Abstract
Cinnamaldehyde (Cin) and citral (Cit) have been studied as antimicrobial agents and natural preservatives, but their action modes are controversial, and the knowledge of their antifungal mechanism against P. expansum is still incomplete. The present study was conducted to evaluate the antifungal mechanism of the combination of Cin and Cit (Cin/Cit) against P. expansum by observing the cellular ultrastructure, fourier transform infrared spectroscopy (FT-IR) fingerprints, plasma membrane, oxidative stress and volatile profile. Cin/Cit caused membrane invaginations, organelles and cytoplasm destruction, as shown by transmission electron microscopy (TEM) observations. The FT-IR spectra and followed principle component analysis (PCA) presented the significant differences in chemical compounds, particularly phospholipid, protein and fatty acids of cells exposed to Cin/Cit. Compared to controls, Cin/Cit induced a decrease of ergosterol by 39.40%, an increase of unsaturated fatty acid, and protein release level (3.5 times). Besides, membrane damage was further verified through the reduction of the membrane integrity by using a flow cytometer. Moreover, the increase of malondialdehyde (MDA) (40.09%) and reactive oxygen species (ROS) accumulation indicated an induction of oxidative stress in cells by Cin/Cit. To resist the unfavorable stress caused by Cin/Cit, P. expansum metabolized Cin or Cit to the predominant detoxification compounds, cinnamic alcohol, nerol, and geraniol. The alterations in volatile profile demonstrated the influences on specific metabolisms in P. expansum caused by Cin/Cit.
Cinnamaldehyde (Cin) and citral (Cit) have been studied as antimicrobial agents and natural preservatives, but their action modes are controversial, and the knowledge of their antifungal mechanism against P. expansum is still incomplete.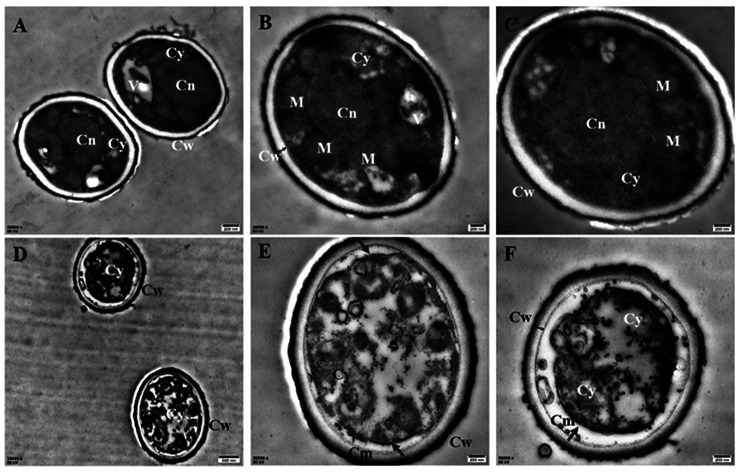
1. Introduction
P. expansum, a widespread fungal pathogen, causes blue mold decay on many types of fruits, resulting in significant economic losses to the fruit industry. P. expansum can spread rapidly under suitable condition and secrete patulin, a secondary metabolite with potential mutagenic, carcinogenic, teratogenic and embryotoxic effects on humans.1 Currently, besides certain prophylactic measures used to prevent the development of fungal pathogen in storage environments, the control of postharvest diseases depends mainly on synthetic fungicides.2 However, loss in the efficiency of fungicides due to the development of pathogen resistance, and concerns regarding chemical residues in food and environment have promoted investigations of alternative strategies to combat fungal decay while increasing food safety and public health.3
Essential oils (EOs) are aromatic and volatile liquids extracted from plant materials and have been proved to possess antimicrobial activities.4–6 Cinnamaldehyde and citral are major components of EOs extracted from Cinnamomum spp. and Cymbopogon citratus, respectively, and have been reported to inhibit microbial growth as a botanical fungicide in fruits. Compared to some compounds used against P. expansum such as potassium phosphite, hydrogen peroxide,2,7 cinnamaldehyde and citral have been legally registered by the Food and Drug Administration (FDA) as flavorings as well as foodstuffs and are generally accepted as safe (GRAS) for human health and environment.8 Moreover, cinnamaldehyde and citral both exhibit direct inhibition of pathogens, as well as indirectly control pathogen growth by inducing the host's defence system.9,10
Researchers have studied the complex action modes of EOs from various perspectives, but some mechanisms are still controversial. Sun et al.11 indicated that cinnamaldehyde inhibited fungal growth and aflatoxin B1 biosynthesis by modulating the oxidative stress response of Aspergillus flavus. Cinnamaldehyde was previously reported to be incapable of disorganizing the outer cell membrane or depleting the intracellular pool of Gram-negative bacteria such as Escherichia coli O157:H7, Salmonella typhimurium, and its biological activity was mainly due to the carbonyl group, which binded to proteins at the bacterial membrane.12,13 However, cinnamon EO, containing 92.4% cinnamaldehyde, effectively inhibited E. coli by causing cell membrane damage.4 Citral exhibited its antifungal activity against Penicillium digitatum by down-regulating ergosterol biosynthesis.14 Shi et al.,5 reported that citral inhibited Cronobacter sakazakii by acting on the cell membrane and intracellular energy production. Besides the direct cell damage caused by EOs, pathogens also display various responses to unfavorable conditions by regulating cell metabolism, such as cell detoxification. Previous studies demonstrated that cinnamaldehyde could be converted to cinnamic alcohol, which showed weaker antifungal activity by Aspergillus ochraceus or E. coli O157:H7.15,16P. expansum produces various volatile organic compounds (VOCs) under different conditions, and some VOCs are considered as biomarkers for P. expansum infection.17 The changes in the volatile profile of P. expansum can reveal the influence of EOs on specific metabolisms pathways and are conducive to analyzing the functional group of the EO component.
The antimicrobial efficacy of EOs combinations has been evaluated.18 However, few studies focused on the combined use of cinnamaldehyde and citral to act on fungal pathogens, particularly P. expansum. Researchers have suggested that the combined effects of some particular EOs would decrease the fungistatic activity, making it difficult for the pathogens to acquire resistance to multiple components of two or more EOs.19 The synergistic antibacterial activity of cinnamaldehyde and nisin has been reported to reduce the problem of Staphylococcus aureus resistance.20 Although cinnamaldehyde and citral have been used against P. expansum, most previous works about their direct antifungal activities focused on the control or application efficacy in food packaging.6,21–24 The knowledge about their action modes on P. expansum is still incomplete. Additionally, the antimicrobial mechanisms of cinnamaldehyde or citral against other bacteria or fungi cannot completely reflect their action modes on P. expansum.
Therefore, the present study aimed to explore the antifungal mechanisms of Cin/Cit against P. expansum through the following aspects: (1) TEM observation of cellular ultrastructure; (2) FT-IR analysis of cellular compounds; (3) plasma membrane (ergosterol, fatty acids, protein release, membrane integrity); (4) oxidative stress (MDA, ROS); (5) volatile metabolites profile, to provide basis for further research against P. expansum.
2. Materials and methods
2.1. Chemicals, strain and growth conditions
Analytical-grade cinnamaldehyde (95%) and citral (96%) were purchased from Jiangxi Xue Song Natural Medicinal Oil Co., Ltd, China. P. expansum F-WY-12-02 was isolated from kiwifruits in our laboratory25 and was maintained in potato dextrose agar (PDA) at 4 °C. The minimum inhibitory concentration (MIC) was determined using a dilution method in PDB (potato dextrose broth), as in a previous work with minor modifications.4 The stock of EOs was dissolved in Tween 80 and added to 10 ml sterile PDB medium. Then, serial EOs dilutions were added to the tubes containing 5 ml of PDB within 107 spores per ml to reach final concentrations ranging from 20 mg l−1 to 200 mg l−1. The same tubes without EOs were set as controls. Samples were incubated for 3 days at 25 °C under continuous shaking and MIC was defined as the lowest concentration of EO with no visible fungal growth. MFC was measured by a subculture of 100 μl from each tube with no visible fungal growth on a PDA plate followed by incubation at 25 °C for 3–5 days. MFC was defined as the lowest concentration of EO with the initial inoculum fungi killed. Each test was performed in triplicate. The Fractional Inhibitory Concentration Index (FICI) of EOs was determined according to a previous study26 to explore the interaction of two EOs. The result showed that the FICI of cinnamaldehyde and citral was 1, indicating their additive effect. Therefore, in the present study, a combined application of Cin/Cit was chosen against P. expansum F-WY-12-02. The FICI, MIC, and MFC of Cin/Cit are shown in Table S1.†
2.2. TEM and FT-IR analysis
Conidia from P. expansum on PDA at 25 °C for 7 days were harvested and suspended in PDB with the concentration of 107 spores per ml. Then 100 μl of different contents of Cin/Cit were added in tubes containing 4.9 ml of PDB within spores reaching the final concentration of 0 and 2 × MIC (90 mg l−1 Cin, 140 mg l−1 Cit). After incubation at 25 °C, 120 rpm for 12 h, the spores were harvested by centrifuging for 5 min at 10 000 × g and washed twice with phosphate buffer solution (PBS). Then the spores were subjected to a series of pretreatments based on Sun et al.11 with slight modifications as follows. The selected spores were added to 1% agarose to avoid losses during the elution process. After solidification of agarose, the agar block was cut into 1 mm3 pieces and subsequently fixed with 2.5% (v/v) glutaraldehyde in 0.1 mol l−1 PBS at 4 °C for 6 h. After that, each of the fixed samples was washed with PBS three times for 10 min and then postfixed with 1% (w/v) osmium tetroxide in PBS at 4 °C for 2 h. Afterwards, the samples were washed with PBS as described above and dehydrated by gradient ethanol solutions (25, 50, 75, 85, 95, and 100%) at 4 °C. The samples were then embedded in LR White Resin and dried for 48 h. At last, the micromorphology was observed using a transmission electron microscope (JEM-1230, JEOL Ltd. Japan).
For the FT-IR analysis, mycelia from 72 h incubation of P. expansum in PDB were collected and washed with PBS (pH 7.4). After that, 0.5 g of wet mycelia were added in tubes containing 8 ml PDB and Cin/Cit at 0, MIC (45 mg l−1 Cin, 70 mg l−1 Cit), 2 × MIC, 6 × MIC. After incubation at 25 °C, 120 rpm for 24 h, the mycelia were harvested by centrifugation for 5 min at 10 000 × g and washed twice with PBS. Subsequently, the mycelia were flash-frozen with liquid nitrogen, ground to fine powder, and pre-freezed at −40 °C for 24 h before vacuum freeze-drying for 24–48 h. After that, the freeze-dried mycelia were ground (100 mesh) to obtain a more homogeneous cell freeze-dried powder. Prior to analysis, samples were prepared by the potassium bromide-disk technique. Each treatment included three replicates.
All spectra were recorded in the reflectance mode using a Bruker Vetex 70 FT-IR spectrometer (Bruker Optics, Germany), and Bruker OPUS 7.0 software was used for data acquisition.27 In addition, FT-IR spectra were recorded between 4000 and 400 cm−1 with a resolution of 4 cm−1 with each spectrum composed of an average of 32 separate scans. Spectral preprocessing and data analysis were performed using the Unscrambler X 10.4 software.
2.3. Plasma membrane analysis
2.3.1. Ergosterol
For ergosterol determination, 500 μl spore suspension (107 spores per ml) was inoculated to each flask containing 50 ml of sterile PDB. After that, Cin/Cit were aseptically added to PDB to obtain the concentrations: 0, 1/4 × MIC-2 × MIC. All flasks were incubated at 25 °C for 5 days under 120 rpm. After incubation, PDB containing P. expansum mycelia was filtered and washed with PBS. Mycelia were dried, weighted and used for ergosterol extraction and determination. Each test was performed in triplicate. Ergosterol was extracted based on a previous study.15 Mycelia were transferred to a clean tube containing 20 ml methanol, 5 ml absolute ethanol, and 2 g potassium hydroxide. The mixed system was shaken for 5 min and placed in a water bath at 70 °C for 40 min. After that, 5 ml of distilled water was added and the solution was centrifuged at 1735 × g for 15 min to remove the mycelia. Then, n-hexane was added in an equal volume to the supernatant and the organic phase was collected and evaporated under a nitrogen flow at 60 °C. The residue was re-suspended in 1 ml of absolute methanol for investigation. High performance liquid chromatography (HPLC, LC-2010A; Shimadzu, Kyoto, Japan) was conducted for ergosterol quantification. The analytical column was Inertsil ODS-SP (250 mm × 4.6 mm i.d., 5 μm, GL Sciences, Japan). Methanol was used as the eluent at a flow rate of 1.0 ml min−1, and the sample injection volume was 20 μl. The detection wavelength was 280 nm.
2.3.2. Fatty acid composition
Mycelia incubated for 5 days in PDB with or without Cin/Cit were collected by filtration and washed with PBS. A chloroform–methanol protocol was used for extraction of lipids and methylation as described previously.28 Fatty acid methyl esters (FAME) were dissolved in hexane for gas chromatography-mass spectrometry (GC-MS) analysis. Then 1 μl from the hexane–FAME sample was injected into the GC-MS system (GCMS-QP2010 Ultra, Shimadzu, Japan), which was fitted with a DB-17MS capillary column (30 m × 0.25 mm i.d., 0.25 μm). Helium was used as a carrier gas with a constant flow rate of 1 ml min−1. The GC-MS conditions were as follows: injector temperature 280 °C, splitless injection mode; starting temperature 150 °C, final temperature 230 °C (8 min), temperature rate 2 °C min−1; electron energy 70 eV; ion source temperature 230 °C. FAME were identified by comparing their retention times and mass fragmentation profiles with those of the standards mix, FAME (Sigma-Aldrich). An alkane (C8–C20) standard was used as a retention time index. The data were expressed as a relative percentage of each fatty acid compared to the total fatty acids area.29 Biological triplicates were used for control and Cin/Cit-treated samples.
2.3.3. Protein release
Mycelia incubated for 5 days in PDB were collected by filtration and washed with PBS. Then PBS containing the same wet weight of mycelia were incubated at 25 °C for 12 h and 24 h under 120 rpm in the presence of Cin/Cit (0, 1/2 × MIC, MIC, 2 × MIC). Then the suspension was centrifuged and the supernatant was collected to determine the concentration of proteins according to Bradford's method.30
2.3.4. Plasma membrane integrity (PMI)
The effect of Cin/Cit on the PMI of P. expansum was evaluated using 1 μg ml−1 of propidium iodide (PI, Sigma-Aldrich). A conidial suspension containing 107 spores per ml was prepared in PBS with 2% (w/v) d-glucose, and the cells were incubated on a rotary shaker with various concentrations of Cin/Cit (0, 1/2 × MIC, 2 × MIC) at 28 °C for 10 h.3 After incubation, the spores were collected, washed with PBS and stained for 15 min at room temperature in the dark with a dye. After washing twice with PBS, the spores were observed by a flow cytometer (BD Biosciences, U.S.A.) with the excitation and emission wavelengths 340 nm and 488 nm, respectively. Unstained cell suspensions were included as autofluorescence controls. Each treatment included three replicates.
2.4. Oxidative stress
2.4.1. MDA content
Mycelia incubated for 5 days in PDB with or without Cin/Cit were collected by filtration and washed with PBS. After that, the mycelia were subjected to MDA determination using a previous method.9 The mycelia of each treatment were homogenized at 0 °C with 12.5 ml of 100 mM PBS (pH 7.8) containing 2% (w/v) polyvinylpyrrolidone, 1 mM EDTA. After centrifugation for 15 min at 10 000 × g and 4 °C, 2 ml supernatant was mixed with 2 ml of 0.5% thiobarbituric acid. Then the mixture was heated at 100 °C for 30 min, cooled, and centrifuged at 10 000 × g for 10 min. Absorbance was recorded at 532 nm, 600 nm and 450 nm by an ultraviolet spectrophotometer. The result is expressed by eqn (1).
| MDA (nmol g−1) = [6.45 × (OD532 − OD600) − 0.56 × OD450] × Vt/(Vs × W) | 1 |
where Vt denotes the total volume of the extract, Vs denotes the volume of the extract used for determination, and W denotes the sample mass.
2.4.2. ROS production
In order to assess the oxidative damage of P. expansum, we determined the production of intracellular ROS using 10 μM 2,7-dichlorodihydrofluorescein diacetate (DCFH-DA; Sigma-Aldrich). After treatment with Cin/Cit (0, 1/2 × MIC, 2 × MIC) for 12 h at 28 °C, the spores were collected, washed with PBS and stained for 15 min at room temperature in the dark. At last, the spores were washed twice with PBS and observed by a flow cytometer (BD Biosciences, U.S.A.) with the excitation and emission wavelengths 488 nm and 535 nm, respectively. Unstained cell suspensions were included as autofluorescence controls. Each treatment included three replicates.
2.5. Volatile profile
After Cin/Cit treatment (0, and 1/2 × MIC) in PDB for 5 days, P. expansum mycelia were collected by filtration, and 10 ml of the filtrate was sterilely taken and transferred to 15 ml glass vials. 2 g NaCl was added to increase the diffusion of volatile organic components (VOCs) from the samples into the headspace. Besides, PDB alone (negative control-1) and PDB/Cin/Cit without fungal inoculation (negative control-2) were also subjected to VOCs determination. Solid phase microextraction (SPME) fiber (100 μm polydimethylsiloxane, Supelco Co., Bellefonte, PA, USA) was used to extract the VOCs. After the enrichment of VOCs, the fiber was automatically inserted into the injection port of the GC-MS system (GCMS-QP2010 Ultra, Shimadzu, Japan), which was fitted with a DB-17MS capillary column (30 m × 0.25 mm i.d., 0.25 μm). Helium was used as carrier gas with a constant flow rate of 1 ml min−1. The GC-MS conditions were as follows: injector temperature 250 °C, splitless injection mode; starting temperature 40 °C (3 min), final temperature 240 °C (9 min), temperature rate 6 °C min−1; electron energy, 70 eV in the range of 35–500 AMU; ion source temperature 230 °C. VOCs were tentatively identified by comparing their mass spectra with those supplied by the National Institute of Standards and Technology (NIST) database (NIST Chemistry WebBook, http://webbook.nist.gov/chemistry/). The retention indice (RI) of VOCs was calculated based on the retention time of a series of n-alkanes (C8–C24) under the same conditions as the analyzed samples. Then the RI of the detected VOCs was compared with the RI provided by the NIST database and published literatures in the same capillary column. The relative contents of components were determined by comparing the peak areas of the volatiles in the sample to the peak area of an internal standard of 3-octanol (0.3273 mg ml−1 in methanol).
2.6. Statistical analysis
Statistical analysis was performed using SPSS version 22.0 (SPSS Inc., IBM, Armonk, NY) and Origin 9.0 (Origin Lab Corporation, USA). Significance was evaluated by one-way ANOVA. Tukey's multiple-range test was performed following a significant (*p < 0.05; **p < 0.01) test. For the VOCs, the relative contents of the components were subjected to log2 transformation prior to the heat map and hierarchical cluster analysis (HCA) using pheatmap R package (http://CRAN.R-project.org/package=pheatmap).
3. Results
3.1. TEM observation
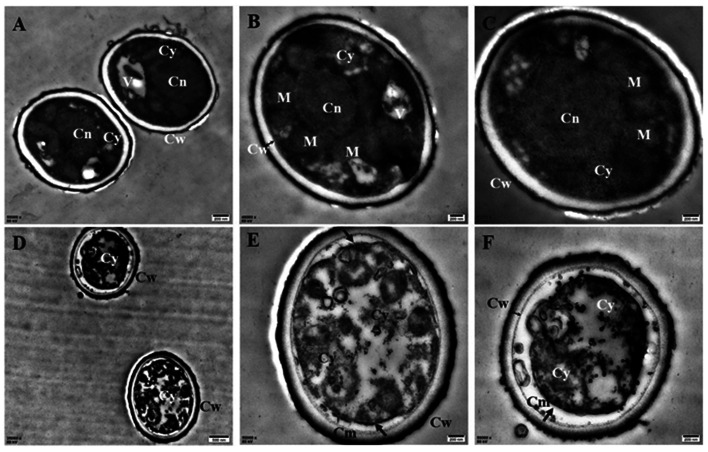
To understand the action mode of Cin/Cit against P. expansum, we started with evaluating the cellular ultrastructure alteration in P. expansum spores by TEM observation (Fig. 1). It can be seen from the control group that organized cell cytoplasm were exhibited and typical organelles e.g., mitochondria, central vacuole, cell nuclear were orderly distributed. Moreover, the cell membrane underlined the cell wall and surrounded the cytoplasm. In contrast, the spores treated by Cin/Cit at 2 × MIC exhibited invaginations of the plasma membrane, destruction of organelles and disorganized cytoplasm, and some cytoplasm even leaked out of the membrane showing the damage of the membrane integrity (Fig. 1D–F).
Fig. 1. Transmission electron microscopy images of P. expansum F-WY-12-02. (A–C) Untreated spores (A, C 50 000×; B 40 000×). (D–F) Treated spores (D 25 000×, E, F 50 000×). V: vacuole; M: mitochondrion; Cy: cytoplasm; Cw: cell wall; Cm: cell membrane; Cn: cell nucleus. Arrows referred to the morphologic changes in spores, two-way arrow referred to the cell wall.
3.2. FT-IR analysis
The FT-IR spectra of untreated P. expansum mycelia and mycelia treated by MIC, 2 × MIC and 6 × MIC of Cin/Cit were recorded and then converted to the second-derivative transformed patterns. The second-derivative transformed FT-IR spectral features were compared to study the mechanism of mycelial injury under conditions of unfavorable stress (Fig. 2). The FT-IR spectra can be roughly divided in wavenumber ranges according to some of the principal biological building blocks: lipids (3000–2800 cm−1), proteins/amides I and II (1700–1500 cm−1), phospholipids/DNA/RNA (1500–1185 cm−1), polysaccharides (1185–900 cm−1) and the fingerprint region (900–600 cm−1).31 Further analysis was conducted among these mainly different spectral regions. Fig. 2A showed the 2nd derivative spectra considering the region between 1800 and 1200 cm−1, particularly the spectra with black dotted rectangles where most differences (spectral intensity, waveform deviation) were observed, showing that Cin/Cit mainly affects phospholipids, nucleic acids, peptides and proteins in cells. While the spectral region between 900 and 1200 cm−1 exhibited no significant differences, suggesting that Cin/Cit did not cause remarkable changes in the content or structure of the polysaccharides of cells. The 2nd derivative spectra between 3100 and 2800 cm−1 were shown in Fig. 2B, and most spectral differences were presented in the region 2950–2850 cm−1, indicating that fatty acid or the hydrophilic molecules of various membrane components were mainly affected by Cin/Cit.
Fig. 2. FT-IR spectra analysis of P. expansum F-WY-12-02 under Cin/Cit treatment. (A and B) FT-IR spectra of the second derivative (900–1800 cm−1, 2800–3100 cm−1, respectively). (C) PCA 3D scores. (D) PCA loading plot. Arrow referred to the greatest contribution to the total variance in the FT-IR spectral data.
PCA clustering analysis was conducted on the 2nd derivative spectra over the wavenumber region 3000–2800 cm−1 and 1800–900 cm−1. The PCA clustering results for P. expansum mycelia exposed to 0, MIC, 2 × MIC, 6 × MIC of Cin/Cit are shown in Fig. 2C and a clear separation was observed between different treatments, suggesting that Cin/Cit affected the chemical compounds of cells. Principal components 1 (PC1), PC2 and PC3 explained 64.88%, 19.41%, and 9.64%, respectively, of the total variability. The first three loading plots from PCA analysis was shown in Fig. 2D. Loading weights indicated the contribution of each variable (wavenumber) to a principal component and provided an indication of which spectral region makes the most significant contributions to data variation.32 Variables between 1800–1500 and 3000–2800 cm−1 with a black arrow provided the greatest contribution to the total variance in the FT-IR spectral data, implying that Cin/Cit caused remarkable changes of phospholipids, nucleic acids, proteins and fatty acids in mycelia.
3.3. Plasma membrane analysis
3.3.1. Ergosterol
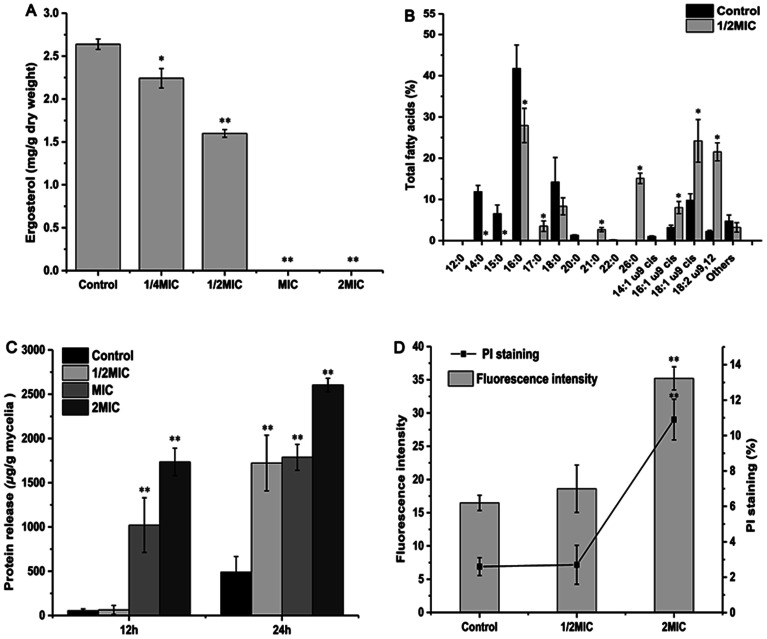
The effects of different concentrations of Cin/Cit on ergosterol accumulation are presented in Fig. 3A. Cin/Cit of MIC and 2 × MIC totally inhibited ergosterol biosynthesis. When P. expansum was treated with 1/4 × MIC and 1/2 × MIC, the reduction in ergosterol ranged from 15.02% to 39.40%.
Fig. 3. Effect of Cin/Cit on ergosterol accumulation (A), fatty acids composition (B), protein release level (C) and PMI (D) of P. expansum F-WY-12-02. PMI was assessed using PI staining by flow cytometry. Bars represent standard deviation of the treatment means.
3.3.2. Fatty acids
Changes in the membrane fatty acids (FAs) composition of P. expansum after Cin/Cit treatment were investigated by GC-MS analysis and the results are shown in Fig. 3B. Compared to the controls, exposure to Cin/Cit induced a significant increase (p < 0.05) of unsaturated fatty acids (UFAs) C16:1 ω9cis, C18:1 ω9cis and C18:2ω9,12, by 60.39%, 59.56%, and 89.58%, respectively. In addition, Cin/Cit treatment increased the relative percentages of some specific long-chain FAs, such as C21:0 and C26:0. However, the levels of some saturated FAs such as C14:0, C15:0, C16:0 and C18:0 were remarkably decreased.
3.3.3. Protein release
In the present study, proteins were evidently released into cell suspension and the levels increased multi-fold after exposure to increasing concentrations of Cin/Cit compared to the controls (p < 0.05) (Fig. 3C). The values of the released proteins increased from 53.84 μg g−1 for the control to 63.12, 1020.19 and 1735.00 μg g−1 mycelia with the presence of Cin/Cit at 1/2 × MIC, MIC and 2 × MIC after 12 h, indicating that the higher content of Cin/Cit caused more serious damage to plasma membrane. With the treatment time prolonged, the level of released proteins increased by 8.09, 26.29, 0.75 and 0.5 times compared to that at 12 h for control, 1/2 × MIC, MIC and 2 × MIC, showing that the higher concentration of Cin/Cit led to a steady state after triggering a rapid and large release of cellular proteins of P. expansum.
3.3.4. Plasma membrane integrity
The results of fluorescence intensity and PI staining cells (%) in the presence of Cin/Cit (0, 1/2 × MIC, 2 × MIC) for 10 h are shown in Fig. 3D. With the increasing concentrations of Cin/Cit, 2 × MIC caused a significant (p < 0.05) increase in fluorescence intensity (35.21) and the percentage of PI-stained cells (10.9%) compared to controls (16.48, 2.6%) and 1/2 × MIC (18.61, 2.7%), indicating a loss of membrane integrity.
3.4. Lipid peroxidation and oxidative stress
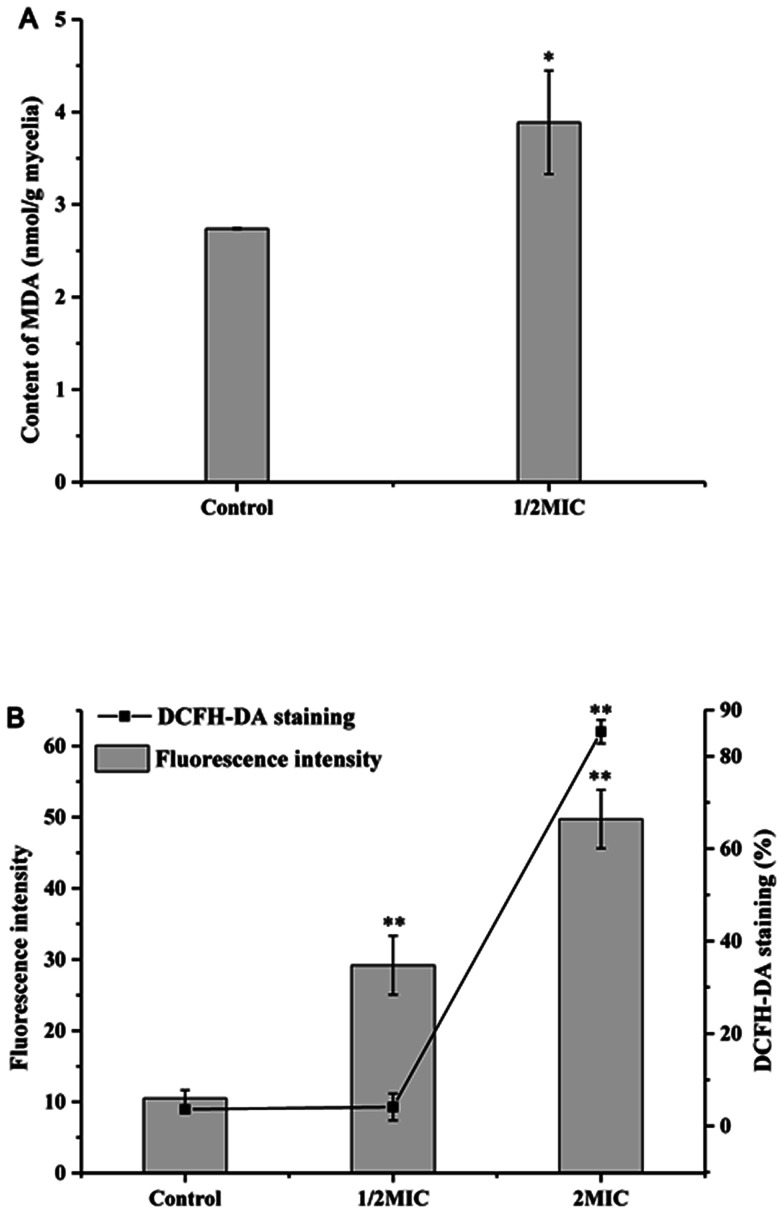
MDA, as the most important lipid peroxidation metabolite, is a crucial parameter reflecting lipid peroxidation level in organisms. As shown in Fig. 4A, Cin/Cit significantly (p < 0.05) induced MDA accumulation in P. expanusm. The intracellular level of MDA was 40.09% more in mycelia treated with 1/2 × MIC, compared with controls.
Fig. 4. Determination of the effect of Cin/Cit on MDA (A) and ROS accumulation (B) of P. expansum F-WY-12-02. ROS was assessed using DCFH-DA staining by flow cytometry. Bars represent standard deviation of the treatment means.
The intracellular ROS level of P. expansum was monitored using a fluorescent dye DCFH-DA (Fig. 4B). A significant (p < 0.05) increase in the fluorescence intensity was detected in P. expansum cells incubated with different concentrations of Cin/Cit for 10 h. Compared with the controls, the promotion of the fluorescence intensity at 1/2 × MIC and 2 × MIC corresponded to 2.78 and 4.74 times, respectively. Besides, the percentage of DCFH-DA staining cells clearly increased with the increase of the Cin/Cit concentration. These data indicated that Cin/Cit induced ROS accumulation in P. expansum cells in a dose-dependent manner.
3.5. Cin/Cit intervented volatile profile of P. expansum
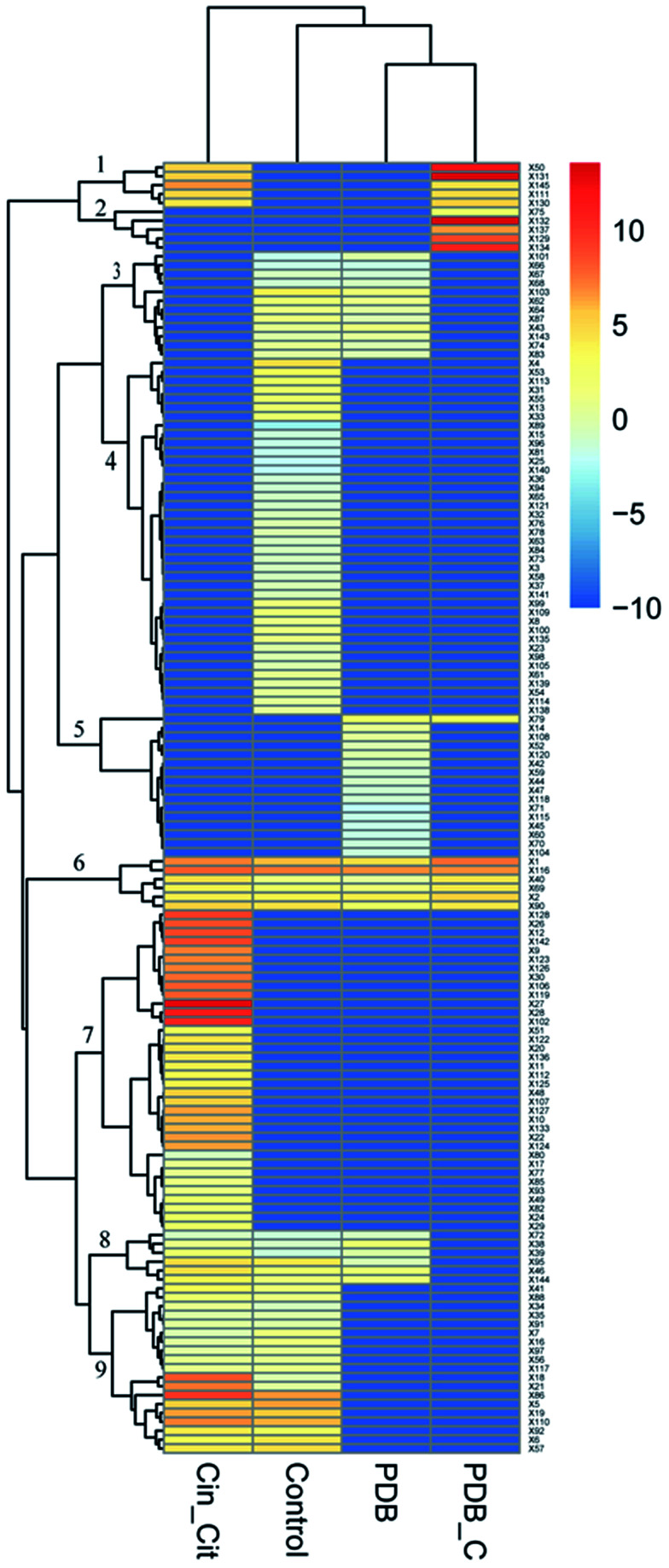
The supplementation of PDB with Cin/Cit provoked marked modifications of the volatile profiles of P. expansum F-WY-12-02. A total of 145 VOCs were detected in four groups, and are listed in Table S2.† These different metabolites were subjected to HCA resulting in 9 clusters (Fig. 5). Clusters 1 and 2 represented VOCs in PDB/Cin/Cit, and most of them were supplemented antimicrobials compounds. VOCs with 41.7% belonging to alkanes in cluster 3 were detected in positive controls and PDB except the treatment group, showing that the metabolism of these compounds might be activated under Cin/Cit stress. Clusters 6 and 8 were comprised of 13 compounds, with ethanol and 2,5-bis(1,1-dimethylethyl)-phenol as predominant compounds. These compounds didn't show significant differences in relative content in PDB with or without fungal growth, showing that they might not be essential nutrients for cells growth. While the compounds, consumed by P. expansum as nutrients, constituted cluster 5, and these substances were metabolized regardless of Cin/Cit treatment. Cluster 9 represented the fungal metabolites with linalool, terpineol, 2-methyl-1,2-ethanediyl-2-propenoic acid ester as predominant compounds, in both treatment group and positive controls, showing that the metabolic pathways related to these VOCs were not influenced significantly under Cin/Cit treatment. Cluster labeled 4 represented volatile metabolites presented only in positive controls, with 31.25% of alcohols and 20% of esters, suggesting that the metabolic pathways related to these VOCs were activated for normal cell growth but depressed under Cin/Cit treatment. By contrast, cluster 7 represented metabolites, with 36.11% of alcohols and 22.22% of terpenes, which occurred only in the treatment group and most belonged to detoxification compounds, indicating an induction of specific metabolism in aromatic substances.
Fig. 5. Effect of Cin/Cit on the volatile profile of P. expansum F-WY-12-02. Columns represent different groups. Control: P. expansum grown in PDB (positive control); Cin_Cit: P. expansum grown in PDB/Cin/Cit (treatment group); PDB: PDB medium alone (negative control-1); PDB_C: PDB/Cin/Cit without fungal inoculation (negative control-2). Rows represent the different VOCs (blue: low abundance; red: high abundance).
4. Discussion
In the present work, in order to study the action modes of Cin/Cit against P. expansum, TEM observation was firstly performed to analyze the changes in the cellular ultrastructure. The results clearly reflected that Cin/Cit caused damage to the intracellular structure. The remarkable separation of the plasma membrane from the cell wall indicated the loss of intracellular substance and membrane damage. Cinnamaldehyde was reported to cause leakage of the cytoplasmic contents of Fusarium verticillioides, as well as the shrink of cytoplasmic substances and mitochondrial destruction of A. flavus, based on TEM observation.11,33 Besides, a phenomenon similar to our results about the membrane damage was obtained from citral against Penicillium italicum.34
After the analysis of the cell structure, FT-IR spectroscopy was conducted to study global macromolecular changes in the cell. FT-IR fingerprints were previously used to assess the metabolomic damage of Listeria innocua and E. coli exerted by N-alkyltropinium bromide surfactants.35 Synchrotron FT-IR spectromicroscopy was used to exam the whole-cell biochemical composition (carbohydrates, proteins and lipids) in fungal cell.27 In this work, the FT-IR spectra of P. expansum treated with different concentrations of Cin/Cit showed significant differences, particularly in the regions 1800–1200 cm−1 and 2950–2850 cm−1, showing that the phospholipids, proteins, fatty acid or hydrophilic molecule of various membrane components were mainly affected by Cin/Cit. The FT-IR spectral region 900–1200 cm−1, containing the information of polysaccharides, exhibited no significant differences (Fig. 2A). A previous report has indicated that limonene treatment caused no significant changes in the cell wall polysaccharide composition of Saccharomyces cerevisiae.28 Besides, the action mode of Cinnamomum verum containing 82.09% of cinnamaldehyde against Candida was also not related to cell wall biosynthesis.36 Based on the TEM observation and FT-IR fingerprints, the cell wall components of P. expansum F-WY-12-02 may be not the main target of Cin/Cit, and they can probably get through the cell wall and act on the cell membrane.
Due to the lipophilic character of EOs, the microbial cytoplasmic membranes have been regarded as the target of bioactive compounds.37,38 Ergosterol is one of the principal sterol components in the fungal membrane and is responsible for regulating cell structure, osmosis, growth, and proliferation.14 In this work, ergosterol biosynthesis of P. expansum F-WY-12-02 was significantly inhibited by Cin/Cit (Fig. 3A). A previous study showed that natural cinnamaldehyde significantly inhibited ergosterol production by 80% to 100% in A. ochraceus.15 Citral inhibited the growth of P. digitatum by down-regulating ergosterol biosynthesis through transcriptional profiling analysis.14 In addition, chemical fungicides, such as triflumizole, were reported to cause the disruption of the P. digitatum cell structure and function, even death by blocking the ergosterol biosynthesis.39 Overall, inhibition of ergosterol biosynthesis is essential for various antifungal agents including Cin/Cit.
Researchers have indicated that microbial cells can respond to environmental stress by modulating the ratio of saturated to unsaturated, cis to trans unsaturation, particular the UFAs. Siroli et al.,29 showed an induction of UFAs by citral and other EOs in L. monocytogenes, Salmonella enteritidis and E. coli, which is similar to our results (Fig. 3B). Based on the above reports and our results, it can be seen that P. expansum modified the membrane FAs composition, mainly by increasing the relative percentages of UFAs, in response to the stress conditions generated by Cin/Cit.
Information on the release of cell constituents can reflect the integrity level of the cell membrane from one side. In the present study, Cin/Cit caused an evident release of proteins into cell suspension (Fig. 3C), which is similar to the findings of Zhang et al.4 Moreover, the FT-IR spectral region between 1700 and 1500 cm−1, containing abundant information on proteins/amides I and II, also exhibited significant differences (Fig. 2A), which may be correlated to the losses of cellular proteins. Our results clearly indicated that the P. expansum plasma membrane integrity was compromised after exposure to Cin/Cit, which could cause cytoplasmic leakage and consequently lead to cell death.
PI is a DNA-staining fluorescent probe that only penetrates dead or dying cells with injured membranes, which has been adopted to detect membrane integrity in previous researches.3,40 Therefore, the effects of Cin/Cit on P. expansum plasma membrane were further examined by monitoring the influx of PI to evaluate the disruption of membrane integrity (Fig. 3D). Once the disturbance of membrane integrity occurred, its functions were compromised not only as a barrier but also as a matrix for enzymes and as an energy transducer, eventually causing the death of P. expansum cells.41
The cellular redox equilibrium can be disturbed by different exogenous stress, leading to enhanced ROS levels called “oxidative stress”.42 Excessive accumulation of ROS can cause oxidative damage to cell components, resulting in compromised cell function and loss of viability.43 Therefore, we determined the levels of MDA, and total ROS to explore whether Cin/Cit caused oxidative stress on P. expanusm.
Our results revealed that Cin/Cit treatment triggered an increase in membrane lipid peroxidation (Fig. 4A). A previous study found that monoterpenes at low concentrations led to lipid peroxidation in fungi.44 Oxidative damage of poly-unsaturated fatty acids (PUFA) caused by oxygen radicals attack is the main reaction in the lipid peroxidation chain, leading to cell membrane damage.45 It should be noted that our data have shown that Cin/Cit treatment induced the accumulation of UFAs in the P. expansum plasma membrane (Fig. 3B), which provided the possibility for membrane lipid peroxidation reaction, and elucidated the increase of MDA reasonably.
In the present work, Cin/Cit induced ROS accumulation in P. expansum cells in a dose-dependent manner (Fig. 4B). Undesirable accumulation of ROS can cause oxidative injury to biological macromolecules; lipid and proteins are particularly vulnerable to oxidative damage, leading to lipid peroxidation and protein carbonylation.7,45 Similar observations demonstrated that the action modes of many antifungal agents were related to the induction of ROS formation such as A. flavus, P. expansum, Botrytis cinerea.7,37,43 Thus, ROS accumulation is the main pathway underlying the cellular damage induced by Cin/Cit.
The observation of volatile metabolites reflected the influence of Cin/Cit on P. expansum from a metabolic point. The VOCs fingerprints of P. expansum were significantly disturbed by Cin/Cit (Fig. 5). Therein, cluster 7 showed only the metabolites occurring in the treatment group and most belonged to detoxification compounds, indicating an induction of specific metabolism of aromatic substances. Our results showed that the relative content of cinnamaldehyde and cinnamic alcohol in PDB/Cin/Cit (negative control-2) and treatment group were 3484, 0 μg l−1 and 41.21, 198.80 μg l−1, respectively. Meanwhile, cinnamic alcohol was undetectable in positive control or PDB (negative control-1). Hua et al.,15 indicated similar results, i.e. cinnamaldehyde was converted to cinnamic alcohol by A. ochraceus. And cinnamic alcohol showed weak antifungal activity compared with cinnamaldehyde, thus the antimicrobial activity of cinnamaldehyde was mainly attributed to its carbonyl aldehyde group. Visvalingam et al.,16 found that E. coli O157:H7 could convert cinnamaldehyde to cinnamic alcohol by alcohol dehydrogenase and 2,5-diketo-d-gluconate reductase A. Therefore, some enzymes in P. expansum such as alcohol dehydrogenase and aldehyde reductase may play important roles in catalyzing cinnamaldehyde to cinnamic alcohol. Additionally, Siroli et al.,29 have shown that citral detoxification molecules, such as nerol, geraniol, β-citronellol, 6-methyl-5-hepten-2-one, etc. characterized the volatilome of cells subjected to citral exposure. Similarly, all these compounds were observed in the volatile metabolites of P. expansum treated by Cin/Cit in the present work, with nerol, geraniol, 6-methyl-5-hepten-2-one as the predominant detoxification compounds. The previous report has attributed the dramatic increase of some alcohols to detoxification of the added antimicrobials, and of aldehydes that are regarded as end products of the breakdown of peroxidated unsaturated fatty acids formed after oxidative stress induced by the growth conditions.29 Except for the added Cin/Cit and their detoxification products, the relative contents of some other VOCs were also remarkably influenced such as heptanal, benzaldehyde, nonanal, 2-methyl-2-propenoic acid, 1,2-ethanediyl ester, etc., suggesting that many other specific response mechanisms or pathways were probably involved, which need further investigations.
5. Conclusions
The present work highlighted the antifungal effect of Cin/Cit on P. expansum F-WY-12-02. Cin/Cit caused membrane invaginations and organelles destruction, as proved by TEM observation. FT-IR spectra reflected remarkable changes of phospholipids and proteins in mycelia. Membrane changes or damages after exposure to Cin/Cit were evidenced by the decrease of ergosterol, an increase in UFAs, protein leakage, and PI staining cells. Moreover, the increase in MDA and ROS accumulation confirmed the oxidative stress in cells. At last, Cin/Cit significantly affected the VOCs profiles of P. expansum with cinnamic alcohol, nerol, and geraniol as the dominant detoxification compounds. The alterations in VOCs profiles may be related to the intervention of some specific mechanism pathway caused by Cin/Cit. These findings indicated that the action mode of Cin/Cit against P. expansum relied on a series of factors, not on any single one of the mechanisms. A variety of factors constituted cascade reaction to induce the death of pathogen. Future researches by applying transcriptomics have been conducted to explain more mechanisms of the inhibitory effect of Cin/Cit on P. expansum and the patulin production.
Conflicts of interest
There are no conflicts of interest to declare.
Supplementary Material
Acknowledgments
The research was supported by the National Basic Research Program of China (2013FY113400), the National Natural Science Foundation of China (31671866), Scientific and Technology Cooperation Project in Hong Kong, Macao, and Taiwan of China (2015DFT30130), and the Science and Technology Research and Development Program of Shaanxi Province, China (2016KTCQ03-12).
Electronic supplementary information (ESI) available. See DOI: 10.1039/c7ra12191a
References
- Moake M. Padillazakour O. I. Worobo R. W. Compr. Rev. Food Sci. Food Saf. 2005;4:8–21. doi: 10.1111/j.1541-4337.2005.tb00068.x. [DOI] [PubMed] [Google Scholar]
- Lai T. Wang Y. Fan Y. Zhou Y. Bao Y. Zhou T. Int. J. Food Microbiol. 2017;244:1–10. doi: 10.1016/j.ijfoodmicro.2016.12.017. [DOI] [PubMed] [Google Scholar]
- Tian J. Wang Y. Zeng H. Li Z. Zhang P. Tessema A. Peng X. Int. J. Food Microbiol. 2015;202:27–34. doi: 10.1016/j.ijfoodmicro.2015.02.022. [DOI] [PubMed] [Google Scholar]
- Zhang Y. B. Liu X. Y. Wang Y. F. Jiang P. P. Quek S. Food Control. 2016;59:282–289. doi: 10.1016/j.foodcont.2015.05.032. [DOI] [Google Scholar]
- Shi C. Song K. Zhang X. Sun Y. Sui Y. Chen Y. Jia Z. Sun H. Sun Z. Xia X. PLoS One. 2016;11:e0159006. doi: 10.1371/journal.pone.0159006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neri F. Mari M. Brigati S. Plant Pathol. 2006;55:100–105. doi: 10.1111/j.1365-3059.2005.01312.x. [DOI] [Google Scholar]
- Qin G. Liu J. Cao B. Li B. Tian S. PLoS One. 2011;6:e21945. doi: 10.1371/journal.pone.0021945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y. Shan T. Yuan Y. Yue T. J. Food Sci. 2016;81:H3043–H3051. doi: 10.1111/1750-3841.13536. [DOI] [PubMed] [Google Scholar]
- Gao J. F., Experimental Guidance for Plant Physiology, World Book Incorporation, Xi'an, 2000 [Google Scholar]
- Fan F. Tao N. Jia L. He X. Postharvest Biol. Technol. 2014;90:52–55. doi: 10.1016/j.postharvbio.2013.12.005. [DOI] [Google Scholar]
- Sun Q. Shang B. Wang L. Lu Z. Liu Y. Appl. Microbiol. Biotechnol. 2016;100:1355–1364. doi: 10.1007/s00253-015-7159-z. [DOI] [PubMed] [Google Scholar]
- Burt S. A. Int. J. Food Microbiol. 2004;94:223–253. doi: 10.1016/j.ijfoodmicro.2004.03.022. [DOI] [PubMed] [Google Scholar]
- Helander I. M. Alakomi H. L. Latvakala K. Mattilasandholm T. De Pol I. V. Smid E. J. Gorris L. G. M. Von Wright A. J. Agric. Food Chem. 1998;46:3590–3595. doi: 10.1021/jf980154m. [DOI] [Google Scholar]
- Ouyang Q. Tao N. Jing G. BMC Genomics. 2016;17:599. doi: 10.1186/s12864-016-2943-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua H. Xing F. Selvaraj J. N. Wang Y. Zhao Y. Zhou L. Liu X. Liu Y. PLoS One. 2014;9:e108285. doi: 10.1371/journal.pone.0108285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visvalingam J. Hernandezdoria J. D. Holley R. A. Appl. Environ. Microbiol. 2013;79:942–950. doi: 10.1128/AEM.02767-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlshoj K. Nielsen P. V. Larsen T. O. J. Agric. Food Chem. 2007;55:4289–4298. doi: 10.1021/jf070134x. [DOI] [PubMed] [Google Scholar]
- Gutierrez J. Barryryan C. Bourke P. Int. J. Food Microbiol. 2008;124:91–97. doi: 10.1016/j.ijfoodmicro.2008.02.028. [DOI] [PubMed] [Google Scholar]
- Stević T. Berić T. Šavikin K. Soković M. Gođevac D. Dimkić I. Stanković S. Ind. Crops Prod. 2014;55:116–122. doi: 10.1016/j.indcrop.2014.02.011. [DOI] [Google Scholar]
- Shi C. Zhang X. Zhao X. Meng R. Liu Z. Chen X. Guo N. Food Control. 2017;71:10–16. doi: 10.1016/j.foodcont.2016.06.020. [DOI] [Google Scholar]
- Manso S. Becerril R. Nerin C. Gomez-Lus R. Food Control. 2015;47:20–26. doi: 10.1016/j.foodcont.2014.06.014. [DOI] [Google Scholar]
- Balaguer M. P. Lopez-Carballo G. Catala R. Gavara R. Hernandez-Munoz P. Int. J. Food Microbiol. 2013;166:369–377. doi: 10.1016/j.ijfoodmicro.2013.08.012. [DOI] [PubMed] [Google Scholar]
- Xing Y. G. Li X. H. Xu Q. L. Yun J. A. Lu Y. Q. Int. J. Food Sci. Technol. 2010;45:1837–1842. doi: 10.1111/j.1365-2621.2010.02342.x. [DOI] [Google Scholar]
- Camele I. Altieri L. De Martino L. De Feo V. Mancini E. Rana G. L. Int. J. Mol. Sci. 2012;13:2290–2300. doi: 10.3390/ijms13022290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y. Yuan Y. Liu B. Zhang Z. Yue T. J. Appl. Microbiol. 2016;121:1384–1393. doi: 10.1111/jam.13208. [DOI] [PubMed] [Google Scholar]
- Iten F. Saller R. Abel G. Reichling J. Planta Med. 2009;75:1231–1236. doi: 10.1055/s-0029-1185541. [DOI] [PubMed] [Google Scholar]
- Jilkine K. Gough K. M. Julian R. Kaminskyj S. G. W. J. Inorg. Biochem. 2008;102:540–546. doi: 10.1016/j.jinorgbio.2007.10.023. [DOI] [PubMed] [Google Scholar]
- Brennan T. C. R. Kromer J. O. Nielsen L. K. Appl. Environ. Microbiol. 2013;79:3590–3600. doi: 10.1128/AEM.00463-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siroli L. Patrignani F. Gardini F. Lanciotti R. Food Chem. 2015;182:185–192. doi: 10.1016/j.foodchem.2015.02.136. [DOI] [PubMed] [Google Scholar]
- Bradford M. M. Anal. Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Maquelin K. Kirschner C. Choosmith L. P. Den Braak N. V. Endtz H. P. Naumann D. Puppels G. J. J. Microbiol. Methods. 2002;51:255–271. doi: 10.1016/S0167-7012(02)00127-6. [DOI] [PubMed] [Google Scholar]
- Kansiz M. Heraud P. Wood B. R. Burden F. R. Beardall J. Mcnaughton D. Phytochemistry. 1999;52:407–417. doi: 10.1016/S0031-9422(99)00212-5. [DOI] [Google Scholar]
- Xing F. Hua H. Selvaraj J. N. Zhao Y. Zhou L. Liu X. Liu Y. Food Control. 2014;46:343–350. doi: 10.1016/j.foodcont.2014.04.037. [DOI] [Google Scholar]
- Tao N. OuYang Q. Jia L. Food Control. 2014;41:116–121. doi: 10.1016/j.foodcont.2014.01.010. [DOI] [Google Scholar]
- Corte L. Tiecco M. Roscini L. De Vincenzi S. Colabella C. Germani R. Tascini C. Cardinali G. PLoS One. 2015;10:1–15. doi: 10.1371/journal.pone.0115275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Essid R. Hammami M. Gharbi D. Karkouch I. Hamouda T. B. Elkahoui S. Limam F. Tabbene O. Appl. Microbiol. Biotechnol. 2017;101:6993–7006. doi: 10.1007/s00253-017-8442-y. [DOI] [PubMed] [Google Scholar]
- Tian J. Ban X. Zeng H. He J. Chen Y. Wang Y. PLoS One. 2012;7:e30147. doi: 10.1371/journal.pone.0030147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabral L. D. C. Pinto V. F. Patriarca A. Int. J. Food Microbiol. 2013;166:1–14. doi: 10.1016/j.ijfoodmicro.2013.05.026. [DOI] [PubMed] [Google Scholar]
- Hamamoto H. Hasegawa K. Nakaune R. Lee Y. J. Makizumi Y. Akutsu K. Hibi T. Appl. Environ. Microbiol. 2000;66:3421–3426. doi: 10.1128/AEM.66.8.3421-3426.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho J. Choi H. Lee J. Kim M.-S. Sohn H.-Y. Lee D. G. Biochim. Biophys. Acta, Biomembr. 2013;1828:1153–1158. doi: 10.1016/j.bbamem.2012.12.010. [DOI] [PubMed] [Google Scholar]
- Jing L. Lei Z. Li L. Xie R. Xi W. Guan Y. Sumner L. W. Zhou Z. J. Agric. Food Chem. 2014;62:3011–3033. doi: 10.1021/jf5006148. [DOI] [PubMed] [Google Scholar]
- Lushchak V. I. Comp. Biochem. Physiol., Part C: Toxicol. Pharmacol. 2011;153:175–190. doi: 10.1016/j.cbpc.2010.10.004. [DOI] [PubMed] [Google Scholar]
- Zhang Z. Qin G. Li B. Tian S. Curr. Microbiol. 2015;71:396–402. doi: 10.1007/s00284-015-0863-1. [DOI] [PubMed] [Google Scholar]
- Lucini E. I. Zunino M. P. López M. L. Zygadlo J. A. J. Phytopathol. 2006;154:441–446. doi: 10.1111/j.1439-0434.2006.01126.x. [DOI] [Google Scholar]
- Repetto M., Semprine J. and Boveris A., Lipid peroxidation: chemical mechanism, biological implications and analytical determination, InTech, New Delhi, 2012 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.