Abstract
Background
Nonalcoholic fatty liver disease (NAFLD) is the leading cause of chronic liver disease, which is associated with features of metabolic syndrome. NAFLD may progress in a subset of patients into nonalcoholic steatohepatitis (NASH) with liver injury resulting ultimately in cirrhosis and potentially hepatocellular carcinoma. Today, there is no approved treatment for NASH due to, at least in part, the lack of preclinical models recapitulating features of human disease. Here, we report the development of a dietary model of NASH in the Göttingen minipig.
Methods
First, we performed a longitudinal characterization of diet-induced NASH and fibrosis using biochemical, histological, and transcriptional analyses. We then evaluated the pharmacological response to Obeticholic acid (OCA) treatment for 8 weeks at 2.5mg/kg/d, a dose matching its active clinical exposure.
Results
Serial histological examinations revealed a rapid installation of NASH driven by massive steatosis and inflammation, including evidence of ballooning. Furthermore, we found the progressive development of both perisinusoidal and portal fibrosis reaching fibrotic septa after 6 months of diet. Histological changes were mechanistically supported by well-defined gene signatures identified by RNA Seq analysis. While treatment with OCA was well tolerated throughout the study, it did not improve liver dysfunction nor NASH progression. By contrast, OCA treatment resulted in a significant reduction in diet-induced fibrosis in this model.
Conclusions
These results, taken together, indicate that the diet-induced NASH in the Göttingen minipig recapitulates most of the features of human NASH and may be a model with improved translational value to prioritize drug candidates toward clinical development
Keywords: NASH, minipig, translational value
Abbreviations: CDAHFD, choline-deficient amino acid-defined high fat diet; FDR, false discovery rate; FFC, fatfructose cholesterol diet; PNPLA3, patatin-like phospholipase domain-containing 3; NAFLD, nonalcoholic fatty liver disease; NAS, NAFLD activity score; NASH, nonalcoholic steatohepatitis
Nonalcoholic fatty liver disease (NAFLD) is a common and progressive disease mainly characterized by hepatic fat accumulation in the absence of alcohol consumption. NAFLD is strongly associated with obesity, Type 2 Diabetes, and dyslipidemia. NAFLD is subdivided into nonalcoholic fatty liver (NAFL) and nonalcoholic steatohepatitis (NASH) based on histological examination of liver biopsy and defined by the presence of lobular inflammation and hepatocyte ballooning with various degrees of fibrosis.1,2 NAFLD is the most common cause of chronic liver disease worldwide with an estimated prevalence of 25%. It is, therefore, considered to be a global health problem associated with a significant socioeconomic burden.3,4 In contrast to NAFL, which is considered as a benign and reversible disease state, NASH accounts for an increased number of patients with cirrhosis, liver failure, and hepatocellular carcinoma.5 NASH patients display increased mortality compared to control population with a high cardiovascular risk. Long-term follow-up studies revealed that fibrosis is the main driver of mortality in NASH.6,7 NAFLD disease phenotype results from the chronic exposure to environmental factors on a susceptible polygenic background comprising multiple independent modifiers.8 Several genes have been associated with the development of NAFLD, and more generally, liver diseases using Genome-Wide Association Studies (GWAS). One of the best characterized single nucleotide polymorphism (SNP) is located within the patatin-like phospholipase domain-containing 3 (PNPLA3) gene, which plays a role in lipid homeostasis.9 This I148M variant is associated with an increase of both NAFLD9,10 and liver fibrosis risk.11 It further confers a significantly increased risk of NAFLD-associated hepatocellular carcinoma.12 There is now a growing list of genetic variants (including but not limited to GCKR, HSD17B13) linked to NALFD (See13 for review).
Despite major progress in the understanding of NASH disease biology and large investments made by the pharmaceutical industry, there is today no approved treatment for NASH.14 There are obvious reasons to explain these failures.15 First, NASH is a very heterogeneous disease with a complex etiology and in most of the cases a late diagnostic. The complex interplay between NASH and fibrosis disease drivers strongly advocates for combination therapy.16 Second, some drugs were moved too quickly into Phase 3 with insufficient efficacy demonstration in small Phase 2 trials or overly optimistic interpretation of equivocal results. Furthermore, positive results on the primary endpoint may not be sometimes strengthened by efficacy on secondary endpoints, which could be equally relevant.15 Third, there are still no noninvasive markers accurately predicting histological outcomes. Finally, the field still suffers from the lack of robust preclinical models both in vitro and in vivo with improved translational value.17 Rodent models (based on genetic or dietary manipulations) have been extensively used to validate targets and to document efficacy upon chronic dosing. These models provide early useful information with respect to target engagement in a pathological setting. Unfortunately, they generally do not capture all the features of human NASH and tend to be overly optimistic in predicting efficacy.15,18 Nonhuman primates have been extensively used for the evaluation of drugs targeting T2D. However, characterization of an obese T2D nonhuman primate model revealed a NAFL phenotype with limited inflammation and mild fibrosis.19 In this study, we have tried to tackle the lack of rodent models fully recapitulating human NASH physiopathology through the development and characterization of a minipig model of NASH and its response to pharmacological intervention. Compared to mice, swine physiology is closer to man, and minipigs have been successfully used to recapitulate both T2D and cardiovascular complications. They are amenable to serial histological examinations on the same set of animals, and therefore, represent an attractive model to perform long-term studies aiming at studying the development of NASH and fibrosis. Indeed, two groups have reported data suggesting that pigs may recapitulate many features of human NASH.20,21 Here, we first analyzed the development of NASH and fibrosis in a cohort of Göttingen minipigs fed with either chow or CDAHFD (Choline-deficient Amino acid-defined High Fat Diet) for up to 6 months. Then, we evaluated the pharmacological response to a drug under late clinical development for NASH, namely Obeticholic acid.
Methods
Animals and Diets
All experimental protocols were approved by the Servier Institutional Animal Care and Use Committee and the French Ministry of Research. Eight-week-old castrated male Göttingen minipigs were provided by Ellegaard (Dalmose, Denmark). Diets were prepared by Special Diet Services (Witham, United Kingdom). Animals were housed in an air-conditioned room with a controlled temperature (22 ± 1 °C) on a 12-h light/dark cycle (light on at 07:00 AM). Pigs were maintained in groups according to their diet and had unlimited access to fresh drinking water. The chow or standard minipig diet contains reduced protein (13%) and carbohydrate (5%) content, crude oil (2.13%), low metabolizable energy. The CDAHFD (Choline-Deficient Amino Acid defined High Fat Diet) contains crude oil (30.05%), protein (20%), 1% cholesterol, 15.8% fructose, no choline but 0.1% methionine. Banana cream flavor was added to the CDAHFD to increase the diet palatability and to help the animal to eat this diet. One week with the progressive introduction of CDAHFD into the meal was required to habituate the pigs to this diet. The amount of CDAHFD given to the animals represents approximately 75% of the given standard chow in order to have isocaloric intake in both groups. Animals were weighed on a weekly basis.
In the first study, animals were placed under CDAHFD (n = 9) or chow diet (n = 6) for 6 months. Liver biopsies, as well as plasma samples, were prepared after a 12 h fast every 2 months for histological and biochemical characterization of the response to the diet. Ultrasound-guided liver biopsy was performed under anesthesia and analgesia using a 14G biopsy device and a vacuum-assisted Vacora Biopsy System. Biopsy samples were fixed in 10% formalin or snap-frozen in liquid nitrogen and stored at −80 °C. At the end of the study, liver samples were collected for gene expression studies, and blood was collected for biochemical determination of different circulating parameters (triglycerides, total cholesterol, glycemia, insulin levels) after a 12 h fast.
In the second study (intervention study), animals were either placed on chow (n = 9 per group) or CDAHFD (n = 18) for 4 weeks. At the end of this induction period, animals fed with CDAHFD were administered Obeticholic acid at 2.5 mg/kg (#AC29217, Purity = 100% from APICHEM, China) (OCA, n = 7) or vehicle (n = 7) for 8 weeks while remaining on CDAHFD. Plasma and liver samples were collected under anesthesia (Zoletil 100, 5 mg/kg, Virbac, France) and analgesia (Buprecare 20 μg/kg, AXIENCE, France) at the end of the study for subsequent analyses.
Biochemical Analyses
Most of the tested plasma parameters were determined with an automatic biochemical analyzer (Indiko Clinical Chemistry Analyzer, Thermofisher). Insulin levels were assessed by Elisa (Mercodia Porcine Insulin ELISA kit)). After KOH and MgCl2 extraction, liver TG levels were measured using a commercially available kit (TRIGLYCERIDES, Thermo Fisher Diagnostics). After extraction using the lipid extraction kit from Biovision, liver total cholesterol levels were determined with the Total cholesterol and Cholesteryl Ester Colorimetric Assay Kit II (Biovision).
Histological Analyses
Formalin-fixed, paraffin-embedded livers were sliced into 3-μm sections. Hematoxylin and Eosin (H&E) staining () was performed to investigate liver histology, and Picro Sirius Red staining was used for liver fibrosis assessment. NAFLD Activity Score (NAS) and fibrosis stage were determined by two double-blinded persons using the NASH CRN scoring system.42 For hepatocellular steatosis, livers were classified into scores 0 to 3 (0: <5% of hepatocytes presenting steatosis, 1: 5–33% of hepatocytes presenting steatosis, 2: 33–66% of hepatocytes presenting steatosis and 3: >66% of hepatocytes presenting steatosis). For lobular inflammation, livers were scored into grades 0 to 3 (0: noninflammatory foci, 1: 1 inflammatory focus, 2: 2 to 4 inflammatory foci, 3: >4 inflammatory foci). Fibrosis was scored into stages from 0 to 4 (0: no fibrosis, 1: perisinusoidal or periportal fibrosis, 2: perisinusoidal and periportal fibrosis, 3: bridging fibrosis or septa, 4: cirrhosis). Analyses were performed at 20 × and 5 × magnification for HE and Picro Sirius Red staining, respectively.
Liver RNA Sequencing
Total RNA was extracted using Qiagen RNA extraction kits following the manufacturer’s instructions. RNA concentrations were obtained using nanodrop or a fluorometric Qubit RNA assay (Life Technologies, Grand Island, New York, USA). The quality of the RNA (RNA integrity number) was determined on the Agilent 2100 Bioanalyzer (Agilent Technologies, Palo Alto, CA, USA) as per the manufacturer’s instructions. To construct the libraries, 400 ng of high-quality total RNA sample (RIN >8) was processed using TruSeq Stranded mRNA kit (Illumina) according to manufacturer instructions. Briefly, after purification of poly-A containing mRNA molecules, mRNA molecules are fragmented and reverse-transcribed using random primers. Replacement of dTTP by dUTP during the second strand synthesis will permit to achieve the strand specificity. The addition of a single A base to the cDNA is followed by ligation of Illumina adapters. Libraries were quantified by qPCR using the KAPA Library Quantification Kit for Illumina Libraries (KapaBiosystems, Wilmington, MA), and library profiles were assessed using the DNA High Sensitivity LabChip kit on an Agilent Bioanalyzer. Libraries were sequenced on an Illumina Nextseq 500 instrument using 75 base-lengths read V2 chemistry in a paired-end mode. After sequencing, a primary analysis based on AOZAN software (ENS, Paris) was applied to demultiplex and control the quality of the raw data (based on FastQC modules/version 0.11.5). Obtained FASTQ files were then aligned using STAR algorithm (version 2.5.2 b) and quality control of the alignment realized with Picard tools (version 2.8.1). Reads were then counted using RSEM, and genes with less than three samples with normalized counts greater than or equal to 10 were filtered out. In the end, our final RNA-Seq data comprises 15,956 genes.
RNA Seq Data Analyses
Data analyses were carried out using R system software (http://www.R-project.org, V4.0.4) packages, including those of Bioconductor or original R code. To identify genes differentially expressed between diets, we performed a linear model (lmFit function from limma R package) on the vst transformed gene expression dataset. The resulting P-values were adjusted for multiple hypothesis testing and filtered to retain DE genes with a False Discovery Rate (FDR) adjusted P-value < 0.05 and a |Fold-Change (FC)| ≥ 1.5. Enrichment analysis was performed by applying the fast gene set enrichment analysis (fgsea function from fgsea R package) against HALLMARK pathways.
Statistical Analysis
For analysis of body weight as a function of time, a two-way ANOVA with repeated measures on time factor was performed for comparison of diet. For liver and body weight, an unpaired t-test was used after verification of the normal distribution of data for comparison of two groups (Chow vs. CDAHFD). For the second study, group comparisons were made by one-way ANOVA followed by Dunnett’s test using the Prism Software v7 (GraphPad Software). A P value less than 0.05 was considered statistically significant. Results are expressed as mean ± SEM.
Results
In the first study, we analyzed the development of NASH and fibrosis in a cohort of Göttingen minipigs fed with either chow or CDAHFD for up to 6 months. Plasma samples and liver biopsies were collected every 2 months in both groups for biochemical and histological analyses, respectively. CDAHFD triggered progressive and significant obesity that became pronounced after 4 months of diet (Figure 1A) despite a similar isocaloric food consumption between groups. CDAHFD induced a marked liver enlargement, as depicted in Figure 1B, which remained significant after normalization to total body weight (Figure 1C–D). Of note, livers from CDAHFD animals were pale, suggesting steatosis (Figure 1B). During the study, two animals in the CDAHFD group had to be euthanized for ethical reasons at weeks 16 and 18. This was due to severe body weight loss secondary to reduced appetite. At necropsy, those animals displayed clear anemia and liver failure. Results from these minipigs were not included in the statistical analyses. Analysis of the blood chemistry revealed a pronounced dyslipidemia with marked hypercholesterolemia in response to CDAHFD (Figure 2B) while triglycerides levels were not largely modified in response to the NASH diet even if they reached statistical significance at the end of the study (Figure 2A). Diet-induced hypercholesterolemia reached its peak as early as 2 months after initiation of the diet. While glucose levels were not modified, CDAHFD resulted in increased plasma insulin levels after 4 and 6 months of diet compared to chow-fed animals (Figure 2D). Moreover, fructosamine levels were also significantly elevated at the end of the study in the CDAHFD group compared to controls (489 ± 37 vs. 371 ± 8, P < 0.001) in line with the insulin results. These results, taken together, results suggest that the CDAHFD diet triggered the development of insulin resistance. Blood chemistry results confirmed severe liver dysfunction with elevated AST, direct and total bilirubin levels as early as 2 months after diet initiation (Figure 3BCD). By contrast, ALT levels were not significantly altered by the diet throughout the study (Figure 3A).
Figure 1.
CDAHFD triggers a significant increase in both liver and total body weight. Body weight follow-up (Panel A), liver morphology (Panel B), total liver weight (Panel C), and liver to body weight ratio (Panel D) at the end of the study (chow n = 6 & CDAHFD = 7 per group). Data shown are mean ± SEM. (∗: P < 0.05; ∗∗∗: P < 0.001 CDAAHFD vs. Chow).
Figure 2.
Marked hypercholesterolemia and insulin resistance in response to CDAHFD. Kinetic analysis of plasma triglycerides (Panel A), cholesterol (Panel B), glucose (Panel C), and insulin levels (Panel D) in Chow (n = 6; white bars), and CDAHFD (n = 7, blue bars) fed minipigs. Data shown are mean ± SEM. (∗∗∗: P < 0.001 CDAHFD vs. Chow).
Figure 3.
Rapid and significant liver dysfunction in response to CDAHFD. Kinetic analysis of plasma liver enzymes (Panel A&B), total and direct bilirubin levels (Panel C&D) in Chow (n = 6; white bars), and CDAHFD (n = 7, blue bars) fed minipigs. Data shown are mean ± SEM. (∗∗: P < 0.01; ∗∗∗: P < 0.001 CDAHFD vs. Chow).
HE staining of liver biopsies revealed a rapid development of steatosis, with more than 60% of the hepatocytes displaying lipid vesicles, as early as 2 months after initiation of CDAHFD with large lipid droplets throughout the liver parenchyma (Figure 4A). There was no further increase in steatosis with time (Figure 4B). Lobular inflammation and limited hepatocellular ballooning (with a time-dependent increase) were also noticed (Figure 4C–D), which translated into a NAS score around 4 after 2 months of CDAHFD. This NAS score, mainly driven by steatosis, did not increase with time. Next, fibrosis was assessed by Sirius Red staining (Figure B). CDAHFD induced a progressive development of both perisinusoidal and portal fibrosis at month 4, reaching fibrotic septa at month 6, with 100% of the animals developing severe fibrosis (Figure 5B–C).
Figure 4.
Pronounced steatosis and limited inflammation in response to CDAHFD. Representative images from HE staining (Panel A), histological analyses (Steatosis Panel B; Inflammation Panel C; Ballooning Panel D), and NAS score determination (Panel E) in Chow (n = 6; white bars), and CDAHFD (n = 7, blue bars) fed minipigs. Data shown are mean ± SEM. (∗∗∗: P < 0.001 CDAHFD vs. Chow).
Figure 5.
Time-dependent increase in liver fibrosis in response to CDAHFD. Representative pictures of Sirius red stained liver samples (Panel A). Fibrosis score (Panel B) and distribution of animals according to score (Panel C) in Chow (n = 6; white bars), and CDAHFD (n = 7, blue bars) fed minipigs. Data shown are mean ± SEM. (∗∗∗: P < 0.001 CDAHFD vs. Chow).
In order to better understand the molecular mechanisms involved in CDAHFD-induced NASH and fibrosis, RNA Seq analysis was performed on liver total RNA at the end of the study. Principal component analysis of RNA Seq data showed a clear separation between groups revealing major transcriptional changes between CDAHFD and chow-fed animals (Supplementary Fig. 1). A total of 5033 genes were found differentially expressed (2747 up and 2286 down) between CDAHFD and chow-fed animals (fold change >1.5, FDR-adjusted P value<0.05) (Figure 6AB). Among the differentially expressed genes, we found major players involved in NASH and fibrosis development, such as regulators of lipid metabolism (cholesterol metabolism, de novo lipogenesis, Supplementary Fig. 2), inflammation (inflammasome, chemotaxis, Supplementary Fig. 3) and fibrosis (Acta2, Collagens, TGFB1, Supplementary Fig. 4). It is noteworthy that these gene modulations were pronounced with very significant P values. These transcriptional changes were further explored by gene set enrichment analysis (Figure 6C). Again, key pathways involved in NASH pathophysiology were identified, such as an inflammatory signature (Interferon response, JAK-STAT3 signaling, TNFA signaling), fibrosis (Epithelium Mesenchymal Transition), lipid (fatty acid, cholesterol, and bile acid metabolism), and mitochondrial dysfunction (oxidative phosphorylation). Ingenuity Pathway Analysis (Qiagen) was then used to determine upstream regulators responsible for these gene modulations (Supplementary Table 1). As expected, key transcription factors/pathways controlling the expression of proinflammatory and profibrotic genes (NFKB and TGFB, respectively) were found activated. By contrast, regulators of lipid metabolism (cholesterol homeostasis, beta-oxidation) and hepatic gene transcription were found inhibited (PPARA, SREBF2) (Supplementary Table 1). Taken together, the results from biochemical, histological, and gene expression analyses indicate that CDAHFD induced in the Göttingen minipig the development of NASH with progressive fibrosis associated with features of the metabolic syndrome.
Figure 6.
RNA Seq analysis of liver samples at the end of the study. Heatmap (Panel A), volcano plot (Panel B), and GSEA (Panel C), of differentially expressed genes from CDAHFD compared to Chow (Fold change cutoff >1.5; FDR-adjusted P-value cutoff: 0.05).
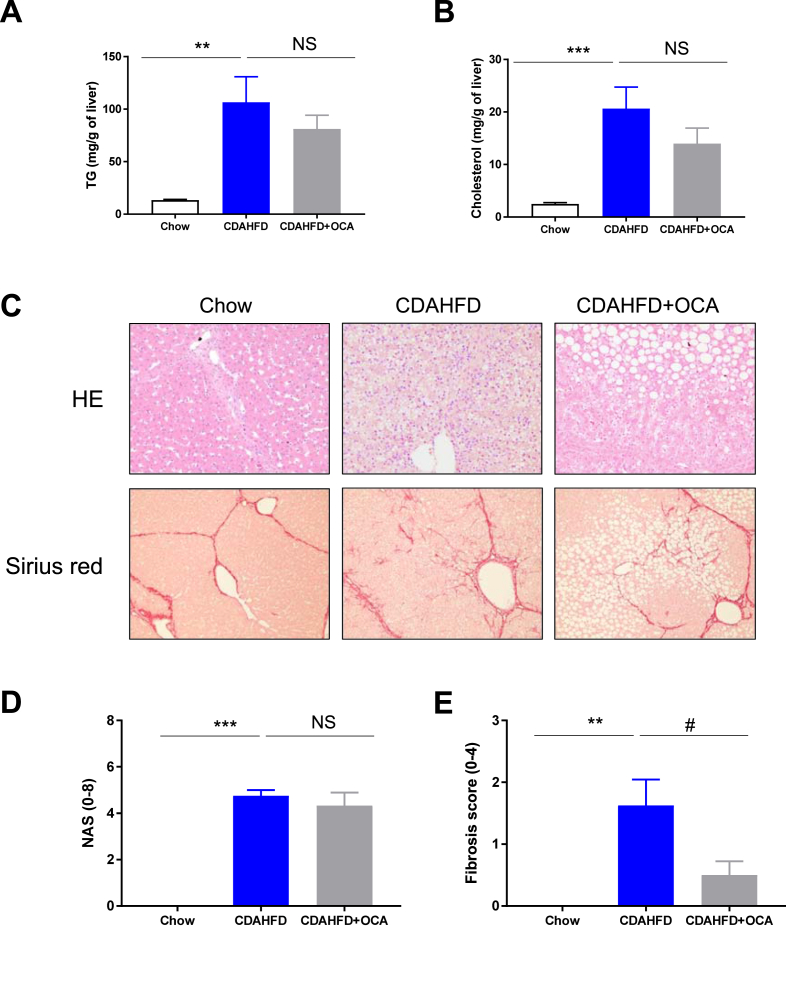
Having characterized the kinetics of development of diet-induced NASH and fibrosis in the minipig fed CDAHFD, we next evaluated the pharmacological response to a drug under late clinical development for NASH. Obeticholic acid (OCA), a bile acid Farnesoid X Receptor (FXR) agonist, is approved for the treatment of Primary Biliary Cholangitis (marketed as OCALIVA) and has demonstrated significant efficacy on fibrosis (improvement >1stage) with no worsening of NASH.22 Preliminary pharmacokinetic analysis led to the selection of 2.5 mg/kg/day as the dose to be tested in the minipig to match clinical exposure obtained at the 25 mg dose,23 a dose resulting in significant improvement of fibrosis in patients with NASH (See Supplementary Table 2). In this intervention study, minipigs were either fed CDAHFD (n = 18) or chow (n = 9) for 4 weeks to initiate NASH development. Then, CDAHFD-fed animals were randomized to receive either OCA (2.5 mg/kg/d) or vehicle for 8 weeks while staying on the very same diet. As expected, CDAHFD induced a significant weight gain compared to chow, in line with our first study at week 12. OCA treatment was well tolerated, although some pigs demonstrated fatigue and digestion problems during few days after treatment induction (side effects that are observed in patients treated with OCA). Interestingly, there was no significant impact of OCA treatment on total body weight nor on liver enlargement (Figure 7). Analysis of blood chemistry confirmed the significant impact of CDAHFD on insulin resistance (Figure 8A–C), dyslipidemia (Figure 8 D-E), and liver dysfunction (Figure 8F–I) compared to chow-fed animals in line with our first study. Again, OCA at the dose tested failed to elicit any meaningful changes on all the analyzed parameters. A small but nonsignificant increase in total cholesterol was nevertheless observed (Figure 8E). CDAHFD triggered a massive accumulation of both triglycerides and cholesterol in the liver (Figure 9 A-B). Nonsignificant reductions of both intrahepatic cholesterol and triglycerides levels were recorded in response to OCA. The trend was clear, but it did not reach statistical significance likely due to the small groups’ size (n = 7 per group).
Figure 7.
OCA treatment had no significant impact on body weight gain nor liver enlargement. Body weight follow-up (Panel A), liver morphology (Panel B), total liver weight (Panel C), and liver to body weight ratio (Panel D) at the end of the study in chow (white bar n = 8); CDAHFD (blue bar n = 8) and CDAHFD + OCA (grey bar n = 6). Data shown are mean ± SEM. (∗∗∗: P < 0.001 CDAHFD vs. Chow; NS: nonsignificant).
Figure 8.
No impact of OCA treatment on insulin resistance, dyslipidemia and liver dysfunction in CDAHFD fed minipigs. Plasma glucose (A), insulin (B), fructosamine (C), TG (D), cholesterol (E), ALT (F), AST (G), ALP (H), and γGT (I) levels were measured at the end of the study. Data shown are means ± SEM (chow white bar n = 8; CDAHFD blue bar n = 8, CDAHFD + OCA grey bar n = 6). ∗: P < 0.05, ∗∗: P < 0.01, ∗∗∗: P < 0.001 CDAHFD vs. Chow.
Figure 9.
Significant impact of OCA on fibrosis but not NASH in minipigs fed with CDAHFD. Liver TG (Panel A) and cholesterol levels (Panel B) were determined at the end of the study. Representative pictures of HE and/or Sirius red stained liver samples at the end of the study (Panel C). NAS (Panel D) and Fibrosis (Panel E) scores. Data shown are mean ± SEM. (chow white bar n = 8; CDAHFD blue bar n = 8, CDAHFD + OCA grey bar n = 6). ∗∗∗: P < 0.001 CDAHFD vs. Chow; #: P < 0.05 CDAHFD + OCA vs. CDAHFD.
Histological analyses confirmed our initial findings, CDAHFD triggering a rapid and significant NASH installation followed by the development of progressive fibrosis. 12 weeks of diet resulted in a NAS score of around 4 and a fibrosis score of around 2 (Figure 9 C-E), in line with our first study (Figure 4, Figure 5). Interestingly OCA treatment significantly reduced the development of fibrosis (>1 point) without affecting NASH progression (no reduction in NAS score). These results, taken together, indicate that OCA treatment reduced fibrosis in a minipig model, thereby validating this new model at the pharmacological level.
Discussion
Here, we report the development, characterization, and pharmacological validation of a preclinical model of NASH associated with fibrosis in the Göttingen minipig. CDAHFD triggered insulin resistance associated with obesity, marked hypercholesterolemia, and liver dysfunction as measured by increased bilirubin and AST levels (Figure 1, Figure 2, Figure 3). However, CDAHFD did not trigger plasma hypertriglyceridemia, a hallmark of human NAFLD likely due to the use of a choline-deficient diet. Histological analyses revealed the rapid installation of NASH mainly driven by steatosis and immune cell infiltration within the lobule. Some hepatocellular ballooning was observed after 2 months of treatment and onward (Figure 4). Last but not least, we observed a time-dependent increase in both perisinusoidal and portal fibrosis (between 2 and 4 months of diet), reaching cirrhosis at month 6, with 100% of the animals developing severe fibrosis (Figure 5). All these results were confirmed at the molecular level by RNA Seq and subsequent GSEA (Figure 6). Our results indicate the CDAHFD-fed Göttingen minipig captures many features of human NASH, or more specifically, Metabolic Associated Fatty Liver Disease (MAFLD).24 Our results are largely in line with previous data generated by Pedersen and colleagues.21 In their work, they compared the ability of several diets to trigger NASH in the Göttingen minipig. Among the tested diets was CDAHFD with either sucrose or fructose. However, animals were placed on the different diets for only 8 weeks, which prevented them from observing obesity and the associated insulin resistance that appeared after more than 10 weeks of diet in our study (Figure 1). Furthermore, histological lesions were logically more pronounced in our model. In general, the phenotype they observed was less marked and likely due to the shorter exposure to the diet21 but highly consistent with respect to results obtained at 8 weeks suggesting the robustness of the model. This choline-deficient diet seems to be the most appropriate to trigger human-like NASH phenotype compared to other tested previously, such as the fat-fructose cholesterol diet (FFC).25 Indeed, prolonged exposure to the FFC diet with or without streptozotocin-induced diabetes failed to trigger the development of steatosis and hepatocellular ballooning, key characteristics of the human disease.25 Other strains of pigs were previously investigated.20 Off note, the Ossabaw miniature swine that shares many features of human metabolic syndrome has been shown to develop NASH in response to a Western diet (fructose-based atherogenic diet)20,26 Interestingly, there was no evidence of macrovesicular steatosis or lobular inflammation in response to this diet (which contains choline and methionine) whatever the duration of the study.20,26 The CDAHFD-fed Göttingen minipig appears therefore as the swine model capturing most of the human NASH features.
One major objective of this work was to develop and characterize the development of NASH and fibrosis in this model but also to try to validate it using a drug. OCA was selected as the drug to be evaluated since it is the most advanced drug in development for NASH with clear efficacy demonstrated in placebo-controlled phase 3 trial in patients with NASH and fibrosis (F2–F3).22 OCA was administered to CDAHFD-fed Göttingen minipigs for 8 weeks at 2.5 mg/kg, a dose selected to match clinical exposure achieved at 25 mg dose tested in the REGENERATE study. OCA treatment was well-tolerated throughout the study. Interestingly, OCA treatment failed to significantly improve CDAHFD-induced liver dysfunction (Figure 7, Figure 8). Furthermore, a nonsignificant increase in total cholesterol (+24%) was observed in line with what was initially reported in patients.27 Nonsignificant reductions in both liver TG and cholesterol levels were noticed (Figure 9). Finally, histological analyses revealed that OCA treatment significantly reduced fibrosis without affecting NASH progression as measured by NAS score (Figure 9). These results are reminiscent of findings of the REGENERATE trial.22 In this trial, OCA (25 mg) met the primary endpoint of improvement in fibrosis with no worsening of NASH in patients with stage F2 or F3 fibrosis at the month-18 interim analysis. It is unclear why OCA failed to improve NASH resolution in this population while early data derived from the FLINT trial were very encouraging.27 These robust antifibrotic properties were documented in rodent preclinical models.28,29 Interestingly, the very same rodent models revealed the major impact of OCA on NASH progression with treatment reducing both steatosis and inflammation in stark contrast to what was observed in our model. It is tempting to speculate that major differences with respect to lipid metabolism such as lack of CETP expression may account for such discrepancies as proposed by Briand and colleagues.30 Indeed, pigs are considered as predictive models for the study of human lipoprotein metabolism and cardiovascular disease.31,32 Nevertheless, more studies are required to determine the overall translational value of the CDAHFD-fed Göttingen minipig.
Transcriptomic analyses revealed major alterations in gene expression in response to CDAHFD (Figure 6) and confirmed the involvement of well-known drivers of human NASH and fibrosis pathophysiology, including but not limited to inflammation, fibrosis, cholesterol metabolism, and mitochondrial dysfunction. It is noteworthy that we found dysregulated in our dataset most of the genes comprised in a recently reported gene signature predicting clinical NAFLD severity.33 For instance, GDF15 that is highly upregulated in the CDAHFD-fed minipig model (Supplementary Fig. 3) has been proposed as a potential biomarker of advanced fibrosis.34 Furthermore, bioinformatic analyses performed using Ingenuity Pathway Analysis led to the identification of upstream regulators that are also well known to play a role in human disease progression (Supplementary Table 1). Among these, PPARα that is known as a protective factor and potential drug target for the treatment of NASH35 is predicted to be inhibited in this study as expected. These transcriptional changes, taken together, underscore the translational value of the minipig model.
The CDAHFD-fed Göttingen minipig model captures many features of MAFLD and offers the possibility to conduct serial histological examination during longitudinal studies, which are key shortcomings of rodent models.18 To further model human disease pathophysiology, it would be interesting to introduce key gene mutations associated with the development of NASH and/or fibrosis and using the Crispr/Cas9 system as recently described by Coutts and colleagues in the Ossabaw swine model for PNPLA3.36 Furthermore, it is even now possible to follow disease progression using noninvasive imaging techniques that are currently used in the clinic. Moreover, this model can also be used to monitor the impact of the disease or the response to a given treatment on cardiovascular readouts. Minipigs are used to study cardiovascular complications and might provide an opportunity to better understand at the preclinical level the impact of liver fibrosis on the development of cardiomyopathy and cardiac arrhythmias (See37 for review). However, such a model presents some limitations such as costs and logistical challenges and requires specific expertise. Rodent models, therefore, remain essential in the drug development process from target validation using genetics to drug evaluation. Recently published models seem to focus much more on human translatability38, 39, 40, 41 and may provide new insights into our understanding of the disease and mechanism of action of drugs.
In conclusion, the CDAHFD-fed Göttingen minipig is a model recapitulating most of the features of human MAFLD and represents an attractive option to assess candidate drug’s efficacy. Further studies, including multiomic approaches, as well as testing drugs with various mechanisms of action and combinations, are required to further establish its translational value.
Credit authorship contribution statement
VD and PD designed experiments and supervised the work. SC, OB, AH, LL, KM, NL, LG, KL, LA, and AH performed all the experiments. JG, FL, VP, NP, VD, and DV contributed to both data and statistical analyses. ELH, TB, NDP, and AB coordinated the work. VD, VP, DV, and PD wrote the manuscript.
Conflicts of interest
VD, SC, OB, AH, LL, KM, NL, LG, KL, NP, ELH, TB, NDP, JG, and PD are employees of Servier. LA, FL, AH, DV, AB, and VP have declared that no conflict of interest exists.
Acknowledgement
We would like to thank the scientific and technical teams at Ellegaard Göttingen Minipigs A/S for their excellent support and scientific guidance.
Funding
This work is part of the QUID-NASH consortium. RHU QUID-NASH is funded by Agence Nationale de la Recherche under the reference QNR-17-RHUS-0009; implemented by Inserm, Universite Paris Descartes, Universite Paris Diderot, CNRS, CEA, Laboratoires Servier, Biopredictive, and AP-HP; and coordinated by Prof. Dominique Valla and Angelique Brzustowski.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jceh.2021.09.001.
Appendix ASupplementary data
The following is the Supplementary data to this article:
References
- 1.Brunt E.M., Wong V.W.S., Nobili V., et al. Nonalcoholic fatty liver disease. Nat Rev Dis Prim. 2015;1 doi: 10.1038/nrdp.2015.80. https://pubmed.ncbi.nlm.nih.gov/27188459/ [Internet]. Nature Publishing Group [cited 2021 Jun 2] Available from: [DOI] [PubMed] [Google Scholar]
- 2.Diehl A.M., Day C. Cause, pathogenesis, and treatment of nonalcoholic steatohepatitis. N Engl J Med. 2017;377:2063–2072. doi: 10.1056/NEJMra1503519. https://pubmed.ncbi.nlm.nih.gov/29166236/ [Internet]. New England Journal of Medicine (NEJM/MMS) [cited 2021 Jun 2] Available from: [DOI] [PubMed] [Google Scholar]
- 3.Younossi Z.M., Golabi P., de Avila L., et al. The global epidemiology of NAFLD and NASH in patients with type 2 diabetes: a systematic review and meta-analysis. J Hepatol. 2019;71:793–801. doi: 10.1016/j.jhep.2019.06.021. https://pubmed.ncbi.nlm.nih.gov/31279902/ [Internet]. Elsevier B.V.[cited 2021 Jun 2] Available from: [DOI] [PubMed] [Google Scholar]
- 4.Younossi Z.M., Koenig A.B., Abdelatif D., Fazel Y., Henry L., Wymer M. Global epidemiology of nonalcoholic fatty liver disease—meta-analytic assessment of prevalence, incidence, and outcomes. Hepatology. 2016;64:73–84. doi: 10.1002/hep.28431. John Wiley and Sons Inc. [DOI] [PubMed] [Google Scholar]
- 5.Loomba R., Friedman S.L., Shulman G.I. Mechanisms and disease consequences of nonalcoholic fatty liver disease. Cell. 2021;184:2537–2564. doi: 10.1016/j.cell.2021.04.015. http://www.ncbi.nlm.nih.gov/pubmed/33989548 [Internet]. Cell [cited 2021 May 23] Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Angulo P., Kleiner D.E., Dam-Larsen S., et al. Liver fibrosis, but no other histologic features, is associated with long-term outcomes of patients with nonalcoholic fatty liver disease. Gastroenterology. 2015;149:389–397. doi: 10.1053/j.gastro.2015.04.043. https://pubmed.ncbi.nlm.nih.gov/25935633/ [Internet]. W.B. Saunders [cited 2021 Jun 2] e10. Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hagström H., Nasr P., Ekstedt M., et al. Risk for development of severe liver disease in lean patients with nonalcoholic fatty liver disease: a long-term follow-up study. Hepatol Commun. 2018;2:48–57. doi: 10.1002/hep4.1124. https://pubmed.ncbi.nlm.nih.gov/29404512/ [Internet]. Wiley [cited 2021 Jun 2] Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Anstee Q.M., Day C.P. The genetics of NAFLD. Nat. Rev. Gastroenterol. Hepatol. Nat Rev Gastroenterol Hepatol. 2013:645–655. doi: 10.1038/nrgastro.2013.182. https://pubmed.ncbi.nlm.nih.gov/24061205/ [Internet] [cited 2021 Jun 2] Available from: [DOI] [PubMed] [Google Scholar]
- 9.Romeo S., Kozlitina J., Xing C., et al. Genetic variation in PNPLA3 confers susceptibility to nonalcoholic fatty liver disease. Nat Genet. 2008;40:1461–1465. doi: 10.1038/ng.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Speliotes E.K., Yerges-Armstrong L.M., Wu J., et al. Genome-wide association analysis identifies variants associated with nonalcoholic fatty liver disease that have distinct effects on metabolic traits. PLoS Genet. 2011;7 doi: 10.1371/journal.pgen.1001324. https://pubmed.ncbi.nlm.nih.gov/21423719/ [ Internet]. Public Library of Science [cited 2021 Jun 2] Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Valenti L., Al-Serri A., Daly A.K., et al. Homozygosity for the patatin-like phospholipase-3/adiponutrin i148m polymorphism influences liver fibrosis in patients with nonalcoholic fatty liver disease. Hepatology. 2010;51:1209–1217. doi: 10.1002/hep.23622. https://pubmed.ncbi.nlm.nih.gov/20373368/ [Internet]. Hepatology [cited 2021 Jun 2] Available from: [DOI] [PubMed] [Google Scholar]
- 12.Liu Y.L., Patman G.L., Leathart J.B.S., et al. Carriage of the PNPLA3 rs738409 C >g polymorphism confers an increased risk of non-alcoholic fatty liver disease associated hepatocellular carcinoma. J Hepatol. 2014;61:75–81. doi: 10.1016/j.jhep.2014.02.030. https://pubmed.ncbi.nlm.nih.gov/24607626/ [Internet]. Elsevier [cited 2021 Jun 2] Available from: [DOI] [PubMed] [Google Scholar]
- 13.Trépo E., Valenti L. Update on NAFLD genetics: from new variants to the clinic. J. Hepatol. Elsevier B.V. 2020:1196–1209. doi: 10.1016/j.jhep.2020.02.020. https://pubmed.ncbi.nlm.nih.gov/32145256/ [Internet] [cited 2021 Jun 2] Available from: [DOI] [PubMed] [Google Scholar]
- 14.Romero F.A., Jones C.T., Xu Y., Fenaux M., Halcomb R.L. The race to bash NASH: emerging targets and drug development in a complex liver disease. J Med Chem. 2020;68:5031–5073. doi: 10.1021/acs.jmedchem.9b01701. http://www.ncbi.nlm.nih.gov/pubmed/31930920 [Internet]. American Chemical Society (ACS) [cited 2020 May 6]; Available from: [DOI] [PubMed] [Google Scholar]
- 15.Ratziu V., Friedman S.L. Why do so many NASH trials fail? Gastroenterology. 2020 doi: 10.1053/j.gastro.2020.05.046. https://pubmed.ncbi.nlm.nih.gov/32439497/ [Internet]. Elsevier BV; [cited 2021 Jun 2]; Available from: S00016-5085 (20) 30680-6. [DOI] [PubMed] [Google Scholar]
- 16.Dufour J.F., Caussy C., Loomba R. Combination therapy for non-alcoholic steatohepatitis: rationale, opportunities and challenges. Gut. 2020;69:1877–1884. doi: 10.1136/gutjnl-2019-319104. https://pubmed.ncbi.nlm.nih.gov/32381514/ [Internet]. BMJ Publishing Group [cited 2021 Jun 2] Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Oseini A.M., Cole B.K., Issa D., Feaver R.E., Sanyal A.J. Translating scientific discovery: the need for preclinical models of nonalcoholic steatohepatitis. Hepatol. Int. Springer. 2018:6–16. doi: 10.1007/s12072-017-9838-6. https://pubmed.ncbi.nlm.nih.gov/29299759/ [Internet] [cited 2021 Jun 2] Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Santhekadur P.K., Kumar D.P., Sanyal A.J. Preclinical models of non-alcoholic fatty liver disease. J. Hepatol. Elsevier B.V. 2018:230–237. doi: 10.1016/j.jhep.2017.10.031. https://pubmed.ncbi.nlm.nih.gov/29128391/ [Internet] [cited 2021 May 23] Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu Y., Gu H., Wang H., et al. 2017. Hepatic Steatosis and Fibrosis in Obese, Dysmetabolic and Diabetic Nonhuman Primates Quantified by Noninvasive Echography. [Google Scholar]
- 20.Lee L., Alloosh M., Saxena R., et al. Nutritional model of steatohepatitis and metabolic syndrome in the Ossabaw miniature swine. Hepatology. 2009;50:56–67. doi: 10.1002/hep.22904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pedersen H.D., Galsgaard E.D., Christoffersen B., et al. NASH-inducing diets in göttingen minipigs. J Clin Exp Hepatol. 2020;10:211–221. doi: 10.1016/j.jceh.2019.09.004. https://pubmed.ncbi.nlm.nih.gov/32405177/ [Internet]. Elsevier B.V. [cited 2021 May 23] Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Younossi Z.M., Ratziu V., Loomba R., et al. Obeticholic acid for the treatment of non-alcoholic steatohepatitis: interim analysis from a multicentre, randomised, placebo-controlled phase 3 trial. Lancet. 2019;394:2184–2196. doi: 10.1016/S0140-6736(19)33041-7. Lancet Publishing Group. [DOI] [PubMed] [Google Scholar]
- 23.Wang M.N., Yu H.T., Li Y.Q., et al. Bioequivalence and pharmacokinetic profiles of generic and branded obeticholic acid in healthy Chinese subjects under fasting and fed conditions. Drug Des Dev Ther. 2021;15:185–193. doi: 10.2147/DDDT.S289016. Dove Medical Press Ltd. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Eslam M., Sanyal A.J., George J., et al. MAFLD: a consensus-driven proposed nomenclature for metabolic associated fatty liver disease. Gastroenterology. 2020;158:1999–2014. doi: 10.1053/j.gastro.2019.11.312. https://pubmed.ncbi.nlm.nih.gov/32044314/ [Internet]. W.B. Saunders [cited 2021 May 23] e1. Available from: [DOI] [PubMed] [Google Scholar]
- 25.Schumacher-Petersen C., Christoffersen B.O., Kirk R.K., et al. Experimental non-alcoholic steatohepatitis in Göttingen Minipigs: consequences of high fat-fructose-cholesterol diet and diabetes. J Transl Med. 2019;17 doi: 10.1186/s12967-019-1854-y. BioMed Central Ltd. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liang T., Alloosh M., Bell L.N., et al. Liver injury and fibrosis induced by dietary challenge in the ossabaw miniature swine. PLoS One. 2015;10 doi: 10.1371/journal.pone.0124173. https://pubmed.ncbi.nlm.nih.gov/25978364/ [Internet]. Public Library of Science [cited 2021 May 23] Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Neuschwander-Tetri B.A., Loomba R., Sanyal A.J., et al. Farnesoid X nuclear receptor ligand obeticholic acid for non-cirrhotic, non-alcoholic steatohepatitis (FLINT): a multicentre, randomised, placebo-controlled trial. Lancet. 2015;385:956–965. doi: 10.1016/S0140-6736(14)61933-4. https://pubmed.ncbi.nlm.nih.gov/25468160/ [Internet]. Lancet Publishing Group [cited 2021 May 23]Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Roth J.D., Feigh M., Veidal S.S., et al. INT-767 improves histopathological features in a dietinduced ob/ob mouse model of biopsy-confirmed nonalcoholic steatohepatitis. World J Gastroenterol. 2018;24:195–210. doi: 10.3748/wjg.v24.i2.195. https://pubmed.ncbi.nlm.nih.gov/29375205/ [Internet]. Baishideng Publishing Group Co [cited 2021 May 25]Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.van den Hoek A.M., Verschuren L., Worms N., et al. A translational mouse model for NASH with advanced fibrosis and atherosclerosis expressing key pathways of human pathology. Cells. 2020;9 doi: 10.3390/cells9092014. https://pubmed.ncbi.nlm.nih.gov/32883049/ [Internet]. NLM (Medline)[cited 2021 May 25]Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Briand F., Brousseau E., Quinsat M., Burcelin R., Sulpice T. Obeticholic acid raises LDL-cholesterol and reduces HDL-cholesterol in the Diet-Induced NASH (DIN) hamster model. Eur J Pharmacol. 2018;818:449–456. doi: 10.1016/j.ejphar.2017.11.021. https://pubmed.ncbi.nlm.nih.gov/29155143/ [Internet]. Elsevier B.V. [cited 2021 May 25] Available from: [DOI] [PubMed] [Google Scholar]
- 31.Olsen AK, Bladbjerg EM, Marckmann P, Larsen LF, Hansen AK. The Gö Ttingen Minipig as a Model for Postprandial Hyperlipidaemia in Man: Experimental Observations. [DOI] [PubMed]
- 32.Ludvigsen T.P., Kirk R.K., Christoffersen B.Ø., et al. Göttingen minipig model of diet-induced atherosclerosis: influence of mild streptozotocin-induced diabetes on lesion severity and markers of inflammation evaluated in obese, obese and diabetic, and lean control animals. J Transl Med. 2015;13:312. doi: 10.1186/s12967-015-0670-2. http://www.translational-medicine.com/content/13/1/312 [Internet]. BioMed Central Ltd. [cited 2021 May 25] Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Govaere O., Cockell S., Tiniakos D., et al. Transcriptomic profiling across the nonalcoholic fatty liver disease spectrum reveals gene signatures for steatohepatitis and fibrosis. Sci Transl Med. 2020;12 doi: 10.1126/scitranslmed.aba4448. American Association for the Advancement of Science. [DOI] [PubMed] [Google Scholar]
- 34.Luo Y., Wadhawan S., Greenfield A., et al. SOMAscan proteomics identifies serum biomarkers associated with liver fibrosis in patients with NASH. Hepatol Commun. 2021;5:760–773. doi: 10.1002/hep4.1670. [Internet]. Wiley [cited 2021 May 25] Available from:/pmc/articles/PMC8122380/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Régnier M., Polizzi A., Smati S., et al. Hepatocyte-specific deletion of Pparα promotes NAFLD in the context of obesity. Sci Rep. 2020;10 doi: 10.1038/s41598-020-63579-3. [Internet]. Nature Research [cited 2021 May 25] Available from:/pmc/articles/PMC7162950/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Coutts A.W., Webster D.A., Chen J., et al. A PNPLA3 I148M gene-edited Ossabaw swine model of Nonalcoholic steatohepatitis (NASH) Faseb J. 2020;34:1. https://faseb.onlinelibrary.wiley.com/doi/full/10.1096/fasebj.2020.34.s1.07238 [Internet]. Wiley [cited 2021 May 25] 1. Available from: [Google Scholar]
- 37.Anstee Q.M., Mantovani A., Tilg H., Targher G. Risk of cardiomyopathy and cardiac arrhythmias in patients with nonalcoholic fatty liver disease. Nat. Rev. Gastroenterol. Hepatol. Nature Publishing Group. 2018:425–439. doi: 10.1038/s41575-018-0010-0. https://pubmed.ncbi.nlm.nih.gov/29713021/ [Internet] [cited 2021 May 25]Available from: [DOI] [PubMed] [Google Scholar]
- 38.Bissig-Choisat B., Alves-Bezerra M., Zorman B., et al. A human liver chimeric mouse model for non-alcoholic fatty liver disease. JHEP Reports. 2021;3:100281. doi: 10.1016/j.jhepr.2021.100281. Elsevier BV. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hansen H.H., Ægidius H.M., Oró D., et al. Human translatability of the GAN diet-induced obese mouse model of non-alcoholic steatohepatitis. BMC Gastroenterol. 2020;20 doi: 10.1186/s12876-020-01356-2. https://pubmed.ncbi.nlm.nih.gov/32631250/ [Internet]. BioMed Central [cited 2021 May 23] Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tsuchida T., Lee Y.A., Fujiwara N., et al. A simple diet- and chemical-induced murine NASH model with rapid progression of steatohepatitis, fibrosis and liver cancer. J Hepatol. 2018;69:385–395. doi: 10.1016/j.jhep.2018.03.011. https://pubmed.ncbi.nlm.nih.gov/29572095/ [Internet]. Elsevier B.V. [cited 2021 May 23] Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Asgharpour A., Cazanave S.C., Pacana T., et al. A diet-induced animal model of non-alcoholic fatty liver disease and hepatocellular cancer. J Hepatol. 2016;65:579–588. doi: 10.1016/j.jhep.2016.05.005. Elsevier B.V. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kleiner D.E., Brunt E.M., Van Natta M., et al. Design and validation of a histological scoring system for nonalcoholic fatty liver disease. Hepatology. 2005;41:1313–1321. doi: 10.1002/hep.20701. https://pubmed.ncbi.nlm.nih.gov/15915461/ [Internet]. Hepatology [cited 2021 May 25] Available from: [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.