Abstract
Many recent Asian clinical Vibrio cholerae E1 Tor O1 and O139 isolates are resistant to the antibiotics sulfamethoxazole (Su), trimethoprim (Tm), chloramphenicol (Cm), and streptomycin (Sm). The corresponding resistance genes are located on large conjugative elements (SXT constins) that are integrated into prfC on the V. cholerae chromosome. We determined the DNA sequences of the antibiotic resistance genes in the SXT constin in MO10, an O139 isolate. In SXTMO10, these genes are clustered within a composite transposon-like structure found near the element's 5′ end. The genes conferring resistance to Cm (floR), Su (sulII), and Sm (strA and strB) correspond to previously described genes, whereas the gene conferring resistance to Tm, designated dfr18, is novel. In some other O139 isolates the antibiotic resistance gene cluster was found to be deleted from the SXT-related constin. The El Tor O1 SXT constin, SXTET, does not contain the same resistance genes as SXTMO10. In this constin, the Tm resistance determinant was located nearly 70 kbp away from the other resistance genes and found in a novel type of integron that constitutes a fourth class of resistance integrons. These studies indicate that there is considerable flux in the antibiotic resistance genes found in the SXT family of constins and point to a model for the evolution of these related mobile elements.
The intercellular spread of the genetic determinants of resistance to antimicrobial agents is facilitated by mobile genetic elements, such as conjugative plasmids and conjugative transposons. The antibiotic resistance genes in these elements are often located within transposons and/or integrons, elements that facilitate the intracellular movement of genes. Two types of transposons have been found to contain resistance genes. Class I transposons, also known as composite transposons, consist of two insertion sequence (IS) elements that flank additional DNA sequences, such as resistance genes. Class II transposons do not contain recognizable IS elements; instead, the genetic information for their transposition and other phenotypes (including antibiotic resistances) is bordered by 35- to 110-bp inverted repeats (reviewed in reference 10). Integrons also play a major role in the spread of antibiotic resistance genes in gram-negative bacteria (32). Integrons are gene-capturing systems that incorporate gene cassettes and convert them to functional genes (31, 32). Integrons characteristically encode an integrase (intI) that mediates recombination between a sequence in the gene cassette (attC) and an integron-associated sequence (attI). This results in integration of the cassette downstream of a resident promoter to permit expression of the encoded protein. While integrons often are found in plasmids and usually contain antibiotic resistance genes, they can also be located on the chromosome and can contain genes that do not specify resistance to antibiotics (4, 26). To date, three classes of resistance integrons have been described based on similarities in the integrase sequences. Class I integrons usually contain the gene sulI, encoding sulfamethoxazole resistance, at their 3′ end (32). Recently, a new type of integron, collectively called chromosomal superintegrons, has been found in the chromosomes of several species belonging to the gamma proteobacteria, including Vibrio cholerae (18, 26, 34).
V. cholerae is the causative agent of the severe and sometimes lethal diarrheal disease cholera. While the genetic bases of resistance to antibiotics in V. cholerae have not been extensively characterized, antibiotic resistance determinants are usually found on plasmids in this organism (13, 17, 40). Historically, only the O1 serogroup of V. cholerae has been associated with epidemic cholera. However, in late 1992 in India and Bangladesh, a novel serogroup designated V. cholerae O139 emerged and gave rise to major cholera outbreaks. Initially, V. cholerae O139 replaced V. cholerae El Tor O1 as the predominant cause of cholera on the Indian subcontinent (5). Microbiologic and genetic characterization of V. cholerae O139 revealed that this serogroup arose from V. cholerae O1 El Tor by horizontal gene transfer and substitution of the genes encoding the O139 serogroup antigen for the genes encoding the O1 serogroup antigen (3, 9, 38, 42). Besides the novel serogroup antigen, the initial O139 isolates could be distinguished from the O1 strains they replaced by characteristic resistances to the antibiotics sulfomethoxazole (Su), trimethoprim (Tm), chloramphenicol (Cm), and low levels of streptomycin (Sm). In MO10, a 1992 clinical O139 isolate, the genes encoding these resistances were found to be located on a novel transmissible genetic element designated the SXT element (referred to here as SXTMO10) (44).
Though it is self-transmissible, an autonomously replicating extrachromosomal form of SXTMO10 has not been isolated; instead, this ∼100-kbp element is always integrated into the 5′ end of the chromosomal gene prfC. SXTMO10 encodes an integrase related to the λ family of site-specific recombinases, and we have shown that the integrase mediates the element's integration and its chromosomal excision, which generates a circular episome (21). This circular but apparently nonreplicating form of the element is believed to be a requisite intermediate for its conjugative transfer between V. cholerae strains, as well as between other gram-negative bacteria. We proposed a new term, constin, an acronym for the element's properties (conjugative, self-transmissible, and integrating) to describe SXTMO10 and other elements with similar features.
After the extensive cholera outbreaks caused by V. cholerae O139 strains, El Tor O1 V. cholerae strains reemerged in 1994 as the predominant cause of cholera on the Indian subcontinent. In contrast to the El Tor O1 strains before the O139 outbreak, these reemerged El Tor strains, like the initial O139 isolates, were resistant to Su, Tm, Cm, and Sm (48). The corresponding resistance genes were found to be located in a constin (designated here SXTET) that is closely related but not identical to SXTMO10 (21, 44). Variation is also evident in more recent O139 isolates from India, as these are generally no longer resistant to Su and Tm (28). However, molecular analyses have revealed the presence of an SXTMO10-like element integrated into prfC in these strains, indicating that they still harbor constins related to SXTMO10 (21).
SXT-like elements are not unique to V. cholerae O139. For example, the IncJ element R391 that mediates kanamycin (Kn) and mercury resistance, originally derived from a South African Providencia retgerii isolate (8), is functionally and genetically related to SXTMO10 (20). Analysis of these two elements suggested that they consist of similar basic building blocks—modules encoding integration and transfer functions—to which have been added genes encoding defining features, such as antibiotic resistance genes (20).
In this study, we determined the sequence and organization of the antibiotic resistance genes in SXTMO10 and compared them to those of other SXT constins. The SXTMO10 resistance genes are embedded in a ∼17.2-kbp composite transposon-like element that interrupts the SXT-encoded rumAB operon. A deletion event, likely mediated by recombination between duplicated sequences in this region, accounts for the Su and Tm sensitivity of recent O139 isolates. In SXTET, unlike in SXTMO10, resistance to Tm is encoded outside the cluster of resistance genes; instead, the Tm resistance determinant is found in a novel class of integrons located far away from the remainder of the antibiotic resistance genes within SXTET. By comparison, the Kn resistance gene in R391 is found to be part of a transposon containing IS26 elements that is located ∼3 kbp 5′ to the R391 rumAB operon. Overall, these studies indicate that the antibiotic resistance determinants on constins are often part of dynamic genetic structures that allow relatively rapid alteration of the properties encoded by a constin.
MATERIALS AND METHODS
Bacterial strains and media.
The bacterial strains used in this study are described in Table 1. Bacterial strains were routinely grown in Luria-Bertani (LB) broth (2) at 37°C and stored at −70°C in LB broth containing 20% (vol/vol) glycerol. Antibiotics were used at the following concentrations: ampicillin (Ap), 100 mg liter−1; Kn, 50 mg liter−1; Su, 160 mg liter−1; Tm, 32 or 250 mg liter−1; tetracycline, 10 mg liter−1; and Cm, 2 mg liter−1 for V. cholerae and 20 mg liter−1 for Escherichia coli.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant genotype or phenotype | Reference or source |
|---|---|---|
| V. cholerae O139 | ||
| MO10 | Toxigenic 1992 clinical isolate from India, SXTMO10+, Sur Tmr Cmr Smr | 41 |
| AS207 | Toxigenic 1997 isolate from Calcutta, Sur TmS Cmr Smr | 36 |
| E712 | Nontoxigenic 1994 isolate from Sri Lanka, Sur Tmr Cmr Smr | 30 |
| 2055 | 1998 clinical isolate from Bangladesh, SuS TmS CmS SmS | 21 |
| HKO139-SXTS | Clinical isolate from Hong Kong, Sus TmS CmS Sms | 47 |
| V. cholerae O1 | ||
| CO943 | El Tor 1994 clinical isolate from India, Sur Tmr Cmr Smr | 44 |
| 1811/98 | El Tor 1998 clinical isolate from Bangladesh, Sur Tmr Cmr Smr | 21 |
| C10488 | El Tor 1999 clinical isolate from Bangladesh, Sur Tmr Cmr Smr | This study |
| E. coli K-12 | ||
| TOP10 | F′ mcrA Δ(mrr-hsdRMS-mcrBC) φ80lacZΔM15 ΔlacX74 recA1 endA1 | Invitrogen |
| DH5α | F−thi-1 ΔlacU196 φ80lacZΔM15 hsdR17 recA1 endA1 | |
| Plasmids | ||
| pSXT1 | pSuperCos1 containing a part of the SXT element encoding Cmr Sur Tmr Smr | 44 |
| pWKS30 | Apr pSC101 derivative | 45 |
| pATMP1 | pWKS30 + 14 kbp from SXTMO10, Cmr Tmr Smr | This study |
| pSUL1 | pWKS30 + 1.7 kbp from SXTMO10, Sur | This study |
| pYL1 | pWKS30 + 2.77 kbp from SXTET, dfrA1+ | This study |
| pYL8 | pWKS30 + 3.8 kbp from SXTET, dfrA1+ | This study |
| pGB2 | Spcr pSC101 derivative | 6 |
| pRLH422 | pGB2 + 11 kbp from R391, Knr | 19, this study |
Molecular biology procedures.
Plasmid DNA was prepared using the Qiaprep Spin Miniprep Kit (Qiagen, Valencia, Calif.), and chromosomal DNA was isolated with the Genome DNA Kit (Bio 101, Vista, Calif.) as described by the manufacturer. Recombinant DNA manipulations were carried out with standard procedures (2). Automated DNA sequencing was carried out as described previously (43) at the Tufts Medical School DNA Sequencing Core Facility. Computer analysis of DNA sequences was performed with the MacVector and AssemblyLIGN programs (Oxford Molecular Group, Campbell, Calif.), the Vector NTI program (InforMax, North Bethesda, Md.), and the BLAST programs (1) available on the web site of the National Center for Biotechnology Information (Bethesda, Md.). Protein sequences were analyzed for the presence of motifs with the SMART program (http://smart.embl-heidelberg.de).
Cloning and sequencing of antibiotic resistance genes of V. cholerae O139 MO10.
The previously described cosmid pSXT1 contains a ∼40-kbp insert of SXTMO10 DNA and mediates resistance to Su, Cm, Tm, and Sm (44). A library of pSXT1 EcoRI fragments was constructed in pWKS30 (45). Subsequently, plasmids mediating resistance to Su, Cm, and Tm were isolated by plating the library on L-agar plates containing the respective antibiotics. One such plasmid, pATMP1, contained a 14-kbp insert that conferred resistance to Cm and Tm; another, pSUL1, contained a 1.7-kbp insert that conferred resistance to Su. Overlapping BamHI, PvuII, and PstI fragments of pATMP1 were subcloned into pUC18, and the DNA sequences of the inserts were determined by primer walking. Additional primer walking using pSXT1 as a template was carried out to determine the sequences flanking the inserts in pATMP1 and pSUL1 on SXTMO10.
Cloning and sequencing of dfrA1 from V. cholerae O1 C10488.
Chromosomal DNA from C10488 was partially digested with Sau3AI, and then fragments of ∼2 to 5 kbp were isolated and ligated with BamHI-digested pWKS30. The ligation mixture was electroporated into E. coli DH5α and plated on L-agar plates containing Tm (250 mg liter−1) and Ap. Two plasmids mediating Tm resistance, pYL1 and pYL8, were isolated. The inserts in these two plasmids (2.77 and 3.8 kbp, respectively) were sequenced and found to overlap.
Cloning and sequencing of aphAI from R391.
As described previously (19, 20), EcoRI fragments of R391 mediating Kn resistance were subcloned into pGB2 (6). One plasmid, called pRLH422, contained a single ∼11-kbp EcoRI fragment and was used for our present studies. The DNA sequence of the ∼11-kbp EcoRI fragment was obtained by nebulizing 20 μg of pRLH422, so as to randomly shear the DNA into fragments of 1 to 2 kbp. These fragments were blunt ended and subsequently cloned into SmaI-digested pUC19; 288 clones were picked and arrayed into three 96-well plates. The DNA sequence of the inserts was obtained using an Applied Biosystems ABI377 sequencer using standard sequencing protocols and primers that were designed to extend from both the 5′ and 3′ ends of the vector into the insert. The sequence data obtained were aligned into a contiguous sequence using the PhredPhrap program, and the correct alignment of the compiled sequence was confirmed by restriction mapping based on the compiled sequence.
PCR amplification.
The primers used in this study are listed in Table 2 and were synthesized by the Tufts Medical School DNA Sequencing Core facility. PCRs were performed using standard reaction conditions in total volumes of 20 μl.
TABLE 2.
DNA sequences of the oligonucleotides used in this study
| Primer | Locus (direction)a | Nucleotide sequence (5′ to 3′) |
|---|---|---|
| INT1 | int (+) | GCTGGATAGGTTAAGGGCGG |
| INT2 | int (−) | CTCTATGGGCACTGTCCACATTG |
| FLOR-F | floR (+) | TTATCTCCCTGTCGTTCCAGCG |
| FLOR-B | floR (−) | TCGTCGAACTCTGCCAAATG |
| SUL2-F | sulII (+) | AGGGGGCAGATGTGATCGAC |
| SUL2-B | sulII (−) | TGTGCGGATGAAGTCAGCTCC |
| STRA-F | strA (+) | TTGATGTGGTGTCCCGCAATGC |
| STRA-B | strA (−) | CCAATCGCAGATAGAAGGCAA |
| TMP-F | dfr18 (+) | TGGGTAAGACACTCGTCATGGG |
| TMP-B | dfr18 (−) | ACTGCCGTTTTCGATAATGTGG |
| TMP3 | orfA (+) | CATGCTGTTTCTCGACGGTG |
| TMP4 | orf6 (−) | GATCCGATCTGTTTGTTCAG |
| LEND4 | orf1 (+) | CCTTTGGTTACACATTCGC |
| LEFTF3 | rum′B (−) | GGTGCCATCTCCTCCAAAGTGC |
| RUMA | Intergenic | CGAGCAATCCCCACATCAAG |
| YL6 | orf73 (+) | TGTGGAACGGCTTTCTGACG |
| YL3 | orfC5A (+) | CGTTGGTTTGGGGTAACACC |
+, oligonucleotides corresponding to the coding strand (forward primer), −, oligonucleotides corresponding to the noncoding strand (backward primer).
Nucleotide sequence accession numbers.
The sequence of the antibiotic resistance gene cluster of SXTMO10 has been deposited in GenBank under accession no. AY034138. The sequence of the integron of SXTET has been deposited under accession no. AY035340. The sequence of the Kn resistance transposon found in R391 has been deposited under accession no. AF375956.
RESULTS AND DISCUSSION
Arrangement of antibiotic resistance genes in V. cholerae O139 strain MO10.
We previously constructed a cosmid library with chromosomal DNA derived from O139 strain MO10, a 1992 clinical isolate from Madras, India (44). pSXT1, one of the cosmids from this library, was found to confer resistance to Su, Cm, Tm, and Sm, indicating that the genes mediating these resistances were not randomly distributed in SXTMO10. Isolation of subclones of the ∼40-kbp insert from pSXT1 (as described in Materials and Methods) revealed that these resistance genes were in fact clustered together in a region of about 9.4 kbp (Fig. 1). Detailed analysis of the DNA sequence of this region along with that of flanking sequences resulted in two major findings. First, the antibiotic resistance genes appear to be part of a large transposon-like element. This element is itself a mosaic composed of other transposon-like elements and DNA sequences found in other mobile elements. Second, SXTMO10 contains previously identified genes (floR, sulII, strA, and strB) encoding resistance to Cm, Su, and Sm, respectively, and a novel gene encoding resistance to Tm.
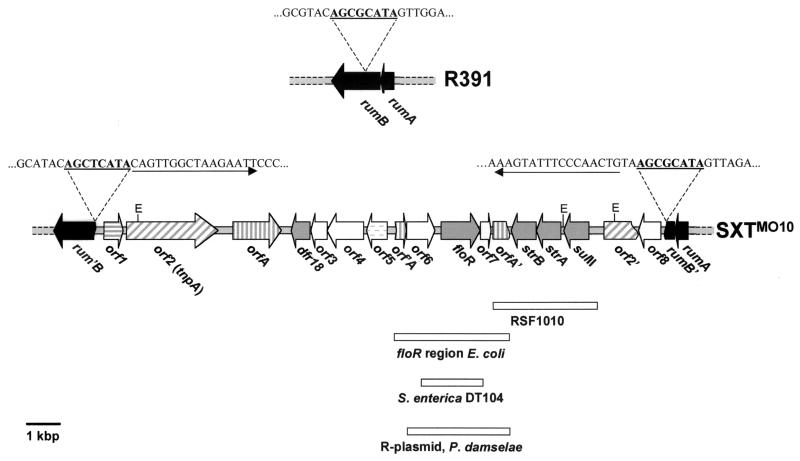
FIG. 1.
Organization of the antibiotic resistance gene cluster in SXTMO10. The SXTMO10 genes mediating resistance to antibiotics, dfr18, floR, strA, strB, and sulII, are represented by gray arrows, and genes with similarity to transposases (orf1, orf2, and orfA) are represented by hatched arrows. Genes encoding hypothetical proteins similar to known proteins are shown as horizontal hatches, and genes encoding hypothetical proteins dissimilar to known proteins are shown in white. Genes rumA and rumB are in black. The rumAB operon of R391 is presented above the SXTMO10 antibiotic resistance gene region. The sequence in rumB which is repeated in SXTMO10 is in bold and underlined; the flanking imperfect repeat (IR) sequences in SXTMO10 are marked by arrows. Also indicated are the EcoRI sites (E) used for construction of pATMP1 and pSUL1. Regions of nucleotide sequence identity to other published nucleotide sequences are represented by boxes.
SXTMO10 appears to have acquired its antibiotic resistance genes and some adjacent sequences via a transposition event(s). This event introduced a 17.2-kbp region containing all five resistance genes into rumB, the second gene of the rumAB operon. This is likely to have been a multistep process, as outlined below. Consistent with this hypothesis, the 17.2-kbp sequence is flanked both by an 8-bp direct repeat (corresponding to amino acids [aa] 76 to 78 of rumB) and by 16-bp imperfect inverted repeats, structures often found at the boundaries of transposons.
A role for transposition is also suggested by the presence of open reading frames (ORFs) with similarity to previous described transposases at the left end of these 17.2 kbp (Fig. 1 and Table 3). The deduced amino acid sequence of orf1 has 39% similarity to the C terminus of a transposase found in Pseudomonas putida (Table 3), and the deduced amino acid sequence of orf2 has 29% identity and 47% similarity to a transposase found in Tn5501 and Tn5502, two cryptic transposons located in P. putida (25). The 5′ end of orf2 is repeated downstream of sulII. However, despite its transposon-like features, the 17.2-kbp sequence is apparently not (or no longer) an autonomously mobile genetic element; all our attempts to mobilize the resistance genes independent of the remainder of SXTMO10 have failed.
TABLE 3.
Gene products with sequence similarity to the antibiotic resistance gene cluster in SXTMO10
| Coding regiona | Gene name | Length (aa) | Closest similarity | Accession no. | % Identity/rangeb |
|---|---|---|---|---|---|
| 1–1044 (−) | rum′B | RumB R391 (C terminus) | 862633 | ||
| 1457–2047 | orf1 | 197 | Transposase, P. putida | 4754812 | 39/188 of 530 |
| 2164–5061 | orf2 (tnpA) | 966 | Transposase, P. putida | 7465523 | 29/956 |
| 5559–7049 | orfA | 497 | Putative transposase, E. coli | 10312101 | 99/496 |
| 7416–7967 (−) | dfr18 | 184 | Dihydrofolate reductase type VIII | 2833495 | 37/157 |
| 7984–8526 (−) | orf3 | 181 | No homology | NAc | NA |
| 8529–9692 (−) | orf4 | 388 | No homology | NA | NA |
| 9710–10435 (−) | orf5 | 294 | Deoxycytidine triphosphate deaminase, P. aeruginosa | 9949625 | 44/208 |
| 10695–11024 | orf′A | 110 | Putative transposase, E. coli (3′ end, aa 388–497) | 10312101 | |
| 11058–11939 | orf6 | 294 | No homology | NA | NA |
| 12159–13370 | floR | 404 | FloR (florfenicol exporter), E. coli | 10312100 | 99/404 |
| 13401–13703 | orf7 | 101 | Putative transcriptional regulator (LysR family), P. aeruginosa | 11352177 | 51/74 |
| 13818–14354 | orfA′ | 179 | Putative transposase, E. coli (5′ end, aa 1–135) | 10312101 | |
| 14332–15165 (−) | strB | 278 | Aminoglycoside phosphotransferase | 420965 | 100/278 |
| 15168–15968 (−) | strA | 267 | Aminoglycoside phosphotransferase | 420964 | 100/267 |
| 16032–16844 (−) | sulII | 271 | Dihydropteroate synthase transposase, P. putida (5′ end) | 1075456 | 100/271 |
| 17315–18674 | orf2′ (tnpA′) | 7465523 | 24 | ||
| 18399–19118 (−) | orfB | 240 | Mutl, V. cholerae | 127554 | 51/173 |
| (217–345 of 563) | |||||
| 19308–19540 (−) | rumB′ | RumB R391 (N terminus) | 862633 | ||
| 19551–19997 (−) | rumA | 149 | RumA R391 | 862632 | 98/149 |
Genes encoded on the minus strand are indicated with (−).
The number of amino acids in a contiguous stretch from which the identity was calculated is shown. The length of the similar protein is also presented.
NA, not applicable.
The Tm resistance gene of SXTMO10 was mapped to subclones of pSXT1 that included a 551-bp ORF. As this ORF's deduced amino acid sequence had 37% identity and 52% similarity (Table 3) to a type VIII dihydrofolate reductase found in some E. coli strains (39), it was named dfr18, for a new gene encoding a Tm-resistant dihydrofolate reductase. dfr18 is preceded by three ORFs, orf3, orf4, and orf5, with the same orientation as dfr18. The deduced amino acid sequences of orf3 and orf4 do not have similarities to any known proteins, but the deduced amino acid sequence of orf5 has 44% identity and 60% similarity (Table 3) to a chromosomal Pseudomonas aeruginosa deoxycytidine triphosphate deaminase (37). Whether orf5 encodes a functional deaminase remains to be studied. These four genes are bracketed by the previously described orfA (7). A complete copy of orfA lies downstream of dfr18, while a 5′-truncated copy of orfA lies upstream of orf5 (Fig. 1 and Table 3). An identical full-length orfA was found by Cloeckaert et al. in a plasmid from an E. coli isolate (7). The predicted OrfA amino acid sequence has been noted to have some similarity to a putative transposase from Pseudomonas pseudoalcaligenes (12). It seems likely that orfA plays some role in promoting the acquisition and loss of antibiotic resistance genes, since orfA or fragments of orfA are closely linked to antibiotic resistance genes in several instances (7, 22, 35). The molecular mechanism(s) by which orfA acts to promote gain or loss of genes remains to be explored.
In two prior cases, orfA or orfA fragments have been found associated with floR (7, 22). This is the case in SXTMO10 as well (Fig. 1). In SXTMO10, floR is found close to the 3′ end of the 5′-truncated orfA, preceded by a putative ORF (orf6) of unknown function. FloR is thought to be an export protein which mediates resistance to Cm and florfenicol. This gene has been found in plasmids derived from E. coli isolates from cattle (7), in the chromosome of the multidrug-resistant Salmonella enterica serovar Typhimurium phagetype DT104 (4) and in an R-plasmid derived from the fish pathogen Photobacterium damselae subsp. piscida (22). As expected, in-frame deletion of floR from SXTMO10 resulted in cells that were no longer resistant to Cm (J. Beaber and B. Hochhut, unpublished observations), confirming that floR is required for resistance to Cm. In SXTMO10, floR is followed by a short putative ORF (orf7) that includes a region with similarity to the helix-turn-helix (HTH) motif of LysR family transcriptional regulators, and another incomplete copy of orfA that is deleted in its 3′ end. The two incomplete copies of orfA that bracket floR together do not constitute a full-length orfA. Comparative DNA sequence analysis revealed extensive nucleotide identity to the floR loci in E. coli isolates and P. damselae (Fig. 1). The genes strA, strB, and sulII, which follow orfA′, are identical to previously described resistance genes and are found on several plasmids, including RSF1010 (35). They encode a sulfonamide-resistant dihydropterate synthase (sulII) and an aminoglycoside phosphotransferase (strAB).
Distribution of SXTMO10 antibiotic resistance genes in related SXT elements from V. cholerae O1 and O139 strains.
Since the discovery of SXTMO10 in isolates from the initial O139 outbreak in 1992, closely related constins have been detected in many V. cholerae isolates of both the O1 and O139 serogroups. These related constins, like SXTMO10, are integrated into prfC (21); however, these elements do not confer the same antibiotic resistances as SXTMO10. For example, many recent O139 clinical isolates from Asia were found to be sensitive to Su and Tm (28). We analyzed the genetic basis for this sensitivity in two O139 clinical isolates, strain 2055 from Bangladesh and strain HKO139-SXTs from Hong Kong. PCR assays designed for amplification of internal sequences of dfr18, floR, strA, and sulII from these strains failed, whereas a PCR amplification of intSXT, a signature sequence of an SXT-related constin, was successful (Table 4). PCR assays utilizing primers that flank the antibiotic resistance genes in SXTMO10 facilitated the mapping of the borders of the DNA missing in strains 2055 and HKO139-SXTS. Using chromosomal DNA from either strain as the template, with primer pair LEND4 and RUMA, we obtained a product of ∼3.3 kbp, and with primer pair LEFTF3 and RUMA, we amplified a 4-kbp product (Fig. 2). In contrast, in MO10 these primer pairs flank sequences of 18.5 and 19.2 kbp.
TABLE 4.
Distribution of antibiotic resistance genes in V. cholerae isolates containing intSXTa
| Strain | Serogroup | Detectionb
|
||||
|---|---|---|---|---|---|---|
| dfr18 | dfrA1 | floR | strA | sulII | ||
| MO10 | O139 | + | − | + | + | + |
| HKO139-SXTS | O139 | − | − | − | − | − |
| 2055 | O139 | − | − | − | − | − |
| AS207 | O139 | − | − | + | + | + |
| E712 | O139 | − | + | + | + | + |
| CO943 | O1 | − | + | + | + | + |
| 1811 | O1 | − | + | + | + | + |
| C10488 | O1 | − | + | + | + | + |
All strains tested were positive for intSXT in a PCR assay.
Symbols: +, listed gene detected by a PCR assay and by a Southern hybridization experiment; −, no detection of the gene.
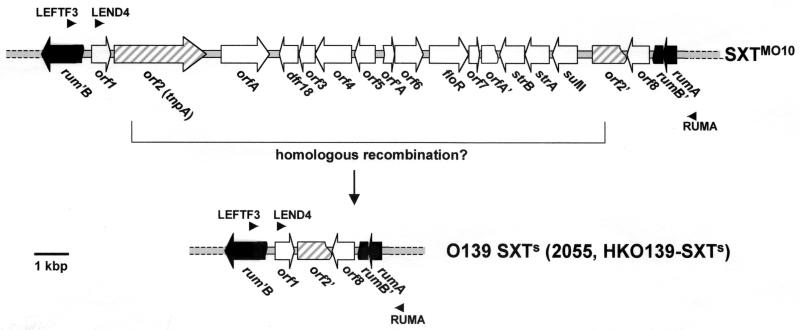
FIG. 2.
Organization of the region containing antibiotic resistance genes in SXTMO10 and in V. cholerae O139 strains sensitive to Tm, Su, and Cm. The gene order found in strains 2055 and HKO139-SXTS (bottom) is compared to that of SXTMO10 (top). Homologous recombination between the identical sequences in orf2 and orf2′ may have resulted in loss of the antibiotic resistance genes. Also shown are the primers (LEFTF3, LEND4, and RUMA) used to amplify this region in 2055 and HKO139-SXTS.
The DNA sequence of the 3.3-kbp fragment was partially determined. As in MO10, the reading frame of rumB is interrupted in these strains, but by a much smaller insert encompassing orf1, orf2′, and orf8. This genetic structure suggests that deletion mediated by homologous recombination between the two identical 5′ ends of orf2 that bracket the resistance gene cluster in SXTMO10 may have rendered these strains sensitive to antibiotics (Fig. 2). An alternative possibility is that these antibiotic-sensitive O139 strains never carried any of the resistance genes and that their constins represent a precursor of SXTMO10. Since the rum operon is interrupted in both types of elements, the latter possibility seems less likely. In either case, the lack of the ∼15.2-kbp fragment from these antibiotic-sensitive O139 strains has not rendered their SXT-like elements deficient for transfer (data not shown).
Other recent intSXT-containing O139 isolates, such as the 1996 Calcutta isolate AS207, have been found to be resistant to Cm, Su, and Sm but sensitive to Tm (21, 27). Using AS207 DNA as the template, we were able to amplify floR, strA, and sulII by PCR (Table 4). Southern hybridization indicated that the arrangement of these genes was similar in AS207 and in MO10 (data not shown). However, both a PCR assay (Table 4) and a Southern hybridization assay (not shown) indicated that AS207 lacked dfr18. The precise borders of the deletion including dfr18 in the AS207 constin are discussed below.
After the initial spread of V. cholerae O139 on the Indian subcontinent in 1993, clinical isolates of V. cholerae O1 El Tor from this region were found to be resistant to the same antibiotics, Su, Sm, Tm, and Cm, as O139 strains. We analyzed the genes encoding these resistances in three El Tor strains, CO943, 1811, and C10488, isolated in different years and from different locations on the Indian subcontinent (Table 1). As in O139 strain MO10, the resistance determinants in these strains were part of a constin designated SXTET, that is very similar but not identical to SXTMO10 (21, 44; data not shown). Using chromosomal DNA from these strains as templates for PCR, products corresponding to internal regions of floR, strA, and sulII were amplified (Table 4). Southern hybridization experiments indicated that the organization of these genes in SXTET is identical to that in SXTMO10 (data not shown). To our surprise, despite their resistance to Tm, these El Tor isolates were found by PCR (Table 4) and Southern hybridization (not shown) not to harbor dfr18.
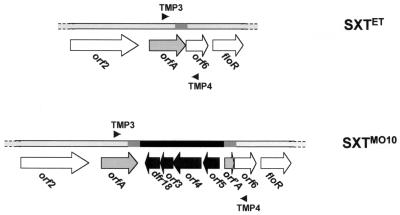
PCR primers (TMP3 and TMP4, Fig. 3) which anneal to sequences that flank dfr18 in SXTMO10 were used to define the extent of the region missing from SXTET. Using these primers and MO10 chromosomal DNA as the template, a PCR product with the expected size of 5.35 kbp was obtained, whereas with C10488 chromosomal DNA as the template, a product of 1.3 kbp was obtained. The DNA sequence of this 1.3-kbp PCR product revealed that in addition to dfr18, orf3, orf4, and orf5 were also absent in C10488 (Fig. 3). Furthermore, in C10488, a complete copy of orfA is followed by orf6 and floR, whereas in MO10, a complete copy of orfA is located next to dfr18 and only a 5′-end-truncated copy of orfA is found next to orf6 (Fig. 3). The 3.34-kbp “insert” that includes the genes dfr18, orf3, orf4, and orf5 and that distinguishes SXTMO10 from the constin present in C10488 is flanked by a 640-bp duplication (Fig. 3). This repeated DNA sequence encompasses the 3′ end of orfA and the first 205 bp of orf6. A PCR showed that the same sequences were also missing from the constins in the other two El Tor Tmr strains, CO943 and 1811/98, as well as in the constin in the Tms O139 strain AS207 discussed above. We have no direct evidence of the mechanism by which these additional genes were acquired by SXTMO10 or, alternatively, lost from the C10488 constin. However, given the presence of the duplicated 640-bp sequence, homologous recombination probably played some role in the loss or acquisition of these four genes.
FIG. 3.
SXTET lacks dfr18, orf3, orf4, and orf5. In El Tor O1 strain C10488, floR is preceded by a complete copy of orf6 and orfA (top). In contrast, in SXTMO10, there is a duplication of 640 bp (dark gray boxes) that flanks the genes dfr18, orf3, orf4, and orf5 (black). The locations of primers (TMP3 and TMP4) used for amplification of this area are also shown.
dfrA1 mediates Tm resistance in V. cholerae O1 constin.
Although the constin in strain C10488 lacked dfr18, we strongly suspected that the determinant of Tm resistance in this strain would be part of SXTET, since the Su, Sm, Cm, and Tm resistance determinants were cotransferred by C10488 (data not shown). We constructed a plasmid library with insert DNA derived from C10488 chromosomal DNA to isolate the Tm resistance determinant(s) from this strain. We identified two recombinant plasmids (pYL1 and pYL8) that allowed their host cells to grow on media containing Tm. Determination of their respective insert DNA sequences revealed that they contained overlapping inserts and that the overlap included an ORF with nucleotide sequence identity to the previously described gene dfrA1 (Fig. 4) (14). dfrA1 encodes a trimethoprim resistance dihydrofolate reductase which until now has been found exclusively as a cassette within class 1 and 2 integrons (11, 32). Instead, dfrA1 from C10488 appears to be part of a novel (class 4) type of integron; 271 bp upstream of the dfrA1 cassette was a gene of 320 codons whose deduced amino acid sequence showed similarity to the site-specific recombinases found in integrons and which has been named intI9. Its predicted product, IntI9, shows 53% identity to IntI2∗ (a 325-amino-acid protein obtained through readthrough of the stop codon at position 178 in intI2 [accession no. NP_065308]), a putative integrase of the class 2 resistance integrons. The paradigm of class 2 integrons is found on Tn7. The second closest relative of IntI9 is SpuIntIA, the Shewanella putrefaciens chromosomal integron integrase (47% identity) (34). dfrA1 and intI9 are oriented in opposite directions, an arrangement characteristic of integrons. Furthermore, the DNA sequence of the dfrA1 cassette is 99.8% identical to the dfrA1 cassette of class 1 and 2 resistance integrons.
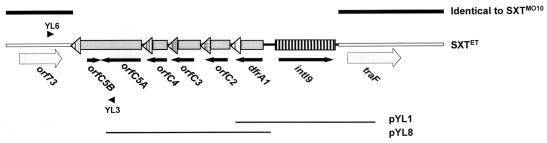
FIG. 4.
Organization of the integron in SXTET constin. The five cassettes found in the SXTET integron are shown. The attC sites are represented by triangles, and ORFs are represented below the cassettes as arrows. Also shown are the genes traF and orf73. DNA sequences identical to SXTMO10 are shown as black lines. The insert DNA in pYL1 and pYL8 is shown below. The positions of primers YL6 and YL3, used to amplify the upstream boundary of the integron insertion in SXTET, are also indicated.
The sequence downstream of the dfrA1 cassette did not show similarity to any known genes. However, analysis of this sequence revealed the presence of four putative consecutive integron cassettes (Fig. 4). Cassettes 2, 3, and 4 each carry a single ORF, while cassette 5 contains two ORFs in opposite orientation. The putative product of orfC2 (142 aa) is predicted to be located in the cytoplasmatic membrane. The deduced amino acid sequence of orfC3 (136 aa) contains a region with similarity to an Xre-type HTH motif and is predicted to the membrane associated. Finally, the product of orfC5A (233 aa) has an AraC-type HTH motif, while the putative product of orfC5B (82 aa) contains a domain conserved among bleomycin resistance proteins. Although integrons were originally described as systems to capture antibiotic resistance genes, analysis of superintegron cassettes has revealed that many of the genes contained therein are of unknown function (32, 34).
As seen for the cassettes carried in the multiresistance integrons, the attC sites carried by the SXTET integron cassettes are extremely different in length (58 to 99 bp) and sequence. Interestingly, the attC site of cassette 2 is almost identical to the attC site of the first cassette of the S. putrefaciens CIP 69.34 superintegron (accession no. AF324211) (34), while the genes carried in both cassettes are unrelated. In contrast, the attC sites of the other three cassettes do not show any significant homology (<50% identity) with the attC sites found either in previously described resistance cassettes or in any of the superintegron cassettes, including those of the V. cholerae superintegron.
In our ongoing research, we are determining the complete nucleotide sequence of SXTMO10. We took advantage of the partially completed sequence to determine where the dfrA1-containing integron is located in the C10488 constin. Downstream of intI9 in the insert of pYL1 was a region with near nucleotide sequence identity to SXTMO10 (Fig. 4). In SXTMO10, this region encodes a putative gene (traF) thought to be required for pilus assembly; it is located about 70 kbp away from the resistance gene cluster. Unlike the insert in pYL1, the sequence of the pYL8 insert did not show any similarity to the sequence of SXTMO10 (Fig. 4).
To identify the upstream boundary of the apparent insertion of the dfrA1-containing integron, we designed PCR primers to amplify the junction between this integron-like element and predicted upstream sequences in SXTMO10 (Fig. 4). With the primer pair YL6/YL3, we amplified a product of ∼1 kbp with C10488 chromosomal DNA as a template. Sequence analysis of this PCR product, combined with the sequences of the pYL1 and pYL8 inserts, revealed that relative to SXTMO10, an insert of 4.77 kbp is present in SXTET between traF and an ORF of unknown function, orf73 (Fig. 4). Examination of the borders of the integron in SXTET did not reveal sequences such as inverted or direct repeats that might suggest the mechanism by which this integron was acquired. However, we noticed that the divergence between the sequences of SXTMO10 and SXTET downstream of orf73 coincided exactly with the core site sequence in attC of cassette 5. In this region the MO10 sequence does not show any of the attC site structural characteristics apart from a conserved CGTT sequence, which is precisely located at the beginning of the identity with the SXTET sequence. Integrase-mediated recombination between attC sites and noncanonical sites, known as secondary sites of consensus GWTMW (15), has been reported several times (15, 16, 33). This suggests that this boundary of the integron likely corresponds to a recombination between the attC site of cassette 5 and the sequence AACGTTCTGC (bases corresponding to bases fitting the secondary site consensus shown above are underlined) of the SXT backbone. To our knowledge, this is the first evidence of such an event to explain the 3′ end of a cassette array in an integron. The only natural case of likely recombination between an attC site and a secondary site described so far was the integration of a single aadB cassette, not an integron, into an RSF1010 plasmid (33).
Like C10488, the other two El Tor strains we studied, CO943 and 1811, also contained a 4.77-kbp sequence inserted between orf73 and traF. Insertion of a dfrA1-containing integron into this locus was not limited to the constins found in El Tor strains. We identified a nontoxigenic O139 isolate, E712, that also contained this insertion (Table 4). In fact, like SXTET, the E712 constin also lacked the 3.34-kbp region containing dfr18, present in SXTMO10 (Table 4), suggesting that the E712 constin was very similar (or identical) to the El Tor constin.
aphAI in R391 is part of a transposon.
Although SXTMO10 and R391 are closely related constins, they encode different sets of antibiotic resistances. Cells carrying R391 are sensitive to Cm, Tm, Su, and Sm but resistant to Kn. As expected, PCR assays and Southern analyses revealed that R391 does not carry any of the resistance genes or putative transposase genes encoded in SXTMO10 (20; data not shown). Also, R391 contains an intact rumAB operon (19, 24). This operon encodes proteins that are phylogenetically related to a superfamily of novel error-prone DNA polymerases found in all three kingdoms of life (46). While R391 complements the DNA repair functions encoded by the umuDC operon in E. coli strains missing these genes (19), SXTMO10 failed to complement a ΔumuDC strain (data not shown), confirming the inactivation of rumB.
DNA sequence analysis of the ∼11-kbp EcoRI fragment carrying the R391 Kn resistance determinant revealed on ORF some ∼4 kbp from the rumABR391 locus that is identical to the previously described aphAI gene, which encodes an aminoglycoside phosphotransferase (29). Immediately 5′ and 3′ of aphAI, we identified two copies of IS26 in opposite orientations (data not shown), indicating that aphAI is part of a novel transposon. Interestingly, linkage of aphAI with IS26 is also found in the multiresistance plasmid pSP9351 from P. damselae (23); however, the organization of IS26 relative to aphAI differs between pSP9351 and R391. Taken together, our data indicate that antibiotic resistance genes can be added to SXT-like constins at several locations and via different mechanisms.
Conclusions.
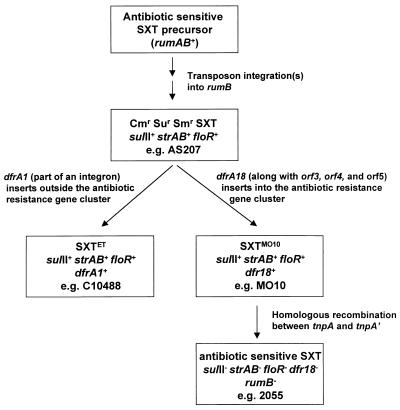
SXT-related constins constitute an important group of transmissible genetic elements that have contributed to the spread of resistance to antimicrobial agents in clinical isolates of V. cholerae from Asia. Our surveys of V. cholerae O139 and O1 clinical isolates from this region indicate that the great majority of post-1993 isolates contain an SXT-related element integrated on the large V. cholerae chromosome at prfC. Thus far, all of the elements tested are self-transmissible and encode IntSXT, the defining features of SXT-related constins. Although the genetic determinants of the transfer and integration functions of these related elements appear to be nearly identical, in the current study we found that the antibiotic resistance genes in these elements differed. In the SXT constin found in the original 1992 O139 outbreak strains (SXTMO10), as exemplified by MO10 (and found in other isolates as well), the antibiotic resistance genes were all clustered together within a ∼17-kbp composite transposon-like structure. In contrast, in the SXTET constin found in the reemerged (post-1993) El Tor O1 strains, this cluster is missing a 3.3-kbp segment that includes the novel dfr18 found in SXTMO10. Instead, SXTET contains a novel integron-like structure that includes dfrA1, located ∼70 kbp away from the other antibiotic resistance genes in this constin. Finally, R391 contains a transposon-associated Kn resistance gene located ∼3.5 kbp away from the site where the composite transposon-like element apparently inserted in SXTMO10. The differences in the antibiotic resistance genes in SXT-related constins suggest that these genes are not intrinsic features of this family of constins; they appear to have inserted themselves on these elements as a way to become transmissible through bacterial populations. Selective pressure to become and remain resistant to antibiotics does not seem to be the only explanation for the dissemination and persistence of SXT-related constins in Asian V. cholerae. This is clear from the absence of antibiotic resistance genes from the SXT-like constins found in many recent O139 isolates, such as strain 2055 analyzed in this study. The advantage(s) conferred by constins lacking resistance genes remains to be elucidated.
A plausible scheme outlining the steps in the acquisition and loss of antibiotic resistance genes in the V. cholerae derived SXT family of constins is shown in Fig. 5. First, in one or several steps, a transposon(s) that included sulII, strAB, and floR inserted into rumB, a gene that is intact in R391, an SXT-related constin. Then, the resulting Sur, Smr, and Cmr constin (such as was found in O139 strain AS207) could have become Tmr by acquiring either the novel integron containing dfrA1, to give rise to SXTET, or dfr18, orf3, orf4, and orf5, to give rise to SXTMO10. This latter event likely depended on orfA (by an unknown mechanism), since orfA is associated with antibiotic resistance genes in several instances. Subsequently, the Sur, Smr, and Cmr constin could have undergone a deletion event, likely mediated by homologous recombination, to give rise to constins that lack antibiotic resistance genes such as those found in O139 strains 2055 and HKO139-SXTS. Even though SXTMO10 was the first SXT-family constin that we identified (from a 1992 O139 isolate) and we did not detect SXTET in O1 strains until 1994, given the differences in the antibiotic resistance genes between these two constins, it seems unlikely that SXTMO10 is an immediate precursor of SXTET. Rather, SXTET, the constin found in most recent O1 isolates, seems to have arisen independently of SXTMO10. We detected an SXTET-like element in an O139 isolate (E712), indicating that SXTET is not limited to the V. cholerae O1 serogroup. Additionally, we found SXTET (or at least very similar elements) in Providencia alcalifaciens isolates from patients in Bangladesh (data not shown). This suggests a recent gene transfer between V. cholerae and P. alcalifaciens. Finally, although SXT family constins are present in virtually all clinical V. cholerae isolates from Asia, these elements are a relatively recent addition to the V. cholerae genome. They are not present in seventh-pandemic V. cholerae isolates, as exemplified by their absence from the genome of N16961, the type strain used for determination of the complete nucleotide sequence of the V. cholerae chromosomes by the Institute for Genome Research. The bacterial species that donated SXT family constins to Asian V. cholerae remains to be determined.
FIG. 5.
Model for antibiotic resistance gene flux in the SXT family of constins.
ACKNOWLEDGMENTS
We are grateful to Michael Bennish for kindly providing us with strain C10488. We thank Brigid Davis and Anne Kane for critical reading of the manuscript.
This work was supported in part by the Deutsche Forschungsgemeinschaft (B.H.), the NIH Intramural program (R.W.), NIH grant AI42347, and a pilot project grant from the NEMC GRASP Center (P30DK-34928). M.K.W. is a PEW scholar and an assistant investigator in the Howard Hughes Medical Institute. D.M. acknowledges the Institut Pasteur and the Programme de Recherche Fondamentale en Microbiologie et Maladies Infectieuses et Parasitaire from the MENRT.
REFERENCES
- 1.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database research programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidmann J G, Smith J A, Struhl K. Current protocols in molecular biology. New York, N.Y: Greene Publishing and Wiley Interscience; 1990. [Google Scholar]
- 3.Bik E M, Bunschoten A E, Gouw R D, Mool F. Genesis of the novel epidemic Vibrio cholerae O139 strain: evidence for horizontal transfer of genes involved in polysaccharide synthesis. EMBO J. 1995;14:209–216. doi: 10.1002/j.1460-2075.1995.tb06993.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Briggs C E, Fratamico P M. Molecular characterization of an antibiotic resistance gene cluster of Salmonella typhimurium DT104. Antimicrob Agents Chemother. 1999;43:846–849. doi: 10.1128/aac.43.4.846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cholera Working Group. Large epidemic of cholera-like disease in Bangladesh caused by Vibrio cholerae O139. Lancet. 1993;342:387–390. [PubMed] [Google Scholar]
- 6.Churchward G, Belin G, Nagamine Y. A pSC101-derived plasmid which shows no sequence homology to other commonly used cloning vectors. Gene. 1984;31:165–171. doi: 10.1016/0378-1119(84)90207-5. [DOI] [PubMed] [Google Scholar]
- 7.Cloeckaert A, Baucheron S, Flaujac G, Schwarz S, Kehrenberg C, Martel J-L, Chaslus-Dancla E. Plasmid-mediated florfenicol resistance encoded by the floR gene in Escherichia coli isolated from cattle. Antimicrob Agents Chemother. 2000;44:2858–2860. doi: 10.1128/aac.44.10.2858-2860.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Coetzee J N, Datta N, Hedges R W. R factors from Proteus retgerri. J Gen Microbiol. 1972;72:543–552. doi: 10.1099/00221287-72-3-543. [DOI] [PubMed] [Google Scholar]
- 9.Comstock L E, Maneval D, Panigrahi P, Joseph A, Levine M M, Kaper J B, Morris J G J, Johnson J A. The capsule and O antigen in Vibrio cholerae O139 Bengal are associated with a genetic region not present in Vibrio cholerae O1. Infect Immun. 1995;63:317–323. doi: 10.1128/iai.63.1.317-323.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Craig N L. Transposition. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C.: ASM Press; 1996. pp. 2339–2362. [Google Scholar]
- 11.Dalsgaard A, Forslund A, Serichantalergs O, Sandvang D. Distribution and content of class 1 integrons in different Vibrio cholerae O-serotype strains isolated in Thailand. Antimicrob Agents Chemother. 2000;44:1315–1321. doi: 10.1128/aac.44.5.1315-1321.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Davis J K, Paoli G C, He Z, Nadeau L J, Somerville C C, Spain J C. Sequence analysis and initial characterization of two isozymes of hydroxylaminobenzene mutase from Pseudomonas pseudoalcaligenes JS45. Appl Environ Microbiol. 2000;66:2965–2971. doi: 10.1128/aem.66.7.2965-2971.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Falbo V, Carattoli A, Tosini F, Pezzella C, Dionisi A M, Luzzi I. Antibiotic resistance conferred by a conjugative plasmid and a class I integron in Vibrio cholerae O1 El Tor strains isolated in Albania and Italy. Antimicrob Agents Chemother. 1999;43:693–696. doi: 10.1128/aac.43.3.693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fling M E, Richards C. Nucleotide sequence of the trimethoprim resistant dihydrofolate reductase harboured by Tn7. Nucleic Acids Res. 1983;11:5147–5158. doi: 10.1093/nar/11.15.5147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Francia M V, Avila P, de la Cruz F, Garcia Lobo J M. A hot spot in plasmid F for site-specific recombination mediated by Tn21 integron integrase. J Bacteriol. 1997;179:4419–4425. doi: 10.1128/jb.179.13.4419-4425.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Francia M V, de la Cruz F, Garcia Lobo J M. Secondary-sites for integration mediated by the Tn21 integrase. Mol Microbiol. 1993;10:823–828. doi: 10.1111/j.1365-2958.1993.tb00952.x. [DOI] [PubMed] [Google Scholar]
- 17.Glass R I, Huq M I, Lee J V, Threlfall E J, Khan M R, Alim A R, Rowe B, Gross R J. Plasmid-borne multiple drug resistance in Vibrio cholerae serogroup O1 biotype El Tor: evidence for a point-source outbreak in Bangladesh. J Infect Dis. 1983;147:204–209. doi: 10.1093/infdis/147.2.204. [DOI] [PubMed] [Google Scholar]
- 18.Heidelberg J F, Eisen J A, Nelson W C, Clayton R A, Gwinn M L, Dodson R J, Haft D H, Hickey E K, Peterson J D, Umayam L, Gill S R, Nelson K E, Read T D, Tettelin H, Richardson D, Ermolaeva M D, Vamathevan J, Bass S, Qin H, Dragoi I, Sellers P, McDonald L, Utterback T, Fleishmann R D, Nierman W C, White O. DNA sequence of both chromosomes of the cholera pathogen Vibrio cholerae. Nature. 2000;406:477–483. doi: 10.1038/35020000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ho C, Kulaeva O I, Levine A S, Woodgate R. A rapid method for cloning mutagenic DNA repair genes: isolation of umu-complementing genes from multidrug resistance plasmids R391, R446b, and R471a. J Bacteriol. 1993;175:5411–5419. doi: 10.1128/jb.175.17.5411-5419.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hochhut B, Beaber J W, Woodgate R, Waldor M K. Formation of chromosomal tandem arrays of the SXT element and R391, two conjugative chromosomally integrating elements that share an attachment site. J Bacteriol. 2001;183:1124–1132. doi: 10.1128/JB.183.4.1124-1132.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hochhut B, Waldor M K. Site-specific integration of the conjugal Vibrio cholerae SXT element into prfC. Mol Microbiol. 1999;32:99–110. doi: 10.1046/j.1365-2958.1999.01330.x. [DOI] [PubMed] [Google Scholar]
- 22.Kim E H, Aoki T. Sequence analysis of the florfenicol resistance gene encoded in the transferable R-plasmid of a fish pathogen, Pasteurella piscida. Microbiol Immunol. 1996;40:397–399. doi: 10.1111/j.1348-0421.1996.tb01125.x. [DOI] [PubMed] [Google Scholar]
- 23.Kim E H, Aoki T. The transposon-like structure of IS26-tetracycline, and kanamycin resistance determinant derived from transferable R plasmid of a fish pathogen, Pasteurella piscida. Microbiol Immunol. 1994;38:31–38. doi: 10.1111/j.1348-0421.1994.tb01741.x. [DOI] [PubMed] [Google Scholar]
- 24.Kulaeva O I, Wootton J C, Levine A S, Woodgate R. Characterization of the umu-complementing operon from R391. J Bacteriol. 1995;177:2737–2743. doi: 10.1128/jb.177.10.2737-2743.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lauf U, Müller C, Herrmann H. The transposable elements resident on the plasmids of Pseudomonas putida strain H, Tn5501 and Tn5502, are cryptic transposons of the Tn3 family. Mol Gen Genet. 1998;259:674–678. doi: 10.1007/s004380050862. [DOI] [PubMed] [Google Scholar]
- 26.Mazel D, Dychinco B, Webb V A, Davies J. A distinctive class of integron in the Vibrio cholerae genome. Science. 1998;280:605–608. doi: 10.1126/science.280.5363.605. [DOI] [PubMed] [Google Scholar]
- 27.Mitra R, Basu A, Dutta D, Nair G B, Takeda Y. Resurgence of Vibrio cholerae O139 Bengal with altered antibiogram in Calcutta, India. Lancet. 1996;348:1181. doi: 10.1016/s0140-6736(05)65326-3. [DOI] [PubMed] [Google Scholar]
- 28.Mukhopadhyay A K, Basu A, Garg P, Bag P K, Ghosh A, Bhattacharya S K, Takeda Y, Nair G B. Molecular epidemiology of re-emergent Vibrio cholerae O139 Bengal in India. J Clin Microbiol. 1998;36:2149–2152. doi: 10.1128/jcm.36.7.2149-2152.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Oka A, Sugisaki H, Takanami M. Nucleotide sequence of the kanamycin resistance transposon Tn903. J Mol Biol. 1981;147:217–226. doi: 10.1016/0022-2836(81)90438-1. [DOI] [PubMed] [Google Scholar]
- 30.Prager R, Streckel R, Stephan J, Bockemuhl T, Shimada T, Tschäpe H. Genomic fingerprinting of Vibrio cholerae O139 from Germany and South Asia in comparison with strains of Vibrio cholerae O1 and other serogroups. Med Microbiol Lett. 1994;5:217–219. [Google Scholar]
- 31.Recchia G D, Hall R M. Origins of the mobile gene cassettes found in integrons. Trends Microbiol. 1997;5:389–394. doi: 10.1016/S0966-842X(97)01123-2. [DOI] [PubMed] [Google Scholar]
- 32.Recchia G D, Hall R M. Gene cassettes: a new class of mobile element. Microbiology. 1995;141:3015–3027. doi: 10.1099/13500872-141-12-3015. [DOI] [PubMed] [Google Scholar]
- 33.Recchia G D, Hall R M. Plasmid evolution by acquisition of mobile gene cassettes: plasmid pIE723 contains the aadB gene cassette precisely inserted at a secondary site in the IncQ plasmid RSF1010. Mol Microbiol. 1995;15:179–187. doi: 10.1111/j.1365-2958.1995.tb02232.x. [DOI] [PubMed] [Google Scholar]
- 34.Rowe-Magnus D A, Guerout A-M, Ploncard P, Dychinco B, Davies J, Mazel D. The evolutionary history of chromosomal super-integrons provides an ancestry for multiresistant integrons. Proc Natl Acad Sci USA. 2001;98:652–657. doi: 10.1073/pnas.98.2.652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Scholz P, Haring V, Wittmann-Liebold B, Ashman K, Bagdasarian M, Scherzinger E. Complete nucleotide sequence and gene organization of the broad-host-range plasmid RSF1010. Gene. 1989;75:271–288. doi: 10.1016/0378-1119(89)90273-4. [DOI] [PubMed] [Google Scholar]
- 36.Sharma C, Maiti S, Mukhopadhyay A K, Basu A, Basu I, Nair G B, Mukhopadhyaya R, Das B, Kar S, Ghosh R K, Ghosh A. Unique organization of the CTX genetic element in Vibrio cholerae O139 strains which reemerged in Calcutta, India, in September 1996. J Clin Microbiol. 1997;95:3348–3350. doi: 10.1128/jcm.35.12.3348-3350.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stover C K, Pham X Q, Erwin A L, Mizoguchi S D, Warrener P, Hickey M J, Brinkman F S, Hufnagle W O, Kowalik D J, Lagrou M, Garber R L, Goltry L, Tolentino E, Westbrock-Wadman S, Yuan Y, Brody L L, Coulter S N, Folger K R, Kas A, Larbig K, Lim R, Smith K, Spencer D, Wong G K, Wu Z, Paulsen I T. Complete genome sequence of Pseudomonas aeruginosa PA01, an opportunistic pathogen. Nature. 2000;406:959–964. doi: 10.1038/35023079. [DOI] [PubMed] [Google Scholar]
- 38.Stroeher U H, Parasivam G, Dredge B K, Manning P A. Novel Vibrio cholerae O139 genes involved in lipopolysaccharide biosynthesis. J Bacteriol. 1997;179:2740–2747. doi: 10.1128/jb.179.8.2740-2747.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sundstrom L, Jansson C, Bemer K, Heikkila E, Olsson-Liljequist B, Skold O. A new dhfrVIII trimethoprim-resistance gene, flanked by IS26, whose product is remote from other dihydrofolate reductases in parsimony analysis. Gene. 1995;154:7–14. doi: 10.1016/0378-1119(94)00905-8. [DOI] [PubMed] [Google Scholar]
- 40.Tabtieng R, Wattanasri S, Echeverria P, Seriwatana J, Bodhidatta L, Chatkaeomorakot A, Rowe B. An epidemic of Vibrio cholerae El Tor Inaba resistant to several antibiotics with a conjugative group C plasmid encoding for type II dihydrofolate reductase in Thailand. Am J Trop Med Hyg. 1989;41:680–686. doi: 10.4269/ajtmh.1989.41.680. [DOI] [PubMed] [Google Scholar]
- 41.Waldor M K, Mekalanos J J. ToxR regulates virulence gene expression in non-O1 strains of Vibrio cholerae that cause epidemic cholera. Infect Immun. 1994;62:72–78. doi: 10.1128/iai.62.1.72-78.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Waldor M K, Mekalanos J J. Vibrio cholerae O139 specific gene sequences. Lancet. 1994;343:1366. doi: 10.1016/s0140-6736(94)92504-6. [DOI] [PubMed] [Google Scholar]
- 43.Waldor M K, Rubin E J, Pearson G D N, Kimsey H, Mekalanos J J. Regulation, replication, and integration functions of the Vibrio cholerae CTXφ are encoded by region RS2. Mol Microbiol. 1997;24:917–926. doi: 10.1046/j.1365-2958.1997.3911758.x. [DOI] [PubMed] [Google Scholar]
- 44.Waldor M K, Tschäpe H, Mekalanos J J. A new type of conjugative transposon encodes resistance to sulfamethoxazole, trimethoprim, and streptomycin in Vibrio cholerae O139. J Bacteriol. 1996;178:4157–4165. doi: 10.1128/jb.178.14.4157-4165.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang R F, Kushner S R. Construction of versatile low-copy-number vectors for cloning, sequencing and gene expression in Escherichia coli. Gene. 1991;100:195–199. [PubMed] [Google Scholar]
- 46.Woodgate R. A plethora of lesion-replicating DNA polymerases. Genes Dev. 1999;13:2191–2195. doi: 10.1101/gad.13.17.2191. [DOI] [PubMed] [Google Scholar]
- 47.Yam W-C, Yuen K-Y, Wong S-S, Que T-L. Vibrio cholerae O139 susceptible to vibriostatic agent O/129 and co-trimoxazole. Lancet. 1994;344:404–405. [PubMed] [Google Scholar]
- 48.Yamamoto T, Nair G B, Alpert M J, Parodi C, Takeda Y. Presented at the Proceedings of the 30th joint conference US-Japan cooperative medical science program for cholera and related diarrheal diseases panel. 1994. Fukuaka, Japan. [Google Scholar]