Abstract
A survey of 1,203 Escherichia coli isolates from 44 hospitals in Taiwan revealed that 136 (11.3%) isolates were resistant to fluoroquinolones and that another 261 (21.7%) isolates had reduced susceptibility. Resistance was more common in isolates responsible for hospital-acquired (mostly in intensive care units) infections (17.5%) than in other adult inpatient (11.4%; P = 0.08) and outpatient isolates (11.9%; P > 0.1). Similarly, reduced susceptibility was more common in isolates responsible for hospital-acquired infections (30.9%) than in other adult inpatient (21.0%; P = 0.04) and outpatient (21.4%; P = 0.06) isolates. Isolates from pediatric patients were less likely to be resistant (1.3 versus 12.0%; P < 0.01) but were nearly as likely to have reduced susceptibility (17.7 versus 21.9%; P > 0.1) as nonpediatric isolates. There was an inverse relationship in the proportion of isolates that were resistant versus the proportion that had reduced susceptibility among isolates from individual hospitals (R = 0.031; P < 0.05). In an analysis of isolates from two hospitals, all 9 resistant strains possessed double point mutations in gyrA and all 19 strains with reduced susceptibility strains had single point mutations; no mutations were found among fully susceptible strains. Risk factors for resistance included underlying cancer (odds ratio [OR], 83; 95% confidence interval [CI95], 7.3 to 2,241; P < 0.001), exposure to a quinolone (OR, undefined; P = 0.02), and exposure to a nonquinolone antibiotic (OR, 20; CI95, 2.2 to 482; P < 0.001); underlying cancer was the only independent risk factor (OR, 83; CI95, 8.6 to 807; P < 0.001). There were no significant associations between any of these factors and reduced susceptibility. Whereas acute and chronic quinolone use in cancer patients is a major selective pressure for resistance, other undetermined but distinct selective pressures appear to be more responsible for reduced susceptibility to fluoroquinolones in E. coli.
The fluoroquinolones are an important class of antibiotics for the treatment of urinary tract infections and other common infections caused by gram-negative bacteria (11, 13, 19). This is largely due to their excellent activities against Escherichia coli, one of the most frequently encountered human bacterial pathogens (27). It is because of their value in human medicine that resistance to fluoroquinolones in E. coli has been viewed with great concern wherever it has begun to emerge, including in parts of Spain (4, 8), other European countries (10, 16, 21, 22), and various countries of East Asia (23, 28, 32). Although there are a few reports of resistance in North America (3), overall rates remain low (1, 26). Recognized clinical risk factors for fluoroquinolone resistance in E. coli include the therapeutic and prophylactic use of fluorquinolones in cancer patients (4, 24, 32) as well as fluoroquinolone use in patients with chronic indwelling urinary catheters or other urinary tract abnormalities (2, 3, 6, 8).
Mutations at the target site appear to be the major mechanism for fluoroquinolone resistance in E. coli (25). In addition, recent work suggests that highly resistant strains of E. coli may also have mutations affecting expression of efflux pumps such as AcrAB (15, 30). Fluoroquinolones possess bacteriocidal activity by the formation of enzyme-DNA complexes involving DNA gyrase and topoisomerase enzymes responsible for unwinding of the bacterial chromosomal during replication; point mutations conferring resistance are localized to particular portions of gyrA, which is a gyrase subunit gene, and parC, which encodes a topoisomerase subunit. In the case of E. coli, mutations in gyrA are predictive of major differences in the level of resistance, irrespective of mutations in parC (12, 23, 31). Whereas high-level resistance in other members of the family Enterobacteriaceae may result from single mutations in gyrA, possibly all E. coli strains with high-level resistance possess double mutations (31).
In contrast, E. coli strains with single mutations in gyrA remain susceptible to the fluoroquinolones. However, the MICs of the fluoroquinolones for these strains are markedly increased (i.e., they have reduced susceptibility) and, like isolates with double mutations, demonstrate resistance to nonfluorinated quinolones such as nalidixic acid. According to studies of mutant E. coli strains selected in vitro, mutations in gyrA result first in a substitution of serine 83 and then in a substitution of aspartate 87 (12). Results of surveys of mutant E. coli found in vivo appear to be consistent with the stepwise occurrence of mutations observed in vitro; nearly all mutants with a single-site mutation have substitutions at serine 83, and most isolates with substitutions at aspartate 87 also have substitutions at serine 83 (7, 23).
Although several reports have described the epidemiology of E. coli with full resistance (i.e., double mutants) (2–4, 6, 8, 24, 32) and a substantial prevalence of E. coli isolates with reduced susceptibility has been reported in Latin America (7), no reports have detailed the epidemiology of reduced susceptibility to fluoroquinolones (i.e., mutants with single-site mutations). Whether risk factors for reduced susceptibility differ from risk factors for resistance and how rates of reduced susceptibility relate to rates of resistance in a community is unknown. Based upon preliminary results of a nationwide survey of antibiotic resistance performed in 1998, we found that over 10% of E. coli isolates in Taiwan were resistant to the fluoroquinolones (14). Moreover, we found another large proportion of E. coli isolates that, although susceptible, possessed reduced susceptibility to fluoroquinolones. These findings prompted us to investigate the epidemiology of emerging fluoroquinolone resistance in Taiwan. Our objectives were to determine the relationship between reduced susceptibility and resistance and, if related, to identify the antibiotic pressure most responsible for each level of resistance. We sought to achieve these objectives through analysis of molecular resistance mechanisms, molecular epidemiology, and clinical risk factors.
MATERIALS AND METHODS
Collection and testing of clinical isolates.
Clinical isolates of E. coli were collected between August and December 1998 from 44 hospitals including 10 medical centers and 34 regional hospitals participating in the Taiwan Surveillance of Antimicrobial Resistance program sponsored by the National Health Research Institutes in Taipei, Taiwan. The medical center status of hospitals is determined and updated regularly by the Taiwan Department of Health on the basis of a larger bed number, an increased proportion of intensive care unit beds, and a medical school affiliation. The hospitals were distributed throughout four geographic regions: 20 in the north, 10 in the south, 10 in the west, and 4 in the sparsely populated east. Each hospital was asked to provide approximately 150 sequential isolates (i.e., isolates were not limited to E. coli) from individual patients (i.e., 1 isolate per patient) in each of four specified categories: 50 from adult inpatients (including at least 25 recovered from blood cultures), 25 from adult outpatients, 25 from adults with documented hospital-acquired infections, and 25 from pediatric patients, if available. Hospital-acquired infections were identified by infection control personnel at each hospital as part of their routine surveillance for nosocomial infections. Surveillance in most participating hospitals was concentrated in the intensive care unit and used definitions based upon the surveillance case definitions for nosocomial infections of the U.S. Centers for Disease Control and Prevention (Atlanta, Ga.) (9).
All isolates were stored frozen (−70°C) in a central laboratory, where the species identification was confirmed by standardized methods. Identification of E. coli was based on Gram staining, colony morphology, spot indole, oxidase, and β-glucoronidase test results and/or results from the Vitek Gram Negative automated identification system (bioMérieux, St. Louis, Mo.). Antimicrobial susceptibility tests were performed with all isolates by the disk diffusion method according to standards developed by the National Committee for Clinical Laboratory Standards (20). In addition to ciprofloxacin, the antimicrobials tested included amikacin, ampicillin, aztreonam, amoxicillin-clavulanate, ceftazidime, cefazolin, cefuroxime, gentamicin, piperacillin, and trimethoprim-sulfamethoxazole. Zones of inhibition were measured with a BIOMIC video reader (Giles Scientific Inc., Santa Barbara, Calif.).
Additional antimicrobial susceptibility testing was performed with a subset of isolates from two hospitals. One hospital, Cathay General Hospital, is a regional hospital located in Taipei, in the northern region of Taiwan. The other, Kaoshiung Medical University Hospital, is a medical center located in the southern city of Kaoshiung. Disk diffusion testing with ofloxacin and nalidixic acid was performed with these isolates, in addition to determination of the MICs of ciprofloxacin and ofloxacin. MIC determinations were performed by gradient agar diffusion by Etests (AB Biodisk, Solna, Sweden).
Molecular assessment of resistance mutations and strain typing.
All resistant isolates, all isolates with reduced susceptibility (see definitions below), and a sample of fully susceptible E. coli isolates from Cathay General and Kaoshiung Medical University Hospitals were assessed for point mutations in the quinolone resistance-determining regions of gyrA and parC. The oligonucleotide primers used for amplification of the gyrA fragment were those described by Weigel et al. (31) Primer gyrA6 (5′-CGACCTTGCGAGAGAAAT-3′) corresponded to nucleotides 6 to 23, and primer gyrA631R (5′-GTTCCATCAGCCCTTCAA-3′) was complementary to nucleotides 631 to 614 of the E. coli gyrA sequence. Primer HJL3 (5′-AATGAGCGATATGGCAGAGC-3′) corresponded to nucleotides −1 to 19 of the E. coli parC sequence, and primer HJL4 (5′-CTGGTCGATTAATGCGATTG-3′) was complementary to nucleotides 594 to 575 of the E. coli parC sequence.
The gyrA and parC gene fragments were amplified from the chromosomal DNA present in crude cell lysates, prepared as follows. A single colony was suspended in 60 μl of distilled water, and the suspension was boiled for 10 min to prepare templates for PCR. The crude cell lysate was diluted 10-fold in distilled water and was either used immediately or stored at −20°C until needed.
Amplifications were carried out with a Peltier Thermal Cycler PTC-200 PCR System (MJ Research Inc., Waltham, Mass.) in 50-μl volumes containing 50 pmol of each primer, 200 μM deoxynucleotide triphosphate, 1× reaction buffer (10 mM Tris-HCl [pH 9], 50 mM KCl, 1.5 mM MgCl2), 1 U of Taq polymerase (Amersham Pharmacia Biotech Inc., Uppsala, Sweden), and 10 μl of diluted cell lysate. An initial 5-min period of denaturation at 95°C was followed by 35 cycles of denaturation (1 min at 94°C), annealing (1 min at 60°C), and extension (1 min at 72°C), followed by a final cycle of 72°C for 15 min. Agarose gel electrophoresis and ethidium bromide staining to confirm the sizes of the gene fragments were used to visualize the amplification products. PCR products were purified on QIA quick spin columns (QIAGEN, Chatsworth, Calif.) according to the manufacturer's instructions. The DNA sequences were determined with a Dye Terminator cycle sequencing kit with a PE Applied Biosystems 377 DNA sequencer (Applera Corporation, Norwalk, Conn.). To eliminate errors caused by amplification artifacts, the forward and reverse sequences of each quinolone resistance-determining region were determined.
Molecular typing of genomic DNA was performed by pulsed-field gel electrophoresis (PFGE) with a temperature-controlled CHEF MAPPER System (Bio-Rad Laboratories, Hercules, Calif.). Extraction and digestion of genomic DNA were done according to the manufacturer's instructions for the GenePath Group 6 Reagent kit (Bio-Rad). The agarose plugs were digested with 50 U of XbaI, and PFGE was performed in 1% Pulsed Field Certified Agarose (Bio-Rad). Gels were electrophoresed for 28 h at 14°C at a constant voltage of 6 V/cm, with pulse times of 6.75 to 44.69 s with linear ramping. Bacteriophage lambda ladders (Bio-Rad) were used as markers.
After staining with ethidium bromide, restriction fragments were imaged with an IS-1000 Digital Imaging System (Alpha Innotech Corporation, San Leandro, Calif.). PFGE pattern analysis was performed with CHEF Mapper XA interactive software (version 1.2; Bio-Rad). Cluster analysis was performed by the unweighted pair group method with arithmetic averages with the Jaccard coefficient; dendrograms were prepared.
Clinical assessment of potential risk factors.
Available medical records of all patients infected or colonized with E. coli submitted from Cathay General and Kaoshiung Medical University Hospitals were reviewed. This review included inpatient and outpatient records from at least 8 weeks before collection of the culture positive for E. coli through 2 weeks after collection. Data collected included patient demographics, hospitalizations, underlying disease, invasive device use, and prior exposure to antimicrobials.
Data analysis and definitions.
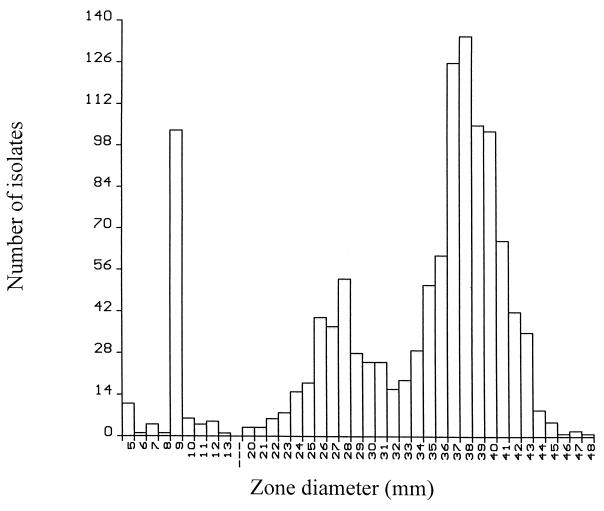
All antimicrobial susceptibility results, including zone diameters, were downloaded from the BIOMIC software, converted in file format, and analyzed with Epi Info software (version 6.04; Centers for Disease Control and Prevention). A histogram of ciprofloxacin zone diameters was produced (see Results) and demonstrated the existence of two distinct populations of ciprofloxacin-susceptible isolates (zone diameters, >15 mm). On the basis of the zone size distributions of the isolates, we defined full susceptibility as a zone diameter of 32 mm and reduced susceptibility as a zone diameter of 16 to 31 mm.
Clinical data were entered into a relational database designed in Access 97 software (Microsoft, Redland, Wash.), converted into a file format, and analyzed with Epi Info software (version 6.04). Associations between various potential risk factors (e.g., underlying diseases, invasive devices, and prior antimicrobials) and outcomes of full or reduced susceptibility or resistance were determined. For the analysis of clinical risk factors, full susceptibility was redefined on the basis of nalidixic acid susceptibility (zone diameter, >16 mm) and the absence of resistance mutations in the case in which an isolate with a ciprofloxacin zone diameter of 31 mm was misclassified as having reduced susceptibility (see Results and Table 2).
TABLE 2.
Quinolone susceptibility and mutations in the gyrA and parC genes of 36 E. coli isolates from two hospitals in Taiwan
| Isolate no.a | Disk zone diam (mm)b
|
MIC (μg/ml)
|
Amino acid change
|
|||||||
|---|---|---|---|---|---|---|---|---|---|---|
| CIP | OFX | NAL | CIP | OFX |
gyrA
|
parC
|
||||
| 83 | 87 | 80 | 81 | 84 | ||||||
| K-12 | Ser | Asp | Ser | Ala | Glu | |||||
| 1 | 6 | 6 | 6 | >32 | >32 | Leu | Asn | Ile | —c | Gly |
| 2 | 6 | 6 | 6 | >32 | >32 | Leu | Asn | — | — | Lys |
| 3 | 6 | 6 | 6 | >32 | >32 | Leu | Asn | Ile | — | — |
| 4 | 6 | 6 | 6 | >32 | >32 | Leu | Asn | Ile | — | — |
| 5 | 6 | 6 | 6 | >32 | >32 | Leu | Asn | Ile | — | — |
| 6 | 6 | 6 | 6 | >32 | >32 | Leu | Asn | Ile | — | — |
| 7 | 6 | 6 | 6 | >32 | >32 | Leu | Asn | Ile | — | — |
| 8 | 6 | 6 | 6 | >32 | >32 | Leu | Asn | Ile | — | — |
| 9 | 12 | 6 | 6 | >32 | >32 | Leu | Asn | Ile | — | — |
| 10 | 18 | 17 | 6 | 0.75 | 2 | Leu | — | — | Pro | — |
| 11 | 22 | 21 | 6 | 0.38 | 1 | Leu | — | Ile | — | — |
| 12 | 23 | 21 | 6 | 0.5 | 1 | Leu | — | Arg | — | — |
| 13 | 24 | 21 | 6 | 0.25 | 0.75 | Leu | — | — | — | — |
| 14 | 24 | 22 | 6 | 0.25 | 1 | Leu | — | — | — | — |
| 15 | 24 | 20 | 6 | 0.19 | 0.75 | Leu | — | — | — | — |
| 16 | 25 | 21 | 6 | 0.38 | 1 | Leu | — | — | — | — |
| 17 | 26 | 22 | 6 | 0.23 | 1 | Leu | — | — | — | — |
| 18 | 26 | 22 | 6 | 0.19 | 0.75 | Leu | — | — | — | — |
| 19 | 26 | 24 | 6 | 0.19 | 0.5 | Leu | — | — | — | — |
| 20 | 26 | 23 | 6 | 0.19 | 0.5 | Leu | — | — | — | — |
| 21 | 26 | 23 | 6 | 0.19 | 0.5 | Leu | — | — | — | — |
| 22 | 26 | 22 | 6 | 0.125 | 0.75 | Leu | — | — | — | — |
| 23 | 27 | 23 | 6 | 0.125 | 0.5 | Leu | — | — | — | — |
| 24 | 28 | 25 | 6 | 0.19 | 0.38 | — | Tyr | — | — | — |
| 25 | 28 | 26 | 6 | 0.094 | 0.38 | — | Asn | — | — | — |
| 26 | 29 | 25 | 6 | 0.19 | 0.38 | Leu | — | — | — | — |
| 27 | 29 | 27 | 6 | 0.094 | 0.38 | — | Gly | — | — | — |
| 28 | 30 | 26 | 6 | 0.125 | 0.38 | — | Tyr | — | — | — |
| 29 | 31 | 27 | 22 | 0.047 | 0.125 | — | — | — | — | — |
| 30 | 32 | 29 | 23 | 0.016 | 0.094 | — | — | — | — | — |
| 31 | 34 | 30 | 29 | 0.016 | 0.094 | — | — | — | — | — |
| 32 | 34 | 30 | 27 | 0.012 | 0.064 | — | — | — | — | — |
| 33 | 35 | 29 | 24 | 0.032 | 0.125 | — | — | — | — | — |
| 34 | 35 | 33 | 27 | 0.016 | 0.064 | — | — | — | — | — |
| 35 | 37 | 32 | 28 | 0.023 | 0.064 | — | — | — | — | — |
| 36 | 38 | 34 | 26 | 0.016 | 0.064 | — | — | — | — | — |
Isolates 1 to 9 are fully resistant, isolates 10 to 28 have reduced susceptibility, and isolates 29 to 36 are fully susceptible, as described in Materials and Methods. Isolate K-12 is a wild-type E. coli strain.
CIP, ciprofloxacin; OFX, ofloxacin; NAL, nalidixic acid.
—, no change.
The statistical significance of the association between categorical variables was assessed by the chi-square test; continuous variables were assessed by analysis of variance, or in cases in which the variance within populations differed significantly, the Kruskal-Wallis one-way analysis of variance test was used. Linear regression of the proportion of resistant isolates and isolates with reduced susceptibility at each hospital was performed by the method of least squares. We used SPSS for Windows (release 10; SPSS, Chicago, Ill.) to perform multivariate analysis of clinical data using binary logistic regression in a stepwise fashion.
RESULTS
Antimicrobial susceptibility tests.
A total of 1,203 E. coli isolates were collected from the 44 hospitals. Overall, 1,067 (88.7%) were susceptible to ciprofloxacin and 136 (11.3%) were resistant to ciprofloxacin. The distribution of ciprofloxacin zone diameters demonstrated three distinct populations (Fig. 1). In addition to resistant isolates (zone diameters, ≤15 mm), there were populations of 261 (21.7%) isolates with reduced susceptibility (zone diameters, 16 to 31 mm) and 806 (67.0%) fully susceptible isolates (zone diameters, ≥32 mm).
FIG. 1.
Distribution of ciprofloxacin inhibition zone diameters for 1,203 E. coli isolates collected throughout Taiwan.
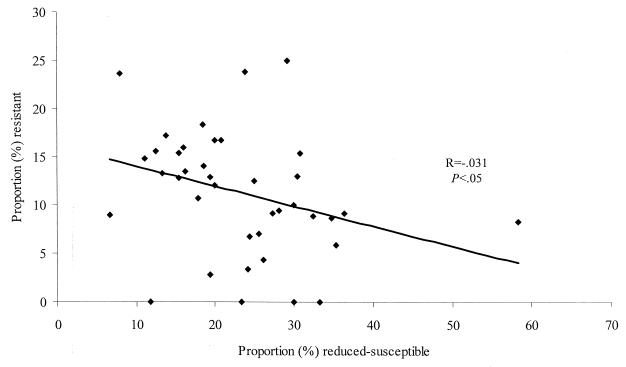
There were no significant differences in the proportion of isolates that had reduced susceptibility or that were resistant by regional hospital versus medical center status, region, geographic location, or site of the body from which the organism was isolated (data not shown). Among isolates from individual hospitals, there was an inverse relationship between the proportions of isolates that were resistant and the proportions of isolates with reduced susceptibility (Fig. 2).
FIG. 2.
Proportion of E. coli isolates with reduced susceptibility to ciprofloxacin versus proportion of E. coli isolates resistant to ciprofloxacin from 40 hospitals that submitted more than 10 isolates.
The rate of resistance tended to be greater among isolates from patients with hospital-acquired (mostly in intensive care units) infections (17 of 97 [17.5%]) than among isolates from other adult inpatients (88 of 775 [11.4%]; P = 0.08) and, although not significantly so, from outpatients (30 of 252 [11.9%]; P > 0.1). Similarly, the rate of reduced susceptibility was greater among isolates from patients with hospital-acquired infections (30.9%) than adult inpatient (163 of 775 [21.0%]; P = 0.04) and outpatient (54 of 252 [21.4%]; P = 0.06) isolates. Isolates from pediatric patients were significantly less likely to be resistant (1 of 79 [1.3%] versus 135 of 1,124 [12.0%]; P < 0.01) but were not significantly less likely to have reduced susceptibility (14 of 79 [17.7%] versus 247 of 1124 [21.9%]; P > 0.1) than isolates from nonpediatric sources.
Other forms of antimicrobial drug resistance and their association with reduced susceptibility or resistance to ciprofloxacin are depicted in Table 1. For each drug tested, resistance was most common in ciprofloxacin-resistant isolates, least common in susceptible isolates, and of intermediate frequency in isolates with reduced susceptibility.
TABLE 1.
Antibiotic resistance in E. coli by degree of ciprofloxacin susceptibility or resistance
| Drug | % Resistant isolates in the following ciprofloxacin susceptibility category:
|
|||
|---|---|---|---|---|
| Fully susceptible (n = 806) | Reduced susceptibility (n = 261) | Resistant (n = 136) | All isolates (n = 1,203) | |
| Amikacin | 1 | 3 | 10 | 3 |
| Ampicillin | 76 | 84 | 97 | 80 |
| Aztreonam | 1 | 6 | 18 | 4 |
| Amoxacillin-clavulanate | 35 | 39 | 73 | 40 |
| Ceftazidime | 1 | 3 | 9 | 2 |
| Cefazolin | 13 | 17 | 48 | 18 |
| Cefuroxime | 4 | 12 | 46 | 10 |
| Gentamicin | 25 | 44 | 72 | 35 |
| Piperacillin | 71 | 79 | 91 | 75 |
| Trimethoprim-sulfamethoxazole | 55 | 83 | 96 | 66 |
Molecular assessment of resistance mutations and strain typing.
A total of 36 isolates from Cathay General and Kaoshiung Medical University Hospitals were analyzed in depth (Table 2). These included all 9 isolates from the hospitals that were fully resistant to ciprofloxacin (zone diameters, ≤15 mm), all 19 strains that had reduced susceptibility (zone diameters, 16 to 31 mm), and 7 randomly chosen fully susceptible strains. Nalidixic acid resistance, as determined by disk diffusion, was strongly predictive of reduced susceptibility and resistance to ciprofloxacin on the basis of either the disk diffusion method or MIC determination. Similarly, mutations in gyrA were found in all isolates with reduced susceptibility and resistance to ciprofloxacin; additional mutations in parC were found in some isolates with reduced susceptibility and all resistant isolates. No parC mutations were found in the absence of gyrA mutations. One isolate (isolate 29) had a ciprofloxacin zone diameter of 31 mm (defined as reduced susceptibility on the basis of the population distribution of all E. coli isolates) but had no mutations in gyrA and parC and was susceptible to nalidixic acid. The MICs of both ciprofloxacin (0.047 μg/ml) and ofloxacin (0.125 μg/ml) for this isolate lay between the usual MICs for other isolates with reduced susceptibility and full susceptibility. For the purpose of further clinical epidemiologic assessment, this isolate was recategorized as fully susceptible.
All 9 resistant isolates and 15 (79%) isolates with reduced susceptibility had codon mutations in the genes encoding the 83rd amino acid of gyrA, leading to a substitution of a leucine (Leu) for the wild-type serine (Ser) at that position. In addition, all resistant isolates had a substitution of asparagine (Asn) for aspartate (Asp) at the 87th amino acid position of gyrA. The nine resistant isolates also had substitutions in the parC subunit of topoisomerase IV. All but one isolate had a substitution of isoleucine (Ile) for serine (Ser) at the 80th amino acid position of parC, and two isolates had substitutions for glutamate at the 84th position (one had a glycine substitution and the other had a lysine substitution). Of the 19 strains with reduced susceptibility, 3 (16%) had mutations that resulted in a single amino acid change of parC, in addition to the 83rd gyrA substitution. Four isolates with reduced susceptibility did not have mutations at the 83rd amino acid position of gyrA; instead, they all had mutations at the 87th amino acid position.
PFGE of the 36 isolates demonstrated three pairs of isolates with reduced susceptibility that were greater than 80% related by dendogram analysis. The first of these pairs (Table 2, isolates 19 and 20) were approximately 90% related, had identical antibiograms, were from two different patients at the same hospital, and were collected 6 days apart. The second pair of isolates (isolates 16 and 23) were just over 80% related, had identical antibiograms, were from different patients at the same hospital at which isolates 19 and 20 were recovered, and were collected 16 days apart. The third isolate pair (isolates 27 and 28) were approximately 95% related and had different antibiograms (one was ampicillin and gentamicin resistant, whereas the other was susceptible to both drugs) and were collected from patients at two different hospitals 21 days apart. Whereas the first two pairs of isolates with greater than 80% homology had codon mutations that resulted in identical amino acid substitutions, the third pair (isolates 27 and 28) had codon mutations that resulted in different amino acid substitutions (Table 2).
Clinical assessment of potential risk factors for reduced susceptibility and resistance.
There were no significant differences in demographics, underlying disease, or potential risk factors for resistance between patients infected with fully susceptible isolates versus patients infected with isolates with reduced susceptibility (Table 3). There were, however, several important differences between patients infected with resistant isolates and patients infected with fully susceptible isolates (Table 4). There was no significant difference between patients infected with resistant isolates or isolates with reduced susceptibility versus patients infected with fully susceptible isolates with regard to the body site of isolation (data not shown). Patients infected with resistant isolates were more likely to have underlying cancer, to have received any antibiotic, or to have received a nonquinolone antibiotic than patients infected with fully susceptible isolates. On the basis of multivariate analysis, only underlying cancer was confirmed to be an independent risk factor for resistance (odds ratio, 83; 95% confidence interval, 8.6 to 807; P < 0.001).
TABLE 3.
Comparison of demographic and clinical factors for patients infected with E. coli isolates with reduced susceptibility to ciprofloxacin versus those infected with fully susceptible E. coli isolates
| Characteristic | Ciprofloxacin susceptibility category
|
Odds ratio (95% CIa) | P | |
|---|---|---|---|---|
| Reduced susceptibility (n = 19) | Fully susceptible (n = 57) | |||
| Median (range) age (yr) | 69 (14–88) | 52 (1–90) | 0.35 | |
| Male (no. [%] of patients) | 7 (37) | 12 (21) | 2.2 (0.6–7.9) | 0.22 |
| No. of days of prior hospitalization (mean [SD]) | 4.8 (7.7) | 5.1 (10.7) | 0.89 | |
| Underlying disease (no. [%] of patients) | ||||
| None | 4 (20) | 17 (30) | 0.6 (0.2–2.5) | 0.67 |
| Renal | 6 (25) | 12 (21) | 1.9 (0.5–7.3) | 0.34 |
| Cancer | 1 (5) | 5 (9) | 0.6 (0.02–5.9) | 1.0 |
| Chronic liver disease | 2 (10) | 5 (9) | 1.2 (0.2–8.3) | 1.0 |
| Invasive device (no. [%] of patients) | ||||
| None | 4 (20) | 6 (11) | 2.3 (0.5–11.0) | 0.25 |
| Peripheral intravenous line | 8 (42) | 38 (67) | 0.4 (0.1–1.2) | 0.1 |
| Urinary catheter | 5 (25) | 13 (23) | 1.2 (0.3–4.6) | 1.0 |
| Central venous catheter | 5 (26) | 7 (12) | 2.6 (0.6–11.1) | 0.28 |
| Receipt of any antibiotic over preceding 2 mo (no. [%] of patients) | 8 (40) | 16 (28) | 1.9 (0.6–6.3) | 0.39 |
| Receipt of quinolone antibiotic over preceding 2 mo (no. [%] of patients) | 1 (5) | 0 | Undefined | 0.25 |
| Receipt of nonquinolone antibiotic over preceding 2 mo (no. [%] of patients) | 7 (35) | 16 (28) | 1.5 (0.4–5.1) | 0.66 |
CI, confidence interval.
TABLE 4.
Comparison of demographic and clinical factors for patients infected with ciprofloxacin-resistant versus fully susceptible E. coli isolates
| Characteristic | Ciprofloxacin susceptibility category
|
Odds ratio (95% CIa) | P | |
|---|---|---|---|---|
| Resistant (n = 9) | Fully susceptible (n = 57) | |||
| Median (range), age (yr) | 68 (11–88) | 52 (1–90) | 0.15 | |
| Male (no. [%] of patients) | 1 (11) | 12 (21) | 0.5 (0.02–4.5) | 0.7 |
| No. of days of prior hospitalization (mean [SD]) | 11.1 (16.3) | 5.1 (10.7) | 0.15 | |
| Underlying disease (no. [%] of patients) | ||||
| None | 0 | 17 (30) | 0 (0–1.6) | 0.01 |
| Renal | 1 (9) | 12 (21) | 0.5 (0.02–5.1) | 1.0 |
| Cancer | 8 (82) | 5 (9) | 83 (7.3–2,241) | <0.001 |
| Chronic liver disease | 2 (22) | 5 (9) | 3.0 (0.3–24) | 0.24 |
| Invasive device (no. [%] of patients) | ||||
| None | 0 | 6 (11) | 0.0 (0–6.6) | 0.59 |
| Peripheral intravenous line | 6 (67) | 38 (67) | 1.0 (0.2–5.8) | 1.0 |
| Urinary catheter | 2 (22) | 13 (23) | 0.97 (0.0–6.3) | 1.0 |
| Central venous catheter | 2 (22) | 7 (12) | 2.0 (0.23–14.9) | 0.6 |
| Receipt of any antibiotic over preceding 2 mo (no. [%] of patients) | 8 (89) | 16 (28) | 20.5 (2.2–482) | <0.001 |
| Receipt of quinolone antibiotic over preceding 2 mo (no. [%] of patients) | 2 (22) | 0 | Undefined | 0.02 |
| Receipt of nonquinolone antibiotic over preceding 2 mo (no. [%] of patients) | 8 (89) | 16 (28) | 20.5 (2.2–482) | <0.001 |
CI, confidence interval.
There was a nonsignificant trend in both nonquinolone antibiotic and overall antibiotic exposure (Tables 3 and 4), with rates of exposure being the lowest in patients infected with fully susceptible isolates (28%), intermediate in patients infected with isolates with reduced susceptibility (35 to 40%), and highest in patients infected with resistant isolates (89%). The one patient who was infected with a strain with reduced susceptibility and who was exposed to a quinolone was exposed only to nalidixic acid, whereas both patients infected with resistant strains were exposed to ciprofloxacin.
DISCUSSION
We found that 11.3% of E. coli isolates in Taiwan were resistant to fluoroquinolones and that another 21.7% had reduced susceptibility. Isolates submitted from two hospitals demonstrated a predictable association between single and double mutations in gyrA and the MICs and zone diameters for fluoroquinolones (Table 2). All isolates with reduced susceptibility from these hospitals possessed single point mutations, a necessary prerequisite for resistance. Therefore, the finding of such a large proportion of E. coli (21.7%) isolates throughout Taiwan with reduced susceptibility suggests that the rate of resistance may rapidly increase further. In addition to increasing the likelihood of evolution toward resistance, this large number of strains with reduced susceptibility may portend an increased risk of clinical failure when infections are treated with fluoroquinolones. For example, it has been demonstrated that infections caused by Salmonella spp. with reduced susceptibility have a higher clinical failure rate than infections caused by fully susceptible strains when treatment is with a fluoroquinolone (18, 29).
The finding that cancer patients were predisposed to infection with fluoroquinolone-resistant E. coli strains has been reported previously (4, 24, 32) and is likely related to the frequent therapeutic and prophylactic use of fluoroquinolones in cancer patients. The fact that the underlying cancer rather than quinolone exposure was an independent risk factor for resistance may be explained by the fact that we recorded only those antibiotic exposures that occurred over the 2 months before the culture date. It is likely that cancer acted as a surrogate marker for previous quinolone exposures that occurred at a higher frequency in cancer patients over an extended time period, even several years.
E. coli strains with reduced susceptibility to fluoroquinolones appeared to be intermediate to fully susceptible and resistant isolates with regard to certain characteristics. Among these were the rates of other forms of resistance in isolates with reduced susceptibility; for each drug tested, the rate of resistance in isolates with reduced susceptibility to fluoroquinolones lay between the rates observed in isolates fully susceptible and resistant to fluoroquinolones (Table 1). In addition, the rates of exposure to both quinolones (5%) and other antibiotics (35%) among patients infected with isolates with reduced susceptibility fluoroquinolones were intermediate to the rates of exposure in patients infected with fully susceptible (0 and 28%, respectively) and resistant (22 and 89%, respectively) isolates (Tables 3 and 4). These findings are consistent with the observations of other investigators suggesting that most resistant strains are selected from a preexisting pool of strains with reduced susceptibility (5, 7, 23).
We found an inverse relationship between the proportions of isolates with reduced susceptibility and the proportions of resistant isolates at different hospitals (Fig. 1). Given the likelihood that resistant strains are selected from strains with preexisting reduced susceptibility (5, 7, 23), this relationship may reflect the presence of distinct selective pressures responsible for each population. Otherwise, if the same selective pressure was responsible for both reduced susceptibility and resistance, one would expect the highest rates of reduced susceptibility to occur among hospitals with the highest rates of resistance.
Other findings support the concept that distinct selective pressures are responsible for reduced susceptibility and resistance. One of these is the proportion of pediatric isolates that had reduced susceptibility (17.7%) and the fact that although fluoroquinolones are not approved for pediatric use in Taiwan, this proportion was similar to the overall proportion of isolates with reduced susceptibility (21.7%). Yet, despite this similar proportion of isolates with reduced susceptibility among pediatric and adult isolates, only 1 of 79 (1.3%) pediatric isolates were resistant, confirming that human fluoroquinolone use is an important selective pressure for the development of full resistance.
In addition, we failed to identify the same patient risk factors for reduced susceptibility as for resistance, suggesting that there may be other distinct but unidentified patient risk factors for reduced susceptibility. However, because our study of patient risk factors involved a small sample size, some of the same risk factors for resistance found by univariate analysis (i.e., underlying cancer, quinolone exposure, and other antibiotic exposure) may in fact be risk factors for reduced susceptibility that were not detected by an underpowered study. Yet, if this is true, these are likely to be much weaker risk factors for reduced susceptibility than for resistance, as we used a slightly larger sample size to study reduced susceptibility versus resistance (Tables 3 and 4).
Because we studied patient quinolone use only over the 2 months prior to isolation of E. coli, one possible risk factor for reduced susceptibility is previous human use of nonfluorinated quinolones in the community. Although the precise amounts used are unknown, claims data submitted to the National Health Insurance Bureau in Taiwan indicate that nalidixic acid and pipemidic acid, both of which are nonfluorinated quinolones, are still used in some practice settings (L. C. McDonald, H. T. Yu, H. C. Yin, C. A. Hsiung, and M. Ho, Proc. 1st Int. Congr. Asia Pacific Soc. Infect. Control, 1999). It is conceivable that nonfluorinated quinolones are dispensed predominantly in small clinics or pharmacies and that this antibiotic pressure selects for reduced susceptibility in the community apart from hospital inpatient and outpatient care areas.
Another possible explanation for such a large proportion of E. coli isolates with reduced susceptibility to fluoroquinolones could be the frequent exposure of humans to bacteria with reduced susceptibility in the food supply. This has been suggested as a possible explanation for the fluoroquinolone-resistant E. coli that have been found to colonize healthy humans in the community in Spain, where fluoroquinolones are used in commercial poultry production and where a large number of flocks are colonized with resistant E. coli strains (8). Although the actual amounts used are unknown, both fluorinated and nonfluorinated quinolones are used in commercial poultry production in Taiwan (17). Moreover, we have found that a large proportion of retail chicken carcasses purchased in both traditional and modern markets around Taipei, Taiwan, are contaminated with E. coli strains with reduced susceptibilities to fluoroquinolones (L. C. McDonald, T. L. Lauderdale, and M. Ho, Abstr. 100th Gen. Meet. Am. Soc. Microbiol. 2000, abstr. Y-7, p. 682, 2000). In addition, we have found that a surprisingly similar proportion of human non-serovar Typhi Salmonella spp. in Taiwan possess reduced susceptibilities to fluoroquinolones (14) and that Salmonella spp. with reduced susceptibilities can also be recovered from retail chicken carcasses (McDonald et al., Abstr. 100th Gen. Meet. Am. Soc. Microbiol. 2000).
There is an urgent need to control the use of fluoroquinolones in certain patient populations, such as cancer patients, in whom resistance is most widespread. However, there is also an urgent need to better understand the relationship between the various steps along the path to resistance, the selective factors involved in these steps, and the best strategies for the prevention and control of resistance. For instance, in settings such as Taiwan, where a very large proportion of isolates with single point mutations already have reduced susceptibility, the use of even relatively limited amounts of fluoroquinolones may result in the rapid development of resistance. We recommend that the prevalence of isolates with reduced susceptibility be investigated in other settings where fluoroquinolone-resistant E. coli strains are emerging. Finally, until there is a better understanding of the precise forms of antibiotic use and overuse most responsible for fluoroquinolone resistance in E. coli, we recommend that the appropriateness and necessity of all quinolone use be carefully reconsidered, including the use of fluorinated and nonfluorinated quinolones in humans and animals. Failure to do so may result in the early senescence of this important class of antibiotics that has already proven so useful to human medicine.
REFERENCES
- 1.Anonymous. Intensive Care Antimicrobial Resistance Epidemiology (ICARE) Surveillance Report, data summary from January 1996 through December 1997: a report from the National Nosocomial Infections Surveillance (NNIS) System. Am J Infect Control. 1999;27:279–284. doi: 10.1053/ic.1999.v27.a98878. [DOI] [PubMed] [Google Scholar]
- 2.Blazquez R, Menasalvas A, Carpena I, Ramirez C, Guerrero C, Moreno S. Invasive disease caused by ciprofloxacin-resistant uropathogenic Escherichia coli. Eur J Clin Microbiol Infect Dis. 1999;18:503–505. doi: 10.1007/s100960050332. [DOI] [PubMed] [Google Scholar]
- 3.Canawati H N, el Farra R, Seymour J, Shimashita J, Dunn D, Montgomerie J Z. Ciprofloxacin-resistant Escherichia coli emerging in a rehabilitation medical center. Diagn Microbiol Infect Dis. 1997;29:133–138. doi: 10.1016/s0732-8893(97)81802-0. [DOI] [PubMed] [Google Scholar]
- 4.Carratala J, Fernandez-Sevilla A, Tubau F, Callis M, Gudiol F. Emergence of quinolone-resistant Escherichia coli bacteremia in neutropenic patients with cancer who have received prophylactic norfloxacin. Clin Infect Dis. 1995;20:557–560. doi: 10.1093/clinids/20.3.557. [DOI] [PubMed] [Google Scholar]
- 5.Conrad S, Oethinger M, Kaifel K, Klotz G, Marre R, Kern W V. gyrA mutations in high-level fluoroquinolone-resistant clinical isolates of Escherichia coli. J Antimicrob Chemother. 1996;38:443–455. doi: 10.1093/jac/38.3.443. [DOI] [PubMed] [Google Scholar]
- 6.Ena J, Amador C, Martinez C, Ortiz V D L T. Risk factors for acquisition of urinary tract infections caused by ciprofloxacin resistant Escherichia coli. J Urol. 1995;153:117–120. doi: 10.1097/00005392-199501000-00040. [DOI] [PubMed] [Google Scholar]
- 7.Gales A C, Gordon K A, Wilke W W, Pfaller M A, Jones R N. Occurrence of single-point gyrA mutations among ciprofloxacin-susceptible Escherichia coli isolates causing urinary tract infections in Latin America. Diagn Microbiol Infect Dis. 2000;36:61–64. doi: 10.1016/s0732-8893(99)00121-2. [DOI] [PubMed] [Google Scholar]
- 8.Garau J, Xercavins M, Rodriguez-Carballeira M, Gomez-Vera J R, Coll I, Vidal D, Llovet T, Ruiz-Bremon A. Emergence and dissemination of quinolone-resistant Escherichia coli in the community. Antimicrob Agents Chemother. 1999;43:2736–2741. doi: 10.1128/aac.43.11.2736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Garner J S, Jarvis W R, Emori T G, Horan T C, Hughes J M. CDC definitions for nosocomial infections, 1988. Am J Infect Control. 1988;16:128–140. doi: 10.1016/0196-6553(88)90053-3. [DOI] [PubMed] [Google Scholar]
- 10.Goettsch W, van Pelt W, Nagelkerke N, Hendrix M G, Buiting A G, Petit P L, Sabbe L J, van Griethuysen A J, de Neeling A J. Increasing resistance to fluoroquinolones in Escherichia coli from urinary tract infections in the netherlands. J Antimicrob Chemother. 2000;46:223–228. doi: 10.1093/jac/46.2.223. [DOI] [PubMed] [Google Scholar]
- 11.Graninger W, Zedtwitz-Liebenstein K, Laferl H, Burgmann H. Quinolones in gastrointestinal infections. Chemotherapy (Basel) 1996;42(Suppl. 1):43–53. doi: 10.1159/000239491. [DOI] [PubMed] [Google Scholar]
- 12.Heisig P, Tschorny R. Characterization of fluoroquinolone-resistant mutants of Escherichia coli selected in vitro. Antimicrob Agents Chemother. 1994;38:1284–1291. doi: 10.1128/aac.38.6.1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hendershot E F. Fluoroquinolones. Infect Dis Clin N Am. 1995;9:715–730. [PubMed] [Google Scholar]
- 14.Ho M, McDonald L C, Lauderdale T L, Yeh L L, Chen P C, Shiau Y R. Surveillance of antibiotic resistance in Taiwan, 1998. J Microbiol Immunol Infect. 1999;32:239–249. [PubMed] [Google Scholar]
- 15.Kern W V, Oethinger M, Jellen-Ritter A S, Levy S B. Non-target gene mutations in the development of fluoroquinolone resistance in Escherichia coli. Antimicrob Agents Chemother. 2000;44:814–820. doi: 10.1128/aac.44.4.814-820.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lehn N, Stower-Hoffmann J, Kott T, Strassner C, Wagner H, Kronke M, Schneider-Brachert W. Characterization of clinical isolates of Escherichia coli showing high levels of fluoroquinolone resistance. J Clin Microbiol. 1996;34:597–602. doi: 10.1128/jcm.34.3.597-602.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McDonald L C, Chen M T, Lauderdale T L, Ho M. The use of antibiotics critical to human medicine in food-producing animals in Taiwan. J Microbiol Immunol Infect. 2001;34:97–102. [PubMed] [Google Scholar]
- 18.Molbak K, Baggesen D L, Aarestrup F M, Ebbesen J M, Engberg J, Frydendahl K, Gerner-Smidt P, Petersen A M, Wegener H C. An outbreak of multidrug-resistant, quinolone-resistant Salmonella enterica serotype Typhimurium DT104. N Engl J Med. 1999;341:1420–1425. doi: 10.1056/NEJM199911043411902. [DOI] [PubMed] [Google Scholar]
- 19.Naber K G. Treatment options for acute uncomplicated cystitis in adults. J Antimicrob Chemother. 2000;46(Suppl. A):23–27. [PubMed] [Google Scholar]
- 20.National Committee for Clinical Laboratory Standards. Performance Standards for Antimicrobial Susceptibility Testing; Ninth Informational Supplement. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 21.Oethinger M, Conrad S, Kaifel K, Cometta A, Bille J, Klotz G, Glauser M P, Marre R, Kern W V. Molecular epidemiology of fluoroquinolone-resistant Escherichia coli bloodstream isolates from patients admitted to European cancer centers. Antimicrob Agents Chemother. 1996;40:387–392. doi: 10.1128/aac.40.2.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Osterlund A, Olsson-Liljequist B. Fluoroquinolone resistance of human pathogenic bacteria. Resistant E. coli now appearing in Sweden. Lakartidningen. 1999;96:1965–1966. . (In Swedish.) [PubMed] [Google Scholar]
- 23.Ozeki S, Deguchi T, Yasuda M, Nakano M, Kawamura T, Nishino Y, Kawada Y. Development of a rapid assay for detecting gyrA mutations in Escherichia coli and determination of incidence of gyrA mutations in clinical strains isolated from patients with complicated urinary tract infections. J Clin Microbiol. 1997;35:2315–2319. doi: 10.1128/jcm.35.9.2315-2319.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Perea S, Hidalgo M, Arcediano A, Ramos M J, Gomez C, Hornedo J, Lumbreras C, Folgueira D, Cortes-Funes H, Rodriguez-Noriega A. Incidence and clinical impact of fluoroquinolone-resistant Escherichia coli in the faecal flora of cancer patients treated with high dose chemotherapy and ciprofloxacin prophylaxis. J Antimicrob Chemother. 1999;44:117–120. doi: 10.1093/jac/44.1.117. [DOI] [PubMed] [Google Scholar]
- 25.Piddock L J. Mechanisms of fluoroquinolone resistance: an update 1994–1998. Drugs. 1999;58(Suppl. 2):11–18. doi: 10.2165/00003495-199958002-00003. [DOI] [PubMed] [Google Scholar]
- 26.Pieroni P, Goodfellow J, Reesor L, Louie M, Simor A E. Antimicrobial susceptibilities of blood culture isolates obtained before and after the introduction of ciprofloxacin. J Antimicrob Chemother. 1997;39:419–422. doi: 10.1093/jac/39.3.419. [DOI] [PubMed] [Google Scholar]
- 27.Thomson C J. The global epidemiology of resistance to ciprofloxacin and the changing nature of antibiotic resistance: a 10 year perspective. J Antimicrob Chemother. 1999;43(Suppl. A):31–40. doi: 10.1093/jac/43.suppl_1.31. [DOI] [PubMed] [Google Scholar]
- 28.Turnidge J. Epidemiology of quinolone resistance. Eastern hemisphere. Drugs. 1995;49(Suppl. 2):43–47. doi: 10.2165/00003495-199500492-00008. [DOI] [PubMed] [Google Scholar]
- 29.Wain J, Hoa N T, Chinh N T, Vinh H, Everett M J, Diep T S, Day N P, Solomon T, White N J, Piddock L J, Parry C M. Quinolone-resistant Salmonella typhi in Viet Nam: molecular basis of resistance and clinical response to treatment. Clin Infect Dis. 1997;25:1404–1410. doi: 10.1086/516128. [DOI] [PubMed] [Google Scholar]
- 30.Wang H, Dzink-Fox J L, Chen M, Levy S B. Genetic characterization of highly fluoroquinolone-resistant clinical Escherichia coli strains from China: role of acrR mutations. Antimicrob Agents Chemother. 2001;45:1515–1521. doi: 10.1128/AAC.45.5.1515-1521.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Weigel L M, Steward C D, Tenover F C. gyrA mutations associated with fluoroquinolone resistance in eight species of Enterobacteriaceae. Antimicrob Agents Chemother. 1998;42:2661–2667. doi: 10.1128/aac.42.10.2661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yoo J H, Huh D H, Choi J H, Shin W S, Kang M W, Kim C C, Kim D J. Molecular epidemiological analysis of quinolone-resistant Escherichia coli causing bacteremia in neutropenic patients with leukemia in Korea. Clin Infect Dis. 1997;25:1385–1391. doi: 10.1086/516132. [DOI] [PubMed] [Google Scholar]