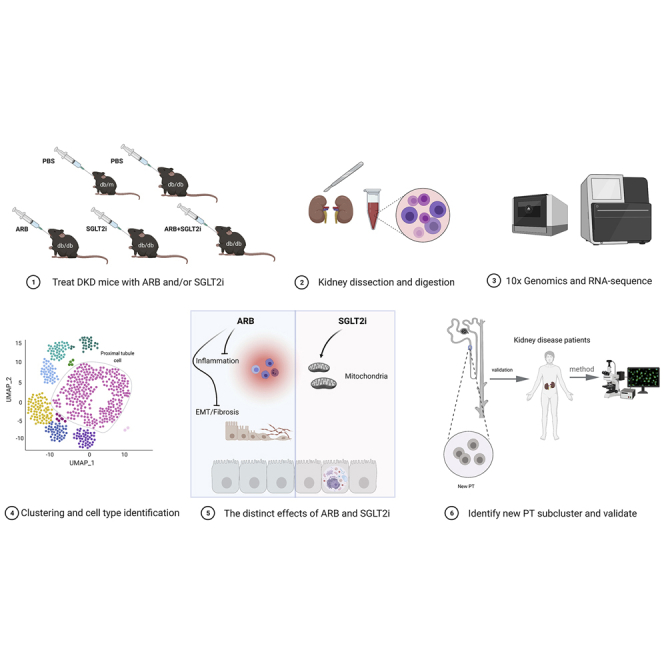
Abstract
Angiotensin receptor blockers (ARBs) and sodium-glucose cotransporter 2 inhibitors (SGLT2i) have been used as the standard therapy for patients with diabetic kidney disease (DKD). However, how these two drugs possess additive renoprotective effects remains unclear. Here, we conducted single-cell RNA sequencing to profile the kidney cell transcriptome of db/db mice treated with vehicle, ARBs, SGLT2i, or ARBs plus SGLT2i, using db/m mice as control. We identified 10 distinct clusters of kidney cells with predominant proximal tubular (PT) cells. We found that ARBs had more anti-inflammatory and anti-fibrotic effects, while SGLT2i affected more mitochondrial function in PT. We also identified a new PT subcluster, was increased in DKD, but reversed by the treatments. This new subcluster was also confirmed by immunostaining of mouse and human kidneys with DKD. Together, our study reveals kidney cell-specific gene signatures in response to ARBs and SGLT2i and identifies a new PT subcluster, which provides new insight into the pathogenesis of DKD.
Keywords: scRNA-seq, diabetic kidney disease, proximal tubular cells, SGLT2i, ARBs
Graphical abstract
Wu and colleagues use scRNA-seq to investigate the mechanisms of combination therapy with ARBs and SGLT2i in diabetic mice and found that ARBs had more anti-inflammatory effects, while SGLT2i affected more mitochondrial function in PT cells. They also identified and validate a new PT subcluster in mouse and human diabetic kidneys.
Introduction
Diabetic kidney disease (DKD) is the major cause of end-stage kidney disease in diabetic patients.1,2 Unfortunately, the treatment options are still limited. Renin-angiotensin system (RAS) blockers, such as angiotensin-converting enzyme inhibitors (ACEi) and angiotensin receptor blockers (ARBs), have been the main treatments for patients with DKD in recent decades.3, 4, 5 Recently, inhibitors of sodium-glucose cotransporter 2 (SGLT2i) have been shown to provide additional renoprotective effects on top of ACEi/ARBs; therefore, combination therapy with RAS blockers and SGLT2i is now considered the standard therapy for DKD patients.6, 7, 8, 9 Several potential renoprotective mechanisms have been proposed for both RAS blockers and SGLT2i; however, the exact mechanisms remain unclear.10,11 In addition, how SGLT2i synergizes with RAS blockade is completely unknown. Understanding this interaction could help us to optimize the current therapy for patients with DKD.
Recently, single-cell RNA sequencing (scRNA-seq) has been used to study kidney development and the pathogenesis of various kidney diseases.12, 13, 14, 15 However, scRNA-seq studies in DKD are limited. To our knowledge, only two studies have been published. We performed a scRNA-seq study of glomeruli isolated from DKD mice and their controls and revealed several glomerular cell-specific differentially expressed genes (DEGs) that play an important role in the progression of DKD.16 Wilson et al. performed single nucleus RNA-seq (snRNA-seq) on three control and three early human DKD samples.17 Analyses of the data demonstrated cell-type-specific changes in gene expression that are important for angiogenesis, ion transport, and immune cell activation. Studies using scRNA-seq to assess drug responses at the single-cell level have not been reported previously for kidney disease.
Here, we used scRNA-seq to assess the response of individual kidney cells to the treatment with ARBs and SGLT2i alone or in combination in type 2 DKD mice (db/db). We were able to dissect specific responses of individual cells to ARBs and/or SGLT2i. We also identified new clusters of proximal tubular (PT) cells in DKD mice.
Results
Effects of ARBs and SGLT2i in diabetic db/db mice
Type 2 diabetic db/db mice at 10 weeks of age were treated with vehicle, SGLT2i (dapagliflozin), ARBs (irbesartan), or both for a total of 8 weeks (Figure S1A). Nondiabetic db/m mice were used as their controls. We found that all diabetic db/db mice had increased blood glucose (BG) compared with nondiabetic db/m mice. Db/db mice treated with SGLT2i or ARBs + SGLT2i had reduced BG levels compared with vehicle-treated and ARB-treated db/db mice (Figure S1B). The systolic blood pressure was not different among the five groups, but a lower diastolic blood pressure was observed in ARB + SGLT2i-treated db/db mice than in vehicle-treated db/db mice (Figures S1C and S1D). Body weight in db/db mice was significantly higher than that in db/m mice, while no difference was found among all groups of db/db mice (Figure S1E). The kidney weight was increased in the db/db mice compared with db/m mice, and a significant reduction was observed in all treatment groups (Figure S1F). db/db mice had increased 24-h urinary albumin excretion (UAE) compared with db/m mice, but a reduction in UAE was observed in all drug-treated groups (Figure S1G). For both kidney weight and UAE, a trend of further reduction was observed in the combination treatment group compared with the individual treatment groups, but this did not reach statistical significance (Figure S1F and S1G). Renal function, as assessed by measurement of blood urea levels, was increased in db/db mice while decreased by SGT2i and ARB + SGLT2i treatment (Figure S1H). Histologically, both glomerular volume and mesangial expansion were increased in db/db mice compared with db/m mice, but these were reduced in all treatment groups (Figures S2A–S2C). A further reduction in glomerular volume was observed in the combination treatment group, but a reduction in the mesangial fraction in the combination group did not reach statistical significance (Figures S2B and S2C). Electronic microscopy confirmed an improvement in podocyte foot process effacement and mitochondrial morphology in all treatment groups compared with the vehicle group (Figure S2D). In addition, we found a significant increase in macrophages in db/db mice compared with db/m mice, but this increase was reduced in all treatment groups (Figures S2E and S2F). These data suggest that SGLT2i and ARBs had individual renoprotective effects in diabetic mice and that additive effects were observed in the combination treatment group at least for some renal parameters.
Single-cell landscape of health and DKD kidney with or without treatment
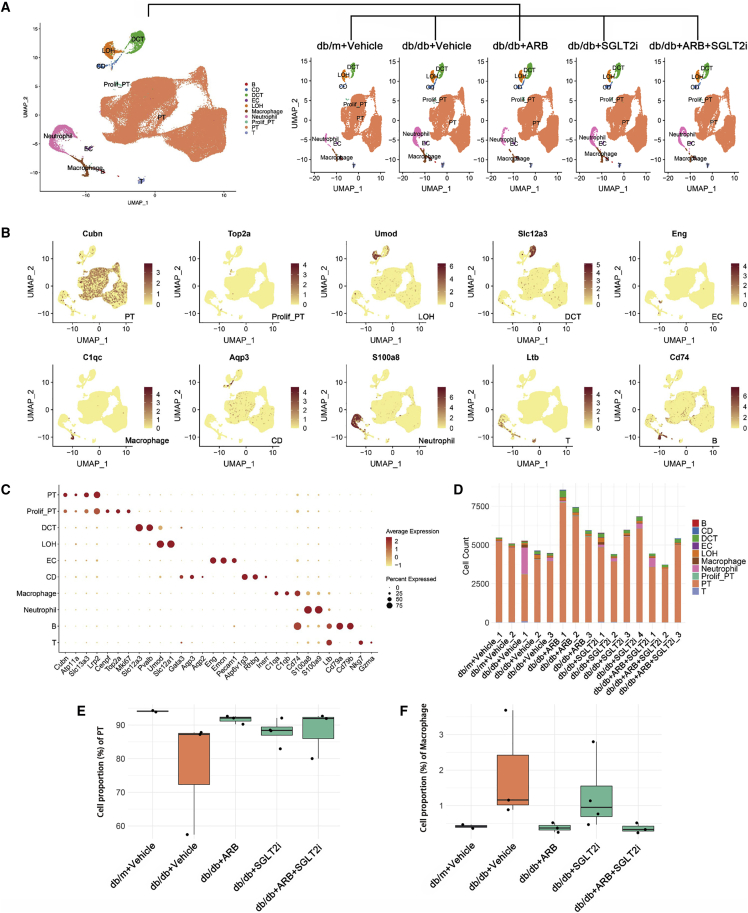
Next, we performed scRNA-seq of the kidneys from these mice as described in the methods and illustrated in Figure S3A. In total, 83,585 single-cell transcriptomes were generated after quality control and filtering (Figure S4). Using unsupervised clustering (uniform manifold approximation and projection [UMAP]), 28 separate cell clusters were identified after pooling all of the samples together (Figure S3B). Ten distinct cell types, including 5 tubular cell types (78,625), 2 lymphoid cell types (525), 2 myeloid cell types (4,201), and one endothelial cell type (234) were annotated based on their known marker gene expression, and they were consistent across all five groups (Figures 1A–1C). One of the cell clusters was annotated as “proliferative PT” since it is characterized by both strong proliferation gene (Mki67) and PT marker gene (Cubn, Atp11a). Represent marker genes are shown in Figure 1B. A full list of marker genes in each cluster is provided in Table S1. Subsequently, to define the major cell-type changes after disease and treatment, the numbers of each cell cluster were compared among the five groups, and PT cells were the dominant component among all of the groups (Figure 1D). In addition, the percentage of PT cells appeared to decrease in db/db mice compared with db/m mice, but was restored in all treatment groups (Figure 1E). In contrast, the proportion of macrophages was increased in the db/db mice compared with controls but suppressed in all treatment groups, with a more prominent suppression in mice treated with ARBs than in mice treated with SGLT2i (Figure 1F). The increase in macrophage infiltration in db/db mice was also confirmed by immunohistochemistry (Figures S2E and S2F).
Figure 1.
Cell diversity in mouse kidney cells delineated by single-cell transcriptomic analysis
(A) All samples with different treatments were integrated into a single dataset and clustered using UMAP. Colors and labels indicate different cell types based on marker gene expression. B, B cells; CD, collecting duct; DCT, distal convoluted tubule; EC, endothelial cell; LOH, loop of henle; PT, proximal tubule; prolif-PT, proliferative PT; T, T cells. (B) Expression of selected marker genes for each cell type projected on UMAP. (C) Dot plot shows the expression of marker genes for the identified cell types. (D) Number of identified cell types in each sample. (E and F) Boxplots of the average proportion of PTs (E) and macrophages (F) in different groups.
PT-specific responses to DKD injury and different treatments
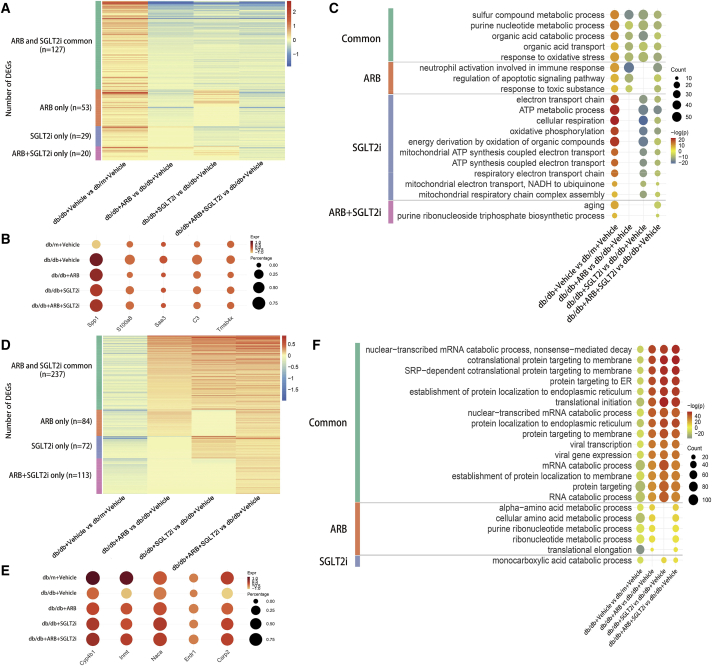
Recent findings suggest that PT injury is not only important for late DKD but also critical for early DKD.18,19 In addition, the primary target of SGLT2i is PT, and it is well known that ARBs also have direct effects on PT.20 Therefore, our analysis focused on the specific responses of PT cells to DKD injury and treatments (ARBs and/or SGLT2i). First, we compared the DEGs between db/db versus db/m and then determined how different treatments with ARBs and/or SGLT2i reversed these DEGs. We analyzed DEGs in two different ways: up-down (upregulated in db/db and downregulated by treatments) and down-up (downregulated in db/db and upregulated by treatments). Among 229 up-down genes, 127 genes were reversed by all treatments, such as Spp1, S100a8, Saa3, C3, and Tmsb4x (Figures 2A and 2B), and Spp1, S100a8, Saa3, and C3 are known pro-inflammatory mediators.15 Tmsb4x, which encodes thymosin β4, an actin sequestering protein, was reported as a modifier of glomerular injury.21 However, 53 DEGs were reversed by ARBs, 29 DEGs were reversed only by SGLT2i and 20 DEGs were affected only by combination treatment. Gene ontology (GO) term enrichment analysis showed that many metabolic pathways were affected by all treatments indicating the critical role of PT in metabolism. Inflammation and apoptosis pathways were enriched in the ARB-treated group and the pathways related to mitochondrial dysfunction were heavily enriched in SGLT2i-treated mice indicating that glucose uptake in PT is a critical driver of mitochondrial dysfunction. A few unique DEGs and pathways were regulated only in the combination treatment group, suggesting that ARBs and SGLT2i likely have additive effects instead of synergistic effects (Figure 2C). We also analyzed 506 genes that followed the down-up manner (Figure 2D). Among these DEGs, 237 genes were downregulated in vehicle-treated db/db mice but reversed in all treatment groups, 84 genes were reversed by ARBs, 72 genes were reversed only by SGLT2i, and 113 genes were affected only in the combination treatment group. The representative genes were shown in Figure 2E including Cyp4b1, Inmt, Naca, Erdr1, and Csrp2, and Cyp4b1 is a key enzyme for drug and lipid metabolism.22,23 Erdr1 is a pivotal regulator of cell apoptosis and its role in kidney disease has not been investigated.24 Csrp2 is a key gene involving in development and cell differentiation.25,26 The GO term analysis showed that many pathways enriched in this group of genes were related to metabolism, suggesting that, in early DKD, metabolism in PT was severely altered and that these metabolic disorders were partially reversed by treatments with either ARBs or SGLT2i. Interestingly, there were no specific GO terms in the combination treatment group, again suggesting additive effects instead of synergistic effects between the two drugs (Figure 2F). The full lists of DEGs and GO terms in each group are provided in Tables S2 and S3. Collectively, these findings suggested that ARBs and SGLT2i treatment regulated many common DEGs and pathways. However, they also had specific effects on PT cells and it seems that SGLT2i preferentially regulates PT mitochondrial function, while ARBs may preferentially protect the kidney by alleviating inflammation. These unique effects of ARBs and SGLT2i may explain their additive renoprotective effects observed in humans with DKD.27
Figure 2.
Differential gene expression analysis reveals PT-specific responses to DKD injury and different treatments
(A) Heatmap shows the number of DEGs that were upregulated in db/db mice compared with db/m mice but downregulated by treatment (up-down pattern). ARBs and SGLT2i Common: DEGs were downregulated by both ARBs and SGLT2i treatment; ARB only: DEGs downregulated specifically by ARB treatment; SGLT2i only: DEGs downregulated specifically by SGLT2i treatment; ARB+SGLT2i only: DEGs downregulated only by combination treatment with ARBs and SGLT2i. The color scale represents the log fold change in the expression levels of genes. (B) Dot plot shows the representative DEGs that follow the up-down pattern in different groups. (C) Dot plot of gene ontology (GO) terms in DEGs that followed the up-down pattern. (D) Heatmap shows the DEGs downregulated in db/db mice compared with db/m mice but upregulated by treatment (down-up pattern). (E) Dot plot shows the representative DEGs that follow the down-up pattern in different groups. (F) Dot plot of GO terms in DEGs that followed the down-up pattern.
Effects of SGLT2i and ARBs in the regulation of fatty acid metabolism, ATP synthesis, inflammation, and fibrosis in PT
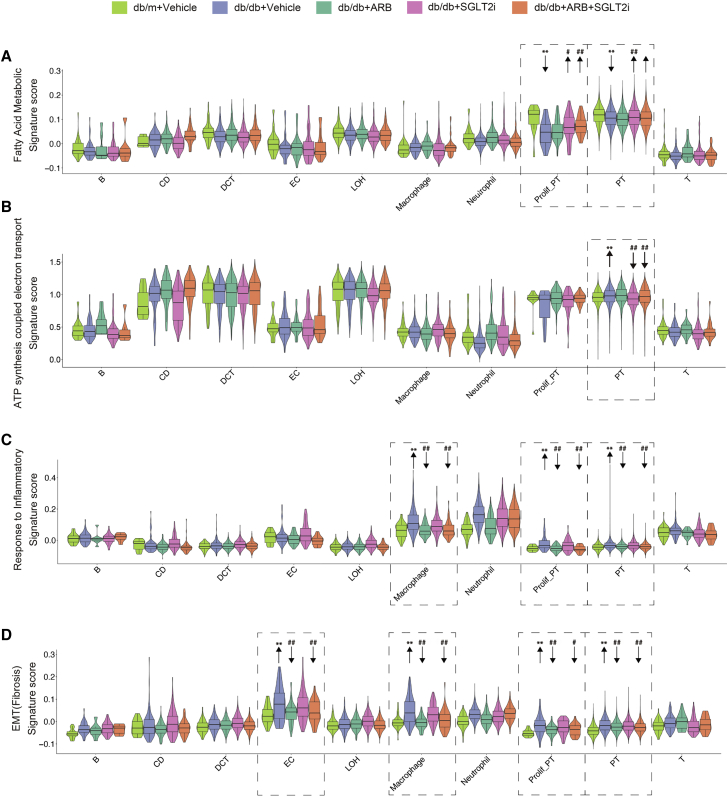
Next, we analyzed the effects of SGLT2i and ARBs specifically on fatty acid (FA) metabolism, ATP synthesis, inflammation, and fibrosis, as these are key events leading to the progression of DKD. We used the scoring gene sets obtained from the MSigDB and MGI database. First, we analyzed the score for FA metabolism, which is critical for PT function under both physiological and pathological conditions. We found that, among all kidney cell clusters, PT had the highest score for FA metabolism (Figure 3A). Furthermore, in PT cells FA metabolism was downregulated in db/db mice but reversed by SGLT2i and combination treatment while ARBs did not have any effects. This observation was more obvious in the proliferative PT cells (Figure 3A). Consistently, the expression levels of genes associated with FA metabolism (Acox1, Lpl, and Cyp2e1) were significantly reduced in db/db mice compared with db/m and reversed by SGLT2i treatment, while ARBs had no effect (Figure S5A). Next, we examined the score for ATP synthesis coupled with electron transport and found that tubule cells had higher scores for ATP synthesis than other cell types. This means that tubule cells require more energy, and this was further increased in db/db mice. In PT cells, SGLT2i reduced the ATP synthesis score, while ARBs had no effect (Figure 3B). Furthermore, expression of ATP synthesis genes (Dld, Sdha, and Ndufs1) was higher in db/db mice than db/m mice and this was reversed by SGLT2i, but not by ARBs (Figure S5A). Then we determined the inflammation score, which was higher in immune cells than in other kidney cell types (Figure 3C). Interestingly, the inflammatory score for both macrophages and PT was increased in db/db mice but reversed mostly by ARB treatment, while SGLT2i had minimal effects. Finally, we examined the epithelial-mesenchymal transition (EMT) score. We are aware that it remains controversial whether EMT occurs during kidney fibrosis process. However, this EMT score, which consists of several profibrotic markers, could be used to assess even relative early fibrosis process in these DKD mice. Interestingly, we found that a high EMT score was observed in endothelial cells and macrophages. The EMT score was increased in these cells as well as in PT cells from db/db mice compared with db/m mice, but this was reversed more by ARB treatment than SGLT2i treatment (Figure 3D). These results were further validated by qPCR analysis of the mRNA levels of marker genes for inflammation (IL-1b and Nfkbia) and EMT (Tnfrsf12a and Jun) (Figure S5B). Collectively, these findings suggest that SGLT2i affects FA metabolism and ATP consumption, whereas ARBs have more anti-inflammatory and antifibrotic effects. This is consistent with our pathway analysis.
Figure 3.
Gene-scoring analysis using the indicated molecular signatures
(A) Gene-scoring analysis of “fatty acid metabolic” in all kidney cell types from different groups of mice. (B) Gene-scoring analysis of “ATP synthesis coupled electron transport” in all kidney cell types from different groups of mice. (C) Gene-scoring analysis of “response to inflammation” across kidney cells in all groups. (D) Gene-scoring analysis of EMT in all kidney cell types from different groups of mice. Arrows indicate comparisons between db/db versus db/m (∗∗p < 0.0001) or db/db + vehicle versus db/db + SGLT2i, db/db + ARB, or db/db + ARB+SGLT2i (#p < 0.01, ##p < 0.0001). p values between groups by one-way ANOVA with Tukey's multiple comparisons test.
Role of PT subclusters in DKD
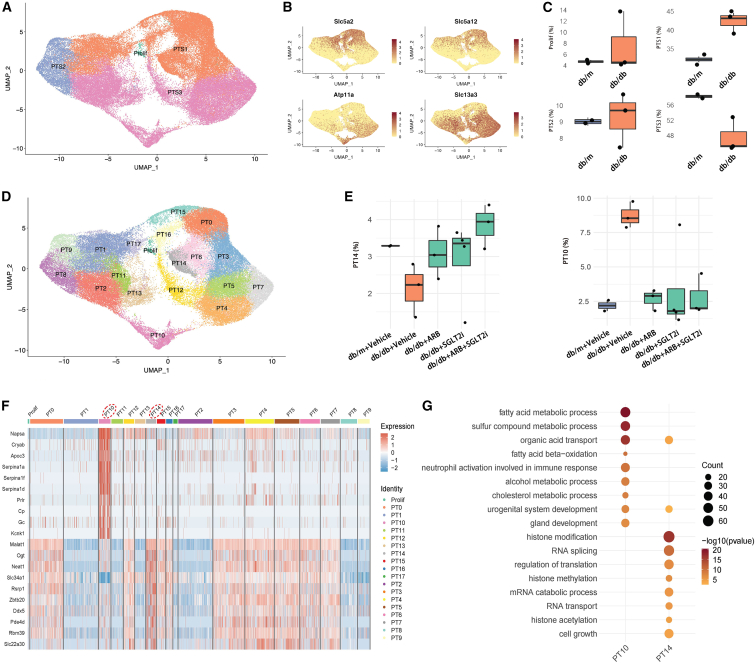
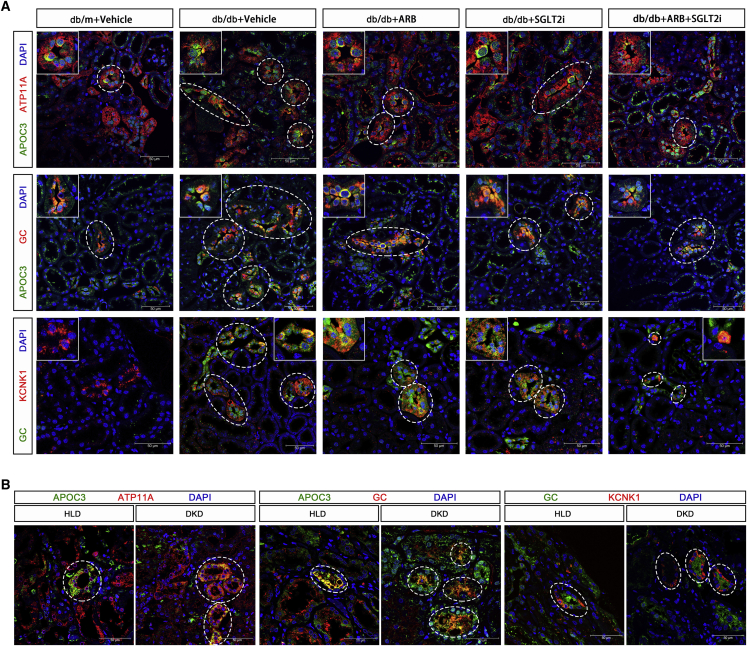
First, we divided PT cells into S1, S2, and S3 segments based on the available specific markers (Figures 4A and 4B). Interestingly, we found that the frequency of PT-S3 cells decreased while the frequency of PT-S1 increased in db/db mice compared with db/m mice (Figure 4C). Then, we further subclustered the PT cells into 19 clusters (Figure 4D). By differential proportion analysis, we found that subcluster 14 from PT-S1 was decreased while subcluster 10 from PT-S3 was increased in db/db mice compared with controls, and these changes were reversed in the treatment groups (Figures 4E and S6). Subcluster 10, which occupied approximately 8%–10% of all PT cells in db/db mice, expressed not only PT-S3 segment markers (Atp11a, Slc13a3) but also a unique set of genes, such as Napsa, Cryab, Apoc3, Gc, and Kcnk1 (Figure 4F). Pathway analysis of the unique gene sets in subclusters 10 and 14 revealed that the pathways enriched in subcluster 10 were absent in subcluster 14, while the pathways enriched in subcluster 14 were absent in subcluster 10, indicating that these two subclusters likely have an opposite function in DKD (Figure 4G). The GO term enrichment analysis also revealed that subcluster 10 was enriched by development-related genes, suggesting that this subcluster could contain potential renal progenitor cells to repair kidney injury or undifferentiated PT. We speculate that this subcluster of the PT-S3 segment plays a major role in kidney injury or repair. To eliminate the possibility of artificial effects, such as enzymatic digestion in the subclustering analysis, we performed several validation analyses and experiments. First, we searched public databases and did not find this PT subcluster in other non-DKD kidney disease models, such as acute kidney injury (Figures S7A and S7B) and unilateral ureter obstruction (Figures S7C and S7D). Second, we did not find increased expression of any known digestion-related genes in this subcluster (data not shown). Most importantly, we validated the expression of the specific markers of subcluster 10 by immunostaining of mouse and human kidneys with DKD and non-DKD glomerular diseases. We confirmed that the staining of APOC3, a specific marker for subcluster 10, was co-localized with the PT-specific marker ATP11A in the mouse kidney. In addition, APOC3 staining was increased in db/db mice compared with db/m mice, but the increased staining was reversed by treatment with either SGLT2i or ARBs, or both (Figure 5A, upper panel). Then, we confirmed that APOC3 staining was co-localized with another specific marker (GC) of subcluster 10, and the staining for both APOC3 and GC was increased in the diabetic mice but reversed by the treatments (Figure 5A middle panel). Moreover, we confirmed another specific marker (KCNK1) of subcluster 10 by co-staining with GC (Figure 5A lower panel). Finally, we stained for ATP11A, APOC3, GC, and KCNK1 in human kidney tissues from normal nephrectomy samples and biopsies from DKD and non-DKD (AKI, IgA nephropathy, and focal segmental glomerulosclerosis) patients (Figures 5B and S8). The pathology and clinical characteristics of these patients are provided in Figure S9 and Table S5. Consistent with the animal data, we found that APOC3 staining was co-localized with ATP11A and increased in DKD patients compared with controls. APOC3 and KCNK1 were also co-localized with GC, and they were increased in DKD patients compared with normal controls. To determine whether this PT-10 cluster is specific for DKD, we also stained these marker genes in the kidneys of non-DKD patients. We found that these genes (APOC3, GC, and KCNK1) were expressed in the kidneys of non-DKD patients but interestingly, they did not co-localize together, suggesting that the PT-10 cluster might be absent in these non-DKD patients (Figure S8). These findings further support the existence of this specific subcluster, and the number of this PT subcluster was increased in DKD kidneys. Further studies are needed to confirm the specificity and the function of this new subcluster in the pathogenesis of DKD.
Figure 4.
Role of PT subclusters in DKD
(A) UMAP plot of PT cells from all samples for identification of S1, S2, and S3 segments. (B) Expression of marker genes for S1 (Slc5a2, Slc5a12) and S3 (Atp11a, Slc13a3) segments. (C) Comparison of the frequency of S1, S2, S3, and prolif-PT cells between db/m and db/db mice. (D) UMAP plot for PT cells from all samples. (E) Boxplot shows the quantitation of subclusters PT14 and PT10 between all groups of mice. (F) Heatmap shows the representative marker gene expression for subclusters PT10 and PT14. (G) GO enrichment analysis of the marker genes in PT14 and PT10.
Figure 5.
Validation of new PT subcluster PT-10
(A) Representative immunofluorescence images from each group for ATP11A (PT-S3 marker, red) and APOC3 (PT-10 marker, green)/GC (PT-10 marker, red/green)/KCNK1 (PT-10 marker, red). (B) Representative immunofluorescence images from healthy living donor (HLD) and DKD patients for ATP11A (red) and APOC3 (green)/GC (red/green)/KCNK1 (red). Original magnification, 63×.
The distinct effects of ARBs and SGLT2i in macrophage
We performed further analysis to examine how macrophages respond differently to ACEi and SGLT2 inhibitors (Figure S10). First, we compared the DEGs between db/db versus db/m and then determined how ARBs and/or SGLT2i reversed these DEGs. We found that the DEG number in macrophage was much smaller than PT. We analyzed DEGs in up-down (upregulated in db/db and downregulated by treatments) and down-up (downregulated in db/db and upregulated by treatments) manners. Among 45 up-down genes, 24 genes were reversed by all treatments, such as Saa3, Ecm1, Wfdc21, and Arg1. 19 DEGs were reversed by ARBs, such as Cxcl2, Slpi, Lcn2, and Fth1 (Figures S10A and S10B). Cxcl2 is a well-known gene involved in inflammatory processes. Slpi, which encodes the secretory leukocyte peptidase inhibitor, was identified as a potential therapy target of kidney injury.28,29 Lcn2 plays an important role in innate immunity and is also an early biomarker of kidney injury.15 Fth1 is the most important iron storage gene and is reported to influence immunity.30 However, only one DEG was reversed by SGLT2i and one DEG was affected only by combination treatment. GO term enrichment analysis showed that ARBs specifically affected response to interleukin-1, regulation of I-κB kinase/NF-κB signaling and cytokine secretion (Figure S10C). We also analyzed 32 genes that followed the down-up manner (Figure S10D). The full list of DEGs and GO terms in macrophage are provided in Tables S6 and S7. Overall, these findings suggest that ARBs has more effects in macrophages than SGLT2i, consistent with its anti-inflammatory effects.
Discussion
Our study provides the first comprehensive analysis of the whole-kidney transcriptome in a type 2 diabetic mouse model with DKD at the single-cell level. We also assessed the response of the individual kidney cells to the two major treatments (ARBs and SGLT2i) of DKD individually or in combination. Our study reveals that ARBs and SGLT2i have distinct effects on PT cells to protect cells from diabetes-induced injury. SGLT2i has more protective effects against mitochondrial dysfunction and FA metabolism while ARBs have more anti-inflammatory and antifibrotic effects. Our study suggests potential mechanisms for the additive renoprotective effects of ARBs and SGLT2i in patients with DKD. In addition, our study reveals a new subcluster of PT cells that increases in DKD. We also validated this new cluster by immunostaining of its specific markers in both mouse and human kidneys with DKD.
Recently, combination therapy with RAS blocker and SGLT2i has been considered the standard therapy for DKD. Therefore, it is critical to understand how these two classes of drugs work together to protect kidney cells from injury. This study could help us to dissect such mechanisms at the single-cell level. To our knowledge, this is the first study to apply scRNA-seq to determine the response of individual kidney cells to treatment. This study helped us not only confirm the drug targets but also to reveal new mechanisms for diabetes-induced kidney cell injury. We used db/db mice here as a type 2 diabetic model with DKD, which mimics early DKD in humans.31 These mice responded to treatment with either ARBs or SGLT2i, and an additive renoprotective effect was also observed with combination therapy. However, it is important to note that the effects of the combination therapy observed in diabetic mice did not completely mimic the human findings. Blood pressure was not suppressed by either ARBs or SGLT2i alone in these mice, which might be different from human studies.32,33 However, the effects of ARBs and SGLT2i that we observed in these db/db mice are unlikely to result from changes in blood pressure. Besides, given that no obvious tubule injury was observed in these animals, the loss of PT cells in diabetic kidney mice could be due to their increased susceptibility to injury during the digestion process.
Our current study focused on PT and, therefore, we used a modified method that allowed us to identify more viable PT cells for scRNA-seq. Therefore, we were not able to have podocyte cluster in our analysis. The percentage of PT cells in our study was higher than that in other scRNA-seq studies.13,15 The reasons for focusing on PT cells are as follows: (1) many studies suggest a critical role of PT cells in the pathogenesis of DKD in both early and late stages,34,35 and (2) PT cells are direct target cells for SGLT2i, and a critical role of RAS in PT cells has also been shown.20 Our study revealed significant alterations of gene expression in PT cells in diabetic mice, and many of these genes were reversed by treatment with either ARBs or SGLT2i or both. We also found that, although ARBs and SGLT2i shared many common pathways, they had distinct effects in PT, indicating the potential mechanisms of additive renal protection. However, there are very few unique genes and pathways regulated by combination therapy, indicating that synergistic effects are unlikely. Further analysis revealed that ARBs have more anti-inflammatory and antifibrotic effects, while SGLT2i improves mitochondrial function. Consistent with our data, it has been shown that RAS blocker has anti-inflammatory effects in tubular cells.36,37 The effects of SGLT2i in the regulation of mitochondrial function in PT cells are likely mediated through its inhibition of glucose uptake. This was further confirmed by analyzing the scores and gene expression for FA metabolism, ATP synthesis, inflammation, and EMT. Indeed, SGLT2i affects more FA metabolism and ATP synthesis, while ARBs regulates inflammation and fibrosis processes. When we compared these scores among different cell types, we confirmed that PT cells had the highest FA score, while immune cells such as macrophages had the highest inflammatory score. Interestingly, a high score for EMT was found in endothelial cells and macrophages, which is consistent with previous reports on potential endothelial to mesenchymal cell38,39 and macrophage to mesenchymal cell transdifferentiation.40 Further analysis of the DEGs involved in these pathways may help us to dissect the detailed mechanisms of the renoprotective effects of these two drugs in individual kidney cells.
In this study, we also identified two novel subclusters of PT named subcluster 10 and subcluster 14. Subcluster 10 has very unique markers that are expressed only in these cells, while the markers for subcluster 14 are also expressed in other cell types but at a much lower level. These two subclusters appeared to have a complementary phenotype because: (1) subcluster 14 decreased while subcluster 10 increased in db/db mice; and (2) GO term enrichment analysis showed that the pathways enriched on one cluster were absent in another cluster. Interestingly, the alteration in the frequency of the cells in these two subclusters was reversed by treatment with either ARBs or SGLT2i, again indicating that these subclusters are related to disease processes. We analyzed published datasets and found that subcluster 10 seems to increase only in diabetic kidneys. We also confirmed the localization and changes of this PT subcluster by immunostaining of mouse and human kidneys with DKD and non-DKD patients. Subcluster 10 has many development-related genes, suggesting that these cells could be renal progenitor cells or undifferentiated cells. Previous scRNA-seq analysis of human kidneys also revealed subclusters of PT that have either a progenitor phenotype or EMT phenotype.41,42 However, subcluster 10 has specific markers that are distinct from other previously reported subclusters. Our clustering analysis also revealed a cluster of cells called proliferative PT, which was previously reported by others.14,43 The number of cells in this cluster also increased in the diabetic kidney but this cluster has distinct markers from the new subcluster 10. Proliferative PT cells are considered as an indicator of the tubular cell injury and repair. Future studies are required to confirm the function of these clusters of PT cells in the pathogenesis of DKD.
We also analyzed how macrophages respond to ARBs and SGLT2i treatments and found that ARBs had more effects in macrophages than SGLT2i, consistent with the anti-inflammatory effects of ARBs. Together with the data in PT cells, our study suggests that ARBs likely have unique renal protective effects independent from SGLT2i. Therefore, our study provides mechanistic support on the use of combination therapy instead of monotherapies with ARBs and SGLT2i in patients with DKD.
Our study have several limitations. Since we used scRNA-seq instead of snRNA-seq, we might not be able to catch all PT cells due to the digestion-related cell injury. To obtain the most viable PT cells for further analysis, we modified the enzyme digestion time, so we were unable to get glomerular cells, such as podocytes. Since we focused on PT cells, we did not perform a detailed analysis of other kidney cell types. For example, it would be interesting to analyze how distal tubular cells respond to SGLT2i with osmotic diuresis caused by the inhibition of sodium-glucose transporters. It would also be interesting to include a control group with similar level of BG control by another hypoglycemic agent in order to distinguish between hyperglycemia-dependent and -independent effects of SGLT2i. The sample sizes are still limited because of the cost and future studies are needed to increase the sample sizes to reduce the variations among the animals. Since uneven drop-out events remain as an issue in the scRNA-seq data, it would be interesting to verify these scRNA-seq findings by using deconvolution of bulk RNA-seq datasets from the same animal models. Finally, animal models of DKD do not fully recapture the features of human DKD, it would be critical to further validate these findings in human DKD.
In summary, we generated a comprehensive kidney transcriptomic profile at the single-cell level in type 2 diabetic mice with DKD. We illustrated how PT cells respond differently to SGLT2i and ARB treatment at the single-cell level. We demonstrated that SGLT2i mostly affects mitochondrial function in PT cells, while ARB have more anti-inflammatory and antifibrotic effects in different kidney cell types. We also identified new clusters of PT cells that likely play an important role in the pathogenesis of DKD. Our study provides new insights into the pathogenesis of DKD and kidney cell-specific responses to ARB and SGLT2i therapy.
Materials and methods
Animals and experiment design
Male db/db mice (BKS.Cg-Dock7m+/+Leprdb/Nju) were purchased from the Model Animal Research Center of Nanjing University. Male age-matched lean db/m mice were purchased from The Jackson Laboratory (stock no. 000642). We used only male mice here because these mice develop DKD more consistently than female mice. Mice were housed in a specific pathogen-free facility with free access to food and water and a 12:12 h night-day cycle. Male db/db mice develop DKD defined by the development of albuminuria at 10 weeks of age. The db/db mice were randomly assigned to four groups as follow: treated with vehicle control (phosphate-buffered saline [PBS]) (n = 8), treated with 3 mg/kg/day of dapagliflozin (n = 8), treated with 20 mg/kg/day of irbesartan (n = 8), and combination therapy with both dapagliflozin and irbesartan (n = 8). Vehicle and drugs were administered daily by oral gavage for 8 weeks and all mice were sacrificed at the age of 18 weeks. All animal studies were performed under the protocol approved by the Animal Care Committee at the Chongqing Medicine University. Body weight and fasting BG levels were monitored weekly by glucometer readings. Urine samples were collected weekly.
Measurement of BG, urinary albumin-to-creatinine ratio, and blood urea
Fasting BG was measured using the Accu-Chek Aviva glucometer (Roche Diabetes care) from tail vein blood samples weekly. The urine albumin concentrations were quantified by ELISA (E80-129; Bethyl Laboratories, Houston, TX). Urine creatinine levels of the same samples were measured by QuantiChrom Creatinine Assay Kit (DICT-500; BioAssay Systems, Hayward, CA) according to the manufacturer's instructions. Blood urea was measured using the QuantiChrom Urea Assay Kit (DIUR-100, BioAssay Systems, Hayward, CA)
Measurement of blood pressure
Systolic and diastolic blood pressure was measured using the CODA noninvasive tail-cuff blood pressure system (Kent Scientific, Torrington, CT). Mice were acclimated to the tail-cuff manometer and restraining device 10–20 min per day for at least 3 days before the day of actual data acquisition. Blood pressure was collected for further analysis.
Kidney histology
Ten percent of formalin-fixed and paraffin-embedded kidney samples were sectioned to 4-μm thickness. Periodic acid-Schiff (PAS)-stained kidney sections were used for the analysis of mesangial matrix expansion and glomerular area. PAS images were taken under 200× and 400× magnification (Leica Microsystems, DM4000B), and quantification was based on an average of 50 glomeruli per section.
Transmission electron microscopy
Tissues were fixed in 2.5% glutaraldehyde with 0.1 M sodium cacodylate (pH 7.4) for 72 h at 4°C. Samples were post-fixed with 1% OsO4 in 0.1 M PBS (pH 7.4) for 2 h at room temperature. After removing OsO4, tissues were rinsed in 0.1 M PBS (pH 7.4) and dehydrated at room temperature. Resin penetration and embedding of samples were performed at 37°C, which was followed by incubation with 2% uranium acetate-saturated alcohol solution. Cuprum grids were put into the grids board and dried overnight at room temperature, and grids were observed under TEM and images taken up to 1,000× magnitude.
Preparation of single cells from kidney
The mice were anesthetized and perfused with chilled 1× PBS via the left heart. Kidneys were removed, minced into 1-mm3 pieces with a razor blade, and incubated in 2 mL of digestion buffer containing 1 mg/mL collagenase type II (Sigma), 1 mg/mL Pronase E (Boster), and 50 μg/mL DNase I (Roche) at 37°C for 10 min. The partially digested tissue was then passed through a 100-μm cell strainer (Falcon) into HBSS (Hank's balanced salt solution) twice on ice and pelleted by centrifugation (200 × g for 5 min). Then, the pellet was resuspended in HBSS and sheared with a 27G needle to remove large clumps. They were further filtered through a 40-μm cell strainer (Falcon) to obtain a single-cell solution. After centrifugation, the cell pellets were incubated with 3 mL of RBC lysis buffer (TIANGEN, RT122-02) to remove red blood cells at room temperature for 3 min. Finally, the cells were washed and resuspended in HBSS three times to remove debris or cell aggregates. Cell viability and number were determined using a Countess II Automated Cell Counter (Invitrogen, C10227) after staining with trypan blue. Using this method, we were able to generate single-cell suspensions with >80% viability.
Sample processing, library generation, and sequencing
Single kidney cells were resuspended in HBSS to a suitable concentration (1,600,000–7,000,000 cells/mL). The viable cells were then loaded into chromium microfluidic chips with 3ʹ (Gel Bead Kit v.2) chemistry and barcoded with a 10x Chromium Controller (10x Genomics). RNA from the barcoded cells was subsequently reverse-transcribed to generate cDNA and sequencing libraries were constructed with reagents from a Chromium Single Cell 3′ v.2 Reagent Kit (10x Genomics) according to the manufacturer's instructions. Sequencing was performed with Illumina (NovaSeq 6000) using 150 bp paired-end sequencing.
scRNA-seq data quality control and analysis
For each sample, the raw unique molecular index (UMI) counts of each gene in each cell were generated using cellRanger count (cell ranger 5.0.0 http://10xgenomics.com) with mm10 as the reference genome. Genes expressed in fewer than 3 cells and cells expressing fewer than 200 genes were removed, while cells expressing more than 5,000 genes were considered potential doublets and removed. Cells were considered injured or apoptotic and removed when mitochondrial genes were >50%. We used a relatively high cutoff mitochondrial gene percentage for matching most PTs in our analysis because the high baseline mitochondria and mitochondrial injury in PTs is well known in DKD (PMID: 29622724). Raw gene UMI counts were further normalized into counts per 10,000 and log transformed using Seurat 3.1.5 (PMID: 31178118). The top 2,000 highly variable genes were selected using Find Variable Features with the default method, “vst”. All of the samples were finally integrated with Seurat Integrate Data using the first 30 canonical correlation analysis (CCAs), with the 2,000 anchors selected by Find Integration Anchors using the first 30 CCAs. The integrated expression profile was then scaled using Scale Data to give each gene equal weights for downstream analysis.
Unsupervised clustering
The first 30 principal components (PCs) identified from principal-component analysis were used to construct the K-nearest neighbor graph using FindNeighbor, and unsupervised clustering analysis was performed by FindClusters with the default method, the Louvain algorithm, with a resolution of 0.8. UMAP coordinates were generated using RunUMAP with the first 30 PCs. Markers of each cell were identified by FindAllMarkers using Wilcoxon rank-sum test.
Identification of differentially expressed genes and GO enrichment analysis
The differentially expressed genes (DEGs) between the cells from groups were identified with FindMarkers and genes with p ≤ 0.01 and log fold change ≥ 0.05 or ≤ −0.05 and expressed in at least 10% of cells in each condition were considered significantly up- or downregulated for further analysis. For each cell type, cells from each condition were pooled together and compared between each condition with the default testing method (Wilcoxon rank-sum test). The GO enrichment for the DEGs between each two groups was analyzed by ClusterProfiler 3.18.0 (PMID: 22455463), and GO terms with p values less than 10−6 were considered significant.
Calculation of the signature score of a gene set
The gene sets of FA metabolic process, inflammatory response, EMT, and ATP synthesis coupled electron transport were generated from MSigDB v.7.4 (PMID 16199517) and MGI 6.16 (PMID 30407599), the full gene list of which is listed in Table S4. The combined signature score was calculated using the AddModuleScore of Seurat.
Immunofluorescence
Kidney tissues from normal nephrectomy samples and biopsies from patients diagnosed with DKD and non-DKD (AKI, IgA nephropathy, and focal segmental glomerulosclerosis) were obtained according to the approved protocol by the Institutional Review Board for Clinical Study at Shanghai Jiao Tong University Affiliated Sixth People's Hospital and The First Affiliated Hospital of Chongqing Medical University. Paraffin-embedded mice kidney sections and human kidney nephrectomy sections were deparaffinized, antigen was retrieved by heat with citric buffer, and endogenous peroxidase was inactivated with 3% hydrogen peroxide (H2O2). Sections were then blocked with 3% bovine serum albumin (BSA) and incubated overnight at 4°C with primary antibody: anti-ATP11A (Invitrogen, catalog PA5-20995), anti-GC (Invitrogen, catalog PA5-20995), or anti-KCNK1 (Novus, NBP2-41301). The next day, sections were washed three times with PBS and then incubated with goat anti-rabbit IgG H&L (HRP) (Abcam, ab136817) and CY3-tyramide (Servicebio, G1223-50UL) for 10 min. Sections were performed at the second antigen retrieval to remove the first primary antibody, and then incubated overnight at 4°C with anti-APOC3 (Servicebio, GB112005). On the third day, sections were again washed three times and incubated with anti-rabbit IgG (Alexa Fluor 488 Conjugate) (CST, 4412S) for 1 h. Finally, kidney sections were stained with DAPI and mounted with reagent (Servicebio, G1225-8).
Immunohistochemistry
Formalin-fixed and paraffin-embedded kidney sections were deparaffinized, and endogenous peroxidase was inactivated with 3% H2O2. Then, sections were blocked by 3% BSA for 1 h and then incubated with anti-F4/80 primary antibody (Abcam, ab6640) at 4°C overnight. The next day, sections were washed three times with 1× PBS, 5 min each, and then incubated with anti-rabbit secondary antibody for 1 h at room temperature and developed using a DAB kit (Abcam). The area of F4/80 was quantified by assessing the positively stained area in 20 high power fields (40×) for each section using ImageJ software.
Quantitative real-time PCR
RNA was isolated from kidney tissues using TRIzol reagent (TaKaRa, 9109), cDNA was reverse-transcribed from 1 μg RNA by using the cDNA Master kit (Roche, 5893151001), and qRT-PCR was run in the Bio-Rad CFX96 machine using SYBR Green Master Mix (Roche, 6924204001) and gene-specific primers. The relative mRNA levels for the specific genes were normalized to the level of Gapdh mRNA expression. The sequences of used primers are shown in Table S8.
Data access
The scRNA-seq data have been deposited in the NCBI Gene Expression Omnibus database under the accession code: GSE181382.
Statistics
Data are presented as mean ± SD. The one-way ANOVA was used for comparisons between multiple groups using the GraphPad software (v.8.0). Two-sided p value less than 0.05 was considered statistically significant.
Data and materials availability
All data needed to analysis and support the conclusions in the paper are available in the main text or the supplementary materials.
Acknowledgments
Q.L. is supported by the National Natural Science Foundation of China (81970720, 81870567) and the Science and Technology Research Program of Chongqing Municipal Education Commission (KJZD-K202000401). J.C.H. is supported by NIH 1R01DK078897, NIH 1R01DK088541, VA Merit Award and NIH P01-DK56492. J.H. is supported by The National Natural Science Foundation of China (81800731). L.M. is supported by The National Natural Science Foundation of China (82000810). K.L. is supported by NIH R01 DK117913.
Author contributions
Q.L. and J.C.H. conceived the project, designed the study, and interpreted the results. J.W. performed the in vivo studies and collected the single cells. Z.S. performed the computational analyses. S.Y. performed the immunostaining imaging. J.F. performed some scRNA-seq analysis. J.H., C.P., and L.M. searched and analyzed the public scRNA-seq database. Y.F. and N.W. provided the kidney samples from non-DKD patients for immunostaining. Z.W. and K.L. provided analytical and experiment support. J.C.H., Q.L., J.W., and Z.S. wrote the manuscript with feedback from all other authors.
Declaration of interests
The authors declare no competing interests.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.ymthe.2021.10.013.
Contributor Information
John Cijiang He, Email: cijiang.he@mssm.edu.
Qifu Li, Email: liqifu@yeah.net.
Supplemental information
References
- 1.Chen T.K., Knicely D.H., Grams M.E. Chronic kidney disease diagnosis and management: a review. JAMA. 2019;322:1294–1304. doi: 10.1001/jama.2019.14745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Reutens A.T. Epidemiology of diabetic kidney disease. Med. Clin. North Am. 2013;97:1–18. doi: 10.1016/j.mcna.2012.10.001. [DOI] [PubMed] [Google Scholar]
- 3.James P.A., Oparil S., Carter B.L., Cushman W.C., Dennison-Himmelfarb C., Handler J., Lackland D.T., LeFevre M.L., MacKenzie T.D., Ogedegbe O., et al. 2014 evidence-based guideline for the management of high blood pressure in adults: report from the panel members appointed to the Eighth Joint National Committee (JNC 8) JAMA. 2014;311:507–520. doi: 10.1001/jama.2013.284427. [DOI] [PubMed] [Google Scholar]
- 4.Palmer S.C., Mavridis D., Navarese E., Craig J.C., Tonelli M., Salanti G., Wiebe N., Ruospo M., Wheeler D.C., Strippoli G.F.M. Comparative efficacy and safety of blood pressure-lowering agents in adults with diabetes and kidney disease: a network meta-analysis. The Lancet. 2015;385:2047–2056. doi: 10.1016/S0140-6736(14)62459-4. [DOI] [PubMed] [Google Scholar]
- 5.Lewis E.J., Hunsicker L.G., Clarke W.R., Berl T., Pohl M.A., Lewis J.B., Ritz E., Atkins R.C., Rohde R., Raz I. Renoprotective effect of the angiotensin-receptor antagonist irbesartan in patients with nephropathy due to type 2 diabetes. N. Engl. J. Med. 2001;345:851–860. doi: 10.1056/NEJMoa011303. [DOI] [PubMed] [Google Scholar]
- 6.Neal B., Perkovic V., Mahaffey K.W., de Zeeuw D., Fulcher G., Erondu N., Shaw W., Law G., Desai M., Matthews D.R., Group C.P.C. Canagliflozin and cardiovascular and renal events in type 2 diabetes. N. Engl. J. Med. 2017;377:644–657. doi: 10.1056/NEJMoa1611925. [DOI] [PubMed] [Google Scholar]
- 7.Wanner C., Inzucchi S.E., Lachin J.M., Fitchett D., von Eynatten M., Mattheus M., Johansen O.E., Woerle H.J., Broedl U.C., Zinman B., Investigators E.-R.O. Empagliflozin and progression of kidney disease in type 2 diabetes. N. Engl. J. Med. 2016;375:323–334. doi: 10.1056/NEJMoa1515920. [DOI] [PubMed] [Google Scholar]
- 8.Wiviott S.D., Raz I., Bonaca M.P., Mosenzon O., Kato E.T., Cahn A., Silverman M.G., Zelniker T.A., Kuder J.F., Murphy S.A., et al. Dapagliflozin and cardiovascular outcomes in type 2 diabetes. N. Engl. J. Med. 2019;380:347–357. doi: 10.1056/NEJMoa1812389. [DOI] [PubMed] [Google Scholar]
- 9.Navaneethan S.D., Zoungas S., Caramori M.L., Chan J.C.N., Heerspink H.J.L., Hurst C., Liew A., Michos E.D., Olowu W.A., Sadusky T., et al. Diabetes management in chronic kidney disease: synopsis of the 2020 KDIGO clinical practice guideline. Ann. Intern. Med. 2021;174:385–394. doi: 10.7326/M20-5938. [DOI] [PubMed] [Google Scholar]
- 10.Alicic R.Z., Neumiller J.J., Johnson E.J., Dieter B., Tuttle K.R. Sodium-glucose cotransporter 2 inhibition and diabetic kidney disease. Diabetes. 2019;68:248–257. doi: 10.2337/dbi18-0007. [DOI] [PubMed] [Google Scholar]
- 11.Heerspink H.J.L., Kosiborod M., Inzucchi S.E., Cherney D.Z.I. Renoprotective effects of sodium-glucose cotransporter-2 inhibitors. Kidney Int. 2018;94:26–39. doi: 10.1016/j.kint.2017.12.027. [DOI] [PubMed] [Google Scholar]
- 12.Wineberg Y., Bar-Lev T.H., Futorian A., Ben-Haim N., Armon L., Ickowicz D., Oriel S., Bucris E., Yehuda Y., Pode-Shakked N., et al. Single-cell RNA sequencing reveals mRNA splice isoform switching during kidney development. J. Am. Soc. Nephrol. 2020;31:2278–2291. doi: 10.1681/ASN.2019080770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Park J., Shrestha R., Qiu C., Kondo A., Huang S., Werth M., Li M., Barasch J., Suszták K. Single-cell transcriptomics of the mouse kidney reveals potential cellular targets of kidney disease. Science (New York, N.Y.) 2018;360:758–763. doi: 10.1126/science.aar2131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dhillon P., Park J., Hurtado Del Pozo C., Li L., Doke T., Huang S., Zhao J., Kang H.M., Shrestra R., Balzer M.S., et al. The nuclear receptor ESRRA protects from kidney disease by coupling metabolism and differentiation. Cell Metab. 2021;33:379–394.e8. doi: 10.1016/j.cmet.2020.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rudman-Melnick V., Adam M., Potter A., Chokshi S.M., Ma Q., Drake K.A., Schuh M.P., Kofron J.M., Devarajan P., Potter S.S. Single-cell profiling of AKI in a murine model reveals novel transcriptional signatures, profibrotic phenotype, and epithelial-to-stromal crosstalk. J. Am. Soc. Nephrol. 2020;31:2793–2814. doi: 10.1681/ASN.2020010052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fu J., Akat K.M., Sun Z., Zhang W., Schlondorff D., Liu Z., Tuschl T., Lee K., He J.C. Single-cell RNA profiling of glomerular cells shows dynamic changes in experimental diabetic kidney disease. J. Am. Soc. Nephrol. 2019;30:533–545. doi: 10.1681/ASN.2018090896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wilson P.C., Wu H., Kirita Y., Uchimura K., Ledru N., Rennke H.G., Welling P.A., Waikar S.S., Humphreys B.D. The single-cell transcriptomic landscape of early human diabetic nephropathy. Proc. Natl. Acad. Sci. U S A. 2019;116:19619–19625. doi: 10.1073/pnas.1908706116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hasegawa K., Wakino S., Simic P., Sakamaki Y., Minakuchi H., Fujimura K., Hosoya K., Komatsu M., Kaneko Y., Kanda T., et al. Renal tubular Sirt1 attenuates diabetic albuminuria by epigenetically suppressing Claudin-1 overexpression in podocytes. Nat. Med. 2013;19:1496–1504. doi: 10.1038/nm.3363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu B.C., Tang T.T., Lv L.L., Lan H.Y. Renal tubule injury: a driving force toward chronic kidney disease. Kidney Int. 2018;93:568–579. doi: 10.1016/j.kint.2017.09.033. [DOI] [PubMed] [Google Scholar]
- 20.Sinha A.D., Agarwal R. Clinical pharmacology of antihypertensive therapy for the treatment of hypertension in CKD. Clin. J. Am. Soc. Nephrol. 2019;14:757–764. doi: 10.2215/CJN.04330418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vasilopoulou E., Kolatsi-Joannou M., Lindenmeyer M.T., White K.E., Robson M.G., Cohen C.D., Sebire N.J., Riley P.R., Winyard P.J., Long D.A. Loss of endogenous thymosin beta4 accelerates glomerular disease. Kidney Int. 2016;90:1056–1070. doi: 10.1016/j.kint.2016.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hsu M.H., Savas U., Griffin K.J., Johnson E.F. Human cytochrome p450 family 4 enzymes: function, genetic variation and regulation. Drug Metab. Rev. 2007;39:515–538. doi: 10.1080/03602530701468573. [DOI] [PubMed] [Google Scholar]
- 23.Lo-Guidice J.M., Allorge D., Cauffiez C., Chevalier D., Lafitte J.J., Lhermitte M., Broly F. Genetic polymorphism of the human cytochrome P450 CYP4B1: evidence for a non-functional allelic variant. Pharmacogenetics. 2002;12:367–374. doi: 10.1097/00008571-200207000-00004. [DOI] [PubMed] [Google Scholar]
- 24.Pinho F.O., de Albuquerque D.M., Olalla Saad S.T., Costa F.F. Reduction of AHSP synthesis in hemin-induced K562 cells and EPO-induced CD34(+) cells leads to alpha-globin precipitation, impairment of normal hemoglobin production, and increased cell death. Exp. Hematol. 2008;36:265–272. doi: 10.1016/j.exphem.2007.11.003. [DOI] [PubMed] [Google Scholar]
- 25.Chen C.H., Ho Y.C., Ho H.H., Chang I.C., Kirsch K.H., Chuang Y.J., Layne M.D., Yet S.F. Cysteine-rich protein 2 alters p130Cas localization and inhibits vascular smooth muscle cell migration. Cardiovasc. Res. 2013;100:461–471. doi: 10.1093/cvr/cvt207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lin D.W., Chang I.C., Tseng A., Wu M.L., Chen C.H., Patenaude C.A., Layne M.D., Yet S.F. Transforming growth factor beta up-regulates cysteine-rich protein 2 in vascular smooth muscle cells via activating transcription factor 2. J. Biol. Chem. 2008;283:15003–15014. doi: 10.1074/jbc.M801621200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Perkovic V., Jardine M.J., Neal B., Bompoint S., Heerspink H.J.L., Charytan D.M., Edwards R., Agarwal R., Bakris G., Bull S., et al. Canagliflozin and renal outcomes in type 2 diabetes and nephropathy. N. Engl. J. Med. 2019;380:2295–2306. doi: 10.1056/NEJMoa1811744. [DOI] [PubMed] [Google Scholar]
- 28.Wu Y., Chen W., Zhang Y., Liu A., Yang C., Wang H., Zhu T., Fan Y., Yang B. Potent therapy and transcriptional profile of combined erythropoietin-derived peptide cyclic helix B surface peptide and caspase-3 siRNA against kidney ischemia/reperfusion injury in mice. J. Pharmacol. Exp. Ther. 2020;375:92–103. doi: 10.1124/jpet.120.000092. [DOI] [PubMed] [Google Scholar]
- 29.Tang Y., Yang X., Shu H., Yu Y., Pan S., Xu J., Shang Y. Bioinformatic analysis identifies potential biomarkers and therapeutic targets of septic-shock-associated acute kidney injury. Hereditas. 2021;158 doi: 10.1186/s41065-021-00176-y. 13.10.1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hu Z.-W., Chen L., Ma R.-Q., Wei F.-Q., Wen Y.-H., Zeng X.-L., Sun W., Wen W.-P. Comprehensive analysis of ferritin subunits expression and positive correlations with tumor-associated macrophages and T regulatory cells infiltration in most solid tumors. Aging (Albany NY) 2021;13:11491–11506. doi: 10.18632/aging.202841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Azushima K., Gurley S.B., Coffman T.M. Modelling diabetic nephropathy in mice. Nat. Rev. Nephrol. 2018;14:48–56. doi: 10.1038/nrneph.2017.142. [DOI] [PubMed] [Google Scholar]
- 32.Scheen A.J. Effect of SGLT2 inhibitors on the sympathetic nervous system and blood pressure. Curr. Cardiol. Rep. 2019;21:70. doi: 10.1007/s11886-019-1165-1. [DOI] [PubMed] [Google Scholar]
- 33.Sternlicht H., Bakris G.L. Blood pressure lowering and sodium-glucose co-transporter 2 inhibitors (SGLT2is): more than osmotic diuresis. Curr. Hypertens. Rep. 2019;21:12. doi: 10.1007/s11906-019-0920-4. [DOI] [PubMed] [Google Scholar]
- 34.Gilbert R.E. Proximal tubulopathy: prime mover and key therapeutic target in diabetic kidney disease. Diabetes. 2017;66:791–800. doi: 10.2337/db16-0796. [DOI] [PubMed] [Google Scholar]
- 35.Zeni L., Norden A.G.W., Cancarini G., Unwin R.J. A more tubulocentric view of diabetic kidney disease. J. Nephrol. 2017;30:701–717. doi: 10.1007/s40620-017-0423-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tylicki L., Lizakowski S., Rutkowski B. Renin-angiotensin-aldosterone system blockade for nephroprotection: current evidence and future directions. J. Nephrol. 2012;25:900–910. doi: 10.5301/jn.5000134. [DOI] [PubMed] [Google Scholar]
- 37.Simoes E.S.A.C., Flynn J.T. The renin-angiotensin-aldosterone system in 2011: role in hypertension and chronic kidney disease. Pediatr. Nephrol. 2012;27:1835–1845. doi: 10.1007/s00467-011-2002-y. [DOI] [PubMed] [Google Scholar]
- 38.Kizu A., Medici D., Kalluri R. Endothelial-mesenchymal transition as a novel mechanism for generating myofibroblasts during diabetic nephropathy. Am. J. Pathol. 2009;175:1371–1373. doi: 10.2353/ajpath.2009.090698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li J., Bertram J.F. Review: endothelial-myofibroblast transition, a new player in diabetic renal fibrosis. Nephrology (Carlton) 2010;15:507–512. doi: 10.1111/j.1440-1797.2010.01319.x. [DOI] [PubMed] [Google Scholar]
- 40.Tang P.M., Nikolic-Paterson D.J., Lan H.Y. Macrophages: versatile players in renal inflammation and fibrosis. Nat. Rev. Nephrol. 2019;15:144–158. doi: 10.1038/s41581-019-0110-2. [DOI] [PubMed] [Google Scholar]
- 41.Lake B.B., Chen S., Hoshi M., Plongthongkum N., Salamon D., Knoten A., Vijayan A., Venkatesh R., Kim E.H., Gao D., et al. A single-nucleus RNA-sequencing pipeline to decipher the molecular anatomy and pathophysiology of human kidneys. Nat. Commun. 2019;10:2832. doi: 10.1038/s41467-019-10861-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang Y., Zhao Y., Zhao Z., Li D., Nie H., Sun Y., Feng X., Zhang T., Ma Y., Nie J., et al. Single-cell RNA-seq analysis identified kidney progenitor cells from human urine. Protein Cell. 2021;12:305–312. doi: 10.1007/s13238-020-00816-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wu H., Kirita Y., Donnelly E.L., Humphreys B.D. Advantages of single-nucleus over single-cell RNA sequencing of adult kidney: rare cell types and novel cell states revealed in fibrosis. J. Am. Soc. Nephrol. 2019;30:23–32. doi: 10.1681/ASN.2018090912. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.