Abstract
It has been reported that penicillin-binding protein 4 (PBP4) activity decreases when a vancomycin-susceptible Staphylococcus aureus isolate is passaged in vitro to vancomycin resistance. We analyzed the PBP profiles of four vancomycin intermediately susceptible S. aureus (VISA) clinical isolates and found that PBP4 was undetectable in three isolates (HIP 5827, HIP 5836, and HIP 6297) and markedly reduced in a fourth (Mu50). PBP4 was readily visible in five vancomycin-susceptible, oxacillin-resistant S. aureus (ORSA) isolates. The nucleotide sequences of the pbp4 structural gene and flanking sequences did not different between the VISA and vancomycin-susceptible isolates. Overproduction of PBP4 on a high-copy-number plasmid in the VISA isolates produced a two- to threefold decrease in vancomycin MICs. Inactivation of pbp4 by allelic replacement mutagenesis in three vancomycin-susceptible ORSA strains (COL, RN450M, and N315) led to a decrease in vancomycin susceptibility, an increase in highly vancomycin-resistant subpopulations, and decreased cell wall cross-linking by high-performance liquid chromatography analysis. Complementation of the COL mutant with plasmid-encoded pbp4 restored the vancomycin MIC and increased cell wall cross-linking. These data suggest that alterations in PBP4 expression are at least partially responsible for the VISA phenotype.
Vancomycin is currently the treatment of choice for serious infections caused by oxacillin-resistant Staphylococcus aureus (ORSA). ORSA isolates recovered from patients with serious infections in both Japan (11) and the United States (23) have recently been described that have reduced susceptibility to vancomycin and other glycopeptides (vancomycin intermediately susceptible S. aureus [VISA]). In addition to having reduced vancomycin susceptibility, these isolates are also resistant to other antimicrobials, leaving few options for effective antimicrobial therapy.
The mechanism of decreased vancomycin susceptibility is poorly understood and still largely speculative (4, 7, 16). VISA isolates demonstrate decreased autolysis, slower growth rates, and thicker cell walls in comparison to vancomycin-susceptible S. aureus (7). In a vancomycin-resistant S. aureus strain derived by in vitro passage, the cell walls of the mutant have a decrease in peptidoglycan cross-bridges and an increase in monomeric muropeptides carrying intact carboxyl-terminal d-alanyl–d-alanine residues (19, 20), the targets of vancomycin, as stem peptide termini. The passage mutant has also been shown to have markedly decreased or absent PBP4, as assessed by radiolabeled penicillin binding (19). PBP4 is a low-molecular-weight (LMW) PBP hypothesized to be involved in secondary cell wall remodeling (9, 10, 26). It has transpeptidase activity and appears also to act as a d,d-carboxypeptidase, cleaving terminal d-alanine residues from un-cross-linked muropeptides (12). It has been proposed that VISA strains, with their thicker cell walls, provide an increased number of vancomycin targets at the cell wall periphery, preventing access of the molecule to its site of lethal action at the cell membrane (19). A decrease in PBP4 activity would increase the number of surrogate vancomycin targets by increasing the total cell wall content of d-alanine–d-alanine-containing muropeptides. However, there has been no rigorous test of this hypothesis. As a first step, we have begun to assess the role of PBP4 in the VISA phenotype among clinical isolates. We have confirmed the absence of PBP4 activity in clinical VISA isolates and have sought to define the role of PBP4 in S. aureus vancomycin susceptibility by pbp4 complementation and deletion mutation. PBP4 activity and regulation appear to be important to the response of S. aureus to vancomycin.
MATERIALS AND METHODS
Bacterial strains and plasmids.
The bacterial strains and plasmids used in the present study are summarized in Table 1.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain, isolate, or plasmid (origin) | Relevant characteristics | Source or reference |
|---|---|---|
| Strains or isolates | ||
| HIP 5836 (New Jersey) | Vancomycin MIC of 8 μg/ml | 23 |
| HIP 6297 (New York) | Vancomycin MIC of 6 μg/ml | 23 |
| HIP 5827 (Michigan) | Vancomycin MIC of 8 μg/ml | 23 |
| Mu50 (Japan) | Vancomycin MIC of 6 μg/ml | 11 |
| Mu3 (Japan) | Vancomycin MIC of 2 μg/ml | 11 |
| RN4220 | Restriction-deficient derivative of 8325-4 | |
| RN450M | Heterogeneous Oxar | 14 |
| N315 | Heterogeneous Oxar | 14 |
| COL | Homogeneous Oxar | 14 |
| 27619 | Homogeneous Oxar | 2 |
| RN450MVR | Passage-derived mutant of RN450M; vancomycin MIC of 8 μg/ml | This study |
| N135VR | Passage-derived mutant of N315; vancomycin MIC of 8 μg/ml | This study |
| COLVR | Passage-derived mutant of COL; vancomycin MIC of 8 μg/ml | This study |
| Plasmids | ||
| pJF3 | 1.8-kb pbp4 gene and 5′ sequences with pRN5543 staphylococcal replicon; Chlr | This study |
| pJF14 | pE194(ts)-based vector with pbp4 interrupted by tetM; Ermr Tetr | This study |
Materials and media.
Mueller-Hinton broth (MHB) and Mueller-Hinton agar (MHA; BBL Microbiology Systems, Cockeysville, Md.) and brain heart infusion (BHI) broth and agar (Difco Laboratories, Detroit, Mich.) with and without selective additives (Sigma, St. Louis, Mo.; United States Biochemicals, Cleveland, Ohio), were used for the subculture and maintenance of Escherichia coli and S. aureus strains. The antibiotics and concentrations used for E. coli strains for initial selection after transformation were as follows: ampicillin, 50 μg/ml; minocycline, 1 μg/ml; chloramphenicol, 25 μg/ml. The antibiotics used for initial selection of S. aureus after electroporation and for subsequent maintenance on agar were as follows: chloramphenicol, 10 μg/ml; erythromycin, 10 μg/ml; minocycline, 1 μg/ml.
Cloning, transformation, and DNA manipulation.
All restriction endonuclease digestions and ligations were performed in accordance with the manufacturer's (New England Biolabs, Beverly, Mass.) specifications. Plasmids were electroporated into E. coli in a Bio-Rad Gene Pulser in accordance with the manufacturer's (Bio-Rad Laboratories, Richmond, Calif.) instructions. Shuttle vectors were moved from E. coli to S. aureus by electroporation (17) into restriction-deficient S. aureus strain RN4220 as previously described (18). VISA isolates were electroporated with plasmid DNA purified from strain RN4220. Plasmids were introduced into other S. aureus strains by transduction with general transduction phage 80α (15). Transductions with phage 80α and isolation of both plasmid and genomic DNAs were performed as previously described (18). PCR of the entire pbp4 gene with its promoter was performed by using primers 20 (5′-ACCCACTGGCCATGATAG-3′) and 40 (5′-TACAGAAGGCATTTCGACG-3′). The resulting 1.8-kb PCR fragment was cloned into pCR2.1 (Invitrogen). To this construct, staphylococcal replicon pRN5543 (3) was added as a BamHI fragment. The resulting plasmid was designated pJF3. This plasmid confers chloramphenicol resistance on S. aureus and ampicillin resistance on E. coli. Construction of pJF14 was accomplished by first cloning the coding region of pbp4 into pUC19. The resulting construct was then digested with SacI, and the tetM cassette was added as a SacI fragment. Temperature-sensitive staphylococcal replicon pE194ts was added as an XbaI fragment. The resulting plasmid (pJF14) confers ampicillin resistance on E. coli and resistance to both minocycline and erythromycin on S. aureus.
Plasmid curing and allelic replacement.
S. aureus isolates harboring plasmid constructs with the pE194ts replicon were cured of their plasmids in order to detect allelic replacement of chromosomal genes by homologous recombination. Briefly, single colonies were inoculated into 5 ml of BHI and allowed to grow for 16 h at the permissive temperature (30°C). Following growth at the nonpermissive temperature for plasmid replication (43°C), colonies were patched to minocycline and erythromycin plates. Colonies were sought that were resistant to minocycline but sensitive to erythromycin, indicating secondary recombination to remove plasmid DNA. Allelic replacement was confirmed by PCR of the pbp4 gene and its promoter, resulting in a fragment 4.7 kb in length compared to the native-size fragment of 1.8 kb. The addition of tetM added roughly 3 kb.
DNA sequencing.
The sequences of pbp4 were determined by direct sequencing of specific amplified PCR products obtained from genomic template DNA prepared with a Genomic Qiagen-tip kit (Qiagen, Valencia, Calif.). Sequencing of the PCR fragments was performed by the dideoxy-chain termination procedure on an ABI 1377 automatic sequencer with an ABI PRISM Dye Terminator Cycle Sequencing Ready Reaction Kit with Ampli-Taq DNA polymerase FS (Perkin-Elmer, Applied Biosystems Division, Foster City, Calif.)
PBP analysis.
PBPs were analyzed by Michael Pucci at the Bristol-Myers Squibb Pharmaceutical Research Institute by methods previously described (14). Briefly, cell membrane protein samples were labeled with [H3]benzylpenicillin (27.2 Ci/mmol; DuPont NEN, Boston, Mass.), resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, fixed, stained, and subjected to fluorography.
Susceptibility testing.
MICs for S. aureus isolates were determined by the broth microdilution method in cation-adjusted MHB in accordance with NCCLS guidelines (13). MICs of vancomycin for isolates containing the multicopy plasmid encoding PBP4 (pJF3) were determined in the presence of chloramphenicol (5 μg/ml). MIC determinations were performed in quadruplicate and read after 18 to 24 h of incubation at 35°C. Susceptibility to vancomycin was also determined by the E-test method (AB Biodisk, Dammartin sur Tigeaux, France) performed in accordance with the manufacturer's recommendations, except that BHI agar was used in addition to MHA.
EOP.
Phenotypic expression of vancomycin resistance was determined by the efficiency-of-plating (EOP) procedure described by Hackbarth et al. (6), except that vancomycin was used instead of methicillin.
Analysis of muropeptide composition.
Isolated cell walls were prepared by the method of Stranden as described previously (25). Lyophilized peptidoglycan was digested with mutanolysin (Sigma), and the resulting muropeptides were reduced to their muramitol derivatives. Separation of muropeptides was achieved by reversed-phase high-pressure liquid chromatography (HPLC) using a Hewlett-Packard 1100 series system. Samples were applied to a Beckman octyldecyl silane column (4.5 by 250 mm) protected by a Altex Ultrasphere- octyldecyl silane precolumn (4.6 by 4.5 mm). The column was eluted with a methanol-NaH2PO4 gradient as previously described (24). Muropeptides were detected at 206 nm. The degree of cross-linking of muropeptides was calculated as described by Stranden et al. (25) by the formula 0.5 × muropeptide dimers (%) + 0.67 × muropeptide trimers (%) + 0.9 × muropeptide oligomers (%).
Generation of passage-derived mutants.
Three methicillin-resistant non-VISA isolates (N315, 450M, and COL) were grown in broth containing no antibiotic to stationary phase. An inoculating loop (10 μl) of each culture was streaked onto vancomycin-containing MHA gradient plates (0 to 8 μg of vancomycin/ml). Gradient plates were incubated overnight at 37°C. Colonies growing on the highest concentration of vancomycin were picked and grown in broth containing a similar vancomycin concentration. This procedure was repeated until mutants with stable vancomycin MICs of 8 to 16 μg/ml were obtained for each isolate.
RESULTS
PBP analysis of VISA isolates.
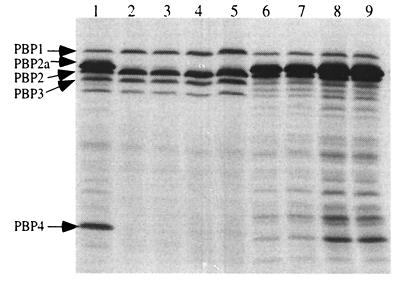
PBP analysis, performed on four clinical VISA isolates and vancomycin-susceptible ORSA isolate 450M (Fig. 1), revealed no or greatly reduced PBP4-binding activity, as assessed by the ability of proteins to bind radiolabeled penicillin, in three of the VISA isolates (HIP5827, HIP5836, and HIP6297). Pre-VISA isolate Mu3 had PBP4 activity. Its intermediately vancomycin-susceptible counterpart, Mu50, had detectable but decreased activity compared to that of Mu3. In addition, each of the VISA isolates was grown in vancomycin prior to PBP analysis and no change in PBP4 activity was detected. An additional five vancomycin-susceptible ORSA isolates have been examined during the course of other studies, and all have been shown to have detectable PBP4 (data not shown).
FIG. 1.
PBP profiles in the presence and absence of vancomycin. Lanes: 1, RN450M without vancomycin; 2, HIP5827 with vancomycin; 3, HIP5827 without vancomycin; 4, HIP5836 with vancomycin; 5, HIP5836 without vancomycin; 6, Mu50 with vancomycin; 7, Mu50 without vancomycin; 8, Mu3 with vancomycin; 9, Mu3 without vancomycin.
PBP4 after passage of VISA isolates in vitro
Three methicillin-resistant non-VISA isolates (COL, RN450M, and N315) were passaged on vancomycin-containing agar until isolates with stable MICs of 8 to 16 μg/ml were produced. None of the three passage-derived isolates, COLVR, RN450MVR, and N315VR, had any difference in PBP4 activity from that seen in the parent.
pbp4 nucleotide sequence in VISA isolates.
The pbp4 structural gene was sequenced in four clinical VISA (HIP5836, HIP5827, HIP6297, and Mu50) and four vancomycin-susceptible, oxacillin-resistant S. aureus (RN450M, 27619, N315, and Mu3) isolates. The predicted amino acid sequences of the pbp4 structural gene were identical among all of the isolates except RN450M, which differed by one amino acid, 409A→T (Table 2). Comparison of the four clinical VISA isolates (Mu50, HIP5836, HIP5827, and HIP6297) and three vancomycin-susceptible isolates (27169, N315, and Mu3) to the previously published sequences of pbp4 demonstrated a two-amino-acid difference from BB938-32 (accession number CAA62899.1), 234H→L and 409A→T; a two-amino-acid difference from 8325-4 (accession number CAA62899.1), 200R→A and 409A→T; and a two-amino-acid difference from SG511-55 (accession number CAA60581.1), 25T→A and 375G→R. None of these amino acid polymorphisms involved known functional motifs or the three predicted penicillin-binding motifs of PBP4. The nucleotide sequence of the 419-bp promoter-operator region 5′ to the start of the pbp4 structural gene was the same among all four VISA isolates (HIP5836, HIP5827, HIP6297, and Mu50), four vancomycin-susceptible S. aureus isolates (RN450M, 27619, N315, and Mu3), and the published sequences of BB938-32 (accession number X91786) and 8325-4 (accession number U29454) but differed from the published sequence of SG511-55 (accession number X87104) by 12 bases.
TABLE 2.
Amino acid polymorphisms in PBP4
| Strain | Amino acid at position:
|
||
|---|---|---|---|
| 200 | 234 | 409 | |
| BB938-32 | R | L | T |
| COL | R | L | T |
| 8325-4 | A | H | T |
| RN450M | R | H | T |
| N315 | R | H | A |
| HIP 5836 | R | H | A |
| HIP 5827 | R | H | A |
| HIP 6297 | R | H | A |
| Mu50 | R | H | A |
Complementation of pbp4 in VISA isolates.
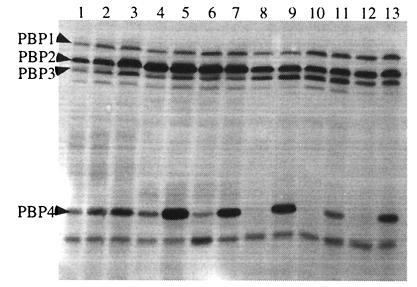
The entire pbp4 gene and 476 bases upstream of the pbp4 translation start site were ligated into an E. coli-staphylococcus shuttle plasmid that replicates in high copy in S. aureus. The resulting plasmid, designated pJF3, was transformed into each of the VISA isolates, and expression of PBP4 was confirmed by PBP analysis (Fig. 2.). All of the VISA isolates showed detectable PBP4 in radiolabeled penicillin-binding assays after transformation by pJF3. Despite the absence of detectable PBP4 among VISA isolates in penicillin-binding assays, Northern analysis demonstrated similar pbp4 transcript levels (data not shown) for both VISA (HIP5827, HIP5836, HIP6297, and Mu50) and vancomycin-susceptible isolates (RN450M, 27619, and Mu3). VISA isolates transformed with high-copy plasmid pJF3 demonstrated large increases in PBP4 transcripts (data not shown).
FIG. 2.
PBP profiles of control strains RN4220 (lane 1), RN4220/pJF3 (lane 2), and RN450M (lane 3) and VISA isolates Mu3 (lane 4), Mu3/pJF3 (lane 5), Mu50 (lane 6), Mu50/pJF3 (lane 7), HIP6297 (lane 8), HIP6297/pJF3 (lane 9), HIP 5836 (lane 10), HIP5836/pJF3 (lane 11), HIP5827 (lane 12), and HIP5827/pJF3 (lane 13).
Determination of vancomycin susceptibilities of pbp4-complemented VISA isolates.
MICs, determined by broth microdilution, were decreased for all of the VISA isolates containing plasmid-encoded pbp4 (Table 3) from threefold to almost fourfold (HIP5827). All strains had no change in vancomycin susceptibility with the plasmid vector alone (pRN5543). COL, RN450M, and Mu3, strains with detectable PBP4 activity, showed no vancomycin MIC change when pbp4 was introduced on a high-copy plasmid (pJF3). Loss of the plasmid restored the parental vancomycin susceptibility to each of the complemented VISA isolates.
TABLE 3.
Effect of PBP4 overproduction on vancomycin susceptibility
| Strain or isolate | Vancomycin MIC (μg/ml)a
|
|||||
|---|---|---|---|---|---|---|
| Broth
microdilutionb
|
E-test
|
|||||
| Parent | Parent + pJF3(pbp4) | Parent + pRN5543 | Parent | Parent + pJF3(pbp4) | Parent + pRN5543 | |
| RN450M | 2 | 2 | 2 | 2 | 2 | 2 |
| Mu3 | 2 | 2 | 2 | 3 | 2 | 3 |
| Mu50 | 6 | 3 | 6 | 8 | 3 | 8 |
| HIP5827 | 12 | 3 | 12 | 12 | 4 | 12 |
| HIP5836 | 8 | 2 | 8 | 8 | 6 | 8 |
| HIP6297 | 6 | 3 | 6 | 6 | 3 | 6 |
Microdilution MICs were determined at 1-μg/ml increments of vancomycin.
In order to keep pressure on plasmid pJF3, 5 μg of chloramphenicol per ml was also added. Failure to do so resulted in loss of the plasmid.
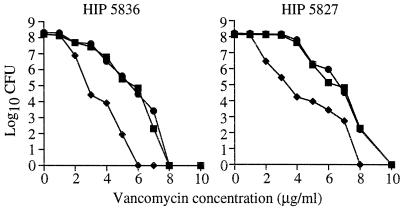
EOP experiments performed with two of the pbp4-complemented VISA isolates (HIP5827 and HIP5836) showed a decrease in highly vancomycin-resistant subpopulations in comparison to the parent or the parent containing the plasmid vector alone (Fig. 3).
FIG. 3.
EOP curves for HIP5836 and HIP5827. Shown are numbers of S. aureus bacteria (in log10 CFU per milliliter) remaining on plates containing various concentrations of vancomycin. Each parental VISA isolate is represented as a solid square. Each parental VISA isolate containing the cloning vector alone (pSK265) is represented as a solid circle. Each VISA isolate expressing pbp4 on high-copy plasmid pJF3 is represented as a solid diamond.
Inactivation of PBP4 in vancomycin-susceptible ORSA.
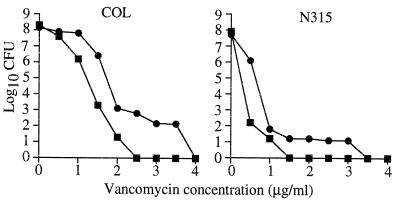
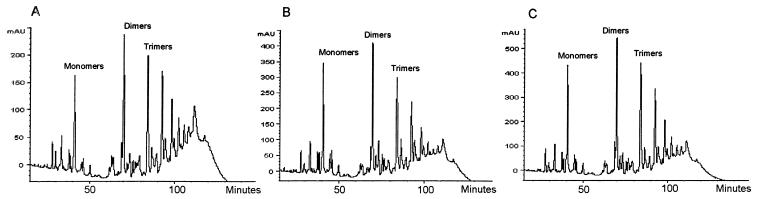
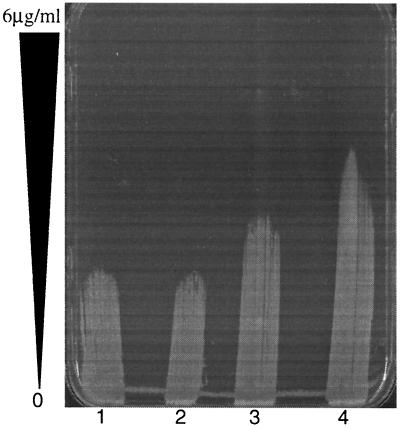
pbp4 was stably inactivated by allelic replacement mutagenesis in three vancomycin-susceptible isolates (N315, RN450M, and COL). Inactivation of PBP4 was confirmed in all three isolates by the absence of a band of the appropriate size in radiolabeled penicillin-binding assays (data not shown). Allelic replacement of pbp4 and secondary recombination to remove integrated plasmid sequences were confirmed by PCR. Following pbp4 inactivation, the vancomycin MIC for neither N315 nor 450M changed in broth microdilution testing using MHB in accordance with NCCLS guidelines (Table 4). However, there were discernible differences in the vancomycin susceptibilities of N315 and RN450M using the vancomycin E-test on BHI agar. Both N315 and RN450M demonstrated a reduction in vancomycin susceptibility following pbp4 inactivation. For COL, the vancomycin MIC increased from 2 to 4 μg/ml following pbp4 inactivation in broth microdilution testing and from 3 to 6 μg/ml in the E-test. Complementation of all three strains with pbp4 inactivated with the high-copy plasmid encoding pbp4 (pJF3) restored the vancomycin susceptibility to that of the parent strains. Subpopulations that were more highly resistant to vancomycin, as determined by EOP, were seen in all three strains following pbp4 inactivation. These subpopulations were most pronounced for COL and COL with pbp4 inactivated (Fig. 4). Inactivation of pbp4 was associated with a lower degree of cross-linking of oligomeric muropeptides, as demonstrated by cell wall analysis by HPLC (Fig. 5). The degrees of cross-linking for COL, 450M, and N315 (78.46, 77.33, and 61.24%, respectively) were substantially decreased in these isolates with pbp4 inactivated (65.06, 63.28, and 54.47%, respectively). Complementation of COL with pbp4 inactivated with high-copy plasmid pJF3 encoding pbp4 led to an increase in muropeptide cross-linking (Fig. 5C). The percentage of cross-linked muropeptide stem peptides increased from 65.06% in COL with pbp4 inactivated to 70.10% following transformation with pJF3 encoding pbp4. Finally, pbp4 inactivation was associated with a greater ability to develop higher levels of vancomycin resistance. Following overnight incubation in vancomycin (1 μg/ml), COL with pbp4 inactivated demonstrated a greater proportion of cells able to grow on higher levels of vancomycin in comparison to its parent with intact pbp4 (Fig. 6).
TABLE 4.
Effect of pbp4 inactivation on vancomycin susceptibility
| Strain | Vancomycin MIC
(μg/ml)a
|
|
|---|---|---|
| Broth microdilution | E-test | |
| RN450M | 2 | 2 |
| RN450MΔPBP4 | 2 | 3 |
| RN450MΔPBP4/pJF3b | 2 | 2 |
| N315 | 1 | 1 |
| N315ΔPBP4 | 1 | 3 |
| N315ΔPBP4/pJF3b | 1 | 1 |
| COL | 2 | 3 |
| COLΔPBP4 | 4.0 | 6 |
| COLΔPBP4/pJF3b | 2.0 | 3 |
MICs were determined by the broth microdilution and E-test methods as described in Materials and Methods.
MIC determination was done in the presence of chloramphenicol (5 μg/ml) to maintain pressure on pJF3.
FIG. 4.
EOP curves for pbp4 knockouts of COL and N315. Shown on the y axis are the numbers of S. aureus bacteria (in log10 CFU per milliliter) remaining on the plates containing various concentrations of vancomycin. Parent strains N315 and COL are represented by solid squares, and their isogenic pbp4 knockouts are represented by solid circles.
FIG. 5.
Analysis of peptidoglycan from S. aureus COL (A), COL with pbp4 inactivated following allelic mutagenesis (B), and COL with pbp4 inactivated complemented with pJF3 (C) by reverse-phase HPLC. COL with pbp4 inactivated demonstrates a lower degree of muropeptide cross-linking, as evidenced by the lower number of oligomeric muropeptides at the end of the chromatogram. Complementation of the inactivated pbp4 gene with pJF3 leads to a higher degree of cross-linking, as evidenced by the increase in trimers and oligomers in panel C. mAU, milliabsorption units.
FIG. 6.
Vancomycin gradient plate of COL and COLΔpbp4 before and after overnight exposure to vancomycin at 1 μg/ml. Lanes: 1, COL with no exposure; 2, COL after overnight exposure to vancomycin; 3, COLΔpbp4 with no exposure; 4, COLΔpbp4 after overnight exposure to vancomycin.
DISCUSSION
PBP4 is the only demonstrated LMW PBP present in S. aureus. (26). In vitro studies have demonstrated that PBP4 has DD-carboxypeptidase, transpeptidase, and β-lactamase activities (12). The extent to which these activities are expressed in vivo is speculative (8, 23, 25). Unlike LMW PBPs of other bacteria that are predominately carboxypeptidases, several lines of evidence indicate that PBP4 in S. aureus functions primarily as a secondary transpeptidase and has little or no in vivo DD-carboxypeptidase activity. First, the high level of cross-linking seen in the S. aureus cell wall makes extensive carboxypeptidase activity unlikely. Increased carboxypeptidase activity decreases cell wall cross-linking due to loss of the free d-Ala–d-Ala termini necessary for transpeptidation. Second, a mutant lacking PBP4 has been shown to have a cell wall with fewer cross-links, indicating decreased transpeptidation (19). In our own experiments, inactivation of PBP4 by allelic mutagenesis in three ORSA isolates (450M, N315, and COL) resulted in a lower degree of cross-linking, as demonstrated by HPLC analysis of muropeptides. Third, a mutant that overproduces PBP4 has been shown to have a more highly cross-linked cell wall than its parent (7), suggesting increased transpeptidation. Lastly, vancomycin-resistant passage mutants that lack PBP4 activity have also been shown to have a cell wall with decreased cross-linking (20).
In the present study, loss of PBP4 activity was a consistent finding in clinical VISA isolates. This loss of activity was shown to be directly related to vancomycin resistance by demonstrating a two- to threefold decrease in vancomycin resistance when PBP4 activity was provided in trans on a high-copy plasmid. Furthermore, pbp4 knockout mutants of vancomycin-susceptible strains COL, RN450M, and N315 had reduced susceptibility to vancomycin, as demonstrated by antimicrobial susceptibility testing. Interestingly, the decrease in vancomycin susceptibility was best demonstrated by the E-test using BHI agar. Previous investigators have also noted an increase in the detection of glycopeptide resistance using BHI-based media in place of MHB-based media in vancomycin susceptibility testing (1). pbp4 knockout mutants also demonstrated an increase in the number of highly vancomycin-resistant subpopulations compared to their parent strains with intact pbp4.
It is unclear how PBP4 activity is related to S. aureus vancomycin susceptibility. Several investigators have suggested that higher levels of resistance to vancomycin are associated with thicker cell walls and a lower degree of cell wall cross-linking, with more d-Ala–d-Ala-terminating cell wall muropeptides (5, 7). These changes could be caused by decreases in PBP4 activity, reducing secondary transpeptidation. This may allow greater vancomycin-binding capacity of the cell wall. Increases in the vancomycin-binding capacity of cell walls have been proposed to occur through three possible mechanisms. First, vancomycin could be bound throughout the cell wall due to increases in the entire cell wall mass. Second, a thicker cell wall could lead to greater binding along the entire periphery of the cell wall. Finally, greater binding of vancomycin at the periphery could lead to reduced passage of additional vancomycin molecules to the interior due to steric hindrance by the large vancomycin complex. However, it is unlikely that the vancomycin-binding capacity of cell walls is the sole explanation for decreases in vancomycin susceptibility. Previous studies with passage-derived glycopeptide-resistant mutants have shown that the vancomycin-binding capacity of cell walls correlates poorly with vancomycin MICs (16, 21).
Loss of PBP4 activity in association with higher levels of vancomycin resistance has been reported previously in passage-derived mutant VM (19, 22). However, several important differences between this laboratory-derived passage mutant and clinical VISA isolates should be noted. Most importantly, there is a large difference between the level of vancomycin resistance seen in clinical VISA isolates (vancomycin MIC, 8 μg/ml) and that seen in VM (vancomycin MIC, 100 μg/ml). During passage to high-level resistance to vancomycin, mutant VM gradually lost PBP4 activity, as measured by radiolabeled penicillin binding (22). Complete loss of detectable PBP4-binding activity was not seen until a vancomycin MIC of 50 μg/ml. The loss of detectable PBP4-binding activity at this level was associated with a mutation of the structural pbp4 gene causing premature translational termination (19). Similarly, the three strains for which the vancomycin MICs are similar to those for VISA isolates (8 to 16 μg/ml) that we generated in the present study by laboratory passage all still demonstrated detectable PBP4-binding activity, a finding similar to that seen in the passage of VM. These studies suggest that passage-derived VISA isolates may have fundamentally different mechanisms of acquired vancomycin resistance than clinical VISA isolates. Since our data indicate that the complete loss of PBP4 activity seen in all of the clinical VISA isolates tested was not associated with changes in the structural gene, the loss of PBP4 in the clinical VISA isolates appears to be posttranscriptional in nature.
The factors responsible for transcriptional regulation of pbp4 are unknown. Immediately 419 bp 5′ to pbp4 is a gene with an unknown function that has an ATP-binding cassette motif in its sequence, abcA. pbp4 and abcA are divergently transcribed and have overlapping promoter regions. Previous studies examining the role of abcA in the regulation of pbp4 have given conflicting results (4, 7). Berger-Bächi et al. found that PBP4 overproduction had no effect on abcA transcription. In that same study, it was also noted that a deletion in the putative promoter region of abcA resulted in increased PBP4 production but no change in abcA transcription (8). These data seemed to indicate the lack of a relationship between pbp4 and abcA. However, Bayles et al. found a sevenfold increase in pbp4 transcription in an abcA knockout mutant compared to the parent with intact abcA (5). It is reasonable to assume that two genes with overlapping and divergent regulatory sequences are coordinately regulated and share a functional pathway. However, any regulatory hypothesis would have to account for the observation that the PBP4 deficiency could be corrected and its quantity could be increased by increasing the pbp4 gene dosage.
In summary, we have shown that detectable binding of radiolabeled penicillin to PBP4 was not observed in any of the clinical VISA strains we examined and both the quantity of radiolabeled PBP4 and the susceptibility of VISA strains to vancomycin were increased by providing the cloned gene on a high-copy-number plasmid. These data suggest that loss of detectable PBP4-binding activity, which appears to result in less cell wall stem peptide cross-linking, is common to all clinical VISA isolates and therefore is likely to be essential for the development of this phenotype. Furthermore, since the altered expression of PBP4 appears to be regulatory or posttranscriptional in nature, finding the factor(s) responsible for this defect may lead to the discovery of other molecules with altered expression as a result of the VISA phenotype.
ACKNOWLEDGMENTS
This work was supported by NIH grant R-37AI3S705 and VA ment grant no. 0010.
We thank Gerithale Cooper, Elizabeth Hanners, and Katrina Williams for their technical assistance.
REFERENCES
- 1.Boyle-Vavra S, Labischinski H, Ebert C C, Ehlert K, Daum R S. A spectrum of changes occurs in peptidoglycan composition of glycopeptide-intermediate clinical Staphylococcus aureusisolates. Antimicrob Agents Chemother. 2001;45:280–287. doi: 10.1128/AAC.45.1.280-287.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Climo M W, Markowitz S, Williams D, Stuart G, Archer G. Comparison of the in vitro and in vivo efficacy of FKO37, vancomycin, imipenem, and nafcillin against staphylococcal species. J Antimicrob Chemother. 1997;40:59–66. doi: 10.1093/jac/40.1.59. [DOI] [PubMed] [Google Scholar]
- 3.Climo, M. W., V. K. Sharma, and G. L. Archer. Identification and characterization of the origin of conjugative transfer (oriT) and a gene (nes) encoding a single-stranded endonuclease on the staphylococcal plasmid pGO1. J. Bacteriol. 178:4975–4983. [DOI] [PMC free article] [PubMed]
- 4.Cui L, Murakami H, Kuwahara-Arai K, Hanaki H, Hiramatsu K. Contribution of a thickened cell wall and its glutamine nonamidated component to the vancomycin resistance expressed by Staphylococcus aureusMu50. Antimicrob Agents Chemother. 2000;44:2276–2285. doi: 10.1128/aac.44.9.2276-2285.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Domanski T L, de Jonge B L, Bayles K W. Transcription analysis of the Staphylococcus aureusgene encoding penicillin-binding protein 4. J Bacteriol. 1997;179:2651–2657. doi: 10.1128/jb.179.8.2651-2657.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hackbarth C J, Miick C, Chambers H F. Altered production of penicillin-binding protein 2a can affect phenotypic expression of methicillin resistance in Staphylococcus aureus. Antimicrob Agents Chemother. 1994;38:2568–2571. doi: 10.1128/aac.38.11.2568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hanaki H, Kuwahara-Arai K, Boyle-Vavra S, Daum R S, Labischinski H, Hiramatsu K. Activated cell-wall synthesis is associated with vancomycin resistance in methicillin-resistant Staphylococcus aureusclinical strains Mu3 and Mu50. J Antimicrob Chemother. 1998;42:199–209. doi: 10.1093/jac/42.2.199. [DOI] [PubMed] [Google Scholar]
- 8.Henze U U, Berger-Bächi B. Penicillin-binding protein 4 overproduction increases beta-lactam resistance in Staphylococcus aureus. Antimicrob Agents Chemother. 1996;40:2121–2125. doi: 10.1128/aac.40.9.2121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Henze U U, Berger-Bächi B. Staphylococcus aureuspenicillin-binding protein 4 and intrinsic beta-lactam resistance. Antimicrob Agents Chemother. 1995;39:2415–2422. doi: 10.1128/aac.39.11.2415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Henze U U, Roos M, Berger-Bächi B. Effects of penicillin-binding protein 4 overproduction in Staphylococcus aureus. Microb Drug Resist. 1996;2:193–199. doi: 10.1089/mdr.1996.2.193. [DOI] [PubMed] [Google Scholar]
- 11.Hiramatsu K. The emergence of Staphylococcus aureuswith reduced susceptibility to vancomycin in Japan. Am J Med. 1998;104:7S–10S. doi: 10.1016/s0002-9343(98)00149-1. [DOI] [PubMed] [Google Scholar]
- 12.Kozarich J W, Strominger J L. A membrane enzyme from Staphylococcus aureuswhich catalyzes transpeptidase, carboxypeptidase, and penicillinase activities. J Biol Chem. 1978;253:1272–1278. [PubMed] [Google Scholar]
- 13.National Committee for Clinical Laboratory Standards. Approved standard M7–A3. 1993. Dilution antimicrobial susceptibility tests for bacteria that grow aerobically. National Committee for Clinical Laboratory Standards, Villanova, Pa. [Google Scholar]
- 14.Niemeyer D M, Pucci M J, Thanassi J A, Sharma V K, Archer G L. Role of mecA transcriptional regulation in the phenotypic expression of methicillin resistance in Staphylococcus aureus. J Bacteriol. 1996;178:5464–5471. doi: 10.1128/jb.178.18.5464-5471.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Novick R. Properties of a cryptic high-frequency transducing phage in Staphylococcus aureus. Virology. 1967;33:155–166. doi: 10.1016/0042-6822(67)90105-5. [DOI] [PubMed] [Google Scholar]
- 16.Pfeltz R F, Singh V K, Schmidt J L, Batten M A, Baranyk C S, Nadakavukaren M J, Jayaswal R K, Wilkinson B J. Characterization of passage-selected vancomycin-resistant Staphylococcus aureusstrains of diverse parental backgrounds. Antimicrob Agents Chemother. 2000;44:294–303. doi: 10.1128/aac.44.2.294-303.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schenk S, Laddaga R A. Improved method for electroporation of Staphylococcus aureus. FEMS Microbiol Lett. 1992;73:133–138. doi: 10.1016/0378-1097(92)90596-g. [DOI] [PubMed] [Google Scholar]
- 18.Sharma V K, Hackbarth C J, Dickinson T M, Archer G L. Interaction of native and mutant MecI repressors with sequences that regulate mecA, the gene encoding penicillin binding protein 2a in methicillin-resistant staphylococci. J Bacteriol. 1998;180:2160–2166. doi: 10.1128/jb.180.8.2160-2166.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sieradzki K, Pinho M G, Tomasz A. Inactivated pbp4 in highly glycopeptide-resistant laboratory mutants of Staphylococcus aureus. J Biol Chem. 1999;274:18942–18946. doi: 10.1074/jbc.274.27.18942. [DOI] [PubMed] [Google Scholar]
- 20.Sieradzki K, Tomasz A. Gradual alterations in cell wall structure and metabolism in vancomycin-resistant mutants of Staphylococcus aureus. J Bacteriol. 1999;181:7566–7570. doi: 10.1128/jb.181.24.7566-7570.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sieradzki K, Tomasz A. Suppression of glycopeptide resistance in a highly teicoplanin-resistant mutant of Staphylococcus aureusby transposon inactivation of genes involved in cell wall synthesis. Microb Drug Resist. 1998;4:159–168. doi: 10.1089/mdr.1998.4.159. [DOI] [PubMed] [Google Scholar]
- 22.Sieradzki K, Wu S W, Tomasz A. Inactivation of the methicillin resistance gene mecA in vancomycin-resistant Staphylococcus aureus. Microb Drug Resist. 1999;5:253–257. doi: 10.1089/mdr.1999.5.253. [DOI] [PubMed] [Google Scholar]
- 23.Smith T L, Pearson M L, Wilcox K R, Cruz C, Lancaster M V, Robinson-Dunn B, Tenover F C, Zervos M J, Band J D, White E, Jarvis W R. Emergence of vancomycin resistance in Staphylococcus aureus. Glycopeptide-Intermediate Staphylococcus aureusWorking Group. N Engl J Med. 1999;340:493–501. doi: 10.1056/NEJM199902183400701. [DOI] [PubMed] [Google Scholar]
- 24.Snowden M A, Perkins H R. Cross-linking and O-acetylation of peptidoglycan in Staphylococcus aureus(strains H and MR-1) grown in the presence of sub-growth-inhibitory concentrations of beta-lactam antibiotics. J Gen Microbiol. 1991;137:1661–1666. doi: 10.1099/00221287-137-7-1661. [DOI] [PubMed] [Google Scholar]
- 25.Stranden A M, Ehlert K, Labischinski H, Berger-Bächi B. Cell wall monoglycine cross-bridges and methicillin hypersusceptibility in a femAB null mutant of methicillin-resistant Staphylococcus aureus. J Bacteriol. 1997;179:9–16. doi: 10.1128/jb.179.1.9-16.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wyke A W, Ward J B, Hayes M V, Curtis N A. A role in vivo for penicillin-binding protein-4 of Staphylococcus aureus. Eur J Biochem. 1981;119:389–393. doi: 10.1111/j.1432-1033.1981.tb05620.x. [DOI] [PubMed] [Google Scholar]