Abstract
Endometrial decidualization is a prerequisite for implantation, and impaired decidualization is associated with recurrent implantation failure (RIF). Coding genes of the HOX family have been clarified as critical regulators in endometrial decidualization, but the role of long non-coding RNAs (lncRNAs) in the HOX gene family has yet to be determined. The aim of this study was to clarify the possible roles of lncRNAs in the HOX gene family in decidualization. In this study, we identified that HOXA11-AS was the most reduced lncRNA in the HOX family in the human endometrium during the window of implantation, and it was elevated in RIF patients. Mechanistically, HOXA11-AS negatively regulated decidualization through competitive interaction with PTBP1, an RNA-binding protein. Binding of PTBP1 to HOXA11-AS limited PTBP1 availability to regulate PKM1/2 alternative splicing, resulting in enhanced PKM1 and diminished PKM2 expression, thus attenuating decidualization. The pattern of high HOXA11-AS expression and impaired PKM2 splicing was consistently observed in RIF patients. Collectively, our study indicates that the increase of HOXA11-AS is detrimental to endometrial decidualization, likely contributing to RIF. Our study may shed light on the pathogenesis and treatment of RIF.
Keywords: HOXA11-AS, endometrial decidualization, recurrent implantation failure, PTBP1, PKM1/2 alternative splicing
Graphical abstract
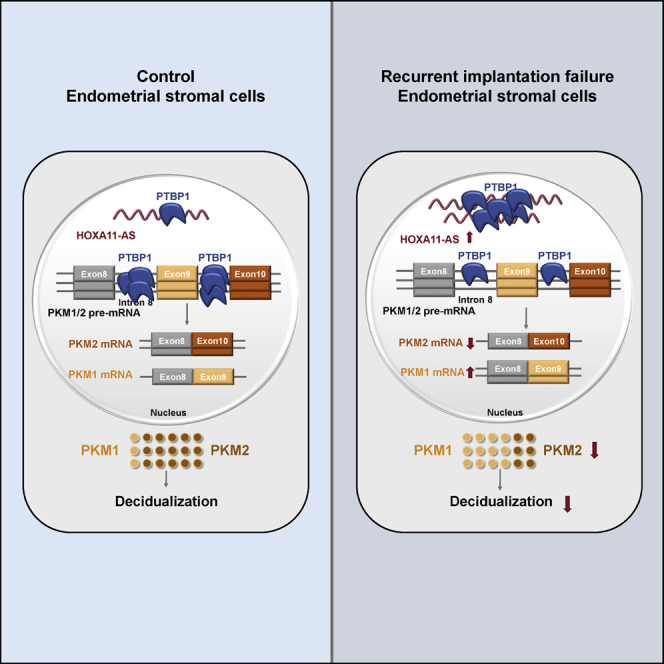
Decidualization of the endometrium is a prerequisite for successful implantation and establishment of pregnancy. Sun et al. showed that increased expression of lncRNA HOXA11-AS attenuated decidualization by competitive binding to PTBP1 and subsequently regulating PKM1/2 alternative splicing both in vitro and in recurrent implantation failure patients
Introduction
In recent years, great advances have been made in assisted reproductive technology, but 15%–20% of patients still experience recurrent implantation failure (RIF).1 RIF refers to failure in achieving pregnancy after three or more embryo transfer cycles with high-quality embryos.1 Successful embryo implantation requires the simultaneous development and interaction between blastocyst and endometrium, which have been proven to be the key factors determining the success of in vitro fertilization-embryo transfer (IVF-ET).2 Although great advances have been made in improving embryo quality, poor endometrial receptivity is still a bottleneck for RIF patients.2,3 Current research reveals that about two-thirds of embryo implantation failures in IVF-ET cycles are due to endometrial receptivity defects.2 During the window of implantation (WOI), human endometrial stromal cells initiate decidualization spontaneously with a series of morphological and functional changes.4 A recent single-cell transcriptomic atlas of human endometrium during the menstrual cycle demonstrated that decidualization is initiated before WOI in a small percentage of human endometrial stromal cells and is widespread at the receptive phase.5 Decidualization is an essential process that determines the establishment and maintenance of endometrial receptivity.6 Impaired decidualization disrupts the invasion of trophoblast cells into the endometrium, causing embryo implantation failure or multiple pregnancy-related diseases, such as preeclampsia7 and premature delivery.8 Although many molecules, such as FOXO1,9 C/EBPβ,10 and HAND2,11 were found to be involved in the spatiotemporal regulation of decidualization, the initiation, maintenance, and regulatory mechanisms of decidualization are still not fully understood.
HOXA10 and HOXA11 belong to the Homeobox gene family (known as HOX genes), which comprise a group of highly conserved genes that encode proteins acting as transcription factors.12 HOXA10 and HOXA11 are expressed in endometrial glands and stroma throughout the menstrual cycle. The expression of HOXA10 and HOXA11 is controlled by estrogen and progesterone, and they are indispensable for embryo implantation in mice and humans because of their vital regulatory roles in decidualization.13, 14, 15 Mutation or deletion of Hoxa10 or Hoxa11 in mice leads to infertility associated with endometrial receptivity and decidualization defects.16,17 Research indicates that HOX family genes also encode many long non-coding RNAs (lncRNAs), most of which are transcribed from the antisense strands of the HOX genes and belong to antisense lncRNAs.18 However, less is known about whether these lncRNAs play a role in the endometrial decidualization and the establishment of endometrial receptivity. HOXA11 antisense (HOXA11-AS) transcript levels change during the menstrual cycle, reaching a peak in the mid-proliferative phase while also varying inversely with HOXA11 mRNA expression levels.19 Nonetheless, HOXA11-AS did not function by binding HOXA11 sense, but by another unknown mechanism.19 The function and regulatory mechanism of HOXA11-AS in endometrial decidualization and receptivity have yet to be elucidated.
In this study, we identified that HOXA11-AS showed the most significant difference in expression between pre-receptive and receptive endometrium among the lncRNA members of HOX family by transcriptomic analysis. The aim of this study was to clarify the possible roles of HOXA11-AS on the pathophysiology of RIF and define the underlying molecular mechanisms of HOXA11-AS in decidualization. We demonstrated that increased HOXA11-AS modulated PKM1/2 alternative splicing by competitive binding to PTBP1, thus attenuating decidualization in RIF patients. Our study, for the first time, demonstrated the function of lncRNA members of the HOX family in decidualization and may shed light on the pathogenesis of RIF.
Results
HOXA11-AS is downregulated during the WOI
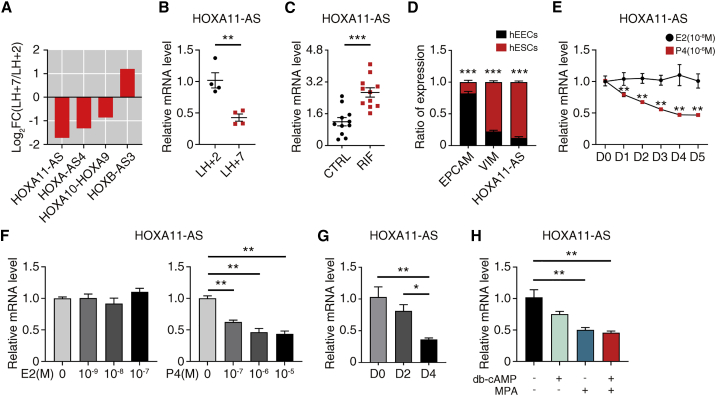
To gain insight into the role of lncRNAs of the HOX family during the WOI, we analyzed their expression changes using our published RNA-seq data for three pre-receptive (LH+2) and three receptive (LH+7) endometria.20 We found that HOXA11-AS was the most significantly changed lncRNA, with a decrease in the receptive phase compared with the pre-receptive phase (Figure 1A). This was validated by qRT-PCR analysis (Figure 1B). Moreover, the expression level of HOXA11-AS was much higher in HCG+7 endometrium of RIF patients (Figure 1C). These data indicate that HOXA11-AS may be a negative regulator in endometrial receptivity.
Figure 1.
HOXA11-AS is downregulated during the window of implantation
(A) Histogram showing differentially expressed lncRNAs in the HOX family between pre-receptive (LH+2) and receptive (LH+7) endometria from patients in the natural cycle (filtered by abs(Log2FC) > 0.5). (B) qRT-PCR assays were conducted to detect the expression of HOXA11-AS in human endometrial samples of LH+2 and LH+7 phases of the natural cycle (n = 4 per group, Student's t test). (C) qRT-PCR assays were performed to examine the expression of HOXA11-AS in human endometrial samples of HCG+7 phase from control (CTRL) and recurrent implantation failure (RIF) patients (n = 11 per group, Student's t test). (D) Expression of HOXA11-AS in primary human endometrial epithelial cells (hEECs) and primary human endometrial stromal cells (hESCs) by qRT-PCR. EPCAM and VIM were used as hEEC and hESC markers (n = 6, Student's t test). (E) Time-dependent effects of estradiol and progesterone at physiological concentration on HOXA11-AS expression level by qRT-PCR (n = 4, one-way ANOVA, Bonferroni test). (F) Dose-dependent effects of estradiol and progesterone on HOXA11-AS expression level on day 4 of treatment by qRT-PCR (n = 4, one-way ANOVA, Bonferroni test). (G) qRT-PCR analysis measuring relative HOXA11-AS level on day 0 (D0), D2, and D4 of in vitro decidualization (n = 4, one-way ANOVA, Bonferroni test). (H) qRT-PCR showing the effects of db-cAMP and MPA, agents for induction of decidualization, on HOXA11-AS expression level on day 4 (n = 4, one-way ANOVA, Bonferroni test). Error bars represent SEM, and the data represent at least three independent experiments. ∗∗∗p < 0.001, ∗∗p < 0.01, ∗p < 0.05.
Human endometrial epithelial cells (hEECs) and human endometrial stromal cells (hESCs) were isolated from proliferative phase endometrium of six healthy women undergoing hysteroscopic endometrial examination, and qRT-PCR analysis showed that HOXA11-AS was predominantly expressed in hESCs (Figure 1D). In addition, we analyzed the expression of HOXA11-AS in different cell types of endometrium using the 10× single-cell dataset from Wang et al.,5 and we found that HOXA11-AS was obviously highly expressed in stromal fibroblasts (Figure S1A), which is consistent with our observation in Figure 1D. We used the 10× dataset to further analyze the expression of HOXA11-AS in stromal fibroblasts across the menstrual cycle. From proliferative phase to late secretory phase, the expression of HOXA11-AS decreased gradually (Figure S1B), which is in accordance with the results in Figure 1B.
Endometrial receptivity is orchestrated by the coordinated actions of ovarian-derived estrogen and progesterone. To further study the effect of estrogen and progesterone on HOXA11-AS expression, hESCs were treated with a physiological concentration of estradiol or progesterone and harvested at different time points. Physiological concentration of progesterone downregulated HOXA11-AS expression in a time-dependent manner, while physiological concentration of estradiol had no effect (Figure 1E). Furthermore, we examined whether HOXA11-AS was regulated by estrogen and progesterone in a dose-dependent manner. We examined HOXA11-AS expression on day 4 because it reaches a plateau after this time point of progesterone treatment (Figure 1E). It turned out that progesterone, but not estradiol, reduced HOXA11-AS in a dose-dependent manner (Figure 1F).
During the WOI, hESCs would initiate spontaneous decidualization, preparing for embryo implantation.6 An in vitro decidualization model was therefore conducted by db-cAMP and MPA treatment. The efficiency of in vitro decidualization was validated by the mRNA and protein levels of two decidualization markers PRL and IGFBP1 (Figures S1C and S1D). We found that HOXA11-AS was significantly lowered in fully decidualized stromal cells on day 4 (Figure 1G). Single-agent treatment with db-cAMP or MPA for 4 days was considered, and MPA displayed a stronger effect on suppressing HOXA11-AS expression than db-cAMP (Figure 1H).
HOXA11-AS attenuates decidualization independent of HOXA10 or HOXA11
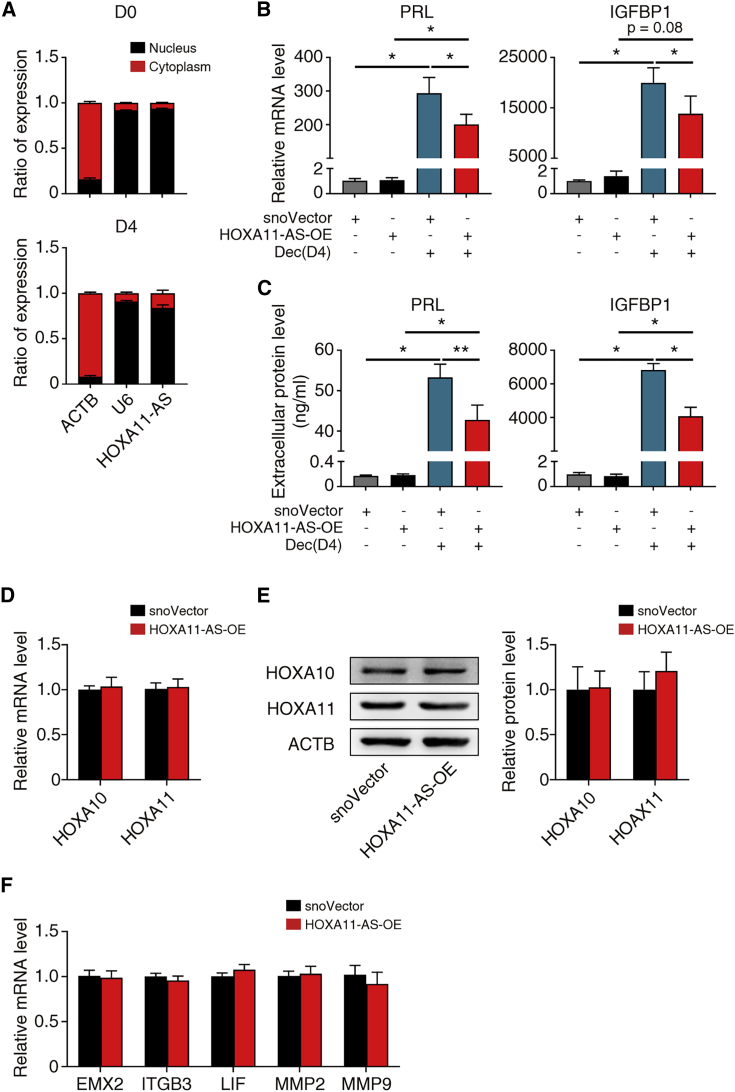
Next, we evaluated whether HOXA11-AS was involved in the regulation of decidualization. Since the subcellular location of lncRNAs is critical,21 we first uncovered the subcellular location of HOXA11-AS in hESCs. The cytoplasmic and nuclear RNA of hESCs from proliferative phase endometrium of healthy women undergoing hysteroscopic endometrial examination were isolated for qRT-PCR analysis, and HOXA11-AS was localized primarily in the nucleus throughout in vitro decidualization (Figure 2A). Thus, we constructed a HOXA11-AS nuclear overexpression plasmid using a snoVector backbone. This construct can stably express sequences of interest and constrain their accumulation to the nucleus without damaging normal nuclear associations and function.22 The overexpression efficiency with or without decidualization and subcellular location of HOXA11-AS were confirmed by qRT-PCR analysis (Figures S2A to S2C). To determine the function of HOXA11-AS on decidualization, we measured the decidualization status of hESCs with or without HOXA11-AS overexpression, after in vitro decidualization or staying untreated for 4 days. HOXA11-AS overexpression significantly decreased PRL and IGFBP1 mRNA and protein levels (Figures 2B and 2C), confirming the hypothesis that HOXA11-AS attenuates decidualization.
Figure 2.
HOXA11-AS attenuates decidualization independent of HOXA10 or HOXA11
(A) qRT-PCR assays were conducted to detect HOXA11-AS expression in the nuclear and cytoplasmic fractions of hESCs before (n = 5) and after (n = 4) in vitro decidualization for 4 days. ACTB and U6 were used as cytoplasmic and nuclear controls, respectively. (B) qRT-PCR showing relative PRL and IGFBP1 mRNA levels in hESCs after transfection with snoVector or HOXA11-AS-OE with or without in vitro decidualization (Dec) for 4 days (n = 4, one-way ANOVA, Bonferroni test). (C) Extracellular PRL and IGFBP1 protein levels after transfection with snoVector or HOXA11-AS-OE with or without in vitro decidualization for 4 days (n = 3, one-way ANOVA, Bonferroni test). (D) qRT-PCR results of relative HOXA10 and HOXA11 mRNA levels in hESCs transfected with snoVector or HOXA11-AS-OE (n = 5, Student's t test). (E) Representative western blotting images and statistical analysis of HOXA10 and HOXA11 in hESCs transfected with snoVector or HOXA11-AS-OE (n = 4, Student's t test). (F) qRT-PCR showing relative mRNA levels of representative downstream genes of HOXA10 and HOXA11 in hESCs transfected with snoVector or HOXA11-AS-OE (n = 5, Student's t test). Error bars represent SEM, and the data represent at least three independent experiments. ∗∗p < 0.01, ∗p < 0.05.
Since HOXA11-AS is the antisense strand of HOXA11, and both HOXA10 and HOXA11 are of immense importance to the regulation of decidualization,15 we wondered whether HOXA11-AS attenuation of decidualization depended on the regulation of HOXA10 or HOXA11. However, overexpression of HOXA11-AS had no influence on the mRNA or protein levels of HOXA10 and HOXA11 (Figures 2D and 2E). In addition, HOXA11-AS overexpression did not change the expression of representative downstream target genes of HOXA10 and HOXA1123,24 (Figure 2F). All of these results suggest that overexpression of HOXA11-AS attenuates decidualization neither by influencing HOXA10 and HOXA11 expression nor by interfering with their function as transcription factors.
HOXA11-AS-attenuated decidualization depends on PTBP1
Interaction with specific proteins is an important mechanism for nuclear lncRNAs to exert their functions.25 Therefore, we conducted a preliminary exploration of the interacting proteins of HOXA11-AS. First, the potential binding proteins of HOXA11-AS were predicted by Annolnc,26 ENCORI,27 RBPDB,28 and starBase29 based on the published CLIP-seq data (Figure S3A). The prediction results are demonstrated in Table S1. Those shared by at least two databases were selected as candidates, with 11 in total (ELAVL1, FUS, SRSF1, YTHDC1, FMR1, PTBP1, CSTF2T, IGF2BP3, DGCR8, TIAL1, and UPF1). Next, we conducted pathway enrichment analysis based on these 11 proteins and found that these proteins were mainly engaged in RNA metabolism, transport, and splicing processes (Figure S3B). Finally, a protein interaction network was constructed using Metascape software,30 and four proteins (ELAVL1, SRSF1, PTBP1, and FUS) were identified to aggregate into one MCODE network (Figure S3C). In particular, the PTBP1 protein played a seed role in the MCODE network (Figure S3C) and it has been reported to bind with pre-mRNA and regulate RNA splicing.31 Hence, PTBP1 was selected for further study as the potential interacting protein of HOXA11-AS had the most predictive value.
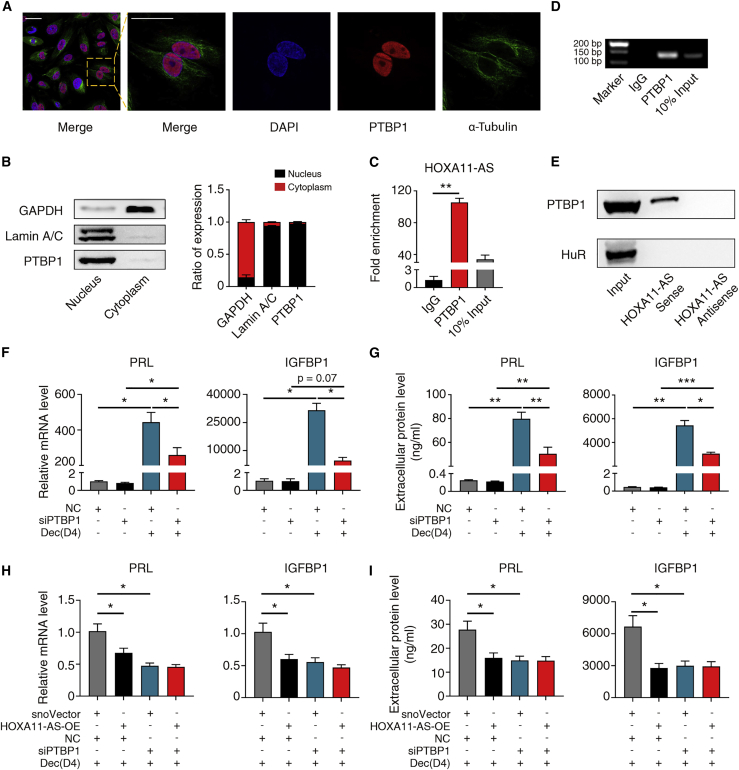
Considering HOXA11-AS is located in the nucleus, we suspected that PTBP1 is also located in the nucleus. Indeed, the nuclear localization of PTBP1 in hESCs was confirmed by western blotting and immunofluorescence (Figures 3A and 3B). To confirm the interaction between PTBP1 and HOXA11-AS in the nucleus, we conducted RIP (RNA immunoprecipitation)-qPCR analysis. A 100-fold enrichment over a non-specific rabbit IgG control was observed (Figures 3C and 3D). In addition, we performed in vitro RNA pull-down assay using in-vitro-transcribed sense or antisense transcripts of HOXA11-AS. HOXA11-AS sense RNA bound to PTBP1 protein while its antisense RNA did not (Figure 3E). At the same time, neither HOXA11-AS sense nor antisense RNA could interact with negative control protein, HuR (Figure 3E). Collectively, these results demonstrate that HOXA11-AS specifically binds to PTBP1.
Figure 3.
HOXA11-AS attenuated decidualization depends on PTBP1
(A) Immunofluorescence images of hESCs. DAPI (blue), PTBP1 (red), and α-tubulin (green). Scale bar, 30 μm. (B) Representative western blotting images and statistical analysis of PTBP1 in the nuclear and cytoplasmic fractions of hESCs. GAPDH and Lamin A/C were used as cytoplasmic and nuclear controls, respectively (n = 3). (C) Enrichment of HOXA11-AS by PTBP1 RIP (RNA immunoprecipitation) in hESCs as determined by qRT-PCR. Normal rabbit immunoglobulin G (IgG) was used as a negative control (n = 3, one-way ANOVA, Bonferroni test). (D) Agarose gel electrophoresis of RIP qRT-PCR products. (E) RNA pull-down assay was performed in hESCs with HOXA11-AS sense and antisense RNA followed by western blotting. HuR was used as a negative control. (F) qRT-PCR showing relative PRL and IGFBP1 mRNA levels after transfection with NC or siPTBP1 with or without in vitro decidualization (Dec) for 4 days (n = 4, one-way ANOVA, Bonferroni test). (G) Extracellular PRL and IGFBP1 protein levels after transfection with NC or siPTBP1 with or without in vitro decidualization for 4 days (n = 4, one-way ANOVA, Bonferroni test). (H) qRT-PCR results of relative PRL and IGFBP1 mRNA levels after PTBP1 knockdown or HOXA11-AS overexpression with in vitro decidualization for 4 days (n = 4, one-way ANOVA, Bonferroni test). (I) Extracellular PRL and IGFBP1 protein levels after PTBP1 knockdown or HOXA11-AS overexpression with in vitro decidualization for 4 days (n = 4, one-way ANOVA, Bonferroni test). Error bars represent SEM, and the data represent at least three independent experiments. ∗∗∗p < 0.001, ∗∗p < 0.01, ∗p < 0.05.
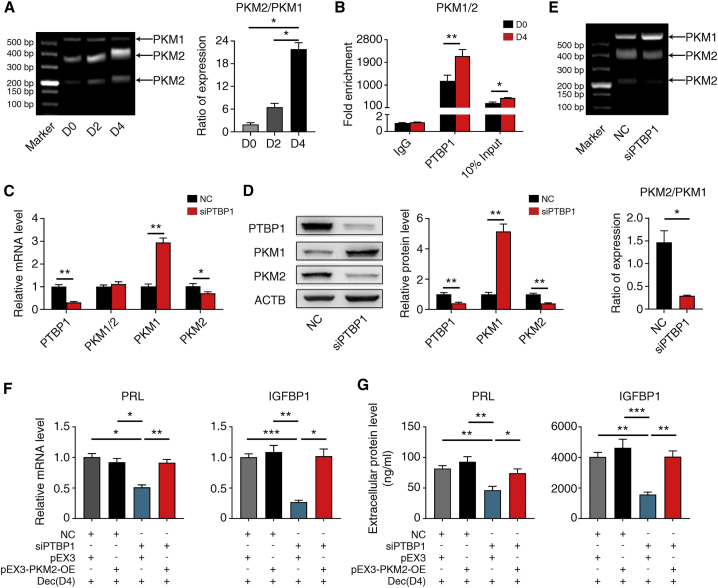
To determine whether PTBP1 affects decidualization, we silenced PTBP1 expression in hESCs with siPTBP1. The knockdown efficiency of PTBP1 was validated by qRT-PCR analysis and western blotting (Figures S3D and S3E). PTBP1 knockdown significantly diminished PRL and IGFBP1 mRNA and protein levels (Figures 3F and 3G), indicating that PTBP1 promoted decidualization. To further investigate whether HOXA11-AS attenuated decidualization depended on PTBP1, we simultaneously overexpressed HOXA11-AS and knocked down PTBP1 in hESCs followed by decidualization for 4 days. PTBP1 knockdown abolished the further attenuated decidualization caused by HOXA11-AS overexpression (Figures 3H and 3I), suggesting that HOXA11-AS attenuates decidualization dependent on PTBP1.
PKM2 is involved in the HOXA11-AS-mediated regulation of decidualization
We next examined how PTBP1 was involved in the HOXA11-AS-mediated regulation of decidualization. The mRNA and protein levels of PTBP1 had no change during decidualization (Figures 4A and 4B), indicating that HOXA11-AS may attenuate decidualization by modulating PTBP1 function rather than its mRNA or protein expression. HOXA11-AS may also modulate some downstream targets of PTBP1 involved in the regulation process.
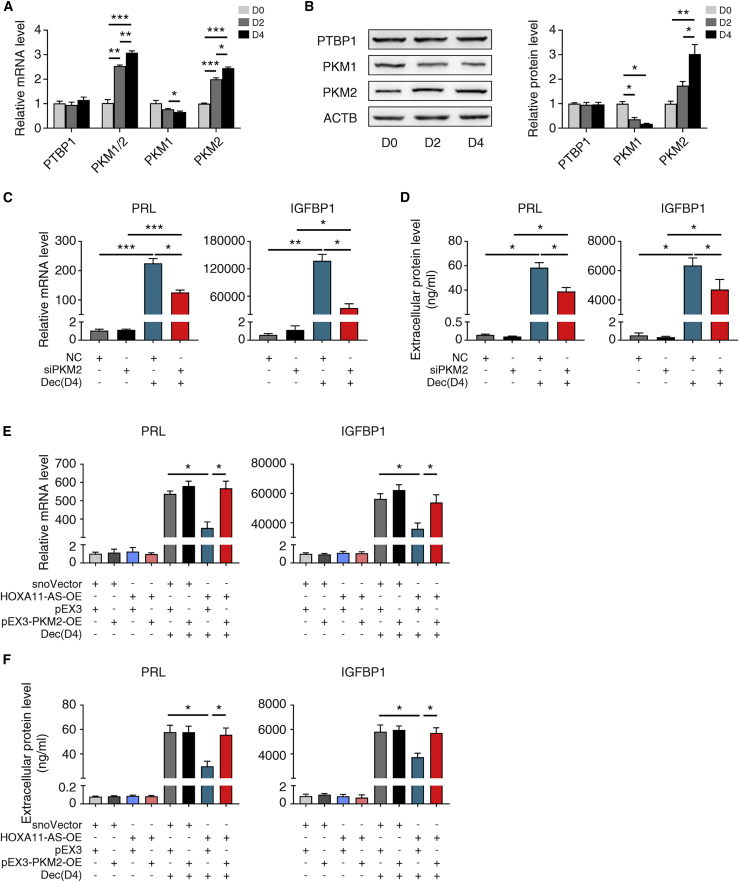
Figure 4.
PKM2 is involved in the HOXA11-AS mediated regulation of decidualization
(A) qRT-PCR showing relative mRNA levels of PTBP1, PKM1/2, PKM1, and PKM2 during in vitro decidualization (n = 4, one-way ANOVA, Bonferroni test). (B) Representative western blotting images and statistical analysis of PTBP1, PKM1, and PKM2 during in vitro decidualization (n = 3, one-way ANOVA, Bonferroni test). (C) qRT-PCR showing relative PRL and IGFBP1 mRNA levels after transfection with NC or siPKM2 with or without in vitro decidualization (Dec) for 4 days (n = 5, one-way ANOVA, Bonferroni test). (D) Extracellular PRL and IGFBP1 protein levels after transfection with NC or siPKM2 with or without in vitro decidualization for 4 days (n = 3, one-way ANOVA, Bonferroni test). (E) qRT-PCR showing relative PRL and IGFBP1 mRNA levels with HOXA11-AS or PKM2 overexpression with or without in vitro decidualization for 4 days (n = 3, one-way ANOVA, Bonferroni test). (F) Extracellular PRL and IGFBP1 protein levels with HOXA11-AS or PKM2 overexpression with or without in vitro decidualization for 4 days (n = 3, one-way ANOVA, Bonferroni test). Error bars represent SEM, and the data represent at least three independent experiments. ∗∗∗p < 0.001, ∗∗p < 0.01, ∗p < 0.05.
Previous studies reveal that PKM2 plays an essential role in mouse decidualization32,33 and that PTBP1 is an important alternative splicing regulator of PKM1/2.34 When PTBP1 expression is low, PKM1 is the dominant isoform and PKM2 is minimal, and vice versa.34 We also found that PTBP1 can interact with PKM1/2 mRNA in hESCs by RIP-qPCR analysis (Figures S4A and S4B). Accordingly, we hypothesized that PKM2 was another mediator downstream of HOXA11-AS to regulate decidualization.
We first evaluated the expression and function of PKM2 through decidualization. PKM2 expression increased and PKM1 expression decreased at both mRNA and protein levels, together with elevated total PKM1/2 mRNA expression (Figures 4A and 4B). Thus, siRNA specifically targeting PKM2 was designed without influencing PKM1 expression (Figures S5A to S5D). Consistent with current evidence from mouse models, PKM2 knockdown significantly lowered PRL and IGFBP1 mRNA and protein levels in hESCs (Figures 4C and 4D), implicating that the modulation of decidualization by PKM2 happened both in mouse and human.
We next transfected hESCs with a pEX3-PKM2-OE plasmid for PKM2 overexpression (Figures S5E–S5H). Intriguingly, PKM2 overexpression completely rescued the suppressed PRL and IGFBP1 mRNA and protein levels caused by HOXA11-AS overexpression (Figures 4E and 4F). These data validate the hypothesis that PKM2 is involved in the HOXA11-AS mediated regulation of decidualization.
PTBP1-regulated decidualization is dependent on PKM2
Both PTBP1 and PKM2 are involved in the regulation of decidualization caused by HOXA11-AS and, as mentioned before, PTBP1 regulates the alternative splicing of PKM1/2.34 However, whether this PTBP1-mediated alternative splicing of PKM1/2 occurs during decidualization has not been reported. Hence, we used the PstI restriction enzyme to distinguish between PKM1 and PKM2 alternative splicing (schematic diagram shown in Figures S6A and S6B) and examined PKM alternative splicing during decidualization. The results showed that PKM1 decreased slightly, while PKM2 increased and became the dominant variant after decidualization (Figure 5A). Although PTBP1 expression did not change during decidualization (Figures 4A and 4B), the enrichment of PKM1/2 mRNA binding to PTBP1 was largely enhanced after decidualization (Figure 5B). This finding suggests that the shift toward the PKM2 variant during decidualization was due to increasing interaction of PTBP1 protein and PKM1/2 mRNA.
Figure 5.
PTBP1-regulated decidualization is dependent on PKM2
(A) Representative gel electrophoresis images and statistical analysis of the proportion of PKM1 and PKM2 during in vitro decidualization (n = 3, one-way ANOVA, Bonferroni test). (B) Enrichment of PKM1/2 mRNA by PTBP1 RIP before and after in vitro decidualization for 4 days was measured by qRT-PCR (n = 3, Student's t test). (C) qRT-PCR showing relative mRNA levels of PTBP1, PKM1/2, PKM1, and PKM2 after transfection with NC or siPTBP1 (n = 4, Student's t test). (D) Representative western blotting images and statistical analysis of PTBP1, PKM1, and PKM2 after transfection with NC or siPTBP1 (n = 4, Student's t test). (E) Representative gel electrophoresis images and statistical analysis of the proportion of PKM1 and PKM2 after transfection with NC or siPTBP1 (n = 3, Student's t test). (F) qRT-PCR showing relative PRL and IGFBP1 mRNA levels after PTBP1 knockdown or PKM2 overexpression as indicated after in vitro decidualization (Dec) for 4 days (n = 4, one-way ANOVA, Bonferroni test). (G) Extracellular PRL and IGFBP1 protein levels after PTBP1 knockdown or PKM2 overexpression as indicated after in vitro decidualization for 4 days (n = 4, one-way ANOVA, Bonferroni test). Error bars represent SEM, and the data represent at least three independent experiments. ∗∗∗p < 0.001, ∗∗p < 0.01, ∗p < 0.05.
To confirm the effect of PTBP1 on PKM1/2 alternative splicing, we examined PKM1 and PKM2 expression and PKM alternative splicing after PTBP1 knockdown. PTBP1 knockdown did not alter PKM1/2 mRNA expression (Figure 5C). However, enhanced PKM1 expression and reduced PKM2 were observed at mRNA and protein levels after PTBP1 knockdown (Figures 5C and 5D). Analysis of PKM alternative splicing also confirmed the proportion change of PKM1 and PKM2 variants (Figure 5E). Concordant with previous results, PKM2 overexpression also completely rescued the attenuated PRL and IGFBP1 mRNA and protein levels caused by PTBP1 knockdown (Figures 5F and 5G). Collectively, the results identify that the mechanism of PTBP1 to regulate PKM alternative splicing without changing total mRNA expression also applies to decidualization.
HOXA11-AS modulates PKM1/2 alternative splicing by competitive binding to PTBP1
Since HOXA11-AS and PKM1/2 mRNA both interacted with PTBP1, and PKM2 served as the common downstream target of HOXA11-AS and PTBP1, we speculated that there was competition between HOXA11-AS and PKM1/2 mRNA to interact with PTBP1 during decidualization, leading to limited PKM2 splicing.
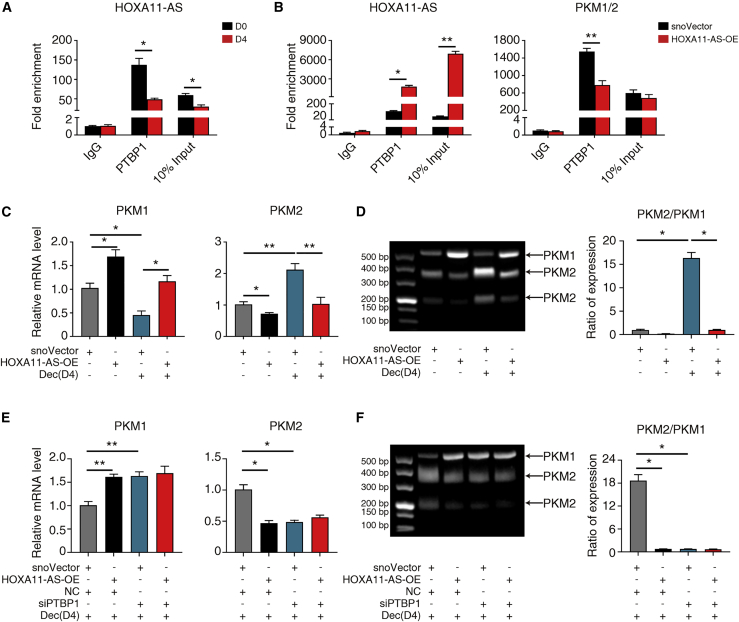
To examine this interesting hypothesis, we tested the PTBP1 interaction with HOXA11-AS and PKM1/2 mRNA with or without HOXA11-AS overexpression. Generally, during decidualization, PTBP1 restricted its interaction with HOXA11-AS (Figure 6A), but promoted its binding to PKM1/2 mRNA (Figure 5B). In contrast, HOXA11-AS overexpression augmented the PTBP1 interaction with HOXA11-AS but mitigated its interaction with PKM1/2 mRNA under decidualization condition (Figure 6B).
Figure 6.
HOXA11-AS modulates PKM1/2 alternative splicing by competitive binding to PTBP1
(A) Enrichment of HOXA11-AS by PTBP1 RIP before and after in vitro decidualization for 4 days was measured by qRT-PCR (n = 3, Student's t test). (B) Enrichment of HOXA11-AS and PKM1/2 mRNA by PTBP1 RIP after HOXA11-AS overexpression under decidualization condition was measured by qRT-PCR (n = 3, Student's t test). (C) qRT-PCR showing relative mRNA levels of PKM1 and PKM2 after transfection with snoVector or HOXA11-AS-OE with or without in vitro decidualization (Dec) for 4 days (n = 7, one-way ANOVA, Bonferroni test). (D) Representative gel electrophoresis images and statistical analysis of the proportion of PKM1 and PKM2 after transfection with snoVector or HOXA11-AS-OE with or without in vitro decidualization for 4 days (n = 3, one-way ANOVA, Bonferroni test). (E) qRT-PCR showing relative mRNA levels of PKM1 and PKM2 after PTBP1 knockdown or HOXA11-AS overexpression as indicated with in vitro decidualization for 4 days (n = 4, one-way ANOVA, Bonferroni test). (F) Representative gel electrophoresis images and statistical analysis of the proportion of PKM1 and PKM2 after PTBP1 knockdown or HOXA11-AS overexpression as indicated with in vitro decidualization for 4 days (n = 3, one-way ANOVA, Bonferroni test). Error bars represent SEM, and the data represent at least three independent experiments. ∗∗p < 0.01, ∗p < 0.05.
Furthermore, HOXA11-AS overexpression did not change total PKM1/2 mRNA (Figure S7A), but instead upregulated PKM1 and downregulated PKM2 at mRNA and protein levels (Figures 6C and S7B). Alternative splicing assay also confirmed the proportion shift of PKM1 and PKM2 variants by HOXA11-AS overexpression (Figure 6D). The data suggest that HOXA11-AS interfered with PKM1/2 alternative splicing.
Finally, we asked whether PKM1/2 splicing regulation by HOXA11-AS overexpression was dependent on PTBP1. Regardless of HOXA11-AS expression, PTBP1 did not regulate PKM1/2 at the transcriptional level (Figures 5C and S7C). Instead, PTBP1 silencing prevented the further alteration of PKM1 and PKM2 mRNA and protein levels caused by HOXA11-AS overexpression (Figures 6E and S7D), consistent with the alternative splicing result that PTBP1 knockdown abrogated the proportion change of PKM1 and PKM2 isoforms caused by HOXA11-AS overexpression (Figure 6F). All these data conclude that HOXA11-AS modulates PKM1/2 alternative splicing by competitive binding to PTBP1.
PKM2 splicing is impaired in RIF patients
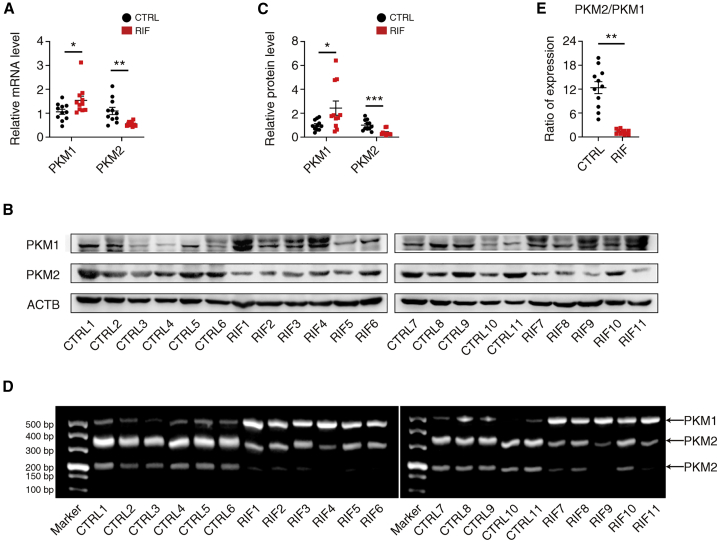
Based on the above results in hESCs and to explore our discovery in RIF patients, we tested PKM1 and PKM2 expression in HCG+7 endometrium of CTRL and RIF patients. Both mRNA and protein levels of PKM2 were lower, whereas those of PKM1 were higher, in HCG+7 endometrium of RIF patients (Figures 7A–7C). In addition, the expression of HOXA11-AS was higher in these RIF patients (Figure 1C). This finding is in accordance with PKM1 and PKM2 mRNA and protein changes caused by HOXA11-AS overexpression in hESCs. We further examined PKM alternative splicing in these CTRL and RIF patients. Consistent with our discovery in hESCs after decidualization, PKM2 splicing was significantly impaired in RIF patients compared with CTRL patients (Figures 7D and 7E). Overall, we found that HOXA11-AS attenuates decidualization by competitive binding to PTBP1 and subsequently regulating PKM1/2 alternative splicing both in vitro and in RIF patients.
Figure 7.
PKM2 splicing is impaired in recurrent implantation failure patients
(A) qRT-PCR showing relative PKM1 and PKM2 mRNA levels in the human endometrial samples of HCG+7 phase from CTRL and RIF patients (n = 11 per group, Student's t test). (B) Western blotting images of PKM1, PKM2, and ACTB in the human endometrial samples of HCG+7 phase from CTRL and RIF patients (n = 11 per group). (C) Statistical analysis of western blotting for PKM1 and PKM2 in the human endometrial samples of HCG+7 phase from CTRL and RIF patients (n = 11 per group, Student's t test). (D) Gel electrophoresis images of the proportion of PKM1 and PKM2 in the human endometrial samples of HCG+7 phase from CTRL and RIF patients (n = 11 per group). (E) Statistical analysis of the proportion of PKM1 and PKM2 in the human endometrial samples of HCG+7 phase from CTRL and RIF patients (n = 11 per group, Student's t test). Error bars represent SEM, and the data represent at least three independent experiments. ∗∗∗p < 0.001, ∗∗p < 0.01, ∗p < 0.05.
Discussion
We for the first time discovered that HOXA11-AS, a lncRNA member of the HOX family, plays a critical regulatory role in decidualization. HOXA11-AS was predominantly expressed in hESCs and the expression of HOXA11-AS was restrained in the WOI and under in vitro decidualization conditions, concordant with the cell type that expressed HOXA11-AS and its expression through menstrual cycle in a recent single-cell transcriptomic dataset.5 Consistently, the abundance of HOXA11-AS was elevated in the endometrium of RIF patients.
LncRNAs are non-coding RNAs with a length of more than 200 nucleotides.35 Increasing studies have shown that lncRNAs play a crucial role in various cellular processes, such as cell cycle, differentiation, proliferation, migration, metabolism, and apoptosis, via interactions with RNA-binding proteins, chromatin modification, and ceRNA (competing endogenous RNA) networks.25,36 Previous studies have demonstrated that lncRNAs may be involved in decidualization.37, 38, 39, 40 Our research group reported that the significantly low expression of lncRNA TUNAR during the WOI promoted decidualization of endometrial stromal cells, and its abnormally high expression was closely related to RIF.41 In this study, we found HOXA11-AS was located predominantly in the nucleus of hESCs and the nuclear overexpression of HOXA11-AS significantly attenuated decidualization. Mechanistically, HOXA11-AS overexpression did not affect HOXA10 and HOXA11 expression or their functions as transcription factors, consistent with the study by Chau et al.19 Furthermore, we clarified that PTBT1, as a novel mediator, was involved in the regulation of decidualization.
PTBP1 is a known regulator of post-transcriptional gene expression, which controls the splicing, translation, stability, and localization of mRNA.31,42 Based on the published sequencing data, PTBP1 was predicted and further confirmed as the most promising binding protein of HOXA11-AS. Binding of PTBP1 in intron 8 of PKM pre-mRNA favors skipping of exon 9 in the mature transcript, thus generating the PKM2 isoform containing exon 10.43 PTBP1-mediated PKM1/2 alternative splicing also occurred in hESCs. Indeed, this regulatory function of PTBP1 rather than its expression mediated the HOXA11-AS attenuated decidualization.
Pkm2 was found to be dramatically increased with the onset of decidualization in mice uteri and Pkm2 knockdown led to abnormal decidualization in mice.32,33 PKM2 overexpression could completely rescue the attenuated decidualization caused by HOXA11-AS overexpression or PTBP1 knockdown. In addition, overexpression of HOXA11-AS resulted in a significant isoform switch from PKM1 to PKM2 without PKM1/2 mRNA change. Specifically, HOXA11-AS competitively bound to PTBP1, making PTBP1 less available for regulating PKM alternative splicing. We also verified this expression correlation of high HOXA11-AS to modest PKM2 in RIF patients.
PTBP1 promotes PKM2 and reduces PKM1 expression, which mainly leads to a metabolic transition from oxidative phosphorylation to glycolysis in cancer cells, and consequently contributes to the Warburg effect and tumorigenesis.34 Salama et al. reported that estradiol induced a Warburg-like glucose metabolism in hESCs and enhanced PKM2 expression.44 In hESCs, knockdown of PKM2 suppresses Warburg-like glucose metabolism and estrogen-induced cell proliferation.44 Although how PKM2 mediated metabolic change contributes to decidualization remains unclear, our data and other reports provide evidence for further investigation of PKM2 mediated metabolic regulation in decidualization.
Lactic acid is the final product of glycolysis. We found that lactic acid was elevated after decidualization (data not shown), which is in accordance with previous studies.33,45 However, we did not observe the enhancement effect of lactic acid on decidualization (data not shown). Kathryn et al. also reported that lactic acid did not change the extent of decidualization.46 These results suggest that lactic acid may not promote decidualization directly. But as an important metabolic product, lactic acid may function at the maternal-fetal interface through other mechanisms, such as promoting trophoblastic invasion, trophoblastic migration, and maternal angiogenesis; providing metabolic fuel; and modulating decidual immune cells to ensure local immune tolerance and defense against any pathogens.47 Arzu et al. performed endometrium spectroscopy on RIF and fertile controls before embryo transfer and used a multi-voxel spectroscopy sequence to detect lactic acid.48 They found that lactic acid was increased in patients who could not conceive.48 So far, the relationship between lactic acid and RIF remains unclear and would be an interesting project to study further.
Admittedly, we cannot exclude that other factors may be involved in downstream of the HOXA11-AS regulatory axis. For example, it has been reported that nuclear HOXA11-AS recruited Ezh2 and Lsd1 protein and regulated RND3 mRNA expression in trophoblast cells of preeclamptic placenta.49 Whether this regulation engages in HOXA11-AS attenuated decidualization remains to be further clarified. Moreover, HOXA11-AS pull-down combined with mass spectrometry will uncover more clues and enrich our understanding of HOXA11-AS functions in decidualization. Based on a recent single-cell RNA sequencing dataset in vitro50 and in vivo,51 decidualization is not a single differentiation response. Examination of HOXA11-AS for 4 days in vitro may not fully recapitulate the whole decidual reaction. There are limitations in only using traditional biomarkers, such as PRL and IGFBP1, to measure decidualization after in vitro decidualization for 4 days. Examining the emergence of anti-inflammatory decidual cells and senescent decidual cells could be another important aspect to evaluate decidualization after in vitro decidualization for 8 days. However, our discovery of the new regulator PTBP1, whose intrinsic function is of greater consequence than its expression level, creates a template for the functional research in decidualization. Examining how many critical PTBP1-like regulators there are and how to dissect their functions during decidualization is interesting and worth investigating.
In summary, our findings reveal that HOXA11-AS is decreased during the WOI and decidualization. Expression of HOXA11-AS is abnormally high in RIF patients. HOXA11-AS attenuates hESC decidualization by interfering with PKM1/2 alternative splicing via competitive binding to PTBP1. Our study provides a new perspective regarding the molecular mechanisms underlying the abnormal decidualization of RIF patients and identifies the HOXA11-AS/PTBP1/PKM2 axis as a novel target pathway for the treatment of RIF.
Materials and methods
Patients and tissue collection
Judged by the urinary luteinizing hormone (LH) surge, endometrial samples were obtained from four women in the LH+2 (pre-receptive) phase and four women in the LH+7 (receptive) phase of the natural cycle. The baseline characteristics of LH+2 and LH+7 patients are shown in Table S2. The endometrium of control (CTRL) (n = 11) and RIF (n = 11) were obtained from patients going through gonadotropin-releasing hormone antagonist stimulation cycle without fresh embryo transfer. Two days after injection of human chorionic gonadotropin (HCG+2), ovum was obtained. From HCG+2, progesterone sustained-release vaginal gel qd and dydrogesterone tablets 10 mg tid were given until HCG+7. Biopsy samples were collected at HCG+7, known as the WOI stage. The control patients referred to those with normal menstrual cycles, who underwent IVF treatment due to fallopian tube factors or male factors, and the first embryo transfer was successful. In this study, RIF is defined as failures after three or more embryo transfer cycles with high-quality embryos.1 Baseline characteristics of CTRL and RIF patients are represented in Table S3. Endometrial biopsies were collected with Pipelle catheters from the uterine fundus, quickly snap-frozen in liquid nitrogen, and stored at −80°C before extraction of protein and total RNA. The proliferative phase endometrium came from six healthy women undergoing hysteroscopic endometrial examination, which was used for the isolation of primary human endometrial epithelial and stromal cells. All the primary endometrial stromal cells for in vitro culture were obtained from healthy women undergoing hysteroscopic endometrial examination. Aside from pathological examination, the remaining endometrium was used for experimental study.
All procedures were performed at the Center for Reproductive Medicine, Ren Ji Hospital, Shanghai Jiao Tong University School of Medicine. Informed consent was obtained from all participants and this project was approved by the Ethics Committee of Ren Ji Hospital (2018072608).
Isolation of primary human endometrial epithelial and stromal cells
Primary hEECs and hESCs were separated from endometrium samples according to a selective attachment strategy based on modifications of previous methods.52 In brief, specimens were first washed several times in Dulbecco's modified Eagle's medium/Hams F12 (DMEM/F12, Gibco, Grand Island, NY) containing 10% (v/v) fetal bovine serum (FBS) (Gibco) and 1% (v/v) penicillin-streptomycin-neomycin (Gibco) to remove excess red blood cells, and then digested with 6 mg/mL collagenase type I at 150 rpm for 40 min and 1 mg/mL deoxyribonuclease I for 15 min without shaking (Sigma-Aldrich, St. Louis, MO). Digested epithelial/stromal mixtures were filtered through 180 and 40 μm cell strainers sequentially. hESCs passed through the 40 μm strainer and were resuspended in DMEM/F12 medium. hEECs retained in the 40 μm strainer were collected by backwashing, and then resuspended in DMEM/F12 medium. Residual hESCs in the epithelial clumps were removed by selective attachment because hEECs attachment were considered to take longer. Cell purity was tested by qRT-PCR for EPCAM and VIM mRNA levels.
Cell culture and treatment
hESCs were cultured in DMEM/F12 medium without phenol red (Gibco) with 2% charcoal/dextran-treated FBS (HyClone, Logan, UT) at 37°C in a humidified 5% CO2 incubator. To establish in vitro decidualization model, hESCs were treated with 1 μM MPA and 0.5 mM db-cAMP (Sigma-Aldrich). After 2 days, the culture medium was renewed with the same treatment. To study the effect of estrogen and progesterone on HOXA11-AS, hESCs were treated with a physiological concentration of estradiol (10−8 M) and progesterone (10−6 M) (Sigma-Aldrich) in a time-dependent manner, and then treated with estradiol and progesterone in a dose-dependent fashion for 4 days.
RNA extraction and reverse transcription
Total RNA was isolated from endometrial tissues and cells using the TRIzol reagent (Invitrogen, Carlsbad, CA) according to the manufacturer's instructions. RNA concentration and quality were determined by NanoDrop ND-2000 spectrophotometer (Thermo-Fisher Scientific, Waltham, MA). cDNA was synthesized from 500 ng of total RNA with the PrimeScript RT reagent kit (TaKaRa, Shiga, Japan) according to the manufacturer's instructions.
Quantitative real-time PCR
Quantitative real-time PCR was performed using SYBR Green Master Mix (TaKaRa) with an ABI Prism System (Applied Biosystems, Carlsbad, CA). All qRT-PCR reactions were performed in duplicate and the final volume of reaction was 10 μL. Relative mRNA or lncRNA levels were calculated by the 2−ΔΔCt method with ACTB as an internal control. Primer sequences are listed in Table S4.
Analysis of PKM alternative splicing
The total PKM1/2 transcripts were amplified from cDNA with Taq DNA Polymerase (CWBIO, Beijing, China) using a pair of universal primers listed in Table S4. After 25 cycles of amplification, PCR products were purified using a TaKaRa MiniBEST DNA Fragment Purification Kit (TaKaRa) according to the manufacturer's instructions, then digested with PstI (New England Biolabs, Ipswich, MA). Digested DNA fragments were examined by agarose gel electrophoresis. The PKM1 variant includes exons 8, 9, 11, and 12, while the PKM2 variant includes exons 8, 10, 11, and 12, and the length of exons 9 and 10 is same. The forward primer is designed in exon 8 and the backward primer is designed in exon 12, so the universal primer can amplify PKM1 and PKM2 variants simultaneously and the PCR products of these two variants have the same length. Because exon 10 of the PKM2 variant has a PstI restriction enzyme cut site while exon 9 of the PKM1 variant does not, so there is one band for PKM1 and two bands for PKM2 after PstI digestion. The schematic diagram was shown in Figures S6A and S6B.
Separation of nucleus and cytoplasm RNA and protein
Nucleus and cytoplasm RNA of hESCs were extracted using a Cytoplasmic & Nuclear RNA Purification Kit (Norgen Biotek, Toronto, Canada) according to the manufacturer's instructions. Nuclear and cytoplasm proteins of hESCs were extracted using a Nuclear/Cytoplasmic Protein Extraction Kit (AmyJet, Wuhan, China) following the manufacturer's instructions.
Transfection of cells
hESCs were seeded into six-well plates and cultured for 1 day before transfection with siRNAs or plasmid DNA using Lipofectamine 3000 Reagent (Invitrogen) following the manufacturer's protocol. The HOXA11-AS nuclear overexpression plasmid was constructed based on snoVector backbone generously provided by the Chinese Academy of Sciences.22 The NC, siPTBP1 and siPKM2 siRNAs were designed by GenePharma (Shanghai, China). NC, a scramble sequence of siRNA, served as a negative control. The siRNAs sequences are provided in Table S5. The PKM2 overexpression plasmid was constructed using a pEX3 vector offered by GenePharma. For one well in a six-well plate, the siRNA transfection complex includes 250 μL opti-MEM, 5 μL 20 μM siRNA, and 3.75 μL lipo3000. For one well in a six-well plate, the plasmid transfection complex includes 250 μL opti-MEM, 2500 ng DNA, 5 μL p3000, and 3.75 μL lipo3000. The transfection complex was mixed well and maintained for 15 min before adding to the well. Twenty-four hours after transfection, the transfection reagents were replaced by DMEM/F12 medium without phenol red (Gibco) with 2% charcoal/dextran-treated FBS (HyClone) in the non-decidualized group, with db-cAMP and MPA also added in the decidualized group. The transfection efficiency was examined 5 days after transfection in the non-decidualized group and 4 days after treatment of db-cAMP and MPA in the decidualized group.
Immunofluorescence
hESCs were seeded in four-well Millicell EZ slides (Millipore, Burlington, MA) with different treatments before immunofluorescence. Then the cells were fixed with 4% paraformaldehyde and permeabilized using 0.5% Triton X-100, followed by blocking in normal goat serum for 1 h at room temperature. After blocking, the cells were incubated with anti-PTBP1 rabbit monoclonal antibody (1:100 diluent, Cell Signaling Technology, Danvers, MA) and Alexa Fluor 488 Anti-alpha Tubulin antibody (1:250 diluent, Abcam, Cambridge, UK) at 4°C overnight, then incubated with Cy3-conjugated goat anti-rabbit IgG antibody (1:100 diluent, Proteintech, Chicago, IL) for 1 h at room temperature, protected from light. Finally, the nucleus was dyed with DAPI (1 μg/mL, Cell Signaling Technology) for 5 min before observation using a Leica SP8 laser scanning confocal microscope (Leica, Weztlar, Germany).
Western blotting
Total protein from hESCs was extracted using an ice-cold radio-immunoprecipitation assay (RIPA) lysis buffer (CWBIO) containing a protease inhibitor cocktail (cOmplete Tablets, Mini EASYpack, one tablet for 10 mL RIPA, Roche, Basel, Switzerland) and a phosphatase inhibitor (PhosSTOP EASYpack, one tablet for 10 mL RIPA, Roche). Protein concentration was quantified using a Bradford assay kit (Beyotime, Shanghai, China), and 30 μg of protein of each sample was mixed with 5× protein loading buffer and denatured at 95°C–100°C for 5 min. The denatured protein was electrophoresed on 10% SDS-polyacrylamide gel and transferred to a nitrocellulose membrane. After blocking, the membranes were incubated with different primary antibodies at 4°C overnight, followed by respective secondary antibody conjugated with horseradish peroxidase for 1 h at room temperature. Relative band intensity was detected using a chemiluminescent detection kit (Epizyme, Shanghai, China), visualized with a G-Box chemiluminescence image capture system (Syngene, Frederick, MD). Primary antibodies are summarized in Table S6.
RIP
Based on the manufacturer's instructions, RIP experiments were conducted using the Magna RIP RNA-Binding Protein Immunoprecipitation Kit (Millipore). PTBP1 antibody (Cell Signaling Technology) and negative control normal rabbit IgG provided in the kit were used for immunoprecipitation. Based on the manufacturer's instructions, 250 μL cell lysis buffer was added to 30 million cells. A total of 100 μL cell lysate was used for IgG and PTBP1 immunoprecipitation, respectively, and 10 μL cell lysate was used as 10% input. Immunoprecipitated RNA was analyzed by qRT-PCR using primers for HOXA11-AS or PKM1/2 mRNA and qRT-PCR products were measured by agarose gel electrophoresis.
RNA pull-down assay
The interaction between HOXA11-AS and PTBP1 protein was determined by RNA pull-down assay. HOXA11-AS sequence was inserted into pSPT19DNA vector (Roche) with SP6 and T7 promoter at either end. Then MEGAscript SP6/T7 Transcription Kit (Invitrogen) was utilized to in vitro transcribe HOXA11-AS sense and antisense RNA. HOXA11-AS sense and antisense RNA were biotin-labeled using a Pierce RNA 3′ End Desthiobiotinylation Kit (Thermo Fisher Scientific), followed by 2U TURBO DNase digestion at 37°C for 15 min without shaking. A pull-down assay was performed using the Pierce Magnetic RNA-Protein Pull-Down Kit (Thermo Fisher Scientific), in accordance with the manufacturer's protocol. Eluted protein was detected by western blotting.
PRL assay
The supernatants of hESCs with different treatments were collected for detecting PRL protein level using a chemiluminescence assay kit (Beckman Coulter, Brea, CA) according to the manufacturer's instructions.
IGFBP1 ELISA
The supernatants of hESCs with different treatments were collected for detecting IGFBP1 protein level by enzyme-linked immunosorbent assay (ELISA) with IGFBP1 ELISA kit (R&D Systems, Minneapolis, MN) according to the manufacturer's instructions.
Statistical analysis
The data are presented as the mean ± standard error of the mean (SEM). Analyses were performed using SPSS 26.0 and GraphPad Prism 9.0. Statistical comparisons between two groups were performed using Student's t test, while differences between three or more groups were assessed by one-way ANOVA followed by Bonferroni tests, with a p value <0.05 considered statistically significant. Statistical significance was evaluated using data from at least three independent experiments.
Acknowledgments
We would like to thank Yichao Han and Fangming Zhu for their valuable support for this work. Especially, we would like to express our sincere thanks to Diego O. Sialer from Weill Cornell Medicine for reviewing the language of this article. This work was generously supported by the National Key R&D Program of China (2019YFA0802604), the National Natural Science Foundation of China (nos. 82130046 and 81771648), the Shanghai Leading Talent Program, Innovative Research Team of High-Level Local Universities in Shanghai (no. SSMU-ZLCX20180401), the Clinical Research Plan of SHDC(SHDC2020CR1046B), the Shanghai Municipal Education Commission-Gaofeng Clinical Medicine grant support (no. 20161413), and the Shanghai Commission of Science and Technology (no. 17DZ2271100).
Author contributions
H.Z. and S.H. performed most of the experiments. J.Q. performed bioinformatic analysis. Y.S. supervised the study and wrote the manuscript. Y.W., Y. Ding, and Q.Z. performed quality control of all data. Y.H., Y.L., Y.Y., S.W., and Y. Du revised the manuscript.
Declaration of interest
The authors declare no competing interests.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.ymthe.2022.01.036.
Supplemental information
References
- 1.Polanski L.T., Baumgarten M.N., Quenby S., Brosens J., Campbell B.K., Raine-Fenning N.J. What exactly do we mean by 'recurrent implantation failure'? A systematic review and opinion. Reprod. Biomed. Online. 2014;28:409–423. doi: 10.1016/j.rbmo.2013.12.006. [DOI] [PubMed] [Google Scholar]
- 2.Timeva T., Shterev A., Kyurkchiev S. Recurrent implantation failure: the role of the endometrium. J. Reprod. Infertil. 2014;15:173–183. [PMC free article] [PubMed] [Google Scholar]
- 3.Lessey B.A., Young S.L. What exactly is endometrial receptivity? Fertil. Steril. 2019;111:611–617. doi: 10.1016/j.fertnstert.2019.02.009. [DOI] [PubMed] [Google Scholar]
- 4.Pan-Castillo B., Gazze S.A., Thomas S., Lucas C., Margarit L., Gonzalez D., Francis L.W., Conlan R.S. Morphophysical dynamics of human endometrial cells during decidualization. Nanomedicine. 2018;14:2235–2245. doi: 10.1016/j.nano.2018.07.004. [DOI] [PubMed] [Google Scholar]
- 5.Wang W., Vilella F., Alama P., Moreno I., Mignardi M., Isakova A., Pan W., Simon C., Quake S.R. Single-cell transcriptomic atlas of the human endometrium during the menstrual cycle. Nat. Med. 2020;26:1644–1653. doi: 10.1038/s41591-020-1040-z. [DOI] [PubMed] [Google Scholar]
- 6.Gellersen B., Brosens J.J. Cyclic decidualization of the human endometrium in reproductive health and failure. Endocr. Rev. 2014;35:851–905. doi: 10.1210/er.2014-1045. [DOI] [PubMed] [Google Scholar]
- 7.Garrido-Gómez T., Castillo-Marco N., Cordero T., Simón C. Decidualization resistance in the origin of preeclampsia. Am. J. Obstet. Gynecol. 2020;S0002-9378:31130–31133. doi: 10.1016/j.ajog.2020.09.039. [DOI] [PubMed] [Google Scholar]
- 8.Romero R., Dey S.K., Fisher S.J. Preterm labor: one syndrome, many causes. Science. 2014;345:760–765. doi: 10.1126/science.1251816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Adiguzel D., Celik-Ozenci C. FoxO1 is a cell-specific core transcription factor for endometrial remodeling and homeostasis during menstrual cycle and early pregnancy. Hum. Reprod. Update. 2021;27:570–583. doi: 10.1093/humupd/dmaa060. [DOI] [PubMed] [Google Scholar]
- 10.Mantena S.R., Kannan A., Cheon Y.P., Li Q., Johnson P.F., Bagchi I.C., Bagchi M.K. C/EBPbeta is a critical mediator of steroid hormone-regulated cell proliferation and differentiation in the uterine epithelium and stroma. Proc. Natl. Acad. Sci. U S A. 2006;103:1870–1875. doi: 10.1073/pnas.0507261103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shindoh H., Okada H., Tsuzuki T., Nishigaki A., Kanzaki H. Requirement of heart and neural crest derivatives-expressed transcript 2 during decidualization of human endometrial stromal cells in vitro. Fertil. Steril. 2014;101:1781–1790e1-5. doi: 10.1016/j.fertnstert.2014.03.013. [DOI] [PubMed] [Google Scholar]
- 12.Taylor H.S., Vanden Heuvel G.B., Igarashi P. A conserved Hox axis in the mouse and human female reproductive system: late establishment and persistent adult expression of the Hoxa cluster genes. Biol. Reprod. 1997;57:1338–1345. doi: 10.1095/biolreprod57.6.1338. [DOI] [PubMed] [Google Scholar]
- 13.Taylor H.S., Arici A., Olive D., Igarashi P. HOXA10 is expressed in response to sex steroids at the time of implantation in the human endometrium. J. Clin. Invest. 1998;101:1379–1384. doi: 10.1172/JCI1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Taylor H.S., Igarashi P., Olive D.L., Arici A. Sex steroids mediate HOXA11 expression in the human peri-implantation endometrium. J. Clin. Endocrinol. Metab. 1999;84:1129–1135. doi: 10.1210/jcem.84.3.5573. [DOI] [PubMed] [Google Scholar]
- 15.Eun Kwon H., Taylor H.S. The role of HOX genes in human implantation. Ann. N. Y. Acad. Sci. 2004;1034:1–18. doi: 10.1196/annals.1335.001. [DOI] [PubMed] [Google Scholar]
- 16.Benson G.V., Lim H., Paria B.C., Satokata I., Dey S.K., Maas R.L. Mechanisms of reduced fertility in Hoxa-10 mutant mice: uterine homeosis and loss of maternal Hoxa-10 expression. Development. 1996;122:2687–2696. doi: 10.1242/dev.122.9.2687. [DOI] [PubMed] [Google Scholar]
- 17.Gendron R.L., Paradis H., Hsieh-Li H.M., Lee D.W., Potter S.S., Markoff E. Abnormal uterine stromal and glandular function associated with maternal reproductive defects in Hoxa-11 null mice. Biol. Reprod. 1997;56:1097–1105. doi: 10.1095/biolreprod56.5.1097. [DOI] [PubMed] [Google Scholar]
- 18.Li L., Wang Y., Song G., Zhang X., Gao S., Liu H. HOX cluster-embedded antisense long non-coding RNAs in lung cancer. Cancer Lett. 2019;450:14–21. doi: 10.1016/j.canlet.2019.02.036. [DOI] [PubMed] [Google Scholar]
- 19.Chau Y.M., Pando S., Taylor H.S. HOXA11 silencing and endogenous HOXA11 antisense ribonucleic acid in the uterine endometrium. J. Clin. Endocrinol. Metab. 2002;87:2674–2680. doi: 10.1210/jcem.87.6.8527. [DOI] [PubMed] [Google Scholar]
- 20.Hu S., Yao G., Wang Y., Xu H., Ji X., He Y., Zhu Q., Chen Z., Sun Y. Transcriptomic changes during the pre-receptive to receptive transition in human endometrium detected by RNA-Seq. J. Clin. Endocrinol. Metab. 2014;99:E2744–E2753. doi: 10.1210/jc.2014-2155. [DOI] [PubMed] [Google Scholar]
- 21.Kopp F., Mendell J.T. Functional classification and experimental dissection of long noncoding RNAs. Cell. 2018;172:393–407. doi: 10.1016/j.cell.2018.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yin Q.F., Hu S.B., Xu Y.F., Yang L., Carmichael G.G., Chen L.L. SnoVectors for nuclear expression of RNA. Nucleic Acids Res. 2015;43:e5. doi: 10.1093/nar/gku1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Du H., Taylor H.S. The role of Hox genes in female reproductive tract development, adult function, and fertility. Cold Spring Harb. Perspect. Med. 2015;6:a023002. doi: 10.1101/cshperspect.a023002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jana S.K., Banerjee P., Mukherjee R., Chakravarty B., Chaudhury K. HOXA-11 mediated dysregulation of matrix remodeling during implantation window in women with endometriosis. J. Assist. Reprod. Genet. 2013;30:1505–1512. doi: 10.1007/s10815-013-0088-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Quinn J.J., Chang H.Y. Unique features of long non-coding RNA biogenesis and function. Nat. Rev. Genet. 2016;17:47–62. doi: 10.1038/nrg.2015.10. [DOI] [PubMed] [Google Scholar]
- 26.Hou M., Tang X., Tian F., Shi F., Liu F., Gao G. AnnoLnc: a web server for systematically annotating novel human lncRNAs. BMC Genomics. 2016;17:931. doi: 10.1186/s12864-016-3287-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li J.H., Liu S., Zhou H., Qu L.H., Yang J.H. starBase v2.0: decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res. 2014;42:D92–D97. doi: 10.1093/nar/gkt1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cook K.B., Kazan H., Zuberi K., Morris Q., Hughes T.R. RBPDB: a database of RNA-binding specificities. Nucleic Acids Res. 2011;39:D301–D308. doi: 10.1093/nar/gkq1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang J.H., Li J.H., Shao P., Zhou H., Chen Y.Q., Qu L.H. starBase: a database for exploring microRNA-mRNA interaction maps from Argonaute CLIP-Seq and Degradome-Seq data. Nucleic Acids Res. 2011;39:D202–D209. doi: 10.1093/nar/gkq1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhou Y., Zhou B., Pache L., Chang M., Khodabakhshi A.H., Tanaseichuk O., Benner C., Chanda S.K. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun. 2019;10:1523. doi: 10.1038/s41467-019-09234-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Oberstrass F.C., Auweter S.D., Erat M., Hargous Y., Henning A., Wenter P., Reymond L., Amir-Ahmady B., Pitsch S., Black D.L., et al. Structure of PTB bound to RNA: specific binding and implications for splicing regulation. Science. 2005;309:2054–2057. doi: 10.1126/science.1114066. [DOI] [PubMed] [Google Scholar]
- 32.Su Y., Guo S., Liu C., Li N., Zhang S., Ding Y., Chen X., He J., Liu X., Wang Y., et al. Endometrial pyruvate kinase M2 is essential for decidualization during early pregnancy. J. Endocrinol. 2020;245:357–368. doi: 10.1530/JOE-19-0553. [DOI] [PubMed] [Google Scholar]
- 33.Zuo R.J., Gu X.W., Qi Q.R., Wang T.S., Zhao X.Y., Liu J.L., Yang Z.M. Warburg-like glycolysis and lactate shuttle in mouse decidua during early pregnancy. J. Biol. Chem. 2015;290:21280–21291. doi: 10.1074/jbc.M115.656629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.He X., Arslan A.D., Ho T.T., Yuan C., Stampfer M.R., Beck W.T. Involvement of polypyrimidine tract-binding protein (PTBP1) in maintaining breast cancer cell growth and malignant properties. Oncogenesis. 2014;3:e84. doi: 10.1038/oncsis.2013.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang K.C., Chang H.Y. Molecular mechanisms of long noncoding RNAs. Mol. Cell. 2011;43:904–914. doi: 10.1016/j.molcel.2011.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Flynn R.A., Chang H.Y. Long noncoding RNAs in cell-fate programming and reprogramming. Cell Stem Cell. 2014;14:752–761. doi: 10.1016/j.stem.2014.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Deng W.B., Liang X.H., Liu J.L., Yang Z.M. Regulation and function of deiodinases during decidualization in female mice. Endocrinology. 2014;155:2704–2717. doi: 10.1210/en.2014-1015. [DOI] [PubMed] [Google Scholar]
- 38.Liang X.H., Deng W.B., Liu Y.F., Liang Y.X., Fan Z.M., Gu X.W., Liu J.L., Sha A.G., Diao H.L., Yang Z.M. Non-coding RNA LINC00473 mediates decidualization of human endometrial stromal cells in response to cAMP signaling. Sci. Rep. 2016;6:22744. doi: 10.1038/srep22744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lv H., Tong J., Yang J., Lv S., Li W.P., Zhang C., Chen Z.J. Dysregulated Pseudogene HK2P1 may contribute to preeclampsia as a competing endogenous RNA for Hexokinase 2 by impairing decidualization. Hypertension. 2018;71:648–658. doi: 10.1161/HYPERTENSIONAHA.117.10084. [DOI] [PubMed] [Google Scholar]
- 40.Jia Y., Cai R., Yu T., Zhang R., Liu S., Guo X., Shang C., Wang A., Jin Y., Lin P. Progesterone-induced RNA Hand2os1 regulates decidualization in mice uteri. Reproduction. 2020;159:303–314. doi: 10.1530/REP-19-0401. [DOI] [PubMed] [Google Scholar]
- 41.Wang Y., Hu S., Yao G., Zhu Q., He Y., Lu Y., Qi J., Xu R., Ding Y., Li J., et al. A novel molecule in human cyclic endometrium: LncRNA TUNAR is involved in embryo implantation. Front. Physiol. 2020;11:587448. doi: 10.3389/fphys.2020.587448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ghetti A., Piñol-Roma S., Michael W.M., Morandi C., Dreyfuss G. hnRNP I, the polypyrimidine tract-binding protein: distinct nuclear localization and association with hnRNAs. Nucleic Acids Res. 1992;20:3671–3678. doi: 10.1093/nar/20.14.3671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Calabretta S., Bielli P., Passacantilli I., Pilozzi E., Fendrich V., Capurso G., Fave G.D., Sette C. Modulation of PKM alternative splicing by PTBP1 promotes gemcitabine resistance in pancreatic cancer cells. Oncogene. 2016;35:2031–2039. doi: 10.1038/onc.2015.270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Salama S.A., Mohammad M.A., Diaz-Arrastia C.R., Kamel M.W., Kilic G.S., Ndofor B.T., Abdel-Baki M.S., Theiler S.K. Estradiol-17β upregulates pyruvate kinase M2 expression to coactivate estrogen receptor-α and to integrate metabolic reprogramming with the mitogenic response in endometrial cells. J. Clin. Endocrinol. Metab. 2014;99:3790–3799. doi: 10.1210/jc.2013-2639. [DOI] [PubMed] [Google Scholar]
- 45.Yang M., Li H., Rong M., Zhang H., Hou L., Zhang C. Dysregulated GLUT1 may be involved in the pathogenesis of preeclampsia by impairing decidualization. Mol. Cell. Endocrinol. 2021;540:111509. doi: 10.1016/j.mce.2021.111509. [DOI] [PubMed] [Google Scholar]
- 46.Gurner K.H., Evans J., Hutchison J.C., Harvey A.J., Gardner D.K. A microenvironment of high lactate and low pH created by the blastocyst promotes endometrial receptivity and implantation. Reprod. Biomed. Online. 2021;S1472-6483:00442–00449. doi: 10.1016/j.rbmo.2021.09.012. [DOI] [PubMed] [Google Scholar]
- 47.Ma L.N., Huang X.B., Muyayalo K.P., Mor G., Liao A.H. Lactic acid: a novel signaling molecule in early pregnancy? Front. Immunol. 2020;11:279. doi: 10.3389/fimmu.2020.00279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yurci A., Dokuzeylul Gungor N., Gurbuz T. Spectroscopy analysis of endometrial metabolites is a powerful predictor of success of embryo transfer in women with implantation failure: a preliminary study. Gynecol. Endocrinol. 2021;37:415–421. doi: 10.1080/09513590.2021.1883584. [DOI] [PubMed] [Google Scholar]
- 49.Xu Y., Wu D., Liu J., Huang S., Zuo Q., Xia X., Jiang Y., Wang S., Chen Y., Wang T., et al. Downregulated lncRNA HOXA11-AS affects trophoblast cell proliferation and migration by regulating RND3 and HOXA7 expression in PE. Mol. Ther. Nucleic Acids. 2018;12:195–206. doi: 10.1016/j.omtn.2018.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lucas E.S., Vrljicak P., Muter J., Diniz-da-Costa M.M., Brighton P.J., Kong C.S., Lipecki J., Fishwick K.J., Odendaal J., Ewington L.J., et al. Recurrent pregnancy loss is associated with a pro-senescent decidual response during the peri-implantation window. Commun. Biol. 2020;3:37. doi: 10.1038/s42003-020-0763-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Garcia-Alonso L., Handfield L.F., Roberts K., Nikolakopoulou K., Fernando R.C., Gardner L., Woodhams B., Arutyunyan A., Polanski K., Hoo R., et al. Mapping the temporal and spatial dynamics of the human endometrium in vivo and in vitro. Nat. Genet. 2021;53:1698–1711. doi: 10.1038/s41588-021-00972-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kirk D., King R.J., Heyes J., Peachey L., Hirsch P.J., Taylor R.W. Normal human endometrium in cell culture. I. Separation and characterization of epithelial and stromal components in vitro. In Vitro. 1978;14:651–662. doi: 10.1007/BF02616162. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.