Abstract
Mutations in several ribosomal proteins are known to be related to antibiotic resistance. For several strains of Escherichia coli, the mutated protein is known but the amino acid actually altered has not been documented. Characterization of these determinants for antibiotic resistance in proteins will further the understanding of the precise mechanism of the antibiotic action as well as provide markers for resistance. Mass spectrometry can be used as a valuable tool to rapidly locate and characterize mutant proteins by using a small amount of material. We have used electrospray and matrix-assisted laser desorption ionization–time of flight (MALDI–TOF) mass spectrometry to map out all 56 ribosomal proteins in E. coli based on intact molecular masses. We used this fingerprinting approach to locate variants of ribosomal proteins displaying a change in mass. In particular we have studied proteins responsible for streptomycin, erythromycin, and spectinomycin resistance in three strains of E. coli, and then we characterized each mutation responsible for resistance by analyzing tryptic peptides of these proteins by using MALDI-TOF and nanoelectrospray tandem mass spectrometry. The results provided markers for antibiotic resistance and demonstrated that mass spectrometry can be used to rapidly investigate changes in individual proteins from a complex with picomole amounts of protein.
Antibiotic resistance presents a significant challenge to scientists in the field of infectious diseases. Identification of protein determinants for resistance will not only provide markers for resistance to a particular drug but will also aid in the understanding of the mechanisms of antibiotic function and resistance. Several antibiotics act by targeting protein biosynthesis, interacting with ribosomal structural proteins, rRNAs, and ribosomal-associated proteins (23). Understanding more about the precise interactions between antibiotics and ribosomal proteins will be especially informative as more detailed information is revealed in higher-resolution structures of the ribosome (4, 9, 11, 15). In particular, mutations in ribosomal proteins L22, S5, and S12 have been observed to confer resistance to erythromycin (10), spectinomycin (7, 14), and streptomycin (1, 24, 25), respectively, in Escherichia coli. As bacteria are exposed to these drugs, the specific changes in ribosomal proteins are incorporated. Patterns of mutations at the particular sites in various strains of E. coli emerge for each drug (10, 14, 25). A method to rapidly screen bacteria for mutations in proteins would be a valuable tool for evaluating antibiotic mechanisms and for determining an appropriate treatment of an infection. Further insights into the antibiotic mechanism could be gleaned from the details of the mutation. For example, knowing which specific amino acid alterations in ribosomal proteins L4 and L22 result in resistance to erythromycin as well as sites in 23S rRNA whose mutations also confer resistance provides information not only on details on the mechanism of erythromycin function but also on details of functional interactions between ribosomal proteins and rRNA (10).
Mass spectrometry (MS) is a powerful tool in protein analysis. Electrospray (13) and matrix-assisted laser desorption ionization (MALDI) (17) time-of-flight (TOF) (8, 12, 27) technologies can be used to precisely detect small changes in the masses of proteins and peptides. These techniques involve the ionization of molecules into products that can be detected. The mass-to-charge ratio of gas phase ions can then be correlated with the molecular structure of the initial species. Electrospray ionization involves an electric field applied to a solution sprayed from a needle. In MALDI, gas phase ions are generated by desorption ionization of the molecule of interest from a layer of crystals formed from volatile matrix molecules. By using these techniques mutated proteins can be detected rapidly, and the precise site of the mutation can be characterized using tandem MS/MS of peptides of the protein. Recently, 55 of the 56 E. coli ribosomal proteins with posttranslational modifications were observed by MALDI-TOF MS (3). We have extended this approach for analysis of antibiotic resistance in E. coli. All 56 ribosomal proteins have been observed in electrospray and/or MALDI-TOF mass spectra. Additionally, three strains of E. coli that exhibit resistance to erythromycin, spectinomycin, and streptomycin due to alterations in the L22, S5, and S12 ribosomal proteins, respectively, have been analyzed. Mass spectral analyses revealed the size of the mass change for the mutated protein associated with resistance, and tandem MS/MS analyses were used to identify the exact site of the mutation by direct analysis of the protein following proteolytic digestion with trypsin. The mutations detected in the previously uncharacterized strains were the same as mutations known to confer resistance in other strains of E. coli. This observation confirmed that the mutations detected were responsible for the resistance observed in these strains. This technique provides a rapid approach to characterize mutations in resistant strains of E. coli that have not been previously characterized.
MATERIALS AND METHODS
Preparation of ribosomal proteins.
The erythromycin- and spectinomycin-resistant strains of E. coli, N281 (rplV281) and E5014 (rpsE2112), respectively, were obtained from Steven T. Gregory and Albert E. Dahlberg at Brown University. For isolation of specific ribosomal proteins, 10-liter fermentations of E. coli strains MRE600, N281, E5014, and a streptomycin-resistant strain generated in-house, called UC15121 (rpsL100), were prepared in Luria broth. A 0.1-ml aliquot of a glycerol stock culture was inoculated into a 100-ml volume of Luria broth, pH 7.5, contained in a 500-ml wide mouth flask. The culture was grown at 37°C, with a shaking rate of 200 rpm. After 16 to 18 h, 4% of this culture was transferred into fresh Luria broth. The culture was grown under the same conditions for an additional 4 to 5 h until late log phase. Cells were then harvested by centrifugation and were stored at −80°C. The 70S ribosomes were prepared similarly to a method described previously (22) with the following alterations. The buffer used consisted of 10 mM Tris, pH 7.5, 5 mM MgCl2, 1 mM dithiothreitol, 60 mM NH4Cl. Lysis was achieved by grinding with alumina, and cells were resuspended in buffer by gentle rocking on ice for at least 30 min. The sucrose gradient used to isolate the ribosomes consisted of 10 ml of 10% sucrose on top of 25 ml of 40% sucrose. Aliquots of ribosomes in suspension were stored at −80°C. Ribosomal proteins were extracted with acetone following acetic acid precipitation of rRNA as described previously (5). Aliquots of the proteins were stored at −20°C.
MALDI-TOF (MS) of intact proteins.
Ribosomal proteins were resuspended in either 0.1 or 1.0% trifluoroacetic acid (TFA) to give 25 mg/ml. A saturated solution of sinapinic acid was prepared in a solution containing 30% acetonitrile and 0.1% TFA for the matrix solution. The sample for MALDI-TOF analysis was then prepared by diluting 1.0 μl of protein solution to 10 μl with matrix solution and was spotted onto a flat, stainless steel target from PerSeptive Biosystems. MALDI-TOF data were collected on a PerSeptive Voyager Elite MALDI-TOF instrument with delayed extraction in positive-ion linear mode. Typical instrument operating settings were 25 kV total accelerating voltage, 91% grid voltage, 0.2% guide wire voltage, delay of 100 to 200 nanoseconds, and 567.2-Da low mass gate. Data were analyzed using GRAMS software (Galactical). The observed masses for two or three ribosomal proteins measured with sufficiently high intensity were used to internally calibrate each mass spectrum.
Electrospray MS of intact proteins.
Ribosomal proteins were resuspended in 0.1% TFA and loaded onto a C4 reversed-phase high-performance liquid chromatography (HPLC) column. Proteins were either analyzed by a Thermoquest LCQ electrospray mass spectrometer in-line or collected in fractions for off-line analysis. For in-line analysis, proteins were eluted in a 60-min gradient from 95% A and 5% B to 100% B, where A was 0.1% TFA and B was 0.1% TFA–90% acetonitrile, directly into the mass spectrometer. For off-line analysis, proteins were eluted in a 120-min gradient from 80% A, 20% B to 15% A, 85% B, where A was 0.1% TFA and B was 0.1% TFA–80% acetonitrile, and collected in 30-s fractions. Fractions near the expected elution time of each mutated protein, based on elution times from MRE600 ribosomal proteins, were then analyzed by electrospray MS. The mass spectral data were analyzed using Xcalibur (Thermoquest).
Tryptic peptide analysis.
HPLC fractions containing separated ribosomal proteins were dried in a speedvac centrifuge to remove the acetonitrile and TFA. Each fraction of proteins was resuspended in 100 μl of 50 mM ammonium bicarbonate. Approximately 0.2 μg of trypsin (sequencing grade modified; Promega) was added to each tube and incubated at 37°C for 2 to 4 h. Digestion was terminated by placing the reaction mixture on ice. The peptides were prepared for MALDI-TOF analysis by being bound to a C18 ZipTip (Millipore) in 0.1% TFA and being eluted in 6 μl of 0.1% TFA– 80% acetonitrile. MALDI-TOF matrix was prepared by dissolving α-cyano-4-hydroxycinnamic acid (CHCA/NC) to a concentration of 20 mg/ml in a solution of (10 mg/ml) nitrocellulose in 50% acetone–50% isopropanol. The matrix solution was spotted onto the MALDI target and allowed to air dry. An equal volume of the peptide mixture was spotted on top of the CHCA/NC matrix and allowed to air dry. MALDI-TOF data were collected on a PerSeptive Biosystems Voyager Elite MALDI-TOF instrument with delayed extraction in positive-ion reflector mode. Typical instrument operating settings were 20 kV total accelerating voltage, 72% grid voltage, 0.055% guide wire voltage, 100nanosecond delay, and 600-Da low mass gate. Data were analyzed using GRAMS software and were used to search the Swiss-Prot database for protein identification using the MS-Fit program of Protein Prospector (P. R. Baker and K. R. Clauser, http://prospector.ucsf.edu). Altered peptides were then analyzed by nanoelectrospray tandem MS (nanospray MS/MS) on a Micromass Q-TOF instrument equipped with a Z-spray ion source using Econo 10 nanospray needles (New Objectives). Peptide parent mass values were recorded in MS mode and were used to acquire MS/MS data on the Q-TOF. Collision energy in the MS/MS experiments was optimized manually for individual peptides being analyzed.
RESULTS
Identification of all 56 ribosomal proteins.
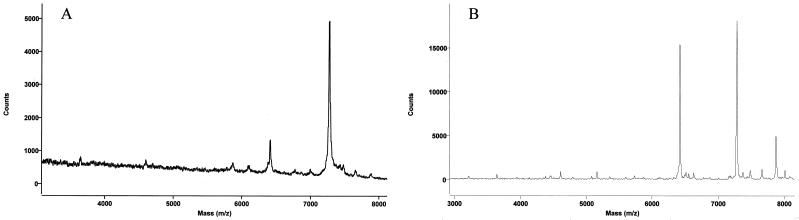
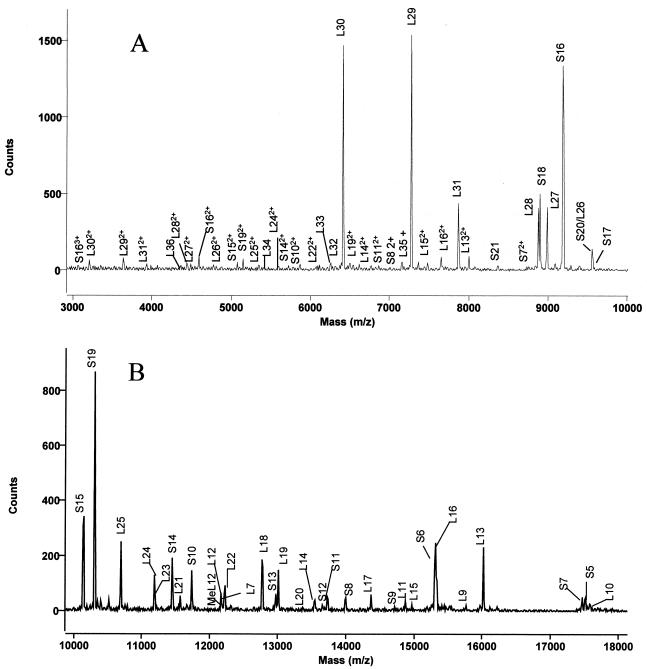
E. coli ribosomal proteins were analyzed by both MALDI-TOF and electrospray MS. The proteins were assigned by comparison with masses calculated from amino acid sequences and posttranslational modifications in the Swiss-Prot database. MALDI-TOF mass spectral data revealed 54 out of 56 MRE600 ribosomal proteins from analyses of two spots on a MALDI target. One spot was prepared using proteins dissolved in 0.1% TFA, and the other was prepared using proteins dissolved in 1.0% TFA. Sinapinic acid was used as the matrix, since it has been shown to selectively ionize large proteins well (6). As can be seen in Fig. 1, ionization of different proteins by this method was dependent upon the amount of TFA used, making it necessary to compile data from spectra obtained in 0.1% TFA and in 1.0% TFA for the most complete intact mass mapping. Two representative spectra, exhibiting many of the ribosomal proteins, are shown in Fig. 2. As can be seen in this figure, several proteins ionize more readily than others do; this method should not be used to quantitate between individual proteins. In general, large proteins are often difficult to observe by MALDI-TOF in the presence of smaller proteins because competitive ionization occurs, whereas small proteins ionize readily compared to larger proteins (18). The only proteins that were not observed by MALDI were the two largest E. coli ribosomal proteins, S1 and L2, with predicted molecular sizes of 61,158 and 29,729 Da, respectively. Many proteins exhibited multiply-charged species in addition to the singly-charged state, e.g., S16. The intact mass can be calculated from these species by multiplying the mass observed, which is actually the mass-to-charge ratio, by the number of charges (20).
FIG. 1.
MALDI-TOF spectra of ribosomal proteins from 3 to 8 kDa, dissolved in 0.1% TFA (A) or 1.0% TFA (B). Ribosomal proteins were dissolved in TFA, mixed with matrix, and spotted onto the stainless steel target. The y axis is the number of counts generated in the MALDI-TOF mass spectrometer, i.e., the intensity of each signal.
FIG. 2.
MALDI-TOF spectra of ribosomal proteins from 3 to 10 kDa (A) or 10 to 18 kDa (B). Ribosomal proteins were dissolved in 1.0% TFA, mixed with matrix, and spotted onto the stainless steel target. The unmodified L12 is indicated as L12, and the methylated form is indicated as MeL12.
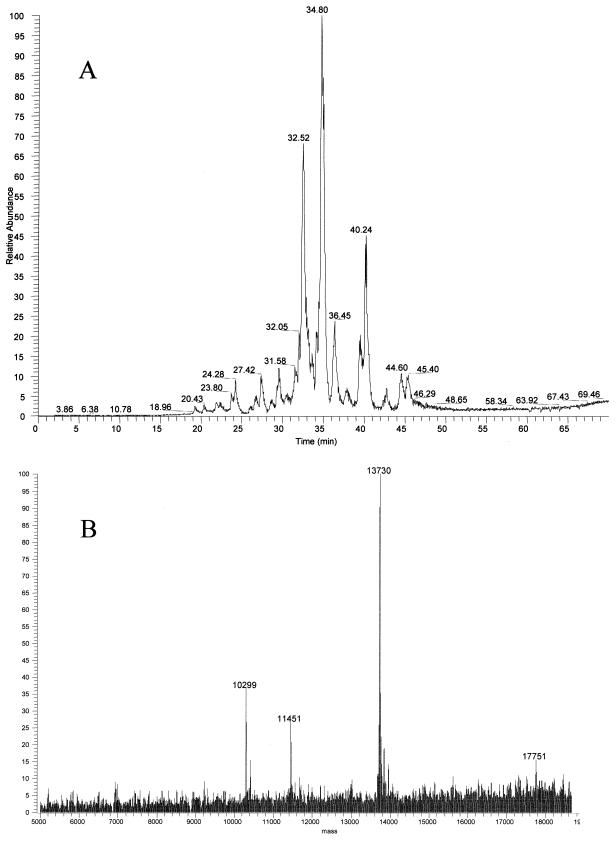
HPLC coupled to electrospray ionization MS was also used in mapping ribosomal proteins by intact mass analysis. Figure 3A displays a representative total ion current (TIC) chromatogram of ribosomal proteins from MRE600 cells from a reversed-phase column by HPLC. As can be seen there are not 56 separated peaks, yet all 56 E. coli ribosomal proteins were observed by electrospray MS. This was possible because the ion signals for several different masses could be deconvoluted from one another within a single peak (Fig. 3B). Therefore, high-resolution HPLC separation was unnecessary, since the intact masses of ribosomal proteins could be determined using a relatively short elution gradient.
FIG. 3.
Reversed-phase HPLC of ribosomal protein separation. (A) Proteins were eluted in a 60-min gradient from 95% A and 5% B to 100% B, where A was 0.1% TFA and B was 0.1% TFA–90% acetonitrile, directly into the LCQ electrospray mass spectrometer. (B) Deconvoluted electrospray mass spectrum demonstrating multiple masses eluting at the same retention time. The y axis of the electrospray spectrum represents the relative intensity of each signal.
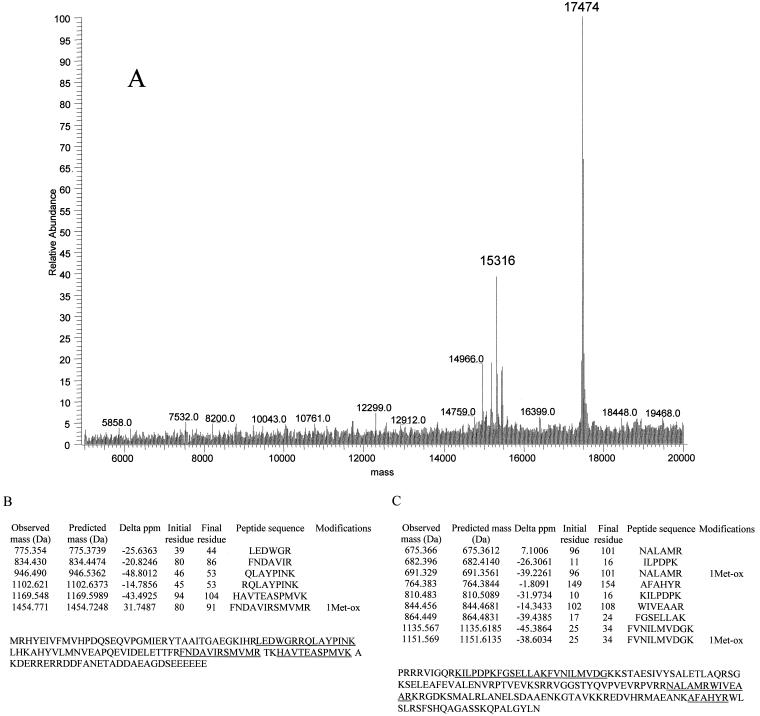
Masses of 15,316 and 17,474 Da (Fig. 4A), which were not reported in an earlier MALDI-TOF study of E. coli ribosomal proteins (3), were observed by both electrospray and MALDI-TOF in this study. These proteins eluted closely in a single HPLC fraction and were digested with trypsin. MALDI-TOF analyses of the tryptic fragments revealed that the 15,316-Da and 17,474-Da proteins were ribosomal proteins S6 (28% coverage) and S7 (24% coverage), respectively (Fig. 4B and C). The gene for the S6 protein encodes two C-terminal Glu residues. Additional Glu residues are known to be added to the C terminus posttranslationally by the enzyme RimK (16). The mass observed at 15,316 Da corresponds well to the predicted mass for S6 with three C-terminal Glu residues (Table 1). Electrospray spectra also reveal the presence of S6 with two C-terminal Glu residues, 15,187 Da, and four C-terminal residues, 15,444 Da, at lower abundance. The protein with a mass of 17,474 Da is a variant form of ribosomal protein S7. E. coli strain B is known to produce a form of S7 missing the C-terminal residues 156 to 178 (21), which would yield a predicted mass of 17,473 Da. Mass assignment to these proteins demonstrated that this technique could be used to detect variants in ribosomal proteins. Table 1 summarizes the average masses observed for E. coli ribosomal proteins by these methods along with the corresponding average masses calculated from the protein sequences.
FIG. 4.
Identification of atypical masses in ribosomal proteins. (A) Deconvoluted electrospray mass spectrum revealed two aberrant signals. Masses of 15,316 Da and 17,474 Da did not match the predicted masses of E. coli ribosomal proteins. (B) Tryptic peptides observed by MALDI-TOF were then used to identify the proteins as ribosomal proteins S6 and S7 (C). The masses of observed and predicted masses are displayed. The protein sequence for each identified protein is indicated, and the covered peptides are underlined.
TABLE 1.
Masses of E. coli ribosomal proteins determined by MALDI-TOF and electrospray MS
| Protein | Predicted mass (Da) | MALDI-TOF mass (Da) | Difference (Da) | Electrospray mass (Da) | Difference (Da) | Modificationsa |
|---|---|---|---|---|---|---|
| S1 | 61,158.2 | 61,159.0 | +0.8 | |||
| S2 | 26,612.5 | 26,616.9 | +4.4 | 26,623.4 | +10.9 | |
| S3 | 25,852.1 | 25,848.1 | −4.0 | 25,853.2 | +1.1 | |
| S4 | 23,338.0 | 23,335.1 | −2.9 | 23,337.4 | −0.6 | |
| S5 | 17,472.2 | 17,515.0 | +42.8 | 17,514.2 | +42.0 | Acetylated |
| S6 | 15,187.1 | 15,187.3 | +0.2 | C-terminal EE | ||
| S6 | 15,316.2 | 15,314.6 | −1.6 | 15,316.5 | +0.3 | C-terminal EEE |
| S6 | 15,445.3 | 15,444.5 | −0.8 | C-terminal EEEE | ||
| S7 | 19,887.9 | 17,474.1 | −2413.8 | 17,474.1 | −2,413.8 | No residues 156 to 178 |
| S8 | 13,995.4 | 13,994.8 | −0.6 | 13,995.1 | −0.3 | |
| S9 | 14,725.0 | 14,725.7 | +0.7 | 14,725.1 | +0.1 | |
| S10 | 11,735.6 | 11,735.6 | 0.0 | 11,734.6 | −1.0 | |
| S11 | 13,713.8 | 13,728.6 | +14.8 | 13,727.8 | +14.0 | Methylated |
| S12 | 13,605.9 | 13,650.7 | −0.2 | 13,651.9 | +1.0 | β-Methylthiolated |
| S13 | 12,968.2 | 12,967.8 | −0.4 | 12,968.1 | −0.1 | |
| S14 | 11,449.3 | 11,449.2 | −0.1 | 11,449.5 | +0.2 | |
| S15 | 10,137.6 | 10,137.4 | −0.2 | 10,137.5 | −0.1 | |
| S16 | 9,190.6 | 9,190.8 | +0.2 | 9,190.4 | −0.2 | |
| S17 | 9,573.3 | 9,573.7 | +0.4 | 9,573.4 | +0.1 | |
| S18 | 8,855.3 | 8,897.2 | +41.9 | 8,897.8 | +42.5 | Acetylated |
| S19 | 10,299.1 | 10,299.1 | 0.0 | 10,298.9 | −0.2 | |
| S20 | 9,553.2 | 9,554.3 | +1.1 | 9,553.2 | 0.0 | |
| S21 | 8,368.8 | 8,370.5 | +1.7 | 8,368.3 | −0.5 | |
| L1 | 24,598.5 | 24,601.1 | +2.6 | 24,598.4 | −0.1 | |
| L2 | 29,729.3 | 29,729.3 | 0.0 | |||
| L3 | 22,243.6 | 22,260.9 | +17.3 | 22,257.6 | +14.0 | Methylated |
| L4 | 22,086.6 | 22,089.2 | +2.6 | 22,086.7 | +0.1 | |
| L5 | 20,170.4 | 20,170.5 | +0.1 | 20,170.5 | +0.1 | |
| L6 | 18,772.6 | 18,772.4 | −0.2 | 18,773.3 | +0.7 | |
| L7 | 12,164.0 | 12,205.4 | +41.4 | 12,204.6 | +40.6 | Acetylated |
| L9 | 15,769.1 | 15,770.7 | +1.6 | 15,770.0 | +0.9 | |
| L10 | 17,580.4 | 17,583.8 | +3.4 | 17,580.5 | +0.1 | |
| L11 | 14,744.2 | 14,870.8 | +126.6 | 14,869.4 | 125.2 | Methylated 9× |
| L12 | 12,164.0 | 12,164.8 | +0.8 | 12,163.7 | −0.3 | |
| L12 | 12,164.0 | 12,180.8 | +16.8 | 12,176.5 | +12.5 | Methylated |
| L13 | 16,018.6 | 16,018.6 | 0.0 | 16,018.4 | −0.2 | |
| L14 | 13,541.1 | 13,541.6 | +0.5 | 13,541.0 | −0.1 | |
| L15 | 14,980.4 | 14,967.5 | −12.9 | 14,966.6 | −13.8 | |
| L16 | 15,281.2 | 15,325.9 | +44.7 | 15,326.3 | +45.1 | Unknown |
| L17 | 14,364.6 | 14,364.9 | +0.3 | 14,364.6 | 0.0 | |
| L18 | 12,769.6 | 12,769.3 | −0.3 | 12,769.8 | +0.2 | |
| L19 | 13,002.1 | 13,001.9 | −0.2 | 13,002.6 | +0.5 | |
| L20 | 13,365.8 | 13,364.7 | −1.1 | 13,364.7 | −1.1 | |
| L21 | 11,564.4 | 11,564.1 | −0.3 | 11,563.1 | −1.0 | |
| L22 | 12,226.3 | 12,226.5 | +0.2 | 12,226.3 | 0.0 | |
| L23 | 11,199.1 | 11,196.5 | −2.6 | 11,198.8 | −0.3 | |
| L24 | 11,185.0 | 11,185.6 | +0.6 | 11,184.9 | −0.1 | |
| L25 | 10,693.5 | 10,693.4 | −0.1 | 10,693.6 | +0.1 | |
| L26 | 9,553.2 | 9,554.3 | +1.1 | 9,553.2 | 0.0 | |
| L27 | 8,993.3 | 8,993.8 | +0.5 | 8,992.5 | −0.8 | |
| L28 | 8,875.3 | 8,875.4 | +0.1 | 8,875.3 | 0.0 | |
| L29 | 7,273.5 | 7,273.2 | −0.3 | 7,272.9 | +0.4 | |
| L30 | 6,410.6 | 6,410.8 | +0.2 | 6,409.8 | −1.0 | |
| L31 | 7,871.1 | 7,871.0 | −0.1 | 7,870.7 | −0.4 | |
| L32 | 6,315.2 | 6,314.9 | −0.3 | 6,314.6 | −0.6 | |
| L33 | 6,240.4 | 6,255.2 | +14.8 | 6,254.0 | +13.6 | Methylated |
| L34 | 5,380.4 | 5,378.9 | −1.5 | 5,379.9 | −0.5 | |
| L35 | 7,157.8 | 7,159.1 | +1.3 | 7,157.4 | −0.4 | |
| L36 | 4,364.4 | 4,367.3 | −0.9 | 4,363.4 | 0.0 |
Characterization of mutated ribosomal proteins.
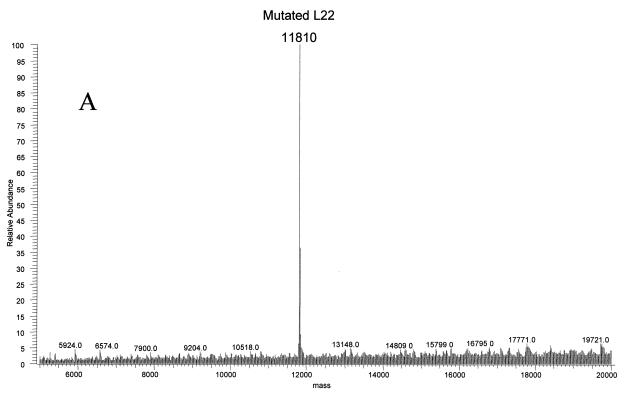
Ribosomal proteins from three strains of E. coli were isolated to investigate antibiotic resistance. The strains N281, UC15121, and E5014 display resistance to erythromycin, streptomycin, and spectinomycin, respectively. Resistance to erythromycin in the N281 strain was known to be conferred by deletion of three residues, MKR at positions 82 to 84, in the L22 ribosomal protein (rplV281) (10). This change should result in a mass decrease of 415.56 Da, yielding an average mass for the protein of 11,810.76 Da. Electrospray analyses of the ribosomal proteins from this strain revealed an average mass of 11,810 Da at the same retention time as that of L22 in MRE600 ribosomal proteins (Fig. 5A). MALDI analyses of tryptic fragments from this protein matched L22, covering 57% of the wild-type sequence (Fig. 5B). A new peptide at residues 74 to 85 was observed with a very strong signal at 1,360.70 Da, matching well with the predicted mass of 1,360.69 Da. The ability to rapidly verify the L22 mutation using this technique provided a basis for using this method to investigate unknown mutations.
FIG. 5.
Ribosomal protein L22 deletion mutant MKR in erythromycin-resistant E. coli. (A) The deconvoluted electrospray mass spectrum was observed to give a mass of 11,810 Da for the mutated L22 protein. This was a decrease of 416 Da from the predicted L22 mass. (B) Analysis of the tryptic peptides observed by MALDI-TOF confirmed that the protein was L22. The observed and predicted masses are displayed. The wild-type protein sequence is indicated, and the covered peptides are underlined. The mutated peptide is double-underlined, with the deleted residues MKR in italics.
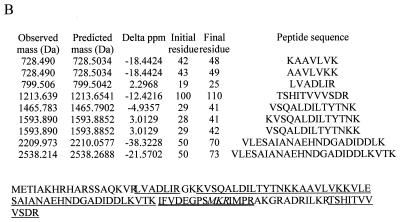
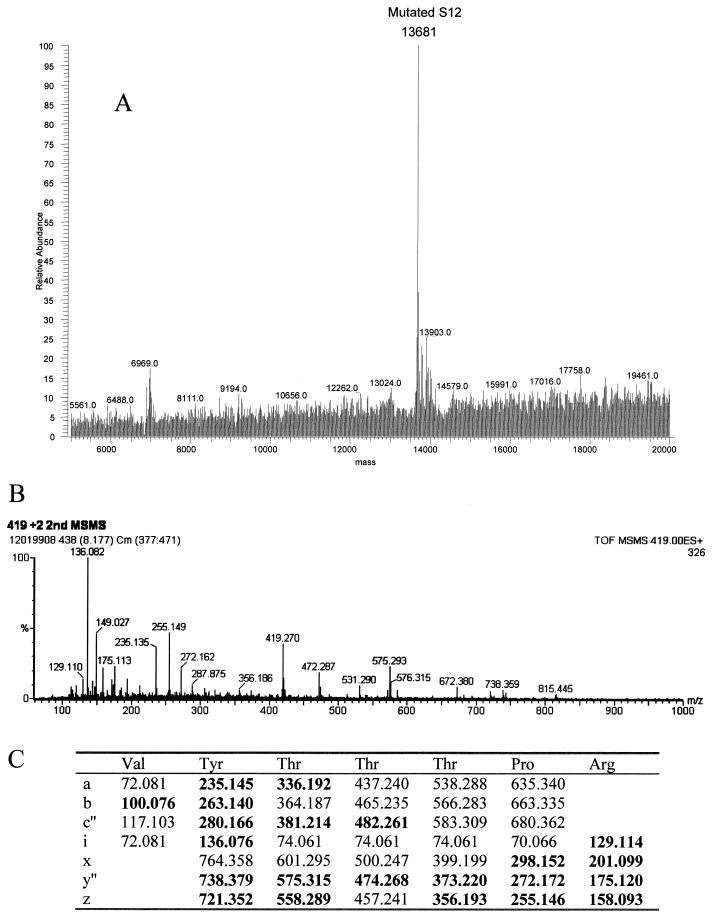
Streptomycin resistance and spectinomycin resistance in E. coli strains UC15121 and E5014 were known to be the result of mutations in ribosomal proteins S12 (rpsL100) and S5 (rpsE2112), respectively. The precise sites of the mutations in the amino acid sequences of these proteins, however, had not been reported. Electrospray analyses of the UC15121 strain revealed a mass of 13,681 Da at the approximate retention time for S12, an increase of 29 ± 2 Da (Fig. 6A). MALDI-TOF analyses of tryptic peptides from this protein confirmed the identity as S12, covering 59% of the wild-type sequence (data not shown). A new peptide with a mass of 837.46 Da was observed. This mass could correspond to the peptide at residues 36 to 42 plus 28.02 Da. Changes at Lys 42, particularly to Arg, have been associated with streptomycin resistance in other strains of E. coli (25). A Lys-to-Arg change corresponds to a mass increase of 28 Da. Tandem MS/MS of the doubly charged species of this peptide was then performed to obtain sequence information on the peptide (Fig. 6B). The masses of the resulting fragments confirmed the sequence VYTTTPR, residues 36 to 42 with the K42R mutation (Fig. 6C).
FIG. 6.
Ribosomal protein S12 mutant K42R in streptomycin-resistant E. coli. (A) The deconvoluted electrospray mass spectrum was observed to give a mass of 13,681 Da for the mutated S12. This was an increase of 29 Da from the predicted S12 mass. (B) Tandem MS/MS analysis of the 837.46-Da peptide in the mutated S12 was then performed. The fragments observed in the spectrum matched well with the predicted fragments (C) of the peptide VYTTTPR. Matches are indicated in bold.
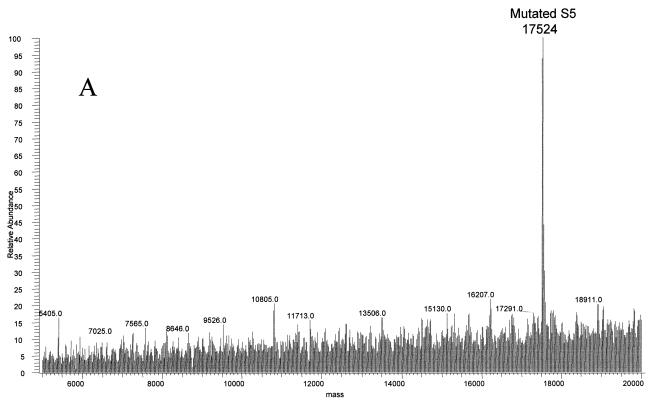
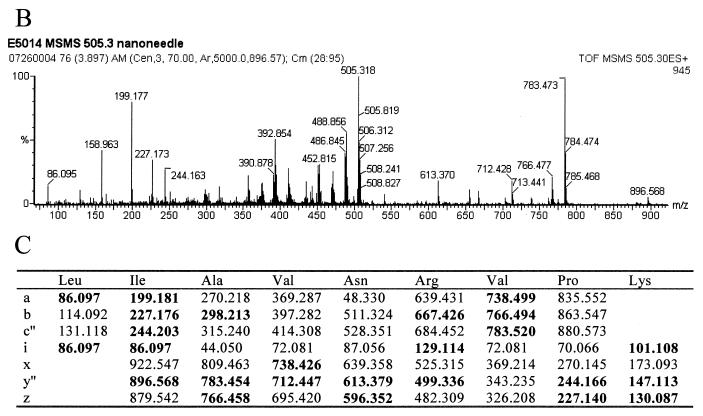
The final strain of E. coli studied by this method was the spectinomycin-resistant E5014. The ribosomal proteins were separated by HPLC, and a mass of 17,524 Da, 10 Da larger than that of the wild type, was observed at the approximate retention time for S5 by electrospray MS (Fig. 7A; see also Table 1). This protein was isolated and digested with trypsin. MALDI-TOF analyses of the resulting peptides confirmed the identification to be ribosomal protein S5, covering 83% of the wild-type sequence (data not shown). Only two regions of the sequence did not contain matches to the peptides observed: residues 20 to 28 and 69 to 85. Previous studies showed that a Ser-to-Pro change in a VSK tripeptide was associated with spectinomycin resistance in a different strain of E. coli (14). Ser to Pro would result in a mass increase of 10 Da. An unmatched peptide in the mutated protein was observed at 1,009.64 Da. This mass could correspond to residues 14 to 22 plus 10.01 Da. Tandem MS/MS was performed on the doubly charged species of this peptide (Fig. 7B). The fragment masses observed matched the sequence LIAVNRVPK, residues 14 to 22 with a P21S mutation (Fig. 7C).
FIG. 7.
Ribosomal protein S5 mutant S21P in spectinomycin-resistant E. coli. (A) The deconvoluted electrospray mass spectrum was observed to give a mass of 17,524 Da for the mutated S5 protein. This was an increase of 10 Da from the predicted S5 mass. (B) Tandem MS/MS analysis of the 1,009.64-Da peptide in the mutated S5 was then performed. The fragments observed in the spectrum matched well with the predicted fragments (C) of the peptide LIAVNRVPK. Matches are indicated in bold.
DISCUSSION
It is well known that MS is a powerful tool for the identification and characterization of peptides and proteins. It can be utilized to identify proteins in a particular biological complex. E. coli ribosomal proteins were evaluated for the feasibility of mapping the proteins by using MS. All 56 intact masses, with posttranslational modifications, were observed by matching them to previously published values. The two methods used in this study, electrospray and MALDI-TOF, provide a balance of speed and detail. MALDI-TOF MS is a rapid method for determining intact masses of nearly all ribosomal proteins in a single analysis. In this study, 54 of 56 proteins were observed without any type of separation in less than 5 min by MALDI-TOF (MS). While MALDI-TOF was very useful, it was incomplete. HPLC separation coupled to electrospray MS determination offers an orthogonal way to establish intact protein average masses within 0.01%, and the separation of the proteins enables more of the proteins to be clearly observed. Deconvolution of electrospray raw data, however, is a manual, somewhat time-consuming process that must be performed to determine each mass. Therefore, it is important to utilize both of these techniques to achieve the best data with the greatest efficiency. Essentially, MALDI-TOF can be used as a first pass to rapidly determine the masses of most of the proteins. Unfortunately, as the mass of the protein increases the accuracy decreases because the peaks begin to significantly broaden. Therefore, electrospray can then be utilized as described as a follow-up approach to determine the remaining masses as well as to more accurately determine the masses of the larger proteins.
Once the wild-type intact protein masses are determined for a given system, alterations in proteins can be rapidly detected by observing mutant intact masses by using MS. Ribosomal proteins are particularly amenable to this approach, since MS is most accurate for relatively small proteins. Only 9 of the 56 E. coli ribosomal proteins are larger than 20 kDa, making small changes in these proteins easily detectable. Alterations can be further characterized by tandem MS/MS analysis of the variant peptide to locate the precise site of the mutation.
In particular, we have shown that this technique can be useful in the characterization of mutated ribosomal proteins related to antibiotic resistance. The three-amino-acid deletion in ribosomal protein L22 from the erythromycin-resistant strain N281, which had been documented in the literature (10), was confirmed by this method. Additionally, two strains in which the specific mutations responsible for resistance had not been identified were characterized. An increase of 29 ± 2 Da was detected in ribosomal protein S12 from the streptomycin-resistant UC15121 strain of E. coli. The K42R mutation, a 28-Da increase, was determined by analyzing tryptic peptides of the mutated S12 protein using MALDI-TOF (MS) in conjunction with tandem MS/MS. This mutation has also been observed in other strains of E. coli that display streptomycin resistance (25). An increase of 10 ± 1 Da was observed for ribosomal protein S5 from spectinomycin-resistant E5014 cells. This mutation was localized to a Ser-to-Pro change, a 10-Da increase, at residue 21 by using the same method as that used to determine the mutation in the S12 protein. This mutation has also been observed in a different strain of spectinomycin-resistant E. coli (14). These findings, coupled with those for the previously characterized strains, reveal the precise mutations responsible for resistance in these strains of E. coli. This technique provides a rapid method for analysis of uncharacterized resistance strains.
As new strains of resistant bacteria emerge, rapid screening of ribosomal proteins by MS may prove valuable. The MS methods described here will provide an alternative approach to sequencing all of the corresponding proteins or genes. In fact, a recent study demonstrated the use of MALDI-TOF MS on whole cells as a tool for fingerprinting strains of E. coli (2). The mutations determined for various strains could also be used as markers of resistance to particular antibiotics. Since several ribosomal protein changes have been documented in the literature to be associated with antibiotic resistance or dependence, a method for quickly identifying these strains would be useful. In one MALDI-TOF experiment requiring a matter of minutes, one could determine if an unknown strain of E. coli exhibited a mass shift correlated with a known mutation and then deduce resistance to a specific antibiotic. Future studies could also use this method to identify novel changes in ribosomal proteins that had previously not been noted.
In addition to a screening technique and to providing protein markers of antibiotic resistance, this type of analysis can provide detailed information on the mechanism of each antibiotic. A specific site of mutation resulting in resistance suggests interaction of the antibiotic with the protein at that site. This data is particularly useful as more detailed structural information about the ribosome is revealed (4,9,11,15). Mutation sites related to resistance in conjunction with the structural information of those sites will likely aid in the understanding of antibiotic function and assist discovery efforts using structure-based drug design.
We have described a method by which we have fingerprinted all 56 E. coli ribosomal proteins. Electrospray MS and MALDI-TOF MS were used together to look for changes in ribosomal proteins. We identified two variant forms of ribosomal proteins in the wild-type strain. We were then able to isolate ribosomal proteins L22, S12, and S5 in strains of E. coli resistant to erythromycin, streptomycin, and spectinomycin, respectively, due to changes in these proteins. Tandem MS/MS was used to characterize the site of the mutation. Each of the mutations revealed had been previously observed in other strains of resistant E. coli. The process provides a rapid means for screening strains of E. coli for mutations known to be associated with antibiotic resistance as well as detecting and characterizing changes in new resistant strains.
ACKNOWLEDGMENTS
We gratefully thank S. T. Gregory and A. E. Dahlberg for providing the N281 and E5014 strains of E. coli, J. I. Cialdella for growing cell cultures, S. M. Swaney and D. L. Shinabarger for instruction in ribosome isolation, and E. Lund for MS/MS technical assistance. We also thank L. D. Adams, E. T. Cecil, W. R. Mathews, and M. C. McCroskey for helpful discussions and critical comments.
REFERENCES
- 1.Allen P N, Noller H F. Mutations in ribosomal proteins S4 and S12 influence the higher order structure of 16S ribosomal RNA. J Mol Biol. 1989;208:457–468. doi: 10.1016/0022-2836(89)90509-3. [DOI] [PubMed] [Google Scholar]
- 2.Arnold R J, Reilly J P. Fingerprint matching of E. coli strains with matrix-assisted laser desorption/ionization time-of-flight mass spectrometry of whole cells using a modified correlation approach. Rapid Commun Mass Spectrom. 1998;12:630–636. doi: 10.1002/(SICI)1097-0231(19980529)12:10<630::AID-RCM206>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- 3.Arnold R J, Reilly J P. Observation of Escherichia coli ribosomal proteins and their posttranslational modifications by mass spectrometry. Anal Biochem. 1999;269:105–112. doi: 10.1006/abio.1998.3077. [DOI] [PubMed] [Google Scholar]
- 4.Ban N, Nissen P, Hansen J, Moore P B, Steitz T A. The complete atomic structure of the large ribosomal subunit at 2.4 Å resolution. Science. 2000;289:905–920. doi: 10.1126/science.289.5481.905. [DOI] [PubMed] [Google Scholar]
- 5.Barritault D, Expert-Benzancon A, Guerin M F, Hayes D. The use of acetone precipitation in the isolation of ribosomal proteins. Eur J Biochem. 1976;63:131–135. doi: 10.1111/j.1432-1033.1976.tb10215.x. [DOI] [PubMed] [Google Scholar]
- 6.Beavis R C, Chait B T. Rapid, sensitive analysis of protein mixtures by mass spectrometry. Proc Natl Acad Sci USA. 1990;87:6873–6877. doi: 10.1073/pnas.87.17.6873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bilgin N, Richter A A, Ehrenberg M, Dahlberg A E, Kurland C G. Ribosomal RNA and protein mutants resistant to spectinomycin. EMBO J. 1990;9:735–739. doi: 10.1002/j.1460-2075.1990.tb08167.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brown R S, Lennon J J. Mass resolution improvement by incorporation of pulsed ion extraction in a matrix-assisted laser desorption/ionization linear time-of-flight mass spectrometer. Anal Chem. 1995;67:1998–2003. doi: 10.1021/ac00109a015. [DOI] [PubMed] [Google Scholar]
- 9.Cate J H, Yusupov M M, Yusupova G Zh, Earnest T N, Noller H F. X-ray crystal structures of 70S ribosome functional complexes. Science. 1999;285:2095–2104. doi: 10.1126/science.285.5436.2095. [DOI] [PubMed] [Google Scholar]
- 10.Chittum H S, Champney W S. Ribosomal protein gene sequence changes in erythromycin-resistant mutants of Escherichia coli. J Bacteriol. 1994;176:6192–6198. doi: 10.1128/jb.176.20.6192-6198.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Clemons W M, Jr, May J L C, Wimberly B T, McCutcheon J P, Capel M S, Ramakrishnan V. Structure of a bacterial 30S ribosomal subunit at 5.5 Å resolution. Nature. 1999;400:833–840. doi: 10.1038/23631. [DOI] [PubMed] [Google Scholar]
- 12.Colby S M, King T B, Reilly J P. Improving the resolution of matrix-assisted laser desorption/ionization time-of-flight mass spectrometry by exploiting the correlation between ion position and velocity. Rapid Commun Mass Spectrom. 1994;8:865–868. [Google Scholar]
- 13.Fenn J B, Mann M, Meng C K, Wong S F, Whitehouse C M. Electrospray ionization for mass spectrometry of large biomolecules. Science. 1989;246:64–71. doi: 10.1126/science.2675315. [DOI] [PubMed] [Google Scholar]
- 14.Funatsu G, Schiltz E, Wittmann H G. Ribosomal proteins XXVII. Localization of the amino acid exchanges in protein S5 from two Escherichia coli mutants resistant to spectinomycin. Mol Gen Genet. 1971;114:106–111. doi: 10.1007/BF00332781. [DOI] [PubMed] [Google Scholar]
- 15.Gabashvili I S, Agrawal R K, Spahn C M T, Grassucci R A, Svergun D I, Frank J, Penczek P. Solution structure of the E. coli 70S ribosome at 11.5 A resolution. Cell. 2000;100:537–549. doi: 10.1016/s0092-8674(00)80690-x. [DOI] [PubMed] [Google Scholar]
- 16.Kang W K, Icho T, Isono S, Kitakawa M, Isono K. Characterization of the gene rimK responsible for the addition of glutamic acid residues to the C-terminus of ribosomal protein S6 in Escherichia coli K12. Mol Gen Genet. 1989;217:281–288. doi: 10.1007/BF02464894. [DOI] [PubMed] [Google Scholar]
- 17.Karas M, Bachmann D, Bahr U, Hillenkamp F. Matrix-assisted ultraviolet laser desorption of non-volatile compounds. Int J Mass Spectrom Ion Processes. 1987;78:53–68. [Google Scholar]
- 18.Kaufmann R. Matrix-assisted laser desorption ionization (MALDI) mass spectrometry: a novel analytical tool in molecular biology and biotechnology. J Biotechnol. 1995;41:155–175. doi: 10.1016/0168-1656(95)00009-f. [DOI] [PubMed] [Google Scholar]
- 19.Kowalak J A, Walsh K A. β-Methylthio-aspartic acid: identification of a novel posttranslational modification in ribosomal protein S12 from Escherichia coli. Protein Sci. 1996;5:1625–1632. doi: 10.1002/pro.5560050816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mann M, Meng C K, Fenn J B. Interpreting mass spectra of multiply charged ions. Anal Chem. 1989;61:1702–1708. [Google Scholar]
- 21.Reinbolt J, Tritsch D, Wittmann-Liebold B. The primary structure of ribosomal protein S7 from E. coli strains K and B. Biochimie. 1979;61:501–522. doi: 10.1016/s0300-9084(79)80207-2. [DOI] [PubMed] [Google Scholar]
- 22.Rheinberger H J, Geigenmuller U, Wedde M, Nierhaus K H. Parameters for the preparation of Escherichia coli ribosomes and ribosomal subunits active in tRNA binding. Methods Enzymol. 1988;164:658–670. doi: 10.1016/s0076-6879(88)64076-6. [DOI] [PubMed] [Google Scholar]
- 23.Spahn C M T, Prescott C D. Throwing a spanner in the works: antibiotics and the translation apparatus. J Mol Med. 1996;74:423–439. doi: 10.1007/BF00217518. [DOI] [PubMed] [Google Scholar]
- 24.Timms A R, Bridges B A. Double, independent mutational events in the rpsL gene of Escherichia coli: an example of hypermutability? Mol Microbiol. 1993;9:335–342. doi: 10.1111/j.1365-2958.1993.tb01694.x. [DOI] [PubMed] [Google Scholar]
- 25.Timms A R, Steingrimsdottir H, Lehmann A R, Bridges B A. Mutant sequences in the rpsL gene of Escherichia coli B/r: mechanistic implications for spontaneous and ultraviolet light mutagenesis. Mol Gen Genet. 1992;232:89–96. doi: 10.1007/BF00299141. [DOI] [PubMed] [Google Scholar]
- 26.Van Buskirk J J, Krisch W M. Gamma-carboxyglutamic acid in eukaryotic and prokaryotic ribosomes. Biochem Biophys Res Commun. 1978;82:1329–1331. doi: 10.1016/0006-291x(78)90334-0. [DOI] [PubMed] [Google Scholar]
- 27.Whittal R M, Li L. High-resolution matrix-assisted laser desorption/ionization in a linear time-of-flight mass spectrometer. Anal Chem. 1995;67:1950–1954. doi: 10.1021/ac00109a007. [DOI] [PubMed] [Google Scholar]
- 28.Wittmann H G. Components of bacterial ribosomes. Annu Rev Biochem. 1982;51:155–183. doi: 10.1146/annurev.bi.51.070182.001103. [DOI] [PubMed] [Google Scholar]