Abstract
The class 1 integron In28, found in the multidrug resistance transposon Tn1403, was found to be located in the res site of the backbone transposon and is flanked by a 5-bp direct duplication, indicating that it reached this position by transposition. In28 has a backbone structure related to that of In4, but has lost internal sequences, including the sul1 gene, due to an IS6100-mediated deletion. In28 also lacks the partial copy of IS6100 found in In4 and contains different gene cassettes, blaP1, cmlA1, and aadA1. In1, the class 1 integron found in the multidrug resistance plasmid R46, is also located in a putative res site and belongs to the In4 group. In1 has a shorter internal deletion than In28 and has also lost one end. Additional integrons with structures related to In4 were also found in databases, and most of them had also lost either one end or internal regions or both. Tn610 belongs to this group.
The integrons found in clinical isolates of gram-negative bacteria generally contain one or more integrated cassettes that each include an antibiotic resistance gene, and a large number of gene cassettes containing different resistance genes have been identified (15, 17, 18, 42). Though integrons belonging to three classes have been found in clinical and environmental strains of multidrug-resistant bacteria (15, 42, 48), class 1 integrons predominate. Despite enormous variation in both the number and the identity of the cassettes associated with any individual integron isolate, integrons belonging to integron class 1 have identical or nearly identical integrase genes (intI1). Furthermore, class 1 integrons are present in many of the earliest multidrug-resistant plasmids isolated. The plasmids NR1 (R100), which is the source of Tn21, and pSa were both isolated in Japan, NR1 in the late 1950s (35, 55) and pSa prior to 1968 (56). Both are now known to contain a class 1 integron (9, 29, 48–50). R46, R751, R1033, and R388, plasmids from resistant strains isolated in Europe during the 1960s and early 1970s, are also known to contain class 1 integrons (9, 20, 36, 41, 48, 50). Indeed, it is possible that that class 1 integrons may have already been present or even widely distributed in bacteria prior to the introduction of antibiotics as human therapeutics (1, 54) and subsequently moved into human pathogens. The variety of backbone structures present in class 1 integrons (8, 16, 36, 41) also suggests an evolutionary history for these elements that is far longer than the 50 years of extensive antibiotic use.
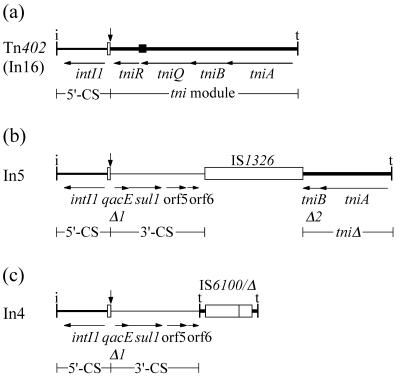
In addition to the intI1 gene, encoding the site-specific recombinase (32) that is responsible for cassette insertion (11), class 1 integrons also include the attI1 site (37, 43) into which the cassettes are incorporated and a promoter, Pc, that directs transcription of the cassette-encoded genes (12). These three features are found in a module of 1.36 kb known as the 5′-conserved segment (5′-CS) (20, 48). The 5′-CS is bounded at the inner end by attI1 and at the outer end by IRi, which is a 25-bp sequence that is found as an inverted repeat, IRt, at the other end of class 1 integrons (8, 20, 36, 41). The 5′-CS or intI1 module is found in all class 1 integrons and is generally followed by a variable region that consists of an array of one or more gene cassettes. To the right of the last cassette, or adjoining the 5′-CS if no cassettes are present, two different sequences have been found (Fig. 1). Most of the class 1 integrons studied to date contain at least part of a region known as the 3′-conserved segment (3′-CS) (8, 16, 48). The longest 3′-CS segment (2,384 bp) is found in In5 (8). It includes the sul1 gene, conferring resistance to sulfonamides (48, 50); the qacEΔ1 gene, conferring marginal resistance to quaternary ammonium compounds such as antiseptics and disinfectants (38); and two open reading frames (ORFs) of unknown function, orf5 and orf6 (36, 48). However, sometimes the 3′-CS is not present. The transposon Tn402 is a class 1 integron that does not include the 3′-CS (41), but is likely to represent the ancestor of integrons that do. Tn402, which is found in plasmid R751, is an active transposon (23, 47) that includes a set of three genes tniA, -B, and -Q, that are required for transposition (24, 41). A fourth gene, tniR, encodes a resolvase that is presumed to be involved in the resolution of the cointegrates that are formed as transpositional intermediates (24).
FIG. 1.
Structure of the backbones of class 1 integrons. Each integron is bounded by 25-bp inverted repeats, designated IRi and IRt (vertical bars labeled i and t), and includes the 5′-CS (medium line), which begins with IRi, contains the intI1 gene, and ends at the vertical arrow in the attI1 site (narrow open box). This arrow marks the position of integrated cassettes, which are not shown. Parts of the 3′-CS (narrow line) and the tni module (thick line) are found to the right of the vertical arrow. Insertion sequences are shown as open boxes. (a) Tn402 (also called In16 or Tn5090) includes the complete tni module that contains a full set of transposition genes (tniA, -B, and -Q), a resolvase gene (tniR), and a res site (solid box). Tn402 contains an array of three cassettes, dfrB3, orfD, and qacE. (b) In5 includes the 3′-CS, containing qacEΔ1, a truncated version of the qacE cassette, the sul1 sulfonamide resistance determinant, orf5 and orf6, and part of the tni module. In5 has a single cassette, aacA(IIa). (c) In4 includes two copies of very short regions of the IRt end of the tni module separated by a complete copy of IS6100 and a partial copy consisting of the last 321 bp of IS6100 (Δ). The cassette array is aacC1-orfE-aadA2-cmlA1.
The class 1 integrons that contain the 3′-CS appear to have arisen from an ancestor with a Tn402-type backbone by incorporation of the genes (qacEΔ1, sul1, orf5, and orf6) that make up the 3′-CS. Indeed, the complete qacE gene is part of a gene cassette (42). This structure has become an integral part of the types of class 1 integrons that are now widely distributed among plasmids, among bacterial species and genera, and geographically. However, the class 1 integrons that include 3′-CS sequences have undergone further rearrangements. All of those studied to date have lost part or all of the tni module and are now transposon derivatives that are defective in self-transposition. Most have also lost part of the 2,384-bp region defined as belonging to the 3′-CS. One group, here called In5-type class 1 integrons (Fig. 1b), were originally identified as containing IS1326 and have lost parts of the 3′-CS and of the tni module, presumably due to IS1326-mediated deletion of DNA adjacent to the insertion sequence (IS) (8). Further members of this group resemble In5 or In0 but have lost IS1326 (27, 41). A second backbone structure (Fig. 1c) has recently been found in integron In4, which constitutes the central region of transposon Tn1696 (GenBank accession no. U12338) (36). In4 contains one complete copy of the insertion sequence IS6100 and an adjacent partial copy in the same orientation. The IS6100 region is flanked by short segments from the outer right-hand end (IRt end) of Tn402 and other class 1 integrons, and these segments are in inverse orientation. The outer and inner copies correspond to the last 152 and 123 bp of Tn402, respectively.
Here, we have examined additional class 1 integrons, In28 and In1. In28 is found within the transposon Tn1403 (28, 53), which was recovered from a clinical strain of Pseudomonas aeruginosa which was isolated in the United States in 1973–1974 (26) and contains the IncP-2 plasmid RPL11. RPL11 transfers resistance to gentamicin, carbenicillin, streptomycin, tetracycline, sulfonamides, chloramphenicol, and mercury (26), while Tn1403 confers resistance to only carbenicillin, streptomycin, spectinomycin, and chloramphenicol (28, 53). In1 is found in the multidrug resistance plasmid R46, originally from a Salmonella enterica var. Typhimurium clinical strain isolated in the United Kingdom prior to 1966 (33). In28 and regions adjacent to both In28 and In1 were sequenced, and both of these integrons were found to be members of the In4 group. The relationships of these integrons to others found in the databases are also reported.
MATERIALS AND METHODS
Bacterial strains and plasmids
Escherichia coli JM109 [(Δlac-proAB) supE thi F′ (traD36 proAB+ lacIq lacZ ΔM15)] was used to propagate plasmid DNA. Plasmids used in this work are shown in Table 1. Fragments from R388::Tn1403 were cloned into either pUC19 (58) or pACYC184 (10) by standard procedures (45). Plasmids containing the appropriate fragments were identified by screening for antibiotic resistance, by restriction mapping, and by sequencing the fragment ends using a universal primer. Subclones were also derived from these primary clones. pRMH519 was constructed by SacI digestion and religation of pRMH518. pRMH527 was constructed by digesting pRMH519 with SalI and religating. Bacteria were routinely cultured at 37°C in Luria-Bertani (LB) medium or on LB agar supplemented as appropriate with ampicillin (100 μg ml−1), chloramphenicol (25 μg ml−1), streptomycin (25 μg ml−1), or sulfamethoxazole (25 μg ml−1). Antibiotics were obtained from Sigma.
TABLE 1.
Plasmids
| Plasmid | Description | Relevant phenotypea | Reference |
|---|---|---|---|
| R388::Tn1403 | Apr Cbr Smr Spr Sur Kmr Tcr Tra+ | G. A. Jacoby | |
| pRMH105 | 10.0-kb SalI-EcoRV fragment of pKM101 (R46) | Apr | 19 |
| pRMH518 | 6.4-kb BamHI-BglII fragment of R388::Tn1403 in pUC19 | Apr | This work |
| pRMH519 | 5.5-kb SacI-BglII fragment of pRMH518 in pUC19 | Apr Smr Spr | This work |
| pRMH527 | 1.5-kb SacI-SalI fragment of pRMH519 in pUC19 | Apr Smr Spr | This work |
| pRMH534 | 10.0-kb HindIII fragment of R388::Tn1403 in pUC19 | Apr Smr | This work |
| pRMH547 | 3.0-kb BamHI fragment of R388::Tn1403 in pACYC184 | Cmr | This work |
Ap, ampicillin; Cb, carbenicillin; Cm, chloramphenicol; Km, kanamycin; Sm, streptomycin; Sp, spectinomycin; Su, sulfonamide; Tc, tetracycline.
DNA isolation and restriction mapping.
Plasmid DNA for restriction analysis and cloning was isolated using an alkaline lysis method (3). Restriction enzymes were used in accordance with the manufacturers' instructions. Fragments were separated by electrophoresis on 1% (wt/vol) agarose gels and visualized by staining with ethidium bromide. An EcoRI digest of bacteriophage SPP1 (Geneworks) was used as size markers. Plasmid DNA for sequencing was purified using the Magic Minipreps DNA purification system (Promega) or Wizard maxiprep kit (Promega).
DNA sequencing and analysis.
The DNA sequence of at least one strand of fragments of Tn1403 cloned in plasmid vectors was determined. Manual DNA sequencing was performed with a Sequenase 2.0 system (51) as recommended by the manufacturer (U.S. Biochemicals) using dITP reaction mixtures followed by a 30-min incubation with a 1 mM deoxynucleoside triphosphate mix and terminal deoxynucleotidyl transferase (Boehringer-Mannheim). Automated sequencing was performed by the Sydney University/Royal Prince Alfred Hospital, Sydney, or by the sequencing facility at the Department of Biological Sciences, Macquarie University, Sydney, on an ABI-Prism 377 sequencer using the Big Dye system. The additional R46 sequence was determined as described by Hall and Vockler (20) using M13 clones isolated in the course of that original study. DNA sequences were assembled using MacVector 6.5 and AssemblyLIGN (Oxford Molecular). GenBank searches were performed using the BlastN and FastA programs available through WebANGIS (Australian National Genomic Information Service). Programs in the GCG Wisconsin package, version 8.1.0, were used via WebANGIS GCG to align and analyze DNA sequences.
Nucleotide sequence accession numbers.
The In28 nucleotide sequence data reported here have been assigned GenBank accession no. AF313472. The additional sequence of R46 reported here has been added as GenBank accession no. M95287.
RESULTS
Structure of In28.
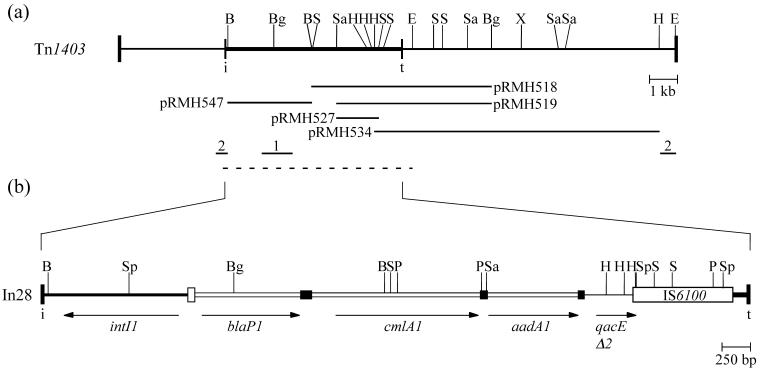
Tn1403 is a transposon isolated from a clinical strain of P. aeruginosa (28, 53) and confers resistance to carbenicillin, streptomycin, spectinomycin, and chloramphenicol, but not to sulfonamides. The most recent published map indicates the presence of blaP1 (PSE-1), cat, aadA, and aphC antibiotic resistance genes (53). An intI1 gene was also shown, and the IRi end of the 5′-CS was sequenced, but the restriction sites characteristic of the 3′-CS are not present, consistent with the absence of sulfonamide resistance. Here restriction fragments that include most of Tn1403 were isolated, and a map of Tn1403 was constructed (Fig. 2). This map differs from the published map, as some restriction sites have been added, removed, or relocated. The sequences of In28, the class 1 integron found in Tn1403, and of the regions adjacent to it were determined (GenBank accession no. AF313472), and the structure of In28 is shown in Fig. 2.
FIG. 2.
Map of Tn1403 and In28. (a) Tn1403. The region shown represents Tn1403, with vertical bars representing the inverted repeats, and In28 is shown as a thicker line ending in bars labeled i and t. The extents of the fragments cloned in various plasmids are shown below. Previously sequenced sections are represented by lines: 1, Huovinen and Jacoby (22); 2, Vézina and Levesque (53); and the extent of the sequence obtained here is indicated by a dotted line. (b) Expanded map of In28. Each cassette is indicated by an open box and adjacent filled box that represents the 59-base element (59-be). Other features are as in Fig. 1. Restriction enzyme sites: B, BamHI; Bg, BglII; E, EcoRI; H, HindIII; P, PstI; S, SalI; Sa, SacI; Sp, SphI; X, XhoI.
The backbone structure is related to that of In4 (36), though the cassettes are different. In28 includes the 1.36-kb 5′-CS that contains the weak variant of the Pc promoter (TGGACA-17 bp-TAAGCT) rather than the strong (TTGACA-17 bp-TAAACT) version found in In4, and does not include the 19-bp duplication in attI1. The 5′-CS is followed by an array of three gene cassettes, blaP1, cmlA1, and aadA1. The presence of the cmlA1 cassette indicates that chloramphenicol resistance is in fact conferred by an efflux mechanism rather than by a chloramphenicol acetyltransferase (Cat). The blaP1 cassette is identical to the previously reported blaP1 (PSE-1) cassette (GenBank accession nos. Z18955 and M69058) (22) and confers resistance to carbenicillin and ampicillin. The cmlA1 cassette sequence differs from the prototype in In4 (GenBank accession no. U12338) at four positions (three amino acid changes). The aadA1 cassette confers resistance to streptomycin and spectinomycin and differs at two positions (G732C and C759T) from the prototype aadA1a sequence (GenBank accession no. X12870), but these differences do not result in any amino acid changes.
To the right of the cassettes, the first 433 bp of the 3′-CS are also present and are adjoined by IS6100. Thus, compared to In4, both the internal 123-bp copy of the IRt end and the region of the 3′-CS that contains orf5, orf6, and the sul1 gene are missing. Beyond IS6100 lies the same short segment of 152 bp from the IRt end of the tni module that is present in In4. In28 does not include the partial copy of IS6100 found in In4. Because this direct duplication is readily lost from cloned fragments (36), its absence was confirmed by DNA-DNA hybridization using digests of R388::Tn1403. The 321-bp PstI fragment created by the presence of the duplication was not detected (data not shown).
The sequence of Tn1403 flanking In28 revealed the presence of a 5-bp duplication (boxed in Fig. 3), indicating that In28 reached its current location by transposition. In28 was located upstream of the tnpR gene in the resI region of the res site of the transposon that forms the backbone of Tn1403 (Fig. 3). The sequence to the left of In28 is identical to that of a short region of Tn1403 published previously (GenBank accession no. M59035) (53), and the regions to the left and right are identical to the corresponding regions of a transposon from a multidrug-resistant Pseudomonas strain isolated from an apple orchard (46). This transposon also contains a class 1 integron in the same position.
FIG. 3.
Sequence surrounding In28. The Tn1403 sequences flanking In28 have been joined together (bases 195 to 73 of GenBank accession no. M59035 plus bases 6357 to 6452 of AF313472) to reconstitute the res site, and the 5 bp duplicated by insertion of In28 are boxed. This sequence is aligned with the res regions of Tn501 (bases 4750 to 4532 in Z00027), Tn1721 (bases 359 to 160 in K01724), and Tn1696 (bases 3562 to 3669 and 12005 to 12116 in U12338) and the extents of the three components of the res site, determined experimentally for Tn1721 by Hall and Halford (21) and Rogowsky et al. (44), are indicated. The 5-bp duplication in Tn1696 is also boxed.
Structure of In1.
We have previously reported that a segment of IS6100 abuts the right-hand end of the region of the 3′-CS present in In1 (16), which is found in the IncN plasmid R46. In In1, only the first 2,197 bp of the 3′-CS are present, as opposed to 2,239 bp in In4, and the internal copy of 123 bp from the IRt end of the tni module is not found. This suggests that a deletion extending from the left-hand boundary of IS6100 to base 2198 of the 3′-CS has occurred. The PstI restriction site expected for the right-hand portion of IS6100 is not present in the map of R46 (7), and IS6100 appears to be interrupted by an IS26 (IS46) element. The sequence of this region of R46 has recently been completed (R. Woodgate, personal communication; GenBank accession no. AF117344). Only the first 298 bp of IS6100 are present, and they are followed by a complete IS26 (Fig. 4b). The remainder of IS6100 and the 152-bp IRt end appear to have been lost, as they are not found elsewhere in R46 (R. Woodgate, personal communication).
FIG. 4.
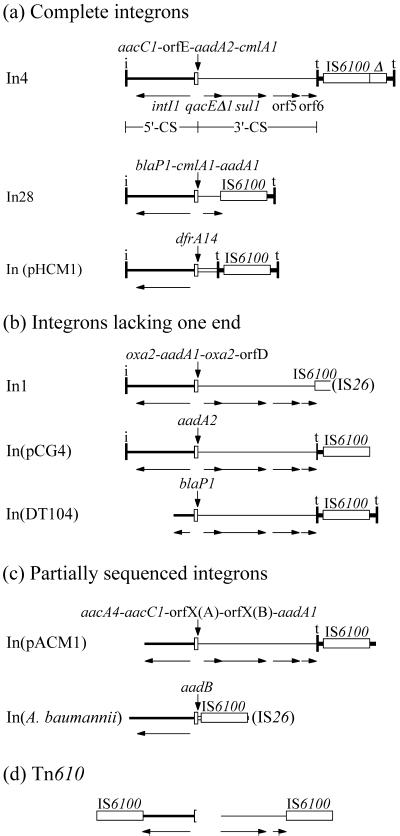
Backbone structures of In4-like class 1 integrons and of Tn610. The structures shown are (a) integrons for which the complete sequence is available and both terminal IRs are present; (b) completely sequenced integrons that lack one of the terminal IRs; (c) integrons for which the sequence does not extend far enough to determine if one or both terminal IRs are present; and (d) Tn610. Features shown are as in Fig. 1, and (IS26) indicates that IS26 is found adjacent to the boundary shown. The sources for the sequences used are: In4, GenBank accession no. U12338; In28, AF313472; pHCM1, http://www.sanger.ac.uk/Projects/S_typhi/; DT104, AF071555 and AF261826; In1, M95287 and AF117344; pCG4, AF164956; pACM1, U90945 and AF107205 plus standard 3′-CS (predicted to be present from available restriction maps); A. baumannii In, AJ289190; Tn610, X53653.
The sequence adjacent to the IRi end of R46 was extended and added to that in GenBank accession no. M95287. It includes a gene, designated resP, encoding a resolvase-like protein. A variant of this resolvase has previously been identified in a derivative of plasmid TP120, which is a relative of R46 (14). In1 is located upstream of resP in the region where the cognate res site would be expected to be found.
Structure of other class 1 integrons.
Searches of bacterial sequences present in the databases revealed several additional examples of class 1 integrons with backbone structures that contain IS6100 and are related to that of In4. Only one of these includes a complete structure extending from an IRi end to an IRt end in the correct orientation (Fig. 4a). It is found in plasmid pHCM1 from a multidrug-resistant Salmonella enterica serovar Typhi strain and is part of a transposon with a backbone structure closely related (greater than 99.9% identical) to that of Tn1696. The integron is in precisely the same position as In4 in Tn1696, with the same 5-bp duplication flanking it. In this integron, the internal 123-bp copy of the IRt end is present, but it abuts an unknown sequence, and none of the 3′-CS is present. A possible explanation for this configuration is that a deletion arising at IRt has extended into a previously unidentified cassette. The partial duplication of IS6100 is not present in this integron.
Two further examples resemble In1 in that one of the ends of the integron has been lost (Fig. 4b). One of these is the integron in pCG4 from Corynebacterium glutamicum (GenBank accession no. AF164956), which has lost the outer 152-bp copy of IRt, presumably due to a deletion arising at the right-hand end of IS6100. The backbone of the In in pCG4 is otherwise identical to the In4 backbone, except that it lacks the partial copy of IS6100.
The second, from the chromosomal multidrug resistance locus of S. enterica var. Typhimurium DT104, was compiled from GenBank accession nos. AF071555 and AF261826 (4, 5). It has lost the IRi end and the first 915 bp of the 5′-CS and the partial copy of IS6100, but is otherwise identical to In4.
In two further cases (Fig. 4c), the available sequence is incomplete. The sequenced region of plasmid pACM1 from Klebsiella oxytoca (40) (compiled from GenBank accession nos. U90945 and AF107205 and the standard 3′-CS sequence) does not extend to either outer end, but the configuration to the right of the cassette array differs from that in In4 only in that the partial copy of IS6100 (Δ in Fig. 4) is missing. In the second case, an integron found in Acinetobacter baumannii, the 3′-CS is missing and the left-hand end of IS6100 abuts the first 53 bp of the oxa3 cassette (39). The right-hand end has been truncated, apparently by insertion of IS26 or an IS26-mediated deletion, and only the first 14 bp of the 152-bp IRt end fragment remain. The available sequence does not extend to the IRi end, and it remains to be established if this is present.
A copy of IS6100 with the 123-bp segment of the tni module adjacent to it is also found in the transposon TnSF1 from Shigella flexneri (GenBank accession no. AF188331). TnSF1 appears to be derived from Tn21 and has a class 1 integron in the same position as In2 in Tn21. This integron seems to have acquired a large block of extra sequence, including the IS6100/IRt feature, resulting in a complex structure (not shown in Fig. 4).
DISCUSSION
We have previously shown (8) that one group of class 1 integrons that contain sequences derived from the 3′-CS have backbone structures related to that of In5. Here, further examples of class 1 integrons that may also contain the 3′-CS but have a different backbone structure related to that of In4 (36) have been identified. In4 has the longest portion of the 3′-CS found in members of this In4-like group and is the only one that includes a partial copy of IS6100 in addition to the complete copy. Though the origin of the complex arrangement of IS6100 and IRt ends found in In4 remains a matter for speculation, In4 appears to be ancestral to the other integrons that contain IS6100, and all other members of the group can be viewed as having derived from it. For example, the direct duplication of IS6100 sequences could have been lost by RecA-dependent homologous recombination. Several further differences are likely to be due to deletions initiated either at the right-hand end of the IS6100, leading to loss of the outer IRt end of the integron, or at the left-hand end of the IS, leading to internal deletions.
Deletions initiated at the left-hand end have led to loss of the internal 123-bp IRt fragment and different lengths of the 3′-CS in In1 and In28, and in the A. baumannii integron, the deletion extends into the cassette array. In the pHCM1 integron, the 3′-CS is also missing, but the deletion appears to have been initiated at the internal IRt end. Insertion of IS26 or IS26-mediated deletion has also played a part in shaping these integrons. Tn610 (Fig. 4d), a transposon isolated from Mycobacterium smegmatis that confers resistance to sulfonamides (31), is another relative. Tn610 is made up of two copies of IS6100 flanking a sequence that consists of bp 334 to 1333 of the 5′-CS joined to bp 454 to 1663 of the 3′-CS. It includes all but the first 4 bp of the sul1 gene. Tn610 may well have arisen from an In4-like integron, but in this case part of the intI1 gene has been lost (presumably due to insertion of the left-hand copy of IS6100) and an internal deletion has led to loss of part of attI1 and the beginning of the 3′-CS. As a consequence, Tn610 is no longer an active integron, as it is unable to incorporate gene cassettes.
Further examples of the In4-like family of class 1 integrons were identified from published restriction maps. For example, one appears to be present in the IncN plasmid N3 (6), and a small amount of available sequence confirms this. Part of IS6100 and the 152-bp IRt fragment are found where they would be expected, adjacent to the EcoRI restriction-modification genes in N3 (2). Another IncN plasmid, R15, includes the appropriate configuration of restriction sites within a transposon, Tn2353 (13), and Tn2353 could be a further relative of Tn1696. Both N3 and R15 are from Shigella strains isolated in Japan prior to 1964 (57).
Most of the In4-like class 1 integrons and integron remnants described here are located on plasmids. A few (In4, In28, and the integron from S. enterica serovar Typhi) are found within a transposon that is located on a plasmid. Only the DT104 integron is located within the bacterial chromosome of the original isolate, and this is presumably a result of the incorporation of extrachromosomal DNA (e.g., a plasmid) into the S. enterica DT104 chromosome (4). Members of this In4-like group have also been recovered from a wide variety of bacterial species. Most are from gram-negative bacteria, but pCG4 is from C. glutamicum and Tn610 was recovered from M. smegmatis. Granted that integrons of this group are found mostly on plasmids, it is perhaps not surprising that they are so broadly disseminated.
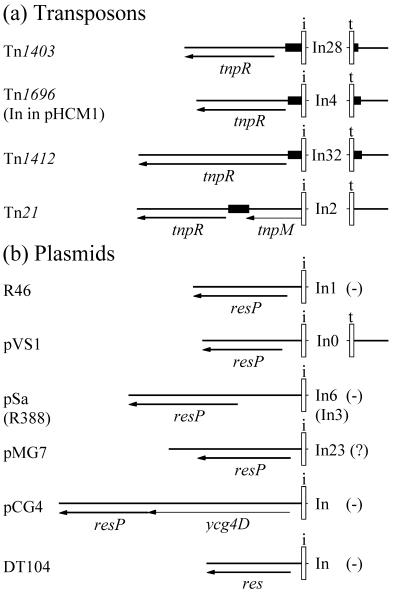
Though integrons belonging to both the In5-like and In4-like groups are unable to mobilize themselves, having lost some or all of the tni genes, they nonetheless appear to move readily. The sequences flanking them are diverse (8, 16), indicating past movement, and only two of the In4-like integrons described here, In4 and the pHCM1 integron, are located in the same DNA context. If the two outer ends (IRi and IRt) are intact and in the correct orientation, movement is presumably accomplished when tni genes are present in the same cell to complement the defect. Transposition of Tn402, and presumably also of the tni-deficient In5-like and In4-like integrons, resembles transposition of the closely related mercury resistance transposon Tn5053 (24, 25) in that it is constrained by stringent target site specificity, with the target being a res site recognized by a resolvase (23, 34). In28, like In4, is located within a res site recognized by the resolvase encoded by the backbone transposon (Fig. 3). This location presumably interferes with the normal transposition process for Tn1403 and Tn1696. Indeed, cointegrates formed on transposition of Tn1403 are not resolved (53). The IRi end of In1 is also adjacent to a plasmid gene, here called resP, encoding a resolvase-like protein and may be located within the cognate res site. A resolvase-like protein is encoded by a gene near the IRi end of the pCG4 integron (Fig. 5). Indeed, genes that encode resolvases are also found adjacent to the IRi end of most class 1 integrons (Fig. 5). Tn402 (not shown in Fig. 5) is an exception (52).
FIG. 5.
Locations of resolvase genes adjacent to class 1 integrons. Integrons (a) found in transposons and (b) found in plasmids. Open bars labeled i and t indicate the outer IRi and IRt ends of the integron. (−) indicates that the outer IRt is not present, and (?) indicates that the sequence of this region has not been determined. The genes encoding resolvases or resolvase-like proteins are shown by thick arrows and are labeled tnpR for transposon genes, resP for plasmid genes, and res for the DT104 gene, whose origin is not known. Other genes and open reading frames are shown by thin arrows. The res sites (solid boxes) are shown only where their locations and extents have been determined experimentally or can be deduced by comparison with an experimentally determined res site (21, 30, 44). The sources of the sequences are: Tn1403/In28, AF313472 and M59035; Tn1696/In4, U12338; Tn1412/In32, L36547; Tn21/In2, AF071413; R46/In1, M95287; pVS1/In0, U10456 and U49101; pSa/In6, U303471; pMG7, S78872; pCG4, AF164956; and DT104, AF071555 and AF261826.
It is clear that class 1 integrons with the In4-like backbone structure described here or with an In5-like backbone described previously (8) are widely distributed. Both types are found among integrons from early clinical isolates of multidrug-resistant strains, indicating that the differences may have arisen prior to the introduction of most antibiotics for extensive therapeutic use. However, sulfonamides were in use for over 10 years before the first use of penicillins, and selection for class 1 integrons containing the 3′-CS, and hence the sul1 gene, may have occurred in that time span. In4-like class 1 integrons are found in a variety of plasmids and transposons as well as in bacterial chromosomes and clearly change their location relatively easily, despite having dispensed with the tni genes. Their location becomes fixed only when one of the outer ends, IRi or IRt, is lost. They are also found in many different bacterial species, presumably as a result of their association with plasmids or transposons.
ACKNOWLEDGMENTS
We thank Cassandra Vockler for determining the R46 sequence.
S.R.P. was supported by a grant from the Australian National Health and Medical Research Council. S. enterica serovar Typhi sequence data were produced by the S. enterica serovar Typhi sequencing group at the Sanger Centre and can be obtained from http://www.sanger.ac.uk/Projects/S_typhi/.
REFERENCES
- 1.Akiba T, Koyama K, Ishiki Y. On the mechanism of the development of multiple-drug-resistant clones of Shigella. Jpn J Microbiol. 1960;4:219–227. doi: 10.1111/j.1348-0421.1960.tb00170.x. [DOI] [PubMed] [Google Scholar]
- 2.Bhagwat A S, Johnson B, Weule K, Roberts R J. Primary sequence of the EcoRII endonuclease and properties of its fusions with β-galactosidase. J Biol Chem. 1990;265:767–773. [PubMed] [Google Scholar]
- 3.Birnboim H C, Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979;7:1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boyd D A, Peters G A, Ng L-K, Mulvey M R. Partial characterization of a genomic island associated with the multidrug resistance region of Salmonella enterica Typhymurium DT104. FEMS Microbiol Lett. 2000;189:285–291. doi: 10.1111/j.1574-6968.2000.tb09245.x. [DOI] [PubMed] [Google Scholar]
- 5.Briggs C E, Fratamico P M. Molecular characterization of an antibiotic resistance gene cluster of Salmonella typhimurium DT104. Antimicrob Agents Chemother. 1999;43:846–849. doi: 10.1128/aac.43.4.846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brown A M C, Coupland G M, Willetts N S. Characterization of IS46, an insertion sequence found in two IncN plasmids. J Bacteriol. 1984;159:472–481. doi: 10.1128/jb.159.2.472-481.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brown A M C, Willetts N S. A physical and genetic map of the IncN plasmid R46. Plasmid. 1981;5:188–201. doi: 10.1016/0147-619x(81)90020-2. [DOI] [PubMed] [Google Scholar]
- 8.Brown H J, Stokes H W, Hall R M. The integrons In0, In2, and In5 are defective transposon derivatives. J Bacteriol. 1996;178:4429–4437. doi: 10.1128/jb.178.15.4429-4437.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cameron F H, Groot Obbink D J, Ackerman V A, Hall R M. Nucleotide sequence of the AAD(2") aminoglycoside adenyltransferase determinant aadB: evolutionary relationship of this region with those surrounding aadA in R538-1 and dhrfII in R388. Nucleic Acids Res. 1986;14:8625–8635. doi: 10.1093/nar/14.21.8625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chang A Y C, Cohen S N. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J Bacteriol. 1978;134:1141–1156. doi: 10.1128/jb.134.3.1141-1156.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Collis C M, Grammaticopoulos G, Briton J, Stokes H W, Hall R M. Site-specific insertion of gene cassettes into integrons. Mol Microbiol. 1993;9:41–52. doi: 10.1111/j.1365-2958.1993.tb01667.x. [DOI] [PubMed] [Google Scholar]
- 12.Collis C M, Hall R M. Expression of antibiotic resistance genes in the integrated cassettes of integrons. Antimicrob Agents Chemother. 1995;39:155–162. doi: 10.1128/aac.39.1.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dobritsa A P. IS8- and Tn2353-mediated cointegration of the plasmids R15 and RP4::Tn1. Mol Gen Genet. 1984;194:206–210. doi: 10.1007/BF00383518. [DOI] [PubMed] [Google Scholar]
- 14.Gigliani F, Sporeno E, Perri S, Battaglia P A. The uvp1 gene of plasmid pR cooperates with mucAB genes in the DNA repair process. Mol Gen Genet. 1989;218:18–24. doi: 10.1007/BF00330560. [DOI] [PubMed] [Google Scholar]
- 15.Hall R M, Brookes D E, Stokes H W. Site-specific insertion of genes into integrons: role of the 59-base element and determination of the recombination crossover point. Mol Microbiol. 1991;5:1941–1959. doi: 10.1111/j.1365-2958.1991.tb00817.x. [DOI] [PubMed] [Google Scholar]
- 16.Hall R M, Brown H J, Brookes D E, Stokes H W. Integrons found in different locations have identical 5′ ends but variable 3′ ends. J Bacteriol. 1994;176:6286–6294. doi: 10.1128/jb.176.20.6286-6294.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hall R M, Collis C M. Antibiotic resistance in gram negative bacteria: the role of gene cassettes and integrons. Drug Resist Updates. 1998;1:109–119. doi: 10.1016/s1368-7646(98)80026-5. [DOI] [PubMed] [Google Scholar]
- 18.Hall R M, Collis C M. Mobile gene cassettes and integrons: capture and spread of genes by site-specific recombination. Mol Microbiol. 1995;15:593–600. doi: 10.1111/j.1365-2958.1995.tb02368.x. [DOI] [PubMed] [Google Scholar]
- 19.Hall R M, Podger D M, Collis C M. An alteration leading to loss of ability to support phleomycin mutagenesis in the pKM101-derived plasmid pGW16 is located in or close to the mucAB genes. Mutat Res. 1985;146:47–53. doi: 10.1016/0167-8817(85)90054-9. [DOI] [PubMed] [Google Scholar]
- 20.Hall R M, Vockler C. The region of the IncN plasmid R46 coding for resistance to β-lactam antibiotics, streptomycin/spectinomycin and sulphonamides is closely related to antibiotic resistance segments found in IncW plasmids and in Tn21-like transposons. Nucleic Acids Res. 1987;15:7491–7501. doi: 10.1093/nar/15.18.7491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hall S C, Halford S E. Specificity of DNA recognition in the nucleoprotein complex for site-specific recombination by Tn21 resolvase. Nucleic Acids Res. 1993;21:5712–5719. doi: 10.1093/nar/21.24.5712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huovinen P, Jacoby G A. Sequence of the PSE-1 β-lactamase gene. Antimicrob Agents Chemother. 1991;35:2428–2430. doi: 10.1128/aac.35.11.2428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kamali-Moghaddam M, Sundström L. Transposon targeting determined by resolvase. FEMS Microbiol Lett. 2000;186:55–59. doi: 10.1111/j.1574-6968.2000.tb09081.x. [DOI] [PubMed] [Google Scholar]
- 24.Kholodii G Y, Mindlin S Z, Bass I A, Yurieva O V, Minakhina S V, Nikiforov V G. Four genes, two ends, and a res region are involved in transposition of Tn5053: a paradigm for a novel family of transposons carrying either a mer operon or an integron. Mol Microbiol. 1995;17:1189–1200. doi: 10.1111/j.1365-2958.1995.mmi_17061189.x. [DOI] [PubMed] [Google Scholar]
- 25.Kholodii G Y, Yurieva O V, Lomovskaya O L, Gorlenko Z M, Mindlin S Z, Nikiforov V G. Tn5053, a mercury resistance transposon with integron's ends. J Mol Biol. 1993;230:1103–1107. doi: 10.1006/jmbi.1993.1228. [DOI] [PubMed] [Google Scholar]
- 26.Korfhagen T R, Loper J C, Ferrel J A. Pseudomonas aeruginosa R factors for gentamicin plus carbenicillin resistance from patients with urinary tract colonizations. Antimicrob Agents Chemother. 1975;7:64–68. doi: 10.1128/aac.7.1.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Laraki N, Galleni M, Thamm I, Riccio M L, Amicosante G, Frère J-M, Rossolini G M. Structure of In31, a blaIMP-containing Pseudomonas aeruginosa integron phyletically related to In5, which carries an unusual array of gene cassettes. Antimicrob Agents Chemother. 1999;43:890–901. doi: 10.1128/aac.43.4.890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Levesque R C, Jacoby G A. Molecular structure and interrelationships of multiresistance β-lactamase transposons. Plasmid. 1988;19:21–29. doi: 10.1016/0147-619x(88)90059-5. [DOI] [PubMed] [Google Scholar]
- 29.Liebert C A, Hall R M, Summers A O. Transposon Tn21, flagship of the floating genome. Microbiol Mol Biol Rev. 1999;63:507–522. doi: 10.1128/mmbr.63.3.507-522.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu C C, Huhne R, Tu J, Lorbach E, Droge P. The resolvase encoded by Xanthomonas campestris transposable element ISXc5 constitutes a new subfamily closely related to DNA invertases. Genes Cells. 1998;3:221–233. doi: 10.1046/j.1365-2443.1998.00182.x. [DOI] [PubMed] [Google Scholar]
- 31.Martin C, Timm J, Rauzier J, Gomez-Lus R, Davies J, Gicquel B. Transposition of an antibiotic resistance element in mycobacteria. Nature. 1990;345:739–743. doi: 10.1038/345739a0. [DOI] [PubMed] [Google Scholar]
- 32.Martinez E, de la Cruz F. Genetic elements involved in Tn21 site-specific integration, a novel mechanism for the dissemination of antibiotic resistance genes. EMBO J. 1990;9:1275–1281. doi: 10.1002/j.1460-2075.1990.tb08236.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Meynell E, Datta N. The relation of resistance transfer factors to the F-factor (sex-factor) of Escherichia coli K12. Genet Res. 1966;7:134–140. doi: 10.1017/s0016672300009538. [DOI] [PubMed] [Google Scholar]
- 34.Minakhina S, Kholodii G, Mindlin S, Yurieva O, Nikiforov V. Tn5053 family transposons are res site hunters sensing plasmidal res sites occupied by cognate resolvases. Mol Microbiol. 1999;33:1059–1068. doi: 10.1046/j.1365-2958.1999.01548.x. [DOI] [PubMed] [Google Scholar]
- 35.Nakaya R, Nakamura A, Murata Y. Resistance transfer agents in Shigella. Biochem Biophys Res Commun. 1960;3:654–659. doi: 10.1016/0006-291x(60)90081-4. [DOI] [PubMed] [Google Scholar]
- 36.Partridge S R, Brown H J, Stokes H W, Hall R M. Transposons Tn1696 and Tn21 and their integrons In4 and In2 have independent origins. Antimicrob Agents Chemother. 2001;45:1263–1270. doi: 10.1128/AAC.45.4.1263-1270.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Partridge S R, Recchia G D, Scaramuzzi C, Collis C M, Stokes H W, Hall R M. Definition of the attI1 site of class 1 integrons. Microbiology. 2000;146:2855–2864. doi: 10.1099/00221287-146-11-2855. [DOI] [PubMed] [Google Scholar]
- 38.Paulsen I T, Littlejohn T G, Rådström P, Sundström L, Sköld O, Swedberg G, Skurray R A. The 3′ conserved segment of integrons contains a gene associated with multidrug resistance to antiseptics and disinfectants. Antimicrob Agents Chemother. 1993;37:761–768. doi: 10.1128/aac.37.4.761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ploy M-C, Denis F, Courvalin P, Lambert T. Molecular characterization of integrons in Acinetobacter baumannii: description of a hybrid class 2 integron. Antimicrob Agents Chemother. 2000;44:2684–2688. doi: 10.1128/aac.44.10.2684-2688.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Preston K, Radomski C C A, Venezia R A. The cassettes and 3′ conserved segment of an integron from Klebsiella oxytca plasmid pACM1. Plasmid. 1999;42:104–114. doi: 10.1006/plas.1999.1418. [DOI] [PubMed] [Google Scholar]
- 41.Rådström P, Sköld O, Swedberg G, Flensburg J, Roy P H, Sundström L. Transposon Tn5090 of plasmid R751, which carries an integron, is related to Tn7, Mu, and the retroelements. J Bacteriol. 1994;176:3257–3268. doi: 10.1128/jb.176.11.3257-3268.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Recchia G D, Hall R M. Gene cassettes: a new class of mobile element. Microbiology. 1995;141:3015–3027. doi: 10.1099/13500872-141-12-3015. [DOI] [PubMed] [Google Scholar]
- 43.Recchia G D, Stokes H W, Hall R M. Characterisation of specific and secondary recombination sites recognised by the integron DNA integrase. Nucleic Acids Res. 1994;22:2071–2078. doi: 10.1093/nar/22.11.2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rogowsky P, Halford S E, Schmitt R. Definition of three resolvase binding sites at the res loci of Tn21 and Tn1721. EMBO J. 1985;4:2135–2141. doi: 10.1002/j.1460-2075.1985.tb03904.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning, a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 46.Schnabel E L, Jones A L. Distribution of tetracycline resistance genes and transposons among phylloplane bacteria in Michigan apple orchards. Appl Environ Microbiol. 1999;65:4898–4907. doi: 10.1128/aem.65.11.4898-4907.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shapiro J A, Sporn P. Tn402: a new transposable element determining trimethoprim resistance that inserts in bacteriophage lambda. J Bacteriol. 1977;129:1632–1635. doi: 10.1128/jb.129.3.1632-1635.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stokes H W, Hall R M. A novel family of potentially mobile DNA elements encoding site-specific gene-integration functions: integrons. Mol Microbiol. 1989;3:1669–1683. doi: 10.1111/j.1365-2958.1989.tb00153.x. [DOI] [PubMed] [Google Scholar]
- 49.Stokes H W, Tomaras C, Parsons Y, Hall R M. The partial 3′-conserved segment duplications in the integrons In6 from pSa and In7 from pDGO100 have a common origin. Plasmid. 1993;30:39–50. doi: 10.1006/plas.1993.1032. [DOI] [PubMed] [Google Scholar]
- 50.Sundström L, Rådström P, Swedberg G, Sköld O. Site-specific recombination promotes linkage between trimethoprim- and sulfonamide resistance genes. Sequence characterization of dhfrV and sulI and a recombination active locus of Tn21. Mol Gen Genet. 1988;213:191–201. doi: 10.1007/BF00339581. [DOI] [PubMed] [Google Scholar]
- 51.Tabor S, Richardson C C. DNA sequence analysis with a modified bacteriophage T7 DNA polymerase. Proc Natl Acad Sci USA. 1987;84:4767–4771. doi: 10.1073/pnas.84.14.4767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Thorsted P B, Macartney D P, Akhtar P, Haines A S, Ali N, Davidson P, Stafford T, Pocklington M J, Pansegrau W, Wilkins B M, Lanka E, Thomas C M. Complete sequence of the IncPβ plasmid R751: implications for evolution and organisation of the IncP backbone. J Mol Biol. 1998;282:969–990. doi: 10.1006/jmbi.1998.2060. [DOI] [PubMed] [Google Scholar]
- 53.Vézina G, Levesque R C. Molecular characterization of the class II multiresistance transposable element Tn1403 from Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1991;35:313–321. doi: 10.1128/aac.35.2.313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Watanabe T. Infective heredity of multiple drug resistance in bacteria. Bacteriol Rev. 1963;27:87–115. doi: 10.1128/br.27.1.87-115.1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Watanabe T, Fukasawa T. “Resistance transfer factor” an episome in Enterobacteriaceae. Biochem Biophys Res Commun. 1960;3:660–665. [Google Scholar]
- 56.Watanabe T, Furuse C, Sakaizumi S. Transduction of various R factors by phage P1 in Escherichia coli and by phage P22 in Salmonella typhimurium. J Bacteriol. 1968;96:1791–1795. doi: 10.1128/jb.96.5.1791-1795.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Watanabe T, Nishida H, Ogata C, Arai T, Sato S. Episome-mediated transfer of drug resistance in Enterobacteriaceae. VII. Two types of naturally occurring R factors. J Bacteriol. 1964;88:716–726. doi: 10.1128/jb.88.3.716-726.1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequence of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]