Abstract
Primary cilia are cell surface, microtubule-based organelles that dynamically extend from cells to receive and process molecular and mechanical signaling cues. In the last decade, this organelle has gained increasing popularity due to its ability to act as a cellular antenna, receive molecular stimuli, and respond to the cell’s environment. A growing field of data suggests that various tissues utilize and interpret the loss of cilia in different ways. Thus, careful examination of the role of cilia on individual cell types and tissues is necessary. Neural crest cells (NCCs) are an excellent example of cells that survey their environment for developmental cues. In this review, we discuss how NCCs utilize primary cilia during their ontogenic development, paying special attention to the role primary cilia play in processing developmental signals required for NCC specification, migration, proliferation, and differentiation. We also discuss how the loss of functional cilia on cranial and trunk NCCs affects the development of various organ systems to which they contribute. A deeper understanding of ciliary function could contribute greatly to understanding the molecular mechanisms guiding NCC development and differentiation. Furthermore, superimposing the ciliary contribution on our current understanding of NCC development identifies new avenues for therapeutic intervention in neurocristopathies.
1. INTRODUCTION
Over 100 years ago, biologists observed a thin, singular eyelash-type extension on various cell types (Zimmermann, 1898). These scientists, without any of the molecular knowledge or tools we have today, hypothesized that this “cilium” had a sensory function within the cell (Zimmermann, 1898). Sadly, after this initial discovery the field of ciliary biology lay essentially dormant for the next 80 years. Now, in the past two decades, the field of ciliary biology has undergone a renaissance due to improved molecular and cellular tools and a better understanding of what these nearly ubiquitous organelles are doing during various cellular processes. This review will discuss what a cilium is, the process of ciliogenesis and ciliary function. We will highlight what is currently known regarding how neural crest cells (NCCs) utilize primary cilia during development and the disorders affecting the NCC derivatives that can arise when ciliary structure or function is disrupted.
2. THE PRIMARY CILIUM: DEFINING THE ORGANELLE
2.1. Structure equals function
A cilium is a microtubule-based cellular projection (Fig. 1). There are several types of cilia, which are typically classified by their microtubule arrangement, capacity for motility, and number per cell. Generally, and for the purposes of this review, a primary cilium is defined by three criteria, (1) a 9+0 microtubule arrangement, (2) lack of motility, and (3) cellular singularity. Establishing how these three criteria define the organelle is important to understand primary cilia function during various cellular processes, including NCC development. Furthermore, understanding how variations in these key characteristics affect ciliary function is also important for gaining a better understanding of how cilia, primary and otherwise, function within a cell.
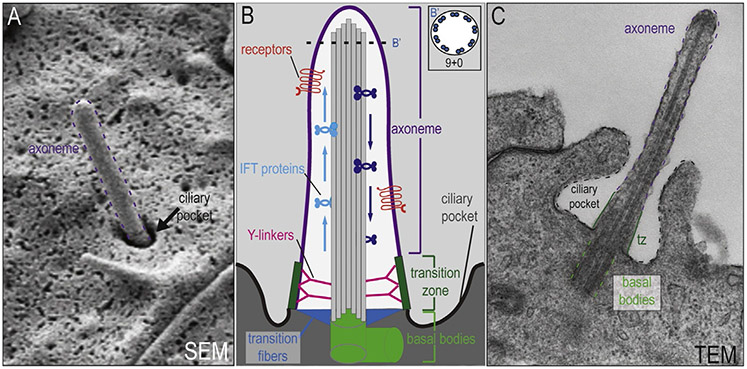
Figure 1.
Structure of the primary cilium. (A) Scanning electron microscopy (SEM) of the primary cilium. The axoneme is outlined in purple and the ciliary pocket is indicated by an arrow. (B) Schematic diagram of the primary cilium. The axoneme (purple) extends out from the basal bodies (light green) into the extracellular space (light gray). Receptors (red) for various signaling pathways localized to the membrane of the axoneme. (B′) Schematic cross-section of the axoneme reveals the microtubules in a 9+0 conformation. Intraflagellar transport proteins travel along microtubules, moving cargo up (light blue) and back down (dark blue) the axoneme. The more proximal portion of the primary cilium contains the transition fibers (blue), transition zone (dark green), and Y-linkers (pink). Collectively, these structures are important for ciliogenesis and act as a selective barrier between the intracellular space (dark gray) and the ciliary compartment (white). (C) Transmission electron microscopy (TEM) of the primary cilium. The axoneme (purple dotted lines), transition zone (tz;dark green lines), basal bodies (light green dotted lines), and ciliary pocket (black dotted lines) have all been highlighted in the image.
The 9+0 conformation of a primary cilium, as the name implies, refers to an arrangement of axonemal microtubules in an outer ring of 9 microtubule doublets with an absence of any inner microtubule pair (Fig. 1). Other types of cilia can have a 9+2 arrangement consisting of an outer ring of 9 microtubule doublets and a central core of 2 microtubules. Even 9+4 cilia have been reported in some species (Feistel & Blum, 2006).
The second feature common to primary cilia is their nonmotile status. Microtubule arrangement broadly correlates with cilium motility: 9+0 cilia are canonically not motile and 9+2 cilia are motile. While this is a general rule to go by, it is more accurate to use the presence or absence of dynein arms to determine motility of the cilium (AfZelius, 1976; Blum, Hayes, Whisnant, & Rosen, 1977). Dyneins are large motor protein complexes that generate force, causing the movement of eukaryotic cilia and flagella. The dynein protein complexes directly connect to the outer ring of microtubules (Neesen et al., 2001). Generally, 9+0 cilia and nonmotile 9+2 cilia lack dynein arms (Bloodgood, 2009).
Finally, primary cilia are reported to be solitary cellular extensions (i.e., each cell only extends one primary cilium). Similar to motility, cilia number has also been linked to microtubule arrangement. Monociliated cells are generally 9+0 and nonmotile, whereas multiciliated cells are generally 9+2 and motile. There are, however, several exceptions to this generality, thus it should not be considered a strict rule. Nodal cilia are perhaps the best example of an exception to this general classification. Current data suggest that the node may possess a mixture of both motile and nonmotile monocilia (McGrath, Somlo, Makova, Tian, & Brueckner, 2003) as well as a mixture of 9+2 and 9+0 monocilia (Caspary, Larkins, & Anderson, 2007).
2.2. Widespread and dynamic
The last 10 years have witnessed increased interest in the primary cilia (Beales & Jackson, 2012). The major causes of this resurgence are twofold; cilia are omnipresent and highly dynamic. First, as with most organelles, they are present in almost all cells rendering their function relevant to just about every tissue and organ system. However, there are a handful of cells and organisms that lack primary cilia. Yeast, fungi, and higher plants do not extend cilia. Of the “model organisms” classically used in biomedical research, the invertebrate C. elegans extend cilia only on sensory neurons, and none of the cilia in the nematode are motile. Similarly, Drosophila also extend cilia on sensory cells, but they are not utilized in the same way as mammalian cilia (Avidor-Reiss et al., 2004; Han, Kwok, & Kernan, 2003; Ray et al., 1999). Primary cilia are much better understood in vertebrates. Still, there are some mammalian cells that lack primary cilia including hepatocytes, acinar cells, and lymphocytes and granulocytes in the hematopoietic lineage (Wheatley, 1969; Wheatley, Wang, & Strugnell, 1996).
Second, unlike many organelles, primary cilia are highly dynamic in their extension, size, and specialization. Primary cilia are extended during times of quiescence (G0) and begin to retract during cell cycle re-entry. This cyclical extension and retraction suggests a mutually exclusive relationship between the cell cycle and ciliogenesis. Indeed, various studies have shown that cell cycle progression requires the continued suppression of primary cilia formation (Goto, Inoko, & Inagaki, 2013). Furthermore, links have been established between ciliary extension and cancer, with some cancers being devoid of primary cilia and some depending on ciliary signaling (Basten & Giles, 2013; Molla-Herman et al., 2010; Wong et al., 2009). Although a bona fide understanding of the exact nature of the relationship between cilia and the cell cycle remains elusive, the current data open the door to the possibility of therapeutic intervention via ciliary targeting, in diseases of aberrant cellular proliferation (Basten & Giles, 2013).
In addition to their dynamic extension and retraction, primary cilia can also vary in length. Under most circumstances, primary cilia are between 1 and 10 μm long, but they can extend to over 20 μm. Various factors can influence the length of cilia including injury/hypoxia (Verghese, Weidenfeld, Bertram, Ricardo, & Deane, 2008; Verghese, Zhuang, Saiti, Ricardo, & Deane, 2011), treatment with monovalent cation chlorides (LiCl, NaCl, and KCl) (Miyoshi, Kasahara, Miyazaki, & Asanuma, 2009; Ou et al., 2009), cytoskeletal dynamics (Miyoshi, Kasahara, Miyazaki, & Asanuma, 2011), and autophagy (Tang et al., 2013; Tang, Zhu, & Zhong, 2014). It is hypothesized that with all other variables being constant, increased length of the cilium is directly related to the sensitivity of the organelle. Understanding the factors that regulate cilium length could determine how sensitive a cell is to the molecular and mechanical stimuli in the surrounding environment. Thus, a feedback system dictating primary cilium length could be an important yet underappreciated factor regulating development (Miyoshi et al., 2011).
2.3. Ciliogenesis: Building the cilium
The cilium itself is composed of various functional domains required for proper ciliary function: basal bodies (centrosomes), transition fibers, transition zone, intraflagellar transport (IFT) machinery, axoneme, and a specialized ciliary membrane (Fig. 1). In cycling cells, two centrioles (mother and daughter) together with pericentriolar material form the centrosome, which serves as microtubule organization center. Once cells enter G1/G0 phase, the mother centriole differentiates into a basal body before ciliogenesis initiates (Kobayashi & Dynlacht, 2011). Similar to the mother centriole, a basal body is comprised of 9 microtubule triplets arranging in a barrel shape, a subdistal appendage (basal foot), and a distal appendage (transition fiber, Fig. 1).
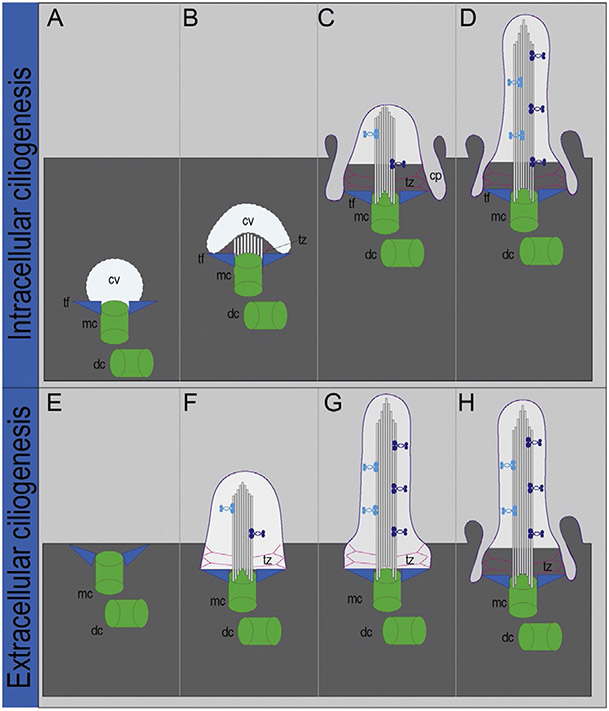
Depending on cell type, ciliogenesis can occur via two distinct mechanisms: (1) extracellular ciliogenesis, where the basal body docks and fuses to the plasma membrane directly or (2) intracellular ciliogenesis, where the basal body first fuses to a ciliary vesicle in the cytoplasm before docking to the plasma membrane (Molla-Herman et al., 2010; Reiter, Blacque, & Leroux, 2012; Sorokin, 1962; Fig. 2). In both pathways, the initial docking is facilitated by the interaction between the membrane and transition fibers. The primary ciliary vesicle adds additional membrane by recruiting secondary vesicles. The enlarged vesicle soon deforms into an invaginated sac, which forms a double membrane sheath (later becoming the ciliary pocket) surrounding the apical end of basal body (Fig. 2). At this point, the transition zone begins to emerge as microtubule doublets start to assemble within the vesicle sac.
Figure 2.
Models of intracellular and extracellular ciliogenesis. Steps of intracellular ciliogenesis. (A) A ciliary vesicle (cv) binds to the distal end of the mother centriole (mc) via associations with transition fibers (tf). (B) Microtubule and transition zone (tz) outgrowth protrudes, causing the cv to invaginate. (C) Docking of the centriolar/cv complex to the plasma membrane. (D) Axonemal outgrowth. Steps of extracellular ciliogenesis. (E) The mother centriole (mc) docks directly to the plasma membrane via associations with transition fibers (tf). (F–G) Axonemal outgrowth occurs. (H) Ciliary pocket forms. dc, daughter centriole;pc, primary cilium; cp, ciliary pocket. Figure modified from Reiter et al. (2012) and Molla-Herman et al. (2010).
While the transition zone is growing, the basal body–ciliary vesicle migrates to the membrane through a process dependent on both the actin cytoskeleton and membrane-associated components in the transition zone (Dawe, Farr, & Gull, 2007). As the basal body–ciliary vesicle complex migrates to, and fuses with, the plasma membrane, the transition zone matures and is now able to function as a ciliary gate. The transition zone is characterized by distinctive Y-shaped fibers, which connect microtubule doublets to the ciliary membrane. Structurally, the transition zone stabilizes the basal body–ciliary vesicle after the initial docking. Functionally, all these components collectively serve as the ciliary gate, a domain required for establishing a distinct ciliary compartment separate from the rest of the cell body.
After formation of the ciliary compartment, the axoneme starts to extend via microtubule lengthening. There is a distinct set of posttranslational modifications on axonemal tubulins as the microtubules are acetylated, detyrosinated, polyglutamylated, and polyglycylated (Konno, Setou, & Ikegami, 2012). These modifications are important for axonemal stability. The microtubule extension occurs through a process called IFT. In IFT, the anterograde transport is carried out by IFT-B complexes and kinesin II motors. IFT builds the microtubule scaffold and carries essential ciliary proteins to the distal tip of the axoneme. Finally, numerous receptors and ion channels localize to the specialized membrane surrounding the fully extended axoneme. With the axoneme extended and functional, cilia are now capable of surveying the cell’s molecular environment. Various cell types utilize these cilia in different ways. In Section 3, we focus on how NCCs utilize primary cilia during their development and differentiation.
3. THE ROLE OF PRIMARY CILIA DURING NCC ONTOGENY
During development, NCCs progress through multiple phases: specification, migration, proliferation, and differentiation. While there are entire fields of study on each phase, little is known regarding primary cilia function during these phases. In this portion of the review, we summarize what is known about primary cilia with respect to each phase.
3.1. Primary cilia and NCC specification
NCC specification occurs at the boundary between the neural plate and the nonneural ectoderm. At this junction Wnt, BMP, and fibroblast growth factor (FGF) signaling initiates a transcriptional program (e.g., Pax3, Pax7, Msx, and Zic1) that will define the neural plate boarder (Garcia-Castro, Marcelle, & Bronner-Fraser, 2002; LaBonne & Bronner-Fraser, 1999; Marchant, Linker, Ruiz, Guerrero, & Mayor, 1998; Mayor, Guerrero, & Martinez, 1997; Monsoro-Burq, Fletcher, & Harland, 2003). Within this region, both NCCs and placodal cells are specified; however, NCCs express a unique combination of transcription factors including Snail2, Sox10, Sox9, and FoxD3 (Bronner & LeDouarin, 2012). While no in-depth study has been conducted specifically examining if primary cilia are required for NCC specification, postmigration data can be used to infer the answer to this question. Various conditional knockouts of ciliary proteins, using NCC drivers, still exhibit the formation of NCCs (Brugmann et al., 2010; Brugmann, Stottmann, unpublished). This suggests that NCCs are specified, to some appreciable extent, in the absence of normal primary cilia function. This conclusion is further supported by data from zebrafish bbs morphant embryos, which exhibit normal domains of foxd3 or sox10 expression suggesting that NCCs are specified properly (Tobin et al., 2008).
While data from both of these model systems support the hypothesis that primary cilia are not absolutely required for proper NCC specification, data from Fuz−/− murine mutants suggests the cilia regulate NCC number. In these mutants, lineage tracing of NCCs indicates an expansion in the domain and number of NCCs in the midbrain, hindbrain, and maxillary process (Gray et al., 2009; Tabler et al., 2013). Taken together, these studies suggest a number of interesting possibilities regarding NCCs and primary cilia. First, perhaps only certain ciliary proteins play a role in NCC specification. Whereas, both Kif3a and the Bardet–Biedl syndrome (BBS) proteins are required for ciliogenesis and localize to the cilia and basal body (Nachury et al., 2007; Yamazaki, Nakata, Okada, & Hirokawa, 1995), respectively, Fuzzy is a cilia-associated protein that functions in the cytoplasm as an effector of the planar cell polarity (PCP) pathway (Brooks & Wallingford, 2012; Park, Haigo, & Wallingford, 2006). Thus, Fuz−/− mutants may have defects in NCC specification due to a cilia-independent mechanism. Second, since Fuz−/− is a null mutant (rather than the conditional knockout targeting ciliary function specifically in NCCs), it is possible that the loss of cilia in surrounding tissues negatively affects the molecular crosstalk required for NCC specification. Continued examination of NCC specification in various ciliary mutants is necessary to clarify this issue.
3.2. Primary cilia and NCC migration
Following specification, NCCs will undergo an epithelial to mesenchymal transition, delaminate from the neural tube and migrate in a highly regulated process. Broadly, NCC migration requires two main types of interactions: (1) between NCCs and the surrounding tissues and (2) between individual migrating NCCs. These interactions are dependent upon NCCs receiving and interpreting either long-range or short-range cellular signals (Teddy & Kulesa, 2004). For migration to efficiently occur, NCCs must be capable of responding to attractants/repellants and creating discrete, directional migration streams (Kuriyama & Mayor, 2008; Teddy & Kulesa, 2004). Chemotaxis, the movement of a cell toward a directive substance, is also a guiding force of NCC migration (Theveneau & Mayor, 2012). Several chemotactic molecules essential for cranial NCC migration have been identified including SDF-1, PDGF-AA, and VEGFA (Theveneau & Mayor, 2012). The receptors for some of these molecules are found on primary cilia (Schneider et al., 2005).
Several ciliary mutants exhibit aberrant NCC cell migration. For example, bbs zebrafish morphants have a significant reduction in the migratory streams of both cranial and trunk NCCs. Through transplantation studies, it was determined that bbs8 is required cell-autonomously for proper NCC migration (Tobin et al., 2008). Fto zebrafish morphants also have NCC migration defects as evidenced by diffuse and reduced Sox10 expression in the head (Osborn et al., 2014). More caudal NCC migration is also severely disrupted in these mutants. NCCs posterior to the seventh somite fail to migrate away from the dorsal neural tube and melanocytes are mislocalized (Osborn et al., 2014). In support of these data from animal models, fibroblasts from ciliopathy patients also fail to migrate normally and exhibit disruptions in actin cytoskeletal architecture (Hernandez-Hernandez et al., 2013; Madhivanan, Mukherjee, & Aguilar, 2012; Tobin et al., 2008).
While, it is clear that aberrant ciliary function can negatively impact NCC migration, we currently lack an understanding of how primary cilia regulate migration. One potential mechanism is platelet-derived growth factor (PDGF)-dependent chemotaxis as PDGF is a known NCC chemoattractant, PDGFRα localizes to the axoneme, and PDGF signaling is transduced through the primary cilium (Schneider et al., 2005). Additionally, studies in both fibroblasts and mouse embryonic fibroblasts (MEFs) demonstrated primary cilia are required for directed migration toward a PDGF-AA source (Schneider et al., 2005, 2010). Interestingly, loss of PDGFRα on cranial NCCs yields aberrant cranial NCC migration (He & Soriano, 2013). Another possible mechanism to affect NCC migration is via regulation of the PCP pathway. The PCP proteins Vangl2 and Dishevelled have been shown to localize to the primary cilium and have genetic interactions with both IFT and basal body proteins (Jones et al., 2008; Ross et al., 2005; Veland et al., 2013). Additionally, ciliopathies such as Lowe, Joubert, and Bardet–Biedl syndromes have defects in RhoA or Rac1 localization or activity, and RhoA has also been show to localize to the basal body (Hernandez-Hernandez et al., 2013; Madhivanan et al., 2012; Valente et al., 2010). All of these parallel findings support the hypothesis that primary cilia play an important role during NCC migration, and we suggest this is an area ripe for continued investigation.
3.3. Primary cilia and NCC proliferation
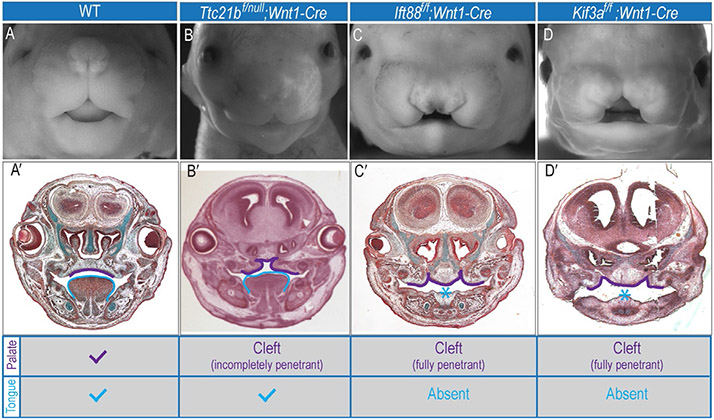
After NCCs have finished migrating, they will continue to proliferate and greatly expand in their target tissues. Primary cilia extension and the cell cycle are tightly linked together. For example, IFT88 is associated with the centrosome throughout the cell cycle (Robert et al., 2007) and is required for spindle orientation in mitosis (Delaval, Bright, Lawson, & Doxsey, 2011). Furthermore, overexpression of IFT88 prevents the G1–S transition and induces apoptotic cell death, whereas knockdown of IFT88 promotes cell cycle progression (Robert et al., 2007). Data from mouse ciliopathic mutants indicate that primary cilia on NCCs are essential for regulating proliferation, especially in the developing face. Kif3af/f;Wnt1-Cre mutants have increased NCC proliferation. This increase is most notable at the facial midline of embryos and correlates with an expansion of Gli1 expression. Phenotypically, the medial increase in proliferation manifests as an expanded midline in these mutants (Brugmann et al., 2010; Fig. 3).
Figure 3.
Craniofacial phenotypes of ciliary mutants. Whole-mount images of e14.5 (A) Wild-type, (B) Ttc21bf/null;Wnt1-Cre, (C) Ift88f/f;Wnt1-Cre, and (D) Kif3af/f;Wnt1-Cre mice. (A′–D′) Frontal sections of (A–D). (A′) Wild-type mice have a fused palatal shelves (purple outline) and a tongue (blue outline). In contrast, all ciliary mutants have (B′–D′) some degree of cleft palate (purple outline) and Ift88f/f;Wnt1-Cre and Kif3af/f;Wnt1-Cre mutants have aglossia (C′, D′). Chart indicates craniofacial phenotype for each mouse.
Although the link to proliferation is clear in Kif3af/f;Wnt1-Cre mutants, the effect that loss of other ciliary proteins has on proliferation is highly variable. Fuz−/− mutants have hyperplastic maxillary prominences that result in a high, arched palate, yet NCC proliferation is actually decreased (Tabler et al., 2013). Furthermore, there are other ciliary mutants that do not show any indication of alterations in NCC proliferation or midline development, such as Ttc21bf/null;Wnt1-Cre (Fig. 3, Stottmann unpublished). These data reiterate that cilia have a role in proliferation, but also suggest that the exact role the cilia play during proliferation is unclear. Currently, the most accurate conclusion to draw from these data is that individual ciliary proteins play specific roles during proliferation.
3.4. Primary cilia and NCC differentiation
NCCs give rise to a wide variety of cell types including bone and cartilage cells in the facial skeleton. Both patients with ciliopathies and ciliopathic animal models often have dysmorphic craniofacial skeletons. In zebrafish, bbs, ift, and ofd1 morphants have dysmorphic craniofacial cartilage (Ferrante et al., 2009; Lunt, Haynes, & Perkins, 2009; Tobin et al., 2008). Murine ciliopathic mutants also frequently have skeletal abnormalities. Mskkrc mutants exhibit misshapen skulls and a decrease in frontal and parietal bone ossification (Weatherbee, Niswander, & Anderson, 2009). Humans with Meckel syndrome, often caused by mutations in the Msk gene, have skeletal malformation of the cranial base (Kjaer, Hansen, Keeling, Nolting, & Kjaer, 1999). Thus, taken together, these data demonstrate that cranioskeletal defects are a common feature of human ciliopathies and further suggest that primary cilia may play an important role in NCC differentiation into skeletal elements.
In addition to the importance of primary cilia for initial skeletal differentiation, they are also critical for postnatal changes in the skeleton. Mechanosensation is another cellular process that helps to shape the architecture of skeletal elements. Primary cilia are able to respond to mechanical signals (Hoey, Downs, & Jacobs, 2012). Specific subsets of ciliary proteins have been shown to be involved in these mechanical-based signaling responses. For example, PKD2 localizes to the primary cilium and is essential for fluid-flow sensation (Nauli et al., 2003). Pkd2f/f;Wnt1-Cre postnatal mutants exhibit dome-shaped skull defects, anterior-posterior snout compression, and abnormal fusion in the face. These abnormalities are not detected in embryos, suggesting that these phenotypes are the result of an inability of NCC derivatives to properly respond to postnatal mechanical forces. These data suggest that primary cilia continue to play a role in development and remodeling of NCC-derived structures even after cell differentiation.
4. CRANIOFACIAL PHENOTYPES IN ANIMAL MODELS AND HUMAN PATIENTS SUPPORT A ROLE FOR PRIMARY CILIA IN NCC DEVELOPMENT
4.1. Insights from animal models
In addition to the ciliary mutants in which early stages of NCC development have been analyzed, there are a significant number of ciliary mouse mutants with craniofacial phenotypes suggesting aberrant NCC behavior (Table 1). For example, several mutants have cleft lip and/or palate, including the Ift172avc1 allele with cleft secondary palate (Friedland-Little et al., 2011), Rpgrip1l with cleft lip and hypoplastic mandible (Delous et al., 2007), Mks1 with cleft lip/palate and pointy snout (Weatherbee et al., 2009), and Kif7 with cleft lip/palate (direct submission by Cecilia Lo to MGI). Other mutants are reported to have phenotypes in the arches, without clefting defects, including Dync2h1 (formerly Dnchc2), which exhibits micrognathia (C Lo direct submission to MGI), ft57 (Hippi), which displays hypertelorism and small arches (Houde et al., 2006), Ift52hypo with fused maxillary prominences (Liu et al., 2005), Tmem67 with mandibular hypoplasia (Abdelhamed et al., 2013), and ft122 mutants with enlarged branchial arches (Cortellino et al., 2009). Furthermore, an in-depth morphometric analysis was performed on BBS mouse models and both Bbs4 and Bbs6 homozygous mutants have larger ratios of mid-face width to height and shortened snouts (Tobin et al., 2008). These craniofacial defects strongly suggest NCC patterning and/or development is frequently affected in ciliary mutants.
Table 1.
Animal models for craniofacial ciliopathies
| Model system |
Mutant/morphant name | Gene | Craniofacial phenotype | References |
|---|---|---|---|---|
| Mouse | Aln | Ttc21b (Ift39) | Delayed eye and forebrain development; neural tube defects | Tran et al. (2008) |
| Bbs4−/− | Bbs4 | Mid-facial shortening due to premaxillary and maxillary hypoplasia; shortened snouts | Tobin et al. (2008) | |
| Bbs6−/− | Bbs6 | Mid-facial shortening due to premaxillary and maxillary hypoplasia; shortened snouts | Tobin et al. (2008) | |
| Dync2h1b2b414Clo | Dync2h1 | Micrognathia | Cecilia Lo to MGI | |
| Evc−/− | Evc | Chondroplasia, hypodontia | Ruiz-Perez et al. (2007) | |
| Fuz−/− | Fuz | Exencephaly; anophthalmia; encephalocoele; mandibular defects; micrognathia; high arched palate; enlarged maxillary processes | Tabler et al. (2013) and Gray et al. (2009) | |
| Hippi−/− | Ift57 | Exencephaly; abnormal cranial flexure; hypertelorism; abnormal maxillary processes; small arches | Houde et al. (2006) | |
| Ift52hypo | Ift52 | Single median nostril; fused maxillary prominences; exencephaly | Liu, Wang, and Niswander (2005) | |
| Ift122−/− | Ift122 | Exencephaly; anencephaly; altered eye and branchial arch development; defects of the ventral portion of the head | Cortellino et al. (2009) | |
| Ift172avc1 | Ift172 | Hydrocephalus; abnormal facies; shortened muzzle; cleft lip and palate | Friedland-Little et al. (2011) | |
| Kif3af/f;Wnt1-Cre | Kif3a in cranial NCCs | Anterior cranium occultum; bifid nasal septum; aglossia; severe hypertelorism; dental abnormalities | Brugmann et al. (2010) | |
| Kipf7−/− | Kif7 | Cleft lip and palate | Cecilia Lo to MGI | |
| Mks−/− | Mks1 | Occipital meningoencephalocele; hypomineralization and/or splitting of the frontal, parietal, and superaoccipital bones; hydrocephaly; cleft palate; pointy snout | Weatherbee et al. (2009) | |
| Ofd1−/− | Ofd1 | Severe cleft palate; shortened skull and facial regions | Ferrante et al. (2006) | |
| Orpk | Ift88 | Dental abnormalities; micrognathia; increased gonial angle; poorly differentiated branchial arches; minor clefting; disorganized/patent frontal sutures | Zhang et al. (2003), Murcia et al., (2000), and Ohazama et al. (2009) | |
| Rpgrip1l−/− | Rpgrip1l/Ftm | Exencephaly, microphthalmia; rounded skull; cleft upper lip; micrognathia | Delous et al. (2007) | |
| Tmem67−/− | Tmem67 | Hypoplastic mandible, semilobar HPE, occipital meningocele, midbrain and/or forebrain exencephaly | Abdelhamed et al. (2013) | |
| Chicken | talpid2 | c2cd3 | Short, wide upper beak; cleft lip/palate; hypo- or aglossia | Chang et al. (2014) |
| talpid3 | talpid3 | Ocular hypotelorism; frontonasal hypoplasia; fused maxillary prominences; micrognathia | Yin et al. (2009) | |
| Zebrafish | Bbs morphants | bbs4, bbs6, bbs8 | Mandibular dysplasia; severe craniofacial reduction; anteroneurocranium hypoplasia | Tobin et al. (2008) |
| Ift mutants | ift57, ift88, ift172 | Mild dysmorphology of craniofacial cartilages | Lunt et al. (2009) | |
| Ift80 morphant | ift80 | Abnormal anterior neurocranial development; midline fusion | Beales et al. (2007) | |
| Ift122 morphant | ift122 | Reduced ocular development; distended cranium with hydrocephalus; otolith defects | Walczak-Sztulpa et al. (2010) | |
| Ofd1 morphant | ofd1 | Abnormal blunting of the jaw; disorganization of Meckel’s cartilage cells | Ferrante et al. (2009) |
Ciliopathic models are grouped according to model system.
Other mutations seem to reveal broader requirements for some ciliary genes. The Ift88Tg737Rpw mice have been described with poorly differentiated branchial arches, minor clefting, supernumerary teeth, and disorganized and patent frontal sutures (Murcia et al., 2000; Zhang et al., 2003). Loss of Ofd1 leads to both severe cleft palate and a shortened skull (Ferrante et al., 2006). Evc mutants have abnormal incisors (Ruiz-Perez et al., 2007). These findings would suggest that ciliary proteins execute a number of roles in craniofacial development, but that loss of particular ciliary genes leads to specific phenotypes. This latter conclusion resonates with our model that cilia in distinct regions of an embryo have possibly unique roles in specific developmental signaling events and not all cilia have the same complement of proteins. However, we must also consider that mouse experiments on different genetic backgrounds may incorporate different modifier genes, which further complicate comparisons between ciliary gene loss-of-function phenotypes. This is consistent with the known multi-allelic effects in ciliopathies (Davis et al., 2011).
Additional insight into the role of cilia in NCC development can be gleaned from a recent study of the Fuzzy mouse (Tabler et al., 2013). This is a unique mutant that exhibits a high arched palate, a phenotype identified in a high proportion of BBS patients (Beales, Elcioglu, Woolf, Parker, & Flinter, 1999). A detailed developmental analysis presented in this study uncovered an enlarged maxillary process and aberrant patterns of NCC migration in E9.5 embryos. Interestingly, a NCC specific ablation of Fuzzy with Wnt1-Cre resulted in a true cleft phenotype without the associated maxillary hypoplasia or hypoplastic tongue (Tabler et al., 2013). Together, these findings are consistent with our model that cilia are actually performing discrete tasks in different regions of the embryo and future studies will need to incorporate this idea to fully explain the role of cilia in craniofacial NCCs.
While mouse models allow for insights into mammalian ciliary function, both fish and chicken models have proven very valuable, especially when evaluating early NCC development. Several fish mutants and morphants exist allowing for rapid evaluation of NCC development, migration, and differentiation (Beales et al., 2007; Ferrante et al., 2009; Lunt et al., 2009; Tobin et al., 2008; Walczak-Sztulpa et al., 2010; Table 1). In addition, two separate, naturally occurring ciliopathic models have been identified in the chicken. The causal genetic elements for both the talpid2 and talpid3 mutants have recently been identified (Chang et al., 2014; Yin et al., 2009). The accessibility of the chicken embryo, and its availability for culturing and transplanting tissues, provides promise for careful analysis of how primary cilia participate in each individual step of NCC development.
4.2. Human craniofacial ciliopathies
Given the data generated in animal models, it is not surprising there is a growing class of human craniofacial ciliopathies with features indicative of aberrant NCC development. Some years ago, Baker and Beales reviewed 102 human conditions of known or possible ciliopathies. Of the 102 conditions, the craniofacial complex was primarily affected in up to 30% of cases (Zaghloul & Brugmann, 2011). Although primary cilia extend from almost all cells in the body, the clinical features observed in patients with ciliopathies highlight the importance of primary cilia in development of certain types of tissue. The phenotypes associated with human craniofacial ciliopathies were similar to those observed in animal models: midline, cephalic, oral, and dental anomalies (Zaghloul & Brugmann, 2011; Table 2). Three such examples of human craniofacial ciliopathies include Meckel–Gruber syndrome (MKS), BBS, and Oro-facial-digital syndrome (OFD). MKS presents with cleft lip in 20% of cases and cleft lip and palate in 45% of cases. Clefting is accompanied by tongue malformations including ankyloglossia, bifid tongue, and anterior marginal hamartomas (Fried, Liban, Lurie, Friedman, & Reisner, 1971; Moerman, Verbeken, Fryns, Goddeeris, & Lauweryns, 1982; Rehder & Labbe, 1981). BBS is associated with oligodontia, hypodontia, and numerous other dental abnormalities (Lofterod, Riise, Skuseth, & Storhaug, 1990). Additional studies have reported characteristic midfacial defects hypothesized to be due to improper hedgehog induced NCC migration (Tobin et al., 2008). A host of other defects related to NCC development/migration including hypopigmentation, heart defects, middle ear defects, choanal atresia, and Hirschsprung disease have also been reported in BBS patients (Gorlin, Cohen, & Levin, 1990). OFD is characterized by a wide nose with a broad, flat nasal bridge, hypertelorism, cleft lip and palate, glossal deformities, hyperplastic frenula, the growth of noncancerous tumors and hypodontia. OFD has long been linked to mutations in OFD1 (Ferrante et al., 2001; Gurrieri, Franco, Toriello, & Neri, 2007). Recently however, a new subtype of OFD has been identified and linked to mutations in C2CD3 (Thauvin-Robinet et al., 2014); the same gene recently found responsible for the talpid2 avian mutant (Chang et al., 2014). The rapid identification of new ciliary genes and ciliopathic models suggests that many yet to be classified syndromes may in fact be ciliopathies. Furthermore, the high frequency of dysmorphic faces suggests NCCs are indeed highly sensitive to the loss of primary cilia.
Table 2.
Human craniofacial ciliopathies and associated phenotypes
| Syndrome | Gene | Craniofacial phenotype |
|---|---|---|
| Bardet–Biedl syndrome | BBS1-12, MKS1, MKS3 | Mid-face shortening and flattening; nasal bridge hypoplasia; reduced length/bulbosity of the nasal tip; mild retrognathia |
| Cranioectodermal dysplasia/Sensenbrenner syndrome | IFT122, IFT144, WDR35 | Sagittal craniosynostosis; epicanthal folds; hypodontia or microdontia; everted lip; multiple oral frenula; high arched palate; skeletal and ectodermal anomalies |
| Ellis–van Creveld syndrome | EVC1, EVC2 | Hypertrophy labiogingival frenulum; upper lip abnormalities; presence of teeth at birth; microdontic teeth |
| Joubert syndrome | MKS3, NPHP6, RPGRIP1L, AH1, ARL13B, CSPP1 | Prominent forehead; high rounded eyebrows; epicanthal folds; ptosis; upturned nose with evident nostrils; hypertelorism; open mouth and tongue protrusion with rhythmic tongue motions |
| Lowe syndrome | OCRL1 | Frontal bossing; retrognathia; high arched palate; deep set eyes; full cheeks; maxillary prognathism; malocclusion |
| Meckel–Gruber syndrome | MKS1, MKS3, TMEM231, TCTN2, CC2D2A | Encephalocele; cleft lip and palate |
| Oro-facial-digital syndrome | OFD1, C2CD3 | Facial asymmetry; hypertelorism; micrognathia; broadened nasal ridge; hypoplasia of the malar bones and nasal alar cartilages; frontal bossing; pseudocleft; cleft palate; hamartomas of the tongue; bifid tongue; hyperplastic oral frenula; up-slanting palpebral fissures |
5. BEYOND THE FACE: TRUNK NCCs ARE ALSO AFFECTED BY THE LOSS OF PRIMARY CILIA
Although NCCs make substantial and vital contributions to the face, they also contribute to many other lineages and primary cilia have been implicated in proper development of many of these populations as well. The best-studied NCC lineage outside the face is arguably the cardiac NCC. Cardiac NCCs normally make a significant contribution to the outflow tract of the heart, which separates the pulmonary and systemic circulations.
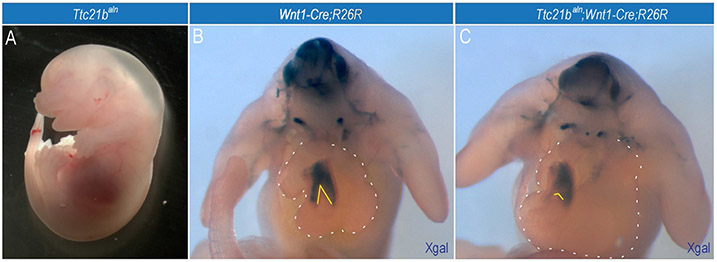
An elegant example of a role for primary cilia in cardiac development is the zebrafish cobblestone (cbs) mutant. Cbs is a hypomorphic allele of Ift88 and homozygous mutants present with a number of the most common congenital heart defects seen in newborn humans including: ventricular and atrial septal defects, and persistent truncus arteriosus (failure of outflow tract to divide). Further study of this mutation showed that the cardiac NCCs migrated normally into the heart, but the NCC-derived outflow tract exhibited a significant reduction in expression of Bmp4 (Willaredt, Gorgas, Gardner, & Tucker, 2012). In our own studies, we have noted frequent edema in late-embryonic stage Ttc21baln mutants and abnormal patterns of NCC migration into the heart (Fig. 4). Thus, primary cilia are important for the molecular signaling that guides outflow tract development.
Figure 4.
Loss of primary cilia affects cardiac NCCs. Ttc21baln mutant has cardiac NCC defects. (A) Whole-mount image of an e14.5 Ttc21baln mutant with systemic edema suggestive of cardiopulmonary dysfunction. (B) Xgal staining in an e11.5 Wnt1-Cre;R26R embryo to trace the NCC lineage. (C) Xgal staining in an e11.5 Ttc21balnWnt1-Cre;R26R embryo. Dotted white lines outline the heart. Solid white lines indicate the degree of cardiac NCC migration into the septating outflow tract.
Another NCC derivative affected by loss of cilia is the enteric nervous system, the main division of the autonomic nervous system that innervates the gut. A failure of NCC migration to the gut or differentiation into enteric ganglia results in Hirschsprung disease (aganglionic megacolon; McKeown, Stamp, Hao, & Young, 2013), a disease in which variable portions of the gut lose the ability to relax. Hirschsprung disease is frequently present in both BBS patients (Amiel et al., 2001) and ciliopathic animal models. Zebrafish bbs8 morphants have decreased vagal NCC and incomplete enteric nervous system formation leading to disordered gut motility (Tobin et al., 2008). Furthermore, reduction of ADP ribosylation factor-like 6 interacting protein 1 (Arl6ip1) in zebrafish produces disorganized cilia, dampened sonic hedgehog (Shh) signaling and subsequent reduction in the number of enteric ganglia (Tu, Yang, Huang, & Tsai, 2012).
Various other NCC derivatives extend cilia and are affected by a loss of primary cilia. Melanocytes extend cilia, both in vitro (Le Coz, Benmerah, & Larue, 2014) and in vivo (Wandel, Steigleder, & Bodeux, 1984) and zebrafish bbs8 morphants exhibit a pigmentation phenotype (Tobin et al., 2008). Primary cilia have been identified on corneal endothelial cells in a number of mammals (Collin & Barry Collin, 2004; Doughty, 1998) and Polaris mutants have disorganized patterning of the corneal endothelium (Blitzer et al., 2011). Although no studies have reported defects in the dorsal root ganglia or sympathetic ganglia, we have noted defects in both of these lineages in the Ttc21baln mutants (Stottmann, unpublished).
6. NCCs UTILIZE PRIMARY CILIA FOR TISSUE-TISSUE INTERACTIONS
6.1. Structures that require reciprocal signaling are disrupted in ciliary mutants
Throughout the course of development NCCs interact with cells from all three germ layers to form epithelial specializations or discrete organs. Many structures that require reciprocal signaling with NCCs for their formation are disrupted in ciliary mutants. Prime examples of structures or tissues that require molecular input from NCCs are teeth, whiskers, and the tongue.
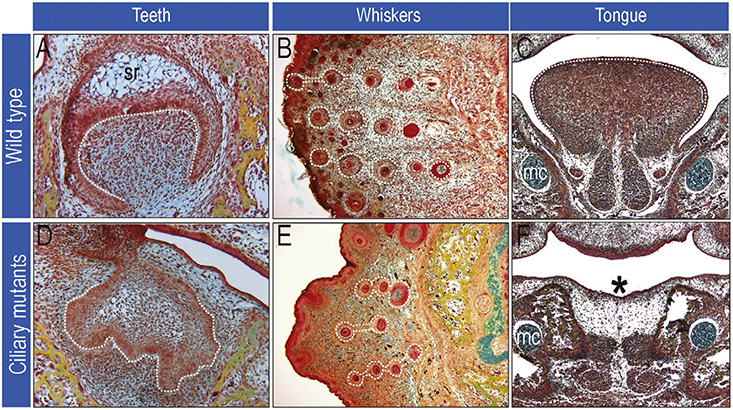
Odontogenesis, or tooth formation, requires reciprocal interactions between the oral ectoderm and the underlying NCC-derived mesenchyme (Thesleff 2006). Many human ciliopathies including Ellis–van Creveld syndrome, OFD syndrome, and Cranioectodermal dysplasia exhibit abnormal dentition (Gurrieri et al., 2007; Mostafa, Temtamy, el-Gammal, & Mazen, 2005; Walczak-Sztulpa et al., 2010; Table 2). Studies in mice provide the genetic advantage of tissue-specific conditional deletions of primary cilia. Ift88 hypomorphs develop supernumerary teeth accompanied by an expansion of Gli1 expression; however, when a keratin (K)-5-Cre driver is used to delete Ift88 from epithelial cells, no dental abnormalities are observed (Ohazama et al., 2009). This suggests that cilia are required specifically on NCCs for proper tooth formation. This hypothesis is supported by data from Kif3af/f;Wnt1-Cre mutants where Shh and Wnt activity in the dental lamina is aberrant (Brugmann et al., 2010), and tooth development is abnormal (Fig. 5). Together, these data suggest that primary cilia are essential mediators of NCC-epithelial signaling during odontogenesis.
Figure 5.
Defects in craniofacial specialization. Images of wild-type (A–C) and ciliary mutant (D–F) craniofacial structures. Ciliary mutants are unable to properly form teeth (compare dotted white lines in A to D), whiskers are abnormally spaced and have aberrant morphology (compare dotted white lines in B to E) and some ciliary mutants can have aglossia (*) (compare dotted white lines in C to asterisk in F). sr, stellate reticulum; mc, Meckel’s cartilage.
A curious example of a ciliopathy causing dental abnormalities is the avian ciliopathic mutant, talpid2, which develops crocodilian-like teeth (Brugmann et al., 2010; Chang et al., 2014; Harris, Hasso, Ferguson, & Fallon, 2006). Modern birds lack teeth. However, some studies suggest that the avian oral ectoderm still possesses instructive signals for forming teeth. Thus, it was hypothesized that avian dentition does not form because the competent NCC-derived mesenchyme is not in a position to receive instructive signaling from the ectoderm (Kollar & Fisher, 1980; Kollar & Mina, 1991). In talpid2, the oral/aboral boundary is shifted such that competent mesenchyme is in a position to receive instructive signal for the ectoderm, promoting specification of dentition (Harris et al., 2006). It is unclear the exact role primary cilia play in this context, as this study was performed prior to the identification of talpid2 as ciliopathic mutant, but these data further suggest that reciprocal signaling between oral ectoderm and NCC-derived mesenchyme require primary cilia.
Similar to ondontogenesis, whisker formation requires reciprocal interactions between NCC-derived mesenchyme and the overlying ectoderm (Chiang et al., 1999). When NCCs lack primary cilia, as in the Kif3af/f;Wnt1-Cre mutants, whiskers still form; however, they are reduced in number and disorganized (Brugmann et al., 2010). Instead of forming in a distinct spatial pattern, they are irregularly spaced (Fig. 5). While an in-depth analysis of whisker formation has not been carried out in cilia mutants, one might expect defects in primary cilia-dependent reciprocal signaling between overlying ectoderm and NCC-derived mesenchyme to at least partially explain this anomaly.
The developing tongue is comprised of three different cell types: epithelial cells encase the tongue, NCC-derived mesenchyme contribute to the vasculature and connective tissue of the tongue, and the mesodermal-derived myoblasts give rise to the muscles of the tongue (Parada, Han, & Chai, 2012). Mesoderm-derived myoblast progenitors associate with NCC-derived mesenchyme once they both populate the tongue anlage (Huang, Zhi, Izpisua-Belmonte, Christ, & Patel, 1999). In order for muscle differentiation to occur, there must be reciprocal interaction between NCCs and the mesoderm. Signaling molecules such as Fgf4, Fgf6, and BMP4 are important for myogenic cell proliferation and differentiation in the tongue and when BMP signaling is reduced intrinsically in NCCs, as is the case for Alk5f/f;Wnt1-Cre mutants, tongue muscles are disrupted (Han, Zhao, Li, Pelikan, & Chai, 2014). Similarly, Shh signaling is also essential for tongue development and both Gli3−/− and Smon/c;Wnt1-Cre mice display abnormal tongue development (Huang, Goudy, Ketova, Litingtung, & Chiang, 2008; Jeong, Mao, Tenzen, Kottmann, & McMahon, 2004).
Several human ciliopathies and animal models encompass tongue malformations. Human ciliopathies that often present with glossal defects include OFD syndrome and Joubert syndrome (Table 2). Patients with OFD syndrome will frequently have a bifid tongue, sometimes accompanied by hamartomas (Gurrieri et al., 2007). With Joubert syndrome, a small percentage of patients will develop soft tissue tumors of the tongue (Pellegrino, Lensch, Muenke, & Chance, 1997). Others exhibit a groove in the midline of the tongue or frequently protrude their tongue rhythmically (Dahlstrom, Cookman, & Jain, 2000; Parisi, 2009). Numerous animal ciliopathic models have glossal defects. The avian talpid2 mutant exhibits hypoglossia/aglossia (Chang et al., 2014) and both the Kif3af/f;Wnt1-Cre and Ift88f/f;Wnt1-Cre mouse mutants display aglossia (Brugmann et al., 2010; Fig. 3). It is tempting to hypothesize that primary cilia on NCCs are required to interpret the Shh signal from the epithelium or other molecular signals from the mesoderm important for specification/patterning of the tongue. It should be noted, however, that other ciliary mutants such as Ttc21bf/f;Wnt1-Cre have normal tongues (Fig. 3). Further investigation is needed to understand why loss of certain ciliary proteins results in aglossia, while others do not.
In addition to craniofacial defects, many ciliopathic patients also have brain defects, including anencephaly, exencephaly, and hydrocephaly (Carter et al., 2012). Similar brain phenotypes are observed in several ciliary mutants including Dynch2GT, Hippi−/−, hennin, Ift122−/−, Msk, and Fuz murine mutants (Caspary et al., 2007; Cortellino et al., 2009; Gray et al., 2009; Houde et al., 2006; May et al., 2005; Weatherbee et al., 2009). Interestingly, brain abnormalities can also occur when primary cilia are lost on NCCs. Kif3af/f;Wnt1-Cre mutants present with agenesis of the corpus callosum (Brugmann et al., 2010). In this case, the expression pattern of Wnt1 in dorsal neural stem cells does not preclude that these phenotypes are due to loss of cilia within the neural plate. However, the possibility remains that loss of cilia affecting NCC derivatives, such as smooth muscle cells or pericytes, could contribute to the aberrant brain development observed in these mutants.
7. THE ROLE FOR PRIMARY CILIA IN MOLECULAR SIGNAL TRANSDUCTION
The data outlined above show NCCs require primary cilia for several developmental processes and intracellular interactions. As previously discussed, primary cilia are essential for the reception and processing of multiple signaling pathways. Below, we briefly summarize what is known regarding the role of primary cilia and transduction of various signaling pathways important in NCC development.
7.1. Sonic hedgehog
NCC survival, proliferation, and patterning are dependent on Shh signaling (Ahlgren & Bronner-Fraser, 1999; Ahlgren, Thakur, & Bronner-Fraser, 2002; Brito, Teillet, & Le Douarin, 2008; Marcucio & Helms, 2002; Wada et al., 2005) and defects in these processes are reflected in the tissues of the face in ciliopathy patients (Brugmann et al., 2010; Tobin et al., 2008). Identification of the primary cilium as a signaling hub for the Hedgehog pathway came from seminal experiments reporting that anterograde and retrograde IFT proteins in the cilium are required for Shh signal propagation (Huangfu & Anderson, 2005; Huangfu et al., 2003). Subsequent analyses showed that Ptch1 and Smo receptors localize to the primary cilium and that in the presence of ligand, Ptch1 is sequestered from the cilium facilitating an accumulation of Smo and activation of signaling (Corbit et al., 2005; Rohatgi, Milenkovic, & Scott, 2007). For a number of years, it was believed that there was a strictly linear relationship between Shh signaling and primary cilia (e.g., the loss of functional cilia would result in a loss of Shh function). In some mutants, however, the exact opposite is the case; loss of cilia causes a gain of Shh signaling (Brugmann et al., 2010; Cortellino et al., 2009; Tran et al., 2008). How might Shh signaling be activated as a consequence of losing ciliary proteins? To begin to hypothesize how this seemingly paradoxical event could occur, we must first understand the relationship between the cilia and components of the Shh pathway.
The most well-known links between primary cilia and the Shh pathway are (1) that the receptors of the pathway are localized to the cilium and (2) that IFT proteins are involved in trafficking and processing of Gli2 and Gli3 from full-length isoforms, into either activator or repressor forms (Haycraft et al., 2005). In the absence of Shh, Ptch1 is present in cilia and Gli3 (and Gli2) may translocate at low levels in and out of cilia, being phosphorylated by PKA. Presumably, IFT proteins are then involved in shuttling Gli3FL to the pericentrosomal proteasome where it is processed into Gli3R and subsequently translocated to the nucleus to inhibit Gli1 and Gli2 (Wen et al., 2010). In the presence of Shh, Smo displaces Ptch1 in cilia, thereby inhibiting PKA phosphorylation and Gli3FL (and Gli2FL) accumulates at the distal tips. Thus, one mechanism by which a ciliary defect could result in a gain of Shh signaling is via aberrant proteolytic processing of Gli3 (the predominant repressor of the pathway). Deregulation of Gli3R production would result in an increased Gli3FL:Gli3R ratio and subsequent increase in Shh activity. This scenario has been observed in numerous IFT mouse models (Haycraft et al., 2005; Huangfu & Anderson, 2005; Liu et al., 2005; May et al., 2005; Tran et al., 2008; Willaredt et al., 2008).
Whereas Gli processing is the most commonly examined aspect in ciliary mutants, the identification of novel ciliary proteins implicates additional mechanisms that could explain how the loss of cilia could cause a gain of Shh activity. For example, recent work identified G-protein-coupled receptor 161 (Gpr161) as a ciliary protein that negatively regulates Shh activity via promoting PKA-mediated generation of GliR (Mukhopadhyay et al., 2013). With respect to Gpr161 function, loss of cilia could alleviate negative regulation of the pathway and thus also cause a gain of Shh activity. Furthermore, some studies have suggested “noncanonical” and ciliary -independent mechanisms for Shh signaling that do not function through Gli activity (Robbins, Fei, & Riobo, 2012). In light of this rapidly emerging data, caution must be taken when making generalizations about the role of primary cilia and a signaling pathway because many of these mechanisms appear to be tissue-specific (Zaghloul & Brugmann, 2011).
7.2. Wnt
Wnt signaling is required throughout NCC development. Various Wnt ligands are expressed in developing neural tissue at stages that strongly suggest a role for Wnt proteins in NCC induction in several species (Rogers, Jayasena, Nie, & Bronner, 2012). Furthermore, Wnts also have reiterative roles later in NCC development during migration (Mayor & Theveneau, 2014), proliferation (Brugmann et al., 2007), and differentiation (Brault et al., 2001). Initial evidence for the involvement of the canonical Wnt pathway in ciliary function came from in vitro experiments demonstrating that Inversin localized to the cilia and physically interacted with Wnt pathway component Dishevelled (Dvl) (Otto et al., 2003; Simons et al., 2005; Watanabe et al., 2003). One study reported that a knockdown of Kif3a led to increased Wnt signaling, suggesting that the primary cilium restricts canonical Wnt signaling in mouse embryos, primary fibroblasts, and embryonic stem cells (Corbit et al., 2008). Other studies focused on different ciliary proteins have supported this hypothesis. MEFs isolated from retrograde protein Ttc21b mutant mice showed an approximately fivefold increase in reporter activity in response to Wnt3a-conditioned media (Tran et al., 2014). Similarly, loss of primary cilia in the mammary ducts of Ift88 mutant mice resulted in increased Wnt signaling (McDermott, Liu, Tlsty, & Pazour, 2010).
Despite this plethora of data suggesting a role for cilia in Wnt signal transduction, there is significant evidence supporting the contrary. Zebrafish ift88 mutants fail to make cilia and display characteristic Shh-related phenotypes, but expression of Wnt targets is unchanged (Huang & Schier, 2009). Another study found that Axin2 expression levels and readouts from Wnt reporters were indistinguishable between wild-type and a number of ciliary mutants (Ocbina, Tuson, & Anderson, 2009). Furthermore, wild-type and ciliary mutant MEFs displayed no difference in response to exposure to both Wnt3a and Wnt5a ligands (Ocbina et al., 2009). In NCCs specifically, the loss of Kif3a did not appear to alter Wnt activity in early facial mesenchyme (Brugmann et al., 2010). Despite these opposing findings, several studies still support a role for ciliary proteins in restraining Wnt signaling. In light of these seemingly contradictory data, one possibility to consider is that the perturbations of the Wnt pathway observed in ciliary mutants may not be direct. Gli proteins can bind to and directly affect transcription of Wnt effector genes. Thus, ciliary-dependent disruptions of posttranslational Gli processing could secondarily affect Wnt activity. Clearly the complexity of the issue requires further investigation regarding Wnt signaling and cilia involvement in NCCs and other cell types.
7.3. Fibroblast growth factor
The FGF signaling pathway has been associated with growth and development of the facial prominences, the facial skeleton, and craniofacial musculature (Creuzet, Schuler, Couly, & Le Douarin, 2004; Macatee et al., 2003; Szabo-Rogers, Geetha-Loganathan, Nimmagadda, Fu, & Richman, 2008; Trumpp, Depew, Rubenstein, Bishop, & Martin, 1999). Similar to Wnts, many FGFs are expressed in spatiotemporal patterns that suggest a role in NCC induction and patterning (Rogers et al., 2012). More recently, a novel role for FGF signaling in ciliary assembly has been hypothesized (Neugebauer, Amack, Peterson, Bisgrove, & Yost, 2009). Fgfr1 morphant zebrafish exhibited reduced expression of the ciliary genes foxj1 and rfx2 as well as diminished ift88 expression, indicating that FGF signaling may exert its effects on cilia by regulating IFT (Neugebauer et al., 2009). Furthermore, cilia length was affected in fgf24/fgf8 double mutants, suggesting that cilia length may be regulated by functional redundancy across FGF ligands. Additional support for this hypothesis comes from studies that report decreased length and number of cilia as a result of knocking down FGF target genes ier2 and fibp1 (Hong & Dawid, 2009). These experiments raise the possibility that FGF-mediated craniofacial malformations could be cilia-related, and in fact, recent studies have begun to substantiate this mechanism (Tabler et al., 2013).
7.4. Platelet-derived growth factor
Aberrant PDGF signaling has been linked with cleft lip/palate in humans (Smith & Tallquist, 2010). Pdgfa/Pdgfc double mutants develop cleft face and cranial bone defects (Ding et al., 2004). Mice deficient in Pdgfra in the neural crest lineage display palatal fusion defects as well as nasal septation and various other anomalies of bone and cartilage (Morrison-Graham, Schatteman, Bork, Bowen-Pope, & Weston, 1992; Soriano, 1997; Tallquist & Soriano, 2003). Primary cilia are required for PDGFα signaling (Schneider et al., 2005, 2009). PDGFα receptor localizes to primary cilium in growth-arrested NIH3T3 cells. Upon signaling activation, the downstream protein Mek1/2 is phosphorylated and localizes to primary cilium. This behavior is lost in cilia mutant Ift88orpk fibroblasts. Moreover, the primary cilium is required for PDGF-AA induced directional cell migration during wound healing (Schneider et al., 2010).
7.5. Notch
Experiments across several species have implicated a role for Notch signaling in NCC induction (Cornell & Eisen, 2002, 2005; Endo, Osumi, & Wakamatsu, 2002, 2003; Ezratty et al., 2011; Glavic, Silva, Aybar, Bastidas, & Mayor, 2004; Leitch, Lodh, Prieto-Echague, Badano, & Zaghloul, 2014; Mead & Yutzey, 2012; Noisa et al., 2014). Notch signaling is required for the induction of NCC-specifier genes in premigratory neural crest-like cells (pNCC) derived from human pluripotent stem cells (hPSC) (Noisa et al., 2014). Activated Notch1 intracellular domain (NICD1) directly binds to and induces expression of NCC-specifier genes. Interestingly, Notch activity is also required for the maintenance of the pNCC fate and the suppression of Notch signaling led to the generation of NCC-derived neurons (Noisa et al., 2014). Furthermore, in avian models, gain and loss of Notch both result in loss of NCC markers (Endo et al., 2002, 2003).
The link between Notch signaling and primary cilia is just beginning to emerge. The Notch3 receptor is enriched in the ciliary membrane while Presenilin-2 (the enzyme responsible for Notch cleavage and activation) is localized to the ciliary basal body. bbs1 and bbs4 morphant zebrafish have increased expression of Notch targets (Leitch et al., 2014), and loss of Bbs1 and Bbs4 caused Notch receptor to accumulate in late endosomes and subsequent lysosomal degradation of Notch receptor was impaired. These experiments strongly suggest that the Notch pathway utilizes the cilium for signal transduction. Uncovering the relationship between Notch signaling, cilia, and NCC development holds great promise for identifying novel molecular mechanism of ciliopathies and possible avenues for therapeutic intervention.
8. CONCLUSIONS
The resurgence of interest in the primary cilium has been impressive and offers new perspectives on how cells sense and respond to their molecular environments. Continued study of primary cilia is vital to both basic science and biomedicine. For basic research, more insight into cilia function will be necessary for truly understanding the molecular mechanisms of signal pathway transduction. For the biomedical community, ciliary research will identify both the basis for ciliopathies as well as potential avenues for therapeutic intervention. Relative to the field of NCC biology, understanding the role primary cilia play during ontogenic development of NCCs could provide novel insights into the etiology of specific neurocristopathies.
Although the field has made significant strides in the last decade, there is still much to learn. For example, why do ciliopathies have differing phenotypes? Why do some ciliopathies have severe craniofacial phenotypes while others do not? Are primary cilia unique depending upon which type of cell they extend from or which signaling pathway they are geared to transduce? In order to address these questions, cilia must be carefully examined both at different time-points during development and on different cell types (Irigoin & Badano, 2011).
The data summarized here emphasize the shift in the current paradigm that primary cilia are homogeneous organelles and that ciliary function can be uniformly defined across multiple tissues and organ systems. Although a plethora of phenotypes have been reported in various ciliopathic animal models (Zaghloul & Brugmann, 2011; Table 1), the perception still exists that disruption of cilia equates simply to a loss of Shh function. Our work, as well as that of many others, suggests that not only will different tissues interpret the loss of cilia in a unique manner, but also the same tissue in different molecular environments will have a unique response to ciliary disruption. It will be of great interest to multiple fields to see what is in store for primary cilia research in the next decade.
REFERENCES
- Abdelhamed ZA, Wheway G, Szymanska K, Natarajan S, Toomes C, Inglehearn C, et al. (2013). Variable expressivity of ciliopathy neurological phenotypes that encompass Meckel-Gruber syndrome and Joubert syndrome is caused by complex de-regulated ciliogenesis, Shh and Wnt signalling defects. Human Molecular Genetics, 22(7), 1358–1372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Afzelius BA (1976). A human syndrome caused by immotile cilia. Science, 193(4250), 317–319. [DOI] [PubMed] [Google Scholar]
- Ahlgren SC, & Bronner-Fraser M (1999). Inhibition of sonic hedgehog signaling in vivo results in craniofacial neural crest cell death. Current Biology, 9(22), 1304–1314. [DOI] [PubMed] [Google Scholar]
- Ahlgren SC, Thakur V, & Bronner-Fraser M (2002). Sonic hedgehog rescues cranial neural crest from cell death induced by ethanol exposure. Proceedings of the National Academy of Sciences of the United States of America, 99(16), 10476–10481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amiel J, Espinosa-Parrilla Y, Steffann J, Gosset P, Pelet A, Prieur M, et al. (2001). Large-scale deletions and SMADIP1 truncating mutations in syndromic Hirschsprung disease with involvement of midline structures. American Journal of Human Genetics, 69(6), 1370–1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avidor-Reiss T, Maer AM, Koundakjian E, Polyanovsky A, Keil T, Subramaniam S, et al. (2004). Decoding cilia function: Defining specialized genes required for compartmentalized cilia biogenesis. Cell, 117(4), 527–539. [DOI] [PubMed] [Google Scholar]
- Basten SG, & Giles RH (2013). Functional aspects of primary cilia in signaling, cell cycle and tumorigenesis. Cilia, 2(1), 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beales PL, Bland E, Tobin JL, Bacchelli C, Tuysuz B, Hill J, et al. (2007). IFT80, which encodes a conserved intraflagellar transport protein, is mutated in Jeune asphyxiating thoracic dystrophy. Nature Genetics, 39(6), 727–729. [DOI] [PubMed] [Google Scholar]
- Beales PL, Elcioglu N, Woolf AS, Parker D, & Flinter FA (1999). New criteria for improved diagnosis of Bardet-Biedl syndrome: Results of a population survey. Journal of Medical Genetics, 36(6), 437–446. [PMC free article] [PubMed] [Google Scholar]
- Beales P, & Jackson PK (2012). Cilia—The prodigal organelle. Cilia, 1(1), 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blitzer AL, Panagis L, Gusella GL, Danias J, Mlodzik M, & Iomini C (2011). Primary cilia dynamics instruct tissue patterning and repair of corneal endothelium. Proceedings of the National Academy of Sciences of the United States of America, 108(7), 2819–2824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloodgood RA (2009). From central to rudimentary to primary: The history of an underappreciated organelle whose time has come. The primary cilium. Methods in Cell Biology, 94, 3–52. [DOI] [PubMed] [Google Scholar]
- Blum JJ, Hayes A, Whisnant CC, & Rosen G (1977). Effect of spin-labeled maleimide on 14S and 30S dyneins in solution and on demembranated ciliary axonemes. Biochemistry, 16(9), 1937–1943. [DOI] [PubMed] [Google Scholar]
- Brault V, Moore R, Kutsch S, Ishibashi M, Rowitch DH, McMahon AP, et al. (2001). Inactivation of the beta-catenin gene by Wnt1-Cre-mediated deletion results in dramatic brain malformation and failure of craniofacial development. Development, 128(8), 1253–1264. [DOI] [PubMed] [Google Scholar]
- Brito JM, Teillet MA, & Le Douarin NM (2008). Induction of mirror-image supernumerary jaws in chicken mandibular mesenchyme by Sonic Hedgehog-producing cells. Development, 135(13), 2311–2319. [DOI] [PubMed] [Google Scholar]
- Bronner ME, & LeDouarin NM (2012). Development and evolution of the neural crest: An overview. Developmental Biology, 366(1), 2–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks ER, & Wallingford JB (2012). Control of vertebrate intraflagellar transport by the planar cell polarity effector Fuz. The Journal of Cell Biology, 198(1), 37–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brugmann SA, Allen NC, James AW, Mekonnen Z, Madan E, & Helms JA (2010). A primary cilia-dependent etiology for midline facial disorders. Human Molecular Genetics, 19(8), 1577–1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brugmann SA, Moore R, Kutsch S, Ishibashi M, Rowitch DH, McMahon AP, et al. (2007). Wnt signaling mediates regional specification in the vertebrate face. Development, 134(18), 3283–3295. [DOI] [PubMed] [Google Scholar]
- Carter CS, Vogel TW, Zhang Q, Seo S, Swiderski RE, Moninger TO, et al. (2012). Abnormal development of NG2+PDGFR-alpha+neural progenitor cells leads to neonatal hydrocephalus in a ciliopathy mouse model. Nature Medicine, 18(12), 1797–1804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caspary T, Larkins CE, & Anderson KV (2007). The graded response to Sonic Hedgehog depends on cilia architecture. Developmental Cell, 12(5), 767–778. [DOI] [PubMed] [Google Scholar]
- Chang CF, Schock EN, O’Hare EA, Dodgson J, Cheng HH, Muir WM, et al. (2014). The cellular and molecular etiology of the craniofacial defects in the avian ciliopathic mutant talpid2. Development, 141(15), 3003–3012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang C, Swan RZ, Grachtchouk M, Bolinger M, Litingtung Y, Robertson EK, et al. (1999). Essential role for Sonic hedgehog during hair follicle morphogenesis. Developmental Biology, 205(1), 1–9. [DOI] [PubMed] [Google Scholar]
- Collin SP, & Barry Collin H (2004). Primary cilia in vertebrate corneal endothelial cells. Cell Biology International, 28(2), 125–130. [DOI] [PubMed] [Google Scholar]
- Corbit KC, Aanstad P, Singla V, Norman AR, Stainier DY, & Reiter JF (2005). Vertebrate smoothened functions at the primary cilium. Nature, 437(7061), 1018–1021. [DOI] [PubMed] [Google Scholar]
- Corbit KC, Shyer AE, Dowdle WE, Gaulden J, Singla V, Chen MH, et al. (2008). Kif3a constrains beta-catenin-dependent Wnt signalling through dual ciliary and non-ciliary mechanisms. Nature Cell Biology, 10(1), 70–76. [DOI] [PubMed] [Google Scholar]
- Cornell RA, & Eisen JS (2002). Delta/Notch signaling promotes formation of zebrafish neural crest by repressing Neurogenin 1 function. Development, 129(11), 2639–2648. [DOI] [PubMed] [Google Scholar]
- Cornell RA, & Eisen JS (2005). Notch in the pathway: The roles of notch signaling in neural crest development. Seminars in Cell & Developmental Biology, 16(6), 663–672. [DOI] [PubMed] [Google Scholar]
- Cortellino S, Wang C, Wang B, Bassi MR, Caretti E, Champeval D, et al. (2009). Defective ciliogenesis, embryonic lethality and severe impairment of the Sonic Hedgehog pathway caused by inactivation of the mouse complex A intraflagellar transport gene Ift122/Wdr10, partially overlapping with the DNA repair gene Med1/Mbd4. Developmental Biology, 325(1), 225–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creuzet S, Schuler B, Couly G, & Le Douarin NM (2004). Reciprocal relationships between Fgf8 and neural crest cells in facial and forebrain development. Proceedings of the National Academy of Sciences of the United States of America, 101(14), 4843–4847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahlstrom JE, Cookman J, & Jain S (2000). Joubert syndrome: An affected female with bilateral colobomata. Pathology, 32(4), 283–285. [PubMed] [Google Scholar]
- Davis EE, Zhang Q, Liu Q, Diplas BH, Davey LM, Hartley J, et al. (2011). TTC21B contributes both causal and modifying alleles across the ciliopathy spectrum. Nature Genetics, 43(3), 189–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawe HR, Farr H, & Gull K (2007). Centriole/basal body morphogenesis and migration during ciliogenesis in animal cells. Journal of Cell Science, 120(Pt. 1), 7–15. [DOI] [PubMed] [Google Scholar]
- Delaval B, Bright A, Lawson ND, & Doxsey S (2011). The cilia protein IFT88 is required for spindle orientation in mitosis. Nature Cell Biology, 13(4), 461–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delous M, Baala L, Salomon R, Laclef C, Vierkotten J, Tory K, et al. (2007). The ciliary gene RPGRIP1L is mutated in cerebello-oculo-renal syndrome (Joubert syndrome type B) and Meckel syndrome. Nature Genetics, 39(7), 875–881. [DOI] [PubMed] [Google Scholar]
- Ding H, Wu X, Boström H, Kim I, Wong N, Tsoi B, et al. (2004). A specific requirement for PDGF-C in palate formation and PDGFR-alpha signaling. Nature Genetics, 36(10), 1111–1116. [DOI] [PubMed] [Google Scholar]
- Doughty MJ (1998). Changes in cell surface primary cilia and microvilli concurrent with measurements of fluid flow across the rabbit corneal endothelium ex vivo. Tissue & Cell, 30(6), 634–643. [DOI] [PubMed] [Google Scholar]
- Endo Y, Osumi N, & Wakamatsu Y (2002). Bimodal functions of Notch-mediated signaling are involved in neural crest formation during avian ectoderm development. Development, 129(4), 863–873. [DOI] [PubMed] [Google Scholar]
- Endo Y, Osumi N, & Wakamatsu Y (2003). Deltex/Dtx mediates NOTCH signaling in regulation of Bmp4 expression in cranial neural crest formation during avian development. Development, Growth & Differentiation, 45(3), 241–248. [DOI] [PubMed] [Google Scholar]
- Ezratty EJ, Stokes N, Chai S, Shah AS, Williams SE, & Fuchs E (2011). A role for the primary cilium in Notch signaling and epidermal differentiation during skin development. Cell, 145(7), 1129–1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feistel K, & Blum M (2006). Three types of cilia including a novel 9+4 axoneme on the notochordal plate of the rabbit embryo. Developmental Dynamics, 235(12), 3348–3358. [DOI] [PubMed] [Google Scholar]
- Ferrante MI, Giorgio G, Feather SA, Bulfone A, Wright V, Ghiani M, et al. (2001). Identification of the gene for oral-facial-digital type I syndrome. American Journal of Human Genetics, 68(3), 569–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrante MI, Romio L, Castro S, Collins JE, Goulding DA, Stemple DL, et al. (2009). Convergent extension movements and ciliary function are mediated by ofd1, a zebrafish orthologue of the human oral-facial-digital type 1 syndrome gene. Human Molecular Genetics, 18(2), 289–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrante MI, Zullo A, Barra A, Bimonte S, Messaddeq N, Studer M, et al. (2006). Oral-facial-digital type I protein is required for primary cilia formation and left-right axis specification. Nature Genetics, 38(1), 112–117. [DOI] [PubMed] [Google Scholar]
- Fried K, Liban E, Lurie M, Friedman S, & Reisner SH (1971). Polycystic kidneys associated with malformations of the brain, polydactyly, and other birth defects in newborn sibs. A lethal syndrome showing the autosomal-recessive pattern of inheritance. Journal of Medical Genetics, 8(3), 285–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedland-Little JM, Hoffmann AD, Ocbina PJ, Peterson MA, Bosman JD, Chen Y, et al. (2011). A novel murine allele of Intraflagellar Transport Protein 172 causes a syndrome including VACTERL-like features with hydrocephalus. Human Molecular Genetics, 20(19), 3725–3737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Castro MI, Marcelle C, & Bronner-Fraser M (2002). Ectodermal Wnt function as a neural crest inducer. Science, 13, 13. [DOI] [PubMed] [Google Scholar]
- Glavic A, Silva F, Aybar MJ, Bastidas F, & Mayor R (2004). Interplay between Notch signaling and the homeoprotein Xiro1 is required for neural crest induction in Xenopus embryos. Development, 131(2), 347–359. [DOI] [PubMed] [Google Scholar]
- Gorlin RJ, Cohen MM, & Levin LS (Eds.), (1990). Syndromes of the head and neck. Oxfordmonographs on medical genetics, 1 (3rd ed.). New York: Oxford University Press. [Google Scholar]
- Goto H, Inoko A, & Inagaki M (2013). Cell cycle progression by the repression of primary cilia formation in proliferating cells. Cellular and Molecular Life Sciences, 70(20), 3893–3905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray RS, Abitua PB, Wlodarczyk BJ, Szabo-Rogers HL, Blanchard O, Lee I, et al. (2009). The planar cell polarity effector Fuz is essential for targeted membrane trafficking, ciliogenesis and mouse embryonic development. Nature Cell Biology, 11(10), 1225–1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurrieri F, Franco B, Toriello H, & Neri G (2007). Oral-facial-digital syndromes: Review and diagnostic guidelines. American Journal of Medical Genetics Part A, 143A(24), 3314–3323. [DOI] [PubMed] [Google Scholar]
- Han YG, Kwok BH, & Kernan MJ (2003). Intraflagellar transport is required in Drosophila to differentiate sensory cilia but not sperm. Current Biology, 13(19), 1679–1686. [DOI] [PubMed] [Google Scholar]
- Han A, Zhao H, Li J, Pelikan R, & Chai Y (2014). ALK5-mediated TGF-beta signaling in neural crest cells controls craniofacial muscle development via tissue-tissue interactions. Molecular and Cellular Biology, 34, 3120–3131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris MP, Hasso SM, Ferguson MW, & Fallon JF (2006). The development of archosaurian first-generation teeth in a chicken mutant. Current Biology, 16(4), 371–377. [DOI] [PubMed] [Google Scholar]
- Haycraft CJ, Banizs B, Aydin-Son Y, Zhang Q, Michaud EJ, & Yoder BK (2005). Gli2 and Gli3 localize to cilia and require the intraflagellar transport protein polaris for processing and function. PLoS Genetics, 1(4), e53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He F, & Soriano P (2013). A critical role for PDGFRalpha signaling in medial nasal process development. PLoS Genetics, 9(9), e1003851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez-Hernandez V, Pravincumar P, Diaz-Font A, May-Simera H, Jenkins D, Knight M, et al. (2013). Bardet-Biedl syndrome proteins control the cilia length through regulation of actin polymerization. Human Molecular Genetics, 22(19), 3858–3868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoey DA, Downs ME, & Jacobs CR (2012). The mechanics of the primary cilium: An intricate structure with complex function. Journal of Biomechanics, 45(1), 17–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong SK, & Dawid IB (2009). FGF-dependent left-right asymmetry patterning in zebrafish is mediated by Ier2 and Fibp1. Proceedings of the National Academy of Sciences of the United States of America, 106(7), 2230–2235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houde C, Dickinson RJ, Houtzager VM, Cullum R, Montpetit R, Metzler M, et al. (2006). Hippi is essential for node cilia assembly and Sonic hedgehog signaling. Developmental Biology, 300(2), 523–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X, Goudy SL, Ketova T, Litingtung Y, & Chiang C (2008). Gli3-deficient mice exhibit cleft palate associated with abnormal tongue development. Developmental Dynamics, 237(10), 3079–3087. [DOI] [PubMed] [Google Scholar]
- Huang P, & Schier AF (2009). Dampened hedgehog signaling but normal Wnt signaling in zebrafish without cilia. Development, 136(18), 3089–3098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang R, Zhi Q, Izpisua-Belmonte JC, Christ B, & Patel K (1999). Origin and development of the avian tongue muscles. Anatomy and Embryology, 200(2), 137–152. [DOI] [PubMed] [Google Scholar]
- Huangfu D, & Anderson KV (2005). Cilia and Hedgehog responsiveness in the mouse. Proceedings of the National Academy of Sciences of the United States of America, 102(32), 11325–11330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huangfu D, Liu A, Rakeman AS, Murcia NS, Niswander L, & Anderson KV (2003). Hedgehog signalling in the mouse requires intraflagellar transport proteins. Nature, 426(6962), 83–87. [DOI] [PubMed] [Google Scholar]
- Irigoin F, & Badano JL (2011). Keeping the balance between proliferation and differentiation: The primary cilium. Current Genomics, 12(4), 285–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong J, Mao J, Tenzen T, Kottmann AH, & McMahon AP (2004). Hedgehog signaling in the neural crest cells regulates the patterning and growth of facial primordia. Genes & Development, 18(8), 937–951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones C, Roper VC, Foucher I, Qian D, Banizs B, Petit C, et al. (2008). Ciliary proteins link basal body polarization to planar cell polarity regulation. Nature Genetics, 40(1), 69–77. [DOI] [PubMed] [Google Scholar]
- Kjaer KW, Hansen BF, Keeling JW, Nolting D, & Kjaer I (1999). Malformations of cranial base structures and pituitary gland in prenatal Meckel syndrome. APMIS, 107(10), 937–944. [DOI] [PubMed] [Google Scholar]
- Kobayashi T, & Dynlacht BD (2011). Regulating the transition from centriole to basal body. The Journal of Cell Biology, 193(3), 435–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kollar EJ, & Fisher C (1980). Tooth induction in chick epithelium: Expression of quiescent genes for enamel synthesis. Science, 207(4434), 993–995. [DOI] [PubMed] [Google Scholar]
- Kollar EJ, & Mina M (1991). Role of the early epithelium in the patterning of the teeth and Meckel’s cartilage. Journal of Craniofacial Genetics and Developmental Biology, 11(4), 223–228. [PubMed] [Google Scholar]
- Konno A, Setou M, & Ikegami K (2012). Ciliary and flagellar structure and function–their regulations by posttranslational modifications of axonemal tubulin. International Review of Cell and Molecular Biology, 294, 133–170. [DOI] [PubMed] [Google Scholar]
- Kuriyama S, & Mayor R (2008). Molecular analysis of neural crest migration. Philosophical Transactions of the Royal Society of London, Series B: Biological Sciences, 363(1495), 1349–1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaBonne C, & Bronner-Fraser M (1999). Molecular mechanisms of neural crest formation. Annual Review of Cell and Developmental Biology, 15, 81–112. [DOI] [PubMed] [Google Scholar]
- Le Coz M, Benmerah A, & Larue L (2014). Quiescent melanocytes form primary cilia. Experimental Dermatology, 23(6), 426–427. [DOI] [PubMed] [Google Scholar]
- Leitch CC, Lodh S, Prieto-Echague V, Badano JL, & Zaghloul NA (2014). Basal body proteins regulate Notch signaling through endosomal trafficking. Journal of Cell Science, 127(Pt. 11), 2407–2419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu A, Wang B, & Niswander LA (2005). Mouse intraflagellar transport proteins regulate both the activator and repressor functions of Gli transcription factors. Development, 132(13), 3103–3111. [DOI] [PubMed] [Google Scholar]
- Lofterod B, Riise R, Skuseth T, & Storhaug K (1990). Laurence-Moon-Bardet-Biedl syndrome. Nordisk Medicin, 105(5), 146–148. [PubMed] [Google Scholar]
- Lunt SC, Haynes T, & Perkins BD (2009). Zebrafish ift57, ift88, and ift172 intraflagellar transport mutants disrupt cilia but do not affect hedgehog signaling. Developmental Dynamics, 238(7), 1744–1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macatee TL, Macatee TL, Hammond BP, Arenkiel BR, Francis L, Frank DU, et al. (2003). Ablation of specific expression domains reveals discrete functions of ectoderm- and endoderm-derived FGF8 during cardiovascular and pharyngeal development. Development, 130(25), 6361–6374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madhivanan K, Mukherjee D, & Aguilar RC (2012). Lowe syndrome: Between primary cilia assembly and Rac1-mediated membrane remodeling. Communicative & Integrative Biology, 5(6), 641–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchant L, Linker C, Ruiz P, Guerrero N, & Mayor R (1998). The inductive properties of mesoderm suggest that the neural crest cells are specified by a BMP gradient. Developmental Biology, 198(2), 319–329. [PubMed] [Google Scholar]
- Marcucio R, & Helms JA (2002). Shh-dependent signals from forebrain direct facial development. Unpublished observations. [Google Scholar]
- May SR, Ashique AM, Karlen M, Wang B, Shen Y, Zarbalis K, et al. (2005). Loss of the retrograde motor for IFT disrupts localization of Smo to cilia and prevents the expression of both activator and repressor functions of Gli. Developmental Biology, 287(2), 378–389. [DOI] [PubMed] [Google Scholar]
- Mayor R, Guerrero N, & Martinez C (1997). Role of FGF and noggin in neural crest induction. Developmental Biology, 189(1), 1–12. [DOI] [PubMed] [Google Scholar]
- Mayor R, & Theveneau E (2014). The role of the non-canonical Wnt-planar cell polarity pathway in neural crest migration. The Biochemical Journal, 457(1), 19–26. [DOI] [PubMed] [Google Scholar]
- McDermott KM, Liu BY, Tlsty TD, & Pazour GJ (2010). Primary cilia regulate branching morphogenesis during mammary gland development. Current Biology, 20(8), 731–737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGrath J, Somlo S, Makova S, Tian X, & Brueckner M (2003). Two populations of node monocilia initiate left-right asymmetry in the mouse. Cell, 114(1), 61–73. [DOI] [PubMed] [Google Scholar]
- McKeown SJ, Stamp L, Hao MM, & Young HM (2013). Hirschsprung disease: A developmental disorder of the enteric nervous system. Wiley Interdisciplinary Reviews Developmental Biology, 2(1), 113–129. [DOI] [PubMed] [Google Scholar]
- Mead TJ, & Yutzey KE (2012). Notch pathway regulation of neural crest cell development in vivo. Developmental Dynamics, 241(2), 376–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyoshi K, Kasahara K, Miyazaki I, & Asanuma M (2009). Lithium treatment elongates primary cilia in the mouse brain and in cultured cells. Biochemical and Biophysical Research Communications, 388(4), 757–762. [DOI] [PubMed] [Google Scholar]
- Miyoshi K, Kasahara K, Miyazaki I, & Asanuma M (2011). Factors that influence primary cilium length. Acta Medica Okayama, 65(5), 279–285. [DOI] [PubMed] [Google Scholar]
- Moerman P, Verbeken E, Fryns JP, Goddeeris P, & Lauweryns JM (1982). The Meckel syndrome. Pathological and cytogenetic observations in eight cases. Human Genetics, 62(3), 240–245. [DOI] [PubMed] [Google Scholar]
- Molla-Herman A, Ghossoub R, Blisnick T, Meunier A, Serres C, Silbermann F, et al. (2010). The ciliary pocket: An endocytic membrane domain at the base of primary and motile cilia. Journal of Cell Science, 123(Pt. 10), 1785–1795. [DOI] [PubMed] [Google Scholar]
- Monsoro-Burq AH, Fletcher RB, & Harland RM (2003). Neural crest induction by paraxial mesoderm in Xenopus embryos requires FGF signals. Development, 130(14), 3111–3124. [DOI] [PubMed] [Google Scholar]
- Morrison-Graham K, Schatteman GC, Bork T, Bowen-Pope DF, & Weston JA (1992). A PDGF receptor mutation in the mouse (Patch) perturbs the development of a non-neuronal subset of neural crest-derived cells. Development, 115(1), 133–142. [DOI] [PubMed] [Google Scholar]
- Mostafa MI, Temtamy SA, el-Gammal MA, & Mazen IM (2005). Unusual pattern of inheritance and orodental changes in the Ellis-van Creveld syndrome. Genetic Counseling, 16(1), 75–83. [PubMed] [Google Scholar]
- Mukhopadhyay S, Wen X, Ratti N, Loktev A, Rangell L, Scales SJ, et al. (2013). The ciliary G-protein-coupled receptor Gpr161 negatively regulates the Sonic hedgehog pathway via cAMP signaling. Cell, 152(1–2), 210–223. [DOI] [PubMed] [Google Scholar]
- Murcia NS, Richards WG, Yoder BK, Mucenski ML, Dunlap JR, & Woychik RP (2000). The Oak Ridge Polycystic Kidney (orpk) disease gene is required for left-right axis determination. Development, 127(11), 2347–2355. [DOI] [PubMed] [Google Scholar]
- Nachury MV, Loktev AV, Zhang Q, Westlake CJ, Peränen J, Merdes A, et al. (2007). A core complex of BBS proteins cooperates with the GTPase Rab8 to promote ciliary membrane biogenesis. Cell, 129(6), 1201–1213. [DOI] [PubMed] [Google Scholar]
- Nauli SM, Alenghat FJ, Luo Y, Williams E, Vassilev P, Li X, et al. (2003). Polycystins 1 and 2 mediate mechanosensation in the primary cilium of kidney cells. Nature Genetics, 33(2), 129–137. [DOI] [PubMed] [Google Scholar]
- Neesen J, Kirschner R, Ochs M, Schmiedl A, Habermann B, Mueller C, et al. (2001). Disruption of an inner arm dynein heavy chain gene results in asthenozoospermia and reduced ciliary beat frequency. Human Molecular Genetics, 10(11), 1117–1128. [DOI] [PubMed] [Google Scholar]
- Neugebauer JM, Amack JD, Peterson AG, Bisgrove BW, & Yost HJ (2009). FGF signalling during embryo development regulates cilia length in diverse epithelia. Nature, 458(7238), 651–654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noisa P, Lund C, Kanduri K, Lund R, Lähdesmäki H, Lahesmaa R, et al. (2014). Notch signaling regulates the differentiation of neural crest from human pluripotent stem cells. Journal of Cell Science, 127(Pt. 9), 2083–2094. [DOI] [PubMed] [Google Scholar]
- Ocbina PJ, Tuson M,& Anderson KV (2009). Primary cilia are not required for normal canonical Wnt signaling in the mouse embryo. PLoS One, 4(8), e6839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohazama A, Haycraft CJ, Seppala M, Blackburn J, Ghafoor S, Cobourne M, et al. (2009). Primary cilia regulate Shh activity in the control of molar tooth number. Development, 136(6), 897–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osborn DP, Roccasecca RM, McMurray F, Hernandez-Hernandez V, Mukherjee S, Barroso I, et al. (2014). Loss of FTO antagonises Wnt signaling and leads to developmental defects associated with ciliopathies. PLoS One, 9(2), e87662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otto EA, Schermer B, Obara T, O’Toole JF, Hiller KS, Mueller AM, et al. (2003). Mutations in INVS encoding inversin cause nephronophthisis type 2, linking renal cystic disease to the function of primary cilia and left-right axis determination. Nature Genetics, 34(4), 413–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou Y, Ruan Y, Cheng M, Moser JJ, Rattner JB, & van der Hoorn FA (2009). Adenylate cyclase regulates elongation of mammalian primary cilia. Experimental Cell Research, 315(16), 2802–2817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parada C, Han D, & Chai Y (2012). Molecular and cellular regulatory mechanisms of tongue myogenesis. Journal of Dental Research, 91(6), 528–535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parisi MA (2009). Clinical and molecular features of Joubert syndrome and related disorders. American Journal of Medical Genetics Part C: Seminars in Medical Genetics, 151C(4), 326–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park TJ, Haigo SL, & Wallingford JB (2006). Ciliogenesis defects in embryos lacking inturned or fuzzy function are associated with failure of planar cell polarity and Hedgehog signaling. Nature Genetics, 38(3), 303–311. [DOI] [PubMed] [Google Scholar]
- Pellegrino JE, Lensch MW, Muenke M, & Chance PF (1997). Clinical and molecular analysis in Joubert syndrome. American Journal of Medical Genetics, 72(1), 59–62. [DOI] [PubMed] [Google Scholar]
- Ray K, Perez SE, Yang Z, Xu J, Ritchings BW, Steller H, et al. (1999). Kinesin-II is required for axonal transport of choline acetyltransferase in Drosophila. The Journal of Cell Biology, 147(3), 507–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehder H, & Labbe F (1981). Prenatal morphology in Meckel’s syndrome. Prenatal Diagnosis, 1(3), 161–171. [DOI] [PubMed] [Google Scholar]
- Reiter JF, Blacque OE, & Leroux MR (2012). The base of the cilium: Roles for transition fibres and the transition zone in ciliary formation, maintenance and compartmentalization. EMBO Reports, 13(7), 608–618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robbins DJ, Fei DL, & Riobo NA (2012). The Hedgehog signal transduction network. Science Signaling, 5(246), re6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert A, Margall-Ducos G, Guidotti JE, Brégerie O, Celati C, Bréchot C, et al. (2007). The intraflagellar transport component IFT88/polaris is a centrosomal protein regulating G1-S transition in non-ciliated cells. Journal of Cell Science, 120(Pt. 4), 628–637. [DOI] [PubMed] [Google Scholar]
- Rogers CD, Jayasena CS, Nie S, & Bronner ME (2012). Neural crest specification: Tissues, signals, and transcription factors. Wiley Interdisciplinary Review Developmental Biology, 1(1), 52–68. [DOI] [PubMed] [Google Scholar]
- Rohatgi R, Milenkovic L, & Scott MP (2007). Patched1 regulates hedgehog signaling at the primary cilium. Science, 317(5836), 372–376. [DOI] [PubMed] [Google Scholar]
- Ross AJ, May-Simera H, Eichers ER, Kai M, Hill J, Jagger DJ, et al. (2005). Disruption of Bardet-Biedl syndrome ciliary proteins perturbs planar cell polarity in vertebrates. Nature Genetics, 37(10), 1135–1140. [DOI] [PubMed] [Google Scholar]
- Ruiz-Perez VL, Blair HJ, Rodriguez-Andres ME, Blanco MJ, Wilson A, Liu YN, et al. (2007). Evc is a positive mediator of Ihh-regulated bone growth that localises at the base of chondrocyte cilia. Development, 134(16), 2903–2912. [DOI] [PubMed] [Google Scholar]
- Schneider L, Cammer M, Lehman J, Nielsen SK, Guerra CF, Veland IR, et al. (2010). Directional cell migration and chemotaxis in wound healing response to PDGF-AA are coordinated by the primary cilium in fibroblasts. Cellular Physiology and Biochemistry, 25(2—3), 279–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider L, Clement CA, Teilmann SC, Pazour GJ, Hoffmann EK, Satir P, et al. (2005). PDGFRalphaalpha signaling is regulated through the primary cilium in fibroblasts. Current Biology, 15(20), 1861–1866. [DOI] [PubMed] [Google Scholar]
- Schneider L, Stock CM, Dieterich P, Jensen BH, Pedersen LB, Satir P, et al. (2009). The Na+/H+ exchanger NHE1 is required for directional migration stimulated via PDGFR-alpha in the primary cilium. The Journal of Cell Biology, 185(1), 163–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simons M, Gloy J, Ganner A, Bullerkotte A, Bashkurov M, Krönig C, et al. (2005). Inversin, the gene product mutated in nephronophthisis type II, functions as a molecular switch between Wnt signaling pathways. Nature Genetics, 37(5), 537–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith CL, & Tallquist MD (2010). PDGF function in diverse neural crest cell populations. Cell Adhesion & Migration, 4(4), 561–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soriano P (1997). The PDGF alpha receptor is required for neural crest cell development and for normal patterning of the somites. Development, 124(14), 2691–2700. [DOI] [PubMed] [Google Scholar]
- Sorokin S (1962). Centrioles and the formation of rudimentary cilia by fibroblasts and smooth muscle cells. The Journal of Cell Biology, 15, 363–377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szabo-Rogers HL, Geetha-Loganathan P, Nimmagadda S, Fu KK,& Richman JM (2008). FGF signals from the nasal pit are necessary for normal facial morphogenesis. Developmental Biology, 318(2), 289–302. [DOI] [PubMed] [Google Scholar]
- Tabler JM, Barrell WB, Szabo-Rogers HL, Healy C, Yeung Y, Perdiguero EG, et al. (2013). Fuz mutant mice reveal shared mechanisms between ciliopathies and FGF-related syndromes. Developmental Cell, 25(6), 623–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tallquist MD, & Soriano P (2003). Cell autonomous requirement for PDGFRalpha in populations of cranial and cardiac neural crest cells. Development, 130(3), 507–518. [DOI] [PubMed] [Google Scholar]
- Tang Z, Lin MG, Stowe TR, Chen S, Zhu M, Stearns T, et al. (2013). Autophagy promotes primary ciliogenesis by removing OFD1 from centriolar satellites. Nature, 502(7470), 254–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Z, Zhu M, & Zhong Q (2014). Self-eating to remove cilia roadblock. Autophagy, 10(2), 379–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teddy JM, & Kulesa PM (2004). In vivo evidence for short- and long-range cell communication in cranial neural crest cells. Development, 131(24), 6141–6151. [DOI] [PubMed] [Google Scholar]
- Thauvin-Robinet C, Lee JS, Lopez E, Herranz-Pérez V, Shida T, Franco B, et al. (2014). The oral-facial-digital syndrome gene C2CD3 encodes a positive regulator of centriole elongation. Nature Genetics, 46(8), 905–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thesleff I (2006). The genetic basis of tooth development and dental defects. American Journal of Medical Genetics Part A, 140(23), 2530–2535. [DOI] [PubMed] [Google Scholar]
- Theveneau E, & Mayor R (2012). Neural crest migration: Interplay between chemorepellents, chemoattractants, contact inhibition, epithelial-mesenchymal transition, and collective cell migration. Wiley Interdisciplinary Reviews Developmental Biology, 1(3), 435–445. [DOI] [PubMed] [Google Scholar]
- Tobin JL, Di Franco M, Eichers E, May-Simera H, Garcia M, Yan J, et al. (2008). Inhibition of neural crest migration underlies craniofacial dysmorphology and Hirschsprung’s disease in Bardet-Biedl syndrome. Proceedings of the National Academy of Sciences of the United States of America, 105(18), 6714–6719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran PV, Talbott GC, Turbe-Doan A, Jacobs DT, Schonfeld MP, Silva LM, et al. (2014). Downregulating hedgehog signaling reduces renal cystogenic potential of mouse models. Journal of the American Society of Nephrology, 25(10), 2201–2212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran PV, Haycraft CJ, Besschetnova TY, Turbe-Doan A, Stottmann RW, Herron BJ, et al. (2008). THM1 negatively modulates mouse sonic hedgehog signal transduction and affects retrograde intraflagellar transport in cilia. Nature Genetics, 40(4), 403–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trumpp A, Depew MJ, Rubenstein JL, Bishop JM, & Martin GR (1999). Cre-mediated gene inactivation demonstrates that FGF8 is required for cell survival and patterning of the first branchial arch. Genes and Development, 13(23), 3136–3148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu CT, Yang TC, Huang HY, & Tsai HJ (2012). Zebrafish arl6ip1 is required for neural crest development during embryogenesis. PLoS One, 7(3), e32899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valente EM, Logan CV, Mougou-Zerelli S, Lee JH, Silhavy JL, Brancati F, et al. (2010). Mutations in TMEM216 perturb ciliogenesis and cause Joubert, Meckel and related syndromes. Nature Genetics, 42(7), 619–625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veland IR, Montjean R, Eley L, Pedersen LB, Schwab A, Goodship J, et al. (2013). Inversin/Nephrocystin-2 is required for fibroblast polarity and directional cell migration. PLoS One, 8(4), e60193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verghese E, Weidenfeld R, Bertram JF, Ricardo SD, & Deane JA (2008). Renal cilia display length alterations following tubular injury and are present early in epithelial repair. Nephrology, Dialysis, Transplantation, 23(3), 834–841. [DOI] [PubMed] [Google Scholar]
- Verghese E, Zhuang J, Saiti D, Ricardo SD, & Deane JA (2011). In vitro investigation of renal epithelial injury suggests that primary cilium length is regulated by hypoxia-inducible mechanisms. Cell Biology International, 35(9), 909–913. [DOI] [PubMed] [Google Scholar]
- Wada N, Javidan Y, Nelson S, Carney TJ, Kelsh RN, & Schilling TF (2005). Hedgehog signaling is required for cranial neural crest morphogenesis and chondrogenesis at the midline in the zebrafish skull. Development, 132(17), 3977–3988. [DOI] [PubMed] [Google Scholar]
- Walczak-Sztulpa J, Eggenschwiler J, Osborn D, Brown DA, Emma F, Klingenberg C, et al. (2010). Cranioectodermal Dysplasia, Sensenbrenner syndrome, is a ciliopathy caused by mutations in the IFT122 gene. American Journal of Human Genetics, 86(6), 949–956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wandel A, Steigleder GK, & Bodeux E (1984). Primary cilia in cells of the epidermis and dermis. Zeitschrift für Hautkrankheiten, 59(6), 389–392. [PubMed] [Google Scholar]
- Watanabe D, Saijoh Y, Nonaka S, Sasaki G, Ikawa Y, Yokoyama T, et al. (2003). The left-right determinant Inversin is a component of node monocilia and other 9+0 cilia. Development, 130(9), 1725–1734. [DOI] [PubMed] [Google Scholar]
- Weatherbee SD, Niswander LA, & Anderson KV (2009). A mouse model for Meckel syndrome reveals Mks1 is required for ciliogenesis and Hedgehog signaling. Human Molecular Genetics, 18(23), 4565–4575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen X, Lai CK, Evangelista M, Hongo JA, de Sauvage FJ, & Scales SJ (2010). Kinetics of hedgehog-dependent full-length Gli3 accumulation in primary cilia and subsequent degradation. Molecular and Cellular Biology, 30(8), 1910–1922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheatley DN (1969). Cilia in cell-cultured fibroblasts. I. On their occurrence and relative frequencies in primary cultures and established cell lines. Journal of Anatomy, 105(Pt. 2), 351–362. [PMC free article] [PubMed] [Google Scholar]
- Wheatley DN, Wang AM, & Strugnell GE (1996). Expression of primary cilia in mammalian cells. Cell Biology International, 20(1), 73–81. [DOI] [PubMed] [Google Scholar]
- Willaredt MA, Gorgas K, Gardner HA, & Tucker KL (2012). Multiple essential roles for primary cilia in heart development. Cilia, 1(1), 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willaredt MA, Hasenpusch-Theil K, Gardner HA, Kitanovic I, Hirschfeld-Warneken VC, Gojak CP, et al. (2008). A crucial role for primary cilia in cortical morphogenesis. The Journal of Neuroscience, 28(48), 12887–12900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong SY, Seol AD, So PL, Ermilov AN, Bichakjian CK, Epstein EH Jr., et al. (2009). Primary cilia can both mediate and suppress Hedgehog pathway-dependent tumorigenesis. Nature Medicine, 15(9), 1055–1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamazaki H, Nakata T, Okada Y, & Hirokawa N (1995). KIF3A/B: A heterodimeric kinesin superfamily protein that works as a microtubule plus end-directed motor for membrane organelle transport. The Journal of Cell Biology, 130(6), 1387–1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Y, Bangs F, Paton IR, Prescott A, James J, Davey MG, et al. (2009). The Talpid3 gene (KIAA0586) encodes a centrosomal protein that is essential for primary cilia formation. Development, 136(4), 655–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaghloul NA, & Brugmann SA (2011). The emerging face of primary cilia. Genesis, 49(4), 231–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q, Murcia NS, Chittenden LR, Richards WG, Michaud EJ, Woychik RP, et al. (2003). Loss of the Tg737 protein results in skeletal patterning defects. Developmental Dynamics, 227(1), 78–90. [DOI] [PubMed] [Google Scholar]
- Zimmermann KW (1898). Beitrage zur Kenntnis einiger Drusen und Epithelien. In 52. Arch. mikr. Anat. u Entwick (p. 552). [Google Scholar]