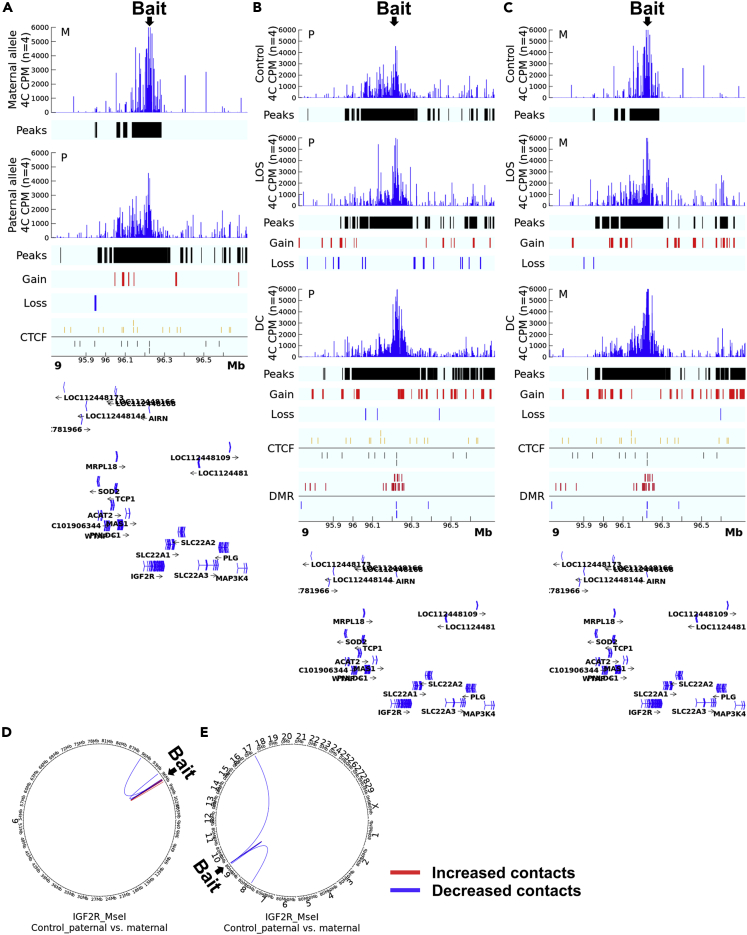
Figure 3.
4C identified allele-specific cis and trans contacts with IGF2R ICR
Shown are data for the IGF2R_MseI assay.
(A) Comparison of cis contacts between the paternal and maternal alleles in controls. Track ‘4C CPM’ shows the mean normalized count of reads aligned to the genome indicating physical contacts with the bait. Track ‘Peaks’show regions with statistically significant contacts with the bait identified by fourSig software within a group. Track ‘Gain’ (red line) and ‘Loss’ (blue line) indicate regions with statistically significant difference in contacts with the bait regions identified by DESeq2 between alleles. Track ‘CTCF’ shows predicted CTCF binding sites on the sense (gold line) or antisense (black line) strand. The gene annotation is at the bottom of the figure. Mb = megabases. CPM = counts per million reads. M = maternal allele. P = paternal allele.
(B and C) Comparison of allele-specific cis contacts between control, LOS, and DC. Shown are the comparison of LOS and DC groups vs controls. Track ‘Gain’ (red line) and ‘Loss’ (blue line) indicate regions with statistically significant difference in contacts with the bait regions identified by DESeq2 between groups. Track ‘DMR’ shows non-allelic differentially methylated regions identified between the LOS and the control group with the red line indicating increased and blue line indicating decreased methylation levels. All other track information as in (A).
(D and E) Comparison of contacts in far-cis and trans between parental alleles in controls. (D) far-cis contacts (chromosome 9) and (E) trans contacts (interchromosomal) in controls. Circos plots showing DESeq2-identified statistically different contacts with the bait in the paternal vs the maternal allele. Red line indicates increased contacts and blue line indicates decreased contacts.