Abstract
Tau proteins aggregate into filaments in brain cells in Alzheimer's disease and related disorders referred to as tauopathies. Here, we used fragments of camelid heavy-chain-only antibodies (VHHs or single domain antibody fragments) targeting Tau as immuno-modulators of its pathologic seeding. A VHH issued from the screen against Tau of a synthetic phage-display library of humanized VHHs was selected for its capacity to bind Tau microtubule-binding domain, composing the core of Tau fibrils. This parent VHH was optimized to improve its biochemical properties and to act in the intra-cellular compartment, resulting in VHH Z70. VHH Z70 precisely binds the PHF6 sequence, known for its nucleation capacity, as shown by the crystal structure of the complex. VHH Z70 was more efficient than the parent VHH to inhibit in vitro Tau aggregation in heparin-induced assays. Expression of VHH Z70 in a cellular model of Tau seeding also decreased the aggregation-reporting fluorescence signal. Finally, intra-cellular expression of VHH Z70 in the brain of an established tauopathy mouse seeding model demonstrated its capacity to mitigate accumulation of pathological Tau. VHH Z70, by targeting Tau inside brain neurons, where most of the pathological Tau resides, provides an immunological tool to target the intra-cellular compartment in tauopathies.
Keywords: VHHs, Nanobodies, Protein-protein interactions, Immunotherapy, Aggregation, Seeding, Alzheimer's disease, Tauopathies, Tau protein
Graphical abstract
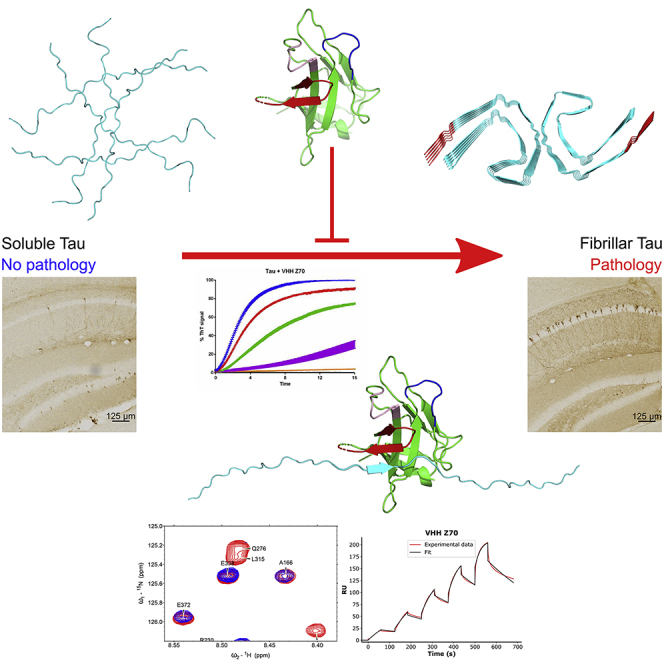
A Tau-specific single-domain antibody fragment targeting Tau aggregation nucleus, consisting of a six-residue peptide called PHF6, can prevent Tau aggregation and seeding in in vitro assays. This Tau-specific nanobody decreases the tauopathy in a mouse model of pathology seeding when expressed in brain cells, demonstrating a therapeutic potential.
Introduction
In neurodegenerative disorders, immunotherapy is currently actively explored as a disease-modifying treatment. Improving immunological tools to treat these disorders is thus a key challenge. Next to classical vaccination or the use of immunoglobulin (Ig) in passive immunotherapies, new approaches are now available. The Fv variable domains, which determine antibody specificity, can now be expressed independently of the Ig framework. These fragments—as single chain fragments of the Fv in scFvs or single domain fragments in VHHs1 (Variable Heavy-chain of the Heavy-chain-only antibodies)—can be engineered to be active intracellularly. Such immunological tools are likely to be useful given that the protein aggregates in neurodegenerative proteinopathy disorders are mostly found within neuronal cells.2 In this work, we have explored the use of VHHs in one group of these disorders, referred to as tauopathies.
Aggregation of the intrinsically disordered neuronal Tau protein to form fibrillar amyloid structures is related to these tauopathies, including the most prevalent, Alzheimer's disease (AD). AD is characterized by both extracellular amyloid deposits made of Aβ (amyloid) peptides and intra-neuronal neurofibrillary tangles (NFTs) formed by Tau protein aggregates.3 In the pathological context, Tau is the principal component of paired helical filaments (PHFs) and straight filaments,4,5 which form the intra-cellular fibrillar deposits leading to the neuropathological lesions. In addition, it has been proposed that extracellular pathological Tau species are taken up in cells, leading to intra-cellular Tau seeding and the polymerization process.6, 7, 8 Intervention strategies based on the amyloid cascade hypothesis had, up to date, limited success despite their being the primary target of clinical assays.9 In AD, the severity of cognitive decline is better correlated with the evolution of NFTs than amyloid deposits.10, 11, 12 In other tauopathies, no amyloid deposition is observed. This emphasizes the need to pursue other biological hypotheses than the amyloid cascade, including Tau-based ones, in search for disease-modifying treatments for tauopathies.
The compelling results of immunotherapies directed against Tau in several transgenic (Tg) mouse models of tauopathies, in decreasing Tau accumulation and in some cases ensuring recovery of cognitive or motor functions,13, 14, 15, 16, 17, 18, 19, 20 have motivated several pharmaceutical companies to launch clinical trials of active and passive immunization, the latter with various Tau-specific monoclonal antibodies.21,22 However, advances in the field still require deciphering key aspects of efficient Tau-specific immunotherapies and to develop their full potential to target tauopathies. The road to anti-Tau immunotherapies is opened based on the evidence of both Tau seeding capacity and Tau propagation.23, 24, 25, 26 Yet, most pathological Tau assemblies remain intra-cellular in the cytoplasm, where it is not the primary target of Tau-specific conventional immunotherapies using Ig. In addition, the extracellular Tau could, at least partly, remain unattainable to Tau-specific antibodies, as would be the case for Tau in extracellular vesicles27,28 or nanotubes.29 Finally, the propagation pattern related to extracellular Tau, clearly defined in AD, could also be less relevant in pure and more acute tauopathies, for which the time frame restrains the propagation.25 In these latter cases, the rational to target extracellular Tau is weaker.
Further exploring the capacity to use immunotherapies targeting the intra-neuronal Tau has thus become an important challenge. With that in mind, we have chosen VHHs, commonly called nanobodies, to target original epitopes of Tau, and to optimize their intra-cellular activity. VHHs consist of a unique heavy chain that corresponds to the variable heavy chain from Camelidae Igs.1 The interest of using the VHHs instead of the classical antibodies stand in their easy generation, from a synthetic library, involving no animal handling, their selection using phage display, their production in periplasm of bacteria, as well as the multiple possibilities offered by modification using protein engineering.30 They can be modified to penetrate into the cytoplasm of cells,31,32 or to be expressed inside the cells,33 and bind specifically to their target epitope. In addition, VHHs showed their potential as diagnostic tools to target NFTs with an affinity and specificity very close to antibodies already used for detecting these pathologic features by immunochemistry, opening the way for new probes in in vivo imaging experiments.34 As demonstrated with scFvs,35,36 we here showed that it is also possible to select, to optimize, and to consider VHHs as therapeutic tools in tauopathies.
Besides the Tau location targeted by the VHHs, the epitope recognition is another parameter of crucial interest to mitigate the seeding and polymerization of the intra-cellular Tau. Tau can be divided into four domains comprising the N-terminal domain (N1-N2), the proline-rich domain (P1-P2), the microtubule-binding domain (MTBD) constituted itself of four partially repeated regions, R1 to R4, and the C-terminal domain (Figure 1B scheme). Two homologous hexapeptides named PHF6∗ (275VQIINK280) and PHF6 (306VQIVYK311), located respectively at the beginning of R2 and R3 repeat regions (Figure 1B scheme) of Tau MTBD, are nuclei of Tau aggregation.37
Figure 1.
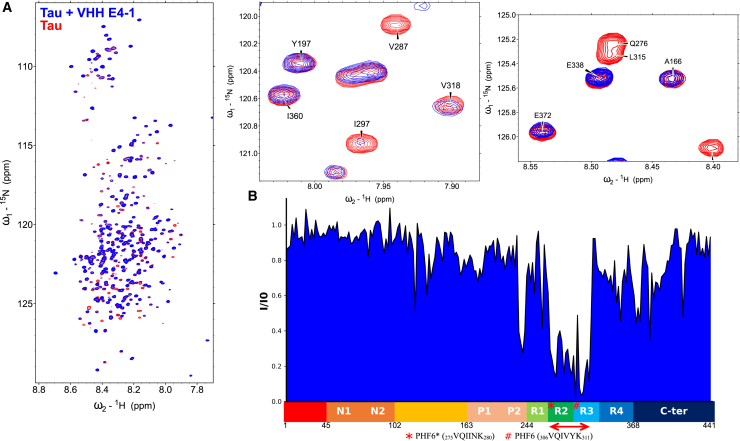
VHH E4-1 binds to the MTBD of Tau
(A) Overlay of 1H, 15N HSQC two-dimensional full spectra and enlargements of free full-length Tau2N4R (in red) or Tau2N4R mixed with non-labeled VHH E4-1 (in blue) (n = 1). In the spectrum of Tau in the presence of VHH E4-1, multiple resonances are broadened beyond detection compared with the Tau control spectrum. (B) Normalized NMR intensities (I/I0) along the Tau sequence with (I0) and (I) corresponding to the resonance intensity when Tau is free in solution or mixed with equimolar quantity of VHH E4-1 (I), respectively. The normalized intensity ratios (I/I0) plot allowed the identification of the Tau MTBD domain as the target of VHH E4-1 interaction. A red double-arrow indicates the region containing the corresponding major broadened resonances, which was mapped to the R2-R3 repeats in the MTBD. N1 and N2 are two inserted sequences in the N-terminal domain (1–163) that are not present in all Tau isoforms (named Tau 0N, Tau 1N or Tau 2N), the proline-rich domain is subdivided in P1 and P2 regions, the MTBD consists of four partially repeated regions, R1 to R4 (in Tau 4R). The R2 repeat is not present in Tau 3R. C-ter is for the C-terminal domain.
Taking that into consideration, a Tau-specific VHH targeting the MTBD was selected from a screen of a humanized naive synthetic library initially performed to obtain VHHs targeting the different Tau domains. In addition, we optimized this parent VHH by a yeast two-hybrid approach to obtain VHH Z70, which efficiently binds Tau once expressed in the intra-cellular environment. VHH Z70 decreased Tau fibrils assembly in vitro and in HEK293 seeding-reporting cellular model. After transduction by lentiviral vectors (LVs) in the hippocampal formation of a tauopathy mouse model, the expressed VHH Z70 significantly reduced Tau seeding induced by human AD brain homogenates. Z70 antibody fragment thus provides an immunological tool to target Tau in cells and to open the way for future gene therapy.
Results
Identification of synthetic VHHs directed against Tau microtubule-binding domain
To generate original VHHs targeting various Tau domains, 20 clones resulting from the screen of a synthetic phage-display library of humanized llama single-domain antibody38 against recombinant Tau protein (Tau 2N4R longest isoform) were initially selected for further analysis. Definition of the region recognized by each of these VHHs was a first step in assessing their properties. Resonance perturbation mapping in 1H, 15N HSQC spectra of 15N-Tau, obtained by nuclear magnetic resonance (NMR) spectroscopy, allowed defining of the various binding regions of the VHHs along Tau sequence. Comparison of the spectra of Tau alone in solution or in the presence of a VHH identified the spectral perturbations that are used to define the binding region. The initial screening of the different clones showed recognition of five different regions of Tau from the proline-rich region to the C-terminus (Figures 1 and S1). We have previously described the series of Tau-specific VHHs targeting the C-terminus of Tau, and parent VHH F8-2, which shows interesting properties as a molecular tool to detect Tau in research experiments. Nevertheless, F8-2 has no capacity to interfere with Tau seeding and polymerization.39
Interestingly, one VHH, named VHH E4-1, affected resonances in the Tau spectrum corresponding to residues within the R2-R3 repeats of the MTBD (Figure 1). The binding mapping was confirmed using a Tau fragment that corresponded to the isolated MTBD. The smaller size of this Tau fragment (124 amino acid residues instead of 441) resulted in fewer resonances and less resonance overlap in the corresponding Tau[244–368] 1H, 15N spectrum, facilitating the identification of the binding region (Figures S2 and S3). The affected resonances corresponded to amino acid residues located in a stretch expanding from residue V275 to K317, including the two aggregation nuclei PHF6∗ and PHF6 (Figure S3B). VHH E4-1 thus binds within the R2-R3 repeats of the MTBD.
Optimization of parent VHH E4-1 into variant VHH Z70
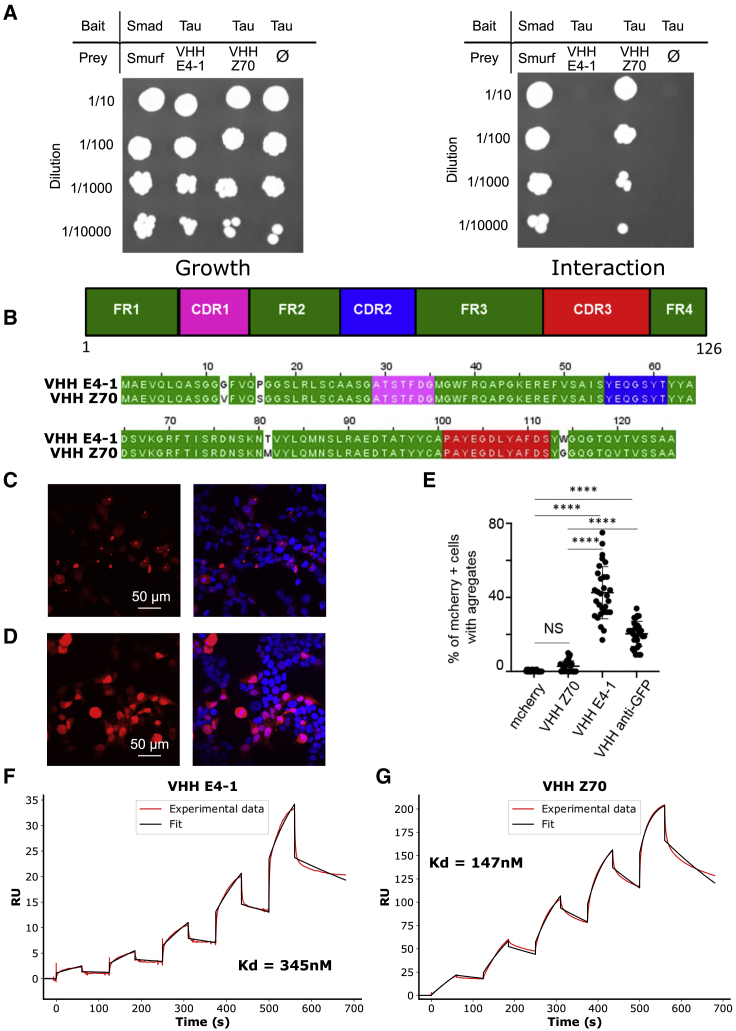
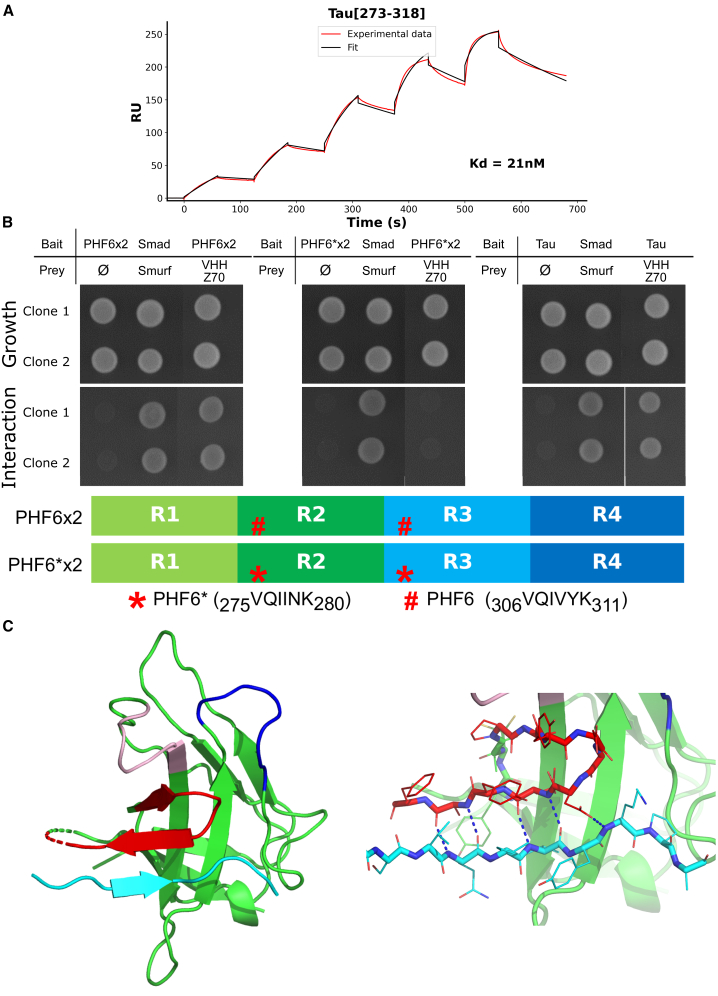
An important property of the VHHs is their capacity to be expressed and to recognize their target in the cellular environment. However, some VHHs might be ineffective in binding once expressed in a cell due to their improper folding leading to aggregation or instability.40 For instance, VHH E4-1 did not bind Tau in yeast two-hybrid assays that require the interaction to take place in the yeast nucleus, providing an evaluation of VHH E4-1 intra-cellular binding capacity41 (Figure 2A). VHH E4-1 was thus next submitted to an original round of optimization, using the yeast two-hybrid system, to maximize its capacity to recognize its target when expressed in a cellular environment. First, we built a cDNA mutant library by random mutagenesis, targeting the whole sequence of VHH E4-1 to produce a variety of VHH preys (VHH-Gal4-activation domain fusion) against the Tau bait (LexA-Tau fusion). The library of 1.5 million variants was transformed in yeast and screen by cell-to-cell mating to get positive colonies under the pressure of selection conditions corresponding to undetected VHH E4-1-Tau interaction (Figure 2A). A single optimized variant named VHH Z70 was obtained, resulting from 4 mutations G12V, P16S, T81M and W114G located in the framework domains (FR) (Figure 2B). Fluorescence imaging of HEK cells expressing mCherry-VHH E4-1 showed focal inclusion bodies illustrated by the detection of fluorescent puncta aggregates (Figures 2C and S4). In contrast, most cells transfected with mCherry-VHH Z70 construct clearly showed the intracellular solubility of VHH Z70 because a homogeneous strong fluorescence signal filled the cells (Figures 2D and S4). The optimization of parent VHH E4-1 for intracellular applications thus succeeded in providing VHH Z70 that had significantly better stability in cells (Figure 2E). Location of the four stabilizing mutations outside the recognition loops (CDR) (Figure 2B) suggests that the epitope recognized by the VHH Z70 mutant is unaltered. Conservation of the binding region was confirmed by NMR perturbation mapping, using labeled Tau and MTBD in the same manner as for the parent VHH E4-1 (Figures S5 and S6). Interactions of VHH E4-1 and VHH Z70 with full-length Tau2N4R were further characterized using surface plasmon resonance spectroscopy (SPR) with biotinylated-Tau immobilized at the surface of a streptavidin-functionalized chip. The assay provided the kinetic parameters of the interactions, characterized by dissociation constants Kd of 345 nM for VHH E4-1 (Figure 2F) and Kd of 147 nM for variant VHH Z70 (Figure 2G). VHH Z70, optimized for intra-cellular activity, had a better affinity for its target than its parent VHH E4-1, the major optimization concerning the association constant (kon) (Figure S7). SPR was additionally performed with biotinylated VHH Z70 immobilized on the chips. A Tau peptide [273–318] corresponding to the NMR-identified VHH binding site, fused to a SUMO domain to improve its solubility, was injected into the flux. VHH Z70 interacted with the fused peptide with a Kd of 21 nM (Figures 3A and S7), confirming that this peptide in Tau sequence was self-sufficient for VHH-Z70 binding.
Figure 2.
VHH Z70 is optimized for intra-cellular binding and has a better affinity for Tau than VHH E4-1
(A) Results from yeast two-hybrid. A growth test on nonselective medium (left panel, lacking only leucine and tryptophan) or on selective medium (right panel, lacking leucine, tryptophan and histidine) was performed with dilution (top to bottom) of the diploid yeast culture expressing both bait and prey constructs. Positive and negative controls of interaction consist respectively of Smad/Smurf interaction42 and Tau alone (empty vector). VHH E4-1 did not interact with Tau in yeast (no growth on selective medium) whereas VHH Z70 did. (B) Domain organization of the VHHs (CDR are for complementarity-determining regions and FR for framework regions) and sequence alignment between VHH E4-1 and VHH Z70 showing four mutations in the FR: G12V, P16S, T81M, and W114G. (C, D) HEK293 cells were transfected with plasmid encoding either (C) mCherry-VHH E4.1 or (D) mCherry-VHH Z70 and mCherry or mCherry VHH anti-GFP (Figure S4). mCherry is visualized in red and nuclei in blue. The scale bar is indicated on the figure. (E) Percentage of mCherry positive cells with puncta is provided for 10 images per group and three independent experiments (30 points). Error bars indicate mean and SD of the data. ∗∗∗∗ correspond to a p value < 0.0001. (F, G) Sensorgrams (reference subtracted data) of single cycle kinetics analysis performed on immobilized biotinylated Tau, with five injections of (F) VHH E4-1 or (G) VHH Z70 at 0.125 μM, 0.25 μM, 0.5 μM, 1 μM, and 2 μM (n = 1). Dissociation equilibrium constant Kd were calculated from the ratio of off-rate and on-rate kinetic constants koff/kon. kon, koff, and Kd are included in the table in Figure S7. Black lines correspond to the fitted curves, red lines to the measurements.
Figure 3.
The PHF6 peptide sequence is essential for VHH Z70 binding to Tau MTBD
(A) Sensorgram (reference subtracted data) of single-cycle kinetics analysis performed on immobilized biotinylated VHH Z70, with five injections of peptide Tau[273–318] fused at its N-terminus with the SUMO protein (n = 1) at 31.25 nM, 62.5 nM, 125 nM, 250 nM, and 500 nM. Tau peptide sequence and, kon, koff, and Kd are included in the table in Figure S7. Black lines correspond to the fitted curves, red lines to the measurements. (B) Results from yeast two-hybrid. The VHH are expressed as preys, with a C-terminal Gal4-activation domain fusion (VHH-Gal4AD) and Tau0N4R/MTBD as bait with a C-terminal fusion with lexA (Tau0N4R/MTBD-LexA). A growth test on nonselective medium (upper panel Growth, lacking only leucine and tryptophane) or on selective medium (lower panel Interaction, lacking leucine, tryptophane, and histidine) was performed of the diploid yeast culture expressing both bait and prey constructs. Positive and negative controls of interaction consist respectively in Smad/Smurf interaction and Tau or chimeric MTBD alone (empty vector). (C) Ribbon representation of the crystal structure of the complex between VHH Z70 and the PHF6 peptide. The CDR1, CDR2, and CDR3 loops of VHH 70 are colored in pink, in dark blue, and in red, respectively. Framework regions of the VHH are represented in green and the PHF6 Tau peptide in cyan. The right panel shows the five intermolecular H-bonds (dashed blue lines) between VHH Z70 and Tau peptide. See Figure S11 for 90°-rotation view and surface representation.
Identification of the minimal tau epitope recognized by VHH Z70
To determine the minimal sequence that VHH Z70 binds within the R2-R3 Tau repeats, an epitope mapping was performed using yeast two-hybrid (267 × 103 tested interactions) with VHH Z70 as bait (LexA-VHH fusion) against a library of Tau fragments as preys (GAL4 activation domain-Tau fragments fusion). Ninety positive clones were selected for their growth in the selection conditions, evidencing binding of VHH Z70 to Tau fragments of various length, from a small-scale cell-to-cell mating screen. Comparison of the Tau prey fragment sequences corresponding to these 90 interactions identified peptide 305SVQIVYKPV313 as the minimal common recognition motif of Tau that VHH Z70 can bind (Figure S8). The sequence is localized in the R3 repeat of the MTBD domain and contains the PHF6 peptide 306VQIVYK311. We next used Tau2N3R isoform that lacks the R2 repeat and so does not contain the PHF6∗ peptide, to confirm that the R3 repeat that contains the PHF6 peptide was sufficient for the interaction. As observed in the NMR intensity profile, the interaction of VHH Z70 with Tau2N3R is maintained, and the most affected resonances in the Tau NMR spectrum corresponded to the PHF6 residues in the R3 repeats (Figure S9), confirming that PHF6∗ is not necessary for VHH Z70 binding to Tau. In the yeast two-hybrid system, we next used chimeric constructs of the MTBD that have been modified to display two PHF6 or two PHF6∗ instead of the wild-type PHF6 and PHF6∗ (Figures 3B and S10A). VHH Z70 did not interact with chimeric MTBD with two PHF6∗ (PHF6∗x2) in yeast (no growth on selective medium), whereas it did interact with Tau and chimeric MTBD with two PHF6 (PHF6x2) (Figure 3B). VHH Z70 was thus not able to bind the double PHF6∗ construct, in conditions corresponding to positive binding to Tau or to the double PHF6 chimeric MTBD. SPR was additionally performed with biotinylated VHH Z70 immobilized on the chip while the MTBD or each of these chimeric constructs was injected in the flux. VHH Z70 interacted with the MTBD or the chimeric MTBD with two PHF6 with Kd of 146 nM and 34 nM, respectively (Figures S10B and S10C), while interaction with the PHF6∗ is characterized by a Kd of 398 nM (Figure S10D). The rate of dissociation (koff) was the major parameter that explained the Kd differences (Figure S7). In conclusion, the epitope of the VHH Z70 was defined as the PHF6 sequence by several concurring approaches and in in vitro conditions, residual interaction of VHH Z70 is detected with the PHF6∗ sequence.
Structure of VHH Z70 in complex with a PHF6 peptide
To obtain further atomic details on the PHF6 recognition by VHH Z70, the structure of the complex between VHH Z70 and a Tau[301–312] peptide, including the PHF6 sequence, was solved by X-ray crystallography at a resolution of 1.7 Å. Structure of this complex clearly demonstrates the direct interaction of residues from the CDR3 loop with each residue of the PHF6 sequence. The complex assembly is characterized by the formation of a three-stranded β-sheet formed by two strands from the CDR3 folded in a β hairpin and one strand formed by the PHF6 sequence that adopts an elongated conformation (Figures 3C and S11A; Video S1). CDR1 and CDR2 loops are not participating directly in the interaction but we cannot exclude that they might be involved in the interaction with full-length Tau. A mean interface area of 480 Å2 (PDBePISA) was calculated from the 526 Å2 of buried accessible surface area of the peptide (38% of total peptide surface, residues 305 to 312) and the 434 Å2 of buried accessible surface area from the VHH (7% of total, residues 39, 47–49, 63–65, 68, 104–110) (Figure S11B). Residues 111 to 115 of VHH Z70 were not resolved, indicating a high degree of flexibility of this region corresponding to the last two residues of CDR3 and the first three residues of FR4. One of the mutations (W114G) that distinguished VHH E4-1 from VHH Z70 was in this segment, indicating that the observed flexibility might be important for CDR3 positioning. The interaction with the CDR3 is stabilized by formation of five intermolecular hydrogen bonds (Figure S11C) involving atoms of the backbone of PHF6 peptide, residues S305 to P312, through a β-augmentation mechanism (Figure 3C). Structure of the complex confirmed that the S305 to P312 sequence was sufficient for VHH Z70 binding, although binding might also be partly context-dependent and optimal when the recognition segment is embedded in the full-length protein (or at least MTBD).
Inhibition of in vitro Tau aggregation
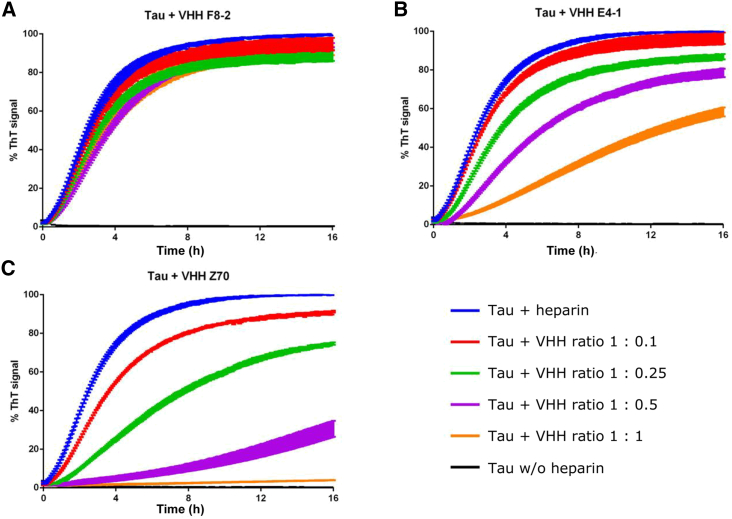
VHH E4-1 and VHH Z70 recognizing Tau peptide PHF6, which is known to nucleate the aggregation process and to form the core of Tau fibers, were assayed for their capacity to interfere with Tau in vitro polymerization. The assays were carried out with recombinant Tau protein in the presence of heparin, using thioflavin T as a dye whose fluorescence is increased in the presence of aggregates (Figure 4). Negative and positive controls consisted of Tau without or with heparin, respectively. An additional control was performed in the presence of VHH F8-2, a VHH issued from the initial phage-library screen (Figure S1), which targets Tau C-terminal domain.39 At 10 μM of Tau, the observed amount of aggregates was maximal (defined as 100%) for the positive control (Tau with heparin, blue line) after 8 h of incubation at 37°C, while no fluorescence change was detected for the negative control (Tau without heparin, black line) (Figure 4). At equimolar concentration of Tau:VHH F8-2, the fluorescence signal reached 91.2% (±3.8%), showing that VHH F8-2 did not affect the aggregation of Tau (Figure 4A, orange line). In contrast, at a molar ratio of 1:0.25 Tau:VHH E4-1, the maximal fluorescence signal reached 86.9% (±2.4%). In addition, about 3.8 h were needed to gain 50% of maximal signal, compared with 2.5 h for the positive control, showing a slower aggregation kinetic in the presence of VHH E4-1 (Figure 4B, green line). At a 1:1 Tau:VHH E4-1 molar ratio, the fluorescence signal only reached 58.3% (±3.9%) and more than 12.8 h were necessary to gain 50% of maximal signal (Figure 4B, orange line). VHH Z70 had an even stronger inhibition effect on the aggregation of Tau than the parent VHH E4-1. At a 1:1 Tau:VHH Z70 molar ratio, the maximal fluorescence signal barely reached above the negative control level, at 4.1% (±0.1%) (Figure 4C, orange line). The link between the thioflavin T fluorescence measurements in our assays and the formation of Tau fibrils at the endpoint of each aggregation assay was confirmed by transmission electron microscopy imaging (Figure S12). Whereas no fibrils were detected without heparin (Figure S12A, negative control), large amounts were observed for Tau in the presence of heparin only (Figure S12B, positive control) or in the additional presence of VHH F8-2 (Figure S12C). Shorter filaments were detected with VHH E4-1 (Figure S12D) and practically none with VHH Z70 (Figure S12E). In conclusion, parent VHH E4-1 and its optimized variant VHH Z70 have both the capacity to interfere with Tau fibrils assembly in vitro.
Figure 4.
VHH E4-1 and VHH Z70 inhibit in vitro Tau aggregation
Aggregation of Tau (10 μM) in the absence of heparin (black curve), in the presence of heparin and of increasing concentration of (A) VHH F8-2, (B) VHH E4-1, and (C) VHH Z70 (0, 1, 2.5, 5, and 10 μM) monitored by Thioflavin T fluorescence at 490 nm (n = 3). Error bars: SEM.
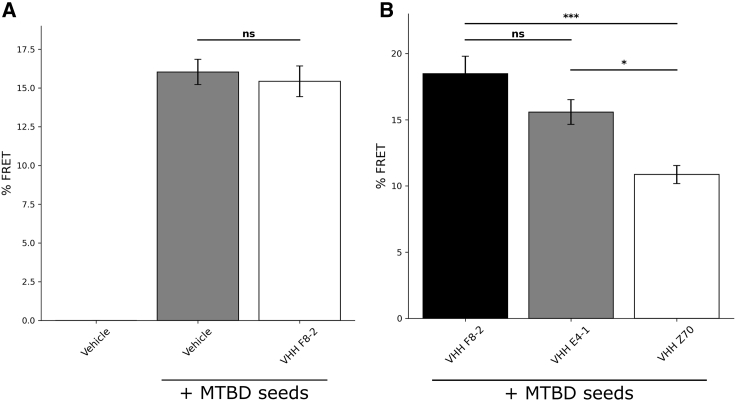
Inhibition of Tau seeding in FRET biosensor reporter cells
The capacity of VHH Z70 and its parent VHH E4-1 to block Tau seeding by using the HEK293 Tau RD P301S FRET Biosensor reporter cell line model was next investigated. This cell line constitutively expresses Tau RD (MTBD), with a P301S mutation, fused to either CFP (Cyan Fluorescent Protein) or YFP (Yellow Fluorescent Protein) that together generate an FRET (Förster Resonance Energy Transfer) signal upon MTBD-P301S polymerization.43 The MTBD-P301S contains the PHF6 peptide 306VQIVYK311 identified as the recognition sequence of VHH-Z70.43 In basal conditions, FRET signal is not detected by flow cytometry (Figure 5A, vehicle). The intra-cellular polymerization of MTBD-P301S protein is induced by treating the cells with Tau seeds (heparin-induced MTBD fibrils),43 leading to an FRET signal (16.0% ± 0.8% FRET-gated positive cells, Figure 5A, gray column). In addition, VHH F8-2 was transfected 1 day prior to MTBD seed treatment as negative control since its binding is outside the MTBD. As expected, VHH F8-2 did not affect the seeding in the reporter cells (15.4% ± 1.0% FRET-gated positive cells, Figure 5A, white column). This negative control also showed that the mCherry fluorophore did not interfere with the seeding, polymerization, and/or FRET signal. We next used the mCherry-VHH fusion proteins to select the transfected cells and to detect FRET signal selectively in mCherry-VHH positive cells (mCherry-gated and FRET-gated positive cells Figure 5B). As expected, given that VHH E4-1 did not bind Tau in cells (Figure 2A), the FRET signal reduction for VHH E4-1 transfected cells was not significant, with a percentage of FRET decreasing to 15.6% ± 0.9%, compared with 18.5% ± 1.3% FRET signal for the VHH F8-2 negative control (Figure 5B). In contrast, VHH Z70 clearly affected the intra-cellular seeding of MTBD-P301S polymerization, as the observed FRET signal for the corresponding transfected cells was significantly decreased to 10.9% ± 0.7% (41% seeding inhibition, p value = 0.0003, Figure 5B). Based on these measurements, we concluded that VHH Z70 has reduced the association of MTBD-P301S CFP and YFP by 40%, showing the efficiency of VHH Z70 to inhibit Tau seeding in this cellular model.
Figure 5.
VHH Z70 blocks intra-cellular seeding of Tau MTBD in HEK293 biosensor cells
(A and B) Analysis of Tau seeding in HEK293 Tau RD P301S FRET Biosensor cells. (A) Percentage of FRET positive cells in the whole population determined from flow cytometry data for cells transfected with vehicle or transfected with vehicle and VHH F8-2 followed by MTBD seeds (n = 3). (B) Percentage of FRET positive cells in the mCherry-gated population determined from flow cytometry data for cells transfected with VHH F8-2, VHH E4-1, and VHH Z70 followed by MTBD seeds (n = 3). A significant decrease of FRET signal, reporting a decrease intra-cellular MTBD aggregation, is observed in the presence of VHH Z70. ∗p value <0.05, ∗∗∗p value < 0.001, ns, not significant. Error bars: SEM.
Inhibition of Tau seeding in the Tg tauopathy mouse model THY-tau 30
The next step was to test the capacity of VHH Z70 to block Tau seeding in a well-characterized model.19 This model consists in the intra-cranial injection of human AD (h-AD) brain-derived materials to robustly induce Tau pathology on a relatively short timescale (1 month), in young Tg THY-tau30 mice (1 month old).19,44 At 1 month old, these mice have low endogenous Tau pathology, which allowed evaluation of the seeding activity associated to the injected human-derived materials. To assay the capacity of VHH Z70 to mitigate the tauopathy in this model, VHH Z70 was expressed as a fusion protein with mCherry inside brain cells, following infection by LVs. LVs were used for their limited diffusion capacity, allowing for evaluation of the local effect of VHH Z70 on Tau seeding. LVs infection of HEK293 cells showed expression of VHH Z70 in the cell fraction, with no detection in the medium, confirming that Z70 VHHs are well-expressed and are not secreted (Figure S13).
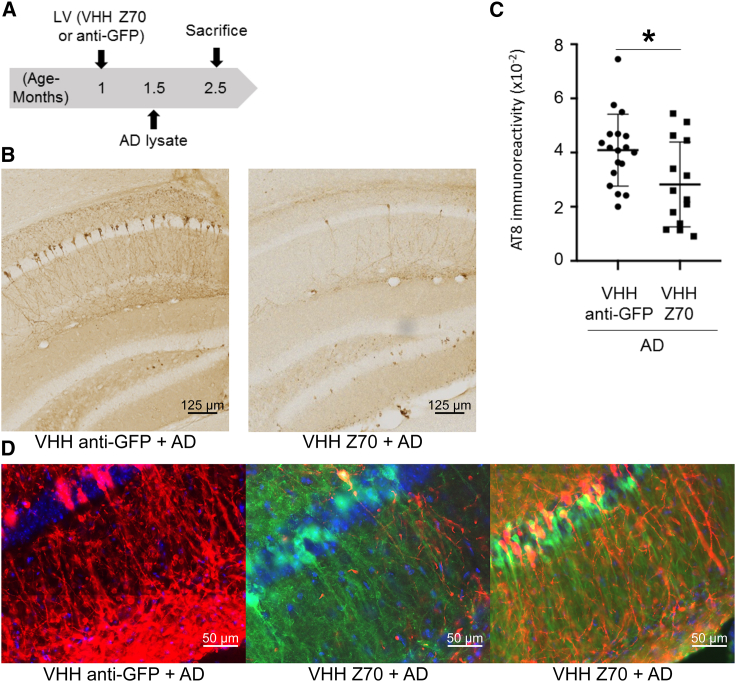
LVs were delivered using intra-cranial injection in each brain hemisphere, 2 weeks prior to the stereotaxic h-AD seed delivery at the same coordinates in the hippocampus region (Figure 6A). Negative control experiments consisted of injection of a VHH directed against GFP (VHH anti-GFP), which is not present in mouse brains, instead of VHH Z70. The expression of VHH Z70 in the brain was monitored in both hemispheres using the mCherry fusion as a reporter (Figure S14). The level of Tau pathology was evaluated, 1 month post-injection of the seeds, by immunohistochemistry with the monoclonal AT8 antibody.10,45 In the animals injected with h-AD brain extract and expressing VHH Z70, a significant decrease of the AT8 labeling compared with the negative control group is observed (Figures 6B and 6C, p value = 0.019, 6D and S15). The decrease detected in the area where the LVs expressing VHH Z70 had diffused and the pathology had spread following injections showed the positive effect of VHH Z70 on mitigating the seeded pathology. Accordingly, when brain tissue sections were double-stained, almost no co-localization of signals was observed in neurons: most neurons expressing VHH Z70 (mCherry positive) had no Tau pathology as defined based on the AT8 signal (Figures 6D and S16). We concluded that intra-cellular immunization with VHH Z70 slows the seeding induced by injection of extracellular h-AD brain extract in THY-tau30.
Figure 6.
VHH Z70 reduces human Tau seeding activity induced by h-AD brain-derived material in THY-tau30 Tg mouse model
(A) Bilateral intra-cranial injection of LVs to express either VHH anti-GFP or VHH Z70 was performed in 1-month-old THY-tau30 mice. Induction of the tauopathy occurred by a second bilateral injection, at the same coordinates, 15 days after with human h-AD. Mice were killed and perfused 1 month later (aged 2.5 months). (B) The whole brain was processed for immunohistochemical analysis using AT8 for the control group (labeled VHH anti-GFP + AD; n = 9) corresponding to injection of LVs VHH anti-GFP followed by h-AD injection and the group (labeled VHH Z70 + AD; n = 7) corresponding to LVs VHH Z70 followed by h-AD injection. Enlarged images are taken from the hippocampus (injection site, AP: −2.46). Scale bars are indicated on the figure. (C) Each data point corresponds to the quantification for one hemisphere. Results are presented as AT8 immunoreactivity (immunoreactive area normalized to the whole area). ∗p value <0.05. Error bars: SD. (D) In a region of the hippocampus (molecular layer) with strong induction of the pathology (detected using AT-8 antibodies, visualized in red, see VHH anti-GFP + AD) and good diffusion of LVs expressing Z70 (detected using mCherry antibodies, visualized in green, see VHH Z70 + AD), there is almost no Tau pathology (red labeling) in neurons positive for VHH-Z70 expression (green labeling). Immunohistochemical analysis is shown for two animals (VHH Z70 + AD) with different levels of pathology. Larger views of the analysis are shown in Figure S16.
Discussion
Tau immunotherapy is an attractive strategy in tauopathies to bind to and to clear extracellular and/or intra-cellular pathological species of the Tau protein to slow disease progression. Indeed, by targeting different Tau epitopes, immunotherapy studies showed a reduction of Tau pathology and cognitive deficit in different mouse models of tauopathy.13, 14, 15, 16, 17, 18, 19, 20 Nevertheless, the mechanistic detail of their mode of action is only recently emerging.
Indeed, it remains difficult to evaluate whether detection of an intra-cellular Ig results from uptake of the Ig by itself or of the immune complex, and thus whether their primary target is Tau in the intra- or extracellular compartment. One study with systematic comparison of intra-neuronal versus extraneuronal-acting equivalent Tau-specific scFvs showed a significant improved effect of the intra-neuronal modified scFvs against the tauopathy, in two tauopathy mouse models.36 Intra-neuronal chimeric scFvs are another successful example, designed to target Tau to the proteasome or lysosomal pathway in human Tau Tg mice.46
We succeeded in selecting a VHH targeted to a specific Tau region in a rather large intrinsically disordered protein, in one single screen. NMR interaction mapping was particularly helpful to select for VHHs binding the MTBD (Figures 1 and S1). Yeast two-hybrid combined with molecular strategies has allowed for the identification of a major binding site 306VQIVYK311 (Figures 3 and S7). In addition, a weaker secondary interaction was detected with PHF6∗ (275VQIINK280) when using sensitive in vitro methods (NMR in Figures 5 and 6). This secondary binding site might contribute to the overall VHH Z70 activity by slowing down dissociation, and by doing so, increasing the apparent affinity for Tau.
The main interest is that VHH Z70 binds in the MTBD, while the majority of epitopes targeted in clinical trial are located in the N- or C-terminal regions.21 Some studies show that targeting N-terminal species could block Tau uptake and its transfer between neurons,47 and could reduce accumulation of Tau in the brain of the mouse model of tauopathy.48 Nevertheless, antibodies targeting N- and C-terminal epitopes bring the risk of binding Tau sequences eliminated by proteolysis.49 They are also unlikely to interfere with seeding and polymerization, other crucial aspects of the pathology. Two phase II trials of passive immunotherapies with monoclonal antibodies targeting the N-terminal part of Tau in supranuclear palsy, gosuranemab50 and tilavonemab,51 reported no clinical treatment effect despite target engagement. Both assay rationales were the removal of the extracellular Tau to slow progression of the disease. Although multiple parameters could explain these results,52 the N-terminal part of Tau remains unlikely to be the mediator of Tau toxicity.22 Conversely, bepranemab, a humanized monoclonal IgG4 antibody binding to the central region of Tau (235–246), near the MTBD, was reported to inhibit seeding in a cellular assay and the THY-tau30 mouse model.19,53 Similarly, treatment with a humanized IgG antibody, binding the R2 and R4 repeats within the MTBD, decreases the level of sarkosyl-insoluble Tau in brain of a mouse model of Tau seeding.54 These antibodies are being proposed for clinical development and their capacity to attenuate the spread of Tau supports the hypothesis that the MTBD provides an efficient target for therapeutic antibodies. VHH Z70 has similar properties but by acting directly intra-cellularly.
Here, we demonstrated the interest of targeting the Tau PHF6 motif, which participates in the aggregation process37 and which is found in the core of Tau fibrils in AD.55, 56, 57, 58 PHF6∗ and PHF6 peptides spontaneously aggregate in solution contrary to the full-length Tau that is a highly soluble protein. Their atomic structures reveal the capacity of these segments to form interdigitated steric-zipper interfaces that seed Tau aggregation.59,60 In addition, the accessibility of the PHF6∗/PHF6 peptides is proposed to be part of the mechanism leading to Tau filament formation. The residues from these peptides are proposed to be shielded in a dynamic hairpin conformation in native Tau, while exposed PHF6 residues would increase Tau sensitivity to aggregation.61 The conversion from an inert Tau monomer to a seed-competent monomer would thus involve an increased accessibility of the PHF motifs.61 Interestingly, several chaperones with Tau anti-aggregation activities, such as Hsc70, Hsp60, DnaJA2, and S100B62 proteins, bind to regions overlapping PHF6, and in a weaker manner PHF6∗. Thus, one major mechanism of the anti-aggregation activities of these chaperones is likely the binding of Tau in the PHF region.63
The cryo-electron microscopy structures of Tau fibers isolated from patient brains affected by various tauopathies: AD,55 corticobasal degeneration,56 Pick's disease,57 chronic traumatic encephalopathy,58 and progressive supranuclear palsy,64 show distinct folds. The common core of these fibrillary structures is nevertheless composed of the subdomains R3 including the PHF6, R4, and a part of the C-terminal domain (V306-F378) that mainly form a β-sheet unit.55 This filament core has the ability to polymerize and form filaments in vitro, and act as seeds to recruit full-length Tau to filaments, in the absence of an inducer such as heparin.65 To form these fibers, a mechanism of β-augmentation is responsible for the stacking of the β-sheet units on top of one another to ensure elongation of the fiber. The PHF6 sequence is thus forming in this process the same type of interactions as those observed with the VHH Z70 (Figure S17). Although it will remain to be demonstrated, the specific β-strand conformation adopted by the PHF6 sequence in the complex suggests that VHH Z70 might not be able to interact with the fibers, except at the free extremity. However, by interacting with the PHF6 at the extremity of Tau oligomers or fibers, VHH Z70 might interfere with the elongation mechanism by competing with the β-sheet augmentation.
In addition, the properties of the parent VHH (E4-1) were improved. VHH Z70 has indeed been selected for its capacity to bind Tau in the intra-cellular compartment, using yeast two-hybrid (Figure 2A). The four mutations differentiating VHH Z70 from the E4-1 are in the framework region and allow the active conformation to be achieved once expressed in cells. This can result either because the cysteine residues are well positioned to form disulfide bridges, despite the reductive environment, or because the mutations allow a disulfide-independent folding. VHH Z70 indeed showed improved solubility when expressed in cells compared with VHH E4-1 that formed aggregates, observed in the experimental conditions as puncta of mCherry fluorescence (Figures 2C–2E and S4). Although we succeeded, the process remains challenging, as a single clone was selected out of the 1.5 million variants screened by two-hybrid. In addition, the affinity reached the 100-nM range and remained an important parameter to optimize as we observed a higher inhibition activity for VHH Z70 compared with VHH-E4-1 in the in vitro aggregation assay (Figure 4). The heparin-induced Tau fibers that are formed in this assay are heterogeneous and have been shown to differ from the structures of the human native fibers.66 Nevertheless, even if not recapitulating all the structural features, the heparin-induced fibers still contain the R3 repeat at the core of a β-sheet unit and fibers are formed by stacking these β-sheet units on one another. Consequently, although the model has limitations, the in vitro heparin-induced aggregation remains of interest when targeting the PHF6 sequence as its key-role in nucleation is conserved in this assay. In addition, VHH Z70 was shown to inhibit seeding in an established cellular model that does not use heparin as inducer of the aggregation. The poor seeding inhibition capacity of VHH-E4-1 in this model (Figure 5) is likely due to its poor intra-cellular activity compared with VHH Z70 (Figures 2A–2C).
Importantly, VHH Z70 decreases the Tau pathology in an established mouse model of tauopathy19 (Figure 6). In this injection model, VHH Z70 mechanism of action likely results from blocking Tau seeding, limiting the accumulation of pathological Tau. Interestingly, the hippocampal neurons expressing VHH Z70 did not show signs of the Tau pathology based on the AT8 labeling (Figures 6D and S16). According to our in vitro results, nucleation is probably blocked by the binding of the VHH Z70 to monomeric Tau, preventing its recruitment by the seeds. We cannot, however, exclude the possibility that VHH Z70 also binds to the seeds, given the lack of a precise definition of the nature of these seeds: seed-competent monomeric, oligomeric, or fibrillar Tau.
VHHs have entered the real world of therapeutics,67 and as they can be delivered as genes, the synergy with the progress in viral vector-mediated gene delivery could open the way for feasible treatments of tauopathies.
Materials and methods
Screening and selection of VHHs directed against Tau protein
The Nali-H1 library of VHHs (3 × 1011 phages) was screened against the recombinant biotinylated-Tau 2N4R as described previously.38,39 EZ-Link Sulfo-NHS-Biotin (Thermo Fisher Scientific) was used for the biotinylation following manufacturer protocol except for a 2-fold molar excess of Sulfo-NHS-Biotin. The unreacted Sulfo-NHS-Biotin was eliminated by desalting on Sepadextran 25 Medium SC (Proteigene). Biotinylated-Tau protein was bound to Dynabeads M-280 Streptavidin (Invitrogen) at a concentration gradually decreasing at each round of selection: 100 nM in first round, 50 nM in the second round, and 10 nM in the third round. Biotinylated-Tau binding was verified by western blot using Streptavidin Protein, HRP (Thermo Fisher Scientific). Non-absorbed Phage ELISA assay using avidin-plates and biotinylated-Tau Antigen (5 μg/ml) was used for cross-validation of 186 randomly picked clones.68
Production and purification of VHHs
Competent Escherichia coli BL21 (DE3) bacterial cells were transformed with the various PHEN2-VHH constructs. Recombinant E. coli cells produced proteins targeted to the periplasm after induction by 1 mM isopropylthiogalactoside (IPTG). Production was pursued for 4 h at 28°C before centrifugation to collect the cell pellet. The pellet was suspended in 200 mM Tris-HCl, 500 mM sucrose, 0.5 mM EDTA, pH 8, and incubated 30 min on ice; 50 mM Tris-HCl, 125 mM sucrose, 0.125 mM EDTA, pH 8, and complete protease inhibitor (Roche) were then added to the cell suspension and incubation continued for 30 min on ice. After centrifugation, the supernatant, corresponding to the periplasmic extract, was recovered. The VHHs were purified by immobilized-metal affinity chromatography (HisTrap HP, 1 mL, Cytiva) followed by size exclusion chromatography (Hiload 16/60, Superdex 75, prep grade, Cytiva) in NMR buffer (50 mM sodium phosphate buffer [NaPi], pH 6.7, 30 mM NaCl, 2.5 mM EDTA, 1 mM DTT).
Production and purification of Tau 2N4R, Tau 2N3R, Tau MTBD, and Tau [208–324]
pET15b-Tau recombinant T7lac expression plasmid was transformed into competent E. coli BL21 (DE3) bacterial cells. A small-scale culture was grown in LB medium at 37°C and was added at 1:10 v/v to 1 L of a modified M9 medium containing MEM vitamin mix 1X (Sigma-Aldrich), 4 g of glucose, 1 g of 15N-NH4Cl (Sigma-Aldrich), 0.5 g of 15N-enriched Isogro (Sigma-Aldrich), 0.1 mM CaCl2, and 2 mM MgSO4. Recombinant 15N Tau production was induced with 0.5 mM IPTG when the culture reached an optical density at 600 nm of 0.8. Proteins were first purified by heating the bacterial extract, obtained in 50 mM NaPi, pH 6.5, 2.5 mM EDTA, and supplemented with complete protease inhibitor cocktail (Sigma-Aldrich), 15 min at 75°C. The resulting supernatant was next passed on a cation exchange chromatography column (Hitrap SP sepharose FF, 5 mL, Cytiva) with 50 mM NaPi, pH 6.5, and eluted with a NaCl gradient. Tau proteins were buffer-exchanged against 50 mM ammonium bicarbonate (Hiload 16/60 desalting column, Cytiva) for lyophilization. The same protocol69 was used to produce and purify Tau 2N3R isoform, Tau[244–368] (designated MTBD, also called K18 fragment), chimeric Tau[244–368] with two PHF6 or PHF6∗ peptide sequences instead of PHF6∗ and PHF6 sequences (Figure S10A) and Tau [208–324].
Production and purification of SUMO-Tau peptides
cDNA encoding peptide Tau[273–318] was amplified from Tau 2N4R cDNA by PCR. cDNA was cloned by a ligation independent protocol into vector pETNKI-HisSUMO3-LIC as described.70 Tau peptide was expressed as a C-terminal SUMO protein fusion with an N-terminal HisTag. His-SUMO-Tau peptide was purified by affinity chromatography on Ni-NTA resin followed by size exclusion chromatography (Hiload 16/60, Superdex 75, prep grade, Cytiva) in SPR buffer (HBS-EP+, GE Healthcare).
Nuclear magnetic resonance spectroscopy experiments
Analysis of the 15N Tau/VHH interactions were performed at 298K on a Bruker Avance Neo 900MHz spectrometer equipped with cryogenic probe. Trimethyl silyl propionate was used as internal reference. Lyophilized 15N Tau were diluted in a buffer containing 50 mM NaPi, 30 mM NaCl, 2.5 mM EDTA, 1 mM DTT, and 10% D2O, pH 6.7 and mixed with VHH at 100 μM final concentration for each protein. A total of 200 μL of each mix in 3-mm tubes was sufficient to obtain the 2D 1H, 15N HSQC spectra with 32 scans. 1H, 15N HSQC were acquired with 3,072 and 416 points in the direct and indirect dimensions, for 12.6 and 25 ppm spectral windows, in the 1H and 15N dimensions, respectively. Each resonance in Tau spectra can be linked to a specific amino acid residue in Tau sequence,71,72 allowing to map the binding region based on the observed differences of chemical shift value and/or intensity for each resonance in the bound versus free condition. Data were processed with Bruker Topspin 3.6 and analyzed with Sparky (T. D. Goddard and D. G. Kneller, SPARKY 3, University of California, San Francisco).
Optimization of VHH E4-1 for intra-cellular expression
VHH E4-1 was amplified from pHEN2 plasmid (oligonucleotides 3390 and 3880 in Figure S18) using Taq polymerase with 14 mM MgCl2 and 0.2 mM MnCl2 and a modified nucleotide pool.73 The amplified cDNAs were transformed in yeast Y187 strain, together with a digested empty derivative of pGADGH vector,74 allowing recombination by gap repair in the vector. The VHH cDNAs are expressed as preys, with a C-terminal Gal4-activation domain fusion (E4-1-Gal4AD). A library of 2.1 million clones was obtained, collected and aliquoted. Tau variant 0N4R isoform (NM_016,834.4) was expressed as bait with a C-terminal fusion with lexA (Tau-LexA) from pB29 vector, which is derived from the original pBTM116.75 The library was screened at saturation, with 20 million tested diploids, using cell-to-cell mating protocol.76 A single clone was obtained, named VHH Z70.
One-to-one interaction by yeast two-hybrid
A one-to-one mating assay was used to test for interaction using a mating protocol with L40ΔGal4 (MATa) transformed with the bait (C-terminal fusion with lexA) and Y187 (MATα) yeast strains transformed with the prey (C-terminal Gal4-activation domain fusion).76 The interaction pairs were tested in triplicate on selective media by streak.
mCherry intracellular aggregation assays
HEK293 cells were seeded in 12-well plates (106 cells per well); 24 h later, cells were transfected with plasmids encoding mCherry, mCherry-VHH Z70, mCherry-VHH E4-1, or mCherry-VHH anti-GFP together with lipofectamine in optiMEM, as recommended by the manufacturer (Invitrogen). Forty-eight hours later, medium was removed, and cells were washed in pre-warmed PBS before 30-min fixation at room temperature (RT) with 4% PFA. After three successive washes in pre-warmed PBS, the nuclei were stained with DAPI (1/10,000) for 15 min at RT. Cells were cover-slipped with VectaMount. Ten images per condition (n = 3 independent experiences) were acquired using a Zeiss AxioObserver Z1 (spinning disk Yokogawa CSU-X1, camera sCMOS Photometrics Prime 95B). The number of mCherry positive cells containing puncta was quantified from the three independent experiments, 10 images per experiment and per group (n = 862 cells for mCherry, n = 932 for VHH-Z70, n = 995 for VHH E4.1 and n = 977 for VHH anti-GFP).
Tau fragment library construction
Tau cDNA (NM_016,834.4) was amplified from Tau-LexA bait vector (oligonucleotides 6690 and 6972 in Figure S18); 5 μg of the PCR product was subjected to Fragmentase treatment (New England Biolab, NEB) until a smear of fragments was detected around 400 to 500 pb by agarose gel electrophoresis. The DNA fragments were purified by phenol/chloroform extraction and ethanol precipitation. The DNA fragments were next subjected to end repair (NEB) and dA-tailing adaptation, using Blunt/TA ligase master mix with NEBNext Adaptor hairpin loop (NEB), followed by AMPure XP bead (Beckman Coulter) purification. After USER enzyme digestion (NEB), DNA fragments were amplified (oligonucleotides 10829 and 10830 in Figure S18) with 15 cycles of PCR using NEBNext Q5 Hot Start HiFi PCR Master Mix (NEB), which allowed addition Gap Repair recombination sequences for the cloning in Gal4-AD prey plasmid pP7. The library comprised 50,000 independent clones.
Tau fragment library screening
The coding sequence for VHH Z70 was PCR-amplified and cloned into pB27 as a C-terminal fusion to LexA (LexA-VHHZ70). The construct was used to produce a bait to screen the Tau fragments library constructed into pP7. pB27 and pP7 derived from the original pBTM11675 and pGADGH74 plasmids, respectively. The Tau fragment library was screened using a mating approach with YHGX13 (Y187 ade2-101:loxP-kanMX-loxP, MATα) and L40ΔGal4 (MATa) yeast strains.76 Ninety His+ colonies corresponding to 267 × 103 tested diploids were selected on a medium lacking tryptophan, leucine, and histidine. The prey fragments of the positive clones were amplified by PCR and sequenced at their 5′and 3′ junctions.
Surface plasmon resonance experiments
Affinity measurements were performed on a BIAcore T200 optical biosensor instrument (Cytiva). Full-length recombinant Tau 2N4R proteins were biotinylated with five molar excess of NHS-biotin conjugates (ThermoFisher) during 4 h at 4°C. Capture of biotinylated Tau was performed on a streptavidin SA sensorchip in HBS-EP + buffer (Cytiva). One flow cell was used as a reference to evaluate nonspecific binding and provide background correction. Biotinylated-Tau was injected at a flow-rate of 30 μL/min, until the total amount of captured Tau reached 500 resonance units (RUs). VHHs were injected sequentially with increasing concentrations ranging between 0.125 and 2 μM in a single cycle, with regeneration (three successive washes of 1M NaCl) between each VHH. On the other hand, a VHH Z70 construct, containing a single C-terminal cysteine, was biotinylated using EZ-Link Maleimide-PEG2-Biotin (Thermo Scientific) and was immobilized on an SA (Streptavidin) chip in HBS-EP + buffer (Cytiva). Increasing concentrations, ranging between 31.25 and 500 nM of the SUMO-Tau peptide, were successively injected. The same functionalized SA chip was also used to inject increasing concentrations of three different MTBD constructs, ranging between 62.5 nM and 1 μM. Single-Cycle Kinetics (SCK) analysis77was performed to determine association kon and dissociation Koff rate constants by curve fitting of the sensorgrams using the 1:1 Langmuir model of interaction of the BIAevaluation software 2.0 (Cytiva). Dissociation equilibrium constants (Kd) were calculated as koff/kon.
VHH Z70/PHF6 Tau peptide complex crystallization and structure determination
VHH Z70 protein solution was dialyzed against 10 mM HEPES pH 7.4, 50 mM NaCl then concentrated to 250 μM and incubated with 1 mM of PHF6 peptide for 30 min before crystallization screening. The PHF6 peptide sequence was 301PGGGSVQIVYKP312KK (Genecust, France), with the last two peptide residues added compared with the native Tau sequence for solubility purposes. From an initial screening of around 600 conditions, optimal crystallization conditions were found to be 0.17 M ammonium sulfate, 25.5% PEG 4000 and 15% glycerol (found in the Cryos Suite, Qiagen). Crystals were evaluated at SOLEIL synchrotron beamline PX1. Crystals belonged to space group P6522 with cell parameters suggesting that the asymmetric unit contains one VHH monomer (98% probability estimated from Matthews coefficient). The best diffraction dataset was obtained at a resolution of 1.7 Å. Structure was solved using molecular replacement (MOLREP)78 with PDB: 01OL0 as template and refined to a Rwork of 0.2 and Rfree of 0.21 using REFMAC579 and COOT.80 Interface of the complex was calculated using PDBePISA (Protein interfaces, surfaces and assemblies' service at the European Bioinformatics Institute, http://www.ebi.ac.uk/pdbe/prot_int/pistart.html).81 The structure was deposited in the worldwide protein databank (wwPDB) with access code PDB: 7QCQ.
In vitro kinetic aggregation assays
Tau 2N4R aggregation assays were performed with 10 μM Tau and with increasing concentrations of VHHs (between 0 and 10 μM) in buffer containing 50 mM MES pH 6.9, 30 mM NaCl, 2.5 mM EDTA, 0.3 mM freshly prepared DTT, 2.5 mM heparin H3 (Sigma-Aldrich), and 50 μM Thioflavin T (Sigma-Aldrich), at 37°C. Experiments were reproduced three times in triplicate for each condition. The resulting fluorescence of Thioflavin T was recorded every 5 min/cycle within 200 cycles using PHERAstar microplate-reader (BMG labtech). The measures were normalized in fluorescence percentage, 100% being defined as the maximum value reached in the positive Tau control, in each experiment.
Transmission electron microscopy
The same samples from the in vitro aggregation assays were recovered and a 10 μL sample of Tau or Tau:VHH ratio 1:1 condition was loaded on a formvar/carbon-coated grid (for 5 min and rinsed twice with water). After drying, the grids were stained with 1% uranyl acetate for 1 min. Tau fibrils were observed under a transmission electron microscope (EM 900 Zeiss equipped with a Gatan Orius 1000 camera).
Seeding assays in HEK293 reporter cell line
Stable HEK293 Tau RD P301S FRET Biosensor cells (ATCC CRL-3275) were plated at a density of 100 k cells/well in 24-well plates. For confocal analysis, cells were plated on glass slides coated with poly-D-lysine and laminin at a density of 100 k cells/well in 24-well plates. At 60% confluency, cells were first transiently transfected with the various pmCherry-N1 plasmid constructs allowing expression of the mCherry-fused VHHs. Transfection complexes were obtained by mixing 500 ng of plasmid diluted in 40 μL of opti-MEM medium, which included 18.5 μL (46.25% v/v) of opti-MEM medium with 1.5 μL (3.75% v/v) Lipofectamine 2000 (Invitrogen). Resulting liposomes were incubated at room temperature for 20 min before addition to the cells. Cells were incubated for 24 h with the liposomes and 1 mL/well of high glucose DMEM medium (ATCC) with fetal bovine serum 1% (Life Technologies). The transfection efficiency was estimated to reach about 46%, for all mCherry-fused VHH plasmids (Figure S19). Eight µM of recombinant MTBD seeds were prepared in vitro, in the presence of 8 μM heparin, as described.43 Cells were then treated with MTBD seeds (10 nM/well) in the presence of transfection reagents forming liposomes as here above described.
FRET flow cytometry
Cells were recovered with trypsin 0.05% and fixed in 2% PFA for 10 min, then suspended in PBS. Flow cytometry was performed on an ARIA SORP BD (acquisition software FACS DIVA V7.0, BD Biosciences). To measure CFP emission fluorescence and FRET, cells were excited with a 405-nm laser. The fluorescence was captured with either a 466/40- or a 529/30-nm filter, respectively. To measure YFP fluorescence, a 488-nm laser was used for excitation and emission fluorescence was captured with a 529/30-nm filter. mCherry cells were excited with a 561-nm laser and fluorescence was captured with a 610/20 nm filter. To selectively detect and quantify FRET, gating was used as described.43,82 The FRET data were quantified using the KALUZA software analyzer v2. Three independent experiments were done in triplicate or quadruplicate, with at least 10,000 cells per replicate analyzed.
Animals
The study was performed in accordance with the ethical standards as laid down in the 1964 Declaration of Helsinki and its later amendments or comparable ethical standards. The experimental research has been performed with the approval of an ethical committee (agreement APAFIS#2264–2015101320441671 from CEEA75, Lille, France) and follows European guidelines for the use of animals. The animals (males and females) were housed in a temperature-controlled (20–22°C) room maintained on a 12 h day/night cycle with food and water provided ad libitum in a specific pathogen-free animal facility (n = 5 mice per cage). Animals were allocated to experimental groups by randomization. AD brain extracts were obtained from the Lille Neurobank (fulfilling criteria of the French law on biological resources and declared to competent authority under the number DC-2008-642) with donor consent, data protection, and ethical committee review. Samples were managed by the CRB/CIC1403 Biobank, BB-0033-00030.
Stereotaxic injection of THY-tau30 Tg mice
THY-tau30 Tg mice express human 1N4R Tau protein with two pathogenic mutations (P301S and G272V) under the control of the neuron-specific Thy1.2 promoter.83,84 One-month-old anesthetized THY-tau30 mice were submitted to stereotaxic intra-cerebrocranial injections (400 ng in 2 μL at 250 nL/min with a Hamilton glass syringe) at the coordinates posterior AP: −2.46, midline ML: −1 and vertical depth DV: −2.3 of both brain hemispheres with LVs expressing either VHH Z70 with an N-terminal mCherry fusion protein (VHH Z70) or a VHH directed against the GFP (VHH anti-GFP).
Two weeks later, these mice were submitted to injections of human AD brain homogenate (h-AD, 15 μg in 2 μL) at the same coordinates of both hemispheres, as previously described in detail.19 The h-AD seeds consisted in a mixture of two postmortem human brain extracts from tissues of patients with confirmed AD (frontal cortex area, Braak stage VI, Brodmann area 10). The injections resulted in two groups of seven and nine mice per group. The mice were killed after a month delay from the injection of the h-AD brain extract. All mice gained weight during the experimental protocol course and no mouse death was recorded, showing no indication of severe toxicity (Figure S20).
Tissue processing, immunohistochemistry, and Tau pathology quantification
THY-tau30 were deeply anesthetized and trans-cardially perfused with ice-cold 0.9% saline solution and subsequently with 4% PFA for 10 min. The brains were immediately removed, fixed overnight in 4% PFA, washed in PBS, placed in 20% sucrose for 16 h, and frozen in isopentane until further use. Free-floating coronal sections (40-μm thickness) were obtained using a cryostat microtome.
Cryostat sections were next used for immunohistochemistry. Nonspecific binding was blocked by using “Mouse in Mouse” reagent (1:100 in PBS; Vector Laboratories). Brain slices were next incubated with the primary monoclonal antibody AT8 (1:500; Thermo MN1020) in PBS- 0.2% Triton X-100, 16 h at 4°C. Labeling was amplified by incubation with an anti-mouse biotinylated IgG (1:400 in PBS-0.2% TritonTM X-100, Vector) followed by the application of the avidin-biotin-HRP complex (ABC kit, 1:400 in PBS, Vector) prior to addition of diaminobenzidine tetrahydrochloride (DAB, Vector) in Tris-HCl 0.1 mol/L, pH 7.6, containing H2O2 for visualization. Brain sections were mounted, air-dried, steadily dehydrated in ethanol (30%, 70%, 95%, 100%), cleared in toluene, and cover-slipped with VectaMount (Vector Laboratories). Mounted brain sections were analyzed using stereology software (Mercator image analysis system; Explora Nova, La Rochelle, France). Threshold was established manually to present a minimum background and remained constant throughout the analysis. The region defined as quantification zone is from bregmas −2.06 to −2.92 (based on the Mouse Atlas, George Paxinos and Keith B.J. Franklin, Second Edition, Academic Press).
ImmunoHistoFluorescence
Brain sections from mice injected with the LVs VHH Z70 with an N-terminal fusion to mCherry were saturated in normal goat serum (1/100; Vector), then were incubated with the primary polyclonal antibodies anti-RFP targeting mCherry protein (1:1,000, rabbit, Polyclonal, Rockland) 16 h at 4°C in PBS-0.2% TritonTM X-100. For the double labeling, incubation was performed in the additional presence of Tau-specific antibody AT8 conjugated with biotin (1:500; Thermo). Labeling was detected using a secondary anti-rabbit antibody (1:500; Invitrogen) functionalized with Alexa 488 and streptavidin functionalized with Alexa 647 (1:500; Invitrogen, visualized in pseudocolor red) for AT8. Section imaging was performed by microscopy using a slide scanner (Axioscan Z1-Zeiss) with a ×20 objective.
Statistical analysis
Data are presented as the means ± SEM for in vitro aggregation assays (Figure 4) and reporter-cell seeding assays (Figure 5), and ±SD for in-cell solubility assays (Figure 2E) and for in vivo experiments (Figure 6C). Experiments were performed at least in triplicate and obtained from three independent experiments. An ordinary one-way nonparametric ANOVA with a Sidak's multiple comparison test has been applied to analyze puncta in mCherry positive cells (Figure 2E). One-way nonparametric ANOVAs (Kruskal-Wallis) with Dunn's multiple comparison test and Mann-Whitney U test were used to analyze data for FRET experiments (Figure 5) and unpaired t test after normality test for in vivo experiments (Figure 6). Statistical analyses were performed with GraphPad Prism 8.0.0.
Acknowledgments
We thank Dr Z. Lens and Dr M. Aumercier for their help on the T200 biacore measurements and Mrs M. Oosterlynck for technical support. We also thank M. Tardivel and A. Bongiovanni for their help on the Zeiss confocal microscope, from the Photonic Microscopy Core BioImaging Center (BiCeL) and N. Jouy for the cytometry experiments, from the Flow Core Facility (BiCel). We would like to thank Tatiana Isabet, Serena Sirigu, and William Shepard for their valuable support during data collection at beamlines PX1 and PX2A at the SOLEIL synchrotron facility (Paris, France). The NMR facilities were funded by the Nord Region Council, CNRS, Institut Pasteur de Lille, European Union, French Research Ministry, and Univ. Lille. Financial support from the IR-RMN-THC FR 3050 CNRS for conducting the research is gratefully acknowledged. We acknowledge SOLEIL for the provision of synchrotron-radiation facilities. This study was supported by the LabEx (Laboratory of Excellence) DISTALZ (Development of Innovative Strategies for a Transdisciplinary approach to Alzheimer's disease ANR-11-LABX-01), by European Union project AgedBrainSYSBIO (Grant Agreement N° 305299), by I-site ULNE (project TUNABLE) and by ANR (project ToNIC, ANR-18-CE44-0016). Our laboratories are also supported by LiCEND (Lille Center of Excellence in Neurodegenerative Disorders), Inserm, Métropole Européenne de Lille, Univ. Lille and FEDER.
Author contributions
J.-C.R., L.B., and I.L. conceptualized the study; C.D., E.D., O.Z., R.C., A.A., S.B., J.M., S.E., A.L., F-X.C. performed the study investigation; C.D., E.D., and M.C. performed the formal analysis; C.D., E.D. and M.C. visualized the study; C.D., E.D., and I.L. wrote the original draft; J.-C.R., M.C., X.H., and L.B. reviewed and edited the manuscript; I.L. and L.B. acquired the funding; E.D., X.H., M.C., I.L., and L.B. supervised the study.
Declaration of interests
A.A. and J.-C.R. are employees of Hybrigenic services.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.ymthe.2022.01.009.
Contributor Information
Luc Buée, Email: luc.buee@inserm.fr.
Isabelle Landrieu, Email: isabelle.landrieu@univ-lille.fr.
Supplemental information
References
- 1.Hamers-Casterman C., Atarhouch T., Muyldermans S., Robinson G., Hamers C., Songa E.B., Bendahman N., Hamers R. Naturally occurring antibodies devoid of light chains. Nature. 1993;363:446–448. doi: 10.1038/363446a0. [DOI] [PubMed] [Google Scholar]
- 2.Messer A.A., Butler D.C. Optimizing intracellular antibodies (intrabodies/nanobodies) to treat neurodegenerative disorders. Neurobiol. Dis. 2020;134:104619. doi: 10.1016/j.nbd.2019.104619. [DOI] [PubMed] [Google Scholar]
- 3.Goedert M., Spillantini M.G. A century of Alzheimer's disease. Science. 2006;314:777–781. doi: 10.1126/science.1132814. [DOI] [PubMed] [Google Scholar]
- 4.Brion J.P., Flament-Durand J., Dustin P. Alzheimer's disease and tau proteins. Lancet. 1986;2:1098. doi: 10.1016/s0140-6736(86)90495-2. [DOI] [PubMed] [Google Scholar]
- 5.Grundke-Iqbal I., Iqbal K., Quinlan M., Tung Y.C., Zaidi M.S., Wisniewski H.M. Microtubule-associated protein tau. A component of Alzheimer paired helical filaments. J. Biol. Chem. 1986;261:6084–6089. [PubMed] [Google Scholar]
- 6.Clavaguera F., Bolmont T., Crowther R.A., Abramowski D., Frank S., Probst A., Fraser G., Stalder A.K., Beibel M., Staufenbiel M., et al. Transmission and spreading of tauopathy in transgenic mouse brain. Nat. Cell Biol. 2009;11:909–913. doi: 10.1038/ncb1901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Frost B., Jacks R.L., Diamond M.I. Propagation of tau misfolding from the outside to the inside of a cell. J. Biol. Chem. 2009;284:12845–12852. doi: 10.1074/jbc.M808759200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mudher A., Colin M., Dujardin S., Medina M., Dewachter I., Alavi Naini S.M., Mandelkow E.M., Mandelkow E., Buée L., Goedert M., Brion J.P. What is the evidence that tau pathology spreads through prion-like propagation? Acta Neuropathol. Commun. 2017;5:99. doi: 10.1186/s40478-017-0488-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cummings J., Lee G., Mortsdorf T., Ritter A., Zhong K. Alzheimer's disease drug development pipeline: 2017. Alzheimers Dement (N Y) 2017;3:367–384. doi: 10.1016/j.trci.2017.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nelson P.T., Alafuzoff I., Bigio E.H., Bouras C., Braak H., Cairns N.J., Castellani R.J., Crain B.J., Davies P., Del Tredici K., et al. Correlation of Alzheimer disease neuropathologic changes with cognitive status: a Review of the literature. J. Neuropathol. Exp. Neurol. 2012;71:362–381. doi: 10.1097/NEN.0b013e31825018f7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schwarz A.J., Yu P., Miller B.B., Shcherbinin S., Dickson J., Navitsky M., Joshi A.D., Devous M.D., Mintun M.S. Regional profiles of the candidate tau PET ligand 18F-AV-1451 recapitulate key features of Braak histopathological stages. Brain. 2016;139:1539–1550. doi: 10.1093/brain/aww023. [DOI] [PubMed] [Google Scholar]
- 12.Wilcock G.K., Esiri M.M. Plaques, tangles and dementia. A quantitative study. J. Neurol. Sci. 1982;56:343–356. doi: 10.1016/0022-510x(82)90155-1. [DOI] [PubMed] [Google Scholar]
- 13.Bi M., Ittner A., Ke Y.D., Götz J., Ittner L.M. Tau-targeted immunization impedes progression of neurofibrillary histopathology in aged P301L tau transgenic mice. PLoS One. 2011;6:e26860. doi: 10.1371/journal.pone.0026860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Asuni A.A., Boutajangout A., Quartermain D., Sigurdsson E.M. Immunotherapy targeting pathological tau conformers in a tangle mouse model reduces brain pathology with associated functional improvements. J. Neurosci. 2007;27:9115–9129. doi: 10.1523/JNEUROSCI.2361-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Boutajangout A., Quartermain D., Sigurdsson E.M. Immunotherapy targeting pathological tau prevents cognitive decline in a new tangle mouse model. J. Neurosci. 2010;30:16559–16566. doi: 10.1523/JNEUROSCI.4363-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dai C.-L., Tung Y.C., Liu F., Gong C.-X., Iqbal K. Tau passive immunization inhibits not only tau but also Aβ pathology. Alzheimers Res. Ther. 2017;9:1. doi: 10.1186/s13195-016-0227-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chai X., Wu S., Murray T.K., Kinley R., Cella C.V., Sims H., et al. Passive immunization with anti-Tau antibodies in two transgenic models: reduction of Tau pathology and delay of disease progression. J. Biol. Chem. 2011;286:34457–34467. doi: 10.1074/jbc.M111.229633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yanamandra K., Kfoury N., Jiang H., Mahan T.E., Ma S., Maloney S.E., Wozniak D.F., Diamond M.I., Holtzman D.M. Anti-tau antibodies that block tau aggregate seeding in vitro markedly decrease pathology and improve cognition in vivo. Neuron. 2013;80:402–414. doi: 10.1016/j.neuron.2013.07.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Albert M., Mairet-Coello G., Danis C., Lieger S., Caillierez R., Carrier S., Skrobala E., Landrieu I., Michel A., Schmitt M., et al. Prevention of tau seeding and propagation by immunotherapy with a central tau epitope antibody. Brain. 2019;142:1736–1750. doi: 10.1093/brain/awz100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Troquier L., Caillierez R., Burnouf S., Fernandez-Gomez F.J., Grosjean M.-E., Zommer N., et al. Targeting phospho-Ser422 by active Tau Immunotherapy in the THYTau22 mouse model: a suitable therapeutic approach. Curr. Alzheimer Res. 2012;9:397–405. doi: 10.2174/156720512800492503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Congdon E.E., Sigurdsson E.M. Tau-targeting therapies for Alzheimer disease. Nat. Rev. Neurol. 2018;14:399–415. doi: 10.1038/s41582-018-0013-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jadhav S., Avila J., Schöll M., Kovacs G.G., Kövari E., Skrabana R., Evans L.D., Kontsekova E., Malawska B., de Silva R., et al. A walk through tau therapeutic strategies. Acta Neuropathol. Commun. 2019;7:22. doi: 10.1186/s40478-019-0664-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kfoury N., Holmes B.B., Jiang H., Holtzman D.M., Diamond M.I. Trans-cellular propagation of Tau aggregation by fibrillar species. J. Biol. Chem. 2012;287:19440–19451. doi: 10.1074/jbc.M112.346072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dujardin S., Lécolle K., Caillierez R., Bégard S., Zommer N., Lachaud C., Carrier S., Dufour N., Aurégan G., Winderickx J., et al. Neuron-to-neuron wild-type Tau protein transfer through a trans-synaptic mechanism: relevance to sporadic tauopathies. Acta Neuropathol. Commun. 2014;2:14. doi: 10.1186/2051-5960-2-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Colin M., Dujardin S., Schraen-Maschke S., Meno-Tetang G., Duyckaerts C., Courade J.-P., Buée L. From the prion-like propagation hypothesis to therapeutic strategies of anti-tau immunotherapy. Acta Neuropathol. 2020;139:3–25. doi: 10.1007/s00401-019-02087-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Clavaguera F., Grueninger F., Tolnay M. Intercellular transfer of tau aggregates and spreading of tau pathology: implications for therapeutic strategies. Neuropharmacology. 2014;76:9–15. doi: 10.1016/j.neuropharm.2013.08.037. [DOI] [PubMed] [Google Scholar]
- 27.Dujardin S., Bégard S., Caillierez R., Lachaud C., Delattre L., Carrier S., Loyens A., Galas M.C., Bousset L., Melki R., et al. Ectosomes: a new mechanism for non-exosomal secretion of tau protein. PLoS One. 2014;9:e100760. doi: 10.1371/journal.pone.0100760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Leroux E., Perbet R., Caillierez R., Richetin K., Lieger S., Espourteille J., et al. Extracellular vesicles: major actors of heterogeneity in tau spreading among human tauopathies. Mol. Ther. J. Am. Soc. Gene Ther. 2021 doi: 10.1016/j.ymthe.2021.09.020. S1525-0016(21)00475–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tardivel M., Bégard S., Bousset L., Dujardin S., Coens A., Melki R., Buée L., Colin M. Tunneling nanotube (TNT)-mediated neuron-to neuron transfer of pathological Tau protein assemblies. Acta Neuropathol. Commun. 2016;4:117. doi: 10.1186/s40478-016-0386-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pain C., Dumont J., Dumoulin M. Camelid single-domain antibody fragments: uses and prospects to investigate protein misfolding and aggregation, and to treat diseases associated with these phenomena. Biochimie. 2015;111:82–106. doi: 10.1016/j.biochi.2015.01.012. [DOI] [PubMed] [Google Scholar]
- 31.Herce H.D., Schumacher D., Schneider A.F.L., Ludwig A.K., Mann F.A., Fillies M., Kasper M.A., Reinke S., Krause E., Leonhardt H., et al. Cell-permeable nanobodies for targeted immunolabelling and antigen manipulation in living cells. Nat. Chem. 2017;9:762–771. doi: 10.1038/nchem.2811. [DOI] [PubMed] [Google Scholar]
- 32.Li T., Bourgeois J.-P., Celli S., Glacial F., Le Sourd A.-M., Mecheri S., Weksler B., Romero I., Couraud P.O., Rougeon F., Lafaye P. Cell-penetrating anti-GFAP VHH and corresponding fluorescent fusion protein VHH-GFP spontaneously cross the blood-brain barrier and specifically recognize astrocytes: application to brain imaging. FASEB J. 2016;26:3969–3979. doi: 10.1096/fj.11-201384. [DOI] [PubMed] [Google Scholar]
- 33.Gormal R.S., Padmanabhan P., Kasula R., Bademosi A.T., Coakley S., Giacomotto J., Blum A., Joensuu M., Wallis T.P., Lo H.P., et al. Modular transient nanoclustering of activated β2-adrenergic receptors revealed by single-molecule tracking of conformation-specific nanobodies. Proc. Natl. Acad. Sci. U S A. 2020;117:30476–30487. doi: 10.1073/pnas.2007443117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li T., Vandesquille M., Koukouli F., Dudeffant C., Youssef I., Lenormand P., Ganneau C., Maskos U., Czech C., Grueninger F., et al. Camelid single-domain antibodies: a versatile tool for in vivo imaging of extracellular and intracellular brain targets. J. Control Release. 2016;243:1–10. doi: 10.1016/j.jconrel.2016.09.019. [DOI] [PubMed] [Google Scholar]
- 35.Vitale F., Giliberto L., Ruiz S., Steslow K., Marambaud P., d'Abramo C. Anti-tau conformational scFv MC1 antibody efficiently reduces pathological tau species in adult JNPL3 mice. Acta Neuropathol. Commun. 2018;6:82. doi: 10.1186/s40478-018-0585-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Goodwin M.S., Sinyavskaya O., Burg F., O’Neal V., Ceballos-Diaz C., Cruz P.E., et al. Anti-tau scFvs targeted to the cytoplasm or secretory pathway variably modify pathology and neurodegenerative phenotypes. Mol. Ther. J. Am. Soc. Gene Ther. 2020 doi: 10.1016/j.ymthe.2020.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.von Bergen M., Friedhoff P., Biernat J., Heberle J., Mandelkow E.M., Mandelkow E. Assembly of tau protein into Alzheimer paired helical filaments depends on a local sequence motif ((306)VQIVYK(311)) forming beta structure. Proc. Natl. Acad. Sci. U S A. 2000;97:5129–5134. doi: 10.1073/pnas.97.10.5129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Moutel S., Bery N., Bernard V., Keller L., Lemesre E., de Marco A., Ligat L., Rain J.C., Favre G., Olichon A., Perez F. NaLi-H1: a universal synthetic library of humanized nanobodies providing highly functional antibodies and intrabodies. eLife. 2016;5:e16228. doi: 10.7554/eLife.16228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dupré E., Danis C., Arrial A., Hanoulle X., Homa M., Cantrelle F.-X., Merzougui H., Colin M., Rain J.C., Buée L., Landrieu I. Single domain antibody fragments as new tools for the detection of neuronal tau protein in cells and in mice studies. ACS Chem. Neurosci. 2019;10:3997–4006. doi: 10.1021/acschemneuro.9b00217. [DOI] [PubMed] [Google Scholar]
- 40.Dingus J., Tang J.C.Y., Cepko C. A general approach for stabilizing nanobodies for intracellular expression. biorxiv. 2021 doi: 10.1101/2021.04.06.438746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vielemeyer O., Nizak C., Jimenez A.J., Echard A., Goud B., Camonis J., Rain J.C., Perez F. Characterization of single chain antibody targets through yeast two hybrid. BMC Biotechnol. 2010;10:59. doi: 10.1186/1472-6750-10-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang Y., Chang C., Gehling D.J., Hemmati-Brivanlou A., Derynck R. Regulation of Smad degradation and activity by Smurf2, an E3 ubiquitin ligase. Proc. Natl. Acad. Sci. U S A. 2001;98:974–979. doi: 10.1073/pnas.98.3.974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Holmes B.B., Furman J.L., Mahan T.E., Yamasaki T.R., Mirbaha H., Eades W.C., Belaygorod L., Cairns N.J., Holtzman D.M., Diamond M.I. Proteopathic tau seeding predicts tauopathy in vivo. Proc. Natl. Acad. Sci. U S A. 2014;111:E4376–E4385. doi: 10.1073/pnas.1411649111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vandermeeren M., Borgers M., Van Kolen K., Theunis C., Vasconcelos B., Bottelbergs A., Wintmolders C., Daneels G., Willems R., Dockx K., et al. Anti-tau monoclonal antibodies derived from soluble and filamentous tau show diverse functional properties in vitro and in vivo. J. Alzheimers Dis. 2018;65:265–281. doi: 10.3233/JAD-180404. [DOI] [PubMed] [Google Scholar]
- 45.Malia T.J., Teplyakov A., Ernst R., Wu S.-J., Lacy E.R., Liu X., Vandermeeren M., Mercken M., Luo J., Sweet R.W., Gilliland G.L. Epitope mapping and structural basis for the recognition of phosphorylated tau by the anti-tau antibody AT8. Proteins. 2016;84:427–434. doi: 10.1002/prot.24988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gallardo G., Wong C.H., Ricardez S.M., Mann C.N., Lin K.H., Leyns C.E.G., Jiang H., Holtzman D.M. Targeting tauopathy with engineered tau-degrading intrabodies. Mol. Neurodegener. 2019;14:38. doi: 10.1186/s13024-019-0340-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nobuhara C.K., DeVos S.L., Commins C., Wegmann S., Moore B.D., Roe A.D., et al. Tau antibody-targeting pathological species block neuronal uptake and interneuron propagation of tau in vitro. Am. J. Pathol. 2017;187:1399–1412. doi: 10.1016/j.ajpath.2017.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Spencer B., Brüschweiler S., Sealey-Cardona M., Rockenstein E., Adame A., Florio J., Mante M., Trinh I., Rissman R.A., Konrat R., Masliah E. Selective targeting of 3 repeat Tau with brain penetrating single chain antibodies for the treatment of neurodegenerative disorders. Acta Neuropathol. 2018;136:69–87. doi: 10.1007/s00401-018-1869-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chen H.-H., Liu P., Auger P., Lee S.-H., Adolfsson O., Rey-Bellet L., Lafrance-Vanasse J., Friedman B.A., Pihlgren M., Muhs A., et al. Calpain-mediated tau fragmentation is altered in Alzheimer's disease progression. Sci. Rep. 2018;8:16725. doi: 10.1038/s41598-018-35130-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dam T., Boxer A.L., Golbe L.I., Höglinger G.U., Morris H.R., Litvan I., Lang A.E., Corvol J.C., Aiba I., Grundman M., et al. Safety and efficacy of anti-tau monoclonal antibody gosuranemab in progressive supranuclear palsy: a phase 2, randomized, placebo-controlled trial. Nat. Med. 2021;27:1451–1457. doi: 10.1038/s41591-021-01455-x. [DOI] [PubMed] [Google Scholar]
- 51.Höglinger G.U., Litvan I., Mendonca N., Wang D., Zheng H., Rendenbach-Mueller B., Lon H.K., Jin Z., Fisseha N., Budur K., et al. Safety and efficacy of tilavonemab in progressive supranuclear palsy: a phase 2, randomised, placebo-controlled trial. Lancet Neurol. 2021;20:182–192. doi: 10.1016/S1474-4422(20)30489-0. [DOI] [PubMed] [Google Scholar]
- 52.Jabbari E., Duff K.E. Tau-targeting antibody therapies: too late, wrong epitope or wrong target? Nat. Med. 2021;27:1341–1342. doi: 10.1038/s41591-021-01465-9. [DOI] [PubMed] [Google Scholar]
- 53.Courade J.-P., Angers R., Mairet-Coello G., Pacico N., Tyson K., Lightwood D., Munro R., McMillan D., Griffin R., Baker T., et al. Epitope determines efficacy of therapeutic anti-Tau antibodies in a functional assay with human Alzheimer Tau. Acta Neuropathol. 2018;136:729–745. doi: 10.1007/s00401-018-1911-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Roberts M., Sevastou I., Imaizumi Y., Mistry K., Talma S., Dey M., Gartlon J., Ochiai H., Zhou Z., Akasofu S., et al. Pre-clinical characterisation of E2814, a high-affinity antibody targeting the microtubule-binding repeat domain of tau for passive immunotherapy in Alzheimer's disease. Acta Neuropathol. Commun. 2020;8:13. doi: 10.1186/s40478-020-0884-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fitzpatrick A.W.P., Falcon B., He S., Murzin A.G., Murshudov G., Garringer H.J., et al. Cryo-EM structures of tau filaments from Alzheimer’s disease. Nature. 2017 doi: 10.1038/nature23002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Arakhamia T., Lee C.E., Carlomagno Y., Duong D.M., Kundinger S.R., Wang K., Williams D., DeTure M., Dickson D.W., Cook C.N., et al. Posttranslational modifications mediate the structural diversity of tauopathy strains. Cell. 2020;180:633–644.e12. doi: 10.1016/j.cell.2020.01.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Falcon B., Zhang W., Murzin A.G., Murshudov G., Garringer H.J., Vidal R., Crowther R.A., Ghetti B., Scheres S.H.W., Goedert M. Structures of filaments from Pick's disease reveal a novel tau protein fold. Nature. 2018;561:137–140. doi: 10.1038/s41586-018-0454-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Falcon B., Zivanov J., Zhang W., Murzin A.G., Garringer H.J., Vidal R., Crowther R.A., Newell K.L., Ghetti B., Goedert M., Scheres S.H.W. Novel tau filament fold in chronic traumatic encephalopathy encloses hydrophobic molecules. Nature. 2019;568:420–423. doi: 10.1038/s41586-019-1026-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sawaya M.R., Sambashivan S., Nelson R., Ivanova M.I., Sievers S.A., Apostol M.I., Thompson M.J., Balbirnie M., Wiltzius J.J., McFarlane H.T., et al. Atomic structures of amyloid cross-beta spines reveal varied steric zippers. Nature. 2007;447:453–457. doi: 10.1038/nature05695. [DOI] [PubMed] [Google Scholar]
- 60.Seidler P.M., Boyer D.R., Rodriguez J.A., Sawaya M.R., Cascio D., Murray K., Gonen T., Eisenberg D.S. Structure-based inhibitors of tau aggregation. Nat. Chem. 2018;10:170–176. doi: 10.1038/nchem.2889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mirbaha H., Chen D., Morazova O.A., Ruff K.M., Sharma A.M., Liu X., Goodarzi M., Pappu R.V., Colby D.W., Mirzaei H., et al. Inert and seed-competent tau monomers suggest structural origins of aggregation. eLife. 2018;7:e36584. doi: 10.7554/eLife.36584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Moreira G.G., Cantrelle F.-X., Quezada A., Carvalho F.S., Cristóvão J.S., Sengupta U., Puangmalai N., Carapeto A.P., Rodrigues M.S., Cardoso I., et al. Dynamic interactions and Ca2+-binding modulate the holdase-type chaperone activity of S100B preventing tau aggregation and seeding. Nat. Commun. 2021;12:6292. doi: 10.1038/s41467-021-26584-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mok S.-A., Condello C., Freilich R., Gillies A., Arhar T., Oroz J., Kadavath H., Julien O., Assimon V.A., Rauch J.N., et al. Mapping interactions with the chaperone network reveals factors that protect against tau aggregation. Nat. Struct. Mol. Biol. 2018;25:384–393. doi: 10.1038/s41594-018-0057-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Shi Y., Zhang W., Yang Y., Murzin A.G., Falcon B., Kotecha A., van Beers M., Tarutani A., Kametani F., Garringer H.J., et al. Structure-based classification of tauopathies. Nature. 2021;598:359–363. doi: 10.1038/s41586-021-03911-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Carlomagno Y., Manne S., DeTure M., Prudencio M., Zhang Y.-J., Hanna Al-Shaikh R., Dunmore J.A., Daughrity L.M., Song Y., Castanedes-Casey M., et al. The AD tau core spontaneously self-assembles and recruits full-length tau to filaments. Cell Rep. 2021;34:108843. doi: 10.1016/j.celrep.2021.108843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhang W., Falcon B., Murzin A.G., Fan J., Crowther R.A., Goedert M., Scheres S.H. Heparin-induced tau filaments are polymorphic and differ from those in Alzheimer's and Pick's diseases. eLife. 2019;8:e43584. doi: 10.7554/eLife.43584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Dutt T., Shaw R.J., Stubbs M.J., Yong J., Bailiff B., Cranfield T., et al. Real-world evidence of caplacizumab use in the management of acute TTP. Blood. 2020;137:1731–1740. doi: 10.1182/blood.2020007599. [DOI] [PubMed] [Google Scholar]
- 68.Matz J., Chames Phage display and selections on purified antigens. Methods Mol. Biol. 2012;907:213–224. doi: 10.1007/978-1-61779-974-7_11. [DOI] [PubMed] [Google Scholar]
- 69.Danis C., Despres C., Bessa L.M., Malki I., Merzougui H., Huvent I. Nuclear magnetic resonance spectroscopy for the identification of multiple phosphorylations of intrinsically disordered proteins. J. Vis. Exp. 2016 doi: 10.3791/55001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Luna-Vargas M.P.A., Christodoulou E., Alfieri A., van Dijk W.J., Stadnik M., Hibbert R.G., Sahtoe D.D., Clerici M., Marco V.D., Littler D., et al. Enabling high-throughput ligation-independent cloning and protein expression for the family of ubiquitin specific proteases. J. Struct. Biol. 2011;175:113–119. doi: 10.1016/j.jsb.2011.03.017. [DOI] [PubMed] [Google Scholar]
- 71.Smet C., Leroy A., Sillen A., Wieruszeski J.M., Landrieu I., Lippens G. Accepting its random coil nature allows a partial NMR assignment of the neuronal Tau protein. Chembiochem. 2004;5:1639–1646. doi: 10.1002/cbic.200400145. [DOI] [PubMed] [Google Scholar]
- 72.Lippens G., Wieruszeski J.M., Leroy A., Smet C., Sillen A., Buée L., Landrieu I. Proline-directed random-coil chemical shift values as a tool for the NMR assignment of the tau phosphorylation sites. Chembiochem. 2004;5:73–78. doi: 10.1002/cbic.200300763. [DOI] [PubMed] [Google Scholar]
- 73.Cadwell R.C., Joyce G.F. Randomization of genes by PCR mutagenesis. PCR Methods Appl. 1992;2:28–33. doi: 10.1101/gr.2.1.28. [DOI] [PubMed] [Google Scholar]
- 74.Bartel P.L., Sternglanz R. In: Cellular Interactions in Development: A Practical Approach Practical Approach Series. Hartley D.A., editor. 1993. Cellular interactions in development: a practical approach; pp. 153–179. [Google Scholar]
- 75.Vojtek A.B., Hollenberg S.M. Ras-Raf interaction: two-hybrid analysis. Methods Enzymol. 1995;255:331–342. doi: 10.1016/s0076-6879(95)55036-4. [DOI] [PubMed] [Google Scholar]
- 76.Fromont-Racine M., Rain J.C., Legrain P. Toward a functional analysis of the yeast genome through exhaustive two-hybrid screens. Nat. Genet. 1997;16:277–282. doi: 10.1038/ng0797-277. [DOI] [PubMed] [Google Scholar]
- 77.Karlsson R., Katsamba P.S., Nordin H., Pol E., Myszka D.G. Analyzing a kinetic titration series using affinity biosensors. Anal. Biochem. 2006;349:136–147. doi: 10.1016/j.ab.2005.09.034. [DOI] [PubMed] [Google Scholar]
- 78.Vagin A., Teplyakov A. Molecular replacement with MOLREP. Acta Crystallogr. D Biol. Crystallogr. 2010;66:22–25. doi: 10.1107/S0907444909042589. [DOI] [PubMed] [Google Scholar]
- 79.Murshudov G.N., Skubák P., Lebedev A.A., Pannu N.S., Steiner R.A., Nicholls R.A., Winn M.D., Long F., Vagin A.A. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr. D Biol. Crystallogr. 2011;67:355–367. doi: 10.1107/S0907444911001314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Emsley P., Lohkamp B., Scott W.G., Cowtan K. Features and development of coot. Acta Crystallogr. D Biol. Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Krissinel E., Henrick K. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 2007;372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 82.Banning C., Votteler J., Hoffmann D., Koppensteiner H., Warmer M., Reimer R., Kirchhoff F., Schubert U., Hauber J., Schindler M. A flow cytometry-based FRET assay to identify and analyse protein-protein interactions in living cells. PLoS One. 2010;5:e9344. doi: 10.1371/journal.pone.0009344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Schindowski K., Bretteville A., Leroy K., Bégard S., Brion J.-P., Hamdane M., Buée L. Alzheimer's disease-like tau neuropathology leads to memory deficits and loss of functional synapses in a novel mutated tau transgenic mouse without any motor deficits. Am. J. Pathol. 2006;169:599–616. doi: 10.2353/ajpath.2006.060002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Leroy K., Bretteville A., Schindowski K., Gilissen E., Authelet M., De Decker R., Yilmaz Z., Buée L., Brion J.P. Early axonopathy preceding neurofibrillary tangles in mutant tau transgenic mice. Am. J. Pathol. 2007;171:976–992. doi: 10.2353/ajpath.2007.070345. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.