Abstract
The methylotrophic yeast Pichia pastoris (syn. Komagataella phaffii) has been extensively engineered for protein production, and is attracting attention as a chassis cell for methanol biotransformation toward production of small molecules. However, the relatively unclear methanol metabolism hampers the metabolic rewiring to improve the biosynthetic efficiency. We here performed a label-free quantitative proteomic analysis of Pichia pastoris when cultivated in minimal media containing methanol and glucose, respectively. There were 243, 158 up-regulated proteins and 244, 304 down-regulated proteins in log and stationary phase, respectively, when cultivated in methanol medium compared with that of glucose medium. Peroxisome enrichment further improved the characterization of more differentially expressed proteins (481 proteins in log phase and 524 proteins in stationary phase). We demonstrated the transaldolase isoenzyme (Tal2, Protein ID: C4R244) was highly up-regulated in methanol medium cultivation, which plays an important role in methanol utilization. Our work provides important information for understanding methanol metabolism in methyltrophic yeast and will help to engineer methanol biotransformation in P. pastoris.
Keywords: Proteomics, Peroxisome, Methanol metabolism, Systems biology
1. Introduction
The methylotrophic yeast Pichia pastoris (syn. Komagataella phaffii) is able to utilize methanol as a sole carbon and energy source, which has been widely used for protein production [1]. Recently, it has attracted great interest for production of chemicals and advanced biofuels from methanol [[2], [3], [4]], which should play an important role toward carbon neutrality. It is required much higher titer, rate and yield in production of small molecules in compared with the protein production due to the relative lower prices. However, the methanol metabolism is very complex and poorly understood, which limits the efficiency of methanol biotransformation toward target molecules. For example, metabolic engineering of P. pastoris only resulted 2.7 g/L malic acid production from sole methanol, while malic acid production reached 8.7 g/L when using glucose as a carbon source [5]. Similarly, β-alanine only reached 5.6 g/L from methanol in engineered P. pastoris evening with supplementation of nutrient-rich ingredients peptone and yeast extract [6]. This low production might attribute to the poorly understanding of methanol metabolism.
The methanol metabolism is localized in different cellular compartments. The initial methanol metabolism is localized in peroxisome [7], which might avoid diffusion of the toxic formaldehyde, the oxidized product of methanol. Thus, the related enzymes such as catalase (Cat), alcohol oxidase (Aox), dihydroxyacetone synthase (Das), S-(hydroxymethyl) glutathione dehydrogenase (Fld), formate dehydrogenase (Fdh) are mainly localized in peroxisome [[8], [9], [10]]. The further steps of methanol assimilation mainly involve glycolysis/gluconeogenesis and pentose phosphate pathway [11]. However, the proteomic response of P. pastoris is not fully understand when using methanol as a carbon source, which might hinder the rational engineering methanol metabolism for production of target compounds.
Here, we performed label-free and comparative quantitative proteomic analysis of whole P. pastoris cells with glucose or methanol as carbon sources. We also enriched peroxisomes by an upgraded density gradient centrifugation for proteomic analysis, considering the importance of peroxisome in methanol metabolism [12]. The proteomic data described here should be helpful for engineering P. pastoris and other methyltrophic yeasts for methanol biotransformation.
2. Materials and methods
2.1. Strains and culture conditions
GS115 (Mut+, his4-) was used for proteome analysis. TAL2 disruption strain TA02 and its parent strain PC111 [13] were used for functional analysis of TAL2 (Table 1). The yeast strains were cultivated at 30 °C, 220 rpm in a shake incubator (Zhichu ZQZY-CS8) to an initial optical density (OD600) of 0.2. The preculture were cultivated in 15 mL tube with a working volume of 2 mL Delft minimal medium (2.5 g/L (NH4)2SO4, 14.4 g/L KH2PO4, 0.5 g/L MgSO4·7H2O, 40 mg/L histidine, 1 mL/L Vitamin solution, 2 mL/L Trace metal solution) [14,15] with 10 g/L glucose. Finally, the preculture cells were inoculated to 20 mL Delft minimal medium containing 10 g/L glucose or methanol as carbon sources.
Table 1.
Strains used in this study.
| Name | Genotype | Resource |
|---|---|---|
| GS115 | Mut+, his4- | Lab stock |
| PC111 | Mut+, his4-, HIS4::PGAP-PpRAD52-TAOX1, PNSI-2::PGAP-hCas9-TDAS1 | [13] |
| TA02 | Mut+, his4-, HIS4::PGAP-PpRAD52-TAOX1, PNSI-2::PGAP-hCas9-TDAS1, tal1-2Δ | This study |
2.2. Reagents
MOPS solution, OptiPrep™ Density Gradient Medium, ethylenediaminetetraacetic acid disodium salt dehydrate (EDTA), potassium hexacyanoferrate (II) trihydrate, cytochrome C, sodium dithionite, tris (hydroxymethy) aminomethane, triton-X 100, lyticase, titanium oxysulfate, titanium (II) oxide, hydrogen peroxide, urea, a protease inhibitor cocktail, dithiothreitol (DTT), iodoacetamide (IAA), formic acid (FA), trifluoroacetic acid (TFA) and dextran coated charcoal were ordered from Sigma-Aldrich. Sucrose was purchased from J&K (Beijing, China). Trypsin was bought from Promega (Madison, WI, USA). Bradford assay kits were purchased from Beyotime (Shanghai, China). D-Sorbitol was purchased from TCI (Shanghai, China). Acetonitrile (ACN) and methanol were purchased from Merck (Darmstadt, Germany). Deionized water was purified using a Milli-Q system (Millipore, Milford, MA, USA). A Microcon filtration device with a relative molecular weight (Mw) cutoff of 10,000 Da (10 K filter) was purchased from Sartorius AG (Gottingen, Germany). All the other chemicals and solvents of analytical-grade were from Kermel (Tianjin, China).
2.3. Peroxisome enrichment
Peroxisomes were enriched by a modified protocol from Cold Spring Harbor [16] with a peroxisome Isolation Kit from Sigma-Aldrich. Approximately 100 mg dry weight of cells with 50-mL tubes were centrifuged for 10 min at 2, 000 × g at 4 °C, washed twice with ice-cold high purity water. And then the cell pellets were incubated with DTT buffer (7 × volume of the wet weight) for 30 min at 30 °C with gentle shaking (80 rpm) and harvested by centrifugation at 1000 g for 15 min at room temperature. Then the cells were washed three times with 10 mL of 1.2 M sorbitol and lysed by incubating in sorbitol buffer (5 × the wet weight) containing 8000 U of lyticase per 100 mg dry weight of cells for 12–16 h. The resulted spheroplasts were washed three times with 5 mL of 1.2 M sorbitol and re-suspended in peroxisome extraction buffer (5 mM MOPS, 0.25 M sucrose, 1 mM EDTA, and 0.1% (v/v) ethanol, 1% (v/v) protease inhibitors, pH 7.65, 2 × the wet weight, 30–60 min at 4 °C). The cell debris were removed by centrifugation at 3000 g (Sigma 1–14 K) for 5 min at 4 °C for twice. The collected supernatants were loaded to the 7 mL cushion of 60% OptiPrep to a 13.2 mL ultra-clear Thinwall tube (Beckman Coulter 344059, USA). Sediments were formed after centrifugation at 25,000 × g (SW 41 Ti, Beckman Coulter Optima™ L-100XP Ultracentrifuge, USA) for 30 min at 4 °C. Collected the organelle particles (OP) carefully in 5-mL tube and dilute OP to 4.5 mL with 22.5% OptiPrep solution (5 mM MOPS, pH 8.0, with 1 mM EDTA and 0.1% (v/v) ethanol). The OP can be used for density gradient centrifugation with a protein load limit of 10 mg. Place 2 ml of the 32% and 27.5% Optiprep solution into a 13.2 ml ultra-clear centrifuge tube successively using an Eppendorf pipette and then overlay with 4.5 ml of the diluted OP sample with 22.5% OptiPrep solution. Overlay the sample with 2 ml 20% Optiprep solution. Centrifuge for 1.5 h at 100,000×g. A light-red ring at the 22.5%/27.5% interface is the enriched peroxisomes. Carefully collected the light-red ring and restored at 4 °C for no more than 24 h.
2.4. Protein solubilization and digestion
Yeast cell pellets were added with 0.2 mL ionic liquid [17] and homogenized with 0.4 mm glass beads by using a FastPrep-24 instrument (MP Biomedicals, USA) for 9 repeated 40 s per cycles at 6.5 m/s, with 60 s pause. The lysate was transferred to new tubes, diluted 10 times with the lysis buffer. The protein concentration was determined by using Bradford Assay Kit. Purified peroxisome samples were sonicated on ice for 120 s (pulse on time 10 s, pulse off time 10 s). All lysis buffers were added with 1% (v/v) protease inhibitor cocktail. Samples of yeast cell pellets and the purified peroxisomes were treated in triplicate using an ionic liquid-based filter-aided sample preparation (i-FASP) method [17]. In brief, samples were added with 0.1 M DTT and incubated at 95 °C for 3 min. The cell debris was removed by centrifugation at 16,000 × g at 4 °C for 5 min, and 100–150 μg of the clarified protein extract was transferred to a 10 kDa filter. After centrifugation on the filter at 14,000 g at 20 °C for 15 min, the extracted proteins were retained, diluted with 200 μL of 50 mM NH4HCO3 and centrifuged again at 14,000 g at 20 °C for 15 min. Subsequently, 200 μL of 50 mM IAA dissolved in 50 mM NH4HCO3 buffer, and the samples were incubated in darkness for 10 min. Then the samples were fileted and washed three times with 200 μL of 50 mM NH4HCO3, then digested for overnight at 37 °C by adding 100 μL of 10 mM NH4HCO3 containing trypsin (3–6 μg). Finally, the tryptic peptides were collected by centrifugation, and 50 μL of water was added to elute the peptide-rich solution twice. Peptides were extracted, vacuum dried, and stored at −80 °C until LC-MS/MS analysis.
2.5. Enzymatic assay of catalase and cytochrome c oxidase
Subcellular localization of organelles can be characterized via measuring the activities of catalase as a peroxisome marker, and cytochrome c oxidase as mitochondrial markers. For measuring the catalase activity, 50 μL of each gradient fraction was mixed with 150 μL reaction buffer (0.33 M sodium phosphate buffer (pH 7.1 at room temperature); 0.1% (v/v) Triton X-100), 500 μL of 1.25 mM hydrogen peroxide solution was added for initiation of the reaction. After 2 min incubation at room temperature, 500 μL of titanium oxysulfate solution was added to stop the reaction. Precipitation was removed by centrifugation at 13, 000 rpm for 10 min at 4 °C and OD410 was recorded. One unit (U) catalase activity is defined as OD410 decrease rate per mg protein.
For quantifying the activity of cytochrome c oxidase, 50 μL of each gradient fraction was added into one well of a 96-well plate containing 200 μL 0.05 M potassium phosphate buffer (pH 7.0 at room temperature). 20 μL of reduced cytochrome c solution was added to start the reaction and 0.2 mg of sodium dithionite. One unit (U) activity of cytochrome c oxidase is defined as OD550 decrease rate per mg protein.
2.6. Nano-RPLC-ESI-MS/MS analysis
A nano-RPLC-ESI-MS/MS system was set up for quantitative proteomic analysis, which consists of Q Exactive mass spectrometer and Ultimate™ 3000 RSLC nano HPLC systems. The experimental conditions of nano-RPLC were as follows: the C18 separation column (ReproSil-Pur C18-AQ, 15 cm × 50 μm i.d., 1.9 μm, 120 Å) was home-made with a C18 trap column (XBP, 3 cm × 75 μm i.d., 5 μm, 150 Å); mobile phase, 2% (v/v) ACN containing 0.1% (v/v) FA (A), 98% (v/v) ACN containing 0.1% (v/v) FA (B); flow rate, 600 nL min−1; separation gradient: 0–10 s, 2–7% B; 10 s–50 min 10 s, 7–23% B; 50 min 10 s–70min 10 s, 23–40% B; 70 min 10 s–72 min 10 s, 40–80% B; 72 min 10 s−85 min 10 s, 80% B.
For Q Exactive analysis, the total ion chromatograms and mass spectra were recorded from m/z 300 to 1800 with Xcalibur software in positive ion mode. The spray voltage was 2.4 kV and the temperature of ion transfer tube was set at 320 °C. Full MS scans were obtained in the orbitrap analyzer with a range of m/z 300–1800 at a mass resolution of 70,000, with an AGC target value of 3E6 and maximum injection time of 60 ms. MS/MS spectra were acquired using a data-dependent top 20 method and precursor selection was based on parent ion intensity from the survey scans. To select peptides for MS/MS analysis, ions with charge states from 2 to 6 were all included, dynamic exclusion was set to 18 s with mass tolerance of 20 ppm, and intensity threshold was set at 1.7E4. Most intense precursor ions were selected and isolated with a window of 2.0 m/z and fragmented by higher-energy collisional dissociation (HCD) with a normalized collision energy of 28. MS/MS spectra were acquired in the ion trap at enhanced scan rate with an AGC target value of 5E4 and maximum injection time at 60 ms.
2.7. MS data analysis
All MS/MS spectra raw files were processed using either two commonly bioinformatics platforms: Proteome Discoverer (version 2.2, Thermo Fisher Scientific) in conjunction with Mascot as a database search engine or MaxLFQ [18] integrated in MaxQuant [19] (version 1.6.17.0), both against the Pichia pastoris GS115 reference proteome database downloaded from UniProt [20] (UP000000314; 5073 sequences; 20210226) and supplemented with GQ68_03479T0(AOA69352.1; NCBI). The parameters were set as follows: MS1 tolerance of 10 ppm; MS2 mass tolerance of 20 ppm; enzyme specificity was set as trypsin with 2 missed cleavages allowed; carbamidomethylation of cysteine was set as a fixed modification (+57.02 Da); acetylation on the protein N-terminal (+42.01 Da) and oxidation of methionine (+15.99 Da) were set as variable modifications. The required false discovery rate (FDR) was set to 1% at both the peptide and the protein level.
2.8. Bioinformatics analysis
All the quantification results of proteins from samples were processed with Z-score normalization and Student's T-test using Perseus (version 1.6.14.0) (http://coxdocs.org/doku.php?id=perseus:start) [21]. Student's T-test was used to calculate the significance of the changes in protein abundance along with their log2 fold changes (log2FC) between the two cultured conditions. To classify the changes in abundance, the Benjamini-Hochberg method for P-value correction was done. If the P-adj < 0.05, it is considered statistically significant. We performed Benjamini-Hochberg correction for P value (P-adj) as previously described [22]: P values are ranked in an ascending array and multiplied by m/k where k is the position of a P value in the sorted vector and m is the number of independent tests. The differentially expressed proteins were picked the log2FC data was greater than 1 or less than −1 in three biological replicates. All the gene ontology (GO) information was obtained from the Uniprot database (http://www.uniprot.org/), the DAVID (version 2021) Knowledgebase (https://david.ncifcrf.gov/summary.jsp), the STRING (version 11.0) analysis database (http://string-db.org/cgi/input.pl) and KEGG mapper (https://www.genome.jp/kegg/mapper/color.html), including biological process (GOBP), cellular component (GOCC), molecular function (GOMF) and KEGG pathway analysis. Only residues significantly enriched (Modified Fisher Exact test; Benjamini-Hochberg corrected P-value: < 0.05) are shown in color.
2.9. Gene deletion
TAL2 deletion in strain PC111 was conducted by using CRISPR-Cas9 method as previously described [13]. The sgRNA plasmid pPICZ-Cas9-gTAL2 was constructed [13] by using ClonExpress® II One Step Cloning Kit (Vazyme, China) with primes gTAL1-2-F (CCTTGACCCTGCAACCTCAGGTTTTAGAGCTAGAAATAGCAAGTTAAAATAAGGC) and gTAL1-2-R (CTGAGGTTGCAGGGTCAAGGGACGAGCTTACTCGTTTCGTCCTC). The underline is sgRNA sequence. The upstream homologues arm was amplified by using primers TA02up-F(GATATCGATCTACACTTAATAGTAGATGACGAGG) and p-R(CAGAAACGTTTGGCCCAATGGCGATATTTCAGCTGAATTTGAAGATACTCATACTATCGCATATTTATAAC), and downstream homologous arm was amplified by using primers TA02dn-F(GTTATAAATATGCGATAGTATGAGTATCTTCAAATTCAGCTGAAATATCGCCATTGGGCCAAACGTTTCTG) and TA02dn-R(AACAATGGTGTCGTAACCGAACTTC). Then these two homologous arms were fused by overlap extension PCR as a donor DNA for TAL2 knockout.
3. Results
3.1. Experiment design and sample preparation
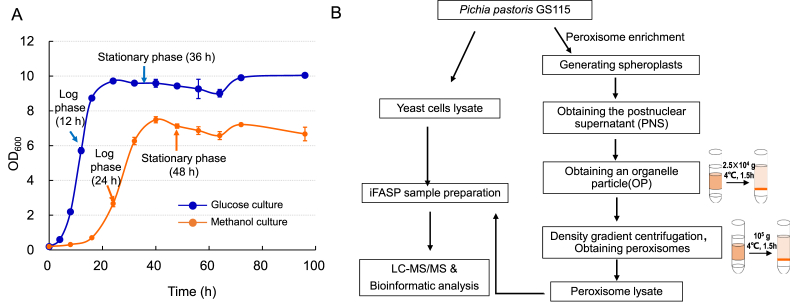
Proteomics analysis of methanol metabolism should be very helpful for engineering methyltrophic yeast for chemical production [[23], [24], [25], [26]]. We here conducted comparative proteomic analysis of Pichia pastoris cultivated in minimal medium containing methanol or glucose by a label-free quantitative proteome strategy (Fig. 1). The middle log phase (12 h glucose culture or 24 h methanol culture) and the stationary phase (36 h glucose culture or 48 h methanol culture) cells were taken for sample preparation (Fig. 1A). With this strategy, we could comprehensively analyze the methanol metabolism by comparing the cellular protein level of methanol cultivated cells with that of glucose cultivated cells. We firstly quantify the total protein level with the whole yeast cell lysate. Then we tried to enrich the peroxisome by using density gradient centrifuge strategy (Fig. 1B), which can help to find peroxisome related proteins and also clearer information on methanol metabolism.
Fig. 1.
Label-free quantitative proteomic analysis of P. pastoris. (A) The growth profiles of P. pastoris GS115 that cultivated in glucose (10 g/L) or methanol (10 g/L) medium and the sample taken time points were indicated as arrows. (B) The workflow for sample preparation. Whole cellular proteome was quantified and the peroxisomes were enriched for proteomic analysis.
3.2. Comparative proteomic analysis of whole cell lysates
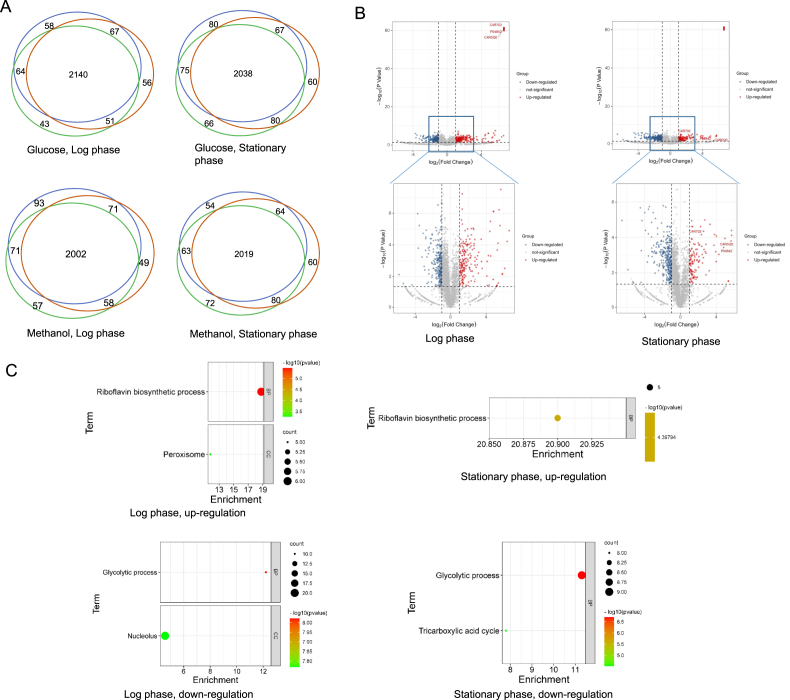
To investigate methanol metabolism of P. pastoris, we comparatively analyzed the proteome of whole yeast cell that cultivated in methanol or glucose medium. There were 487 proteins differentially expressed in log phase (P-adj < 0.05, log2FC > 1 or < −1) with 243 up-regulation and 244 down-regulation under methanol medium, in compared with that of glucose medium (Fig. 2A, Supplementary material). While in stationary phase, there were 462 differentially expressed protein (P-adj < 0.05, log2FC > 1 or < −1) with 158 up-regulation and 304 down-regulation, respectively (Fig. 2A). As expected, the methanol utilization related enzymes are highly up-regulated. The key enzymes of primary methanol assimilation are alcohol oxidase 2 (gene ID Aox2, uniport C4R702), dihydroxyacetone synthase 1 and 2 (Das1, C4R5P8; Das2, C4R5Q0) and dihydroxyacetone kinase (Dak; C4R5Q6), were dramatically up-regulated in methanol cultured cells (Fig. 2B). We further performed GO-term enrichment analysis on biological processes (BP), cellular components (CC) and molecular functions (MF) in DAVID (Benjamini-Hochberg correct P values: < 0.05) analysis for the up- and down-regulated proteins (Fig. 2C), which showed that proteins located in peroxisome were dominantly enriched in methanol than in glucose while proteins related with glycolytic process were dominantly enriched in glucose than methanol. We also observed the significant enrichment of the riboflavin biosynthetic process (Fig. 2C) both in log and stationary phase on up-regulated proteins.
Fig. 2.
Proteomic analysis on Methanol/glucose of whole cell lysate of P. pastoris in log and stationary phase, respectively. Each experimental condition was independently repeated three times and in each of these three biological repetitions, three technical replicas were made. (A) Overlap of identified proteins from three biological repetitions, which are the proteome obtained from the cells that cultured in methanol or glucose. (B) Volcanic maps for differentially expressing proteins. Log-transformed P value (Student's T-test, Benjamini-Hochberg corrected P value: < 0.05) are plotted against log-transform fold change. Red dots (log2FC > 1), significantly upregulation. Blue dots (log2FC < −1), significantly downregulation. Gray dots, non-differentially expressed proteins. Aox1, methanol oxidase 1 (uniprot: P04842), Aox2, methanol oxidase 2 (C4R702) and Das2, dihydroxyacetone synthase 2(C4R5Q0) were dramatically higher up-regulated in log phase than in stationary phase. (C) GO enrichment analysis represented in bubble diagram, including the biological processes (BP), cellular components (CC), and molecular functions (MF). The left and right column charts represent GO enrichment in log phase and in stationary phase, respectively. Only significantly enriched proteins (Modified Fisher Exact test; Benjamini-Hochberg corrected P value: < 0.05) are shown.
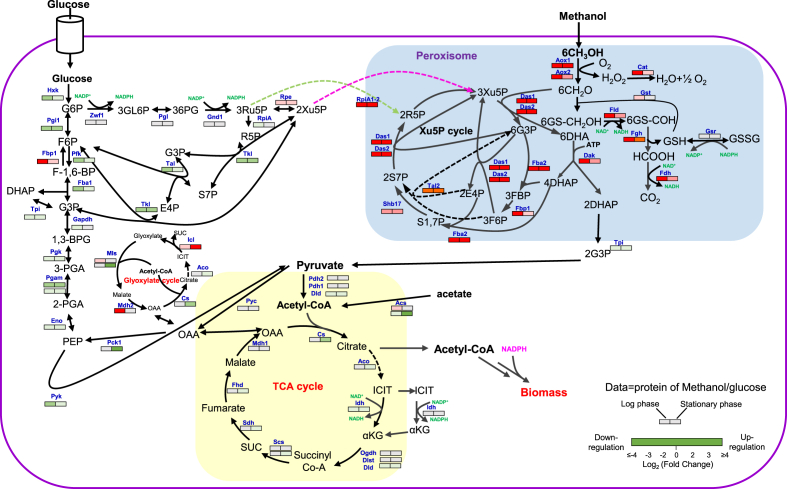
We then mapped the differentially enzyme to the central metabolic pathways (Fig. 3). The xylulose-5 phosphate (Xu-5-P) cycle [11] was active for methanol assimilation and cell growth in methanol medium. The methanol catabolism pathway is also highly activated for providing the NADH and ATP for growth with generating CO2 and the toxic compound H2O2 [27]. Cat was up-regulated to 47.3-fold in log phase, which was assumed to be beneficial for alleviating the toxicity by degrading H2O2. The enzymes related to glyoxylate cycle were up-regulated in methanol medium compared with that of glucose medium (Fig. 3), which was similar with a previous report [11]. The up-regulated glyoxylate cycle might be helpful for the biosynthesis of TCA cycle-derived amino acids when using methanol as a substrate. Interesting, the key gluconeogenesis enzymes fructose-1,6-bisphosphate aldolase (Fba2, C4QWS2) and fructose-1,6-bisphosphatase (Fbp1, C4R5T8) were up-regulated, and the ribulose-5-phosphate 3-epimerase (Rpe) of the pentose-phosphate-pathway (PPP) was also up-regulated (Fig. 3). These results suggested that methanol utilization requires the redirecting metabolic flux toward gluconeogenesis and PPP to provide xylulose-5-phosphate for assimilating formaldehyde, the methanol oxidized metabolite. The enzymes related to TCA cycle had been slightly down-regulated in methanol medium.
Fig. 3.
Comparing the expression of protein related to central metabolism of P. pastoris that cultivated on methanol or glucose media. The heat map shows the protein expression level of methanol cultured cells compared with glucose cultured cells. Statistical analysis was performed using Student's T-test (Benjamini-Hochberg corrected P value: < 0.05). Red lanes (log2FC > 1), significantly upregulation. Green lanes (log2FC < −1), significantly downregulation. Gray lanes, nondifferentially expressed proteins. Methanol metabolism: Aox1, methanol oxidase 1 (uniprot: P04842; EC: 1.1.3.13); Aox2, methanol oxidase 2 (C4R702; 1.1.3.13); Cat, catalase (C4R2S1; 1.11.1.6); Das1, dihydroxyacetone synthase 1(C4R5P8; 2.2.1.3); Das2, dihydroxyacetone synthase 2(C4R5Q0; 2.2.1.3); Dak, dihydroxyacetone kinase (C4R5Q6; 2.7.1.29); Fba2, fructose 1,6-bisphosphate aldolase (C4QWS2; 4.1.2.13); Fbp1, fructose-bisphosphates (C4R5T8; 3.1.3.11); Shb17, sedoheptulose 1,7-bisphosphatase (C4R2M0; 3.1.3.37); Tal2, transaldolase 2 (C4R244; 2.2.1.2); Gst, glutathione transferase (C4R2T6; 2.5.1.18); Gsr, glutathione reductase (C4R686; 1.8.1.7); Fld, S-hydroxymethyl dehydrogenase (C4R6A5; 1.1.1.284); Fgh, S-formyl glutathione hydrolase (C4R5T5; 3.1.2.12); Fdh, formate dehydrogenase (C4R606; 1.17.1.9); RpiA1-2, d-ribose-5-phosphate ketol-isomerase (C4R763; 5.3.1.6). Pentose phosphate pathway (PPP): Zwf1, glucose-6-phosphate 1-dehydrogenase (C4R099; 1.1.1.49); Pgl, 6-phosphogluconolactonase (C4QWM6; 3.1.1.31); Gnd1, 6-phosphogluconate dehydrogenase (C4R430; 1.1.1.343); Tkl, transketolase (C4QXK7; 2.2.1.1); Tal1, transaldolase 1 (C4R245; 2.2.1.2); RpiA, d-ribose-5-phosphate ketol-isomerase (C4R764; 5.3.1.6); Rpe, ribulose-5-phosphate-3-epimerase (AOA69352; 5.1.3.1). Glycolysis/gluconeogenesis pathway: Hkx, phosphotransferase (C4R8F9; 2.7.1.1); Pgi1, glucose-6-phosphate isomerase (C4R4L7; 5.3.1.9); Fbp1, fructose-bisphosphatase (C4R5T8; 3.1.3.11); Pfk, 6-phosphofructokinase (C4QXA5, Q92448; 2.7.1.11); Fba1, fructose-bisphosphate aldolase (C4QW09; 4.1.2.13); Tpi, triosephosphate isomerase(C4R626; 5.3.1.1); Gapdh, glyceraldehydes-3-phosphate dehydrogenase(C4R0P1; 1.2.1.12); Pgk, phosphoglycerate kinase(C4QY07; 2.7.2.3); Pgam, phosphoglycerate mutase(C4R5A6, C4R5P4; 5.4.2.11); Eno, enolase(C4R3H8; 4.2.1.11); Pyk, pyruvate kinase(C4R1P9; 2.7.1.40); Pck1, phosphoenolpyruvate carboxykinase (C4QZB8; 4.1.1.49); Pyc, pyruvate carboxylase (C4R339; 6.4.1.1). Tricarboxylic acid cycle (TCA): Pdh1, dihydrolipoyllysine-residue acetyltransferase (C4QVY5; 2.3.1.12); Pdh2, pyruvate dehydrogenase (acetyl-transferring) (C4QYX8; 1.2.4.1); Dld, dihydrolipoyl dehydrogenase (C4R312; 1.8.1.4); Cs, citrate synthase (C4QW60; 2.3.3.1); Aco, aconitate hydratase (C4QV90; 4.2.1.3); Idh, isocitrate dehydrogenase (NAD+) (C4QZQ0, C4R8B4; 1.1.1.41); Idh, isocitrate dehydrogenase (NADP+) (C4QWH9, C4R142; 1.1.1.42); Ogdh, oxoglutarate dehydrogenase (succinyl-transferring) (C4QZL6; 1.2.4.2); Dlst, dihydrolipoyllysine-residue succinyltransferase (C4QV80; 2.3.1.61); Scs, succinate—CoA ligase (C4R1X8, C4R5P6; 6.2.1.4); Sdh, succinate dehydrogenase (quinone) (C4R8S1, C4R6J2; 1.3.5.1); Fhd, fumarate hydratase (C4R559; 4.2.1.2); Mdh1, malate dehydrogenase 1 (C4R024; 1.1.1.37); Acs, acetate-CoA ligase (C4R1P7,C4R4G6; 6.2.1.1); Glyoxylic acid cycle: Mdh2, malate dehydrogenase 2 (C4R911; 1.1.1.37); Mls, malate synthase (C4QYI8, C4R741; 2.3.3.9); Icl, isocitrate lyase (C4QY57; 4.1.3.1).
3.3. Enriching peroxisomes for proteomic analysis of methanol metabolism
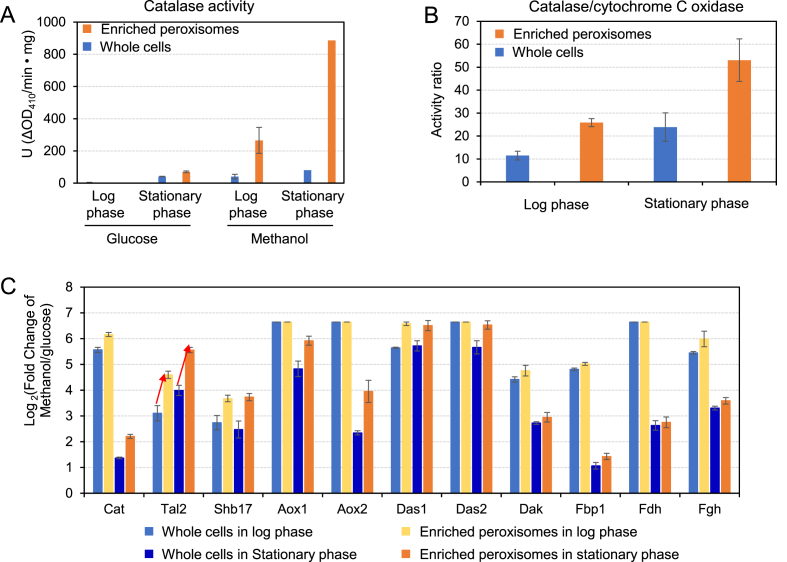
Considering the methanol assimilation is mainly localized in peroxisome, we then enriched peroxisomes by density gradient centrifugation. Peroxisome enrichment further improved the characterization of more differentially expressed proteins (481 proteins in log phase and 524 proteins in stationary phase). The peroxisome marker enzyme catalase was enhanced significantly after enrichment in both glucose and methanol cultivated cells (Fig. 4A). Furthermore, the activity ratio of catalase/cytochrome c oxidase was significantly higher in the enriched fraction (Fig. 4B), which suggested that the peroxisome was successfully enriched with partially removing the mitochondria.
Fig. 4.
Enriching peroxisomes for comparative proteomics analysis. Statistical analysis was performed using Student's T-test (Benjamini-Hochberg corrected P value: < 0.05). (A) The catalase activity of the whole lysate and the peroxisome enriched fraction. (B) The enzyme activity ratio of catalase/cytochrome c oxidase showed that the peroxisome was enriched and also partially purified from mitochondria. (C) The enzyme up-regulation fold of whole cell lysate and enriched peroxisome fraction.
We then investigated the enriched peroxisome proteomics for exploring the peroxisome function and methanol metabolism of P. pastoris. We found that a group of up-regulated proteins were enriched (Fig. 4C), suggesting that these proteins were related to peroxisomes. In particular, transaldolase 2 (Tal2), sedoheptulose 1,7-bisphosphatase (Shb17) and Fba2 were had much higher up-regulation after peroxisome enrichment in both log phase and stationary phase cells. All these proteins contain a type 1 peroxisome targeting signal, which suggested that these enzymes were localized in peroxisome and played an important role in methanol utilization. Among these proteins, Tal2 was largely enriched (24.2- or 47.1-fold change) in the peroxisome enriched fraction (Fig. 4C), which suggested that Tal2 participated the Xu-5-P cycle for methanol assimilation.
3.4. TAL2 deletion
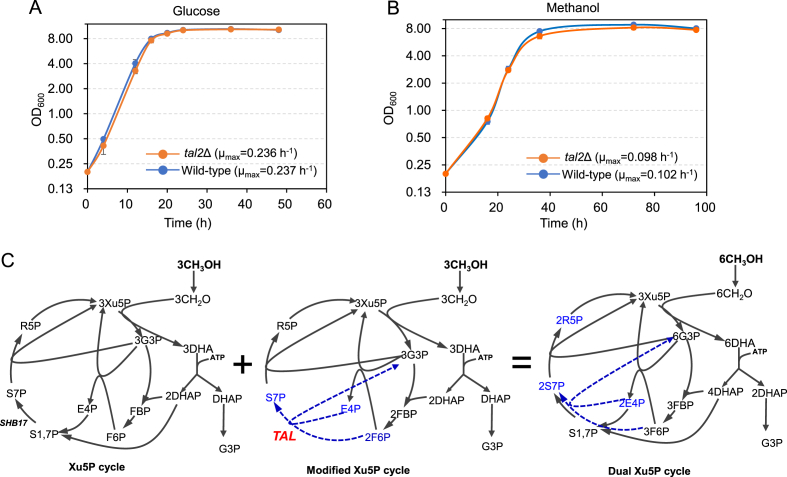
Since Tal2 was significantly up-regulated in methanol medium and this up-regulation was significantly strengthened in enriched peroxisomes, we thus tried to verify its function in methanol metabolism. Deletion of TAL2 had marginal effect on cell growth when cultured in glucose medium (Fig. 5A), while slightly retarded cell growth when cultured in methanol medium (Fig. 5B). These results suggested that TAL2 was beneficial for methanol metabolism by participating in a modified Xu-5-P cycle and the main Xu-5-P cycle was sufficient to support efficient methanol utilization in P. pastoris (Fig. 5C). This proposed dual Xu-5-P cycle might enabled efficient methanol utilization of P. pastoris, which could be used a robust cell chassis for methanol biotransformation.
Fig. 5.
TAL2 deletion retarded cell growth in methanol medium. Cell growth of wild-type strain and tal2Δ strain in glucose (A) or methanol medium (B). The cells were cultivated in Delft minimal medium containing 10 g/L glucose or methanol at 30 °C, 220 rpm. (C) The proposed Xu-5-P cycle for methanol utilization.
4. Discussion
We here conducted a comparative proteomic analysis of methyltrophic yeast P. pastoris under glucose and methanol minimal medium by using the whole cells and peroxisome enriched systems. As observed previously [11], methanol assimilation-related enzymes that localized in peroxisome were highly regulated in methanol medium compared with that in glucose medium. The TCA cycle was relatively stable in both media, suggesting that the energy generation was equivalent for cell growth in those conditions. The enzymes of PPP cycle were slightly repressed except Rpe, which was supposed to be helpful in providing Xu-5-P for methanol assimilation. We would like to mention that the different sample taking time might have slight variation of protein expression profile in batch culture, alternatively, chemo-state culture should be more stable for proteomic analysis.
Tal2, an enzyme supposed to be localized in peroxisome and involved in methanol assimilation, was up-regulated in methanol medium. Disruption of TAL2 slightly retarded the cell growth in methanol medium, suggesting that Tal2 was helpful for methanol utilization. We suppose that the Tal2 catalyzes a peroxisomal transaldolation that involved in an alternate Xu-5-P cycle, which could complement the main Xu-5-P cycle (Fig. 5). With the development of genetic engineering tools [13], clearer genetic function could be characterized in methyltrophic yeasts.
In summary, we investigated the global protein expression file in P. pastoris by using a comparative proteomic analysis, which should provide many insights for engineering this methyltrophic yeast for production of chemicals from methanol.
Availability of data and material
Data that supports the finding of this study are available in the main text and the supplementary materials.
Ethical approval
This article does not contain studies with human participants or animals performed by any of the authors.
Consent for publication
All listed authors have approved the manuscript before submission, including the names and order of authors.
CRediT authorship contribution statement
Rui Hou: Methodology, Investigation, Writing – original draft. Linhui Gao: Methodology, Resources. Jianhui Liu: Methodology. Zhen Liang: Methodology, Resources. Yongjin J. Zhou: Conceptualization, Writing – review & editing, Supervision, Project administration, Funding acquisition. Lihua Zhang: Conceptualization, Writing – review & editing, Supervision, Project administration, Funding acquisition. Yukui zhang: Conceptualization, Supervision.
Declaration of competing interest
The authors declare that there is no conflict of interests.
Acknowledgement
The work was financially supported by National Natural Science Foundation of China (22161142008, M − 0246), DMTO research grant (grant no. DICP DMTO201701) from Dalian institute of Chemical Physics, CAS.
Footnotes
Peer review under responsibility of KeAi Communications Co., Ltd.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.synbio.2022.04.005.
Contributor Information
Yongjin J. Zhou, Email: zhouyongjin@dicp.ac.cn.
Lihua Zhang, Email: lihuazhang@dicp.ac.cn.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Brady J.R., Whittaker C.A., Tan M.C., II Kristensen D.L., Ma D., Dalvie N.C., et al. Comparative genome-scale analysis of Pichia pastoris variants informs selection of an optimal base strain. Biotechnol Bioeng. 2020;117:543–555. doi: 10.1002/bit.27209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gao J., Zhou Y.J. Advances in methanol bio-transformation. Syn. Biol. J. 2020;1:158–173. [Google Scholar]
- 3.Duan X., Gao J., Zhou Y.J. Advances in engineering methylotrophic yeast for biosynthesis of valuable chemicals from methanol. Chin Chem Lett. 2018;29:681–686. [Google Scholar]
- 4.Zhou Y.J., Kerkhoven E.J., Nielsen J. Barriers and opportunities in bio-based production of hydrocarbons. Nat Energy. 2018;3:925–935. [Google Scholar]
- 5.Guo F., Dai Z., Peng W., Zhang S., Zhou J., Ma J., et al. Metabolic engineering of Pichia pastoris for malic acid production from methanol. Biotechnol Bioeng. 2021;118:357–371. doi: 10.1002/bit.27575. [DOI] [PubMed] [Google Scholar]
- 6.Miao T., Li Y., Zhu T. Metabolic engineering of methylotrophic Pichia pastoris for the production of β-alanine. Bioresour Bioprocess. 2021;8:89. doi: 10.1186/s40643-021-00444-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.van der Klei I.J., Yurimoto H., Sakai Y., Veenhuis M. The significance of peroxisomes in methanol metabolism in methylotrophic yeast. Biochim Biophys Acta. 2006;1763:1453–1462. doi: 10.1016/j.bbamcr.2006.07.016. [DOI] [PubMed] [Google Scholar]
- 8.Yurimoto H., Sakai Y. Methanol-inducible gene expression and heterologous protein production in the methylotrophic yeast Candida boidinii. Biotechnol Appl Biochem. 2009;53:85–92. doi: 10.1042/BA20090030. [DOI] [PubMed] [Google Scholar]
- 9.Purdue P.E., Lazarow P.B. Peroxisome biogenesis. Annu Rev Cell Dev Biol. 2001;17:701–752. doi: 10.1146/annurev.cellbio.17.1.701. [DOI] [PubMed] [Google Scholar]
- 10.Sakai Y., Oku M., van der Klei I.J., Kiel J.A. Pexophagy: autophagic degradation of peroxisomes. Biochim Biophys Acta. 2006;1763:1767–1775. doi: 10.1016/j.bbamcr.2006.08.023. [DOI] [PubMed] [Google Scholar]
- 11.Russmayer H., Buchetics M., Gruber C., Valli M., Grillitsch K., Modarres G., et al. Systems-level organization of yeast methylotrophic lifestyle. BMC Biol. 2015;13:80. doi: 10.1186/s12915-015-0186-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.van der Klei I.J., Yurimoto H., Sakai Y., Veenhuis M. The significance of peroxisomes in methanol metabolism in methylotrophic yeast. Bba-Mol Cell Res. 2006;1763:1453–1462. doi: 10.1016/j.bbamcr.2006.07.016. [DOI] [PubMed] [Google Scholar]
- 13.Cai P., Duan X., Wu X., Gao L., Ye M., Zhou Y.J. Recombination machinery engineering facilitates metabolic engineering of the industrial yeast Pichia pastoris. Nucleic Acids Res. 2021;49:7791–7805. doi: 10.1093/nar/gkab535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhou Y.J., Buijs N.A., Zhu Z., Qin J., Siewers V., Nielsen J. Production of fatty acid-derived oleochemicals and biofuels by synthetic yeast cell factories. Nat Commun. 2016;7:11709. doi: 10.1038/ncomms11709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhou Y.J., Buijs N.A., Zhu Z., Gomez D.O., Boonsombuti A., Siewers V., et al. Harnessing yeast peroxisomes for biosynthesis of fatty-acid-derived biofuels and chemicals with relieved side-pathway competition. J Am Chem Soc. 2016;138:15368–15377. doi: 10.1021/jacs.6b07394. [DOI] [PubMed] [Google Scholar]
- 16.Cramer J., Effelsberg D., Girzalsky W., Erdmann R. Small-Scale purification of peroxisomes for analytical applications. Cold Spring Harb Protoc. 2015;(9):838–845. doi: 10.1101/pdb.prot083717. [DOI] [PubMed] [Google Scholar]
- 17.Fang F., Zhao Q., Chu H., Liu M., Zhao B., Liang Z., et al. Molecular dynamics simulation-assisted ionic liquid screening for deep coverage proteome analysis. Mol Cell Proteomics. 2020;19:1724–1737. doi: 10.1074/mcp.TIR119.001827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cox J., Hein M.Y., Luber C.A., Paron I., Nagaraj N., Mann M. Accurate proteome-wide label-free quantification by delayed normalization and maximal peptide ratio extraction, termed MaxLFQ. Mol Cell Proteomics. 2014;13:2513–2526. doi: 10.1074/mcp.M113.031591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tyanova S., Temu T., Cox J. The MaxQuant computational platform for mass spectrometry-based shotgun proteomics. Nat Protoc. 2016;11:2301–2319. doi: 10.1038/nprot.2016.136. [DOI] [PubMed] [Google Scholar]
- 20.Bateman A., Martin M.J., Orchard S., Magrane M., Alpi E., Bely B., et al. UniProt: a worldwide hub of protein knowledge. Nucleic Acids Res. 2019;47:D506–D515. doi: 10.1093/nar/gky1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tyanova S., Temu T., Sinitcyn P., Carlson A., Hein M.Y., Geiger T., et al. The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat Methods. 2016;13:731–740. doi: 10.1038/nmeth.3901. [DOI] [PubMed] [Google Scholar]
- 22.Jafari M., Ansari-Pour N. Why, when and how to adjust your P values? Cell J. 2019;20:604–607. doi: 10.22074/cellj.2019.5992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liang S., Wang B., Pan L., Ye Y., He M., Han S., et al. Comprehensive structural annotation of Pichia pastoris transcriptome and the response to various carbon sources using deep paired-end RNA sequencing. BMC Genom. 2012;13:738. doi: 10.1186/1471-2164-13-738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Baumann K., Carnicer M., Dragosits M., Graf A.B., Stadlmann J., Jouhten P., et al. A multi-level study of recombinant Pichia pastoris in different oxygen conditions. BMC Syst Biol. 2010;4:141. doi: 10.1186/1752-0509-4-141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Love K.R., Shah K.A., Whittaker C.A., Wu J., Bartlett M.C., Ma D., et al. Comparative genomics and transcriptomics of Pichia pastoris. BMC Genom. 2016;17:550. doi: 10.1186/s12864-016-2876-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.van Zutphen T., Baerends R.J., Susanna K.A., de Jong A., Kuipers O.P., Veenhuis M., et al. Adaptation of Hansenula polymorpha to methanol: a transcriptome analysis. BMC Genom. 2010;11:1. doi: 10.1186/1471-2164-11-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Austin R.J., Kuestner R.E., Chang D.K., Madden K.R., Martin D.B. SILAC compatible strain of Pichia pastoris for expression of isotopically labeled protein standards and quantitative proteomics. J Proteome Res. 2011;10:5251–5259. doi: 10.1021/pr200551e. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data that supports the finding of this study are available in the main text and the supplementary materials.