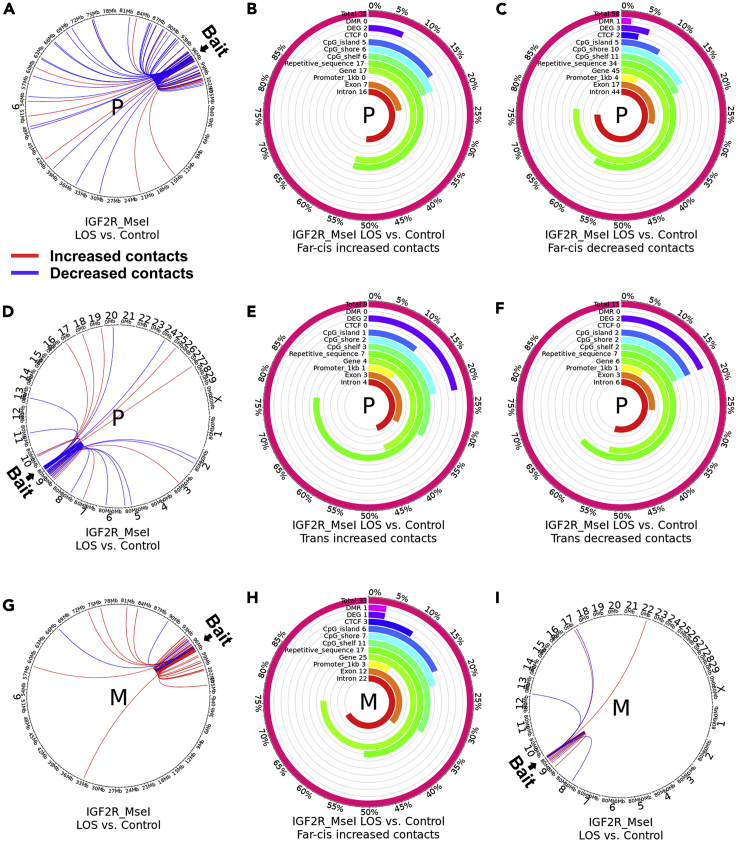
Figure 5.
Distribution of altered IGF2R ICR far-cis and trans contact across various genomic contexts
Shown are data for the IGF2R_MseI assay. (A, D, G, and I) Circos plots showing DESeq2-identified statistically different contacts with the bait in LOS vs. controls. Red line indicates increased contacts and blue line indicates decreased contacts. (A and D) Paternal and (G and I) maternal allele-specific comparisons. (A and G) far-cis contacts (chromosome 9) and (D and I) trans contacts (interchromosomal). P = paternal allele. M = maternal allele. (B-C, E-F, and H) Figures show the total number of altered far-cis (B-C and H) or trans (E-F) contacts identified and the number and percent of increased (B, E, and H) and decreased (C and F) contacts over each genomic context. For example, ∼55% of increased far-cis contacts in the paternal allele overlap with repetitive sequences (n = 17) and genes (n = 17). In addition, the figures include the number and percent of altered contacts that overlap differentially methylated regions (DMR) and within 100kb of differentially expressed genes (DEG) reported in this work. Analyses were only conducted for conditions with greater than five altered contacts.