Summary
Background
We aimed to develop a deep learning-based segmentation system for rapid on-site cytopathology evaluation (ROSE) to improve the diagnostic efficiency of endoscopic ultrasound-guided fine-needle aspiration (EUS-FNA) biopsy.
Methods
A retrospective, multicenter, diagnostic study was conducted using 5345 cytopathological slide images from 194 patients who underwent EUS-FNA. These patients were from Nanjing Drum Tower Hospital (109 patients), Wuxi People's Hospital (30 patients), Wuxi Second People's Hospital (25 patients), and The Second Affiliated Hospital of Soochow University (30 patients). A deep convolutional neural network (DCNN) system was developed to segment cell clusters and identify cancer cell clusters with cytopathological slide images. Internal testing, external testing, subgroup analysis, and human–machine competition were used to evaluate the performance of the system.
Findings
The DCNN system segmented stained cells from the background in cytopathological slides with an F1-score of 0·929 and 0·899–0·938 in internal and external testing, respectively. For cancer identification, the DCNN system identified images containing cancer clusters with AUCs of 0·958 and 0·948–0·976 in internal and external testing, respectively. The generalizable and robust performance of the DCNN system was validated in sensitivity analysis (AUC > 0·900) and was superior to that of trained endoscopists and comparable to cytopathologists on our testing datasets.
Interpretation
The DCNN system is feasible and robust for identifying sample adequacy and pancreatic cancer cell clusters. Prospective studies are warranted to evaluate the clinical significance of the system.
Funding
Jiangsu Natural Science Foundation; Nanjing Medical Science and Technology Development Funding; National Natural Science Foundation of China.
Keywords: Rapid on-site cytopathology evaluation, EUS-FNA, Deep convolutional neural network, Pancreatic mass
Research in context.
Evidence before the study
Rapid on-site cytopathology evaluation (ROSE) is an effective modality to improve the efficiency of endoscopic ultrasound-guided fine-needle aspiration (EUS-FNA) biopsy for the pathological diagnosis of pancreatic masses, but the lack of cytopathologists has limited the application of this modality. Deep convolutional neural network (DCNN) models have shown promising results in analyzing medical images, and several studies used deep learning for the classification of pancreatic lesions, including pancreatic cystic lesions, autoimmune pancreatitis, pancreatic benign tumors, and malignant tumors, under endoscopic ultrasound. Three recent conference abstracts reported deep learning-based cytopathic evaluation for EUS-FNA of pancreatic masses. However, no original studies have reported the utilization of deep learning-based segmentation algorithms for ROSE of EUS-FNA samples of pancreatic masses.
Added value of the study
In the present study, we attempted to develop a deep learning-based segmentation system for ROSE. To the best of our knowledge, we included the largest sample size of cytopathological slide images from multiple institutions. The deep learning-based system could evaluate the adequacy of stained cell clusters and identify pancreatic cancer clusters with outstanding generalization and robustness on our testing datasets. The study also demonstrated that performance of the deep learning-based system can be superior to that of the trained endoscopists, and comparable to the cytopathologists on our testing datasets.
Implications of all the available evidence
The well-established deep learning-based system may assist endoscopists in conducting rapid on-site cytopathology evaluations during EUS-FNA of pancreatic masses without the presence of cytopathologists. The system could potentially improve the diagnostic yields of EUS-FNA of pancreatic masses by reducing diagnostic variation among endoscopists and may promote the coverage of EUS-FNA in various hospitals, especially in community hospitals.
Alt-text: Unlabelled box
Introduction
An accurate pathological diagnosis of pancreatic masses is essential to establish an optimal treatment strategy.1,2 The therapeutic plans and prognoses of pancreatic malignancies (pancreatic cancer) are quite different from those of benign pancreatic lesions (such as pancreatic neuroendocrine tumors and solid pseudopapillary tumors).2,3 However, it is a major challenge to make a correct pathological diagnosis of pancreatic masses under conventional imaging techniques.
Endoscopic ultrasound-guided fine-needle aspiration (EUS-FNA) has been demonstrated to be an integral modality for the diagnosis and staging of pancreatic masses.4 Recent meta-analyses showed that EUS-FNA could achieve a reliable pathological diagnosis of pancreatic masses with an accuracy of 80–86·2%.5,6 However, substantial variance was observed in the diagnostic yield of EUS-FNA, with a sensitivity for pancreatic malignancies ranging from 64% to 96%.7 Several factors, such as the number of needle passes, needle type and size, and the experience of endoscopists, were associated with the diagnostic efficiency of EUS-FNA.5,6 Rapid on-site cytopathology evaluation (ROSE) has been considered as an effective modality to increase the diagnostic efficiency of EUS-FNA by providing immediate feedback on the characteristics and adequacy of the tissue samples.8,9 Previous reports demonstrated that ROSE could improve the diagnostic yield of EUS-FNA by 10–30% by increasing the adequacy of tissue samples for cytopathological evaluation.10, 11, 12 However, the lack of on-site cytopathologists and suboptimal agreement among cytopathologists have limited the widespread use of EUS-FNA in most hospitals.
Artificial intelligence (AI) systems based on deep convolutional neural network (DCNN) models have shown promising results in analyzing medical images.13 Previous studies have found that DCNN shows satisfactory diagnostic performance based on pathological images and cytopathological slides in prostate cancer, cervical cancer, and breast cancer.14, 15, 16 Three preliminary conference abstracts have implemented deep learning in the pathological classification of pancreatic solid masses using a relatively small sample size of cytopathological slides from EUS-FNA and achieved limited diagnostic performance in single-center validation.17, 18, 19 However, it is more clinically applicable for a deep learning-based system to be capable of segmenting the cell clusters and be validated on datasets from multiple hospitals. To the best of our knowledge, no study has adopted DCNN-based segmentation algorithms for evaluating sample adequacy and identifying cancer cell clusters in cytopathological slides from EUS-FNA.
In this study, we developed a DCNN-based segmentation model to differentiate stained cells from the background to ensure the adequacy of the samples and to identify pancreatic malignant cell clusters in cytopathological slides. We validated the DCNN system in the identification of pancreatic cancer cell clusters with internal and external validation datasets at per-image and per-patient levels. We also compared the diagnostic outcomes of the DCNN system with those of trained endoscopists and cytopathologists to assess the transformative potential of the DCNN system in clinical application.
Methods
Study design and participants
This retrospective multicenter diagnostic study was performed in four institutions in China: Nanjing Drum Tower Hospital (NJDTH), Wuxi People's Hospital (WXPH), Wuxi Second People's Hospital (WXSPH), and The Second Affiliated Hospital of Soochow University (SAHSU). The participants included in this study and the clinical baseline characteristics of the patients are shown in Table 1. For pancreatic cancer group, we included the following patients from January 2016 to January 2019: patients who underwent EUS-FNA, had histologically confirmed adenocarcinoma based on evaluation of resected specimens or cytopathologically confirmed adenocarcinoma based on evaluation of EUS-FNA specimens, and cytopathological slides with at least 1 cell cluster were available. For patients who underwent more than one EUS-FNA, only the data from the latest surgery with or before the final diagnosis was included. For the non-cancer group, patients who underwent EUS-FNA with cytopathological diagnosis of mild atypical lesions, other tumors (including pancreatic neuroendocrine tumor and solid pseudopapillary tumor), or no tumors (including chronic pancreatitis and autoimmune pancreatitis) from January 2016 to January 2019 were included. All the patients in the non-cancer group were confirmed with no malignant findings by two experienced certificated cytopathologists and underwent at least 12 months of follow-up with no rapid progression of the pancreatic disease. After selection, a total of 110 malignant cases and 84 non-cancer patients were included. Among the included patients, 109 patients (64 pancreatic cancer patients and 45 non-cancer patients) were from NJDTH, 30 patients (14 pancreatic cancer patients and 16 non-cancer patients) were from WXPH, 25 patients (16 pancreatic cancer patients and 9 non-cancer patients) were from WXSPH, and the rest 30 patients (16 pancreatic cancer patients and 14 non-cancer patients) were from SAHSU.
Table 1.
Baseline characteristics of the training, validation, and testing datasets.
| Total, n = 194 | Training dataset, n = 66 | Validation dataset, n = 16 | Internal testing dataset, n = 27 | External testing datasets |
|||
|---|---|---|---|---|---|---|---|
| WXPH, n = 30 | WXSPH, n = 25 | SAHSC, n = 30 | |||||
| Age (year), mean±sd | 62·1 ± 11·6 | 63·8 ± 10·2 | 59·5 ± 14·4 | 62·1 ± 12·5 | 60·0 ± 11·2 | 64·6 ± 10·5 | 59·9 ± 13·2 |
| Sex, n (%) | |||||||
| Male | 116 (59·8%) | 34 (51·5%) | 12 (75·0%) | 14 (51·9%) | 21 (70·0%) | 17 (68·0%) | 18 (60·0%) |
| Female | 78 (40·2%) | 32 (48·5%) | 4 (25·0%) | 13 (48·1%) | 9 (30·0%) | 8 (32·0%) | 12 (40·0%) |
| Size (cm), mean±sd | 3·3 ± 1·1 | 3·2 ± 1·1 | 3·3 ± 0·9 | 3·2 ± 1·1 | 3·3 ± 1·0 | 3·7 ± 1·7 | 3·2 ± 1·0 |
| Location, n (%) | |||||||
| Head | 114 (58·8%) | 41 (62·1%) | 6 (37·5%) | 16 (59·3%) | 19 (63·3%) | 13 (52·0%) | 19 (63·3%) |
| Body | 45 (23·2%) | 14 (21·2%) | 7 (43·8%) | 7 (25·9%) | 7 (23·3%) | 5 (20·0%) | 5 (16·7%) |
| Tail | 35 (18·0%) | 11 (16·7%) | 3 (18·8%) | 4 (14·8%) | 4 (13·3%) | 7 (28·0%) | 6 (20·0%) |
| Cytopathological diagnosis, n (%) | |||||||
| No tumorsa | 47 (24·2%) | 17 (25·8%) | 3 (18·8%) | 5 (18·5%) | 9 (30·0%) | 4 (16·0%) | 9 (30·0%) |
| Mild atypia | 23 (11·9%) | 7 (10·6%) | 2 (12·5%) | 3 (11·1%) | 4 (13·3%) | 4 (16·0%) | 3 (10·0%) |
| Cancer | 113 (58·2%) | 40 (60·6%) | 10 (62·5%) | 17 (63·0%) | 14 (46·7%) | 16 (64·0%) | 16 (53·3%) |
| Other tumorsb | 11 (5·7%) | 2 (3·0%) | 1 (6·2%) | 2 (7·4%) | 3 (10·0%) | 1 (4·0%) | 2 (6·7%) |
WXPH, Wuxi People's Hospital; WXSPH, Wuxi Second People's Hospital; SAHSC, The Second Affiliated Hospital of Soochow University.
No tumors included chronic pancreatitis (n = 37) or autoimmune pancreatitis (n = 10).
Other tumors included pancreatic neuroendocrine tumors (n = 7) and solid pseudopapillary tumors (n = 4).
Datasets distribution
All the available cytopathological slides of the included patients which were stained with hematoxylin and eosin (H&E) staining or Liu's (modified Romanowsky stain) staining were collected and photographed with a camera (Olympus DP73, Olympus Corporation, Tokyo, Japan) mounted on the microscope (Olympus BX40, Olympus Corporation) at 400 × magnification by an experienced certificated cytopathologist (JY.Z). All the images were delineated along the margin of the cell clusters, and the regions were labeled (annotated) to be cancer, non-cancer and background at pixel level by the experienced certificated cytopathologist (JY.Z). After data annotation, the dataset from NJDTH was randomly split into training, validation, and internal testing datasets (Fig. S1). The datasets from three other centers were considered as external testing datasets. Patients were independent in different datasets.
(1) Training dataset: 1434 images from 37 patients with pancreatic cancer and 732 images from 29 patients in the non-cancer group from NJDTH were used to train the DCNN system.
(2) Validation dataset: 384 images from 10 patients with pancreatic cancer and 311 images from 6 patients in the non-cancer group from NJDTH were used to validate the model and select the optimal hyperparameters.
(3) Internal testing dataset: 865 images from 17 patients with pancreatic cancer and 297 images from 10 patients in the non-cancer group from NJDTH were used to test the model internally.
(4) The external testing dataset: 737 images from 46 patients with pancreatic cancer and 585 images from 39 patients in the non-cancer group from three other hospitals were used to test the model externally. Specifically, 238 images from 14 patients with pancreatic cancer and 290 images from 16 non-cancer patients were contributed by WXPH, 266 images from 16 patients with pancreatic cancer and 99 images from 9 non-cancer patients were contributed by WXSPH, and 233 images from 16 patients with pancreatic cancer and 196 images from 14 non-cancer patients were from SAHSU.
(5) Human–machine competition dataset 1 (randomly selected from the internal testing dataset): 422 images from 6 patients with pancreatic cancer and 158 images from 4 patients in the non-cancer group from NJDTH were used to compare the performance of the DCNN system and the reports by trained endoscopists and cytopathologists.
(6) Human–machine competition dataset 2 (randomly selected from the external testing dataset): 200 images from 45 patients with pancreatic cancer and 200 images from 36 patients in the non-cancer group from the external testing datasets were used to compare the performance of the DCNN system and the reports by trained endoscopists and cytopathologists.
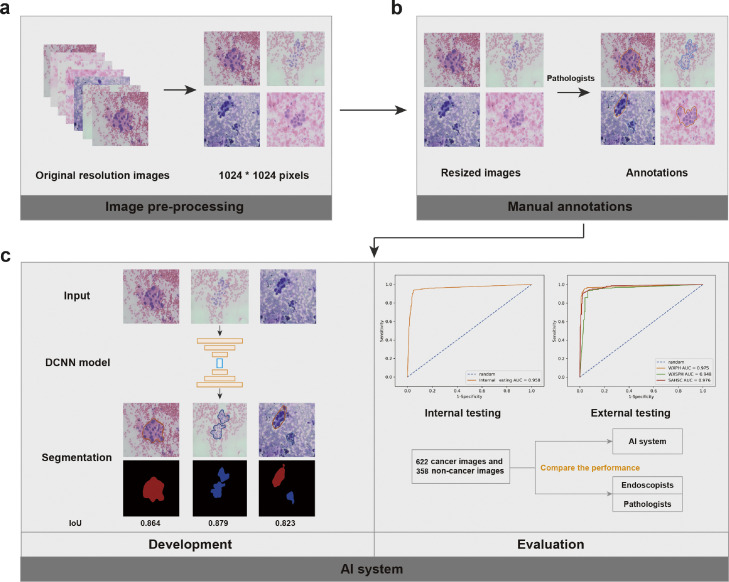
Figure 1 shows the flow chart of our study design.
Figure 1.
The flow chart of study design. (a) All the images of cytopathological slides with original resolutions were resized to a standard resolution (1024 × 1024 pixels). (b) The resized images were annotated with delineation along the margin of the cell clusters and addition of the corresponding labels on the delineated cell clusters. (c) A deep convolutional neural network model was developed using the training dataset and optimal hyperparameters were selected with the validation dataset. (d) The performance of the established deep-learning system was evaluated using the internal and external testing datasets and then compared with those of endoscopists and cytopathologists. DCNN=Deep convolutional neural network. AI=Artificial intelligence.
Training DCNN for cell image segmentation
All the images were resized to 1024 × 1024 pixels before feeding into a UNet-based DCNN system.20 The UNet architecture consisted of an encoder and a decoder to extract and combine different levels of features for prediction. For the encoder, the ResNet101 network was chosen as the backbone, which consisted of 4 convolutional blocks with 9, 12, 69 and 9 convolutional layers.20 For the decoder, 4 convolutional blocks and 4 bilinear interpolation upsampling layers were used. Skip connections were used for merging features from different layers. Finally, the model mapped each pixel to a vector, and the predicted class of each pixel was defined by the index of the highest value (Figure S2). An early stopping strategy was used to prevent overfitting (Figure S3a & S3b). Several data augmentation strategies were used to improve the diversity of the training dataset and overcome the overfitting. Specifically, random rotations, horizontal flips, vertical flips, brightness changes, and hue transformations were performed on the training images.
Testing of the DCNN system and comparison with trained endoscopists and cytopathologists
Firstly, the internal and external testing datasets were used for the assessment. Secondly, sensitivity analysis was conducted to evaluate the DCNN system for cancer identification according to sex, age, lesion location and lesion size on the internal testing dataset. Thirdly, the human–machine competition datasets were used to compare the performance of the system in cancer identification with those of five endoscopists with six-month cytopathological training and three cytopathologists with two-year experience. The endoscopists and cytopathologists were asked to differentiate between cancer and non-cancer images in the slide images from the human–machine competition datasets, which are randomly shown on the computer screen. The endoscopists and cytopathologists were not involved in the selection and annotation of all the datasets, and were masked to the clinical characteristics, endoscopic features, and pathological results of patients on the testing datasets.
Outcomes
The primary outcome was the area under the receiver operating characteristic curve (AUC) in cancer identification. The optimal proportion of segmented cancer in the entire segmented areas was determined by the highest Youden index (Figure S3c). For the diagnosis of malignant lesions in each patient, we used three positive images as the threshold. The accuracy, sensitivity, specificity, positive predictive value (PPV) and negative predictive value (NPV) were also calculated to assess the performance in distinguishing cancer images from non-cancer images.
In addition, we evaluated the diagnostic adequacy of the slides according to the results of stained cell segmentation. The segmentation performance of the DCNN system was assessed using intersection over union (IoU), accuracy, precision, recall, and F1-score. We then calculated precision and recall at per-patient level by setting an optimal IoU and accuracy of segmented images to imitate clinical scenarios. All the details and formula involved were listed in supplementary materials.
Statistical analysis
A two-sided McNemar test was conducted to compare the differences in accuracy, sensitivity, and specificity. Generalized score statistics were utilized to compare the discrepancies in PPV and NPV. The kappa value was calculated to evaluate the diagnostic consistency of the DCNN system, endoscopists, and cytopathologists. All statistical analyses were conducted using R (version 4·1·2; https://www.r-project.org).
Ethics
The study protocol was reviewed and approved by the Medical Ethics Committee of Nanjing Drum Tower Hospital (IRB no. 2020-044-01). Informed consent was waived since only deidentified data were retrospectively included. The study was registered in the WHO Registry Network's Primary Registries (ChiCTR2000032664).
Role of the funding source
The funders played no role in study design, data collection, data analyses, interpretation, or writing of the article. The corresponding authors had full access to all the data generated in the study.
Results
Performance of cell cluster segmentation
Several deep-learning algorithms were used to segment all stained cell clusters and cancer cell clusters. The results showed that UNet along with the ResNet101 encoder achieved the best segmentation performance compared with other deep-learning algorithms on the validation dataset, with a mean IoU of 0·875 for all stained cell clusters (Figure S4, Table S1). Moreover, the results showed that the IoU of the DCNN in segmenting all stained cell clusters (irrespective of cell types) was 0·867 on the internal testing dataset and 0·817–0·883 on the external testing datasets (Table S2). The accuracy, precision, recall, and F1-score of the DCNN system in segmenting all cell clusters were 0·964, 0·927, 0·931, and 0·929, respectively, on the internal testing dataset (Table S2). On the external testing datasets, the accuracy, precision, recall, and F1-score of the DCNN system in segmenting all cell clusters were 0·947–0·969, 0·889–0·940, 0·891–0·936, and 0·899–0·938, respectively (Table S2).
To evaluate the DCNN system in assessing cell adequacy, we calculated recall (cell adequacy rate) by setting an optimal threshold of IoU at per-image and per-patient level. The results showed that the recall was 0·842 in internal testing and 0·821 in external testing at per-image level (Table S3). Notably, the recall of the DCNN system could reach 0·963 in internal testing and 0·906 in external testing at per-patient level (Table S4). Collectively, these results indicated that the DCNN system can delineate the margins of cell clusters in cytopathological slides with high accuracy and substantial robustness.
Performance of the DCNN system in identifying pancreatic cancer
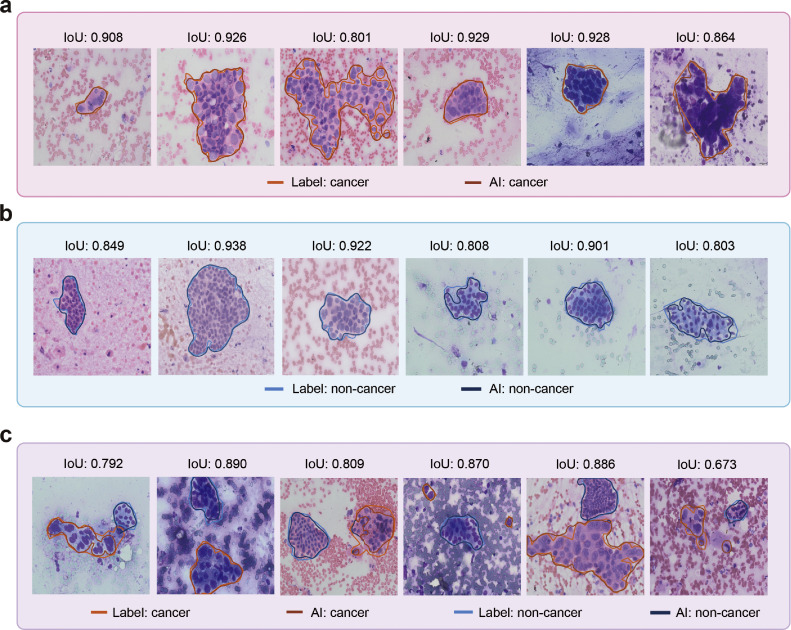
We next evaluated the performance of the DCNN system in identifying pancreatic cancer in cytopathological slides at per-image level. Representative predictions of the DCNN system are shown in Figure 2. The results showed that the DCNN system identified pancreatic cancer with an accuracy of 0·944 (95% CI, 0·929–0·956), sensitivity of 0·940 (95% CI, 0·917–0·963), specificity of 0·946 (95% CI, 0·930–0·962), PPV of 0·901 (95% CI, 0·873–0·930), and NPV of 0·968 (95% CI, 0·955–0·980) on the internal testing dataset (Table S5). Moreover, the DCNN system also showed satisfactory performance in pancreatic cancer identification on the external testing datasets with an accuracy of 0·912–0·958, sensitivity of 0·928–0·944, specificity of 0·875–0·971, PPV of 0·897–0·928, and NPV of 0·930–0·971 (Table S5). The AUC of the DCNN system on the internal testing dataset was 0·958, and the AUCs on the external testing datasets were 0·948–0·976 (Table S5, Figure 3a & b).
Figure 2.
Visualization of DCNN performance in segmenting pancreatic cancer cell clusters and noncancer cell clusters. (a) Representative prediction results of the DCNN system for pancreatic cancer cell cluster segmentation. (b) Representative prediction results of the DCNN system for noncancer cell cluster segmentation. (c) Representative prediction results of the DCNN system for pancreatic cancer and noncancer cell cluster segmentation in a single visual field. DCNN=Deep convolutional neural network. IoU=Intersection over Union.
Figure 3.
DCNN performance in identifying pancreatic cancer on internal and external testing datasets. (a) Image-level ROC curves for pancreatic cancer on the internal testing dataset. (b) Image-level receiver operating characteristic curves for pancreatic cancer on the external testing datasets. (c) Patient-level receiver operating characteristic curves for pancreatic cancer on the internal testing dataset. (d) Patient-level receiver operating characteristic curves for pancreatic cancer on the external testing datasets. Image-level receiver operating characteristic curves for pancreatic cancer in the subgroup analysis of the internal testing dataset according to age (e), sex (f), lesion location (g), and lesion size (h). DCNN= Deep convolutional neural network. AUC=Area under the receiver operating characteristic curve. NJDTH=Nanjing Drum Tower Hospital. WXPH=Wuxi People's Hospital. WXSPH=Wuxi Second People's Hospital. SAHSC=The Second Affiliated Hospital of Soochow University.
Since one patient may have more than two slides in the datasets, we evaluated the performance of our DCNN system in pancreatic cancer identification at per-patient level to further verify the effectiveness of our DCNN system. On the internal testing dataset, the accuracy, sensitivity, specificity, PPV, and NPV of the DCNN system in distinguishing cancer from noncancer cytopathology were 0·889 (95% CI, 0·719–0·961), 1·000 (95% CI, 1·000–1·000), 0·700 (95% CI, 0·416–0·984), 0·850 (95% CI, 0·694–1·000), and 1·000 (95% CI, 1·000–1·000), respectively (Table S5). Notably, the DCNN system also showed generalizable and robust performance on the external testing datasets with an accuracy of 0·920–0·967, a sensitivity of 0·929–1·000, a specificity of 0·778–0·938, a PPV of 0·889–0·941, and an NPV of 0·938–1·000 (Table S5). The AUC of the DCNN system on the internal testing dataset was 0·982, and the AUCs on the external testing datasets were 0·993–1·000 (Table S5, Figure 3c & d).
To further validate the promising performance of the DCNN system, we performed sensitivity analysis according to different ages, sexes, lesion locations, and lesion sizes. The results showed that the AUCs of the model were 0·949–0·969 for different ages, 0·936–0·982 for different sexes, 0·937–0·958 for different locations and 0·936–0·993 for different sizes (Table S6, Figure 3e–h).
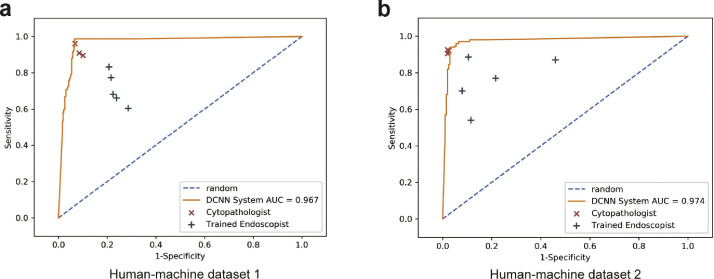
Performance comparison among the DCNN system, endoscopists, and cytopathologists
Subsequently, we compared the performance between the DCNN system and the cytopathologists. The results showed that the DCNN system achieved a higher accuracy (0·945, 95% CI, 0·933–0·955) than the cytopathologists (0·917, 95% CI, 0·903–0·930, P = 0·001) on the human-machine competition dataset 1 (Figure 4a, Tables S7–S9). Notably, the sensitivity (0·987, 95% CI, 0·969–1·000) and NPV (0·995, 95% CI, 0·988–1·000) of the DCNN system were significantly higher than those of the cytopathologists (sensitivity: 0·922, 95% CI, 0·898–0·947, P < 0·001; NPV: 0·970, 95% CI, 0·961–0·980, P < 0·001) (Table S7–S9). The specificity (0·930, 95% CI, 0·905–0·954) and PPV (0·835, 95% CI, 0·781–0·889) of the DCNN system were comparable to those of the cytopathologists (specificity: 0·915, 95% CI, 0·900–0·931, P = 0·175; PPV: 0·798, 95% CI, 0·764–0·832, P = 0·064) (Table S7–S9). Consistently, the DCNN system also showed a comparable performance with the cytopathologists with relatively higher sensitivity and NPV (P < 0.01), and a lower specificity and PPV (P < 0.001) in the human-machine competition dataset 2 (Figure 4b, Tables S10–S12).
Figure 4.
Comparison of the diagnostic performance of the DCNN system and doctors. (a) Comparison of the diagnostic performance of the DCNN system, trained endoscopists, and cytopathologists on human-machine competition dataset 1. (b) Comparison of the diagnostic performance of the DCNN system, trained endoscopists, and cytopathologists on human-machine competition dataset 2. DCNN=Deep convolutional neural network. AUC=Area under the receiver operating characteristic curve.
Since cytopathologists were not available in the field of EUS-FNA for most hospitals, several studies trained endoscopists to increase the diagnostic performance of EUS-FNA.21,22 Thus, we compared the performance of the DCNN system and trained endoscopists. Interestingly, the accuracy, sensitivity, specificity, PPV, and NPV of the DCNN system were all significantly better than those of trained endoscopists on both human-machine competition datasets (P < 0·001) (Figure 4, Table S7–S12). Similarly, the diagnostic consistency of cytopathologists (κ: 0·616–0·670) was higher than that of endoscopists (κ: 0·162–0·333) (Table S13, Figure S5a). Moreover, the consistency between the DCNN system and cytopathologists (κ: 0·683–0·754) was also higher than that between the DCNN system and endoscopists (κ: 0·286–0·505) (Table S13, Figure S5b).
Discussion
In this study, we developed a novel DCNN system to render ROSE during EUS-FNA. The DCNN system showed promising performance in differentiating stained cells from the background to ensure the adequacy of the samples and in identifying pancreatic malignant cell clusters from benign cell clusters with cytopathological slides. The system also showed generalizable and robust performance on the internal dataset, external datasets, and subgroup analysis. The diagnostic performance of the DCNN system could be comparable to that of cytopathologists and was superior to that of trained endoscopists. To the best of our knowledge, this study was the first and largest to establish a deep learning-based system for segmenting stained cell clusters and identifying pancreatic cancer in ROSE during EUS-FNA, which might be used to improve the diagnostic performance of EUS-FNA.
It is pivotal to evaluate the diagnostic adequacy of EUS-FNA samples since only sufficient high-quality samples can be concluded with a preliminary diagnosis.23 Several studies have demonstrated the efficacy of ROSE in improving the diagnostic adequacy of EUS-FNA.24 However, the lack of cytopathologists in many hospitals, especially rural hospitals, has refrained the wide use of ROSE. Although on-site cytotechnicians could improve the diagnostic adequacy of EUS-FNA, cytotechnicians without sufficient training could only achieve moderate diagnostic adequacy (70%).25 Several studies assessed the performance of endoscopists or endosonographers in assessing the diagnostic adequacy of EUS-FNA samples and demonstrated a higher diagnostic adequacy among these endoscopists or endosonographers.21,26,27 However, the training curriculums varied in different studies, leading to a considerable inconsistency between endoscopists and cytopathologists.28 To the best of our knowledge, no studies have implemented deep learning in assessing the diagnostic adequacy of EUS-FNA. Here, our DCNN system achieved IoUs of 0·817–0·883 and F1-scores of 0·899–0·938 in segmenting all the stained cell clusters on the internal and external testing datasets. In clinical practice, the DCNN system could be used to evaluate the adequacy of cytopathological samples by setting a suitable threshold of IoU to achieve satisfactory performance.
Three preliminary studies have implemented deep learning in pancreatic cancer identification. Hashimoto et al. established a deep learning model with 800 adenocarcinoma and 400 benign images and found that the deep learning model achieved a slightly lower accuracy than the cytopathologists.17 Patel et al. found that their deep learning model achieved an accuracy of 0·87 in pancreatic cancer identification and ranked 4th among the included 19 observers.18 Notably, they found that the deep learning model outperformed all 6 interventional gastroenterologists. Thosani et al. also developed a deep learning-based classification model for pancreatic cancer diagnosis.19 However, the specificity of the DCNN system was relatively low, which indicated substantial false positive classifications and might confuse endoscopists in real clinical scenarios. All the previous DCNN systems could only classify pancreatic malignancy images from non-malignancy images but could not show the exact location of the stained malignant cells. Here, our DCNN system showed generalizable and robust performance in identifying pancreatic cancer cell clusters in EUS-FNA cytopathology at both per-image level and per-patient level. Moreover, we also found that the DCNN system showed stable diagnostic performance in several subgroups, including different ages, sexes, lesion locations, and lesion sizes. Based on the outstanding performance of our DCNN system, we developed a workstation that integrated the DCNN system into the diagnostic workflow of ROSE during EUS-FNA (Figure S6, Video S1). Prospective studies will be conducted to further evaluate the assistant value of the DCNN system in the diagnostic yield of EUS-FNA in real clinical practice.
The present DCNN system was set with a relatively high sensitivity to reduce potential missing lesions. In comparison with trained endoscopists and cytopathologists, the performance of the DCNN system was significantly higher than that of the trained endoscopists, and was comparable to that of the cytopathologists on our testing datasets. This indicated that the system could assist the EUS-FNA process reliably.
Hikichi et al. conducted a comparative study on the diagnostic accuracy of ROSE by trained endoscopists and cytopathologists.21 The results showed that the diagnostic accuracies of endoscopists and cytopathologists were 94·7% and 94·3%, respectively. However, Savoy et al. compared trained endoscopists and cytotechnologists in determining the sample adequacy and diagnosing a malignancy of EUS-FNA specimens.22 They found that endoscopists achieved a very low sensitivity (56–60%) in the diagnosis of EUS-FNA specimens. This indicated that although endoscopists trained with cytopathology may be capable of establishing an accurate diagnosis of EUS-FNA specimens, the performance of the endoscopists showed obvious variance. Here, the diagnostic performance of our DCNN system was comparable and concordant with that of cytopathologists and was much better than that of trained endoscopists. Combined with high sensitivity and PPV, our DCNN system could be deployed in clinical scenes to assist endoscopists for increasing the diagnostic yield of EUS-FNA.
Although this study has achieved promising results, there still exist several limitations in this study. First, this was a retrospective study, which may contain selection bias. The excellent performance of the DCNN system cannot reflect its clinical application in the real world. We will collect more data from variant sources to enhance the generalizability of the DCNN system. We will design a multicenter prospective validation trial, and a randomized controlled trial to further validate the applicability of this DCNN system. Second, only high-quality images were used in this study. Although it is easy to control the quality of images in real clinical applications, we still plan to collect more images of different qualities to further enhance the generalization of our DCNN system.
In conclusion, we developed a DCNN system to assist endoscopists in rendering ROSE during EUS-FNA. The DCNN system showed outstanding performance in evaluating diagnostic adequacy and identifying pancreatic cancer cell clusters, and the performance of the DCNN system was superior to the trained endoscopists, and comparable to the cytopathologists on our testing datasets. Prospective validation and intervention studies are needed to provide high-level evidence for the clinical significance of the DCNN system in real clinical practice. We believe that this system can assist endoscopists in performing ROSE by themselves when cytopathologists are not available, with increased diagnostic yield during EUS-FNA for pancreatic cancer.
Contributors
X.P.Z., Y.L., L.W. and W.J.L.: conceived and supervised this study. Y.L. and C.Y.P. supported the study. S.Z. and D.H.T.: designed the experiments. S.Z., Y.F.Z., D.H.T., and M.H.N.: conducted the experiments. G.F.X., S.S.S., C.Y.P., Q.Z., X.Y.W. and D.M.H.: collected the images. Y.F.Z., W.J.L. and L.W.: established the deep learning model. J.Y.Z.: labelled the images. S.Z., Y.F.Z., D.H.T., and M.H.N.: analysed the data. D.H.T., S.Z., and Y.F.Z.: wrote the manuscript. W.J.L. and Y.L.: modified the language of the manuscript. All authors reviewed and approved the final version of the manuscript. All authors had full access to all the data in the study, and had final responsibility for the decision to submit for publication.
Data sharing statement
Due to patient privacy, all the data sets generated or analyzed in this study are available from the corresponding author upon reasonable request (Xiaoping Zou, zouxp@nju.edu.cn). To ensure the transparency and reproducibility of the study, we have provided a free access to the model on the website (http://39.100.80.45/ROSE/).
Funding
Jiangsu Natural Science Foundation; Nanjing Medical Science and Technology Development Funding; National Natural Science Foundation of China.
Declaration of interests
All the authors have no conflicts of interest to declare.
Acknowledgments
We thank Tao Bai, Wei Li, Qi Sun, and Li Gao for contributions to this study. This work was supported by the General Program of Jiangsu Natural Science Foundation (SBK2019022491), the Nanjing Medical Science and Technology Development Fund (ZKX18022) and National Natural Science Foundation of China (Grant No. 62006113). The funders had no role in the study design, data collection, data analyses, interpretation, or writing of the report.
Footnotes
Supplementary material associated with this article can be found in the online version at doi:10.1016/j.ebiom.2022.104022.
Contributor Information
Wu-Jun Li, Email: liwujun@nju.edu.cn.
Lei Wang, Email: leiwang9631@nju.edu.cn.
Ying Lv, Email: lvying@njglyy.com.
Xiaoping Zou, Email: zouxp@nju.edu.cn.
Appendix. Supplementary materials
References
- 1.Mizrahi J.D., Surana R., Valle J.W., Shroff R.T. Pancreatic cancer. Lancet. 2020;395(10242):2008–2020. doi: 10.1016/S0140-6736(20)30974-0. [DOI] [PubMed] [Google Scholar]
- 2.Cives M., Strosberg J.R. Gastroenteropancreatic neuroendocrine tumors. CA Cancer J Clin. 2018;68(6):471–487. doi: 10.3322/caac.21493. [DOI] [PubMed] [Google Scholar]
- 3.Grossberg A.J., Chu L.C., Deig C.R., et al. Multidisciplinary standards of care and recent progress in pancreatic ductal adenocarcinoma. CA Cancer J Clin. 2020;70(5):375–403. doi: 10.3322/caac.21626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang A.Y., Yachimski P.S. Endoscopic management of pancreatobiliary neoplasms. Gastroenterology. 2018;154(7):1947–1963. doi: 10.1053/j.gastro.2017.11.295. [DOI] [PubMed] [Google Scholar]
- 5.Bang J.Y., Hawes R., Varadarajulu S. A meta-analysis comparing ProCore and standard fine-needle aspiration needles for endoscopic ultrasound-guided tissue acquisition. Endoscopy. 2016;48(4):339–349. doi: 10.1055/s-0034-1393354. [DOI] [PubMed] [Google Scholar]
- 6.van Riet P.A., Erler N.S., Bruno M.J., Cahen D.L. Comparison of fine-needle aspiration and fine-needle biopsy devices for endoscopic ultrasound-guided sampling of solid lesions: a systemic review and meta-analysis. Endoscopy. 2021;53(4):411–423. doi: 10.1055/a-1206-5552. [DOI] [PubMed] [Google Scholar]
- 7.Hawes R.H. The evolution of endoscopic ultrasound: improved imaging, higher accuracy for fine needle aspiration and the reality of endoscopic ultrasound-guided interventions. Curr Opin Gastroenterol. 2010;26(5):436–444. doi: 10.1097/MOG.0b013e32833d1799. [DOI] [PubMed] [Google Scholar]
- 8.Polkowski M., Larghi A., Weynand B., et al. Learning, techniques, and complications of endoscopic ultrasound (EUS)-guided sampling in gastroenterology: European Society of Gastrointestinal Endoscopy (ESGE) technical guideline. Endoscopy. 2012;44(2):190–206. doi: 10.1055/s-0031-1291543. [DOI] [PubMed] [Google Scholar]
- 9.Schmidt R.L., Walker B.S., Howard K., Layfield L.J., Adler D.G. Rapid on-site evaluation reduces needle passes in endoscopic ultrasound-guided fine-needle aspiration for solid pancreatic lesions: a risk-benefit analysis. Dig Dis Sci. 2013;58(11):3280–3286. doi: 10.1007/s10620-013-2750-6. [DOI] [PubMed] [Google Scholar]
- 10.Matynia A.P., Schmidt R.L., Barraza G., Layfield L.J., Siddiqui A.A., Adler D.G. Impact of rapid on-site evaluation on the adequacy of endoscopic-ultrasound guided fine-needle aspiration of solid pancreatic lesions: a systematic review and meta-analysis. J Gastroenterol Hepatol. 2014;29(4):697–705. doi: 10.1111/jgh.12431. [DOI] [PubMed] [Google Scholar]
- 11.Harada R., Kato H., Fushimi S., et al. An expanded training program for endosonographers improved self-diagnosed accuracy of endoscopic ultrasound-guided fine-needle aspiration cytology of the pancreas. Scand J Gastroenterol. 2014;49(9):1119–1123. doi: 10.3109/00365521.2014.915051. [DOI] [PubMed] [Google Scholar]
- 12.Kong F., Zhu J., Kong X., et al. Rapid on-site evaluation does not improve endoscopic ultrasound-guided fine needle aspiration adequacy in pancreatic masses: a meta-analysis and systematic review. PLoS One. 2016;11(9) [Google Scholar]
- 13.Bi W.L., Hosny A., Schabath M.B., et al. Artificial intelligence in cancer imaging: clinical challenges and applications. CA Cancer J Clin. 2019;69(2):127–157. doi: 10.3322/caac.21552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Song Y., Zhang L., Chen S., Ni D., Lei B., Wang T. Accurate segmentation of cervical cytoplasm and nuclei based on multiscale convolutional network and graph partitioning. IEEE Trans Biomed Eng. 2015;62(10):2421–2433. doi: 10.1109/TBME.2015.2430895. [DOI] [PubMed] [Google Scholar]
- 15.Dey P., Logasundaram R., Joshi K. Artificial neural network in diagnosis of lobular carcinoma of breast in fine-needle aspiration cytology. Diagn Cytopathol. 2013;41(2):102–106. doi: 10.1002/dc.21773. [DOI] [PubMed] [Google Scholar]
- 16.Momeni-Boroujeni A., Yousefi E., Somma J. Computer-assisted cytologic diagnosis in pancreatic FNA: an application of neural networks to image analysis. Cancer Cytopathol. 2017;125(12):926–933. doi: 10.1002/cncy.21915. [DOI] [PubMed] [Google Scholar]
- 17.Hashimoto Y. Prospective comparison study of eus-fna onsite cytology diagnosis by pathologist versus our designed deep learning algorhythm in suspected pancreatic cancer. Gastroenterology. 2020;158(6):17. [Google Scholar]
- 18.Patel J., Bhakta D., Elzamly S., et al. Artificial intelligence based rapid onsite cytopathology evaluation (rose-aidtm) vs. physician interpretation of cytopathology images of endoscopic ultrasound-guided fine-needle aspiration (eus-fna) of pancreatic solid lesions. Gastrointest Endosc. 2021;93(6):AB193–ABAB4. [Google Scholar]
- 19.Thosani N., Patel J., Moreno V., et al. Development and validation of artificial intelligence based rapid onsite cytopathology evaluation (rose-aidtm) for endoscopic ultrasound-guided fine-needle aspiration (eus-fna) of pancreatic solid lesions. Gastroenterology. 2021;160(6):S–17. [Google Scholar]
- 20.He K., Zhang X., Ren S., Sun J. Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR) 2016. Deep residual learning for image recognition; pp. 770–778. 2016 27-30 June 2016. [Google Scholar]
- 21.Hikichi T., Irisawa A., Bhutani M.S., et al. Endoscopic ultrasound-guided fine-needle aspiration of solid pancreatic masses with rapid on-site cytological evaluation by endosonographers without attendance of cytopathologists. J Gastroenterol. 2009;44(4):322–328. doi: 10.1007/s00535-009-0001-6. [DOI] [PubMed] [Google Scholar]
- 22.Savoy A.D., Raimondo M., Woodward T.A., et al. Can endosonographers evaluate on-site cytologic adequacy? A comparison with cytotechnologists. Gastrointest Endosc. 2007;65(7):953–957. doi: 10.1016/j.gie.2006.11.014. [DOI] [PubMed] [Google Scholar]
- 23.Harewood G.C., Wiersema M.J. Endosonography-guided fine needle aspiration biopsy in the evaluation of pancreatic masses. Am J Gastroenterol. 2002;97(6):1386–1391. doi: 10.1111/j.1572-0241.2002.05777.x. [DOI] [PubMed] [Google Scholar]
- 24.Iglesias-Garcia J., Dominguez-Munoz J.E., Abdulkader I., et al. Influence of on-site cytopathology evaluation on the diagnostic accuracy of endoscopic ultrasound-guided fine needle aspiration (EUS-FNA) of solid pancreatic masses. Am J Gastroenterol. 2011;106(9):1705–1710. doi: 10.1038/ajg.2011.119. [DOI] [PubMed] [Google Scholar]
- 25.Petrone M.C., Arcidiacono P.G., Carrara S., Mezzi G., Doglioni C., Testoni P.A. Does cytotechnician training influence the accuracy of EUS-guided fine-needle aspiration of pancreatic masses? Dig Liver Dis. 2012;44(4):311–314. doi: 10.1016/j.dld.2011.12.001. [DOI] [PubMed] [Google Scholar]
- 26.Hayashi T., Ishiwatari H., Yoshida M., et al. Rapid on-site evaluation by endosonographer during endoscopic ultrasound-guided fine needle aspiration for pancreatic solid masses. J Gastroenterol Hepatol. 2013;28(4):656–663. doi: 10.1111/jgh.12122. [DOI] [PubMed] [Google Scholar]
- 27.Nebel J.A., Soldan M., Dumonceau J.M., et al. Rapid on-site evaluation by endosonographer of endoscopic ultrasound fine-needle aspiration of solid pancreatic lesions: a randomized controlled trial. Pancreas. 2021;50(6):815–821. doi: 10.1097/MPA.0000000000001846. [DOI] [PubMed] [Google Scholar]
- 28.Assef M., Rossini L., Nakao F., Araki O., Bueno F., Sayeg M. Interobserver concordance for endoscopic ultrasonography-guided fine-needle aspiration on-site cytopathology. Endosc Ultrasound. 2014;3(1):S15. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.