Abstract
Background and aims
The COVID-19 pandemic has prompted researchers to look for effective therapeutic targets. The effect of endocannabinoid system against infectious diseases is investigated for several years. In this study, we evaluated the expression level of CNR1 and CNR2 genes in patients with COVID-19 with and without diabetes to provide new insights regarding these receptors and their potential effect in COVID-19 disease.
Methods
In this study, peripheral blood monocytes cells (PBMCs) were isolated from eight different groups including COVID-19 patients, diabetic patients, and healthy individuals. RNA were extracted to evaluate the expression level of CNR1 and CNR2 genes using real-time PCR. The correlation between the expression levels of these genes in different groups were assessed.
Results
A total of 80 samples were divided into 8 groups, with each group consisting of ten samples. When comparing severe and moderate COVID-19 groups to healthy control group, the expression levels of the CNR1 and CNR2 genes were significantly higher in the severe and moderate COVID-19 groups. There were no significant differences between the mild COVID-19 group and the healthy control group. It was found that the expression levels of these genes in patients with diabetes who were infected with SARS-COV-2 did not differ across COVID-19 groups with varying severity, but they were significantly higher when compared to healthy controls.
Conclusion
Our study suggests the possible role of endocannabinoid system during SARS-COV-2 pathogenicity as the expression of CNR1 and CNR2 were elevated during the disease.
1. Introduction
In late 2019, the world has encountered a new coronavirus, SARS-COV-2, which has led to a worldwide pandemic. The virus results in a wide range of clinical presentations (most commonly associated with respiratory symptoms) causing COVID-19 disease [1].
According to reports, the elderly and comorbid patients are the most sensitive to the infection as compared to healthy people [2,3]. Several studies have suggested that the disease is associated with severe manifestations in patients with diabetes and uncontrolled blood glucose level [4,5]. Although the exact difference between diabetes types 1 and 2 outcomes in COVID-19 patients has yet to be determined, both groups have been found to have a higher risk of disease progression than the general population, [6].
While the immune system response (particularly the innate immune system) is critical in controlling viral infections, the exacerbated immune response and cytokine storm are two of the most common consequences in COVID-19 patients, which have been linked to disease severity [7]. Several COVID-19 therapies have been proposed to address this issue, and their efficacy is still being assessed [8].
Since the emergence of COVID-19, a number of studies have discussed the potential effect of cannabinoid use in these patients as a potent immune modulator [[9], [10], [11]]. The endocannabinoid system is found in several systems in human body, including CNS and the immune system. It is comprised of three key elements: 1) cannabinoid receptor 1 (CNR1), which is mostly expressed in the CNS, and cannabinoid receptor 2 (CNR2), which is expressed peripherally by the immune cells; 2) endogenous cannabinoid ligand (anandamide and 2-arachidonoylglycerol); and 3) metabolic enzymes [12]. Furthermore, multiple studies have highlighted the possible effect of targeting the endocannabinoid system in diabetic patients, as this system has been hypothesized to have a significant role in metabolic disorders [[13], [14], [15]]. To develop successful strategies to prevent or reduce SARS-COV-2 morbidity, a greater understanding of cannabinoid receptors and their role throughout the virus's pathogenicity is required. Therefore, in this study, we evaluated the expression level of CNR1 and CNR2 genes in patients with COVID-19 with and without diabetes to provide new insights regarding these receptors and their potential effect in COVID-19 disease.
2. Methods
2.1. -Study population
Present study was conducted in “Imam Ali” hospital, Alborz province, Iran, in June–December 2021. A trained infectious disease specialist confirmed the diagnosis of COVID-19 disease based on clinical manifestations, a computed tomography (CT) scan, and SARS-CoV-2 molecular detection (RT-qPCR). The status of COVID-19 patients is defined as: 1) severe: patients with O2 saturation less than 90%, 2) moderate: patients with respiratory rate≥24 per minute and O2 saturation between 90 and 93%, 3) mild: patients with respiratory rate <24 per minute and O2 saturation between≥94% (out-patient). All groups were matched on age and sex. Participants with a history of hypertension were excluded due to potential bias that can arise in the expression of endocannabinoid receptors in these individuals. Eligible participants were included in eight groups: A) Diabetic patients with severe COVID-19 disease, B) Non-diabetic patients with severe COVID-19 disease, C) Diabetic patients with moderate COVID-19 disease, D) Non-diabetic patients with moderate COVID-19 disease, Diabetic patients with mild COVID-19 disease, F) Non-diabetic patients with mild COVID-19 disease, G) Diabetic patients without COVID-19 disease and H) Non-diabetic patients (without COVID-19 disease). This study was approved by the ethics committee of Alborz university of medical sciences, Alborz, Karaj, Iran. (IR.ABZUMS.REC.1399.042).
2.2. -Sampling, RNA extraction and reverse-transcription polymerase chain reaction (RT - PCR) amplification of the CNR1 and CNR2 genes
Blood sample were collected from the participants after written informed consent was taken using EDTA tubes. Peripheral blood mononuclear cells (PBMCs) were isolated from the blood samples using Ficoll density gradient medium (Lympholyte H, Cedarlane, CANADA). Total RNA was extracted from PBMCs using RNA Extraction RNJia PB Kit (Roje, Iran) based on the manufacturers' guidelines. The RNA was then reverse transcribed using oligo dT primer as recommended by the RT-ROSET kit (ROJE, Iran). Gene expression of CNR1 and CNR2 was evaluated using SYBR Green-based RT-qPCR (TaKaRa, Otsu, Japan), and the Q-6000 machine (Qiagen, Germany) was used to perform relative 5 standard curves real-time PCR on the cDNA samples. The RPLP0 gene was used as a housekeeping gene for data normalization (Table 1 ).
Table 1.
Forward and reverse primers used for determining CNR1 and CNR2 gene expression.
| Forward | Reverse | |
|---|---|---|
| CNR1 | 5ʹ-CACCTTCCGCACCATCACCACT -3ʹ | 5ʹ-ACCTGGTCTGCTGGGACTAGCT-3ʹ |
| CNR2 | 5ʹ-CCTGTTCATTGGCAGCTTGGCT-3ʹ | 5ʹ-CGCAGGCAGAGGTATCGGTCAA-3ʹ |
| RPLP0 | 5-GACAAAGTGGGAGCCAGCGA-3 | 5-ACACCCTCCAGGAAGCGAGA-3 |
2.3. -Outcomes
The primary outcome was to compare CNR1 and CNR2 gene expression levels in healthy individuals and patients with different COVID-19 status. The secondary outcome was to address whether diabetes could change the expression level of these genes across the groups.
2.4. -Statistical analyses
All the statistical analyses were performed using Graph-Pad Prism software version 8 (GraphPad Software, Inc., San Diego, CA, USA). Kruskal–Wallis test was applied to determine between group differences. P-value < 0.05 was considered statistically significant.
3. Results
3.1. Study population
Total 80 participants included in this study, of them 43 (53.75%) were women and 37 were man (46.25%), with median age [IQR] 48 [21.25–54.75] years. A total of 10 participants were included in each group.
3.2. CNR1 gene expression
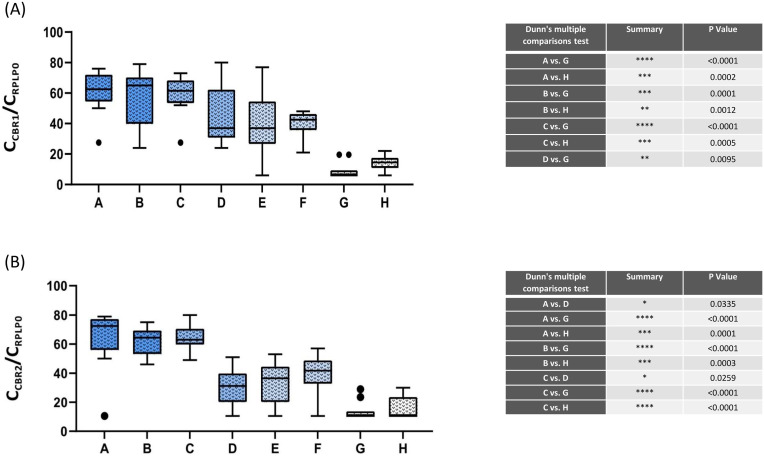
Based on the Kruskal–Wallis test, a significant difference was observed in CNR1 expression across groups (p < 0.0001). The mean (±SE) expression of CNR1 (Diabetic + severe COVID-19: 95.63 ± 28.26; non-diabetic + severe COVID-19: 115.3 ± 54.73; Diabetic + moderate COVID-19: 67.45 ± 17.01; Diabetic + moderate COVID-19: 126.7 ± 82.71; Diabetic + mild COVID-19: 58.85 ± 50.22; non-diabetic + mild COVID-19: 3.63 ± 0.95; Diabetic + without COVID-19: 0.002 ± 0.001; non-diabetic + without COVID-19: 0.004 ± 0.001) was compared pairwise using Dunn's multiple comparisons test. Significant statistical differences were observed between the following groups: 1) Diabetic + severe COVID-19 and Diabetic + without COVID-19 (p < 0.0001); 2) Diabetic + severe COVID-19 and non-diabetic + without COVID-19 (p = 0.0002); 3) non-diabetic + severe COVID-19 and Diabetic + without COVID-19 (p = 0.0001); 4) non-diabetic + severe COVID-19 and non-diabetic + without COVID-19 (p = 0.0012); 5) Diabetic + moderate COVID-19 and Diabetic + without COVID-19 (p < 0.0001); 6) Diabetic + moderate COVID-19 and non-diabetic + without COVID-19 (p = 0.0005); 7) Non diabetic + moderate COVID-19 and Diabetic + without COVID-19 (p = 0.0095). (Fig. 1 A).
Fig. 1.
The expression levels of cannabinoid receptor 1(A)and cannabinoid receptor 2(B) in COVID-19 and healthy individuals. Overall, a significant difference was observed in cannabinoid receptor expression level between the SARS-CoV-2 infected group compared with non-infected individuals, suggesting the possible role of the endocannabinoid system during the infection. Error bars represent minimum and maximum values in each group. Outliers were shown by black dots in the figure. Abbreviations: Group A: Diabetic patients with severe COVID-19 disease, Group B: Non-diabetic patients with severe COVID-19 disease, Group C: Diabetic patients with moderate COVID-19 disease, Group D: Non-diabetic patients with moderate COVID-19 disease, Group E: Diabetic patients with mild COVID-19 disease, Group F: Non-diabetic patients with mild COVID-19 disease, Group G: Diabetic without COVID-19 disease, Group H: Non-diabetic patients without COVID-19 infection.
3.3. CNR2 gene expression
Based on the Kruskal–Wallis test, a significant difference was observed in CNR2 expression across groups (p < 0.0001). The mean (±SE) expression of CNR2 (Diabetic + severe COVID-19: 61.07 ± 13.61; non-diabetic + severe COVID-19: 38.49 ± 9.64; Diabetic + moderate COVID-19: 44.35 ± 12.27; non-diabetic + moderate COVID-19: 1.11 ± 0.50; Diabetic + mild COVID-19: 1.74 ± 0.74; non-diabetic + mild COVID-19: 2.75 ± 0.94; Diabetic + without COVID-19: 0.006 ± 0.004; non-diabetic + without COVID-19: 0.010 ± 0.006) was compared pairwise using Dunn's multiple comparisons test. Significant statistical differences were observed between the following groups: 1) Diabetic + severe COVID-19 and non-diabetic + moderate COVID-19 (p = 0.033); 2) Diabetic + severe COVID-19 and Diabetic + without COVID-19 (p < 0.0001); 3) Diabetic + severe COVID-19 and non-diabetic + without COVID-19 (p = 0.0001); 4) non-diabetic + severe COVID-19 and Diabetic + without COVID-19 (p < 0.0001); 5) non-diabetic + severe COVID-19 and non-diabetic + without COVID-19 (p = 0.0003); 6) Diabetic + moderate COVID-19 and non-diabetic + moderate COVID-19 (p = 0.025); 7) Diabetic + moderate COVID-19 and Diabetic + without COVID-19 (p < 0.0001); 8) Diabetic + moderate COVID-19 and non-diabetic + without COVID-19 (p < 0.0001). (Fig. 1B).
4. Discussion
To our knowledge, this is the first study investigating the expression levels of cannabinoid receptors (CNR) in COVID-19 patients with and without diabetes mellitus. The precise role of this receptor implicated in the pathogenicity of COVID-19 patients is still unknown. Herein, we tried to shed a light on the differences between the receptor expressions in 8 different groups.
The result obtained from this study revealed elevated both CNR1 and CNR2 expression in patients with COVID-19 in comparison with healthy individuals. This difference was statistically significant primarily between COVID-19 patients with severe and moderate disease and healthy individuals who did not have the disease. Generally, there was no significant difference in CNR1 and CNR2 expression across the COVID-19 groups, except for diabetes with the severe COVID-19 group and non-diabetic with moderate COVID-19, and diabetes with moderate COVID-19 group and non-diabetic with moderate COVID-19, where the difference was statistically significant in CNR2 expression.
Till now, many treatments have been proposed for treating COVID-19. A number of these drugs deliver their efficacy through regulating the over-reacted immune system. A massive release of cytokines that contribute to the cytokine storm might cause extensive lung inflammation in some people causing fatal COVID-19 cases [16]. Based on this fact, corticosteroids, which operate as an immune suppressor, are one of the most beneficial treatments for severe patients [17]. Because of the limited results available, additional therapeutic approaches must be considered until certain medications are developed. One of these approaches was the use of cannabinoids (CBDs) in treating COVID-19 patients based on their modulation function in immune system [[9], [10], [11]]. As previously stated, the endocannabinoid system is comprised of endocannabinoids such as anandamide (AEA) and 2-arachidonoylglycerol (2-AG), enzymes that degrade ligands, and cannabinoid receptors (mostly CNR1 and CNR2, but other receptors such as GPR55, GPR18, PPAR-a, PPAR-b, and TRPV1 are also activated by ligands, although they are not considered as an individual cannabinoid receptor) [10,18]. Exogenous cannabinoids are mainly derived from Cannabis sativa L [19]. Some of which is Δ9-tetrahydrocannabinol (THC) and cannabidiol, which are partial CNR1 and CNR2 agonists, respectively [20,21].
CNRs' increased expression suggests that they may play a role in immune system functions [22]. Until now, few research has been done when it comes to the effect of cannabinoids in viral infections. An in-vitro study recently publisehed by Breeman et al. indicates the possible effect of Cannabinoids in blocking SARS-COV-2 cell entry. The use of cannabigerolic and cannabidiolic acids prevented infection by blocking pseudoviruses producing spike protein and the live SARS-CoV-2 virus from entering the living cell [11]. A study done by Krishnan et al. investigated the role of endocannabinoids on modulating innate immune response during HIV-1 Tat cytotoxicity [23]. By eliciting a cytokine response in CNS cells, the HIV-1 Tat protein has been linked to neurodegeneration. The results of the study showed that both endocannabinoids had a potent role in regulating cytokine production by altering the NF–B complex, which includes IRAK1BP1 and TAB2, at the transcriptional level [23]. Another study by Mestre et al. showed the AEA effect on Theiler's virus (a murine virus of the picornavirus family) induced VCAM-1 in CNS endothelial cells. Their findings imply that VCAM-1 suppression mediated by the CNR1 is an unique mechanism for AEA-reduced leukocyte transmigration suggesting the immune modulatory effect of the cannabinoid system [24]. Several studies on respiratory syncytial virus (RSV) infection also revealed the CNR1 and CNR2 activation could potentially reduce the signs of infection [25,26]. Furthermore, a CNR2 gene polymorphism (Q63R, a missense mutation that reduces the endocannabinoid system response) has recently been associated with a greater risk of COVID-19 disease severity [27]. According to the present study, patients with COVID-19 disease had a higher level of CNR gene expression than healthy people. It's worth noting that the rate of expressing these receptors increases as the disease advances, which could be attributed to the increased necessity for immunomodulatory effects of this system during disease pathogenicity. However, the differences between COVID-19 groups were not statistically significant and are just based on observational findings.
Many studies have been conducted to determine the exact role of the endocannabinoid system in energy homeostasis and metabolic diseases such as diabetes [[13], [14], [15],28]. Despite the numerous debates in this sector, it has been widely accepted that the use of CB1 receptor antagonists and CB2 receptor agonists have potential therapeutic effects due to the signaling disruption caused by metabolic disorders. [29]. Therefore, in addition to our primary outcomes, we also investigated whether COVID-19 patients with diabetes have different CNR expression compared with non-diabetics. As the study findings show, there were no notable differences in the expression levels of CNR1 and CNR2 between diabetic and non-diabetic COVID-19 patients. The scope of this article extends beyond a detailed explanation of the endocannabinoid system's function and its pathogenic role during diabetes.
In conclusion, the outcomes of this research supports the possible role of endocannabinoid system during SARS-COV-2 pathogenicity as the expression of CNR1 and CNR2 were elevated during the disease. Moreover, despite their limitations due to psychiatric side effects, the regulated use of cannabinoids should be examined by researchers to identify their potential effectiveness as a therapeutic target in COVID-19 disease.
Financial Support
This study was funded by Alborz University of Medical Sciences (Grant number: 3770).
Declaration of competing interest
No Conflict of interest.
Acknowledgement
Not applicable.
References
- 1.Guo Y.-R., Cao Q.-D., Hong Z.-S., Tan Y.-Y., Chen S.-D., Jin H.-J., et al. vol. 7. 2020. pp. 1–10. (The origin, transmission and clinical therapies on coronavirus disease 2019 (COVID-19) outbreak–an update on the status). 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen Y., Klein S.L., Garibaldi B.T., Li H., Wu C., Osevala N.M., et al. Aging in COVID-19: vulnerability, immunity and intervention. Ageing Res Rev. 2021;65 doi: 10.1016/j.arr.2020.101205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Perrotta F., Corbi G., Mazzeo G., Boccia M., Aronne L., D'Agnano V., et al. COVID-19 and the elderly: insights into pathogenesis and clinical decision-making. Aging Clin Exp Res. 2020;32(8):1599–1608. doi: 10.1007/s40520-020-01631-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhu L., She Z.-G., Cheng X., Qin J.-J., Zhang X.-J., Cai J., et al. vol. 31. 2020. pp. 1068–1077. (Association of blood glucose control and outcomes in patients with COVID-19 and pre-existing type 2 diabetes). 6.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Petrilli C.M., Jones S.A., Yang J., Rajagopalan H., O'Donnell L., Chernyak Y., et al. vol. 369. 2020. (Factors associated with hospital admission and critical illness among 5279 people with coronavirus disease 2019 in New York City: prospective cohort study). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barron E., Bakhai C., Kar P., Weaver A., Bradley D., Ismail H., et al. Associations of type 1 and type 2 diabetes with COVID-19-related mortality in England: a whole-population study. Lancet Diabetes Endocrinol. 2020;8(10):813–822. doi: 10.1016/S2213-8587(20)30272-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jiang Y., Rubin L., Peng T., Liu L., Xing X., Lazarovici P., et al. Cytokine storm in COVID-19: from viral infection to immune responses, diagnosis and therapy. Int J Biol Sci. 2022;18(2):459–472. doi: 10.7150/ijbs.59272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Siemieniuk R.A., Bartoszko J.J., Ge L., Zeraatkar D., Izcovich A., Kum E., et al. 2020. Drug treatments for covid-19: living systematic review and network meta-analysis; p. 370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.El Biali M., Broers B., Besson M., Demeules J. Cannabinoids and COVID-19. Medical cannabis and cannabinoids. 2020;3(2):111–115. doi: 10.1159/000510799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lucaciu O., Aghiorghiesei O., Petrescu N.B., Mirica I.C., Benea H.R.C., Apostu D. In quest of a new therapeutic approach in COVID-19: the endocannabinoid system. Drug Metab Rev. 2021;53(4):478–490. doi: 10.1080/03602532.2021.1895204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.van Breemen R.B., Muchiri R.N., Bates T.A., Weinstein J.B., Leier H.C., Farley S., et al. Cannabinoids block cellular entry of SARS-CoV-2 and the emerging variants. J Nat Prod. 2022;85(1):176–184. doi: 10.1021/acs.jnatprod.1c00946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Farquhar-Smith W.P., Egertová M., Bradbury E.J., McMahon S.B., Rice A.S., Elphick M.R.J.M., et al. vol. 15. 2000. pp. 510–521. (Cannabinoid CB1 receptor expression in rat spinal cord). 6. [DOI] [PubMed] [Google Scholar]
- 13.Borowska M., Czarnywojtek A., Sawicka-Gutaj N., Woliński K., Płazińska M.T., Mikołajczak P., et al. The effects of cannabinoids on the endocrine system. Endokrynol Pol. 2018;69(6):705–719. doi: 10.5603/EP.a2018.0072. [DOI] [PubMed] [Google Scholar]
- 14.Gonçalves E.D., Dutra R.C. Cannabinoid receptors as therapeutic targets for autoimmune diseases: where do we stand? Drug Discov Today. 2019;24(9):1845–1853. doi: 10.1016/j.drudis.2019.05.023. [DOI] [PubMed] [Google Scholar]
- 15.Silvestri C., Di Marzo V. The endocannabinoid system in energy homeostasis and the etiopathology of metabolic disorders. Cell Metabol. 2013;17(4):475–490. doi: 10.1016/j.cmet.2013.03.001. [DOI] [PubMed] [Google Scholar]
- 16.Onaivi E.S., Sharma V. Cannabis for COVID-19: can cannabinoids quell the cytokine storm? Future science OA. 2020;6(8) doi: 10.2144/fsoa-2020-0124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Siemieniuk R.A., Bartoszko J.J., Ge L., Zeraatkar D., Izcovich A., Kum E., et al. vol. 370. 2020. (Drug treatments for covid-19: living systematic review and network meta-analysis). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hernández-Cervantes R., Méndez-Díaz M., Prospéro-García Ó., Morales-Montor J. Immunoregulatory role of cannabinoids during infectious disease. Neuroimmunomodulation. 2017;24(4–5):183–199. doi: 10.1159/000481824. [DOI] [PubMed] [Google Scholar]
- 19.Hanuš L.O., Meyer S.M., Muñoz E., Taglialatela-Scafati O., Appendino G. Phytocannabinoids: a unified critical inventory. Nat Prod Rep. 2016;33(12):1357–1392. doi: 10.1039/c6np00074f. [DOI] [PubMed] [Google Scholar]
- 20.Pertwee R.G. Pharmacological actions of cannabinoids. Handb Exp Pharmacol. 2005;(168):1–51. doi: 10.1007/3-540-26573-2_1. [DOI] [PubMed] [Google Scholar]
- 21.Pisanti S., Malfitano A.M., Ciaglia E., Lamberti A., Ranieri R., Cuomo G., et al. Cannabidiol: state of the art and new challenges for therapeutic applications. Pharmacol Therapeut. 2017;175:133–150. doi: 10.1016/j.pharmthera.2017.02.041. [DOI] [PubMed] [Google Scholar]
- 22.Rieder S.A., Chauhan A., Singh U., Nagarkatti M., Nagarkatti P. Cannabinoid-induced apoptosis in immune cells as a pathway to immunosuppression. Immunobiology. 2010;215(8):598–605. doi: 10.1016/j.imbio.2009.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Krishnan G., Chatterjee N. Endocannabinoids affect innate immunity of Muller glia during HIV-1 Tat cytotoxicity. Molecular and cellular neurosciences. 2014;59:10–23. doi: 10.1016/j.mcn.2014.01.001. [DOI] [PubMed] [Google Scholar]
- 24.Mestre L., Iñigo P.M., Mecha M., Correa F.G., Hernangómez-Herrero M., Loría F., et al. Anandamide inhibits Theiler's virus induced VCAM-1 in brain endothelial cells and reduces leukocyte transmigration in a model of blood brain barrier by activation of CB(1) receptors. J Neuroinflammation. 2011;8:102. doi: 10.1186/1742-2094-8-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tahamtan A., Samieipoor Y., Nayeri F.S., Rahbarimanesh A.A., Izadi A., Rashidi-Nezhad A., et al. Effects of cannabinoid receptor type 2 in respiratory syncytial virus infection in human subjects and mice. Virulence. 2018;9(1):217–230. doi: 10.1080/21505594.2017.1389369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tahamtan A., Tavakoli-Yaraki M., Shadab A., Rezaei F., Marashi S.M., Shokri F., et al. The role of cannabinoid receptor 1 in the immunopathology of respiratory syncytial virus. Viral Immunol. 2018;31(4):292–298. doi: 10.1089/vim.2017.0098. [DOI] [PubMed] [Google Scholar]
- 27.Rastegar M., Samadizadeh S., Yasaghi M., Moradi A., Tabarraei A., Salimi V., et al. Functional variation (Q63R) in the cannabinoid CB2 receptor may affect the severity of COVID-19: a human study and molecular docking. Arch Virol. 2021;166(11):3117–3126. doi: 10.1007/s00705-021-05223-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Deeba F., Kumar A., Mukherjee M., Sharma A.K., Sharma M. Targeting the endocannabinoid system in diabesity: fact or fiction? Drug Discov Today. 2021;26(7):1750–1758. doi: 10.1016/j.drudis.2021.03.022. [DOI] [PubMed] [Google Scholar]
- 29.Gruden G., Barutta F., Kunos G., Pacher P. vol. 173. 2016. pp. 1116–1127. (Role of the endocannabinoid system in diabetes and diabetic complications). 7. [DOI] [PMC free article] [PubMed] [Google Scholar]