Abstract
In a large-scale radiological catastrophe, it is expected that hundreds and thousands of people could be exposed to radiation. A rapid method is required for triage of casualties to determine proper medical treatment. In this article, mice were exposed to different radiation doses and sera of mice were investigated by surface-enhanced Raman spectroscopy (SERS) and orthogonal projections to latent structure discriminant analysis (OPLS-DA) after total body irradiation (TBI). The results of the present study indicated that differences have widened over time. The different radiation groups showed a slight overlap at 24 h and 72 h but were completely distinct at the 10th day after TBI. The SERS spectrum between the normal group and the irradiated group showed a significant difference at 24 hours. The same trend was depicted in scatting score plots. Significant differences in Raman peaks were found, such as 744 and 1495 cm−1 corresponding to riboflavin and 593 and 1204 cm−1 corresponding to l-tryptophan. The lack of riboflavin and l-tryptophan will influence metabolism levels. Above all, these results bear potential in the development of label-free and rapid tools for on-site detection and screening of irradiation injuries.
A rapid and on-site detection method would be needed to triage of casualties and proper medical treatment in a large scale radiological catastrophe. The detection method of SERS would be a effective approach.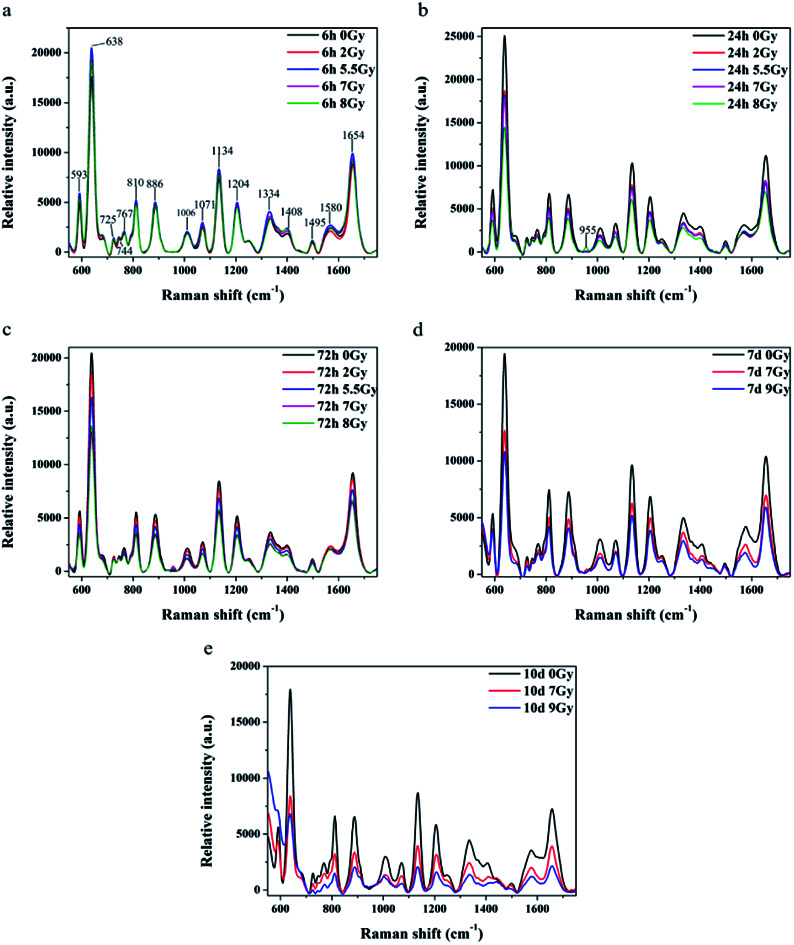
Introduction
During a large-scale radiological event, hundreds of thousands of individuals may be exposed to radiation and need emergency treatment, so a rapid biodosimetric method is required for triage of casualties to determine proper medical treatment. Classical biodosimetric methods, including chromosomal dicentrics (ring) or micronuclei, require at least 48 h for lymphocyte culture and experienced researchers, who must be trained for long periods for credible results.1,2 Electron paramagnetic resonance spectroscopy, which detects radiation-induced radicals in teeth and fingernails, requires large devices which are difficult to move.3 Thus, a more convenient and rapid method is necessary for triage of casualties in a large scale radiation event.
Surface-enhanced Raman spectra (SERS) was widely employed in biological medical research.4 Because of its “fingerprint” characteristic, Raman spectroscopy can provide information on structure and conformation of macromolecules, such as proteins, nucleic acids, and lipids. In recent years, label-free SERS detection was widely used in different serum studies.4–7 Patients were separated from normal individuals using statistical formulas. And variations of components posing significant effects on diseases8,9 were also detected by using this method.
Ag (Au) NPs has been reported as an optimum nanoparticles material in antibacterial.10 Ag NPs could interact with peptidoglycan and lipopolysaccharide (LPS) to damage bacterial cell wall and membrane in various bacterial cell models.11 Zheng K. et al. reported that chemically integrated daptomycin–AgNCs showed improved bacterial killing efficiency over the physically mixed daptomycin and Ag nanocluster (Ag NCs).12 Ultra small Au nanoclusters (Au NCs) could interact with bacteria and induce metabolic imbalance to kill Gram-positive and Gram-negative bacteria.13 There were also reports on Au NPs/NCs as promising radiosensitizer for cancer radiotherapy.14 Hainfeld J. F. et al. reported that mice bearing subcutaneous EMT-6 mammary carcinomas showed one-year survival of 86% versus 20% with X-rays alone and 0% with gold alone after receiving Au NPs injection and X-ray therapy.15 Ag (Au) in SERS has been used in gastric cancer detection based on blood plasma and circulation miRNA detection in human serum.9,16
Structural changes induced by radiation in cell membranes have been studied by Raman spectroscopy to illustrate lipid and protein changes on stretching regions.17–19 Brain and tissue of mice irradiated have been studied to identify the biochemical changes occurring as a result of radiation injury.20 Moreover, SERS detection method for DNA has been carried out to achieve various goals. DNA damage after X-ray have been analyzed using SERS technique to provide information for radiation-induced the DNA damage at the molecular level.21
In this paper, SERS was introduced to detect the sera of mice exposed to different dose γ-ray. The results revealed that the normal, sub lethal, and lethal groups could be separated much clearly with time. Significant difference in SERS spectrum between normal group and radiation groups emerged at 24 h. And same results were depicted in scattering score plots. Furthermore, significant differences in Raman peaks were found, such as 744 and 1495 cm−1 corresponding to riboflavin and 593 and 1204 cm−1 corresponding to l-tryptophan. The lack of them will influence level of metabolism. Results also indicated that SERS detection maybe a useful tool for triage casualties of radiation accidents.
Material and methods
Mice, total body irradiation (TBI) radiation, and serum collection
C57BL/6J male mice (6–8 weeks old) were obtained from and bred in the Academy of Military Medical Sciences (Beijing, China). Mice were irradiated in 0, 2, 5.5, 7, and 8 Gy (or 9 Gy) by using 60Co source with a dose rate of 101.90 or 91.11 cGy min−1. Eight mice were included in each dose group, with 352 animals in total. For blood counts, 20 μL blood was obtained from the tail vein at 6 h and at 1, 3, 7, 11, 15, 20, 25, and 30 days after radiation. NIHON KOHDEN MEK-722K peripheral blood-cell analyzer was used for analysis. Mice body weights were measured at 1, 3, 7, 11, 15, 20, 25, and 30 days. Mice survival was observed and recorded. Blood of mice were collected by removing the eye at different time after radiation and incubated at room temperature (RT) for 1 h to allow clotting. Blood samples were then centrifuged at 3000 rpm for 5 min at RT. Supernatant was collected and re-spun at the above conditions to remove any remaining cellular contaminants. The resulting supernatant (serum) was stored in aliquots at −80 °C. The animal care and handling were performed in accordance with ‘Guide for the Care and Use of Laboratory Animal of AMMS in China’ and all animal experiments were approved by Animal Care and Use Committee of Beijing Institute of Radiation Medicine (Beijing, China).
Preparation of silver colloids and SERS measurement
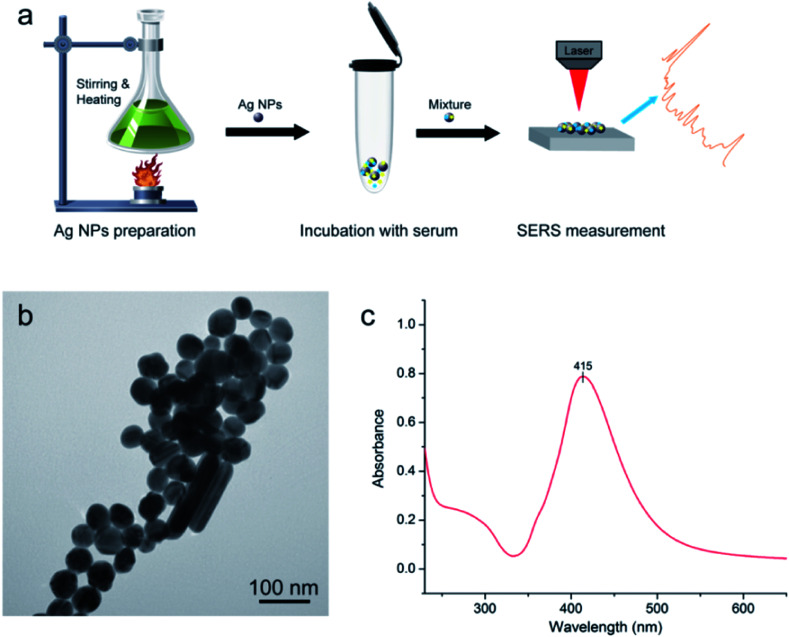
Silver (Ag) colloids were prepared using a standard citrate reduction procedure.22,23Fig. 1a shows the procedure for SERS-based diagnosis. The first step is preparation of Ag NPs. The second step involved mixture of serum and Ag colloid at 1 : 2 ratio. Then, 2.5 μL mixtures was dropped onto a silicon wafer and aired for further measurements.
Fig. 1. (a) Schematic illustration of the procedure for SERS-based diagnosis; (b) transmission electron microscopy (TEM) image of Ag nanoparticles (NPs) of silver colloid; (c) UV/visible absorption spectrum of Ag NPs.
SERS spectra were recorded using a portable Raman system B&W Teki-Raman Plus BWS465-785H spectrometer equipped with a back-illuminated CCD detector cooled at −2 °C. The spectrum was measured at the range of 550–1750 cm−1 under a 785 nm laser excitation with a power 25 mW. And SERS spectra were obtained through a microscope with a 20× objective with 10 s integration time. The spectrum of each sample was obtained from the average of three spectra measured at random spots.
Statistical analysis
Origin8.5 software was used to generate line charts of animal models and mean spectrum of each radiation group at 550–1750 cm−1 spectral range after background subtraction and spectral smoothing. SIMCA14.1 software was applied to create orthogonal projections to latent structure discriminant analysis (OPLS-DA) score plots using normalized data and goodness of fit (R2Y) and predictive ability (Q2)24,25 were obtained. Significant differences in SERS peaks were analyzed by SIMCA14.1 software for variable importance plot (VIP), and SPSS 16.0 was used for analysis of variance (ANOVA).
Results
Quality of silver (Ag) colloids
Fig. 1b displays results of Ag nanoparticles (NPs) and transmission electron microscopy (TEM) micrograph with 100 nm bar. The quality of these Ag NPs was shown through the UV absorption spectrum (Fig. 1c), with maximum absorption peak at 415 nm. The quality of Ag NPs in this measurement was high.10,26
Mice model of acute radiation injury
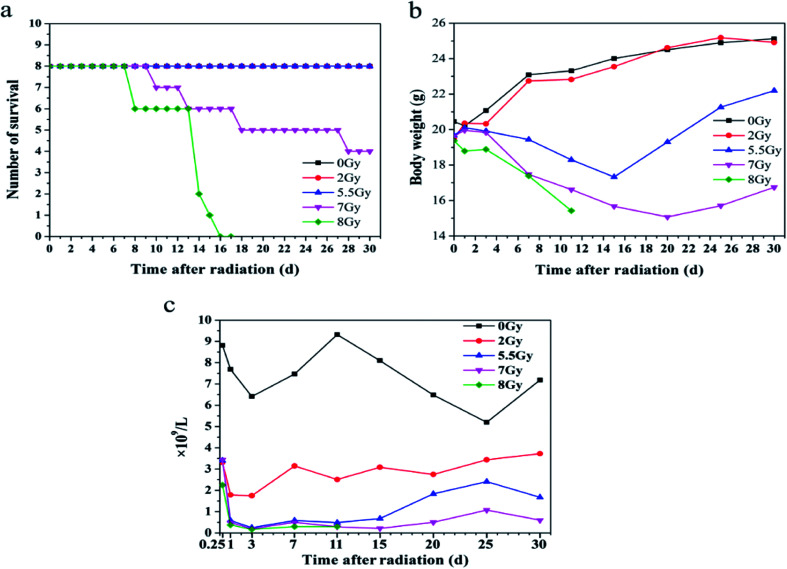
To establish mice models induced by radiation, mice were divided into five groups and exposed to 0, 2, 5.5, 7, or 8 Gy. Survival, body weight, and blood counts were obtained after radiation, as shown in Fig. 2. Lymphocytes decreased dramatically at 6 h in all mice exposed to different doses of radiation compared with the lymphocytes number of 8.8 × 109 L−1 of 0 Gy group mice. And 8 Gy group presented the lowest count at 2.25 × 109 L−1. On the first day after TBI, lymphocyte counts of 2, 5.5, 7, and 8 Gy groups decreased to 3.3, 0.6, 0.5, and 0.4 × 109 L−1, respectively. Thirty days after TBI, lymphocyte counts of 2, 5.5, and 7 Gy groups recovered and reached 3.7, 1.7, and 0.6 × 109 L−1, respectively. Lymphocyte count of 8 Gy group decreased to 0.3 × 109 L−1 in two days after TBI until all mice died. The data indicate that sera from mice exposed to 2, 5.5, 7, and 8 Gy can be used in the following experiments.
Fig. 2. Model of laboratory animal, C57BL/6J, included normal control (n = 8) and total body irradiation (TBI) with four different doses (each group n = 8). (a) Comparison of survival numbers from 0 day to 30 days; (b) comparison of body weights from 0 day to 30 days; (c) comparison of lymphocyte counts from 0 day to 30 days.
SERS detection spectrum for different mice sera with different irradiation doses
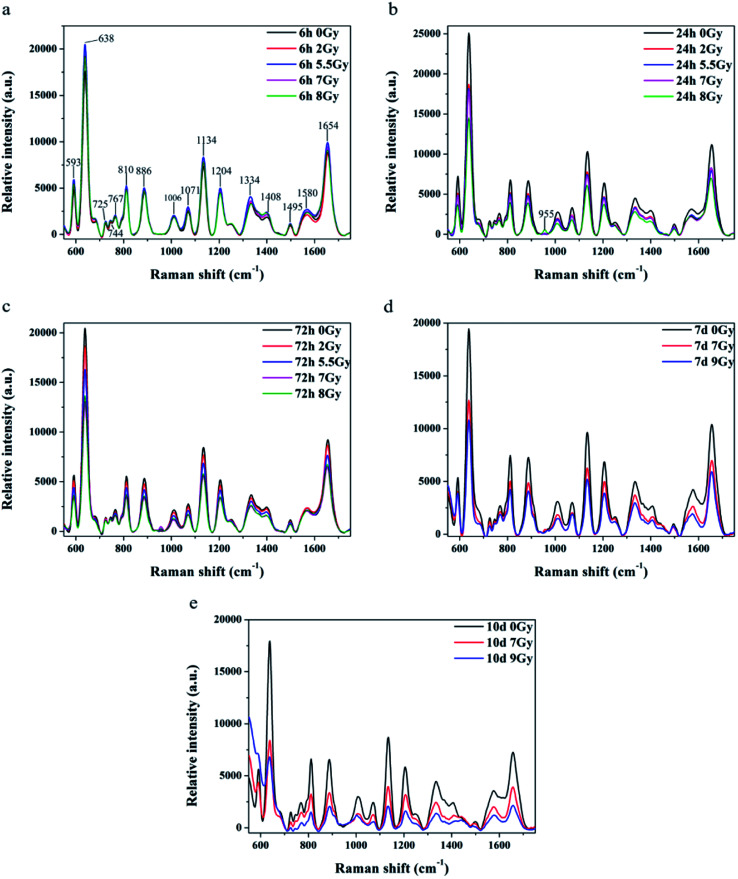
All spectra shown in Fig. 3a–c were measured under the same instrumentation set-up. Fluorescence background of original SERS data was deducted prior to page creation. Fig. 3a–c display SERS spectra of normal control (0 Gy) versus other dose groups (2, 5.5, 7 and 8 Gy) at 6, 24, and 72 h, respectively, after TBI. All of Raman spectra featured similar spectral shapes, whereas peaks were observed at 593, 638, 725, 744, 767, 810, 886, 1006, 1071, 1134, 1204, 1334, 1408, 1495, 1580, and 1654 cm−1. Different doses groups cannot be distinguished at 6 h after TBI in Fig. 3a. Normal group and 8 Gy group were distinctly separated to other doses (2, 5.5 and 7 Gy) groups, respectively, at 24 h in Fig. 3b. And the doses groups of 2, 5.5 and 7 Gy cannot be distinguished to each other at 24 h. Normal group, 2 Gy and 5.5 Gy dose group was distinguished to half lethal (7 Gy) and lethal groups (8 Gy), respectively, at 72 h in Fig. 3c. But the half lethal (7 Gy) and lethal groups (8 Gy) cannot be separated clearly in SERS spectrum at 72 h. Thus, other experiments were necessary to determine time points for recognizing half lethal and lethal doses. Another model was divided into the normal control, half lethal and lethal group and exposed to 0, 7, and 9 Gy, respectively. As shown in ESI Fig. S1,† results for body weight and survival were similar to previous findings. Fig. 3d and e are SERS spectra of mice sera collected at days 7 and 10 after TBI. Fig. 3d revealed that higher dose presented lower intensity, whereas Fig. 3e showed more distinction on differences. And the results of SERS spectrums indicated that different doses of radiation were distinguished more clearly by the Raman spectrum with increasing time.
Fig. 3. Comparison of mean SERS spectra for relative intensities of normal serum (black line, n = 8) versus those under different irradiation doses and time points. Bars represent color of each curve. Comparison of relative intensities under five irradiation doses at (a) 6 h, (b) 24 h, and (c) 72 h after TBI. Comparison of relative intensities under three irradiation doses at (d) 7 days and (e) 10 days after TBI.
Peaks were labeled in Fig. 3a and b. Table 1 shows SERS peak positions and vibrational mode assignments. Analysis of VIP and SPSS ANOVA were used to determine significant difference in SERS peaks in different exposure dose groups. In metabonomics, VIPs > 1 possibly represent between-group variance of metabolin. Significant differences in peaks were defined by SPSS ANOVA. Given these results, peaks at 593, 744, 767, 810, 886, 955, 1006, 1071, 1134, 1204, 1334, 1408, and 1495 cm−1 showed significant differences (P < 0.05).
SERS peak positions and vibrational mode assignments.9,27–30.
| Peak position (cm−1) | Vibrational mode | Major assignments |
|---|---|---|
| 593 | l-Tryptophan | |
| 638 | ν(C–S) | l-Tyrosine, lactose, acetoacetate |
| 725 | C–H | Adenine, acetyl coenzyme A |
| 744 | Riboflavin | |
| 767 | Thymine | |
| 810 | l-Serine | |
| 886 | δ(C–O–H) | d-(C)-galactosamine, glutathione |
| 955 | ν(C–C) | α-Helix, proline |
| 1006 | ν s(C–C) | l-Phenylalanine, l-tryptophan, acetoacetate |
| 1071 | β-d-glucose, lactose | |
| 1134 | ν(C–N) | l-Arginine, d-mannose |
| 1204 | l-Tryptophan | |
| 1334 | ν(C–H) | Nucleic acid bases |
| 1408 | l-Histidine, l-alanine, coenzyme A | |
| 1495–1500 | Glycine, riboflavin | |
| 1580 | δ(C C) | Acetoacetate |
| 1654 | ν(C O) | α-Helix, collagen, triolein |
OPLS-DA score plot for different irradiation doses in mice
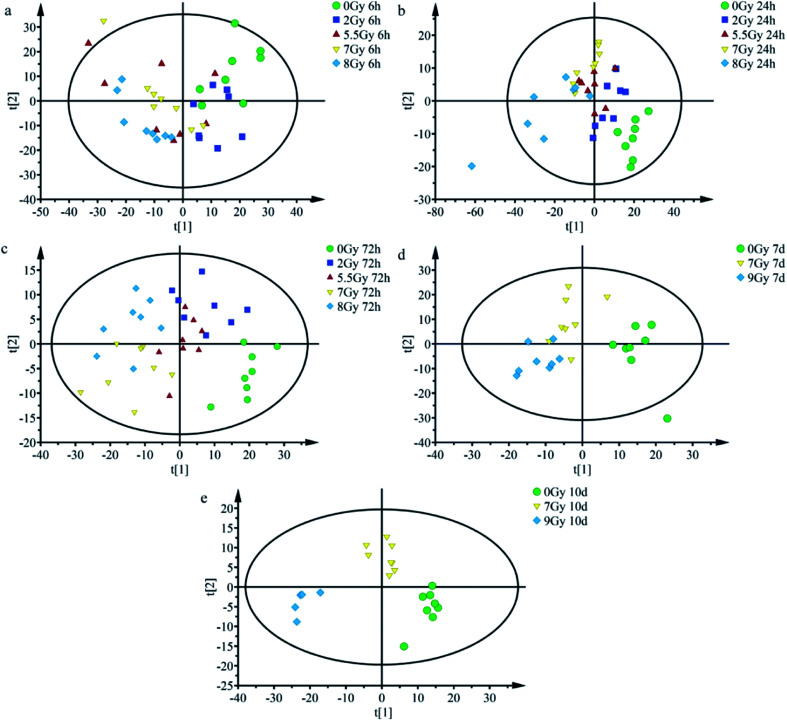
OPLS-DA serves as good strategy for distinguishing and discerning individual differences, whereas score plots can intuitively reflect effects of classification. OPLS-DA could be applied to obtain better discrimination among normal control group (0 Gy) and other radiation dose groups (2, 5.5, 7, 8 or 9 Gy) with increasing time (6, 24, and 72 h and 7 and 10 days), as indicated in Fig. 4. As shown in Fig. 4a, at 6 h, all groups overlapped with one another and yielded R2Y = 0.225; Q2 = 0.111. Fig. 4b (24 h) and 4c (72 h) showed that the normal control group (0 Gy) was completely divided from radiation groups (2, 5.5, 7, and 8 Gy). Fig. 4b and c presented R2Y = 0.473 and Q2 = 0.302; R2Y = 0.602 and Q2 = 0.258, respectively, at indicated times. As shown in Fig. 4b, at 24 h, each group was preliminarily concentrated and different radiation dose groups started separating.
Fig. 4. Scatter plots for OPLS-DA scores of mice irradiated under different doses after normalizing at different time points. Scatter plots for OPLS-DA scores of different radiation doses at (a) 6 h (R2Y = 0.225; Q2 = 0.111), (b) 24 h (R2Y = 0.473, Q2 = 0.302), and (c) 72 h (R2Y = 0.602; Q2 = 0.258); scatter plots for OPLS-DA scores for different radiation doses at (d) 7th days (R2Y = 0.624, Q2 = 0.413) and (e) 10th days (R2Y = 0.844, Q2 = 0.644).
Different irradiation dose groups showed better central tendency, as shown in Fig. 4c (72 h). As depicted in Fig. 4d, 7th day, 7 and 9 Gy groups still presented a slight overlap but separated more, yielding values of R2Y = 0.624 and Q2 = 0.413. Separation was more remarkable at the 10th day. Fig. 4e (10th days) showed complete difference between half lethal (7 Gy) and lethal (9 Gy) groups, whereas it yielded R2Y = 0.844 and Q2 = 0.644. As presented in Fig. 4a–e, after TBI, half lethal and lethal groups separated more clearly as time progressed. Dispersion degrees of each spot accord with results in Fig. 3. These findings show much clear distinction among different irradiated groups with increasing time and radiation dose.
Discussion
Radiation/nuclear accidents, such as nuclear leakage events in Chernobyl and Fukushima threatened hundreds of thousands of people. Existing radiation biodosimeter has disadvantages, such as time consumption and professional limitations, and cannot rapidly detect huge numbers of exposed population. It is necessary to develop rapid and effective biodosimetric methods for estimating unexpected and accidental exposure to radiation.31
Radiation causes serious and potentially lethal lesions of nuclear DNA,32 potential anomalies in mitochondrial membrane, DNA damage in mitochondria,33 and changes in enzyme activity. Damage in mitochondria particularly leads to oxidation and reduction imbalance.33 Radiation also generates multiple variations, such as reactive oxygen, cytokines, and intercellular communication.34 Finally, all submicroscopic changes lead to metabolic disorders and nutrient scarcity in organisms.35 In this experiment, all mice featured bone marrow type of ARS.
Fig. 2a–c and S1a and b† showed all mice models after TBI. The models had differences at lethal dose level. Fig. S1a and b,† the later model, featured high dose of 9 Gy, whereas lethal dose for the previous one measured 8 Gy because of declining rate of 60Co γ source. Fig. 2c revealed injury in mice hematopoietic system based on lymphocyte counts. Lymphocyte is one of the most radiation-sensitive cells. Sensitive cells include hematopoietic, germ (especially spermatogenic and follicular cells), intestinal nest cells (especially those of small intestine), and lymphocytes.36 Compared with other radiation-sensitive cells, lymphocytes in peripheral blood are collected and detected much easily than the others. Thus, lymphocyte counts were used to estimate injury after radiation exposure.
Raman shift of SERS peaks of serum reflected vibration of various biomolecules, such as amino acid, lipids, and nucleic acids.29 Variations of the molecular in serum reveal that metabolism of immune, biochemistry, signal communication and nutriment have been changed.37Table 1 lists tentative assignments for the observed SERS bands according to literature.9,27–30 Spectra indicated that SERS intensity decreased with increasing time and stimulated radiation dose. According to the Raman peaks shown in Table 1, riboflavin, carbohydrates, amino acids, and nucleic acids in sera declined after TBI. Radiation will cause redox and energy metabolism disorder in cell to impact the organism.35 SERS peaks at 744 and 1495 cm−1 correspond to riboflavin, which participates in oxidation and energy metabolism in organisms. This observation is related to metabolism of carbohydrates, proteins, nucleic acids, and lipids. Lack of riboflavin may cause metabolic disorders.38,39 SERS peak at 1071 cm−1 corresponds to glucose, lactose, and fructose.30 These sugars are closely related to energy metabolism, and their reduction indicated lack of nutrition in body of mice. SERS peaks at 593 and 1204 cm−1 correspond to l-tryptophan, whereas that at 1495 cm−1 corresponds to glycine. Lack of tryptophan leads to weight loss, growth retardation, and reduction of fat accumulation.40 Both glycine and tryptophan are related to the immune system. Deficiencies in these amino acids result in decline in humoral immunity. Compared with mice models in Fig. 2, results of SERS detection agree with weight loss, sharp decline in lymphocytes, and reduced immune ability after TBI.
Multivariate statistical analysis method which combined with SERS spectral data analysis is used much frequently in detection of cancer serum, environmental microorganisms, and viruses.41–43 OPLS-DA obtains principal components by transformation of independent and dependent variables to generate score plots.24,44Fig. 4 shows goodness of fit (R2Y) and predictive ability (Q2) at different time points. R2Y and Q2 all increased at considered time ranges.44 Results indicated enhanced predictive ability and recognition effect of OPLS-DA classification after TBI.
Total body irradiation impacts the whole body organ and system, and damages serum nucleic acids and proteins. Score plots in Fig. 4 display better separation between normal control and radiation groups; especially under severe radiation at 24 h and 72 h. And the results indicated that exposed mice can be easily distinguished from the normal population at 24 h, and different irradiated groups were preliminarily recognized before 72 h. When normal control was considered in classification, complete separation of half lethal and lethal groups was recognized 10 days after TBI. Thus, SERS detection technology is an appropriate rapid method for screening exposure at early time periods. OPLS-DA results revealed potential of label-free SERS detection technology for rapid diagnosis of radiation injury. With increasing time and dosage, detection and diagnostic abilities were enhanced.
Our study suggested that normal group was separated from radiation groups at 24 h. Furthermore, we applied label-free SERS detection technology combined with OPLS-DA multivariate statistical analysis method to screening for different exposure doses of mice sera. Our findings also demonstrate that SERS bears potential as a technique for label-free, non-invasive detection, and diagnosis of radiation. All results provide possibility for development of a rapid and on-site detection technology.
Conclusion
In recent years, many studies focused on rapid diagnosis of diseases, such as cancer, for early detection and treatment. ARS is an event requiring rapid diagnosis and treatment. A rapid on-site detection technology for ARS will distinguish high- or non-exposed individuals and subject them to appropriate treatments, thus saving people's lives.
This paper applied label-free SERS detection technology combined with OPLS-DA multivariate statistical analysis method to realize rapid on-site detection and screening for different exposure doses of mice sera. At 24 h after TBI, concentrated tendency in each dose group and separated between groups started to emerge. Separation effect became more prominent and improved with increasing time between different irradiated groups.
This detection method features fast speed, obtaining results 30 min after sample preparation. Thus, this method may be effective for on-site screening in large-scale radiological events. In conclusion, for the first time, this study reveals that SERS bears potential as a label-free, non-invasive detection and diagnosis technique for radiation biodosimeter.
Author contributions
P. K. Z., S. Q. W. and Z. D. W. designed the study. H. J. F. and X. Q. Z. collected peripheral blood of mice. Q. W. and Z. R. analyzed significant of peaks. L. S. L., Z. D. W. and R. X. experimental, collection, analysis of data, creation images and manuscript writing. All authors reviewed the manuscript.
Conflicts of interest
The authors declare no competing financial interests.
Supplementary Material
Acknowledgments
This work was supported by Grants from Applied Basic Research Project (No. 15QNP089 and No. 16QNP118), the Major Projects (No. AWS14C014 and No. AWS13C007), and Beijing Municipal Science & Technology Commission (No. Z161100000116040).
Electronic supplementary information (ESI) available. See DOI: 10.1039/c7ra12238a
References
- Ainsbury E. A. Vinnikov V. A. Maznyk N. A. Lloyd D. C. Rothkamm K. Radiat. Prot. Dosim. 2013;155:253–267. doi: 10.1093/rpd/ncs335. [DOI] [PubMed] [Google Scholar]
- Tucker J. D. Ramsey M. J. Lee D. A. Minkler J. L. Int. J. Radiat. Biol. 1993;64(1):27–37. doi: 10.1080/09553009314551081. [DOI] [PubMed] [Google Scholar]
- Williams B. B. Dong R. Flood A. B. Grinberg O. Kmiec M. Lesniewski P. N. Matthews T. P. Nicolalde R. J. Raynolds T. Salikhov I. K. Swartz H. M. Radiat. Meas. 2011;46:772–777. doi: 10.1016/j.radmeas.2011.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vo-Dinh T. Yan F. Wabuyele M. B. J. Raman Spectrosc. 2005;36:640–647. doi: 10.1002/jrs.1348. [DOI] [Google Scholar]
- Wang H. N. Vo-Dinh T. Nanotechnology. 2009;20:065101. doi: 10.1088/0957-4484/20/6/065101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G. Lipert R. J. Jain M. Kaur S. Chakraboty S. Torres M. P. Batra S. K. Brand R. E. Porter M. D. Anal. Chem. 2011;83:2554–2561. doi: 10.1021/ac102829b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J. Chen R. Feng S. Pan J. Li Y. Chen G. Cheng M. Huang Z. Yu Y. Zeng H. Nanomedicine. 2011;7:655–663. doi: 10.1016/j.nano.2011.01.012. [DOI] [PubMed] [Google Scholar]
- Lin D. Pan J. Huang H. Chen G. Qiu S. Shi H. Chen W. Yu Y. Feng S. Chen R. Sci. Rep. 2014;4:4751. doi: 10.1038/srep04751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng S. Chen R. Lin J. Pan J. Wu Y. Li Y. Chen J. Zeng H. Biosens. Bioelectron. 2011;26:3167–3174. doi: 10.1016/j.bios.2010.12.020. [DOI] [PubMed] [Google Scholar]
- Fayaz A. M. Balaji K. Girilal M. Yadav R. Kalaichelvan P. T. Venketesan R. Nanomedicine. 2010;6:103–109. doi: 10.1016/j.nano.2009.04.006. [DOI] [PubMed] [Google Scholar]
- Zheng K. Setyawati M. I. Leong D. T. Xie J. Coord. Chem. Rev. 2018;357:1–17. doi: 10.1016/j.ccr.2017.11.019. [DOI] [Google Scholar]
- Zheng K. Setyawati M. I. Lim T.-P. Leong D. T. Xie J. ACS Nano. 2016;10:7934–7942. doi: 10.1021/acsnano.6b03862. [DOI] [PubMed] [Google Scholar]
- Kaiyuan Zheng M. I. S. Tai Leong D. Xie J. ACS Nano. 2017;11:6904–6910. doi: 10.1021/acsnano.7b02035. [DOI] [PubMed] [Google Scholar]
- Goswami N. Luo Z. Yuan X. Leong D. T. Xie J. Mater. Horiz. 2017;4:817–831. doi: 10.1039/C7MH00451F. [DOI] [Google Scholar]
- Hainfeld J. F. Slatkin D. N. Smilowitz H. M. Phys. Med. Biol. 2004;49:N309–N315. doi: 10.1088/0031-9155/49/18/N03. [DOI] [PubMed] [Google Scholar]
- Zheng J. Ma D. Shi M. Bai J. Li Y. Yang J. Yang R. Chem. Commun. 2015;51:16271–16274. doi: 10.1039/C5CC06549F. [DOI] [PubMed] [Google Scholar]
- Verma S. P. Singhal A. Sonwalkar N. Int. J. Radiat. Biol. 1993;63:297–288. doi: 10.1080/09553009314550381. [DOI] [PubMed] [Google Scholar]
- Verma S. Radiat. Res. 1986;107:183–193. doi: 10.2307/3576806. [DOI] [PubMed] [Google Scholar]
- Verma S. P. Sonwalkar N. Radiat. Res. 1991;126:27–35. doi: 10.2307/3578167. [DOI] [PubMed] [Google Scholar]
- Lakshmi R. J. Kartha V. B. Murali Krishna C. Solomon J. G. R. Ullas G. Uma Devi P. Radiat. Res. 2002;157:175–182. doi: 10.1667/0033-7587(2002)157[0175:TRSFTS]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- Ou L. Chen Y. Su Y. Zou C. Chen Z. Appl. Spectrosc. 2016;70:1821–1830. doi: 10.1177/0003702816671073. [DOI] [PubMed] [Google Scholar]
- Lee P. C. Meisel D. J. Phys. Chem. 1982;86:3391–3395. doi: 10.1021/j100214a025. [DOI] [Google Scholar]
- Wang C. Xiao R. Wu X. Dong P. Rong Z. Chen J. Wang S. Laser Phys. 2014;24:045807. doi: 10.1088/1054-660X/24/4/045807. [DOI] [Google Scholar]
- Boccard J. Rutledge D. N. Anal. Chim. Acta. 2013;769:30–39. doi: 10.1016/j.aca.2013.01.022. [DOI] [PubMed] [Google Scholar]
- Chen X. Huang Z. Feng S. Chen J. Wang L. Lu P. Zeng H. Chen R. Int. J. Nanomed. 2012;7:6115–6121. doi: 10.2147/IJN.S37782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nanda A. Saravanan M. Nanomedicine. 2009;5:452–456. doi: 10.1016/j.nano.2009.01.012. [DOI] [PubMed] [Google Scholar]
- Feng S. Chen R. Lin J. Pan J. Chen G. Li Y. Cheng M. Huang Z. Chen J. Zeng H. Biosens. Bioelectron. 2010;25:2414–2419. doi: 10.1016/j.bios.2010.03.033. [DOI] [PubMed] [Google Scholar]
- Huang Z. Chen X. Chen Y. Chen J. Dou M. Feng S. Zeng H. Chen R. J. Biomed. Opt. 2011;16:110501. doi: 10.1117/1.3650310. [DOI] [PubMed] [Google Scholar]
- De Gelder J. De Gussem K. Vandenabeele P. Moens L. J. Raman Spectrosc. 2007;38:1133–1147. doi: 10.1002/jrs.1734. [DOI] [Google Scholar]
- Pichardo-Molina J. L. Frausto-Reyes C. Barbosa-Garcia O. Huerta-Franco R. Gonzalez-Trujillo J. L. Ramirez-Alvarado C. A. Gutierrez-Juarez G. Medina-Gutierrez C. Lasers Med. Sci. 2007;22:229–236. doi: 10.1007/s10103-006-0432-8. [DOI] [PubMed] [Google Scholar]
- Flood A. B. Nicolalde R. J. Demidenko E. Williams B. B. Shapiro A. Wiley Jr A. L. Swartz H. M. Radiat. Meas. 2011;46:916–922. doi: 10.1016/j.radmeas.2011.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teoule R. Int. J. Radiat. Biol. Relat. Stud. Phys., Chem. Med. 1987;51(4):573–589. doi: 10.1080/09553008414552111. [DOI] [PubMed] [Google Scholar]
- Kim G. J. Chandrasekaran K. Morgan W. F. Mutagenesis. 2006;21:361–367. doi: 10.1093/mutage/gel048. [DOI] [PubMed] [Google Scholar]
- Klammer H. Mladenov E. Li F. Iliakis G. Cancer Lett. 2015;356:58–71. doi: 10.1016/j.canlet.2013.12.017. [DOI] [PubMed] [Google Scholar]
- Azzam E. I. Jay-Gerin J. P. Pain D. Cancer Lett. 2012;327:48–60. doi: 10.1016/j.canlet.2011.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heylmann D. Rodel F. Kindler T. Kaina B. Biochim. Biophys. Acta. 2014;1846:121–129. doi: 10.1016/j.bbcan.2014.04.009. [DOI] [PubMed] [Google Scholar]
- Zeng J. Yin P. Tan Y. Dong L. Hu C. Huang Q. Lu X. Wang H. Xu G. J. Proteome Res. 2014;13:3420–3431. doi: 10.1021/pr500390y. [DOI] [PubMed] [Google Scholar]
- Hoppel C. DiMarco J. P. Tandler B. J. Biol. Chem. 1979;254:4164–4170. [PubMed] [Google Scholar]
- van der Beek E. J. van Dokkum W. Wedel M. Schrijver J. van den Berg H. J. Am. Coll. Nutr. 1994;13:629–640. doi: 10.1080/07315724.1994.10718459. [DOI] [PubMed] [Google Scholar]
- Ray P. D. Foster D. O. Lardy H. A. J. Biol. Chem. 1966;241:3904–3908. [PubMed] [Google Scholar]
- Wang M. Cao X. Lu W. Tao L. Zhao H. Wang Y. Guo M. Dong J. Qian W. RSC Adv. 2014;4:64225–64234. doi: 10.1039/C4RA07603F. [DOI] [Google Scholar]
- Driskell J. D. Zhu Y. Kirkwood C. D. Zhao Y. Dluhy R. A. Tripp R. A. PLoS One. 2010;5:e10222. doi: 10.1371/journal.pone.0010222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li-Chan E. C. Y. Trends Food Sci. Technol. 1996;7(11):361–370. doi: 10.1016/S0924-2244(96)10037-6. [DOI] [Google Scholar]
- Bylesjö M. Rantalainen M. Cloarec O. Nicholson J. K. Holmes E. Trygg J. J. Chemom. 2006;20:341–351. doi: 10.1002/cem.1006. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.