Abstract
Bacterial small RNAs (sRNAs) play a pivotal role in post-transcriptional regulation of gene expression and participate in many physiological circuits. An ~80-nt-long RyjB was earlier identified as a novel sRNA, which appeared to be accumulated in all phases of growth in Escherichia coli. We have taken a comprehensive approach in the current study to understand the regulation of ryjB expression under normal and pH stress conditions. RpoS was not necessary for ryjB expression neither at normal condition nor under acid stress. Hfq also emerged to be unnecessary for RyjB accumulation. Interestingly, RyjB was detected as a novel acid stress induced sRNA. A DNA binding protein PhoP, a component of PhoP/Q regulon, was found to regulate ryjB expression at low pH, as the elimination of phoP allele in the chromosome exhibited a basal level of RyjB expression under acid stress. Ectopic expression of PhoP in ΔphoP cells restored the overabundance of RyjB in the cell. Overexpression of RyjB increased the abundance of sgcA transcripts, with which RyjB shares a 4-nt overlap. The current study increases our knowledge substantially regarding the regulation of ryjB expression in E. coli cell.
Keywords: acid stress, Hfq, small RNAs, sRNA-mediated gene regulation
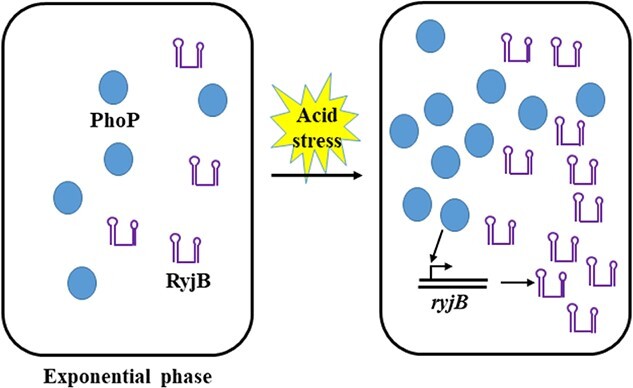
Graphical Abstract
Graphical Abstract.
Introduction
A wide range of physiological circuits in bacteria is regulated by small RNAs (sRNAs) (1–5). These sRNA regulators are primarily induced in response to changing environmental cues (5–12) and play a key role in modulating gene expressions essentially at the post-transcriptional level. Numerous physiological processes like stress response, virulence and quorum sensing (2–5) have been reported to be regulated by sRNAs. sRNAs with a lower degree of complementarity to their target RNAs require a chaperon action by Hfq. Function of some sRNAs in Escherichia coli have been discovered, but many of them have been reported with unidentified role in the cell. RyjB in E. coli is one such sRNA; the expression of which was never systematically investigated.
Although RyjB in E. coli was not detected in the Hfq-immunoprecipitate, a lower abundance of RyjB was reported in hfq-mutant strain. Initially, it was validated as an ~130-nt-long sRNA (12), subsequent sequence inspection of RyjB and 5′ and 3′ RACE analysis demonstrated that RyjB is an ~80-nt-long sRNA, which is located in the intergenic region of sgcA and sgcQ genes in reverse polarities (13). A 4-nt overlap exists between ryjB and a putative phosphotransferase system transporter protein encoding gene sgcA (14). Hence, RyjB was speculated to act as a cis-acting antisense RNA. A σ70 promoter was predicted to be within the sgcA gene for RyjB expression; however, it was never experimentally validated (14). RyjB was detected in all the growth phases of the cell, but its relative abundance was never quantitatively estimated at different phases of growth. RyjB has interestingly been reported as an inducible sRNA under high Mg2+ concentration and thus, how RyjB expression is related to PhoP/PhoQ, a two-component regulatory system controlling Mg2+ homeostasis in E. coli, is a long-standing question to the researchers (14).
Aside from the aforementioned features of RyjB, regulation of its expressions and physiological role are unknown until now. In the current study, we have taken a systematic approach to investigate the underlying regulation of RyjB expression in E. coli. We have found that the accumulation of RyjB did not alter significantly in a 24-h growth cycle. Neither RpoS nor Hfq affects its expression dramatically in any phases of cell growth. Interestingly, RyjB was found to be an acid stress inducible sRNA, but its expression in alkaline condition remains unaltered. RyjB induction under acid stress conditions appeared to be regulated by PhoP as the elimination of the chromosomal copy of phoP gene resulted in a constant low level RyjB expression. Ectopic expression of PhoP in ΔphoP strain was found to increase RyjB abundance confirming its regulatory role in ryjB expression in E. coli cells. Overexpression of RyjB increased the intracellular accumulation of sgcA suggesting its role as an antisense RNA.
Materials and Methods
Materials
Genomic DNA isolation kit was purchased from Promega Co. (USA). Total RNA isolation kits were procured from KPC Life Sc. (India) and Qiagen (Germany). SYBR green master mix and cDNA synthesis kit (iScript™) were purchased from Bio-Rad (USA). Growth media was obtained from Hi-Media (India). MEGAshortscript™ transcription kit was purchased from Ambion (USA). GeneElute™ PCR clean-up kit was procured from Sigma Chemical Co. (USA). DNaseI was procured from Fermentas, USA. Oligonucleotide primers were synthesized by Integrated DNA Technologies, Inc. (USA). Anti-FLAG M2 mAbs and homopiperazine-1,4-bis(2-ethanesulfonic acid) (Homopipes) were purchased from Sigma chemical Co. (USA). Anti-mouse IgG HRP conjugate was from Santa Cruz Biotechnology (USA). ECL solution was procured from PerkinElmer (USA). All other chemicals were molecular biology grade reagents.
Bacterial strain construction and growth condition
Wild type E. coli K-12 strain MG1655 was obtained from laboratory stock. Deletion of rpoS, hfq, phoP genes were introduced into MG1655 strain through phage P1vir-mediated transduction (15). N-terminus of chromosomal copy of phoP gene was fused with a 2 × FLAG sequence by recombineering using primers F1 and F2 (see oligonucleotide sequences in Supplemental Table T2), and the resulting gene fusion was confirmed by DNA sequencing. Cells were grown overnight in 5 ml of LB medium with appropriate antibiotic when needed at 37°C under shaking at 180 rpm. Primary cultures were diluted 1:100 into fresh LB medium. Portions were removed at different time points and instantly mixed with ice-cold stop solution (95% ethanol, 5% phenol). Antibiotics, when present, were at the following concentrations: kanamycin, 25 μg/ml; ampicillin, 100 μg/ml.
For pH stress, cells were grown up to mid-exponential phase (OD600 = 0.4) and then the pH of the medium was adjusted either to 5.0 by buffering the medium with 10 mM HOMOPIPES or sodium acetate (pH 5.0) or to 9.0 by 50-mM Tris–HCl (pH 9.0). Sodium phosphate buffer (pH 7.0) was used to maintain the cells at neutral pH. Cells were withdrawn at regular intervals after they were exposed to stress followed by total RNA/protein extraction from each sample.
RNA extraction and real-time PCR
Wild type and mutant E. coli cells were grown overnight in LB media with relevant antibiotics as required. Primary cultures were diluted 1:100 in fresh LB broth and were grown continuously at 37°C in an orbital shaker at 180 rpm. Equal volumes of culture were taken away at different time points. Cells were pelleted down and total RNA was extracted from each of those cell pellets using RNeasy kit (Qiagen). Each RNA sample was treated with DNaseI (Fermentas, USA) to eliminate DNA contamination. UV spectrophotometry (Jasco V-730) was used to quantitate RNA concentration. Complementary DNA was synthesized from 1 μg of total RNA utilizing iScript™ cDNA synthesis kit (Bio-Rad) with random hexamer (Bio-Rad C1000, USA) as a primer. Synthesized cDNA was diluted 100-fold by nuclease-free water and 1 μL of that diluted cDNA was employed in qPCR reaction in a 96-well microtiter PCR plate using SYBR green master mix (Bio-Rad) in CFX96 Touch™ Real-Time PCR Detection System (Bio-Rad). A typical Real-time PCR reaction mix contained 20 ng of cDNA, 1 μM of each sRNA primer (Supplementary Table T2), 10 μl of SYBR Green Master Mix and 8-μl nuclease-free water in 20-μl reaction volume. Each sample was tested in triplicate. The 5S rRNA gene was taken as an internal reference as it has been widely used as a control to examine the sRNA expression (16). Relative gene expression was calculated using 2-ΔΔCt method. All experiments were performed according to the manufacturer’s instructions.
In vitro transcription
Escherichia coli RyjB RNA was synthesized from DNA templates of ryjB genes by in vitro transcription reactions using the MEGAshortscript™ transcription kit (Ambion). Genomic DNA was utilized to synthesize the template for transcription reaction by PCR with the forward primer containing the T7 RNA polymerase promoter sequence. PCR products were purified using the GeneElute™ PCR clean-up kit (Sigma). Transcribed RyjB RNA was purified by phenol:chloroform:isoamylalcohol (25:24:1) extraction followed by ethanol precipitation.
Northern blotting
Fifteen micrograms of total RNA samples, extracted at the indicated time point after the cells were exposed to pH 5.0, were mixed with gel loading buffer and resolved on a 2% denaturing agarose gel in 1X MOPS (4-morpholinepropanesulfonic acid) buffer. Deionized water was used to wash the gel and RNA was transferred on nylon membrane (GE Healthcare) by downward capillary transfer for 15 h at room temperature. RNA transferred on the membrane was UV cross-linked (Herolab, Germany) to it. A DNA oligonucleotide probe labelled with Cy5 at 5′-end was utilized to detect RyjB RNA. The probe was allowed to anneal to the cross-linked RNAs on the membrane by incubating overnight for hybridization (Thermo Fisher Scientific, USA) at 58°C in ExpressHyb hybridization solution (Clontech). Membrane was then washed thrice with pre-warmed SSC buffer and the bands were visualized by high-resolution gel imaging for fluorescence system (G:Box Chemi-XX9, SYNGENE). Bands were quantitated by ImageJ.
Western blotting
Fresh LB medium was used to dilute 24-h grown wild type MG1655 cells (1:100). Cells were grown afterwards constantly until the mid-exponential phase (OD600 = 0.4) was reached. The pH of the growth medium was then adjusted to 5.0. Cells were removed at indicated time points after acidifications. Cell pellets were gently resuspended in 250 μl of lysis buffer (25-mM Tris pH 7.4, 150-mM KCl, 1-mM MgCl2 and 1-mM DTT). A total of 250 μl of glass beads (0.1 mm) was added to the mixture and it was then vortexed for 30 s followed by 15 s cooling on ice. This step was repeated for 5 min. Mixture was then treated with an additional 250-μl lysis buffer and centrifuged at 15000 rpm for 10 min at 4°C to separate beads and insoluble materials (16). Protein concentration in the cell-free extract was estimated through Bradford assay (Bio-rad). Protein samples (15 μg) were resolved in a 10% SDS-PAGE and were transferred onto PVDF membranes, which were then treated with 3% nonfat milk, and probed with anti-FLAG M2 mAbs (1:1000 dilution). PhoP was detected with HRP-conjugated anti-mouse IgG antibody and visualized with ECL solutions as described (17).
Cloning and overexpression of RyjB and PhoP
Genomic DNA from wild type MG1655 cells was isolated using genomic DNA extraction Kit (Promega Co.) and used as a template for the amplification of ryjB and phoP gene (primers are in Supplementary Table T1) in a PCR reaction utilizing KOD hot start DNA polymerase (Invitrogen). The template DNA was eliminated by DpnI digestion and the amplicon was purified by gel extraction. Both the PCR product and the vector pUC19 were digested with HindIII and BamHI at 37°C for 2 h. Digested products were purified by phenol: chloroform: isoamyl alcohol (25:24:1) extraction. T4 DNA ligase was used to ligate the digested PCR product in plasmid within HindIII and BamHI sites. The resulting recombinant plasmid was chemically transformed into E. coli DH5α cells, which were then grown overnight on LB agar plates (Amp+) containing 100 μg/ml X-gal, and 1-mM IPTG. White colonies thus observed were grown in (Amp+) LB broth and recombinant plasmids (pRyjB or pPhoP) were isolated using QIAprep Spin Kit (Qiagen). Cloning of ryjB and phoP in pUC19 was confirmed by DNA sequencing.
Recombinant plasmid (pRyjB and pPhoP), isolated from DH5α cells, was transformed into E. coli cells. Cells were then grown in LB media until mid-exponential phase (OD600 = 0.4). IPTG (1 mM) was added to that culture and allowed to grow further for an hour. Cells were then collected through centrifugation at 5000 rpm at 4°C and total RNA was isolated from that sample for further analysis.
Statistical analysis
Experimental data, collected from three different experiments, were presented as mean ± standard deviation (SD). One-way analysis of variance was applied to analyse the differences between multiple groups. Statistically, a significant difference was considered as P < 0.05.
Results
Regulation of RyjB expression
RyjB was initially found to be transcribed from a gene flanked between sgcA and sgcQ genes (Fig. 1A) (12). RyjB expression was reported to be higher in the exponential phase than the stationary phase of E. coli cells (13). However, a systematic approach has never been taken to analyse how RyjB expression fluctuates over a period of continuous growth of E. coli cells. Herein, we have taken a comprehensive approach to examine RyjB expression in a 24-h growth cycle of E. coli. Wild type E. coli MG1655 cells grown for 24-h were diluted (1:100) in a fresh LB medium and were continuously grown at 37°C under shaking conditions. Cells were withdrawn at a regular time span and total RNA was isolated from the cells collected at each time point. One microgram of the total RNA was employed in a cDNA synthesis reaction. RyjB expression was analysed by RT-qPCR utilizing the cDNA appearing at each time point.
Figure 1.
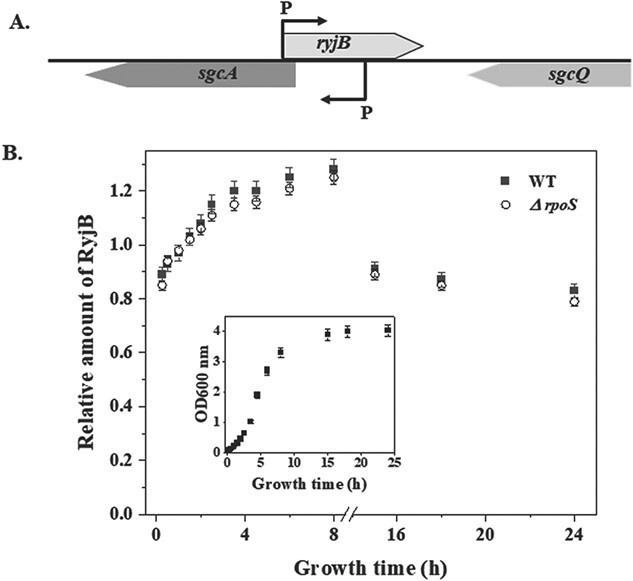
Accumulation of RyjB at different growth phases of E. coli cells. A. Schematic illustration of genomic location and orientation of ryjB gene. B. Relative expression of ryjB gene was analysed through RT-qPCR analysis in a 24-h growth cycle. RyjB quantity at each point was normalized by comparison to RT-qPCR of 5S rRNA. Each point in the graph represents the relative amount of RyjB when compared with 1.5-h sample. Data at each point were presented as the average ± SD from three separate experiments. Inset, growth curve of MG1655 cells. Each point on the curve represents the point of sample withdrawal.
Figure 1B shows ryjB expression in a 24-h growth cycle (growth curve was shown in the inset of Fig. 1B) of E. coli. Although RyjB has been found to be present in all phases of growth, but its abundance was more prevalent in the exponential phase. RyjB transcript was found to be depleted by ~40% in the mid-stationary phase compared with the late exponential phase of E. coli growth. Moreover, a detailed investigation of RyjB status in the exponential phase revealed that the RyjB population increased by ~30% from early to late exponential phase and culminated prior to the entry to the stationary phase. Although it was predicted that RyjB expression is controlled by the presence of Eσ70 promoter (12, 13), the reason of increment in the population from an early to late exponential phase is still unknown.
To ascertain that the stationary phase specific σ-factor, RpoS, does not play any role in RyjB expression in wild type cells, we tested the growth-phase dependent expression of ryjB in wild type and ΔrpoS cells (Fig. 1B). Profile of ryjB expression in both the cells emerged to be unaltered, which confirmed that RpoS was not necessary for RyjB synthesis under normal physiological conditions.
Role of Hfq
RyjB was found to be less abundant in Δhfq cells; however, it was not detected in the Hfq-immunoprecipitate by northern analysis (12,13). Thus, a detailed investigation on the role of Hfq, if any, on RyjB stability is required to understand its functional mechanism. Hfq plays a central role to protect sRNA from cellular ribonucleases and also promotes base pairing between sRNA and its target. Binding of sRNA to the Hfq is mediated through a stable interaction of 3′-poly(U) rich region of sRNA to the proximal face of Hfq (18). Thus, the presence of a poly(U) rich region in the sRNA structure is essential for its binding to Hfq. RyjB structure was determined by Mfold web server (19) and the predicted structure of lowest energy was utilized for its structural analysis. Two adjacent stem-loops of different length were present in its structure. A 5-nt 3′-single stranded region in the RyjB structure was devoid of any ‘U’ residue (Fig. 2A). Hence, it was presumed that RyjB lacks the ability to bind Hfq.
Figure 2.
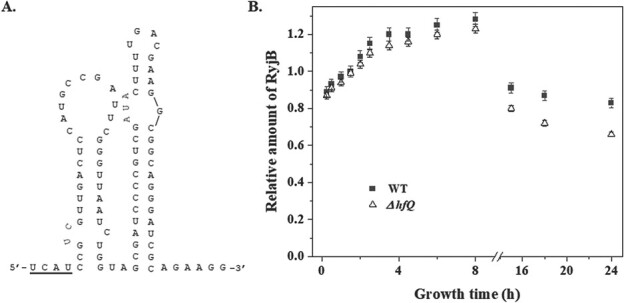
Role of Hfq in RyjB expression. A. Structure of RyjB predicted by Mfold web server. The sequence above the bold line in the structure is complementary to the 5′-end of sgcA mRNA. B. Comparison of the accumulation of RyjB in wild type and Δhfq cells in a 24-h growth cycle analysed by RT-qPCR analysis. 5S rRNA was taken as internal control. Relative amount of RyjB was presented at each point compared to the RyjB accumulation at 1.5 h, which was set as 1. Data were presented as mean ± S.D. from three independent experiments.
To test our hypothesis that RyjB is an Hfq-independent sRNA, we compared ryjB accumulation in wild type and Δhfq cells in a 24-h growth cycle. Overnight grown cells were diluted (1:100) in fresh LB medium and were constantly grown at 37°C. Cells were removed at the indicated times. Total RNA was isolated from each sample and was subjected to RT-qPCR study. Figure 2B shows the relative amount of RyjB accumulated in wild type and Δhfq cells over a period of 24 h. RyjB abundance in the exponential phase of both wild type and Δhfq cells are identical, indicating that Hfq does not play any role for RyjB accumulation in the exponential phase. However, RyjB build-up in the stationary phase is ~20% less in Δhfq cells than that in wild type cells. These data are also in line with the observation of Kawano et al. (13) and establishes that Hfq generally is not necessary for RyjB expression; however, why it is decreased in the stationary phase in hfq-mutant compared with wild type cells still has to be elucidated.
RyjB is an acid stress inducible sRNA
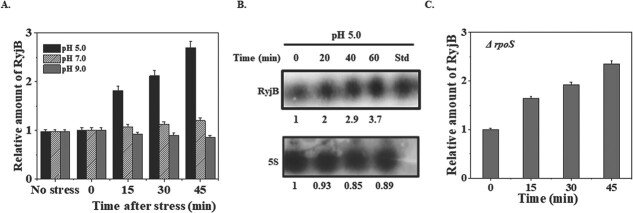
RpoS independent expression of RyjB and its lower abundance in the stationary phase instigated us to examine whether RyjB is a stress sensitive sRNA or not. Since the stationary phase of growth is a condition of multiple stresses, and E. coli cells also exposed to low pH transiently during its passage through the human stomach, we desired to investigate the effect of acid stress on RyjB expression. A 15-h grown E. coli MG1655 culture was diluted (1:100) in a fresh LB medium and was allowed to grow until mid-exponential phase (OD600 = 0.4). The growth medium was acidified to bring down pH to 5.0. Cells were withdrawn at indicated time points after acidification and total RNA was isolated from each sample. RyjB accumulation in each RNA sample was determined by RT-qPCR. Figure 3A shows RyjB expression profile upon pH stress as a function of time. A dramatic result, which was not anticipated earlier, revealed that RyjB population increased with time and nearly 2.5-fold upregulation was observed after 45 min of the acid stress. An identical experiment carried out at pH 7.0 and 9.0 revealed an unaltered expression of RyjB at neutral and alkaline conditions (Fig. 3A).
Figure 3.
Amount of RyjB in WT and ΔrpoS cells under pH stress. A. Wild type cells were grown up to mid-exponential phase (OD600–0.4) at 37°C and pH of the medium was adjusted to either 5.0 (adjusted by 50-mM HOMOPIPES) or 9.0 (adjusted by 50 mM Tris–HCl). Samples were removed at the indicated time and total RNA was isolated for RT-qPCR analysis. 5S rRNA gene was taken as internal control. RyjB accumulation at 0 min was set as 1. B. 15 μg of total RNAs isolated from the samples withdrawn at indicated time after acid stress (pH adjusted by 50-mM sodium acetate) were analysed by northern blot. RyjB transcribed in vitro was run in the last lane (Std) as size standard. The number under each lane represents the relative amount of RyjB when compared with 0-min sample, which was set as 1. 5S rRNA gene was taken as loading control. C. Accumulation of RyjB in ΔrpoS cells under acid stress analysed by RT-qPCR. 5S rRNA gene was used as internal control.
To further confirm the induction of RyjB at low pH, northern blot analysis (Fig. 3B) was performed with the RNA samples extracted at the indicated time points from the cells after they were exposed to pH 5.0. RyjB transcribed in vitro was also utilized in the northern blot as a standard. Quantitation of the bands in the northern blot revealed that RyjB expression was induced by ~2.9-fold after 40 min exposure of the cells to pH 5.0. Continued acid stress on the cells upto 60 min resulted in about 4-fold higher accumulation of RyjB. 5S rRNA expression was unaltered under acid stress and thus was taken as loading control. These data corroborate well with the result obtained in the above-mentioned RT-qPCR analysis. Although RyjB is upregulated at low pH, the reason why RyjB was elevated under that condition is yet to be known. An independent experiment conducted with the aforementioned experimental condition using ΔrpoS cells exhibited an identical RyjB profile as in wild type cells under acid stress (Fig. 3C). These data clearly indicated that global stress regulator RpoS is not necessary for RyjB induction under acid stress.
PhoP upregulates RyjB under acid stress
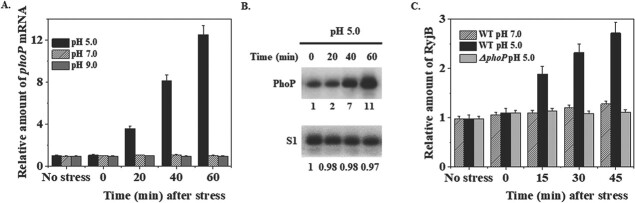
The question how RyjB was upregulated under acid stress drew immense attention for investigation. PhoP, an integrant of two component regulatory system PhoPQ, has previously been reported to control a large number of genes involved in major cellular functions such as Mg2+-homeostasis, cell envelope composition, bacterial virulence and acid resistance (20–22). To test the effect of acid stress on PhoP accumulation, mid-exponential phase cells (OD600 = 0.4) were exposed to pH 5.0 and were persistently grown at 37°C. Cells were removed at indicated time and total RNA was isolated. Status of phoP mRNA at the low pH condition was analysed by RT-qPCR method. Figure 4A represents the relative amount of phoP mRNA as a function of time after acid stress and revealed that phoP was induced by 12.3-fold after 1 h of acid stress. Examination of PhoP protein under the same condition through western blot analysis demonstrated that PhoP protein level is elevated by approximately 11-fold after 1 h of acid stress (Fig. 4B). These data confirmed that PhoP is an acid stress inducible protein, which is induced as much as 11-fold under the above-mentioned experimental condition. These data are also in line with the previous reports of low pH induction of PhoP in both Salmonella sp. (23) and E. coli (24).
Figure 4.
RyjB expression is regulated by PhoP. A. Amount of phoP mRNA upon acid stress (pH adjusted by 50-mM HOMOPIPES) was estimated by RT-qPCR analysis. 5S rRNA gene was taken as internal control for the normalization of the amount of phoP mRNA at each time point. Accumulation of phoP mRNA at 0 min was set as 1 and relative amount of phoP at other time point was calculated. B. PhoP accumulation after acid stress (pH adjusted by 50 mM sodium acetate) in wild type cells was measured by western blotting. The number under each lane represents the relative amount of PhoP protein when compared with 0-min sample, which was set as 1. Ribosomal protein S1 was taken as loading control. C. Acid stress was given to mid-exponential phase (OD600 = 0.4) wild type and ΔphoP cells. Total RNA was isolated from the cells at indicated times after acid stress and was utilized for RT-qPCR analysis. Expression of 5S rRNA gene was utilized as internal control.
Acid stress induction of PhoP apprised us to examine whether PhoP plays any role for RyjB elevation at low pH. Cells grown overnight were diluted in fresh LB medium (1:100). Cells were then continuously grown until the mid-exponential phase (OD600 = 0.4) and the growth medium was adjusted with pH 5.0. Equal volumes were removed from the growth medium in the post acidification phase and total RNA was isolated from each of the samples. RyjB expression was investigated by RT-qPCR. Comparison of RyjB accumulation in wild type and ΔphoP cells under acid stress (Fig. 4C) revealed that even though RyjB is induced by about 2.5-fold under acid stress, the elimination of phoP allele from the chromosome of E. coli cells resulted in a constant basal-level expression of RyjB. These data clearly indicates that PhoP directly regulates RyjB expression under acid stress and hence, RyjB appears to be a PhoP-dependent acid stress inducible sRNA in E. coli cells.
Ectopic expression of PhoP in ΔphoP cells restores RyjB induction
It is evident from the above experiment that PhoP is required for RyjB induction under acid stress. Basal level expression of ryjB in ΔphoP cells at low pH encouraged us to look over whether the deficiency in ryjB induction in ΔphoP strain can be complemented by the ectopic expression of PhoP. To demonstrate that, phoP gene was cloned in the pUC19 vector under the control of Lac-promoter (Plac) and the resulting recombinant plasmid (pPhoP) was chemically transformed into ΔphoP cells. Cells were then grown for 15 h and diluted in fresh LB medium (1:100). Cells grown up to mid-exponential phase (OD600 = 0.4) were treated with 1-mM IPTG and grown for an additional hour. Total RNA was isolated from the cells and used in RT-qPCR analysis. Wild type and ΔphoP cells harbouring empty plasmid were also treated in an identical manner. Figure 5A shows the accumulation of phoP transcript in wild type and ΔphoP strains. It is evident from Fig. 5A that phoP transcript was absent in ΔphoP cells containing empty plasmid but was induced about 7.5-fold when expressed from Plac compared to its accumulation in wild type cells.
Figure 5.
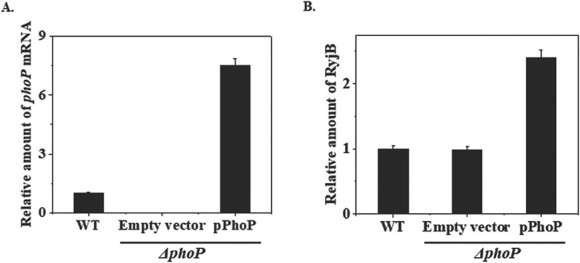
Ectopic expression of PhoP upregulates ryjB expression. A. RT-qPCR analysis was performed to estimate the amount of phoP mRNA in wild type (WT) and ΔphoP cells harbouring empty vector or pPhoP recombinant plasmid. 5S rRNA gene was taken as internal control for the normalization of the amount of phoP mRNA at each time point. Accumulation of phoP mRNA in WT cells was set as 1 and relative amount of phoP at other time point was calculated. B. RyjB abundance was quantified through RT-qPCR analysis in WT and ΔphoP cells harbouring either empty vector or pPhoP recombinant plasmid. 5S rRNA gene was taken as reference gene for the normalization of the amount of RyjB at each time point. Accumulation of RyjB in WT cells was set as 1 and relative amount of RyjB at other time point was calculated.
To investigate the effect of ectopically expressed phoP on ryjB expression, exponentially growing ΔphoP cells (OD600 = 0.4) harbouring recombinant pPhoP plasmid were treated with 1 mM IPTG for an hour. Cells were then harvested and total RNA was isolated from the cells. RT-qPCR analysis was performed to measure RyjB accumulation in the sample. Wild type and ΔphoP cells containing empty plasmid were also processed in an identical method. Figure 5B shows a 2.4-fold induction of RyjB in ectopically expressed PhoP in ΔphoP cells. These data indicate a direct role of PhoP in ryjB regulation in E. coli.
Overexpression of RyjB induces sgcA expression
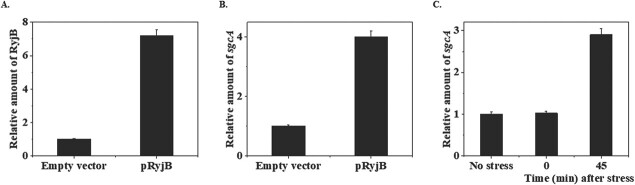
RyjB is synthesized from a strand complementary to one from which sgcA is transcribed. Presence of 4-nt complementary region between ryjB and sgcA transcripts persuaded us to investigate the effect of elevated plenitude of RyjB on sgcA transcript. Since ryjB and sgcA genes are located in opposite directions, deletion of ryjB will eventually eliminate the regulatory region of sgcA gene and vice versa. Thus, RyjB was decided to be overexpressed in wild type cells to examine the effect of its overabundance on sgcA expression. To examine that fact, wild type MG1655 cells harbouring recombinant pRyjB plasmid were grown until mid-exponential phase (OD600 = 0.4). IPTG (1 mM) was added to the growth medium and cells were further grown for an hour. Cells were pelleted down and total RNA was isolated from them. RT-qPCR analysis was performed to estimate the accumulation of ryjB and sgcA transcripts. Cells with empty plasmid were also treated in an identical manner. Figure 6A shows that the overexpression of the ryjB gene resulted in approximately 7-fold higher accumulation of intracellular RyjB. Cells overexpressing RyjB was found to concomitantly increase sgcA expression by at least about 4-fold (Fig. 6B), which indicates the direct interaction of RyjB with sgcA transcript.
Figure 6.
Effect of RyjB overexpression on sgcA accumulation in the cell. Cells with empty vector or pRyjB growing at mid-exponential phase (OD600 = 0.4) were treated with 1-mM IPTG for an hour. Cells were harvested and total RNA was isolated from each sample. Accumulation of RyjB (A) or sgcA (B) was measured by RT-qPCR analysis. 5S rRNA gene was taken as internal control for the normalization of the amount of ryjB or sgcA transcript at each point. Accumulation of RyjB or sgcA in the cells with empty vector was set as 1 and relative amount of RyjB or sgcA in the cells with pRyjB was calculated. C. RT-qPCR analysis was performed to measure the accumulation of sgcA mRNA under acid stress as described earlier. Expression of 5S rRNA gene was utilized as internal control.
Since overexpressed RyjB enhances intracellular accumulation of sgcA and RyjB is induced under acid stress, we wanted to examine whether sgcA is also induced under acid stress. To test that hypothesis, cells were grown till mid-exponential phase (OD600 = 0.4) and pH of the medium was reduced to 5.0 using 10-mM homopipes buffer as described in the ‘Materials and Methods’ section. Cells exposed to low pH were collected after 45 min and total RNA was isolated, which was then used to measure sgcA transcript by RT-qPCR analysis. Figure 6C shows the sgcA expression was induced by about 3-fold under acid stress. This fact indicates that higher accumulation of sgcA under acid stress is possibly driven by higher accumulation of RyjB at low pH.
Discussion
We have taken a comprehensive approach in the current study to investigate the regulation of expression of a novel sRNA RyjB. Stationary phase sigma factor EσS was shown to be unnecessary for RyjB transcription under both normal and acid stress conditions. RyjB accumulation in Δhfq cells is identical to that in wild type cells, except in the stationary phase, where the RyjB population is ~30% less in Δhfq cells than in wild type cells. Thus, it was believed that Hfq was not necessary for the post-transcriptional abundance of RyjB at least in the exponential phase. Interestingly, RyjB was found to be an acid stress inducible sRNA; however, alkaline pH did not show any effect on RyjB expression. A comprehensive investigation of RyjB induction under acid stress revealed that the higher abundance of PhoP upregulates RyjB expression at low pH. RyjB overexpression resulted in higher sgcA accumulation in the cells indicating its role as a cis-acting sRNA.
Enteric bacteria like E. coli naturally colonize in mammalian digestive tract and their passage to intestine is achieved by their transmission through stomach (pH 1.5–3.0). Escherichia coli utilizes diverse metabolic and proton consuming mechanisms to survive at external pHs, which are way beyond the cytoplasmic pH range (25, 26). Numerous sRNAs participate in pH adaptation mechanisms and promote cell survival at extreme external pHs (27). RyjB induction at low pH indicated its possible involvement in pH homeostasis in the cell. Clearly, more investigation is required to settle this issue. sRNAs were reported to improve cell survival under acid stress. GcvB has been demonstrated to improve the survival of E. coli cells at low pH by stimulating the acid resistance system through upregulating the levels of RpoS (28). An EσS-dependent antisense RNA GadY protects as well as stabilizes gadX transcript under acid stress condition, which in turn increases the acid tolerance of E. coli (29).
Low abundance of RyjB in the stationary phase led us to perceive that this sRNA might be sensitive to stress as the stationary phase provides a condition of multiple stresses originating from nutrient deprivation, accumulation of secondary metabolites, alteration in pH of the medium, increased population density, etc. Measurement of pH at different growth phases of E. coli cells revealed that pH of the medium gradually drops to acidic range from early exponential to early stationary phase and increases afterwards to alkaline range (Siddiqui and Dutta, unpublished observation). These data also corroborate previously published reports (30). Thus, it was anticipated that lowering of pH in the exponential phase might be the reason for higher abundance of RyjB in that phase than the stationary phase of growth.
The pleiotropic two-component regulatory system PhoP-PhoQ, although acts as a response regulator of low Mg2+ adaptation (16, 31), has also been conferred to be involved in acid resistance regulatory network (20). PhoP/Q was found to control the expression of numerous sRNAs like MgrR, SokX and IsrC in either Mg2+ dependent or independent manner (14, 32). RyjB appeared to be another PhoP-regulated sRNA, which is induced in response to acid stress. RyjB was also identified as the sRNA, which is induced under high Mg2+ concentration (14). Since PhoP is activated by PhoQ-phosphorylation at low level of extracellular Mg2+ (33); therefore, PhoP seemed to be unnecessary for RyjB induction under high Mg2+ level. However, it has been explicated that the PhoP/PhoQ system controls acid resistance genes in E. coli and a class of such genes including dps, gadA, hdeA and hdeB has been delineated to be highly induced at low pH under the control of PhoP (20). Thus, PhoP-mediated induction of RyjB under acid stress is a new addition to that list.
Cells contain diverse mechanisms of sRNA induction under acid stress. Majority of acid inducible sRNAs have been demonstrated to activate global stress responsive transcription factor RpoS for the acquisition of acid stress tolerance in E. coli (34). It has been shown that overexpression of Hfq interacting sRNAs like RprA, DsrA and ArcZ, which are translational activators of RpoS, increased the acid-tolerance of E. coli cells (35). Wherefore, it was suggested that the enhanced RpoS level through the action of the aforementioned sRNAs possibly produce an effectual response to acid stress (34). GcvB was also found to rescue E. coli cells from acid stress vulnerability by upregulating rpoS expression (28). RpoS-independent activation of a sRNA RyeA was demonstrated (36, 37). PhoP-mediated activation RyjB is a new regulatory pathway of sRNA induction under acid stress.
Although RyjB is a conserved sRNA in E. coli and Salmonella, quite a few differences in its properties have been observed in both the enterobacteria. Growth-dependent expression of RyjB in Salmonella (38) is somewhat identical to its expression pattern in E. coli; nonetheless, the role of Hfq in RyjB abundance differs moderately in both microorganisms. Hfq in Salmonella has been implicated to be nonessential for RyjB stability (39), but RyjB appeared to be significantly unstable in the stationary phase of Δhfq E. coli cells. The distinctive attribute of RyjB, which lacks in Salmonella, is its induction under acid stress. PhoP is essential for its acid induction as the elimination of phoP allele from E. coli chromosome precluded its elevation at low pH. Even though PhoPQ has been reported to regulate RyjB expression in Salmonella (40), but RyjB was never perceived to be acid stress inducible sRNA in Salmonella (39). Difference in RyjB profile in E. coli and Salmonella under acid stress might stem from the differences in natural habitats of bacteria. Unlike Salmonella, which is primarily pathogenic, E. coli, a commonly nonpathogenic bacterium, colonizes in human intestine and frequently experiences low pH exposure of the stomach. This difference in habitats might have evolved E. coli RyjB to be an acid induced sRNA. A comprehensive work will elucidate the reason for the aforementioned differences.
Although the regulation of ryjB expression is now known (current study), but the key role of this sRNA has yet to be discerned. A 4-nt overlap of ryjB gene with sgcA evinces its probable function as an antisense RNA. In the current study, it has been shown that the overexpression of RyjB at normal condition increases sgcA accumulation. Expression of sgcA was also found to be induced under acid stress, which was believed to be attributed by RyjB. However, additional work has to be performed to establish that RyjB directly interacts with sgcA transcript and regulates its abundance. Moreover, small length of the overlapping region and the presence of mostly AU sequence in the overlapping nucleotides of ryjB and sgcA genes call for a systematic study to demonstrate the antisense action of RyjB on sgcA expression. Low pH induction of RyjB points to its role in pH homeostasis in E. coli. Like other acid stress inducible sRNAs, e.g. RprA, DsrA and ArcZ, whether it acts as the activator of global stress responsive transcription factor RpoS or it directly interacts to protein involved in glutamic/arginine dependent acid resistance systems (41) has to be investigated. A recent finding identified the intergenic region of gene encoding isocitrate dehydrogenase kinase/phosphatase (aceK) as a target for RyjB through RIL-seq analysis (42). This suggests that RyjB might be involved in regulating gene in multiple regulatory network. A rigorous study is required to settle this issue.
RyjB expression under different conditions and its regulation known thus far have potentially established itself as an acid stress inducible sRNA, which is regulated by PhoP at the transcriptional level. However, the complete transcriptional regulation of RyjB at low pH is still unknown. Whether any other transcription factor participates in its induction mechanism has yet to be unveiled. Results presented in this study add substantially to our knowledge and will help to identify its mechanism of action under acid stress condition.
Funding
We acknowledge Science and Engineering Research Board, Department of Science and Technology, Government of India, Indian Council for Medical Research, Government of India and Board of Research in Nuclear Sciences, Government of India for funding the research (ECR/2016/000178, 52/07/2019-BMS and 58/14/11/2019-BRNS/103871 to T.D.).
Authors’ contribution statement
TD conceptualized and designed the experiments. NS and AKG performed the experiments and TD wrote the paper.
Supplementary Data
Supplementary Data are available at JB Online.
Acknowledgement
We sincerely thank Prof. J. Gowrishankar of the Centre for DNA Fingerprinting and Diagnostics, Hyderabad, India, and Prof. Umesh Varshney of the Indian Institute of Science, Bengaluru, India, for providing mutant strains. We also thank Prof. Christopher S. Hayes, University of California, Santa Barbara, for providing us plasmids. Both N.S. and A.K.G. acknowledge Indian Institute of Technology Delhi for fellowship.
Conflict of Interest
Authors do not have any potential conflict of interest.
Supplementary Material
Contributor Information
Namra Siddiqui, RNA Biology Laboratory, Department of Chemistry, Indian Institute of Technology Delhi, Hauz Khas, New Delhi 110016, India.
Amit Kumar Gupta, Center for Cancer Research, National Cancer Institute, Frederick, MD 21702-1201, USA.
References
- 1. Dutta, T., and Srivastava, S. (2018) Small RNA-mediated regulation in bacteria: a growing palette of diverse mechanisms. Gene 656, 60–72 [DOI] [PubMed] [Google Scholar]
- 2. Storz, G., Vogel, J., and Wassarman, K.M. (2011) Regulation by small RNAs in bacteria: expanding frontiers. Mol. Cell. 43, 880–891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Wagner, E.G.H., and Romby, P. (2015) Small RNAs in bacteria and archaea: who they are, what they do, and how they do it. Adv. Genetics 90, 1–76 [DOI] [PubMed] [Google Scholar]
- 4. Mandin, P., and Gottesman, S. (2009) Regulating the regulators: an RNS decoy acts as an OFF switch for the regulation of a sRNA. Gene Dev. 23, 1981–1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Water, L.S., and Storz, G. (2009) Regulatory RNAs in bacteria. Cell 136, 615–628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Papenfort, K., and Vanderpool, C.K. (2015) Target activation of regulatory RNAs in bacteria. FEMS Microbiol. Rev. 39, 362–378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Prevost, K., Desnoyers, G., Jacques, J.F., Lavoie, F., and Masse, E. (2011) Small RNA-induced mRNA degradation achieved through both translation block and activated cleavage. Genes Dev. 25, 385–396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Massé, E., Escorcia, F.E., and Gottesman, S. (2003) Coupled degradation of a small regulatory RNA and its mRNA targets in Escherichia coli. Genes Dev. 17, 2374–2383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wassarman, K.M., Zhang, A., and Storz, G. (1999) Small RNAs in Escherichia coli. Trends Microbiol 7, 37–45 [DOI] [PubMed] [Google Scholar]
- 10. Masse, E., and Gottesmen, S. (2002) A small RNA regulates the expression of genes involved in iron metabolism in Escherichia coli. Proc. Natl. Acad. Sci. USA 99, 4620–4625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Papenfort, K., Sun, Y., Miyakoshi, M., Vanderpool, C.K., and Vogel, J. (2013) Small RNA-mediated activation of sugar phosphate mRNA regulates glucose homeostasis. Cell 153, 426–437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Zhang, A., Wasserman, K.M., Rosenow, C., Tjaden, B.C., Storz, G., and Gottesman, S. (2003) Global analysis of small RNA and mRNA targets of Hfq. Mol. Microbiol. 50, 1111–1124 [DOI] [PubMed] [Google Scholar]
- 13. Kawano, M., Reynolds, A.A., Miranda-Rios, J., and Storz, G. (2005) Detection of 5′- and 3′-UTR-derived small RNAs and cis-encoded antisense RNAs in Escherichia coli. Nucleic Acids Res. 33, 1040–1050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Reizer, J., Charbit, A., Reizer, A., and Saier, M.H., Jr. (1996) Novel phosphotransferase system genes revealed by bacterial genome analysis: operons encoding homologues of sugar-specific permease domains of the phosphotransferase system and pentose catabolic enzymes. Genome Sci. Technol. 1, 53–75 [Google Scholar]
- 15. Lennox, E.S. (1955) Transduction of linked genetic characters of the host by bacteriophage P1. Virology 1, 190–206 [DOI] [PubMed] [Google Scholar]
- 16. Raghavan, R., Groisman, E., and Ochman, H. (2011) Genome-wide detection of novel regulatory RNAs in E. coli. Genome Res. 21, 1487–1497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Chen, H., Dutta, T., and Deutscher, M.P. (2016) Growth phase-dependent variation of RNase BN/Z affects small RNAs: REGULATION OF 6S RNA. J. Biol. Chem. 291, 26435–26442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Sauer, E., and Weichenrieder, O. (2011) Structural basis for RNA 3′-end recognition by Hfq. Proc. Natl. Acad. Sci. USA 108, 13065–13070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zuker, M. (2003) Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 31, 3406–3415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zwir, I., Shin, D., Kato, A., Nishino, K., Latifi, T., Solomon, F., Hare, J.M., Huang, H., and Groisman, E.A. (2005) Dissecting the PhoP regulatory network of Escherichia coli and Salmonella enterica. Proc. Natl. Acad. Sci. USA 102, 2862–2867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Coornaert, A., Chiaruttini, C., Springer, M., and Guillier, M. (2013) Post-transcriptional control of the Escherichia coli PhoQ-PhoP two-component system by multiple sRNAs involves a novel pairing region of GcvB. PLOS Genet. 9, e1003156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Xu, J., Li, T., Gao, Y., Deng, J., and Gu, J. (2019) MgrB affects the acid stress response of Escherichia coli by modulating the expression of iraM. FEMS Microbiol. Lett. 366, 1–9 [DOI] [PubMed] [Google Scholar]
- 23. Bearson, B.L., Wilson, L., and Foster, J.W. (1998) A low pH-inducible, PhoPQ-dependent acid tolerance response protects Salmonella typhimurium against inorganic acid stress. J. Bacteriol. 180, 2409–2417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kanda, T., Abiko, G., Kanesaki, Y., Iwai, N., and Wachi, M. (2020) RNase E dependent degradation of tnaA mRNA encoding tryptophanase is prerequisite for the induction of acid resistance in Escherichia coli. Sci. Rep. 10, 7128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Krulwich, T.A., Sachs, G., and Padan, E. (2011) Molecular aspects of bacterial pH sensing and homeostasis. Nat. Rev. Microbial. 9, 330–344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kanjee, U., and Houry, W.A. (2013) Mechanism of acid resistance in Escherichia coli. Annu. Rev. Microbiol. 67, 65–81 [DOI] [PubMed] [Google Scholar]
- 27. Aiso, T., Kamiya, S., Yonezawa, H., and Gamou, S. (2014) Overexpression of an antisense RNA, ArrS, increases the acid resistance of Escherichia coli. Microbiology 160, 954–961 [DOI] [PubMed] [Google Scholar]
- 28. Jin, Y., Watt, R.M., Danchin, A., and Huang, J. (2009) Small noncoding RNA GcvB is a novel regulator of acid resistance in Escherichia coli. BMC Genomics 10, 165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Opdyke, J.A., Kang, J.G., and Storz, G. (2004) A small RNA regulator of acid response genes in Escherichia coli. J. Bacteriol. 186, 6698–6705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Clemente, R.S., Igeno, M.I., Poblacion, A.G., Guijo, M.I., Merchan, F., and Blasco, R. (2018) Study of pH changes in the media during growth of several environmental strains. Proceedings 2, 1297 [Google Scholar]
- 31. Groisman, E.A., Hollands, K., Kriner, M.A., Lee, E.J., Park, S.Y., and Pontes, M.H. (2013) Bacterial Mg2+ homeostasis, transport, and virulence. Annu. Rev. Genet. 47, 625–646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Moon, K., and Gottesman, S. (2009) A PhoQ/P-regulated small RNA regulates sensitivity of Escherichia coli to antimicrobial peptides. Mol. Microbiol. 74, 1314–1330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kato, A., Tanabe, H., and Utsumi, R. (1999) Molecular characterization of the PhoP-PhoQ two component system in Escherichia coli K-12: identification of extracellular Mg2+-responsive promoter. J. Bacteriol. 181, 5516–5520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. De Bruijn, F.J. (2016) Stress and Environmental Regulation of Gene Expression and Adaptation in Bacteria. pp 402–420 John Wiley & Sons, USA [Google Scholar]
- 35. Gaida, S.M., Al-Hinai, M.A., Indurthi, D.C., Nicolaou, S.A., and Papoutsakis, E.T. (2013) Synthetic tolerance: three noncoding small RNAs, DsrA, ArcZ and RprA, acting supra-additively against acid stress. Nucleic Acids Res. 41, 8726–8737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Gupta, A.K., Siddiqui, N., and Dutta, T. (2020) A novel mechanism of RyeA/SraC induction under acid stress. Biochem. Biophys. Res. Commun. 525, 298–302 [DOI] [PubMed] [Google Scholar]
- 37. Gupta, A.K., Siddiqui, N., and Dutta, T. (2019) Regulation of RyeA/SraC expression in Escherichia coli. Biochem. Biophys. Res. Commun. 516, 661–665 [DOI] [PubMed] [Google Scholar]
- 38. Perez, E., Samper, S., Bordas, Y., Guilhot, C., Gicquel, B., and Martin, C. (2001) An essential role for phoP in Mycobacterium tuberculosis virulence. Mol. Microbiol. 41, 179–187 [DOI] [PubMed] [Google Scholar]
- 39. Kröger, C., Colgan, A., Srikumar, S., Handler, K., Sivasankaran, K. S., Hammarlof, D. L., Canals, R., Grissom, J. E., Conway, T., Hokamp, K., and Hinton, J. C. D. (2013) An infection-relevant transcriptomic compendium for Salmonella enterica serovar typhimurium. Cell Host Microbe 14, 683–695 [DOI] [PubMed] [Google Scholar]
- 40. Colgan, A.M., Kröger, C., Diard, M., Hardt, W.D., Puente, J.L., Sivasankaran, S.K., Hokamp, K., and Hinton, J. C. D. (2016) The impact of 18 ancestral and horizontally-acquired regulatory proteins upon the transcriptome and sRNA landscape of Salmonella enterica serovar typhimurium. PLoS Genet. 12, e1006258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Foster, J.W. (2004) Escherichia coli acid resistance: tales of an amateur acidophile. Nat. Rev. Microbiol. 2, 898–907 [DOI] [PubMed] [Google Scholar]
- 42. Bar, A., Argaman, L., Altuvia, Y., and Margalit, H. (2021) Prediction of novel bacterial small RNAs from RIL-Seq RNA-RNA interaction data. Front. Microbiol. 12, 635070. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.