Abstract
Mutations in the Plasmodium falciparum gene (dhfr) encoding dihydrofolate reductase are associated with resistance to antifols. Plasmodium vivax, the more prevalent malaria parasite in Asia and the Americas, is considered antifol resistant. Functional polymorphisms in the dhfr gene of P. vivax (pvdhfr) were assessed by PCR-restriction fragment length polymorphism using blood samples taken from 125 patients with acute vivax malaria from three widely separated locations, Thailand (n = 100), India (n = 16), and Madagascar and the Comoros Islands (n = 9). Upon evaluation of the three important codons (encoding residues 57, 58, and 117) of P. vivax dhfr (pvdhfr), double- or triple-mutation genotypes were found in all but one case from Thailand (99%), in only three cases from India (19%) and in no cases from Madagascar or the Comoros Islands (P < 0.0001). The dhfr PCR products of P. vivax from 32 Thai patients treated with the antifolate sulfadoxine-pyrimethamine (S-P) were investigated. All samples showed either double (53%) or triple (47%) mutations. Following treatment, 34% of the patients had early treatment failures and only 10 (31%) of the patients cleared their parasitemias for 28 days. There were no significant differences in cure rates, but parasite reduction ratios at 48 h were significantly lower for patients whose samples showed triple mutations than for those whose samples showed double mutations (P = 0.01). The three mutations at the pvdhfr codons for residues 57, 58, and 117 are associated with high levels of S-P resistance in P. vivax. These mutations presumably arose from selection pressure.
Drugs which inhibit plasmodial folate biosynthesis represent a major component of the antimalarial armamentarium. The sulfonamides and sulfones inhibit dihydropteroate synthase, and pyrimethamine and cycloguanil (the metabolite of proguanil) inhibit dihydrofolate reductase (DHFR). Sequential inhibition results in a synergistic antimalarial effect (6). Unfortunately, resistance to these drugs develops relatively quickly if they are widely used. Although Plasmodium falciparum acquires resistance readily (10, 11), it has been considered that P. vivax is intrinsically resistant to pyrimethamine (27). However, the initial sensitivity of this parasite to proguanil shortly after its initial deployment in peninsular Malaya in 1947 would suggest that acquired resistance is a more likely explanation for treatment failure (11). P. vivax in Thailand is considered highly resistant to sulfadoxine-pyrimethamine (S-P) (15, 20).
Molecular studies associated with detailed clinical and epidemiological observations of falciparum malaria have demonstrated that the major mechanism of resistance to pyrimethamine and sulfadoxine results from specific point mutations in the parasite's dhfr and dhps genes, respectively (6, 8, 13, 19, 23). These mutations result in the nonsynonymous replacement of key amino acid residues and thus a much reduced affinity of the mutant enzyme for the respective drug. Simple and sensitive methods for the detection of these point mutations have been devised for the P. falciparum dhfr and dhps genes (1, 2, 5, 12–14, 17–19, 21, 25, 28). Recently, the gene coding for DHFR-thymidylate synthase of P. vivax has been cloned and sequenced (3). The presence of mutations in the residues of the P. vivax enzyme (at positions 15, 50, 58, 117, and 173) which were predicted by secondary-structure analysis and amino acid homology to correspond to the five key positions on the P. falciparum gene (positions 16, 51, 59, 108, and 164, respectively) was determined for 30 P. vivax isolates of various geographical origins. Although a total of 12 nonsynonymous mutations were observed for these P. vivax dhfr (pvdhfr) gene sequences, only 3 corresponded to the positions mentioned above. Double mutations at residues 117 and 58 were observed most frequently (14 of the 30 samples), but a mutation at residue 173 was found in only 1 of these samples. No mutations were found at residue 15 or 50. The only other mutations encountered more than once were at residues 33 and 57, which were detected in three and in two samples, respectively. These results suggested that residues 58 and 117 are linked to pyrimethamine resistance, but the number of samples analyzed was too small to provide a firm conclusion (4).
In this article, we have extended these observations by analyzing the pvdhfr gene using a large number of P. vivax samples which were obtained from three areas which differ substantially in their current and previous use of antimalarial drugs: Madagascar and the Comoros Islands, where S-P is not and has not been deployed; India, where S-P has not been used widely until recently; and Thailand, where S-P has been used extensively and where high-level resistance in P. falciparum has developed. We have correlated the pvdhfr mutation pattern with clinical and parasitological responses in 32 P. vivax-infected Thai patients who were monitored for 28 days following treatment with S-P. A sensitive nested-PCR–restriction fragment length polymorphism (RFLP) protocol was developed for the study of mutations in the pvdhfr gene. We present an analysis of mutation frequencies at the five positions which have previously been suggested to be involved in resistance to S-P. Our epidemiological and clinical data suggest that of these mutations, only those occurring at positions 57, 58, and 117 of the P. vivax DHFR enzyme sequence are implicated in resistance to pyrimethamine.
MATERIALS AND METHODS
Blood samples.
Admission blood samples were collected in three separate geographical regions from a total of 125 symptomatic patients who acquired P. vivax infections between 1995 and 1998. Samples from Madagascar or the Comoros Islands (n = 9) were obtained from the Centre Hospitalier de Dunkerque, Rosendael, France; samples from the Indian subcontinent (n = 16) were obtained from Northwick Park Hospital, Harrow, United Kingdom; and samples from Thailand (n = 100) were collected from adult patients admitted to the Bangkok Hospital for Tropical Diseases. All samples were collected on admission before the start of treatment and stored frozen at −30 or −70°C until DNA extraction. The diagnoses were made by microscopic examination of Field- or Giemsa-stained thin and thick blood smears. The presence of P. vivax was confirmed by duplicate PCR analysis of a 5-ul aliquot from each sample.
Patients.
A subset of 32 adult patients admitted to the Hospital for Tropical Diseases in Bangkok, who gave fully informed consent, were treated with a combination of 1,500 mg of sulfadoxine and 75 mg of pyrimethamine and were kept under daily parasitological and clinical observation for 28 days. Details of these studies, which were approved by the ethical committee of the Faculty of Tropical Medicine of Mahidol University, are to be reported in full elsewhere. These patients were subdivided into three groups according to the clinical outcome as follows: the cured group, whose parasites and fevers cleared within 7 days and who showed no further symptoms or detectable parasitemia; the recurrence group, whose parasites reappeared in the blood circulation between days 7 and 28 following the initial clearance of the parasitemia; and the failure group, whose parasites failed to clear during the first 7 days following treatment. Those patients whose primary treatment failed received standard retreatment with chloroquine. Parasite counts were determined using microscopy, by counting infected red blood cells per 1,000 red blood cells in thin smears or by calculating the parasite count per 200 white blood cells in thick smears. Parasite clearance time was calculated as the time taken for the parasitemia at the time of admission to fall below detectable levels. Fever clearance time (FCT) was the duration of time that elapsed before the patient's temperature returned to <37.5°C following admission and remained below this temperature for >48 h. The parasite reduction ratio at 48 h (PRR48) is the ratio of the parasite count before treatment to that observed at 48 h (26).
DNA template preparation and pvdhfr amplification.
Template DNA was purified from 1 ml of infected blood using the QIAamp DNA kit (Qiagen, Hilden, Germany). The DNA was eluted with Tris-EDTA buffer (10 mM Tris-HCl [pH 8.0], 0.1 mM EDTA [pH 8.0]) such that 1 μl of the solution corresponded to 5 μl of whole blood.
The oligonucleotides used to amplify a fragment of the pvdhfr gene were designed using a published sequence of the dhfr-ts gene of P. vivax (GenBank accession no. X98123). Nested- or seminested-PCR amplification strategies were adopted. In the primary reaction, the whole of the pvdhfr-ts gene, 1,872 bp excluding the stop codon, was amplified using the oligonucleotide pair VDT-OF (5′-ATGGAGGACCTTTCAGATGTATTTGACATT-3′) and VDT-OR (5′-GGCGGCCATCTCCATGGTTATTTTATCGTG-3′). The product of this reaction was then used to initiate a second round of amplification in which either the first 611 bp of the 711 bp of pvdhfr, using the oligonucleotide pair VDT-OF and VDF-NR (5′-TCACACGGGTAGGCGCCGTTGATCCTCGTG-3′), or the first 238 bp of pvdhfr, using the oligonucleotide pair VDT-OF and VDF-NR58 (5′-GGTACCTCTCCCTCTTCCACTTTAGCTTCT-3′), were amplified. A fragment (ca. 250 bp) spanning the repeat region of pvdhfr was amplified using the oligonucleotide pair VDF-N2F (5′-CGGTGACGACCTACGTGGATGAGTCAAAGT-3′) and N2R (5′-TAGCGTCTTGGAAAGCACGACGTTGATTCT-3′).
All amplification reactions were carried out in a final volume of 20 μl, which included 1 μl of template in the form of genomic DNA for the primary reactions or of the product of the primary reaction for the secondary amplification. Oligonucleotide primers were each used at final concentrations of 125 nM in the primary reactions and 250 nM in the secondary reactions. The reaction mixture contained 10 mM Tris-HCl (pH 8.3), 50 mM KCl, a 125 μM concentration of each of the four deoxynucleoside triphosphates, and 0.4 U of AmpliTaq polymerase (Perkin-Elmer Cetus, Mass.). The reactions were carried out in the presence of 2 mM MgCl2 for all oligonucleotide combinations except the VDT-OF–VDF-NR58 pair, for which a concentration of 3 mM MgCl2 was used. The cycling parameters for the PCR were as follows: an initial denaturation step at 95°C for 5 min was followed by 25 (primary reaction) or 30 (secondary reaction) cycles of annealing at 64°C (62°C for VDF-N2F–N2R) for 2 min, extension at 72°C for 2 min, and denaturation at 94°C for 1 min. After a final annealing step followed by 5 min of extension, the reaction was stopped. PCR products were stored at 4°C until analysis.
Analysis of pvdhfr.
The DNA fragments obtained following PCR amplification or RFLP analysis were analyzed following electrophoresis on 3% MetaPhor agarose gels (performed in Tris-borate-EDTA buffer). Digestion of 10 μl of the PCR product was performed using 10 U of each restriction enzyme (New England Biolabs Inc.) for 3 h at 37°C in a total volume of 20 μl.
RESULTS
Amplification of pvdhfr fragments.
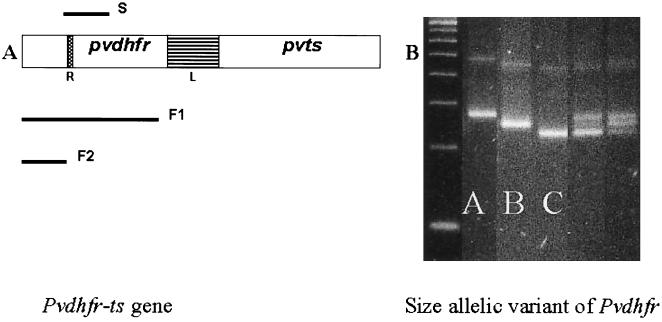
In order to improve on the previously published method, where the detection of the pvdhfr gene required the presence of 100 or more parasites in the aliquot to be analyzed, a nested- or seminested-PCR amplification strategy was developed. Specific primers, designed from the previously reported sequence and used in a primary amplification reaction, generated a single fragment (1,872 bp of the whole dhfr-ts gene) from P. vivax genomic DNA (Fig. 1). Three separate secondary amplification reactions were then carried out. The primer S spanned the repeat region (product of ca. 250 bp), and the primers F1 and F2 amplified, respectively, the first 611 bp of the pvdhfr gene and a fragment of 238 bp which included the codon for residue 58.
FIG. 1.
(A) Schematic representation of the pvdhfr-ts gene, with the linker region (L) and the repeat region (R) indicated. Three fragments were obtained by nested-PCR amplifications (S, F1, and F2) for further analysis. (B) Three S fragments of different sizes were observed and designated A, B, and C. In some samples, mixed infections were observed. Electrophoresis was performed in MetaPhor agarose in Tris-borate-EDTA, and the product was visualized by UV transillumination following ethidium bromide staining.
The protocol was optimized using 5-μl aliquots of uninfected blood spiked with known numbers of P. vivax-infected red blood cells. Successful nested-PCR amplification using the three secondary-reaction primer pairs was obtained reproducibly from samples containing as little as 1 to 10 P. vivax genomes (ca. 0.00001%). The specificity of the reaction was confirmed by the absence of amplification products when genomic DNA from P. falciparum, P. malariae, or P. ovale parasites or genomic DNA from human blood was used as a template. Specific amplification products were obtained from all the samples collected in this study.
Size polymorphism of pvdhfr genes at repeat region.
The pvdhfr gene contains a tandem repeat region between nucleotides 262 and 309 (3). Four allelic variants that differed with respect to the repeat motifs were found previously. The variant without the deletion was found in 15 of the 30 samples for which the pvdhfr gene product was sequenced; the sequence THGGD (15 bp) was missing in 12 of 15 samples, TSGGDN (18 bp) was missing in 2 of 15 samples, and GGDNAD (18 bp) was missing in the last 15 samples (4). In the present study, oligonucleotide primers were designed to amplify a small fragment spanning this region. Three size variants, designated A, B, and C from the largest to the smallest in base pairs, respectively (range, ca. 230 to 280 bp), were found in the 125 blood samples (Fig. 1). The percentages of distribution of pvdhfr genotypes for vivax malaria patients from all three geographic regions were 17% for the type A variant, 58% for the type B variant, and 45% for the type C variant. The genotypes for the majority of Thai samples were type C (43%), but those for most of the parasites from the other two regions were type B (87% of those from India and 100% of those from Madagascar and the Comoros islands). Mixed-genotype infections were observed only in Thai isolates (20%).
RFLP determination of specific mutations in pvdhfr.
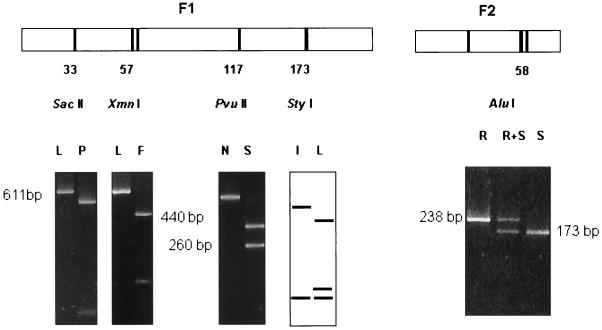
The presence of mutations in the codons for residues 33, 57, 58, 117, and 173 of the product of the pvdhfr gene (numbered according to the sequence available under EMBL GeneBank accession number X98123) was assessed by RFLP analysis using the enzymes indicated (Fig. 2): the 611-bp product was digested with SacII (residue 33, P→L), XmnI (residue 57, F→L), and PvuII (residue 117, S→N), for which digestion was observed only for the wild-type sequence. The mutation at residue 173 (I→L) results in the generation of a second StyI site in the amplified 611-bp fragment. The mutation at residue 58 (S→R) also abolishes the recognition sequence of AluI; however, as this enzyme has multiple sites in the 611-bp fragment, the analysis was carried out on the 238-bp amplified fragment spanning this position (Fig. 2), which is cut twice by AluI for the wild-type sequence but only once for the mutant sequence.
FIG. 2.
MetaPhor electrophoresis of PCR-RFLP products specific for F1 and F2 fragments showing size allelic variants of wild and mutant types at codons corresponding to pvdhfr residues 33, 57, 58, 117, and 173. The restriction sites of SacII, XmnI, PvuII, StyI, and AluI are indicated. F, phenylalanine; I, isoleucine; L, leucine; N, asparagine; P, proline; S, serine; R, arginine.
Mutation frequencies of pvdhfr genes.
Mutations at residue 173 (I→L) were not detected in any of the the samples analyzed. The overall frequencies of the mutations found for the four other positions are presented in Table 1. The overall frequencies of mutant pvdhfr genotypes were 99% for the samples from Thailand, 25% for those from India, and 44% for those from Madagascar and the Comoros Islands. Mutations at residue 33 (P→L) were observed only in 4 of the 9 samples originating from Madagascar and the Comoros Islands. These proved to be the only mutations detected in the pvdhfr gene from these parasites. The mutation at residue 57 (F→L) was detected in 54 of the 100 Thai samples but only 1 of the 16 Indian samples. Nearly all of the parasites from Thailand carried mutations at residues 58 (S→R) (99 of the 100 samples) and 117 (S→N) (98 of the 100 samples), but these mutations were found in only 3 of the 16 Indian samples (P < 0.001).
TABLE 1.
Distribution of pvdhfr mutations at five specific codons
| Countries | Total no. of samples | No. of wild-type samples | No. of samples with mutation at codons for residue:
|
||||
|---|---|---|---|---|---|---|---|
| 33 | 57 | 58 | 117 | 173 | |||
| Thailand | 100 | 1 | 0 | 54a | 99b | 98 | 0 |
| India | 16 | 12 | 0 | 1c | 3d | 3 | 0 |
| Madagascar and Comoros islands | 9 | 5 | 4 | 0 | 0 | 0 | 0 |
Included 40 with mutant genotypes and 14 with mixed infections of wild-type and mutant genotypes.
Included 98 with mutant genotypes and 1 with mixed infections of wild-type and mutant genotypes.
Both wild-type and mutant genotypes present.
Included two with mutant genotypes and one with mixed infections of wild-type and mutant genotype.
Thus, with respect to amino acid positions 57, 58, and 117, the majority (12 of the 16) of Indian samples had parasites with the wild-type genotype (F-S-S at the respective positions), 2 had parasites with a genotype with a single mutation (L-S-S in one case and F-S-N in the other), and 3 had parasites that harbored a double-mutation genotype (F-R-N). Mixed-genotype infections were found in 2 of the 16 Indian samples (F-S-S plus L-S-S and F-S-N plus F-R-N).
The situation was markedly different for the parasites in the 100 Thai samples, where only 1 sample contained the wild-type genotype, 59 contained a double-mutation genotype (L-R-S in 1 case and F-R-N in 58 cases), and 53 carried a triple mutation (L-R-N) (P < 0.0001). Mixed-genotype infections were found in 14 of the 100 samples from Thailand, with F-R-N plus L-R-N observed 13 times, while in the remaining sample the presence of mixed mutations at residues 57 and 58 did not allow us to categorize the genotypes unambiguously.
The relationships of the genotypes of PCR-RFLP products and allelic size variations of all studied samples are shown in Table 2. Evaluations of genotypes using both the PCR-RFLP products and allelic size variations showed a high prevalence of type B in triple mutations and type C in double mutations. The percentage of the type B genotype (either on its own or mixed with the C genotype) in triple-mutation pvdhfr (47 of 53 samples; 87%) was significantly higher than those of the other genotypes of the PCR-RFLP product (P < 0.01). The percentage of the type C genotype in double-mutation pvdhfr isolates (either alone or mixed) (45 of 49 samples; 92%) was also significantly higher than those of the other genotypes of the PCR-RFLP products (P < 0.01). The majority of mixed genotypes (A, B, and C) per sample had triple mutations (18 of 20 samples; 90%).
TABLE 2.
pvdhfr allelic size variants classified according to number of point mutations
| No. of point mutations | No. of samples with indicated allelic variants
|
Total no. of samples | |||||
|---|---|---|---|---|---|---|---|
| A | B | C | A+B | A+C | A+B+C | ||
| None | 17 | 1 | 18 | ||||
| Single | 1 | 4 | 5 | ||||
| Double | 4 | 43 | 1 | 1 | 49 | ||
| Triple | 35 | 7 | 6 | 5 | 53 | ||
| Total | 1 | 60 | 44 | 7 | 7 | 6 | 125 |
Relationship of pvdhfr genetic mutations to clinical responses following S-P treatment.
Of the P. vivax-infected Thai patients whose samples were analyzed in the present study, 32 were treated with S-P. These patients were kept under observation for 28 days at the Hospital for Tropical Diseases in Bangkok as part of chemotherapy studies to be reported elsewhere. The patients can be divided into three groups by their clinical and parasitological responses to S-P treatment. The parasites did not clear in 11 patients (high-grade resistance), recurrence of P. vivax was observed after initial clearance in a further 11 patients, and cure for up to 28 days was obtained in only 10 (31%) patients. Samples from these patients were analyzed with respect to mutations at residues 57, 58, and 117 encoded by the pvdhfr gene. All the patients harbored parasites carrying mutations at residues 58 and 117, and an additional mutation at residue 57 was observed in 15 of the 32 patients. Thus, double mutations of the pvdhfr gene (resulting in F-R-N) alone were seen in samples from 17 patients, triple mutations were seen in those from 11 patients, and mixed-genotype infections (double mutations plus triple mutations) were seen in those from the remaining 4 patients.
There were no significant differences in age or parasitemia (geometric mean, 11,761 parasites/μl range, 900 to 107,765 per μl) at time of admission whether the patients were grouped by clinical outcome or by number of observed pvdhfr mutations. As expected, FCT correlated with the overall cure rate. There was a significant difference in FCT between the cured and the recurrence groups (28 and 52 h, respectively) and the failure group (100 h) (P = 0.001). PRR48 also corresponded to overall treatment efficacy, with median values of 3.3 (range, 0.33 to 15.7) for the failure group, 10.4 (range, 2 to 75.8) for the recurrence group, and 28.9 (range, 2 to 1,500) for the cured group.
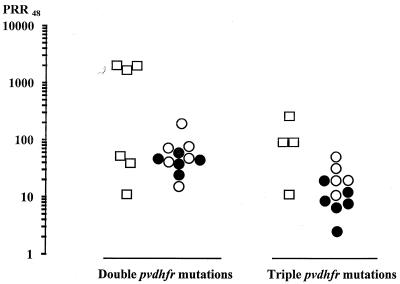
Parasites with pvdhfr genes carrying the double or triple mutation were equally distributed between the three clinical-outcome groups. Although parasite clearance time and FCT did not differ significantly between the patients harboring parasites with double or triple mutations (P > 0.05), there was a significant difference in the PRR48s (P = 0.01) (Fig. 3). The median PRR48 for the patients with parasites with double mutations was 13 (range, 2 to 1,500), and that for those with triple mutations was 3.3 (range, 0.3 to 108). The interval from treatment to reappearance of parasitemia in the recurrence group was shorter for patients with parasites with triple mutations (13 days; range, 11 to 21) than that for those admitted with infections with parasites with double mutations (18 days; range, 11 to 28), but this difference was not statistically significant (P > 0.05).
FIG. 3.
PRR48 of 32 patients with vivax malaria treated with S-P. The clinical responses were classified as cure (□), recurrence (●), or treatment failure (○).
The association between allelic size variants (repeat region) and the specific mutant genotypes was evaluated in all 32 monitored patients (Table 3). Of the triple- and double-mutations PCR-RFLP genotypes, type B (either on its own or mixed with other genotypes) was significantly more commonly associated with the triple mutations (13 of 15 [87%] versus 2 of 15 [13%]) (P < 0.05) while type C (either on its own or mixed with the type A genotype) was significantly more commonly associated with the double mutations (16 of 17 [94%] versus 1 of 17 [6%]) (P < 0.05). There were no significant differences in the incidence of these associated genotypes (triple mutants and type B variants or double mutants and type C variants) between patients with different clinical outcomes (cure, recurrence, or treatment failure).
TABLE 3.
Distribution of allelic size varients of samples with either double or triple mutations of pvfdhr with respect to clinical outcome
| Group of patients | No. of patients with samples showing allelic mutations in codons 57, 58, and 117 (repeat region[s]) resulting in amino acids:
|
||
|---|---|---|---|
| L-R-Na | L/F-R-Na | F-R-Na | |
| Treatment failure (n = 11) | 5 (B), 1 (A+B) | 5 (C) | |
| Recurrence (n = 11) | 2 (B), 2 (A+B) | 1 (A+C) | 6 (C) |
| Cure (n = 10) | 1 (B) | 1 (A+C), 2 (A+B+C) | 5 (C), 1 (A+B+C) |
L, leucine; R, arginine; N, asparagine; F, phenylalanine.
DISCUSSION
The dhfr gene of P. falciparum has been studied extensively. Elegant molecular, epidemiological, and modeling studies have led to the identification of defined residues, most of which are closely associated with the active site, where mutation results in the loss of the parasite's sensitivity to the DHFR inhibitor pyrimethamine (2, 5, 6, 12, 13, 16–18, 23, 24, 28). Analysis of the predicted amino acid sequence of the pvdhfr gene product revealed potential analogous polymorphic residues in this enzyme. Sequence analysis of 30 P. vivax isolates has provided preliminary evidence that residues 58 and 117 are implicated in pyrimethamine resistance, since nonsynonymous mutations at these positions were noted in 13 of the 16 isolates originating from Southeast Asia (3, 4).
In this report we present a highly sensitive nested-PCR–RFLP detection method which we used to analyze the frequency of mutations at defined residues of the gene product in isolates from 125 P. vivax-infected blood samples. These were collected from three geographical regions which differ historically in their use of antimalarial drugs. Chloroquine is the first-line treatment of P. vivax throughout the Indian subcontinent and the Indian Ocean islands (9). S-P use is rare in Madagascar and the Comoros Islands and has been relatively infrequent in India. Although chloroquine is also used to treat vivax malaria in Thailand (7, 22), the prevalence of highly chloroquine-resistant falciparum malaria resulted for a time in the widespread use of S-P. This compound is still available widely in areas where malaria is endemic and is still a first-line treatment in the adjacent countries Laos and Myanmar. Our samples therefore represent a decreasing East-to-West change in drug pressure, and we reasoned that this decrease might be reflected in the accumulation of mutations in the pvdhfr gene of parasites from these regions. Five residues were targeted for analysis in this study, of which three (residues 58, 117, and 173) are predicted to correspond to residues 59, 108, and 164 of the P. falciparum gene product.
Mutations of pvdhfr were significantly more common in Thai parasites (99 of the 100 samples) than in those (8 of 25 samples) found in patients from India, Madagascar, and the Comoros Islands (P < 0.0001). Nearly all the parasites from Thailand had mutations at both residues 58 and 117, whereas these mutations were detected only in 19% of the Indian samples. A similar, albeit less-pronounced, trend was observed for mutations at residue 57, which were noted in 2 of the 30 samples studied previously (4). Triple mutations were found exclusively in Thai parasites. None of the 125 samples showed evidence of a mutation at residue 173. These results provide strong supportive evidence that residues 58 and 117 are involved in pyrimethamine resistance, and residue 57 is then added to this list. Although the data seem to indicate that residue 173 might not play a role in this resistance, it is possible that the fixation of mutations at this site occurs only after sustained drug pressure, as is thought to be the case for residue 164 of the dhfr gene product of P. falciparum. Alternatively, these triple mutations in P. vivax may confer such high levels of resistance that there is no additional selection for residue 173. The fact that mutations at residue 33 were not found in any of the samples except those from Madagascar and the Comoros Islands supports the suggestion of Eldin de Pécoulas et al. (4) that this mutation is idiosyncratic. The existence of “neutral” genetic variants of P. vivax in different regions is consistent with the observation that the frequency of the deletion variants in a region of pvdhfr, which is thought not to be of catalytic importance, differs between genetically isolated parasites.
Many factors affect the therapeutic response in malaria patients. In this study, parasites harboring triple mutations were cleared significantly more slowly than those with double mutations following S-P treatment, irrespective of subsequent clinical outcome. The lower fractional reduction in parasite numbers in samples with triple mutations suggests that these parasites are intrinsically less susceptible to antifolate treatment.
In conclusion, we provide epidemiological and clinical data which are strongly suggestive of a role of mutations in three defined residues of the pvdhfr gene in resistance to S-P. The PCR-RFLP methodology to detect these mutations is practical and simple and can be applied in most research laboratories in countries of P. vivax malaria endemicity. Confirmation that defined genetic mutations result in the resistance of P. vivax to pyrimethamine would be much helped by a routine in vitro test. Further sequencing of samples would also be desirable in determining whether there are other key residues. Finally, molecular studies similar to those performed for the enzyme of P. falciparum, using recombinant proteins with a known pattern of mutations, will ultimately demonstrate to what extent each residue contributes to the alteration of the inhibition constant of the drug and whether synergistic interactions between mutations are a feature of pyrimethamine resistance in P. vivax.
ACKNOWLEDGMENTS
We are grateful to the Royal Golden Jubilee Program of the Thailand Research Fund and the Wellcome Trust for supporting the study.
This study was part of the Wellcome Trust-Mahidol University, Oxford Tropical Medicine Research Programme, funded by the Wellcome Trust of Great Britain.
REFERENCES
- 1.Basco L K, Eldin de Pécoulas P, Wilson C M, Le Bras J, Mazabraud A. Point mutations in dihydrofolate reductase-thymidylate synthase gene and pyrimethamine and cycloguanil resistance in Plasmodium falciparum. Mol Biochem Parasitol. 1995;69:135–138. doi: 10.1016/0166-6851(94)00207-4. [DOI] [PubMed] [Google Scholar]
- 2.Cowman A F, Morry M J, Biggs B A, Cross G A, Foote S J. Amino acid changes linked to pyrimethamine resistance in the dihydrofolate reductase-thymidylate synthase gene of Plasmodium falciparum. Proc Natl Acad Sci USA. 1988;85:9109–9113. doi: 10.1073/pnas.85.23.9109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Eldin de Pecoulas P, Basco L K, Tahar R, Ouatas T, Mazabraud A. Analysis of the Plasmodium vivax dihydrofolate reductase-thymidylate synthase gene sequences. Gene. 1998;211:177–185. doi: 10.1016/s0378-1119(98)00118-8. [DOI] [PubMed] [Google Scholar]
- 4.Eldin de Pécoulas P, Tahar R, Ouatas T, Mazabraud A, Basco L K. Sequences variations in the Plasmodium vivax dihydrofolate reductase-thymidylate synthase gene and their relationship with pyrimethamine resistance. Mol Biochem Parasitol. 1998;92:265–273. doi: 10.1016/s0166-6851(97)00247-8. [DOI] [PubMed] [Google Scholar]
- 5.Foote S J, Galatis D, Cowman A F. Amino acids in the dihydrofolate reductase-thymidylate synthase gene of Plasmodium falciparum involved in cycloguanil resistance differ from those involved in pyrimethamine resistance. Proc Natl Acad Sci USA. 1990;87:3014–3017. doi: 10.1073/pnas.87.8.3014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Foote S J, Cowman A F. The mode of action and the mechanism of resistance to antimalarial drugs. Acta Trop. 1994;56:157–171. doi: 10.1016/0001-706x(94)90061-2. [DOI] [PubMed] [Google Scholar]
- 7.Luxemburger C, van Vugt M, Jonathan S, McGready R, Looareesuwan S, White N J, Nosten F. Treatment of vivax malaria in an endemic area on the western border of Thailand. Trans R Soc Trop Med Hyg. 1993;93:433–438. doi: 10.1016/s0035-9203(99)90149-9. [DOI] [PubMed] [Google Scholar]
- 8.Matthews D A, Alden R A, Bolin J T, Freer S T, Hamlin R, Xuong N, Kraut J, Poe M, Williams M, Hoogsteen K. Dihydrofolate reductase: X-ray structure of the binary complex with methotrexate. Science. 1977;197:452–455. doi: 10.1126/science.17920. [DOI] [PubMed] [Google Scholar]
- 9.Most H, London I M, Kane A, Lavietes P H, Schroeder E F. Chloroquine for treatment of acute attacks of vivax malaria. JAMA. 1946;131:963–967. doi: 10.1001/jama.1946.02870290013005. [DOI] [PubMed] [Google Scholar]
- 10.Onyiorah E, van Hensbroek M B, Jah M S, Greenwood B. Early clinical failures after pyrimethamine-sulfadoxine treatment of complicated falciparum malaria. Trans R Soc Trop Med Hyg. 1996;90:307–308. doi: 10.1016/s0035-9203(96)90265-5. [DOI] [PubMed] [Google Scholar]
- 11.Peters W. Chemotherapy and drug resistance in malaria. 2nd ed. London, England: Academic Press; 1987. [Google Scholar]
- 12.Peterson D S, Walliker D, Wellems T E. Evidence that a point mutation in dihydrofolate reductase-thymidylate synthase confers resistance to pyrimethamine in falciparum malaria. Proc Natl Acad Sci USA. 1988;85:9114–9118. doi: 10.1073/pnas.85.23.9114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Peterson D S, Milhous W K, Wellems T E. Molecular basis of differential resistance to cycloguanil and pyrimethamine in Plasmodium falciparum malaria. Proc Natl Acad Sci USA. 1990;87:3018–3022. doi: 10.1073/pnas.87.8.3018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Peterson D S, Di Santi S M, Povoa M, Calvosa V S, de Rosario V E, Wellems E E. Prevalence of the dihydrofolate reductase Asn-108 mutation as the basis for pyrimethamine-resistant falciparum malaria in the Brazilian Amazon. Am J Trop Med Hyg. 1991;45:492–497. doi: 10.4269/ajtmh.1991.45.492. [DOI] [PubMed] [Google Scholar]
- 15.Pukrittayakamee S, Chantra A, Simpson J A, Vanijanonta S, Clemens R, Looareesuwan S, White N J. Therapeutic responses to different antimalarial drugs in vivax malaria. Antimicrob Agents Chemother. 2000;44:1680–1685. doi: 10.1128/aac.44.6.1680-1685.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sirawaraporn W, Sirawaraporn R, Cowman A F, Yuthavong Y, Santi D V. Heterologous expression of active thymidylate synthase-dihydrofolate reductase from Plasmodium falciparum. Biochemistry. 1990;29:10779–10785. doi: 10.1021/bi00500a009. [DOI] [PubMed] [Google Scholar]
- 17.Sirawaraporn W, Prapunwattana P, Sirawaraporn R, Yuthavong Y, Santi D V. The dihydrofolate reductase domain of Plasmodium falciparum thymidylate synthase-dihydrofolate reductase. Gene synthesis, expression, and anti-folate-resistant mutants. J Biol Chem. 1993;268:21637–21644. [PubMed] [Google Scholar]
- 18.Sirawaraporn W, Sathikul T, Sirawaraporn R, Yuthavong Y, Santi D V. Antifolate-resistant mutants of Plasmodium falciparum dihydrofolate reductase. Proc Natl Acad Sci USA. 1997;94:1124–1129. doi: 10.1073/pnas.94.4.1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Snewin V A, England S M, Sims P F G, Hyde J E. Characterisation of the dihydrofolate reductase-thymidylate synthetase gene from human malaria parasites highly resistant to pyrimethamine. Gene. 1989;76:41–52. doi: 10.1016/0378-1119(89)90006-1. [DOI] [PubMed] [Google Scholar]
- 20.Thaithong S, Chan S W, Songsomboon S, Wilairat P, Seesod N, Sueblingwong T, Goman M, Ridley R, Beale G. Pyrimethamine resistant mutations in Plasmodium falciparum. Mol Biochem Parasitol. 1992;52:149–158. doi: 10.1016/0166-6851(92)90047-n. [DOI] [PubMed] [Google Scholar]
- 21.Triglia T, Menting J G T, Wilson C, Cowman A F. Mutations in dihydropteroate synthase are responsible for sulfone and sulfonamide resistance in Plasmodium falciparum. Proc Natl Acad Sci USA. 1997;94:13944–13949. doi: 10.1073/pnas.94.25.13944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.van Vugt M, White N J. The treatment of chloroquine-resistant malaria. Trop Dr. 1999;29:176–179. doi: 10.1177/004947559902900321. [DOI] [PubMed] [Google Scholar]
- 23.Volz K W, Matthews D A, Alden R A, Freer S T, Hansch C, Kaufman B T, Kraut J. Crystal structure of avian dihydrofolate reductase containing phenyltriazine and NADPH. J Biol Chem. 1982;257:2528–2536. [PubMed] [Google Scholar]
- 24.Walter R D. Folate metabolism as a target for chemotherapy of malaria. In: Coombs G H, North M J, editors. Biochemical protozoology. London, England: Taylor and Francis; 1991. [Google Scholar]
- 25.Wang P, Read M, Sims P F, Hyde J E. Sulfadoxine resistance in the human malaria parasite Plasmodium falciparum is determined by mutations in dihydropteroate synthase and an additional factor associated with folate utilization. Mol Microbiol. 1997;23:979–986. doi: 10.1046/j.1365-2958.1997.2821646.x. [DOI] [PubMed] [Google Scholar]
- 26.White N J. Assessment of the pharmacodynamic properties of antimalarial drugs in vivo. Antimicrob Agents Chemother. 1997;41:1413–1422. doi: 10.1128/aac.41.7.1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Young M D, Burgess R W. Pyrimethamine resistance in Plasmodium vivax malaria. Bull W H O. 1959;20:27–36. [PMC free article] [PubMed] [Google Scholar]
- 28.Zolg J W, Plitt J R, Chen G X, Palmer S. Point mutations in the dihydrofolate reductase-thymidylate synthase gene as the molecular basis for pyrimethamine resistance in Plasmodium falciparum. Mol Biochem Parasitol. 1989;36:253–262. doi: 10.1016/0166-6851(89)90173-4. [DOI] [PubMed] [Google Scholar]