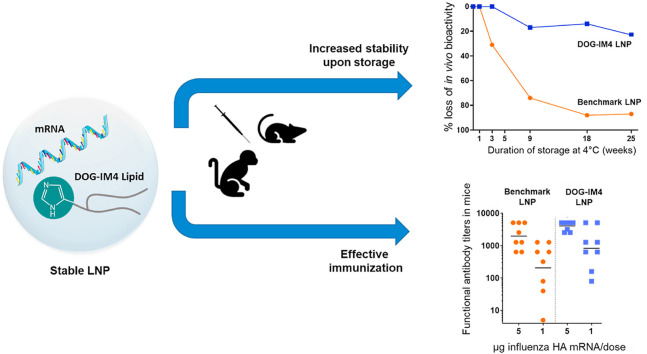
Abstract
The mRNA vaccine technology has promising applications to fight infectious diseases as demonstrated by the licensing of two mRNA-based vaccines, Comirnaty® (Pfizer/BioNtech) and Spikevax® (Moderna), in the context of the Covid-19 crisis. Safe and effective delivery systems are essential to the performance of these vaccines and lipid nanoparticles (LNPs) able to entrap, protect and deliver the mRNA in vivo are considered by many as the current “best in class”. Nevertheless, current mRNA/LNP vaccine technology has still some limitations, one of them being thermostability, as evidenced by the ultracold distribution chain required for the licensed vaccines. We found that the thermostability of mRNA/LNP, could be improved by a novel imidazole modified lipid, DOG-IM4, in combination with standard helper lipids. DOG-IM4 comprises an ionizable head group consisting of imidazole, a dioleoyl lipid tail and a short flexible polyoxyethylene spacer between the head and tail. Here we describe the synthesis of DOG-IM4 and show that DOG-IM4 LNPs confer strong immunization properties to influenza HA mRNA in mice and macaques and a remarkable stability to the encapsulated mRNA when stored liquid in phosphate buffered saline at 4 °C. We speculate the increased stability to result from some specific attributes of the lipid's imidazole head group.
Keywords: mRNA vaccine, Lipid nanoparticles, Storage stability, Ionizable lipid, Imidazole lipid
Graphical abstract
1. Introduction
mRNA-based vaccines stimulate strong immune responses by mimicking natural infection and are advantageous from an industrial standpoint by enabling standardization of vaccine design and manufacturing, quality control, safety assessment and regulatory approval processes. They are effective weapons to rapidly respond to emerging pandemic threats [[1], [2], [3], [4], [5], [6]] as perfectly illustrated by the rapid development of mRNA vaccines against SARS-CoV-2 [[7], [8], [9], [10], [11]].
Delivery systems are essential to the performance of mRNA vaccines. Lipid nanoparticles (LNPs) composed of ionizable cationic lipids, phospholipids, cholesterol and lipid-anchored polyethylene glycol (PEG) are potent vectors for in vivo RNA vaccine delivery [6,[12], [13], [14], [15]] and are used in the marketed Covid-19 vaccines, Comirnaty™ (Pfizer/BioNtech) and Spikevax™ (Moderna). Ionizable cationic lipids with a pKa of seven or lower are most critical to the in vivo potency of LNPs [[16], [17], [18], [19], [20], [21], [22]]. Such lipids contain pH-sensitive amines which maintain a close to neutral surface charge at physiological pH, thereby reducing nonspecific lipid–protein interactions and turn into cationic groups within the acidic environment of the endosomes after cellular uptake, thus facilitating endosomal destabilization and mRNA release into the cytosol [23,24]. Examples of such ionizable cationic lipids comprise MC3 [17] that is present in the marketed product Onpattro™ (Patisiran from Alnylam) [25,26], L319 that was devised as a biodegradable analogue of MC3 [27] and more recent compounds that were selected from library screens and rational design [[20], [21], [22],[28], [29], [30], [31]] as can be found in Comirnaty™ and Spikevax™. The development and evolution of cationic lipids for LNP formulation has been the topic of some excellent review articles [14,15,[32], [33], [34], [35], [36], [37]].
Although LNP-formulated mRNA vaccines have now reached the market in the context of the Covid-19 crisis, there remain some hurdles to overcome to enable global access and widespread applications. One of them is the challenging ultracold distribution chain required for both Comirnaty™ and Spikevax™ [38] indicating that mRNA vaccine formulations could still benefit from improvements, especially in terms of thermostability. From a formulation perspective, there are essentially two ways for improving the stability of these vaccines. The most straightforward one is to use drying technologies and to introduce excipients able to protect the formulation during drying to generate a stable dry product. However, this approach has limitations in terms of volumes and could significantly increase the cost of the final vaccine product. The second one is to act directly on the LNP composition itself by using for instance ionizable lipids interacting with the mRNA in a way conferring increased stability [39,40].
In the present study we developed such an ionizable cationic lipid for the formulation of LNPs displaying increased stability at 4 °C in liquid form and promoting robust mRNA expression and strong immune responses. The lipid was synthesized by assembling three building blocks: i) a dioleoyl lipid tail comprising stable ether linkages, ii) a flexible spacer arm made of three polyoxyethylene units, and iii) an ionizable cationic head group composed of a simple imidazole cycle attached at position 4 to the spacer arm. This new lipid was termed DOG-IM4. A chemical isomer, DOG-IM2, with position 2 of the imidazolium ring attached to the spacer arm was also synthesized but was found less effective.
2. Materials & methods
2.1. Lipids
All reagents and solvents used for the lipid synthesis were obtained from Sigma-Aldrich. 1H and 13C NMR spectra were recorded at room temperature on a Bruker Avance 400 spectrometer (NMR 1H: 400 MHz and NMR 13C: 75 MHz). Recorded shifts were reported in parts per million (δ) and calibrated using traces of undeuterated solvent (CHCl3: 1H 7.26 ppm; 13C 77.16 ppm, MeOH 1H 3.31 ppm; 13C: 49.0 ppm). Data was represented as follows: chemical shift, multiplicity (s = singlet, d = doublet, t = triplet, q = quartet and m = multiplet), coupling constant (J in Hz), integration and attribution. High-resolution mass spectra (HRMS) were obtained using an Agilent Q-TOF (time of flight) 6520 and Low-resolution mass spectra (LCMS) using an Agilent MSD 1200 SL (ESI/APCI) with an Agilent HPLC 1200 SL.
DLin-MC3-DMA (MC3) and L319 were synthesized as respectively described by Jayaraman M et al. [17] and Maier MA et al. [27] and obtained from SAI Life Sciences (Hyderabad, India). 1,2-distearoyl-sn-glycero-3-phosphocholine (DSPC), 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine (DOPE), 1,2-dimyristoyl-rac-glycero-3-methoxypolyethylene glycol-2000 (DMG-PEG2000), 1,2-dimyristoyl-sn-glycero-3-phosphoethanolamine-N-[methoxy(polyethylene glycol)-2000 (PEG2000-PE) and Cholesterol were obtained from Avanti Polar Lipids (Alabaster, AL, USA).
The novel synthetic dioleoyl lipids bearing an imidazolium head group, DOG-IM4 and DOG-IM2, were synthesized respectively by coupling 4-imidazolcarboxylic acid or 2-imidazolcarboxylic acid to dioleylglycero-ethoxy-ethoxy-ethoxy-ethylamine via acyl chloride formation. The starting dioleylglycero-ethoxy-ethoxy-ethoxy-ethylamine was prepared by reacting dioleoyl glycerol, synthesized according to Espuelas et al. [41], with methanesulfonyloxy-ethoxy-ethoxy-ethoxy-ethyl-azoture followed by treatment with triphenylphosphine [42]. Details of the lipid synthesis and analytical characterization of the synthetic intermediates and final products are provided in supplementary information.
2.2. mRNA
High purity, custom-synthesized mRNAs encoding the hemagglutinin (HA) of the influenza strain A/Netherlands/602/2009 (H1N1) were obtained from TriLink (San Diego, CA) and from AmpTec GMBH (Hamburg, Germany). Codon optimization and 5′ and 3’ UTRs in these constructs were specific to the suppliers. AmpTec supplied one HA mRNA containing only natural nucleotides and another one containing 1-methyl-pseudouridine (1MpU) in place of uridine. The 5-methoxyuridine-modified mRNAs encoding firefly luciferase (FLuc) and human erythropoietin (hEPO) were obtained from TriLink (respective catalogue reference numbers: L-7202 and L-7209).
2.3. Formulation of LNPs
LNPs were prepared by microfluidic mixing as described in Belliveau NM et al. [43]. Briefly, lipids were dissolved in ethanol at molar ratios of 50:10:38.5:1.5 (ionizable lipid/Phospholipid/cholesterol/PEGLipid). The lipids in ethanol solution at 20 mg/mL and the mRNA in 50 mM citrate buffer (pH 4.0) at 0.265 mg/mL for the preparation of DOG-IM4 and DOG-IM2 LNPs or at 0.305 mg/mL or 0.314 mg/mL for the preparation of respectively L319 LNPs or MC3 LNPs, were then injected into a microfluidic mixer (NanoAssemblr™, Precision Nanosystems, Vancouver, BC) at a flow rate ratio of 1:3 with a combined final flow rate of 4 mL/min. The selected lipid/mRNA concentrations yielded a constant N/P ratio of 6 (cationic nitrogen groups from the ionizable lipid over anionic phosphate groups from the mRNA) corresponding to lipid/mRNA ratios of 25 in DOG-IM4 and DOG-IM2 LNPs or 22 in L319 and MC3 LNPs. Formulations were then dialyzed against 50 mM citrate buffer, pH 4.0, for at least 4 h followed by phosphate buffered saline, pH 7.4 (PBS), for 24 h by using 10 kDa MWCO dialysis cassettes (Spectrum Labs, Rancho Dominguez, CA). Final LNPs were filtered on a 0.22 μm PES filter (Merck-Millipore) and stored liquid at 4 °C in PBS under nitrogen atmosphere.
2.4. LNP characterization
LNP size determined by dynamic light scattering (DLS) and zeta potential were measured using the Malvern Zetasizer NanoZS (Malvern, Worcestershire, UK). mRNA encapsulation efficiency was determined by mRNA accessibility to Ribogreen using the QuantiTRibogreen RNA assay (Life Technologies, Burlington, ON). Briefly, LNPs were incubated at 37 °C for 10 min in the presence or absence of 1% Triton X-100 (Sigma). Upon the addition of the Ribogreen reagent, fluorescence intensities (Ex: 480 nm; Em: 520 nm) for untreated samples (representing non-encapsulated mRNA) and samples treated with Triton X-100 (representing total mRNA) were determined.
2.4.1. Apparent pKa measurement
Apparent pKa was measured by the 6-(p-toluidino)-2-naphthalenesulfonic acid (TNS) fluorescence method as described in Hassett et al. [22]. In brief, in a black-bottom 96-well plate, 5 μL of LNPs at 50 μg mRNA/mL were mixed with 5 μL of 100 mM TNS reagent (Sigma) and 190 μL of various buffers (citrate, phosphate or borate) at pH ranging from 3 to 11. Each pH unit of buffer was repeated in triplicate. TNS fluorescence intensity (Ex: 325 nm; Em: 435 nm) was read on a SpectraMax® i3k Microplate reader and was plotted as a function of pH. Apparent pKa of the LNP was assigned to the log of the inflection point.
2.4.2. UHPLC-CAD-MS method for LNP lipid analysis
The separation and analysis of the LNP lipids was performed by UHPLC (Vanquish® Thermo Scientific) using an Acquity Premier CSH C18 column, 1.7 μm, 2.1 × 50 mm (Waters; Ref. 186009460) maintained at 55 °C and eluted with a discontinuous gradient of solvent system B [Isopropanol/Acetonitrile (90:10) containing 10 mM ammonium formate and 0.1%formic acid] in A [Acetonitrile/Water (60:40) containing 10 mM ammonium formate and 0.1%formic acid] at a flow rate of 0.4 mL/min. The gradient was as follows: T0.0: 40% B, T15.0 min: 70% B, T23.0 min: 99% B, T23.1 min: 40% B, T25.0 min: 40% B. A charged aerosol detector (Corona® CAD; Thermo Scientific) set at 50 °C and power function 1 was used to detect the lipids eluted from the column. Mass spectrometry was performed using the Orbitrap mass spectrometry system (Q Focus Exactive Thermo Scientific) and the workstation was Chromelon® 7.3. Electrospray ionization in positive/negative ion mode was used, and full scan mass spectra were measured in the mass range of m/z 133.4 to 2000 using the sensitivity mode. For positive ESI, spray voltage was set to 3 kV. For negative ESI, spray voltage was set to 2.5 kV. The S-lens RF Level was at 50, the sheath gas flow was set to 60 arbitrary units, auxiliary gas flow to 20 arbitrary units, sweep gas flow to 0 arbitrary units, and the capillary temperature to 380 °C.
2.4.3. Extraction of mRNA from DOG-IM4 LNPs and integrity analysis
The following procedure allowed a reproducible extraction of 75 ± 15% of total mRNA from DOG-IM4 LNPs. In brief, LNPs were incubated under agitation for 10 min at 50 °C in the presence of 1% Triton X100 and successively extracted with a mixture of Phenol/Chloroform/Isoamyl alcohol 25:24:1 (Sigma; Ref. 77617) and then with Chloroform/Isoamyl alcohol 24:1 (Acros Organics; Ref. 327155000) to get rid of the lipids. The mRNA was then precipitated from the extracted solution by addition of 0.1 volume of 3 M sodium acetate, pH 5.2 (Molecular biologics; Ref. R1181) and 2.5 vol of 100% ethanol. After 12 h of incubation at −20 °C, the mRNA was pelleted by centrifugation at 12000g for 10 min at 4 °C (Eppendorf tabletop microcentrifuge). The mRNA pellet was washed with 70% ethanol, dried by using a Speedvac vacuum concentrator and resuspended in RNAse-free water to enable quantification by UV absorbance at 260 nm with a Nanodrop spectrophotometer (Thermo Scientific) and integrity analysis by percentage of main peak on an Agilent 5200 Fragment Analyzer.
2.4.4. LNP morphology
The morphology of LNPs was observed by cryo-electron microscopy. In brief, 4 μL of concentrated LNPs were deposited on Quantifoil R2/2 cupper 300mesh grids (Quantifoil Instruments GmbH, Germany) after 90 s of glow discharge on an ELMO ionizer (Cordouan, France). Grids were blotted and frozen using a Vitrobot MARK IV (Thermo Scientific) and transferred for observation onto a TEM Tecnai-G20 (Thermo Scientific) operated at 200 kV using a 910 cryo-holder (Gatan Inc., USA). Images were recorded at 4 μm defocus and at low-dose mode (electron doses between 10 and 15 e−/Å2) using an ssCCD Ultrascan 4000 (Gatan Inc., USA). Pixel size of recorded images was estimated to 0.221 nm after TEM calibration using a cross line grid (EMS, USA) with pitch spacing of 500 nm and 2000 lines/mm.
2.4.5. Stability of DOG-IM4, MC3 and L319 LNPs
hEPO mRNA (Trilink) was formulated in LNPs composed of MC3, L319 or DOG-IM4/DSPC/Chol/DMG-PEG 50:10:38.5:1.5 mol/mol and dialyzed into PBS, as described above and diluted to 20 μg/mL with PBS. The product was filled into 3 mL borosilicate type I clear glass vials under nitrogen (850 μL/vial) and stored at 4 °C, 25 °C or 37 °C. At different time points, vials were removed from storage and the LNPs were injected intramuscularly to Balb/c mice (1 μg mRNA/50 μL dose) to determine the expression of hEPO in the mouse serum by using the Human Erythropoietin Quantikine IVD ELISA Kit (R&D Systems; Ref. DEP00) with human serum standards (R&D Systems; Ref. CEP01 and CEP03) to control and standardize the assay. LNPs were further characterized for stability of particle sizes and PDI by DLS, mRNA encapsulation by Ribogreen accessibility assay, mRNA integrity by fragment analyzer after extraction of the mRNA and lipid integrity by UHPLC-CAD-MS.
2.5. In vivo studies
2.5.1. Animals and ethics statements
BALB/c ByJ mice (Balb/c) were purchased from Charles River (Saint-Germain-Nuelles, France) and cynomolgus macaques (Macaca fascicularis) from Noveprim (Camarney, Spain). Animals were housed at Sanofi Pasteur in an animal facility accredited by AAALAC International since 2011. All the studies were prepared from an Animal Use Protocol reviewed by the Ethics Committee of Sanofi Pasteur. The macaque experiment was included in the project number APAFIS#16801–2018092011168040 v2 and the mouse experiment in the project AFAPIS#16508–2018081313297024 v1, both approved by the French Ministry of Higher Education, Research and Innovation (MHERI). All experiments were conducted in accordance with the European Directive 2010/63/UE as published in the French Official Journal of February 7th, 2013.
2.6. Immunization studies
2.6.1. Immunization of mice
Groups of 8 female Balb/c mice (8 weeks old at the time of the first immunization) were immunized twice three weeks apart (D0, D21) with 0.5–5 μg of mRNA in LNPs injected intramuscularly (IM) into the quadriceps with a volume of 50 μL. A positive control group of 8 mice, received 10 μg of monovalent A/California/07/2009 (H1N1) split vaccine (MIV) derived from Vaxigrip® process (Sanofi Pasteur). Blood samples were collected on D20 (post-1) and D42 (post-2) for functional antibody response analysis by hemagglutination inhibition (HI) assay. Clinical observations were performed daily over a period of 3 days after each injection.
2.6.2. Immunization of non-human primates (NHPs)
Groups of 4 or 5 female cynomolgus macaques (2.4–3.5 kg body weight) were immunized twice four weeks apart (D0, D28) with 10–50 μg of mRNA in LNPs injected IM into the quadriceps or deltoid with a volume of 500 μL. Blood samples were collected pre-immunization (D-33, D-21, D-13) and at different time points following immunization for the analysis of functional antibody responses by HI titration (D28, D56, D84, D111 and D140), HA-specific T cell responses by ELISPOT (D-33 and D42), innate cytokines by Meso Scale Discovery (MSD) assay (D-21 and D1) and standard blood parameters (D-21, D-13, D1, D2, D6, D29, D30 and D34). Clinical observations were performed daily over a period of 8 days after each injection.
2.6.3. Hemagglutination inhibition (HI) assay
The level of functional antibodies was determined by HI titration of individual sera against 4 hemagglutination units (HAU) of a A/California/07/09 (H1N1) influenza virus (strain closely related to A/Netherlands/602/2009 encoded by the mRNA vaccine) by using chicken red blood cells (cRBCs). Individual sera collected at defined time-points were first treated with Receptor Destroying Enzyme (RDE) and absorbed on 10% cRBCs in PBS. The HI titer of a given serum was defined as the reciprocal of its last dilution preventing hemagglutination. A value of 5, corresponding to half of the initial dilution (1/10), was arbitrarily given to a serum with no HI activity to perform statistical analysis.
2.6.4. T cell ELISPOT assay
ELISPOT plates (Millipore; Ref. S5EJ104I07) were coated with purified anti-monkey IFN-γ (Mabtech; Ref. MT126L) or anti-human IL-4 (Mabtech; Ref. IL4 I). After blocking with RPMI medium supplemented with glutamine, streptomycin, penicillin and 10% heat-inactivated fetal calf serum, peripheral blood mononuclear cells (PBMCs) were added and stimulated for 24 h with recombinant influenza hemagglutinin (rHA) from A/California/07/2009 strain (Sanofi - Protein Sciences) or medium alone for negative control wells. IFN-γ and IL-4 secreting cells were detected using respectively BAM-conjugated anti-monkey/human IFN-γ (Mabtech; Ref. 7-B6-1-BAM) and biotinylated anti-human IL-4 (Mabtech; Ref. IL4 II), followed by anti-BAM 490 (Mabtech; Ref. anti-BAM 490) and streptavidin-550 (Mabtech; Ref. SA-550). IFN-γ and IL-4 secreting cells were counted using a fluorospot plate reader (Microvision Instruments, Evry, France). Data shown were corrected for background by subtracting counts from negative control wells containing PBMCs collected from the same animal prior immunization.
2.6.5. Innate cytokine assay
Ten cytokines (Eotaxin, IFN-α2a; IL-1β; IL-1RA, IL-6, IL-8, IL-17 A; I-TAC, MCP-1, TNFα) were tested by using MSD U-plex kit (MSD; Ref. K15068L-1) by following the instructions from the kit.
2.6.6. Blood biomarkers
Plasma levels of alkaline phosphatase (ALP), alanine aminotransferase (ALT), aspartate aminotransferase (AST), creatinine, and lipase were determined by colorimetry on a Pentra C400 (Horiba, Palaiseau, France). The determination of C reactive protein (CRP) levels by immunoturbidimetry was performed by Biovelys (Marcy l’Etoile, France).
2.7. Bioimaging studies
Eight-week-old female Balb/c mice were anesthetized with 2% isoflurane in oxygen (Alcyon, France; Ref. 1818290) and injected intramuscularly in the right quadriceps with 5 μg of FLuc mRNA in L319 or DOG-IM4 LNPs under 50 μL of PBS. At 6-, 24-, 48- and 72-h post-injection, mice were anesthetized again and imaged 15 min after intraperitoneal injection of D-luciferin (Invitrogen) at a dose of 150 mg/kg. Bioluminescence imaging was performed using an IVIS Spectrum imaging system (Perkin Helmer) using an auto exposure time to ensure the acquired signal was within effective detection range (above noise levels and below CCD saturation limit). Bioluminescence values were quantified by measuring Radiance (photons/second) in the region of interest where bioluminescence signal emanated using the Living Image software provided by PerkinElmer.
2.8. Statistical analysis
HI titers were log10 transformed prior to statistical analysis. To compare LNPs, a model of analysis of variances with two factors (LNPs and doses) was applied with a Turkey adjustment for multiple comparisons. Statistical analyses were performed on groups with more than 50% of responders.
The model's residuals were studied to test the model's validity (normality, extreme individuals, etc.). All analyses were done on SAS v9.4®. A margin of error of 5% was used for effects of the main factors.
3. Results
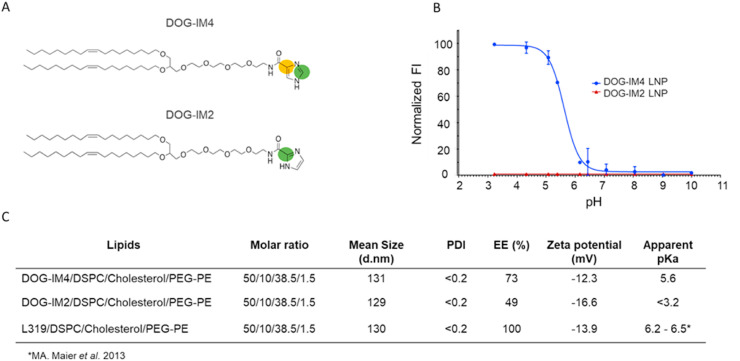
DOG-IM4 and DOG-IM2 were synthesized with a purity >95%. The two lipids were readily soluble in ethanol, which was important for LNP formulation by the ethanol dilution method using the NanoAssemblr®. LNPs were formed by combining DOG-IM4 (or DOG-IM2), a neutral co-lipid (DSPC or DOPE), cholesterol, and a PEGylated lipid (DMPE-PEG2000 or DMG-PEG2000) with mRNA encoding influenza HA (H1N1), hEPO or FLuc by using a NanoAssemblr®. LNPs comprising L319 instead of DOG-IM4 were used as a benchmark in the present studies as we found that L319 LNPs outperformed DLin-MC3-DMA (MC3) [17] and DLin-KC2-DMA (KC2) [16] LNPs for the induction of immune responses in preliminary mouse and cynomolgus macaques studies using A/Netherland/602/2009H1N1 influenza HA mRNA constructs (manuscript in preparation). In terms of formulation, all LNPs used in here were formulated with standard helper lipids: DSPC, cholesterol and PEG2000Lipids at a molar ratio of Ionizable lipid/DSPC/Chol/PEG2000Lipid 50:10:38.5:1.5, and at a ionizable lipid to RNA charge ratio (N/P) of 6, knowing from the literature and from our preliminary studies that these conditions were appropriate for our reference lipid, L319 [27]. DOG-IM4, DOG-IM2 and L319 LNPs containing unmodified mRNA expressing influenza A/Netherland/602/2009 (H1N1) HA obtained from Trilink, were thus prepared, and characterized for particle size and polydispersity index by DLS, surface charge by zetametry, pKa by TNS dye binding assay and mRNA entrapment by Ribogreen accessibility assay. As shown on Fig. 1 , DOG-IM4 and DOG-IM2 LNPs had hydrodynamic diameters of around 130 nm both and a negative surface charge as indicated by zeta potentials of respectively −12.3 and −16.6 mV, when formulated at N/P ratio of 6. The substitution of DOG-IM4 with DOG-IM2 decreased the mRNA encapsulation efficiency by about 25% as assessed by measuring the mRNA accessible to the Ribogreen dye and had a drastic effect on the LNP pKa, which in turn negatively affected the in vivo potency (Fig. 3). In comparison, L319 LNPs also displayed an average particle size of around 130 nm and a negative surface charge with a zeta potential of −13.9 mV but a higher encapsulation efficiency of nearly 100%. The apparent pKa of the L319 LNPs was taken from the literature and was comprised between 6.2 and 6.5 [27].
Fig. 1.
Comparison of DOG-IM4, DOG-IM2 and L319 LNPs characteristics.DOG-IM4, DOG-IM2 and L319 were synthesized (A) and combined with standard helper lipids for the preparation of mRNA-containing LNPs as described in Materials and Methods. The LNPs were characterized for pKa by TNS dye binding assay (B), particle size and surface charge by using a Zetasizer (Malvern instrument), and mRNA entrapment by Ribogreen accessibility assay (C). FI stands for fluorescence intensity, EE for encapsulation efficiency and PDI for polydispersity index.
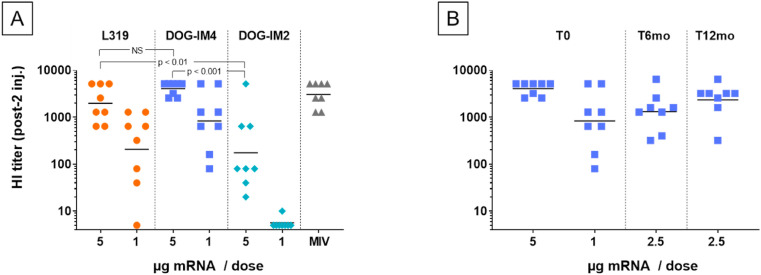
Fig. 3.
DOG-IM4 LNPs induce higher functional antibody titers than DOG-IM2 LNPs in immunogenicity studies with mice and are stable at 4°C.PanelA: Individual and mean hemagglutination inhibiting antibody titers (HI titers) measured in sera collected at D42 from Balb/c mice (n = 8) immunized at D0 and D21 with 10 μg of a monovalent A/California/07/2009 (H1N1) split influenza vaccine (Vaxigrip®; MIV) or with DOG-IM4, DOG-IM2 and L319 LNPs loaded with indicated amounts of mRNA encoding full-length hemagglutinin (HA) of closely related influenza virus strain A/Netherlands/602/2009 (H1N1). The vaccines in PBS were injected into the mouse quadriceps under a final volume of 50 μL. PanelB: Samples from the lot of DOG-IM4 LNPs used in (A) were retested after 6 and 12 months of storage in PBS under nitrogen at 4°C. Individual and mean HI titers are shown. The study comprised also a group of mice injected with MIV (10 μg Vaxigrip®) at each timepoint to confirm the consistency of HI titration test (not shown). NS = not statistically significant.
The DOG-IM4 LNPs were also analyzed by cryo-TEM (Fig. 2 ) and the obtained images indicated a majority of bilayer structures forming blebs emanating from small electron dense bodies as already observed by others [44,45]. The average particle sizes measured by cryoTEM was in the range of 100–130 nm, corroborating the DLS measurements.
Fig. 2.
Cryo EM micrograph for DOG-IM4 LNPs; scale = 100 nm.
Next, the HA mRNA-containing-LNPs were tested directly for the immunization of Balb/c mice. Mice (8 per group) were immunized with 1–5 μg of mRNA in either DOG-IM4, DOG-IM2 or L319 LNPs. After two IM injections of 5 μg, mice responded with high HI titers, which were in the same range as those obtained with 10 μg of a monovalent A/California/07/2009 split influenza vaccine (MIV; Vaxigrip®, Sanofi Pasteur) (Fig. 3 ). DOG-IM4 LNPs were at least as potent as L319 LNPs and significantly more potent than DOG-IM2 LNPs at inducing functional HI titers in this study (p-value <0.007). The HI response induced by the different LNPs was dose dependent, with undetectable titers in the DOG-IM2 group at 1 μg dose of mRNA, while HI titers remained high at this dosage in the DOG-IM4 group (average titer: 830). In this study, the different LNPs all comprised the same helper lipids at the same molar ratio: ionizable lipid/DSPC/Chol/DMPE-PEG2000, 50:10:38.5:1.5 mol/mol and a N/P ratio of 6 was used for formulation. To note, the replacement of DSPC with the fusogenic lipid DOPE and the replacement of DMPE-PEG2000 with DMG-PEG2000 had no impact on the HI titers induced by DOG-IM4 LNPs as observed in a separate mouse study (not shown). To test for stability, the DOG-IM4 LNPs used in this study, which were stored in PBS under nitrogen at 4 °C, were retested after 6 and 12 months of storage following the same immunization regimen. The same Vaxigrip® control (10 μg dose) was used as a reference in each of these separate studies to confirm the consistency of the HI titrations. Unfortunately, due to the limited lot volume, the product could only be tested at a 2.5 μg dose and not at the initial 1 and 5 μg doses. In this study, we found that the ability of DOG-IM4 LNPs to induce high HI titers did not decrease over time with average comparable HI titers of 1312 and 2334, obtained after respectively 6 and 12 months of storage under nitrogen in PBS at 4 °C (Fig. 3, Panel B). We also found that the DOG-IM4 LNP particle size and encapsulation efficiency remained stable in these storage conditions. These results provided a first indication suggesting that DOG-IM4 LNPs could be advantageous in terms of stability and motivated additional studies of DOG-IM4 LNPs with reporter mRNAs, including an accelerated stability study using hEPO mRNA depicted in Fig. 6.
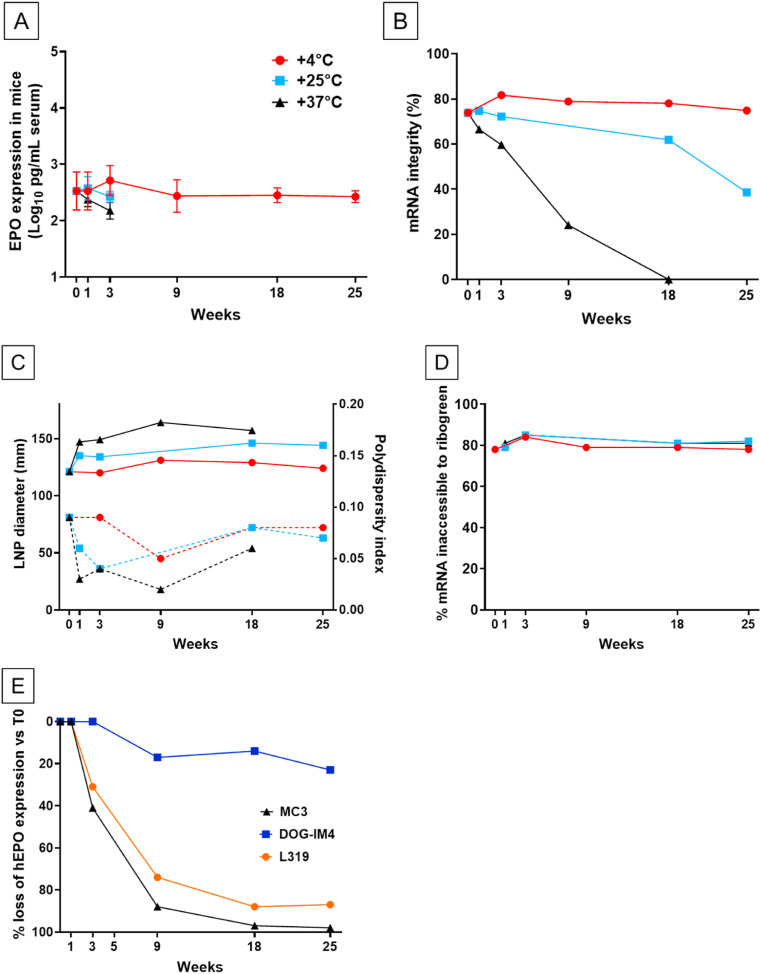
Fig. 6.
Stability of DOG-IM4 LNPs over time at different storage temperatures.DOG-IM4 LNPs containing hEPO mRNA were stored in PBS under nitrogen at 4°C, 25°C or 37°C, and after different periods of time (indicated in weeks) the LNPs were tested for: PanelA: In vivo bioactivity via circulating hEPO determination 6h after IM injection of 1 μg mRNA into Balb/c mice (4 mice/group injected with 50 μL LNP in the quadriceps); PanelB: mRNA integrity by fragment analyzer after extraction from the DOG-IM4 LNPs; PanelC: Average particle size (z-Average and PDI in dotted lines) by dynamic light scattering (DLS); PanelD: mRNA encapsulation (Ribogreen assay). PanelE: The hEPO expression level from DOG-IM4 LNPs stored at 4°C is compared to that of L319 and MC3 LNPs stored in the same conditions. Loss of hEPO expression is expressed as a percent of hEPO measured at T0.
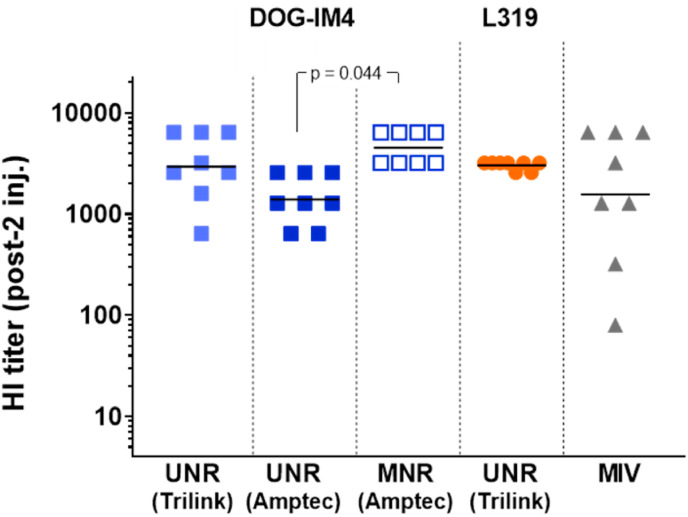
The potency of DOG-IM4 LNPs was then further studied with 1-methyl pseudouridine (1MpU)-modified mRNA (MNR) encoding A/Netherland/602/2009H1N1 influenza HA obtained from AmpTec since our preliminary study with this mRNA construct indicated that some LNPs could benefit more than others from the use of MNR (manuscript in preparation). Studies with MNR always comprised groups immunized with the same construct containing non-modified uridine (UNR) obtained from the same supplier (AmpTec in this case). To compare this study with the previous ones, the same immunization regimen was used, and groups immunized with UNR obtained from Trilink as in Fig. 3 and a group immunized with monovalent split vaccine (MIV) were introduced as references. Results in Fig. 4 indicate that comparable HI titers could be obtained from DOG-IM4 and L319 LNPs with UNR obtained from Trilink, confirming that the results of Fig. 3 were reproducible. When comparing the titers obtained with UNR and MNR from AmpTec, we observed that MNR induced significantly higher HI titers than its UNR counterpart when delivered using DOG-IM4 LNPs (respective average titers 4525 vs 1396; p = 0.044).
Fig. 4.
DOG-IM4 LNPs induce high functional antibody titers with both UNR and MNR in immunogenicity studies with Balb/c mice.Balb/c mice (n = 8) were immunized as inFig. 3with 10 μg of a monovalent A/California/07/2009 (H1N1) split influenza vaccine (Vaxigrip®; MIV) or with DOG-IM4 LNPs loaded with 5 μg of unmodified (UNR) or 1MpU-modified mRNA (MNR) encoding full-length hemagglutinin (HA) of closely related influenza virus strain A/Netherlands/602/2009 (H1N1). Individual and mean HI titers measured three weeks after the second injection (D42) are shown. UNR was either from Trilink (as inFig. 3) or from AmpTec and MNR was from AmpTec. A group immunized with L319 LNPs loaded with 5 μg of UNR from Trilink (as inFig. 3) was included as a reference.
In these different studies, DOG-IM4 LNPs induced mild to moderate injection site swelling, especially after the second dose, in some mice injected with 5 μg UNR that was seen also in mice injected with L319 LNPs but at a lower frequency. In all cases, this local reactogenicity was transient and returned to normal after a maximum of 5 days post injection without affecting the animal's behavior and normal weight gain.
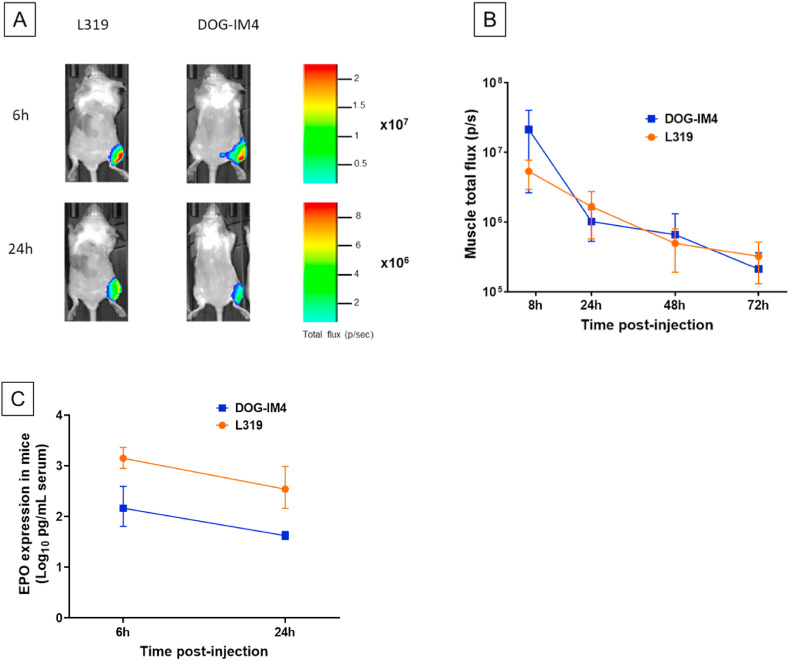
To further characterize the in vivo transfection activity and stability of the LNPs, DOG-IM4 and L319 were used to prepare LNPs with hEPO mRNA and FLuc mRNA. Both mRNAs were obtained from TriLink and contained 5-methoxy uridine instead of uridine. The lipid composition and N/P ratio in these LNPs were maintained the same, i.e., DOG-IM4 or L319/DSPC/Chol/DMG-PEG2000 (50:10:38.5:1.5 mol/mol) with an N/P ratio of 6, and the LNPs were prepared and dialyzed into PBS pH 7.4, as for the immunization studies. In mice injected IM (right quadriceps) with 5 μg FLuc mRNA, the levels of luciferase expression detected in the injected muscles was essentially the same for L319 and DOG-IM4 LNPs and followed similar kinetics (Fig. 5 , Panels A and B). In this study, luciferase expression peaked at 6 h and declined progressively to return to background levels after about 4 days for both LNPs. Conversely, in mice injected IM with 1 μg hEPO mRNA, the systemic expression of hEPO after 6 and 24 h was about 10-fold higher with L319 LNPs as compared to DOG-IM4 LNPs (Fig. 5, Panel C), indicating that LNP in vivo performance assessment may vary depending on the reporter system used.
Fig. 5.
Reporter protein expression in BALB/c mice after IM injection of hEPO and FLuc mRNAs loaded in DOG-IM4 or L319 LNPs. Panel A: In vivo imaging of Balb/c mice 6h and 24h after IM administration of a 5 μg dose of Fluc mRNA in DOG-IM4 or L319 LNPs under 50 μL of PBS. One representative mouse from groups of 4 is shown for each formulation.Panel B: Luciferase expression from the injected muscles was quantified and average Radiance (total photon flux/second) (±SD) from groups of 4 injected mice is represented as a function of time.Panel C: Average (±SD) hEPO expression in the serum of Balb/c mice (4/group) determined by ELISA 6h and 24h after IM administration of 1 μg of hEPO mRNA in either DOG-IM4 or L319 LNPs under 50 μl of PBS.
Due to its convenience the hEPO reporter system was then further used to assess the heat stability of DOG-IM4 LNPs. In the heat stability study depicted in Fig. 6 , DOG-IM4 LNPs loaded with hEPO mRNA were stored under nitrogen in PBS at different temperatures: 4 °C, 25 °C and 37 °C. L319 LNPs containing the same mRNA and stored under nitrogen in PBS at 4 °C were used as a benchmark along with MC3 LNPs, which comprise the L319 analogue, MC3, devoid of hydrolysable ester bonds [17,27]. At different time points, the LNPs were characterized for bioactivity (hEPO expression after IM injection of 1 μg into mice), particle size and PDI (DLS), mRNA encapsulation (Ribogreen assay) and integrity (fragment analyzer), and lipid integrity by HPLC analysis with CAD-MS detection.
Fig. 6 indicates that DOG-IM4 LNPs retained most of their in vivo bioactivity (<20% loss) after 25 weeks of storage at 4 °C, while in the same conditions, L319 and MC3 LNPs lost more than 80% of their bioactivity (Fig. 6, Panel E). At 25 °C and 37 °C, hEPO expression from the DOG-IM4 LNPs declined rapidly over 3 weeks but appeared to remain stable for at least one week at 25 °C (Fig. 6, Panel A). The limitation of these observations being the inherent variability of a bioactivity assay performed in different mice at different points in time, we tried to correlate these observations with an evolution of the physicochemical characteristics of the LNPs. The loss of bioactivity upon storage at 25 °C and 37 °C was found to correlate with an accelerated mRNA degradation at these temperatures as assessed by performing fragment analysis on the mRNA after extraction from the LNPs (Fig. 6, Panel B). An apparent increase in DOG-IM4 LNP average particle size from 130 to 150 nm, which correlated with mRNA degradation when the LNPs were stored at 25 °C and 37 °C was also observed (Fig. 6, Panel C). There was no apparent change in mRNA encapsulation (Fig. 6, Panel D) and LNP lipid integrity (Fig. 7 ) during this accelerated stability study. Note that mRNA integrity analysis could not be performed on L319 and MC3 LNPs due to poor mRNA extraction yields from these LNPs. The other physicochemical parameters of the L319 and MC3 LNPs (particle sizes, mRNA encapsulation and lipid integrity), however, remained stable over the 25-week storage period at 4 °C (data not shown).
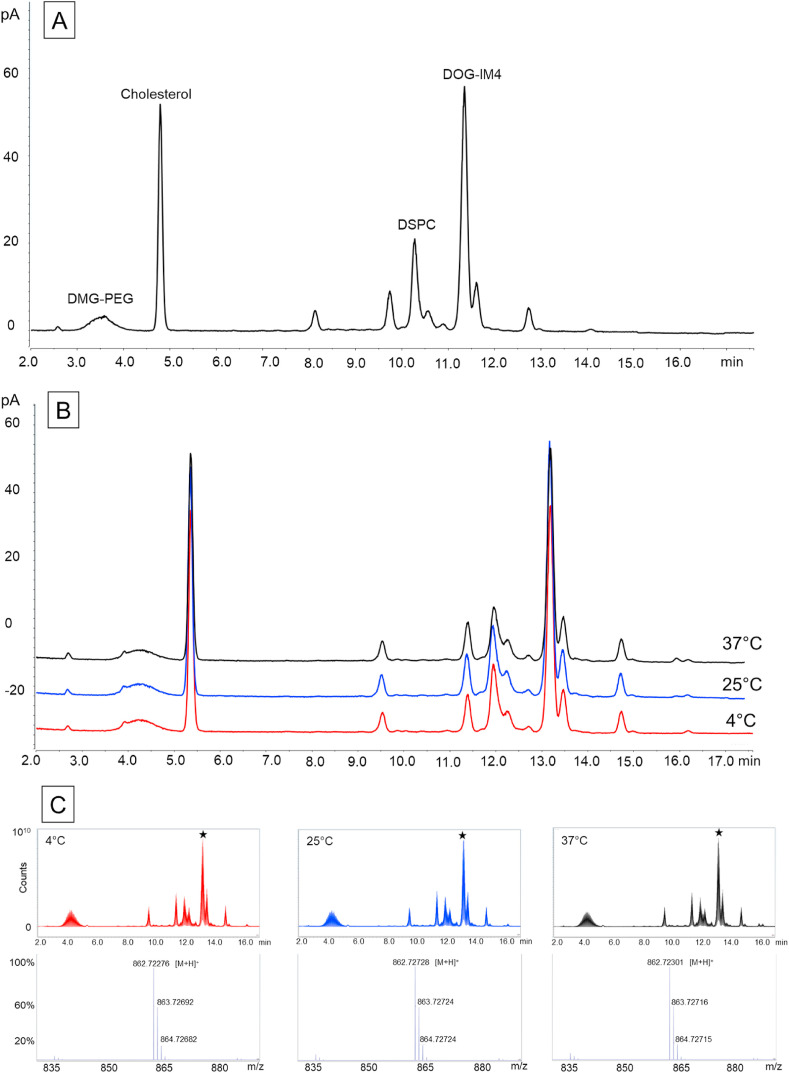
Fig. 7.
Lipid analysis of DOG-IM4 LNPs by UHPLC-CAD-MS at T0 and after 18 weeks of storage at different temperatures. The LNPs used in the stability study described inFig. 7were analyzed for lipid integrity by UHPLC-CAD-MS. PanelA: CAD chromatogram at T0; PanelB: CAD chromatograms after 18 weeks at 4°C (in red), 25°C (in blue) or 37°C (in black). The slight shift in retention times observed between T0 and T18 weeks chromatograms is due to the replacement of the HPLC column by a new one. PanelC: Total Ion Current and mass spectra after 18 weeks at 4°C (in red), 25°C (in blue) or 37°C (in black). The mass spectra correspond to the retention time of DOG-IM4 (13.1 min; indicated by the black star).
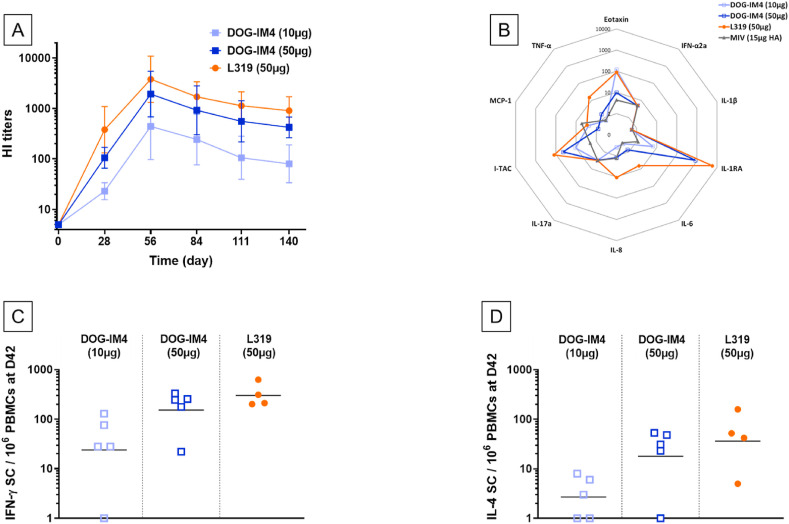
Based on the outstanding stability of DOG-IM4 LNPs and the high and homogeneous HI titers obtained with DOG-IM4 and MNR in mice, this formulation was then tested in non-human primates. In this study, groups of 5 cynomolgus macaques were immunized twice with 10 or 50 μg of MNR in DOG-IM4 LNPs at an N/P ratio of 6. A group of 4 macaques immunized with 50 μg of the same mRNA in L319 LNPs was used as a control group. HI titers obtained with DOG-IM4 LNPs were similar to the titers obtained with L319 LNPs at the 50 μg dose and were significantly higher than titers obtained with a 10 μg dose (p = 0.002). In the 50 μg dose groups, HI titers peaked four weeks after the second immunization with average peak values of 1931 and 3805 for DOG-IM4 and L319 LNPs, respectively, and declined progressively to reach respective average values of 422 and 905 at the end of the study five months later (Fig. 8 , Panel A). The ability of the immunization regimen to induce IFN-γ and IL-4 secreting T cells was monitored by antigen specific ELISPOT assays performed on PBMCs collected prior immunization (baseline) and two weeks after the second injection. In the macaques immunized with DOG-IM4 LNPs, which responded to the mRNA vaccine with high HI titers, a high frequency of IFN-γ secreting cells (20–400 cells/106 PBMCs on average) and a lower frequency of IL-4 secreting cells (1–50 cells/106 PBMCs) was observed (Fig. 8, Panel C&D). A similar trend reflective of Th-1 biased immunity was observed in the macaques immunized with L319 LNPs.
Fig. 8.
DOG-IM4 LNPs induce high functional antibody titers, innate cytokines and Th-1 biased T cell responses in immunogenicity studies with NHPs in a dose-dependent manner.PanelA: Time course of mean (±SD) hemagglutination inhibiting antibody titers (HI titers) measured in sera collected from cynomolgus macaques (N = 5 in DOG-IM4 groups and N = 4 in L319 group) immunized at D0 and D28 with DOG-IM4 or L319 LNPs loaded with 1MpU-modified mRNA (MNR from AmpTec) encoding full-length hemagglutinin (HA) of influenza virus strain A/Netherlands/602/2009 (H1N1). DOG-IM4 LNPs contained either 10 μg or 50 μg of MNR and L319 LNPs contained 50 μg of MNR. The LNPs in PBS were injected into the quadriceps under a final volume of 500 μL. PanelB: Twenty-four hours following the first administration, plasma samples from the immunized monkeys were tested for ten chemokines/cytokines (Eotaxin, IFN-α2a; IL-1β; IL-1RA, IL-6, IL-8, IL-17A; I-TAC, MCP-1, TNF-α) by MSD. Levels are shown in pg/mL on a spider diagram. The levels induced by an injection of 15 μg of monovalent split vaccine (Vaxigrip; MIV) come from a previous study and were plotted as a comparator. PanelC & D: Two weeks following the second administration (D42), PBMCs were collected from the immunized macaques and analyzed for IFN-γ (PanelC) and IL-4 (PanelD) secreting cells by ELISPOT assay after stimulation for 24h with recombinant influenza hemagglutinin (rHA) from A/California/07/2009 strain (H1N1). Cell counts from each individual monkey are shown after subtracting counts obtained with PBMCs collected from the same animal pre-immunization.
None of the immunized macaques experienced clinical symptoms, except one in the 50μg/DOG-IM4 group which developed injection site redness 6 days after the second injection, which resolved within 3 days. The animals were also tested all over the study duration for a typical set of blood biomarkers (CRP, Lipase, AST, ALT, ALP and creatinine) and for innate cytokines and chemokines prior and 24 h post-first injection, by using a multiplexing assay for IL-1RA, IL-1β, IL-6, IL-8 (CXCL8), IL-17, IFN-α2a, I-TAC (CXCL11), MCP-1 (CCL2), TNF-α and eotaxin. The induction of innate chemokines/cytokines in the serum of immunized macaques is depicted on Spider diagram of Fig. 8, Panel B. Both DOG-IM4 and L319 LNPs induced similar moderate to high levels of IL-1RA (1000–10000 pg/mL), I-TAC (100 pg/mL) and eotaxin (100 pg/mL) at a 50 μg dose. These levels were higher than those induced by IM injection of 15 μg of monovalent Vaxigrip® (measured in a previous macaque study). The level of all other cytokines/chemokines remained around or below 10 pg/mL; there was no increase for IFN-α2a, IL-1β and IL-17. At the 10 μg mRNA dose, the DOG-IM4-induced cytokine/chemokine levels were low and comparable to that of the Vaxigrip® reference, except for eotaxin, which reached 100 pg/mL. Importantly, none of the major inflammatory cytokines, i.e. TNFα, IL-1β and IL-6, was increased in the serum 24 h after the first injection as compared to baseline levels of the Vaxigrip® benchmark.
Immunization induced a transient elevation of CRP in 4/5 and 3/5 animals in the 50 μg DOG-IM4 group post first and second injection, respectively, and a more pronounced elevation in 4/4 animals in the L319 group post each injection. The CRP increased one day following each immunization and decreased from two days post-injection to return to normal levels six days post-injection. This observation suggests that the 50 μg doses could induce transient systemic inflammation in the vaccinated monkeys (Supp. Figure S1). Concerning the other biochemical parameters, despite some inter individual variability, especially for lipase, ALP and creatinine, the levels of these biomarkers remained essentially unaffected by the immunization regimen, except for two monkeys that experienced a transient increase in lipase after the second injection in the L319 group (Supp. Figure S1).
4. Discussion
This study describes a novel ionizable lipid, DOG-IM4, for the formulation of LNPs that remain active upon long term storage in PBS under nitrogen at 4 °C. DOG-IM4 comprises an ionizable head group consisting of imidazole, a dioleoyl lipid tail and a short flexible polyoxyethylene spacer between the head and tail. The imidazole head group was inspired by pioneering works on polymers, peptides and lipids containing histidine or imidazole as nucleic acid carriers [[46], [47], [48], [49]]. This head group was selected for its pKa in the range of 5.5–6.5, enabling a neutral charge in the extracellular space at pH 7.4 and a cationic charge after endocytosis into endosomal vesicles (pH < 5.5). The pH-sensitivity was shown to be a critical factor for the efficacy of in vivo mRNA delivery and release of the mRNA into the cytosol after endocytosis. Furthermore, the imidazole group, which is found in the natural amino acid histidine, may be susceptible to increase LNP/mRNA stability by enabling π interactions [50] and providing antioxidant properties [51,52]. A flexible spacer arm consisting of three polyoxyethylene units was introduced to facilitate electrostatic and π−π stacking interactions between the imidazolium head groups and mRNA. To maintain in vivo activity, it is critical to attach the spacer arm at position 4 of the imidazole head group as shown by comparing DOG-IM4 with DOG-IM2, where the spacer arm was attached at position 2 of the imidazole head group. The difference in potency between DOG-IM4 and DOG-IM2 is an illustration of how subtle differences in the chemical structure of the ionizable cationic lipid can affect potency, as already observed by others [[29], [30], [31],[53], [54], [55], [56]].
In terms of LNP formulation, this initial work was performed by using standard helper lipids and a ionizable lipid to RNA charge ratio (N/P) of 6 [27]. We did not try to modify formulation parameters at this stage but DOG-IM4 LNP formulation optimization through a design of experiment approach is currently underway in our laboratory. LNP formulation was performed by using a NanoAssemblr™, a microfluidic mixing device enabling monodispersed particle populations and batch-to-batch reproducibility at laboratory scale and potential scale-up possibilities [43,57,58]. By proceeding this way, robust and potent LNP formulations with consistent particle sizes and encapsulation rates could be rapidly produced. While a small particle size below 200 nm was known to be important for lymphatic uptake [[59], [60], [61]], the effect of LNP particle size on the quality of immune responses induced by mRNA vaccines was only recently studied by Hassett and coworkers [62]. By using a fixed lipid composition and controlled process parameters, these authors formulated various LNP particle sizes ranging from 60 to 150 nm and observed that larger particle sizes of 80 nm and above were optimal for the induction of strong immune responses in mice and monkeys. With an average particle size around 130 nm, both DOG-IM4 and L319 LNPs should therefore be in the appropriate size range.
While biodegradability was the main driver for the design of L319 and other more recent cationic lipids [22], biodegradability was not the main driver for the design of DOG-IM4. Rather, stable ether bonds and a heterocyclic amino head group enabling π−π stacking interactions were selected to provide increased stability in a liquid formulation at 4 °C, as this may represent an important asset for mRNA vaccine development [38]. In our mouse immunization study using influenza HA expressing UNR from Trilink, the potency of DOG-IM4 LNPs remained essentially the same over one year of storage in PBS at 4 °C, based on their capacity to induce high functional HI antibodies. This initial observation was confirmed in a temperature stress study using hEPO reporter mRNA to provide a rapid readout for in vivo bioactivity. The functional in vivo assay was accompanied by a set of biochemical and biophysical assays including mRNA integrity and encapsulation, particle sizing, and lipid integrity by HPLC-CAD-MS. The hEPO results confirmed that DOG-IM4 LNPs would be stable for at least one week at 25 °C and for at least 6 months at 4 °C. In this temperature stress study, the decline of potency of the LNPs over time could not be attributed to a degradation of the LNP lipid components as assessed by RP-HPLC-MS analysis but rather to a degradation of mRNA as assessed by fragment analyzer on a fraction of mRNA extracted from the LNPs. The mRNA degradation was accompanied with an evolution of LNP particle size from 130 nm to 150 nm, suggesting an evolution of the LNP structure. RNA degradation could occur through in-line hydrolytic cleavage and attack by nucleases, oxidizers, and chemical modifiers that may be present in the RNA's environment [63]. In-line hydrolytic cleavage refers to mRNA backbone cleavage by inline attack of ribose 2′OH on the neighboring phosphodiester bond. This transesterification reaction, which may be catalyzed by amino lipids able to decrease the pKa of 2′OH, relies on the RNA backbone being able to adopt a specific conformation where the 2′OH is in line with the leaving 5′ oxyanion of the phosphodiester [64]. DOG-IM4 was designed with an imidazole group attached to a flexible spacer arm as to enable π − π interactions within the LNPs, not only between neighboring imidazole head groups but also with nitrogenous base rings on mRNA on top of the electrostatic interactions with the mRNA phosphate backbone. By doing so, the imidazole lipid may block the mRNA conformation and protect the mRNA backbone from self-cleaving by preventing inline attack of ribose 2′OH on neighboring phosphodiester bond. A similar mechanism of protection has been proposed in mRNA secondary structures where base pairing by hydrogen bonding and π-stacking is thought to protect from inline cleavage and enzymatic degradation [65,66]. Furthermore, the antioxidant properties of the imidazole group in DOG-IM4 may protect mRNA from degrading by oxidation [63]. Finally, a novel mechanism for the loss of mRNA activity in lipid nanoparticle delivery systems was recently described, involving cationic lipid tertiary amine oxidation, hydrolysis and mRNA-lipid adduct formation [67]. Contrarily to the tertiary amino group of L319, MC3 or of SM-102 and ALC-0315, the ionizable amino lipids used respectively in Spikevax and Comirnaty [11], the imidazole head group in DOG-IM4 would not be compatible with this mechanism, adding rational to explain the retention of mRNA activity in DOG-IM4 LNPs. Obviously, more work is necessary to study LNP stability, to correlate changes in biophysical characteristics with activity and understand the underlying mechanisms of mRNA stabilization in DOG-IM4 LNPs at refrigerated temperature. In this regard, molecular modeling and electron microscopy work was initiated in our group to study the partition of mRNA between the aqueous cavities and dense bodies of the DOG-IM4 LNPs observed by electron microscopy (Fig. 2) and to better understand the interactions taking place between mRNA and DOG-IM4 in these structures.
In terms of potency, the DOG-IM4 LNPs used with an mRNA encoding the influenza HA of A/Netherlands 2009 strain (H1N1 strain) were found capable of inducing high functional HI antibody titers in mice and macaques upon intramuscular injection. The immune responses obtained with DOG-IM4 LNPs were in the same range and displayed the same Th-1 bias (based on the ratio of IFN-γ/IL-4 producing T cells), as those obtained with the L319 LNPs. The induction of a Th-1 type immunity observed with these formulations was in accordance with previous observations and with the general attributes of mRNA vaccines [68]. When comparing natural influenza HA mRNA to 1MpU-modified HA mRNA from the same supplier (AmpTec), meaning that the same UTRs and production and purification methods were used for both constructs, the 1MpU mRNA outperformed the natural mRNA for the induction of high functional antibody titers, which was in accordance with previous reports indicating that 1MpU modification could increase mRNA translation [[69], [70], [71], [72], [73]] and mRNA vaccine efficacy [74]. The superiority of the modified nucleotide mRNA vaccines is typically attributed to weaker stimulation of innate immune pathways, such as 2′-5′-oligoadenylate synthetase and TLR3, TLR7 and TLR8 activation capable of impeding mRNA expression [71,[75], [76], [77]]. However, as indicated by comparing natural mRNAs from two different suppliers (Trilink vs AmpTec), the efficacy of mRNA vaccines encoding the same antigen depends also largely on mRNA design, sequence optimization and purity as already demonstrated by others [12,78,79].
Although the in vivo studies described in this report were not designed for safety assessment, the administration of DOG-IM4 LNPs to mice and monkeys did not induce any reactogenicity of concern. In fact, despite mild to moderate injection site reactions observed in most of the mice at the 5 μg dose (but not at the 1 μg dose) and in one of the monkeys at the 50 μg dose (but not at the 10 μg dose), these reactions remained transient and disappeared within 3–6 days and did not affect the animal normal behavior and weight gain. The determination of the innate cytokine/chemokine profile induced by the injection of the DOG-IM4 mRNA vaccine in macaques and the monitoring of standard blood biomarkers over the study course, also suggested that this formulation would be tolerated at least as well as L319 LNPs when used with 1MpU modified mRNA.
Noteworthy, upon IM administration of 1 μg hEPO mRNA into mice, about ten-fold more hEPO was measured in the circulation from L319 LNPs as compared to DOG-IM4 LNPs (Fig. 5, panel C), while the latter brought about the same immune responses as L319 LNPs when assessed with influenza HA expressing mRNAs. Interestingly, with 5 μg of FLuc mRNA, DOG-IM4 and L319 LNPs appeared to induce a similar pattern of mRNA expression at the site of injection in mice, as assessed by using two-dimensional whole-body imaging (Fig. 5, Panel A&B). It is our observation however, that, in general, there is a poor correlation between the level of mRNA expression and the induced immune responses from a given LNP formulation, as already reported by others [22]. This could be due to multiple factors including specific biodistribution and delivery patterns and intrinsic immunostimulatory capacity of the LNP. In the present case, L319 LNPs may have driven higher levels of hEPO expression in general or reached organs/cells that are more susceptible to secrete the expressed hEPO into the circulation.
5. Conclusion
In summary we have designed, synthesized, and evaluated a new ionizable lipid, DOG-IM4, that has potential for mRNA LNP stabilization. DOG-IM4 LNPs brought about the same immune responses as L319 LNPs when assessed with natural or 1MpU-modified influenza HA mRNA in mice and monkeys. When assessed in the hEPO reporter system, the potency of DOG-IM4 LNPs was found stable (<20% activity loss) for at least 6 months, when LNPs were stored in PBS under nitrogen at 4 °C and for one week at 25 °C. Results of this study were in accordance with immunogenicity results suggesting that influenza HA mRNA in DOG-IM4 LNPs could be stable long term when stored in PBS under nitrogen at 4 °C. The long-term liquid stability of DOG-IM4 LNPs when stored at 4 °C and 25 °C warrants further development of this lipid family for the formulation of mRNA vaccines. More work is currently underway in our group to study if potency of DOG-IM4 LNPs could be further improved by introducing changes in the lipid structure (lead optimization) and by changing formulation parameters such as molar ratios of the LNP constituents and/or the N/P charge ratio.
Credit author statement
Conceptualization and methodology : M.R., M-C.B., B.F. and J.H. Investigations: M.R., M-C.B., C.V., E.B., S.C., V.P., K.L., M-C.N., P.B., A.K. Writing – Original Draft: M.R., M-C.B. and J.H. Writing – Review & Editing: C.V., A.K., B.F. and J.H. Supervision: M-C.B., A.K., B.F. and J.H.
Disclaimer
Certain commercial materials, equipment and suppliers are identified in this manuscript for the sake of clarity. This does not imply that the materials, equipment, and suppliers identified are necessarily the best available for the purpose.
Funding
This work was funded by Sanofi.
Data availability statement
Research data are not shared.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
We gratefully thank Nadine Petiot and Stéphanie Guinamand for expert technical assistance, Julie Piolat for statistical analyses, Luc Even for helpful discussions and Bachra Rokbi for supporting this work and critical review of the manuscript.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.biomaterials.2022.121570.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Sahin U., Karikó K., Türeci Ö. mRNA-based therapeutics--developing a new class of drugs. Nat. Rev. Drug Discov. 2014;13:759–780. doi: 10.1038/nrd4278. [DOI] [PubMed] [Google Scholar]
- 2.Thran M., Mukherjee J., Pönisch M., Fiedler K., Thess A., Mui B.L., Hope M.J., Tam Y.K., Horscroft N., Heidenreich R., Fotin-Mleczek M., Shoemaker C.B., Schlake T. mRNA mediates passive vaccination against infectious agents, toxins, and tumors. EMBO Mol. Med. 2017;9:1434–1447. doi: 10.15252/emmm.201707678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pardi N., Hogan M.J., Porter F.W., Weissman D. mRNA vaccines - a new era in vaccinology. Nat. Rev. Drug Discov. 2018;17:261–279. doi: 10.1038/nrd.2017.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pardi N., Hogan M.J., Weissman D. Recent advances in mRNA vaccine technology. Curr. Opin. Immunol. 2020;65:14–20. doi: 10.1016/j.coi.2020.01.008. [DOI] [PubMed] [Google Scholar]
- 5.Zhang C., Maruggi G., Shan H., Li J. Advances in mRNA vaccines for infectious diseases. Front. Immunol. 2019;10:594. doi: 10.3389/fimmu.2019.00594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jackson N.A.C., Kester K.E., Casimiro D., Gurunathan S., DeRosa F. The promise of mRNA vaccines: a biotech and industrial perspective. NPJ Vaccines. 2020;5:11. doi: 10.1038/s41541-020-0159-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Krammer F. SARS-CoV-2 vaccines in development. Nature. 2020;586:516–527. doi: 10.1038/s41586-020-2798-3. [DOI] [PubMed] [Google Scholar]
- 8.Wang F., Kream R.M., Stefano G.B. An evidence based perspective on mRNA-SARS-CoV-2 vaccine development. Med. Sci. Mon. Int. Med. J. Exp. Clin. Res. 2020;26 doi: 10.12659/MSM.924700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bettini E., Locci M. SARS-CoV-2 mRNA vaccines: immunological mechanism and beyond. Vaccines (Basel) 2021;9 doi: 10.3390/vaccines9020147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Verbeke R., Lentacker I., De Smedt S.C., Dewitte H. The dawn of mRNA vaccines: the COVID-19 case. J. Contr. Release. 2021;333:511–520. doi: 10.1016/j.jconrel.2021.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schoenmaker L., Witzigmann D., Kulkarni J.A., Verbeke R., Kersten G., Jiskoot W., Crommelin D.J.A. mRNA-lipid nanoparticle COVID-19 vaccines: structure and stability. Int. J. Pharm. 2021;601:120586. doi: 10.1016/j.ijpharm.2021.120586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lutz J., Lazzaro S., Habbeddine M., Schmidt K.E., Baumhof P., Mui B.L., Tam Y.K., Madden T.D., Hope M.J., Heidenreich R., Fotin-Mleczek M. Unmodified mRNA in LNPs constitutes a competitive technology for prophylactic vaccines. NPJ Vaccines. 2017;2:29. doi: 10.1038/s41541-017-0032-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zeng C., Zhang C., Walker P.G., Dong Y. Formulation and delivery technologies for mRNA vaccines. Curr. Top. Microbiol. Immunol. 2020 doi: 10.1007/82_2020_217. https://link.springer.com/content/pdf/10.1007%2F82_2020_217.pdf?msclkid=268492fbcf8611ec8ea331b451db98d1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Buschmann M.D., Carrasco M.J., Alishetty S., Paige M., Alameh M.G., Weissman D. Nanomaterial delivery systems for mRNA vaccines. Vaccines (Basel) 2021;9 doi: 10.3390/vaccines9010065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim J., Eygeris Y., Gupta M., Sahay G. Self-assembled mRNA vaccines. Adv. Drug Deliv. Rev. 2021;170:83–112. doi: 10.1016/j.addr.2020.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Semple S.C., Akinc A., Chen J., Sandhu A.P., Mui B.L., Cho C.K., Sah D.W.Y., Stebbing D., Crosley E.J., Yaworski E., Hafez I.M., Dorkin J.R., Qin J., Lam K., Rajeev K.G., Wong K.F., Jeffs L.B., Nechev L., Eisenhardt M.L., Jayaraman M., Kazem M., Maier M.A., Srinivasulu M., Weinstein M.J., Chen Q., Alvarez R., Barros S.A., De S., Klimuk S.K., Borland T., Kosovrasti V., Cantley W.L., Tam Y.K., Manoharan M., Ciufolini M.A., Tracy M.A., de Fougerolles A., MacLachlan I., Cullis P.R., Madden T.D., Hope M.J. Rational design of cationic lipids for siRNA delivery. Nat. Biotechnol. 2010;28:172–176. doi: 10.1038/nbt.1602. [DOI] [PubMed] [Google Scholar]
- 17.Jayaraman M., Ansell S.M., Mui B.L., Tam Y.K., Chen J., Du X., Butler D., Eltepu L., Matsuda S., Narayanannair J.K., Rajeev K.G., Hafez I.M., Akinc A., Maier M.A., Tracy M.A., Cullis P.R., Madden T.D., Manoharan M., Hope M.J. Maximizing the potency of siRNA lipid nanoparticles for hepatic gene silencing in vivo. Angew Chem. Int. Ed. Engl. 2012;51:8529–8533. doi: 10.1002/anie.201203263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tam Y.Y.C., Chen S., Cullis P.R. Advances in lipid nanoparticles for siRNA delivery. Pharmaceutics. 2013;5:498–507. doi: 10.3390/pharmaceutics5030498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sato Y., Hatakeyama H., Sakurai Y., Hyodo M., Akita H., Harashima H. A pH-sensitive cationic lipid facilitates the delivery of liposomal siRNA and gene silencing activity in vitro and in vivo. J. Contr. Release. 2012;163:267–276. doi: 10.1016/j.jconrel.2012.09.009. [DOI] [PubMed] [Google Scholar]
- 20.Sato Y., Hashiba K., Sasaki K., Maeki M., Tokeshi M., Harashima H. Understanding structure-activity relationships of pH-sensitive cationic lipids facilitates the rational identification of promising lipid nanoparticles for delivering siRNAs in vivo. J. Contr. Release. 2019;295:140–152. doi: 10.1016/j.jconrel.2019.01.001. [DOI] [PubMed] [Google Scholar]
- 21.Sabnis S., Kumarasinghe E.S., Salerno T., Mihai C., Ketova T., Senn J.J., Lynn A., Bulychev A., McFadyen I., Chan J., Almarsson Ö., Stanton M.G., Benenato K.E. A novel amino lipid series for mRNA delivery: improved endosomal escape and sustained pharmacology and safety in non-human primates. Mol. Ther. 2018;26:1509–1519. doi: 10.1016/j.ymthe.2018.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hassett K.J., Benenato K.E., Jacquinet E., Lee A., Woods A., Yuzhakov O., Himansu S., Deterling J., Geilich B.M., Ketova T., Mihai C., Lynn A., McFadyen I., Moore M.J., Senn J.J., Stanton M.G., Almarsson Ö., Ciaramella G., Brito L.A. Optimization of lipid nanoparticles for intramuscular administration of mRNA vaccines. Mol. Ther. Nucleic Acids. 2019;15:1–11. doi: 10.1016/j.omtn.2019.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xu Y., Szoka F.C. Mechanism of DNA release from cationic liposome/DNA complexes used in cell transfection. Biochemistry. 1996;35:5616–5623. doi: 10.1021/bi9602019. [DOI] [PubMed] [Google Scholar]
- 24.Hafez I.M., Maurer N., Cullis P.R. On the mechanism whereby cationic lipids promote intracellular delivery of polynucleic acids. Gene Ther. 2001;8:1188–1196. doi: 10.1038/sj.gt.3301506. [DOI] [PubMed] [Google Scholar]
- 25.Akinc A., Maier M.A., Manoharan M., Fitzgerald K., Jayaraman M., Barros S., Ansell S., Du X., Hope M.J., Madden T.D., Mui B.L., Semple S.C., Tam Y.K., Ciufolini M., Witzigmann D., Kulkarni J.A., van der Meel R., Cullis P.R. The Onpattro story and the clinical translation of nanomedicines containing nucleic acid-based drugs. Nat. Nanotechnol. 2019;14:1084–1087. doi: 10.1038/s41565-019-0591-y. [DOI] [PubMed] [Google Scholar]
- 26.Rizk M., Tüzmen Ş. Update on the clinical utility of an RNA interference-based treatment: focus on Patisiran. Pharmgenomics Pers Med. 2017;10:267–278. doi: 10.2147/PGPM.S87945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Maier M.A., Jayaraman M., Matsuda S., Liu J., Barros S., Querbes W., Tam Y.K., Ansell S.M., Kumar V., Qin J., Zhang X., Wang Q., Panesar S., Hutabarat R., Carioto M., Hettinger J., Kandasamy P., Butler D., Rajeev K.G., Pang B., Charisse K., Fitzgerald K., Mui B.L., Du X., Cullis P., Madden T.D., Hope M.J., Manoharan M., Akinc A. Biodegradable lipids enabling rapidly eliminated lipid nanoparticles for systemic delivery of RNAi therapeutics. Mol. Ther. 2013;21:1570–1578. doi: 10.1038/mt.2013.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fenton O.S., Kauffman K.J., McClellan R.L., Appel E.A., Dorkin J.R., Tibbitt M.W., Heartlein M.W., DeRosa F., Langer R., Anderson D.G. Bioinspired alkenyl amino alcohol ionizable lipid materials for highly potent in vivo mRNA delivery. Adv Mater. 2016;28:2939–2943. doi: 10.1002/adma.201505822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Carrasco M.J., Alishetty S., Alameh M.-G., Said H., Wright L., Paige M., Soliman O., Weissman D., Cleveland T.E., Grishaev A., Buschmann M.D. Ionization and structural properties of mRNA lipid nanoparticles influence expression in intramuscular and intravascular administration. Commun Biol. 2021;4:1–15. doi: 10.1038/s42003-021-02441-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hajj K.A., Ball R.L., Deluty S.B., Singh S.R., Strelkova D., Knapp C.M., Whitehead K.A. Branched-tail lipid nanoparticles potently deliver mRNA in vivo due to enhanced ionization at endosomal pH. Small. 2019;15 doi: 10.1002/smll.201805097. [DOI] [PubMed] [Google Scholar]
- 31.M. Cornebise, E. Narayanan, Y. Xia, E. Acosta, L. Ci, H. Koch, J. Milton, S. Sabnis, T. Salerno, K.E. Benenato, Discovery of a Novel Amino Lipid That Improves Lipid Nanoparticle Performance through Specific Interactions with mRNA, Adv. Funct. Mater.. n/a (n.d.) 2106727. 10.1002/adfm.202106727. [DOI]
- 32.Cullis P.R., Hope M.J. Lipid nanoparticle systems for enabling gene therapies. Mol. Ther. 2017;25:1467–1475. doi: 10.1016/j.ymthe.2017.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Guan S., Rosenecker J. Nanotechnologies in delivery of mRNA therapeutics using nonviral vector-based delivery systems. Gene Ther. 2017;24:133–143. doi: 10.1038/gt.2017.5. [DOI] [PubMed] [Google Scholar]
- 34.Hajj K.A., Whitehead K.A. Tools for translation: non-viral materials for therapeutic mRNA delivery. Nat. Rev. Mater. 2017;2:1–17. doi: 10.1038/natrevmats.2017.56. [DOI] [Google Scholar]
- 35.Kaczmarek J.C., Kowalski P.S., Anderson D.G. Advances in the delivery of RNA therapeutics: from concept to clinical reality. Genome Med. 2017;9:60. doi: 10.1186/s13073-017-0450-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kowalski P.S., Rudra A., Miao L., Anderson D.G. Delivering the messenger: advances in technologies for therapeutic mRNA delivery. Mol. Ther. 2019;27:710–728. doi: 10.1016/j.ymthe.2019.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Samaridou E., Heyes J., Lutwyche P. Lipid nanoparticles for nucleic acid delivery: current perspectives. Adv. Drug Deliv. Rev. 2020;154–155:37–63. doi: 10.1016/j.addr.2020.06.002. [DOI] [PubMed] [Google Scholar]
- 38.Crommelin D.J.A., Anchordoquy T.J., Volkin D.B., Jiskoot W., Mastrobattista E. Addressing the cold reality of mRNA vaccine stability. J Pharm Sci. 2021;110:997–1001. doi: 10.1016/j.xphs.2020.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Suzuki Y., Hyodo K., Tanaka Y., Ishihara H. siRNA-lipid nanoparticles with long-term storage stability facilitate potent gene-silencing in vivo. J. Contr. Release. 2015;220:44–50. doi: 10.1016/j.jconrel.2015.10.024. [DOI] [PubMed] [Google Scholar]
- 40.S. Alishetty, M. Carrasco, M.-G. Alameh, M. Paige, H. Said, L. Wright, A. Narayanan, F. Alem, K. Hernandez, P. Gillevet, O. Soliman, P. Hicks, T. Manzoni, P. Bates, A. Stephens-Shields, T. Cleveland, A. Grishaev, D. Weissman, M. Buschmann, Novel lipid nanoparticle provides potent SARS-CoV-2 mRNA vaccine at low dose with low local reactogenicity, high thermostability and limited systemic biodistribution, 17 August 2021, PREPRINT (Version 1) available at: Research Square [ 10.21203/rs.3.rs-798453/v1]. [DOI]
- 41.Espuelas S., Haller P., Schuber F., Frisch B. Synthesis of an amphiphilic tetraantennary mannosyl conjugate and incorporation into liposome carriers. Bioorg. Med. Chem. Lett. 2003;13:2557–2560. doi: 10.1016/s0960-894x(03)00472-4. [DOI] [PubMed] [Google Scholar]
- 42.Vittoria Spanedda M., Salomé C., Hilbold B., Berner E., Heurtault B., Fournel S., Frisch B., Bourel-Bonnet L. Smart tools and orthogonal click-like reactions onto small unilamellar vesicles: additional molecular data. Data Brief. 2015;5:145–154. doi: 10.1016/j.dib.2015.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Belliveau N.M., Huft J., Lin P.J., Chen S., Leung A.K., Leaver T.J., Wild A.W., Lee J.B., Taylor R.J., Tam Y.K., Hansen C.L., Cullis P.R. Microfluidic synthesis of highly potent limit-size lipid nanoparticles for in vivo delivery of siRNA. Mol. Ther. Nucleic Acids. 2012;1:e37. doi: 10.1038/mtna.2012.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Brader M.L., Williams S.J., Banks J.M., Hui W.H., Zhou Z.H., Jin L. Encapsulation state of messenger RNA inside lipid nanoparticles. Biophys. J. 2021;120:2766–2770. doi: 10.1016/j.bpj.2021.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Leung A.K.K., Tam Y.Y.C., Chen S., Hafez I.M., Cullis P.R. Microfluidic mixing: a general method for encapsulating macromolecules in lipid nanoparticle systems. J. Phys. Chem. B. 2015;119:8698–8706. doi: 10.1021/acs.jpcb.5b02891. [DOI] [PubMed] [Google Scholar]
- 46.Kumar V.V., Pichon C., Refregiers M., Guerin B., Midoux P., Chaudhuri A. Single histidine residue in head-group region is sufficient to impart remarkable gene transfection properties to cationic lipids: evidence for histidine-mediated membrane fusion at acidic pH. Gene Ther. 2003;10:1206–1215. doi: 10.1038/sj.gt.3301979. [DOI] [PubMed] [Google Scholar]
- 47.Midoux P., Pichon C., Yaouanc J.-J., Jaffrès P.-A. Chemical vectors for gene delivery: a current review on polymers, peptides and lipids containing histidine or imidazole as nucleic acids carriers. Br. J. Pharmacol. 2009;157:166–178. doi: 10.1111/j.1476-5381.2009.00288.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ripoll M., Neuberg P., Kichler A., Tounsi N., Wagner A., Remy J.-S. pH-responsive nanometric polydiacetylenic micelles allow for efficient intracellular siRNA delivery. ACS Appl. Mater. Interfaces. 2016;8:30665–30670. doi: 10.1021/acsami.6b09365. [DOI] [PubMed] [Google Scholar]
- 49.Lointier M., Aisenbrey C., Marquette A., Tan J.H., Kichler A., Bechinger B. Membrane pore-formation correlates with the hydrophilic angle of histidine-rich amphipathic peptides with multiple biological activities. Biochim. Biophys. Acta Biomembr. 1862:183212. doi: 10.1016/j.bbamem.2020.183212. 2020. [DOI] [PubMed] [Google Scholar]
- 50.Corley M., Burns M.C., Yeo G.W. How RNA-binding proteins interact with RNA: molecules and mechanisms. Mol Cell. 2020;78:9–29. doi: 10.1016/j.molcel.2020.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wade A.M., Tucker H.N. Antioxidant characteristics of L-histidine. J. Nutr. Biochem. 1998;9:308–315. doi: 10.1016/s0955-2863(98)00022-9. [DOI] [Google Scholar]
- 52.Sorrenti V., Salerno L., Di Giacomo C., Acquaviva R., Siracusa M.A., Vanella A. Imidazole derivatives as antioxidants and selective inhibitors of nNOS. Nitric Oxide. 2006;14:45–50. doi: 10.1016/j.niox.2005.09.005. [DOI] [PubMed] [Google Scholar]
- 53.Heyes J., Palmer L., Bremner K., MacLachlan I. Cationic lipid saturation influences intracellular delivery of encapsulated nucleic acids. J. Contr. Release. 2005;107:276–287. doi: 10.1016/j.jconrel.2005.06.014. [DOI] [PubMed] [Google Scholar]
- 54.Jarzębińska A., Pasewald T., Lambrecht J., Mykhaylyk O., Kümmerling L., Beck P., Hasenpusch G., Rudolph C., Plank C., Dohmen C. A single methylene group in oligoalkylamine-based cationic polymers and lipids promotes enhanced mRNA delivery. Angew Chem. Int. Ed. Engl. 2016;55:9591–9595. doi: 10.1002/anie.201603648. [DOI] [PubMed] [Google Scholar]
- 55.Li B., Luo X., Deng B., Giancola J.B., McComb D.W., Schmittgen T.D., Dong Y. Effects of local structural transformation of lipid-like compounds on delivery of messenger RNA. Sci. Rep. 2016;6:22137. doi: 10.1038/srep22137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Fenton O.S., Kauffman K.J., Kaczmarek J.C., McClellan R.L., Jhunjhunwala S., Tibbitt M.W., Zeng M.D., Appel E.A., Dorkin J.R., Mir F.F., Yang J.H., Oberli M.A., Heartlein M.W., DeRosa F., Langer R., Anderson D.G. Synthesis and biological evaluation of ionizable lipid materials for the in vivo delivery of messenger RNA to B lymphocytes. Adv Mater. 2017;29 doi: 10.1002/adma.201606944. [DOI] [PubMed] [Google Scholar]
- 57.Zhigaltsev I.V., Belliveau N., Hafez I., Leung A.K.K., Huft J., Hansen C., Cullis P.R. Bottom-up design and synthesis of limit size lipid nanoparticle systems with aqueous and triglyceride cores using millisecond microfluidic mixing. Langmuir. 2012;28:3633–3640. doi: 10.1021/la204833h. [DOI] [PubMed] [Google Scholar]
- 58.Shepherd S.J., Issadore D., Mitchell M.J. Microfluidic formulation of nanoparticles for biomedical applications. Biomaterials. 2021;274:120826. doi: 10.1016/j.biomaterials.2021.120826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Swartz M.A. The physiology of the lymphatic system. Adv. Drug Deliv. Rev. 2001;50:3–20. doi: 10.1016/s0169-409x(01)00150-8. [DOI] [PubMed] [Google Scholar]
- 60.Oussoren C., Zuidema J., Crommelin D.J., Storm G. Lymphatic uptake and biodistribution of liposomes after subcutaneous injection. II. Influence of liposomal size, lipid compostion and lipid dose. Biochim. Biophys. Acta. 1997;1328:261–272. doi: 10.1016/s0005-2736(97)00122-3. [DOI] [PubMed] [Google Scholar]
- 61.Carstens M.G., Camps M.G.M., Henriksen-Lacey M., Franken K., Ottenhoff T.H.M., Perrie Y., Bouwstra J.A., Ossendorp F., Jiskoot W. Effect of vesicle size on tissue localization and immunogenicity of liposomal DNA vaccines. Vaccine. 2011;29:4761–4770. doi: 10.1016/j.vaccine.2011.04.081. [DOI] [PubMed] [Google Scholar]
- 62.Hassett K.J., Higgins J., Woods A., Levy B., Xia Y., Hsiao C.J., Acosta E., Almarsson Ö., Moore M.J., Brito L.A. Impact of lipid nanoparticle size on mRNA vaccine immunogenicity. J. Contr. Release. 2021;335:237–246. doi: 10.1016/j.jconrel.2021.05.021. [DOI] [PubMed] [Google Scholar]
- 63.Pogocki D., Schöneich C. Chemical stability of nucleic acid-derived drugs. J Pharm Sci. 2000;89:443–456. doi: 10.1002/(SICI)1520-6017(200004)89:4<443::AID-JPS2>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 64.Li Y., Breaker R.R. Kinetics of RNA degradation by specific base catalysis of transesterification involving the 2‘-hydroxyl group. J. Am. Chem. Soc. 1999;121:5364–5372. doi: 10.1021/ja990592p. [DOI] [Google Scholar]
- 65.Wayment-Steele H.K., Kim D.S., Choe C.A., Nicol J.J., Wellington-Oguri R., Watkins A.M., Parra Sperberg R.A., Huang P.-S., Participants E., Das R. Theoretical basis for stabilizing messenger RNA through secondary structure design. Nucleic Acids Res. 2021;49:10604–10617. doi: 10.1093/nar/gkab764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mikkola S., Kaukinen U., Lönnberg H. The effect of secondary structure on cleavage of the phosphodiester bonds of RNA. Cell Biochem. Biophys. 2001;34:95–119. doi: 10.1385/CBB:34:1:95. [DOI] [PubMed] [Google Scholar]
- 67.Packer M., Gyawali D., Yerabolu R., Schariter J., White P.A. A novel mechanism for the loss of mRNA activity in lipid nanoparticle delivery systems. Nat Commun. 2021;12:6777. doi: 10.1038/s41467-021-26926-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Liang F., Lindgren G., Lin A., Thompson E.A., Ols S., Röhss J., John S., Hassett K., Yuzhakov O., Bahl K., Brito L.A., Salter H., Ciaramella G., Loré K. Efficient targeting and activation of antigen-presenting cells in vivo after modified mRNA vaccine administration in rhesus macaques. Mol. Ther. 2017;25:2635–2647. doi: 10.1016/j.ymthe.2017.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Karikó K., Muramatsu H., Welsh F.A., Ludwig J., Kato H., Akira S., Weissman D. Incorporation of pseudouridine into mRNA yields superior nonimmunogenic vector with increased translational capacity and biological stability. Mol. Ther. 2008;16:1833–1840. doi: 10.1038/mt.2008.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Karikó K., Muramatsu H., Keller J.M., Weissman D. Increased erythropoiesis in mice injected with submicrogram quantities of pseudouridine-containing mRNA encoding erythropoietin. Mol. Ther. 2012;20:948–953. doi: 10.1038/mt.2012.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Karikó K., Buckstein M., Ni H., Weissman D. Suppression of RNA recognition by Toll-like receptors: the impact of nucleoside modification and the evolutionary origin of RNA. Immunity. 2005;23:165–175. doi: 10.1016/j.immuni.2005.06.008. [DOI] [PubMed] [Google Scholar]
- 72.Kormann M.S.D., Hasenpusch G., Aneja M.K., Nica G., Flemmer A.W., Herber-Jonat S., Huppmann M., Mays L.E., Illenyi M., Schams A., Griese M., Bittmann I., Handgretinger R., Hartl D., Rosenecker J., Rudolph C. Expression of therapeutic proteins after delivery of chemically modified mRNA in mice. Nat. Biotechnol. 2011;29:154–157. doi: 10.1038/nbt.1733. [DOI] [PubMed] [Google Scholar]
- 73.Svitkin Y.V., Cheng Y.M., Chakraborty T., Presnyak V., John M., Sonenberg N. N1-methyl-pseudouridine in mRNA enhances translation through eIF2α-dependent and independent mechanisms by increasing ribosome density. Nucleic Acids Res. 2017;45:6023–6036. doi: 10.1093/nar/gkx135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Knudson C.J., Alves-Peixoto P., Muramatsu H., Stotesbury C., Tang L., Lin P.J.C., Tam Y.K., Weissman D., Pardi N., Sigal L.J. Lipid-nanoparticle-encapsulated mRNA vaccines induce protective memory CD8 T cells against a lethal viral infection. Mol. Ther. 2021;29:2769–2781. doi: 10.1016/j.ymthe.2021.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Anderson B.R., Muramatsu H., Jha B.K., Silverman R.H., Weissman D., Karikó K. Nucleoside modifications in RNA limit activation of 2’-5’-oligoadenylate synthetase and increase resistance to cleavage by RNase L. Nucleic Acids Res. 2011;39:9329–9338. doi: 10.1093/nar/gkr586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Pollard C., De Koker S., Saelens X., Vanham G., Grooten J. Challenges and advances towards the rational design of mRNA vaccines. Trends Mol. Med. 2013;19:705–713. doi: 10.1016/j.molmed.2013.09.002. [DOI] [PubMed] [Google Scholar]
- 77.Lokugamage M.P., Gan Z., Zurla C., Levin J., Islam F.Z., Kalathoor S., Sato M., Sago C.D., Santangelo P.J., Dahlman J.E. Mild innate immune activation overrides efficient nanoparticle-mediated RNA delivery. Adv Mater. 2020;32 doi: 10.1002/adma.201904905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Karikó K., Muramatsu H., Ludwig J., Weissman D. Generating the optimal mRNA for therapy: HPLC purification eliminates immune activation and improves translation of nucleoside-modified, protein-encoding mRNA. Nucleic Acids Res. 2011;39:e142. doi: 10.1093/nar/gkr695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Gebre M.S., Rauch S., Roth N., Yu J., Chandrashekar A., Mercado N.B., He X., Liu J., McMahan K., Martinot A., Martinez D.R., Giffin V., Hope D., Patel S., Sellers D., Sanborn O., Barrett J., Liu X., Cole A.C., Pessaint L., Valentin D., Flinchbaugh Z., Yalley-Ogunro J., Muench J., Brown R., Cook A., Teow E., Andersen H., Lewis M.G., Boon A.C.M., Baric R.S., Mueller S.O., Petsch B., Barouch D.H. Optimization of non-coding regions for a non-modified mRNA COVID-19 vaccine. Nature. 2022;601:410–414. doi: 10.1038/s41586-021-04231-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Research data are not shared.