SCC7 cells using the CRISPR/Cas9 technique (Figure S3A, C), and Nt5e was rescued by CD73‐GFP lentivirus transfection (Figure S3D).
Intratumoral injection of sEVs from SCC7, SCC7‐Nt5eKO, or SCC7‐Nt5eOE cells was performed in a tumour‐bearing mouse model (Figure 4a). Compared with the SCC7 control group, the growth rate was slower, and size of tumours was significantly smaller in SCC7 Rab27a KO group, whereas Rab27aKO did not affect SCC7 proliferation in vitro (Figure S3B). Furthermore, compared with control group, the infiltration of CD8+ T cells increased, whereas TAMs, CD4+ T cells, and Tregs decreased in the SCC7 Rab27a KO group, expressing lower levels of CD73 and PD‐1 (Figures 4e–g and S4A–D). Thus, sEVs may be important in forming the immunosuppressive microenvironment during neoplasia. Injection of sEVs from SCC7 (sEVsSCC7) significantly accelerated tumour growth compared to sEVs from SCC7 Nt5e KO (sEVsSCC7‐ Nt5e KO) (Figure 4b–d). Exogenous sEVsCD73 recruited more TAMs, CD4+ T cells, and Tregs and accelerate the exhaustion of CD8+ T cells, attenuating the immune defence (Figures 4e–g and S4D). In addition, a rescue experiment involving injection of sEVsSCC7‐ Nt5e OE showed similar results as observed for the sEVsCD73 group (Figures 4e–g and S4C,D), suggesting that CD73 in HNSCC‐derived sEVs play a main role in this model. The Lat A‐treated group exhibited a smaller tumour size and less immunosuppression than control group (Figures 4b–g and S4C,D), suggesting the importance of sEVs internalized by TAMs.
FIGURE 4.
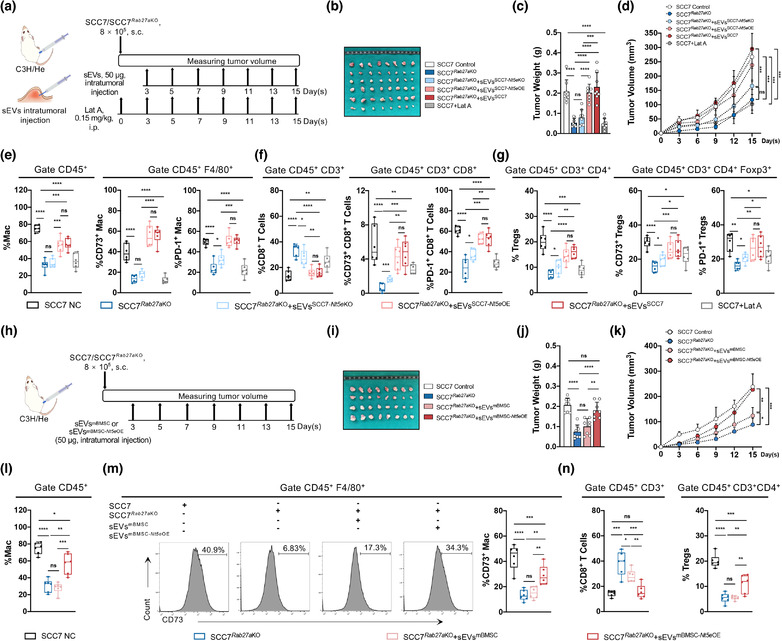
Absence of CD73 in sEVs rescues immune suppression and restrains tumour growth in vivo. (a) Schematic of subcutaneous tumorigenesis in vivo experiment, followed with intratumoral injection of sEVs which were collected from SCC7, SCC7‐Nt5eKO or SCC7‐Nt5eOE cells grown in vitro. (b) The exhibition of isolated tumours. (c and d) The tumour weight and the time course of tumour growth in grams for 15 days postinjection with SCC7 or SCC7 Rab27a KO cells with or without CD73 in sEVs. Lat A was used as inhibitor of sEVs uptaken. (e) Flow cytometry analysis for infiltration of macrophages and percentage of CD73+/PD‐1+macrophages in tumours. (f). Flow cytometry analysis for infiltration of CD8+ T cells and percentage of CD73+/PD‐1+ CD8+ T cells in tumours. (g) Flow cytometry analysis for infiltration of Tregs and percentage of CD73+/PD‐1+ Tregs in tumours. (h) Schematic of subcutaneous tumorigenesis followed with intratumoral injection of engineered sEVs from mBMSC or mBMSC Nt5e OE. (i) The exhibition of dissected tumours. (j and k) The tumour weight and tumour growth curve of SCC7 control or SCC7 Rab27a KO with indicated treatment. (l and m) Flow cytometry analysis for infiltration of macrophages and percentage of CD73+ macrophages in tumours. (n) Flow cytometry analysis for infiltration of CD8+ T cells and Tregs in tumours. Data were analysed by Mann‐Whitney test (ns, no significant difference, *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001)
sEVs derived from mBMSC cells overexpressing CD73 by lentivirus transfection (sEVsmBMSC‐ Nt5e OE) were used as engineered sEVs to determine the in vivo function of sEVsCD73 (Figure 4h). The sEVsmBMSC enhanced tumour progression but did not increase the tumour size (p = 0.1788) (Figure 4i–k). However, overexpression of sEVsCD73 prominently promoted tumorigenesis in the sEVsmBMSC‐ Nt5e OE group, similar to the effects of SCC7‐derived sEVs carrying abundant CD73 (Figure 4i–k). Compared with the sEVsmBMSC group, infiltration of TAMs increased significantly (p = 0.0004), and CD8+ T cells were remarkably decreased (p = 0.0024), whereas significant difference in CD4+ T (p = 0.0138) and Tregs (p = 0.0044) in the sEVsmBMSC‐ Nt5e OE group (Figures 4l–n and S4G) was also observed. In addition, the percentage of CD73 increased more prominently in TAMs (p = 0.0038) than in CD8+ T cells (p = 0.027), CD4+ T cells (p = 0.0288), and Tregs (p = 0.0198) (Figures 4m and S4F,H). The results suggested that CD73 in sEVs was mainly phagocytosed by TAMs, whereas engineered sEVs with high levels of CD73 further converted TAMs into TAMsCD73high, which exhibited a more malignant phenotype.
3.5. sEVs carrying CD73 functionally regulate TAMs via NF‐κB pathway
HNSCC sEVs‐treated M2 macrophages were subjected to RNA sequencing. The sEVs were harvested from HN6, HN6 NT5E KO, SCC25, SCC25 NT5E KO, CAL27 and CAL27 NT5E KO. The 511 upregulated genes from the comparison between the NC group and M2+sEVs group were screened, and 204 downregulated genes were selected from the comparison between the M2+sEVs NT5E KO and M2+sEVs groups. The 143 common genes in these two groups are shown in a Venn diagram (Figure 5a), and differentially expressed genes were found to be involved in macrophage regulation by sEVsCD73 (Table S5). Using the Metascape platform, NFκB1 was identified as the most closely related transcription factor among 143 differentially expressed genes (Figure 5b). NFκB1 activates the classic NF‐κB pathway, which is involved in inflammation, the immune response, and tumour progression (Cartwright et al., 2016); NF‐κB‐related transcription factors such as RELA, SP1, and STAT3 were also identified. The Circos‐gram including pathways and gene shown in Figure 5c also indicate the highest enrichment for the NF‐κB pathway. The Gene Set Enrichment Analysis results showed that IκB phosphorylation and NIK were promoted when HNSCC sEVs were mixed with macrophages (Figure 5d); this trend was reversed when macrophages were treated with sEVs NT5E KO (Figure 5e). The regulatory flow of sEVsCD73, NF‐κB pathway, and macrophages was studied. Macrophages were cocultured with sEVs for 3 h to assess the effect of sEVsCD73 on translocation of p65 into the nucleus, which represented NF‐κB pathway activation. Immunofluorescence revealed that complete nuclear accumulation of p65 was only observed when exogenous sEVs carrying CD73 were added (Figure 5f). To evaluate the role of the CD73‐p65‐NF‐κB axis in this process, M2 macrophages were treated with 100 μM PDTC for 1 h, which inhibited sEVCD73‐induced p65 nuclear translocation (Figure 5f). Therefore, CD73 carried by sEVs activated the NF‐κB pathway in macrophages.
FIGURE 5.
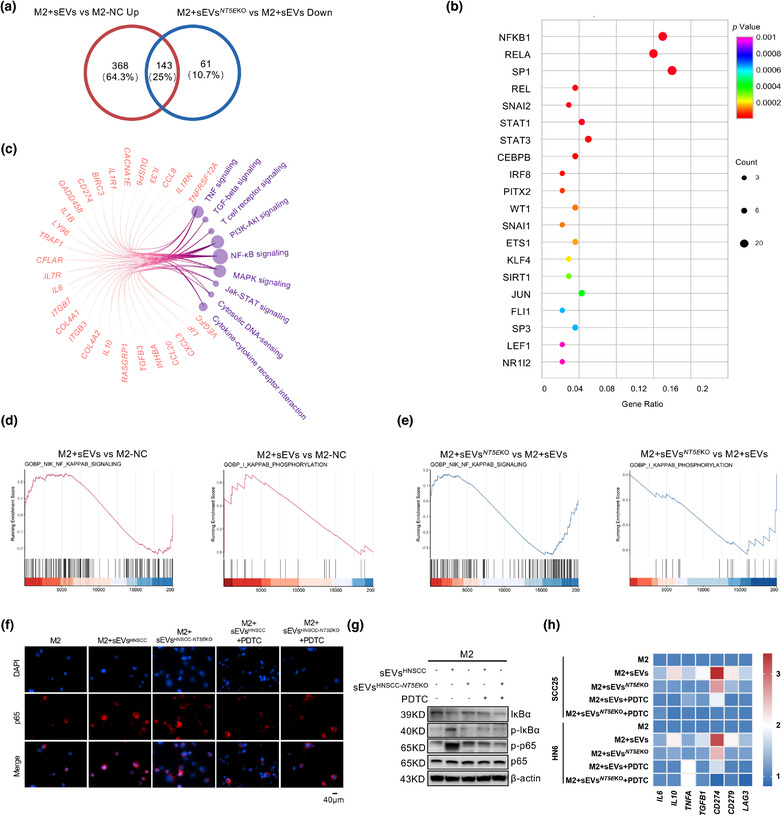
CD73 in sEVs regulates the immune functions of TAMs through NF‐κB pathway. (a) Venn diagram showing the differential expressed gene (DEG) of M2 macrophages depending on the regulation by sEVsCD73 or sEVs NT5E KO, the numbers of shared and exclusive genes were exhibited. (b) Bubble plot showing the predicted transcription factors of shared DEGs from AC. The shared DEGs and the signalling pathways which they belonged to were shown in circos gram, the number of genes was represented by node size. (d and e) GSEA analysis of up‐stream events of NF‐κB regulation. IκB phosphorylation and NIK signaling activity was measure after sEVsCD73 (up regulated) or sEVs NT5E KO (down regulated) incubation. (f) M2 macrophages were treated by HNSCC cell lines‐derived sEVs with or without CD73 for 3 h, with or without pretreatment of 100 μM PDTC for 1h. IF was applied for assessing the translocation of p65 in M2 macrophages (Scale bar = 40 μm). (g) Western blot was performed to validate the status of IκBα degradation, IκBα phosphorylation, p65 and phosphorylated p65 in M2 macrophages following treatment with sEVs or sEVs NT5E KO from HNSCC cell lines. (h) The heatmap showed the mRNA levels of IL6, IL10, TNFA, TGFB1, CD274, CD279, and LAG3 in M2 macrophages which were stimulated by HNSCC derived sEVs, the expressions were partially downregulated when sEVs NT5E KO were added, and further inhibited when pretreated with PDTC
The NF‐κB‐activating effect of sEVCD73 also stimulated the phosphorylation of p65 and IκBα, further terminating the degradation of IκBα (Figure 5g). Among the 143 differentially expressed genes selected, IL6, IL10, TNFA, TGFB1, and CD274 have been reported as downstream genes of the NF‐κB pathway (Antonangeli et al., 2020; Rong et al., 2020; Zheng et al., 2020). Cytokines such as IL6, IL10, TNFA, and TGFB1 were upregulated by sEVsCD73, but the effect was partially reversed when macrophages were treated with sEVs NT5E KO or PDTC (Figure 5h). The immune checkpoint molecules LAG3, CD274, and CD279 exhibited similar trends (Figure 5h). Thus, sEVsCD73 influenced macrophages to promote the expression of immunosuppressive molecules (LAG3, CD279, and CD274) and related cytokines (IL6, IL10, TNFA, and TGFB1) and activated the NF‐κB pathway, resulting in immune escape and tumour progression.
3.6. sEVsCD73 is a potential checkpoint and therapeutic target for enhancing the antitumour efficacy of anti‐PD‐1 therapy
The proportion of CD73high macrophages was higher in PBMCs from patients with HNSCC than in healthy donors (Figures 6b and S5A). To further explore the potential of CD73 as a target for clinical diagnosis and treatment, CD73 in circulating sEVs from patients with HNSCC was evaluated using ELISA (Figure 6a). sEVs were purified from the serum. Detection of circulating sEVsCD73 using ELISA demonstrated that the concentration of CD73 in sEVs was significantly higher in patients with HNSCC than normal people (Figure 6c). In addition, high levels of circulating sEVsCD73 may predict poorer lymph node metastasis and larger tumour size (Figure 6d; Table 2).
FIGURE 6.
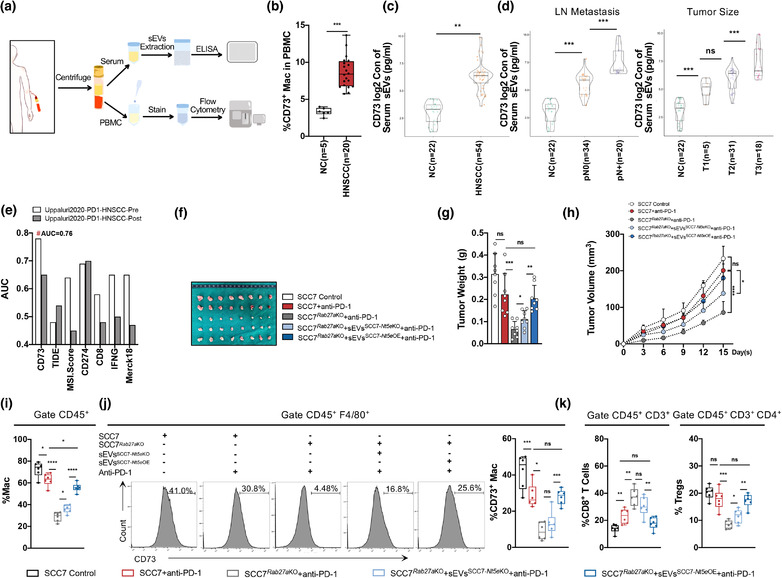
The sEVsCD73 predicts HNSCC metastasis and targeting sEVsCD73 abolishes immunotherapy resistance. (a) Schematic of the circulating sEVs from HNSCC patients processing. (b) Percentage of CD73+ macrophages in PBMC. (c) The concentration of CD73 in serum sEVs (log2 TPM) detected by ELISA from HNSCC patients. (d) Level of CD73 (log2 TPM) on serum sEVs predicts higher risk of lymph node metastasis and larger tumour size. (e) CD73 indicated as a potential candidate to predict anti‐PD‐1 therapy response for HNSCC patients compared to other existed biomarkers, analysis by TIDE framework (http://tide.dfci.harvard.edu), TIDE, tumour immune dysfunction and exclusion. (f) The exhibition of isolated tumours 15 days after injection of SCC7 or SCC7 Rab27a KO cells with or without CD73 absent in sEVs combined with anti‐PD‐1 therapy. (g and h) The tumour weight and tumour growth curve of SCC7 control or SCC7 Rab27a KO with indicated treatment combined with anti‐PD‐1 therapy. (j) Flow cytometry analysis for infiltration of macrophages and percentage of CD73+ macrophages in tumours. (k) Flow cytometry analysis for infiltration of CD8+ T cells and Tregs in tumours. Data were analyzed by Mann–Whitney test (ns, no significant difference, *: p < 0.05, **: p < 0.01, ***: p < 0.001, ****: p < 0.0001)
TABLE 2.
Relationship between sEVs CD73 in serum and clinicopathologic features (n = 54)
| Characteristics | Number of patients | % proportion | CD73 expression of serum sEVs (pg/ml) | p‐Value |
|---|---|---|---|---|
| Age (years) | 0.2974 | |||
| ≥60 | 22 | 40.74 | 140.76 | |
| <60 | 32 | 59.26 | 138.24 | |
| Gender | 0.2251 | |||
| Male | 37 | 68.52 | 142.39 | |
| Female | 17 | 31.48 | 138.61 | |
| Lymph node metastasis | 0.0003 | |||
| Non‐metastasis | 34 | 62.96 | 67.14 | |
| Metastasis | 20 | 37.04 | 262.97 | |
| Tumour size | 0.0043 | |||
| I | 5 | 9.26 | 35.22 | |
| II | 31 | 57.41 | 85.60 | |
| III | 18 | 33.33 | 262.62 | |
| IV | 0 | 0 | 0 |
The TIDE analysis platform was used to predict the response to ICB (P. Jiang, Gu, et al., 2018). CD73 was further validated as a marker for predicting the response to immune checkpoint therapy based on different indices, which effectively predicted the sensitivity to anti‐PD‐1 therapy in HNSCC (pre‐PD‐1 treatment AUC = 0.76). Data on the indices as TMB, T. Clonality, and B. Clonality were unavailable and thus, were omitted from the chart. Moreover, CD73, a single molecular, predicted the outcomes of patients with HNSCC treated with first‐line anti‐PD‐1 more accurately than other common biomarkers such as the microsatellite instability score, PD‐L1 and IFNR levels, CD8, Merck18, and TIDE indices (Figure 6e). Because CD73 is released through sEVs and facilitates tumour immune evasion, we investigated whether targeting of sEVsCD73 could reverse the resistance to anti‐PD‐1 therapy in patients with HNSCC.
To explore the therapeutic potency of sEVsCD73 in anti‐PD‐1 therapy, an anti‐PD‐1 agent (BioXcell, USA) was injected into mice with SCC7 or SCC7 Rab27a KO cells, along with sEVs with or without CD73 (Figure S5B). After 15 days of anti‐PD‐1 therapy, the growth rate and weight of the tumour were not significantly different from those in the SCC7 control group (p = 0.0709) (Figure 6f–h). However, the resistance to anti‐PD‐1 therapy was reversed when CD73 in sEVs was knocked out (Figure 6f–h), demonstrating the important function of tumour cell‐derived sEVsCD73. In addition, for anti‐PD‐1 therapy without the participation of sEVsCD73, infiltration of TAMs, CD4+ T cells and Tregs decreased remarkably, whereas the number of CD8+ T cells increased, with decreased expression of CD73 and PD‐1 (Figures 6i–k and S5C–F). Compared with sEVs Nt5e KO, the presence of CD73 in sEVs significantly reduced the therapeutic effect of Rab27aKO combined with anti‐PD‐1 and aggravated suppression of the immune microenvironment. Among these immunocytes, TAMs and CD8+ T cells were more sensitive to sEVsCD73, promoting the immunosuppressive process, as PD‐1 and CD73 were upregulated in CD8+ T cells, Tregs, and TAMs (Figures 6i–k and S5C–F).
Overall, CD73 in tumour cell‐derived sEVs attenuated the therapeutic effect of anti‐PD‐1 by exacerbating immunosuppression. The level of sEVsCD73 predicts HNSCC metastasis and is a potential combined immunotherapeutic target with anti‐PD‐1 therapy in HNSCC.
4. DISCUSSION
Due to TME complexity, the efficiency of immune therapy is highly heterogeneous among patients. As important immunomodulators of tumorigenesis, sEVs contribute to various hallmarks of cancer, particularly in TME remodelling to trigger tumour metastasis and immune escape (Henrich et al., 2020). Studies have shown that cancer‐ and TME‐derived sEVs carrying cargoes are closely linked to immunosuppression, which may further modulate the response to ICB therapy (Marar et al., 2021). For example, cancer‐derived sEVs potently mediate immunosuppression by elevating the levels of the immunoregulatory molecule PD‐L1. PD‐L1+ sEVs in the plasma are potential predictors of the response to anti‐PD1 therapy, highlighting valuable therapeutic strategies for depleting cancer‐derived sEVs. In addition, immunocyte‐derived sEVs directly or indirectly suppress T cell by antigen‐presenting cell‐mediated cross‐priming, including antigen‐presenting cell‐derived sEVs that contain miR‐142‐3p to induce apoptosis in T cells (O' Reilly et al., 2009). Tregs can inhibit the activity of other immunocytes by secreting tRNA fragments containing sEVs (Sakaguchi et al., 1995). In our study, HNSCC‐derived sEVs carried abundant CD73, primarily taken up by TAMs to facilitate formation of an immunosuppressive niche. Further elimination of sEVs secretion from primary tumour cells decreased CD73 expression in macrophages, increased effector T cell infiltration, and improved the anti‐PD‐1theraputic response. These results suggest a potential target of immunotherapy, as CD73+ sEVs aggravate the instability of the TME.
CD73 is an ectonucleotidase that, along with its upstream signalling molecule CD39, converts extracellular ATP to extracellular adenosine (eADO), which can be induced by inflammation, hypoxia, and specific oncogenic pathways (D. Allard et al., 2019). Increasing evidence suggests the involvement of CD73‐eADO is important for innate and adaptive immune responses regulation, as well as immunosuppression (Bareche et al., 2021). Studies have linked high expression of CD73 with poor prognosis in patients with cancer, such as melanoma, breast cancer, lung cancer, and glioblastoma (T. Jiang, Xu, et al., 2018; H. Wang et al., 2012; Zhi et al., 2007). CD73 promotes the hijacking of NK cells for immune escape (Neo et al., 2020), while its upregulation in Tregs generates an immunosuppressive phenotype and promotes tumour growth (Romio et al., 2011). Goswami et al. (2020) found that CD73 was highly expressed in macrophages of glioblastoma, and the absence of CD73 provided better ICB therapeutic efficiency. The importance of CD73 in sEVs has also been reported, which affected immunocytes and caused immune suppression. Human effector CD8+ T cells release sEVs containing CD73, which would promote the production of adenosine and mediate immune suppression (Schneider et al., 2021). CD73 in sEVs from TME has also been reported to enhance the expression of CD73 in DC cells, thereby inhibiting CD8+ T cell responses and causing immunosuppression in an adenosine‐dependent manner (Salimu et al., 2017). Not only did we explore the influence of tumour cell‐derived sEVs on immune microenvironment, but also demonstrated that CD73 was highly expressed in sEVs derived from HNSCC cells, which predicted a worse prognosis and poorer response to anti‐PD‐1 therapies in HNSCC. To highlight the function of sEVsCD73, two independent strategies were applied. Endocytosis was inhibited by Lat A, while the production of sEVs was blocked by Rab27a knockout (Johnson et al., 2016; Mulcahy et al., 2014). Considering that Lat A was also reported to have antitumour effects, these two strategies supported the tumour‐promoting effect of sEVsCD73 to some extent (Konishi et al., 2009). Following intratumoral injection of sEVsCD73, TAMs showed the most significant correlation with sEVsCD73, with CD8+ T cells obviously exhausted when the immune checkpoint receptors were upregulated, including PD‐1, PD‐L1, and LAG3, whereas CD4+ T cells and Tregs were rather insensitive. As a membrane protein, CD73 reshaped TAMs mainly through HNSCC tumour cell‐derived sEVs. In addition, engineered sEVs from BMSC NT5E OE cells were introduced, which overexpressed CD73 in released sEVs. Data on whether BMSC‐derived sEVs promote or inhibit tumour growth are conflicting (Vakhshiteh et al., 2019). BMSC‐derived sEVs have been reported to induce macrophages to the M2 subtype and enhance the levels of IL‐10 and TGF‐β1 to accelerate Treg proliferation. Our results indicate that overexpression of CD73 in BMSC‐derived sEVs imitated the immunosuppressive effect of HNSCC tumour cell‐derived sEVs, emphasizing the tumorigenic ability of sEVsCD73.
Moreover, the expression of coinhibitory receptors, including LAG3, VISTA, PD‐1, and PD‐L1, was significantly upregulated in CD73‐positive NK cells compared to in CD73‐negative NK cells, suggesting that CD73 defined regulatory NK cells in the immune‐suppressive TME (Neo et al., 2020). A unique population of CD73high macrophages in glioblastomas has been identified as a specific immunotherapeutic target (Goswami et al., 2020). In addition, sEVs derived from immunocytes, such as Tregs, show outstanding tumour immunosuppressive ability in inducing effector T cell exhaustion by expressing IL‐35, and accelerating the expression of PD‐1, T cell immunoglobulin, and mucin domain‐containing protein 3 (TIM3), and LAG3 (Sullivan et al., 2020). These findings suggest that CD73 from cancer‐derived sEVs can be taken up by TAMs to define a specific subpopulation, CD73high TAMs, which possess a more malignant phenotype than M2 macrophages. We examined TCGA database, focusing on cohorts of patients with HNSCC, and showed that high infiltration of CD73high macrophages was closely associated with poor prognosis and a high risk of metastasis. However, another HNSCC cohort of single‐cell RNA‐sequencing data showed that the mRNA level of NT5E in macrophages was extremely low (Cillo et al., 2020), suggesting that TAMs in HNSCC are reprogramed by tumour‐derived CD73+ sEVs toward a more malignant phenotype, CD73high TAMs. Co‐culture experiments with sEVsCD73 showed that the expression of PD‐1, PD‐L1, and LAG3 in TAMs was enhanced by sEVsCD73 via activation of the NF‐κB pathway. The associated mechanism includes inflammatory cytokines, such as TGF‐β, IL‐6, IL‐10, and IL‐8, which further induce immunosuppression (Hugo et al., 2016; Mcdermott et al., 2018). Our study showed that sEVsCD73 also remodelled the TME to support an immunosuppressive state by increasing the secretion of IL‐6, IL‐10, TNF‐α, and TGF‐β1 in HNSCC TAMs. The attenuated sensitivity to anti‐PD‐1 therapy caused by CD73 was validated by in vivo findings. These results suggest that the absence of CD73 in sEVs can reverse the immunosuppressive state and predict the efficacy of combined anti‐PD‐1 with anti‐CD73 therapy.
Tumour‐derived sEVs can be taken up by organ‐specific cells to form the premetastatic niche (Hoshino et al., 2015; Kang et al., 2021). Lymphatic vessels are among the most common routes for the dissemination of HNSCC, which is important for treatment design and is a factor for prognosis (Pascual et al., 2017). Tumour‐derived sEVs drain into lymph nodes through lymph vessels and exchange molecules and information with immune cells in the lymph nodes to further remodel the microenvironment (Page et al., 2014; Poggio et al., 2019). However, constant stimulation of tumour sEVs causes immunosuppression (Frey, 2015). The sEVs can suppress the activation and proliferation of T cells and promote metastasis (Poggio et al., 2019). A similar phenotype was observed in DLNs following CD73+ sEVs injection: the proportion of CD8+ T cells decreased while the infiltration of CD73+ macrophages increased in lymph nodes with high expression of PD‐1 to form an environment favourable for metastasis. Tumour cell‐derived CD73+ sEVs may be critical for reprogramming of the premetastatic niche in lymph nodes (Sleeman, 2015). However, in‐depth mechanistic studies are needed to explore the role of CD73 in sEVs in lymph node metastasis.
Circulating sEVs are useful for the early diagnosis of tumours and can be obtained noninvasively (De Abreu et al., 2020; Möhrmann et al., 2018; T. T. Tang et al., 2020). Considering the role of CD73+ sEVs in immunosuppressive TME and ICB therapeutic resistance, we measured the level of circulating sEVsCD73 in patients with HNSCC and showed that CD73 was more enriched in serum sEVs from patients with HNSCC than healthy people, which was closely related to a late TNM stage and lymph node metastasis. These findings suggest that sEVsCD73 is a marker for prognosis and metastasis risk in patients with HNSCC. Moreover, we used TIDE, as reported by P. Jiang, Gu, et al. (2018), which reflected the power of biomarkers to predict the sensitivity to immune therapy in different clinical trials. Compared with the diagnostic methods from TIDE, CD73 could better predict the sensitivity of anti‐PD‐1 treatment in HNSCC. We also found that higher levels of HNSCC tumour cell‐derived sEVsCD73 hinders anti‐PD‐1 treatment. More CD73high TAMs were induced by HNSCC tumour cell‐derived sEVsCD73, which exhibited malignant phenotypes and affected the TME, leading to resistance to anti‐PD‐1 therapy. Thus, suppressing sEVsCD73 in HNSCC tumour cells can reverse immunosuppression and improve the sufficiency of anti‐PD‐1 therapy. These results provide insight into combined treatment with anti‐CD73 and anti‐PD‐1, particularly for patients with high levels of sEVsCD73. However, the potential of circulating sEVsCD73 to predict the sensitivity of anti‐PD‐1 therapy in patients with HNSCC should be further evaluated.
In conclusion, our study illustrated that HNSCC tumour cell‐derived sEVsCD73 mediate phenotypic and functional changes in TAMs to induce immune tolerance, and proposes that sEVsCD73 as a potential predictive marker and combined target for dual blockade therapy with PD‐1 against HNSCC.
CONFLICTS OF INTEREST
The authors declare no competing interests.
AUTHOR CONTRIBUTIONS
Ming Yan, Wantao Chen designed the project. Tingwei Lu and Zhen Zhang performed the experiments, data statistics and bioinformatics analysis. Jianjun Zhang, Xu Wang kindly provided suggestions for the project. Zhihui Li, Xinhua Pan, Min Ruan and Tingwei Lu collected the clinical samples. Xueqin Zhu and Huasheng Li kindly provided support during animal experiments and cytologic assays. Tingwei Lu and Zhen Zhang wrote the manuscript. All the authors read and approved the final manuscript.
Supporting information
Supporting Information
Supporting Information
Supporting Information
Supporting Information
Supporting Information
FIGURE S1. CD73 was riched in HNSCC cells derived sEVs. A. Quantitative proteome analysis for cells released sEVs from HNSCC adjacent normal tissues, shown by venn diagram. B. Quantitative proteome for sEVs from HNSCC tissues with or without metastasis, shown by venn diagram. C. Immunoblotting for CD73 in HNSCC cell lines derived sEVs. D. Quantitative proteome results of sEVs from HN6, SCC25 and CAL27, shown by venn diagram. E. The mass spectrum of CD73 in sEVs from HN6, SCC25 and CAL27.
FIGURE S2. CD73 were closely related to macrophages in HNSCC. A. NT5E expression level in different types of tumors utilizing TCGA datasets (log2 TPM). B. The overall survival rate of HNSCC patients from TCGA datasets. C‐F. Survival curves regarding NT5E expression based on TCGA HNSCC cohort. (C, D) Log‐rank Mantel‐Cox test was used to assess significance. Patients with high macrophages and low macrophages gene signature were shown. (E) Patients with M2high and M2low gene signature were shown. (F) Data were classified into low NT5E signature (NT5Elow), high NT5E signature (NT5Ehigh), low macrophage/M2 macrophage signature (Maclow/M2low) and high macrophage/M2 macrophage signature (Machigh/M2high). G. Flow cytometry analysis for percentage of sEVsSCC7‐GFP uptaken in DLNs after injecting sEVsSCC7‐GFP (10 ng‐50 μg) for 30min. H. Flow cytometry analysis about DLNs for percentage of CD4+ T cells, CD8+ T cells, M2 macrophages and Tregs after injecting sEVs (25 μg) every day. I. Gating strategy for identification of immunocytes subsets in DLNs by manual gating. J. Gating strategy for identification of immunocytes in DLNs by manual gating. K. Schematic of SCC7 subcutaneous tumorigenesis. After 21 days, DLNs were dissected. L. Flow cytometry analysis for percentage of macrophages/CD73+ macrophages/PD‐1+ macrophages in DLNs. L. Flow cytometry analysis for percentage of CD8+ T cells/CD73+ CD8+ T cells/PD‐1+ CD8+ T cells in DLNs. Lymph nodes from untreated mice were used as normal control. Data were analyzed by Mann‐Whitney test. (ns, no significant difference, *: p < 0.05, **: p < 0.01, ***: p < 0.001, ****: p < 0.0001).
FIGURE S3. The role of HNSCC‐derived sEVsCD73 in macrophages function. A. Western blot for CD73 and CD63 in sEVs to measure the efficiency of CRISPR/Cas9‐mediated RAB27A/ Rab27a disruption. B. Cell proliferation over time for HN6, SCC25, SCC7 with or without RAB27A/ Rab27a KO. C. Western blot for CD73 in sEVs to detect the efficiency of CRISPR/Cas9‐mediated NT5E/Nt5e disruption. D. Western blot for CD73 in sEVs after NT5E/Nt5e overexpression on NT5E/Nt5eKO cells mediated by lentivirus transfection. E. Representative graphs of CD73+ macrophages in different co‐culture systems. F. Gating strategy for identification of CD73+ macrophages subsets by manual gating. G. Macrophages (1x106) co‐cultured with or without sEVsHNSCC (25 μg‐100 μg) for 24h. Flow cytometry analysis for CD73 expression in macrophages. H. Western blot for CD73 expression levels in sEVs derived from HN6, SCC25, hBMSC, hBMSC‐ NT5EOE, SCC7, mBMSC and mBMSC‐Nt5eOE. Data were analyzed by Mann‐Whitney test. (ns, no significant difference, *: p < 0.05, **: p < 0.01, ***: p < 0.001, ****: p < 0.0001).
FIGURE S4. Absence of CD73 in sEVs restrains tumor growth in vivo. A. Gating strategy for identification of immunocytes subsets by manual gating. B. Representative graphs of CD73+ macrophages in different groups. C. Quantification of PD‐L1+ cells in macrophages. D. Flow cytometry analysis for infiltration of CD4+T cells and percentage of CD73+/PD‐1+ CD4+T cells in tumors. E. Quantification of PD‐1+/PD‐L1+ cells in macrophages. F. Flow cytometry analysis for infiltration of CD4+T cells and percentage of CD73+/PD‐1+ CD4+T cells in tumors. G. Percentage of CD73+/PD‐1+ Tregs in tumors. Data were analyzed by Mann‐Whitney test. (ns, no significant difference, *: p < 0.05, **: p < 0.01, ***: p < 0.001, ****: p < 0.0001).
FIGURE S5. Targeting CD73 in sEVs abolishes immunotherapy resistance in vivo. A. Representative graphs of CD73+ macrophages gating from PBMC in normal people or HNSCC patients. B. Schematic of subcutaneous tumorigenesis 15 days after injection of SCC7 or SCC7Rab27aKO cells with or without CD73 absent in sEVs combined with anti‐PD‐1 therapy. C. Quantification of PD‐1+/PD‐L1+ cells in macrophages. D. Quantification of CD73+/PD‐1+ cells in macrophages. E. Flow cytometry analysis for infiltration of CD4+ T cells and percentage of CD73+/PD‐1+ CD4+ T cells in tumors. F. Quantification of CD73+/PD‐1+ cells in Tregs. Data were analyzed by Mann‐Whitney test. (ns, no significant difference, *: p < 0.05, **: p < 0.01, ***: p < 0.001, ****: p < 0.0001).
ACKNOWLEDGEMENTS
This work was supported by the National Natural Science Foundation of China grant 82173217, 81672829 by Ming Yan, and grant 81874126 by Wantao Chen. We thank funds for the Shanghai Committee of Science and Technology: Shanghai Oral and Maxillofacial Tumor Tissue Samples and Bioinformatics Database Professional Technical Service Platform (21DZ2292000), and SJTU Trans‐med Awards Research (20210103). We thank Chuwen Li, Chao Lou, Changsheng Lu, Ran Yan, Fang Xia and Yuan Dai, who kindly provided help. We thank Mr. Qidong Zu (OE Biotech, Inc., Shanghai, China) for assistance with the bioinformatics analysis of RNA‐seq. We thank Caihua Zhu from Shanghai Applied Protein Technology for technical support.
Lu, T. , Zhang, Z. , Zhang, J. , Pan, X. , Zhu, X. , Wang, X. , Li, Z. , Ruan, M. , Li, H. , Chen, W. , & Yan, M. (2022). CD73 in small extracellular vesicles derived from HNSCC defines tumor‐associated immunosuppression mediated by macrophages in the microenvironment. Journal of Extracellular Vesicles, 11, e12218. 10.1002/jev2.12218
Tingwei Lu and Zhen Zhang contributed equally to this work.
Contributor Information
Wantao Chen, Email: chenwantao196323@sjtu.edu.cn.
Ming Yan, Email: yanming8012@126.com.
REFERENCES
- Allard, B. , Allard, D. , Buisseret, L. , & Stagg, J. (2020). The adenosine pathway in immuno‐oncology. Nature Reviews Clinical Oncology, 17(10), 611–629. [Google Scholar]
- Allard, D. , Chrobak, P. , Allard, B. , Messaoudi, N. , & Stagg, J. (2019). Targeting the CD73‐adenosine axis in immuno‐oncology. Immunology Letters, 205, 31–39. [DOI] [PubMed] [Google Scholar]
- Antonangeli, F. , Natalini, A. , Garassino, M. C. , Sica, A. , Santoni, A. , & Di Rosa, F. (2020). Regulation of PD‐L1 expression by NF‐κB in cancer. Frontiers in Immunology, 11, 584626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bareche, Y. , Pommey, S. , Carneiro, M. , Buisseret, L. , Cousineau, I. , Thebault, P. , Chrobak, P. , Communal, L. , Allard, D. , Robson, S. C. , Mes‐Masson, A. M. , Provencher, D. , Lapointe, R. , & Stagg, J. (2021). High‐dimensional analysis of the adenosine pathway in high‐grade serous ovarian cancer. Journal for ImmunoTherapy of Cancer, 9(3), e001965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biray Avci, C. , Sezgin, B. , Goker Bagca, B. , Karci, H. B. , & Gode, S. (2020). PI3K/AKT/mTOR pathway and autophagy regulator genes in paranasal squamous cell carcinoma metastasis. Molecular Biology Reports, 47(5), 3641–3651. [DOI] [PubMed] [Google Scholar]
- Brown, M. C. , Mosaheb, M. M. , Mohme, M. , Mckay, Z. P. , Holl, E. K. , Kastan, J. P. , Yang, Y. , Beasley, G. M. , Hwang, E. S. , Ashley, D. M. , Bigner, D. D. , Nair, S. K. , & Gromeier, M. (2021). Viral infection of cells within the tumor microenvironment mediates antitumor immunotherapy via selective TBK1‐IRF3 signaling. Nature Communication, 12(1), 1858. [Google Scholar]
- Cartwright, T. , Perkins, N. D. , & Wilson, C. L. (2016). NFKB1: A suppressor of inflammation, ageing and cancer. Febs Journal, 283(10), 1812–1822. [Google Scholar]
- Chen, G. , Huang, A. C. , Zhang, W. , Zhang, G. , Wu, M. , Xu, W. , Yu, Z. , Yang, J. , Wang, B. , Sun, H. , Xia, H. , Man, Q. , Zhong, W. , Antelo, L. F. , Wu, B. , Xiong, X. , Liu, X. , Guan, L. , Li, T. , … Guo, W. (2018). Exosomal PD‐L1 contributes to immunosuppression and is associated with anti‐PD‐1 response. Nature, 560(7718), 382–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow, L. Q. M. (2020). Head and neck cancer. New England Journal of Medicine, 382(1), 60–72. [Google Scholar]
- Cillo, A. R. , Kürten, C. H. L. , Tabib, T. , Qi, Z. , Onkar, S. , Wang, T. , Liu, A. , Duvvuri, U. , Kim, S. , Soose, R. J. , Oesterreich, S. , Chen, W. , Lafyatis, R. , Bruno, T. C. , Ferris, R. L. , & Vignali, D. A. A. (2020). Immune landscape of viral‐ and carcinogen‐driven head and neck cancer. Immunity, 52(1), 183–199.e9.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen, E. E. W. , Bell, R. B. , Bifulco, C. B. , Burtness, B. , Gillison, M. L. , Harrington, K. J. , Le, Q. ‐. T. , Lee, N. Y. , Leidner, R. , Lewis, R. L. , Licitra, L. , Mehanna, H. , Mell, L. K. , Raben, A. , Sikora, A. G. , Uppaluri, R. , Whitworth, F. , Zandberg, D. P. , & Ferris, R. L. (2019). The society for immunotherapy of cancer consensus statement on immunotherapy for the treatment of squamous cell carcinoma of the head and neck (HNSCC). Journal for ImmunoTherapy of Cancer, 7(1), 184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong, L. , & Zhang, F. (2015). Genome engineering using CRISPR‐Cas9 system. Methods in Molecular Biology, 1239, 197–217. [DOI] [PubMed] [Google Scholar]
- De Abreu, R. C. , Fernandes, H. , Da Costa Martins, P A. , Sahoo, S. , Emanueli, C. , & Ferreira, L. (2020). Native and bioengineered extracellular vesicles for cardiovascular therapeutics. Nature Reviews Cardiology, 17(11), 685–697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frey, A. B. (2015). Suppression of T cell responses in the tumor microenvironment. Vaccine, 33(51), 7393–7400. [DOI] [PubMed] [Google Scholar]
- Goswami, S. , Walle, T. , Cornish, A. E. , Basu, S. , Anandhan, S. , Fernandez, I. , Vence, L. , Blando, J. , Zhao, H. , Yadav, S. S. , Ott, M. , Kong, L. Y. , Heimberger, A. B. , De Groot, J. , Sepesi, B. , Overman, M. , Kopetz, S. , Allison, J. P. , Pe'er, D. , & Sharma, P. (2020). Immune profiling of human tumors identifies CD73 as a combinatorial target in glioblastoma. Nature Medicine, 26(1), 39–46. [Google Scholar]
- Greening, D. W. , Xu, R. , Ji, H. , Tauro, B. J. , & Simpson, R. J. (2015). A protocol for exosome isolation and characterization: Evaluation of ultracentrifugation, density‐gradient separation, and immunoaffinity capture methods. Methods in Molecular Biology, 1295, 179–209. [DOI] [PubMed] [Google Scholar]
- Gu, L. , Li, H. , Chen, L. , Ma, X. , Li, X. , Gao, Y. , Zhang, Y. , Xie, Y. , & Zhang, X. (2016). Prognostic role of lymphocyte to monocyte ratio for patients with cancer: Evidence from a systematic review and meta‐analysis. Oncotarget, 7(22), 31926–31942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henrich, S. E. , Mcmahon, K. M. , Plebanek, M. P. , Calvert, A. E. , Feliciano, T. J. , Parrish, S. , Tavora, F. , Mega, A. , De Souza, A. , Carneiro, B. A. , & Thaxton, C. S. (2020). Prostate cancer extracellular vesicles mediate intercellular communication with bone marrow cells and promote metastasis in a cholesterol‐dependent manner. Journal of Extracellular Vesicles, 10(2), e12042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoshino, A. , Costa‐Silva, B. , Shen, T. L. , Rodrigues, G. , Hashimoto, A. , Tesic Mark, M. , Molina, H. , Kohsaka, S. , Di Giannatale, A. , Ceder, S. , Singh, S. , Williams, C. , Soplop, N. , Uryu, K. , Pharmer, L. , King, T. , Bojmar, L. , Davies, A. E. , Ararso, Y. , … Lyden, D. (2015). Tumour exosome integrins determine organotropic metastasis. Nature, 527(7578), 329–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hugo, W. , Zaretsky, J. M. , Sun, L. , Song, C. , Moreno, B. H. , Hu‐Lieskovan, S. , Berent‐Maoz, B. , Pang, J. , Chmielowski, B. , Cherry, G. , Seja, E. , Lomeli, S. , Kong, X. , Kelley, M. C. , Sosman, J. A. , Johnson, D. B. , Ribas, A. , & Lo, R. S. (2016). Genomic and transcriptomic features of response to anti‐PD‐1 therapy in metastatic melanoma. Cell, 165(1), 35–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hume, D. A. , & Macdonald, K. P. A. (2012). Therapeutic applications of macrophage colony‐stimulating factor‐1 (CSF‐1) and antagonists of CSF‐1 receptor (CSF‐1R) signaling. Blood, 119(8), 1810–1820. [DOI] [PubMed] [Google Scholar]
- Jiang, P. , Gu, S. , Pan, D. , Fu, J. , Sahu, A. , Hu, X. , Li, Z. , Traugh, N. , Bu, X. , Li, B. , Liu, J. , Freeman, G. J. , Brown, M. A. , Wucherpfennig, K. W. , & Liu, X. S. (2018). Signatures of T cell dysfunction and exclusion predict cancer immunotherapy response. Nature Medicine, 24(10), 1550–1558. [Google Scholar]
- Jiang, T. , Xu, X. , Qiao, M. , Li, X. , Zhao, C. , Zhou, F. , Gao, G. , Wu, F. , Chen, X. , Su, C. , Ren, S. , Zhai, C. , & Zhou, C. (2018). Comprehensive evaluation of NT5E/CD73 expression and its prognostic significance in distinct types of cancers. BMC Cancer, 18(1), 267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson, J. L. , Ramadass, M. , He, J. , Brown, S. J. , Zhang, J. , Abgaryan, L. , Biris, N. , Gavathiotis, E. , Rosen, H. , & Catz, S. D. (2016). Identification of neutrophil exocytosis inhibitors (Nexinhibs), small molecule inhibitors of neutrophil exocytosis and inflammation: Druggability of the small GTPase Rab27a. Journal of Biological Chemistry, 291(50), 25965–25982. [Google Scholar]
- Kang, M. , Jordan, V. , Blenkiron, C. , & Chamley, L. W. (2021). Biodistribution of extracellular vesicles following administration into animals: A systematic review. Journal of Extracellular Vesicles, 10(8), e12085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konishi, H. , Kikuchi, S. , Ochiai, T. , Ikoma, H. , Kubota, T. , Ichikawa, D. , Fujiwara, H. , Okamoto, K. , Sakakura, C. , Sonoyama, T. , Kokuba, Y. , Sasaki, H. , Matsui, T. , & Otsuji, E. (2009). Latrunculin a has a strong anticancer effect in a peritoneal dissemination model of human gastric cancer in mice. Anticancer Research, 29(6), 2091–2097. [PubMed] [Google Scholar]
- Laviron, M. , & Boissonnas, A. (2019). Ontogeny of Tumor‐Associated Macrophages. Frontiers in immunology, 10, 1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, H. , Lee, S. , & Heo, Y. S. (2019). Molecular interactions of antibody drugs targeting PD‐1, PD‐L1, and CTLA‐4 in immuno‐oncology. Molecules (Basel, Switzerland), , 24(6), 1190. [Google Scholar]
- Lee, H. H. , Wang, Y. N. , Xia, W. , Chen, C. H. , Rau, K. M. , Ye, L. , Wei, Y. , Chou, C. K. , Wang, S. C. , Yan, M. , Tu, C. Y. , Hsia, Te‐C. , Chiang, S. F. , Chao, K. S. C. , Wistuba, I. I. , Hsu, J. L. , Hortobagyi, G. N. , & Hung, M. C. (2019). Removal of N‐linked glycosylation enhances PD‐L1 detection and predicts anti‐PD‐1/PD‐L1 therapeutic efficacy. Cancer Cell, 36(2), 168–178.e4.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leemans, C. R. , Braakhuis, B. J. M. , & Brakenhoff, R. H. (2011). The molecular biology of head and neck cancer. Nature Reviews Cancer, 11(1), 9–22. [DOI] [PubMed] [Google Scholar]
- Li, M. , Soder, R. , Abhyankar, S. , Abdelhakim, H. , Braun, M. W. , Trinidad, C. V. , Pathak, H. B. , Pessetto, Z. , Deighan, C. , Ganguly, S. , Dawn, B. , Mcguirk, J. , Dunavin, N. , & Godwin, A. K. (2021). WJMSC‐derived small extracellular vesicle enhance T cell suppression through PD‐L1. Journal of Extracellular Vesicles, 10(4), e12067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, T. , Fu, J. , Zeng, Z. , Cohen, D. , Li, J. , Chen, Q. , Li, B. , & Liu, X. S. (2020). TIMER2.0 for analysis of tumor‐infiltrating immune cells. Nucleic Acids Research, 48(W1), W509–W514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, W. , Zhang, X. , Wu, F. , Zhou, Y. , Bao, Z. , Li, H. , Zheng, P. , & Zhao, S. (2019). Gastric cancer‐derived mesenchymal stromal cells trigger M2 macrophage polarization that promotes metastasis and EMT in gastric cancer. Cell Death & Disease, 10(12), 918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Q. , Wu, H. , Li, Y. , Zhang, R. , Kleeff, J. , Zhang, X. , Cui, M. , Liu, J. , Li, T. , Gao, J. , Pan, B. , Wu, W. , Wang, W. , Zhou, L. , Guo, J. , Dai, M. , Zhang, T. , Liao, Q. , Lu, Z. , & Zhao, Y. (2020). Combined blockade of TGf‐β1 and GM‐CSF improves chemotherapeutic effects for pancreatic cancer by modulating tumor microenvironment. Cancer Immunology, Immunotherapy, 69(8), 1477–1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, X. , Ory, V. , Chapman, S. , Yuan, H. , Albanese, C. , Kallakury, B. , Timofeeva, O A. , Nealon, C. , Dakic, A. , Simic, V. , Haddad, B R. , Rhim, J S. , Dritschilo, A. , Riegel, A. , Mcbride, A. , & Schlegel, R. (2012). ROCK inhibitor and feeder cells induce the conditional reprogramming of epithelial cells. American Journal of Pathology, 180(2), 599–607. [Google Scholar]
- Lu, T. , Shao, Z. , Liu, B. , & Wu, T. (2020). Recent advance in patient‐specific 3D printing templates in mandibular reconstruction. Journal of the Mechanical Behavior of Biomedical Materials, 106, 103725. [DOI] [PubMed] [Google Scholar]
- Mantovani, A. , Marchesi, F. , Malesci, A. , Laghi, L. , & Allavena, P. (2017). Tumour‐associated macrophages as treatment targets in oncology. Nature Reviews Clinical Oncology, 14(7), 399–416. [Google Scholar]
- Marar, C. , Starich, B. , & Wirtz, D. (2021). Extracellular vesicles in immunomodulation and tumor progression. Nature Immunology, 22(5), 560–570. [Google Scholar]
- Mariathasan, S. , Turley, S. J. , Nickles, D. , Castiglioni, A. , Yuen, K. , Wang, Y. , Kadel Iii, E. E. , Koeppen, H. , Astarita, J. L. , Cubas, R. , Jhunjhunwala, S. , Banchereau, R. , Yang, Y. , Guan, Y. , Chalouni, C. , Ziai, J. , Şenbabaoğlu, Y. , Santoro, S. , Sheinson, D. , … Powles, T. (2018). TGFβ attenuates tumour response to PD‐L1 blockade by contributing to exclusion of T cells. Nature, 554(7693), 544–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mcdermott, D. F. , Huseni, M. A. , Atkins, M. B. , Motzer, R. J. , Rini, B. I. , Escudier, B. , Fong, L. , Joseph, R. W. , Pal, S. K. , Reeves, J. A. , Sznol, M. , Hainsworth, J. , Rathmell, W. K. , Stadler, W. M. , Hutson, T. , Gore, M. E. , Ravaud, A. , Bracarda, S. , Suárez, C. , … Powles, T. (2018). Clinical activity and molecular correlates of response to atezolizumab alone or in combination with bevacizumab versus sunitinib in renal cell carcinoma. Nature Medicine, 24(6), 749–757. [Google Scholar]
- Mei, J. , Xiao, Z. , Guo, C. , Pu, Q. , Ma, L. , Liu, C. , Lin, F. , Liao, H. , You, Z. , & Liu, L. (2016). Prognostic impact of tumor‐associated macrophage infiltration in non‐small cell lung cancer: A systemic review and meta‐analysis. Oncotarget, 7(23), 34217–34228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng, F. , Guo, Z. , Hu, Y. , Mai, W. , Zhang, Z. , Zhang, B. , Ge, Q. , Lou, H. , Guo, F. , Chen, J. , Duan, S. , & Gao, Z. (2019). CD73‐derived adenosine controls inflammation and neurodegeneration by modulating dopamine signalling. Brain, 142(3), 700–718. [DOI] [PubMed] [Google Scholar]
- Möhrmann, L. , Huang, H. J. , Hong, D. S. , Tsimberidou, A. M. , Fu, S. , Piha‐Paul, S. A. , Subbiah, V. , Karp, D. D. , Naing, A. , Krug, A. , Enderle, D. , Priewasser, T. , Noerholm, M. , Eitan, E. , Coticchia, C. , Stoll, G. , Jordan, L. M. , Eng, C. , Kopetz, E. S. , … Janku, F. (2018). Liquid biopsies using plasma exosomal nucleic acids and plasma cell‐free DNA compared with clinical outcomes of patients with advanced cancers. Clinical Cancer Research, 24(1), 181–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison, A. C. , Wilson, C. B. , Ray, M. , & Correll, P. H. (2004). Macrophage‐stimulating protein, the ligand for the stem cell‐derived tyrosine kinase/RON receptor tyrosine kinase, inhibits IL‐12 production by primary peritoneal macrophages stimulated with IFN‐gamma and lipopolysaccharide. Journal of Immunology, 172(3), 1825–1832. [Google Scholar]
- Mounir, M. , Lucchetta, M. , Silva, T. C. , Olsen, C. , Bontempi, G. , Chen, X. , Noushmehr, H. , Colaprico, A. , & Papaleo, E. (2019). New functionalities in the TCGAbiolinks package for the study and integration of cancer data from GDC and GTEx. Plos Computational Biology, 15(3), e1006701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulcahy, L. A. , Pink, R. C. , & Carter, D. R. (2014). Routes and mechanisms of extracellular vesicle uptake. Journal of Extracellular Vesicles, 3. [Google Scholar]
- Neo, S. Y. , Yang, Y. , Record, J. , Ma, R. , Chen, X. , Chen, Z. , Tobin, N. P. , Blake, E. , Seitz, C. , Thomas, R. , Wagner, A. K. , Andersson, J. , De Boniface, J. , Bergh, J. , Murray, S. , Alici, E. , Childs, R. , Johansson, M. , Westerberg, L. S. , … Lundqvist, A. (2020). CD73 immune checkpoint defines regulatory NK cells within the tumor microenvironment. Journal of Clinical Investigation, 130(3), 1185–1198. [Google Scholar]
- Newman, A. M. , Liu, C. L. , Green, M. R. , Gentles, A. J. , Feng, W. , Xu, Y. , Hoang, C. D. , Diehn, M. , & Alizadeh, A. A. (2015). Robust enumeration of cell subsets from tissue expression profiles. Nature Methods, 12(5), 453–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ngambenjawong, C. , Gustafson, H. H. , & Pun, S. H. (2017). Progress in tumor‐associated macrophage (TAM)‐targeted therapeutics. Advanced Drug Delivery Reviews, 114, 206–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O' Reilly, L. A. , Tai, L. , Lee, L. , Kruse, E. A. , Grabow, S. , Fairlie, W. D. , Haynes, N. M. , Tarlinton, D. M. , Zhang, J.‐G. , Belz, G. T. , Smyth, M. J. , Bouillet, P. , Robb, L. , & Strasser, A. (2009). Membrane‐bound Fas ligand only is essential for Fas‐induced apoptosis. Nature, 461(7264), 659–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page, D. B. , Postow, M. A. , Callahan, M. K. , Allison, J. P. , & Wolchok, J. D. (2014). Immune modulation in cancer with antibodies. Annual Review of Medicine, 65, 185–202. [Google Scholar]
- Park, E. K. , Jung, H. S. , Yang, H. I. , Yoo, M. C. , Kim, C. , & Kim, K. S. (2007). Optimized THP‐1 differentiation is required for the detection of responses to weak stimuli. Inflammation Research, 56(1), 45–50. [DOI] [PubMed] [Google Scholar]
- Pascual, G. , Avgustinova, A. , Mejetta, S. , Martín, M. , Castellanos, A. , Attolini, C. S. O. , Berenguer, A. , Prats, N. , Toll, A. , Hueto, J. A. , Bescós, C. , Di Croce, L. , & Benitah, S. A. (2017). Targeting metastasis‐initiating cells through the fatty acid receptor CD36. Nature, 541(7635), 41–45. [DOI] [PubMed] [Google Scholar]
- Poggio, M. , Hu, T. , Pai, C. C. , Chu, B. , Belair, C. D. , Chang, A. , Montabana, E. , Lang, U. E. , Fu, Q. , Fong, L. , & Blelloch, R. (2019). Suppression of exosomal PD‐L1 induces systemic anti‐tumor immunity and memory. Cell, 177(2), 414–427.e13.e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu, Z. , Jiang, C. , Wu, J. , & Ding, Y. (2015). Exosomes as potent regulators of HCC malignancy and potential bio‐tools in clinical application. International Journal of Clinical and Experimental Medicine, 8(10), 17088–17095. [PMC free article] [PubMed] [Google Scholar]
- Quail, D. F. , & Joyce, J. A. (2013). Microenvironmental regulation of tumor progression and metastasis. Nature Medicine, 19(11), 1423–1437. [Google Scholar]
- Quail, D. F. , & Joyce, J. A. (2017). Molecular Pathways: Deciphering Mechanisms of Resistance to Macrophage‐Targeted Therapies. Clinical Cancer Research, 23(4), p. 876–884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raposo, G. , & Stoorvogel, W. (2013). Extracellular vesicles: Exosomes, microvesicles, and friends. Journal of Cell Biology, 200(4), 373–383. [Google Scholar]
- Roh, M. , Wainwright, D. A. , Wu, J. D. , Wan, Y. , & Zhang, B. (2020). Targeting CD73 to augment cancer immunotherapy. Current Opinion in Pharmacology, 53, 66–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romio, M. , Reinbeck, B. , Bongardt, S. , Hüls, S. , Burghoff, S. , & Schrader, J. (2011). Extracellular purine metabolism and signaling of CD73‐derived adenosine in murine Treg and Teff cells. American Journal of Physiology. Cell Physiology, 301(2), C530–C539. [DOI] [PubMed] [Google Scholar]
- Rong, C. , Shi, Y. , Huang, J. , Wang, X. , Shimizu, R. , Mori, Y. , Murai, A. , & Liang, J. (2020). The effect of Metadherin on NF‐κB activation and downstream genes in ovarian cancer. Cell Transplantation, 29, 096368972090550. [Google Scholar]
- Ruan, S. , Lin, M. , Zhu, Y. , Lum, L. , Thakur, A. , Jin, R. , Shao, W. , Zhang, Y. , Hu, Y. , Huang, S. , Hurt, E. M. , Chang, A. E. , Wicha, M. S. , & Li, Q. (2020). Integrin β4‐targeted cancer immunotherapies inhibit tumor growth and decrease metastasis. Cancer Research, 80(4), : P. 771–783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruffell, B. , Affara, N. I. , & Coussens, L. M. (2012). Differential macrophage programming in the tumor microenvironment. Trends in Immunology, 33(3), 119–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakaguchi, S. , Sakaguchi, N. , Asano, M. , Itoh, M. , & Toda, M. (1995). Immunologic self‐tolerance maintained by activated T cells expressing IL‐2 receptor alpha‐chains (CD25). Breakdown of a single mechanism of self‐tolerance causes various autoimmune diseases. Journal of Immunology, 155(3), 1151–1164. [Google Scholar]
- Salimu, J. , Webber, J. , Gurney, M. , Al‐Taei, S. , Clayton, A. , & Tabi, Z. (2017). Dominant immunosuppression of dendritic cell function by prostate‐cancer‐derived exosomes. Journal of Extracellular Vesicles, 6(1), 1368823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider, E. , Winzer, R. , Rissiek, A. , Ricklefs, I. , Meyer‐Schwesinger, C. , Ricklefs, F. L. , Bauche, A. , Behrends, J. , Reimer, R. , Brenna, S. , Wasielewski, H. , Lauten, M. , Rissiek, B. , Puig, B. , Cortesi, F. , Magnus, T. , Fliegert, R. , Müller, C. E. , Gagliani, N. , & Tolosa, E. (2021). CD73‐mediated adenosine production by CD8 T cell‐derived extracellular vesicles constitutes an intrinsic mechanism of immune suppression. Nature Communication, 12(1), 5911. [Google Scholar]
- Sleeman, J. P. (2015). The lymph node pre‐metastatic niche. Journal of Molecular Medicine (Berl), 93(11), 1173–1184. [Google Scholar]
- Sturm, G. , Finotello, F. , Petitprez, F. , Zhang, J. D. , Baumbach, J. , Fridman, W. H. , List, M. , & Aneichyk, T. (2019). Comprehensive evaluation of transcriptome‐based cell‐type quantification methods for immuno‐oncology. Bioinformatics, 35(14), i436–i445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan, J. A. , Tomita, Y. , Jankowska‐Gan, E. , Lema, D. A. , Arvedson, M. P. , Nair, A. , Bracamonte‐Baran, W. , Zhou, Y. , Meyer, K. K. , Zhong, W. , Sawant, D. V. , Szymczak‐Workman, A. L. , Zhang, Q. , Workman, C. J. , Hong, S. , Vignali, D. A. A. , & Burlingham, W. J. (2020). Treg‐cell‐derived IL‐35‐coated extracellular vesicles promote infectious tolerance. Cell reports, 30(4), 1039–1051.e5.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang, T. T. , Wang, B. , Wu, M. , Li, Z. L. , Feng, Y. , Cao, J. Y. , Yin, D. , Liu, H. , Tang, Ri‐N. , Crowley, S. D. , Lv, L. ‐L. , & Liu, Bi‐C. (2020). Extracellular vesicle‐encapsulated IL‐10 as novel nanotherapeutics against ischemic AKI. Science Advances, 6(33), eaaz0748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang, Y. , Li, C. , Zhang, Y. ‐. J. , & Wu, Z.‐H. (2021). Ferroptosis‐related long non‐coding RNA signature predicts the prognosis of Head and neck squamous cell carcinoma. International Journal of Biological Sciences, 17(3), 702–711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Théry, C. , Witwer, K. W. , Aikawa, E. , Alcaraz, M. J. , Anderson, J. D. , Andriantsitohaina, R. , Antoniou, A. , Arab, T. , Archer, F. , Atkin‐Smith, G. K. , Ayre, D. C. , Bach, J. M. , Bachurski, D. , Baharvand, H. , Balaj, L. , Baldacchino, S. , Bauer, N. N. , Baxter, A. A. , Bebawy, M. , … Zuba‐Surma, E. K. (2018). Minimal information for studies of extracellular vesicles 2018 (MISEV2018): A position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. Journal of Extracellular Vesicles, 7(1), 1535750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tjin, M. S. , Chua, A. W. C. , Moreno‐Moral, A. , Chong, Li Y. , Tang, Po Y. , Harmston, N. P. , Cai, Z. , Petretto, E. , Tan, B. K. , & Tryggvason, K. (2018). Biologically relevant laminin as chemically defined and fully human platform for human epidermal keratinocyte culture. Nature Communication, 9(1), 4432. [Google Scholar]
- Vakhshiteh, F. , Atyabi, F. , & Ostad, S. N. (2019). Mesenchymal stem cell exosomes: A two‐edged sword in cancer therapy. International Journal of Nanomedicine, 14, 2847–2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan, S. , Zhao, E. , Kryczek, I. , Vatan, L. , Sadovskaya, A. , Ludema, G. , Simeone, D. M. , Zou, W. , & Welling, T. H. (2014). Tumor‐associated macrophages produce interleukin 6 and signal via STAT3 to promote expansion of human hepatocellular carcinoma stem cells. Gastroenterology, 147(6), 1393–1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, H. , Lee, S. , Nigro, C. L. o. , Lattanzio, L. , Merlano, M. , Monteverde, M. , Matin, R. , Purdie, K. , Mladkova, N. , Bergamaschi, D. , Harwood, C. , Syed, N. , Szlosarek, P. , Briasoulis, E. , Mchugh, A. , Thompson, A. , Evans, A. , Leigh, I. , Fleming, C. , … Crook, T. (2012). NT5E (CD73) is epigenetically regulated in malignant melanoma and associated with metastatic site specificity. British Journal of Cancer, 106(8), 1446–1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, H. W. , & Joyce, J. A. (2010). Alternative activation of tumor‐associated macrophages by IL‐4: Priming for protumoral functions. Cell Cycle, 9(24), 4824–4835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whiteside, T. L. (2016). Exosomes and tumor‐mediated immune suppression. Journal of Clinical Investigation, 126(4), 1216–1223. [Google Scholar]
- Willingham, S. B. , Volkmer, J. P. , Gentles, A. J. , Sahoo, D. , Dalerba, P. , Mitra, S. S. , Wang, J. , Contreras‐Trujillo, H. , Martin, R. , Cohen, J. D. , Lovelace, P. , Scheeren, F. A. , Chao, M. P. , Weiskopf, K. , Tang, C. , Volkmer, A. K. , Naik, T. J. , Storm, T. A. , Mosley, A. R. , … Weissman, I. L. (2012). The CD47‐signal regulatory protein alpha (SIRPa) interaction is a therapeutic target for human solid tumors. Proceedings of the National Academy of Sciences of the United States of America, 109(17), 6662–6667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witwer, K. W. , Buzás, E. I. , Bemis, L. T. , Bora, A. , Lässer, C. , Lötvall, J. , Nolte‐'t Hoen, E. N. , Piper, M. G. , Sivaraman, S. , Skog, J. , Théry, C. , Wauben, M. H. , & Hochberg, F. (2013). Standardization of sample collection, isolation and analysis methods in extracellular vesicle research. Journal of Extracellular Vesicles, 2. [Google Scholar]
- Xiao, C. , Song, F. , Zheng, Yu L. , Lv, J. , Wang, Q. F. , & Xu, N. (2019). Exosomes in head and neck squamous cell carcinoma. Frontiers in Oncology, 9, 894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, M. , Li, J. , Gu, P. , & Fan, X. (2021). The application of nanoparticles in cancer immunotherapy: Targeting tumor microenvironment. Bioactive Materials, 6(7), 1973–1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yegutkin, G. G. (2008). Nucleotide‐ and nucleoside‐converting ectoenzymes: Important modulators of purinergic signalling cascade. Biochimica Et Biophysica Acta, 1783(5), 673–694. [DOI] [PubMed] [Google Scholar]
- Yu, G. , Wang, Li‐G. , Han, Y. , & He, Q.‐Y. (2012). clusterProfiler: An R package for comparing biological themes among gene clusters. OMICS, 16(5), 284–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, H. , Li, R. , Cao, Y. , Gu, Y. , Lin, C. , Liu, X. , Lv, K. , He, X. , Fang, H. , Jin, F. , Fei, Y. , Chen, Y. , Wang, J. , Liu, H. , Li, H. , Zhang, H. , He, H. , & Zhang, W. (2020). Poor clinical outcomes and immunoevasive contexture in intratumoral IL‐10‐producing macrophages enriched gastric cancer patients. Annals of Surgery, 275(4), e626–e635. [Google Scholar]
- Zhang, Y. , Yu, G. , Chu, H. , Wang, X. , Xiong, L. , Cai, G. , Liu, R. , Gao, H. , Tao, B. , Li, W. , Li, G. , Liang, J. , & Yang, W. (2018). Macrophage‐associated PGK1 phosphorylation promotes aerobic glycolysis and tumorigenesis. Molecular Cell, 71(2), 201–215.e7.e7. [DOI] [PubMed] [Google Scholar]
- Zheng, Z. C. , Zhu, W. , Lei, L. , Liu, X. ‐Q. , & Wu, Y. ‐. G. (2020). Wogonin ameliorates renal inflammation and fibrosis by inhibiting NF‐κB and TGF‐β1/Smad3 signaling pathways in diabetic nephropathy. Drug Design, Development and Therapy, 14, 4135–4148. [Google Scholar]
- Zhi, X. , Chen, S. , Zhou, P. , Shao, Z. , Wang, L. , Ou, Z. , & Yin, L. (2007). RNA interference of ecto‐5'‐nucleotidase (CD73) inhibits human breast cancer cell growth and invasion. Clinical & Experimental Metastasis, 24(6), 439–448. [DOI] [PubMed] [Google Scholar]
- Zhou, L. , Mudianto, T. , Ma, X. , Riley, R. , & Uppaluri, R. (2020). Targeting EZH2 enhances antigen presentation, antitumor immunity, and circumvents anti‐PD‐1 resistance in head and neck cancer. Clinical Cancer Research, 26(1), 290–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, Y. , Zhou, B. , Pache, L. , Chang, M. , Khodabakhshi, A. H. , Tanaseichuk, O. , Benner, C. , & Chanda, S. K. (2019). Metascape provides a biologist‐oriented resource for the analysis of systems‐level datasets. Nature Communication, 10(1), 1523. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information
Supporting Information
Supporting Information
Supporting Information
Supporting Information
FIGURE S1. CD73 was riched in HNSCC cells derived sEVs. A. Quantitative proteome analysis for cells released sEVs from HNSCC adjacent normal tissues, shown by venn diagram. B. Quantitative proteome for sEVs from HNSCC tissues with or without metastasis, shown by venn diagram. C. Immunoblotting for CD73 in HNSCC cell lines derived sEVs. D. Quantitative proteome results of sEVs from HN6, SCC25 and CAL27, shown by venn diagram. E. The mass spectrum of CD73 in sEVs from HN6, SCC25 and CAL27.
FIGURE S2. CD73 were closely related to macrophages in HNSCC. A. NT5E expression level in different types of tumors utilizing TCGA datasets (log2 TPM). B. The overall survival rate of HNSCC patients from TCGA datasets. C‐F. Survival curves regarding NT5E expression based on TCGA HNSCC cohort. (C, D) Log‐rank Mantel‐Cox test was used to assess significance. Patients with high macrophages and low macrophages gene signature were shown. (E) Patients with M2high and M2low gene signature were shown. (F) Data were classified into low NT5E signature (NT5Elow), high NT5E signature (NT5Ehigh), low macrophage/M2 macrophage signature (Maclow/M2low) and high macrophage/M2 macrophage signature (Machigh/M2high). G. Flow cytometry analysis for percentage of sEVsSCC7‐GFP uptaken in DLNs after injecting sEVsSCC7‐GFP (10 ng‐50 μg) for 30min. H. Flow cytometry analysis about DLNs for percentage of CD4+ T cells, CD8+ T cells, M2 macrophages and Tregs after injecting sEVs (25 μg) every day. I. Gating strategy for identification of immunocytes subsets in DLNs by manual gating. J. Gating strategy for identification of immunocytes in DLNs by manual gating. K. Schematic of SCC7 subcutaneous tumorigenesis. After 21 days, DLNs were dissected. L. Flow cytometry analysis for percentage of macrophages/CD73+ macrophages/PD‐1+ macrophages in DLNs. L. Flow cytometry analysis for percentage of CD8+ T cells/CD73+ CD8+ T cells/PD‐1+ CD8+ T cells in DLNs. Lymph nodes from untreated mice were used as normal control. Data were analyzed by Mann‐Whitney test. (ns, no significant difference, *: p < 0.05, **: p < 0.01, ***: p < 0.001, ****: p < 0.0001).
FIGURE S3. The role of HNSCC‐derived sEVsCD73 in macrophages function. A. Western blot for CD73 and CD63 in sEVs to measure the efficiency of CRISPR/Cas9‐mediated RAB27A/ Rab27a disruption. B. Cell proliferation over time for HN6, SCC25, SCC7 with or without RAB27A/ Rab27a KO. C. Western blot for CD73 in sEVs to detect the efficiency of CRISPR/Cas9‐mediated NT5E/Nt5e disruption. D. Western blot for CD73 in sEVs after NT5E/Nt5e overexpression on NT5E/Nt5eKO cells mediated by lentivirus transfection. E. Representative graphs of CD73+ macrophages in different co‐culture systems. F. Gating strategy for identification of CD73+ macrophages subsets by manual gating. G. Macrophages (1x106) co‐cultured with or without sEVsHNSCC (25 μg‐100 μg) for 24h. Flow cytometry analysis for CD73 expression in macrophages. H. Western blot for CD73 expression levels in sEVs derived from HN6, SCC25, hBMSC, hBMSC‐ NT5EOE, SCC7, mBMSC and mBMSC‐Nt5eOE. Data were analyzed by Mann‐Whitney test. (ns, no significant difference, *: p < 0.05, **: p < 0.01, ***: p < 0.001, ****: p < 0.0001).
FIGURE S4. Absence of CD73 in sEVs restrains tumor growth in vivo. A. Gating strategy for identification of immunocytes subsets by manual gating. B. Representative graphs of CD73+ macrophages in different groups. C. Quantification of PD‐L1+ cells in macrophages. D. Flow cytometry analysis for infiltration of CD4+T cells and percentage of CD73+/PD‐1+ CD4+T cells in tumors. E. Quantification of PD‐1+/PD‐L1+ cells in macrophages. F. Flow cytometry analysis for infiltration of CD4+T cells and percentage of CD73+/PD‐1+ CD4+T cells in tumors. G. Percentage of CD73+/PD‐1+ Tregs in tumors. Data were analyzed by Mann‐Whitney test. (ns, no significant difference, *: p < 0.05, **: p < 0.01, ***: p < 0.001, ****: p < 0.0001).
FIGURE S5. Targeting CD73 in sEVs abolishes immunotherapy resistance in vivo. A. Representative graphs of CD73+ macrophages gating from PBMC in normal people or HNSCC patients. B. Schematic of subcutaneous tumorigenesis 15 days after injection of SCC7 or SCC7Rab27aKO cells with or without CD73 absent in sEVs combined with anti‐PD‐1 therapy. C. Quantification of PD‐1+/PD‐L1+ cells in macrophages. D. Quantification of CD73+/PD‐1+ cells in macrophages. E. Flow cytometry analysis for infiltration of CD4+ T cells and percentage of CD73+/PD‐1+ CD4+ T cells in tumors. F. Quantification of CD73+/PD‐1+ cells in Tregs. Data were analyzed by Mann‐Whitney test. (ns, no significant difference, *: p < 0.05, **: p < 0.01, ***: p < 0.001, ****: p < 0.0001).


