Abstract
Background
Previous studies have confirmed the oncogenic role of HMGB2 in various cancers, but the biological functions of HMGB2-derived circRNAs remain unknown. Thus, we intended to investigate the potential role of HMGB2-derived circRNAs in lung adenocarcinomas (LUAD) and squamous cell carcinomas (LUSC).
Methods
The expression profiles of HMGB2-derived circRNAs in LUAD and LUSC tissues and matched normal tissues were assessed using qRT–PCR. The role of circHMGB2 in the progression of the LUAD and LUSC was determined in vitro by Transwell, CCK-8, flow cytometry and immunohistochemistry assays, as well as in vivo in an immunocompetent mouse model and a humanized mouse model. In addition, in vivo circRNA precipitation assays, luciferase reporter assays and RNA pulldown assays were performed to explore the underlying mechanism by which circHMGB2 promotes anti-PD-1 resistance in the LUAD and LUSC.
Results
The expression of circHMGB2 (hsa_circ_0071452) was significantly upregulated in NSCLC tissues, and survival analysis identified circHMGB2 as an independent indicator of poor prognosis in the LUAD and LUSC patients. We found that circHMGB2 exerted a mild effect on the proliferation of the LUAD and LUSC cells, but circHMGB2 substantially reshaped the tumor microenvironment by contributing to the exhaustion of antitumor immunity in an immunocompetent mouse model and a humanized mouse model. Mechanistically, circHMGB2 relieves the inhibition of downstream CARM1 by sponging miR-181a-5p, thus inactivating the type 1 interferon response in the LUAD and LUSC. Moreover, we found that the upregulation of circHMGB2 expression decreased the efficacy of anti-PD-1 therapy, and we revealed that the combination of the CARM1 inhibitor EZM2302 and an anti-PD-1 antibody exerted promising synergistic effects in a preclinical model.
Conclusion
circHMGB2 overexpression promotes the LUAD and LUSC progression mainly by reshaping the tumor microenvironment and regulating anti-PD-1 resistance in the LUAD and LUSC patients. This study provides a new strategy for the LUAD and LUSC treatment.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12943-022-01586-w.
Keywords: circHMGB2, CARM1, NSCLC, TME, Anti-PD-1 treatment
Introduction
Lung cancer remains one of the most common malignancies, and it is the leading cause of cancer-related death in the world; non-small cell lung cancer (NSCLC) mainly including lung adenocarcinomas (LUAD) and squamous cell carcinomas (LUSC) accounts for approximately 85% of all cases of lung cancer [1]. While curative resection by lobectomy or segmentectomy has achieved promising effects in patients with early NSCLC, the overall prognosis of NSCLC remains poor; the overall 5-year survival rate is only 16% since most patients are diagnosed at an advanced stage due to the lack of symptoms of early NSCLC [1, 2]. Thus, further investigation of the oncogenesis and progression of NSCLC could have potential clinical value and may facilitate the development of new therapeutic strategies for patients with NSCLC.
Circular RNAs (circRNAs) are a class of regulatory RNAs that are characterized by a covalently closed loop structure; circRNAs are generated by the backsplicing of exons in precursor mRNA [3]. Previous studies have revealed the potential effects of circRNAs in various biological processes, such as cardiovascular disease [4], immunity [5] and cancer [6]. The biological functions and relevant mechanisms of circRNAs in various cancers have been thoroughly investigated, and the findings have provided new directions for the development of treatment strategies and cancer biomarkers. For instance, circRanGAP1 facilitates the progression of gastric cancer via the miR-887-3p/VEGFA axis [7]. Circ-cRAPGEF5 inhibits the invasion of renal cell carcinoma cells by sponging miR-27a-3p to block the inhibition of the downstream target TXNIP [8]. While some studies have reported a role of circRNAs in the oncogenesis and progression of NSCLC, the potential roles of most circRNAs in the development of NSCLC remain unclear and require further investigation [6].
High-mobility group box 2 (HMGB2) is a ubiquitous nuclear protein in humans that is responsible for the activation of chromatin domains [9]. HMGB2 is highly conserved and has universal biological characteristics, such as its ability to bind to DNA without sequence specificity [10]. Previous studies have identified HMGB2 as an oncogene that is associated with the poor prognosis of various cancers, including hepatocellular carcinoma, breast cancer and NSCLC [11–13]. Thus, we wondered whether the circRNAs transcribed by HMGB2 contribute to the progression of NSCLC (here indicated LUAD and LUSC). In this study, we compared the expression profiles of circRNAs transcribed by HMGB2 in NSCLC tissues and paired normal tissues and observed a significant upregulation of circHMGB2 (hsa_circ_0071452) in NSCLC tissues. Interestingly, the high expression of circHMGB2 indicated a poor prognosis of NSCLC patients and contributed to the malignant properties of NSCLC by supporting an immunosuppressive microenvironment. Mechanistically, HMGB2 sponged miR-181a-5p and further blocked the inhibition of the downstream molecule CARM1, which is responsible for the inactivation of the type 1 interferon (IFN) response; thus, HMGB2 led to the immune evasion of NSCLC. Therefore, this study presents circHMGB2 as a potential molecular target for immunotherapy in NSCLC.
Methods
Cell lines
The cell lines used in this study included the human NSCLC cell lines A549, PC-9, NCI-H460, NCI-H1299, NCI-H1703 and human bronchial epithelial (HBE) cells. The mouse lung cancer cell lines LLC and HEK-293 T were obtained from the Cell Bank of the Chinese Academy of Sciences (Shanghai, China). The cells were cultivated in DMEM and RPMI-1640 (Gibco, USA) supplemented with 10% fetal bovine serum (Gibco, USA) and 1% penicillin–streptomycin (Yeasen, Shanghai, China). The environmental conditions were 37 °C and 5% CO2.
Patients and follow-up
A total of 120 pairs of NSCLC tissues that were identified by two experienced pathologists were collected from patients who underwent lobectomy or segmentectomy at the Second Affiliated Hospital of Nanchang University from 2013 to 2014. These tissues were used to construct a tissue microarray (TMA). The TMA was used to perform immunohistochemistry (IHC) staining. The last follow-up occurred in June 2019. The Ethics Committee of the Second Affiliated Hospital of Nanchang University approved the human ethics-related protocols of this study, and informed consent was obtained from all the patients.
Agarose gel electrophoresis, Western blotting, quantitative real-time polymerase chain reaction (qRT–PCR), IHC, fluorescence in situ hybridization (FISH), wound healing assays, Matrigel Transwell assays, cell counting Kit-8 (CCK-8) assays and colony formation assays
These experiments were performed following protocols described in previous studies [14], and the details are provided in the Supplementary Methods and Materials. Information about the antibodies and primers is presented in supplementary Tables 1 and 2. The probe sequences for circHMGB2 and miR-181a-5p are listed in Supplementary Table 3.
Transfection of lentiviral vectors and generation of CRISPR Cas9-edited LLC and A549 cells
The lentiviral vectors carrying circHMGB2 and shcircHMGB2 were purchased from Genomeditech (Shanghai, China) and used to establish stably transfected cell lines. The transfection efficiency was assessed by qRT–PCR. Knockout of the CARM1 gene in the LLC and A549 cell lines via the CRISPR Cas9-gDNA system was accomplished by Genomeditech, and western blotting was used to determine the efficacy. The sequences of shcircHMGB2 and sgCARM1 are listed in Supplementary Tables 4 and 5.
In vivo circRNA precipitation (circRIP) and RNA immunoprecipitation (RIP)
The experimental procedures for the circRIP and RIP assays were described in a previous study [14]. For the circRIP assay, the biotin-labeled circHMGB2 probe was purchased from Gene-Chem (Shanghai, China). Briefly, after transfection with the biotin-circHMGB2 and NC probes, H1299 cells were fixed with 4% formaldehyde, lysed, sonicated and centrifuged. Next, the supernatant was incubated with M280 streptavidin Dynabeads (Invitrogen) for 12 h. Then, the mixture was washed and suspended in lysis buffer. The total RNA was extracted from the mixture with TRIzol reagent (Invitrogen).
For the RIP assay, the Magna RIP kit (Millipore, USA) was used to enrich circHMGB2 and miRNA. Total RNA was extracted with TRIzol reagent, and the levels of the target circRNAs and miRNAs were measured by qRT–PCR.
RNA pull-down assay and co-IP combined with MS
The pull-down assay was performed according to a previous study [15]. First, the biotinylated miR-181a-5p mimics and negative control (NC) mimics were mixed with M-280 streptavidin magnetic beads (Invitrogen) and incubated for approximately 2 h. Then, H1299 cells were lysed and incubated with the mixture for 12 h. Finally, the RNAs sponged by the beads were extracted and analyzed by qRT–PCR.
The immunoprecipitation was performed in H1299, A549 and PC9 cells, and the CARM1 antibody preabsorbed protein A- and G-Sepharose beads was used as the primary antibody. The 2D-LC-MS/MS protocol was performed as previously described [16].
Dual-luciferase reporter gene assay
pGL3-LUC-circHMBG2, pGL3-LUC-CARM1, mutant pGL3-LUC-circHMGB2, and mutant-pGL3-LUC-CARM1 were co-transfected with miR-181a-5p mimics or NC mimics into HEK-293 T cells for 48 h. Then, the cells were lysed and centrifuged, and the supernatants were collected. The luciferase activities in the supernatants were measured using a dual-luciferase reporter assay system (Promega). The activation of the target gene was calculated according to the ratio of firefly luciferase activity/Renilla luciferase activity.
Flow cytometry analysis
The immune cell profiles in subcutaneous tumors were investigated by flow cytometry. Fresh tumor tissues were homogenized into single cell suspensions, and immune cells were isolated with Percoll. For surface marker staining, 1 × 106 cells were washed with 2 ml staining buffer (PBS with 1% FBS and 0.2% EDTA) and centrifuged at 350 g × 6 min. Then, the cells were incubated with Fc blocker and antibodies for 30 min. For the staining of Foxp3 in the nucleus, after surface marker staining for 30 min, the cells were washed with 1 ml staining buffer, fixed with 4% paraformaldehyde for 15 min and permeabilized with 0.1% Triton X-100 for 30 min. Next, the cells were washed with 1 ml staining buffer and incubated with Fc blocker and anti-Foxp3 antibodies for 90 min. Flow cytometry analysis was performed with a Fortessa flow cytometer (BD Biosciences, USA). Information about the antibodies is listed in Supplementary Table 1, and the CD45+CD3+CD8+ indicates CD8 cells; CD45+CD3+CD4+, CD4 cells; CD45+CD3−NK1.1/CD56+, NK; CD45+CD3+CD4+FOXP3+, Treg; CD45+CD11b+F4/80+ and DC, CD45+CD11b−CD11c+ TAM.
Exosome extraction and electron microscopy
Exosomes in the supernatants of NSCLC cells were extracted by ExoQuick Exosome Precipitation Solution (SBI System Biosciences) according to the manufacturer’s protocol. The exosomes were further identified by electron microscopy and western blotting.
Humanized mouse generation
The huHSC-NOG-EXL mice were purchased from Beijing Vital River Laboratory. New-born NOG-EXL mice were irradiated with 2Gy and subsequently injected with 5 × 104 CD34+ human hematopoietic stem cells (HSCs) through the tail vein. After 7–8 weeks of HSC differentiation, the reconstitution of human immune system components in the peripheral blood of humanized NOG-EXL mice was analyzed. A proportion of hCD45+ cells higher than 45% was considered to indicate the successful establishment of huHSC-NOG-EXL mice. Then, tumor engraftment was performed in weeks 8–11 (Fig. 4E).
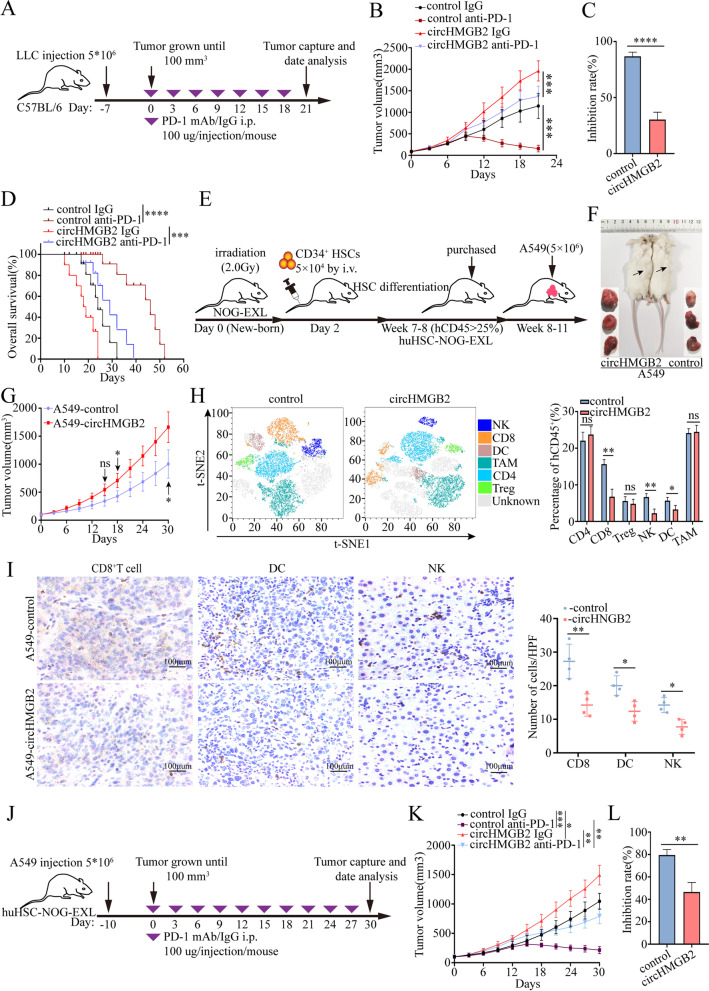
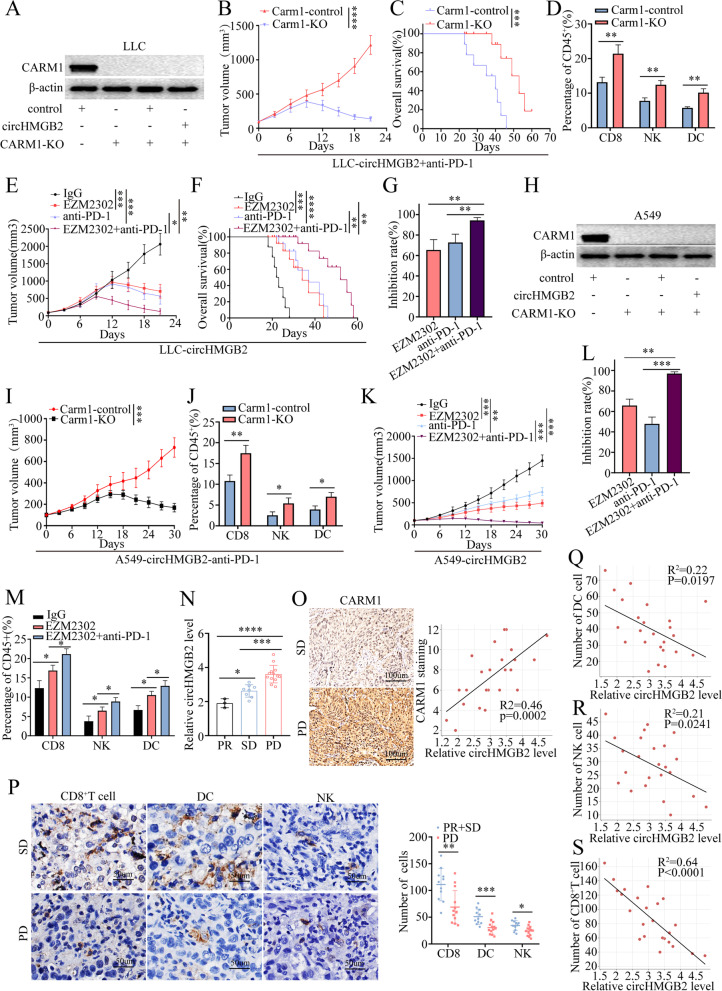
Fig. 4.
CircHMGB2 decreased the efficacy of PD-1 blockade in NSCLC treatment. A A schematic diagram of the treatment plan in C57BL/6 mice. B Growth of subcutaneous tumors in C57BL/6 mice inoculated with LLC-control or LLC-circHMGB2 cells and administered an anti-PD-1 antibody. C The efficacy of PD-1 therapy after the overexpression of circHMGB2. D The prognosis of C57BL/6 mice subcutaneously implanted with LLC-circHMGB2 cells and administered an anti-PD-1 antibody. E A schematic diagram of the establishment of huHSC-NOG-EXL mice. F and G huHSC-NOG-EXL mice were subcutaneously implanted with A549-control and A549-circHMGB2 cells, and the tumor size was assessed every 3 days. H The immune cell profiles of subcutaneous tumors derived from A549-control and A549-circHMGB2 cells were detected by flow cytometry. I IHC staining of CD8+ T cells, DCs, and NK cells in subcutaneous A549-control and A549-circHMGB2 tumors. J A schematic view of the treatment plan in huHSC-NOG-EXL mice. K. Growth of subcutaneous tumors in huHSC-NOG-EXL mice inoculated with A549-control or A549-circHMGB2 cells and administered an anti-PD-1 antibody. L The efficacy of PD-1 therapy after the overexpression of circHMGB2. Data are presented as the means ± SD; n = 3, *P < 0.05, **P < 0.01, ***P < 0.001, ***P < 0.0001, ns: not significant
In vivo tumor growth and anti-PD-1 therapy
C57BL/6 mice were purchased from Jiesijie (Shanghai, China). The C57BL/6 mice and huHSC-NOG-EXL mice were fed in a pathogen-free environment in the Center for Experimental Animals of Zhongshan Hospital. The animal experiments were approved by the Ethics Committee of Zhongshan Hospital. Approximately 5 × 106 cells were resuspended in 150 μl DMEM medium and subcutaneously injected into the right flanks of the C57BL/6 mice or huHSC-NOG-EXL mice. When the tumor size reached 100mm3, the PD-1 antibody or IgG was administered i.p. at a dose of 100 μg/injection every three days. The CARM1 inhibitor EZM2302 was administered orally at a dose of 150 mg/kg twice a day for 14 days [17]. The tumor size was assessed every 3 days.
Statistical analysis
Data analysis was performed with 23.0 SPSS (Chicago, IL). The values are shown as the mean ± standard deviation. Student’s t test was chosen to compare differences in measurement data between two groups, while categorical variables were compared via chi-squared or Fisher’s exact tests. Spearman correlation analysis was used to analyze correlations between the levels of circHMGB2, miR-181a-5p, CARM1, CD8, CD56, NK1.1 (CD161), CD11C, p-STAT1, ISG15 and IFIT1. The Kaplan–Meier method and the log-rank test were used to analyze differences in the prognosis of NSCLC patients. Cox’s regression model was used to investigate independent prognostic factors. All the p values were two tailed, and differences with p < 0.05 were considered statistically significant.
Results
Clinical significance of circHMGB2 in NSCLC patients
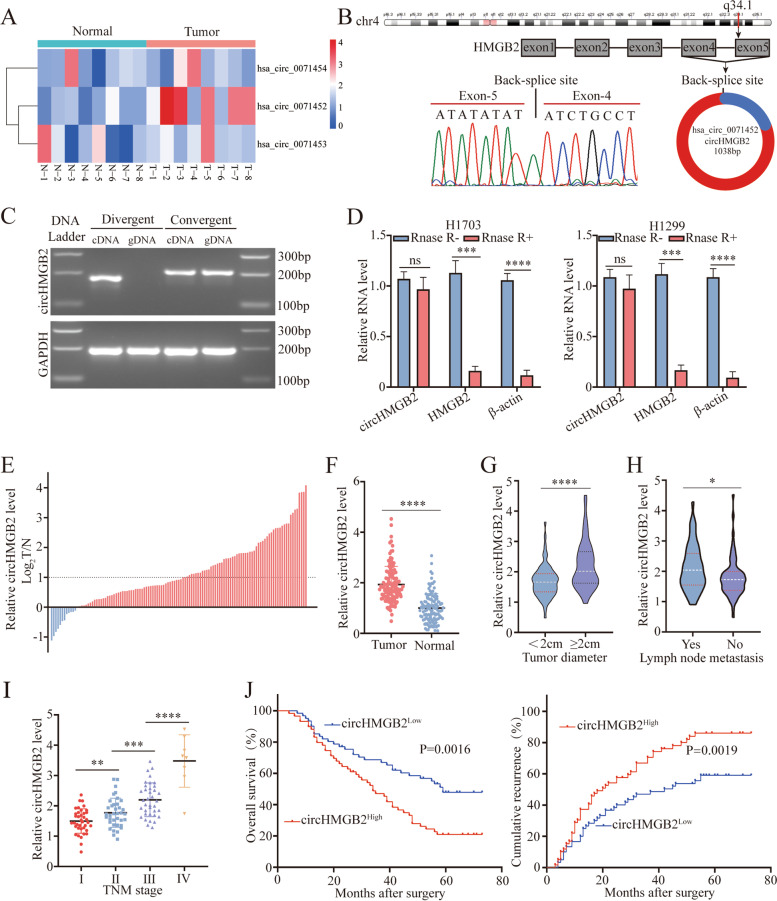
HMGB2 is reported to be involved in the progression of many types of cancers, including NSCLC [11–13, 18]. In general, the expression of circRNAs is closely related to the expression of the corresponding mRNAs since circRNAs are derived from the back-splicing of precursor mRNAs [3]. Thus, the expression of three circRNAs derived from HMGB2 was measured in 8 pairs of NSCLC tissues and paired normal tissues via qRT–PCR. The results showed that hsa_circ_0071452 (circHMGB2) was the most significantly upregulated circRNA in NSCLC tissues (Fig. 1A and Supplementary Fig. 1A). Sanger sequencing with divergent primers also identified the loop structure of circHMGB2 (Fig. 1B). In addition, PCR results showed that circHMGB2 could be transcribed with divergent primers only from cDNA but not from gDNA (Fig. 1C). After incubation with RNase R for 30 min, the qRT–PCR results showed that circHMGB2 was resistant to RNase R, while the mRNA of HMGB2 and β-actin as positive control was substantially degraded (Fig. 1D).
Fig. 1.
The clinical significance of circHMGB2 in NSCLC patients. A The expression of HMGB2-derived circRNAs was measured in 8 pairs of NSCLC tissues and matched normal tissues. B Schematic diagram of circHMGB2 and Sanger sequencing results of the site of circHMGB2 backsplicing. C PCR results of circHMBG2 transcribed with divergent or convergent primers from cDNA and gDNA. D circHMGB2 and HMGB2 in H1703 and H1299 cell lines were detected by qRT–PCR after digestion by RNase. E and F The expression of circHMGB2 was detected by qRT–PCR in 120 pairs of NSCLC tissues and matched normal tissues. G and H The expression of circHMGB2 was analyzed according to tumor diameter (< 2 cm vs. ≥ 2 cm) and lymph node metastasis status (yes vs. no). I The expression of circHMGB2 was analyzed according to TNM stage. J Survival analysis of the recurrence and OS of 120 NSCLC patients divided into groups according to circHMGB2 expression (circHMGB2high vs. circHMBG2low) was performed using Kaplan–Meier and log rank analysis. Data are presented as the means ± SD of three independent experiments. *P < 0.05, **P < 0.01, *** P < 0.001, ***P < 0.0001, ns: not significant
To further evaluate the clinical role of circHMGB2 in NSCLC, the level of circHMGB2 was measured in 120 NSCLC tissues and matched normal tissues using qRT–PCR. CircHMGB2 was highly expressed in the majority of tumor tissues, and the expression of circHMGB2 in 57 NSCLC tissues was more than twofold higher than that in normal tissues (Fig. 1E and F). The relationships between the level of circHMGB2 and the clinical characteristics of 120 NSCLC patients are presented in Table 1. These results suggested that high expression of circHMGB2 was related to large tumor size and lymph node metastasis in NSCLC patients (Fig. 1G and H). We also observed that circHMGB2 expression was increased as clinical stage increased (Fig. 1I). Moreover, Kaplan–Meier analysis revealed that patients with high circHMGB2 expression had lower overall survival (OS) and higher postoperative recurrence rates (Fig. 1J) than those with lower circHMGB2 expression. Furthermore, multivariate Cox analysis confirmed that circHMGB2 was an independent factor in the prognosis of NSCLC patients (Tables 2 and 3). Additionally, we further analyzed the clinical implication of circHMGB2 in LUAD (Supplementary Fig. 1B and C). In short, these results suggest that high expression of circHMGB2 indicates a poor prognosis in NSCLC patients and is involved in the progression of NSCLC.
Table 1.
Correlations between circRNA HMGB2 and clinical characteristics in 120 NSCLC patients
| Clinicopathological parameters | No. of cases | CircHMGB2 expression level | P value | |
|---|---|---|---|---|
| Low | High | |||
| Age | ||||
| ≥60 | 53 | 28 | 25 | |
| <60 | 67 | 32 | 35 | 0.581 |
| Gender | ||||
| Male | 67 | 36 | 31 | |
| Female | 53 | 24 | 29 | 0.358 |
| Smoking history | ||||
| Smokers | 54 | 24 | 30 | |
| Nonsmokers | 66 | 36 | 30 | 0.271 |
| Histological type | ||||
| Adenocarcinoma | 78 | 41 | 37 | |
| Squamous | 42 | 19 | 23 | 0.444 |
| Tumor stage | ||||
| I–II | 72 | 31 | 41 | |
| III–IV | 48 | 29 | 19 | 0.062 |
| Lymph node metastasis | ||||
| Yes | 50 | 19 | 31 | |
| NO | 70 | 41 | 29 | 0.026 |
| Tumor size | ||||
| >2 cm | 55 | 21 | 34 | |
| ≤2 cm | 65 | 39 | 26 | 0.017 |
| Differentiation | ||||
| Well and moderate | 57 | 23 | 34 | |
| Poor | 63 | 37 | 26 | 0.044 |
Table 2.
Univariate and multivariate analyses of factors associated with overall survival
| Factors | OS | |||||
|---|---|---|---|---|---|---|
| Univariate | Multivariate | |||||
| HR | 95% CI | P value | HR | 95% CI | P value | |
| Age(≥60 VS <60) | 0.856 | 0.683–1.073 | 0.178 | NA | ||
| Gender(Male VS Female) | 0.914 | 0.726–1.151 | 0.446 | NA | ||
| Smoking history (smokers VS Nonsmokers) | 0.946 | 0.755–1.186 | 0.631 | NA | ||
| Histological type (adenocarcinoma VS squamous) | 1.082 | 0.676–1.733 | 0.741 | NA | ||
| Tumor stage (III–IV VS I–II) | 0.79 | 0.630–0.991 | 0.041 | 0.692 | 0.544–0.882 | 0.003 |
| Lymph node metastasis (Yes VS No) | 0.716 | 0.571–0.898 | 0.004 | 0.735 | 0.581–0.929 | 0.01 |
| Tumor size (>2 cm VS ≤2 cm) | 0.732 | 0.582–0.919 | 0.007 | 0.811 | 0.639–1.030 | 0.086 |
| Differentiation (poor VS well and moderate) | 0.75 | 0.597–0.943 | 0.014 | 0.664 | 0.517–0.851 | < 0.001 |
| CircHMGB2 expression (High vs. Low) | 0.709 | 0.562–0.894 | 0.004 | 0.600 | 0.459–0.783 | <0.001 |
Table 3.
Univariate and multivariate analyses of factors associated with cumulative recurrence
| Factors | OS | |||||
|---|---|---|---|---|---|---|
| Univariate | Multivariate | |||||
| HR | 95% CI | P value | HR | 95% CI | P value | |
| Age(≥60 VS <60) | 0.957 | 0.773–1.185 | 0.687 | NA | ||
| Gender(Male VS Female) | 0.931 | 0.750–1.156 | 0.518 | NA | ||
| Smoking history (smokers VS Nonsmokers) | 0.919 | 0.743–1.138 | 0.44 | NA | ||
| Histological type (adenocarcinoma VS squamous) | 0.985 | 0.629–1.544 | 0.949 | NA | ||
| Tumor stage (III–IV VS I–II) | 0.77 | 0.622–0.955 | 0.017 | 0.68 | 0.540–0.856 | 0.001 |
| Lymph node metastasis (Yes VS No) | 0.722 | 0.582–0.895 | 0.003 | 0.749 | 0.598–0.939 | 0.012 |
| Tumor size (>2 cm VS ≤2 cm) | 0.778 | 0.627–0.965 | 0.022 | 0.854 | 0.681–1.070 | 0.17 |
| Differentiation (poor VS well and moderate) | 0.782 | 0.630–0.970 | 0.025 | 0.711 | 0.564–0.896 | 0.004 |
| CircHMGB2 expression (High vs. Low) | 0.713 | 0.573–0.888 | 0.002 | 0.617 | 0.479–0.795 | <0.001 |
CircHMGB2 promotes the proliferation of NSCLC and reshapes the tumor microenvironment (TME)
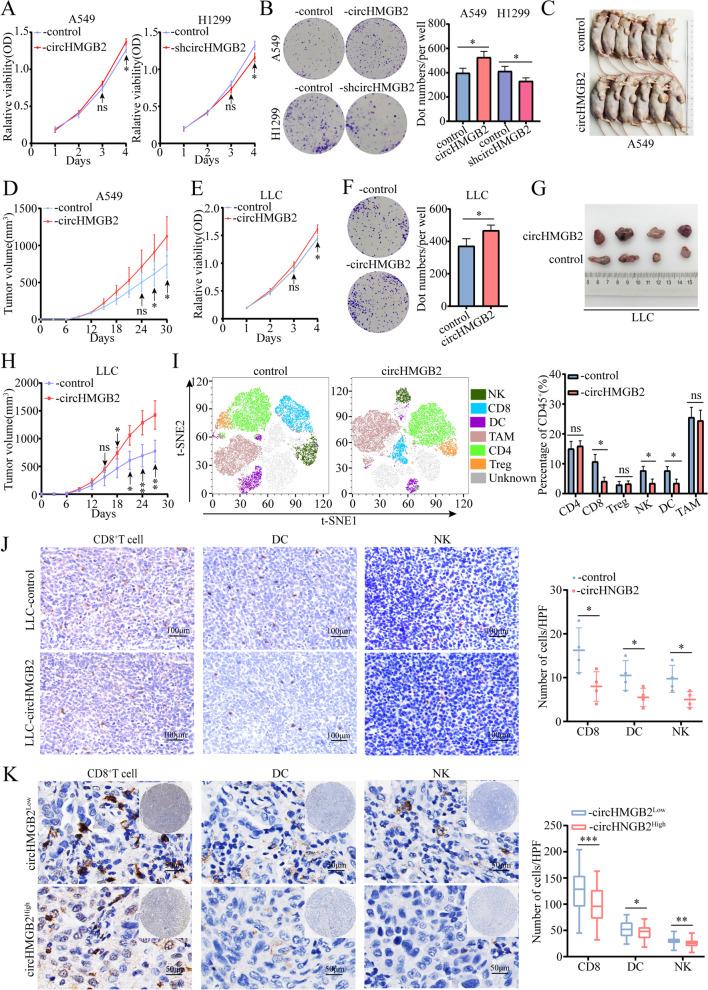
Considering the potential effect of circHMGB2 on the prognosis of NSCLC patients, the impact of circHMGB2 on the biological functions of NSCLC was further investigated. The expression of circHMGB2 in 5 NSCLC cell lines and LLC cells was measured by qRT–PCR, and the results showed that circHMGB2 was expressed at the highest level in H1299 cells and the lowest level in A549 cells (here only including LUAD and LUSC cell lines) (Supplementary Fig. 2A). Then, circHMGB2 expression was knocked down in H1299 cells (H1299-shcircHMGB2) and overexpressed in A549 and LLC cells (A549-circHMGB2, LLC-circHMGB2) by lentivirus transfection, and three stable cell lines were generated (Supplementary Fig. 2B). A matrigel transwell assay revealed no significant effect of circHMGB2 on the invasion of NSCLC cells (Supplementary Fig. 2C). Consistently, a wound healing assay showed no effect of circHMGB2 in inhibiting or promoting the migration of A549-circHMGB2 or H1299-shHMGB2 cells (Supplementary Fig. 2D). However, CCK-8 and colony formation assays showed that proliferation was decreased after the knocked-down of circHMGB2 and increased after the elevated circHMGB2 expression in NSCLC cells (Fig. 2A and B). In addition, the in vitro analysis of nude mice subcutaneously implanted with A549-circHMGB2 and control cells further showed that the overexpression of circHMGB2 could accelerate the growth of NSCLC (Fig. 2C and D).
Fig. 2.
CircHMGB2 promotes the proliferation of NSCLC and reshapes the tumor TME. A The proliferation of A549-circHMGB2 and H1299-shcircHMGB2 cells was measured by CCK-8 assay. B The viability of A549-circHMGB2 and H1299-shcircHMGB2 cells was measured by colony formation assays. C and D Nude mice were subcutaneously implanted with A549-control and A549-circHMGB2 cells, and the tumor size was assessed every 3 days. E The proliferation of LLC-circHMGB2 cells was measured by CCK-8 assay. F The viability of LLC-circHMGB2 cells was measured by colony formation assays. G and H C57BL/6 mice were subcutaneously implanted with LLC-control and LLC-circHMGB2 cells, and the tumor size was assessed every 3 days. I The immune cell profiles of subcutaneous tumors derived from LLC-control and LLC-circHMGB2 cells were assessed by flow cytometry. J IHC staining of CD8+ T cells, DCs, and NK cells in subcutaneous tumors derived from LLC-control and LLC-circHMGB2 cells. K The infiltration of CD8+ T cells, DCs and NK cells in 120 NSCLC tissues divided into circHMGB2low and circHMGB2high groups was assessed by IHC. Data are presented as the means ± SD; n = 3, *P < 0.05, **P < 0.01, ***P < 0.001, ns: not significant
Additionally, further studies were performed to determine whether circHMGB2 could potentially impact the TME. Firstly, CCK-8 and colony formation assays showed that proliferation was increased after circHMGB2 expression was upregulated in LLC cells (Fig. 2E and F). Then, we performed an in vivo assay in C57BL/6 mice via subcutaneously inject with LLC-circHMGB2 and LLC-control cells, and found that circHMGB2 could dramatically potentiate the growth of subcutaneous tumors in immunocompetent mice (Fig. 2G and H). Moreover, the immune profiles in the tumors were evaluated using flow cytometry. The results revealed the presence of exhausted of CD8+ T cells, NK cells and DCs in the subcutaneous tumors derived from LLC-circHMGB2 cells (Fig. 2I). The IHC staining of CD8+ T cells, CD4+T cells, NK1.1+ NK cells, CD68+TAM, FOXP3+Treg cells and CD11C+ DCs in the tumor tissues also showed a similar trend (Fig. 2J and Supplementary Fig. 2E). To further verify the in vivo results, IHC staining of CD8, CD56 and CD11C expression in 120 NSCLC tissues was performed, and the results revealed similar trends. The infiltration of CD8+ T cells, CD56+ NK cells and CD11C+ DCs was remarkably lower in NSCLC samples with high circHMGB2 expression (Fig. 2K). Considering the evidently effects of circHMGB2 on the NSCLC TME, we hypothesized that circHMGB2 induces the progression of NSCLC mainly by limiting antitumor immunity in the TME.
CircHMGB2 upregulated the expression of the downstream molecule CARM1 by sponging miR-181a-5p
Further investigation of the relationship between circHMGB2 and the TME led to two possible hypotheses: (1) circHMGB2 is directly transmitted into the TME and affects the biological functions of immune cells or (2) the upregulation of circHMGB2 expression inhibits the immune response to NSCLC. Since exosomes have been reported to be a critical mechanism by which circRNAs are transmitted between cells [15, 19, 20], exosomes were extracted from the supernatants of H460-circHMGB2 cells to test the first hypothesis (Supplementary Fig. 3A and B). However, no markedly enrichment in the exosomes derived from H460-circHMGB2 cells was observed (Supplementary Fig. 3C). Therefore, further studies focused on the function of circHMGB2 in NSCLC.
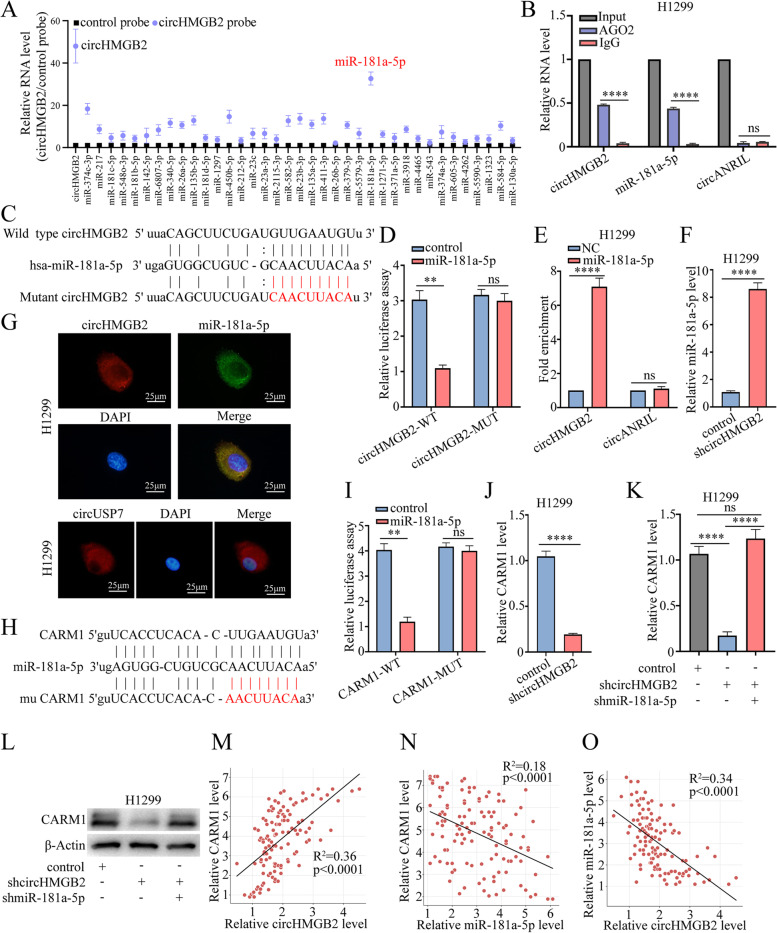
Since circRNAs have been widely reported to act as competing endogenous RNAs (ceRNAs) for miRNAs, potential miRNAs that could be sponged by circHMGB2 were predicted using StarBase 3.0. Then, circRIP with a circHMGB2 probe was performed in H1299 cells, and the significant enrichment of miR-181a-5p was observed via qRT–PCR (Fig. 3A). Further verification of the interaction between circHMGB2 and miR-181a-5p using RIP revealed the remarkable enrichment of circHMGB2 and miR-181a-5p by an anti-AGO2 antibody (Fig. 3B). Moreover, miR-181a-5p mimics were co-transfected with a luciferase plasmid carrying the wild-type circHMGB2 sequence and a mutant sequence into HEK-293 T cells. The miR-181a-5p mimics obviously weakened the luciferase activity of the wild-type circHMGB2 sequence but not the luciferase activity of the mutant circHMGB2 sequence (Fig. 3C and D). The miR-181a-5p pulldown assay also revealed significant enrichment of circHMGB2 (Fig. 3E). In addition, the qRT–PCR results showed that the level of miR-181a-5p was elevated after the knockdown of circHMGB2 expression in H1299 cells (Fig. 3F). FISH staining of circHMGB2 and miR-181a-5p in H1299 cells showed that these molecules colocalized in the cytoplasm (Fig. 3G). Thus, circHMGB2 may perform its biological function by sponging miR-181a-5p. To identify the downstream mRNAs, StarBase 3.0, miRanda and PITA were used to predict the potential targets of miR-181a-5p, and the results showed that the immune-related genes SIRT1 [21], CDK8 [22], IRS2 [23], BHLHE40 [24] and CARM1 [25] contained potential binding sites for miR-181a-5p. Furthermore, the CARM1 mRNA was enriched by biotin-miR-181a-5p in NSCLC cells (Supplementary Fig. 4A). Subsequently, a luciferase reporter assay showed that the luciferase activity of the wild-type CARM1 sequence was reduced compared to that of the mutant CARM1 sequence (Fig. 3H, I and Supplementary Fig. 4B-E). qRT–PCR also showed that the expression of CARM1 was reduced in H1299-shcircHMGB2 cells (Fig. 3J), while the knockdown of miR-181a-5p expression in H1299-shcircHMGB2 cells significantly restored the expression of CARM1 (Fig. 3K and L). Moreover, the levels of circHMGB2, miR-181a-5p and CARM1 in 120 NSCLC patients were measured, and the results showed that the expression of miR-181a-5p was negatively associated with that of circHMGB2 and CARM1 level, while the expression of circHMGB2 was positively correlated with the level of CARM1 (Fig. 3M-O). Thus, circHMGB2 relieves the inhibition of the downstream molecule CARM1 by sponging miR-181a-5p in NSCLC.
Fig. 3.
CircHMGB2 upregulates the expression of the downstream molecule CARM1 by sponging miR-181a-5p. A A circRIP assay was performed with a circHMGB2 probe in H1299 cells. B The RIP assay was performed using the anti-AGO2 antibody in H1299 cells. C The putative binding sites of circHMGB2 and miR-181a-5p. D The luciferase activity of circHMGB2 in HEK-293 T cells transfected with miR-181a-5p. E The RNA pulldown assay was performed with H1299 cells transfected with biotinylated miR-181a-5p. F The expression of miR-181a-5p was measured via qRT–PCR after the knockdown of circHMGB2 in H1299 cells. G FISH staining of circHMGB2 and miR-181a-5p in H1299 cells. circUSP7 as positive control H The putative binding sites of CARM1 and miR-181a-5p. I The luciferase activity of CARM1 was measured in HEK-293 T cells transfected with miR-181a-5p. J The expression of CARM1 was measured via qRT–PCR after the overexpression of circHMGB2. K The expression of CARM1 was measured using qRT–PCR after the dual knockdown of circHMGB2 and miR-181a-5p expression. L The expression of CARM1 was measured using western blotting after the dual knockdown of circHMGB2 and miR-181a-5p expression. M-O The relationships among the expression levels of circHMGB2, miR-181a-5p and CARM1 in 120 NSCLC tissues were assessed using qRT–PCR. Data are presented as the means ± SD; n = 3, **P < 0.01, ****P < 0.001, ns: not significant
CircHMGB2 limits the efficacy of PD-1 blockade in NSCLC treatment
A previous study reported that the inactivation of CARM1 could sensitize tumors to T cell-dependent immune attack [25]. Thus, we hypothesized that high expression of circHMGB2 might limit the immune response to NSCLC and inhibit the efficacy of anti-PD-1 therapy. Given the potential difference in the miR-181a-5p and CARM1 sequences between humans and mice, the predicted binding sites were compared between the two species; the results showed that humans and mice shared the same sequence of miR-181a-5p and that the predicted binding sites of Carm1 were compatible (Supplementary Fig. 5A and B). miR-181a-5p substantially decreased the luciferase activity of the wild-type Carm1 sequence but not that of the mutant Carm1 sequence (Supplementary Fig. 5C). The overexpression of circHMGB2 in LLC cells enhanced the expression of Carm1, while transfection of miR-181a-5p mimics could reverse this phenomenon (Supplementary Fig. 5D and E). Thus, circHMGB2 could also regulate the expression of the downstream molecule Carm1 in LLC cells. Moreover, the potential effects of circHMGB2 on the efficacy of anti-PD-1 treatment were evaluated in C57BL/6 mice bearing circHMGB2-overexpressing or control tumors. The PD-1 blockade or IgG was administered i.p. at a dose of 100 μg/injection every 3 days when the tumor size reached 100mm3 (Fig. 4A). The in vivo results showed that the overexpression of circHMGB2 significantly minimized the efficacy of anti-PD-1 treatment after 2 weeks (Fig. 4B and C). In addition, upregulated circHMGB2 expression reduced the survival of immunocompetent mice (Fig. 4D).
In order to further confirm that circHMGB2 can reshape the tumor immune microenvironment and limit the efficacy of anti-PD-1 in NSCLC, we performed in vivo experiments in mice with a humanized immune system. NOG-EXL mice can be reconstituted with human lymphoid cells and myeloid cells, and can better simulate the cellular composition of the human immune system. Consequently, these mice have been widely used in the study of tumor immunity [26]. We established huHSC-NOG-EXL mice (Fig. 4E). These mice were subcutaneously inoculated with A549-circHMGB2 and A549-control cells, and we found that circHMGB2 could dramatically potentiate the growth of subcutaneous tumors in humanized immune system mice (Fig. 4F and G). Then, the immune profiles in the tumors were assessed using flow cytometry. The results revealed the presence of exhaustion of CD8+ T cells, NK cells and DCs in the subcutaneous tumors derived from A549-circHMGB2 cells (Fig. 4H). The IHC staining of CD8+ T cells, CD56+ NK cells and CD11C+ DCs in the tumor tissues also showed a similar trend (Fig. 4I). We also verified the efficacy of anti-PD-1 treatment in humanized mice models inoculated with A549-circHMGB2 or A549-control cells, and a dosing schedule for immunotherapy was developed (Fig. 4J). The results confirmed that circHMGB2 overexpression obviously limited the efficacy of anti-PD-1 treatment (Fig. 4K and L).
Collectively, these results showed that the efficacy of anti-PD-1 treatment was limited in both a mouse model with a humanized immune system and a xenograft model of circHMGB2-overxpressing NSCLC.
Knockout of the CARM1 gene sensitizes NSCLC cells with high circHMGB2 expression to anti-PD-1 antibody treatment
Since the above results revealed that high circHMGB2 expression inhibited the efficacy of anti-PD-1 therapy in NSCLC, we hypothesized that the inactivation of CARM1 might sensitize the treatment of anti-PD-1. Thus, the Carm1 gene was completely knocked out with the CRISPR-Cas9 system in LLC-circHMGB2 cells (Fig. 5A). To verify the synergetic effects of Carm1 knockout and PD-1 immunotherapy, a xenograft model was established by subcutaneously implanting mice with LLC-Carm1-KO-circHMGB2 and LLC-Carm1-control-circHMGB2 cell lines. The results showed that the knockout of Carm1 significantly delayed the growth of circHMGB2-overexpressing tumors after the administration of anti-PD-1 and greatly improved the survival of the mice in this group (Fig. 5B and C). Flow cytometry analysis revealed that anti-PD-1 therapy increased the infiltration of CD8+ T cells, NK cells and DCs into subcutaneous circHMGB2-overexpresing tumors in C57BL/6 mice after the knockout of Carm1 (Fig. 5D). Importantly, the administration of EZM2302 (a small molecule inhibitor of CARM1) and an anti-PD-1 antibody achieved similar results (Fig. 5E-G). We knock out CARM1 gene in A549 cells (Fig. 5H), and established A549-CARM1-control-circHMGB2 and A549-CARM1-KO-circHMGB2 stably transfected cell lines. Then, the subcutaneous tumor models in humanized mice with A549-CARM1-control-circHMGB2 and A549-CARM1-KO-circHMGB2 cells were established, and those mice were administrated anti-PD-1 mAb. The in vivo experiments verified that the knockout of CARM1 significantly delayed the growth of tumors overexpressing circHMGB2 after the administration of anti-PD-1 and promoted the infiltration of CD8+ T cells, NK cells and DCs in the subcutaneous tumors tissue (Fig. 5I and J). Moreover, the administration of EZM2302 and anti-PD-1 mAb achieved similar results (Fig. 5K-M). The in vivo experiments showed that the EZM2302 treatment could increase the infiltration of CD8+ T cells, NK cells and DCs (Fig. 5M), which indicate that EZM2302 may have synergistic effect with anti PD-1 Ab to improve the anti-tumor response in murine cancer models.
Fig. 5.
Knockout of CARM1 sensitized NSCLC cells with high expression of circHMGB2 to treatment with an anti-PD-1 antibody. A The knockout of the Carm1 gene in LLC cells by the CRISPR-Cas9 system was verified by western blotting. B The growth of subcutaneous tumors in C57BL/6 mice after the knockout of the Carm1 gene in LLC-circHMGB2 cells and the administration of an anti-PD-1 antibody. C The prognosis of C57BL/6 mice after subcutaneous implantation of LLC-circHMGB2 cells with Carm1 knockout and treatment with PD-1 antibody administration. D The infiltration of CD8+ T cells, NK cells and DCs in tumors overexpressing circHMGB2 and Carm1 knockout after the administration of anti-PD-1 antibody was assessed by flow cytometry. E The growth of subcutaneous tumors in C57BL/6 mice implanted with LLC-circHMGB2 cells after the synergistic usage of EZM2302 and an anti-PD-1 antibody. F The prognosis of C57BL/6 mice subcutaneously implanted with LLC-circHMGB2 cells after treatment with the synergistic combination of EZM2302 and an anti-PD-1 antibody. G The efficacy of the synergistic combination of EZM2302 and an anti-PD-1 antibody. H The knockout of the CARM1 gene in A549 cells by the CRISPR-Cas9 system was verified by western blotting. I The growth of subcutaneous tumors in huHSC-NOG-EXL mice after the knockout of the CARM1 gene in A549-circHMGB2 cells and the administration of an anti-PD-1 antibody. J The infiltration of CD8+ T cells, NK cells and DCs in tumors was assessed by flow cytometry which derived from A549-CARM1-control-circHMGB2 or A549-CARM1-KO-circHMGB2 cells after the administration of an anti-PD-1 antibody. K The efficacy of the synergistic usage of EZM2302 and an anti-PD-1 antibody in huHSC-NOG-EXL mice subcutaneously implanted with A549-circHMGB2 cells. L The efficacies of anti-PD-1 treatment, EZM2302 treatment or the combination anti-PD-1 and EZM2302 treatment. M The infiltration of CD8+ T cells, NK cells and DCs into tumors derived from A549-circHMGB2 cells was detected by flow cytometry after the administration of an anti-PD-1 antibody and EZM2302. N The expression of circHMGB2 in 24 NSCLC patients who received anti-PD-1 immunotherapy and who were stratified into three groups: the PR, SD and PD groups. O The relationship between CARM1 and circHMGB2 expression in NSCLC patients who received anti-PD-1 immunotherapy was assessed by IHC. P The infiltration of CD8+ T cells, DCs and NK cells in the PR + SD group and PD group who received anti-PD-1 immunotherapy. Q-S The relationship between the expression of circHMGB2 and the infiltration of CD8+ T cells, DCs and NK cells in 24 NSCLC patients who received anti-PD-1 immunotherapy. Data are presented as the means ± SD; n = 3, *P < 0.05, **P < 0.01, ***P < 0.001, ***P < 0.0001, ns: not significant, SD: stable disease, PR: partial remission, PD: progressive disease
To further study the expression of circHMGB2 and the efficacy of PD-1 blockade therapy, we also analyzed the expression patterns in 24 patients who received anti-PD-1 therapy. According to the RECIST1.1 standard, 13 patients achieved progressive disease (PD) status, 8 patients achieved stable disease (SD) status, and only 3 patients were considered to have achieved partial remission (PR) status. The qRT–PCR results showed that the expression of circHMGB2 in the PD group was significantly higher than that in the PR and SD groups (Fig. 5N). Spearman analysis showed that the expression of circHMGB2 was positively correlated with the expression of CARM1 in the PD, PR and SD groups (Fig. 5O). Moreover, IHC staining showed that the infiltration of CD8+ T cells, NK cells and DCs in the PR and SD groups was significantly higher than that in the PD group (Fig. 5P). Spearman analysis showed that the expression of circHMGB2 was positively correlated with the expression of CARM1 but negatively correlated with the infiltration of CD8+ T cells, DCs and NK cells in 24 NSCLC patients received anti-PD-1 therapy (Fig. 5Q-S). Thus, these data show that the inhibition of CARM1 can improve the efficacy of anti-PD-1 therapy in NSCLC patients with high circHMGB2 expression.
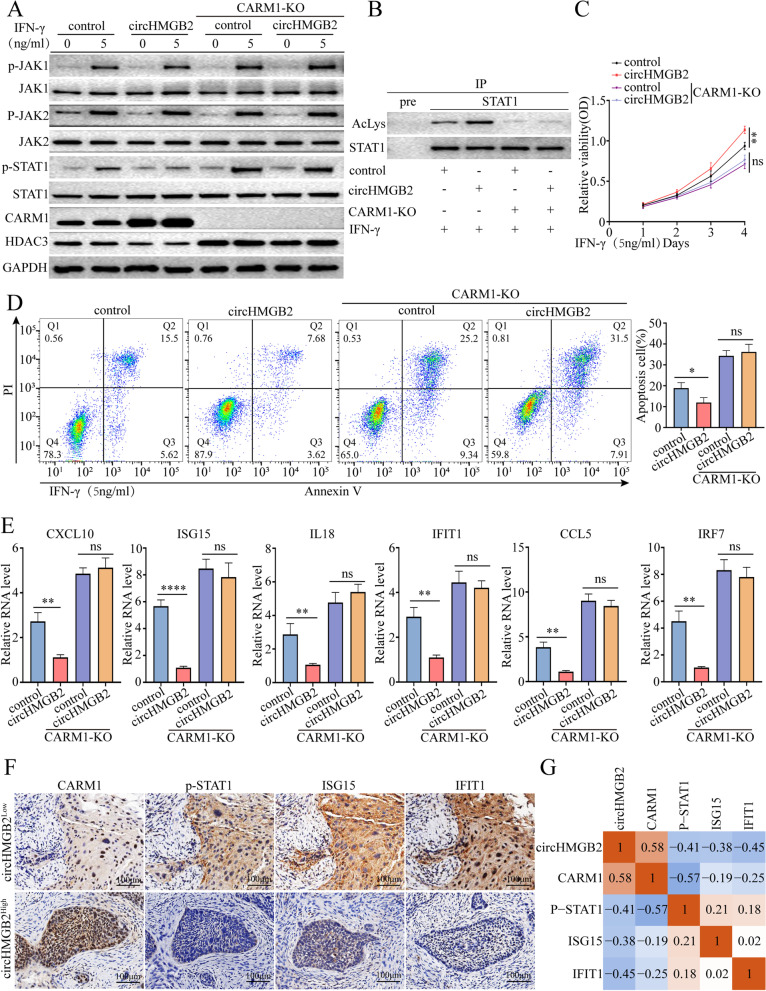
CircHMGB2 inactivates the type 1 IFN response to NSCLC via CARM1
We had showed that the high level of circHMGB2 was associated with the exhaustion of CD8+ T, NK and DC cells in C57BL/6 mice and human NSCLC samples. Moreover, elevated circHMGB2 was identified to upregulate the expression of the downstream molecule CARM1 by sponging miR-181a-5p, which indicated that high level of circHMGB2 could reset the tumor immune microenvironment by CARM1. Coincidently, a previous study identified that CARM1 could inhibit the type 1 IFN response in tumors and desensitize tumors to the cytotoxic T cell-mediated immune response [25]. Thus, we hypothesized that high expression of circHMGB2 might inactivate the type 1 IFN response via CARM1 to enhance NSCLC resistance to cytotoxic T cells and foster an immunosuppressive environment in NSCLC. The knockout of the CARM1 gene in A549-control or A549-circHMGB2 cells also led to a significantly increased response to IFN-γ compared to that of A549-circHMGB2 cells. Mechanistically, stimulation with IFN-γ (5 ng/ml) upregulated the expression of IFN-activated genes and activated JAK and STAT1, which are involved in a pathway that is important for the type 1 IFN response (Fig. 6A) [27, 28]. In order to further demonstrate the mechanism of CRAM1 in IFN-γ signal, Co-IP combined with MS was used employed to isolate and identify the interactome of CRAM1 in A549, PC9 and H1299 cells, and HDAC3 was found and further identified to interact with CRAM1(Supplementary Fig. 6A and B). HDAC3 levels did not increase after stimulation with IFN-γ; however, overexpression of circHMGB2 induced a reduction in the HDAC3 levels. Moreover, CARM1 KO significantly increased HDAC3 expression (Fig. 6A). HDAC3 is involved in the regulation of the balance between STAT1 phosphorylation and acetylation [29]. In Co-IP experiments, overexpression of circHMGB2 inhibited STAT1 deacetylation, and CARM1 KO reversed this phenomenon (Fig. 6B). These findings confirmed that overexpression of circHMGB2 inhibited STAT1 deacetylation, thereby reducing the phosphorylation of STAT1 and inhibiting IFN-γ signal transduction. The proliferation of CARM1-knockout A549 cells was significantly impaired compared to that of A549-circHMGB2 or control cells, while the apoptosis of CARM1-KO A549 cells was remarkably enhanced after stimulation with IFN-γ (Fig. 6C and D). In addition, the expression of IFN-activated genes, including CXCL10, ISG15, IL18, IFIT1, CCL5 and IFR7, was measured using qRT–PCR. The results showed that the expression of these genes were significantly increased after the knockout of CARM1 but inhibited after the overexpression of circHMGB2 (Fig. 6E, Supplementary Fig. 6C). To further verify the role of circHMGB2 in regulating the type 1 IFN response, the phosphorylation of STAT1, the expression of CARM1 and the expression of the IFN response genes ISG15 and IFIT were evaluated in tumor tissues from 120 NSCLC patients using IHC. The results suggested that the expression of circHMGB2 was positively correlated with the staining of CARM1 but negatively correlated with the phosphorylation of STAT1 and the expression of ISG15 and IFIT (Fig. 6F and G). Thus, these results indicate that circHMGB2 suppresses the type 1 IFN response via CARM1, which induces resistance to the cytotoxic immune response in NSCLC.
Fig. 6.
CircHMGB2 inhibited the type 1 IFN response via CARM1 in NSCLC. A Activation of the STAT1 pathway in A549-circHMGB2 cells after stimulation with IFN-γ for 48 h was verified by western blotting. B STAT1 acetylation in circHMGB2-overexpressing A549 cells or CARM1-KO A549 cells was analyzed by STAT1-IP and anti-acetyl lysine Western blotting. C The effect of CARM1 on the proliferation of A549-circHMGB2 cells stimulated with IFN-γ was measured by CCK-8 assay. D The effect of CARM1 on the apoptosis of A549-HMGB2 cells stimulated with IFN-γ was measured by flow cytometry. E Expression levels of type 1 IFN response-related genes were measured by qRT–PCR after the overexpression of circHMGB2 and the knockout of CARM1 in A549 cells. F and G The relationship between the expression of circHMGB2 and the staining of CARM1, p-STAT1, ISG15 and IFIT1 in 120 NSCLC patients was assessed by IHC and qRT–PCR. Data are presented as the means ± SD; n = 3, **P < 0.01, ***P < 0.001, ns: not significant
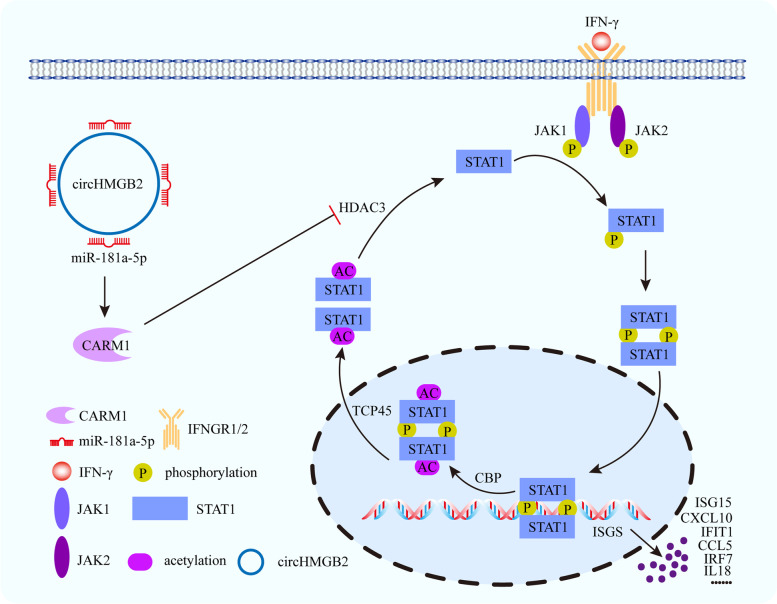
Figure 7 summarizes the main findings of this study. In NSCLC, the upregulation of circHMGB2 expression shapes the immunosuppressive microenvironment and leads to resistance to immunotherapy. Mechanistically, circHMGB2 relieves the inhibition of the downstream molecule CARM1 by sponging miR-181a-5p, which inhibits the type 1 IFN response by HDAC3 and promotes resistance to cytotoxic T cells. In addition, we found that the synergistic combination of EZM2302 and anti-PD-1 treatment substantially increased the susceptibility to immunotherapy in NSCLC patients with high circHMGB2 expression.
Fig. 7.
Schematic diagram illustrates the mechanism of circHMGB2 relieves the inhibition of the downstream molecule CARM1 by sponging miR-181a-5p, which induces a reduction in the HDAC3 levels. Then, STAT1 deacetylation is inhibited, thereby reducing the phosphorylation of STAT1 and inhibiting IFN-γ signal transduction. Subsequently, the expression of IFN-activated genes, including CXCL10, ISG15, IL18, IFIT1, CCL5 and IFR7, were inhibited. Those drive immunosuppression and anti-PD-1 resistance in lung adenocarcinomas and squamous cell carcinomas
Discussion
Since circRNAs were first discovered in the cytoplasm of eukaryotic cells in 1976, various circRNAs have been identified by high-throughput deep sequencing [30, 31]. Previous studies have shown the differential expression of many circRNAs in various cells and reported the potential biological functions of circRNAs in various pathophysiological processes, such as cancer, heart failure, atherosclerosis and immunity [32–35]. In addition, abnormally expressed circRNAs are closely related to cancer development, progression and treatment resistance. However, the underlying molecular mechanisms are still unclear [35, 36].
Previous studies have confirmed the oncogenic role of HMGB2 in various cancers. After the detection of 3 HMGB2-derived circRNAs in 8 pairs of NSCLC tissues and matched normal tissues, circHMGB2 (hsa_circ_0071452) was found to be highly expressed in tumor tissues, and high circHMGB2 expression was identified as an indicator of poor prognosis in NSCLC patients. Moreover, we further showed that circHMGB2 not only modulated the proliferation of NSCLC cells but also restricted the immune response by fostering the immunosuppressive cancer microenvironment. Notably, the overexpression of circHMGB2 was found to inhibit the type 1 IFN response in NSCLC and enhance the exhaustion of cytotoxic T cells via the miR-181a-5p/CARM1 axis. Moreover, the synergistic combination of the CARM1 inhibitor EZM2302 and an anti-PD-1 antibody significantly inhibited the growth of circHMGB2-overexpressing tumors in mice with a humanized immune system and in immunocompetent mice; these results indicated a promising strategy for immunotherapy for the subgroup of NSCLC patients with high circHMGB2 expression. Therefore, this study provides a new oncogene that induces the formation of an immunosuppressive microenvironment and impairs the efficacy of anti-PD-1 therapy in NSCLC.
The biological functions of circRNAs have been thoroughly investigated in various cancers. Competitive endogenous RNAs (ceRNAs) are a unique feature of circRNAs and have been widely reported in numerous studies [6]. CircRNAs contain nucleotide sequences that interact with miRNAs, and the sponging of miRNAs by circRNAs interrupts the binding of miRNAs to the 3′-UTR regions of target mRNAs, thus eliminating the inhibition of downstream mRNAs and regulating the biological function of cancers [37]. In addition, circRNAs can interact with RNA-binding proteins, which are vital for the transcription and translation of genes or encoded proteins that regulate the progression of cancers [38, 39]. Moreover, circRNAs can directly regulate the transcription of genes, bypassing the sponging of miRNAs [40]. In this study, circHMGB2 was shown to function as a ceRNA for miR-181a-5p and further induce the upregulation of the downstream molecule CARM1. Thus, we conclude that the circHMGB2/miR-181a-5p/CARM1 axis reshapes the TME in NSCLC.
The communication network between cancer cells and the TME is intricate, and understanding the underlying mechanisms is vital to preventing immune evasion and developing effective therapeutic strategies. Previous studies have confirmed the critical roles of circRNAs in the regulation of tumor immunity by both enhancing and inhibiting antitumor immunity [41]. Mechanistically, circRNAs can regulate the expression of immune-related genes in cancer cells at the posttranscriptional level and impact the TME. For instance, circRNAs can modulate the expression of molecules related to the immune checkpoint PD-1/PD-L1 axis and influence the cytotoxic function of effector T lymphocytes [42]. In addition, since exosomes and extracellular vesicles facilitate communication between cells, circRNAs can be transmitted from host cells to the TME via these vehicles to regulate biological functions in immune cells [37]. Furthermore, the levels of circRNAs in the plasma of cancer patients reflect immune infiltration into the TME [43]. In this study, we found that circHMGB2 had a slight effect on the proliferation, but not on the invasion, of NSCLC; it inhibited IFN response-related gene expression and enhanced tumor resistance to cytotoxic T cells, further inducing an immunosuppressive microenvironment in NSCLC.
The type 1 IFN response was first shown to interfere with the antiviral immune response, which protects the host from a second virus attack after primary viral infection [44]. In addition to its antiviral effects, accumulating evidence has confirmed that the type 1 IFN response enhances host immunity and plays a critical role in modulating the activation, differentiation and apoptosis of various subgroups of immune cells [45]. Moreover, previous studies showed the potential effects of type 1 IFNs on host immunosurveillance in the TME and antitumor therapy. The efficacy of antitumor chemotherapy, radiotherapy, targeted therapy and immunotherapy greatly depends on the activation of the type 1 IFN response [44, 46]. In this study, the overexpression of circHMGB2 inhibited the type 1 IFN response and decreased the efficacy of anti-PD-1 therapy in NSCLC. The transcriptional coactivator CARM1 has been reported to be widely involved in biological processes such as cell differentiation, autophagy, metabolism and cancer [47]. Recently, CARM1 was discovered to be a negative regulator of tumor immunity that contributes to resistance to checkpoint blockade therapy [25]. Thus, we found that circHMGB2 induces the formation of an immunosuppressive TME via the miR-181a-5p/CARM1 axis, which inhibits the type 1 IFN response in cancer cells and enhances cancer cell resistance to the cytotoxic effects of T cells.
The discovery of the immune checkpoint PD-1 was a breakthrough in cancer immunotherapy, and drugs targeting this checkpoint have been rapidly commercialized in recent years. Interruption of the PD-1/PD-L1 axis reverses the dysfunctional status of cytotoxic T cells and enhances the immune surveillance of cancer cells. Nonetheless, a large number of patients have no obvious response to PD-1 blockade, which is attributed to the complicated biological regulatory network in most advanced cancers [48]. Therefore, an increasing number of studies have investigated the mechanism underlying resistance to anti-PD-1 therapy and explored the dual blockade of the PD-1/PD-L1 axis and other targets that led to the failure of PD-1 blockade [49]. Since circHMGB2 overexpression upregulated the expression of the downstream target CARM1, which facilitated the formation of an immunosuppressive TME and decreased the efficacy of anti-PD-1 monotherapy, we explored the feasibility of the dual blockade of PD-1 and CARM1. The preclinical results revealed that EZM2302 evidently improved the efficacy of PD-1 blockade in circHMGB2-overexpressing NSCLC in a synergistic manner.
Conclusion
This study revealed a critical role of circHMGB2 in the TME of LUAD and LUSC, and provided a new strategy for improving the efficacy of PD-1 immunotherapy in LUAD and LUSC.
Supplementary Information
Additional file 2: Supplementary Fig. 1. A. The expression of circUSP7 was measured in 3 pairs of NSCLC tissues and matched normal tissues. CircUSP7 was used as a positive control in this study. B. The expression of circHMGB2 was analyzed according to tumor diameter (< 2 cm vs. ≥ 2 cm), lymph node metastasis status (yes vs. no), and TNM stage in 78 LUAD patients. C, Survival analysis of the recurrence and OS of 78 LUAD patients divided into groups according to circHMGB2 expression (circHMGB2high vs. circHMBG2low) was performed using Kaplan–Meier and log rank analysis. Data are presented as the means ± SD; n = 3, *P < 0.05, **P < 0.01, ***P < 0.0001. Supplementary Fig. 2. A. The expression of circHMGB2 was measured in HBE, LLC and 5 NSCLC cell lines (NCI-H460, A549, PC9, H1703, and NCI-H1299 cells) using qRT–PCR. B. The transfection efficiency of three stable cell lines, A549-circHMGB2, H1299-shcircHMGB2 and LLC-circHMGB2, was validated by qRT–PCR. C. The invasion of A549-circHMGB2 and H1299-shcircHMGB2 cells was assessed by Matrigel Transwell assay. D. The migration of A549-circHMGB2 and H1299-shcircHMGB2 cells was assessed by wound healing assay. E, IHC staining of CD4+ T cells, Tregs, and TAM in subcutaneous tumors derived from LLC-control and LLC-circHMGB2 cells. Data are presented as the means ± SD; n = 3, ****P < 0.0001, ns: not significant. Supplementary Fig. 3. A. Electron microscopy image of exosomes in the supernatants of H460 cells. B. The levels of biomarkers of exosomes from the supernatants of NSCLC cells were measured by western blotting. C. The expression of circHMGB2 in exosomes derived from the supernatants of H460-control and H460-circHMGB2 cells was measured by qRT–PCR. Data are presented as the means ± SD; n = 3, ns: not significant. Supplementary Fig. 4. A. The RNA pulldown assay was performed with PC9, A549 and H1299 cells transfected with biotinylated miR-181a-5p. B, The luciferase activity of SIRT1 was measured in HEK-293 T cells transfected with miR-181a-5p. C. The luciferase activity of CDK8 was measured in HEK-293 T cells transfected with miR-181a-5p. D. The luciferase activity of IRS2 was measured in HEK-293 T cells transfected with miR-181a-5p. E. The luciferase activity of BHLHE40 was measured in HEK-293 T cells transfected with miR-181a-5p. Data are presented as the means ± SD; n = 3, *P < 0.05, **P < 0.01, ***P < 0.001, ***P < 0.0001, ns: not significant. Supplementary Fig. 5. A. Comparison of the nucleotide sequences between hsa-miR-181a-5p and mmu-miR-181a-5p. B. The putative binding sites of murine Carm1 and miR-181a-5p. C. The luciferase activity of Carm1 was measured in HEK-293 T cells transfected with miR-181-5p. D. The expression of Carm1 was measured in LLC cells overexpressing both circHMGB2 and miR-181a-5p using qRT–PCR. E. The expression of Carm1 was measured in LLC cells overexpressing both circHMGB2 and miR-181a-5p using western blotting. Data are presented as the means ± SD; n = 3, ****P < 0.0001, ns: not significant. Supplementary Fig. 6. A, MS (mass spectrometry) was performed in PC9, A549 and H1299 cells respectively. B, The Co-IP assay was performed in A549 cells to confirm the interaction between CARM1 and HDAC3. C, The expression of type 1 IFN response-related genes was measured in circHMGB2-overexpressing or CARM1-knockout LLC cells after stimulation with IFN-γ for 48 h. Data are presented as the means ± SD; n = 3, ***P < 0.001, ***P < 0.0001, ns: not significant.
Acknowledgments
None.
Abbreviations
- NSCLC
non-small-cell lung cancer
- LUAD
lung adenocarcinomas
- LUSC
squamous cell carcinomas
- circRNA
circular RNA
- HMGB2
high-mobility group box 2
- qRT–PCR
quantitative real-time polymerase chain reaction
- PD-1
programmed cell death 1
- TMA
tissue microarray
- IHC
immunohistochemistry
- FISH
fluorescence in situ hybridization
- CCK-8
cell counting kit-8
- circRIP
circRNA precipitation
- miRNA
microRNA
- NC
negative control
- OS
overall survival
- RFS
recurrence-free survival
- TME
tumor microenvironment
- PD
progressive disease
- SD
stable disease
- PR
partial remission
Authors’ contributions
Y-B W, L-X Z and J G conceived and designed the experiments; L-X Z, J G, L X and P-F Z performed the experiments; X Y, S-Q Z, X P, B-Q Q, S-W C, F L, K L and J-J X analyzed the data; L-X Z and J G wrote the paper. All authors have read and approved the final manuscript.
Funding
This work was supported by the following grants: National Natural Science Foundation of China (81860520 and 82072575).
Availability of data and materials
All data in our study are available upon request.
Declarations
Ethics approval and consent to participate
The ethical approval was provided by the Ethics Committee of the Second Affiliated Hospital of Nanchang University, and written informed consent was obtained from each patient.
Consent for publication
Not applicable.
Competing interests
None.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Ling-Xian Zhang, Jian Gao, Xiang Long and Peng-Fei Zhang contributed equally to this work.
References
- 1.Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer statistics, 2021. CA Cancer J Clin. 2021;71(1):7–33. doi: 10.3322/caac.21654. [DOI] [PubMed] [Google Scholar]
- 2.Duggan MA, Anderson WF, Altekruse S, Penberthy L, Sherman ME. The surveillance, epidemiology, and end results (SEER) program and pathology: toward strengthening the critical relationship. Am J Surg Pathol. 2016;40(12):e94–e102. doi: 10.1097/PAS.0000000000000749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kristensen LS, Andersen MS, Stagsted L, Ebbesen KK, Hansen TB, Kjems J. The biogenesis, biology and characterization of circular RNAs. Nat Rev Genet. 2019;20(11):675–691. doi: 10.1038/s41576-019-0158-7. [DOI] [PubMed] [Google Scholar]
- 4.Li H, Xu JD, Fang XH, Zhu JN, Yang J, Pan R, Yuan SJ, Zeng N, Yang ZZ, Yang H, Wang XP, Duan JZ, Wang S, Luo JF, Wu SL, Shan ZX. Circular RNA circRNA_000203 aggravates cardiac hypertrophy via suppressing miR-26b-5p and miR-140-3p binding to Gata4. Cardiovasc Res. 2020;116(7):1323–1334. doi: 10.1093/cvr/cvz215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu CX, Li X, Nan F, Jiang S, Gao X, Guo SK, Xue W, Cui Y, Dong K, Ding H, Qu B, Zhou Z, Shen N, Yang L, Chen LL. Structure and degradation of circular RNAs regulate PKR activation in innate immunity. Cell. 2019;177(4):865–880. doi: 10.1016/j.cell.2019.03.046. [DOI] [PubMed] [Google Scholar]
- 6.Goodall GJ, Wickramasinghe VO. RNA in cancer. Nat Rev Cancer. 2021;21(1):22–36. doi: 10.1038/s41568-020-00306-0. [DOI] [PubMed] [Google Scholar]
- 7.Lu J, Wang YH, Yoon C, Huang XY, Xu Y, Xie JW, Wang JB, Lin JX, Chen QY, Cao LL, Zheng CH, Li P, Huang CM. Circular RNA circ-RanGAP1 regulates VEGFA expression by targeting miR-877-3p to facilitate gastric cancer invasion and metastasis. Cancer Lett. 2020;471:38–48. doi: 10.1016/j.canlet.2019.11.038. [DOI] [PubMed] [Google Scholar]
- 8.Chen Q, Liu T, Bao Y, Zhao T, Wang J, Wang H, Wang A, Gan X, Wu Z, Wang L. CircRNA cRAPGEF5 inhibits the growth and metastasis of renal cell carcinoma via the miR-27a-3p/TXNIP pathway. Cancer Lett. 2020;469:68–77. doi: 10.1016/j.canlet.2019.10.017. [DOI] [PubMed] [Google Scholar]
- 9.Zirkel A, Nikolic M, Sofiadis K, Mallm JP, Brackley CA, Gothe H, Drechsel O, Becker C, Altmüller J, Josipovic N, Georgomanolis T, Brant L, Franzen J, Koker M, Gusmao EG, Costa IG, Ullrich RT, Wagner W, Roukos V, Nürnberg P, Marenduzzo D, Rippe K, Papantonis A. HMGB2 loss upon senescence entry disrupts genomic organization and induces CTCF clustering across cell types. Mol Cell. 2018;70(4):730–744. doi: 10.1016/j.molcel.2018.03.030. [DOI] [PubMed] [Google Scholar]
- 10.Hock R, Furusawa T, Ueda T, Bustin M. HMG chromosomal proteins in development and disease. Trends Cell Biol. 2007;17(2):72–79. doi: 10.1016/j.tcb.2006.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kwon JH, Kim J, Park JY, Hong SM, Park CW, Hong SJ, Park SY, Choi YJ, Do IG, Joh JW, Kim DS, Choi KY. Overexpression of high-mobility group box 2 is associated with tumor aggressiveness and prognosis of hepatocellular carcinoma. Clin Cancer Res. 2010;16(22):5511–5521. doi: 10.1158/1078-0432.CCR-10-0825. [DOI] [PubMed] [Google Scholar]
- 12.Fu D, Li J, Wei J, Zhang Z, Luo Y, Tan H, Ren C. HMGB2 is associated with malignancy and regulates Warburg effect by targeting LDHB and FBP1 in breast cancer. Cell Commun Signal. 2018;16(1):8. doi: 10.1186/s12964-018-0219-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lou N, Zhu T, Qin D, Tian J, Liu J. High-mobility group box 2 reflects exacerbated disease characteristics and poor prognosis in non-small cell lung cancer patients. Ir J Med Sci. 2022;191(1):155–162. doi: 10.1007/s11845-021-02549-8. [DOI] [PubMed] [Google Scholar]
- 14.Zhang PF, Pei X, Li KS, Jin LN, Wang F, Wu J, Zhang XM. Circular RNA circFGFR1 promotes progression and anti-PD-1 resistance by sponging miR-381-3p in non-small cell lung cancer cells. Mol Cancer. 2019;18(1):179. doi: 10.1186/s12943-019-1111-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang PF, Gao C, Huang XY, Lu JC, Guo XJ, Shi GM, Cai JB, Ke AW. Cancer cell-derived exosomal circUHRF1 induces natural killer cell exhaustion and may cause resistance to anti-PD1 therapy in hepatocellular carcinoma. Mol Cancer. 2020;19(1):110. doi: 10.1186/s12943-020-01222-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Peng R, Zhang PF, Yang X, Wei CY, Huang XY, Cai JB, Lu JC, Gao C, Sun HX, Gao Q, Bai DS, Shi GM, Ke AW, Fan J. Overexpression of RNF38 facilitates TGF-β signaling by Ubiquitinating and degrading AHNAK in hepatocellular carcinoma. J Exp Clin Cancer Res. 2019;38(1):113. doi: 10.1186/s13046-019-1113-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Drew AE, Moradei O, Jacques SL, Rioux N, Boriack-Sjodin AP, Allain C, Scott MP, Jin L, Raimondi A, Handler JL, Ott HM, Kruger RG, McCabe MT, Sneeringer C, Riera T, Shapiro G, Waters NJ, Mitchell LH, Duncan KW, Moyer MP, Copeland RA, Smith J, Chesworth R, Ribich SA. Identification of a CARM1 inhibitor with potent in vitro and in vivo activity in preclinical models of multiple myeloma. Sci Rep. 2017;7(1):17993. doi: 10.1038/s41598-017-18446-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fang J, Ge X, Xu W, Xie J, Qin Z, Shi L, Yin W, Bian M, Wang H. Bioinformatics analysis of the prognosis and biological significance of HMGB1, HMGB2, and HMGB3 in gastric cancer. J Cell Physiol. 2020;235(4):3438–3446. doi: 10.1002/jcp.29233. [DOI] [PubMed] [Google Scholar]
- 19.Ding C, Yi X, Wu X, Bu X, Wang D, Wu Z, Zhang G, Gu J, Kang D. Exosome-mediated transfer of circRNA CircNFIX enhances temozolomide resistance in glioma. Cancer Lett. 2020;479:1–12. doi: 10.1016/j.canlet.2020.03.002. [DOI] [PubMed] [Google Scholar]
- 20.Chen W, Quan Y, Fan S, Wang H, Liang J, Huang L, Chen L, Liu Q, He P, Ye Y. Exosome-transmitted circular RNA hsa_circ_0051443 suppresses hepatocellular carcinoma progression. Cancer Lett. 2020;475:119–128. doi: 10.1016/j.canlet.2020.01.022. [DOI] [PubMed] [Google Scholar]
- 21.Yu Q, Dong L, Li Y, Liu G. SIRT1 and HIF1α signaling in metabolism and immune responses. Cancer Lett. 2018;418:20–26. doi: 10.1016/j.canlet.2017.12.035. [DOI] [PubMed] [Google Scholar]
- 22.Steinparzer I, Sedlyarov V, Rubin JD, Eislmayr K, Galbraith MD, Levandowski CB, Vcelkova T, Sneezum L, Wascher F, Amman F, Kleinova R, Bender H, Andrysik Z, Espinosa JM, Superti-Furga G, Dowell RD, Taatjes DJ, Kovarik P. Transcriptional responses to IFN-γ require mediator kinase-dependent pause release and mechanistically distinct CDK8 and CDK19 functions. Mol Cell. 2019;76(3):485–499. doi: 10.1016/j.molcel.2019.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zarling AL, Obeng RC, Desch AN, Pinczewski J, Cummings KL, Deacon DH, Conaway M, Slingluff CJ, Engelhard VH. MHC-restricted phosphopeptides from insulin receptor substrate-2 and CDC25b offer broad-based immunotherapeutic agents for cancer. Cancer Res. 2014;74(23):6784–6795. doi: 10.1158/0008-5472.CAN-14-0043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li C, Zhu B, Son YM, Wang Z, Jiang L, Xiang M, Ye Z, Beckermann KE, Wu Y, Jenkins JW, Siska PJ, Vincent BG, Prakash YS, Peikert T, Edelson BT, Taneja R, Kaplan MH, Rathmell JC, Dong H, Hitosugi T, Sun J. The transcription factor Bhlhe40 programs mitochondrial regulation of resident CD8(+) T cell fitness and functionality. Immunity. 2019;51(3):491–507. doi: 10.1016/j.immuni.2019.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kumar S, Zeng Z, Bagati A, Tay RE, Sanz LA, Hartono SR, Ito Y, Abderazzaq F, Hatchi E, Jiang P, Cartwright A, Olawoyin O, Mathewson ND, Pyrdol JW, Li MZ, Doench JG, Booker MA, Tolstorukov MY, Elledge SJ, Chédin F, Liu XS, Wucherpfennig KW. CARM1 inhibition enables immunotherapy of resistant tumors by dual action on tumor cells and T cells. Cancer Discov. 2021;11(8):2050–2071. doi: 10.1158/2159-8290.CD-20-1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Maser IP, Hoves S, Bayer C, Heidkamp G, Nimmerjahn F, Eckmann J, Ries CH. The tumor milieu promotes functional human tumor-resident Plasmacytoid dendritic cells in humanized mouse models. Front Immunol. 2020;11:2082. doi: 10.3389/fimmu.2020.02082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Guarda G, Braun M, Staehli F, Tardivel A, Mattmann C, Förster I, Farlik M, Decker T, Du Pasquier RA, Romero P, Tschopp J. Type I interferon inhibits interleukin-1 production and inflammasome activation. Immunity. 2011;34(2):213–223. doi: 10.1016/j.immuni.2011.02.006. [DOI] [PubMed] [Google Scholar]
- 28.Michalska A, Blaszczyk K, Wesoly J, Bluyssen H. A positive feedback amplifier circuit that regulates interferon (IFN)-stimulated gene expression and controls type I and type II IFN responses. Front Immunol. 2018;9:1135. doi: 10.3389/fimmu.2018.01135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Krämer OH, Knauer SK, Greiner G, Jandt E, Reichardt S, Gührs KH, Stauber RH, Böhmer FD, Heinzel T. A phosphorylation-acetylation switch regulates STAT1 signaling. Genes Dev. 2009;23(2):223–235. doi: 10.1101/gad.479209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhou R, Wu Y, Wang W, Su W, Liu Y, Wang Y, Fan C, Li X, Li G, Li Y, Xiong W, Zeng Z. Circular RNAs (circRNAs) in cancer. Cancer Lett. 2018;425:134–142. doi: 10.1016/j.canlet.2018.03.035. [DOI] [PubMed] [Google Scholar]
- 31.Sanger HL, Klotz G, Riesner D, Gross HJ, Kleinschmidt AK. Viroids are single-stranded covalently closed circular RNA molecules existing as highly base-paired rod-like structures. Proc Natl Acad Sci U S A. 1976;73(11):3852–3856. doi: 10.1073/pnas.73.11.3852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Devaux Y, Creemers EE, Boon RA, Werfel S, Thum T, Engelhardt S, Dimmeler S, Squire I. Circular RNAs in heart failure. Eur J Heart Fail. 2017;19(6):701–709. doi: 10.1002/ejhf.801. [DOI] [PubMed] [Google Scholar]
- 33.Chen X, Yang T, Wang W, Xi W, Zhang T, Li Q, Yang A, Wang T. Circular RNAs in immune responses and immune diseases. Theranostics. 2019;9(2):588–607. doi: 10.7150/thno.29678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cao Q, Guo Z, Du S, Ling H, Song C. Circular RNAs in the pathogenesis of atherosclerosis. Life Sci. 2020;255:117837. doi: 10.1016/j.lfs.2020.117837. [DOI] [PubMed] [Google Scholar]
- 35.Lei M, Zheng G, Ning Q, Zheng J, Dong D. Translation and functional roles of circular RNAs in human cancer. Mol Cancer. 2020;19(1):30. doi: 10.1186/s12943-020-1135-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ng WL, Mohd MT, Shukla K. Functional role of circular RNAs in cancer development and progression. RNA Biol. 2018;15(8):995–1005. doi: 10.1080/15476286.2018.1486659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang Q, Wang W, Zhou Q, Chen C, Yuan W, Liu J, Li X, Sun Z. Roles of circRNAs in the tumour microenvironment. Mol Cancer. 2020;19(1):14. doi: 10.1186/s12943-019-1125-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Conn SJ, Pillman KA, Toubia J, Conn VM, Salmanidis M, Phillips CA, Roslan S, Schreiber AW, Gregory PA, Goodall GJ. The RNA binding protein quaking regulates formation of circRNAs. Cell. 2015;160(6):1125–1134. doi: 10.1016/j.cell.2015.02.014. [DOI] [PubMed] [Google Scholar]
- 39.Zheng X, Chen L, Zhou Y, Wang Q, Zheng Z, Xu B, Wu C, Zhou Q, Hu W, Wu C, Jiang J. A novel protein encoded by a circular RNA circPPP1R12A promotes tumor pathogenesis and metastasis of colon cancer via hippo-YAP signaling. Mol Cancer. 2019;18(1):47. doi: 10.1186/s12943-019-1010-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yang Q, Du WW, Wu N, Yang W, Awan FM, Fang L, et al. A circular RNA promotes tumorigenesis by inducing c-myc nuclear translocation. Cell Death Differ. 2017;24(9):1609-1620. [DOI] [PMC free article] [PubMed]
- 41.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 42.Zhang XL, Xu LL, Wang F. Hsa_circ_0020397 regulates colorectal cancer cell viability, apoptosis and invasion by promoting the expression of the miR-138 targets TERT and PD-L1. Cell Biol Int. 2017;41(9):1056–1064. doi: 10.1002/cbin.10826. [DOI] [PubMed] [Google Scholar]
- 43.Weng Q, Chen M, Li M, Zheng YF, Shao G, Fan W, Xu XM, Ji J. Global microarray profiling identified hsa_circ_0064428 as a potential immune-associated prognosis biomarker for hepatocellular carcinoma. J Med Genet. 2019;56(1):32–38. doi: 10.1136/jmedgenet-2018-105440. [DOI] [PubMed] [Google Scholar]
- 44.Zitvogel L, Galluzzi L, Kepp O, Smyth MJ, Kroemer G. Type I interferons in anticancer immunity. Nat Rev Immunol. 2015;15(7):405–414. doi: 10.1038/nri3845. [DOI] [PubMed] [Google Scholar]
- 45.Fuertes MB, Woo SR, Burnett B, Fu YX, Gajewski TF. Type I interferon response and innate immune sensing of cancer. Trends Immunol. 2013;34(2):67–73. doi: 10.1016/j.it.2012.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Montoya M, Schiavoni G, Mattei F, Gresser I, Belardelli F, Borrow P, Tough DF. Type I interferons produced by dendritic cells promote their phenotypic and functional activation. Blood. 2002;99(9):3263–3271. doi: 10.1182/blood.V99.9.3263. [DOI] [PubMed] [Google Scholar]
- 47.Suresh S, Huard S, Dubois T. CARM1/PRMT4: making its mark beyond its function as a transcriptional Coactivator. Trends Cell Biol. 2021;31(5):402–417. doi: 10.1016/j.tcb.2020.12.010. [DOI] [PubMed] [Google Scholar]
- 48.Xu-Monette ZY, Zhang M, Li J, Young KH. PD-1/PD-L1 blockade: have we found the key to unleash the antitumor immune response? Front Immunol. 2017;8:1597. doi: 10.3389/fimmu.2017.01597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Melero I, Berman DM, Aznar MA, Korman AJ, Pérez GJ, Haanen J. Evolving synergistic combinations of targeted immunotherapies to combat cancer. Nat Rev Cancer. 2015;15(8):457–472. doi: 10.1038/nrc3973. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 2: Supplementary Fig. 1. A. The expression of circUSP7 was measured in 3 pairs of NSCLC tissues and matched normal tissues. CircUSP7 was used as a positive control in this study. B. The expression of circHMGB2 was analyzed according to tumor diameter (< 2 cm vs. ≥ 2 cm), lymph node metastasis status (yes vs. no), and TNM stage in 78 LUAD patients. C, Survival analysis of the recurrence and OS of 78 LUAD patients divided into groups according to circHMGB2 expression (circHMGB2high vs. circHMBG2low) was performed using Kaplan–Meier and log rank analysis. Data are presented as the means ± SD; n = 3, *P < 0.05, **P < 0.01, ***P < 0.0001. Supplementary Fig. 2. A. The expression of circHMGB2 was measured in HBE, LLC and 5 NSCLC cell lines (NCI-H460, A549, PC9, H1703, and NCI-H1299 cells) using qRT–PCR. B. The transfection efficiency of three stable cell lines, A549-circHMGB2, H1299-shcircHMGB2 and LLC-circHMGB2, was validated by qRT–PCR. C. The invasion of A549-circHMGB2 and H1299-shcircHMGB2 cells was assessed by Matrigel Transwell assay. D. The migration of A549-circHMGB2 and H1299-shcircHMGB2 cells was assessed by wound healing assay. E, IHC staining of CD4+ T cells, Tregs, and TAM in subcutaneous tumors derived from LLC-control and LLC-circHMGB2 cells. Data are presented as the means ± SD; n = 3, ****P < 0.0001, ns: not significant. Supplementary Fig. 3. A. Electron microscopy image of exosomes in the supernatants of H460 cells. B. The levels of biomarkers of exosomes from the supernatants of NSCLC cells were measured by western blotting. C. The expression of circHMGB2 in exosomes derived from the supernatants of H460-control and H460-circHMGB2 cells was measured by qRT–PCR. Data are presented as the means ± SD; n = 3, ns: not significant. Supplementary Fig. 4. A. The RNA pulldown assay was performed with PC9, A549 and H1299 cells transfected with biotinylated miR-181a-5p. B, The luciferase activity of SIRT1 was measured in HEK-293 T cells transfected with miR-181a-5p. C. The luciferase activity of CDK8 was measured in HEK-293 T cells transfected with miR-181a-5p. D. The luciferase activity of IRS2 was measured in HEK-293 T cells transfected with miR-181a-5p. E. The luciferase activity of BHLHE40 was measured in HEK-293 T cells transfected with miR-181a-5p. Data are presented as the means ± SD; n = 3, *P < 0.05, **P < 0.01, ***P < 0.001, ***P < 0.0001, ns: not significant. Supplementary Fig. 5. A. Comparison of the nucleotide sequences between hsa-miR-181a-5p and mmu-miR-181a-5p. B. The putative binding sites of murine Carm1 and miR-181a-5p. C. The luciferase activity of Carm1 was measured in HEK-293 T cells transfected with miR-181-5p. D. The expression of Carm1 was measured in LLC cells overexpressing both circHMGB2 and miR-181a-5p using qRT–PCR. E. The expression of Carm1 was measured in LLC cells overexpressing both circHMGB2 and miR-181a-5p using western blotting. Data are presented as the means ± SD; n = 3, ****P < 0.0001, ns: not significant. Supplementary Fig. 6. A, MS (mass spectrometry) was performed in PC9, A549 and H1299 cells respectively. B, The Co-IP assay was performed in A549 cells to confirm the interaction between CARM1 and HDAC3. C, The expression of type 1 IFN response-related genes was measured in circHMGB2-overexpressing or CARM1-knockout LLC cells after stimulation with IFN-γ for 48 h. Data are presented as the means ± SD; n = 3, ***P < 0.001, ***P < 0.0001, ns: not significant.
Data Availability Statement
All data in our study are available upon request.