Abstract
Multiplexed tissue imaging facilitates the diagnosis and understanding of complex disease traits. However, the analysis of such digital images heavily relies on the experience of anatomical pathologists for the review, annotation, and description of tissue features. In addition, the wider use of data from tissue atlases in basic and translational research and in classrooms would benefit from software that facilitates the easy visualization and sharing of the images and the results of their analyses. In this Perspective, we describe the ecosystem of software available for the analysis of tissue images and discuss the need for interactive online guides that help histopathologists make complex images comprehensible to non-specialists. We illustrate this idea via a software interface (Minerva), accessible via web browsers, that integrates multi-omic and tissue-atlas features. We argue that such interactive narrative guides can effectively disseminate digital histology data and aid their interpretation.
Diagnostic pathology relies on the microscopic analysis of histological specimens stained with colorimetric dyes or antibodies. Multiplexed high-resolution imaging1–4 can provide information on the expression levels and subcellular localization of 20–60 proteins, facilitating the study of cell states, tissue biomarkers and cell–cell interactions in normal and diseased conditions, and enabling the molecular profiling of tissues while preserving the native tissue architecture5. A common application of tissue imaging in oncology involves the identification and quantification of immune cell types and the mapping of their locations relative to the tumour boundaries and to stromal cells5. This type of spatially resolved data is particularly relevant to the study of the mechanisms of action of immunotherapies, such as the immune checkpoint inhibitors targeting the programmed cell death protein 1 (PD-1) or its ligand PD-L16, which function by blocking juxtracrine signalling between immune cells and tumour cells. However, images of tissues can be complex owing to the range of sizes of the biologically relevant structures: from the micrometre scale of subcellular vesicles and nuclear granules to the millimetre scale of cell configurations in blood and lymphatic vessels, to the centimetre scale required to visualize the interactions among endothelia, epithelia, muscle and the other cell types forming functional tissues.
There are a number of multiplexed tissue-imaging methods in current use. Imaging mass cytometry (IMC)1 and multiplexed ion beam imaging (MIBI)2 detect antigens via antibodies labelled with metal isotopes, followed by tissue ablation and atomic mass spectrometry. Methods such as multiplexed immunofluorescence (MxIF)3, co-detection by indexing (CODEX)7, tissue-based cyclic immunofluorescence (t-CyCIF)4, multiplexed immunohistochemistry (mIHC)8,9, and immunostaining with signal amplification by exchange reaction (immuno-SABER)10 use fluorescently labelled (or enzyme-linked) antibodies followed by microscopy. These methods differ in the number of antigens that they can detect on a single tissue section (at present, ~12 for multiplexed IHC and ~40–60 for IMC, MxIF, CODEX and t-CyCIF). Some methods are restricted to selected fields of view (in particular, ~1 mm2 for MIBI and IMC), whereas others can perform whole-slide imaging (WSI) on areas ~100–400 times larger (this is the case for MxIF, t-CyCIF, CODEX and mIHC). Most multiplexed tissue-imaging methods are in active development, and hence their strengths and limitations regarding imaging speed, sensitivity, resolution and other performance parameters are in flux. However, they all generate multiplexed two-dimensional (2D) images of cells and supporting tissue structures in situ. It is also possible to generate three-dimensional (3D) images via optical-sectioning and tissue-clearing methods11.
Tissue and tumour atlases
International projects are currently underway to create publicly accessible atlases of normal human tissues and tumours (Table 1). These include the Human Cell Atlas12, the Human BioMolecular Atlas Program (HuBMAP)13, and the Human Tumor Atlas Network (HTAN)14. For example, HTAN is envisioned as the spatially resolved counterpart of the well-established Cancer Genome Atlas15 (TCGA) (Figure 1). HTAN atlases aim to integrate genetic and molecular information from dissociative single-cell methods, such as single-cell RNA sequencing, with morphological and spatial details obtained from tissue imaging and spatial transcriptomics13,14. These atlases will combine images from many specimens into common reference systems to enable systematic inter-patient and cross-disease analysis of the data13. Conceptually, integration across samples and data types is easiest to achieve at the level of derived features, such as a census of cell types and positions (from imaging data) or transcript levels (from single-cell RNA sequencing). Adding mesoscopic-scale information from images—such as the arrangement of supporting stroma, membranes, blood and lymphatic vessels—is more challenging.
Table 1.
Available atlases of normal and diseased human tissue
| Atlas | Year Established | Microscopy | Lead Country | Atlas Link | Description/Goals |
|---|---|---|---|---|---|
| Human Genome Project | 1990 | − | International | https://www.genome.gov/human-genome-project | Map all genes of the human genome |
| FANTOM (Functional ANnoTation Of the Mammalian genome) | 2000 | − | International | https://fantom.gsc.riken.jp/ | Assign functional annotations to full-length cDNAs collected in Mouse Encyclopedia Project at RIKEN |
| ENCODE (Encyclopedia of DNA Elements) | 2003 | + | US | https://www.encodeproject.org/ | Identify functional elements in the human genome |
| Human Protein Atlas (HPA) | 2003 | ++ | Sweden | https://www.proteinatlas.org/ | Map all human proteins in cells, tissues, and organs |
| Wellcome Trust Case Control Consortium (WTCCC) | 2005 | + | UK | https://www.wtccc.org.uk/ | Understand patterns of human genome sequence variation to explore the utility, design and analyses of genome-wide association studies (GWAS) |
| The Cancer Genome Atlas (TCGA) | 2005 | + | US | https://portal.gdc.cancer.gov/ | Obtain a comprehensive understanding of the genomic alterations that underlie all major cancers |
| International Cancer Genome Consortium (ICGC) | 2007 | − | International | https://icgc.org/ | Unravel genomic changes present in many forms of cancer that contribute to the burden of disease |
| European Bioinformatics Institute (EMBL-EBI) Expression Atlas | 2009 | − | International/EBI | https://www.ebi.ac.uk/gxa/home | Database for querying differential gene expression across tissues, cell types and cell lines under various biological conditions |
| The Genotype-Tissue Expression (GTEx) project | 2010 | + | US | https://gtexportal.org/home/ | Study how inherited changes in genes lead to common diseases |
| LungMAP | 2010 | +++ | US | https://www.lungmap.net/ | Generate structural and molecular data on perinatal and postnatal lung development |
| BLUPRINT | 2011 | − | Netherlands/EU | https://www.blueprint-epigenome.eu/ | Understand how genes are activated or repressed in both healthy and diseased haematopoietic cells |
| Cancer Cell Line Encyclopedia (CCLE) | 2012 | − | US | https://depmap.org/portal/ccle/ | Compile gene expression, copy number and sequencing data from 947 human cancer cell lines |
| GenitoUrinary Development Molecular Anatomy Project (GUDMAP) | 2015 | ++ | US | https://www.gudmap.org/ | Develop tools to facilitate research on the genito-urinary (GU) tract |
| Accelerating Medicine Partnership (AMP) | 2014 | + | US | https://www.nih.gov/research-training/accelerating-medicines-partnership-amp | Identify and validate biological targets for diagnostics & drug development |
| Human Cell Atlas | 2016 | ++ | International | https://www.humancellatlas.org/ | Develop reference maps of all human cells as a basis for diagnosing, monitoring, treating disease |
| Stimulating Peripheral Activity to Relieve Conditions (SPARC) | 2016 | ++ | US | https://sparc.science/ | Accelerate development of devices that modulate neuronal activity to improve organ function |
| Pancreatlas | 2016 | +++ | US | https://pancreatlas.org/ | Generate comprehensive images of the human pancreas |
| Human Tumor Atlas Network (HTAN) | 2017 | ++ | US | https://humantumoratlas.org/ | Multidimensional molecular, cellular, and morphological mapping of human cancers |
| 4D Nucleome (4DN) | 2017 | ++ | US | https://www.4dnucleome.org/ | Study the three-dimensional organization of the nucleus in space and time (the 4th dimension) |
| Kidney Precision Medicine Project/Kidney Tissue Atlas | 2017 | ++ | US | https://atlas.kpmp.org/repository | Study kidney biopsies to map heterogeneity and molecular pathways for drug discovery |
| Human BioMolecular Atlas Program (HuBMAP) | 2017 | ++ | US | https://hubmapconsortium.org/ | Develop an open and global platform to map healthy cells in the human body |
| Single Cell Expression Atlas | 2018 | − | International/EBI | https://www.ebi.ac.uk/gxa/sc/home | Display gene expression in single cells from 12 species |
| BRAIN Initiative Cell Census Network (BICCN) | 2018 | +++ | US | https://biccn.org/ | Identify and provide experimental access to different brain cell types to determine their roles in health and disease |
| Oncobox Atlas of Normal Tissue Expression (ANTE) | 2019 | − | Russia | https://www.nature.com/articles/s41597–019–0043–4 | Atlas of RNA sequencing profiles for normal human tissues |
| LifeTime Initiative | 2019 | +++ | International/EU | https://lifetime-initiative.eu/ | Single-cell multi-omics & imaging, AI and patient-derived experimental disease models of health & disease |
| Pediatric Cell Atlas | 2019 | ++ | US | https://www.humancellatlas.org/pca/ | Cytogenomic framework for study of pediatric health and disease. |
| Japanese cancer genome atlas (JCGA) | 2019 | − | Japan | https://pubmed.ncbi.nlm.nih.gov/31863614/ | The Japanese TCGA |
| Gut Cell Atlas | 2019 | ++ | International | https://www.gutcellatlas.helmsleytrust.org/ | Catalogue cell types in small, large intestines |
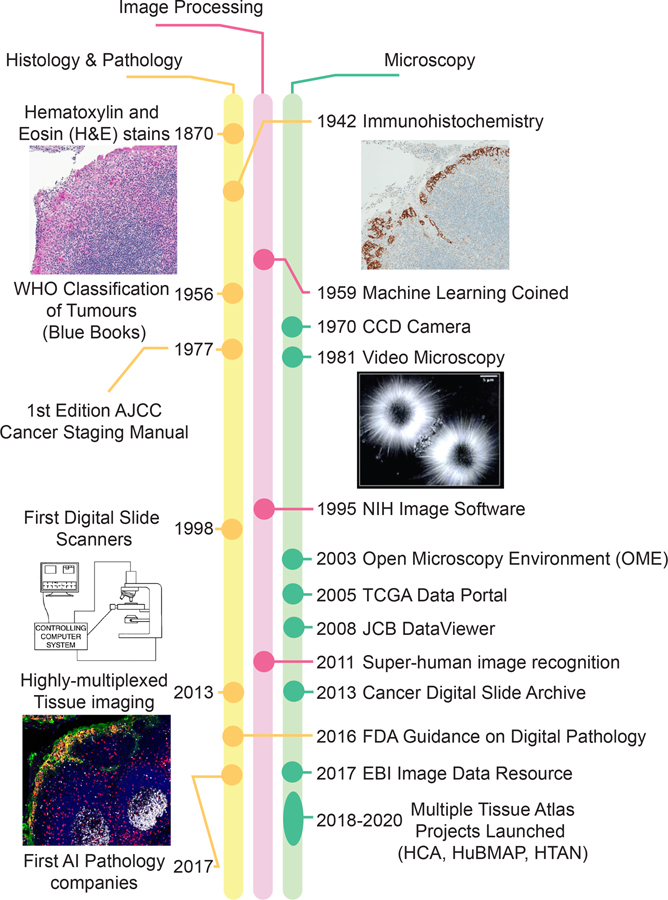
Figure 1. Milestones in the development of histopathology, image processing and microscopy.
Methods used for histology and anatomical pathology have evolved towards digitalization in parallel with the growth of image-processing capabilities, multiplexed imaging modalities and the launch of multiple tissue-atlas projects. The first use of the term “machine learning”, in 1959, is widely credited to A. Samuel (ref. 73). Publicly available atlases of human tissues, tumours and cell types are listed in Table 1. Video microscopy image reproduced with permission from ref. 74. Slide scanner diagram reproduced with permission from ref. 75. AI, artificial intelligence; AJCC, American Joint Committee on Cancer; CCD, charge-coupled device; WHO, World Health Organization.
Computational methods that automatically extract spatial information from images are still under development; therefore, the practice of annotating slides by anatomical pathologists remains essential. Ongoing research to better understand how pathologists make diagnoses from tissue specimens16 and to quantify connections between features computed from cellular neighbourhoods and clinical outcome17 will provide useful insight into the development of automated image-analysis pipelines. However, human oversight of tissue images will remain essential for relating morphology to pathophysiology and for assessing the quality of image-processing algorithms and training machine-learning classifiers. Therefore, any software-based solution aimed at users of digital atlases must enable free exploration of image data while also allowing users to benefit from the expertise of pathologists who currently work almost entirely with physical specimens on glass slides. In this Perspective, we describe software interfaces that meet these considerations. The software interfaces allow users working in research or in clinical practice to browse increasingly large image datasets at low cost and without the need for complex software. The interfaces accomplish this by using ‘digital docents’ and narration so that users can benefit from the expert description of a complex specimen, while also facilitating access to quantitative information derived from computational analysis. They can also augment reports and publications so that reported imaging data are not restricted to showing selected small fields of view.
Tissue imaging in a clinical setting
An expansion in the use of multiplexed tissue-imaging methods in research settings is happening at the same time as a transition to digital technologies in clinical histopathology18; the two developments are, however, not optimally linked or coordinated (Figure 1). Although genetics is increasingly pertinent to disease diagnosis, particularly in cancer and in inherited diseases, histology and cytology remain the central pillars of routine clinical work for the confirmation or diagnosis of many diseases. In current practice, tissue samples recovered by biopsy or surgical resection are formaldehyde-fixed and paraffin-embedded, sliced into 5-μm sections, and stained with haematoxylin and eosin (H&E). Samples for immediate study (used when a patient is undergoing surgery, for example) are frozen, sectioned and stained (these are often called OCT samples, because of the ‘optimal cutting temperature’ medium in which they are embedded). Liquid haematological samples are spread on a slide to create blood smears, which are also stained with colorimetric dyes (Romanowsky–Giemsa staining). H&E staining imparts a characteristic pink-blue colour on cells and other structures, and pathologists review these specimens using simple bright-field microscopes; other stains are analysed in a similar way. Some clinical samples are also subjected to IHC to obtain information on the expression of one or few protein biomarkers per slide19. Although cost-effective and widely used, many histopathology methods were developed over a century ago, and IHC is itself 75 years old20 (Figure 1). Moreover, diagnoses based on these techniques generally do not capture the depth of molecular information needed to optimally select targeted therapies. The latter remains the purview of mRNA and DNA sequencing, which, in a clinical setting, often involves exome sequencing of several hundred selected genes.
Recently, pathologists have started to leverage the pattern-recognition capabilities of machine-learning algorithms to extract deeper diagnostic insight into histological data. The digital analysis of histological specimens first became possible with the introduction of bright-field WSI instruments 20 years ago21,22, but it was not until 2016 that the United States Food and Drug Administration (FDA) released guidance on the technical requirements for the use of digital imaging in diagnosis23. Digital-pathology instruments, software and start-ups have proliferated over the past few years, fuelled in large part by the development of machine-learning algorithms that assist in the interpretation of H&E-stained slides24, which histopathology laboratories need to process at high volumes (often more than 1 million slides per year in a single hospital). Machine learning of images has shown its worth in several areas of medicine25,26, and promises to assist practitioners by increasing the efficiency and reproducibility of pathological diagnoses24. The pathology departments at several comprehensive cancer centres (institutions that are part of a network coordinated by the United States National Cancer Institute) have recently introduced multiplexed image-based immunoprofiling services for the identification of patients most likely to benefit from immune checkpoint inhibitors. Nonetheless, in most hospitals, the vast majority of diagnostic pathology still involves the visual inspection of physical specimens; only a minority of slides are scanned and digitized for concurrent or subsequent review on a computer screen. This is widely anticipated to change over the next decade; in fact, several European countries have national digital-pathology programs. As clinical pathology incorporates new measurement modalities and becomes increasingly digital, compatible and interoperable software and standards for clinical and research purposes need further development, and this is likely to be particularly important in the conduct of clinical trials. From a technical perspective, standards for digital image files (such as the OME-TIFF format) developed for preclinical research can work well with multiplexed images from clinical services. However, software for clinical use also requires security, workflows and billing features that are not necessary components of software used for research.
Sharing imaging data from tissue atlases
Algorithms, software and standards for high-dimensional image data27,28 remain underdeveloped in comparison to the tools avail- able for almost all types of genomic information. Moreover, with sequencing data, the information present in primary data files (such as FASTQ files) are fully retained (or enhanced) when reads are aligned or when count tables are generated; it is rarely necessary to re-access the primary data. In contrast, methods for the extraction of features from tissue images are immature, and their visual inspection by experts for diagnostic or scientific purposes, or for the testing of new algorithms, requires repeated access to the original images at their native resolution.
As far back as 2008, the Journal of Cell Biology worked with the founders of the Open Microscopy Environment (OME)29 to deploy a JCB DataViewer30, which provided direct access to primary high-resolution microscopy data (much of it from tissue culture cells and model organisms). Economic pressures ended this ambitious effort31, which emphasizes that funds have long been available to purchase expensive microscopes but not to distribute the resulting data. Currently, most H&E, IHC and multiplexed tissue images are shared only as figure panels in manuscripts, a form that typically provides access to a few selected fields of view at a single (and often downscaled) resolution. In the best case, these data might be available via public data repositories such as figshare32. The European Bioinformatics Institute (EBI) Image Data Resource (IDR) is a notable exception; IDR uses the OME-compatible OMERO33 server to provide full-resolution access to selected microscopy data34. The importance of sharing image data is particularly pronounced in the case of research biopsies, including those being used to assemble tumour atlases. These biopsies are obtained to expand scientific knowledge rather than to inform the treatment of individual patients, and there is an ethical obligation for the resulting data (appropriately anonymized) to be made widely available in an open and useful form for research purposes35. Open-source data and code are widely recognized as valuable tools in genomics36,37 and in basic cell-and-developmental-biology research, but availability has only recently been flagged as a significant barrier to scientific progress38. More generally, digital pathology and medical imaging are disciplines in which the goal of making research findable, accessible, interoperable and reusable (FAIR)39 is desirable, but the computational infrastructure is insufficient to meet these aspirations.
Software for image analysis and interpretation
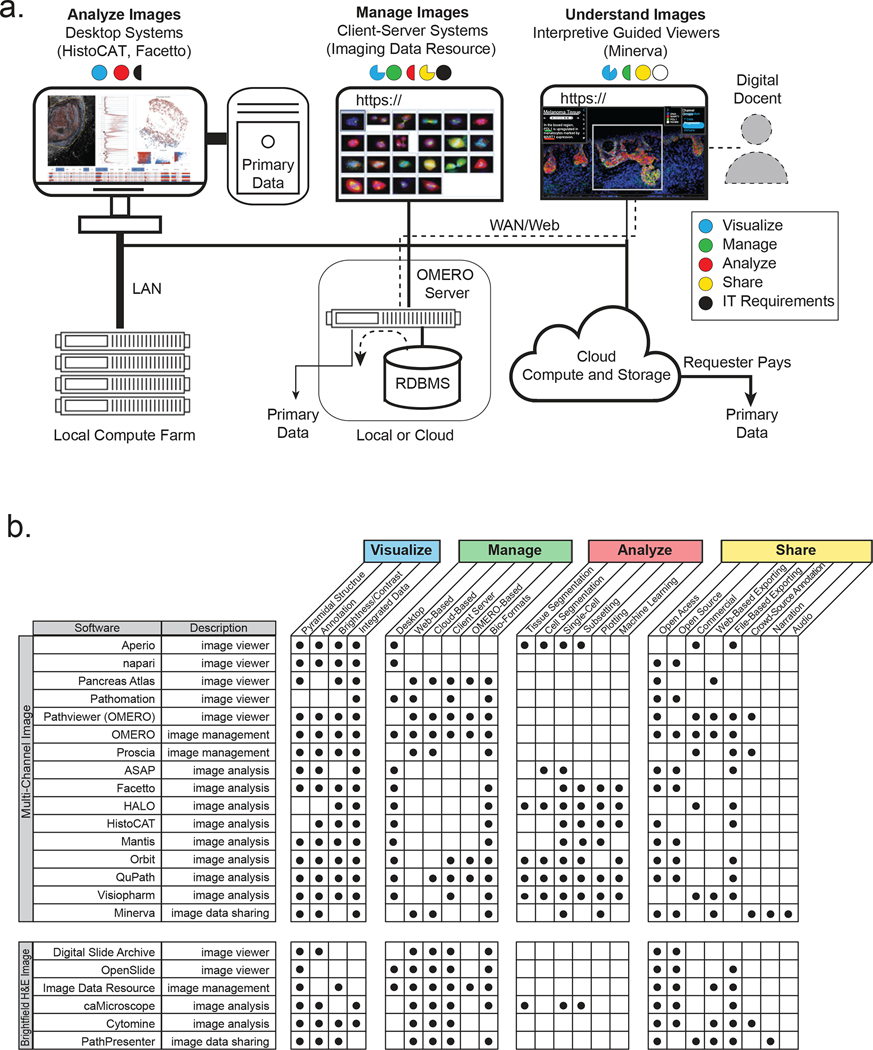
A wide variety of academic and commercial microscopy software systems are available either as a desktop system, for data analysis, or as a client–server relational database management system (RDBMS), for image management (Figure 2a and Boxes 1 and 2). Desktop software is particularly good for interactive image analysis because it exploits graphics cards for rapid image rendering as well as high-bandwidth connections to local data for computation. RDBMSs are instead ideal for data management because they enable relational queries, support multiple simultaneous users, ensure data integrity and effectively manage access to large-scale local and cloud-based computer resources (Figure 2b provides a detailed comparison of available software).
Figure 2. Software used to visualize, analyse, manage and share tissue images.
Software systems for processing tissue images are typically optimized to perform a select number of tasks and to co-evolve to complement each other’s strengths and weaknesses as part of an ecosystem held together by common data standards and interoperable application programming interfaces. a, Desktop applications such as histoCat provide sophisticated tools for the interactive quantification and analysis of primary image data, and effectively exploit embedded image-rendering capabilities. Many desktop applications can be run in a ‘headless’ configuration on computer farms to accelerate the analysis of large images. They can also fetch images from three-tier client–server systems such as OMERO. OMERO involves a RDBMS, an image server and one or more browser-based clients and desktop clients; such systems provide a full-featured approach to organizing, visualizing and sharing large numbers of images, but they typically support limited data processing. The Minerva application discussed in this Perspective is optimized for data sharing and interpretive image viewing and enables a detailed narration of image features and derived data. Pre-rendering of stories with multiple waypoints in an image mimics the insight provided by a human expert or guide (a ‘docent’). Public access to large-scale primary data is typically limited with all of these systems because of the costs of data transfer. However, in some OMERO configurations (in particular, the IDR), primary data can be downloaded from the same file system. Another approach to primary-data access being used by HTAN involves ‘requester pays’ cloud-based storage (for instance, an AWS S3 bucket). Minerva Author may also be extended to enable direct access to primary data stored in OMERO or these S3 buckets. b, The key features of commercial and academic software tools suitable for viewing image data; each tool has its strengths and weaknesses, but none comprehensively satisfies all of the functions needed for complete image analysis. Data generators and consumers rely on a suite of interoperable desktop-and-server or cloud-based software systems. Panel a reproduced with permission from ref. 57, IEEE (left) and ref. 33 (right).
Box 1: Software for managing and visualizing image data.
The OME-based OMERO33 server is the most widely used image-informatics system for microscopy data in a research setting. It is the foundation of the IDR-based resource at the EBI34, a prominent large-scale publicly accessible image repository, and is part of more specialized repositories such as Pancreatlas76. OMERO has a client–server three-tier architecture involving a relational database, an image server and one or more interoperable user interfaces. OMERO is well suited to managing image data and metadata and to organizing images so that they can be queried using a visual index or via search77 (Figure 2a). In its current form, it does not perform sophisticated image analysis and it is not specialized for the creation of narrative guides.
A range of other software tools are available for the static visualization or partially interactive visualization of H&E and IHC images in a web browser. In particular, caMicroscope78 is used to organize IHC and H&E images for TCGA15. Other publicly available image interfaces include the Cancer Imaging Archive79, the Digital Slide Archive38, PathPresenter80 and Aperio81 (Figure 2b). A white paper from the Digital Pathology Association provides a description of the tools being developed to view bright-field digital-pathology data52. However, such H&E and IHC viewers are not generally compatible with multichannel images or with the integration of different types of omics data.
Box 2: Software for the analysis of image data.
Multiple software suites—CellProfiler82, histoCAT56, Facetto57, QuPath83, Orbit84, Mantis85 and ASAP86, among others—have been developed for the analysis of high-dimensional image data from tissues or cultured cells. Many of these software systems are desktop-based and run locally (Figure 2b). They generally perform image segmentation to identify individual cells or tissue-level features, determine cell centroids and shape parameters (such as area and eccentricity), and compute staining intensities in designated regions of an image and across all channels. The resulting vectors can then be processed using standard tools for high-dimensional data analysis such as supervised and unsupervised clustering, t-SNE87 or uniform manifold approximation and projection (UMAP)88 to identify cell types and to study cell–cell interactions89. Although some tools support segmentation, others require pre-generated segmentation masks and single-cell feature tables. As an alternative, other approaches can analyse tissues at the level of individual pixels via the use of machine-learning algorithms and convolutional neural networks90. This approach potentially bypasses the need to segment individual cells from densely packed tissues, in which cells can dramatically vary in size. A key feature of software such as histoCat56 or Facetto57 is the integration of an image viewer with feature-based representations of the same data. This is essential for training and testing classifiers, for the quality control of image-processing routines and for obtaining insight into spatial characteristics. Pipelines that integrate and simplify the processing of high-plex images have started to appear60 and are based on technologies used in existing pipelines for the processing of sequencing data.
Several cloud-based computing platforms for digital pathology are being commercially developed, such as HALO (Indica Labs), Visiopharm (Visiopharm) and PathViewer (Glencoe Software, the commercial developer of OMERO). Academic efforts such as the Allen Cell Explorer91 and napari92 build on highly successful open-source software platforms such as ImageJ93. Napari is particularly attractive to computational biologists because it is written entirely using the programming language Python and has user-interface elements and a console. Commercial tools often strive for an all-in-one approach to analysis and visualization, but this comes at the cost of complexity, proprietary implementations, and licensing fees. It also ignores one of the primary lessons from genomics: progress in rapidly developing research fields rarely involves the use of a single integrated software suite; rather, it relies on an ecosystem of interoperable tools specialized for specific tasks.
However, with the introduction of high-plex and whole-slide tissue images, new software is required to guide users through the extraordinary complexity of images that encompass multiple square centimetres of tissue, 105–107 cells and upwards of 100 channels. Images of this size are not only difficult to process and share but they are also difficult to understand. We envision a key role for ‘interpretive guides’ or ‘digital docents’ that help walk users through a series of human-provided and machine-generated annotations about an image in much the same way that the results section of a published article guides users through a multi-panel figure. Genomic science faced an analogous need for efficient and intuitive visualization tools a decade ago, and this led to the development of the influential Integrative Genomics Viewer40 and its many derivatives.
Interactive guides of pictures and images have proven highly successful in other scientific fields, as exemplified by Project Mirador (https://projectmirador.org). Project Mirador focuses on the development of web-based interpretive tours of cultural resources such as art museums, illuminated manuscripts and maps of culturally or historically significant cities. In these online tours, a series of waypoints (reference points used to help navigate the information displayed) and accompanying text direct users to areas of interest while also allowing free exploration (the user can then return to the narrative when needed). This type of interactive narration—also known as digital storytelling or visual storytelling—mimics some of the benefits provided by tour guides and functions as a pedagogical tool that enhances comprehension41 and memory formation42. Multiple studies have identified benefits associated with receiving complex information in a narrative manner, and digital storytelling has been applied to several areas of medicine and research, including oncology43, mental health44, health equity45 and science communication46,47.
How might these lessons be applied to tissue images? At the very least, they suggest that simply making gigapixel-sized images available for download and analysis on desktop software is insufficient to optimally advance scientific discovery and to improve diagnostic outcomes. Instead, an easy approach that guides users through the salient features of an image with associated annotation and commentary would be beneficial to both specialists and non-specialists. Currently, pathologists share insights with colleagues using a multi-head microscope to pan across an image and to switch between high and low-power fields (magnifications), thereby studying cells in detail while also placing them in the context of the overall tissue. In this process, key features are often highlighted using a pointer. In the case of multiplex images, time-tested pathology practice must be built into software along with the means to toggle channels on and off (so that the contribution of specific antibodies to the final image can be ascertained) and display quantitative data arising from image analysis.
Software-based interpretive guides
How might interpretive guides for tissue images be implemented? One possibility involves the use of an OMERO33 client. OMERO is the most widely deployed open-source image-informatics system, and it is compatible with a range of software clients. However, an OMERO client requires access to an OMERO database and, as images get larger, the server becomes substantially loaded, which limits the number of concurrent sessions. Database-independent viewers are used by Project Mirador and are based on the open-source OpenSeadragon48 platform. OpenSeadragon makes it easy to zoom and pan across images (in a manner similar to Google Maps49). Minerva, which is customized for guided viewing of multiplexed images, has taken this approach50. Minerva is a web application that uses a client-side JavaScript (and thus requires no downloaded software), and it is easily deployable on standard commercial clouds such as Amazon Web Services (AWS) (Figure 3) or on local computing servers that support the static site generator Jekyll51. OpenSeadragon has been previously used for displaying H&E images52, but in Minerva, it is paired with narrative features, interactive views of derived single-cell features within the image space, lightweight implementation and the ability to accommodate both bright-field (H&E and IHC) and multiplexed immunofluorescence images. Minerva can also display images and annotations on smartphones.
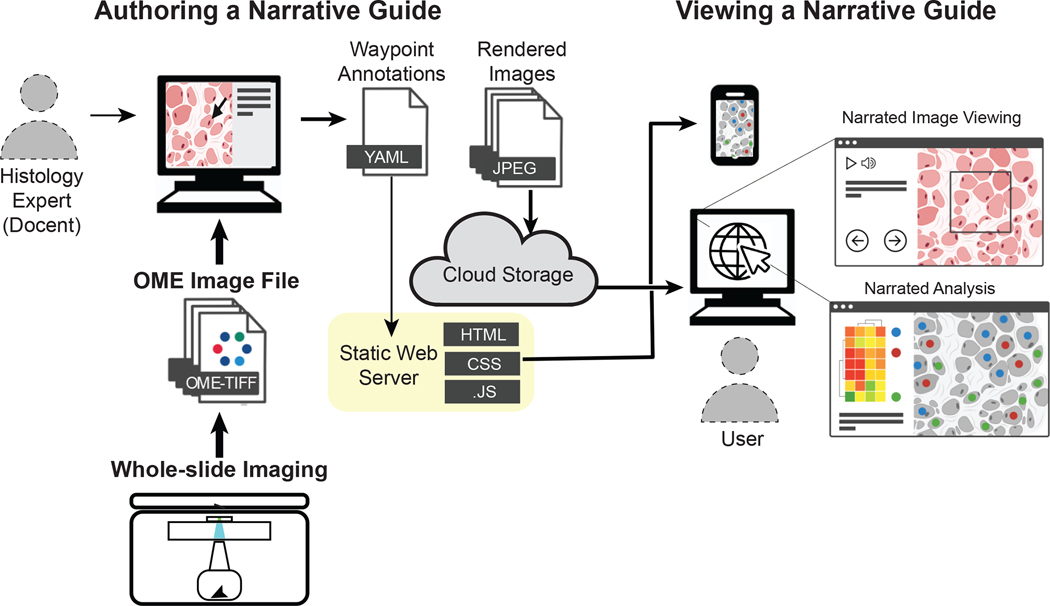
Figure 3. A system for generating and viewing online narrative guides for histopathology tissue images.
The illustrated architecture is based on the OpenSeadragon viewer Minerva Story and the narration tool Minerva Author. The thickness of the arrows indicates the amount of data transferred. Whole-slide images, including bright-field and multi-channel microscopy images, in the standard OME-TIFF format are imported into Minerva Author by a user with expertise in tissue biology and image interpretation (the ‘docent’); in many cases this individual is a pathologist, histologist or cell biologist. The docent uses tools in Minerva Author to pan across the image and to then set channel-rendering values (background levels and intensity scale), specify waypoints and add text and graphical annotations. Minerva Author then renders image pyramids for all channel groupings as JPEG files and generates a YAML configuration file that specifies waypoints and associated information. The rendered images are stored on a cloud host (such as AWS S3) and accessed via a static web server supporting Jekyll (on GitHub, for example). A user simply opens a web browser, selects a link, and Minerva Story launches the necessary client-side JavaScript (.JS), making it possible to follow the story and to freely explore the image by panning, zooming and selecting channels. Because interactivity is handled on the client side, no special backend software or server is needed. Images rendered by Minerva Story are compatible with multiple devices, including cell phones. Source code for the Minerva Story can be found at https://github.com/labsyspharm/minerva-story, and detailed documentation and user guide at https://github.com/labsyspharm/minerva-story/wiki.
Minerva is OME-compatible and BioFormats-compatible and is therefore usable with images from virtually any existing microscope or slide scanner. There is no practical limit to the number of users who can concurrently access narrative guides developed in Minerva and there is no requirement for specialized servers or a relational database, thereby keeping complexity and costs low. Anyone familiar with GitHub and AWS (or similar cloud services) can deploy a Minerva story in a few minutes, and new stories can then be generated by individuals with little expertise in software and computational biology. Developers can also build on the Minerva framework to develop narration tools for other applications (for example, for interpreting protein structures or complex high-dimensional datasets). Software viewers such as Minerva are not intended to be all-in-one solutions to the many computational challenges associated with processing and analysing tissue data; rather, they are specialized browsers that perform one task well. In the case of Minerva, it provides an intuitive and interpretive approach to images that have already been analysed using other tools. Genome browsers are similar: they do not perform alignment and data analysis; instead, they make it possible to interact with the processed sequences effectively.
Within a Minerva window on a standard web browser, a narration panel directs the user’s attention to particular regions of an image and to specific channel groupings, and is accompanied by a text description (which Minerva can read aloud) and by image annotations involving overlaid geometric shapes or text (Figure 4). Each image can be associated with more than one narrated story and with different ways of viewing the same type of data. A fundamental aspect of the narrative guides is that individuals with expertise in a particular disease or tissue (in particular, a pathologist) can create stories for use by others to assist them in the understanding of the morphology of a specimen. Creating narrations requires an authoring tool; for Minerva, this is the Minerva Author. Minerva Author is a desktop application (in JavaScript React with a Python Flask backend) that converts images in OME-TIFF format53 to pyramid images (layered images with different resolution, with the largest one at the bottom and the smallest one on the top) and that assists with the addition of waypoints and text annotations. Minerva Author supports RGB images, such as bright-field, H&E and IHC, as well as multichannel images such as immunofluorescence, CODEX and CyCIF. After specifying the rendering settings and writing the waypoints in Minerva Author, a user receives a configuration file and image pyramids to deploy to AWS S3 or to another web-based storage location (Figure 3). Stories can be as simple as a single panel with a short introduction or a multi-panel narration enriched with a series of views with detailed descriptions, changes in zoom level and associated data analysis. It takes users a few hours to learn the software and then from about 30 min to a few hours to create a story; this is about the same time required to create a good static figure panel for a published article, and much less time than that needed for data collection, image registration, segmentation and data analysis.
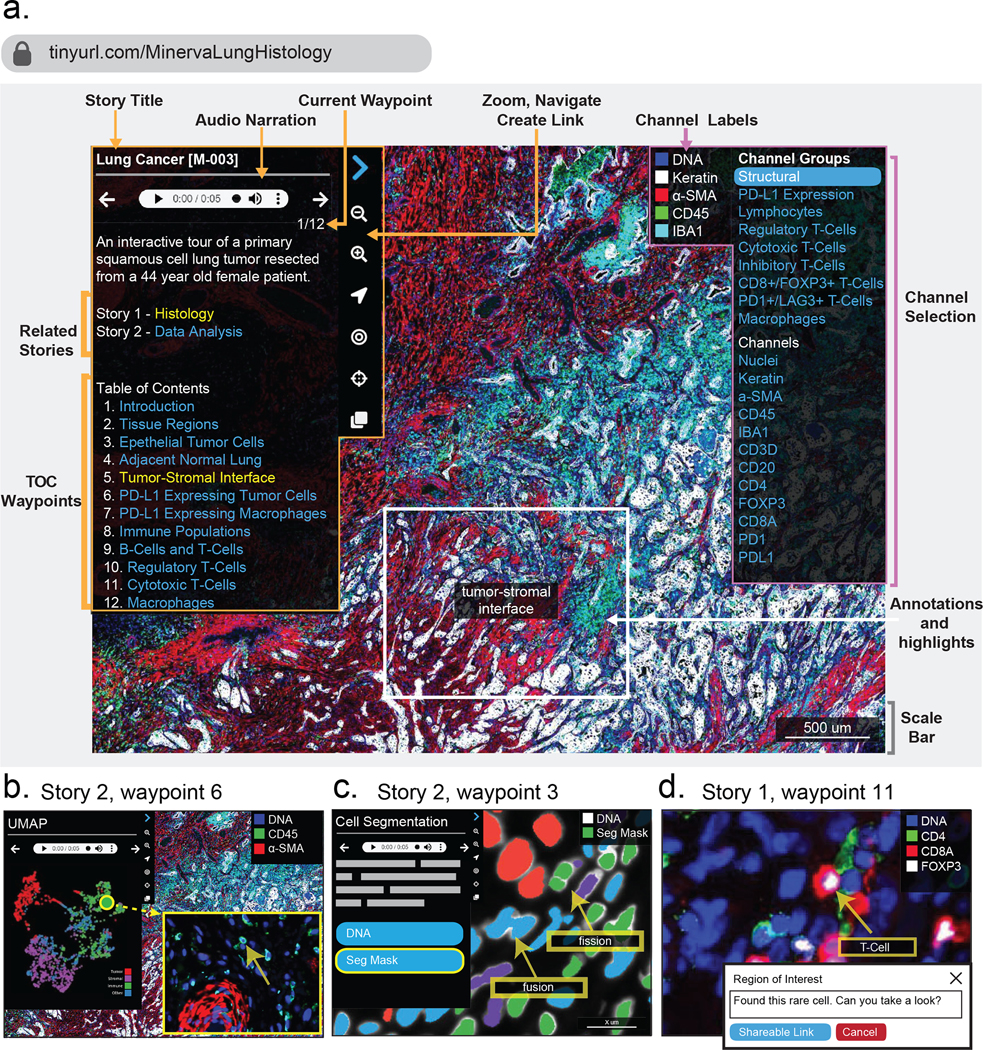
Figure 4. The key features of the user interface of Minerva Story.
Two stories exemplify the use of Minerva for a multiplexed image of a specimen of lung adenocarcinoma. Story 1 focuses on histology and immune populations and can be accessed at https://www.cycif.org/MinervaLungHistology. Story 2 focuses on data generation and analysis and can be accessed at https://www.cycif.org/MinervaLungData. a, The home screen for story 1. A narration panel on the left-hand side shows the title and narrative text, which can be read aloud by the software (through the use of the audio panel). Also, the left panel has links to related stories for the specimen and a table of contents (TOC) listing each waypoint in the story (outlined in orange). Users can select the navigation arrows to step through each waypoint or skip to a waypoint of interest by using the table of contents. On the right-hand side, the channel-selection panel allows users to change which channels are rendered (outlined in pink). At any point, users can depart from a story and freely pan and zoom in or out of the image by using a mouse, trackpad or the magnification icons in the navigation panel. Arrowheads, concentric circles and other graphical elements can be used to annotate images and to generate web URLs specific to the current rendering. The viewer also contains a scale bar that grows and shrinks as a user navigates across zoom levels (grey). The screenshot shown includes only a subset of the waypoints and channel groups. Extensive information on these features is available at https://github.com/labsyspharm/minerva-story/wiki. b, Left: waypoint 6 from story 2 displays an interactive plot of a UMAP performed on a random sample of 2,000 cells from a tissue. Users can select any data point in the plot (each data point represents a single cell), and the browser will zoom to the position of that cell in the image and place an arrow on it. Middle: waypoint 3 from story 2 shows each cell in the tissue overlaid with a segmentation mask in which the colour denotes the cell type, as determined by quantitative k-means clustering. This makes it possible for users to assess the accuracy and the effects of segmentation on downstream cell-type-calling. The user can toggle image data and segmentation masks for different immunofluorescence channels on and off, and also overlay the clustering results and the image data in the same view. Waypoint 3 annotates two types of common segmentation errors: fission and fusion. Right: on-the-fly annotation tool. Users can add annotations and enter text and then hit the blue button to generate a URL that allows anyone to render the same image and location, preserving the panning and zoom settings, the marker group and the annotations. The screenshots displayed in this figure contain text of size and rendering that are optimized for presentation in this figure panel.
Interactive guides for exploring human tumours
As a case study of an interactive tissue guide, we used Minerva Story to mark up a large specimen of human lung adenocarcinoma (https://www.cycif.org/MinervaLungHistology). This primary tumour measured ~5 × 3.5 mm and was imaged at subcellular resolution using 44-plex t-CyCIF4. Multiple fields were then stitched into a single image (described in ref. 54; the image is referred to within that paper as LUNG-3-PR). Because there is no single best way to analyse an image containing several hundred thousands of cells, we created two guides: a guide focused on the histological review of regions of interest and on specific types of immune cells and tumour cells, and a guide focused on the presentation of quantitative data analysis in the context of the original image (Figure 4a). A panel to the left of the Minerva Story window (outlined in orange in Figure 4a) shows the name of the samples, the links to its related stories, a table of contents and the navigation tools. On the right of the window (outlined in pink), another navigation panel allows for the selection of channels of interest. Channels and channel groups, as well as the cell types that they define, can be pre-specified. Each protein (antigen) is linked to an explanatory source of information (such as the GeneCards55 database; a more customized and tissue-specific annotation of markers is under development).
Stories progress from one waypoint to the next (analogous to the numbered system used by museum audio guides), and each waypoint can involve a different field of view, magnification and set of channels, as well as arrows and text describing specific features of interest (marked in grey). At any point, users can diverge from a story by panning and zooming around the image or by selecting different channels, and then return to the narrative by selecting the appropriate waypoint on the table of contents. In Figure 4a, ‘waypoint one’ shows pan-cytokeratin-positive tumour cells growing in chords and clusters at the tumour–stroma interface. This region of the tumour is characterized by an inflammatory microenvironment, as evidenced by the presence of a variety of lymphocyte and macrophage populations distinguishable by the expression of their cell surface markers. By using the panel on the right, users can toggle these markers on and off to explore the images and data, and to evaluate the accuracy of the classification. The remainder of the story explores the expression of PD-L1 and the localization of populations of lymphocytes and macrophages.
Narrative guides are also useful for showing the results of quantitative data analyses in the context of the original image (as described in the lung adenocarcinoma story). For example, the data analysis of high-plex tissue images typically involves measuring the co-expression of multiple cell type markers (such as immune lineage markers) for the identification of individual cell types. The analysis of cell states, morphologies and neighbourhood relationships are also common. Within Minerva Story, it is possible to link representations of quantitative data directly to the image space. For example, when data are captured in a 2D plot, such as a UMAP for dimension reduction (Figure 4b), selecting a data point takes the user directly to the corresponding position of the cell in the image (denoted with an arrow). This is a standard feature in desktop software such as histoCat56 and Facetto57, and greatly enhances a user’s understanding of the relationship between images and image-derived features. The display of segmentation masks is similarly useful for troubleshooting and for assessing data quality (for example, in Figure 4b, middle, unwanted fusion and fission events are highlighted by arrows, both of which result from errors in segmentation). Additionally, users can interactively highlight areas of interest, add notes and generate sharable links that allow others to navigate to the same position in the image and to view any added annotations and text (for example, a high-magnification view with a labelled CD8+ Foxp3+ regulatory T cell in Figure 4b, right).
Narrative guides for medical education
Narrative tissue guides can be used for teaching. Histology is challenging to teach in an undergraduate setting58 and changes in curriculum have reduced the amount of time that medical students and residents spend in front of a microscope. Online collections of tissue images are a frequent substitute. In fact, pairing classroom instruction with dynamic viewing and flexible interaction with image data is essential for learning59. We therefore created a narrative guide of H&E images obtained from specimens of heart tissue from a patient who experienced multiple episodes of myocardial infarction (Figure 5). An introductory panel depicts the overall structure of the heart and the positions from which various specimens were resected. These images display the histological hallmarks of ischaemic heart disease: severe coronary artery atherosclerosis, plaque rupture, stunned myocardium, reperfusion injury, as well as the early, intermediate and late features of myocardial tissue infarction. Rather than looking at snippets of histological data in a textbook or in poorly annotated online images, the interactive narrations of tissue-image data provide context that facilitates the more nuanced understanding of common cardiac pathophysiology.
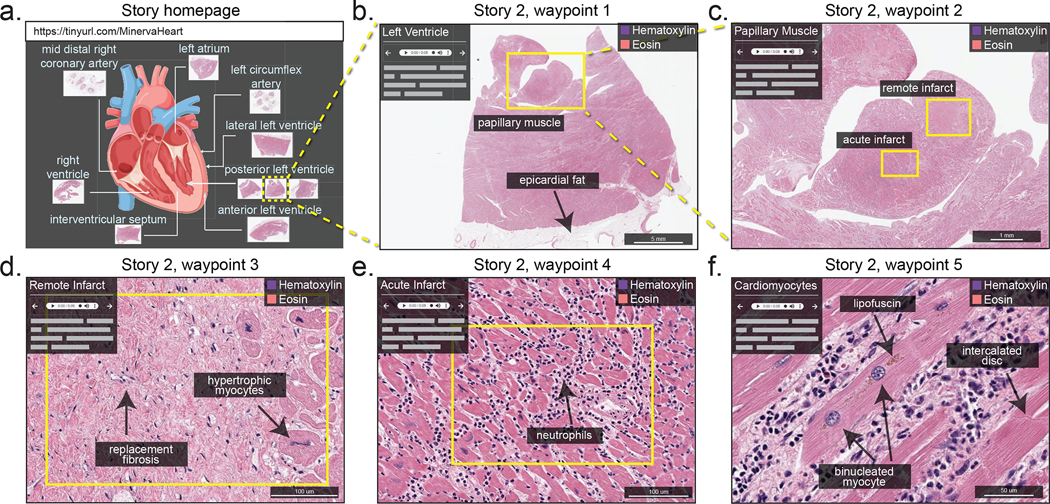
Figure 5. Minerva Story for medical education.
Minerva Story can be used for the guided viewing of conventional H&E-stained sections. For example, a Minerva story was created to guide students through tissue specimens from different anatomical regions of the heart of a patient who suffered multiple myocardial infarcts. The Story homepage shows an anatomical schematic of the heart, indicating the regions from which tissue specimens in the Minerva story were collected. Waypoint 1 shows a specimen sliced with a posterior view of the left ventricle, with box, arrow and text annotations of a few histological features. Waypoint 2 of the story shows a zoomed-in view of a papillary muscle, characteristic of the left ventricle, with annotations indicating regions of tissue showing acute (~12 days old) and remote (6 weeks to years old) myocardial infarction. Waypoint 3 and Waypoint 4 guide the user to the regions of remote infarct and acute infarct in the papillary muscle, describing histological hallmarks to distinguish the two types of infarcts. Waypoint 5 depicts microscopic cellular structures of cardiomyocytes at high resolution in a region of the tissue with a late acute infarct. This Minerva story is available at https://www.cycif.org/MinervaHeart.
Software such as Minerva Story should be useful for medical education and could enhance interactive textbooks. With relatively little effort, text can be hyperlinked to stories and waypoints, and students could use image-annotation features to take notes and to ask questions. Audio-based narration is advantageous in this context, as it could allow students to receive lecture content while simultaneously concentrating on a relevant image.
Outlook
Multiplexed tissue-imaging methods generate images that are data-rich; however, their potential to inform basic, translational and clinical research remains largely untapped, which is in large part because the complexity and size of these images makes it difficult to process and share them. As tissue atlases become increasingly available, software that enables interactive data exploration with cloud-based storage of pre-rendered narrative guides could dramatically increase the number of people who can access a pathologist’s expertise and view an image. Ideal systems combine intuitive panning, zooming and data-exploration interfaces, guidance from digital docents and low costs for the data providers. These possibilities are illustrated by the Minerva software and the OpenSeadragon platform on which it is based. Ideally, publication of high-plex images could in the future be tied to the release of the primary data and to software viewers having digital docents to make the data easier to interpret; this would meet the key goals of the JCB DataViewer project30 and be a step towards realizing FAIR data standards.
With respect to data analysis, a long-term goal of computational biology is to have a single copy of the data in the cloud so that computational algorithms can be more easily used across cloud datasets. At present, however, the dominant approach of local data analysis remains, in particular for interactive tasks such as optimizing segmentation, training machine-learning algorithms, and performing and validating data clustering, which all require local access to full-resolution primary data. With commercial cloud services, data download has become the primary expense and can be substantial for multi-terabyte datasets. A solution could be the use of ‘requester pays’ buckets, available on cloud services from Amazon, Google and other providers of these services. These would allow a data generator to make even large datasets publicly available by requiring the requester to cover the cost of data transfer. In the short term, we envision an ecosystem in which web clients for databases such as OMERO provide interactive viewing of primary data in private datasets for which demand on the OMERO server can be better managed. Lightweight viewers such as Minerva would provide unrestricted access to published and processed data, making the data easy to access and understand by non-experts, and requester-pays cloud buckets would allow experts (in particular, computational biologists) to access primary data and to perform their own analyses locally. In the longer term, these functions are likely to merge, with OMERO providing sophisticated cloud-based processing of data and Minerva serving as a means to access data virtually cost-free, either in OMERO or in a static file system.
An obvious extension of Minerva would be the addition of tools that facilitate supervised machine learning; in such an application, a Minerva variant would provide an efficient way of adding the needed labels to data to train classifiers and neural networks. The results of the machine-learning-based classification could also be checked in Minerva, as its lightweight implementation facilitates crowdsourcing. With these possibilities in mind, a key question is whether tools such as Minerva should also expand to include sophisticated image-processing functions. We do not think so; instead, we believe that narrative guides should be optimized for image review, publication and description, and other interoperable software should continue to be used for data analysis. These tools can be joined together into efficient workflows (such as MCMICRO60) that use software containers (such as Docker)61 and pipeline frameworks (such as Nextflow62). This approach is not perfect63 (Box 2), but it cannot reasonably be replaced by all-in-one commercial or academic software. Recent interest in single-cell tissue biology derives not only from advances in microscopy but also from the widespread adoption of single-cell sequencing64,65. Comprehensive characterization of normal and diseased tissues will almost certainly involve the integration of data from multiple analytical modalities, including imaging, spatial transcriptomic profiling66,67, mass spectrometry imaging of metabolites and drugs68,69, and computational registration of dissociated single-cell RNA sequencing70 with spatial features. Minerva cannot currently handle all of these tasks, but it can be readily combined with other tools to create the multi-omic viewers that are needed for tissue atlases. An attractive possibility is to add narrative tissue guides to widely used genomics platforms such as cBioPortal for Cancer Genomics71 to create environments in which genomic and multiplexed tissue histopathology can be viewed simultaneously. Better visualization could also help with the more conceptually challenging task of integrating spatiomolecular features in multiplex images with gene expression and mutational data. With regards to clinical applications, the biomedical community needs to ensure that digital-pathology systems do not become locked behind proprietary data formats based on non-interoperable software. OME and BioFormats for microscopy, and DICOM for radiology72, exemplify that it is possible for academic developers, commercial instrument manufactures and software vendors to work together for mutual benefit. Easy-to-use visualization software based on permissive open-source licensing could readily be deployed by research groups, instrument makers and scientific publishers to make high-plex image data widely accessible and easy to understand. This would help to realize a FAIR future for tissue imaging and digital pathology.
Acknowledgements
This work was funded by NIH grants U54-CA225088 to P.K.S. and S.S., and by the Ludwig Center at Harvard. The Dana-Farber/Harvard Cancer Center is supported in part by NCI Cancer Center Support Grant P30-CA06516.
Competing interests
P.K.S. is a member of the SAB and BOD member of Applied Biomath, RareCyte Inc., and Glencoe Software, which distributes a commercial version of the OMERO database. P.K.S. is also a member of the NanoString SAB. In the past 5 years, the Sorger Laboratory has received research funding from Novartis and Merck. P.K.S. declares that none of these relationships have influenced the content of this manuscript. S.S. is a consultant for RareCyte Inc. The remaining authors declare no competing interests.
Data availability
Source code for Minerva Story can be found at https://github.com/labsyspharm/minerva-story and detailed documentation and user guide at https://github.com/labsyspharm/minerva-story/wiki.
References
- 1.Giesen C et al. Highly multiplexed imaging of tumor tissues with subcellular resolution by mass cytometry. Nat. Methods 11, 417–422 (2014). [DOI] [PubMed] [Google Scholar]
- 2.Angelo M et al. Multiplexed ion beam imaging of human breast tumors. Nat. Med. 20, 436–442 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gerdes MJ et al. Highly multiplexed single-cell analysis of formalin-fixed, paraffin-embedded cancer tissue. Proc. Natl Acad. Sci. USA 110, 11982–11987 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lin J-R et al. Highly multiplexed immunofluorescence imaging of human tissues and tumors using t-CyCIF and conventional optical microscopes. eLife 10.7554/eLife.31657 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bodenmiller B Multiplexed epitope-based tissue imaging for discovery and healthcare applications. Cell Syst. 2, 225–238 (2016). [DOI] [PubMed] [Google Scholar]
- 6.Coy S et al. Multiplexed immunofluorescence reveals potential PD-1/ PD-L1 pathway vulnerabilities in craniopharyngioma. Neuro Oncol. 20, 1101–1112 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Goltsev Y et al. Deep profiling of mouse splenic architecture with CODEX multiplexed imaging. Cell 174, 968–981.e15 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stack EC, Wang C, Roman KA & Hoyt CC Multiplexed immunohistochemistry, imaging, and quantitation: a review, with an assessment of tyramide signal amplification, multispectral imaging and multiplex analysis. Methods 70, 46–58 (2014). [DOI] [PubMed] [Google Scholar]
- 9.Akturk G, Sweeney R, Remark R, Merad M & Gnjatic S Multiplexed immunohistochemical consecutive staining on single slide (MICSSS): multiplexed chromogenic IHC assay for high-dimensional tissue analysis. Methods Mol. Biol. 2055, 497–519 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Saka SK et al. Immuno-SABER enables highly multiplexed and amplified protein imaging in tissues. Nat. Biotechnol. 37, 1080–1090 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li W, Germain RN & Gerner MY Multiplex, quantitative cellular analysis in large tissue volumes with clearing-enhanced 3D microscopy (Ce3D). Proc. Natl Acad. Sci. USA 114, E7321–E7330 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Regev A et al. The Human Cell Atlas. eLife 10.7554/eLife. 27041 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Consortium HuBMAP. The human body at cellular resolution: the NIH Human Biomolecular Atlas Program. Nature 574, 187–192 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rozenblatt-Rosen O et al. The Human Tumor Atlas Network: charting tumor transitions across space and time at single-cell resolution. Cell 181, 236–249 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.The Cancer Genome Atlas Research Network et al. The Cancer Genome Atlas Pan-Cancer analysis project. Nat. Genet. 45, 1113–1120 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shin D et al. PathEdEx—uncovering high-explanatory visual diagnostics heuristics using digital pathology and multiscale gaze data. J. Pathol. Inf. 8, 29 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wagner J et al. A single-cell atlas of the tumor and immune ecosystem of human breast cancer. Cell 177, 1330–1345.e18 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bui MM et al. Digital and computational pathology: bring the future into focus. J. Pathol. Inform. 10, 10 (2019). [Google Scholar]
- 19.Brown M & Wittwer C Flow cytometry: principles and clinical applications in hematology. Clin. Chem. 46, 1221–1229 (2000). [PubMed] [Google Scholar]
- 20.Coons AH, Creech HJ, Jones RN & Berliner E The demonstration of pneumococcal antigen in tissues by the use of fluorescent antibody. J. Immunol. 45, 159–170 (1942). [Google Scholar]
- 21.Pantanowitz L et al. Twenty years of digital pathology: an overview of the road travelled, what is on the horizon, and the emergence of vendor-neutral archives. J. Pathol. Inf. 9, 40 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gurcan MN et al. Histopathological image analysis: a review. IEEE Rev. Biomed. Eng. 2, 147–171 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Technical Performance Assessment of Digital Pathology Whole Slide Imaging Devices (US Food and Drug Administration, 2016); http://www.fda.gov/regulatory-information/search-fda-guidance-documents/technical-performance-assessment-digital-pathology-whole-slide-imaging-devices [Google Scholar]
- 24.Ehteshami Bejnordi B et al. Diagnostic assessment of deep learning algorithms for detection of lymph node metastases in women with breast cancer. JAMA 318, 2199–2210 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gulshan V et al. Development and validation of a deep learning algorithm for detection of diabetic retinopathy in retinal fundus photographs. JAMA 316, 2402–2410 (2016). [DOI] [PubMed] [Google Scholar]
- 26.Greenspan H, van Ginneken B & Summers RM Guest editorial deep learning in medical imaging: overview and future promise of an exciting new technique. IEEE Trans. Med. Imaging 35, 1153–1159 (2016). [Google Scholar]
- 27.Abels E et al. Computational pathology definitions, best practices, and recommendations for regulatory guidance: a white paper from the Digital Pathology Association. J. Pathol. 249, 286–294 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schapiro D et al. MITI Minimum Information guidelines for highly multiplexed tissue images. Preprint at https://arxiv.org/abs/2108.09499 (2021). [DOI] [PMC free article] [PubMed]
- 29.Goldberg IG et al. The Open Microscopy Environment (OME) data model and XML file: open tools for informatics and quantitative analysis in biological imaging. Genome Biol. 6, R47 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hill E Announcing the JCB DataViewer, a browser-based application for viewing original image files. J. Cell Biol. 183, 969–970 (2008). [Google Scholar]
- 31.Swedlow JR, Goldberg I, Brauner E & Sorger PK Informatics and quantitative analysis in biological imaging. Science 300, 100–102 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Singh J FigShare. J. Pharm. Pharmacother. 2, 138–139 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Allan C et al. OMERO: flexible, model-driven data management for experimental biology. Nat. Methods 9, 245–253 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Williams E et al. The Image Data Resource: a bioimage data integration and publication platform. Nat. Methods 14, 775–781 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Levit LA et al. Ethical framework for including research biopsies in oncology clinical trials: American Society of Clinical Oncology research statement. J. Clin. Oncol. 37, 2368–2377 (2019). [DOI] [PubMed] [Google Scholar]
- 36.Kaye J, Heeney C, Hawkins N, de Vries J & Boddington P Data sharing in genomics—reshaping scientific practice. Nat. Rev. Genet. 10, 331–335 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Reardon J et al. Bermuda 2.0: reflections from Santa Cruz. Gigascience 5, 1–4 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gutman DA et al. The Digital Slide Archive: a software platform for management, integration, and analysis of histology for cancer research. Cancer Res. 77, e75–e78 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wilkinson MD et al. The FAIR Guiding Principles for scientific data management and stewardship. Sci. Data 3, 160018 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Robinson JT et al. Integrative genomics viewer. Nat. Biotechnol. 29, 24–26 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stephens GJ, Silbert LJ & Hasson U Speaker–listener neural coupling underlies successful communication. Proc. Natl Aacd. Sci. USA 107, 14425–14430 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rieger KL et al. Digital storytelling as a method in health research: a systematic review protocol. Syst. Rev. 7, 41 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wilson DK, Hutson SP & Wyatt TH Exploring the role of digital storytelling in pediatric oncology patients’ perspectives regarding diagnosis: a literature review. SAGE Open 10.1177/2158244015572099 (2015). [DOI] [Google Scholar]
- 44.De Vecchi N, Kenny A, Dickson-Swift V & Kidd S How digital storytelling is used in mental health: a scoping review. Int J. Ment. Health Nurs. 25, 183–193 (2016). [DOI] [PubMed] [Google Scholar]
- 45.Lee H, Fawcett J & DeMarco R Storytelling/narrative theory to address health communication with minority populations. Appl. Nurs. Res 30, 58–60 (2016). [DOI] [PubMed] [Google Scholar]
- 46.Botsis T, Fairman JE, Moran MB & Anagnostou V Visual storytelling enhances knowledge dissemination in biomedical science. J. Biomed. Inf. 107, 103458 (2020). [DOI] [PubMed] [Google Scholar]
- 47.ElShafie SJ Making science meaningful for broad audiences through stories. Integr. Comp. Biol. 58, 1213–1223 (2018). [DOI] [PubMed] [Google Scholar]
- 48.OpenSeadragon v.2.4.2 (OpenSeadragon contributors, 2013); https://openseadragon.github.io/ [Google Scholar]
- 49.Jianu R & Laidlaw DH What Google Maps can do for biomedical data dissemination: examples and a design study. BMC Res. Notes 6, 179 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hoffer J et al. Minerva: a light-weight, narrative image browser for multiplexed tissue images. J. Open Source Softw. 5, 2579 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jekyll v.4.2.0 (The Jekyll Team; 2021); https://jekyllrb.com/ [Google Scholar]
- 52.Aeffner F et al. Introduction to digital image analysis in whole-slide imaging: a white paper from the Digital Pathology Association. J. Pathol. Inf. 10, 9 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hiner MC, Rueden CT & Eliceiri KW SCIFIO: an extensible framework to support scientific image formats. BMC Bioinformatics 17, 521 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rashid R et al. Highly multiplexed immunofluorescence images and single-cell data of immune markers in tonsil and lung cancer. Sci. Data 6, 323 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rebhan M, Chalifa-Caspi V, Prilusky J & Lancet D GeneCards: integrating information about genes, proteins and diseases. Trends Genet. 13, 163 (1997). [DOI] [PubMed] [Google Scholar]
- 56.Schapiro D et al. histoCAT: analysis of cell phenotypes and interactions in multiplex image cytometry data. Nat. Methods 14, 873–876 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Krueger R et al. Facetto: combining unsupervised and supervised learning for hierarchical phenotype analysis in multi-channel image data. IEEE Trans. Vis. Comput. Graph. 26, 227–237 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.García M, Victory N, Navarro-Sempere A & Segovia Y Students’ views on difficulties in learning histology. Anat. Sci. Educ. 12, 541–549 (2019). [DOI] [PubMed] [Google Scholar]
- 59.Mione S, Valcke M & Cornelissen M Remote histology learning from static versus dynamic microscopic images. Anat. Sci. Educ. 9, 222–230 (2016). [DOI] [PubMed] [Google Scholar]
- 60.Schapiro D et al. MCMICRO: a scalable, modular image-processing pipeline for multiplexed tissue imaging. Preprint at bioRxiv 10.1101/2021.03.15.435473 (2021). [DOI] [PMC free article] [PubMed]
- 61.O’Connor BD et al. The Dockstore: enabling modular, community-focused sharing of Docker-based genomics tools and workflows. F1000Res 6, 52 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Di Tommaso P et al. Nextflow enables reproducible computational workflows. Nat. Biotechnol. 35, 316–319 (2017). [DOI] [PubMed] [Google Scholar]
- 63.Siepel A Challenges in funding and developing genomic software: roots and remedies. Genome Biol. 20, 147 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Macaulay IC, Ponting CP & Voet T Single-cell multiomics: multiple measurements from single cells. Trends Genet. 33, 155–168 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Trapnell C Defining cell types and states with single-cell genomics. Genome Res. 25, 1491–1498 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ståhl PL et al. Visualization and analysis of gene expression in tissue sections by spatial transcriptomics. Science 353, 78–82 (2016). [DOI] [PubMed] [Google Scholar]
- 67.Schulz D et al. Simultaneous multiplexed imaging of mRNA and proteins with subcellular resolution in breast cancer tissue samples by mass cytometry. Cell Syst. 6, 25–36.e5 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Van de Plas R, Yang J, Spraggins J & Caprioli RM Image fusion of mass spectrometry and microscopy: a multimodality paradigm for molecular tissue mapping. Nat. Methods 12, 366–372 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Liu X et al. Molecular imaging of drug transit through the blood–brain barrier with MALDI mass spectrometry imaging. Sci. Rep. 3, 2859 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Moncada R et al. Integrating microarray-based spatial transcriptomics and single-cell RNA-seq reveals tissue architecture in pancreatic ductal adenocarcinomas. Nat. Biotechnol. 38, 333–342 (2020). [DOI] [PubMed] [Google Scholar]
- 71.Cerami E et al. The cBio Cancer Genomics Portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2, 401–404 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Mildenberger P, Eichelberg M & Martin E Introduction to the DICOM standard. Eur. Radio. 12, 920–927 (2002). [DOI] [PubMed] [Google Scholar]
- 73.Samuel AL Some studies in machine learning using the game of checkers. IBM J. Res. Dev. 3, 210–229 (1959). [Google Scholar]
- 74.Inoué S & Spring K Video Microscopy: The Fundamentals (Springer US, 1997). [Google Scholar]
- 75.Bacus JV & Bacus JW Method and apparatus for acquiring and reconstructing magnified specimen images from a computer-controlled microscope. US patent 6,101,265 (2000).
- 76.HANDEL-P (Pancreatlas, 2021); https://pancreatlas.org/ [Google Scholar]
- 77.Rubin DL, Greenspan H & Brinkley JF in Biomedical Informatics: Computer Applications in Health Care and Biomedicine (eds Shortliffe EH & Cimino JJ) 285–327 (Springer, 2014). [Google Scholar]
- 78.caMicroscope (GitHub, 2021); https://github.com/camicroscope [Google Scholar]
- 79.Clark K et al. The Cancer Imaging Archive (TCIA): maintaining and operating a public information repository. J. Digit. Imaging 26, 1045–1057 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.PathPresenter (Singh R et al. , 2021); https://public.pathpresenter.net/#/login
- 81.Olson AH Image Analysis Using the Aperio ScanScope (Quorum Technologies, 2006). [Google Scholar]
- 82.McQuin C et al. CellProfiler 3.0: next-generation image processing for biology. PLoS Biol. 16, e2005970 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Bankhead P et al. QuPath: open source software for digital pathology image analysis. Sci. Rep. 7, 16878 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Stritt M, Stalder AK & Vezzali E Orbit Image Analysis: an open-source whole slide image analysis tool. PLoS Comput. Biol. 16, e1007313 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Mantis Viewer (Parker Institute for Cancer Immunotherapy, 2021); https://mantis.parkerici.org/ [Google Scholar]
- 86.ASAP—Automated Slide Analysis Platform (Computation Pathology Group—Radboud University Medical Center, 2021); https://computationalpathologygroup.github.io/ASAP/#home [Google Scholar]
- 87.Maaten Lvander & Hinton G Visualizing data using t-SNE. J. Mach. Learn. Res. 9, 2579–2605 (2008). [Google Scholar]
- 88.McInnes L, Healy J & Melville J UMAP: uniform manifold approximation and projection for dimension reduction. Preprint at https://arxiv.org/abs/1802.03426 (2018).
- 89.Arnol D, Schapiro D, Bodenmiller B, Saez-Rodriguez J & Stegle O Modeling cell–cell interactions from spatial molecular data with spatial variance component analysis. Cell Rep. 29, 202–211.e6 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Berg S et al. ilastik: interactive machine learning for (bio)image analysis. Nat. Methods 16, 1226–1232 (2019). [DOI] [PubMed] [Google Scholar]
- 91.Chen J et al. The Allen Cell Structure Segmenter: a new open source toolkit for segmenting 3D intracellular structures in fluorescence microscopy images. Preprint at bioRxiv 10.1101/491035 (2018). [DOI] [Google Scholar]
- 92.Sofroniew N et al. napari/napari: 0.3.6rc2. Zenodo; 10.5281/zenodo.3951241 (2020). [DOI] [Google Scholar]
- 93.Schindelin J, Rueden CT, Hiner MC & Eliceiri KW The ImageJ ecosystem: an open platform for biomedical image analysis. Mol. Reprod. Dev. 82, 518–529 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Source code for Minerva Story can be found at https://github.com/labsyspharm/minerva-story and detailed documentation and user guide at https://github.com/labsyspharm/minerva-story/wiki.